ABSTRACT
Pyrazinamide (PZA) is a widely used antitubercular chemotherapeutic. Typically, PZA resistance (PZA-R) emerges in Mycobacterium tuberculosis strains with existing resistance to isoniazid and rifampin (i.e., multidrug resistance [MDR]) and is conferred by loss-of-function pncA mutations that inhibit conversion to its active form, pyrazinoic acid (POA). PZA-R departing from this canonical scenario is poorly understood. Here, we genotyped pncA and purported alternative PZA-R genes (panD, rpsA, and clpC1) with long-read sequencing of 19 phenotypically PZA-monoresistant isolates collected in Sweden and compared their phylogenetic and genomic characteristics to a large set of MDR PZA-R (MDRPZA-R) isolates. We report the first association of ClpC1 mutations with PZA-R in clinical isolates, in the ClpC1 promoter (clpC1p−138) and the N terminus of ClpC1 (ClpC1Val63Ala). Mutations have emerged in both these regions under POA selection in vitro, and the N-terminal region of ClpC1 has been implicated further, through its POA-dependent efficacy in PanD proteolysis. ClpC1Val63Ala mutants spanned 4 Indo-Oceanic sublineages. Indo-Oceanic isolates invariably harbored ClpC1Val63Ala and were starkly overrepresented (odds ratio [OR] = 22.2, P < 0.00001) among PZA-monoresistant isolates (11/19) compared to MDRPZA-R isolates (5/80). The genetic basis of Indo-Oceanic isolates’ overrepresentation in PZA-monoresistant tuberculosis (TB) remains undetermined, but substantial circumstantial evidence suggests that ClpC1Val63Ala confers low-level PZA resistance. Our findings highlight ClpC1 as potentially clinically relevant for PZA-R and reinforce the importance of genetic background in the trajectory of resistance development.
KEYWORDS: pyrazinamide, pyrazinamide resistance, clpC1, mode of action, antimicrobial resistance, low-level resistance, monoresistance, Mycobacterium tuberculosis, antibiotic resistance, pncA
INTRODUCTION
The primary mechanism of resistance to pyrazinamide (PZA) is inactivation or diminished expression of pyrazinamidase/nicotinamidase (PZase) (1), which converts PZA to its active form, pyrazinoic acid (POA). Dozens of unique mutations in the gene encoding PZase, pncA, reportedly confer resistance (2) through this mechanism, though not all appear in clinical isolates (3). A more limited number of pncA mutations have been described not to influence the susceptibility to PZA (3). The mode of POA action is less clear. Several models have been proposed for the mode of action of and alternative mechanisms of Mycobacterium tuberculosis POA/PZA (1). These include disruption of ribosomal salvage via trans-translation inhibition (4) due to POA binding of the 30S ribosomal subunit S1 (RpsA) and disrupting coenzyme A (CoA) synthesis (5) by inhibiting the rate-limiting enzyme of the pathway (6), l-aspartate decarboxylase (PanD) (7). Two groups have identified a connection between POA resistance and ClpC1 (8, 9), an AAA+ ATPase that acts as the chaperone for the cytoplasmic Clp protease (10). Recent work (11) proposed a mechanism for ClpC1-mediated POA resistance based on the finding that altered Clp protease degradation of PanD functions as part of mechanism of POA action and that mutation in ClpC1 of the Clp complex (Clp protease plus ClpC1) mitigates POA-driven efficiency of PanD proteolysis. Particularly in cases where pncA mutations are absent, the targets mediating resistance against these proposed modes of action are candidates for conferring phenotypic resistance.
A recent analysis of 26 isolates from Sweden with PZA-resistant (PZA-R) but wild-type (WT) pncA was conducted to understand the nature of this noncanonical genetic basis of PZA-R (12). That study was able to attribute resistance to false resistance in 8/26 isolates through replicate drug-susceptibility testing (DST) at two concentrations and resistance in another seven isolates to heteroresistance by pncA genotyping following drug pressure in a Bactec mycobacterial growth indicator tube (MGIT) PZA test. To potentially explain the 11 remaining unexplained isolates, the authors of that study genotyped proposed alternative PZA resistance genes (rpsA and panD) with Ion Torrent sequencing. All 11 appeared to be explained by either mixed populations with naturally PZA-R Mycobacterium avium (n = 3), pncA mutation not captured by their primer (n = 1), or alternative mechanisms involving mutation in panD (n = 4) or rpsA (n = 3). However, they found that 17/400 PZA-S isolates harbored nonsynonymous rpsA mutations, including five with the same mutation seen in two of their three PZA-R rpsA mutants (12).
Here, we used long-read sequencing data to genotype 19 PZA-monoresistant isolates from Sweden, including nine of the WT-pncA PZA-R isolates described above. We contrast the genetic basis of PZA resistance in these isolates with that of a large set of multidrug-resistant (MDRPZA-R) isolates isolated in Sweden during a similar time period to identify aspects of PZA resistance that distinguish PZA monoresistance from MDRPZA-R. We analyzed pncA and genes suggested or shown to mediate alternative resistance mechanisms (panD, rpsA, and clpC1), focusing on how the prevalence of mutations in these genes differs between PZA-monoresistant isolates and MDRPZA-R isolates.
RESULTS AND DISCUSSION
PZA-monoresistant clinical isolates.
Isolates were selected from samples collected in Swedish clinical tuberculosis (TB) labs (from patients in Sweden who were most likely infected primarily in Asia and Africa) and processed at the Public Health Agency of Sweden between 2000 and 2015 on the basis of PZA-R in initial phenotypic drug susceptibility testing (DST) and phenotypic susceptibility to isoniazid, rifampin, ethambutol, and amikacin. Genotypic DST corroborated the phenotypic results and implied susceptibility to fluoroquinolones and second-line injectable drugs, with only established lineage markers observed (Table 1). PZA-monoresistant isolates span three of the four major M. tuberculosis lineages and multiple sublineages within the Indo-Oceanic and Euro-American lineages (Table 1).
TABLE 1.
Lineage and genotypic resistance to first- and second-line drugs for PZA-monoresistant clinical isolatesa
| Isolate | Phylogenetic typing |
Mutationb |
||||||
|---|---|---|---|---|---|---|---|---|
| INH |
RIF | FQ |
SLIDs | |||||
| Lineagec | Spoligotypingd | katG | fabG1p | rpoB | gyrA | gyrB | rrs | |
| SEA12126 | IO | EAI5 (SIT 1082) | Arg463Leu | WT | Synonymous | Glu21Gln, Ser95Thr, Ala384Val, Gly668Asp | Ala130Ser, Met291Ile | WT |
| SEA12334 | IO | EAI6_BGD1 (SIT 43) | Arg463Leu | WT | Synonymous | Glu21Gln, Ser95Thr, Ala384Val, Gly668Asp | Met291Ile | WT |
| SEA13111 | IO | EAI3-IND (SIT 414) | Arg463Leu | WT | Synonymous | Glu21Gln, Ser95Thr, Ala384Val, Gly668Asp | Met291Ile | WT |
| SEA12202 | IO | NA | Arg463Leu | WT | Synonymous | Glu21Gln, Ser95Thr, Ala384Val, Gly668Asp | Met291Ile | WT |
| SEA00042 | EAM | NA | WT | WT | Synonymous | Glu21Gln, Ser95Thr, Gly668Asp | WT | WT |
| SEA06535 | EAM | U (SIT 1142) | WT | WT | Synonymous | Glu21Gln, Ser95Thr, Gly668Asp | WT | WT |
| SEA08151 | IO | EAI1_SOM (SIT 48) | Arg463Leu | WT | Synonymous | Glu21Gln, Ser95Thr, Ala384Val, Gly668Asp | Met291Ile | G883A |
| SEA08162 | EAM | LAM4 (SIT 1530) | WT | WT | Glu21Gln, Ser95Thr, Gly247Ser, Gly668Asp | WT | WT | |
| SEA10007 | IO | EAI1_SOM (SIT 48) | Arg463Leu | WT | Synonymous | Glu21Gln, Ser95Thr, Ala384Val, Gly668Asp | Met291Ile | G883A |
| SEA11278 | Ethiopian | SIT 910 | Arg463Leu | WT | Synonymous | Glu21Gln, Ser95Thr, Gly668Asp | WT | WT |
| SEA14117 | EAS | Beijing-like (SIT 269) | Arg463Leu | WT | Synonymous | Glu21Gln, Ser95Thr, Gly668Asp | WT | WT |
| SEA14318 | IO | EAI5 (SIT 138) | Arg463Leu | WT | Synonymous | Ser95Thr, Glu214Asp, Ala384Val, Gly668Asp | Met291Ile | WT |
| SEA15209 | M. canettii | NA | Arg463Leu | C-183G | Synonymous | Glu21Gln, Ser95Thr, Asp504Glu, Gly668Asp, -ATCAGGCTC2518 | WT | T6G |
| SEA10470 | EAM | H3 (SIT 50) | WT | WT | Synonymous | Glu21Gln, Ser95Thr, Gly668Asp | WT | WT |
| SEA09167 | EAM | LAM11_ZWE (SIT 59) | WT | WT | WT | Glu21Gln, Ser95Thr, Gly668Asp | Val301Leu | WT |
| SEA14333 | IO | EAI1-SOM (SIT 735) | Arg463Leu | WT | Synonymous | Glu21Gln, Ser95Thr, Ala384Val, Gly668Asp | Met291Ile | G1454C |
| SEA15228 | IO | EAI1-SOM (SIT 48) | Arg463Leu | WT | Synonymous | Glu21Gln, Ser95Thr, Ala384Val, Gly668Asp | Met291Ile | WT |
| SEA15229 | IO | EAI3-IND (SIT 11) | Arg463Leu | WT | Synonymous | Glu21Gln, Ser95Thr, Ala384Val, Gly668Asp | Met291Ile | WT |
| SEA15230 | IO | EAI5 (SIT 299) | Arg463Leu | WT | Synonymous | Glu21Gln, Ser95Thr, Ala384Val, Gly668Asp | Met291Ile | WT |
Isolates were collected from TB patients in Sweden between 2000 and 2015.
Mutations in known resistance genes. Genes with a wild-type allele (WT) are those matching reference and virulent type strain H37Rv. INH, isoniazid; RIF, rifampin; FQ, fluoroquinolones; SLIDs, second-line injectable drugs.
Defined according to large sequence polymorphism classification (34). IO, Indo-Oceanic; EAM, Euro-American; EAS, East Asian.
NA, not available (spoligotyping data unavailable).
With a single exception, PacBio sequencing calls were concordant with initial Sanger genotyping of pncA and with genotyping of alternative resistance targets by Sanger and Ion Torrent sequencing. The sole exception was an isolate (SEA10470) (Table 2) initially identified as WT/Ser65Ser (a minor population of Ser65Ser) but had a large deletion spanning the beginning of pncA and its upstream region in the PacBio data (Table 2). In their recent work (which included SEA10470), upon observing that their pncA primer for Sanger sequencing from the MGIT PZA tube did not return anything to sequence, Werngren and colleagues sequenced the PZA-containing MGIT-derived sample on Ion Torrent (12) and discovered the same large deletion that we found in single-molecule real-time (SMRT) sequencing data following several weeks of growth in Löwenstein-Jensen (LJ) medium, implying that WT-pncA was ultimately outcompeted by the pncA deletion in both the presence (MGIT PZA tube) and absence of PZA (growth in LJ medium preceding SMRT sequencing). Thus, we conclude SEA10470 is a heteroresistant pncA mutant with limited or no fitness cost in LJ medium.
TABLE 2.
Genotypic and phenotypic profiles of 19 monoresistant PZA samples isolated in Sweden
| Isolate | Lineagea | PZA DST result |
pncA | Resistance conferringb | Genotype |
Resistance explanationc | |||
|---|---|---|---|---|---|---|---|---|---|
| Initial | MGIT (100/200 mg/liter) | panD | rpsA | clpC1 | |||||
| SEA12126 | IO | R | S/S | WT | WT | Val260Ile | Val63Ala | DST flipped | |
| SEA12334 | IO | R | S/S | WT | WT | WT | Val63Ala | DST flipped | |
| SEA13111 | IO | R | S/S | WT | WT | WT | Val63Ala | DST flipped | |
| SEA12202 | IO | R | R/R | Leu172Pro | Y | WT | WT | Val63Ala | pncA mediated |
| SEA00042 | EAM | R | R/R | His51Gln | Y | WT | WT | WT | pncA mediated |
| SEA06535 | EAM | R | R/R | Arg123Pro | Y | WT | WT | WT | pncA mediated |
| SEA08151 | IO | R | R/R | -A416 | Yd | WT | WT | Val63Ala | pncA mediated |
| SEA08162 | EAM | R | R/R | Leu172Pro | Y | WT | WT | Asn806Asn | pncA mediated |
| SEA10007 | IO | R | R/R | -A416 | Yd | WT | WT | Val63Ala | pncA mediated |
| SEA12178 | Ethiopian | R | R/R | Cys138Tyr | Y | Asp133Ala | Thr459Pro | WT | pncA mediated |
| SEA14117 | EAS | R | R/R | Ile90Ser | Y | WT | Arg212Arg | Tyr389Tyr, promoter: −C138 | pncA mediated |
| SEA14318 | IO | R | R/R | DVVG129-132G | Ye | WT | WT | Thr241Thr, Val63Ala | pncA mediated |
| SEA15209 | M. canettii | R | R/R | Ala46Ala | NA | Met117Thr, Ala13Ala | Thr5Ala, Pro9Pro, Thr210Ala, Glu457Glu | promoter: +A-170, A-177G, 49 synonymous mutations | Intrinsic PZA resistance |
| SEA10470 | EAM | R | R/R | deleted nucleotides 264–158 | Yf | WT | WT | WT | Heteroresistanceg |
| SEA09167 | EAM | R | R/R | WT | Ile115Thr | WT | Asn806Asn | Alternative mechanism | |
| SEA14333 | IO | R | R/R | WT | WT | Ile55Val, Val260Ile | Val63Ala | Alternative mechanism | |
| SEA15228 | IO | R | R/R | WT | WT | Val260Ile | Val63Ala | Alternative mechanism | |
| SEA15229 | IO | R | R/S | WT | WT | WT | Val63Ala | Alternative mechanism | |
| SEA15230 | IO | R | R/R | WT | Ile49Val | WT | Val63Ala | Alternative mechanism | |
IO, Indo-Oceanic (lineage 1); EAM, Euro-American (lineage 2); EAS, East-Asian (lineage 4). Ethiopian is lineage 7.
Resistance mutations described by Yadon et al. (2) and/or Ramirez-Busby and Valafar (3). NA, not applicable (M. canetti is intrinsically PZA resistant).
“DST flipped” indicates that no genotyping of MGIT from PZA tube was performed.
Other frameshifts in this region have been reported to confer resistance (3).
Other deletions affecting these residues have been reported to confer resistance (3).
Certainly would disrupt PncA expression and ostensibly confer resistance.
Complex case of heteroresistance described in the text.
panD, rpsA, and clpC1 mutations in PZA-monoresistant isolates with wild-type pncA.
To identify the genetic basis of our PZA-monoresistant isolates, we first checked for pncA mutations. PncA alleles for all 10 pncA mutants had been reported previously for clinical isolates (Table 3), substantiating their role in conferring PZA-R to these isolates. After accounting for isolates with pncA mutations (Table 3), PZA-R in nine WT-pncA isolates remained unexplained. One isolate is Mycobacterium canettii and is therefore naturally PZA-R (13), leaving 8 PZA-R WT-pncA M. tuberculosis isolates. This prevalence of noncanonical PZA resistance among monoresistant M. tuberculosis isolates (8/18) is remarkably higher (P < 0.00001, odds ratio = 32.2, 95% confidence interval [CI] = 35.47 to 351, two-tailed Fisher’s exact test) than in MDRPZA-R Swedish isolates (2/80), suggesting that the nature of PZA monoresistance is different from that of PZA resistance in MDR isolates. All eight had at least one nonsynonymous mutation in panD, rpsA, or clpC1. Three of the eight were susceptible upon repeated phenotypic DST, and one exhibited low-level resistance (resistant at 100 μg/ml but not 200 μg/ml). All four isolates that remained unexplained, with neither PncA mutations nor inconsistent phenotyping, had multiple mutations in alternative PZA resistance genes. Two had nonsynonymous mutations in panD and the other two in rpsA (Table 2), and all had the missense mutation ClpC1Val63Ala. However, the rpsA mutations also occurred in many Swedish isolates susceptible to PZA (12), suggesting either that these mutations confer low-level resistance around the critical concentration or that other genetic elements are at play.
TABLE 3.
Sublineage diversity of PZA-monoresistant clpC1Val63Ala mutants within the Indo-Oceanic lineagea
| Sublineage | Isolate | Spoligotype |
|---|---|---|
| 1.1.1 | SEA12126 | EAI5 (SIT 1082) |
| SEA15228 | EAI1 SOM (SIT 48) | |
| SEA14333 | EAI1 SOM (SIT 735) | |
| 1.1.2 | SEA15230 | EAI5 (SIT 299) |
| SEA15229 | EAI3 IND (SIT 11) | |
| SEA13111 | EAI3 IND (SIT 414) | |
| 1.2.1 | SEA12334 | EAI6 BGD1 (SIT 43) |
| SEA12202 | NA | |
| 1.2.2 | SEA10007 | EAI1_SOM (SIT 48) |
| SEA08151 | EAI1 SOM (SIT 48) | |
| SEA14318 | EAI5 (SIT 138) |
Sublineage membership was determined using the lineage-specific SNPs described by Coll et al. (19). NA, not available (spoligotyping data are unavailable for this isolate).
Novel mutations implicate regions mediating PanD proteolysis by ClpC1.
Recent work demonstrated that POA increases PanD proteolysis by the Clp complex and proposed this as its mode of action (11). This proteolysis requires a C-terminal degradation tag (PanD127–139) for recognition by the N terminus of ClpC1. While PZA-R clpC1 mutants have not been previously reported among clinical isolates, they have been isolated following selection under PZA (14) and POA (9) pressure in vitro and POA pressure in an in vivo mouse model (8). Here, we report the first account of clpC1 mutations in PZA-R clinical isolates with WT-pncA and a panD mutation (panDAsp133Ala) in its C-terminal recognition tag novel to clinical isolates.
The first clpC1 mutation is a deletion in the clpC1 promoter (clpC1p), 138 bp upstream of its start codon (clpC1p−C138). The isolate harboring clpC1p−C138 (SEA14117) (Table 2) has a pncA mutation (pncAIle90Ser) previously reported in PZA-R clinical isolates. However, pncAIle90Ser is rare and—unlike most clinically impactful pncA mutations (2)—is not enriched following selection under PZA pressure in vitro or in mice, and neither were any other codon 90 pncA mutations. While the possibility that pncAIle90Ser accounts fully for PZA-R in SEA14117 cannot be dismissed, the irreproducibility of pncAIle90Ser conferring resistance on its own in vitro leaves open the possibility that clpC1p−C138 may contribute to resistance either alternatively to pncAIle90Ser or perhaps additively. While clpC1p−C138 has not been described previously, other clpC1 promoter mutations have emerged under POA selection pressure in vitro (9), suggesting that clpC1 promoter mutations can confer POA resistance. Viewing this observation through the model of Clp proteolysis of PanD as the POA mode of action (11), clpC1p mutations could confer PZA/POA resistance by weakening the promoter, reducing ClpC1 expression and, in turn, Clp-mediated PanD degradation.
The second clpC1 mutant was a missense mutation in the ClpC1 N terminus, Val63Ala (clpC1Val63Ala). Eleven of 19 PZA-monoresistant isolates were clpC1Val63Ala mutants, exclusively and invariably of the Indo-Oceanic lineage. These 11 isolates span four Indo-Oceanic sublineages (each with multiple isolates) and comprise seven distinct sublineage-spoligotype combinations (Table 3). Thus, PZA-monoresistant clpC1Val63Ala mutants are neither a clonal expansion nor monophyletic. Rather, they are spread throughout the Indo-Oceanic lineage. The effects of the variants clpC1Val63Ala and panDAsp133Ala converge on the interaction between the ClpC1 N-terminal and PanD127-139 C-terminal degradation tags (9). Though it is only speculation at present, the functional effects implied by these mutations’ location fits well into the mode of POA action proposed recently by Gopal and colleagues (11).
Indo-Oceanic strains in this study (11/19), which selected isolates on the basis of PZA monoresistance, were far more prevalent (P < 0.000001, odds ratio = 22.2, 95% CI = 6.05 to 121, two-tailed Fisher’s exact test) than PZA-resistant Indo-Oceanic isolates from Sweden in a study (35) (5/80) in which isolates were selected on the basis of MDR phenotype. The frequency of ClpC1 mutants that we observed here stands in contrast to a recent study of 269 PZA-resistant isolates in China where no isolates harbored a nonsynonymous ClpC1 mutation (16). However, that study had mostly Beijing lineage isolates (lineage was not indicated for the remaining isolates). This overrepresentation of Indo-Oceanic isolates in monoresistant PZA-R strains relative to MDRPZA-R strains has been observed previously in the United States (17) and Iran (18), suggesting that it is a phenomenon not isolated to samples derived from patients in Sweden. This repeatedly observed trend of Indo-Oceanic isolates developing PZA monoresistance, when considered alongside the mechanistic plausibility of ClpC1 N-terminal mutations affecting POA resistance (8, 9, 11, 14) and the vacillating of these ClpC1 mutants around the resistance threshold (Table 2), suggests that clpC1Val63Ala may confer low-level PZA resistance to Indo-Oceanic isolates. The missense mutation clpC1Val63Ala is specific to and invariably present in Indo-Oceanic isolates (15, 19) and thus would represent a lineage-specific resistance phenotype akin to the intrinsic PZA resistance of the ancestral mycobacteria M. canettii (13) and Mycobacterium bovis (1), but at a lower level. However, confirmatory mutagenesis is needed to verify this possibility.
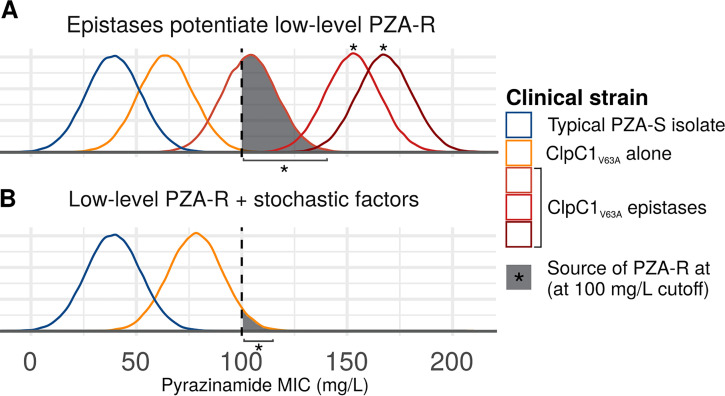
How might one reconcile the proposed low-level PZA resistance conferred by the clpC1Val63Ala allele with the observation that Indo-Oceanic isolates often register as phenotypically susceptible to PZA? It is important to consider that the Bactec MGIT 960 critical concentration (100 μg/ml) has been calibrated to detect pncA-conferred resistance, which is severalfold higher than the 2- to 8-fold MIC increases observed for POA-resistant mutants harboring panD and clpC1 mutations (8, 9). This could explain the variable and discrepant phenotypic results we observe in Indo-Oceanic isolates upon replicating DST (Table 2; Fig. 1). This scenario has been reported for PZA/POA-resistant panD mutants isolated following selection under POA pressure; despite a 2- to 8-fold MIC increase for G139* panD mutants relative to the wild type, seven of eight clinical isolates that Gopal and colleagues found with the mutation were classified as susceptible to PZA (8). Similarly, rpsAΔ438Ala and rpsAAsp123Ala mutations were recently shown to confer 2-fold increases in MIC relative to H37Rv (20). Low-level resistance conferred by the clpC1Val63Ala allele would simultaneously explain the prevalence of Indo-Oceanic lineage and WT-PncA PZA-R among PZA-monoresistant isolates (17, 18) as well as the variable phenotypic results for these isolates. Epistasis (Fig. 1A) and resistance near the critical concentration vacillating above it due to nongenetic factors (Fig. 1B) could also produce the bias toward Indo-Oceanic lineage isolates and inconsistent DST results among monoresistant PZA isolates resulting from clpC1Val63Ala. In either case, together, these considerations suggest clpC1Val63Ala low-level resistance, though confirmation is required. Future work with a larger set of PZA-monoresistant and MDRPZA-R isolates and susceptibility testing across a range of lower MICs would more precisely delineate the role of clpC1Val63Ala in PZA resistance.
FIG 1.
Phenomena potentially underlying low-level PZA-R monoresistance conferred by clpC1Val63Ala. Curves depict theoretical probability density functions of MICs obtained for a given isolate. (A) Epistasis. In the epistatic scenario, clpC1Val63Ala alone confers low-level resistance, invariably below the 100-mg/liter critical concentration, but some isolates (reds) harbor additional mutations that confer higher-level PZA-R through additive or synergistic interaction with clpC1Val63Ala. (B) Near-critical concentration MIC. In this scenario, low-level PZA resistance conferred by clpC1Val63Ala alone sometimes exceeds the 100-mg/liter cutoff, due to nongenetic factors contributing to MIC variance (30, 31), such as inoculum size (32), growth medium, or interlaboratory variation (33). The phenomena depicted in panels A and B are not mutually exclusive and could both be at work.
Concluding remarks.
By analyzing monoresistant PZA isolates collected in Sweden, we found that monoresistant isolates have mutational profiles distinct from those of MDRPZA-R isolates and identified clpC1Val63Ala as a potential low-level resistance marker common to Indo-Oceanic isolates. Despite this mutation’s being common to Indo-Oceanic isolates (19) and POA/PZA-resistant ClpC1 mutants isolated from in vitro screens (8, 9), this is, to our knowledge, the first report of PZA-resistant ClpC1 mutants in clinical isolates. Further study of the hypothesized link between clpC1Val63Ala and low-level resistance warrants mechanistic study, as does evaluation of its effect on PZA/POA resistance in vitro and in the context of infection. The functional effects implicated by clpC1Val63Ala and panDAsp133Ala converge on the interaction between the ClpC1 N-terminal and PanD127-139 C-terminal degradation tags (9), consistent with the emerging model of POA mode of action where PanD degradation by Clp proteolysis is enhanced by POA (11). Functional studies assessing the effect of these two mutations on PanD degradation natively and in the presence of POA would be of great interest. More broadly, the bias of PZA monoresistance toward Indo-Oceanic isolates underlines the relevance of genetic background in evolution of resistance (21–23), a factor that is often precluded from analysis in molecular epidemiology and genome-wide association studies on the assumption that they do not influence resistance. Our findings suggest otherwise.
MATERIALS AND METHODS
Isolate selection.
PZA-monoresistant isolates were selected from TB patients seen in Sweden between 2003 and 2015, irrespective of lineage, TB presentation, or geographic origin. The geographic origin of the 19 TB strains isolated in Sweden was typical of the general TB epidemiology in Sweden, with roughly half of the patients coming from Africa, a fifth from Asia, and the rest from Sweden or other parts of the globe (24).
Culturing and DNA extraction.
M. tuberculosis samples were prepared and extracted at the World Health Organization Supranational Reference Laboratory in Stockholm, Sweden. Isolates growing on LJ medium were received from clinical TB labs and subsequently Sanger sequenced. The genotypic DST result (pncA-Sanger) was then compared to the phenotypic DST result (PZA Bactec 460 until 2008, MGIT 960 thereafter) from the clinical lab, and discordant cases were retested in PZA MGIT. In cases of verified PZA resistance, Sanger sequencing was repeated on the growth from the PZA MGIT tube. Samples still lacking a pncA mutation were then subjected to whole-genome sequencing (WGS). Preparation and extraction were performed as previously described (12).
SMRT sequencing.
DNA sequencing was performed at the Institute for Genomic Medicine at the University of California, San Diego, CA. DNA libraries for PacBio (Pacific Biosciences, Menlo Park, CA) were prepared using PacBio’s DNA template prep kit with no follow-up PCR amplification. Briefly, sheared DNA was end repaired, and hairpin adapters were ligated using T4 DNA ligase. Incompletely formed SMRTbell templates were degraded with a combination of exonuclease III and exonuclease VII. The resulting DNA templates were purified using solid-phase reversible-immobilization (SPRI) magnetic beads (AMPure; Agencourt Bioscience, Beverly, MA) and annealed to a 2-fold molar excess of a sequencing primer that specifically bound to the single-stranded loop region of the hairpin adapters. SMRTbell templates were subjected to standard SMRT sequencing using an engineered phi29 DNA polymerase on the PacBio RS II system according to the manufacturer’s protocol.
Sanger and Ion Torrent sequencing.
DNA sequencing with Sanger and Ion Torrent was carried out at World Health Organization Supranational Reference Laboratory in Stockholm, Sweden, as previously described (12).
Genotyping from SMRT sequencing data.
Raw Pacific Biosciences SMRT sequencing reads were aligned to the reference genome of the M. tuberculosis virulent type strain H37Rv reference strain (GenBank accession number NC_000962.3) using BLASR (25) (v1.3) with default parameters. PBHoover (26) (https://gitlab.com/LPCDRP/pbhoover) corrected aligned reads and called variants based on a maximum-likelihood criterion. VCF formatted files were further annotated with Variant Effect Predictor (27) (VEP) (v87) to determine the consequence of each variant. Variants within or proximally upstream of clpC1, panD, rpsA, and pncA were screened for using a custom python script (https://gitlab.com/LPCDRP/drug-resistance/-/blob/master/src/known-resistance-association.py).
Lineage determination and spoligotyping.
Lineage was determined by in silico mycobacterial interspersed repetitive unit (MIRU) typing and spoligotyping with MiruHero (https://gitlab.com/LPCDRP/miru-hero). Spoligotyping was also carried out in Stockholm, Sweden, as described previously (28). Sublineage membership for Indo-Oceanic isolates was determined by the lineage-specific single nucleotide polymorphisms (SNPs) defined by Coll and colleagues (19).
Drug susceptibility testing.
For cases with discordance between initial Sanger sequencing of pncA and the PZA Bactec 460/MGIT result from the clinical lab, PZA DST was performed in the Bactec MGIT system (Becton Dickinson) according to instructions from the manufacturer. The DST inoculum was prepared from bacterial growth on Löwenstein-Jensen egg medium at 37°C. Briefly, two 1-μl loops of bacteria were suspended in 3 ml phosphate-buffered saline (PBS) in a small glass tube with glass beads. The bacterial suspension was homogenized using vortexing or an ultrasound water bath to disperse any clumps. The suspension was then left to sediment for 20 min, and the upper 2 ml was transferred to a new tube and left to sediment for another 15 min. Before inoculation of the MGIT PZA medium culture tubes (pH 5.9), the bacterial suspension was adjusted to a McFarland turbidity of 0.5 and diluted in PBS per the manufacturer’s PZA test protocol.
Statistical tests.
Two-tailed Fisher’s exact test was used to assess independence and implemented in R statistical programming language (29).
Data availability.
All code is available on GitLab at https://gitlab.com/LPCDRP/miru-hero.
ACKNOWLEDGMENTS
We acknowledge Derek Conkle-Gutierrez and Bria Gorman for careful review and helpful feedback during manuscript revision.
This work was funded through a grant (R01AI105185) by the National Institute for Allergy and Infectious Diseases (NIAID) awarded to FV. The funding bodies had no role in the design of the study, in collection, analysis, and interpretation of data, or in writing the manuscript.
Conceptualization, methodology, and supervision: S.J.M., F.V., and S.E.H.; data curation: S.J.M. and S.E.H.; formal analysis: T.M. and S.J.M.; funding acquisition: F.V.; investigation: S.J.M., T.M., M.M., and J.W.; project administration: F.V.; visualization: S.J.M.; writing – original draft: S.J.M. and T.M.; writing – review and editing: S.J.M., T.M., S.E.H., J.W., M.M., and F.V. All authors reviewed and approved the final manuscript.
REFERENCES
- 1.Lamont EA, Dillon NA, Baughn AD. 2020. The bewildering antitubercular action of pyrazinamide. Microbiol Mol Biol Rev 84:e00070-19. 10.1128/MMBR.00070-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yadon AN, Maharaj K, Adamson JH, Lai YP, Sacchettini JC, Ioerger TR, Rubin EJ, Pym AS. 2017. A comprehensive characterization of PncA polymorphisms that confer resistance to pyrazinamide. Nat Commun 8:588. 10.1038/s41467-017-00721-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ramirez-Busby S, Valafar F. 2015. Systematic review of mutations in pyrazinamidase associated with pyrazinamide resistance in Mycobacterium tuberculosis clinical isolates. Antimicrob Agents Chemother 59:5267–5277. 10.1128/AAC.00204-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shi W, Zhang X, Jiang X, Yuan H, Lee JSJS, Barry CECE, Wang H, Zhang W, Zhang Y. 2011. Pyrazinamide inhibits trans-translation in Mycobacterium tuberculosis. Science 333:1630–1632. 10.1126/science.1208813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang S, Chen J, Shi W, Liu W, Zhang W, Zhang Y. 2013. Mutations in panD encoding aspartate decarboxylase are associated with pyrazinamide resistance in Mycobacterium tuberculosis. Emerg Microbes Infect 2:e34. 10.1038/emi.2013.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jackowski S, Rock CO. 1981. Regulation of coenzyme A biosynthesis. J Bacteriol 148:926–932. 10.1128/JB.148.3.926-932.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chopra S, Pai H, Ranganathan A. 2002. Expression, purification, and biochemical characterization of Mycobacterium tuberculosis aspartate decarboxylase, PanD. Protein Expr Purif 25:533–540. 10.1016/s1046-5928(02)00039-6. [DOI] [PubMed] [Google Scholar]
- 8.Gopal P, Tasneen R, Yee M, Lanoix JP, Sarathy J, Rasic G, Li L, Dartois V, Nuermberger E, Dick T. 2017. In vivo-selected pyrazinoic acid-resistant Mycobacterium tuberculosis strains harbor missense mutations in the aspartate decarboxylase PanD and the unfoldase ClpC1. ACS Infect Dis 3:492–501. 10.1021/acsinfecdis.7b00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yee M, Gopal P, Dick T. 2017. Missense mutations in the unfoldase ClpC1 of the caseinolytic protease complex are associated with pyrazinamide resistance in Mycobacterium tuberculosis. Antimicrob Agents Chemother 61:e02342-16. 10.1128/AAC.02342-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Akopian T, Kandror O, Raju RM, Unnikrishnan M, Rubin EJ, Goldberg AL. 2012. The active ClpP protease from M. tuberculosis is a complex composed of a heptameric ClpP1 and a ClpP2 ring. EMBO J 31:1529–1541. 10.1038/emboj.2012.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gopal P, Sarathy JP, Yee M, Ragunathan P, Shin J, Bhushan S, Zhu J, Akopian T, Kandror O, Lim TK, Gengenbacher M, Lin Q, Rubin EJ, Grüber G, Dick T. 2020. Pyrazinamide triggers degradation of its target aspartate decarboxylase. Nat Commun 11:1661. 10.1038/s41467-020-15516-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Werngren J, Alm E, Mansjö M. 2017. Non-pncA gene-mutated but pyrazinamide-resistant Mycobacterium tuberculosis: why is that? J Clin Microbiol 55:1920–1927. 10.1128/JCM.02532-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Supply P, Marceau M, Mangenot S, Roche D, Rouanet C, Khanna V, Majlessi L, Criscuolo A, Tap J, Pawlik A, Fiette L, Orgeur M, Fabre M, Parmentier C, Frigui W, Simeone R, Boritsch EC, Debrie AS, Willery E, Walker D, Quail MA, Ma L, Bouchier C, Salvignol G, Sayes F, Cascioferro A, Seemann T, Barbe V, Locht C, Gutierrez MC, Leclerc C, Bentley SD, Stinear TP, Brisse S, Médigue C, Parkhill J, Cruveiller S, Brosch R. 2013. Genomic analysis of smooth tubercle bacilli provides insights into ancestry and pathoadaptation of Mycobacterium tuberculosis. Nat Genet 45:172–179. 10.1038/ng.2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang S, Chen J, Shi W, Cui P, Zhang J, Cho S, Zhang W, Zhang Y. 2017. Mutation in clpC1 encoding an ATP-dependent ATPase involved in protein degradation is associated with pyrazinamide resistance in Mycobacterium tuberculosis. Emerg Microbes Infect 6:1–2. 10.1038/emi.2017.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rose G, Cortes T, Comas I, Coscolla M, Gagneux S, Young DB. 2013. Mapping of genotype–phenotype diversity among clinical isolates of Mycobacterium tuberculosis by sequence-based transcriptional profiling. Genome Biol Evol 5:1849–1862. 10.1093/gbe/evt138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hameed HMA, Tan Y, Islam MM, Lu Z, Chhotaray C, Wang S, Liu Z, Fang C, Tan S, Yew WW, Zhong N, Liu J, Zhang T. 2020. Detection of novel gene mutations associated with pyrazinamide resistance in multidrug-resistant Mycobacterium tuberculosis clinical isolates in southern China. Infect Drug Resist 13:217–227. 10.2147/IDR.S230774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kurbatova E, Cavanaugh J, Dalton T, Click ES, Cegielski JP, TD-C infectious. 2013. Epidemiology of pyrazinamide-resistant tuberculosis in the United States, 1999–2009. Clin Infect Dis 57:1081–1093. 10.1093/cid/cit452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doustdar F, Khosravi AD, Farnia P. 2009. Mycobacterium tuberculosis genotypic diversity in pyrazinamide-resistant isolates of Iran. Microb Drug Resist 15:251–256. 10.1089/mdr.2009.0066. [DOI] [PubMed] [Google Scholar]
- 19.Coll F, McNerney R, Guerra-Assunção JA, Glynn JR, Perdigão J, Viveiros M, Portugal I, Pain A, Martin N, Clark TG. 2014. A robust SNP barcode for typing Mycobacterium tuberculosis complex strains. Nat Commun 5:4–8. 10.1038/ncomms5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shi W, Cui P, Niu H, Zhang S, Tønjum T, Zhu B, Zhang Y. 2019. Introducing RpsA point mutations 438A and D123A into the chromosome of mycobacterium tuberculosis confirms their role in causing resistance to pyrazinamide. Antimicrob Agents Chemother 63:e02681-18. 10.1128/AAC.02681-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kavvas ES, Catoiu E, Mih N, Yurkovich JT, Seif Y, Dillon N, Heckmann D, Anand A, Yang L, Nizet V, Monk JM, Palsson BO. 2018. Machine learning and structural analysis of Mycobacterium tuberculosis pan-genome identifies genetic signatures of antibiotic resistance. Nat Commun 9:4306. 10.1038/s41467-018-06634-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Merker M, Kohl TA, Barilar I, Andres S, Fowler PW, Chryssanthou E, Ängeby K, Jureen P, Moradigaravand D, Parkhill J, Peacock SJ, Schön T, Maurer FP, Walker T, Köser C, Niemann S. 2020. Phylogenetically informative mutations in genes implicated in antibiotic resistance in Mycobacterium tuberculosis complex. Genome Med 12:27. 10.1186/s13073-020-00726-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Castro RAD, Ross A, Kamwela L, Reinhard M, Loiseau C, Feldmann J, Borrell S, Trauner A, Gagneux S. 2020. The genetic background modulates the evolution of fluoroquinolone-resistance in Mycobacterium tuberculosis. Mol Biol Evol 37:195–207. 10.1093/molbev/msz214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Folkhälsomyndigheten. 2019. Tuberculosis. Folkhälsomyndigheten, Solna, Sweden. https://www.folkhalsomyndigheten.se/folkhalsorapportering-statistik/statistik-a-o/sjukdomsstatistik/tuberkulos/?p=18610#statistics-nav.
- 25.Chaisson MJ, Tesler G. 2012. Mapping single molecule sequencing reads using basic local alignment with successive refinement (BLASR): application and theory. BMC Bioinformatics 13:238. 10.1186/1471-2105-13-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramirez-Busby SM, Elghraoui A, Bin Kim Y, Kim K, Valafar F. 2018. PBHoover and CigarRoller: a method for confident haploid variant calling on Pacific Biosciences data and its application to heterogeneous population analysis. bioRxiv 10.1101/360370. [DOI]
- 27.McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GRS, Thormann A, Flicek P, Cunningham F. 2016. The Ensembl Variant Effect Predictor. Genome Biol 17:122. 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kamerbeek J, Schouls L, Kolk A, Van Agterveld M, Van Soolingen D, Kuijper S, Bunschoten A, Molhuizen H, Shaw R, Goyal M, Van Embden J. 1997. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J Clin Microbiol 35:907–914. 10.1128/JCM.35.4.907-914.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.R Core Team. 2018. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/. [Google Scholar]
- 30.Farhat MR, Freschi L, Calderon R, Ioerger T, Snyder M, Meehan CJ, de Jong B, Rigouts L, Sloutsky A, Kaur D, Sunyaev S, van Soolingen D, Shendure J, Sacchettini J, Murray M. 2019. GWAS for quantitative resistance phenotypes in Mycobacterium tuberculosis reveals resistance genes and regulatory regions. Nat Commun 10:2128. 10.1038/s41467-019-10110-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mouton JW, Meletiadis J, Voss A, Turnidge J. 2018. Variation of MIC measurements: the contribution of strain and laboratory variability to measurement precision. J Antimicrob Chemother 73:2374–2379. 10.1093/jac/dky232. [DOI] [PubMed] [Google Scholar]
- 32.Alexander HK, Craig MacLean R. 2020. Stochastic bacterial population dynamics restrict the establishment of antibiotic resistance from single cells. Proc Natl Acad Sci U S A 117:19455–19464. 10.1073/pnas.1919672117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Juréen P, Ängeby K, Sturegård E, Chryssanthou E, Giske CG, Werngren J, Nordvall M, Johansson A, Kahlmeter G, Hoffner S, Schön T. 2010. Wild-type MIC distributions for aminoglycoside and cyclic polypeptide antibiotics used for treatment of Mycobacterium tuberculosis infections. J Clin Microbiol 48:1853–1858. 10.1128/JCM.00240-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gagneux S, DeRiemer K, Van T, Kato-Maeda M, de Jong BC, Narayanan S, Nicol M, Niemann S, Kremer K, Gutierrez MC, Hilty M, Hopewell PC, Small PM. 2006. Variable host-pathogen compatibility in Mycobacterium tuberculosis. Proc Natl Acad Sci U S A 103:2869–2873. 10.1073/pnas.0511240103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mansjö M, Werngren J, Hoffner S. 2017. Characterization of pyrazinamide resistance in consecutive multidrug-resistant mycobacterium tuberculosis isolates in Sweden between 2003 and 2015. Int J Mycobacteriol 6:156–161. 10.4103/ijmy.ijmy_23_17. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All code is available on GitLab at https://gitlab.com/LPCDRP/miru-hero.