ABSTRACT
Helicobacter pylori infection is the leading cause of chronic gastritis, which can develop into gastric cancer. Eliminating H. pylori infection with antibiotics achieves the prevention of gastric cancer. Currently, the prevalence of H. pylori resistance to clarithromycin and metronidazole, and the dual resistance to metronidazole and clarithromycin (C_R, M_R, and C/M_R, respectively), remains at a high level worldwide. As a means of exploring new candidate proteins for the management of H. pylori infection, secreted proteins from antibiotic-susceptible and antibiotic-resistant H. pylori-associated gastritis strains were obtained by in-solution tryptic digestion coupled with nano-liquid chromatography tandem mass spectrometry (nano-LC-MS/MS). A total of 583, 582, 590, and 578 differential expressed proteins were identified from C_R, M_R, C/M_R, and antibiotic-sensitive strain (S_S) samples, respectively. Of these, 23 overlapping proteins were found by Venn diagram analysis. Based on heat map analyses, the most and least differing protein expressions were observed from C/M_R strains and S_S strains, respectively. Of the proteins secreted by the S_S strain, only nine were found. After predicting the protein interaction with metronidazole and clarithromycin via the STITCH database, the two most interesting proteins were found to be rpoBC and FBPAII. After quantitative real-time reverse transcription PCR (qRT-PCR) analysis, a downregulation of rpoB from M_R strains was observed, suggesting a relationship of rpoB to metronidazole sensitivity. Inversely, an upregulation of fba from C_R, M_R, and C/M_R strains was noticed, suggesting the paradoxical expression of FBPAII and the fba gene. This report is the first to demonstrate the association of these two novel secreted proteins, namely, rpoBC and FBPAII, with antibiotic-sensitive H. pylori-associated gastritis strains.
KEYWORDS: Helicobacter pylori-associated gastritis, antibiotic-sensitive, antibiotic resistance, clarithromycin, metronidazole, dual resistance to metronidazole and clarithromycin, in-solution digestion proteomic study
INTRODUCTION
Helicobacter pylori, a spiral-shaped Gram-negative bacterium, is an important clinical pathogen that causes gastritis and peptic ulcers and, in the worst case, leads to gastric mucosa-associated lymphoid tissue lymphoma (MALT) and gastric adenocarcinoma (1–3). The eradication of H. pylori with proton-pump inhibitors in combination with clarithromycin (Cla) or metronidazole (Mtz)/amoxicillin is not only helpful in alleviating peptic ulcers, but also helps prevent the occurrence of cancer (4). However, the efficacy of this therapeutic regimen is in decline because of increasing of antibiotic resistance of H. pylori (5). The prevalence of H. pylori resistance to Cla and Mtz (C_R and M_R, respectively) over the past 10 years in the Asia Pacific region has significantly increased (from 7% to 21% and 36% to 45%, respectively) (6), while in the Association of Southeast Asian Nations (ASEAN) countries, the prevalence of Cla resistance and Mtz resistance is still common (7). In Thailand, the prevalence of C_R and M_R was moderate (14% and 36%, respectively) (7). In addition, a 10% rate of concurrent resistance to Cla and Mtz (dual resistance, C/M_R) has been reported (8). Therefore, a greater understanding of the resistance mechanism is likely to lead to the development of better treatment in H. pylori-resistance patients.
Cla is a macrolide derived from erythromycin. Its antimicrobial mechanism consists of the inhibition of protein synthesis by binding with 23S rRNA, which then blocks the translocation process. The World Health Organization (WHO) published a report in 2017 stating that C_R H. pylori is on the global list of antibiotic resistance crises (9). The most common resistance mechanism is associated with the A2142G or A2143G mutation in domain V of the 23S mRNA (10). Another possible mechanism of C_R is the reduction of intracellular antimicrobial concentrations through H. pylori resistance-nodulation-division (RND) efflux of antimicrobial compounds (11–13), whereas Mtz is a nitroimidazole derivative drug. Its antimicrobial activity is based on toxic free-radical formation of the nitro group on the Mtz-prodrug by the oxygen-insensitive NADPH nitroreductase or NAD(P)H-flavin oxidoreductase, which is encoded by the rdxA gene or the frxA gene, respectively (14–16). The Mtz-active drug then induces several strong oxidizing agents which ultimately damage the DNA, thus inhibiting nucleic acid synthesis. The most reported mutations in M_R H. pylori is from insertions and deletions of transposons or missense and frameshift mutations in the rdxA and frxA genes (17–20). In addition to the most common resistance mechanisms, other mechanisms detected by genomic tools have been clarified recently. For example, mutation in the gene HP1027 (Fur) or Fur downregulation causes an overexpression of superoxide dismutase, suggesting the association of H. pylori M_R (21). In practice, most of the information about this resistance mechanism is known to be derived from molecular techniques (22, 23). However, the use of proteomics techniques to investigate protein expression is another effective method which is currently popular in studies of antibiotic resistance of H. pylori (24–28).
Proteomic analysis is the tool for identifying and quantifying proteins in protein mixtures under defined conditions by using mass spectrometry (MS) (29, 30). Outlines of typical procedures for protein identification by MS-based proteomics include cell extraction, one-dimensional (1-D) or 2-D gel electrophoresis, and protein digestion. The peptides are then analyzed using liquid chromatography tandem mass spectrometry (LC-MS/MS), followed by peptide identification and analysis, and are then compared with a standardized database (31, 32). Nonetheless, the top-down proteomic approach has the advantage of providing a complete protein sequence as well as a reliable and comprehensive analysis of all types of posttranslation modification (33). Furthermore, because of the easy adaptation to the high-throughput analysis of the bottom-up approach, bottom-up proteomics is currently the most common approach in use (34). Moreover, to date, in-gel digestion (after 1-D or 2-D gel electrophoresis) and in-solution digestion are the two most popular approaches for digestion of the protein sample in preparing it for mass spectrometry analysis (30, 35), while efficiency and reproducibility from prefractionation of the sample are a major advantage for in-gel digestion. In addition, simpler sample preparation, less labor, and less sample demand are also advantages of in-solution digestion (36, 37). In this study, in order to identify the secreted proteins of H. pylori-associated gastritis strains, experiments were therefore designed using the bottom-up proteomics with in-solution digestion protocols.
However, a proteome analysis study of H. pylori can explore the protein under a variety of conditions, such as under oxidative stress, under a spiral and coccoid form, or even from a clinical strain (38, 39). Also, as mentioned above, it has been found that antibiotic resistance in H. pylori infection is increasing from year to year worldwide. Additionally, gastritis caused by H. pylori infection is an important first cause in promoting tissue transformation, cellular alterations, and cell morphological changes until it eventually becomes cancerous. However, only a few studies have been conducted of the proteome analysis of H. pylori-associated gastritis (40–42). Even fewer studies were found of the proteome analysis of antibiotic-sensitive and antibiotic-resistant H. pylori (25, 26, 43), whereas no proteomic studies were found of antibiotic-sensitive and antibiotic-resistant H. pylori-associated gastritis strains. Therefore, a study describing protein secreted from antibiotic-sensitive strains compared with the antibiotic-resistant clinical strains (C_R, M_R, and C/M_R) that have been associated with H. pylori-associated gastritis is still necessary. This information would provide evidence of the secreted proteins associated with H. pylori-resistant and -sensitive strains and be a notable benefit for the understanding of the resistance mechanism.
In the present study, the secreted proteins from antibiotic-sensitive (S_S) and antibiotic-resistant (C_R, M_R and C/M_R) H. pylori-associated gastritis strains were examined using proteomic tools via in-solution digestion and quantitative analysis through nano-LC-MS/MS. A Venn diagram of the identified protein and prediction of protein to protein, including chemical and drug interactions, was created through the STITCH database. This report is the first to demonstrate the proteomic profiles of secreted proteins from S_S, C_R, M_R, and C/M_R H. pylori-associated gastritis strains.
RESULTS
Protein identification from MS/MS.
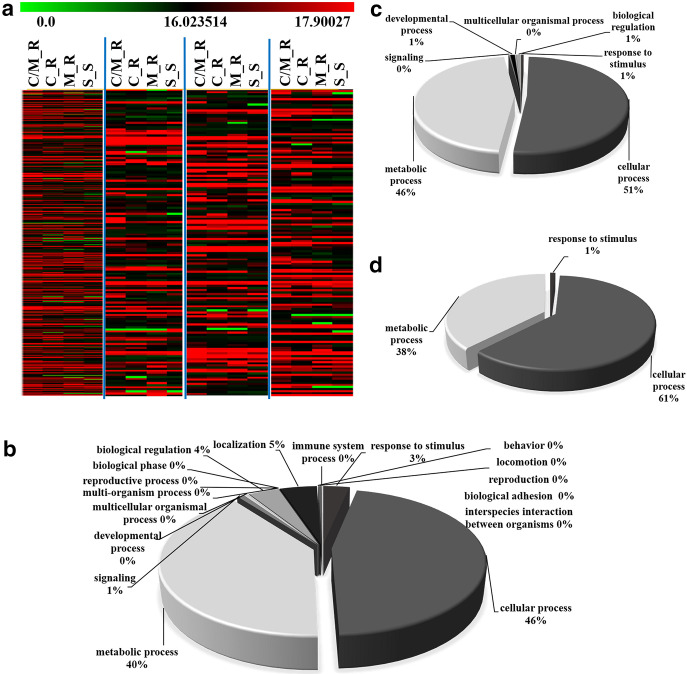
The aim of this study was to compare the differential expression of the protein secreted from H. pylori-associated gastritis strains among four antibiotic response groups (C_R, M_R, C/M_R, and S_S strains). The decisive parameter was achieved using the intensity of the maximum protein represented as log2 intensities, which was indicated by DeCyder (DeCyder MS; GE Healthcare, Amersham, UK). The data were filtered to show different expression changes with a statistically significant value of only P < 0.05 (one-way analysis of variance [ANOVA] and standard t test) as mentioned above. From this criterion, 592 differentially expressed proteins were detected from nano-LC-MS/MS analysis, 583, 582, 590, and 578 of which were identified from C_R, M_R, C/M_R, and S_S samples, respectively. All of the differentially expressed proteins were imported into PANTHER (http://pantherdb.org/) for biological process annotation. These proteins are involved in various cellular processes (Fig. 1b), such as in the cellular process (46%), the metabolic process (40%), process localization (5%), etc. Heat map analyses of 583, 582, 590, and 578 different proteins were also shown, which gave a significant overall picture of the up- and downregulated proteins in each of the strains (Fig. 1a). In addition, biological process annotation of the 23 overlapping proteins from the four strains and 9 identified proteins from the S_S strains were also shown (Fig. 1c and d, respectively). The results showed that the secreted proteins consisted predominantly of a group of proteins involved in cellular processes and metabolic processes.
FIG 1.
Heat map analysis (a) and biological function of 592 differentially expressed proteins (b), 23 overlapping proteins from the four strains (c), and 9 identified proteins from the S_S strains (d). (a) A total of 583, 582, 590, and 578 different proteins are displayed as a fold change in heat map analyses. Biological function is based on the PANTER database classifications of (b) 592 secreted proteins, (c) 23 overlapping proteins and (d) 9 identified proteins from the S_S strains. These classifications demonstrate the first two functions—the cellular process (46%, 51%, and 61%, respectively) and the metabolic process (40%, 46%, and 38%, respectively).
Group of the secreted proteins by the Venn diagram.
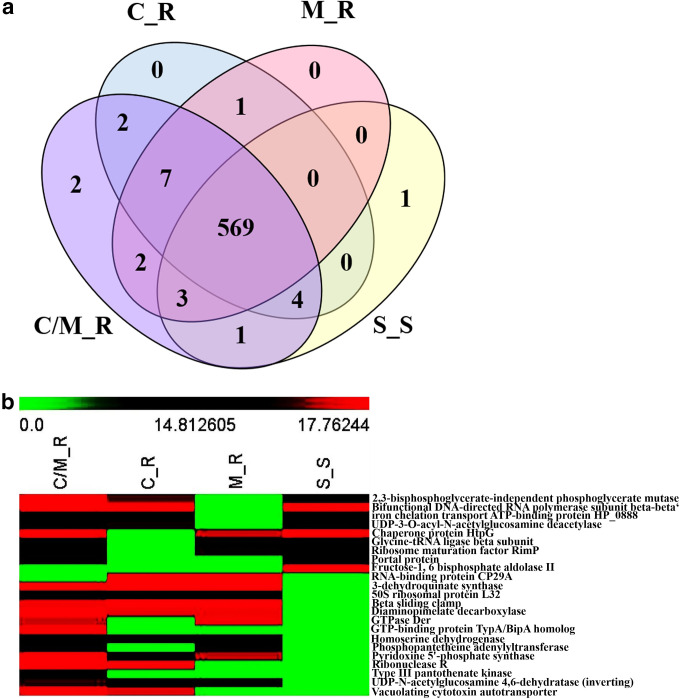
From the distribution of the proteins as shown by the Venn diagram, the 569 overlapping proteins from the four groups were found (Fig. 2a). On the other hand, 23 proteins overlapping between each group were observed at the intersection of each group. Alternatively, when considering the secreted proteins found in the S_S strain, nine proteins (numbers 1 to 9 in Table 1) were identified (2,3-bisphosphoglycerate-independent phosphoglycerate mutase, bifunctional DNA-directed RNA polymerase subunit beta-beta′, UDP-3-O-acyl-N-acetylglucosamine deacetylase, iron chelation transport ATP-binding protein HP_0888, chaperone protein HtpG, glycine-tRNA ligase beta subunit, ribosome maturation factor RimP, portal protein, and fructose-1, 6 bisphosphate aldolase II [FBPAII]). Among these proteins, the first four and next three proteins (numbers 1 to 4 and numbers 5 to 7 in Table 1) were found explicitly in the S_S and C/M_R strains, but not in M_R or in C_R strains, respectively. The next protein (number 8 in Table 1), meanwhile, is specifically identified in the S_S and C/M_R strains, but not in the M_R or C_R strains. In particular, the last protein, fructose-bisphosphate aldolase (FBPAII), was found only in the S_S strain. Additionally, two unique proteins (GTP-binding protein TypA/BipA homolog and type III pantothenate kinase) and one unique protein (FBPAII) were found in C/M_R and S_S strains, respectively (numbers 22 to 23 and number 9, respectively, in Table 1). In contrast, no unique proteins were detected in the M_R and C_R strains (Table 1). Moreover, after heat map analysis of the 23 overlapping secreted proteins (Fig. 2b), C/M_R strains showed the most differentially expressed proteins, whereas less differential expression was found in S_S strains.
FIG 2.
Venn diagram of 592 differentially expressed proteins from (a) nano-LC-MS/MS analysis and (b) heat map of 23 overlapping proteins among C_R, M_R, C/M_R, and S_S strains. (a) A total of 583, 582, 590, and 578 unique proteins were identified from the C_R, M_R, C/M_R, and S_S samples, respectively. (b) A higher than average abundance of a protein (indicated by log2) is displayed in shades of red, whereas reduced abundance is displayed in shades of green. The heat map analysis revealed that FBPAII is the only secreted protein found exclusively in the S_S strain.
TABLE 1.
The overlapping secreted proteins differentially expressed in each group of antibiotic-resistant strains, as identified by nano-LC-MS/MS analysis and classification of the protein functions according to the UniProt database (http://www.uniprot.org/)
| Protein no. | GenBank accession no. | Protein name | Gene name | Function | Peptide sequence | MOWSE score | Log 2 abundance |
|||
|---|---|---|---|---|---|---|---|---|---|---|
| C/M_R | C_R | M_R | S_S | |||||||
| 1 | gi|123373726 | 2,3-bisphosphoglycerate-independent phosphoglycerate mutase (2,3-BPGM) | gpmI | Glucose catabolic process (GO:0006007); glycolytic process (GO:0006096) | VLIPSPK | 2.62 | 18.11 | 16.56 | 0 | 15.87 |
| 2 | gi|41017590 | Bifunctional DNA-directed rNA polymerase subunit beta-beta′ (RpoBC) | rpoB | Transcription, DNA-templated (GO:0006351) | VGAGQIIADGPSMDR | 4.6 | 19.98 | 19.66 | 0 | 18.75 |
| 3 | gi|14285572 | UDP-3-O-acyl-N-acetylglucosamine deacetylase | lpxC | lipid A biosynthetic process (GO:0009245) | EFALQK | 7.23 | 14.66 | 16.19 | 0 | 15.14 |
| 4 | gi|2492572 | Iron chelation transport ATP-binding protein HP_0888 | HP_0889 | Transmembrane ABC‐transporter components FecE, ion transport (GO:0006811); iron ion homeostasis (GO:0055072) | QMVLLAR | 4.54 | 14.89 | 14.29 | 0 | 14.47 |
| 5 | gi|118575192 | Chaperone protein HtpG | htpG | Protein folding (GO:0006457) | KTLELNPNHAILQK | 1.24 | 18.03 | 0 | 17.04 | 18.08 |
| 6 | gi|238064852 | Glycine-tRNA ligase beta subunit | glyS | Glycyl-tRNA aminoacylation (GO:0006426) | YFATFCQK | 2.18 | 15.22 | 0 | 13.04 | 13.23 |
| 7 | gi|6648012 | Ribosome maturation factor RimP | rimP | Ribosomal small subunit biogenesis (GO:0042274) | VEVKLINK | 2.08 | 15.32 | 0 | 16.10 | 14.67 |
| 8 | gi|426021105 | Portal protein | AC1LGJY | Head-to-tail connector in H. pylori bacteriophage KHP30 (GO:0099002) | QLLRLLAGLNDESLGMAVNR | 2.34 | 15.95 | 0 | 0 | 14.74 |
| 9 | gi|9789722 | Fructose-1, 6 bisphosphate aldolase II (FBPAII) | fba | Fructose 1,6-bisphosphate metabolic process (GO:0030388); glycolytic process (GO:0006096) | KFFSPAQLALK | 5.23 | 0 | 0 | 0 | 22.40 |
| 10 | gi|2492962 | 3-dehydroquinate synthase | aroB | Aromatic amino acid family biosynthetic process (GO:0009073); chorismate biosynthetic process (GO:0009423) | MQEILIPLKEK | 23.44 | 17.76 | 17.74 | 18.26 | 0 |
| 11 | gi|2500328 | 50S ribosomal protein L32 | rpmF | Translation (GO:0006412) | MAVPDRR | 3.47 | 14.12 | 15.71 | 15.67 | 0 |
| 12 | gi|3913505 | Beta sliding clamp | dnaN | DNA strand elongation involved in DNA replication (GO:0006271) | RELAGILMQFDQK | 2.89 | 17.44 | 18.71 | 18.67 | 0 |
| 13 | gi|8134398 | Diaminopimelate decarboxylase | lysA | Lysine biosynthetic process via diaminopimelate (GO:0009089) | FGVEEK | 1.4 | 20.33 | 17.22 | 17.36 | 0 |
| 14 | gi|11132649 | Homoserine dehydrogenase | hom | Isoleucine biosynthetic process (GO:0009097); methionine biosynthetic process (GO:0009086); threonine biosynthetic process (GO:0009088) | AMLAYHRYELEQIAK | 8.18 | 14.32 | 14.02 | 15.55 | 0 |
| 15 | gi|226700973 | Pyridoxine 5′-phosphate synthase | pdxJ | Pyridoxine biosynthetic process (GO:0008615) | RHIQNEDVLR | 6.69 | 18.71 | 13.73 | 16.99 | 0 |
| 16 | gi|81341467 | UDP-N-acetylglucosamine 4,6-dehydratase (inverting)b | pseB | VLDTTNAK | 3.65 | 16.41 | 15.47 | 16.01 | 0 | |
| 17 | gi|7674337 | Ribonuclease Rb | rnr | EALQSNKDR | 6.11 | 19.16 | 20.50 | 0 | 0 | |
| 18 | gi|2499107 | Vacuolating cytotoxin autotransporter | vacA | Pathogenesis (GO:0009405) | NDKNESAK | 9.96 | 18.00 | 17.19 | 0 | 0 |
| 19 | gi|238058975 | GTPase Der | der | Ribosome biogenesis (GO:0042254) | NTSPKTLK | 3.29 | 17.01 | 0 | 17.50 | 0 |
| 20 | gi|226709008 | Phosphopantetheine adenylyltransferase | coaD | Coenzyme A biosynthetic process (GO:0015937) | MMQLATKSFK | 1.25 | 13.71 | 0 | 15.61 | 0 |
| 21 | gi|30316379 | RNA-binding protein CP29A | CP29A | Chloroplast rRNA processing (GO:1901259); cold acclimation (GO:0009631); mRNA processing (GO:0006397); RNA stabilization (GO:0043489) | SSYGSGSGSGSGSGSGNR | 4.11 | 0 | 19.37 | 18.80 | 0 |
| 22 | gi|8134781 | GTP-binding protein TypA/BipA homolog | typA | Ribosome biogenesis (GO:0042254) | CEEMGEGK | 5.8 | 22.84 | 0 | 0 | 0 |
| 23 | gi|81555831 | Type III pantothenate kinase | coaX | Coenzyme A biosynthetic process (GO:0015937) | SAKILEQPFK | 4.63 | 15.47 | 0 | 0 | 0 |
MOWSE, molecular weight search.
Denotes a protein not previously reported as the biological function.
Protein-protein interactions.
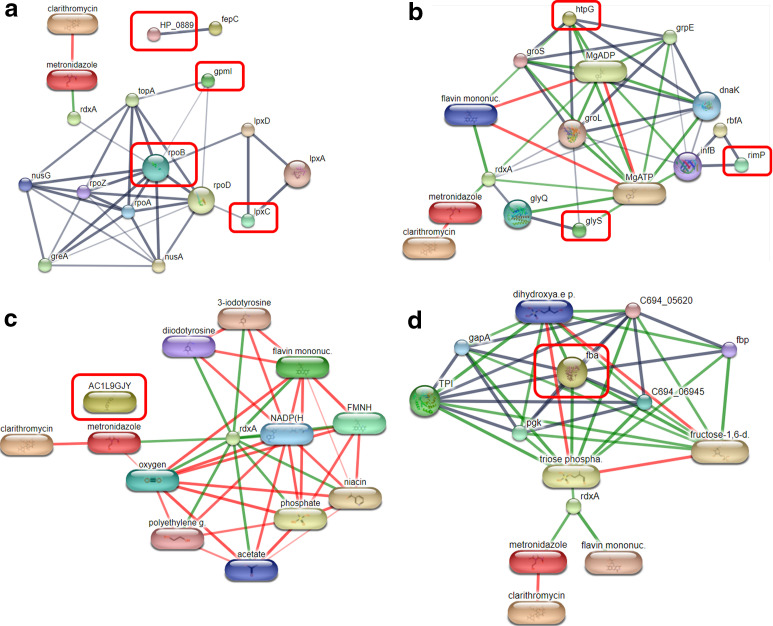
Since we are interested in identifying proteins secreted by antibiotic-susceptible strains (numbers 1 to 9 in Table 1), a network of nine proteins and other known proteins, as well as two antibiotics, Cla and Mtz, was generated using the STITCH database (Fig. 3). The STITCH prediction of 4 proteins secreted from the S_S strains, but not in the M_R strains (number 1 to 4 in Table 1), is shown in Fig. 3a. The interaction of 2,3-BPGM (2,3-bisphosphoglycerate-independent phosphoglycerate mutase) (gpmI) and UDP-3-O-acyl-N-acetylglucosamine deacetylase (lpxC) with bifunctional DNA-directed RNA polymerase subunit beta-beta′ (rpoB) with both of the antibiotics via oxygen-insensitive NADPH nitroreductase (rdxA) was predicted. Meanwhile, the three proteins were secreted by the S_S strain but not by the C_R strain (numbers 5 to 7 in Table 1), and the chaperone proteins HtpG (htpG), glycine-tRNA ligase beta subunit (glyS), and ribosome maturation factor RimP (rimP) were also associated with both of the antibiotics via rdxA (Fig. 3b). In addition, the STITCH analysis predicts that the links are interconnected through other proteins found in the 569 secreted proteins identified in the study (e.g., 60-kDa chaperonin [groL], glycine-tRNA ligase alpha subunit [glyQ], ribosome-binding factor A [rbfA], and translation initiation factor IF-2 [infB]). Ultimately, fructose-1,6 bisphosphate aldolase II (fba), a unique protein from the S_S strain (number 9 in Table 1), was found to be linked with trios phosphate, which was directly related to the two antibiotics (Fig. 3d). This interaction also occurs through rdxA. Moreover, our study found two proteins that were not associated with either antibiotic, HP_0888, an iron chelation transport ATP-binding protein (HP_0889) (Fig. 3a), and portal protein (AC1LGJY) (numbers 4 and 8, respectively, in Table 1) (Fig. 3c). Based on the predictive results from the STITCH database of nine proteins secreted by the S_S strain, the antibiotic association was predicted directly through the bifunctional DNA-directed RNA polymerase subunit beta-beta′ (rpoB) and fructose-1,6 bisphosphate aldolase II (fba). To verify the differential expressions of rpoB and fba, their mRNA levels were quantified using quantitative real-time reverse transcription-PCR (RT-PCR) analysis. The results are shown in the next session.
FIG 3.
The interaction of the chemical and proteins network of 9 proteins secreted by antibiotic-susceptible strains (numbers 1 to 9 in Table 1) and other known proteins, including clarithromycin and metronidazole, by STITCH 4.0. Protein-protein interactions are shown in gray, chemical-protein interactions are shown in in green, and interactions between chemicals are shown in in red. (a) Interaction between four secreted proteins from the S_S strains but not the M_R strains (numbers 1 to 4 in Table 1). (b) Interaction between three secreted proteins from the S_S strains but not the C_R strains (numbers 5 to 7 in Table 1). (c) Interaction between 1 secreted protein from S_S strains but not from the M_R and C_R strains (number 8 in Table 1). (d) Interaction between one unique protein found only in the S_S strain (number 9 in Table 1).
Quantitative real-time RT-PCR of rpoB and fba.
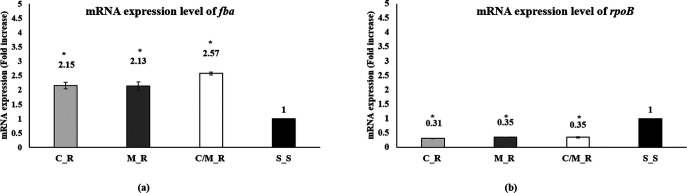
The mRNA expression level of rpoB and fba was analyzed using RT-PCR (their primers are shown in Table 2). Using the S_S strain expression level as a reference, the rpoB mRNA expression was significantly downregulated across all antibiotic-resistant strains (Fig. 4b). Meanwhile, a 2-fold increase in the fba mRNA expression levels in all three antibiotic-resistant strains (C_R, M_R, and C/M_R) was observed (Fig. 4a).
TABLE 2.
Oligonucleotide primers for fba and rpoB genes
| Name | Product size (bpb) | Directiona | 5′→3′ | Length |
|---|---|---|---|---|
| Fructose-bisphosphate aldolase (fba) | 161 | F | GGTGGGGGCGTTTAATTTCG | 20 |
| R | AGCGTTCGCACATGGTTTTC | 20 | ||
| Bifunctional DNA-directed RNA polymerase subunit beta-beta′ (rpoB) | 190 | F | ATATGCGCTACAGGAGTGGC | 20 |
| R | AACGAGACGGCTTGTTTTGC | 20 |
F, forward; R, reverse.
bp, base pair.
FIG 4.
mRNA expression pattern of fba and rpoB. (a and b) The fold change values in mRNA expression of (a) fba and (b) rpoB were analyzed by qRT-PCR from the four strains used in this study; the C_R, M_R, C/M_R, and S_S samples (*, P < 0.05, versus S_S as the control). Error bars indicate the standard deviation values from three independent experiments. The fold changes represented in this figure were calculated by using the expression level at the S_S strains as a reference.
DISCUSSION
It is well known that an untreated H. pylori infection is the major cause of chronic gastritis, a condition that can develop into gastric cancer. An earlier H. pylori eradication by triple therapy including two antibiotics (Cla and Mtz) and a proton-pump inhibitor is the treatment of choice for preventing incidences of gastric cancer. The current prevalence of H. pylori infection that is C_R, M_R, and C/M_R has continued at a higher rate in recent years (5, 44, 45). To date, there is growing interest in studies of proteins secreted by clinical strains of H. pylori because of their direct contact with tissues and because they may mediate important interactions between the pathogen and the host (46–48). However, there are limited reports of studies comparing proteins secreted by antibiotic-susceptible and antibiotic-resistant H. pylori clinical strains (25, 26, 43). From our point of view, if we identify proteins secreted only from S_S compared to those from antibiotic-resistant strains, the results will be useful in developing new drug targets or new serological tools for better diagnosis of H. pylori infection. Therefore, this study aims to investigate the new candidate proteins secreted from S_S and to compare them to those secreted from C_R, M_R, and C/M_R H. pylori-associated gastritis strains by the proteomic approach (in-solution tryptic digestion coupled with nano-LC-MS/MS).
A triple replication of samples was decided upon in order not only to minimize biological and technical errors, but also to limit the possibility of missing values that might have occurred during the experiments. Our results from the Venn diagram show that 569 different proteins were identified in all of the strains. Of these, UreI and HopB, which are inner membrane and outer membrane protein markers, were not detected (49). The absence of these markers indicated that the procedure used in this study minimized the contamination from both inner membrane proteins and outer membrane proteins. Moreover, several proteins have been previously reported as H. pylori-secreted proteins. Included among these were the flagella component proteins (flagellin A, flagellin B, flagella P-ring protein, flagellar hook protein FlgE, etc.), antioxidant enzymes (superoxide dismutase, catalase, thioredoxin reductase, etc.), cag-PAI proteins (CAG pathogenicity island proteins 12, 13, and 23), plasminogen-binding protein PgbA, carbonic anhydrase flavodoxin FldA, and VacA (50, 51). In addition, the results of this study revealed more than 500 secreted proteins, compared to 26 secreted proteins as published by Bumann et al. (51), which is the original publication we refer to in the experimental methods. The difference may be the result of differences in protein preparations. We prepared our sample with in-solution digestion, which not only has a higher throughput than in-gel digests, but can also identify all peptides in solution (52), although not all of the secreted proteins were found by the original researchers. In addition to differences in the experimental methods mentioned above, this variation may be the result of the genetic diversity of the H. pylori clinical strain (53). Although a small sample size (n = 3 per strain) may be a limitation in this study, these findings ultimately show that our experimental procedures are being performed to determine the protein secreted by H. pylori bacteria.
The biological activity of both the 592 secreted proteins and the 23 overlapping proteins, including 9 secreted proteins of the S_S strain, was identified through a protein database search. The results showed that its primary function was related to cellular processes (46%, 51%, and 61%, respectively) and metabolic processes (40%, 46%, and 38%, respectively) (Fig. 1b to d), where these functional proteins are most often found in the cytoplasm. The question of how the nonclassical secretion protein is secreted from the cytoplasm of the cell has been extensively studied. From the evidence, it includes the activation of a cellular lysis mechanism through the adhesion protein group, the attack of a membrane autolysin pathway, and the change in osmotic stress from the environment, as well as the activation of channels from antibiotic treatment, which then increases permeabilization of the membranes and the formation of outer membrane vesicles (OMVs) (54, 55). Recently, OMVs have been described as a releasing mechanism that plays a role in the process of delivering virulence factors of Gram-negative bacteria, including H. pylori. Well-known H. pylori proteins that are exported via OMVs include vacuolating cytotoxin (VacA) (56). In addition, the cell secretion system responsible for CagA transport from the cytoplasm to enable an interaction with host cells is identified as type IV secretion system (T4SS)-dependent (57, 58). It may be possible to hypothesize that the H. pylori-associated gastritis strains in this study secrete proteins by mediation through the generation of OMVs and/or a T4SS-dependent secretion system. However, the exact mechanisms and detailed information need to be examined further.
The interaction network of the secreted proteins, as well as Cla and Mtz via the STITCH database, predicted an interaction of 2,3-bisphosphoglycerate-independent phosphoglycerate mutase (gpmI) and UDP-3-O-acyl-N-acetylglucosamine deacetylase (lpxC) with the bifunctional DNA-directed RNA polymerase subunit beta-beta′ (rpoB) in S_S strains, although no such interaction was predicted within the M_R strains (Fig. 3a). The detection of RNA polymerase in the secreted protein was consistent with the previous report (59). Moreover, it is well known that the rpoB mutations play an important role in the resistance to rifampicin, a third-line antibiotic used in the eradication of H. pylori (60). However, Nishizawa et al. reported on the rpoB mutations in rifampicin-sensitive H. pylori clinical strains as well (61). Additionally, synonymous mutations for rpoB were also reported as an association with antibiotic resistance against several different antibiotic medications (62, 63). This information might support our results, since we found rpoBC in the secreted proteins from S_S and C_R, as well as C/M_R H. pylori strains. It is, however, not identified from M_R strains (Table 1). Additionally, by qRT-PCR analysis, a downregulation of rpoB from all antibiotic-resistant strains, including M_R strains, was observed (Fig. 4b). Moreover, it is normally a cytoplasmic protein, but was previously reported as a fibronectin-binding protein in group B streptococci and MUC7-binding adhesion in Streptococcus gordonii (64, 65). Nonetheless, there has been no previous evidence reporting any detection of rpoBC in secreted proteins from H. pylori S_S, although as based on our findings. It is possible that this secreted protein may be associated with dual resistance or a second antibiotic resistance mechanism, as well as Mtz susceptibility. However, to summarize this hypothesis, rpoB cloning to detect its mutation and a Western blot analysis of this protein from all strains are going to be required, together with evidence for a total amino acid sequence (to determine whether it has any mutations between any strains) and a three-dimensional structure prediction.
In addition, the other interesting secreted protein is FBPAII. FBPAII in H. pylori played a role of primarily catalyzing the condensation reaction to convert substrates such as pyruvate back into sugars, which are the essential substrates for nucleic acid and peptidoglycan synthesis (66). Beyond its cytoplasmic enzyme that catalyzed glycolytic/gluconeogenesis, its plasminogen-binding proteins and host cell adhesion have been recently reported in Paracoccidioides and Neisseria meningitides, respectively (67, 68). Apart from its role in hosting plasminogen-binding proteins and host cell adhesion, FBPA class II was also studied as (i) a protein vaccine candidate for Streptococcus pneumoniae (69), (ii) an immunogenic surface target for Listeria genus detection (70), and (iii) a promising target of new antibiotics (71). Moreover, its function as a transcriptional regulator has been recently reported in Francisella novicida, as described by Ziveri et al. (72). Based on our evidence, this protein was secreted only from S_S (Table 1). The results thus greatly promote FBPAII as a new antibiotic target for the treatment of Helicobacter pylori, as reported by Fonvielle et al. (73). In contrast, qRT-PCR analyses showed an upregulation of the fba gene in all of the antibiotic-resistant strains (Fig. 4a). It indicated that the absence of any release was not the result of decreased expression. It may be possible, instead, to explain this paradoxical event as a result of a delay in protein synthesis, as described by Liu et al. (74). In addition, it may also be explained by the fact that the correlation of cellular protein levels with mRNA levels is not 100% interrelated but depends on the process of transcription, postlocalization, and protein degradation, as previously reviewed by Vogel and Marcotte (75). To account for these inconsistent results, a quantitative proteomic analysis of the cell lysate using a protein expression-measurement improvement technique, such as SILAC, should be performed (76). While the links of the exact molecular mechanisms of FBPAII in H. pylori-susceptible strains still need to be investigated further, our results might be expanding our knowledge of this protein in H. pylori-associated gastritis strains.
Lastly, among the nine proteins secreted from the S_S strains, two identified proteins, iron chelation transport ATP-binding protein HP_0888 and portal protein (head-to-tail connector gp8), showed no association with Cla and Mtz from the STITCH prediction (Fig. 3a and c). HP0888, also known as the transmembrane ABC‐transporter component FecE, was previously reported for its ability to provoke an immune response (77). If it is proven to be present in the blood during infection or is identified for its antigenic pattern with patient serum as previously described by Kimmel et al. (78), HP0888 might become a new serological marker for antibiotic susceptibility testing. The portal protein, on the other hand, is the protein that plays a critical role for the head-to-tail connector in H. pylori bacteriophage KHP30. Recently, various studies have shown that the existence of the KHP30 phage bacteria is associated with H. pylori genetic diversity (79–81). This association may support the explanation of the genetic variants of the H. pylori clinical strains used in this study.
In summary, this study reports the use of a proteomic tool to explore the secreted protein profile from antibiotic-sensitive and antibiotic-resistant Helicobacter pylori-associated gastritis strains. The results obtained from this study supply fundamental information and provide an effective tool to expand knowledge about the protein secreted by this gastrointestinal (GI)-infecting bacterium. To our knowledge, this report is the first to have elucidated the fact that FBPAII and rpoBC are two newly secreted proteins identified in S_S strains. They are both of interest in our further study, as we seek a new target of Helicobacter pylori eradication. In addition, HP0888 may be useful for serological markers; however, the validity of the antigen pattern should be checked beforehand.
MATERIALS AND METHODS
H. pylori strain and culture conditions.
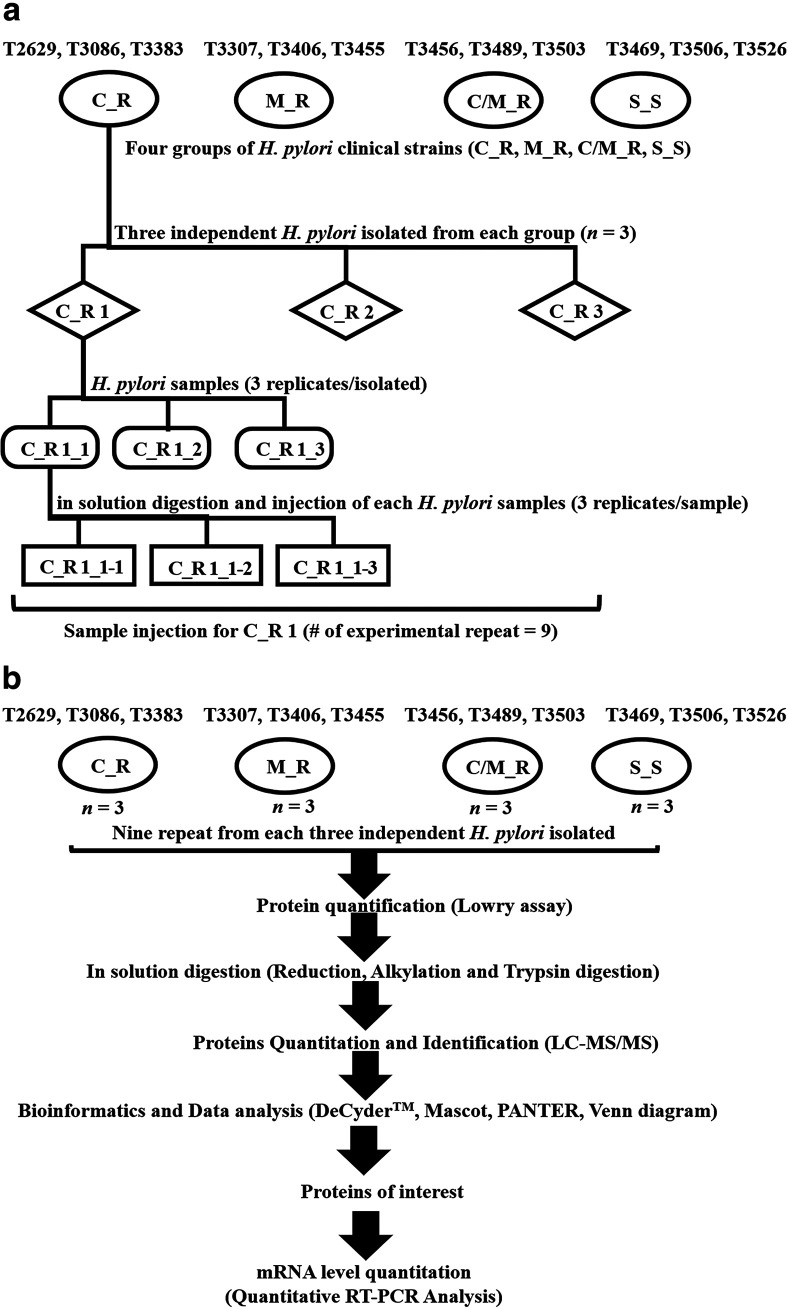
The H. pylori strains used in this study were T2629, T3086, and T3383 (C_R strains), T3307, T3406, and T3455 (M_R strains), T3456, T3489, and T3503 (C/M_R strains), and T3456, T3489, and T3503 (S_S strains). They were all isolated from patients with gastritis (4 males and 8 females). The average age of each group was 50.7, 56, 45.7, and 51.3 years, respectively (Table 3). They were kindly provided from the clinical stock culture of the Thammasat University Center of Excellence in Digestive Diseases. Each of the strains was isolated from three antral gastric biopsy specimens from each patient as previously reported (82, 83). In brief, after homogenization of the biopsy specimen in the saline solution, H. pylori was inoculated onto selective medium as a brain heart infusion (BHI) agar (Becton, Dickinson, Sparks, MD, USA). The plates were grown under microaerophilic conditions (5% O2, 10% CO2, and 85% N2) for 5 to 7 days at 37°C. The whole H. pylori-like colonies were subcultured in a BHI agar supplemented with 7% horse blood (Nippon Biotest, Tokyo, Japan) without antibiotics. H. pylori was confirmed by Gram staining, microscopic identification, and assay of urease, catalase, and oxidase activity. To identify the MICs of antibiotic susceptibility, viable H. pylori colonies were suspended in Mueller-Hinton II broth (Becton, Dickinson) with 7% horse blood. The culture suspension equivalent to a 2 McFarland standard was applied to inoculate the plates. They were categorized as susceptible or resistant to antibiotics based on the EUCAST recommendations, i.e., Cla (R > 0.5 μg/ml) and Mtz (R > 8 μg/ml) (84). Isolated strains were then collected and stored at −80°C. After ethics approval from the Research Ethics Subcommittee in People Thammasat University Series 1 (approval number 236/2019), a triplicate set of H. pylori samples (numbers C_R 1-3, M_R 4-6, C/M_R 7-9, and S_S 10–12,) was independently restored in BHI broth and incubated at 37°C for 3 days. Before using the strains for a proteomic study, they were also microscopically confirmed and analyzed using all specific enzymes (urease, catalase, and oxidase activity), as well as being tested with antibiotics as susceptible or drug-resistant strains according to EUCAST recommendations, as described previously (Fig. 5a). All procedures were performed in a class II biosafety cabinet (BSC), and the use of safety equipment was in accordance with the guidelines and regulations for standard and special microbiological practices for biosafety level II. This study was initiated after approval from Thammasat University’s Institution Biosafety Committee (IBC 126/2019). A schematic overview of the sample preparation for LC/MS-MS analysis in this study is shown in Fig. 5b.
TABLE 3.
Clinical and demographic data of the patient samples with endoscopic diagnosis of gastritis
| Antibiotic susceptibility | Code | Sex | Age |
|---|---|---|---|
| C_R | T2629 | F | 61 |
| T3086 | F | 49 | |
| T3383 | M | 42 | |
| M_R | T3307 | F | 68 |
| T3406 | F | 50 | |
| T3455 | M | 50 | |
| C/M_R | T3456 | F | 51 |
| T3489 | M | 33 | |
| T3503 | F | 53 | |
| S_S | T3469 | F | 58 |
| T3506 | F | 56 | |
| T3526 | M | 40 |
FIG 5.
(a and b) Schematic diagram of bacterial strain cultures with C_R clinical strain as an example (a) and overview of the experimental workflow in this study (b). The number of experimental repeats in this study was 9 for each of the H. pylori samples (numbers C_R 1–3, M_R 4-6, C/M_R 7-9, and S_S 10-12). The oval enclosures represent H. pylori clinical strains (C_R, M_R, C/M_R, and S_S); the diamond enclosures represent the patient number (e.g., C_R 1 = T2629, C_R 2 = T3086, C_R 2 = T3383); the rounded rectangular and rectangular enclosures represent the triplicate in each experimental workflow.
Secreted protein preparation for the proteome study.
The proteins secreted by each strain of H. pylori were prepared as previously described (51). In summary, after growing the selected sample from each of the strains in BHI broth overnight at 37°C at 150 rpm in a microaerobic atmosphere until reaching an optical density at 600 nm (OD600) of 0.5 to 1, the bacteria were recovered by centrifugation and then washed with BHI. The bacteria were then inoculated into a new BHI broth culture (triplicate for each sample, 20 ml each) until an OD600 of 0.01 was reached. The bacterium sample was then grown at 37°C and 150 rpm for 20 to 24 h until the cultures typically reached the mid-exponential growth phase with an OD600 of 0.3 to 0.5. The cultures were then collected and separated into two parts, a supernatant and an H. pylori cell pellet, by centrifugation at 5,000 × g for 10 min (4°C). The supernatant was subsequently centrifuged at 10,000 × g for 10 min (4°C) and filtered through a 0.2-µm-pore-size membrane filter to remove cell debris. The H. pylori cell pellets were then used for RNA isolation and reverse transcription to quantify the genes of interest by a real-time PCR.
Protein determination.
The protein concentration was determined with a Lowry protein assay with bovine serum albumin (BSA) as a standard (85). In summary, 5 µl of diluted sample (1:25) or standard (BSA, 2 to 10 mg/ml) was mixed with 0.2 ml of freshly prepared alkaline copper solution (0.4% CuSO4.7H20 in tartaric acid, 5% SDS, 0.8 M NaOH, and 20% sodium carbonate) in a separate 96-well microtiter plate. After incubation at room temperature for 30 min, 0.05 ml 20% Folin-Ciocalteu phenol reagent was added, mixed vigorously, and then allowed to stand at room temperature in the dark for 30 min. Absorbance at 690 nm was read with a microplate reader (Rayto Life and Analytical Sciences Co., Ltd., Shenzhen, China).
In-solution digestion.
After determining the protein concentration, protein digestion was carried out via the in-solution method with a minor modification as described previously (86, 87). In summary, after resuspending 50 μg of prepared protein sample in 10 µl of 10 mM AMBIC solution (NH4HCO3 in double-distilled water [ddH2O]) (Sigma, USA); 5 mM dithiothreitol (DTT) (GE Healthcare, UK) in AMBIC solution was then added and incubated at 60°C for 1 h to cleave disulfide bonds. After allowing the sample to settle to an ambient temperature, 15 mM freshly prepared iodoacetamide (GE Healthcare, UK) was added to the AMBIC solution and incubated at room temperature in the dark for 45 min. Trypsinization was achieved by adding 20 μl of sequencing-grade trypsin (1:20 wt/wt) (Promega, Walldorf, Germany) to the sample and incubated overnight at 37°C. The tryptic peptides were dried (SpeedVac system; Thermo Fisher) and kept at −20°C until use.
Nano-liquid chromatography and tandem mass-spectrophotometry (nano-LC-MS/MS) analysis.
After suspending the tryptic peptides with 0.1% formic acid (FA) (Merck, Germany), 1 µl of sample was injected in triplicate into an ion-trap mass spectrometer (HCT Ultra ion trap; Bruker Daltonics, Germany) coupled to a nano-LC system (Ultimate 3000 LC system; Thermo Fisher Scientific, Waltham, MA, USA). The peptide mixture for each sample was fractionated by using a reverse-phase high-performance liquid-chromatography column (Acclaim PepMap 100 Å, 75 µm by 5 cm; Thermo Scientific, UK, and PepSwift monolithic trap column 200 µm by 5 cm; Thermo Scientific, UK) coupled to an ion-trap mass spectrometer (HCT Ultra ion trap; Bruker Daltonics). Peptides were eluted with a mobile phase consisting of a linear gradient of 4% to 70% buffer B (80% acetonitrile, 19.9% water, and 0.1% formic acid [vol/vol/vol]) in buffer A (99.9% water and 0.1% formic acid [vol/vol/vol]) over a 20-min period. Mass spectra (MS) and MS/MS spectra were obtained in the data-dependent acquisition (DDA) mode over a range (m/z) of 400 to 1,500 (MS) and 200 to 2,800 Hz (MS/MS).
Bioinformatics and data analysis.
The protein intensity was measured based on the peptide MS signal of the individual LC-MS. Data were analyzed with the DeCyder MS 2.0 differential analysis instrument (GE Healthcare, Amersham, UK) (88, 89). The highest-intensity control and log2 intensity were selected and calculated with the DeCyder MS program as a default. An average abundance ratio of more than 2-fold was determined as an overexpressed protein with a significant standard t test and one-way ANOVA P value of <0.05. All MS/MS spectra from the DeCyder MS analysis were searched against the NCBI protein databases (https://www.ncbi.nlm.nih.gov/) with Helicobacter pylori proteins (2,306 sequences; downloaded on 19 December 2019) by using the Mascot software search engine tool (Matrix Science, London, UK) with the following parameters: fixed modification of carbamidomethyl (C), a variable modification of oxidation (M), the peptide charge state (1+, 2+, and 3+), a peptide mass tolerance ±1.2 Da, and a fragment mass toleranceof ±0.6 Da. To identify matching peptides, these spectra were then searched against the NCBI protein databases (https://www.ncbi.nlm.nih.gov/) with the Helicobacter pylori genome (2,306 sequences; downloaded on 19 December 2019) by using the Mascot software search engine tool (Matrix Science, London, UK). The identified proteins were normalized with 200 fmol bovine serum albumin (BSA) and filtered with a one-way ANOVA (P < 0.05). Protein function classification, annotations, and subcellular location were identified using the PANTHER web tool (http://pantherdb.org/), which classified based on the UniProt database (http://www.uniprot.org/).
Quantitative real-time reverse transcription-PCR of differential protein analysis.
To examine the different expressions of fba and rpoB from all of the H. pylori strains used in this study, a quantitative real-time reverse transcription-PCR (RT-PCR) technique was performed. DNA-free total RNA was isolated from the H. pylori cell pellet using a GenUP total RNA kit (Biotechrabbit, Germany) in accordance with the manufacturer’s instructions. Genomic DNA was physically eliminated by binding to the DNA filter without any DNase treatment during the isolation process. Reverse transcription was performed by adding 2 μg of total RNA, 1 μg of oligo(dT), and 10 μl of SuPrimeScript RT-PCR premix (2×) (GeNet Bio, South Korea), and the mixture was incubated at 42°C for 60 min. PCR amplification quantitation of both genes was performed with an Exicycler 96 real-time PCR instrument (Bioneer, South Korea). The sequences of the primers (Table 2) were designed from Primer3 software (https://bioinfo.ut.ee/primer3-0.4.0/). To ensure their specificity, the designed primers were then compared with an existing database at GenBank (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Each PCR mixture consisted of 1 μl of cDNA, 0.4 μl of forward and reverse primers, 5 μl of 5× HOT FIREPol EvaGreen qPCR master mix (Solis BioDyne), and 3.2 μl of nuclease-free water in a total volume of 10 μl. The PCR cycling conditions were as follows: 95°C for 5 min, followed by 40 cycles of denaturation at 95°C for 30 sec, annealing at 60°C for 30 sec, and elongation at 72°C for 30 s. Positive controls (DNA), a negative control (distilled water), and RT-negative controls (total RNA sample) were also included in each run. For each of the RNA extractions, measurements of gene expression were obtained in triplicate. The result of the qPCR was determined as the cycle threshold (CT) value. The relative expression for the qPCR results of both genes were normalized by the CT values of 16S RNA for each sample. The relative transcript levels of fba and rpoBC were calculated by 2−ΔΔCT (90). The data were analyzed using Student’s t test with a statistical significance of P < 0.05.
ACKNOWLEDGMENTS
We appreciate the Proteomics Research Laboratory, National Center for Genetic Engineering and Biotechnology (BIOTEC), for their assistance in using the reagents and proteomics instruments, particularly for the nano-LC-MS/MS and data analysis. We thank the Thammasat University Center of Excellence in Digestive Diseases for kindly providing all of the clinical stock culture used in this study.
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
We declare no competing interest.
Contributor Information
Panadda Rojpibulstit, Email: panadda@tu.ac.th.
Denise Monack, Stanford University.
REFERENCES
- 1.Peek RM Jr, Crabtree JE. 2006. Helicobacter infection and gastric neoplasia. J Pathol 208:233–248. doi: 10.1002/path.1868. [DOI] [PubMed] [Google Scholar]
- 2.Moss SF. 2017. The clinical evidence linking Helicobacter pylori to gastric cancer. Cell Mol Gastroenterol Hepatol 3:183–191. doi: 10.1016/j.jcmgh.2016.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang XY, Zhang PY, Aboul-Soud MA. 2017. From inflammation to gastric cancer: role of Helicobacter pylori. Oncol Lett 13:543–548. doi: 10.3892/ol.2016.5506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chiang TH, Chang WJ, Chen SL, Yen AM, Fann JC, Chiu SY, Chen YR, Chuang SL, Shieh CF, Liu CY, Chiu HM, Chiang H, Shun CT, Lin MW, Wu MS, Lin JT, Chan CC, Graham DY, Chen HH, Lee YC. 2020. Mass eradication of Helicobacter pylori to reduce gastric cancer incidence and mortality: a long-term cohort study on Matsu Islands. Gut 70:243–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Savoldi A, Carrara E, Graham DY, Conti M, Tacconelli E. 2018. Prevalence of antibiotic resistance in Helicobacter pylori: a systematic review and meta-analysis in world health organization regions. Gastroenterology 155:1372–1382.e1317. doi: 10.1053/j.gastro.2018.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuo YT, Liou JM, El-Omar EM, Wu JY, Leow AHR, Goh KL, Das R, Lu H, Lin JT, Tu YK, Yamaoka Y, Wu MS, Asian Pacific Alliance on Helicobacter andMicrobiota. 2017. Primary antibiotic resistance in Helicobacter pylori in the Asia-Pacific region: a systematic review and meta-analysis. Lancet Gastroenterol Hepatol 2:707–715. doi: 10.1016/S2468-1253(17)30219-4. [DOI] [PubMed] [Google Scholar]
- 7.Vilaichone RK, Quach DT, Yamaoka Y, Sugano K, Mahachai V. 2018. Prevalence and pattern of antibiotic resistant strains of Helicobacter pylori Infection in ASEAN. Asian Pac J Cancer Prev 19:1411–1413. doi: 10.22034/APJCP.2018.19.5.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu HH, Lai FP, Lo HY, Sheu BS, Yang YJ. 2019. Increasing antimicrobial resistance to clarithromycin and metronidazole in pediatric Helicobacter pylori infection in southern Taiwan: a comparison between two decades. Helicobacter 24:e12633. doi: 10.1111/hel.12633. [DOI] [PubMed] [Google Scholar]
- 9.WHO. 2017. WHO global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics. https://www.who.int/medicines/publications/WHO-PPL-Short_Summary_25Feb-ET_NM_WHO.pdf?ua=1.
- 10.Oleastro M, Menard A, Santos A, Lamouliatte H, Monteiro L, Barthelemy P, Megraud F. 2003. Real-time PCR assay for rapid and accurate detection of point mutations conferring resistance to clarithromycin in Helicobacter pylori. J Clin Microbiol 41:397–402. doi: 10.1128/jcm.41.1.397-402.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bina JE, Alm RA, Uria-Nickelsen M, Thomas SR, Trust TJ, Hancock RE. 2000. Helicobacter pylori uptake and efflux: basis for intrinsic susceptibility to antibiotics in vitro. Antimicrob Agents Chemother 44:248–254. doi: 10.1128/aac.44.2.248-254.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu ZQ, Zheng PY, Yang PC. 2008. Efflux pump gene hefA of Helicobacter pylori plays an important role in multidrug resistance. World J Gastroenterol 14:5217–5222. doi: 10.3748/wjg.14.5217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marques AT, Vitor JMB, Santos A, Oleastro M, Vale FF. 2020. Trends in Helicobacter pylori resistance to clarithromycin: from phenotypic to genomic approaches. Microb Genom 6:e000344. doi: 10.1099/mgen.0.000344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Olekhnovich IN, Goodwin A, Hoffman PS. 2009. Characterization of the NAD(P)H oxidase and metronidazole reductase activities of the RdxA nitroreductase of Helicobacter pylori. FEBS J 276:3354–3364. doi: 10.1111/j.1742-4658.2009.07060.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sisson G, Jeong JY, Goodwin A, Bryden L, Rossler N, Lim-Morrison S, Raudonikiene A, Berg DE, Hoffman PS. 2000. Metronidazole activation is mutagenic and causes DNA fragmentation in Helicobacter pylori and in Escherichia coli containing a cloned H. pylori RdxA(+) (nitroreductase) gene. J Bacteriol 182:5091–5096. doi: 10.1128/jb.182.18.5091-5096.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jeong JY, Mukhopadhyay AK, Akada JK, Dailidiene D, Hoffman PS, Berg DE. 2001. Roles of FrxA and RdxA nitroreductases of Helicobacter pylori in susceptibility and resistance to metronidazole. J Bacteriol 183:5155–5162. doi: 10.1128/jb.183.17.5155-5162.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goodwin A, Kersulyte D, Sisson G, Veldhuyzen van Zanten SJ, Berg DE, Hoffman PS. 1998. Metronidazole resistance in Helicobacter pylori is due to null mutations in a gene (rdxA) that encodes an oxygen-insensitive NADPH nitroreductase. Mol Microbiol 28:383–393. doi: 10.1046/j.1365-2958.1998.00806.x. [DOI] [PubMed] [Google Scholar]
- 18.Jenks PJ, Ferrero RL, Labigne A. 1999. The role of the rdxA gene in the evolution of metronidazole resistance in Helicobacter pylori. J Antimicrob Chemother 43:753–758. doi: 10.1093/jac/43.6.753. [DOI] [PubMed] [Google Scholar]
- 19.Kwon DH, Hulten K, Kato M, Kim JJ, Lee M, El-Zaatari FA, Osato MS, Graham DY. 2001. DNA sequence analysis of rdxA and frxA from 12 pairs of metronidazole-sensitive and -resistant clinical Helicobacter pylori isolates. Antimicrob Agents Chemother 45:2609–2615. doi: 10.1128/aac.45.9.2609-2615.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moore JM, Salama NR. 2005. Mutational analysis of metronidazole resistance in Helicobacter pylori. Antimicrob Agents Chemother 49:1236–1237. doi: 10.1128/AAC.49.3.1236-1237.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsugawa H, Suzuki H, Satoh K, Hirata K, Matsuzaki J, Saito Y, Suematsu M, Hibi T. 2011. Two amino acids mutation of ferric uptake regulator determines Helicobacter pylori resistance to metronidazole. Antioxid Redox Signal 14:15–23. doi: 10.1089/ars.2010.3146. [DOI] [PubMed] [Google Scholar]
- 22.Albert TJ, Dailidiene D, Dailide G, Norton JE, Kalia A, Richmond TA, Molla M, Singh J, Green RD, Berg DE. 2005. Mutation discovery in bacterial genomes: metronidazole resistance in Helicobacter pylori. Nat Methods 2:951–953. doi: 10.1038/nmeth805. [DOI] [PubMed] [Google Scholar]
- 23.Chua EG, Debowski AW, Webberley KM, Peters F, Lamichhane B, Loke MF, Vadivelu J, Tay CY, Marshall BJ, Wise MJ. 2019. Analysis of core protein clusters identifies candidate variable sites conferring metronidazole resistance in Helicobacter pylori. Gastroenterol Rep (Oxf) 7:42–49. doi: 10.1093/gastro/goy048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McAtee CP, Hoffman PS, Berg DE. 2001. Identification of differentially regulated proteins in metronidozole resistant Helicobacter pylori by proteome techniques. Proteomics 1:516–521. doi:. [DOI] [PubMed] [Google Scholar]
- 25.Smiley R, Bailey J, Sethuraman M, Posecion N, Showkat Ali M. 2013. Comparative proteomics analysis of sarcosine insoluble outer membrane proteins from clarithromycin resistant and sensitive strains of Helicobacter pylori. J Microbiol 51:612–618. doi: 10.1007/s12275-013-3029-5. [DOI] [PubMed] [Google Scholar]
- 26.Hanafi A, Lee WC, Loke MF, Teh X, Shaari A, Dinarvand M, Lehours P, Megraud F, Leow AH, Vadivelu J, Goh KL. 2016. Molecular and proteomic analysis of levofloxacin and metronidazole resistant Helicobacter pylori. Front Microbiol 7:2015. doi: 10.3389/fmicb.2016.02015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sugiyama N, Miyake S, Lin MH, Wakabayashi M, Marusawa H, Nishiumi S, Yoshida M, Ishihama Y. 2019. Comparative proteomics of Helicobacter pylori strains reveals geographical features rather than genomic variations. Genes Cells 24:139–150. doi: 10.1111/gtc.12662. [DOI] [PubMed] [Google Scholar]
- 28.Khodadadi E, Zeinalzadeh E, Taghizadeh S, Mehramouz B, Kamounah FS, Khodadadi E, Ganbarov K, Yousefi B, Bastami M, Kafil HS. 2020. Proteomic applications in antimicrobial resistance and clinical microbiology studies. Infect Drug Resist 13:1785–1806. doi: 10.2147/IDR.S238446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Link AJ, Eng J, Schieltz DM, Carmack E, Mize GJ, Morris DR, Garvik BM, Yates JR III.. 1999. Direct analysis of protein complexes using mass spectrometry. Nat Biotechnol 17:676–682. doi: 10.1038/10890. [DOI] [PubMed] [Google Scholar]
- 30.Aebersold R, Mann M. 2003. Mass spectrometry-based proteomics. Nature 422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 31.Eng JK, McCormack AL, Yates JR. 1994. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom 5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 32.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. 1999. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20:3551–3567. doi:. [DOI] [PubMed] [Google Scholar]
- 33.Catherman AD, Skinner OS, Kelleher NL. 2014. Top Down proteomics: facts and perspectives. Biochem Biophys Res Commun 445:683–693. doi: 10.1016/j.bbrc.2014.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y, Fonslow BR, Shan B, Baek MC, Yates JR III.. 2013. Protein analysis by shotgun/bottom-up proteomics. Chem Rev 113:2343–2394. doi: 10.1021/cr3003533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li N, Shaw AR, Zhang N, Mak A, Li L. 2004. Lipid raft proteomics: analysis of in-solution digest of sodium dodecyl sulfate-solubilized lipid raft proteins by liquid chromatography-matrix-assisted laser desorption/ionization tandem mass spectrometry. Proteomics 4:3156–3166. doi: 10.1002/pmic.200400832. [DOI] [PubMed] [Google Scholar]
- 36.Kim SC, Chen Y, Mirza S, Xu Y, Lee J, Liu P, Zhao Y. 2006. A clean, more efficient method for in-solution digestion of protein mixtures without detergent or urea. J Proteome Res 5:3446–3452. doi: 10.1021/pr0603396. [DOI] [PubMed] [Google Scholar]
- 37.Choksawangkarn W, Edwards N, Wang Y, Gutierrez P, Fenselau C. 2012. Comparative study of workflows optimized for in-gel, in-solution, and on-filter proteolysis in the analysis of plasma membrane proteins. J Proteome Res 11:3030–3034. doi: 10.1021/pr300188b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang CH, Chiou SH. 2011. Proteomic analysis of upregulated proteins in Helicobacter pylori under oxidative stress induced by hydrogen peroxide. Kaohsiung J Med Sci 27:544–553. doi: 10.1016/j.kjms.2011.06.019. [DOI] [PubMed] [Google Scholar]
- 39.Muller SA, Pernitzsch SR, Haange SB, Uetz P, von Bergen M, Sharma CM, Kalkhof S. 2015. Stable isotope labeling by amino acids in cell culture based proteomics reveals differences in protein abundances between spiral and coccoid forms of the gastric pathogen Helicobacter pylori. J Proteomics 126:34–45. doi: 10.1016/j.jprot.2015.05.011. [DOI] [PubMed] [Google Scholar]
- 40.Repetto O, Zanussi S, Casarotto M, Canzonieri V, De Paoli P, Cannizzaro R, De Re V. 2014. Differential proteomics of Helicobacter pylori associated with autoimmune atrophic gastritis. Mol Med 20:57–71. doi: 10.2119/molmed.2013.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang YN, Ding SG, Huang LH, Zhang J, Shi YY, Zhong LJ. 2011. Comparative proteome analysis of Helicobacter pylori clinical strains by two-dimensional gel electrophoresis. J Zhejiang Univ Sci B 12:820–827. doi: 10.1631/jzus.B1000445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Govorun VM, Moshkovskii SA, Tikhonova OV, Goufman EI, Serebryakova MV, Momynaliev KT, Lokhov PG, Khryapova EV, Kudryavtseva LV, Smirnova OV, Toropyguine IY, Maksimov BI, Archakov AI. 2003. Comparative analysis of proteome maps of Helicobacter pylori clinical isolates. Biochemistry (Mosc) 68:42–49. doi: 10.1023/A:1022189200944. [DOI] [PubMed] [Google Scholar]
- 43.Chen J, Ye L, Jin L, Xu X, Xu P, Wang X, Li H. 2018. Application of next-generation sequencing to characterize novel mutations in clarithromycin-susceptible Helicobacter pylori strains with A2143G of 23S rRNA gene. Ann Clin Microbiol Antimicrob 17:10. doi: 10.1186/s12941-018-0259-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Suzuki S, Esaki M, Kusano C, Ikehara H, Gotoda T. 2019. Development of Helicobacter pylori treatment: how do we manage antimicrobial resistance? World J Gastroenterol 25:1907–1912. doi: 10.3748/wjg.v25.i16.1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yousefi-Avarvand A, Vaez H, Tafaghodi M, Sahebkar AH, Arzanlou M, Khademi F. 2018. Antibiotic resistance of Helicobacter pylori in Iranian children: a systematic review and meta-analysis. Microb Drug Resist 24:980–986. doi: 10.1089/mdr.2017.0292. [DOI] [PubMed] [Google Scholar]
- 46.Tavares R, Pathak SK. 2017. Helicobacter pylori secreted protein HP1286 triggers apoptosis in macrophages via TNF-independent and ERK MAPK-dependent pathways. Front Cell Infect Microbiol 7:58. doi: 10.3389/fcimb.2017.00058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kao JY, Rathinavelu S, Eaton KA, Bai L, Zavros Y, Takami M, Pierzchala A, Merchant JL. 2006. Helicobacter pylori-secreted factors inhibit dendritic cell IL-12 secretion: a mechanism of ineffective host defense. Am J Physiol Gastrointest Liver Physiol 291:G73–G81. doi: 10.1152/ajpgi.00139.2005. [DOI] [PubMed] [Google Scholar]
- 48.Carlsohn E, Nystrom J, Karlsson H, Svennerholm AM, Nilsson CL. 2006. Characterization of the outer membrane protein profile from disease-related Helicobacter pylori isolates by subcellular fractionation and nano-LC FT-ICR MS analysis. J Proteome Res 5:3197–3204. doi: 10.1021/pr060181p. [DOI] [PubMed] [Google Scholar]
- 49.Kim N, Weeks DL, Shin JM, Scott DR, Young MK, Sachs G. 2002. Proteins released by Helicobacter pylori in vitro. J Bacteriol 184:6155–6162. doi: 10.1128/jb.184.22.6155-6162.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zanotti G, Cendron L. 2014. Structural and functional aspects of the Helicobacter pylori secretome. World J Gastroenterol 20:1402–1423. doi: 10.3748/wjg.v20.i6.1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bumann D, Aksu S, Wendland M, Janek K, Zimny-Arndt U, Sabarth N, Meyer TF, Jungblut PR. 2002. Proteome analysis of secreted proteins of the gastric pathogen Helicobacter pylori. Infect Immun 70:3396–3403. doi: 10.1128/iai.70.7.3396-3403.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Leon IR, Schwammle V, Jensen ON, Sprenger RR. 2013. Quantitative assessment of in-solution digestion efficiency identifies optimal protocols for unbiased protein analysis. Mol Cell Proteomics 12:2992–3005. doi: 10.1074/mcp.M112.025585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Blaser MJ, Berg DE. 2001. Helicobacter pylori genetic diversity and risk of human disease. J Clin Invest 107:767–773. doi: 10.1172/JCI12672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lekmeechai S, Su YC, Brant M, Alvarado-Kristensson M, Vallstrom A, Obi I, Arnqvist A, Riesbeck K. 2018. Helicobacter pylori outer membrane vesicles protect the pathogen from reactive oxygen species of the respiratory burst. Front Microbiol 9:1837. doi: 10.3389/fmicb.2018.01837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ebner P, Gotz F. 2019. Bacterial excretion of cytoplasmic proteins (ECP): occurrence, mechanism, and function. Trends Microbiol 27:176–187. doi: 10.1016/j.tim.2018.10.006. [DOI] [PubMed] [Google Scholar]
- 56.Chmiela M, Walczak N, Rudnicka K. 2018. Helicobacter pylori outer membrane vesicles involvement in the infection development and Helicobacter pylori-related diseases. J Biomed Sci 25:78. doi: 10.1186/s12929-018-0480-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Backert S, Tegtmeyer N, Fischer W. 2015. Composition, structure and function of the Helicobacter pylori cag pathogenicity island encoded type IV secretion system. Future Microbiol 10:955–965. doi: 10.2217/fmb.15.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Backert S, Tegtmeyer N. 2017. Type IV secretion and signal transduction of Helicobacter pylori caga through interactions with host cell receptors. Toxins (Basel) 9:115. doi: 10.3390/toxins9040115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lasserre JP, Beyne E, Pyndiah S, Lapaillerie D, Claverol S, Bonneu M. 2006. A complexomic study of Escherichia coli using two-dimensional blue native/SDS polyacrylamide gel electrophoresis. Electrophoresis 27:3306–3321. doi: 10.1002/elps.200500912. [DOI] [PubMed] [Google Scholar]
- 60.Hays C, Burucoa C, Lehours P, Tran CT, Leleu A, Raymond J. 2018. Molecular characterization of Helicobacter pylori resistance to rifamycins. Helicobacter 23:e12451. doi: 10.1111/hel.12451. [DOI] [PubMed] [Google Scholar]
- 61.Nishizawa T, Suzuki H, Matsuzaki J, Muraoka H, Tsugawa H, Hirata K, Hibi T. 2011. Helicobacter pylori resistance to rifabutin in the last 7 years. Antimicrob Agents Chemother 55:5374–5375. doi: 10.1128/AAC.05437-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hoeksema M, Jonker MJ, Brul S, Ter Kuile BH. 2019. Effects of a previously selected antibiotic resistance on mutations acquired during development of a second resistance in Escherichia coli. BMC Genomics 20:284. doi: 10.1186/s12864-019-5648-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lai CC, Chen CC, Lu YC, Chuang YC, Tang HJ. 2018. The clinical significance of silent mutations with respect to ciprofloxacin resistance inMRSA. Infect Drug Resist 11:681–687. doi: 10.2147/IDR.S159455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Beckmann C, Waggoner JD, Harris TO, Tamura GS, Rubens CE. 2002. Identification of novel adhesins from Group B streptococci by use of phage display reveals that C5a peptidase mediates fibronectin binding. Infect Immun 70:2869–2876. doi: 10.1128/iai.70.6.2869-2876.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kesimer M, Kilic N, Mehrotra R, Thornton DJ, Sheehan JK. 2009. Identification of salivary mucin MUC7 binding proteins from Streptococcus gordonii. BMC Microbiol 9:163. doi: 10.1186/1471-2180-9-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Doig P, de Jonge BL, Alm RA, Brown ED, Uria-Nickelsen M, Noonan B, Mills SD, Tummino P, Carmel G, Guild BC, Moir DT, Vovis GF, Trust TJ. 1999. Helicobacter pylori physiology predicted from genomic comparison of two strains. Microbiol Mol Biol Rev 63:675–707. doi: 10.1128/MMBR.63.3.675-707.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chaves EG, Weber SS, Bao SN, Pereira LA, Bailao AM, Borges CL, Soares CM. 2015. Analysis of Paracoccidioides secreted proteins reveals fructose 1,6-bisphosphate aldolase as a plasminogen-binding protein. BMC Microbiol 15:53. doi: 10.1186/s12866-015-0393-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tunio SA, Oldfield NJ, Berry A, Ala’Aldeen DA, Wooldridge KG, Turner DP. 2010. The moonlighting protein fructose-1, 6-bisphosphate aldolase ofNeisseria meningitidis: surface localization and role in host cell adhesion. Mol Microbiol 76:605–615. doi: 10.1111/j.1365-2958.2010.07098.x. [DOI] [PubMed] [Google Scholar]
- 69.Elhaik Goldman S, Dotan S, Talias A, Lilo A, Azriel S, Malka I, Portnoi M, Ohayon A, Kafka D, Ellis R, Elkabets M, Porgador A, Levin D, Azhari R, Swiatlo E, Ling E, Feldman G, Tal M, Dagan R, Mizrachi Nebenzahl Y. 2016. Streptococcus pneumoniae fructose-1,6-bisphosphate aldolase, a protein vaccine candidate, elicits Th1/Th2/Th17-type cytokine responses in mice. Int J Mol Med 37:1127–1138. doi: 10.3892/ijmm.2016.2512. [DOI] [PubMed] [Google Scholar]
- 70.Mendonca M, Moreira GM, Conceicao FR, Hust M, Mendonca KS, Moreira AN, Franca RC, da Silva WP, Bhunia AK, Aleixo JA. 2016. Fructose 1,6-bisphosphate aldolase, a novel immunogenic surface protein on Listeria species. PLoS One 11:e0160544. doi: 10.1371/journal.pone.0160544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Daher R, Coincon M, Fonvielle M, Gest PM, Guerin ME, Jackson M, Sygusch J, Therisod M. 2010. Rational design, synthesis, and evaluation of new selective inhibitors of microbial class II (zinc dependent) fructose bis-phosphate aldolases. J Med Chem 53:7836–7842. doi: 10.1021/jm1009814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ziveri J, Tros F, Guerrera IC, Chhuon C, Audry M, Dupuis M, Barel M, Korniotis S, Fillatreau S, Gales L, Cahoreau E, Charbit A. 2017. The metabolic enzyme fructose-1,6-bisphosphate aldolase acts as a transcriptional regulator in pathogenic Francisella. Nat Commun 8:853. doi: 10.1038/s41467-017-00889-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fonvielle M, Coincon M, Daher R, Desbenoit N, Kosieradzka K, Barilone N, Gicquel B, Sygusch J, Jackson M, Therisod M. 2008. Synthesis and biochemical evaluation of selective inhibitors of class II fructose bisphosphate aldolases: towards new synthetic antibiotics. Chemistry 14:8521–8529. doi: 10.1002/chem.200800857. [DOI] [PubMed] [Google Scholar]
- 74.Liu Y, Beyer A, Aebersold R. 2016. On the dependency of cellular protein levels on mRNA abundance. Cell 165:535–550. doi: 10.1016/j.cell.2016.03.014. [DOI] [PubMed] [Google Scholar]
- 75.Vogel C, Marcotte EM. 2012. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat Rev Genet 13:227–232. doi: 10.1038/nrg3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. 2002. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. MolCell Proteomics 1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 77.Spreng S, Gentschev I, Goebel W, Mollenkopf H, Eck M, Muller-Hermelink HK, Schmausser B. 2000. Identification of immunogenic antigens of Helicobacter pylori via the Escherichia coli hemolysin secretion system(1). FEMS Microbiol Lett 186:251–256. doi: 10.1111/j.1574-6968.2000.tb09113.x. [DOI] [PubMed] [Google Scholar]
- 78.Kimmel B, Bosserhoff A, Frank R, Gross R, Goebel W, Beier D. 2000. Identification of immunodominant antigens from Helicobacter pylori and evaluation of their reactivities with sera from patients with different gastroduodenal pathologies. Infect Immun 68:915–920. doi: 10.1128/iai.68.2.915-920.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Takeuchi H, Kira M, Konishi S, Uchiyama J, Matsuzaki S, Matsumura Y. 2018. Polymorphisms in the Helicobacter pylori NY43 strain and its prophage-cured derivatives. Microbiology (Reading) 164:877–882. doi: 10.1099/mic.0.000665. [DOI] [PubMed] [Google Scholar]
- 80.Vale FF, Lehours P. 2018. Relating phage genomes to Helicobacter pylori population structure: general steps using whole-genome sequencing data. Int J Mol Sci 19:1831. doi: 10.3390/ijms19071831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Uchiyama J, Takeuchi H, Kato S, Gamoh K, Takemura-Uchiyama I, Ujihara T, Daibata M, Matsuzaki S. 2013. Characterization of Helicobacter pylori bacteriophage KHP30. Appl Environ Microbiol 79:3176–3184. doi: 10.1128/AEM.03530-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Vilaichone RK, Mahacahai V, Tumwasorn S, Kachintorn U. 2011. CagA genotype and metronidazole resistant strain of Helicobacter pylori in functional dyspepsia in Thailand. J Gastroenterol Hepatol 26 Suppl 3:46–48. doi: 10.1111/j.1440-1746.2011.06652.x. [DOI] [PubMed] [Google Scholar]
- 83.Vilaichone RK, Ratanachu-Ek T, Gamnarai P, Chaithongrat S, Uchida T, Yamaoka Y, Mahachai V. 2016. Extremely high prevalence of metronidazole-resistant Helicobacter pylori strains in mountain people (Karen and Hmong) in Thailand. Am J Trop Med Hyg 94:717–720. doi: 10.4269/ajtmh.15-0449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.EUCAST. 2015. The European Committee on Antimicrobial Susceptibility Testing. Breakpoint tables for interpretation of MICs and zone diameters. Version 5.0. https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_5.0_Breakpoint_Table_01.pdf.
- 85.Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. 1951. Protein measurement with the Folin phenol reagent. J Biol Chem 193:265–275. doi: 10.1016/S0021-9258(19)52451-6. [DOI] [PubMed] [Google Scholar]
- 86.Losuwannarak N, Maiuthed A, Kitkumthorn N, Leelahavanichkul A, Roytrakul S, Chanvorachote P. 2019. Gigantol targets cancer stem cells and destabilizes tumors via the suppression of the PI3K/AKT and JAK/STAT pathways in ectopic lung cancer xenografts. Cancers (Basel) 11:2032. doi: 10.3390/cancers11122032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wongin S, Narkbunnam R, Waikakul S, Chotiyarnwong P, Aresanasuwan T, Roytrakul S, Viravaidya-Pasuwat K. 2020. Construction and evaluation of osteochondral-like tissue using chondrocyte sheet and cancellous bone. Tissue Eng Part A 27:282–295. doi: 10.1089/ten.TEA.2020.0107. [DOI] [PubMed] [Google Scholar]
- 88.Johansson C, Samskog J, Sundstrom L, Wadensten H, Bjorkesten L, Flensburg J. 2006. Differential expression analysis of Escherichia coli proteins using a novel software for relative quantitation of LC-MS/MS data. Proteomics 6:4475–4485. doi: 10.1002/pmic.200500921. [DOI] [PubMed] [Google Scholar]
- 89.Thorsell A, Portelius E, Blennow K, Westman-Brinkmalm A. 2007. Evaluation of sample fractionation using micro-scale liquid-phase isoelectric focusing on mass spectrometric identification and quantitation of proteins in a SILAC experiment. Rapid Commun Mass Spectrom 21:771–778. doi: 10.1002/rcm.2898. [DOI] [PubMed] [Google Scholar]
- 90.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]