ABSTRACT
Long disregarded as junk DNA or genomic dark matter, endogenous retroviruses (ERVs) have turned out to represent important components of the antiviral immune response. These remnants of once-infectious retroviruses not only regulate cellular immune activation, but may even directly target invading viral pathogens. In this Gem, we summarize mechanisms by which retroviral fossils protect us from viral infections. One focus will be on recent advances in the role of ERVs as regulators of antiviral gene expression.
KEYWORDS: endogenous retroviruses, antiviral immunity, sensing
INTRODUCTION
Viruses do not have a good reputation. They are generally perceived as harmful pathogens causing acute or chronic diseases that need to be well controlled or, ideally, eradicated. Thus, it may at first glance seem worrisome that about 8% of the human genome consists of retroviral DNA sequences. These so-called endogenous retroviruses (ERVs) represent remnants of once infectious exogenous retroviruses that became fixed in our DNA and are now inherited in a Mendelian manner. While many of them are usually dormant, they can be reactivated by several stimuli, including viral infections. For example, dengue, herpes simplex, influenza, and human immunodeficiency viruses have all been shown to induce the transcription and translation of endogenous retroviral elements (1–5). Although human ERVs (HERVs) are not known to produce infectious viral particles in vivo, this resurgence of virus-derived RNAs and proteins in our cells may seem alarming. However, many ERVs are not detrimental and have even been coopted for important physiological functions in the host. Besides well-known examples, such as syncytins that regulate placental development (6, 7), ERVs have become integral parts of immune defense mechanisms and help to fight off invading viral pathogens.
To date, hundreds of thousands of HERVs have been identified, ranging from full-length proviruses to short gene fragments (8). Thus, it is not surprising that they support the antiviral immune response via numerous mechanisms. These include the enhancement of cellular sensing pathways, regulation of viral gene expression, blockade of entry receptors, and direct restriction of virion assembly. In this Gem, we summarize recent advances in the interplay of endogenous and exogenous retroviruses. We present important principles underlying the antiviral activities of ERVs and illustrate how the host cell has coopted fossils of possibly once harmful retroviruses to limit the spread of current viral pathogens.
ERV-DERIVED NUCLEIC ACIDS TRIGGER INNATE SENSING CASCADES
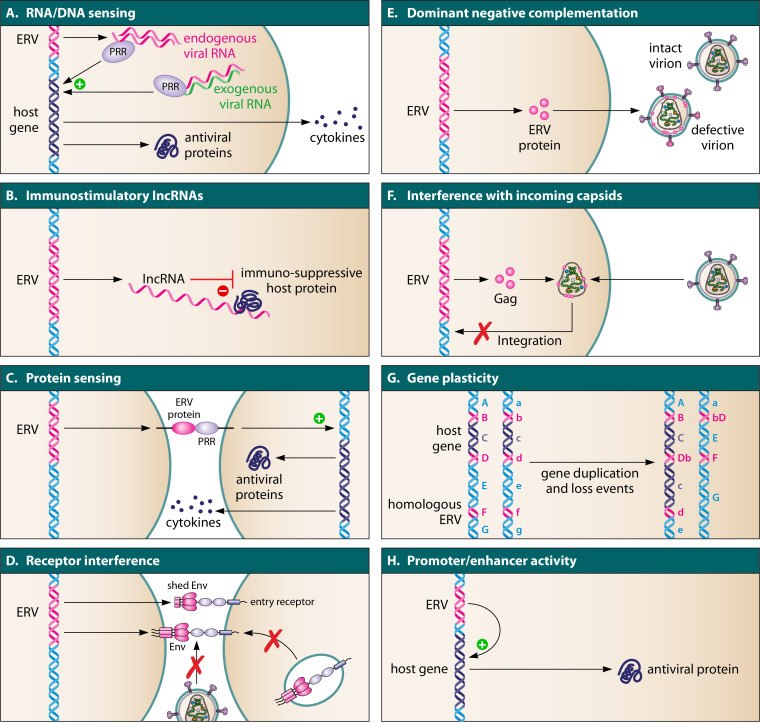
Host cells are equipped with a variety of pattern recognition receptors (PRRs) that sense virus-derived nucleic acids and induce antiviral gene expression upon infection. These include cytosolic sensors such as RIG-I or MDA5, as well as Toll-like receptors (TLRs) such as TLR3 or TLR9 that recognize double-stranded RNA (dsRNA), single-stranded RNA (ssRNA), and/or RNA-DNA hybrids (9–13). Usually, these sensors efficiently distinguish between self and nonself to prevent undesirable activation in the absence of infection (14). In a process of viral mimicry, however, the expression and subsequent sensing of endogenous retroviral nucleic acids may be beneficial and boost the induction of an antiviral state in the infected host (Fig. 1A). In particular, dsRNA seems to play an important role in ERV sensing. Upon ERV activation, dsRNA species may be formed by pairing of ERV RNA with cellular antisense transcripts or complementary exogenous viral RNA. In addition, intramolecular pairing of ERV transcripts can also activate PRRs that detect dsRNA.
FIG 1.
ERV-mediated mechanisms of antiviral immunity. (A) Enhancement of innate sensing by ERV-derived nucleic acids. (B) Regulation of antiviral immune responses by ERV-derived long noncoding RNAs (lncRNAs). (C) Enhancement of innate sensing by ERV-derived proteins. (D) Receptor interference by endogenous retroviral envelope (Env) proteins. (E) Dominant negative complementation of exogenous viral particles by endogenous retroviral proteins. (F) Interference with incoming viral capsids. (G) Increased gene plasticity due to ERV-mediated recombination events. (H) Regulation of antiviral gene expression by ERV-derived promoter and enhancer elements.
One striking example of ERV sensing was recently described by the group of Benjamin Hale (15). The authors showed that influenza A virus (IAV) and influenza B virus (IBV) infection triggers a loss of SUMO-modified TRIM28/KAP1. RIG-I, MDA5, and alpha interferon (IFN-α) were not required for this loss, and the viral and/or cellular pathways triggering this SUMO switch remain to be discovered. Since TRIM28 acts as a restriction factor that limits retroviral transcription (16, 17), this derepression resulted in an increased expression of endogenous retroviruses. PRR knockout and inhibitor experiments suggested that the sensing of HERV-derived dsRNA via RIG-I triggers the expression of type I interferons (IFNs) and IFN-stimulated genes (ISGs), thereby limiting IAV replication (15). While the correlation of increased HERV expression with ISG induction is very suggestive, a causal link has not been proven in a definitive manner.
Another ERV-derived nucleic acid that is sensed and may contribute to antiviral immune responses has been described in birds. In 2019, Chen and colleagues demonstrated that an antisense long noncoding RNA (lncRNA) derived from an endogenous avian leucosis virus (ALVE1) activates the dsRNA sensor TLR3 (18). As a result, this lncRNA (termed lnc-ALVE1-AS1) induces the expression of beta interferon (IFN-β) and ISGs in chicken embryonic fibroblasts and suppresses replication of avian leukosis virus subgroup J (ALVJ) (Table 1). While expression of this lncRNA could be induced by the DNA methylation inhibitor 5-Aza-dC, its expression upon viral infection remains to be determined.
TABLE 1.
Examples for endogenous retroviruses involved in immunity
| Mode of action | ERVb | Species (examples) | Molecular mechanisma | Target virus(es) | Reference(s) |
|---|---|---|---|---|---|
| Induction of sensing via nucleic acids | Multiple ERVs (e.g., ERV3-2, ERV4700) | Humans | Sensing of ERV-derived nucleic acids by MDA5, RIG-I, and TLR3 | Most likely several viruses | 15, 20–23 |
| ALVE1 | Chickens | Induction of TLR3 signaling by ERV-derived lncRNA (lnc-ALVE1-AS1) | Avian leukosis virus subgroup J, possibly others | 18 | |
| Regulation of immune signaling by lncRNAs | lnc-EPAV | Mice | lncRNA-mediated sequestration of SFPQ, a repressor of Rela expression | VSV, possibly others | 25 |
| MER9a2, LTR5A, MLT2A1 | Humans | Binding of SFPQ; induction of Rela expression (?) | Most likely several viruses | 25 | |
| Modulation of immune activation by ERV proteins | HERV-W Env | Humans | Activation of TLR4/CD14; induction of IL-1β, IL-6, TNF-α, and IL-12p40 | Most likely several viruses | 26 |
| HERV-K(HML2) dUTPase | Humans | Activation of NF-κB via TLR2; induction of IL-1β, IL-6, IL-8, IL-10, IL-12p40, IL-23, IL-17, TNF-α, RANTES, IFN-γ, and CCL20 | Most likely several viruses | 28 | |
| Receptor interference | ev3, ev6, ev9 | Chickens | Receptor blockade | Avian leukosis virus, subgroup E | 40 |
| Fv-4r (Akvr-1) | Mice | Blockade of the receptor CAT-1 | Ecotropic murine leukemia viruses | 41, 42, 46 | |
| Rcmf, Rcmf2 | Mice | Blockade of the receptor XPR1 | Polytropic murine leukemia viruses | 47–51 | |
| en-JSRV env loci | Sheep | Blockade of the receptor hyaluronidase 2 (HYAL2) | Jaagsiekte sheep retrovirus, enzootic nasal tumor virus (?) | 52 | |
| Refrex-1 (loci ERV-DC7 and –DC16) | Cats | Secreted truncated Env; extracellular receptor interference | Endogenous and exogenous feline retroviruses (ERV-DC genotype I, FeLV-D) | 54 | |
| FeLIX | Cats | Secreted truncated Env; extracellular receptor interference | FeLV-B | 55 | |
| HERV-T env locus | Primates | Depletion of MCT-1 from the cell surface | Extinct HERV-T ancestor | 56 | |
| Suppressyn | Primates | Blockade of the receptor ASCT2 | RD114/mammalian type-D retroviruses | 57 | |
| Dominant negative virion complementation | Fv-4r | Mice | Virion incorporation of Env with inactive fusion peptide | Ecotropic murine leukemia viruses | 58 |
| HERV-K gag locus | Humans | Coassembly with exogenous viral Gag; generation of malformed and poorly infectious virions; reduced virion release | HIV-1 | 62, 63 | |
| enJS56A1, enJSRV-20 (?) | Sheep | Dominant negative interference of a defective Gag protein with viral exit | Jaagsiekte sheep retrovirus | 64–66 | |
| Interference with incoming viral particles | Fv1 | Mice | Binding to incoming capsids | Murine leukemia viruses | 67–70 |
| Increased plasticity of immune genes | Multiple ERVs | Several species | Gene conversion and duplication events increasing the diversity of MHC II genes and potentially others | Most likely several viruses | 85 |
| Regulation of antiviral gene expression | X-MLV LTR | Mice | Enhancer of mA3; increase in the levels and antiviral activity of APOBEC3 | MMTV, potentially others | 91, 92 |
| HERVP71A LTR/enhancer L | Humans | Enhancer of HLA-G | Unclear | 93, 94 | |
| MER41 | Humans | IFN-γ-inducible enhancer of AIM2 expression | Most likely several viruses | 97 | |
| MER41 | Humans | IFN-γ-inducible enhancer of IFI6 expression | Several flaviviruses | 97 | |
| MER41 | Humans | IFN-γ-inducible enhancer of SECTM1 expression | Most likely several viruses | 97 | |
| ERV9 | Apes | Transcription start site for IRGM | Active against mycobacteria, antiviral activity unclear | 105 | |
| LTR12C | Humans | IFN-γ-inducible promoter of GBP2 | HIV, measles virus, Zika virus, and other furin-dependent viral pathogens | 107 | |
| LTR12C | Humans | Promoter of GBP5 | HIV, measles virus, Zika virus, and other furin-dependent viral pathogens | 107 | |
| LTR12C | Humans | Promoter of SEMA4D (?) | Multiple viruses? | 107 |
TLR, Toll-like receptor; IL, interleukin; TNF-α, tumor necrosis factor alpha; IFN-γ, gamma interferon; lncRNA, long noncoding RNA.
LTR, long terminal repeat.
It has also been proposed that more specific immune activation may be achieved by ERV-derived antisense transcripts that form dsRNA molecules upon binding to complementary RNA (cRNA) molecules derived from exogenous retroviruses (13, 19). In this pathway, the ERV RNA not only acts as a pathogen-associated molecular pattern (PAMP), but may also be considered a kind of PRR itself, since it recognizes complementary exogenous RNA and subsequently triggers the activation of downstream dsRNA-sensing cascades.
Additional evidence for a contribution of viral mimicry to cellular immune responses comes from cancer research. For example, several studies reported a beneficial effect of DNA demethylating drugs in tumor therapy that involves the induction and sensing of endogenous retroviruses and other transposable elements (20–22). In all cases, the induction of antitumor immune responses required the presence of dsRNA sensors, such as TLR3, MDA5, or RIG-I, and may result in increased T cell-mediated killing of tumor cells. Similarly, Lee and colleagues suggested that MDA5-mediated sensing of ERVs contributes to the beneficial effects of tumor radiotherapy (23). Intriguingly, additional knockout of the ERV-repressor TRIM28/KAP1 further boosted the beneficial effects of radiotherapy in two tumor models (23).
Further evidence for a role of ERVs in immune activation comes from congenital diseases, in which the accumulation of retroviral nucleic acids may result in a detrimental chronic IFN response. For example, patients suffering from Aicardi-Goutières syndrome (AGS) show aberrant immune activation and lack cellular factors such as SAMHD1 or TREX1 that reduce the levels of endogenous retroviral RNA/DNA species (24).
Together, these observations highlight the potential of ERV-derived nucleic acids to enhance cellular immune responses. While translational approaches that take advantage of this sensing pathway are already pursued in tumor research, the induction of ERVs in the treatment of viral infections also deserves further attention.
ERV-DERIVED lncRNAs REGULATE ANTIVIRAL IMMUNE RESPONSES
ERV-derived nucleic acids may not only act as PAMPs triggering cellular sensing cascades, but also regulate immune signaling in a more direct manner (Fig. 1B). One interesting example is a murine ERV-derived lncRNA termed lnc-EPAV (for ERV-derived lncRNA positively regulating antiviral responses) that boosts antiviral gene expression by increasing the levels of the transcription factor NF-κB (25). More specifically, lnc-EPAV is expressed upon Sendai virus and vesicular stomatitis virus (VSV) infection and induces the expression of NF-κB/RelA by sequestering its repressor SFPQ (splicing factor, proline and glutamine rich) in the nucleus. As a result, the production of antiviral cytokines such as IFN-β or interleukin 6 (IL-6) is increased. Since lnc-EPAV itself is also NF-κB responsive, this ERV pathway may act as a positive feedback loop enhancing antiviral immune responses (25). Notably, the human ortholog of SFPQ also interacts with multiple HERV-derived RNAs (e.g., MER9a2, LTR5A, and MLT2A1), suggesting that a similar immune-regulatory mechanism exists in humans (25).
ENDOGENOUS RETROVIRAL PROTEINS MODULATE IMMUNE ACTIVATION
Besides endogenous retroviral nucleic acids, ERV-derived proteins may also be sensed and/or regulate immune activation (Fig. 1C). For example, plasma membrane-associated TLRs have been shown to detect different retroviral proteins. This includes sensing of the HERV-W envelope (Env) protein by TLR4 and its coreceptor CD14 (26). As a consequence, proinflammatory and antivirally active cytokines such as IL-1β, IL-6, TNF-α, and IL-12p40 are released (26). The TLR4-dependent induction of cytokines by HERV-W Env was confirmed in an independent study (27) that additionally described an increased expression of inducible nitric oxide synthase (iNOS) in the presence of this retroviral envelope protein. While the production of nitric oxide by iNOS is an important component of the innate immune response, the authors speculate that HERV-W Env-mediated iNOS induction may also contribute to brain damage in multiple sclerosis (27). Another TLR-stimulating retroviral protein is the dUTPase of HERV-K (HML2) that activates NF-κB and induces the release of cytokines (i.e., IL-1β, IL-6, IL-8, IL-10, IL-12p40, IL-23, IL-17, TNF-α, RANTES, IFN-γ, and CCL20) in a TLR2-dependent manner (28).
Exogenous retroviral Envs can also directly activate canonical NF-κB signaling, independently of PRR activation (29). This process involves recruitment of transforming growth factor beta (TGF-β)-activated kinase 1 (TAK1) to the cytoplasmic domain of Env. Thus, it is tempting to speculate that endogenous retroviral Envs with similar NF-κB-activating activity remain to be discovered. Notably, several HERV Envs with immunosuppressive activity have already been described. These include human and murine syncytins, which are expressed in placental cells and may allow maternal immune tolerance to the fetus and placenta (30, 31). Finally, endogenous retroviral Envs of HERV-K18 (32, 33) and possibly HERV-W (34) have been suggested to contribute to the pathogenesis of auto-immune disorders because they act as superantigens that induce excessive activation of T cells. However, this hypothesis remains controversial. While some studies observed an association of HERV-K18 polymorphisms with type I and II diabetes (33, 35), others found no evidence for a contribution of this HERV to the pathogenesis of autoimmune diseases (36, 37). Nevertheless, the above-mentioned examples illustrate that transcription and translation of ERVs need to be tightly regulated in order to prevent detrimental effects on the host.
ENDOGENOUS RETROVIRAL ENVELOPE PROTEINS BLOCK VIRUS ENTRY RECEPTORS
Apart from rather broad and unspecific immune-modulatory activities, endogenous retroviral Env proteins can also directly inhibit viral infection via a process termed receptor interference. This mechanism involves direct binding of cell-associated or cell-free endogenous retroviral Env proteins to cellular entry receptors (Fig. 1D). As a result, these receptors are blocked and cannot mediate attachment of exogenous viruses using the same entry pathway. Furthermore, ERV-derived Envs may bind newly synthesized entry receptors intracellularly and prevent their transport to the cell surface. Since exogenous and endogenous retroviruses frequently share the same receptors, endogenous Envs offer a large repertoire for broadly active antiviral factors.
Early reports of receptor interference date back to the 1960s, when several groups demonstrated that Env proteins of avian leukosis viruses (ALV) induce resistance to exogenous viruses of the same subgroup (38, 39). In 1981, Robinson and colleagues identified three Env-expressing endogenous retroviruses in chickens (ev3, ev6, and ev9) that reduce the susceptibility to and entry of subgroup E ALVs (40). The authors already speculated that additional examples of receptor interference will be found and hypothesized that the previously identified antiviral genes Fv-4r (41) and Akvr-1 (42) “represent defective endogenous viruses” in mice (40). Indeed, Fv-4r and Akvr-1 subsequently turned out to represent a single transcriptionally active but truncated provirus (43–45). Experiments in transgenic mice revealed that the endogenous retroviral Env protein expressed from the Fv-4r (Akvr-1) locus is responsible for the observed restriction of murine leukemia viruses (MLV) (46). Similarly, the murine loci Rmcf and Rmcf2 also express endogenous retroviral Env proteins blocking MLV entry (47–51). Apart from chickens and mice, protective retroviral env genes have also been identified in sheep (52, 53) and cats (54, 55). Notably, the antivirally active Env proteins Refrex-1 and FeLIX in cats are C-terminally truncated and released into the extracellular space, where they can block receptors and thus entry of feline leukemia viruses (FeLV) (54, 55).
More recently, Env-derived restriction factors have also been discovered in primates. Blanco-Melo et al. identified a retroviral envelope protein (HERV-T Env) in humans that fails to mediate infection, but abrogates susceptibility of human cells to infection with reconstituted HERV-T-like viral particles (56). This inhibitory effect was associated with decreased surface levels of the virus entry receptor monocarboxylate transporter-1 (MCT-1). Similarly, the Env-derived protein suppressyn may protect humans and other hominoids from retroviral infections, as it blocks the surface receptor ASCT2 that mediates attachment of RD114/mammalian type-D retroviruses (57). Members of this virus group circulate in nonhuman primates and other mammalian species. Thus, cooption of retroviral envelope genes enables efficient restriction of (zoonotic) transmission events and may have contributed to the extinction of the corresponding exogenous retroviruses.
ERV PROTEINS COMPLEMENT VIRIONS IN A DOMINANT NEGATIVE MANNER
Intriguingly, certain endogenous Env proteins suppress viral replication at several steps of the viral life cycle. For example, murine Fv-4 not only inhibits MLV entry via receptor interference, but also reduces virion infectivity (Fig. 1E). More specifically, Fv-4 is incorporated into budding virions, but fails to mediate infection as it harbors an inactivating mutation in its fusion peptide (58). Similarly, Env proteins of the evolutionarily young HERV-K family of human endogenous retroviruses can be incorporated into HIV-1 particles in vitro (59, 60). However, it remains to be determined whether HERV-K Env is also incorporated into HIV-1 virions and reduces particle infectivity in infected individuals. HERV-K108 Env has also been shown to negatively interfere with HIV-1 replication (61). However, the underlying mechanisms may be independent of virion incorporation, as this Env reduces HIV-1 protein levels and overall virion production (61).
Besides Env, endogenous retroviral Gag proteins can also restrict retroviral replication. This includes the Gag protein of HERV-K, which coassembles with HIV-1 Gag, thereby reducing the production of fully infectious HIV-1 particles (62). Subsequent mechanistic analyses revealed that the inhibitory activity of HERV-K Gag depends on its N-terminal capsid domain and results in the formation of malformed HIV-1 virions (63). Another interesting example comes from sheep that harbor the endogenous Jaagsiekte sheep retrovirus enJS56A1, which expresses a nonfunctional Gag (64). This defective protein directly associates with Gag proteins of exogenous JSRV. As a result, egress of JSRV is suppressed in a dominant negative fashion, and viral particles accumulate in intracellular compartments (64, 65). enJSRV-20, a related endogenous retrovirus, may restrict JSRV replication in a similar manner, as it harbors the same inactivating mutation (Trp21) in its Gag protein (66).
ERV PROTEINS INTERFERE WITH INCOMING VIRAL PARTICLES
The best-characterized Gag-derived restriction factor is probably Fv1 in mice, which inhibits MLV replication (67, 68) and interacts with exogenous retroviral capsid proteins (69). In contrast to the Gag proteins mentioned above, however, Fv1 acts in viral target cells and restricts MLV replication before integration by directly interacting with incoming retroviral capsids (69, 70) (Fig. 1F). Thus, the ability of endogenous viral Gag proteins to interact with their exogenous counterparts may confer blocks at several steps of the retroviral replication cycle. The importance of Fv1 in antiviral immune responses is further highlighted by signatures of positive selection in the Fv1 gene in the genus Mus that are indicative of a long-lasting coevolution with exogenous viral pathogens (71, 72).
Notably, the description of additional coopted gag genes such as wucaishi1 (wcs1) and wucaishi2 (wcs2) (73) suggests that more antiviral Gag proteins remain to be discovered. Moreover, other products of ERVs (e.g., RNA, polymerase, integrase, and reverse transcriptase) may in principle also be incorporated into exogenous viral particles and interfere with viral replication. Laderoute and colleagues reported an interesting association in this regard; they observed an increased number of genomic HERV-K102 pol copy numbers in highly exposed HIV-1-seronegative sex workers compared to those in HIV-1-infected individuals (74). However, the molecular mechanisms underlying this association have remained unclear. Notably, the incorporation of ERV-derived gene products into exogenous viral particles may not always negatively interfere with their assembly or infectivity. For example, certain HERV-derived protease (75, 76), integrase (77), or Env (78, 79) proteins may at least partially complement HIV-1 mutants with defects in the respective genes and sometimes even expanded the tropism to CD4-negative cells (78, 79). Nevertheless, the cooption of retroviral genes to negatively interfere with virion production represents a simple yet effective antiviral mechanism that has evolved several times independently during mammalian evolution.
REPETITIVE ERV ELEMENTS INCREASE PLASTICITY OF IMMUNITY GENES
All of the antiviral mechanisms described above depend on the transcription and/or translation of endogenous retroviral sequences. Nevertheless, transcriptionally silent ERVs can also provide a selection advantage to the host. For example, the presence of repetitive ERV elements may promote recombination processes that increase the number and/or allelic variety of host genes (80–82) (Fig. 1G). This mechanism of gene plasticity might usually be selected against in monogenic systems, since homologous recombination events can also result in the loss of genes or gene fragments (83, 84). In contrast, the risk-benefit ratio of ERV-dependent recombination is most likely lower in multigene families, where the insertion of repetitive ERV sequences may provide a fitness advantage and facilitate adaptation to viral pathogens. One notable example is the MHC locus in primates, where the integration of different ERVs may have promoted the diversity of alleles and thereby broadened the spectrum of viral peptides that can be presented to T helper cells (85, 86).
ERV-DERIVED PROMOTERS AND ENHANCERS REGULATE ANTIVIRAL GENE EXPRESSION
Retroviral elements may also contribute to the regulation of host gene expression (Fig. 1H). One key characteristic of retroviruses is the presence of promoters, enhancers, transcription start sites, and other gene regulatory elements is their long terminal repeats (LTRs). Exogenous retroviruses harbor multiple binding sites for cellular transcription factors in their LTRs and exploit them to regulate the expression of their own genes. This frequently includes immune-regulatory transcription factors such as NF-κB, interferon regulatory factors (IRFs), or STATs, since they are activated in response to viral infection. Upon endogenization, the ability of LTRs to recruit transcription factors can be exploited by the host to regulate the expression of cellular genes. This cooption of regulatory elements is not a rare phenomenon, and it has been estimated that about 20% of all transcription factor binding sites in humans are found in HERVs and other transposable elements (87). In line with this, a meta-analysis of chromatin immunoprecipitation sequencing (ChIP-Seq) data sets identified about 800,000 transcription factor binding sites within HERVs (88).
Intriguingly, almost 90% of all HERVs represent so-called solo LTRs (89). These HERVs lost the prototypical retroviral genes gag, pol, and env due to homologous recombination of their flanking LTR sequences, leaving single LTR promoters in the genome. Due to their activation upon immune stimulation, ERV LTRs have already been termed “landing strips for inflammatory transcription factors” (90), and evidence for their role in regulating cellular immune responses is growing. One of the earlier reports comes from mice, where the insertion of a retroviral LTR increases the transcription of APOBEC3 (91). This deaminase introduces hypermutations in retroviral genomes, thereby restricting replication of mouse mammary tumor virus (MMTV) and potentially that of other viruses (92). The endogenized LTR is derived from a xenotropic mouse gammaretrovirus and harbors typical regulatory elements, including CAT and TATA boxes and an enhancer region. Mice harboring the LTR insertion show about 4- to 20-fold higher APOBEC3 mRNA levels than in those lacking it (91). These findings strongly suggest that the fixation of a retroviral LTR in the resistance gene APOBEC3 confers a selection advantage in virally infected mice, although direct in vivo evidence is missing.
Another example of an endogenous retroviral LTR modulating the expression of an immunity gene is the HERVP71A LTR (termed enhancer L), which acts as a distal enhancer for HLA-G in trophoblast cells (93, 94). This nonclassical MHC protein has been suggested to primarily prevent killing of placenta cells by NK cells, but it may also play a role in antiviral immune responses, as it is able to present intracellular peptides (95, 96).
In 2016, Chuong and colleagues performed a seminal study, deciphering the role of ERVs in IFN-mediated immune responses in a global and unbiased manner (97). Taking advantage of available ChIP-Seq data, they searched the human genome for transposable elements that show an increase in STAT1 and/or IRF1 binding upon IFN-γ stimulation and which may therefore contribute to the expression of ISGs upon viral infection. This approach identified 20 HERV families that are enriched for IRF1/STAT1 binding peaks upon IFN-γ stimulation. One notable example is the MER41 family of endogenous gammaretroviruses, whose members frequently harbor a so-called gamma-activated site (GAS) that is bound by STAT1. Notably, STAT1-binding MER41 elements were found in the vicinity of immunity genes such as AIM2, APOL1, IFI6, and SECTM1 (97). While AIM2 triggers an inflammasome-mediated immune response upon sensing of cytosolic dsDNA (98), APOL1 lyses intracellular parasites (99), IFI6 inhibits replication of several flaviviruses (100–103), and SECTM1 contributes to T-cell activation (104). In agreement with an important immune-regulatory activity of MER41 elements in vivo, their CRISPR/Cas9-mediated deletion reduced or abrogated the responsiveness of the adjacent immunity genes to IFN-γ (97).
The endogenous LTRs mentioned above all act as enhancers and may increase antiviral gene expression in response to viral infection and/or IFN stimulation. In some cases, however, ERVs have also been coopted as the primary promoters and/or transcription start sites (TSS) for immunity genes. One example is an ERV9 element harboring the TSS for the immunity-related GTPase family M protein (IRGM) (105). This large IFN-inducible GTPase eliminates mycobacteria by inducing autophagy (106). In this case, the insertion of an Alu retrotransposon initially disrupted the open reading frame of IRGM in the common anthropoid ancestor, but was subsequently resurrected by the insertion of the ERV9 transcription start site (TSS) (105). Interestingly, expression of two additional members of the family of large IFN-inducible GTPases is also regulated by ERV9 promoters. Using an RNA sequencing approach, we recently found that guanylate-binding proteins 2 and 5 (GBP2 and GBP5) are expressed under the control of ERV9 solo-LTRs, termed LTR12C, and can be further increased by IFN-γ stimulation and HIV-1 infection (107, 108). These two GTPases exert broad antiviral activity by inhibiting the virus dependency factor furin (109). Intriguingly, numerous LTR12C and related LTR12D elements are activated upon HIV-1 infection (107, 110), suggesting that they may be part of a larger network regulating antiviral immune responses. In line with this, expression of an LTR12C-SEMA4D fusion transcript is also increased in HIV-1-infected cells (107). SEMA4D (also called CD100) is a regulator of B-cell activation and may contribute to T-cell senescence in HIV-1-infected individuals (111, 112).
In the context of the current COVID-19 pandemic, the report of a retroelement (MIRb) that acts as promoter for an unusual isoform of ACE2 has received some attention (113). Since ACE2 is the primary receptor of SARS-CoV-2, its expression pattern and inducibility are important determinants of the tissue tropism and pathogenicity of this pandemic pathogen. However, the MIRb-ACE2 transcript turned out to be translated into a truncated and unstable form of ACE2 that most likely fails to mediate viral entry, as it lacks domains required for SARS-CoV-2 binding (113). In contrast to the canonical ACE2 isoform, the unusual MIRb-ACE2 transcript is strongly IFN inducible. However, its exact role in viral infections remains unclear.
In summary, retroviral LTRs have been coopted independently several times to regulate the expression of antiviral host factors, and it is very likely that additional examples remain to be discovered.
IMPORTANCE AND FUTURE DIRECTIONS
Our knowledge about endogenous retroviral elements involved in cellular immune responses is constantly increasing. Recent key findings include the discovery of retroviral Envs restricting retroviral infection in primates (56, 57), the identification of a SUMO-dependent pathway of antiviral immunity that involves sensing of ERV nucleic acids (15), and the description of additional retroviral promoter and enhancer elements regulating antiviral gene expression (93, 97, 107). These reports not only provide important insights into innate antiviral defense mechanisms, but also help to assess the role of ERV activation that is observed in a variety of infectious and noninfectious diseases. Furthermore, they strongly suggest that the host accepts possible risks associated with the cooption and activation of endogenous retroviruses (e.g., detrimental immune activation), and that the positive effects of ERVs usually prevail.
Nevertheless, several open questions remain. Although many studies report interesting associations of increased ERV activity with immune activation, causality is frequently not shown, and the relative contribution of ERVs to antiviral immune responses still remains unclear. Furthermore, several studies characterize antiviral mechanisms of individual ERVs in vitro, but do not investigate whether these antivirally active ERVs are activated upon infection in vivo.
Another important aspect of future studies will be to decipher the pathways that trigger the activation of specific ERV repeats or families upon viral infection. Are ERVs primarily activated by the cytokine storm in response to viral infection (97)? Are they derepressed because exogenous viruses counteract host restriction factors that suppress retroviral gene expression, such as TRIM28 (15, 23)? Why do viruses such as HIV-1 primarily activate specific families of ERVs (107, 110)? Answering these questions will shed light on the ERV immune network that is activated in response to infection and may uncover strategies that viruses have evolved to evade ERV-mediated immunity. Finally, cancer research has already demonstrated that artificial induction of ERV expression can boost antitumor immune responses, and it will be important to investigate whether similar beneficial effects can be achieved for the therapy of viral diseases.
ACKNOWLEDGMENTS
We thank Jumpei Ito, Frank Kirchhoff, Dorota Kmiec, and Kei Sato for fruitful discussions and for critically reading the manuscript.
D.S. was supported by the Heisenberg Programme (grant SA 2676/3-1) and Priority Programme SPP1923 of the German Research Foundation. S.S.B. was supported by the International Graduate School in Molecular Medicine Ulm (IGradU) and the Research Training Group “Cellular and Molecular Mechanisms in Aging” (CEMMA).
Contributor Information
Daniel Sauter, Email: daniel.sauter@med.uni-tuebingen.de.
Michaela Ulrike Gack, Cleveland Clinic.
REFERENCES
- 1.Wang M, Qiu Y, Liu H, Liang B, Fan B, Zhou X, Liu D. 2020. Transcription profile of human endogenous retroviruses in response to dengue virus serotype 2 infection. Virology 544:21–30. 10.1016/j.virol.2020.01.014. [DOI] [PubMed] [Google Scholar]
- 2.Nellåker C, Yao Y, Jones-Brando L, Mallet F, Yolken RH, Karlsson H. 2006. Transactivation of elements in the human endogenous retrovirus W family by viral infection. Retrovirology 3:44. 10.1186/1742-4690-3-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhardwaj N, Maldarelli F, Mellors J, Coffin JM. 2014. HIV-1 infection leads to increased transcription of human endogenous retrovirus HERV-K (HML-2) proviruses in vivo but not to increased virion production. J Virol 88:11108–11120. 10.1128/JVI.01623-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Contreras-Galindo R, López P, Vélez R, Yamamura Y. 2007. HIV-1 infection increases the expression of human endogenous retroviruses type K (HERV-K) in vitro. AIDS Res Hum Retroviruses 23:116–122. 10.1089/aid.2006.0117. [DOI] [PubMed] [Google Scholar]
- 5.Vincendeau M, Göttesdorfer I, Schreml JMHH, Wetie AGNG, Mayer J, Greenwood AD, Helfer M, Kramer S, Seifarth W, Hadian K, Brack-Werner R, Leib-Mösch C. 2015. Modulation of human endogenous retrovirus (HERV) transcription during persistent and de novo HIV-1 infection. Retrovirology 12:27. 10.1186/s12977-015-0156-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mi S, Lee X, Li X, Veldman GM, Finnerty H, Racie L, LaVallie E, Tang XY, Edouard P, Howes S, Keith JC, McCoy JM. 2000. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature 403:785–789. 10.1038/35001608. [DOI] [PubMed] [Google Scholar]
- 7.Blaise S, de Parseval N, Bénit L, Heidmann T. 2003. Genomewide screening for fusogenic human endogenous retrovirus envelopes identifies syncytin 2, a gene conserved on primate evolution. Proc Natl Acad Sci U S A 100:13013–13018. 10.1073/pnas.2132646100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paces J, Pavlícek A, Paces V. 2002. HERVd: database of human endogenous retroviruses. Nucleic Acids Res 30:205–206. 10.1093/nar/30.1.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, Taira K, Akira S, Fujita T. 2004. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol 5:730–737. 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 10.Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, Uematsu S, Jung A, Kawai T, Ishii KJ, Yamaguchi O, Otsu K, Tsujimura T, Koh C-S, Reis e Sousa C, Matsuura Y, Fujita T, Akira S. 2006. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 441:101–105. 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 11.Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. 2001. Recognition of double-stranded RNA and activation of NF-κB by Toll-like receptor 3. Nature 413:732–738. 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- 12.Rigby RE, Webb LM, Mackenzie KJ, Li Y, Leitch A, Reijns MAM, Lundie RJ, Revuelta A, Davidson DJ, Diebold S, Modis Y, MacDonald AS, Jackson AP. 2014. RNA:DNA hybrids are a novel molecular pattern sensed by TLR9. EMBO J 33:542–558. 10.1002/embj.201386117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hurst TP, Magiorkinis G. 2015. Activation of the innate immune response by endogenous retroviruses. J Gen Virol 96:1207–1218. 10.1099/jgv.0.000017. [DOI] [PubMed] [Google Scholar]
- 14.Sauter D, Kirchhoff F. 2021. Evolutionary conflicts and adverse effects of antiviral factors. Elife 10:e65243. 10.7554/eLife.65243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schmidt N, Domingues P, Golebiowski F, Patzina C, Tatham MH, Hay RT, Hale BG. 2019. An influenza virus-triggered SUMO switch orchestrates co-opted endogenous retroviruses to stimulate host antiviral immunity. Proc Natl Acad Sci U S A 116:17399–17408. 10.1073/pnas.1907031116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rowe HM, Jakobsson J, Mesnard D, Rougemont J, Reynard S, Aktas T, Maillard PV, Layard-Liesching H, Verp S, Marquis J, Spitz F, Constam DB, Trono D. 2010. KAP1 controls endogenous retroviruses in embryonic stem cells. Nature 463:237–240. 10.1038/nature08674. [DOI] [PubMed] [Google Scholar]
- 17.Wolf D, Goff SP. 2007. TRIM28 mediates primer binding site-targeted silencing of murine leukemia virus in embryonic cells. Cell 131:46–57. 10.1016/j.cell.2007.07.026. [DOI] [PubMed] [Google Scholar]
- 18.Chen S, Hu X, Cui IH, Wu S, Dou C, Liu Y, Sun Z, Xue S, Geng T, Liu Z, Qin A, Cui H. 2019. An endogenous retroviral element exerts an antiviral innate immune function via the derived lncRNA lnc-ALVE1-AS1. Antiviral Res 170:104571. 10.1016/j.antiviral.2019.104571. [DOI] [PubMed] [Google Scholar]
- 19.Gürtler C, Bowie AG. 2013. Innate immune detection of microbial nucleic acids. Trends Microbiol 21:413–420. 10.1016/j.tim.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chiappinelli KB, Strissel PL, Desrichard A, Li H, Henke C, Akman B, Hein A, Rote NS, Cope LM, Snyder A, Makarov V, Budhu S, Buhu S, Slamon DJ, Wolchok JD, Pardoll DM, Beckmann MW, Zahnow CA, Merghoub T, Mergoub T, Chan TA, Baylin SB, Strick R. 2015. Inhibiting DNA methylation causes an interferon response in cancer via dsRNA including endogenous retroviruses. Cell 162:974–986. 10.1016/j.cell.2015.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Cubas AA, Dunker W, Zaninovich A, Hongo RA, Bhatia A, Panda A, Beckermann KE, Bhanot G, Ganesan S, Karijolich J, Rathmell WK. 2020. DNA hypomethylation promotes transposable element expression and activation of immune signaling in renal cell cancer. JCI Insight 5:e137569. 10.1172/jci.insight.137569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roulois D, Loo Yau H, Singhania R, Wang Y, Danesh A, Shen SY, Han H, Liang G, Jones PA, Pugh TJ, O’Brien C, De Carvalho DD. 2015. DNA-demethylating agents target colorectal cancer cells by inducing viral mimicry by endogenous transcripts. Cell 162:961–973. 10.1016/j.cell.2015.07.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee AK, Pan D, Bao X, Hu M, Li F, Li CY. 2020. Endogenous retrovirus activation as a key mechanism of anti-tumor immune response in radiotherapy. Radiat Res 193:305–317. 10.1667/RADE-20-00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Crow YJ, Manel N. 2015. Aicardi-Goutières syndrome and the type I interferonopathies. Nat Rev Immunol 15:429–440. 10.1038/nri3850. [DOI] [PubMed] [Google Scholar]
- 25.Zhou B, Qi F, Wu F, Nie H, Song Y, Shao L, Han J, Wu Z, Saiyin H, Wei G, Wang P, Ni T, Qian F. 2019. Endogenous retrovirus-derived long noncoding RNA enhances innate immune responses via derepressing RELA expression. mBio 10:e00937-19. 10.1128/mBio.00937-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rolland A, Jouvin-Marche E, Viret C, Faure M, Perron H, Marche PN. 2006. The envelope protein of a human endogenous retrovirus-W family activates innate immunity through CD14/TLR4 and promotes Th1-like responses. J Immunol 176:7636–7644. 10.4049/jimmunol.176.12.7636. [DOI] [PubMed] [Google Scholar]
- 27.Kremer D, Schichel T, Förster M, Tzekova N, Bernard C, van der Valk P, van Horssen J, Hartung H-P, Perron H, Küry P. 2013. Human endogenous retrovirus type W envelope protein inhibits oligodendroglial precursor cell differentiation. Ann Neurol 74:721–732. 10.1002/ana.23970. [DOI] [PubMed] [Google Scholar]
- 28.Ariza M-E, Williams MV. 2011. A human endogenous retrovirus K dUTPase triggers a TH1, TH17 cytokine response: does it have a role in psoriasis? J Invest Dermatol 131:2419–2427. 10.1038/jid.2011.217. [DOI] [PubMed] [Google Scholar]
- 29.Postler TS, Desrosiers RC. 2012. The cytoplasmic domain of the HIV-1 glycoprotein gp41 induces NF-κB activation through TGF-β-activated kinase 1. Cell Host Microbe 11:181–193. 10.1016/j.chom.2011.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mangeney M, Renard M, Schlecht-Louf G, Bouallaga I, Heidmann O, Letzelter C, Richaud A, Ducos B, Heidmann T. 2007. Placental syncytins: genetic disjunction between the fusogenic and immunosuppressive activity of retroviral envelope proteins. Proc Natl Acad Sci U S A 104:20534–20539. 10.1073/pnas.0707873105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tolosa JM, Schjenken JE, Clifton VL, Vargas A, Barbeau B, Lowry P, Maiti K, Smith R. 2012. The endogenous retroviral envelope protein syncytin-1 inhibits LPS/PHA-stimulated cytokine responses in human blood and is sorted into placental exosomes. Placenta 33:933–941. 10.1016/j.placenta.2012.08.004. [DOI] [PubMed] [Google Scholar]
- 32.Conrad B, Weissmahr RN, Böni J, Arcari R, Schüpbach J, Mach B. 1997. A human endogenous retroviral superantigen as candidate autoimmune gene in type I diabetes. Cell 90:303–313. 10.1016/s0092-8674(00)80338-4. [DOI] [PubMed] [Google Scholar]
- 33.Marguerat S, Wang WYS, Todd JA, Conrad B. 2004. Association of human endogenous retrovirus K-18 polymorphisms with type 1 diabetes. Diabetes 53:852–854. 10.2337/diabetes.53.3.852. [DOI] [PubMed] [Google Scholar]
- 34.Emmer A, Staege MS, Kornhuber ME. 2014. The retrovirus/superantigen hypothesis of multiple sclerosis. Cell Mol Neurobiol 34:1087–1096. 10.1007/s10571-014-0100-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dickerson F, Rubalcaba E, Viscidi R, Yang S, Stallings C, Sullens A, Origoni A, Leister F, Yolken R. 2008. Polymorphisms in human endogenous retrovirus K-18 and risk of type 2 diabetes in individuals with schizophrenia. Schizophr Res 104:121–126. 10.1016/j.schres.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 36.Nyegaard M, Demontis D, Thestrup BB, Hedemand A, Sørensen KM, Hansen T, Werge T, Hougaard DM, Yolken RH, Mortensen PB, Mors O, Børglum AD. 2012. No association of polymorphisms in human endogenous retrovirus K18 and CD48 with schizophrenia. Psychiatr Genet 22:146–148. 10.1097/YPG.0b013e328353953c. [DOI] [PubMed] [Google Scholar]
- 37.Ramos-Lopez E, Ghebru S, Van Autreve J, Aminkeng F, Herwig J, Seifried E, Seidl C, Van der Auwera B, Badenhoop K. 2006. Neither an intronic CA repeat within the CD48 gene nor the HERV-K18 polymorphisms are associated with type 1 diabetes. Tissue Antigens 68:147–152. 10.1111/j.1399-0039.2006.00637.x. [DOI] [PubMed] [Google Scholar]
- 38.Vogt PK, Ishizaki R. 1966. Patterns of viral interference in the avion leukosis and sarcoma complex. Virology 30:368–374. 10.1016/0042-6822(66)90115-2. [DOI] [PubMed] [Google Scholar]
- 39.Steck FT, Rubin H. 1966. The mechanism of interference between an avian leukosis virus and Rous sarcoma virus. II. Early steps of infection by RSV of cells under conditions of interference. Virology 29:642–653. 10.1016/0042-6822(66)90288-1. [DOI] [PubMed] [Google Scholar]
- 40.Robinson HL, Astrin SM, Senior AM, Salazar FH. 1981. Host susceptibility to endogenous viruses: defective, glycoprotein-expressing proviruses interfere with infections. J Virol 40:745–751. 10.1128/JVI.40.3.745-751.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Suzuki S. 1975. FV-4: a new gene affecting the splenomegaly induction by Friend leukemia virus. Jpn J Exp Med 45:473–478. [PubMed] [Google Scholar]
- 42.Gardner MB, Rasheed S, Pal BK, Estes JD, O’Brien SJ. 1980. Akvr-1, a dominant murine leukemia virus restriction gene, is polymorphic in leukemia-prone wild mice. Proc Natl Acad Sci U S A 77:531–535. 10.1073/pnas.77.1.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.O’Brien SJ, Berman EJ, Estes JD, Gardner MB. 1983. Murine retroviral restriction genes Fv-4 and Akvr-1 are alleles of a single locus. J Virol 47:649–651. 10.1128/JVI.47.3.649-651.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gardner MB, Kozak CA, O’Brien SJ. 1991. The Lake Casitas wild mouse: evolving genetic resistance to retroviral disease. Trends Genet 7:22–27. 10.1016/0168-9525(91)90017-K. [DOI] [PubMed] [Google Scholar]
- 45.Ikeda H, Laigret F, Martin MA, Repaske R. 1985. Characterization of a molecularly cloned retroviral sequence associated with Fv-4 resistance. J Virol 55:768–777. 10.1128/JVI.55.3.768-777.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Limjoco TI, Dickie P, Ikeda H, Silver J. 1993. Transgenic Fv-4 mice resistant to Friend virus. J Virol 67:4163–4168. 10.1128/JVI.67.7.4163-4168.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jung YT, Lyu MS, Buckler-White A, Kozak CA. 2002. Characterization of a polytropic murine leukemia virus proviral sequence associated with the virus resistance gene Rmcf of DBA/2 mice. J Virol 76:8218–8224. 10.1128/jvi.76.16.8218-8224.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu T, Yan Y, Kozak CA. 2005. Rmcf2, a xenotropic provirus in the Asian mouse species Mus castaneus, blocks infection by polytropic mouse gammaretroviruses. J Virol 79:9677–9684. 10.1128/JVI.79.15.9677-9684.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Buller RS, Ahmed A, Portis JL. 1987. Identification of two forms of an endogenous murine retroviral env gene linked to the Rmcf locus. J Virol 61:29–34. 10.1128/JVI.61.1.29-34.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bassin RH, Ruscetti S, Ali I, Haapala DK, Rein A. 1982. Normal DBA/2 mouse cells synthesize a glycoprotein which interferes with MCF virus infection. Virology 123:139–151. 10.1016/0042-6822(82)90301-4. [DOI] [PubMed] [Google Scholar]
- 51.Ruscetti S, Davis L, Feild J, Oliff A. 1981. Friend murine leukemia virus-induced leukemia is associated with the formation of mink cell focus-inducing viruses and is blocked in mice expressing endogenous mink cell focus-inducing xenotropic viral envelope genes. J Exp Med 154:907–920. 10.1084/jem.154.3.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Spencer TE, Mura M, Gray CA, Griebel PJ, Palmarini M. 2003. Receptor usage and fetal expression of ovine endogenous betaretroviruses: implications for coevolution of endogenous and exogenous retroviruses. J Virol 77:749–753. 10.1128/jvi.77.1.749-753.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Varela M, Spencer TE, Palmarini M, Arnaud F. 2009. Friendly viruses. Ann N Y Acad Sci 1178:157–172. 10.1111/j.1749-6632.2009.05002.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ito J, Watanabe S, Hiratsuka T, Kuse K, Odahara Y, Ochi H, Kawamura M, Nishigaki K. 2013. Refrex-1, a soluble restriction factor against feline endogenous and exogenous retroviruses. J Virol 87:12029–12040. 10.1128/JVI.01267-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McDougall AS, Terry A, Tzavaras T, Cheney C, Rojko J, Neil JC. 1994. Defective endogenous proviruses are expressed in feline lymphoid cells: evidence for a role in natural resistance to subgroup B feline leukemia viruses. J Virol 68:2151–2160. 10.1128/JVI.68.4.2151-2160.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Blanco-Melo D, Gifford RJ, Bieniasz PD. 2017. Co-option of an endogenous retrovirus envelope for host defense in hominid ancestors. Elife 6:e22519. 10.7554/eLife.22519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Frank JA, Singh M, Cullen HB, Kirou RA, Coyne CB, Feschotte C. 2020. Antiviral activity of a human placental protein of retroviral origin. bioRxiv https://www.biorxiv.org/content/10.1101/2020.08.23.263665v1 [DOI] [PMC free article] [PubMed]
- 58.Taylor GM, Gao Y, Sanders DA. 2001. Fv-4: identification of the defect in Env and the mechanism of resistance to ecotropic murine leukemia virus. J Virol 75:11244–11248. 10.1128/JVI.75.22.11244-11248.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brinzevich D, Young GR, Sebra R, Ayllon J, Maio SM, Deikus G, Chen BK, Fernandez-Sesma A, Simon V, Mulder LCF. 2014. HIV-1 interacts with human endogenous retrovirus K (HML-2) envelopes derived from human primary lymphocytes. J Virol 88:6213–6223. 10.1128/JVI.00669-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dewannieux M, Blaise S, Heidmann T. 2005. Identification of a functional envelope protein from the HERV-K family of human endogenous retroviruses. J Virol 79:15573–15577. 10.1128/JVI.79.24.15573-15577.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Terry SN, Manganaro L, Cuesta-Dominguez A, Brinzevich D, Simon V, Mulder LCF. 2017. Expression of HERV-K108 envelope interferes with HIV-1 production. Virology 509:52–59. 10.1016/j.virol.2017.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Monde K, Contreras-Galindo R, Kaplan MH, Markovitz DM, Ono A. 2012. Human endogenous retrovirus K Gag coassembles with HIV-1 Gag and reduces the release efficiency and infectivity of HIV-1. J Virol 86:11194–11208. 10.1128/JVI.00301-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Monde K, Terasawa H, Nakano Y, Soheilian F, Nagashima K, Maeda Y, Ono A. 2017. Molecular mechanisms by which HERV-K Gag interferes with HIV-1 Gag assembly and particle infectivity. Retrovirology 14:27. 10.1186/s12977-017-0351-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mura M, Murcia P, Caporale M, Spencer TE, Nagashima K, Rein A, Palmarini M. 2004. Late viral interference induced by transdominant Gag of an endogenous retrovirus. Proc Natl Acad Sci U S A 101:11117–11122. 10.1073/pnas.0402877101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Murcia PR, Arnaud F, Palmarini M. 2007. The transdominant endogenous retrovirus enJS56A1 associates with and blocks intracellular trafficking of Jaagsiekte sheep retrovirus Gag. J Virol 81:1762–1772. 10.1128/JVI.01859-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Arnaud F, Caporale M, Varela M, Biek R, Chessa B, Alberti A, Golder M, Mura M, Zhang Y, Yu L, Pereira F, DeMartini JC, Leymaster K, Spencer TE, Palmarini M. 2007. A paradigm for virus–host coevolution: sequential counter-adaptations between endogenous and exogenous retroviruses. PLoS Pathog 3:e170. 10.1371/journal.ppat.0030170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stoye JP. 1998. Fv1, the mouse retrovirus resistance gene. Rev Sci Tech 17:269–277. 10.20506/rst.17.1.1080. [DOI] [PubMed] [Google Scholar]
- 68.Best S, Le Tissier P, Towers G, Stoye JP. 1996. Positional cloning of the mouse retrovirus restriction gene Fvl. Nature 382:826–829. 10.1038/382826a0. [DOI] [PubMed] [Google Scholar]
- 69.Hilditch L, Matadeen R, Goldstone DC, Rosenthal PB, Taylor IA, Stoye JP. 2011. Ordered assembly of murine leukemia virus capsid protein on lipid nanotubes directs specific binding by the restriction factor, Fv1. Proc Natl Acad Sci U S A 108:5771–5776. 10.1073/pnas.1100118108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Passerini LD, Keckesova Z, Towers GJ. 2006. Retroviral restriction factors Fv1 and TRIM5α act independently and can compete for incoming virus before reverse transcription. J Virol 80:2100–2105. 10.1128/JVI.80.5.2100-2105.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Boso G, Buckler-White A, Kozak CA. 2018. Ancient evolutionary origin and positive selection of the retroviral restriction factor Fv1 in muroid rodents. J Virol 92:e00850-18. 10.1128/JVI.00850-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yan Y, Buckler-White A, Wollenberg K, Kozak CA. 2009. Origin, antiviral function and evidence for positive selection of the gammaretrovirus restriction gene Fv1 in the genus Mus. Proc Natl Acad Sci U S A 106:3259–3263. 10.1073/pnas.0900181106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang J, Gong Z, Han G-Z. 2019. Convergent co-option of the retroviral gag gene during the early evolution of mammals. J Virol 93:e00542-19. 10.1128/JVI.00542-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Laderoute MP, Larocque LJ, Giulivi A, Diaz-Mitoma F. 2015. Further evidence that human endogenous retrovirus K102 is a replication competent foamy virus that may antagonize HIV-1 replication. Open AIDS J 9:112–122. 10.2174/1874613601509010112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Towler EM, Gulnik SV, Bhat TN, Xie D, Gustschina E, Sumpter TR, Robertson N, Jones C, Sauter M, Mueller-Lantzsch N, Debouck C, Erickson JW. 1998. Functional characterization of the protease of human endogenous retrovirus, K10: can it complement HIV-1 protease? Biochemistry 37:17137–17144. 10.1021/bi9818927. [DOI] [PubMed] [Google Scholar]
- 76.Padow M, Lai L, Fisher RJ, Zhou YC, Wu X, Kappes JC, Towler EM. 2000. Analysis of human immunodeficiency virus type 1 containing HERV-K protease. AIDS Res Hum Retroviruses 16:1973–1980. 10.1089/088922200750054701. [DOI] [PubMed] [Google Scholar]
- 77.Ogata T, Okui N, Sakuma R, Kobayashi N, Kitamura Y. 1999. Integrase of human endogenous retrovirus K-10 supports the replication of replication-incompetent Int− human immunodeficiency virus type 1 mutant. Jpn J Infect Dis 52:251–252. [PubMed] [Google Scholar]
- 78.An DS, Xie Y, Chen ISY. 2001. Envelope gene of the human endogenous retrovirus HERV-W encodes a functional retrovirus envelope. J Virol 75:3488–3489. 10.1128/JVI.75.7.3488-3489.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tang Y, Woodward BO, Pastor L, George AM, Petrechko O, Nouvet FJ, Haas DW, Jiang G, Hildreth JEK. 2020. Endogenous retroviral envelope syncytin induces HIV-1 spreading and establishes HIV reservoirs in placenta. Cell Rep 30:4528–4539.e4. 10.1016/j.celrep.2020.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hughes JF, Coffin JM. 2005. Human endogenous retroviral elements as indicators of ectopic recombination events in the primate genome. Genetics 171:1183–1194. 10.1534/genetics.105.043976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Feschotte C, Gilbert C. 2012. Endogenous viruses: insights into viral evolution and impact on host biology. Nat Rev Genet 13:283–296. 10.1038/nrg3199. [DOI] [PubMed] [Google Scholar]
- 82.Hughes JF, Coffin JM. 2001. Evidence for genomic rearrangements mediated by human endogenous retroviruses during primate evolution. Nat Genet 29:487–489. 10.1038/ng775. [DOI] [PubMed] [Google Scholar]
- 83.Sun C, Skaletsky H, Rozen S, Gromoll J, Nieschlag E, Oates R, Page DC. 2000. Deletion of azoospermia factor a (AZFa) region of human Y chromosome caused by recombination between HERV15 proviruses. Hum Mol Genet 9:2291–2296. 10.1093/oxfordjournals.hmg.a018920. [DOI] [PubMed] [Google Scholar]
- 84.Campbell IM, Gambin T, Dittwald P, Beck CR, Shuvarikov A, Hixson P, Patel A, Gambin A, Shaw CA, Rosenfeld JA, Stankiewicz P. 2014. Human endogenous retroviral elements promote genome instability via non-allelic homologous recombination. BMC Biol 12:74. 10.1186/s12915-014-0074-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Doxiadis GGM, de Groot N, Bontrop RE. 2008. Impact of endogenous intronic retroviruses on major histocompatibility complex class II diversity and stability. J Virol 82:6667–6677. 10.1128/JVI.00097-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kulski JK, Gaudieri S, Martin A, Dawkins RL. 1999. Coevolution of PERB11 (MIC) and HLA class I genes with HERV-16 and retroelements by extended genomic duplication. J Mol Evol 49:84–97. 10.1007/pl00006537. [DOI] [PubMed] [Google Scholar]
- 87.Sundaram V, Cheng Y, Ma Z, Li D, Xing X, Edge P, Snyder MP, Wang T. 2014. Widespread contribution of transposable elements to the innovation of gene regulatory networks. Genome Res 24:1963–1976. 10.1101/gr.168872.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ito J, Sugimoto R, Nakaoka H, Yamada S, Kimura T, Hayano T, Inoue I. 2017. Systematic identification and characterization of regulatory elements derived from human endogenous retroviruses. PLoS Genet 13:e1006883. 10.1371/journal.pgen.1006883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Belshaw R, Watson J, Katzourakis A, Howe A, Woolven-Allen J, Burt A, Tristem M. 2007. Rate of recombinational deletion among human endogenous retroviruses. J Virol 81:9437–9442. 10.1128/JVI.02216-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Manghera M, Douville RN. 2013. Endogenous retrovirus-K promoter: a landing strip for inflammatory transcription factors? Retrovirology 10:16. 10.1186/1742-4690-10-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sanville B, Dolan MA, Wollenberg K, Yan Y, Martin C, Yeung ML, Strebel K, Buckler-White A, Kozak CA. 2010. Adaptive evolution of Mus Apobec3 includes retroviral insertion and positive selection at two clusters of residues flanking the substrate groove. PLoS Pathog 6:e1000974. 10.1371/journal.ppat.1000974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Okeoma CM, Lovsin N, Peterlin BM, Ross SR. 2007. APOBEC3 inhibits mouse mammary tumour virus replication in vivo. Nature 445:927–930. 10.1038/nature05540. [DOI] [PubMed] [Google Scholar]
- 93.Ferreira LMR, Meissner TB, Mikkelsen TS, Mallard W, O’Donnell CW, Tilburgs T, Gomes HAB, Camahort R, Sherwood RI, Gifford DK, Rinn JL, Cowan CA, Strominger JL. 2016. A distant trophoblast-specific enhancer controls HLA-G expression at the maternal–fetal interface. Proc Natl Acad Sci U S A 113:5364–5369. 10.1073/pnas.1602886113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Grandi N, Tramontano E. 2018. Human endogenous retroviruses are ancient acquired elements still shaping innate immune responses. Front Immunol 9:2039. 10.3389/fimmu.2018.02039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Diehl M, Münz C, Keilholz W, Stevanović S, Holmes N, Loke YW, Rammensee H-G. 1996. Nonclassical HLA-G molecules are classical peptide presenters. Curr Biol 6:305–314. 10.1016/s0960-9822(02)00481-5. [DOI] [PubMed] [Google Scholar]
- 96.Schmidt CM, Orr HT. 1993. Maternal/fetal interactions: the role of the MHC class I molecule HLA-G. Crit Rev Immunol 13:207–224. [PubMed] [Google Scholar]
- 97.Chuong EB, Elde NC, Feschotte C. 2016. Regulatory evolution of innate immunity through co-option of endogenous retroviruses. Science 351:1083–1087. 10.1126/science.aad5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hornung V, Ablasser A, Charrel-Dennis M, Bauernfeind F, Horvath G, Caffrey DR, Latz E, Fitzgerald KA. 2009. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature 458:514–518. 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Vanhamme L, Paturiaux-Hanocq F, Poelvoorde P, Nolan DP, Lins L, Van Den Abbeele J, Pays A, Tebabi P, Van Xong H, Jacquet A, Moguilevsky N, Dieu M, Kane JP, De Baetselier P, Brasseur R, Pays E. 2003. Apolipoprotein L-I is the trypanosome lytic factor of human serum. Nature 422:83–87. 10.1038/nature01461. [DOI] [PubMed] [Google Scholar]
- 100.Meyer K, Kwon Y-C, Liu S, Hagedorn CH, Ray RB, Ray R. 2015. Interferon-α inducible protein 6 impairs EGFR activation by CD81 and inhibits hepatitis C virus infection. Sci Rep 5:9012. 10.1038/srep09012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Qi Y, Li Y, Zhang Y, Zhang L, Wang Z, Zhang X, Gui L, Huang J. 2015. IFI6 Inhibits apoptosis via mitochondrial-dependent pathway in dengue virus 2 infected vascular endothelial cells. PLoS One 10:e0132743. 10.1371/journal.pone.0132743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Richardson RB, Ohlson MB, Eitson JL, Kumar A, McDougal MB, Boys IN, Mar KB, De La Cruz-Rivera PC, Douglas C, Konopka G, Xing C, Schoggins JW. 2018. A CRISPR screen identifies IFI6 as an ER-resident interferon effector that blocks flavivirus replication. Nat Microbiol 3:1214–1223. 10.1038/s41564-018-0244-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Dukhovny A, Lamkiewicz K, Chen Q, Fricke M, Jabrane-Ferrat N, Marz M, Jung JU, Sklan EH. 2019. A CRISPR activation screen identifies genes that protect against Zika virus infection. J Virol 93:e00211-19. 10.1128/JVI.00211-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wang T, Huang C, Lopez-Coral A, Slentz-Kesler KA, Xiao M, Wherry EJ, Kaufman RE. 2012. K12/SECTM1, an interferon-γ regulated molecule, synergizes with CD28 to costimulate human T cell proliferation. J Leukoc Biol 91:449–459. 10.1189/jlb.1011498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bekpen C, Marques-Bonet T, Alkan C, Antonacci F, Bruna ML, Ventura M, Kidd JM, Siswara P, Howard JC, Eichler E. 2009. Death and resurrection of the human IRGM gene. PLoS Genet 5:e1000403. 10.1371/journal.pgen.1000403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Singh SB, Davis AS, Taylor GA, Deretic V. 2006. Human IRGM induces autophagy to eliminate intracellular mycobacteria. Science 313:1438–1441. 10.1126/science.1129577. [DOI] [PubMed] [Google Scholar]
- 107.Srinivasachar Badarinarayan S, Shcherbakova I, Langer S, Koepke L, Preising A, Hotter D, Kirchhoff F, Sparrer KMJ, Schotta G, Sauter D. 2020. HIV-1 infection activates endogenous retroviral promoters regulating antiviral gene expression. Nucleic Acids Res 48:10890–10908. 10.1093/nar/gkaa832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Krapp C, Hotter D, Gawanbacht A, McLaren PJ, Kluge SF, Stürzel CM, Mack K, Reith E, Engelhart S, Ciuffi A, Hornung V, Sauter D, Telenti A, Kirchhoff F. 2016. Guanylate binding protein (GBP) 5 is an interferon-inducible inhibitor of HIV-1 infectivity. Cell Host Microbe 19:504–514. 10.1016/j.chom.2016.02.019. [DOI] [PubMed] [Google Scholar]
- 109.Braun E, Hotter D, Koepke L, Zech F, Groß R, Sparrer KMJ, Müller JA, Pfaller CK, Heusinger E, Wombacher R, Sutter K, Dittmer U, Winkler M, Simmons G, Jakobsen MR, Conzelmann K-K, Pöhlmann S, Münch J, Fackler OT, Kirchhoff F, Sauter D. 2019. Guanylate-binding proteins 2 and 5 exert broad antiviral activity by inhibiting furin-mediated processing of viral envelope proteins. Cell Rep 27:2092–2104.e10. 10.1016/j.celrep.2019.04.063. [DOI] [PubMed] [Google Scholar]
- 110.Sherrill-Mix S, Ocwieja KE, Bushman FD. 2015. Gene activity in primary T cells infected with HIV89.6: intron retention and induction of genomic repeats. Retrovirology 12:79. 10.1186/s12977-015-0205-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kuklina ЕМ, Nekrasova IV, Valieva YV. 2017. Involvement of semaphorin (Sema4D) in T-dependent activation of B cells. Bull Exp Biol Med 163:447–450. 10.1007/s10517-017-3825-8. [DOI] [PubMed] [Google Scholar]
- 112.Eriksson EM, Milush JM, Ho EL, Batista MD, Holditch SJ, Keh CE, Norris PJ, Keating SM, Deeks SG, Hunt PW, Martin JN, Rosenberg MG, Hecht FM, Nixon DF. 2012. Expansion of CD8+ T cells lacking Sema4D/CD100 during HIV-1 infection identifies a subset of T cells with decreased functional capacity. Blood 119:745–755. 10.1182/blood-2010-12-324848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ng KW, Attig J, Bolland W, Young GR, Major J, Wrobel AG, Gamblin S, Wack A, Kassiotis G. 2020. Tissue-specific and interferon-inducible expression of nonfunctional ACE2 through endogenous retroelement co-option. Nat Genet 52:1294–1302. 10.1038/s41588-020-00732-8. [DOI] [PMC free article] [PubMed] [Google Scholar]