ABSTRACT
Human immunodeficiency virus (HIV) persistence in tissue reservoirs is a major barrier to HIV cure. While antiretrovirals (ARVs) suppress viral replication, antiretroviral therapy (ART) interruption results in rapid rebound viremia that may originate from lymphoid tissues. To understand the relationship between anatomic distribution of ARV exposure and viral expression in lymph nodes, we performed mass spectrometry imaging (MSI) of 6 ARVs, RNAscope in situ hybridization for viral RNA (vRNA), and immunohistochemistry of collagen in mesenteric lymph nodes from 8 uninfected and 10 reverse transcriptase simian/human immunodeficiency virus (RT-SHIV)-infected rhesus macaques dosed to steady state with combination ART. MATLAB-based quantitative imaging analysis was used to evaluate spatial and pharmacological relationships between these ARVs, viral RNA (both vRNA+ cells and follicular dendritic cell [FDC]-bound virions), and collagen deposition. Using MSI, 31% of mesenteric lymph node tissue area was found to be not covered by any ARV. Additionally, 28% of FDC-trapped virions and 21% of infected cells were not exposed to any detected ARV. Of the 69% of tissue area that was covered by cumulative ART exposure, nearly 100% of concentrations were greater than in vitro 50% inhibitory concentration (IC50) values; however, 52% of total tissue coverage was from only one ARV, primarily maraviroc. Collagen covered ∼35% of tissue area but did not influence ARV distribution heterogeneity. Our findings are consistent with our hypothesis that ARV distribution, in addition to total-tissue drug concentration, must be considered when evaluating viral persistence in lymph nodes and other reservoir tissues.
KEYWORDS: antiretroviral agents, drug distribution, human immunodeficiency virus, mass spectrometry
INTRODUCTION
Antiretroviral (ARV) therapy (ART) has been an effective treatment for human immunodeficiency virus (HIV) for over 30 years (1). Combination ART successfully suppresses plasma viral load (pVL), contributing to increased life expectancy and fewer comorbidities for people living with HIV (2). However, despite ART’s success, there is still no cure for HIV, which necessitates lifelong drug adherence to avoid the viral rebound and subsequent immune sequelae that occur when ART is discontinued (3, 4).
One major barrier to HIV cure is the persistence of virus within the body’s tissues, even during ART (5, 6). These anatomical reservoirs include tissues like the lymph nodes, which contain large populations of HIV-targeted CD4+ T cells (7–12). There are several possible mechanisms of viral persistence within the lymph node during ART; some hypothesize that active, low-level HIV replication maintains the reservoir (13, 14), while others contest that latently infected cells undergo clonal expansion and actively produce virus only after treatment is discontinued (15, 16). Both mechanisms may be influenced by ARV pharmacology in the lymph node; if ARVs are not proximate to infected cells, or if drug concentrations are subtherapeutic, HIV persistence may occur.
Liquid chromatography-tandem mass spectrometry (LC-MS/MS) is the gold standard for measuring ARV concentrations in tissue, but due to different methods of tissue processing before analysis, even LC-MS/MS can produce conflicting results for ARV lymph node penetration. Our lab has previously measured ARV concentrations in homogenized lymph node tissue from nonhuman primates (NHPs) and humans and found that lymph node concentrations ranged from 1- to 100-fold higher than plasma concentrations (17). However, others analyzed ARV concentrations in isolated lymph node mononuclear cells—a method which may underestimate ARV exposure due to drug loss during cell isolation—and reported that ARV concentrations in these cells were 66 to 100% lower than those in peripheral blood (18). In both studies, ARV penetration was highly variable between tissue compartments, and the LC-MS/MS approach did not account for the spatial distribution of drug in lymph nodes. To understand this component of ARV pharmacology, novel quantitative mass spectrometry imaging (MSI) methods have been used to reveal heterogeneous ARV distribution in other reservoir tissues such as the ileum, rectum, brain, and axillary lymph nodes (19–24).
Many studies in NHPs and humans have shown that chronic immune activation and inflammation caused by simian immunodeficiency virus (SIV) or HIV infection can lead to extensive tissue fibrosis (8, 25–30), which may impair ARV distribution in the lymph node. Immunohistochemistry (IHC) imaging of collagen deposition, combined with MSI of ARVs, may help explain heterogeneous ARV distribution in tissue. Furthermore, when MSI results are combined with RNAscope in situ hybridization (ISH) for viral RNA distribution, the potential efficacy of ARVs in tissue may be elucidated.
In this study, we performed infrared matrix-assisted laser desorption electrospray ionization (IR-MALDESI) MSI of 6 ARVs, RNAscope ISH of viral RNA, and IHC imaging of collagen in mesenteric lymph nodes (MLNs) from uninfected NHPs and NHPs infected with reverse transcriptase simian-human immunodeficiency virus (RT-SHIV). We utilized quantitative imaging analysis methods to understand the spatial and pharmacological relationships between these targets.
RESULTS
MSI analysis of ARVs.
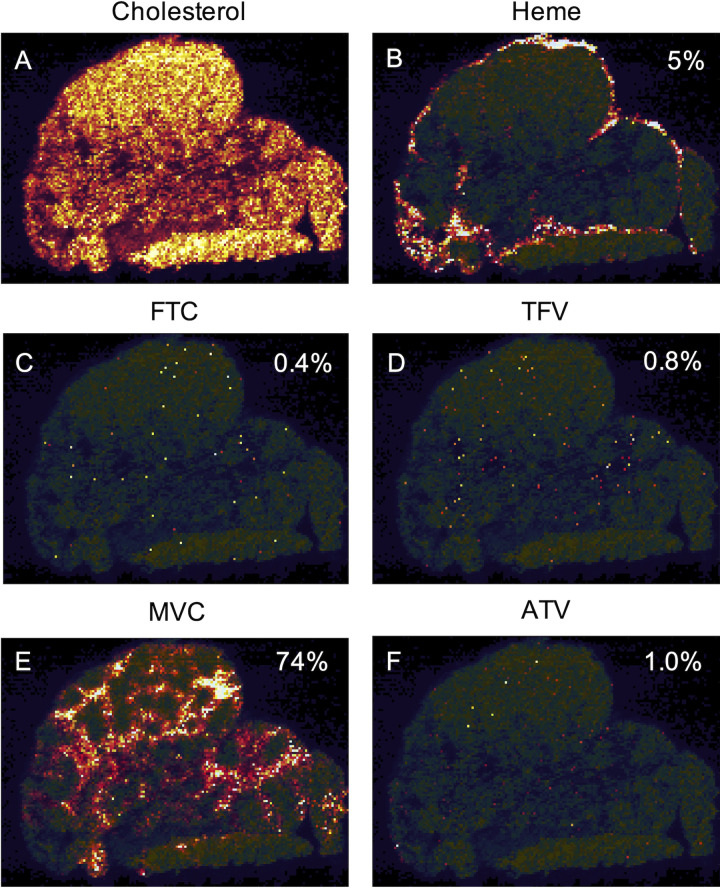
Representative images of ARV distribution are shown for RT-SHIV+ NHP 42966 in Fig. 1, reflecting FTMA dosing (see Materials and Methods). MLN drug concentrations determined through quantitative MSI were generally well correlated with those found by gold standard LC-MS/MS methods (Table 1). Across all 17 MLNs (NHP 40422 was excluded due to liver failure, as described in Materials and Methods), the heme-corrected cumulative (full 4-drug regimen) ARV response covered a median 69% (range, 5 to 98%) of MLN tissue area (Table 2). Excluding samples with drug concentrations that were below the limit of quantitation (BLQ), nearly 100% of the heme-corrected ARV response was at concentrations greater than in vitro 50% inhibitory concentration (IC50) estimates (Table 2). Compared to raw ARV response, heme correction did not significantly reduce cumulative drug coverage or individual drug coverage for either regimen (Table 2), suggesting minimal blood contamination of the tissues. ARV distribution was primarily localized to the medullary and paracortical regions of the MLN tissue.
FIG 1.
Representative intensity-scaled MSI images of the distribution of cholesterol (A), heme (B), emtricitabine (FTC) (C), tenofovir (TFV) (D), maraviroc (MVC) (E), and atazanavir (ATV) (F) in RT-SHIV+ NHP 42966. The percentage of MLN area covered by each target is indicated in each image. Brighter colors (white and yellow) represent higher MSI signal intensity, while darker colors (black and red) represent lower MSI signal intensity.
TABLE 1.
Median (range) MLN drug concentrations quantified by LC-MS/MS and MSI
| Drug |
n (n BLQa) |
LC-MS/MS concn (ng/g of tissue) |
MSI concn (ng/g of tissue) |
r | P |
|---|---|---|---|---|---|
| FTC | 17 (11) | 55 (10–135) | BLQ (BLQ–348) | 0.24 | 0.35 |
| TFV | 17 (3) | 1,477 (109–7,140) | 520 (BLQ–2,741) | 0.64 | 0.006 |
| EFV | 8 (0) | 157 (43–1,231)b | 152 (15–2,457)b | 0.62 | 0.11 |
| RAL | 8 (7) | 20 (11–62) | BLQ (BLQ–71) | 0.74 | 0.03 |
| MVC | 9 (0) | 4,792 (1,054–14,622) | 7,439 (1,221–39,711) | 0.75 | 0.02 |
| ATV | 9 (3) | 12 (1.7–784)b | 50 (12–825)b | 0.90 | 0.001 |
BLQ, below limit of quantitation.
Protein binding adjusted; Pearson correlation test.
TABLE 2.
MLN area covered by raw and heme-corrected drug response and heme-corrected drug response at concentrations greater than the IC50a
| Drug |
n (n BLQ) |
Median (range) % of MLN area covered by ARV |
Median (range) % of heme-corrected ARV area >IC50 (BLQs excluded) |
IC50 value (ng/g of tissue) |
IC50 reference | ||
|---|---|---|---|---|---|---|---|
| Raw | Heme corrected | P | |||||
| FTC | 17 (11) | 1.6 (0.09–11) | 1.6 (0.07–11) | NS | 100 (95–100) | 149 | 35 |
| TFV | 17 (3) | 8.4 (0.1–71) | 7.8 (0.1–71) | NS | 97 (23–100) | 2,303 | 34 |
| EFV | 8 (0) | 22 (5.2–77) | 22 (4.9–76) | NS | 100 (100–100) | 11.9b | 41 |
| RAL | 8 (7) | 0.10 (0.03–0.42) | 0.08 (0.03–0.42) | NS | 100 (100–100) | 2.9 | 42 |
| MVC | 9 (0) | 79 (56–97) | 79 (55–96) | NS | 100 (100–100) | 2.2 | 43 |
| ATV | 9 (3) | 3.3 (1.2–97) | 3.2 (1–96) | NS | 100 (100–100) | 16.6b | 44 |
| Cumulative | 17 | 70 (5.8–99) | 69 (5.4–98) | NS | |||
Mann-Whitney rank sum test. NS, not significant.
Protein binding adjusted.
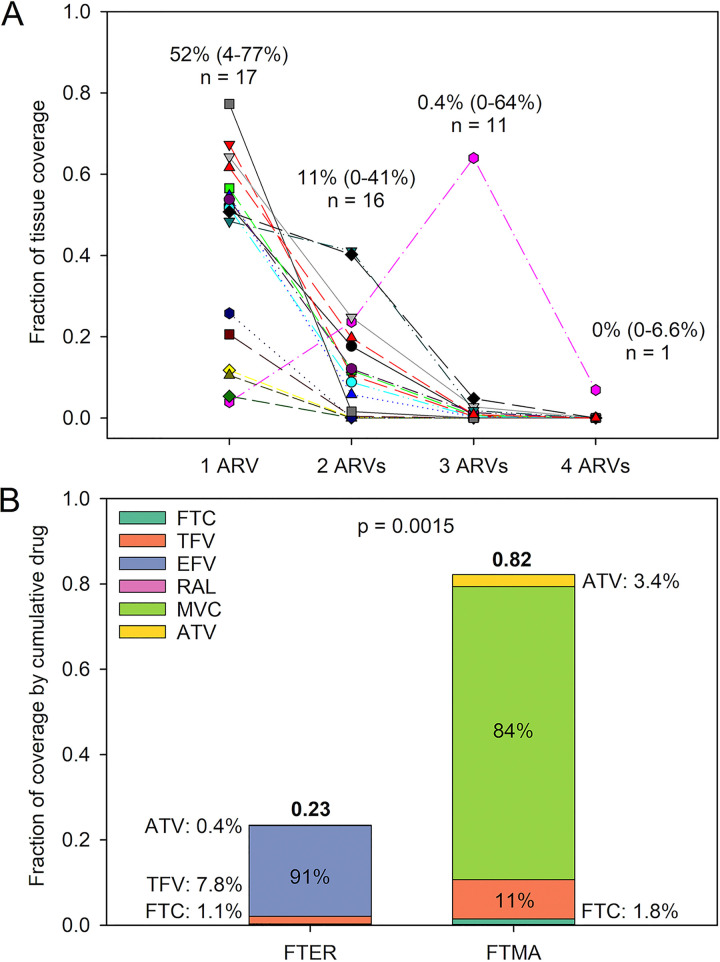
In evaluating ARV coverage in lymph node slices, a median of 52% of tissue area was covered by one ARV in all 17 MLNs. However, in assessing colocalization of multiple drugs, only 11% of tissue area in 16 MLNs were found to contain an overlap of 2 ARVs, 0.4% of tissue area in 11 MLNs contained an overlap of 3 ARVs, and only 1 MLN had overlap of all 4 ARVs, covering 6.9% of tissue area (Fig. 2A). There was no effect of RT-SHIV infection on cumulative regimen or individual drug coverage in MLNs (P > 0.05). Cumulative coverage differed by drug regimen; the FTER regimen (see Materials and Methods) resulted in less tissue area coverage than the FTMA regimen (P = 0.015 [Fig. 2B]). Efavirenz (EFV) and maraviroc (MVC) had the greatest contribution to each regimen, accounting for 91% and 84% of each regimen’s measured tissue exposure, respectively (Fig. 2B).
FIG 2.
(A) Fraction of MLN area covered by some or all of the ARVs in the 4-drug regimen. The median (range) percentage of coverage and the sample size are given for each number of ARVs. (B) Median fraction of MLN coverage by each of the 4-drug regimens compared by Student’s t test. The percent contribution of each ARV to the regimen’s cumulative coverage is also shown.
Quantitative imaging analysis of FDC-trapped virions and cell-associated viral RNA and colocalization with ARVs.
RT-SHIV follicular dendritic cell (FDC)-bound virions were identified in 4 of the 6 infected MLNs analyzed and accounted for a median 0.11% (range, 0 to 6%) of the analyzed tissue area. The median vRNA+ infected cell viral load (copies per square millimeter of tissue) was 106.4 (range, 27 to 171), which accounted for 0.007% (0.002 to 0.014%) of the tissue area. Due to the short duration of ART following 6 weeks of RT-SHIV infection, FDC-trapped virions were found within B cell follicles (BCFs) as expected, while vRNA+ infected cells were commonly present within and just outside BCFs in the surrounding T cell paracortical zones. Quantitative image analysis of virions and vRNA+ infected cell viral load (copies per square millimeter) in MLN tissue were compared to more traditional measurements (Table 3); our estimates of virion viral load were confirmed by strong correlation to necropsy MLN viral load (r = 0.99; P = 0.012). Virion viral load was also strongly correlated with peak pVL (r = 0.99; P = 0.006), while viral load based on only vRNA+ infected cells was not significantly correlated with other measures of viral burden (r ≤ 0.66; P ≥ 0.15) (Table 3).
TABLE 3.
Pearson correlation between different measures of viral load in MLNs
| Viral load measure | n | ART day 0 pVL (copies/ml) |
MLN VL at necropsy (copies/106 cells) |
Virion imaging (copies/mm2) |
caRNA imaging (copies/mm2) |
|---|---|---|---|---|---|
| Peak pVL (copies/ml) |
6 | r = 0.33; P = 0.52 | r = 0.97; P = 0.001 | r = 0.99; P = 0.006 | r = 0.62; P = 0.19 |
| ART day 0 pVL (copies/ml) |
6 | r = 0.48; P = 0.33 | r = 0.16; P = 0.84 | r = 0.66; P = 0.15 | |
| MLN VL at necropsy (copies/106 cells) |
6 | r = 0.99; P = 0.012 | r = 0.60; P = 0.21 | ||
| Virion imaging (copies/mm2) |
4 | r = 0.57; P = 0.43 |
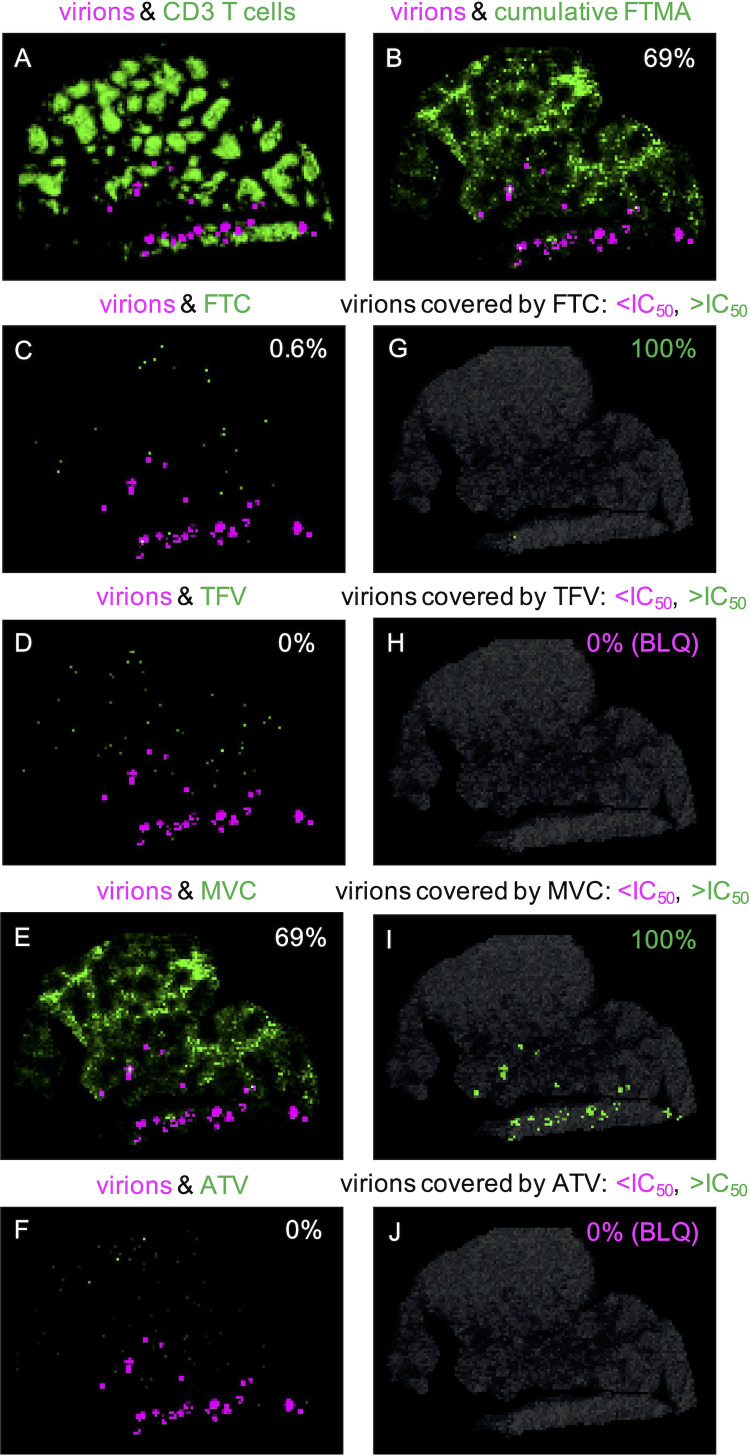
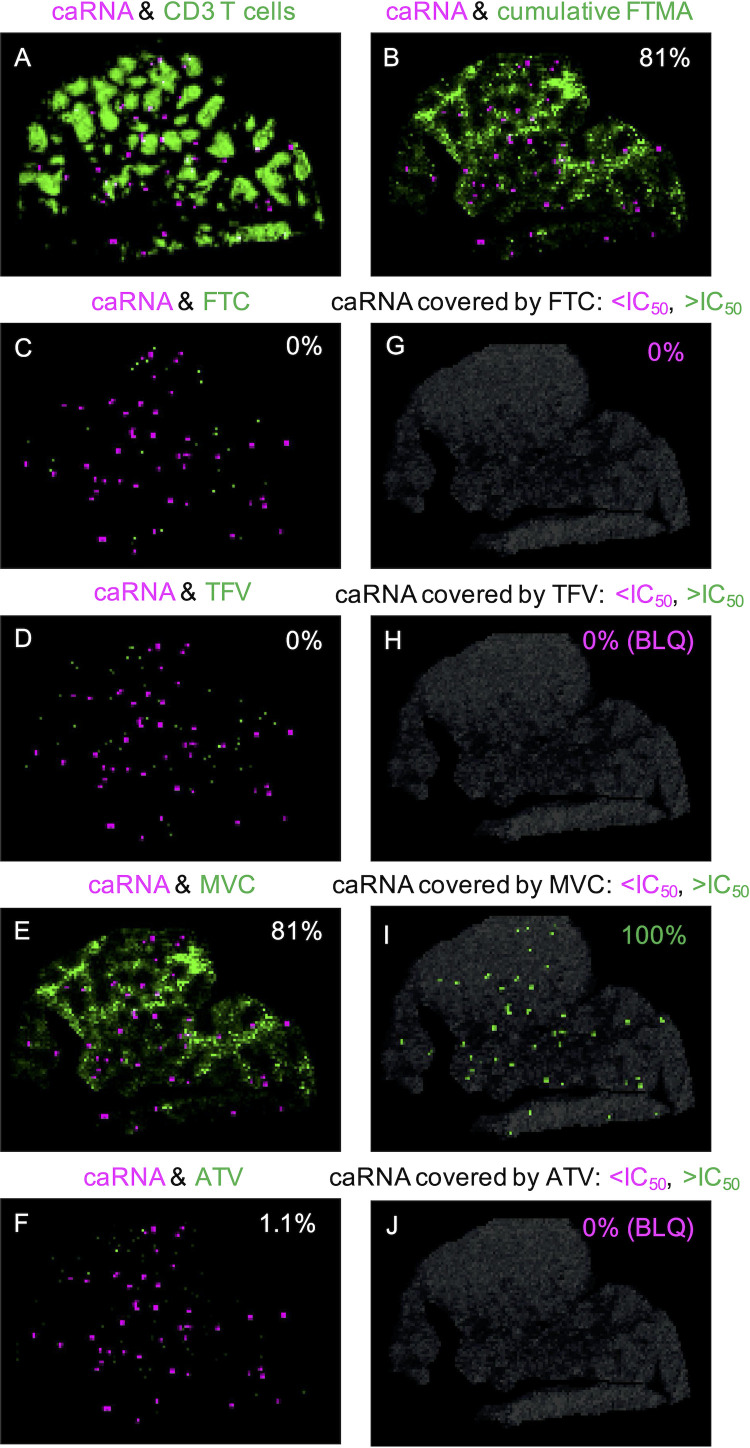
Representative images of FDC-trapped virions and vRNA+ infected cell (cell-associated RNA [caRNA]) colocalization with ARVs are shown for NHP 42966 in Fig. 3 and 4, respectively. Seventy-two percent (range, 11 to 100%) of virions and 79% (range, 13 to 81%) of caRNA colocalized with any ARV (Table 4). Once again, MVC had the largest contribution to overall coverage, colocalizing with 69% (range, 68 to 83%) of virions and 74% (range, 51 to 81%) of caRNA. We evaluated viral colocalization with inhibitory ARV coverage—excluding samples with drug concentrations that were BLQ—and found that 100% of the virus-colocalized drug response was at concentrations greater than the in vitro IC50 estimates (Table 4).
FIG 3.
(A to F) Representative images of the colocalization between FDC-trapped virions (magenta) and other targets (green) in RT-SHIV+ NHP 42966: virions and CD3+ T cells (A), virions and cumulative FTMA (B), and virions and individual ARVs, with the percentage of colocalization indicated in each image (C to F). (G to J) Images of only drug-covered virions, with the percentages below (magenta) and above (green) the IC50 indicated in each image. BLQ, below the limit of quantitation.
FIG 4.
(A to F) Representative images of the colocalization between caRNA (magenta) and other targets (green) in RT-SHIV+ NHP 42966: caRNA and CD3+ T cells (A), caRNA and cumulative FTMA (B), and caRNA and individual ARVs, with the percentage of colocalization indicated in each image (C to F). (G to J) Images of only drug-covered caRNA, with the percentages below (magenta) and above (green) the IC50 indicated in each image.
TABLE 4.
Virions and caRNA area covered by any drug response and ARV-covered virus at concentrations greater than the IC50
| Drug | FDC-trapped virions |
Cell-associated RNA |
||||
|---|---|---|---|---|---|---|
|
n (n BLQ) |
Median (range) % of virus area covered by ARV | Median (range) % of ARV-covered virus >IC50 (BLQs excluded) |
n (n BLQ) |
Median (range) % of virus area covered by ARV |
Median (range) % of ARV-covered virus >IC50 (BLQs excluded) |
|
| FTC | 4 (3) | 0.34 (0–2.5) | 100 (100–100) | 6 (3) | 0 (0–3.1) | 0 (0–0) |
| TFV | 4 (2) | 8.3 (0–19) | 100 (100–100) | 6 (2) | 8.6 (0–29) | 100 (100–100) |
| EFV | 1 (0) | 10 | 100 (100–100) | 1 (0) | 13 (13–13) | 100 (100–100) |
| RAL | 1 (1) | 0.4 | 1 (1) | 0.7 (0.7–0.7) | ||
| MVC | 3 (0) | 69 (68–83) | 100 (100–100) | 5 (0) | 74 (51–81) | 100 (100–100) |
| ATV | 3 (2) | 2.6 (0–17) | 100 (100–100) | 5 (2) | 2.0 (0–32) | 100 (0–100) |
| Cumulative | 4 | 72 (11–100) | 6 | 79 (13–81) | ||
Quantitative imaging analysis of collagen and colocalization with ARVs.
Across all 17 MLNs, collagen covered a median 35% (range, 8% to 68%) of tissue area, with greater collagen in uninfected NHPs (48%) than RT-SHIV+ animals (28%; P = 0.01). Representative images of collagen colocalization with ARVs are shown for NHP 42966 in Fig. 5. Based on morphological assessment, there were no apparent trends in the location of collagen relative to ARVs. Quantitative imaging analysis found that a median of 61% (range, 5.5 to 89%) of collagen was colocalized with ARVs (Table 5). The majority of collagen colocalization was with EFV (19% [range, 4.9 to 66%]) and MVC (73% [range, 45 to 88%]), driven by the large contribution of these ARVs to each regimen’s tissue coverage.
FIG 5.
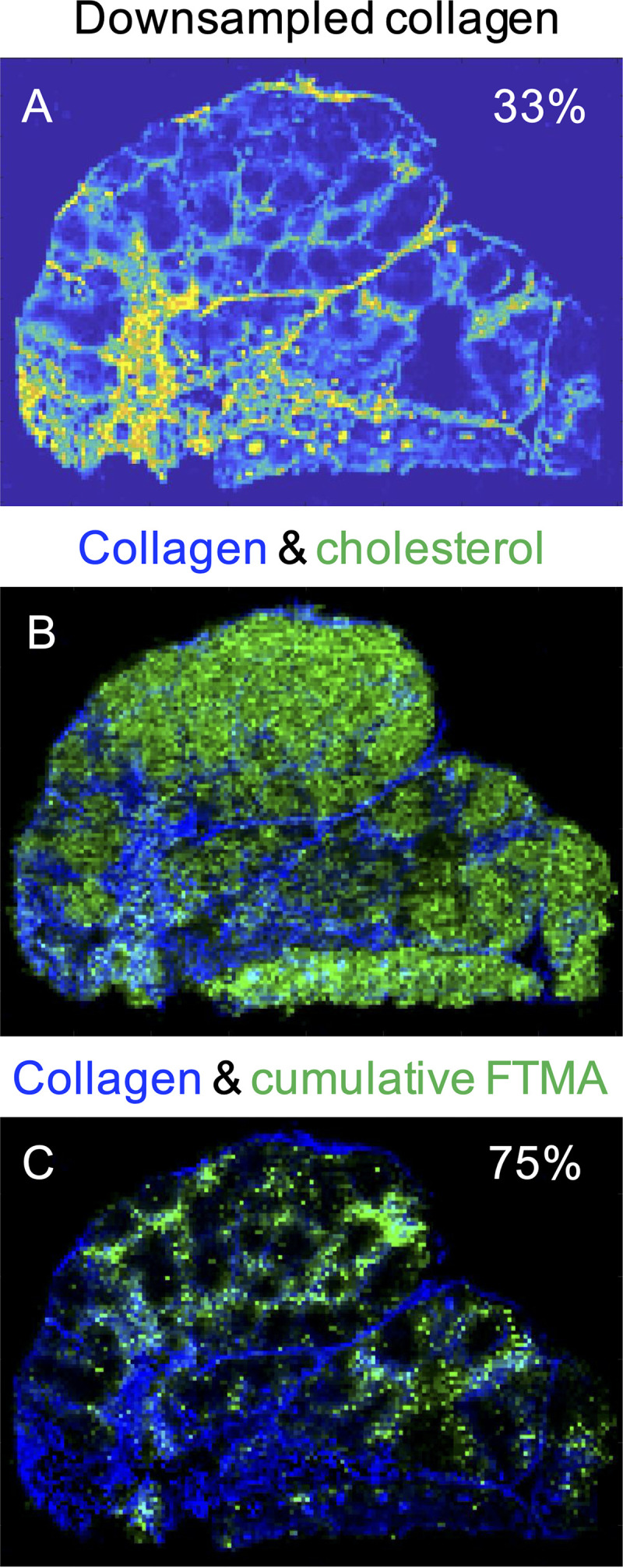
Representative images of the distribution of downsampled collagen (A), coregistered collagen (blue) and cholesterol (green) (B), and collagen and cumulative FTMA regimen coverage (C) in RT-SHIV+ NHP 42966. The percentage of MLN area covered by collagen is indicated in panel A, and the percentage of overlap between collagen and FTMA is indicated in panel C.
TABLE 5.
Collagen area covered by drug response
| Drug | n | Median (range) % of collagen area covered by ARV |
|---|---|---|
| FTC | 17 | 0.99 (0.03–10) |
| TFV | 17 | 7.1 (0.09–60) |
| EFV | 8 | 19 (4.9–66) |
| RAL | 8 | 0.10 (0.04–0.36) |
| MVC | 9 | 73 (45–88) |
| ATV | 9 | 2.6 (0.99–79) |
| Cumulative | 17 | 61 (5.5–89) |
DISCUSSION
This study assessed the spatial relationships of ARVs, collagen, and RT-SHIV RNA in the MLNs of NHPs using multiple quantitative imaging modalities. As with previous investigations (19, 20, 23, 24), we found heterogeneous drug distribution throughout this lymphoid tissue, with 31% of MLN area uncovered by any ARV. However, of the tissue covered by quantifiable drug, almost 100% of this coverage was above IC50 estimates. We assessed the colocalization of viral RNA with ARVs and found that 79% of caRNA and 72% of FDC-trapped virions were exposed to drug—though this was primarily just MVC—and again, 100% of quantifiable drug concentrations that colocalized with virus exceeded IC50 estimates. Drug heterogeneity did not appear to be caused by fibrosis, as collagen represented a median of 35% of tissue area, and there was 61% overlap between collagen and drug.
Out of 17 animals, only in 1 NHP could we demonstrate any tissue overlap of all 4 drugs, which covered 6.9% of the tissue area. We found that 69% of MLN tissue area is covered by cumulative ARV exposure and 52% of that coverage is from one drug alone, primarily EFV or MVC. Ongoing MSI analysis of axillary lymph nodes from these NHPs also found the greatest drug response from EFV and MVC (24). Interestingly, there was no significant difference in FTMA regimen coverage between MLNs (82%) and axillary lymph nodes (74%), but MVC concentrations were 2.5- to 4-fold higher in MLNs (see Fig. S1 in the supplemental material), demonstrating that drug concentrations are not necessarily correlated with extent of drug distribution in tissue. High MVC concentrations in MLNs may be a result of physicochemical properties that favor lymphatic absorption following oral dosing (31–36). After crossing into the enterocyte, soluble drugs with logP values greater than 5 (Table S2) are more likely to be incorporated into lipoproteins and transported out of the enterocyte and into the mesenteric lymphatics (37). MVC has a logP of 5.1 (34) and is highly soluble across the physiological pH range from 1.0 to 7.5 (38), so it is possible that these properties account for increased distribution to nearby draining MLNs compared to axillary lymph nodes. ATV also has a logP estimate of 5.6 (Table S2), but its solubility is variable and highly dependent on pH (39); the wide range of pH in NHP stomachs compared to humans (40) may be the cause of low ATV concentrations across all lymph nodes sampled (Fig. S1).
Using RNAscope, we identified virions distributed throughout the MLNs but predominantly within follicles, where they are expected to be trapped on FDCs (8, 18, 41, 42). When we quantified lymph node viral load based on virion signal, our estimate strongly correlated with peak pVL in weeks 2 to 3 after RT-SHIV infection, but not with pVL on day 0 of ART. This result is expected given that the size of the FDC reservoir is a marker of all previous virus production, rather than recently produced virus (18, 43). The size of the FDC reservoir is also known to be correlated with the number of productively infected cells in lymphoid tissue (44); however, we did not find this relationship in our MLNs. This may be due to underprediction of the infected cell viral load caused by saturated RNA signal intensity in infected cells; the signal saturation results in a high threshold for a single vRNA copy and therefore lowers the number of copies per square millimeter that meet that threshold. To truly confirm that the infected cells we quantified were transcriptionally active, we attempted to perform duplexed RNA/DNAscope to identify RNA+/DNA+ cells. However, our fresh-frozen tissue sections were too fragile for this method and resulted in extensive tissue damage and nonspecific HIV DNA staining. Despite this limitation, our designation of RNA+ T cells as infected cells is consistent with previous imaging analyses of lymphoid tissues (8, 18, 45).
Our quantitative imaging analyses demonstrate the utility of combining multiple imaging modalities to understand the pharmacology of ARVs within tissue reservoirs of HIV. We found that 72% of FDC-trapped virions and 79% of caRNA in MLNs were colocalized with any detectable ARV, though the majority of that coverage (82% and 84%, respectively) was by MVC alone. We also found that 100% of quantifiable, virus-colocalized ARV concentrations exceeded IC50 estimates. Prior work by our group has shown that in NHP ileal and rectal tissues, up to 80% of vRNA was colocalized with ARV concentrations exceeding IC90 thresholds (20). Here, we show that even with lower drug penetration in lymph nodes than in the gut (18, 46, 47), the extents of colocalization of virus and inhibitory drug exposure are similar between these tissues. While these results indicate significant ARV colocalization with vRNA in tissue, they do not guarantee complete inhibitory coverage of the large, inducible, replication-competent reservoir that exists in lymphoid tissues (45, 48–50). Indeed, many studies have shown that lymph nodes and gut-associated lymphoid tissue are the largest reservoirs of viral RNA and DNA during ART (9, 11, 45) and are likely sites of viral rebound (45, 51–53). We believe that taken together, these data support our hypothesis that heterogeneous ARV distribution—in this case, the 31% of MLN area unexposed to inhibitory drug concentrations—may contribute to viral persistence in reservoir tissues. Analyzing and improving ARV distribution in HIV reservoirs are important not only for HIV treatment (and adherence “forgiveness”) but also for the development of “kick-and-kill” cure strategies, since the successful use of these therapies is predicated on protective ARV coverage of uninfected cells during latency reversal.
To understand what causes heterogeneous drug distribution, we investigated the role of fibrosis in limiting drug distribution. We used IHC imaging and quantitative analysis and found that one-third of the MLN area represented collagen, which was primarily located in the medullary, sinusoidal, and capsule regions, where connective tissue is expected to be found for structural support. If fibrosis was preventing drug permeation, we would expect collagen and ARVs to have little overlap and occupy distinct areas within the MLNs; however, 61% of collagen colocalized with drug, mainly in the medulla and sinuses, where lymphatic fluid flow is prominent. This demonstrates that collagen was not restricting ARV distribution but was providing the normal structural support through which drug diffusion and cell trafficking may occur. There is also no evidence that this colocalization represents collagen binding to ARVs; in fact, collagen drug delivery systems often have difficulty retaining small molecules unless the drug is synthetically modified to include a collagen-binding domain (54).
Previous studies have reported that during SIV/HIV infection, disruptive collagen deposition occurs in the T cell zone of the lymph node (8, 25–30); however, as stated above, we observed collagen primarily in the medulla, sinuses, and capsule. We also saw more collagen deposition in uninfected MLNs, in contrast to existing evidence that T cell zone lymphoid tissue fibrosis increases significantly with infection (26, 28–30). To compare with these previous data, we performed additional imaging analyses to quantify collagen in only the T cell zone (data not shown), but the fraction of collagen coverage was still higher in uninfected MLNs (8.3% of T cell zone area) than infected MLNs (4.4%). These contradicting results may be explained by differences in viral kinetics between the RT-SHIV used in our study and SIV strains used in previous collagen analyses. One study investigating the viral kinetics of different simian HIV strains found that NHPs infected with SHIV89.6P had significantly higher peak viral loads and lower viral load set points than animals with SIVmac251 (55). Lower viral load set points in our RT-SHIV+ NHPs could lead to lower levels of immune activation, decreased transforming growth factor β1 (TGF-β1) signaling, and therefore reduced collagen deposition (25, 27, 56). Furthermore, the lack of collagen in the infected lymph nodes could simply be due to the short period of infection (6 weeks) of these NHPs, which may not have been long enough for appreciable fibrotic damage to occur.
Our prior work in lymph nodes from the same NHPs found that there was little influence of drug transporters on ARV penetration in these tissues; transporter protein and gene expression was low, and multiple linear regression analysis found no predictive relationships between transporter expression and ARV concentrations (17). This was also the case in the ileum and rectum tissue from the same NHPs (20), indicating that variability in drug distribution may be caused by more physiological mechanisms, such as changes in local pH, capillary permeability, lymphocyte trafficking, and lymph flow rates and patterns (57, 58). Current drug development research aims to increase ARV retention in lymph nodes using nanoparticle/lipid conjugate formulations (37, 59–61); MSI will be a useful tool to evaluate whether these macromolecule therapies improve drug distribution throughout tissue (62).
Imaging analyses have some inherent limitations. These analyses represent only one point in time, and a tissue section represents only one two-dimensional (2D) view of a 3D lymph node. This can have implications for the extent of viral RNA or collagen staining found in that section; a section containing many follicles will likely show increased viral RNA response due to FDC-trapped virions, while a section containing a large portion of the medulla might show increased collagen coverage. To account for this limitation, we made every attempt to section the MLNs along the medial plane, so that the sections we analyzed for viral RNA, collagen, and ARVs were representative of a full cross-section of MLN morphology and physiology.
MSI analysis of both MLN and gut tissues from these NHPs showed minimal detectable concentrations/coverage of emtricitabine (FTC) and tenofovir (TFV), contradicting LC-MS/MS analyses (20, 63). This discrepancy is a limitation of MSI sensitivity to detect and quantify these compounds (lower limits of quantitation: FTC, 116.13 fmol/voxel, and TFV, 122.01 fmol/voxel) compared to LC-MS/MS (FTC, 0.002 ng/g, and TFV, 0.02 ng/g). For stability reasons, we were unable to visualize and quantify the distribution of the intracellular active metabolites tenofovir diphosphate (TFVdp) and emtricitabine triphosphate (FTCtp). However, in our previous study, we performed LC-MS/MS analysis of TFVdp and FTCtp in the same NHP MLNs and found that the concentrations of TFVdp were well above its IC90 value (1.7 × 105 fmol/g of tissue [64], or 76 ng/g of tissue), and ratios of active metabolite to endogenous nucleotide (TFVdp:dATP and FTCtp:dCTP) exceeded predicted 90% effective concentration (EC90) targets (0.29 and 0.07, respectively [65]) by 1 to 2 logs (17). It is likely that TFVdp and FTCtp exhibit heterogeneous distribution within tissue reservoirs, since these intracellular metabolites are trafficked by T cells; indeed, studies have shown that penetration of these metabolites and their parent compounds differs between tissues and results in varied prevention of HIV infection (65, 66). Future work aims to increase IR-MALDESI sensitivity to TFV, FTC, TFVdp, and FTCtp to improve our understanding of spatial patterns between parent compounds, active metabolites, and T cell distribution.
Resolution of MSI images (100 μm/pixel) is much lower than for IHC/ISH images (0.5 μm/pixel) (67, 68), requiring a downsampling of ISH images to match MSI resolution and avoiding an underestimation of drug-virus colocalization. However, the downsampling process can limit location specificity of viral RNA. In this case, the size of a virion or infected cell becomes artificially inflated, and consequently, colocalization reflects proximity to ARVs within the dimension of a voxel. However, this voxel margin of error does help visualize possible in vivo drug or cell movement in the static MSI and ISH images, which otherwise would not be accounted for. Further work is ongoing to optimize the balance between IR-MALDESI sensitivity and resolution.
The second complication of our imaging analysis is the quantitation of infected cells. As previously discussed, we were unable to utilize duplexed RNA/DNAscope to identify productively infected, replication-competent cells in favor of snap-freezing tissue to avoid any shifting of diluting of drug concentrations in tissue. Rather, we relied on several techniques to arrive at an objective estimate of infected cell count. First, images were annotated to distinguish the visual appearance of FDC-trapped virions compared to infected cells. Then, we utilized these annotations within our MATLAB code to isolate and create separate images of infected cells and FDC-trapped virions, to ensure that these two measures of virus were analyzed separately. Furthermore, we also limited our RNA analysis to regions of the tissue that were intact and undamaged by the ISH process, to ensure that we did not include tissue artifacts in our analysis. All methods were automated within our MATLAB code, and all MLN viral RNA images were processed by the same code, eliminating investigator bias.
Using multiple imaging modalities to assess ARV, viral RNA, and collagen distribution in NHP MLNs, we demonstrated that ARV distribution is heterogeneous in tissue, and this heterogeneity was not caused by fibrosis at this stage of infection. We found that about one-quarter of RT-SHIV virions and infected cells were not exposed any ARV, providing evidence to support viral persistence mechanisms within MLNs. Continued ARV distribution and imaging work will be critical for effective kick-and-kill cure strategies aimed at eliminating virus within tissue reservoirs like the lymph nodes.
MATERIALS AND METHODS
NHP study and lymph node collection.
The NHP study was conducted at the California National Primate Research Center in partnership with the University of California, Davis, and was performed in accordance with Institutional Animal Care and Use Committee protocol 18345 from the University of California, Davis. Further details on animal dosing, infection, and tissue collection can be found in a previous report (17). Eighteen rhesus macaques approximately 3 to 4 years old and weighing 4 to 6 kg were used in this study. Eight NHPs (2 females and 6 males) were left uninfected, while 10 NHPs (4 females and 6 males) were infected for 6 weeks with RT-SHIV (11). pVL was monitored during the infection period; peak- and pre-ART pVL is reported in Table S1. After the 6-week infection period, all animals were dosed to steady state (8 to 10 days) with ARV doses based on previously reported effective treatment regimens in this animal model (69, 70). All NHPs received daily subcutaneous doses of 30 mg/kg of body weight of tenofovir (TFV) and 16 mg/kg of emtricitabine (FTC) plus one of the following oral combinations: 200 mg of efavirenz (EFV) daily plus 100 mg of raltegravir (RAL) twice daily (FTER regimen; n = 9) or 150 mg/kg of maraviroc (MVC) twice daily plus 270 mg/kg of atazanavir (ATV) twice daily (FTMA regimen; n = 9). One day after the final dose, NHPs were euthanized by phenobarbital overdose and necropsy was performed. At necropsy, MLNs were collected from each NHP and snap-frozen. MLN viral load at the time of necropsy is reported in Table S1. Tissue was stored at −80°C until analysis.
Tissue sectioning.
One MLN from each NHP (n = 18) was mounted on a specimen disc using O.C.T. (Tissue-Tek; Sakura Finetek, Inc., Torrance, CA) and cryosectioned at −24°C using a Leica CM1950 cryomicrotome (Leica Biosystems, Buffalo Grove, IL) at a thickness of 10 μm. Lymph node sections were thaw-mounted onto positively charged glass slides (IHC and ISH) or plain glass slides uniformly sprayed with internal standards (IR-MALDESI). Serial sections were collected from each MLN in the following order and quantity for each imaging method: IHC, 5 sections; IR-MALDESI, 3 sections; ISH, 2 sections; and LC-MS/MS, 2 sections.
LC-MS/MS and IR-MALDESI MSI analyses.
NHP 40422, on the FTER regimen, developed liver failure, resulting in plasma drug concentrations 17- to 260-fold higher than for other animals; this NHP was excluded from further analysis. LC-MS/MS methods for MLN samples have been detailed previously (17). Rapid equilibrium dialysis was performed on MLN homogenate following previous protocols (19, 71), and the extent of MLN protein binding of EFV (64% bound) and ATV (40% bound) was used to correct LC-MS/MS and MSI drug concentrations. Protein binding of TFV and FTC is negligible (<1% bound) (72, 73), and our rapid equilibrium analysis found minimal binding of RAL and MVC (17% and 10% bound, respectively).
Tissue sections taken for MSI analysis were mounted onto glass slides uniformly coated with internal standards using an HTX TM-Sprayer (HTX Technologies, LLC, Chapel Hill, NC). ARV standards were also spotted on undosed MLN tissues to generate calibration curves for drug quantification. The IR-MALDESI method of MSI has been described previously (20, 23, 74) and is briefly summarized here. Each mounted tissue section was cooled to −9°C in the source chamber, covered with a thin matrix of ice by increasing the relative humidity in the chamber, and ablated using an IR laser pulse (IR-Opolette 2371; Opotek, Carlsbad, CA) at a 100-μm spot-to-spot distance. Molecules from each ablated tissue volumetric pixel (voxel) were ionized by an orthogonal electrospray plume and collected into a Q-Exactive mass spectrometer (Thermo Fisher Scientific, Bremen, Germany). Raw signal intensity data from each voxel were converted to .imzML files using MSConvert (75) and imzMLConverter (76) and loaded into MSiReader (MATLAB application; Mathworks, Natick, MA) (77) to produce ion maps of ARV distribution throughout the MLN section (78). In addition to ARVs, spatial signal intensities were also acquired for cholesterol (an endogenous marker of overall tissue shape) and heme (to correct for blood contamination). Data from all ions of interest were exported into an Excel file for each of the tissue sections analyzed. For each MLN, the total concentration of each ARV in “ng/slice” was quantified using the calibration curve and normalized by the internal standards abacavir (29 μg/ml; FTC and MVC) and chloro-dihydro-pentynyl-trifluoromethyl benzoxazinone (32 μg/ml; TFV, EFV, RAL, and ATV). Drug concentrations were reported on a per-mass basis (nanograms per gram of tissue) by dividing the total concentration per slice by the volume of the tissue slice (area × thickness) and the tissue density (1.06 g/cm3). ARV maps were further thresholded by in vitro 50% inhibitory concentration (IC50) values to assess the distribution of what might be considered an “efficacious” ARV concentration in the MLNs. IC50 values for each ARV can be found in Table 4 (72, 73, 79–82).
RNAscope ISH analysis and IHC multiplex.
ISH processing of the MLN sections from NHPs 42226, 42971, and 40437 resulted in tissue damage that made further image analysis unreliable. These NHPs were excluded from viral RNA imaging but were included in drug and collagen distribution analyses. ISH analysis of viral RNA was performed on mounted MLN sections using a modification of the previously published RNAscope protocol (8). Slides were incubated for 30 min in Prefer fixative at room temperature and then rinsed twice in double-distilled water (ddH2O). Heat-induced epitope retrieval was performed by boiling slides in 1× target retrieval (ACD, Newark, CA; 322000) for 10 min, followed by peroxidase blocking for 10 min at room temperature. Slides were incubated with the target probe SIVmac239 sense (ACD; 312811) overnight at 40°C, and amplification was performed with an RNAscope 2.5 HD detection kit (Brown; ACD; 322310) using 0.5× wash buffer (ACD; 310091). Slides were developed with Alexa Fluor 647 (AF647)-conjugated tyramide.
To remove/inactivate the in situ amplification tree/horseradish peroxidase (HRP) complexes, slides were gently boiled in ACD retrieval buffer for 10 min. Slides were incubated with the target probe SIVmac239 antisense (ACD; 314071) for 2 h at 40°C, and amplification was performed with an RNAscope 2.5 HD detection kit (Brown; ACD; 322310) using 0.5× wash buffer (ACD; 310091). Slides were developed with Alexa Fluor 568-conjugated tyramide.
Slides were then stained for CD3 (Thermo Fisher; RM-9107-S [rabbit]) and developed with anti-mouse-AF488 (Invitrogen; A21202) and anti-rabbit-AF755 (Invitrogen; SA5-10043). Slides were counterstained with 4′,6-diamidino-2-phenylindole (DAPI; Invitrogen; D1306) at a dilution of 0.5 μg/ml in ddH2O for 10 min, coverslipped using Prolong Gold antifade mounting medium, and scanned using a Zeiss AxioScan Z1.
IHC analysis for collagen type I.
Mounted MLN sections taken for IHC analysis were dried in a fume hood at room temperature for 30 min, baked at 60°C for 30 min, fixed in 10% neutral buffered formalin for 15 min, and then washed 3 times in 1× phosphate-buffered saline (PBS). After fixation, each set of serial tissue sections underwent IHC analysis for collagen type I (Sigma-Aldrich, St. Louis, MO; catalog number C2456; dilution, 1:500), followed by chromogenic detection using 3,3′-diaminobenzidine (DAB)-Brown/horseradish peroxidase. Staining was performed in a Bond fully automated slide stainer, and images were digitized using an Aperio ScanScope XT slide scanner with a 20× objective (Leica Biosystems).
Quantitative image analysis and colocalization.
Quantitative imaging analysis of ARVs, collagen, T cells, and viral RNA was performed using MATLAB (Mathworks, Natick, MA). Collagen and multiplexed CD3 T cell and RNA microscopy images were imported into MATLAB, resized to reduce the image file size, and then flipped and/or rotated to match the orientation of ARV images. Bright-field RGB collagen images were color thresholded to isolate the DAB-Brown collagen stain, while grayscale CD3 T cell and RNA images derived from immunofluorescence were adjusted for brightness and intensity-thresholded to isolate cells and RNA while removing background. Follicular dendritic cell (FDC)-trapped virions and cell-associated RNA (caRNA) were isolated by hand and annotated using xy coordinates, and those annotations were used to create two separate images: one of the FDC-trapped virion response and one of the caRNA response. Binary masks of the thresholded FDC-trapped virions, caRNA, and collagen were generated. CD3+ T cell masks were also used to perform initial colocalization alignment steps. Lymph node viral load (copies per square millimeter) was determined separately for FDC-trapped virions and infected cells by analyzing the ISH signal associated with a single virion and dividing that signal by 2 to obtain an average estimate of the signal for a single copy of vRNA in each case.
MSI signal abundances were read into MATLAB from the exported Excel files, and scaled heat maps were generated for each ion. Cholesterol and heme maps were overlaid on each ARV and used as masks to eliminate off-tissue and blood-related drug responses, respectively. Annotations provided by RNAscope collaborators were also used to mask off regions of tissue damaged during ISH analysis. Due to the differences in resolution between IHC/ISH (∼0.5 μm/pixel) and IR-MALDESI (100 μm/pixel), virion, caRNA, collagen, and CD3 masks were downsampled to match the resolution and dimensions of the MSI data.
ARV distributions were then coregistered with isolated virion, caRNA, and collagen masks to assess spatial relationships between each target. The coregistration process has been previously reported (20) and is summarized here for ARVs and viral RNA: since lymph node shape and detail could be seen most clearly in the cholesterol and CD3 images, these were used to select corresponding control points based on matched section features. The cholesterol map was then transformed to match the CD3 mask using the MATLAB image warp function. This transformation was then applied to all heme-corrected ARV maps to ensure consistent alignment. Finally, the aligned, heme-corrected ARV maps were overlaid with viral RNA masks to evaluate colocalization, including IC50-thresholded ARV maps to assess virus colocalization with inhibitory drug concentrations. This process was repeated between collagen and ARVs.
Statistical analysis.
Data are presented as medians (ranges). Relationships between LC-MS/MS and MSI drug concentrations were assessed using a Pearson correlation test. Differences in raw and heme-corrected individual ARV responses were evaluated by one-way analysis of variance (ANOVA) with Dunn’s method for multiple comparisons; cumulative drug regimen raw response versus heme-corrected response was evaluated by a Mann-Whitney rank sum test. The difference in the fraction of tissue covered by the FTER regimen versus the FTMA regimen was assessed with Student’s t test. Variance in the extent of collagen coverage between uninfected and RT-SHIV+ MLNs was evaluated by a Mann-Whitney rank sum test. A Pearson correlation test was used to examine the relationships between various measures of viral load. All statistical tests were performed using SigmaPlot 13.0 (Systat Software, Inc., San Jose, CA). A P value of <0.05 was considered statistically significant.
Data availability.
Initial MSI data processing was performed using the free, open-source MSiReader software (https://www.msireader.wordpress.ncsu.edu/) (77), and quantitative imaging analyses were performed using MATLAB software accessed through a university license (https://www.mathworks.com/products/matlab.html). MSI data are available online through METASPACE database (https://www.metaspace2020.eu) (83) under the project name “Scholz_NHP_LN_ARV.”
ACKNOWLEDGMENTS
This work was supported by the National Institutes of Health, National Institute of Allergy and Infectious Diseases, grants R01 AI111891, R01 AI143411-01A1, and R01 AI149672, The University of North Carolina at Chapel Hill Center for AIDS Research (UNC CFAR) grant P30 AI50410, The Bill and Melinda Gates Foundation grant INV-002704, and the Base Grant to the California National Primate Research Center (P51 OD011107) and Oregon National Primate Research Center (P51 OD011092).
Footnotes
Supplemental material is available online only.
aac.00019-21-s000s1.pdf (154.2KB, pdf)
REFERENCES
- 1.Palmisano L, Vella S. 2011. A brief history of antiretroviral therapy of HIV infection: success and challenges. Ann Ist Super Sanita 47:44–48. 10.4415/ANN_11_01_10. [DOI] [PubMed] [Google Scholar]
- 2.Samji H, Cescon A, Hogg RS, Modur SP, Althoff KN, Buchacz K, Burchell AN, Cohen M, Gebo KA, Gill MJ, Justice A, Kirk G, Klein MB, Korthuis PT, Martin J, Napravnik S, Rourke SB, Sterling TR, Silverberg MJ, Deeks S, Jacobson LP, Bosch RJ, Kitahata MM, Goedert JJ, Moore R, Gange SJ, North American AIDS Cohort Collaboration on Research and Design (NA-ACCORD) of IeDEA. 2013. Closing the gap: increases in life expectancy among treated HIV-positive individuals in the United States and Canada. PLoS One 8:e81355. 10.1371/journal.pone.0081355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chun TW, Davey RT, Engel D, Lane HC, Fauci AS. 1999. Re-emergence of HIV after stopping therapy. Nature 401:874–875. 10.1038/44755. [DOI] [PubMed] [Google Scholar]
- 4.Lewin SR, Rouzioux C. 2011. HIV cure and eradication: how will we get from the laboratory to effective clinical trials? AIDS 25:885–897. 10.1097/QAD.0b013e3283467041. [DOI] [PubMed] [Google Scholar]
- 5.Natarajan V, Bosche M, Metcalf JA, Ward DJ, Lane HC, Kovacs JA. 1999. HIV-1 replication in patients with undetectable plasma virus receiving HAART. Highly active antiretroviral therapy. Lancet 353:119–120. 10.1016/s0140-6736(05)76156-0. [DOI] [PubMed] [Google Scholar]
- 6.Buzón MJ, Codoñer FM, Frost SDW, Pou C, Puertas MC, Massanella M, Dalmau J, Llibre JM, Stevenson M, Blanco J, Clotet B, Paredes R, Martinez-Picado J. 2011. Deep molecular characterization of HIV-1 dynamics under suppressive HAART. PLoS Pathog 7:e1002314. 10.1371/journal.ppat.1002314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wong JK, Yukl SA. 2016. Tissue reservoirs of HIV. Curr Opin HIV AIDS 11:362–370. 10.1097/COH.0000000000000293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deleage C, Wietgrefe SW, Del Prete G, Morcock DR, Hao XP, Piatak M, Bess J, Anderson JL, Perkey KE, Reilly C, McCune JM, Haase AT, Lifson JD, Schacker TW, Estes JD. 2016. Defining HIV and SIV reservoirs in lymphoid tissues. Pathog Immun 1:68–106. 10.20411/pai.v1i1.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Horiike M, Iwami S, Kodama M, Sato A, Watanabe Y, Yasui M, Ishida Y, Kobayashi T, Miura T, Igarashi T. 2012. Lymph nodes harbor viral reservoirs that cause rebound of plasma viremia in SIV-infected macaques upon cessation of combined antiretroviral therapy. Virology 423:107–118. 10.1016/j.virol.2011.11.024. [DOI] [PubMed] [Google Scholar]
- 10.Schrager LK, D’Souza MP. 1998. Cellular and anatomical reservoirs of HIV-1 in patients receiving potent antiretroviral combination therapy. JAMA 280:67–71. 10.1001/jama.280.1.67. [DOI] [PubMed] [Google Scholar]
- 11.North TW, Higgins J, Deere JD, Hayes TL, Villalobos A, Adamson L, Shacklett BL, Schinazi RF, Luciw PA. 2010. Viral sanctuaries during highly active antiretroviral therapy in a nonhuman primate model for AIDS. J Virol 84:2913–2922. 10.1128/JVI.02356-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pantaleo G, Graziosi C, Butini L, Pizzo PA, Schnittman SM, Kotler DP, Fauci AS. 1991. Lymphoid organs function as major reservoirs for human immunodeficiency virus. Proc Natl Acad Sci U S A 88:9838–9842. 10.1073/pnas.88.21.9838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lorenzo-Redondo R, Fryer HR, Bedford T, Kim E-Y, Archer J, Pond SLK, Chung Y-S, Penugonda S, Chipman J, Fletcher CV, Schacker TW, Malim MH, Rambaut A, Haase AT, McLean AR, Wolinsky SM. 2016. Persistent HIV-1 replication maintains the tissue reservoir during therapy. Nature 530:51–56. 10.1038/nature16933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rose R, Lamers SL, Nolan DJ, Maidji E, Faria NR, Pybus OG, Dollar JJ, Maruniak SA, McAvoy AC, Salemi M, Stoddart CA, Singer EJ, McGrath MS. 2016. HIV maintains an evolving and dispersed population in multiple tissues during suppressive combined antiretroviral therapy in individuals with cancer. J Virol 90:8984–8993. 10.1128/JVI.00684-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McManus WR, Bale MJ, Spindler J, Wiegand A, Musick A, Patro SC, Sobolewski MD, Musick VK, Anderson EM, Cyktor JC, Halvas EK, Shao W, Wells D, Wu X, Keele BF, Milush JM, Hoh R, Mellors JW, Hughes SH, Deeks SG, Coffin JM, Kearney MF. 2019. HIV-1 in lymph nodes is maintained by cellular proliferation during antiretroviral therapy. J Clin Invest 129:4629–4642. 10.1172/JCI126714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kearney MF, Wiegand A, Shao W, McManus WR, Bale MJ, Luke B, Maldarelli F, Mellors JW, Coffin JM. 2017. Ongoing HIV replication during ART reconsidered. Open Forum Infect Dis 4:ofx173. 10.1093/ofid/ofx173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burgunder E, Fallon JK, White N, Schauer AP, Sykes C, Remling-Mulder L, Kovarova M, Adamson L, Luciw P, Garcia JV, Akkina R, Smith PC, Kashuba ADM. 2019. Antiretroviral drug concentrations in lymph nodes: a cross-species comparison of the effect of drug transporter expression, viral infection, and sex in humanized mice, nonhuman primates, and humans. J Pharmacol Exp Ther 370:360–368. 10.1124/jpet.119.259150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fletcher CV, Staskus K, Wietgrefe SW, Rothenberger M, Reilly C, Chipman JG, Beilman GJ, Khoruts A, Thorkelson A, Schmidt TE, Anderson J, Perkey K, Stevenson M, Perelson AS, Douek DC, Haase AT, Schacker TW. 2014. Persistent HIV-1 replication is associated with lower antiretroviral drug concentrations in lymphatic tissues. Proc Natl Acad Sci U S A 111:2307–2312. 10.1073/pnas.1318249111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Srinivas N, Rosen EP, Gilliland WM, Kovarova M, Remling-Mulder L, De La Cruz G, White N, Adamson L, Schauer AP, Sykes C, Luciw P, Garcia JV, Akkina R, Kashuba ADM. 2019. Antiretroviral concentrations and surrogate measures of efficacy in the brain tissue and CSF of preclinical species. Xenobiotica 49:1192–1201. 10.1080/00498254.2018.1539278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thompson CG, Rosen EP, Prince HMA, White N, Sykes C, de la Cruz G, Mathews M, Deleage C, Estes JD, Charlins P, Mulder LR, Kovarova M, Adamson L, Arora S, Dellon ES, Peery AF, Shaheen NJ, Gay C, Muddiman DC, Akkina R, Garcia JV, Luciw P, Kashuba ADM. 2019. Heterogeneous antiretroviral drug distribution and HIV/SHIV detection in the gut of three species. Sci Transl Med 11:eaap8758. 10.1126/scitranslmed.aap8758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ntshangase S, Mdanda S, Naicker T, Kruger HG, Baijnath S, Govender T. 2019. Spatial distribution of elvitegravir and tenofovir in rat brain tissue: application of MALDI-MSI and LC-MS/MS. Rapid Commun Mass Spectrom 33:1643–1651. 10.1002/rcm.8510. [DOI] [PubMed] [Google Scholar]
- 22.Ntshangase S, Mdanda S, Singh SD, Naicker T, Kruger HG, Baijnath S, Govender T. 2019. Mass spectrometry imaging demonstrates the regional brain distribution patterns of three first-line antiretroviral drugs. ACS Omega 4:21169–21177. 10.1021/acsomega.9b02582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thompson CG, Bokhart MT, Sykes C, Adamson L, Fedoriw Y, Luciw PA, Muddiman DC, Kashuba ADM, Rosen EP. 2015. Mass spectrometry imaging reveals heterogeneous efavirenz distribution within putative HIV reservoirs. Antimicrob Agents Chemother 59:2944–2948. 10.1128/AAC.04952-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rosen E. 2018. Mass spectrometry imaging reveals associations between antiretrovirals, SHIV, and cells in lymph nodes, abstr 475. 2018 CROI.
- 25.Estes JD, Wietgrefe S, Schacker T, Southern P, Beilman G, Reilly C, Milush JM, Lifson JD, Sodora DL, Carlis JV, Haase AT. 2007. Simian immunodeficiency virus-induced lymphatic tissue fibrosis is mediated by transforming growth factor beta 1-positive regulatory T cells and begins in early infection. J Infect Dis 195:551–561. 10.1086/510852. [DOI] [PubMed] [Google Scholar]
- 26.Estes JD, Haase AT, Schacker TW. 2008. The role of collagen deposition in depleting CD4+ T cells and limiting reconstitution in HIV-1 and SIV infections through damage to the secondary lymphoid organ niche. Semin Immunol 20:181–186. 10.1016/j.smim.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zeng M, Smith AJ, Wietgrefe SW, Southern PJ, Schacker TW, Reilly CS, Estes JD, Burton GF, Silvestri G, Lifson JD, Carlis JV, Haase AT. 2011. Cumulative mechanisms of lymphoid tissue fibrosis and T cell depletion in HIV-1 and SIV infections. J Clin Invest 121:998–1008. 10.1172/JCI45157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schacker TW, Nguyen PL, Beilman GJ, Wolinsky S, Larson M, Reilly C, Haase AT. 2002. Collagen deposition in HIV-1 infected lymphatic tissues and T cell homeostasis. J Clin Invest 110:1133–1139. 10.1172/JCI0216413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schacker TW, Brenchley JM, Beilman GJ, Reilly C, Pambuccian SE, Taylor J, Skarda D, Larson M, Douek DC, Haase AT. 2006. Lymphatic tissue fibrosis is associated with reduced numbers of naive CD4+ T cells in human immunodeficiency virus type 1 infection. Clin Vaccine Immunol 13:556–560. 10.1128/CVI.13.5.556-560.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schacker TW, Reilly C, Beilman GJ, Taylor J, Skarda D, Krason D, Larson M, Haase AT. 2005. Amount of lymphatic tissue fibrosis in HIV infection predicts magnitude of HAART-associated change in peripheral CD4 cell count. AIDS 19:2169–2171. 10.1097/01.aids.0000194801.51422.03. [DOI] [PubMed] [Google Scholar]
- 31.National Center for Biotechnology Information. 2021. PubChem compound summary for CID 464205, tenofovir. https://pubchem.ncbi.nlm.nih.gov/compound/Tenofovir.
- 32.National Center for Biotechnology Information. 2021. PubChem compound summary for CID 60877, emtricitabine. https://pubchem.ncbi.nlm.nih.gov/compound/Emtricitabine.
- 33.Moss DM, Siccardi M, Murphy M, Piperakis MM, Khoo SH, Back DJ, Owen A. 2012. Divalent metals and pH alter raltegravir disposition in vitro. Antimicrob Agents Chemother 56:3020–3026. 10.1128/AAC.06407-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.National Center for Biotechnology Information. 2021. PubChem compound summary for CID 3002977, maraviroc. https://pubchem.ncbi.nlm.nih.gov/compound/Maraviroc.
- 35.National Center for Biotechnology Information. 2021. PubChem compound summary for CID 148192, atazanavir. https://pubchem.ncbi.nlm.nih.gov/compound/Atazanavir.
- 36.National Center for Biotechnology Information. 2021. PubChem compound summary for CID 64139, efavirenz. https://pubchem.ncbi.nlm.nih.gov/compound/Efavirenz.
- 37.Trevaskis NL, Kaminskas LM, Porter CJH. 2015. From sewer to saviour—targeting the lymphatic system to promote drug exposure and activity. Nat Rev Drug Discov 14:781–803. 10.1038/nrd4608. [DOI] [PubMed] [Google Scholar]
- 38.Food and Drug Administration. SELZENTRY (maraviroc) package insert. Food and Drug Administration, Silver Spring, MD. [Google Scholar]
- 39.Berlin M, Ruff A, Kesisoglou F, Xu W, Wang MH, Dressman JB. 2015. Advances and challenges in PBPK modeling—analysis of factors contributing to the oral absorption of atazanavir, a poorly soluble weak base. Eur J Pharm Biopharm 93:267–280. 10.1016/j.ejpb.2015.03.031. [DOI] [PubMed] [Google Scholar]
- 40.Hatton GB, Yadav V, Basit AW, Merchant HA. 2015. Animal farm: considerations in animal gastrointestinal physiology and relevance to drug delivery in humans. J Pharm Sci 104:2747–2776. 10.1002/jps.24365. [DOI] [PubMed] [Google Scholar]
- 41.Bronnimann MP, Skinner PJ, Connick E. 2018. The B-cell follicle in HIV infection: barrier to a cure. Front Immunol 9:20. 10.3389/fimmu.2018.00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Heesters BA, Lindqvist M, Vagefi PA, Scully EP, Schildberg FA, Altfeld M, Walker BD, Kaufmann DE, Carroll MC. 2015. Follicular dendritic cells retain infectious HIV in cycling endosomes. PLoS Pathog 11:e1005285. 10.1371/journal.ppat.1005285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Haase AT. 1999. Population biology of HIV-1 infection: viral and CD4+ T cell demographics and dynamics in lymphatic tissues. Annu Rev Immunol 17:625–656. 10.1146/annurev.immunol.17.1.625. [DOI] [PubMed] [Google Scholar]
- 44.Haase AT, Henry K, Zupancic M, Sedgewick G, Faust RA, Melroe H, Cavert W, Gebhard K, Staskus K, Zhang ZQ, Dailey PJ, Balfour HH, Erice A, Perelson AS. 1996. Quantitative image analysis of HIV-1 infection in lymphoid tissue. Science 274:985–989. 10.1126/science.274.5289.985. [DOI] [PubMed] [Google Scholar]
- 45.Estes JD, Kityo C, Ssali F, Swainson L, Makamdop KN, Del Prete GQ, Deeks SG, Luciw PA, Chipman JG, Beilman GJ, Hoskuldsson T, Khoruts A, Anderson J, Deleage C, Jasurda J, Schmidt TE, Hafertepe M, Callisto SP, Pearson H, Reimann T, Schuster J, Schoephoerster J, Southern P, Perkey K, Shang L, Wietgrefe SW, Fletcher CV, Lifson JD, Douek DC, McCune JM, Haase AT, Schacker TW. 2017. Defining total-body AIDS-virus burden with implications for curative strategies. Nat Med 23:1271–1276. 10.1038/nm.4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Devanathan AS, Pirone JR, Akkina R, Remling-Mulder L, Luciw P, Adamson L, Garcia JV, Kovarova M, White NR, Schauer AP, Blake K, Sykes C, Burgunder EM, Srinivas N, Rosen EP, Kashuba ADM. 2019. Antiretroviral penetration across three preclinical animal models and humans in eight putative HIV viral reservoirs. Antimicrob Agents Chemother 64:e01639-19. 10.1128/AAC.01639-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee SA, Telwatte S, Hatano H, Kashuba ADM, Cottrell ML, Hoh R, Liegler TJ, Stephenson S, Somsouk M, Hunt PW, Deeks SG, Yukl S, Savic RM. 2020. Antiretroviral therapy concentrations differ in gut vs. lymph node tissues and are associated with HIV viral transcription by a novel RT-ddPCR assay. J Acquir Immune Defic Syndr 83:530–537. 10.1097/QAI.0000000000002287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sanyal A, Mailliard RB, Rinaldo CR, Ratner D, Ding M, Chen Y, Zerbato JM, Giacobbi NS, Venkatachari NJ, Patterson BK, Chargin A, Sluis-Cremer N, Gupta P. 2017. Novel assay reveals a large, inducible, replication-competent HIV-1 reservoir in resting CD4(+) T cells. Nat Med 23:885–889. 10.1038/nm.4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ho Y-C, Shan L, Hosmane NN, Wang J, Laskey SB, Rosenbloom DIS, Lai J, Blankson JN, Siliciano JD, Siliciano RF. 2013. Replication-competent noninduced proviruses in the latent reservoir increase barrier to HIV-1 cure. Cell 155:540–551. 10.1016/j.cell.2013.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bruner KM, Murray AJ, Pollack RA, Soliman MG, Laskey SB, Capoferri AA, Lai J, Strain MC, Lada SM, Hoh R, Ho Y-C, Richman DD, Deeks SG, Siliciano JD, Siliciano RF. 2016. Defective proviruses rapidly accumulate during acute HIV-1 infection. Nat Med 22:1043–1049. 10.1038/nm.4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vibholm LK, Lorenzi JCC, Pai JA, Cohen YZ, Oliveira TY, Barton JP, Garcia Noceda M, Lu C-L, Ablanedo-Terrazas Y, Del Rio Estrada PM, Reyes-Teran G, Tolstrup M, Denton PW, Damsgaard T, Søgaard OS, Nussenzweig MC. 2019. Characterization of intact proviruses in blood and lymph node from HIV-infected individuals undergoing analytical treatment interruption. J Virol 93:e01920-18. 10.1128/JVI.01920-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rothenberger MK, Keele BF, Wietgrefe SW, Fletcher CV, Beilman GJ, Chipman JG, Khoruts A, Estes JD, Anderson J, Callisto SP, Schmidt TE, Thorkelson A, Reilly C, Perkey K, Reimann TG, Utay NS, Nganou Makamdop K, Stevenson M, Douek DC, Haase AT, Schacker TW. 2015. Large number of rebounding/founder HIV variants emerge from multifocal infection in lymphatic tissues after treatment interruption. Proc Natl Acad Sci U S A 112:E1126–E1134. 10.1073/pnas.1414926112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chaillon A, Gianella S, Dellicour S, Rawlings SA, Schlub TE, De Oliveira MF, Ignacio C, Porrachia M, Vrancken B, Smith DM. 2020. HIV persists throughout deep tissues with repopulation from multiple anatomical sources. J Clin Invest 130:1699–1712. 10.1172/JCI134815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wallace DG, Rosenblatt J. 2003. Collagen gel systems for sustained delivery and tissue engineering. Adv Drug Deliv Rev 55:1631–1649. 10.1016/j.addr.2003.08.004. [DOI] [PubMed] [Google Scholar]
- 55.Parker RA, Regan MM, Reimann KA. 2001. Variability of viral load in plasma of rhesus monkeys inoculated with simian immunodeficiency virus or simian-human immunodeficiency virus: implications for using nonhuman primate AIDS models to test vaccines and therapeutics. J Virol 75:11234–11238. 10.1128/JVI.75.22.11234-11238.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Estes JD, Li Q, Reynolds MR, Wietgrefe S, Duan L, Schacker T, Picker LJ, Watkins DI, Lifson JD, Reilly C, Carlis J, Haase AT. 2006. Premature induction of an immunosuppressive regulatory T cell response during acute simian immunodeficiency virus infection. J Infect Dis 193:703–712. 10.1086/500368. [DOI] [PubMed] [Google Scholar]
- 57.Jafarnejad M, Woodruff MC, Zawieja DC, Carroll MC, Moore JE. 2015. Modeling lymph flow and fluid exchange with blood vessels in lymph nodes. Lymphat Res Biol 13:234–247. 10.1089/lrb.2015.0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Müller M, dela Peña A, Derendorf H. 2004. Issues in pharmacokinetics and pharmacodynamics of anti-infective agents: distribution in tissue. Antimicrob Agents Chemother 48:1441–1453. 10.1128/aac.48.5.1441-1453.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shao J, Kraft JC, Li B, Yu J, Freeling J, Koehn J, Ho RJ. 2016. Nanodrug formulations to enhance HIV drug exposure in lymphoid tissues and cells: clinical significance and potential impact on treatment and eradication of HIV/AIDS. Nanomedicine (Lond) 11:545–564. 10.2217/nnm.16.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ali Khan A, Mudassir J, Mohtar N, Darwis Y. 2013. Advanced drug delivery to the lymphatic system: lipid-based nanoformulations. Int J Nanomedicine 8:2733–2744. 10.2147/IJN.S41521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Perazzolo S, Shireman LM, Koehn J, McConnachie LA, Kraft JC, Shen DD, Ho RJY. 2018. Three HIV drugs, atazanavir, ritonavir, and tenofovir, coformulated in drug-combination nanoparticles exhibit long-acting and lymphocyte-targeting properties in nonhuman primates. J Pharm Sci 107:3153–3162. 10.1016/j.xphs.2018.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stevens DM, Adiseshaiah P, Dasa SSK, Potter TM, Skoczen SL, Snapp KS, Cedrone E, Patel N, Busman-Sahay K, Rosen EP, Sykes C, Cottrell M, Dobrovolskaia MA, Estes JD, Kashuba ADM, Stern ST. 2020. Application of a scavenger receptor A1-targeted polymeric prodrug platform for lymphatic drug delivery in HIV. Mol Pharm 17:3794–3812. 10.1021/acs.molpharmaceut.0c00562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thompson CG, Fallon JK, Mathews M, Charlins P, Remling-Mulder L, Kovarova M, Adamson L, Srinivas N, Schauer A, Sykes C, Luciw P, Garcia JV, Akkina R, Smith PC, Kashuba ADM. 2017. Multimodal analysis of drug transporter expression in gastrointestinal tissue. AIDS 31:1669–1678. 10.1097/QAD.0000000000001554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cherrington JM, Allen SJW, Bischofberger N, Chen MS. 1995. Kinetic interaction of the diphosphates of 9–(2-phosphonylmethoxyethyl)adenine and other anti-HIV active purine congeners with HIV reverse transcriptase and human DNA polymerases α, β and γ. Antivir Chem Chemother 6:217–221. 10.1177/095632029500600403. [DOI] [Google Scholar]
- 65.Cottrell ML, Yang KH, Prince HMA, Sykes C, White N, Malone S, Dellon ES, Madanick RD, Shaheen NJ, Hudgens MG, Wulff J, Patterson KB, Nelson JAE, Kashuba ADM. 2016. A translational pharmacology approach to predicting outcomes of preexposure prophylaxis against HIV in men and women using tenofovir disoproxil fumarate with or without emtricitabine. J Infect Dis 214:55–64. 10.1093/infdis/jiw077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Patterson KB, Prince HA, Kraft E, Jenkins AJ, Shaheen NJ, Rooney JF, Cohen MS, Kashuba ADM. 2011. Penetration of tenofovir and emtricitabine in mucosal tissues: implications for prevention of HIV-1 transmission. Sci Transl Med 3:112re4. 10.1126/scitranslmed.3003174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Robichaud G, Barry JA, Garrard KP, Muddiman DC. 2013. Infrared matrix-assisted laser desorption electrospray ionization (IR-MALDESI) imaging source coupled to a FT-ICR mass spectrometer. J Am Soc Mass Spectrom 24:92–100. 10.1007/s13361-012-0505-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chaurand P, Schriver KE, Caprioli RM. 2007. Instrument design and characterization for high resolution MALDI-MS imaging of tissue sections. J Mass Spectrom 42:476–489. 10.1002/jms.1180. [DOI] [PubMed] [Google Scholar]
- 69.Massud I, Aung W, Martin A, Bachman S, Mitchell J, Aubert R, Solomon Tsegaye T, Kersh E, Pau C-P, Heneine W, García-Lerma JG. 2013. Lack of prophylactic efficacy of oral maraviroc in macaques despite high drug concentrations in rectal tissues. J Virol 87:8952–8961. 10.1128/JVI.01204-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shytaj IL, Norelli S, Chirullo B, Della Corte A, Collins M, Yalley-Ogunro J, Greenhouse J, Iraci N, Acosta EP, Barreca ML, Lewis MG, Savarino A. 2012. A highly intensified ART regimen induces long-term viral suppression and restriction of the viral reservoir in a simian AIDS model. PLoS Pathog 8:e1002774. 10.1371/journal.ppat.1002774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Thermo Scientific. RED device systems brochure. Thermo Fisher, Waltham, MA. [Google Scholar]
- 72.Food and Drug Administration. VIREAD (tenofovir disoproxil fumarate) new drug application. Food and Drug Administration, Silver Spring, MD. [Google Scholar]
- 73.Food and Drug Administration. EMTRIVA (emtricitabine) new drug application. Food and Drug Administration, Silver Spring, MD. [Google Scholar]
- 74.Bokhart MT, Rosen E, Thompson C, Sykes C, Kashuba ADM, Muddiman DC. 2015. Quantitative mass spectrometry imaging of emtricitabine in cervical tissue model using infrared matrix-assisted laser desorption electrospray ionization. Anal Bioanal Chem 407:2073–2084. 10.1007/s00216-014-8220-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chambers MC, Maclean B, Burke R, Amodei D, Ruderman DL, Neumann S, Gatto L, Fischer B, Pratt B, Egertson J, Hoff K, Kessner D, Tasman N, Shulman N, Frewen B, Baker TA, Brusniak M-Y, Paulse C, Creasy D, Flashner L, Kani K, Moulding C, Seymour SL, Nuwaysir LM, Lefebvre B, Kuhlmann F, Roark J, Rainer P, Detlev S, Hemenway T, Huhmer A, Langridge J, Connolly B, Chadick T, Holly K, Eckels J, Deutsch EW, Moritz RL, Katz JE, Agus DB, MacCoss M, Tabb DL, Mallick P. 2012. A cross-platform toolkit for mass spectrometry and proteomics. Nat Biotechnol 30:918–920. 10.1038/nbt.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Race AM, Styles IB, Bunch J. 2012. Inclusive sharing of mass spectrometry imaging data requires a converter for all. J Proteomics 75:5111–5112. 10.1016/j.jprot.2012.05.035. [DOI] [PubMed] [Google Scholar]
- 77.Bokhart MT, Nazari M, Garrard KP, Muddiman DC. 2018. MSiReader v1.0: evolving open-source mass spectrometry imaging software for targeted and untargeted analyses. J Am Soc Mass Spectrom 29:8–16. 10.1007/s13361-017-1809-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Robichaud G, Garrard KP, Barry JA, Muddiman DC. 2013. MSiReader: an open-source interface to view and analyze high resolving power MS imaging files on Matlab platform. J Am Soc Mass Spectrom 24:718–721. 10.1007/s13361-013-0607-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Acosta EP, Limoli KL, Trinh L, Parkin NT, King JR, Weidler JM, Ofotokun I, Petropoulos CJ. 2012. Novel method to assess antiretroviral target trough concentrations using in vitro susceptibility data. Antimicrob Agents Chemother 56:5938–5945. 10.1128/AAC.00691-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Temesgen Z, Siraj DS. 2008. Raltegravir: first in class HIV integrase inhibitor. Ther Clin Risk Manag 4:493–500. 10.2147/tcrm.s2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dorr P, Westby M, Dobbs S, Griffin P, Irvine B, Macartney M, Mori J, Rickett G, Smith-Burchnell C, Napier C, Webster R, Armour D, Price D, Stammen B, Wood A, Perros M. 2005. Maraviroc (UK-427,857), a potent, orally bioavailable, and selective small-molecule inhibitor of chemokine receptor CCR5 with broad-spectrum anti-human immunodeficiency virus type 1 activity. Antimicrob Agents Chemother 49:4721–4732. 10.1128/AAC.49.11.4721-4732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.European Medicines Agency. REYATAZ (atazanavir) new drug application, scientific discussion. European Medicines Agency, Amsterdam, the Netherlands. [Google Scholar]
- 83.Palmer A, Phapale P, Chernyavsky I, Lavigne R, Fay D, Tarasov A, Kovalev V, Fuchser J, Nikolenko S, Pineau C, Becker M, Alexandrov T. 2017. FDR-controlled metabolite annotation for high-resolution imaging mass spectrometry. Nat Methods 14:57–60. 10.1038/nmeth.4072. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Initial MSI data processing was performed using the free, open-source MSiReader software (https://www.msireader.wordpress.ncsu.edu/) (77), and quantitative imaging analyses were performed using MATLAB software accessed through a university license (https://www.mathworks.com/products/matlab.html). MSI data are available online through METASPACE database (https://www.metaspace2020.eu) (83) under the project name “Scholz_NHP_LN_ARV.”