ABSTRACT
Lipoprotein diacylglyceryl transferase (Lgt) catalyzes the first step in the biogenesis of Gram-negative bacterial lipoproteins which play crucial roles in bacterial growth and pathogenesis. We demonstrate that Lgt depletion in a clinical uropathogenic Escherichia coli strain leads to permeabilization of the outer membrane and increased sensitivity to serum killing and antibiotics. Importantly, we identify G2824 as the first-described Lgt inhibitor that potently inhibits Lgt biochemical activity in vitro and is bactericidal against wild-type Acinetobacter baumannii and E. coli strains. While deletion of a gene encoding a major outer membrane lipoprotein, lpp, leads to rescue of bacterial growth after genetic depletion or pharmacologic inhibition of the downstream type II signal peptidase, LspA, no such rescue of growth is detected after Lgt depletion or treatment with G2824. Inhibition of Lgt does not lead to significant accumulation of peptidoglycan-linked Lpp in the inner membrane. Our data validate Lgt as a novel antibacterial target and suggest that, unlike downstream steps in lipoprotein biosynthesis and transport, inhibition of Lgt may not be sensitive to one of the most common resistance mechanisms that invalidate inhibitors of bacterial lipoprotein biosynthesis and transport.
IMPORTANCE As the emerging threat of multidrug-resistant (MDR) bacteria continues to increase, no new classes of antibiotics have been discovered in the last 50 years. While previous attempts to inhibit the lipoprotein biosynthetic (LspA) or transport (LolCDE) pathways have been made, most efforts have been hindered by the emergence of a common mechanism leading to resistance, namely, the deletion of the gene encoding a major Gram-negative outer membrane lipoprotein lpp. Our unexpected finding that inhibition of Lgt is not susceptible to lpp deletion-mediated resistance uncovers the complexity of bacterial lipoprotein biogenesis and the corresponding enzymes involved in this essential outer membrane biogenesis pathway and potentially points to new antibacterial targets in this pathway.
KEYWORDS: Lgt, Lpp, antibiotic resistance, lipoproteins
INTRODUCTION
The cell envelope of a typical Gram-negative bacterium consists of two membranes, namely, a phospholipid inner membrane (IM) and an asymmetrical outer membrane (OM), the latter of which is composed of a phospholipid inner leaflet and a lipopolysaccharide (LPS) outer leaflet. The IM and OM are separated by the periplasm, which contains a peptidoglycan (PGN) cell wall (reviewed in detail in reference 1). Escherichia coli harbors >90 lipoproteins, many of which are localized to the inner leaflet of the OM but can also be exposed on the bacterial cell surface (2, 3). Bacterial lipoproteins play critical roles in adhesion, nutrient uptake, antibiotic resistance, virulence, invasion, and immune evasion (4), making the lipoprotein biosynthetic and transport pathways attractive targets for novel antibacterial drug discovery.
Lipoprotein biosynthesis in Gram-negative bacteria is mediated by three IM-localized enzymes: Lgt, LspA, and Lnt (Fig. 1). All preprolipoproteins contain a signal peptide followed by a conserved four-amino-acid sequence, [LVI][ASTVI][GAS]C, also known as a lipobox (5). After secretion of preprolipoproteins through the IM via the Sec or Tat pathways, Lgt catalyzes the attachment of a diacylglyceryl moiety from phosphatidylglycerol to the thiol group of the conserved +1-position cysteine via a thioether bond (6). The second enzyme, prolipoprotein signal peptidase (LspA), is an aspartyl endopeptidase which cleaves off the signal peptide N-terminal to the conserved diacylglyceryl-modified +1 cysteine (7) and is the molecular target of the Gram-negative-specific natural-product antibiotics globomycin and myxovirescin (8–11). In Gram-negative and high-GC-content Gram-positive bacteria, a third enzyme, lipoprotein N-acyl transferase (Lnt), catalyzes the addition of a third acyl chain to the amino group of the N-terminal cysteine via an amide linkage. Mature triacylated lipoproteins destined for the OM are extracted from the IM by the LolCDE ATP-binding cassette (ABC) transporter and transported to the OM via the periplasmic chaperone protein LolA and the OM lipoprotein LolB (12, 13) (Fig. 1).
FIG 1.
Lipoprotein biosynthesis and transport in Gram-negative bacteria. Prolipoprotein substrates translocate through the IM via the Sec or Tat pathway and are sequentially modified by Lgt, LspA, and Lnt. Triacylated lipoproteins that are destined for the OM are recognized by the Lol system (LolABCDE) and transported to the OM. Lpp and Pal are two OM lipoproteins that tether the OM to the PGN layer. Pal also binds to TolB, which also can interact with Lpp and OmpA, an OM β-barrel protein that can also associate with PGN (not shown).
Two OM lipoproteins, Lpp (also known as murein lipoprotein or Braun’s lipoprotein) and Pal (peptidoglycan-associated lipoprotein), mediate tethering of the PGN layer to the OM in Escherichia coli. Lpp is a small ∼8-kDa lipoprotein that is the most abundant OM protein in E. coli (∼500,000 molecules per cell), and a third of all Lpp is covalently linked to PGN (2, 14). E. coli mutants deficient in Lpp exhibit increased sensitivity to ethylenediaminetetraacetic acid and SDS, leakage of periplasmic components, increased outer membrane vesicle (OMV) release, and increased sensitivity to complement-mediated lysis (15–17). Mislocalization and accumulation of PGN-linked Lpp in the IM upon inhibition of LspA (11, 18) and LolCDE (19, 20) is believed to lead to bacterial cell death (21–23). In addition to Lpp, Pal binds PGN and interacts with OmpA, Lpp, and the Tol complex and is crucial for maintaining OM integrity in E. coli (24–27). While LspA and LolCDE inhibitors were previously identified (19, 28), no inhibitors of the first committed step in bacterial lipoprotein biosynthesis have been described. Since many natural product antibiotics, including those that inhibit LspA, are cyclic (29, 30), we screened a macrocyclic peptide library to identify Lgt inhibitors. In this study, we identify and characterize G2824 as the first-described inhibitor of Lgt that inhibits growth of wild-type E. coli and Acinetobacter baumannii strains. In comparison to the downstream inhibition of LspA by globomycin, we demonstrate that either Lgt depletion or pharmacologic inhibition of Lgt by G2824 does not lead to significant accumulation of PGN-linked Lpp in the IM and, as such, is not sensitive to rescue of bacterial growth mediated by deletion of lpp.
RESULTS
Modest depletion of Lgt leads to loss of bacterial viability that is insensitive to lpp deletion.
Previous investigations into the role of Lgt in E. coli have focused on laboratory strains, specifically, those lacking the O-antigen of LPS. Here, we engineered the uropathogenic E. coli clinical isolate CFT073 so that the only copy of lgt was under the control of an arabinose-inducible promoter (CFT073 Δlgt) and hence requires arabinose for Lgt expression. As expected, genetic depletion of Lgt was lethal in vitro, and growth was rescued after complementation with lgt from either E. coli, Pseudomonas aeruginosa PA14, or A. baumannii ATCC 17978 (which have 51.6% and 48.6% sequence identity to E. coli lgt, respectively) (Fig. 2A; see Fig. S1A in the supplemental material). thyA, the gene that encodes thymidylate synthase, is downstream of lgt, and its ribosome binding site overlaps the lgt stop codon. We confirmed that thyA expression, which is regulated by transcription from the lgt promoter and translational coupling (31), was unaffected after Lgt depletion (Fig. S1B). Overexpression of the E. coli genes encoding the downstream enzymes in lipoprotein biosynthesis (lspA and lnt) and transport (lolCDE) did not rescue growth of CFT073 Δlgt in spite of detectable levels of LspA, Lnt, and LolCDE (Fig. S1C to G). We next determined how much depletion of Lgt was needed to cause cell death and lead to accumulation of unmodified pro-Lpp (UPLP), a major substrate of Lgt. Incubating cells with various arabinose concentrations led to a dose-dependent depletion of Lgt, UPLP accumulation, and decreased bacterial viability (Fig. 2B and C). While depletion of ∼25% of Lgt was sufficient for loss of cell viability (Fig. 2B and C), CFT073 Δlgt cells expressing as high as ∼90% of normal levels of Lgt were significantly more sensitive to complement-mediated killing of the normally serum-resistant E. coli CFT073 (Fig. 2D) and showed increased incorporation of SYTOX green, a dye that normally does not penetrate an intact OM (Fig. 2E). Depletion of Lgt also resulted in an increase in cell size and an Lpp-dependent IM contraction due to osmotic stress (Fig. 2F and S2), as previously reported (32–34). Consistent with these results, partial depletion of Lgt led to increased sensitivity to antibiotics that are normally excluded by the impermeable Gram-negative OM in spite of normal growth in vitro (Table 1). Depletion of Lgt also resulted in significant attenuation in a mouse E. coli bacteremic infection model (Fig. 2G). Cumulatively, these data suggest that Lgt could be an attractive antibiotic target, since partial inhibition of Lgt may be sufficient to lead to significant attenuation of growth and cellular morphology.
FIG 2.

Lgt is essential for in vitro growth, membrane integrity, serum resistance and virulence. (A) CFT073 Δlgt cells were grown in the presence of 4% arabinose (red) or 0.2% glucose (gray), and CFU were enumerated over 7 h posttreatment. CFT073 Δlgt cultured in the presence of 0.2% glucose was complemented with empty pLMG18 plasmid (green) or pLMG18 plasmids expressing lgt from E. coli (blue), A. baumannii (magenta), or P. aeruginosa (orange). The dashed line represents the limit of detection (200 CFU/ml) of the experiment. (B and C) A modest reduction in Lgt levels results in a significant loss in viability over time with a concurrent accumulation of the unmodified pro-Lpp (ø; UPLP). CFT073 Δlgt cells were treated with a range of arabinose concentrations, and CFU were enumerated over 20 h. CFU growth data are from experiments performed in duplicates. Western blot analysis for expression of Lgt and Lpp was performed using WT CFT073 and CFT073 Δlgt total cell lysates harvested at 3 h post-arabinose treatment. To quantitate Lgt expression levels, Lgt levels were normalized to GroEL and quantitated as fold change relative to that of WT CFT073 (FC versus WT). Lpp forms are denoted as follows: *, triacylated free Lpp; §, PGN-linked diacylglyceryl pro-Lpp (DGPLP); ø, unmodified pro-Lpp (UPLP). Data are representative of two independent experiments. (D) Lgt depletion leads to increased serum sensitivity. WT CFT073 and CFT073 Δlgt cells grown in the presence of a range of arabinose concentrations (2%, magenta; 0.2%, light blue; 0.1%, green, and 0.05%, orange) were incubated with 50% normal human serum (nHS), heat-inactivated human serum (HIHS), or medium (no serum) for 1 h, and CFU were enumerated. Data are representative of at least three independent experiments, each performed in duplicates. (E) Lgt depletion leads to increased OM permeability. WT CFT073 and CFT073 Δlgt cells were incubated with the same range of arabinose concentrations as in panel D and incubated with the nucleic acid dye SYTOX green; flow cytometry was performed to determine the level of dye incorporation. While SYTOX green does not efficiently incorporate in bacterial cells with an intact OM (CFT073 Δlgt treated with 2% arabinose, magenta), SYTOX green incorporation in bacterial cells increases after Lgt depletion. Intact CFT073 (WT; gray) or CFT073 treated with 70% ethanol (WT+ETOH; black), which permeabilizes the cells, were used as controls. Data are representative of two independent experiments. (F) Lgt depletion results in a globular cellular phenotype and membrane blebbing. WT CFT073 or CFT073 Δlgt cells were grown in either arabinose or glucose for 4 h, fixed, and incubated with FM-64 dye (red) and DAPI (blue) to detect OM and nucleic acids, respectively. Cells were visualized by confocal microscopy. Arrows represent membrane blebs. Bars, 1 μm. Quantitation of cell size was performed using ImageJ software. (G) Lgt depletion leads to significant attenuation in virulence. Intravenous infection of neutropenic A/J mice with WT CFT073 (black) or CFT073 Δlgt (red) cells. At 0.5 h and 24 h postinfection, bacterial burden in the liver and spleen was enumerated. Overall P value for the analysis of variance (ANOVA) is <0.0001. Pairwise comparisons were analyzed using unpaired Mann-Whitney test (**, P = 0.0079). The dashed line represents the limit of detection (200 CFU/ml) for this experiment. (H) Deletion of lpp does not rescue growth after Lgt depletion. E. coli MG1655 (WT, black) or inducible deletion strains for lgt (Δlgt), lspA (ΔlspA) and lolCDE (ΔlolCDE) that either contained lpp or had lpp deleted were grown under conditions that allowed normal growth (AraWT, 2% arabinose) or decreased growth (AraLow, 0.0125% arabinose), and CFU were enumerated at 5 h posttreatment. Data are representative of two independent experiments, each performed in duplicates. ns, not significant; *, P < 0.05; **, P < 0.01.
TABLE 1.
Antibiotic sensitivity of WT CFT073 versus CFT073 Δlgt cells expressing wild-type (4% Ara) or low (0.25% Ara) levels of Lgt
| Antibiotic | MIC (μM)a for: |
||
|---|---|---|---|
| WT CFT073 | CFT073 Δlgt |
||
| Lgt4% Ara | Lgt0.25% Ara | ||
| Vancomycin | >100 | >100 | 12.5 |
| Rifamycin | 6.3 | 6.3 | 0.8 |
| Penicillin G | >50 | >50 | 0.8 |
| Oxacillin | >100 | >100 | 12.5 |
| Zeocin | 12.5 | 12.5 | 0.8 |
| Norfloxacin | 0.4 | 0.6 | 0.2 |
MIC values are averages from two independent experiments, each performed in duplicates.
Since bactericidal activity of LspA and LolCDE inhibitors is sensitive to deletion of the gene encoding the major OM lipoprotein, Lpp (18, 19), we wanted to determine if lpp deletion rescued bacterial cell growth after Lgt depletion. To compare growth to lspA and lolCDE inducible deletion strains, we constructed a lgt inducible deletion strain in E. coli MG1655 with and without lpp (MG1655 Δlgt and MG1655 Δlgt Δlpp). Expectedly, lpp deletion rescued the growth of the lspA and lolCDE inducible deletion strains after depletion of LspA and LolCDE, respectively (Fig. 2H). In contrast, the lpp mutant was more sensitive to Lgt depletion, leading to a greater loss of CFU. Since the loss of lpp is a primary mechanism of resistance to inhibitors of LspA and LolCDE, identification of Lgt inhibitors would serve to confirm these genetic data and aid in identifying novel antibacterials.
Identification and characterization of G2824 as a macrocyclic peptide inhibitor of Lgt.
Since many launched drugs are cyclic natural products or derivatives thereof (29), we screened a macrocyclic peptide library to identify specific and high-affinity binders of Lgt. We used a genetically reprogrammed in vitro translation system combined with mRNA affinity selection methods to generate large macrocycle peptide libraries with sizes ranging from 8 to 14 amino acids in length (35–37) (Fig. 3A). The variable sequence (6 to 12 amino acids) of the macrocycle libraries encoded the random incorporation of 11 natural amino acids (Ser, Tyr, Trp, Leu, Pro, His, Arg, Asn, Val, Asp, and Gly) and 5 nonnatural amino acids (Fig. 3B). The library screening is schematically depicted in Fig. 3C. Biotinylated Lgt was solubilized in 0.02% n-dodecyl β-d-maltoside (DDM), immobilized on streptavidin magnetic beads, and incubated with the macrocyclic library. Iterative rounds of affinity selection were performed to identify Lgt-binding macrocycles. After five rounds of enrichment, two additional rounds of off-rate selections were performed by increasing the stringency of the washes before high-affinity binders were eluted. Hit macrocycles were identified using next-generation sequencing (NGS) on the last four rounds of selection followed by a frequency analysis calculation. Two macrocycles, 692 and 693 (see Fig. S3A), were significantly enriched in the final round of selection, accounting for 19.4% and 10.1%, respectively, of peptide sequences as measured by NGS. Macrocycles 692 and 693 each contain 7 amino acids, are related to one another with a charge swap at position 2, and have molecular weights (MWs) of 1428.66 and 1259.55, respectively. The calculated values for LogP (cLogP), which is the logarithm of the compound’s partition coefficient between n-octanol and water and a measure of a molecule’s hydrophilicity, were 1.8 and 1.7 for 692 and 693, respectively. Macrocycle 692 was synthesized with a Gly off the C terminus and renamed G9066 (Fig. 3D). Macrocycle 693 was synthesized with a Gly-Lys-Lys tail off the C terminus to aid in solubility and was renamed G2824 (Fig. 3D).
FIG 3.

G9066 and G2824 macrocyclic peptides inhibit Lgt enzymatic activity. (A) Representation of macrocycle peptide libraries ranging in length from 8 to 14 amino acids. The variable region (Xn) of the macrocycle libraries was encoded to allow the random incorporation of 11 natural amino acids and 5 nonnatural amino acids. (B) The 5 nonnatural amino acids used in the generation of the libraries were N-α-methyl-l-phenylalanine (MeF), N-α-methyl-l-glycine (MeG, sarcosine), (S)-2-aminoheptanoic acid (Ahp), 4-phenyl-l-phenylalanine (Bph), and (S)-1,2,3,4-tetrahydroisoquinoline-3-carboxylic acid (Tic). (C) Schematic representation of affinity-based selections using recombinant biotinylated Lgt immobilized on streptavidin magnetic beads. As discussed in Materials and Methods, Lgt-DDM was incubated with the macrocycle library and Lgt binders were eluted, amplified and translated to generate new libraries enriched for Lgt binders. Iterative rounds of affinity selection and washing were performed against recombinant Lgt and macrocycles that bound to Lgt were identified using next-generation sequencing. (D) Structure of the macrocyclic peptides G9066 and G2824 identified in this study. (E) Schematic of the in vitro Lgt biochemical Lgt reaction and coupled luciferase reaction. Lgt was incubated with phosphatidylglycerol and the Pal-IAAC peptide substrate for 60 min at RT, as described in Materials and Methods. As phosphatidylglycerol substrate used in our biochemical assay contains a racemic glycerol moiety at the end of phosphatidyl group, transfer of diacylglyceryl from phosphatidylglycerol to Pal-IAAC leads to the release of both G1P and G3P. In the coupling reaction, G3P is quantitatively converted to dihydroxyacetone phosphate (DHAP) with concomitant formation of an equivalent amount of NADH by the action of glycerol-3-phosphate dehydrogenase (G3PDH). Newly formed NADH, in turn, quantitatively reacts with proluciferin by the action of proluciferin reductase to generate equivalent amounts of luciferin, which ultimately results in proportional luminescence by luciferase. (F) Mass spectrometric confirmation of diacylglyceryl modification of the Pal-IAAC peptide by Lgt. Spectra depict the base peak chromatograms of the Pal-IAAC peptide ± Lgt. (G) MS1 spectra of the Pal-IAAC peptide in absence (2.59-min peak, top) or presence (5.61-min peak, bottom) of 100 nM Lgt. (H) Lgt reactions in the presence or absence of Lgt were analyzed by SDS-PAGE followed by staining with Coomassie blue. The slower migrating form, corresponding to the diacylglyceryl-modified Pal-IAAC peptide, was detected only in the presence of Lgt. (I) Dose-dependent inhibition of Lgt biochemical activity. Lgt reaction mixtures containing phosphatidylglycerol and Pal-IAAC were incubated in the presence or absence of G9066 (red), G2824 (blue), or a mutant Pal-IAAA peptide (black), which has the conserved cysteine mutated to alanine. Luminescence values were normalized to those of DMSO controls (0% inhibition) and no enzyme controls (100% inhibition). Negative-control reactions for each inhibitor were run in the absence of Lgt (open symbols). Data are representative of at least two independent experiments, each performed in triplicates.
We then tested the ability of G9066 and G2824 to inhibit E. coli Lgt enzymatic activity in vitro by measuring the release of glycerol phosphate, which is a by-product released from the Lgt-catalyzed transfer of diacylglyceryl from phosphatidylglycerol to a peptide substrate via formation of a thioether bond (Fig. 3E). The peptide substrate was derived from the Pal lipoprotein (Pal-IAAC, where C is the conserved cysteine that is modified by Lgt). While glycerol-1-phosphate (G1P) is the expected by-product of the Lgt enzymatic activity (6), the phosphatidylglycerol substrate used in our biochemical assay contains a racemic glycerol moiety at the end of the phosphatidyl group; hence, both G1P and glycerol-3-phosphate (G3P) are released from phosphatidylglycerol as Lgt catalyzes the reaction (Fig. 3E). The detection of G3P is based on a coupled luciferase reaction (described in Materials and Methods and Fig. 3E). Using mass spectrometry, we determined that the addition of 552 Da to the Pal-IAAC peptide, which corresponds to the diacylglyceryl moiety, is Lgt dependent (Fig. 3F and G and S3B and C). Furthermore, SDS-PAGE analysis of the Lgt reaction confirmed that detection of a slower-migrating Pal-IAAC species, corresponding to diacylglyceryl Pal-IAAC, is Lgt dependent (Fig. 3H). G9066 and G2824 potently inhibit Lgt biochemical activity (50% inhibitory concentrations [IC50s] of 0.24 μM and 0.18 μM, respectively) (Fig. 3I). In comparison, a mutant Pal peptide substrate with the conserved cysteine mutated to alanine (Pal-IAAA), which cannot be modified and acts as a Lgt-binding nonreactive, substrate-based competitive inhibitor (Fig. S3D), inhibits Lgt with an IC50 of 4.4 μM (Fig. 3F). Generation of G3P was dependent on Lgt, as reactions performed in the absence of Lgt (Fig. 3F) or using heat-inactivated Lgt (Fig. S3E) did not yield a luciferase signal. Furthermore, G9066 and G2824 did not inhibit steps involved in the coupling reaction (Fig. S3F) or the background signal (Fig. S3G), demonstrating that the inhibitory effects observed with G9066 and G2824 were specifically due to inhibition of the Lgt-catalyzed enzymatic reaction.
G9066 and G2824 were then tested against bacterial cells in MIC growth assays. While G9066 and G2824 only modestly inhibited growth of WT E. coli MG1655 and CFT073 (MIC, ∼100 μM), both molecules showed greater growth inhibition of the lpp-deleted strains (MG1655 Δlpp and CFT073 Δlpp) (Table 2). In contrast, the lpp-deleted MG1655 and CFT073 strains were resistant to the natural product inhibitor of LspA, globomycin, as expected. Both G9066 and G2824 also inhibited growth of wild-type (WT) A. baumannii 19606, with a MIC of 37.5 μM. Using bacterial cells treated with EDTA or expressing the imp4213 allele of lptD, which leads to permeabilization of the OM (38), we demonstrate that G9066 and G2824 inhibit growth of OM-permeabilized E. coli, P. aeruginosa PA14, and A. baumannii 19606 (Table 2). Consistent with data from the WT strains, lpp deletion in the context of the imp4213 or EDTA treatment led to a modest increase in G9066 and G2824 antibacterial activity, unlike that seen with globomycin (Table 2). G2824 showed minimal nonspecific activity against eukaryotic cells and the Gram-positive Staphylococcus aureus strain USA300, consistent with data demonstrating lgt is dispensable for Gram-positive bacterial growth in vitro (39). In contrast, G9066 inhibited growth of USA300 to a greater extent, suggesting G9066 may have additional targets or nonspecific cellular effects. Given G9066 and G2824 are structurally very similar, we decided to focus on G2824 for the remainder of this study.
TABLE 2.
Growth inhibition of a panel of bacterial strains and eukaryotic cells by G9066, G2824, globomycin, and vancomycin
| Bacterium or effect | Strain and treatment | MIC or EC50 (μM)a |
|||
|---|---|---|---|---|---|
| G9066 | G2824 | Globomycin | Vancomycin | ||
| E. coli | MG1655 | >100 | 93.8 | 25 | >100 |
| MG1655 Δlpp | 31.3 | 62.5 | 50 | >100 | |
| MG1655 + EDTA | 3.1 | 3.1 | 0.8 | 3.1 | |
| MG1655 Δlpp + EDTA | 3.1 | 3.1 | 12.5 | 0.8 | |
| CFT073 | 100 | 92 | 29.2 | >100 | |
| CFT073 Δlpp | 31.3 | 62.5 | >100 | >100 | |
| CFT073 + EDTA | 3.1 | 3.1 | 1.1 | 0.8 | |
| CFT073 Δlpp + EDTA | 1.6 | 2.3 | 4.7 | 0.8 | |
| CFT073 imp4213 | 5.7 | 8.4 | 0.5 | 0.6 | |
| CFT073 imp4213 Δlpp | 6.8 | 8.9 | 11.5 | 0.7 | |
| A. baumannii | 19606 | 37.5 | 37.5 | 25 | >100 |
| 19606 + EDTA | 3.1 | 6.3 | 0.6 | 0.2 | |
| P. aeruginosa | PA14 | >100 | >100 | >100 | >100 |
| PA14 imp4213 | 6.3 | 6.3 | 50 | 12.5 | |
| PA14 + EDTA | 6.3 | 6.3 | 50 | 6.3 | |
| S. aureus | USA300 | 6.3 | >100 | >100 | 0.4 |
| Mammalian cytotoxicityb | HepG2 | >100 | >100 | >100 | >100 |
| Hela | >100 | >100 | >100 | >100 | |
| 293T | >100 | >100 | >100 | >100 | |
All E. coli MIC values represent averages from at least four independent experiments, each performed in duplicates. For other bacterial strains, MIC values represent averages from two independent experiments each performed in duplicates.
Mammalian cytotoxicity (EC50) values are representative of three independent replicates.
G2824 targets Lgt function in E. coli, leading to growth inhibition.
While G2824 inhibited both Lgt enzymatic function and bacterial growth, it was unclear whether inhibition of bacterial cell growth was mediated by specific inhibition of Lgt function. While G2824 solubility issues prevented performing agar plate-based resistance selections, multistep resistance selections did not identify on-target resistant mutants to G2824 in CFT073 imp4213. Resistant mutations in lptD (D322V or N274I) were identified, and since N274I was previously demonstrated to be a suppressor of the imp4213 phenotype (38), we believe the mutations identified were nonspecific suppressors leading to increased OM integrity. Therefore, multiple experimental approaches were undertaken to determine if inhibition of bacterial growth was indeed Lgt dependent. As the accumulation of Lpp intermediates detected by Western blotting has been successfully used to confirm inhibition or deletion of enzymes in lipoprotein biosynthesis or transport (20, 21), we asked if Lgt treatment led to the accumulation of UPLP, the pro-Lpp substrate of Lgt. We initially sought to verify the various Lpp forms by leveraging a previously validated protocol using SDS fractionation (17, 40, 41), which separates the SDS-insoluble PGN-associated proteins (PAP) and SDS-soluble non-PGN-associated proteins (non-PAP) (Fig. 4A). Lysozyme was added to allow for the identification of PGN-linked Lpp forms, as previously demonstrated (42). As expected, the fastest migrating form representing the triacylated mature form of Lpp (*) was enriched in the non-PAP fraction, and the PGN-linked Lpp forms (†) were enriched in the PAP fraction (Fig. 4B). We also detected a form corresponding the diacylglyceryl modified pro-Lpp (DGPLP; §) enriched in the PAP fraction, which was previously reported to be PGN linked (42). We then asked if we could detect UPLP in total cell lysates after Lgt depletion in the lgt inducible deletion strain. We confirmed that specific depletion of Lgt led to the accumulation of the UPLP (Fig. 4C, ø), consistent with previous results (43). Depletion of LspA but not Lgt led to the accumulation of DGPLP (§) and other PGN-linked Lpp forms (†) (Fig. 4C). These results now enabled us to determine whether accumulation of UPLP was also detected after treatment with G2824.
FIG 4.

G2824 inhibits Lgt enzymatic activity in bacterial cells. (A) Schematic representing the isolation of PAP and non-PAP fractions. Bacterial cultures were resuspended in 6 ml of PAP extraction buffer and centrifuged at 100,000 × g for 60 min. Six milliliters of supernatants was collected, and pellets were resuspended in 200 μl of PAP extraction buffer. (B) SDS fractionation of WT CFT073 cells to distinguish PGN-associated versus non-PGN-associated forms of Lpp. CFT073 imp4213 cells were treated with SDS to enrich for PAP and non-PAP fractions as discussed in Materials and Methods, and Western blot analysis was performed to detect levels of Lpp. GroEL was used as a control for enrichment of the PAP fraction. While triacylated free Lpp (*) was enriched in the SDS-soluble non-PAP fraction, higher-molecular-weight Lpp species (§ and †) were enriched in the SDS-insoluble PAP fraction (§, DGPLP; †, other PGN-linked Lpp forms). Molecular weight markers (in kilodaltons) are denoted on the left of the blots. (C) Detection of Lpp intermediates in WT MG1655, MG1655 Δlgt and MG1655 ΔlspA inducible deletion strains by Western blotting. WT or inducible deletion strains were treated with 2% arabinose (A) or 0.2% glucose (G), and total cell lysates were harvested at 3 h posttreatment (Ø, unmodified pro-Lpp [UPLP]). PGN-linked DGPLP (§) and other higher-molecular-weight Lpp species (†) accumulated after LspA depletion. (D) G2824 treatment leads to accumulation of UPLP. CFT073 imp4213 cells were treated with 0.5× MICs of G2824 or globomycin for 30 min, and Western blot analysis was performed to measure levels of Lpp forms, which are denoted as described above and in the figure. (E) CRISPRi knockdown of lgt gene expression sensitized cells to G2824 but not globomycin. E. coli BW25113 cells expressing dCas9 and gRNAs specific to lgt or lspA were untreated (black bars) or treated (white bars) with 2 μM G2824, 0.05 μM globomycin or 0.016 μM ciprofloxacin. A scrambled (scr) gRNA and gRNA specific to folA (dihydrofolate reductase) were used as negative controls. Bacterial growth was measured by OD600, and values were normalized to the untreated sample for each gRNA, which was set at 100%. P values were determined comparing to scr gRNA-expressing cells treated with the specific inhibitor (***, P < 0.001). Data are representative of at least two independent experiments, each performed in triplicates. (F) G2824 treatment leads to cell morphology changes and membrane blebs. CFT073 imp4213 cells were left untreated or treated with G2824 for 30 min, fixed, and incubated with FM-64 dye (red) and DAPI solution (blue) to stain membranes and nucleic acid, respectively, and visualized by confocal microscopy. Arrows represent membrane blebs. Bars, 3 μm. (G) Quantitation of cell size after treatment with G2824. A total of 104 ± 4 cells per treatment were quantitated using ImageJ. *, P = 0.04; ***, P = 0.002.
Since G2824 has moderate activity against WT bacterial strains, we performed the mechanistic studies in CFT073 imp4213, as Lgt depletion in CFT073 imp4213 (CFT073 imp4213 Δlgt) leads to a similar extent of Lgt depletion, loss in viable CFU, and accumulation of UPLP as seen in CFT073 Δlgt (compare Fig. S4A and B and Fig. 2B and C), confirming that CFT073 imp4213 cells are not more sensitive to Lgt inhibition. G2824 treatment of CFT073 imp4213 led to an accumulation of UPLP (Fig. 4D, ø), consistent with what was observed with the lgt inducible deletion strains in both the WT background and imp4213 backgrounds. As expected, treatment with globomycin led to an accumulation of DGPLP and other PGN-linked Lpp forms. Levels of triacylated free Lpp were not significantly decreased after either depletion or inhibition of Lgt or LspA (Fig. 4C and D), likely due to the presence of high levels of mature triacylated Lpp expressed in the OM. We engineered CFT073 imp4213 to express Lpp in an arabinose-inducible manner (CFT073 imp4213 lppAra) to demonstrate that G2824, like globomycin, prevented the formation of mature triacylated Lpp. Lpp expression was induced prior to treatment with G2824 or globomycin to minimize the level of preexisting triacylated Lpp. G2824 treatment of CFT073 imp4213 lppAra resulted in accumulation of UPLP as seen in CFT073 imp4213 and, like globomycin, led to decreased mature Lpp (Fig. 4D, right, *). These data demonstrate that G2824 identified in this study leads to the accumulation of Lgt substrate, UPLP, and inhibits the generation of mature triacylated Lpp.
Lgt-specific G2824 activity in bacterial cells was further confirmed using two additional methods. First, we asked whether cells expressing reduced levels of Lgt would be specifically sensitized to G2824 compared to the other inhibitors. To test this hypothesis, we utilized CRISPR interference (CRISPRi) technology to decrease gene expression of the enzymes involved in lipoprotein biosynthesis and transport. BW25113 cells containing plasmids expressing dCas9 and guide RNAs (gRNAs) specific to lgt and lspA were treated with G2824, globomycin, and a control antibiotic, ciprofloxacin, and bacterial growth was measured. gRNA-mediated downregulation of target gene expression (Fig. S4C and D) was consistent with published reports for CRISPRi in bacterial cells (44). Decreased lgt expression specifically sensitized cells to G2824 but not globomycin compared to that of the negative-control gRNAs targeting a scrambled sequence (scr) or the folA gene (Fig. 4E). As expected, decreased expression of lspA specifically led to enhanced growth inhibition by globomycin and not G2824 (Fig. 4E). In contrast, decreased lgt or lspA expression did not lead to increased sensitivity to ciprofloxacin compared to scr or folA gRNAs. Although not specific to targeting lipoprotein biosynthesis, G2824 treatment also led to the expected increase in cell size, as seen with inhibitors of LspA and LolCDE, and OM blebbing (Fig. 4F and G), as seen with the inducible deletion strain (Fig. 4F) and previously demonstrated in a Pal-deficient E. coli strain (45). Cumulatively, our data demonstrate that the novel Lgt-binding macrocycle G2824 interferes with Lgt activity, leading to inhibition of E. coli growth.
Loss of peptidoglycan-linked Lpp enhances the rate of G2824 antibacterial activity.
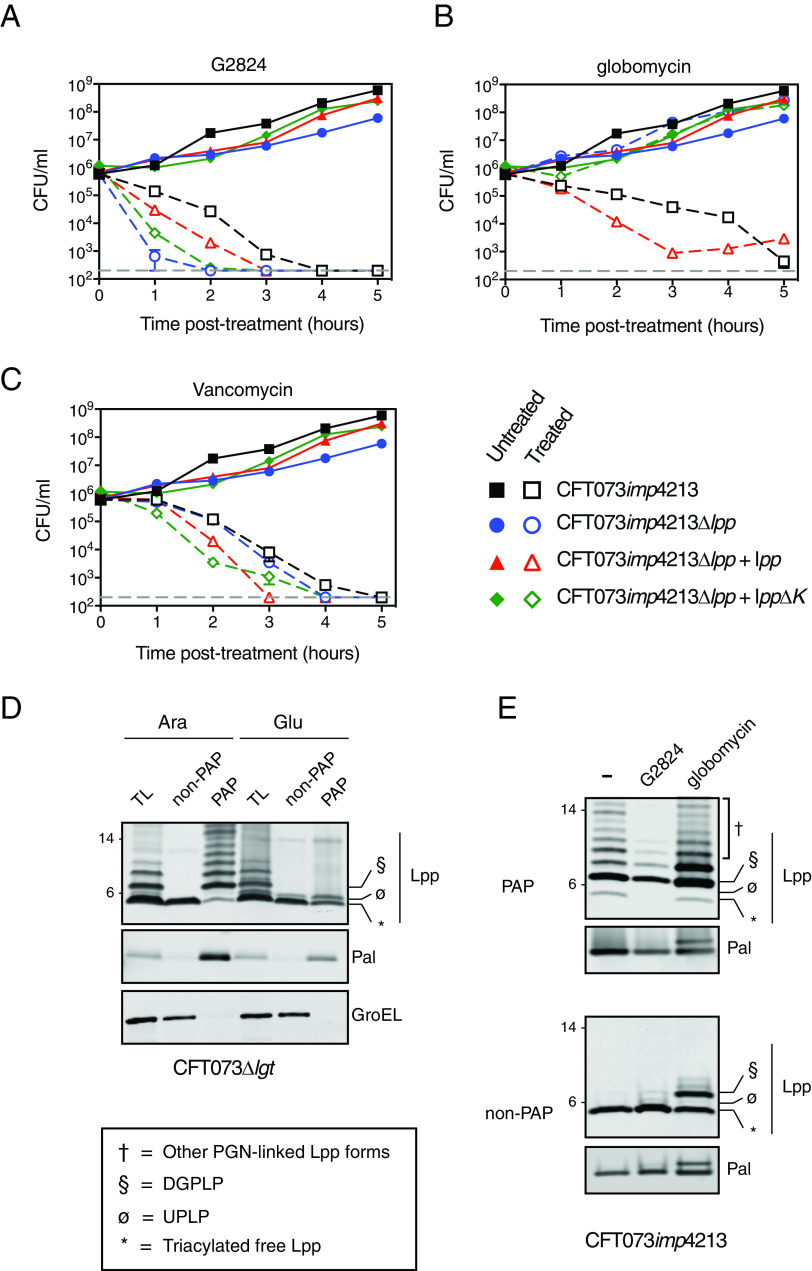
Our data with the inducible deletion strains (Fig. 2H) as well as MIC data (Table 2) suggested that lpp-deleted cells are more sensitive to Lgt depletion or Lgt inhibition, distinguishing it from the mechanism of cell death involved in the later steps of lipoprotein biosynthesis and transport. To compare the rate of bactericidal killing of G2824 with that of globomycin, we treated CFT073 imp4213 and CFT073 imp4213 Δlpp cells with G2824 and globomycin at 2× MIC of the respective inhibitors against CFT073 imp4213 and enumerated viable CFU counts. Consistent with our previous data, CFT073 imp4213 Δlpp cells were not protected from G2824, and, in fact, G2824 led to a more rapid loss of CFT073 imp4213 Δlpp viability than that of CFT073 imp4213 (Fig. 5A). As expected, inhibition of bacterial growth by globomycin was lost in the absence of lpp (Fig. 5B). In contrast, vancomycin showed equivalent killing of CFT073 imp4213 and CFT073 imp4213 Δlpp at 5 h posttreatment (Fig. 5C), confirming the vancomycin MIC data (Table 2) that the lack of lpp does not lead to nonspecific increases in OM permeability. These data confirm that lpp deletion is not a mechanism of resistance to G2824 and that loss of lpp leads to increased bactericidal activity of G2824.
FIG 5.
lpp deletion does not rescue growth after G2824 treatment. CFT073 imp4213 (black), CFT073 imp4213 Δlpp (blue), or CFT073 imp4213 Δlpp complemented with pBAD24 plasmids encoding WT lpp (red) or lppΔK (green) were untreated or treated with 12.5 μM G2824 (A), 3.2 μM globomycin (B), or 1.6 μM vancomycin (C). CFU were enumerated at various times posttreatment. Data are representative of two independent experiments, each performed in triplicates. (D) Lgt depletion leads to loss of PGN-linked Lpp and Pal. CFT073 Δlgt inducible deletion cells were grown in arabinose (Ara) or glucose (Glu), and Lpp and Pal expression was determined in total cell lysates (TL), SDS-insoluble (PAP), and SDS-soluble (non-PAP) fractions 5 h postincubation. GroEL was used as a control for fractionation. (E) G2824 treatment leads to decreased PGN-associated Lpp and Pal. CFT073 imp4213 cells were untreated or treated with G2824 or globomycin for 60 min at 0.5× MIC, and levels of Lpp and Pal were measured in PAP and non-PAP fractions. These data are representative of two independent experiments. Lpp forms are denoted as described in the figure: *, triacylated free Lpp; §, DGPLP; ø, UPLP; †, other PGN-linked Lpp forms.
To determine if loss of PGN linkage of Lpp leads to increased G2824 bactericidal activity, we treated CFT073 imp4213 Δlpp cells complemented with either WT lpp or a mutant form that is unable to covalently link to PGN (lppΔK). Using the previously described SDS fractionation protocol, we confirmed that while WT Lpp localized to both PAP and non-PAP fractions, the LppΔK mutant was primarily detected in the non-PAP fraction (see Fig. S5A). As we noted earlier, PGN-linked DGPLP (§) and other PGN-linked Lpp forms (†) were primarily detectable in the PAP fraction (Fig. S5A). Bacterial viability of cells not expressing lpp or only expressing the mutant lppΔK was lost at a higher rate than cells expressing WT lpp after treatment with G2824 (Fig. 5A). In comparison, cells expressing lppΔK were completely resistant to globomycin, similar to the lpp-deleted cells (Fig. 5B). These data suggest that PGN-linked Lpp is important for OM integrity after treatment with an Lgt inhibitor.
Lgt depletion or inhibition leads to decreased PGN-linked Lpp.
Unlike with the LspA inhibitors globomycin and myxovirescin, lpp deletion does not rescue growth after treatment with G2824, suggesting that the PGN-linkage state and/or localization of Lpp must differ after G2824 treatment. Using SDS fractionation of CFT073 Δlgt inducible deletion cells, we found that Lgt depletion led to significantly lower levels of PGN-linked Lpp, including DGPLP, in the PAP fraction (Fig. 5D). Lgt depletion also led to decreased PGN-associated Pal, likely due to decreased overall levels of triacylated Pal. We next tested if G2824 treatment also led to a similar loss of PGN-associated Lpp. As seen with the lgt inducible deletion strain, treatment with G2824 led to decreased levels of PGN-linked Lpp and Pal (Fig. 5E). Globomycin treatment led to an expected accumulation of PGN-linked DGPLP (§) and other PGN-linked Lpp forms (†). These data suggest that after Lgt inhibition, the accumulated Lgt substrate, UPLP, either is not sufficiently PGN-linked or does not accumulate in the IM to levels needed to cause cell lysis.
Inhibition of Lgt does not lead to significant accumulation of PGN-linked Lpp in the IM.
To determine if Lpp accumulated in the IM after G2824 treatment, we utilized sucrose gradient centrifugation to separate the IM and OM and measured levels of OM lipoproteins (Lpp, Pal, and BamB) and outer membrane proteins (OMPs) (BamA and OmpA) by Western blotting analyses. Separation of IM and OM from MG1655 imp4213 was more efficient than that seen with CFT073 imp4213 (Fig. 6A and S5B), as determined by probing sucrose gradient centrifugation fractions for MsbA, OmpA, and Lpp levels by Western blotting, and was therefore used to test the various inhibitors. In comparison to untreated cells, treatment with globomycin or G2824 led to increases in total protein levels in the IM fraction and a decrease in OM density (Fig. 6). This was further validated by demonstrating IM accumulation of OM lipoproteins (Pal and BamB) and OMPs (BamA and OmpA) after treatment with G2824 or globomycin (Fig. 6B and C). While globomycin treatment also led to a significant IM accumulation of PGN-linked Lpp, including DGPLP, no such accumulation of PGN-linked Lpp was detected after G2824 treatment. We did observe IM accumulation of UPLP after G2824 treatment (Fig. 6B). Since the DGPLP form of Lpp is PGN linked, our data suggest that the lack of significant IM accumulation of PGN-linked Lpp forms, including DGPLP, after G2824 treatment could explain why lpp deletion does not mediate resistance to Lgt inhibitors.
FIG 6.

Inhibition of Lgt leads to depletion of essential OM lipoproteins and OMPs and minimal IM accumulation of PGN-linked DGPLP. MG1655 imp4213 cells were left untreated (A) or treated with G2824 (B) or globomycin (C) for 60 min at 1× MIC and subjected to isopycnic sucrose gradient ultracentrifugation as described in Materials and Methods. Total protein in each fraction was quantitated. Total protein from G2824-treated (red) and globomycin-treated (blue) samples were compared to that in untreated cells (black in panel A; gray in panels B and C). IM and OM fractions were assigned based on the total protein peaks and expression of MsbA and OmpA. These data are representative of at least two independent experiments. *, triacylated free Lpp; §, DGPLP; ø, UPLP; †, other PGN-linked Lpp forms.
DISCUSSION
Lipoprotein biosynthesis is a critical pathway involved in the biogenesis and maintenance of the Gram-negative bacterial OM, and disruption of any step in this pathway leads to loss of cell viability. Lpp maintains the integrity of the Gram-negative bacterial cell surface by covalent interaction between the C-terminal lysine and the meso-diaminopimelic acid residue of the PGN layer (15, 46–49). Published data suggest that lpp deletion leads to rescue of growth after inhibition of LspA and LolCDE (11, 18–20, 23) as well as rescue of the temperature-sensitive Salmonella enterica serovar Typhimurium lgt and lnt mutants (50, 51). While E. coli lnt is essential in the absence of lpp (22), Lpp overexpression in the E. coli lnt mutant leads to bacterial cell growth arrest (21). Following up on data from Pailler et al., who demonstrated that lgt is essential in BW25113, a derivative of E. coli K-12 strain BD792, and that Lgt depletion leads to increased DNA leakage from the cell pole (43), we demonstrate that Lgt depletion in the clinical E. coli strain CFT073 leads to significant perturbations to the bacterial cell envelope, leading to increased sensitivity to antibiotics, increased serum killing, and attenuated virulence in vivo.
Based on these published reports, we had expected that deletion of lpp would also lead to rescue of growth after Lgt depletion or pharmacologic inhibition of the enzyme. However, our data demonstrate that pharmacologic inhibition of Lgt by G2824 or Lgt depletion leads to cell death that is not rescued by lpp deletion in two E. coli strain backgrounds, CFT073 and MG1655. Furthermore, neither genetic depletion nor enzymatic inhibition of Lgt leads to significant accumulation of PGN-linked Lpp forms. While a previous report demonstrated that UPLP and DGPLP are both linked to a single muropeptide unit (42), our data suggest that the level of complete PGN linkage is likely less efficient in the absence of diacylglyceryl modification of pro-Lpp. While our findings suggest the lack of diacylglyceryl modification by Lgt may generate a less optimal substrate for the l,d-transpeptidases that covalently link Lpp to PGN, they do not rule out the possibility that PGN linkage occurs at or after modification by Lgt. Our data are in contrast to those generated by Zhang et al. (49), who immunoprecipitated [35S]methionine-labeled Lpp from JA221 E. coli cells to demonstrate that lipid modification of Lpp is not essential for linkage to PGN. While the explanation for this is unclear, this could be due, in part, to differences in E. coli strains used in both studies, as the same mutation generated in JA221 and MG1655, both K-12-derived E. coli strains, can lead to strikingly different sensitivities (∼4,000-fold) to bacterial cell stress (52). Both G2824 and globomycin treatment lead to mislocalization of the β-barrel protein, OmpA (Fig. 6B and C), which is likely due to decreased OM localization of BamB and other Bam lipoproteins that are crucial for optimal OM localization of BamA in E. coli. Since most of the studies with G2824 were performed in cells expressing the lptD imp4213 mutant allele, identifying more-potent Lgt inhibitors will be needed to determine the effects of Lgt inhibition in WT bacterial cells. However, given that Lgt depletions in CFT073 and CFT073 imp4213 show similar kinetics of cell viability loss, Lgt depletion, and accumulation of pro-Lpp (Fig. 2; see Fig. S4 in the supplemental material), we believe our results in the imp4213 background should translate to WT E. coli. It is conceivable that Lpp normally protects cells against inhibitors of lipoprotein biosynthesis and transport by maintaining optimal OM structure and integrity but that this is masked in the case of LspA and LolCDE inhibitors, as they lead to the accumulation of PGN-linked Lpp, which itself induces cell death.
Using a combination of biochemical and genetic strategies, we believe the Lgt inhibitor G2824 identified in this study inhibits bacterial cell growth through inhibition of the diacylglyceryl transferase activity of Lgt. First, G2824 was identified using a specific Lgt binding assay and confirmed to inhibit Lgt enzymatic function in vitro. Second, the results generated from pharmacologic inhibition of Lgt by G2824 in both CFT073 and MG1655 E. coli strains are highly consistent with the genetic depletion of Lgt, strongly arguing against off-target effects as a main cause of G2824 bactericidal activity. Third, our sucrose gradient centrifugation data clearly demonstrate that G2824, like globomycin, leads to IM accumulation of OM lipoproteins (Pal and BamB) and OMPs that are dependent on lipoproteins for their localization (BamA). While the sucrose gradients clearly separate IM and OM in untreated cells, some overlap of the IM fractions is detected for G2824 and globomycin-treated cells. This is not unexpected and is detected in most published reports when using sucrose gradient centrifugation to examine membrane localization of Lpp in WT E. coli membranes (22, 23, 53), suggesting this is unlikely due to the use of lptD imp4213-expressing cells. Interestingly, separation of IM and OM in globomycin-treated cells was more challenging, likely explained by the significant accumulation of membranes as detected by electron microscopy after inhibition of LspA (54). While we were unable to raise on-target resistant mutants to G2824, we cannot fully rule out that G2824 does not have additional targets in bacterial cells. However, one could speculate that if G2824 binds to the conserved phosphatidylglycerol binding site in Lgt, mutations disrupting G2824 binding might be incompatible with bacterial viability. In fact, no on-target resistance mutations have ever been identified against the LspA inhibitor globomycin (55) or its improved analog, G0790 (54), which is believed to act as a substrate mimic and bind the highly conserved LspA active site (56). As recent publications have revealed significant insights into the potential mechanisms of diacylglyceryl modification by Lgt (57, 58), further studies aimed at determining if G2824 competitively inhibits binding of the phosphatidylglycerol or prolipoprotein substrates would be needed to better understand the mechanism by which G2824 interferes with this critical OM biogenesis pathway. Although we do not detect MIC shifts with G2824 in cells overexpressing lgt (data not shown), drug resistance in E. coli after target overexpression is known to vary (increase, remain unchanged, or decrease) depending on the balance between bacterial fitness costs and inhibition of enzymatic activity (59). One could speculate that even a modest inhibition of Lgt could lead to significant defects in OM integrity and cellular fitness, which may counteract any resistance arising from lgt overexpression.
In summary, our study is the first to systematically differentiate the role of Lpp in targeting multiple steps of bacterial lipoprotein biosynthesis. The lack of significant PGN association of Lpp, resulting from Lgt depletion or pharmacologic inhibition of the enzyme, eliminates a major mechanism of resistance mediated by deletion of lpp and, as such, could inform future antibacterial strategies to inhibit this essential OM biogenesis pathway.
MATERIALS AND METHODS
Ethics statement.
All mice used in this study were housed and maintained at Genentech in accordance with American Association of Laboratory Animal Care guidelines. All experimental studies were conducted under protocol 13-0979A and approved by the Institutional Animal Care and Use Committee of Genentech Lab Animal Research and performed in an Association for Assessment and Accreditation of Laboratory Animal Care International (AAALAC)-accredited facility in accordance with the Guide for the Care and Use of Laboratory Animals and applicable laws and regulations.
Antibodies.
The anti-Pal antibody was a generous gift from Shaw Warren (Massachusetts General Hospital). The anti-OmpA (Antibody Research Corporation), anti-GroEL (Enzo Life Sciences), anti-ThyA (GeneTex, Inc.) and anti-His (Cell Signaling Technology) antibodies were obtained from commercial sources. Generation of anti-Lpp and anti-BamA antibodies was previously described (17, 60, 61). Recombinant Lgt was used to generate rabbit polyclonal antibodies. Rabbit immunizations, generation of antisera, and purification of rabbit polyclonal antibodies were performed as previously described for Lpp (17).
Generation of bacterial strains and plasmids.
Bacterial strains and plasmids used in this study are listed in Table S1 in the supplemental material. E. coli strain CFT073 (ATCC 700928) (62) and MG1655 (ATCC 700926) were purchased from ATCC. Gene disruption in CFT073 was performed as previously described (17, 63). CFT073 Δlgt and MG1655 ΔlspA were generated using λ Red recombination (17, 63). The primers used to generate the CFT073 and MG1655 mutants are listed in Table S2. Plasmids pKD46 for the λ Red recombinase (17, 63), pKD4 or pSim18 for the integration construction (17, 63), and pCP20 (64) for the FLP recombinase were used in this study. The 3′ primers (Table S2) used to generate the integration construct from pKD4 contain a juxtaposed ribosome binding site downstream of the kanamycin (Kan) cassette to allow for the creation of nonpolar gene deletions within operons (63), which is critical when generating the lgt inducible deletion strain given that the ribosomal binding site of the thyA gene downstream of lgt is within the lgt coding region. The inducible deletion strains (MG1655 Δlgt and MG1655 ΔlolCDE) in either the WT and/or Δlpp backgrounds were generated using similar methods as previously described (17, 65). To delete lpp in the lgt, lspA, and lolCDE inducible deletion strains, the Kan cassette was removed by transforming the FLP recombinase-expressing pCP20 plasmid followed by deletion of the lpp gene using λ Red recombinase as discussed above. Similar protocols were used to introduce the imp4213 mutant allele. The PA14 imp4213 strain was generated based on published protocols (66–68). Briefly, the lptD imp4213 mutant allele was generated by PCR amplifying PA14 genomic DNA using primer pairs PA14imp4213_up-F/PA14imp4213_up-R (upstream region) and PA14imp4213_dn-F/PA14imp4213_dn-R (downstream region) to create lptD missing amino acids 425 to 447. PCR products were combined with the PCR-amplified vector pEX18gm with primers pLMG18_GA-F/pLMG18_GA-R and assembled using Gibson assembly (NEB). The resultant suicide vector pEX18gm was confirmed by sequence analysis (ELIM Biopharm). Single recombination mutants were selected on LB containing 10 μg/ml gentamicin and 25 μg/ml irgasan. Double recombination mutants were selected on LB without NaCl plates containing 10% sucrose and confirmed by PCR and sequence analysis. For expression under the isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible promoter, DNA encoding the full-length sequences of lgt, lspA, lnt, and lolCDE were cloned into pLMG18 and induced using 2.5 mM IPTG.
In vitro growth inhibition and serum sensitivity assays.
Unless stated otherwise, E. coli cells were grown in Luria-Bertani (LB) medium (0.5% yeast extract, 1% tryptone, 0.5% NaCl) at 37°C. Where indicated, kanamycin (Kan) was added to culture media at a 50-μg/ml final concentration. MIC assays were performed based on Clinical and Laboratory Standards Institute (CLSI) guidelines. For in vitro growth curves, overnight cultures of WT CFT073, CFT073 Δlgt, and CFT073 Δlgt complemented with lgt from E. coli (lgtEc) or P. aeruginosa (lgtPa) were grown to mid-exponential phase (optical density at 600 nm [OD600] of 0.6) and then diluted to an OD600 of 0.1 to initiate growth curves. At various times, culture aliquots were diluted and plated in dilutions on LB+Kan agar, and CFU were enumerated in duplicates. Growth of MG1655 inducible deletion strains was measured by culturing in the presence of 2-fold dilutions of arabinose (starting arabinose concentrations for CFT073 and MG1655 inducible deletion strains were 4% and 0.8%, respectively). While 2% arabinose was sufficient for WT growth of MG1655 Δlgt, 4% arabinose was used for CFT073 Δlgt based on comparing its growth to that of WT CFT073 as measured by CFU. OD600 growth measurements were performed using an EnVision 2101 multilabel reader plate reader (PerkinElmer) linked with Echo liquid handler (Labcyte). For time-kill experiments, bacteria were harvested in mid-exponential phase and treated with 12.5 μM G2824, 3.2 μM globomycin, and 1.6 μM vancomycin. CFU were enumerated at various times posttreatment. Bacterial culture medium containing 2% arabinose was used to induce lpp or lppΔK expression from pBad24 plasmids. Bacterial viability at different time points during the treatment was measured by enumerating CFU. Serum killing assays were carried out as previously described (17).
Detection of membrane permeability using SYTOX green incorporation.
To determine the effect of Lgt depletion on membrane permeability, WT CFT073 and CFT073 Δlgt strains were streaked onto LB agar plates containing 4% arabinose and cultured at 37°C for 18 h. From a single colony, bacteria were cultured in LB broth containing 4% arabinose and cultured at 37°C to an OD of 0.5. One-milliliter cultures at an OD of 0.5 for both strains were harvested, washed, and resuspended in LB broth or medium containing a range of arabinose (2, 0.2, 0.1, 0.05, and 0%) or glucose (0.2%) concentrations and incubated at 37°C for 2 h. Cells were harvested by centrifugation at 4,000 × g at 4°C for 5 min. Intact CFT073 or CFT073 treated with 70% ethanol at room temperature (RT) for 15 min to permeabilize the cells were used as controls. Cells were incubated with SYTOX green according to the manufacturer’s recommendations, washed with phosphate-buffered saline (PBS; 3 times) and fixed in 2% paraformaldehyde. SYTOX green incorporation was measured by flow cytometry using a FACSAria II (Becton Dickenson) and analyzed using FlowJo software.
Mouse infections.
Overnight bacterial cultures were back diluted 1:100 in M9 medium and grown to an OD600 of 0.8 to 1 at 37°C. Cells were harvested, washed once with PBS, and resuspended in PBS containing 10% glycerol. Cells were frozen in aliquots, and thawed aliquots were measured for CFU prior to mouse infections. Virulence of WT CFT073 and CFT073 Δlgt was measured using the neutropenic E. coli infection model (69). Seven-week-old female A/J mice (Jackson Laboratory) were rendered neutropenic by peritoneal injections of 2 doses of cyclophosphamide (150 mg/kg body weight on day −4 and 100 mg/kg body weight on day −1). On day 0, mice were infected with 5 × 105 CFU of mid-exponential-phase bacteria diluted in PBS by intravenous injection through the tail vein. At 30 min and 24 h postinfection, bacterial burdens in the liver and spleen were determined by serial dilutions of tissue homogenates on LB plates.
Expression and purification of recombinant Lgt.
DNA encoding full-length E. coli Lgt fused to a C-terminal Flag tag was transformed into Rosetta 2(DE3) Gold cells (Agilent). Starter cultures were grown in Terrific broth (TB) medium with carbenicillin (50 μg/ml) and chloramphenicol (12.5 μg/ml) at 37°C for 3 h. The starter cultures were diluted 1:50 in TB medium with carbenicillin (50 μg/ml), chloramphenicol (12.5 μg/ml), and glycerol (1%) and grown at 37°C for 2 h with shaking at 200 rpm. The temperature of the culture was reduced to 30°C, and they were grown for an additional 2 h before the temperature of the culture was reduced to 16°C for growth for 64 h. The cells were harvested by centrifugation, resuspended in lysis buffer (20 mM Tris [pH 8.0], 300 mM NaCl, protease inhibitor cocktail, and Lysonase), and stirred at 4°C for 30 min before being passed through a microfluidizer 3 times. The membrane fraction was solubilized by adding DDM directly to the lysate to a final concentration of 1% and stirring at 4°C for 2 h before centrifugation at 40,000 rpm for 1 h. Preequilibrated Flag resin was added to the supernatant and incubated with rotation at 4°C for 2 h. The slurry was added to a gravity column, and the column was washed with 10 culture volumes (CV) buffer A (20 mM Tris [pH 8.0], 300 mM NaCl, 5% glycerol, 1% DDM) and 10 CV buffer B (20 mM Tris [pH 8.0], 300 mM NaCl, 5% glycerol, 0.05% DDM). The bound fraction was eluted by the addition of 5 CV of buffer C (20 mM Tris [pH 8.0], 300 mM NaCl, 5% glycerol, 0.05% DDM, 100 μg/ml Flag peptide). The peak fractions were collected, concentrated to less than 5 ml, and loaded onto a Superdex 200 16/60 column equilibrated with buffer C [20 mM Tris (pH 8.0), 300 mM NaCl, 5% glycerol, 0.05% DDM, 1 mM Tris(2-carboxyethyl)phosphine hydrochloride (TCEP)]. The peak fractions were collected, analyzed by SDS-PAGE, and stored at −80°C.
Macrocyclic peptide library design and selection of Lgt-binding molecules.
A thioether-macrocyclic peptide library was constructed by using N-chloroacetyl d-phenylalanine (ClAc-f) as an initiator in a genetically reprogrammed in vitro translation system (36). The genetic code was designed with the addition of two N-methyl amino acids, N-methyl-l-phenylalanine (MeF) and N-methyl-l-glycine (MeG), and three unnatural amino acids, (S)-2-aminoheptanoic acid (Ahp), (S)-3-([1,1′-biphenyl]-4-yl)-2-aminopropanoic acid (Bph), and (S)-1,2,3,4-tetrahydroisoquinoline-3-carboxylic acid (Tic) in addition to 11 natural amino acids (Ser, Tyr, Trp, Leu, Pro, His, Arg, Asn, Val, Asp, and Gly). After in vitro translation, a thioether bond formed spontaneously between the N-terminal ClAc group of the initiator d-phenylalanine residue and the sulfhydryl group of a downstream cysteine residue to generate the macrocyclic peptides.
Lgt was biotinylated by the addition of 400 μl of a freshly prepared solution of 20 mM N-hydroxysuccinimide (NHS)-polyethylene glycol 4 (PEG 4)-biotin (Thermo) to 1.375 ml of 10 mg/ml Lgt, as per the manufacturer’s instructions. The reaction mixture was incubated for 30 min at RT, and free NHS-PEG 4-biotin was removed by gel filtration chromatography on a Superdex 200 16/60 column that had been preequilibrated with 20 mM Tris (pH 8.0), 250 mM NaCl, 5% glycerol, and 0.02% DDM. The final biotinylated sample was concentrated to 1 mg/ml. Affinity selection of macrocyclic peptides binding to Lgt was performed using E. coli Lgt-biotin in 0.02% n-dodecyl β-d-maltoside (DDM). Briefly, 10 μM mRNA library was hybridized with a peptide linker (11 μM) at RT for 3 min. The mRNA library was translated at 37°C for 30 min in the reprogrammed in vitro translation system to generate the peptide-mRNA fusion library (35, 37). Each reaction mixture contained 2 μM mRNA-peptide-linker conjugate, 12.5 μM initiator tRNA (tRNAfMet aminoacylated with ClAc-d-Phe), and 25 μM each elongator tRNA aminoacylated with the specified noncanonical/canonical amino acids. In the first round of selections, translation was performed at a 20-μl scale. After the translation, the reaction was quenched with 17 mM EDTA. The product was subsequently reverse transcribed using RNase H minus reverse transcriptase (Promega) at 42°C for 30 min, and buffer was exchanged for DDM buffer: 50 mM Tris (pH 8), 5 mM EDTA, 200 mM NaCl, 0.02% DDM, and 1 mM glutathione. For affinity selection, the peptide-mRNA/cDNA solution was incubated with 250 nM biotinylated E. coli Lgt for 60 min at 4°C, and the streptavidin-coated beads (Dynabeads M-280 streptavidin; Thermo) were further added and incubated for 10 min to isolate Lgt binders. The beads were washed once with cold DDM buffer, the cDNA was eluted from the beads by heating for 5 min at 95°C, and fractional recovery from the affinity selection step was assessed by quantitative PCR using Sybr green I on a LightCycler thermal cycler (Roche). After five rounds of affinity maturation, two additional rounds of off-rate selections were performed by increasing the wash stringency before elution to identify high-affinity binders. Sequencing of the final enriched cDNA was carried out using a MiSeq next-generation sequencer (Illumina).
Peptide synthesis.
Thioether macrocyclic peptides were synthesized using standard 9-fluorenylmethoxy carbonyl (Fmoc) solid-phase peptide synthesis (SPPS). Following coupling of all amino acids, the deprotected N terminus was chloroacetylated on resin followed by global deprotection using a trifluoroacetic acid (TFA) deprotection cocktail. The peptides were then precipitated from the deprotection solution by adding >10-fold excess diethyl ether. Crude peptide pellets were then dissolved and repelleted 3 times using diethyl ether. After the final wash, the pellet was left to dry, and then the pellet was resuspended in dimethyl sulfoxide (DMSO) followed by the addition of triethylamine for intramolecular cyclization via formation of a thioether bond between the thiol of the cysteine and N-terminal chloroacetyl group. Upon completion of cyclization, the reaction was quenched with acetic acid (AcOH), and the cyclic peptide was purified using standard reverse-phase high-pressure liquid chromatography (HPLC) methods. For the Lgt biochemical assay, the Pal-IAAC and Pal-IAAA peptides were synthesized by CPC Scientific.
SDS-PAGE and Western immunoblotting.
Bacterial cell samples normalized for equivalent OD600 and resuspended in BugBuster lysis buffer (Fischer Scientific) with the addition of sample buffer (LI-COR), and proteins were separated by SDS-PAGE using 16% Tricine protein gels or NuPAGE 4% to 20% bis-Tris gels (Thermo Fisher Scientific), transferred to nitrocellulose membranes using the iBlot 2 dry blotting system (Invitrogen), and blocked using LI-COR blocking buffer for 30 min. Unless stated otherwise, loading buffer with reducing agents was added and samples were not boiled prior to SDS-PAGE. For the sucrose gradient centrifugation, samples were boiled prior to running the SDS-PAGE. Primary antibodies were used at a final concentration of 1 μg/ml with some exceptions: rabbit anti-Lpp polyclonal antibody (0.1 μg/ml), murine anti-Pal 6D7 antibody (0.5 μg/ml), rabbit anti-GroEL antibody (1:10,000 final dilution), and rabbit anti-OmpA antibody (1:50,000 final dilution). The secondary antibodies were all obtained from LI-COR and used as per manufacturer’s instructions. Images were collected using the Odyssey CLx imaging system (LI-COR) and analyzed by Image Studio Lite.
Development of the Lgt biochemical assay.
The Lgt enzymatic activity was measured by specific detection of G3P. Both G3P and G1P are released from phosphatidylglycerol as Lgt catalyzes the transfer of diacylglyceryl from phosphatidylglycerol to the preprolipoprotein substrate, since the PGN substrate used in the assay contains a racemic glycerol moiety at the end of phosphatidyl group. The standard assay consists of a 6-μl reaction mixture with 3 nM Lgt-DDM, 50 μM phosphatidylglycerol (1,2-dipalmitoyl-sn-glycero-3-phospho-[1′-rac-glycerol]; Avanti), and 12.5 μM Pal-IAAC peptide substrate derived from the Pal lipoprotein (MQLNKVLKGLMIALPVMAIAACSSNKN) in 50 mM Tris (pH 8), 200 mM NaCl, 5 mM EDTA, 0.02% DDM, 0.05% bovine skin gelatin, and 1 mM glutathione. As a control, we used a mutant nonmodifiable Pal substrate peptide containing a cysteine-to-alanine mutation (Pal-IAAA), which served as a competitive nonmodifiable inhibitor. The reaction was quenched after 60 min at RT with 0.5 μl of 4.8% lauryl dimethylamine-N-oxide (Anatrace), followed by addition of 6 μl detection solution. After incubation for 120 min at RT, the luminescence signal was read. The detection solution was modified based on an NAD Glo protocol (Promega, G9072), per the manufacturer’s instructions. Specifically, 10 ml detection solution consists of 3-fold dilution of luciferin detection reagents, supplemented with 10 μl reductase, 2.5 μl reductase substrate, 1 mM NAD, and 4.25 U of glycerol-3-phosphate dehydrogenase (G3PDH) (Roche Diagnostics, 10127779001). The luciferin detection reagents, reductase, and reductase substrate were all from the NAD Glo kit (Promega). Luminescence values were normalized to those of DMSO controls (0% inhibition) and no enzyme controls (100% inhibition). IC50 values were calculated using a 4-parameter logistic model using GraphPad Prism software.
Mass spectrometry.
Ammonium acetate, acetic acid, formic acid, and trifluoroacetic acid were purchased from Sigma-Aldrich (St. Louis, MO). Acetonitrile (ACN) was purchased from Fisher Scientific (Hampton, NH).
For liquid chromatography UV mass spectrometry (LC-UV-MS) analysis and top down sequencing, 1 μl corresponding to 12.5 μM Pal peptide, MQLNKVLKGLMIALPVMAIAACSSNKN (2859.6 Da), was injected onto a Q Exactive UHMR Orbitrap (70) coupled to a Vanquish liquid chromatograph (Thermo Fisher Scientific) through a 2.1 mm by 50 mm MAbPac reverse-phase (RP) HPLC column (80°C). Ions of interest, representing the most abundant charge state of the peptide with or without Lgt treatment, m/z 953.86 (3+) and 1,137.69 (3+), respectively, were isolated in the quadrupole and selected for fragmentation in the high-energy collisional dissociation (HCD) cell at a gas pressure of 3 × 10−5 to 7 × 10−5 Pa, with UHMR mode “on.” The MS parameters used were spray voltage of 1.2 to 1.3 V, source temperature of 300°C, collision energy of 60 V, source dissociation energy of 5 V, and resolution (at m/z 200) of 35,000. The instrument was mass calibrated as described previously using a solution of CsI (71).
Theoretical fragment ions were generated using MS-Product in Protein Prospector (https://prospector.ucsf.edu) and annotated manually to determine the exact mass addition on Cys. Spectra were subsequently analyzed and fragmentation map generated in ProSightLite (http://prosightlite.northwestern.edu) with the assigned mass addition to Cys for the treated peptide. Fragment ions were matched to within 10 ppm of theoretical b-type and y-type ions.
Transmission electron microscopy, time-lapse microcopy, and confocal microcopy.
Electron microscopy was performed as previously described (65). For time-lapse microscopy, WT CFT073, CFT073 Δlgt, and CFT073 Δlgt Δlpp cells were grown overnight in LB medium containing 4% arabinose, back diluted to a final OD600 of 0.1, and immediately placed between a coverslip and 1% agarose pad containing 0.2% glucose for imaging. Cells were maintained at 37°C during imaging in a stage-top chamber (Okolab Inc.). Cells were imaged on a Nikon Eclipse Ti inverted confocal microscope (Nikon Instruments Inc.) coupled with an UltraVIEW VoX (PerkinElmer Inc.) and a 100× (numerical aperture [NA], 1.40) oil immersion lens objective. Images were captured at various times using an ORCA-Flash 4.0 complementary metal oxide semiconductor (CMOS) camera (Hamamatsu Photonics), collected using Volocity software (Quorum Technologies), and processed using Fiji (72). For confocal microscopy, images were acquired on a Leica SP8 STED 3× platform using a 100× white-light, NA 1.4 oil immersion lens objective. Cells were treated with arabinose or glucose (CFT073 Δlgt) or G2824 or globomycin at 1× MIC for 30 min (CFT073 imp4213), fixed with 4% paraformaldehyde, and incubated with 1 μg/ml FM-64 dye and 1 μg/ml 4′,6-diamidino-2-phenylindole (DAPI) solution. Quantitation of bacterial cell area was performed using the ImageJ program by measuring at least ∼100 bacterial cells from two independent experiments.
Downregulation of gene expression by CRISPRi.
The two-plasmid bacterial CRISPRi system pdCas9-bacteria_GNE and PGNRNA-bacteria_GNE are based on the AddGene plasmids 44249 and 44251 (73), respectively. The plasmid was synthesized in smaller DNA fragments (500 bp to 3 kb) (IDT gBlocks) and assembled by Gibson assembly (NEB) according to the manufacturer’s protocols. Plasmids were confirmed by sequencing (ELIM Bio). gRNAs were designed to target the 5′ end of the gene on the nontemplate strand using Benchling CRISPR software (74). gRNAs were cloned into PGNRNA-bacteria using Gibson assembly (NEB) according to the manufacturer’s protocols and sequence confirmed (ELIM Bio).
Bacterial cultures were grown overnight on LB agar supplemented with carbenicillin (50 μg/ml) and chloramphenicol (12.5 μg/ml) to maintain both plasmids, pdCas9-bacteria and PGNRNA-bacteria with each gRNA as appropriate. Cells were scraped from the plate into fresh medium. The OD600 was measured, and the culture was subsequently diluted to an OD600 0.001 in the presence or absence of G2824, globomycin, or ciprofloxacin. Two hundred microliters was transferred to a 96-well plate (Corning) and monitored for growth by measuring the OD600 (EnVision multimode plate reader; PerkinElmer). All treatments were performed in triplicates. The specificity of CRISPRi downregulation was measured using reverse transcriptase quantitative PCR (RT-qPCR).
Purification of peptidoglycan-associated proteins.
Purification of PGN-associated proteins (PAP) was performed according to published methods (17, 40, 41) with some modifications. Briefly, bacteria were harvested in mid-exponential phase for treatment and then subjected to PAP extraction by resuspending the cell pellets from an OD (A600) of 10 in 6 ml of PAP extraction buffer containing 2% (wt/vol) SDS in 100 mM Tris-HCl (pH 8.0) with 100 mM NaCl, 10% glycerol, and cOmplete, mini, EDTA-free protease inhibitor cocktail (Sigma-Aldrich). After 60 min at RT, the extraction was subjected to centrifugation at 100,000 × g for 60 min at 22°C, and the pellet, containing PGN and associated proteins, was washed once with the same PAP extraction buffer with centrifugation at 100,000 × g for 30 min and resuspended in 200 μl of PAP extraction buffer (referred to as the SDS-insoluble on PAP fraction). The supernatant containing the SDS-soluble fraction was aliquoted and frozen (referred to as non-PAP fraction). Both fractions were treated with equal volumes of BugBuster buffer prior to the addition of sample buffer for Western immunoblotting as described above. It should be noted that the final PAP fractions were ∼30-fold more concentrated than the non-PAP fractions.
Isolation of E. coli IM and OM using sucrose gradient centrifugation.
Bacterial inner and outer membranes were separated by isopycnic sucrose gradient centrifugation as described previously (75) and by the Hancock Laboratory (http://cmdr.ubc.ca/bobh/method/outer-membrane-preparation-one-step-sucrose-gradient-procedure/), with some modifications. Briefly, bacteria were grown in Luria broth at 37°C to mid-exponential phase (OD600 of 0.8 to 0.9) and then treated with 1× MIC of designated inhibitors for 1 h. Cells representing equivalents of an OD600 of 10 were harvested by centrifugation at 4,000 × g for 15 min, washed once, and resuspended in cold 10 mM Tris, pH 8.0, containing 20% (wt/vol) sucrose, cOmplete EDTA-free protease inhibitor cocktail (Roche), and 1,000 U/ml nuclease (Benzonase nuclease; EMD Millipore). The cell suspension was disrupted by two passages through an LV1 Microfluidizer (Microfluidics). Unbroken cells were removed by centrifugation at 4,000 × g, and membranes were collected by ultracentrifugation at 100,000 × g for 1 h and washed once with cold 10 mM Tris, pH 8.0. The final membrane preparation was resuspended in 10 mM Tris, pH 8.0, containing 10% sucrose, cOmplete EDTA-free protease inhibitor cocktail (Roche), and 1,000 U/ml nuclease and then applied to the top of the sucrose gradients (73%, 60%, and 20%). The loaded gradients were spun at 39,000 rpm for 22 h at 4°C in a Beckman SW 41 Ti rotor. Fractions were collected from the top and analyzed by SDS-PAGE and immunoblotting with appropriate antibodies.
Statistical analyses.
All statistical analyses were performed using GraphPad Prism 6.0 software. The data were tested to ensure appropriate use in parametric statistics, and statistical analyses were performed on log-transformed data. All graphs present the means ± the standard errors of the means (SEMs). Unless stated otherwise, P values for all data were determined using a Mann-Whitney unpaired t test. Bonferroni’s correction was applied to control for multiple comparisons for CRIPSRi data shown in Fig. 4G.
ACKNOWLEDGMENTS
We thank Shaw Warren (Massachusetts General Hospital) for the anti-Pal antibody, Scott Stawicki for assistance with generating the rabbit polyclonal antibodies, and Eric Brown for helpful comments and suggestions for the manuscript.
This study was supported by internal Genentech funds.
All authors except R.K., T.S., H.I., H.O., H.Y., J.N., and P.C.R. are employees of Genentech, a member of the Roche Group, and are shareholders of Roche. R.K., T.S., H.I., H.O., H.Y., J.N., and P.C.R. are employees of PeptiDream, Inc.
Footnotes
Supplemental material is available online only.
Contributor Information
Christian N. Cunningham, Email: cunningham.christian@gene.com.
Sharookh B. Kapadia, Email: kapadia.sharookh@gene.com.
Laurie E. Comstock, Brigham and Women's Hospital/Harvard Medical School
REFERENCES
- 1.Silhavy TJ, Kahne D, Walker S. 2010. The bacterial cell envelope. Cold Spring Harb Perspect Biol 2:a000414. doi: 10.1101/cshperspect.a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cowles CE, Li Y, Semmelhack MF, Cristea IM, Silhavy TJ. 2011. The free and bound forms of Lpp occupy distinct subcellular locations in Escherichia coli. Mol Microbiol 79:1168–1181. doi: 10.1111/j.1365-2958.2011.07539.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wilson MM, Bernstein HD. 2016. Surface-exposed lipoproteins: an emerging secretion phenomenon in Gram-negative bacteria. Trends Microbiol 24:195–208. doi: 10.1016/j.tim.2015.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kovacs-Simon A, Titball RW, Michell SL. 2011. Lipoproteins of bacterial pathogens. Infect Immun 79:548–561. doi: 10.1128/IAI.00682-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schlesinger MJ. 1992. Lipid modifications of proteins. CRC Press, Boca Raton, FL. [Google Scholar]
- 6.Sankaran K, Wu HC. 1994. Lipid modification of bacterial prolipoprotein. Transfer of diacylglyceryl moiety from phosphatidylglycerol. J Biol Chem 269:19701–19706. doi: 10.1016/S0021-9258(17)32077-X. [DOI] [PubMed] [Google Scholar]
- 7.Tokunaga M, Tokunaga H, Wu HC. 1982. Post-translational modification and processing of Escherichia coli prolipoprotein in vitro. Proc Natl Acad Sci U S A 79:2255–2259. doi: 10.1073/pnas.79.7.2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Olatunji S, Yu X, Bailey J, Huang C-Y, Zapotoczna M, Bowen K, Remškar M, Müller R, Scanlan EM, Geoghegan JA, Olieric V, Caffrey M. 2020. Structures of lipoprotein signal peptidase II from Staphylococcus aureus complexed with antibiotics globomycin and myxovirescin. Nat Commun 11:140. doi: 10.1038/s41467-019-13724-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dev IK, Harvey RJ, Ray PH. 1985. Inhibition of prolipoprotein signal peptidase by globomycin. J Biol Chem 260:5891–5894. doi: 10.1016/S0021-9258(18)88911-6. [DOI] [PubMed] [Google Scholar]
- 10.Gerth K, Irschik H, Reichenbach H, Trowitzsch W. 1982. The myxovirescins, a family of antibiotics from Myxococcus virescens (Myxobacterales). J Antibiot 35:1454–1459. doi: 10.7164/antibiotics.35.1454. [DOI] [PubMed] [Google Scholar]
- 11.Xiao Y, Gerth K, Müller R, Wall D. 2012. Myxobacterium-produced antibiotic TA (myxovirescin) inhibits type II signal peptidase. Antimicrob Agents Chemother 56:2014–2021. doi: 10.1128/AAC.06148-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Narita S-I, Tokuda H. 2010. Sorting of bacterial lipoproteins to the outer membrane by the Lol system. Methods Mol Biol 619:117–129. doi: 10.1007/978-1-60327-412-8_7. [DOI] [PubMed] [Google Scholar]
- 13.Narita S-I. 2011. ABC transporters involved in the biogenesis of the outer membrane in Gram-negative bacteria. Biosci Biotechnol Biochem 75:1044–1054. doi: 10.1271/bbb.110115. [DOI] [PubMed] [Google Scholar]
- 14.Neidhardt FC. 1996. Chemical composition of Escherichia coli, p 1035–1063. Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, DC. [Google Scholar]
- 15.Suzuki H, Nishimura Y, Yasuda S, Nishimura A, Yamada M, Hirota Y. 1978. Murein-lipoprotein of Escherichia coli: a protein involved in the stabilization of bacterial cell envelope. Mol Gen Genet 167:1–9. doi: 10.1007/BF00270315. [DOI] [PubMed] [Google Scholar]
- 16.Yem DW, Wu HC. 1978. Physiological characterization of an Escherichia coli mutant altered in the structure of murein lipoprotein. J Bacteriol 133:1419–1426. doi: 10.1128/JB.133.3.1419-1426.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Diao J, Bouwman C, Yan D, Kang J, Katakam AK, Liu P, Pantua H, Abbas AR, Nickerson NN, Austin C, Reichelt M, Sandoval W, Xu M, Whitfield C, Kapadia SB. 2017. Peptidoglycan association of murein lipoprotein is required for KpsD-dependent group 2 capsular polysaccharide expression and serum resistance in a uropathogenic Escherichia coli isolate. mBio 8:e00603-17. doi: 10.1128/mBio.00603-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zwiebel LJ, Inukai M, Nakamura K, Inouye M. 1981. Preferential selection of deletion mutations of the outer membrane lipoprotein gene of Escherichia coli by globomycin. J Bacteriol 145:654–656. doi: 10.1128/JB.145.1.654-656.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McLeod SM, Fleming PR, MacCormack K, McLaughlin RE, Whiteaker JD, Narita S-I, Mori M, Tokuda H, Miller AA. 2015. Small molecule inhibitors of Gram-negative lipoprotein trafficking discovered by phenotypic screening. J Bacteriol 197:1075–1082. doi: 10.1128/JB.02352-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nickerson NN, Jao CC, Xu Y, Quinn J, Skippington E, Alexander MK, Miu A, Skelton N, Hankins JV, Lopez MS, Koth CM, Rutherford S, Nishiyama M. 2018. A novel inhibitor of the LolCDE ABC transporter essential for lipoprotein trafficking in Gram-negative bacteria. Antimicrob Agents Chemother 62:e02151-17. doi: 10.1128/AAC.02151-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Narita S-I, Tokuda H. 2011. Overexpression of LolCDE allows deletion of the Escherichia coli gene encoding apolipoprotein N-acyltransferase. J Bacteriol 193:4832–4840. doi: 10.1128/JB.05013-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robichon C, Vidal-Ingigliardi D, Pugsley AP. 2005. Depletion of apolipoprotein N-acyltransferase causes mislocalization of outer membrane lipoproteins in Escherichia coli. J Biol Chem 280:974–983. doi: 10.1074/jbc.M411059200. [DOI] [PubMed] [Google Scholar]
- 23.Yakushi T, Tajima T, Matsuyama S, Tokuda H. 1997. Lethality of the covalent linkage between mislocalized major outer membrane lipoprotein and the peptidoglycan of Escherichia coli. J Bacteriol 179:2857–2862. doi: 10.1128/jb.179.9.2857-2862.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mizuno T. 1979. A novel peptidoglycan-associated lipoprotein found in the cell envelope of Pseudomonas aeruginosa and Escherichia coli. J Biochem 86:991–1000. doi: 10.1093/oxfordjournals.jbchem.a132631. [DOI] [PubMed] [Google Scholar]
- 25.Leduc M, Ishidate K, Shakibai N, Rothfield L. 1992. Interactions of Escherichia coli membrane lipoproteins with the murein sacculus. J Bacteriol 174:7982–7988. doi: 10.1128/JB.174.24.7982-7988.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cascales E, Bernadac A, Gavioli M, Lazzaroni J-C, Lloubes R. 2002. Pal lipoprotein of Escherichia coli plays a major role in outer membrane integrity. J Bacteriol 184:754–759. doi: 10.1128/JB.184.3.754-759.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clavel T, Germon P, Vianney A, Portalier R, Lazzaroni JC. 1998. TolB protein of Escherichia coli K-12 interacts with the outer membrane peptidoglycan-associated proteins Pal, Lpp and OmpA. Mol Microbiol 29:359–367. doi: 10.1046/j.1365-2958.1998.00945.x. [DOI] [PubMed] [Google Scholar]
- 28.Kitamura S, Owensby A, Wall D, Wolan DW. 2018. Lipoprotein signal peptidase inhibitors with antibiotic properties identified through design of a robust in vitro HT platform. Cell Chem Biol 25:301–308.e12. doi: 10.1016/j.chembiol.2017.12.011. [DOI] [PubMed] [Google Scholar]
- 29.Igarashi M. 2019. New natural products to meet the antibiotic crisis: a personal journey. J Antibiot (Tokyo) 72:890–898. doi: 10.1038/s41429-019-0224-6. [DOI] [PubMed] [Google Scholar]
- 30.Rossiter SE, Fletcher MH, Wuest WM. 2017. Natural products as platforms to overcome antibiotic resistance. Chem Rev 117:12415–12474. doi: 10.1021/acs.chemrev.7b00283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gan K, Sankaran K, Williams MG, Aldea M, Rudd KE, Kushner SR, Wu HC. 1995. The umpA gene of Escherichia coli encodes phosphatidylglycerol:prolipoprotein diacylglyceryl transferase (lgt) and regulates thymidylate synthase levels through translational coupling. J Bacteriol 177:1879–1882. doi: 10.1128/jb.177.7.1879-1882.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Inukai M, Enokita R, Torikata A, Nakahara M, Iwado S, Arai M. 1978. Globomycin, a new peptide antibiotic with spheroplast-forming activity. I. Taxonomy of producing organisms and fermentation. J Antibiot (Tokyo) 31:410–420. doi: 10.7164/antibiotics.31.410. [DOI] [PubMed] [Google Scholar]
- 33.Inukai M, Nakajima M, Osawa M, Haneishi T, Arai M. 1978. Globomycin, a new peptide antibiotic with spheroplast-forming activity. II. Isolation and physico-chemical and biological characterization. J Antibiot (Tokyo) 31:421–425. doi: 10.7164/antibiotics.31.421. [DOI] [PubMed] [Google Scholar]
- 34.Rojas ER, Billings G, Odermatt PD, Auer GK, Zhu L, Miguel A, Chang F, Weibel DB, Theriot JA, Huang KC. 2018. The outer membrane is an essential load-bearing element in Gram-negative bacteria. Nature 559:617–621. doi: 10.1038/s41586-018-0344-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goto Y, Katoh T, Suga H. 2011. Flexizymes for genetic code reprogramming. Nat Protoc 6:779–790. doi: 10.1038/nprot.2011.331. [DOI] [PubMed] [Google Scholar]
- 36.Kashiwagi K, Reid CP. 28 April 2011. Rapid display method in translational synthesis of peptide. International patent WO2011049157.
- 37.Ishizawa T, Kawakami T, Reid PC, Murakami H. 2013. TRAP display: a high-speed selection method for the generation of functional polypeptides. J Am Chem Soc 135:5433–5440. doi: 10.1021/ja312579u. [DOI] [PubMed] [Google Scholar]
- 38.Ruiz N, Falcone B, Kahne D, Silhavy TJ. 2005. Chemical conditionality: a genetic strategy to probe organelle assembly. Cell 121:307–317. doi: 10.1016/j.cell.2005.02.014. [DOI] [PubMed] [Google Scholar]
- 39.Stoll H, Dengjel J, Nerz C, Götz F. 2005. Staphylococcus aureus deficient in lipidation of prelipoproteins is attenuated in growth and immune activation. Infect Immun 73:2411–2423. doi: 10.1128/IAI.73.4.2411-2423.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakae T, Ishii J, Tokunaga M. 1979. Subunit structure of functional porin oligomers that form permeability channels in the other membrane of Escherichia coli. J Biol Chem 254:1457–1461. doi: 10.1016/S0021-9258(17)37790-6. [DOI] [PubMed] [Google Scholar]
- 41.Whitfield C, Hancock RE, Costerton JW. 1983. Outer membrane protein K of Escherichia coli: purification and pore-forming properties in lipid bilayer membranes. J Bacteriol 156:873–879. doi: 10.1128/JB.156.2.873-879.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Suzuki M, Hara H, Matsumoto K. 2002. Envelope disorder of Escherichia coli cells lacking phosphatidylglycerol. J Bacteriol 184:5418–5425. doi: 10.1128/jb.184.19.5418-5425.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pailler J, Aucher W, Pires M, Buddelmeijer N. 2012. Phosphatidylglycerol::prolipoprotein diacylglyceryl transferase (Lgt) of Escherichia coli has seven transmembrane segments, and its essential residues are embedded in the membrane. J Bacteriol 194:2142–2151. doi: 10.1128/JB.06641-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rousset F, Cui L, Siouve E, Becavin C, Depardieu F, Bikard D. 2018. Genome-wide CRISPR-dCas9 screens in E. coli identify essential genes and phage host factors. PLoS Genet 14:e1007749. doi: 10.1371/journal.pgen.1007749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kowata H, Tochigi S, Kusano T, Kojima S. 2016. Quantitative measurement of the outer membrane permeability in Escherichia coli lpp and tol-pal mutants defines the significance of Tol-Pal function for maintaining drug resistance. J Antibiot (Tokyo) 69:863–870. doi: 10.1038/ja.2016.50. [DOI] [PubMed] [Google Scholar]
- 46.Hirota Y, Suzuki H, Nishimura Y, Yasuda S. 1977. On the process of cellular division in Escherichia coli: a mutant of E. coli lacking a murein-lipoprotein. Proc Natl Acad Sci U S A 74:1417–1420. doi: 10.1073/pnas.74.4.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Braun V, Wolff H. 1970. The murein-lipoprotein linkage in the cell wall of Escherichia coli. Eur J Biochem 14:387–391. doi: 10.1111/j.1432-1033.1970.tb00301.x. [DOI] [PubMed] [Google Scholar]
- 48.Zhang WY, Wu HC. 1992. Alterations of the carboxyl-terminal amino acid residues of Escherichia coli lipoprotein affect the formation of murein-bound lipoprotein. J Biol Chem 267:19560–19564. doi: 10.1016/S0021-9258(18)41811-X. [DOI] [PubMed] [Google Scholar]
- 49.Zhang WY, Inouye M, Wu HC. 1992. Neither lipid modification nor processing of prolipoprotein is essential for the formation of murein-bound lipoprotein in Escherichia coli. J Biol Chem 267:19631–19635. doi: 10.1016/S0021-9258(18)41821-2. [DOI] [PubMed] [Google Scholar]
- 50.Gan K, Gupta SD, Sankaran K, Schmid MB, Wu HC. 1993. Isolation and characterization of a temperature-sensitive mutant of Salmonella Typhimurium defective in prolipoprotein modification. J Biol Chem 268:16544–16550. doi: 10.1016/S0021-9258(19)85453-4. [DOI] [PubMed] [Google Scholar]
- 51.Gupta SD, Gan K, Schmid MB, Wu HC. 1993. Characterization of a temperature-sensitive mutant of Salmonella Typhimurium defective in apolipoprotein N-acyltransferase. J Biol Chem 268:16551–16556. doi: 10.1016/S0021-9258(19)85454-6. [DOI] [PubMed] [Google Scholar]
- 52.Herman A, Serfecz J, Kinnally A, Crosby K, Youngman M, Wykoff D, Wilson JW. 2016. The bacterial iprA gene is conserved across Enterobacteriaceae, is involved in oxidative stress resistance, and influences gene expression in Salmonella enterica serovar Typhimurium. J Bacteriol 198:2166–2179. doi: 10.1128/JB.00144-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ito H, Ura A, Oyamada Y, Yoshida H, Yamagishi J-I, Narita S-I, Matsuyama S-I, Tokuda H. 2007. A new screening method to identify inhibitors of the Lol (localization of lipoproteins) system, a novel antibacterial target. Microbiol Immunol 51:263–270. doi: 10.1111/j.1348-0421.2007.tb03906.x. [DOI] [PubMed] [Google Scholar]
- 54.Pantua H, Skippington E, Braun M-G, Noland CL, Diao J, Peng Y, Gloor SL, Yan D, Kang J, Katakam AK, Reeder J, Castanedo GM, Garland K, Komuves L, Sagolla M, Austin CD, Murray J, Xu Y, Modrusan Z, Xu M, Hanan EJ, Kapadia SB. 2020. Unstable mechanisms of resistance to inhibitors of Escherichia coli lipoprotein signal peptidase. mBio 11:e02018-20. doi: 10.1128/mBio.02018-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lehman KM, Grabowicz M. 2019. Countering Gram-negative antibiotic resistance: recent progress in disrupting the outer membrane with novel therapeutics. Antibiotics (Basel) 8:163. doi: 10.3390/antibiotics8040163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vogeley L, Arnaout El T, Bailey J, Stansfeld PJ, Boland C, Caffrey M. 2016. Structural basis of lipoprotein signal peptidase II action and inhibition by the antibiotic globomycin. Science 351:876–880. doi: 10.1126/science.aad3747. [DOI] [PubMed] [Google Scholar]
- 57.Mao G, Zhao Y, Kang X, Li Z, Zhang Y, Wang X, Sun F, Sankaran K, Zhang XC. 2016. Crystal structure of E. coli lipoprotein diacylglyceryl transferase. Nat Commun 7:10198. doi: 10.1038/ncomms10198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Singh W, Bilal M, McClory J, Dourado D, Quinn D, Moody TS, Sutcliffe I, Huang M. 2019. Mechanism of phosphatidylglycerol activation catalyzed by prolipoprotein diacylglyceryl transferase. J Phys Chem B 123:7092–7102. doi: 10.1021/acs.jpcb.9b04227. [DOI] [PubMed] [Google Scholar]
- 59.Palmer AC, Kishony R. 2014. Opposing effects of target overexpression reveal drug mechanisms. Nat Commun 5:4296. doi: 10.1038/ncomms5296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Storek KM, Auerbach MR, Shi H, Garcia NK, Sun D, Nickerson NN, Vij R, Lin Z, Chiang N, Schneider K, Wecksler AT, Skippington E, Nakamura G, Seshasayee D, Koerber JT, Payandeh J, Smith PA, Rutherford ST. 2018. Monoclonal antibody targeting the β-barrel assembly machine of Escherichia coli is bactericidal. Proc Natl Acad Sci U S A 115:3692–3697. doi: 10.1073/pnas.1800043115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Storek KM, Chan J, Vij R, Chiang N, Lin Z, Bevers J, Koth CM, Vernes J-M, Meng YG, Yin J, Wallweber H, Dalmas O, Shriver S, Tam C, Schneider K, Seshasayee D, Nakamura G, Smith PA, Payandeh J, Koerber JT, Comps-Agrar L, Rutherford ST. 2019. Massive antibody discovery used to probe structure-function relationships of the essential outer membrane protein LptD. Elife 8:e46258. doi: 10.7554/eLife.46258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mobley HL, Green DM, Trifillis AL, Johnson DE, Chippendale GR, Lockatell CV, Jones BD, Warren JW. 1990. Pyelonephritogenic Escherichia coli and killing of cultured human renal proximal tubular epithelial cells: role of hemolysin in some strains. Infect Immun 58:1281–1289. doi: 10.1128/IAI.58.5.1281-1289.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cherepanov PP, Wackernagel W. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9–14. doi: 10.1016/0378-1119(95)00193-a. [DOI] [PubMed] [Google Scholar]
- 65.Noland CL, Kattke MD, Diao J, Gloor SL, Pantua H, Reichelt M, Katakam AK, Yan D, Kang J, Zilberleyb I, Xu M, Kapadia SB, Murray JM. 2017. Structural insights into lipoprotein N-acylation by Escherichia coli apolipoprotein N-acyltransferase. Proc Natl Acad Sci U S A 114:E6044–E6053. doi: 10.1073/pnas.1707813114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hmelo LR, Borlee BR, Almblad H, Love ME, Randall TE, Tseng BS, Lin C, Irie Y, Storek KM, Yang JJ, Siehnel RJ, Howell PL, Singh PK, Tolker-Nielsen T, Parsek MR, Schweizer HP, Harrison JJ. 2015. Precision-engineering the Pseudomonas aeruginosa genome with two-step allelic exchange. Nat Protoc 10:1820–1841. doi: 10.1038/nprot.2015.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Balibar CJ, Grabowicz M. 2016. Mutant alleles of lptD increase the permeability of Pseudomonas aeruginosa and define determinants of intrinsic resistance to antibiotics. Antimicrob Agents Chemother 60:845–854. doi: 10.1128/AAC.01747-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schweizer HP, Hoang TT. 1995. An improved system for gene replacement and xylE fusion analysis in Pseudomonas aeruginosa. Gene 158:15–22. doi: 10.1016/0378-1119(95)00055-B. [DOI] [PubMed] [Google Scholar]
- 69.Cross AS, Siegel G, Byrne WR, Trautmann M, Finbloom DS. 1989. Intravenous immune globulin impairs anti-bacterial defences of a cyclophosphamide-treated host. Clin Exp Immunol 76:159–164. [PMC free article] [PubMed] [Google Scholar]
- 70.van de Waterbeemd M, Fort KL, Boll D, Reinhardt-Szyba M, Routh A, Makarov A, Heck AJR. 2017. High-fidelity mass analysis unveils heterogeneity in intact ribosomal particles. Nat Methods 14:283–286. doi: 10.1038/nmeth.4147. [DOI] [PubMed] [Google Scholar]
- 71.Rosati S, Yang Y, Barendregt A, Heck AJR. 2014. Detailed mass analysis of structural heterogeneity in monoclonal antibodies using native mass spectrometry. Nat Protoc 9:967–976. doi: 10.1038/nprot.2014.057. [DOI] [PubMed] [Google Scholar]
- 72.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez J-Y, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods 9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA. 2013. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Peters JM, Colavin A, Shi H, Czarny TL, Larson MH, Wong S, Hawkins JS, Lu CHS, Koo B-M, Marta E, Shiver AL, Whitehead EH, Weissman JS, Brown ED, Qi LS, Huang KC, Gross CA. 2016. A comprehensive, CRISPR-based functional analysis of essential genes in bacteria. Cell 165:1493–1506. doi: 10.1016/j.cell.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cian MB, Giordano NP, Mettlach JA, Minor KE, Dalebroux ZD. 2020. Separation of the cell envelope for Gram-negative bacteria into inner and outer membrane fractions with technical adjustments for Acinetobacter baumannii. J Vis Exp 2020:e60517. doi: 10.3791/60517. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Kashiwagi K, Reid CP. 28 April 2011. Rapid display method in translational synthesis of peptide. International patent WO2011049157.
Supplementary Materials
Fig. S1 to S5 and Tables S1 and S2. Download JB.00149-21-s0001.pdf, PDF file, 1.11 MB (1.1MB, pdf)