ABSTRACT
Salmonella enterica can exist in food animals as multiserovar populations, and different serovars can harbor diverse antimicrobial resistance (AMR) profiles. Conventional Salmonella isolation assesses AMR only in the most abundant members of a multiserovar population, which typically reflects their relative abundance in the initial sample. Therefore, AMR in underlying serovars is an undetected reservoir that can readily be expanded upon antimicrobial use. CRISPR-SeroSeq profiling demonstrated that 60% of cattle fecal samples harbored multiple serovars, including low levels of Salmonella serovar Reading in 11% of samples, which were not found by culture-based Salmonella isolation. An in vitro challenge revealed that Salmonella serovar Reading was tetracycline resistant, while more abundant serovars were susceptible. This study highlights the importance of AMR surveillance in multiserovar populations.
KEYWORDS: Salmonella, tetracycline, CRISPR-SeroSeq, cattle, serovar Reading, antimicrobial resistance
INTRODUCTION
Salmonella enterica is responsible for more than a million human salmonellosis cases each year in the United States, with 212,500 cases attributed to antimicrobial-resistant Salmonella (1–3). Salmonella surveillance and isolation rely on culture methodology that typically concludes with serotyping one or a few colonies per sample (4). These colonies usually reflect those serovars that were most abundant in the original sample or were favored by the chosen culture methods. A previous study reported that in order to detect two serovars that are in equal proportion in a population with a 95% probability, six colonies must be selected per sample (5). Since it is not feasible to routinely pick several colonies, in samples with mixed serovars of unequal relative frequencies (or where some serovars or strains are outcompeted during Salmonella isolation), the serovar(s) present at lower frequencies remains undetected due to effective masking by more abundant serovars. As a result, the antimicrobial resistance (AMR) profiles of these low-abundance serovars also remain unknown. Since Salmonella serovars can exhibit different AMR profiles (6), it is possible that multiserovar populations contribute to a more diverse AMR reservoir. Cattle can harbor multiserovar Salmonella populations (7, 8); however, high-resolution analysis of these populations is almost impossible to discern using conventional Salmonella culture methodology.
Salmonella clustered regularly interspaced short palindromic repeat (CRISPR) spacer content is tractable with serovar identity, and these sequences have been employed effectively for molecular serotyping (9–13). CRISPR-SeroSeq is an amplicon-based sequencing tool that uses Salmonella CRISPR identities to quantify the relative frequency of multiple serovars in a single sample, down to serovars comprising as little as 0.003% of the population (14, 15). In a complementary but targeted approach, quantitative PCR (qPCR) assays can detect a single serovar at low quantities within a mixed Salmonella culture (13).
In a previous study, the effects of injectable ceftiofur crystalline-free acid (CCFA) and in-feed chlortetracycline (CTC) administration on Salmonella in feedlot cattle were explored (16). Culturing of fecal samples identified selection of multidrug-resistant (MDR) (including resistance to tetracycline) Salmonella serovar Reading within 4 days of CTC treatment. Importantly, Salmonella serovar Reading was never detected in fecal samples collected prior to antibiotic administration. We hypothesized that Salmonella serovar Reading was present in the pretreatment samples at lower levels than other serovars and was not detected in the initial study because of the low resolution provided by culture-based methods. Given the profiling capabilities and high-resolution detection of underlying serovars in a sample, we hypothesized that CRISPR-SeroSeq could be used to reveal the presence of Salmonella serovar Reading in pretreated cattle. This hypothesis is further supported by a subsequent study that showed most resistant Salmonella were below the limit of quantification prior to antibiotic treatment (17). Using that study as a framework, we used CRISPR-SeroSeq analysis to retrospectively reveal a high prevalence of multiserovar Salmonella populations in cattle feces, including the presence of Salmonella serovar Reading at low levels in fecal samples collected prior to antibiotic treatment. A subsequent in vitro challenge showed these Salmonella serovar Reading bacteria were resistant to tetracycline, while other more abundant serovars were tetracycline susceptible.
Cattle fecal samples from our previous study were stored at −80°C in glycerol. Since this was a retrospective study, Salmonella was reisolated for this study from the Salmonella-positive fecal samples that were collected on day 0 and day 20 of the original study (16) by preenrichment and subsequent selective enrichment and plating, as previously described. Total genomic DNA was isolated from 1 ml of the enriched broth culture as described previously (14). An additional 5 ml of the enrichment culture was centrifuged at 5,000 rpm for 5 min, and the bacterial pellet was resuspended in tryptic soy broth (TSB) with 20% glycerol and stored at −80°C for the tetracycline challenge experiments. CRISPR-SeroSeq was performed using 2 μl of DNA as the template as described previously (14). Thirty-six samples were multiplexed on a single MiSeq (Illumina Inc., San Diego, CA) run, including one positive (S. enterica serovar Enteritidis) and two negative controls. A CRISPR-SeroSeq Python script that scans sequence reads and then uses BLAST to match sequence reads to a database of more than 130 serovars was used to profile serovars, and the output was written directly to a spreadsheet (14, 15). Serovars were counted if they contained multiple CRISPR spacers unique to that serovar and if the cumulative number of reads for all the spacers in that serovar constituted a relative frequency of at least 0.02% of the population. CRISPR-SeroSeq data were visualized via graphs and Sankey plots built using SankeyMATIC (www.sankeymatic.com).
A 200-μl aliquot of thawed enriched cattle fecal sample was inoculated into 10 ml LB broth and Salmonella were allowed to recover for 5 h at 37°C. Two 4-ml aliquots were removed from the culture, and 16 μg/ml tetracycline (MilliporeSigma, Burlington, MA) was added to one aliquot. Following incubation at 37°C for 19 h, the samples were subcultured 1:100 into fresh medium, maintaining tetracycline selection in the one culture, and incubated for an additional 24 h at 37°C. Genomic DNA was isolated as described above from cultures directly after the 5-h recovery incubation (but before antibiotic addition) and again after 48 h. The tetracycline challenge experiments were performed in biological triplicates on separate days with fresh aliquots of the frozen enrichment culture. Each qPCR was performed in triplicate with 2 μl genomic DNA as a template as described previously (13, 18) on the qTower3 platform and analyzed using qPCRsoft 4.0 software (Analytik Jena, Jena, Germany). The primer and probe sequences are shown in Table 1. The fold change in target DNA between 48-h-treated and untreated samples was calculated as the log2 difference in threshold cycle (CT) values.
TABLE 1.
qPCR primer and probe sequences used in this study
| Serovar | Forward primer (5′–3′) | Reverse primer (5′–3′) | Probe (5′–3′)a | CRISPR locusb |
|---|---|---|---|---|
| Give | GCGGCAGCGGTGGCTAATATA | GCGGGGAACACATGGTCTGAAA | CGGATCATGTCCATGTGCGGTTTATCCCC | CRISPR2, sp 17-18 |
| Mbandaka | ACCGGTACGGAAATTTGTGTCAGA | GGGAACACTATCCTGCGCAATTC | CGAACTGTGGGCACGGTTTATCCCC | CRISPR1, sp 8-9 |
| Montevideo | CCCTGGTTAATGATGGTTGTCAGCTT | CGGGGAACACCACCGGATA | CCGGGTTCTCAGCTGCCACC | CRISPR1, sp 34-35 |
| Reading | GCTAACAGAAACATAGCTGATAGTTGGCG | CGGGGAACACACTGGTCTG | ACGGTCAGTCCTGCAAACGGTTTATCCC | CRISPR1, sp 31-32 |
Probes were labeled with a 6-carboxyfluorescein (FAM) fluorophore and contained an IOWA-Black quencher.
sp, spacers; refers to spacer location within the designated CRISPR array.
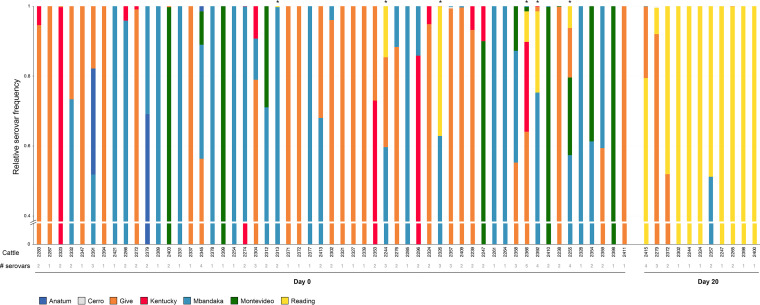
To establish whether cattle fecal samples from the Ohta et al. (16) study harbored multiple Salmonella serovars, we performed CRISPR-SeroSeq on 55 enriched fecal samples collected before CCFA and CTC were administered (day 0) and 11 enriched fecal samples collected after antibiotic treatment was completed (day 20). The samples from this study are listed in Table S1 in the supplemental material and show which cattle were positive for Salmonella (and the serovar) at three different time points in the original study. Day 20 samples were selected as they represented the completion of both antimicrobial treatment regimens (i.e., CCFA and CTC) and because data from Ohta et al. (16) suggested that by the end of the experiment (day 26), the effects of the antimicrobials were waning, and Salmonella populations were reverting back to “normal” (Table S1). Overall, 60% (44/60) fecal samples contained two or more serovars (Fig. 1). We found up to five serovars in a single sample (sample 2366, day 0), though there was no difference in the number of serovars found between day 0 and day 20 samples (two-tailed t test, P = 0.65). There was, however, a drastic difference in the serovar identity between the two sets of fecal samples: on day 0, Salmonella serovars Give and Mbandaka were the most frequently detected serovars, each found in 62% of samples, while in day 20 samples, Salmonella serovar Reading was found in all samples (100%). These data are consistent with earlier work (16) and with serotyping performed in that original study, which was not previously published (Table S1). Importantly, we detected Salmonella serovar Reading in six day 0 fecal samples, with a relative frequency ranging from 0.2% of the population (sample 2313) to 37% (sample 2326) (Fig. 2A). In every instance, other serovars had a greater relative frequency than Salmonella serovar Reading, explaining why it was not detected in our earlier work, which relied on characterizing individual colonies.
FIG 1.
Salmonella populations in feedlot cattle feces are diverse and consist of multiple serovars. CRISPR-SeroSeq identified multiple serovars in 60% and 45% of Salmonella enriched fecal samples from day 0 (55 total samples) and day 20 (11 total samples), respectively. Day 0 samples that contain serovar Reading (yellow) are noted with an asterisk above the bar. Each color represents a different serovar as indicated, and the colors used are the same colors used by Ohta et al. (16). At the bottom of the figure, the gray numbers show the numbers of different serovars per sample. The cattle identification numbers are shown as black numbers; each unique number corresponds to a single steer. All cattle in this study were treated with in-feed CTC.
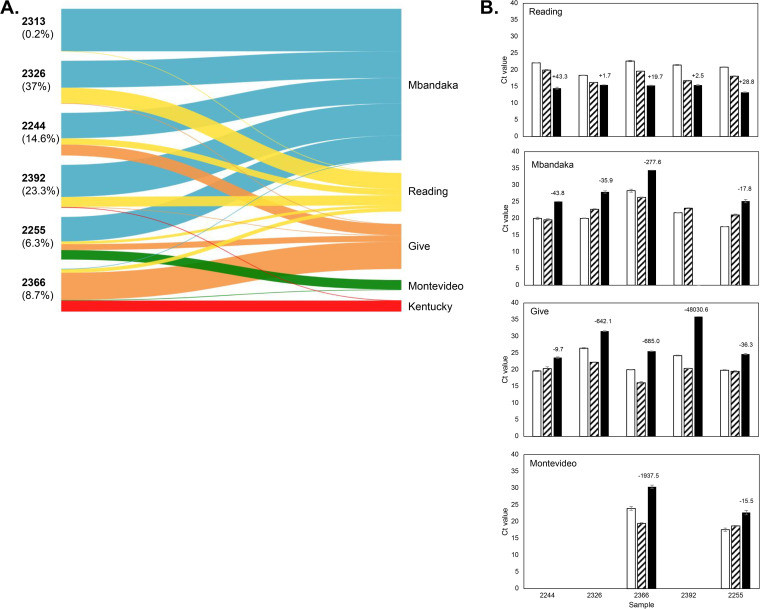
FIG 2.
Salmonella serovar Reading that is present at low levels in untreated cattle is resistant to tetracycline. (A) CRISPR-SeroSeq identified six fecal samples collected before CCFA and CTC treatment that contained low levels of Salmonella serovar Reading. The relative Salmonella serovar frequency is represented by the thickness of the line in the Sankey plots, and each serovar is represented by a different color as labeled on the right and matches the color used by Ohta et al. (16). Each node on the left side of the Sankey plots represents an enriched fecal sample from a single steer; the cattle identification number is shown in bold type, and each number is unique to a single steer. The percentage values represent the percentages of Salmonella serovar Reading found in the samples. (B) Fecal samples enriched from cattle at day 0 were recovered from frozen glycerol stocks by culturing for 5 h in LB broth (white bars) and then cultured in the presence (black bars) or absence (diagonal bars) of 16 μg/ml tetracycline for 48 h. TaqMan-based qPCR assays targeting CRISPR sequences unique to each serovar were used to assess relative changes in serovar abundance. The numbers above the black bars represent the fold differences in DNA between treated and untreated samples at 48 h, calculated as the log2 of the difference in CT values. This experiment is representative of three independent experiments performed on separate days with different aliquots of the frozen culture. Sample 2313 was not included in these experiments as we could not reproducibly detect Salmonella serovar Reading at the 5-h time point. We suspect this is because its relative frequency was very low (0.2%), and the necessitated repeated freezing and thawing of the glycerol cultures damaged the integrity of that sample. For sample 2392, Salmonella serovar Mbandaka was undetectable after 40 cycles when cultured in tetracycline.
There is some discordance between Salmonella serovars that have the greatest relative frequency as defined by CRISPR-SeroSeq (Fig. 1) and the colonies that were serotyped in the first study (Table S1). In previous studies where we have directly compared CRISPR-SeroSeq frequencies to cultured isolates that are serotyped, we find extremely high concordance (14, 15). We expect that the discrepancies found here are due to differences that occurred as a result of reenriching a different aliquot of the same fecal sample, which may have had an unequal distribution of Salmonella. Nonetheless, the serovars found by CRISPR-SeroSeq were concordant with those enriched in a particular pen (Table S1).
The population analysis presented here provides evidence that Salmonella serovar Reading was present in a small number of cattle on day 0. Of the six cattle that were positive for Salmonella serovar Reading at the beginning of the study, only two were positive for Salmonella on day 20 in the earlier study (samples 2244 and 2366; Table S1). Given the intermittent nature of Salmonella shedding in cattle, this is expected. Of these two samples, only fecal sample 2244 yielded Salmonella when the enrichment was repeated for this study; therefore, we have only one paired sample. CRISPR-SeroSeq provides a high resolution of Salmonella serovar frequency, and in this study, we were able to detect serovars contributing to as low as 0.02% of the Salmonella population. Therefore, the possibility that other cattle at day 0 carried Salmonella serovar Reading but that we did not capture it is low. Rather, the transient nature of Salmonella transmission within the dense feedlot pen environments and between different cattle within and between pens likely contributed to identifying Salmonella serovar Reading in multiple different cattle by the end of the study.
To determine whether the Salmonella serovar Reading detected at day 0 was in fact resistant to tetracycline, we treated the enriched day 0 cultures that contained Salmonella serovar Reading with or without tetracycline for 48 h and analyzed changes in Salmonella serovar Reading levels by qPCR. The CT values from samples collected after a 5-h recovery were congruent with the relative serovar frequencies detected using CRISPR-SeroSeq in day 0 fecal samples (Fig. 2B). In all five tetracycline-treated cultures, there was an increase of Salmonella serovar Reading after 48 h in comparison to the untreated samples, with a maximum 43-fold increase in sample 2244. The smallest change was in sample 2326, which was expected as it had a higher initial relative amount of Salmonella serovar Reading. Consistent with an increase in Salmonella serovar Reading, we observed a decrease of Salmonella serovars Give, Mbandaka, and Montevideo (when present), confirming their susceptibility to tetracycline.
Cattle are an important Salmonella reservoir, and using a high-resolution amplicon sequencing approach, we have revealed that nearly two thirds of the Salmonella-positive cattle fecal samples analyzed contained multiple serovars. The population analysis presented here provides evidence that Salmonella serovar Reading was present in a small number of cattle before CTC treatment. The transient nature of Salmonella transmission within feedlots and the selection pressure of antibiotic administration, as observed in the first study (16), likely contributed to identifying Salmonella serovar Reading in multiple cattle by the end of the study, including those where Salmonella serovar Reading was not present at the beginning of the study. The contracted list of Salmonella-positive cattle on day 20 (Fig. 1 and Table S1) reflects not only the antibiotic-driven expansion of resistant Salmonella serovar Reading but also the elimination of susceptible Salmonella. Our data also suggest that in the absence of selective pressure, Salmonella serovar Reading is outcompeted by other serovars, including Salmonella serovars Mbandaka and Give. This is supported by other studies showing that MDR carriage can incur a fitness cost in Salmonella and other members of the Enterobacteriaceae family (19, 20). This is the first study to precisely reveal the composition of multiserovar Salmonella populations in cattle, demonstrating how they can shift in response to antimicrobial treatment, and to use the population information to detect less abundant AMR serovars. With respect to AMR in low-abundance serovars, this study highlights the importance of a high-resolution surveillance platform that can detect multiserovar populations. This study was designed based on the hindsight provided by our earlier work, and we strongly suspected that we would find Salmonella serovar Reading present in the pretreated cattle. Future applications would be performing this blind, where the presence of an AMR phenotype is unknown. Here, we expect that this approach would work in two steps. First, CRISPR-SeroSeq would be used to reveal serovar profiles in a sample. Second, enriched, mixed cultures would be treated with an antibiotic. Using the information from CRISPR-SeroSeq, serovar-specific qPCR assays could then be used to rapidly screen the antibiotic-treated aliquots to determine AMR serovars. This approach would be faster than the current alternative, which involves streaking multiple samples onto agar and then serotyping individual colonies. Additionally, further characterization of isolates would need to be conducted to identify AMR. The two-step approach suggested could be scaled up to treat aliquots of a sample with a panel of different antibiotics and could potentially be used to define the antimicrobial profile of all serovars in a population. Collectively, the work presented here underscores the importance of being able to analyze phenotypes of clinical importance, including AMR, within entire Salmonella serovar populations and provides a powerful framework with which to assess the dynamics of antimicrobial resistance among bacterial populations.
ACKNOWLEDGMENTS
We thank David Medina and Cameron Thompson for their assistance in preparing CRISPR-SeroSeq libraries, Erin Schroeder for helping to design the qPCR assays, Javier Vinasco for curating and preparing samples and isolates from previous work, and Edgar Morales for his help in isolating Salmonella from fecal samples.
We are grateful for funding support from USDA-NIFA (award number 2016-69003-24615 to N.W.S. for development of CRISPR-SeroSeq and qPCR assays), USDA-NIFA-AFRI (formerly CSREES; award numbers 2008-35201-30235 and 2008-35201-04682 to B.N., G.H.L., and H.M.S. for the cattle field trial), and USDA-NIFA-NIFSI (award number 2010-51110-21083 to G.H.L. and H.M.S. for phenotypic analysis). Any recommendations, opinions, findings or conclusions expressed in this publication are those of the publishing authors and do not necessarily represent those of the U.S. Department of Agriculture.
Footnotes
Supplemental material is available online only.
aac.00048-21-s000s1.pdf (98.9KB, pdf)
REFERENCES
- 1.Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson MA, Roy SL, Jones JL, Griffin PM. 2011. Foodborne illness acquired in the United States—major pathogens. Emerg Infect Dis 17:7–15. 10.3201/eid1701.p11101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tack DM, Marder EP, Griffin PM, Cieslak PR, Dunn J, Hurd S, Scallan E, Lathrop S, Muse A, Ryan P, Smith K, Tobin‐D’Angelo M, Vugia DJ, Holt KG, Wolpert BJ, Tauxe R, Geissler AL. 2019. Preliminary incidence and trends of infections with pathogens transmitted commonly through food − Foodborne Diseases Active Surveillance Network, 10 U.S. sites, 2015−2018. MMWR Morb Mortal Wkly Rep 68:369–373. 10.15585/mmwr.mm6816a2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Centers for Disease Control and Prevention. 2019. Antibiotic resistance threats in the United States, 2019. Centers for Disease Control and Prevention, Atlanta, GA. [Google Scholar]
- 4.US Department of Agriculture. 2017. Laboratory Guidebook Notice of Change Title: Isolation and identification of Salmonella from meat, poultry, pasteurized egg, and siluriformes (fish) products and carcass and environmental sponges. US Department of Agriculture, Washington, DC.
- 5.Cason J, Cox N, Buhr R, Bourassa D, Richardson L. 2011. Probability of identitying different Salmonella serotypes in poultry samples, abstr 262P, p 75–76. In International poultry scientific forum. Southern Poultry Science Society, Mississippi State, MS. [Google Scholar]
- 6.Shah DH, Paul NC, Sischo WC, Crespo R, Guard J. 2017. Population dynamics and antimicrobial resistance of the most prevalent poultry-associated Salmonella serotypes. Poult Sci 96:687–702. 10.3382/ps/pew342. [DOI] [PubMed] [Google Scholar]
- 7.Gragg SE, Loneragan GH, Nightingale KK, Brichta-Harhay DM, Ruiz H, Elder JR, Garcia LG, Miller MF, Echeverry A, Ramírez PR, Brashears MM. 2013. Substantial within-animal diversity of Salmonella isolates from lymph nodes, feces, and hides of cattle at slaughter. Appl Environ Microbiol 79:4744–4750. 10.1128/AEM.01020-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vohra P, Bugarel M, Turner F, Loneragan GH, Hope JC, Hopkins J, Stevens MP. 2017. Quantifying the survival of multiple Salmonella enterica serovars in vivo via massively parallel whole-genome sequencing to predict zoonotic risk. Appl Environ Microbiol 84:e02262-17. 10.1128/AEM.02262-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Touchon M, Rocha EPC. 2010. The small, slow and specialized CRISPR and anti-CRISPR of Escherichia and Salmonella. PLoS One 5:e11126. 10.1371/journal.pone.0011126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fabre P-H, Hautier L, Dimitrov D, Douzery EJ. 2012. A glimpse on the pattern of rodent diversification: a phylogenetic approach. BMC Evol Biol 12:88. 10.1186/1471-2148-12-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shariat N, Timme RE, Pettengill JB, Barrangou R, Dudley EG. 2015. Characterization and evolution of Salmonella CRISPR-Cas systems. Microbiology (Reading) 161:374–386. 10.1099/mic.0.000005. [DOI] [PubMed] [Google Scholar]
- 12.Bugarel M, den Bakker H, Grout J, Vignaud M-L, Loneragan GH, Fach P, Brisabois A. 2018. CRISPR-based assay for the molecular identification of highly prevalent Salmonella serotypes. Food Microbiol 71:8–16. 10.1016/j.fm.2017.03.016. [DOI] [PubMed] [Google Scholar]
- 13.Richards AK, Hopkins BA, Shariat NW. 2020. Conserved CRISPR arrays in Salmonella enterica serovar Infantis can serve as qPCR targets to detect Infantis in mixed serovar populations. Lett Appl Microbiol 71:138–145. 10.1111/lam.13296. [DOI] [PubMed] [Google Scholar]
- 14.Thompson CP, Doak AN, Amirani N, Schroeder EA, Wright J, Kariyawasam S, Lamendella R, Shariat NW. 2018. High-resolution identification of multiple Salmonella serovars in a single sample by using CRISPR-SeroSeq. Appl Environ Microbiol 84:e01859-18. 10.1128/AEM.01859-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cox N, Berrang M, House S, Medina D, Cook K, Shariat N. 2019. Population analyses reveal preenrichment method and selective enrichment media affect Salmonella serovars detected on broiler carcasses. J Food Prot 82:1688–1696. 10.4315/0362-028X.JFP-19-166. [DOI] [PubMed] [Google Scholar]
- 16.Ohta N, Norman KN, Norby B, Lawhon SD, Vinasco J, den Bakker H, Loneragan GH, Scott HM. 2017. Population dynamics of enteric Salmonella in response to antimicrobial use in beef feedlot cattle. Sci Rep 7:14310. 10.1038/s41598-017-14751-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ohta N, Norby B, Loneragan GH, Vinasco J, den Bakker HC, Lawhon SD, Norman KN, Scott HM. 2019. Quantitative dynamics of Salmonella and E. coli in feces of feedlot cattle treated with ceftiofur and chlortetracycline. PLoS One 14:e0225697. 10.1371/journal.pone.0225697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tracy LM, Hicks JA, Grogan KB, Nicholds JA, Morningstar-Shaw BR, Shariat NW. 2020. Molecular detection of Salmonella enterica subsp. arizonae by quantitative PCR. Avian Dis 64:305–309. 10.1637/aviandiseases-D-19-00197. [DOI] [PubMed] [Google Scholar]
- 19.Bythwood TN, Soni V, Lyons K, Hurley-Bacon A, Lee MD, Hofacre C, Sanchez S, Maurer JJ. 2019. Antimicrobial resistant Salmonella enterica Typhimurium colonizing chickens: the impact of plasmids, genotype, bacterial communities, and antibiotic administration on resistance. Front Sustain Food Syst 3:20. 10.3389/fsufs.2019.00020. [DOI] [Google Scholar]
- 20.Johnson T, Singer R, Isaacson R, Danzeisen J, Lang K, Kobluk K, Rivet B, Borewicz K, Frye J, Englen M, Anderson J, Davies P. 2015. In vivo transmission of an IncA/C plasmid in Escherichia coli depends on tetracycline concentration, and acquisition of the plasmid results in a variable cost of fitness. Appl Environ Microbiol 81:3561–3570. 10.1128/AEM.04193-14. [DOI] [PMC free article] [PubMed] [Google Scholar]