ABSTRACT
Escherichia coli is the most commonly identified human pathogen and a prominent microorganism of the gut microbiota. Acquired resistance to antibiotics in this species is driven mainly by horizontal gene transfer and plasmid acquisition. Currently, the main concern is the acquisition of extended-spectrum β-lactamases of the CTX-M type in E. coli, a worldwide-observed phenomenon. Plasmids encoding CTX-M enzymes have different scaffolds and conjugate at different frequencies. Here, we show that the conjugation rates of several plasmid types encoding broad-spectrum β-lactamases are increased when the E. coli donor strain is exposed to subinhibitory concentrations of diverse orally given antibiotics, including fluoroquinolones, such as ciprofloxacin and levofloxacin, but also trimethoprim and nitrofurantoin. This study provides insights into underlying mechanisms leading to increased plasmid conjugation frequency in relation to DNA synthesis inhibitor-type antibiotics, involving reactive oxygen species (ROS) production and probably increased expression of genes involved in the SOS response. Furthermore, we show that some antioxidant molecules currently approved for unrelated clinical uses, such as edaravone, p-coumaric acid, and N-acetylcysteine, may antagonize the ability of antibiotics to increase plasmid conjugation rates. These results suggest that several antioxidative molecules might be used in combination with these “inducer” antibiotics to mitigate the unwanted increased resistance plasmid dissemination.
KEYWORDS: Escherichia coli, ROS, SOS, antibiotic, antioxidant, inducer, plasmid
INTRODUCTION
The concomitant increase in resistance to antibiotics and shortage of newly marketed antimicrobial drugs is raising significant concerns, necessitating alternative and innovative approaches to mitigate the spread of resistant bacteria but also of the corresponding resistance genes (1, 2). There are many factors that may influence the emergence and dissemination of antimicrobial resistance, among which the use, misuse, and overuse of antibiotics are important.
Even though acquisition of resistance to antibiotics may occur through chromosomal mutations, it is well established that horizontal gene transfer (HGT) is the leading cause of its dissemination. Another worrying aspect of HGT is the phenomenon of accumulation that is frequently observed, with genetic structures involved in HGT (mostly plasmids, transposons, and insertion sequences [IS]) often gathering multiple resistance genes, leading to a multidrug resistance pattern (3).
Currently, among the most clinically relevant resistance traits in Gram-negative bacteria are the extended-spectrum-β-lactamase (ESBL) genes, encoding enzymes capable of hydrolyzing most β-lactam substrates, with the exception of cephamycins and carbapenems. Among those genes, blaCTX-M-15 is the most commonly identified in Enterobacterales (and particularly in Escherichia coli) (4–6). Its emergence and extensive spread worldwide have been hypothesized to be possibly related to the overall increased usage of broad-spectrum cephalosporins (7).
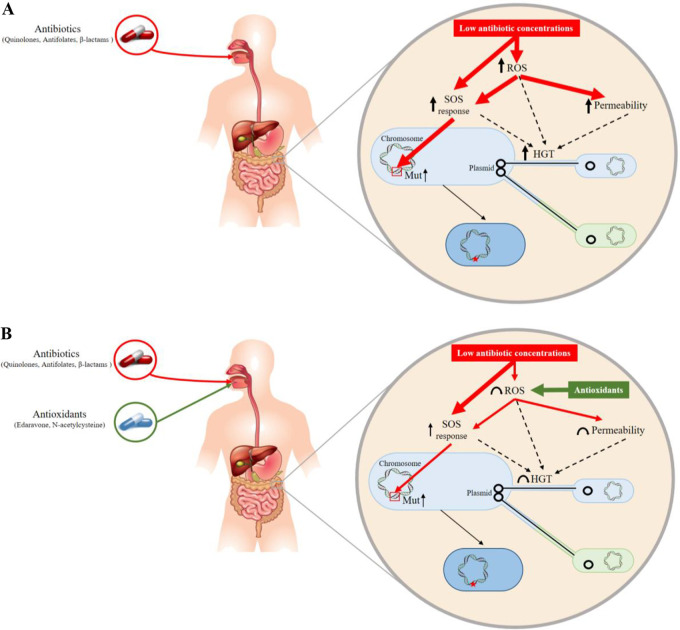
Noteworthily, antibiotics, apart from their antimicrobial function, also have side effects on bacterial populations, stimulating their genetic evolution and their dissemination. In some instances, antibiotics may enhance HGT and therefore complexify the whole picture of resistance dissemination, with the curative agent itself promoting the occurrence of corresponding resistance. This is especially true within the gut microbiota, which represents a rich source of mobile antibiotic resistance genes and on which subinhibitory concentrations of orally given antibiotics might exert significant selective pressure (8, 9) (Fig. 1). The most commonly prescribed and orally given antibiotics are fluoroquinolones, trimethoprim (TMP), nitrofurantoin (NIT), β-lactams (penicillins and cephalosporins), and fosfomycin (FOS), particularly for treating urinary tract infections due to Escherichia coli (10). Those antibiotic molecules have diverse modes of action. Fluoroquinolones (ciprofloxacin [CIP] and levofloxacin [LEV]) inhibit DNA replication by binding to the enzyme-DNA complex, blocking movement of the DNA replication fork, and stabilizing DNA strand breaks created by DNA gyrase and topoisomerase IV, eventually leading to cell death (11). TMP is an analogue of folic acid able to inhibit the action of the dihydrofolate reductase enzyme by fitting well into its nucleotide-binding site (12). NIT is activated intracellularly via the flavoprotein NIT reductase and interferes with enzymes involved in DNA, RNA, and protein synthesis (13). FOS is a peptidoglycan synthesis inhibitor that acts by blocking the MurA enzyme, involved in the first steps of the peptidoglycan biosynthetic pathway (14). β-Lactams act by inhibiting the synthesis of the peptidoglycan layer of bacterial cell walls by interfering with the dd-transpeptidases, also known as penicillin-binding proteins (15).
FIG 1.
Possible mechanisms through which stress conditions could accelerate the evolution and spread of resistance genes. Drugs present at subinhibitory concentrations in body compartments may act as inducers, leading to increases (arrows) in mutagenesis (Mut) and horizontal gene transfer (HGT) by means of an ROS/SOS response.
Several antibiotics such as fluoroquinolones, cephalosporins, and penicillins, when present at subinhibitory concentrations, have been previously reported to increase the frequency of plasmid conjugation in E. coli (16–18). Moreover, other studies suggested that this increased rate of plasmid conjugation was likely related to induction of the SOS response, production of reactive oxygen species (ROS), and an increase in membrane permeability (19, 20). Furthermore, a single study showed that CIP could enhance the transfer of multidrug resistance plasmids between commensal E. coli strains and fluoroquinolone-resistant Shigella sonnei strains (21). Increased mutational rates have been observed in many studies upon exposure to fluoroquinolones either in Enterobacterales or in Pseudomonas aeruginosa, in relation to an SOS-mediated mutagenesis phenomenon (22, 23).
Bacteria commonly respond to stress conditions through the activation of the SOS system, which may also result from increased production of ROS. The SOS response is a coordinated cellular response to DNA damage and replication blockage. In E. coli, the SOS regulon is composed of at least 40 genes, many of them coding for DNA repair functions, such as nucleotide excision repair, translation synthesis, and homologous recombination (24). Responding to environmental stress may induce a significant SOS response at the bacterial level (25). Such activation may lead to an increase in expression of genes related to genetic plasticity, which is actually one pathway by which the bacteria respond to the stress. Hence, the SOS response promotes horizontal gene transfer and mutagenesis (26, 27).
Apart from the SOS response, which can be antibiotic induced, oxidative metabolism (which is a process in which oxygen is used to generate energy from nutrients) is also known to be regulated upon stress conditions. In bacteria growing under aerobic conditions, as for E. coli, most energy is produced by respiration using molecular oxygen as the final electron acceptor, although unwanted by-products of respiration are ROS. Bacteria possess defense mechanisms just sufficient to protect themselves against endogenous ROS. However, a stress-induced increase in ROS production has deleterious consequences, including increased mutation rates. A series of studies have shown that some antibiotic selective pressure may increase ROS production and subsequently mutation rates in E. coli (28, 29).
With respect to this bacterial response to stress conditions that may be deleterious, a series of molecules have been shown to contribute to a reduction of the mutation rate, such as N-acetylcysteine (NAC), which acts as a blocker of the SOS system and ROS production (28, 29), and edaravone (EDA; 3-methyl-1-phenyl-2-pyrazolin-5-one), which acts as a ROS scavenger. NAC is a glutathione precursor used as a mucolytic agent and used for the treatment of numerous disorders, including oral paracetamol intoxication. It has well-known protective effects against oxidative stress, due to its antioxidant properties (30, 31). EDA is a potent free-radical scavenger approved by the U.S. Food and Drug Administration for use in humans to treat acute stroke caused by cerebral thrombosis and embolism and to treat amyotrophic lateral sclerosis (32). This synthetic pyrazolone derivative was recently shown to modulate oxidative stress and the production of ROS in bacteria (33, 34). Another interesting molecule is p-coumaric acid (pCA), a phenolic compound present in many food products and possessing various potential properties, including antioxidant, anti-inflammatory, and immunomodulatory effects (35–38).
Here, we aimed to investigate two aspects of antibiotic-related selective pressure in E. coli and their potential inhibition. We focused on the most commonly identified ESBL-encoding gene, namely, blaCTX-M-15, as the most prevalent ESBL gene worldwide among human isolates but also on other clinically relevant β-lactamase genes, such as blaOXA-48 and blaKPC-2, encoding widespread carbapenemases. First, we evaluated whether any orally given antibiotic, when present at subinhibitory concentrations, could increase the frequency of conjugation of plasmids carrying those β-lactamase genes, thus contributing to the dissemination of the corresponding resistance determinant. A particular focus was on the effect of CIP, since this antibiotic is used for treating infections caused by Gram-negative and Gram-positive bacteria and is one of the main antibiotics prescribed worldwide (39, 40). Then, several drugs possessing antioxidant properties were evaluated with the object of preventing or reducing the spread of resistance genes. The ultimate goal of our work was to elaborate strategies mitigating the effect of antibiotics as inducers of antibiotic resistance gene dissemination.
RESULTS
Increased conjugation frequencies in the presence of subinhibitory concentrations of oral antibiotics.
Orally administered antibiotics, including the β-lactams amoxicillin, amoxicillin-clavulanic acid (AMC), and cefalexin, but also CIP, LEV, TMP, NIT, FOS, azithromycin, erythromycin, and tetracycline, and the intravenously given drug amikacin as a representative of the aminoglycoside antibiotic class were tested at subinhibitory concentrations to evaluate their potential impact on plasmid conjugation frequencies. E. coli RZ211 harboring the self-conjugative and gentamicin resistance-encoding F-plasmid derivative pOX38 bearing the blaCTX-M-15 gene was used as the donor for conjugation assays. Before it was mixed with the E. coli J53 recipient strain, cultures of E. coli RZ211 were performed in the presence of 1/2 and 1/4 of the MICs of different antibiotics (preculture steps).
Preliminary evaluation of the relative number of donors and recipients in term of CFU showed that the number of donors was indeed affected by the exposure to subinhibitory antibiotic concentrations. For example, the concentration of the E. coli pOX38 donor strain after 4 h growth was 8 × 107 CFU/ml in the absence of any antibiotic, while it was 1 × 107 CFU/ml when supplemented with 1/2 the MIC of CIP. Since the conjugation frequencies were calculated with respect to the number of donors, the bias created by the exposition to antibiotics was subsequently considered in the provided values.
Conjugation assays performed at a 1:4 volume ratio showed a significant increase in plasmid pOX38 conjugation frequencies in the presence of subinhibitory concentrations of TMP, CIP, LEV, and NIT, with changes of ca. 10-, 20-, 40-, and 100-fold, respectively, compared to the control experiment performed in the absence of antibiotic during the preculture step (Table 1).
TABLE 1.
Conjugation frequency and fold change in mating assays for E. coli RZ211(pOX38)a
| Antibiotic | MIC (μg/ml) | Control |
1/4 MIC |
1/2 MIC |
1/2 MIC + EDA |
||||
|---|---|---|---|---|---|---|---|---|---|
| CF | Fold change | CF | Fold change | CF | Fold change | CF | Fold change | ||
| Ciprofloxacin | 0.004 | 1.1 × 100 ± 1.2 × 100 | 1 | 3.3 × 100 ± 2.8 × 100 | 3.3 | 2.1 × 101 ± 1.9 × 101 | 21 | 3.2 × 100 ± 3.7 × 100 | 3.2 |
| Levofloxacin | 0.032 | 7 × 10−1 ± 5 × 10−1 | 1 | 4.3 × 100 ± 4.8 × 100 | 6 | 2.6 × 101 ± 9.8 × 101 | 37 | 5.3 × 100 ± 3.7 × 100 | 7.5 |
| Trimethoprim | 0.06 | 4 × 10−1 ± 1.2 × 10−1 | 1 | 4 × 100 ± 2.8 × 100 | 10 | 4.4 × 100 ± 2.3 × 100 | 11 | 2.5 × 100 ± 2 × 10−1 | 6 |
| Fosfomycin | 4 | 6 × 10−1 ± 1.1 × 10−1 | 1 | 1.6 × 100 ± 9 × 10−1 | 2.7 | 4.4 × 100 ± 1.8 × 100 | 7.3 | 1.3 × 100 ± 4.5 × 10−1 | 2.2 |
| Amoxicillin | >512 | 7.5 × 10−1 ± 3.4 × 10−1 | 1 | 2 × 100 ± 9.8 × 10−1 | 2.7 | 2.3 × 100 ± 7.5 × 10−1 | 3 | 9.5 × 10−1 ± 7 × 10−2 | 1.3 |
| Amoxicillin/clavulanic acidb | 4 | 1.8 × 100 ± 1 × 100 | 1 | 2.2 × 101 ± 1.6 × 101 | 12.2 | 2.4 × 101 ± 1.8 × 101 | 13 | 1.3 × 101 ± 1 × 101 | 7.2 |
| Nitrofurantoin | 4 | 2.8 × 10−1 ± 2.5 × 10−1 | 1 | 1.4 × 101 ± 1.1 × 101 | 50 | 3 × 101 ± 9.8 × 100 | 107 | 1.5 × 101 ± 4.2 × 100 | 50 |
| Cefalexin | 512 | 7.6 × 10−1 ± 2.7 × 10−1 | 1 | 1.6 × 100 ± 1.2 × 100 | 2.1 | 5 × 100 ± 2.5 × 100 | 6.6 | 1.5 × 100 ± 1.2 × 100 | 2 |
| Azithromycin | 2 | 2.3 × 100 ± 6.6 × 10−1 | 1 | 3.3 × 100 ± 1.4 × 100 | 1.4 | 3.2 × 100 ± 1.5 × 100 | 1.4 | 2.9 × 100 ± 9.8 × 10−1 | 1.3 |
| Sulfonamides | 32 | 9 × 10−1 ± 3 × 10−1 | 1 | 1.1 × 100 ± 1.3 × 100 | 1.2 | 1.7 × 100 ± 1.7 × 100 | 1.8 | 9 × 10−1 ± 9 × 10−1 | 1 |
| Tetracycline | 2 | 9 × 10−1 ± 3 × 10−1 | 1 | 1.3 × 100 ± 1.3 × 100 | 1.4 | 1.2 × 100 ± 1.3 × 100 | 1.3 | 1.2 × 100 ± 1.1 × 100 | 1.3 |
| Erythromycin | 32 | 4.4 × 100 ± 3.6 × 100 | 1 | 2.4 × 100 ± 2.2 × 100 | 0.5 | 4.1 × 100 ± 9.1 × 10−1 | 0.9 | 2.5 × 100 ± 2.8 × 10−1 | 0.6 |
| Amikacin | 6 | 7.5 × 10−1 ± 3.4 × 10−1 | 1 | 1 × 100 ± 5.2 × 10−1 | 1.3 | 1.5 × 100 ± 7 × 10−2 | 1.9 | 8 × 10−1 ± 1.4 × 10−1 | 1 |
EDA, edaravone (0.1 mM); CF, conjugation frequency. Statistically significant fold changes are in bold (P < 0.05).
Fixed concentration of clavulanic acid at 2 μg/ml.
A statistically significant, albeit more moderate, increase in conjugation frequency was also observed with subinhibitory concentrations of amoxicillin, cefalexin, and FOS, with ca. 3-fold, 7-fold, and 7-fold changes, respectively (Table 1). Noteworthily, the latter antibiotics act on bacterial growth by interfering with cell wall synthesis (15, 41).
In contrast, inhibitors of protein synthesis targeting ribosomes, such as azithromycin, erythromycin, amikacin, and tetracycline, did not modify the conjugation rate of plasmid pOX38 (Table 1). Interestingly, while the effect observed with amoxicillin was moderate and not statistically significant, a much higher induction effect was observed when amoxicillin was combined with clavulanic acid at a concentration of 2 μg/ml (Table 1).
Natural plasmids may disseminate at higher frequencies after CIP exposure.
Since significant increases in plasmid conjugation were observed with CIP in our preliminary E. coli RZ211(pOX38) model, CIP was further retained as an inducer for subsequent conjugation assays performed with natural plasmids and using clinical isolates as donors. A total of five clinical isolates (a single E. coli isolate and four K. pneumoniae isolates) were thus selected as donors harboring the plasmid types most commonly identified as sources of acquisition of the most commonly identified and clinically relevant ESBLs or carbapenemases, namely, IncFIB and IncN plasmids bearing blaKPC-3, IncI1 and IncFII plasmids bearing the blaCTX-M-15 gene, and IncL bearing the blaOXA-48 gene (Table 2). Of note, none of these plasmids carried any other known resistance gene interfering with the activity of quinolone molecules (data not shown). Induction experiments were performed by adding subinhibitory concentrations of CIP to the culture broth of each donor, before mating-out assays using the same protocol as for the model strain. A significant increase in plasmid conjugation frequencies was observed for plasmids IncL (blaOXA-48), IncI1 (blaCTX-M-15), IncFII (blaCTX-M-15), and IncFIB (blaKPC-3) (ca. 5-, 10-, 20-, and 25-fold, respectively) in the presence of half of the MIC of CIP (Table 3). This increased rate was actually lower with concentrations corresponding to 1/4 the MIC of CIP (Table 3). In contrast, no increased frequency was observed for plasmid IncN-blaKPC-3.
TABLE 2.
Strains used in the study
| Strain | Plasmid or description | Reference |
|---|---|---|
| E. coli RZ211 | pOX38 (Genr blaCTX-M-15) | This work |
| K. pneumoniae KPM26 | IncFIB (blaKPC-3) | 56 |
| K. pneumoniae KPM2 | IncN (blaKPC-3) | 56 |
| K. pneumoniae KPI1 | IncI1 (blaCTX-M-15) | This work |
| K. pneumoniae KP11978 | IncL (blaOXA-48) | 57 |
| E. coli EC1010 | IncFII (blaCTX-M-15) | This work |
| E. coli J53 | Recipient strain (azide resistant) |
TABLE 3.
Conjugation frequency and fold change in conjugation frequencies for natural plasmids after ciprofloxacin exposure with or without EDAa
| Strains | Plasmid | Resistance determinant | MIC (μg/ml) | Control |
1/2 MIC |
1/2 MIC + EDA |
|||
|---|---|---|---|---|---|---|---|---|---|
| CF | Fold change | CF | Fold change | CF | Fold change | ||||
| RZ211 | pOX38 | bla CTX-M-15 | 0.004 | 1.1 × 100 ± 8.4 × 10−1 | 1 | 2.3 × 101 ± 5.8 × 100 | 19.2 | 5.4 × 100 ± 2.6 × 100 | 4.5 |
| KPM26 | IncFIB | bla KPC-3 | 1 | 4 × 10−6 ± 5.5 × 10−8 | 1 | 1 × 10−4 ± 1.7 × 10−5 | 24.6 | 1.2 × 10−5 ± 1 × 10−5 | 3 |
| KPM2 | IncN | bla KPC-3 | 0.016 | 6.9 × 10−6 ± 7 × 10−6 | 1 | 5 × 10−6 ± 8 × 10−6 | 0.7 | 2.5 × 10−6 ± 3.5 × 10−6 | 0.4 |
| KPI1 | IncI1 | bla CTX-M-15 | 0.032 | 4.7 × 10−7 ± 1 × 10−7 | 1 | 4.8 × 10−6 ± 1.6 × 10−6 | 10.2 | 6.2 × 10−7 ± 2.9 × 10−7 | 1.3 |
| KP11978 | IncL/M | bla OXA-48 | 1.5 | 2.1 × 10−4 ± 5.3 × 10−5 | 1 | 9.2 × 10−4 ± 1.2 × 10−4 | 4.1 | 1.7 × 10−5 ± 7.3 × 10−6 | 0.1 |
| EC1010 | IncFII | bla CTX-M-15 | 0.032 | 5.7 × 10−5 ± 5.5 × 10−5 | 1 | 1 × 10−3 ± 5.5 × 10−4 | 17.5 | 5 × 10−4 ± 2 × 10−4 | 8.7 |
EDA, edaravone (0.1 mM); CF, conjugation frequency. Statistically significant changes are in bold (P < 0.05).
Increased plasmid transfer in the presence of subinhibitory CIP concentration using S-sS medium.
In order to mirror the potential impact of antibiotics on genetic plasticity in the gut flora, mating-out assays were then performed in semisolid stool medium (S-sS medium). E. coli RZ211(pOX38) and E. coli J53 were used as donor and recipient strains, respectively. In contrast to former experiments during which CIP was added only during the growth of the donor before mixing both donor and recipient, the incubation of both donor and recipient was performed in the presence of subinhibitory concentrations of CIP. Concentrations of 1/2 and 1/4 the MIC of CIP (with respect to the recipient strain) raised the frequency of conjugation of plasmid pOX38, with the number of transconjugants obtained being increased by 2- and 4.3-fold, respectively (see Table S1 and Fig. S1 in the supplemental material). Hence, the induction of the plasmid conjugation frequency by CIP could be further demonstrated in the artificial S-sS medium.
EDA mitigates the increased rate of conjugation frequency.
Since the conjugation rates of reference plasmid pOX38 and the natural plasmids used in our experiments were shown to be induced by subinhibitory concentrations of CIP or other antibiotic molecules (Table 1), and since it was speculated that this phenomenon was related to the production of ROS, the effect of EDA as a possible inhibitor of ROS production was tested.
First, at the concentration of 0.1 mM used for mating-out experimental assays, incubation with EDA did not affect the growth rate of the wild-type E. coli donor strain RZ210. This fixed concentration of 0.1 mM EDA was chosen in accordance with the reported serum concentration measured upon clinical use, which is ca. 90 μM upon oral delivery (28, 42). When EDA concentrations were increased 100-fold, still no growth inhibition could be observed (Fig. S2). In parallel, an assay was performed to evaluate the putative impact of EDA on CIP antibacterial activity as described elsewhere (28), and the results showed that no synergistic antibacterial activity actually occurred and that this therefore would not bias the subsequent observations (Table S2).
Results of the mating-out assays showed that addition of EDA reduced or even suppressed the augmentation of plasmid conjugation frequency observed with antibiotics for which induction properties were observed. At 0.1 mM EDA, reduction of the increased transfer rate was estimated to be between 2- and 6.5-fold, depending on the antibiotics used as inducers. However, in a context of noninducibility, EDA did not modify the conjugation rate baseline for the tested plasmids (Table 1). These results showed that the effect of antioxidant addition was more significant when an SOS-ROS-related induction had actually occurred. Nevertheless, the fosfomycin-induced plasmid conjugation observed in the absence of ROS-SOS-related induction was also alleviated by EDA (Table 1).
CIP-induced conjugation rates of natural plasmids from clinical isolates were reduced between 2- and 8-fold, depending on the plasmids tested, when the exposure to CIP was combined with 0.1 mM EDA (Table 3). Moreover, a decrease by 8.6 ± 2.1-fold of the rate of pOX38 conjugation by CIP was also observed when S-sS medium was used.
Incubation with antibiotic targeting DNA or DNA metabolism showed a correlation between SOS response and ROS generation.
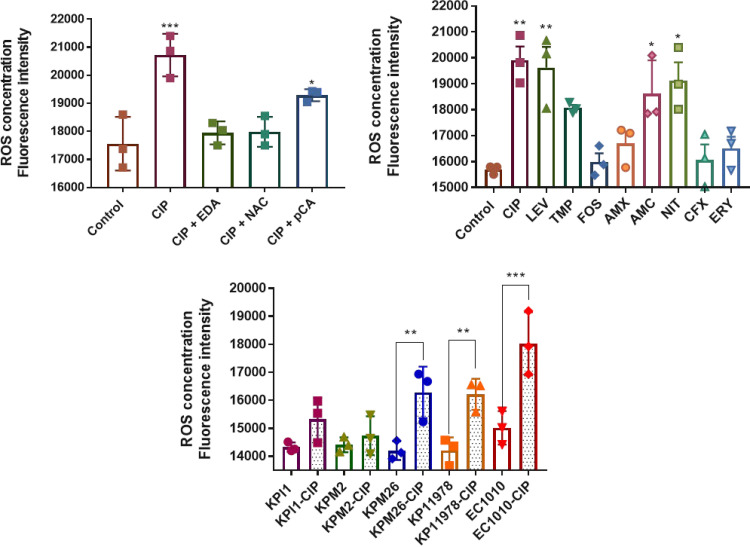
ROS production and SOS-related genes expression were evaluated in E. coli RZ211(pOX38), under conditions shown to induce plasmid conjugation, namely, exposure to CIP, LEV, TMP, FOS, amoxicillin, AMC, NIT, and cefalexin. In addition, erythromycin was added as a control, since no induction of the plasmid conjugation process has been observed with this antibiotic. Overall, CIP, LEV, TMP, NIT (whose respective mechanisms of actions interfere with DNA synthesis), and AMC were found to significantly induce the production of ROS (Fig. 2). In contrast, no significant increase in ROS concentration was observed in the presence of drugs acting by other mechanisms of action, such as FOS, amoxicillin, cephalexin, and erythromycin (control) (Fig. 2B).
FIG 2.
ROS measurement by fluorimeter. (A) Comparative analysis between E. coli RZ211 ROS measurement after incubation with ciprofloxacin and incubation with ciprofloxacin plus an antioxidant. (B) ROS concentration in E. coli RZ211 after induction with several antibiotics. (C) Production of ROS by clinical strains with and without exposure to ciprofloxacin. CIP, ciprofloxacin; LEV, levofloxacin; TMP, trimethoprim; FOS, fosfomycin; AMX, amoxicillin; NIT, nitrofurantoin; CFX, cephalexin; ERY, erythromycin; EDA, edaravone; NAC, N-acetylcysteine; pCA, p-coumaric acid. Antibiotics at 1/2 the MIC and antioxidant molecules at 0.1 mM were used for all the experiments. Three independent replicates were performed. The data are means and standard deviations (SD). *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001.
Results of SOS gene expression measurements showed that only antibiotics which are capable of significantly inducing ROS production and whose mechanism of action is actually related to DNA synthesis inhibition, such as CIP, LEV, TMP, and NIT, increased the expression of the recA and/or sfiA genes, in a range between 2.5- and 12.5-fold (Table 4). In contrast, a significant increase in ROS production but no increase of the expression of the recA and sfiA genes was observed in the presence of subinhibitory concentrations of AMC (Table 4). Compared to the control, a slight increase in ROS concentration was observed in the presence of subinhibitory concentrations of amoxicillin and cephalexin; however, it did not result in an induction of SOS-related-gene expression (Table 4 and Fig. 2B), as expected. Those data therefore showed that induction of the SOS-related-gene expression may occur upon exposure to certain amounts of ROS, as described previously (29), and that antibiotics targeting DNA metabolism are more prone to induce the ROS/SOS system.
TABLE 4.
Expression of two chromosomal SOS-related genes in E. coli RZ211(pOX38) at sub-MIC of molecules representatives of different classes of antibioticsa
| Gene | 2−ΔΔCT |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C | CIP | EDA | CIP+E | LEV | TMP | FOS | AMX | AMC | NIT | CFX | ERY | |
| recA | 1 | 12.5 ± 1.6 | 0.6 ± 0.2 | 4.0 ± 0.5 | 2.6 ± 0.4 | 2.7 ± 0.2 | 1.75 ± 0.7 | 0.9 ± 0.1 | 0.9 ± 0.1 | 2.5 ± 0.3 | 2.2 ± 0.1 | 1.2 ± 0.4 |
| sfiA | 1 | 6 ± 1.7 | 0.4 ± 0.2 | 3.0 ± 0.3 | 3.5 ± 0.4 | 1.3 ± 0.1 | 1.3 ± 0.3 | 0.7 ± 0.2 | 0.7 ± 0.2 | 2.7 ± 0.3 | 1.9 ± 0.8 | 1.2 ± 0.3 |
C, control; CIP, ciprofloxacin (0.002 mg/liter); EDA, edaravone (0.1 mM); CIP+E: ciprofloxacin (0.002 mg/liter) plus edaravone (0.1 mM); LEV, levofloxacin (0.016 mg/liter); TMP, trimethoprim (0.03 mg/liter); FOS, fosfomycin (2 mg/liter); AMX, amoxicillin (512 mg/liter); AMC, amoxicillin-clavulanic acid (2/4 mg/liter); NIT, nitrofurantoin (2 mg/liter); CFX, cephalexin (256 mg/liter); ERY, erythromycin (16 mg/liter). Fold change in mRNA levels were determined by RT-qPCR. The data were normalized to the 16S rRNA gene. Three independent replicates were performed. Bold indicates significant results (P < 0.05).
Activation of the ROS/SOS route in clinical isolates after exposure to subinhibitory concentrations of CIP.
In order to identify whether some specific plasmid scaffolds are more prompt to have their transfer rate induced by some antibiotics (keeping in mind that our “model” plasmid is an IncF-type plasmid), ROS and SOS genes expression levels were measured using clinical isolates as plasmid donors (Table 2). CIP was again selected as a model of induction of the ROS/SOS system, and different subinhibitory concentrations were tested. Compared with the untreated counterpart strain, an increased production of ROS products was observed for isolates KPI1 (IncI1), EC1010 (IncFII), KP11978 (IncL/M), and KPM26 (IncFIB) in the presence of CIP (Fig. 2C). By measuring the expression of two SOS-related genes as described above, namely, recA and sfiA, we observed that at least one of the two genes was overexpressed ca. 3.3- to 10.5-fold compared with the untreated sample in all but one strain (Table 5). Notably, clinical strains for which the highest increase in ROS production was observed were not those for which the highest increased expressions of the SOS-related genes were observed, indicating a lack of directly proportional interplay between those two pathways activated upon CIP exposure. A similar observation was made with respect to the conjugation rate, with the increased conjugation rates in response to the antibiotic-related induction being not systematically directly proportional to the increased production of ROS or overexpression of SOS-related genes. In contrast, no increase in ROS production and no induction of the expression of the recA and sfiA genes was observed for isolate KPM2 (IncN-blaKPC-3), as expected, since no increased rate of conjugation had been observed.
TABLE 5.
Expression of two chromosomally encoded SOS-related genes for clinical E. coli isolates harboring different types of plasmids after exposure to sub-MIC of ciprofloxacin
| Gene | 2−ΔΔCT for strain at indicated CIP concn (μg/ml)a |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| KPI1 (IncI1, blaCTX-M-15) |
KPM2 (IncN, blaKPC-3) |
KPM26 (IncFIB, blaKPC-3) |
KP11978 (IncL/M, blaOXA-48) |
EC1010 (IncFII, blaCTX-M-15) |
||||||
| 0 | 0.016 | 0 | 0.008 | 0 | 0.5 | 0 | 0.75 | 0 | 0.016 | |
| recA | 1 | 8.6 ± 0.4 | 1 | 0.7 ± 0.15 | 1 | 4.5 ± 0.9 | 1 | 4.1 ± 0.15 | 1 | 3.3 ± 0.7 |
| sfiA | 1 | 0.6 ± 0.1 | 1 | 0.2 ± 0.05 | 1 | 10.5 ± 0.1 | 1 | 1.1 ± 0.07 | 1 | 0.8 ± 0.25 |
Fold change in mRNA levels were determined by RT-qPCR. The data were normalized to the mRNA levels of the 16S rRNA gene. Three independent replicates were performed. Bold indicates significant results (P < 0.05).
Testing of other antioxidant molecules.
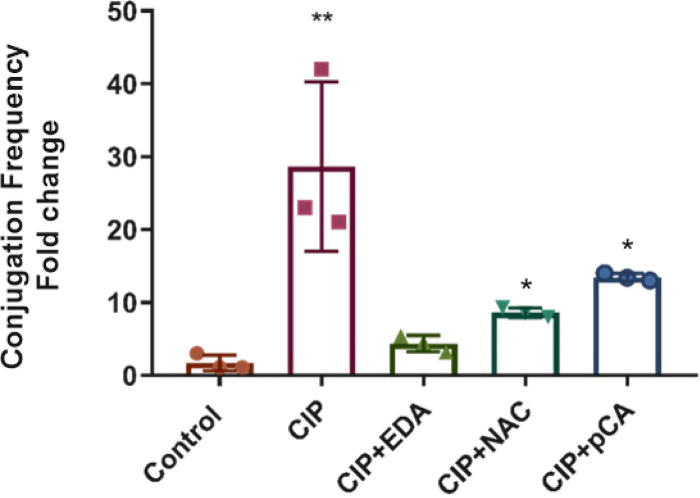
NAC and pCA antioxidant molecules were additionally tested as conjugation-mitigating molecules, using concentrations mirroring their serum concentrations: 170 μM and 160 μM, respectively (43, 44). Those molecules together were used in the mating-out assays by using E. coli RZ211(pOX38) and CIP as the inducer. The pOX38 conjugation rate was increased by 21-fold in the presence of CIP compared with the standard assay. Adding EDA (0.1 mM) and NAC (0.1 mM) reduced the CIP-induced transfer rate by 6.5- and 3-fold, respectively, under CIP-supplemented conditions, while pCA (0.1 mM) reduced the transfer rate by only 2-fold under the same conditions (Fig. 3). Measurement of the ROS levels in those different assay conditions performed in the presence of CIP revealed a significant reduction of the ROS amounts in the presence of EDA and NAC. In contrast, no significative reduction was observed after incubation with pCA at the tested concentrations (Fig. 2A). Additionally, measurement of the expression of genes involved in SOS response in the presence of subinhibitory concentrations of CIP revealed significant reductions of expression of the recA and sfiA genes in the presence of EDA, 3- and 2-fold, respectively (Table 4).
FIG 3.
Fold change in plasmid pOX38 conjugation frequency from E. coli RZ211 under ciprofloxacin (1/2 the MIC) and several antioxidants (0.1 mM) exposure. Control, no exposure to antibiotic; CIP, ciprofloxacin; EDA, edaravone; NAC, N-acetylcysteine; pCA, p-coumaric acid. Three independent replicates were performed. The data are means and SD. *, P ≤ 0.05; **, P ≤ 0.01.
DISCUSSION
Emergence and dissemination of resistant bacteria are known to be related to antibiotic consumption (45, 46). The term “emergence of resistance” refers to selection of a resistant strain deriving from a susceptible one, consequent upon acquisition of a resistance mechanism whose genetic basis mainly corresponds to acquisition of chromosomal mutations or of foreign genetic material through HGT. We focused our study on plasmid-mediated acquired resistance, with a specific focus on plasmid conjugation frequencies, which basically constitutes the most common transfer mechanism in Gram-negative bacteria. Considering that bacterial strains are frequently exposed to nonlethal concentrations of drugs, especially in the gut in the presence of antibiotics, we evaluated the extent to which orally given antibiotics may induce the spread of resistance genes in the gut flora and how this transfer could be partially prevented.
We showed that a series of antibiotics may increase plasmid conjugation rates in E. coli, the further consequence being to increase the rate of acquisition of plasmid-borne resistance genes. Our study showed that antibiotics acting as DNA synthesis inhibitors, such as CIP, levofloxacin, trimethoprim, and nitrofurantoin, had the potential to enhance the plasmid conjugation rate at a higher rate than cell wall inhibitor antibiotics, such as amoxicillin, cefalexin, and FOS.
Previous studies showed that cefotaxime and fluoroquinolones might be inducers of plasmid conjugation frequency, which was confirmed here for CIP and levofloxacin (16, 17). Interestingly, Møller et al. (17) showed that the increased rate observed upon selective pressure from high-dose cefotaxime was related not to induction of the SOS response but rather to an increased expression of genes involved in plasmid conjugation, i.e., those involved in the pilus synthesis and assembly system, the DNA transfer system, and also structure of the pilus. This was confirmed in our experiments, even though the induction impact may vary significantly depending on the class of antibiotics tested.
Noteworthily, other nonantibiotic drugs, such as the anticonvulsant carbamazepine, the anticancer molecule cisplatin, and the selective serotonin reuptake inhibitor fluoxetine (used as antiepileptic, cancer therapy, and antidepressant treatments, respectively) have been reported to induce HGT and to modulate the SOS system (47–49). It is therefore tempting to speculate that such molecules could also have an impact on plasmid dissemination, and further experiments will be required to measure any potential undesired effect.
Our study identified antibiotic molecules promoting higher conjugation frequencies likely associated with the ROS/SOS response (namely, CIP, LEV, NIT, and TMP) and ROS production (AMC). Previous studies showed that quinolones (CIP) and antifolates (TMP) may be inducers of the SOS system by means of increased recA expression levels, which was further proven by the observation of a lack of antibiotic-induced mutagenesis upon inactivation of this gene (27). Conversely, induction of the SOS response during the conjugation process has been reported (50). Altogether, those observations and ours evidenced a striking interaction between the antibiotic-induced SOS response and HGT, particularly that related to plasmid conjugation. In contrast, supplementation with amoxicillin, cefalexin, and FOS led to a moderate increase in conjugation rate that was not significantly related to increased ROS concentration. The lack of significant increase in ROS concentration in the presence of amoxicillin and cefalexin might be related to the presence of the blaCTX-M-15 gene in the model we used, with CTX-M-15 conferring high MICs of amoxicillin and cefalexin. Accordingly, a previous study showed that an increase in ROS levels was observed only for strains susceptible to the corresponding bactericidal drug (51). The lack of induction of HGT by antibiotics inhibiting bacterial protein synthesis (aminoglycosides, macrolides, and tetracyclines) may be related to inhibition of a series of proteins, including those involved in the conjugation machinery.
The relevance of our study is emphasized by the choice to test clinically relevant and orally given antibiotics as inducers of plasmid transfer, considering that these molecules are subsequently largely diffused within the gut flora, where many genetic exchanges between bacteria may occur. Many clinical studies have highlighted the in vivo transfer of resistance plasmids within the gut microbiome and, in particular, plasmids carrying ESBL- or carbapenemase-encoding genes (21, 52). Our data further strengthen the observation that subinhibitory concentrations of several classes of antibiotics in the gut have a significant impact on antibiotic resistance dissemination.
Here, as a proof of concept, we confirmed through mating-out using E. coli strains as plasmid donors performed in S-sS medium simulating the gut flora that induction of plasmid conjugation frequencies also occurred upon antibiotic administration. Then, the model was reproduced by using clinical strains as donors, including diverse enterobacterial species (E. coli and K. pneumoniae), diverse β-lactamase genes (blaKPC-3, blaCTX-M-15, and blaOXA-48), and diverse plasmid types (IncFII, IncFIB, IncN, IncI1, and IncL/M).
Then, we aimed to identify a “plasmid antidissemination” strategy based on the principle of counteracting the CIP-induced SOS/ROS response. The antioxidant molecule EDA was shown to significantly reduce the ROS levels of E. coli clinical isolates being cultured at subinhibitory CIP concentrations, and a subsequent reduction of the normally observed conjugation rate was obtained. This phenomenon was also observed using the model of E. coli/pOX38 treated with CIP on S-sS medium, which simulates the gut flora environment. EDA could also reduce ROS levels and expression of SOS-related genes in CIP-treated samples. The correlation between the reduction of the CIP-increased transfer rate and recA gene expression (SOS response) was evidenced. Furthermore, edaravone was able to reduce the induced expression of recA, but not up to the baseline of recA expression, as observed with the conjugation frequency. This suggested that edaravone only partially inhibited the pathway by which the SOS response is actually induced by CIP. When it was compared with the efficacy of other anti-oxidative molecules such as NAC and pCA, a further reduction of the ROS levels was indeed observed with EDA.
The significant action of antioxidant molecules, used in combination with the “inducer” antibiotic, indicates their capacity to mitigate the effects of this antibiotic-related induction on plasmid dissemination. Considering that these molecules have already been developed and approved for human use, we might consider a repurposing strategy based on a combination of antibiotics and antioxidants to prevent the dissemination of antibiotic resistance genes. For further confirmation of the potential interest of such combinations, experimental models of gut colonization will be required and are already planned. Such animal models might, however, present some limitations, such as the difficulty of extrapolating results from experimental conditions with respect to the antioxidant concentrations in the gut, that remain unknown, since only plasma concentrations are available so far for some of these molecules.
Finally, in the current One Health concept of emerging resistance, identification of antibiotic classes that are prone to induce plasmid transfer as well as molecules that can mitigate such plasmid transfer may be relevant. Those findings may be used to limit the spread of antibiotic resistance in animals and later to its transfer to humans.
MATERIALS AND METHODS
Chemicals.
The antimicrobial agents were obtained as standard laboratory powders and were used immediately after solubilization. The agents and their sources were as follows: amoxicillin, AMC, TMP, CIP, FOS, tetracycline, sulfamethoxazole, cefalexin, NIT, EDA, N-acetyl-l-cysteine, and p-coumaric acid were from Sigma (Saint-Quentin Falavier, France); amikacin and erythromycin were from Acros Organic (Geel, Belgium); LEV, gentamicin, and azithromycin were from Apollo Scientific (Denton, United Kingdom); and clavulanic acid was from TCR Canada (Zurich, Switzerland). The probe 2,7-dichlorodihydrofluorescein diacetate (DCFH-DA; Sigma-Aldrich) was dissolved in dimethyl sulfoxide (DMSO; Sigma-Aldrich) and used as a probe in the ROS detection assay. Sodium hydroxide (NaOH; 0.01 N) and phosphate-buffered saline (PBS; 1 M) were used for probe pretreatment as described elsewhere (53).
Bacterial strains and plasmids.
E. coli RZ211 harboring plasmid pOX38, which encodes resistance to gentamicin and harbors the blaCTX-M-15 gene, was used as the donor in our experiments. In addition, several E. coli and K. pneumoniae clinical isolates harboring natural plasmids encoding a series of different β-lactamase genes were also used as donor strains in further mating-out assays (Table 2). The azide-resistant E. coli strain J53 was used as the recipient in conjugation experiments.
Antimicrobial susceptibility testing and culture medium.
MICs were determined by the microdilution method in Mueller-Hilton medium (MH) (Bio-Rad, Cressier, Switzerland) according to the EUCAST guidelines (54). Clinical and recombinants strains were inoculated in Luria-Bertani medium (LB) (Bio-Rad) with sub-MIC antibiotic concentrations and incubated at 37°C for 4 h with shaking. Selective medium was used for selection of transconjugant colonies. The antibiotic concentrations in solid selective medium were amoxicillin (50 and 100 mg/liter), gentamicin (7 and 15 mg/liter), and sodium azide (100 mg/liter). Semisolid stool medium (S-sS medium) was prepared by adding phosphate buffer (pH 7) to stool sample in a 1:5 ratio (1 g/5 ml) until a viscous texture compatible with our experiment was achieved.
Conjugation assays.
E. coli RZ211 and E. coli J53 were used as donor and recipient strains, respectively, in the conjugation experiments performed in the presence or absence of potential conjugation inhibitors and of subinhibitory antibiotic concentrations. In order to remove antibiotics from the donor strain culture, cells were washed with sterile sodium chloride and centrifugation at 3,000 × g for 2 min at room temperature and then resuspended in fresh LB medium. The recipient was always grown without antibiotic. Conjugations were performed on LB medium at 37°C. Donors (grown with or without antibiotic) and recipient were mixed in a 1:4 volume ratio, respectively, and then the mix was incubated at 37°C for 4 h without agitation. Then, serial dilutions of that mixed culture were made, and 100 μl of the respective mixtures was plated onto LB agar plates containing 15 mg/liter gentamicin (to quantify donor plus transconjugant cells) and plates containing100 mg/liter azide and 15 mg/liter gentamicin (to quantify respective transconjugants only). The conjugation frequency (CF) was calculated as the number of transconjugants divided by the number of donors. EDA, NAC, and pCA were tested by mating assay in combination with CIP against E. coli RZ211.
Several E. coli and K. pneumoniae isolates which harbored different natural plasmids were used as donors and E. coli J53 was the recipient strain in the conjugation experiments, which used standard conditions or CIP as the inducer. Conjugation assays were performed on LB agar plates (Carl Roth, Karlsruhe, Germany) with filters (0.22 μm; Merck Millipore, Cork, Ireland) at 37°C. Donor and recipient were mixed in a 1:4 volume ratio on the filters. Mixtures were incubated at 37°C without shaking, and conjugation was performed for 4 h. The bacterial material was washed off the filters using isotonic NaCl, and the mixture was vortexed to stop the conjugation process. A dilution series was made, and the mixture was plated on LB agar plates containing 100 mg/liter ampicillin (for quantifying donors and transconjugants) and 100 mg/liter ampicillin and 100 mg/liter azide (for quantifying transconjugants) and incubated overnight at 37°C. The CF was calculated as the number of transconjugants divided by the number of donors.
E. coli RZ211 and E. coli J53 were used as donor and recipient strains, respectively, in the conjugation experiments on S-sS medium with and without edaravone and sub-MIC of CIP. The recipient and the donor were grown with the molecules on the medium. The conjugations were performed on S-sS medium at 37°C. Donor and recipient were mixed in a 1:1 volume ratio. S-sS medium with a mixture of bacteria was incubated at 37°C for 8 h. A dilution series was made, and the mixture was plated onto LB agar plates containing 15 mg/liter gentamicin (for quantifying donors and transconjugants) and 100 mg/liter azide and 15 mg/liter gentamicin (for quantifying transconjugants). The CF was calculated as the number of transconjugants divided by the number of donors. S-sS medium was tested by using LB agar plates containing 100 mg/liter azide and 15 mg/liter gentamicin to confirm the absence of resistant bacteria on the medium.
Fluorescence ROS detection assay.
Measurement of the ROS production level was performed as reported by Castro-Alférez et al. (53), by measuring the chemical hydrolysis of DCFH-DA, which is an indicator of hydroxyl radicals, hydrogen peroxide, and peroxyl radicals. Briefly, the DCFH-DA substrate is naturally hydrolyzed by endogenous cell esterases, subsequently forming dichlorodihydrofluorescein (DCFH). Then, nonfluorescent DCFH gives rise to 2,7-dichlorofluorescein (DCF) by direct interactions with different compounds, such as iron, NO2, and H2O2. DCF is a fluorescent compound that was detected by fluorescence spectroscopy at 522- and 498-nm emission and excitation wavelengths, respectively. Addition of DCFH to bacterial suspension was performed after an incubation period of 6 h in culture broths that were supplemented with antibiotics or unsupplemented. After 15 min of incubation with DCFH, fluorescence was detected using a fluorimeter (TECAN 200Pro).
mRNA extraction.
mRNA samples were obtained after 4 h of bacterial growth, performed with or without antibiotic. Isolation of total RNA was performed by using the TRIzol Max bacterial RNA isolation kit (Thermo Fisher Scientific, Waltham, MA, USA) as specified by the manufacturer. RNA samples were subsequently treated with DNase to remove any contaminating DNA by using the DNA-free kit (Thermo Fisher Scientific, Waltham, MA, USA). Finally, DNA-free RNA samples were cleaned by using the RNeasy Power Clean Pro cleanup kit (Qiagen, Hilden, Germany) and measured using the NanoDrop 2000 spectrophotometer (Thermo Scientific, Littau, Switzerland).
RT-qPCR.
A 200-ng portion of each RNA sample was reverse transcribed with a PrimeScript RT reagent kit (TaKaRa, Saint-Germain-en-Laye, France) according to the manufacturer’s instructions. Quantitative real-time PCR (RT-qPCR) experiments were performed using the Rotor-Gene Q cycler (Qiagen, Hilden, Germany). Primer sequences were designed to amplify partial sequences (<150 bp) of intrinsic genes of E. coli and K. pneumoniae, namely, the 16S rRNA-encoding gene (reference) (16SFW, 5′-GTG CAA TAT TCC CCA CTG CT-3′; 16SRV, 5′-CGA TCC CTA GCT GGT CTG AG-3′), the recombinase-encoding gene recA (recAFW, 5′-ACA CCG GCG AGC AGG CAC TGG AAA-3′; recARV, 5′-ACG TGC CGC AAG GCC CAT GTG A-3′), and the cell division inhibitory gene sfiA (sfiAFW, 5′-CGS GAA TGG GTT CAG KCM KCY GGK C-3′; sfiARV, 5′- TRC CCG TGC GYA RRG CRC GRA YCA-3′). Reactions were set up in a total volume of 25 μl with a Rotor-Gene SYBR green PCR kit (Qiagen, Hilden, Germany). Three independent replicates were performed. The obtained cycle threshold (CT) values were analyzed by the 2−ΔΔCT method (55). Relative expression levels were calculated by comparison with control samples. The condition values were corrected with the appropriate reference gene.
Statistical analysis.
Conjugation frequencies were calculated by dividing the number of transconjugants by the number of donor bacteria. Significant differences were analyzed by one-way analysis of variance (ANOVA) in GraphPad Prism 7 software.
Data availability.
The complete nucleotide sequence of K. pneumoniae KPM2 was deposited in GenBank with accession number SRP308093.
ACKNOWLEDGMENTS
This work was funded by the University of Fribourg, by the Swiss National Science Foundation (projects JPI-AMR FNS-31003A_163432 and PNR72-40AR40_173686), and by the INSERM, Paris, France.
Footnotes
Supplemental material is available online only.
aac.02658-20-s0001.pdf (1.3MB, pdf)
REFERENCES
- 1.O'Neill J. 2014. Antimicrobial resistance. Tackling a crisis for the health and wealth of nations. The Wellcome Trust, London, United Kingdom. [Google Scholar]
- 2.Clatworthy AE, Pierson E, Hung DT. 2007. Targeting virulence: a new paradigm for antimicrobial therapy. Nat Chem Biol 3:541–548. 10.1038/nchembio.2007.24. [DOI] [PubMed] [Google Scholar]
- 3.Soucy SM, Huang J, Gogarten JP. 2015. Horizontal gene transfer: building the web of life. Nat Rev Genet 16:472–482. 10.1038/nrg3962. [DOI] [PubMed] [Google Scholar]
- 4.Poirel L, Gniadkowski M, Nordmann P. 2002. Biochemical analysis of the ceftazidime-hydrolysing extended-spectrum ß-lactamase CTX-M-15 and of its structurally related beta-lactamase CTX-M-3. J Antimicrob Chemother 50:1031–1034. 10.1093/jac/dkf240. [DOI] [PubMed] [Google Scholar]
- 5.Bevan ER, Jones AM, Hawkey PM. 2017. Global epidemiology of CTX-M β-lactamases: temporal and geographical shifts in genotype. J Antimicrob Chemother 72:2145–2155. 10.1093/jac/dkx146. [DOI] [PubMed] [Google Scholar]
- 6.Stoesser N, Sheppard AE, Pankhurst L, De Maio N, Moore CE, Sebra R, Turner P, Anson LW, Kasarskis A, Batty EM, Kos V, Wilson DJ, Phetsouvanh R, Wyllie D, Sokurenko E, Manges AR, Johnson TJ, Price LB, Peto TEA, Johnson JR, Didelot X, Walker AS, Crook DW, Modernizing Medical Microbiology Informatics Group. 2016. Evolutionary history of the global emergence of the Escherichia coli epidemic clone ST131. mBio 7:e02162-15. 10.1128/mBio.02162-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rossolini GM, D'Andrea MM, Mugnaioli C. 2008. The spread of CTX-M-type extended-spectrum beta-lactamases. Clin Microbiol Infect 14:33–41. 10.1111/j.1469-0691.2007.01867.x. [DOI] [PubMed] [Google Scholar]
- 8.Sommer MOA, Dantas G, Church GM. 2009. Functional characterization of the antibiotic resistance reservoir in the human microflora. Science 325:1128–1131. 10.1126/science.1176950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kintses B, Méhi O, Ari E, Számel M, Györkei Á, Jangir PK, Nagy I, Pál F, Fekete G, Tengölics R, Nyerges A, Likó I, Bálint A, Molnár T, Bálint B, Vásárhelyi BM, Bustamante M, Papp B, Pál C. 2019. Phylogenetic barriers to horizontal transfer of antimicrobial peptide resistance genes in the human gut microbiota. Nat Microbiol 4:447–458. 10.1038/s41564-018-0313-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Novelli A, Rosi E. 2017. Pharmacological properties of oral antibiotics for the treatment of uncomplicated urinary tract infections. J Chemother 29:10–18. 10.1080/1120009X.2017.1380357. [DOI] [PubMed] [Google Scholar]
- 11.Naeem A, Badshah SL, Muska M, Ahmad N, Khan K. 2016. The current case of quinolones: synthetic approaches and antibacterial activity. Molecules 21:268. 10.3390/molecules21040268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sköld O. 2010. Sulfonamides and trimethoprim. Expert Rev Anti Infect Ther 8:1–6. 10.1586/eri.09.117. [DOI] [PubMed] [Google Scholar]
- 13.Huttner A, Verhaegh EM, Harbarth S, Muller AE, Theuretzbacher U, Mouton JW. 2015. Nitrofurantoin revisited: a systematic review and meta-analysis of controlled trials. J Antimicrob Chemother 70:2456–2464. 10.1093/jac/dkv147. [DOI] [PubMed] [Google Scholar]
- 14.Falagas ME, Vouloumanou EK, Samonis G, Vardakas KZ. 2016. Fosfomycin. Clin Microbiol Rev 29:321–347. 10.1128/CMR.00068-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kohanski MA, Dwyer DJ, Collins JJ. 2010. How antibiotics kill bacteria: from targets to networks. Nat Rev Microbiol 8:423–435. 10.1038/nrmicro2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shun-Mei E, Zeng JM, Yuan H, Lu Y, Cai RX, Chen C. 2018. Sub-inhibitory concentrations of fluoroquinolones increase conjugation frequency. Microb Pathog 114:57–62. 10.1016/j.micpath.2017.11.036. [DOI] [PubMed] [Google Scholar]
- 17.Møller TS, Liu G, Boysen A, Thomsen LE, Lüthje FL, Mortensen S, Møller-Jensen J, Olsen JE. 2017. Treatment with cefotaxime affects expression of conjugation associated proteins and conjugation transfer frequency of an IncI1 plasmid in Escherichia coli. Front Microbiol 8:2365. 10.3389/fmicb.2017.02365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu G, Bogaj K, Bortolaia V, Olsen JE, Thomsen LE. 2018. Antibiotic-induced, increased conjugative transfer is common to diverse naturally occurring ESBL plasmids in Escherichia coli. Front Microbiol 10:2119. 10.3389/fmicb.2019.02119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu J, Wang Y, Jin M, Yuan Z, Bond P, Guo J. 2020. Both silver ions and silver nanoparticles facilitate the horizontal transfer of plasmid-mediated antibiotic resistance genes. Water Res 169:115229. 10.1016/j.watres.2019.115229. [DOI] [PubMed] [Google Scholar]
- 20.Zhang Y, Gu AZ, He M, Li D, Chen J. 2017. Subinhibitory concentrations of disinfectants promote the horizontal transfer of multidrug resistance genes within and across genera. Environ Sci Technol 51:570–580. 10.1021/acs.est.6b03132. [DOI] [PubMed] [Google Scholar]
- 21.Duy PT, Nguyen TNT, Thuy DV, The HC, Alcock F, Boinett C, Thanh HND, Tuyen HT, Thwaites GE, Rabaa MA, Baker S. 2020. Commensal Escherichia coli are a reservoir for the transfer of XDR plasmids into epidemic fluoroquinolone-resistant Shigella sonnei. Nat Microbiol 5:256–264. 10.1038/s41564-019-0645-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blázquez J, Rodríguez-Beltrán J, Matic I. 2018. Antibiotic-induced genetic variation: how it arises and how it can be prevented. Annu Rev Microbiol 72:209–230. 10.1146/annurev-micro-090817-062139. [DOI] [PubMed] [Google Scholar]
- 23.Blázquez J, Couce A, Rodríguez-Beltrán J, Rodríguez-Rojas A. 2012. Antimicrobials as promoters of genetic variation. Curr Opin Microbiol 15:561–569. 10.1016/j.mib.2012.07.007. [DOI] [PubMed] [Google Scholar]
- 24.Sutton MD, Smith BT, Godoy VG, Walker GC. 2000. The SOS response: recent insights into umuDC-dependent mutagenesis and DNA damage tolerance. Annu Rev Genet 34:479–497. 10.1146/annurev.genet.34.1.479. [DOI] [PubMed] [Google Scholar]
- 25.Baharoglu Z, Mazel D. 2014. SOS, the formidable strategy of bacteria against aggressions. FEMS Microbiol Rev 38:1126–1145. 10.1111/1574-6976.12077. [DOI] [PubMed] [Google Scholar]
- 26.Beaber JW, Hochhut B, Waldor MK. 2004. SOS response promotes horizontal dissemination of antibiotic resistance genes. Nature 427:72–74. 10.1038/nature02241. [DOI] [PubMed] [Google Scholar]
- 27.Thi TD, López E, Rodríguez-Rojas A, Rodríguez-Beltrán J, Couce A, Guelfo JR, Castañeda-García A, Blázquez J. 2011. Effect of recA inactivation on mutagenesis of Escherichia coli exposed to sublethal concentrations of antimicrobials. J Antimicrob Chemother 66:531–538. 10.1093/jac/dkq496. [DOI] [PubMed] [Google Scholar]
- 28.Pribis JP, García-Villada L, Zhai Y, Lewin-Epstein O, Wang AZ, Liu J, Xia J, Mei Q, Fitzgerald DM, Bos J, Austin RH, Herman C, Bates D, Hadany L, Hastings PJ, Rosenberg SM. 2019. Gamblers: an antibiotic-induced evolvable cell subpopulation differentiated by reactive-oxygen-induced general stress response. Mol Cell 74:785–800.E7. 10.1016/j.molcel.2019.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rodríguez-Rosado AI, Valencia EY, Rodríguez-Rojas A, Costas C, Galhardo RS, Rodríguez-Beltrán J, Blázquez J. 2019. N-acetylcysteine blocks SOS induction and mutagenesis produced by fluoroquinolones in Escherichia coli. J Antimicrob Chemother 74:2188–2196. 10.1093/jac/dkz210. [DOI] [PubMed] [Google Scholar]
- 30.Dhouib IE, Jallouli M, Annabi A, Gharbi N, Elfazaa S, Lasram MM. 2016. A minireview on N-acetylcysteine: an old drug with new approaches. Life Sci 151:359–363. 10.1016/j.lfs.2016.03.003. [DOI] [PubMed] [Google Scholar]
- 31.Samuni Y, Goldstein S, Dean OM, Berk M. 2013. The chemistry and biological activities of N-acetylcysteine. Biochim Biophys Acta 1830:4117–4129. 10.1016/j.bbagen.2013.04.016. [DOI] [PubMed] [Google Scholar]
- 32.Watanabe K, Tanaka M, Yuki S, Hirai M, Yamamoto Y. 2018. How is edaravone effective against acute ischemic stroke and amyotrophic lateral sclerosis? J Clin Biochem Nutr 62:20–38. 10.3164/jcbn.17-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pérez-González A, Galano A. 2011. OH radical scavenging activity of Edaravone: mechanism and kinetics. J Phys Chem B 115:1306–1314. 10.1021/jp110400t. [DOI] [PubMed] [Google Scholar]
- 34.Bailly C. 2019. Potential use of edaravone to reduce specific side effects of chemo-, radio- and immuno-therapy of cancers. Int Immunopharmacol 77:105967. 10.1016/j.intimp.2019.105967. [DOI] [PubMed] [Google Scholar]
- 35.Kong CS, Jeong CH, Choi JS, Kim KJ, Jeong JW. 2013. Antiangiogenic effects of p-coumaric acid in human endothelial cells. Phytother Res 27:317–323. 10.1002/ptr.4718. [DOI] [PubMed] [Google Scholar]
- 36.Shen Y, Song X, Li L, Sun J, Jaiswal Y, Huang J, Liu C, Yang W, Williams L, Zhang H, Guan Y. 2019. Protective effects of p-coumaric acid against oxidant and hyperlipidemia—an in vitro and in vivo evaluation. Biomed Pharmacother 111:579–587. 10.1016/j.biopha.2018.12.074. [DOI] [PubMed] [Google Scholar]
- 37.Pragasam SJ, Venkatesan V, Rasool M. 2013. Immunomodulatory and anti-inflammatory effect of p-coumaric acid, a common dietary polyphenol on experimental inflammation in rats. Inflammation 36:169–176. 10.1007/s10753-012-9532-8. [DOI] [PubMed] [Google Scholar]
- 38.Ferguson LR, Lim IF, Pearson AE, Ralph J, Harris PJ. 2003. Bacterial antimutagenesis by hydroxycinnamic acids from plant cell walls. Mutat Res 542:49–58. 10.1016/j.mrgentox.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 39.Redgrave LS, Sutton SB, Webber MA, Piddock LJ. 2014. Fluoroquinolone resistance: mechanisms, impact on bacteria, and role in evolutionary success. Trends Microbiol 22:438–445. 10.1016/j.tim.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 40.Hooper DC, Jacoby GA. 2015. Mechanisms of drug resistance: quinolone resistance. Ann N Y Acad Sci 1354:12–31. 10.1111/nyas.12830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Giske CG. 2015. Contemporary resistance trends and mechanisms for the old antibiotics colistin, temocillin, fosfomycin, mecillinam and nitrofurantoin. Clin Microbiol Infect 21:899–905. 10.1016/j.cmi.2015.05.022. [DOI] [PubMed] [Google Scholar]
- 42.Parikh A, Kathawala K, Tan CC, Garg S, Zhou XF. 2016. Development of a novel oral delivery system of edaravone for enhancing bioavailability. Int J Pharm 515:490–500. 10.1016/j.ijpharm.2016.10.052. [DOI] [PubMed] [Google Scholar]
- 43.Greene SC, Noonan PK, Sanabria C, Peacock WF. 2016. Effervescent N-acetylcysteine tablets versus oral solution N-acetylcysteine in fasting healthy adults: an open-label, randomized, single-dose, crossover, relative bioavailability study. Curr Ther Res Clin Exp 83:1–7. 10.1016/j.curtheres.2016.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Konishi Y, Hitomi Y, Yoshioka E. 2004. Intestinal absorption of p-coumaric and gallic acids in rats after oral administration. J Agric Food Chem 52:2527–2532. 10.1021/jf035366k. [DOI] [PubMed] [Google Scholar]
- 45.Van Boeckel TP, Gandra S, Ashok A, Caudron Q, Grenfell BT, Levin SA, Laxminarayan R. 2014. Global antibiotic consumption 2000 to 2010: an analysis of national pharmaceutical sales data. Lancet Infect Dis 14:742–750. 10.1016/S1473-3099(14)70780-7. [DOI] [PubMed] [Google Scholar]
- 46.Wellington EM, Boxall AB, Cross P, Feil EJ, Gaze WH, Hawkey PM, Johnson-Rollings AS, Jones D, Lee NM, Otten W, Thomas CM, Williams AP. 2013. The role of the natural environment in the emergence of antibiotic resistance in gram-negative bacteria. Lancet Infect Dis 13:155–165. 10.1016/S1473-3099(12)70317-1. [DOI] [PubMed] [Google Scholar]
- 47.Wang Y, Lu J, Mao L, Li J, Yuan Z, Bond PL, Guo J. 2019. Antiepileptic drug carbamazepine promotes horizontal transfer of plasmid-borne multi-antibiotic resistance genes within and across bacterial genera. ISME J 13:509–522. 10.1038/s41396-018-0275-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chistyakov VA, Prazdnova EV, Mazanko MS, Churilov MN, Chmyhalo VK. 2018. Increase in bacterial resistance to antibiotics after cancer therapy with platinum-based drugs. Mol Biol (Mosk) 52:270–276. 10.7868/S002689841S020106. [DOI] [PubMed] [Google Scholar]
- 49.Jin M, Lu J, Chen Z, Nguyen SH, Mao L, Li J, Yuan Z, Guo J. 2018. Antidepressant fluoxetine induces multiple antibiotics resistance in Escherichia coli via ROS-mediated mutagenesis. Environ Int 120:421–430. 10.1016/j.envint.2018.07.046. [DOI] [PubMed] [Google Scholar]
- 50.Baharoglu Z, Bikard D, Mazel D. 2010. Conjugative DNA transfer induces the bacterial SOS response and promotes antibiotic resistance development through integron activation. PLoS Genet 6:e1001165. 10.1371/journal.pgen.1001165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hoeksema M, Brul S, Ter Kuile BH. 2018. Influence of reactive oxygen species on de novo acquisition of resistance to bactericidal antibiotics. Antimicrob Agents Chemother 62:e02354-17. 10.1128/AAC.02354-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Aires-de-Sousa M, Ortiz de la Rosa JM, Goncalves ML, Costa A, Nordmann P, Poirel L. 2020. Occurrence of NDM-1-producing Morganella morganii and Proteus mirabilis in a single patient in Portugal: probable in vivo transfer by conjugation. J Antimicrob Chemother 75:903–906. 10.1093/jac/dkz542. [DOI] [PubMed] [Google Scholar]
- 53.Castro-Alférez M, Polo-López MI, Fernández-Ibáñez P. 2016. Intracellular mechanisms of solar water disinfection. Sci Rep 6:38145. 10.1038/srep38145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.European Committee on Antimicrobial Susceptibility Testing. 2020. Breakpoint tables for interpretation of MICs and zone diameters. Version 10.0. https://www.eucast.org/ast_of_bacteria/mic_determination/?mo_cache=1.
- 55.Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29:e45. 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Aires-de-Sousa M, Ortiz de la Rosa JM, Gonçalves ML, Pereira AL, Nordmann P, Poirel L. 2019. Epidemiology of carbapenemase-producing Klebsiella pneumoniae in a hospital, Portugal. Emerg Infect Dis 25:1632–1638. 10.3201/eid2509.190656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Potron A, Poirel L, Nordmann P. 2014. Derepressed transfer properties leading to the efficient spread of the plasmid encoding carbapenemase OXA-48. Antimicrob Agents Chemother 58:467–471. 10.1128/AAC.01344-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The complete nucleotide sequence of K. pneumoniae KPM2 was deposited in GenBank with accession number SRP308093.