ABSTRACT
Bartonella spp., mostly Bartonella quintana and B. henselae, are a common cause of culture-negative endocarditis. Serology using immunofluorescence assay (IFA) and PCR performed on cardiac tissues are the mainstays of diagnosis. We developed an enzyme immunoassay (EIA) and a novel multiplex real-time PCR assay, utilizing Bartonella genus-specific, B. henselae-specific, and B. quintana-specific SimpleProbe probes, for diagnosis of Bartonella endocarditis. We aimed to evaluate the performance of these assays. Thirty-seven patients with definite endocarditis, 18 with B. henselae, 18 with B. quintana, and 1 with B. koehlerae, were studied. Diagnosis was confirmed by conventional PCR and DNA sequencing of surgical cardiac specimens. Similar to the case with IFA, anti-Bartonella IgG titers of ≥1:800 were found in 94% of patients by EIA; cross-reactivity between B. henselae and B. quintana precluded species-specific serodiagnosis, and frequent (41%) but low-titer cross-reactivity between Coxiella burnetii antibodies and B. henselae antigen was found in patients with Q fever endocarditis. Low-titer (1:100) cross-reactivity was uncommonly found also in patients with brucellosis and culture-positive endocarditis, particularly Enterococcus faecalis endocarditis. Real-time PCR performed on explanted heart valves/vegetations was in complete agreement with results of sequence-based diagnosis with characteristic melting curves. The genus-specific probe identified five additional endocarditis-associated Bartonella spp. at the genus level. In conclusion, EIA coupled with a novel real-time PCR assay can play an important role in Bartonella endocarditis diagnosis and expand the diagnostic arsenal at the disposal of the clinical microbiologist. Since serology remains a major diagnostic tool, recognizing its pitfalls is essential to avoid incorrect diagnosis.
KEYWORDS: Bartonella sp., Bartonella henselae, Bartonella quintana, B. koehlerae, immunofluorescence assay, IFA, enzyme immunoassay, EIA, ELISA, real-time PCR, endocarditis, development, diagnosis, human, Bartonella spp., diagnostics
INTRODUCTION
Following Q fever, Bartonella spp. are the second most common cause of culture-negative endocarditis, accounting for up to 28% of cases (1–3). Most reported cases have been attributed to Bartonella quintana, followed by B. henselae, while other Bartonella spp. have been implicated in single cases of endocarditis, including B. elizabethae (4), Bartonella vinsonii subsp. berkhoffii (1, 5, 6), Bartonella vinsonii subsp. arupensis (7), B. koehlerae (8), B. alsatica (1, 9, 10), and “Candidatus Bartonella mayotimonensis” (11). Bartonella spp. are fastidious bacteria with notoriously poor sensitivity of axenic cultures performed on clinical specimens of human blood or tissue. Laboratory diagnosis of Bartonella endocarditis is therefore based mainly on serological assays and PCR performed on infected tissues. Microscopic immunofluorescence assay (IFA) is the most commonly used serological method utilized for diagnosis of Bartonella infections, although other methods, such as enzyme immunoassay (EIA), are used as well (12–16). Compared with EIA, IFA is laborious and time-consuming, and interpretation is subjective. A limitation of all serological assays for the diagnosis of Bartonella infection is cross-reactivity between B. henselae and B. quintana. Bartonella IFA has also been reported to have cross-reactivity with Coxiella burnetii and Chlamydia pneumoniae (17–19). The Bernard Pridan Laboratory for Molecular Biology of Infectious Disease at Tel Aviv Sourasky Medical Center serves as the national reference laboratory for human Bartonella infections in Israel and has developed over the past 30 years serological and molecular tools for the diagnosis of Bartonella infection, including an EIA and various conventional and real-time PCR assays. Although we have occasionally used these assays for non-henselae Bartonella-associated infections, they have been developed and utilized primarily for the diagnosis of cat scratch disease, which is caused predominantly by B. henselae (12, 14, 15, 20–24). A systematic evaluation of the performance of our assays for the diagnosis of Bartonella endocarditis has not been conducted thus far. The aim of the current study was to evaluate an EIA and molecular assays, including a novel multiplex real-time PCR assay, for the diagnosis of Bartonella endocarditis.
MATERIALS AND METHODS
Patients and clinical samples.
As part of a surveillance study of Bartonella infections ongoing in Israel since 1991, all clinical specimens from patients with suspected Bartonella endocarditis, including serum samples and tissue for PCR assays and cultures, are referred to a single laboratory at Tel Aviv Sourasky Medical Center, thus allowing identification of essentially all laboratory-confirmed cases of Bartonella endocarditis in Israel. The referring hospital-based physicians are requested to fill out a questionnaire providing data on patients’ demographic and clinical features and are contacted to obtain additional information. For the purpose of the current study, we included patients with culture-negative endocarditis who met the following two criteria: (i) they have undergone valve surgery for endocarditis-associated valve dysfunction complicated by heart failure and/or the presence of intracardiac abscess, and (ii) Bartonella was identified in the valvular/paravalvular tissue or vegetation by PCR and DNA sequencing. The study included 37 patients with Bartonella endocarditis diagnosed in hospitals throughout Israel in 2001 to 2020: 18 patients with B. henselae, 18 with B. quintana, and 1 with B. koehlerae. All patients had definite endocarditis based on at least two major criteria of the Duke score: (i) echocardiogram positive for endocarditis as defined in the modified Duke criteria (25) and (ii) Bartonella-positive PCR from infected cardiac tissue corroborated by DNA sequencing, that is, if one accepts Bartonella-positive PCR as a finding with equal significance to a major criterion of the modified Duke criteria (25, 26). Patients with Bartonella endocarditis without PCR-based diagnosis for species identification were excluded from the study. The study was approved by the institutional review board (Helsinki Committee) of the Tel Aviv Sourasky Medical Center.
Bacterial strains and cultures.
Bacterial strains used in this study are described in Table 1, including 7 Bartonella spp. that have been implicated in human endocarditis and non-Bartonella bacterial species that have been associated with culture-negative endocarditis (27). All bacterial strains were obtained from known reference collections (American Type Culture Collection [ATCC], USA, National Collection of Type Cultures [NCTC], United Kingdom, Collection Nationale de Cultures de Microorganismes [CNCM], Institute Pasteur, France, and Collection of the Institute Pasteur, France [CIP]). Exceptions included Nine Mile phase I Coxiella burnetii DNA (Vircell, Spain), B. henselae isolate BhTA-4, cultured in our laboratory from lymph node biopsy specimen of a patient with typical cat scratch disease, and B. koehlerae isolate C-508, cultured in our laboratory from a bacteremic cat and found to have ribC and gltA DNA sequences identical to those of B. koehlerae amplified from a valve of an endocarditis patient reported by our group (8). Bartonella spp. and cardiac tissue specimens, whenever sufficient material was available, were cultured on chocolate agar plates as previously described (12). Bartonella spp. were identified by PCR targeting various genes with amplicon sequencing (Table 2).
TABLE 1.
Results of the real-time PCR assay performed on Bartonella spp. and other bacterial species associated with culture-negative endocarditis
| Bacterial species/subsp. | Strain designation(s)b | Probe Tma (°C) or status |
||
|---|---|---|---|---|
| Bartonella genus | Bartonella henselae | Bartonella quintana | ||
| B. henselae | BhTA-4, ATCC 49793 (87-66) | 58–61 | 47–51 | Negative |
| B. quintana | CIP 103739 (Toulouse), CIP 107027 (Fuller) | 57–61 | Negative | 68–71 |
| B. koehlerae | ATCC 700693 | 57–59 | Negative | Negative |
| B. vinsonii subsp. berkhoffii | ATCC 51672 | 57 | Negative | Negative |
| B. vinsonii subsp. arupensis | ATCC BAA-2250 | 58 | Negative | Negative |
| B. elizabethae | ATCC 49927 | 43 | Negative | Negative |
| B. washoensis | ATCC BAA-2254 | 50 | Negative | Negative |
| Coxiella burnetii | ATCC VR-615 (Nine Mile phase I) | Negative | Negative | Negative |
| Tropheryma whipplei | CNCM I-2202T (Twist-Marseille) | Negative | Negative | Negative |
| Legionella pneumophila subsp. pneumophila | ATCC 33152 | Negative | Negative | Negative |
| Aggregatibacter aphrophilus | ATCC 33389 | Negative | Negative | Negative |
| Aggregatibacter paraphrophilus | NCTC 10557 | Negative | Negative | Negative |
| Aggregatibacter actinomycetemcomitans | ATCC 700685 | Negative | Negative | Negative |
| Aggregatibacter segnis | NCTC 10977 | Negative | Negative | Negative |
| Cardiobacterium hominis | NCTC 10426 | Negative | Negative | Negative |
| Eikenella corrodens | ATCC 23834 | Negative | Negative | Negative |
| Kingella kingae | NCTC 10529 | Negative | Negative | Negative |
| Abiotrophia defectiva | ATCC 49176 | Negative | Negative | Negative |
| Granulicatella adiacens | ATCC 49175 | Negative | Negative | Negative |
| Granulicatella elegans | ATCC 700633 | Negative | Negative | Negative |
| Gemella bergeriae | ATCC 700627 | Negative | Negative | Negative |
| Gemella haemolysans | ATCC 10379 | Negative | Negative | Negative |
| Gemella morbillorum | NCTC 11323 | Negative | Negative | Negative |
| Gemella sanguinis | ATCC 700632 | Negative | Negative | Negative |
Tm, melting temperature.
ATCC, American Type Culture Collection, USA; CIP, Collection of the Institute Pasteur, France; CNCM, Collection Nationale de Cultures de Microorganismes, Institut Pasteur, France; NCTC, National Collection of Type Cultures, Central Public Health Laboratory, United Kingdom.
TABLE 2.
Results of the real-time PCR assay (expressed as melting temperatures) and the enzyme immunoassay (expressed as the reciprocal titers) among 37 patients with Bartonella endocarditis
| Patient no. | Age (yrs)/sexa | Molecular diagnosisb | Conventional PCR target gene(s) | Real-time PCR Tm (°C)c |
Maximal reciprocal titer (anti-Bartonella IgG antibodies) |
||||
|---|---|---|---|---|---|---|---|---|---|
| Tm-1 | Tm-2 | Tm-3 | B. henselae | B. quintana | B. koehlerae | ||||
| 1 | 60/M | B. koehlerae | gltA, ribC | 59 | 400 | 800 | 1,600 | ||
| 2 | 63/M | B. henselae | gltA | 49 | 59 | 1,600 | 400 | 800 | |
| 3 | 71/M | B. henselae | gltA | 49 | 59 | 3,200 | 1,600 | 3,200 | |
| 4 | 37/M | B. henselae | gltA | 49 | 59 | 3,200 | 1,600 | 1,600 | |
| 5 | 58/M | B. henselae | ribC, pap31 | NAd | NA | NA | 12,800 | 6,400 | NDe |
| 6 | 63/M | B. henselae | gltA | 49 | 60 | 400 | NA | ND | |
| 7 | 22/F | B. henselae | gltA | 49 | 59 | 3,200 | 1,600 | ND | |
| 8 | 71/M | B. henselae | pap31 | NA | NA | NA | 3,200 | 3,200 | ND |
| 9 | 74/F | B. henselae | ribC | 49 | 59 | 3,200 | 800 | ND | |
| 10 | 75/F | B. henselae | ribC | 49 | 59 | 12,800 | 12,800 | ND | |
| 11 | 73/M | B. henselae | ribC | 49 | 59 | 1,600 | 800 | ND | |
| 12 | 43/M | B. henselae | pap31, 16S rRNA | 51 | 61 | NA | NA | ND | |
| 13 | 56/M | B. henselae | gltA | 49 | 60 | 1,600 | ND | ND | |
| 14 | 71/F | B. henselae | gltA, 16S rRNA | 49 | 59 | 6,400 | 6,400 | ND | |
| 15 | 20/M | B. henselae | ribC | 51 | 60 | 6,400 | 3,200 | ND | |
| 16 | 56/F | B. henselae | 16S rRNA | 49 | 59 | 12,800 | 12,800 | ND | |
| 17 | 50/M | B. henselae | gltA | 49 | 59 | 3,200 | 1,600 | ND | |
| 18 | 42/F | B. henselae | 16S rRNA | 50 | 60 | 12,800 | 6,400 | ND | |
| 19 | 56/M | B. henselae | gltA | 49 | 59 | 1,600 | 800 | ND | |
| 20 | 32/M | B. quintana | gltA, groEL, rpoB, ftsZ | 59 | 70 | 400 | 1,600 | 400 | |
| 21 | 20/M | B. quintana | gltA, groEL, rpoB, ftsZ | 59 | 70 | 400 | 1,600 | 200 | |
| 22 | 56/M | B. quintana | gltA, groEL, rpoB, ftsZ | 59 | 69 | 1,600 | 6,400 | 800 | |
| 23 | 43/M | B. quintana | 16S rRNA, gltA, pap31 | NA | NA | NA | 800 | 1,600 | ND |
| 24 | 72/M | B. quintana | gltA | NA | NA | NA | 3,200 | 3,200 | 1,600 |
| 25 | 56/M | B. quintana | ribC | 59 | 70 | 200 | 1,600 | ND | |
| 26 | 48/F | B. quintana | gltA, groEL, rpoB, ftsZ | 59 | 70 | 1,600 | 6,400 | ND | |
| 27 | 12/F | B. quintana | gltA, groEL, rpoB, ftsZ | 58 | 69 | 100 | 1,600 | ND | |
| 28 | 7/F | B. quintana | gltA, groEL, rpoB, ftsZ | 61 | 71 | 1,600 | NA | ND | |
| 29 | 28/M | B. quintana | gltA, groEL, rpoB, ftsZ | 61 | 71 | 3,200 | 6,400 | ND | |
| 30 | 52/M | B. quintana | 16S rRNA | 58 | 69 | 12,800 | 25,600 | ND | |
| 31 | 16/F | B. quintana | gltA, groEL, rpoB, ftsZ | 58 | 68 | 12,800 | 6,400 | ND | |
| 32 | 9/F | B. quintana | gltA, groEL, rpoB, ftsZ | 59 | 70 | 3,200 | 6,400 | ND | |
| 33 | 75/M | B. quintana | gltA, groEL, rpoB, ftsZ | 58 | 69 | 25,600 | 25,600 | ND | |
| 34 | 65/M | B. quintana | gltA, groEL, rpoB, ftsZ | 57 | 68 | 3,200 | 6,400 | ND | |
| 35 | 20/F | B. quintana | ribC | 59 | 70 | 200 | 200 | ND | |
| 36 | 11/M | B. quintana | gltA, groEL, rpoB, ftsZ | 59 | 70 | 12,800 | 25,600 | ND | |
| 37 | 75/F | B. quintana | 16S rRNA | 58 | 69 | 3,200 | 6,400 | ND | |
M, male; F, female.
Molecular diagnosis was determined by sequencing of the conventional PCR products and by real-time PCR melting-curve analysis. All B. henselae strains were Huston-1 type as determined by DNA sequencing.
Tm-1, melting temperature generated by the B. henselae-specific probe; Tm-2, melting temperature generated by the Bartonella genus probe; Tm-3, melting temperature generated by the B. quintana-specific probe.
NA, not available. Specimen was not available for further testing.
ND, not done.
Serology.
EIA for the detection of anti-Bartonella antibodies was performed essentially as reported previously, with some modifications (12, 14, 23). The EIA antigen was prepared as sarcosyl-insoluble, presumably outer membrane protein extracts of agar-derived B. henselae strain 87-66 (ATCC 49793), B. quintana Fuller strain (CIP 107027), or B. koehlerae strain C-508 (8), as needed. Sera were initially screened at a 1:100 dilution, and positive sera at this dilution, as determined previously (12), were tested at incremental dilutions to determine their endpoints. To assess assay specificity, serum specimens from the following control groups were subjected to Bartonella EIA. (i) The first group consisted of patients with blood culture-positive endocarditis (n = 62), including 31 cases of Enterococcus faecalis endocarditis and 31 cases with other pathogens, including Streptococcus viridans, Streptococcus sanguinis, Streptococcus mutans, Streptococcus mitis, Streptococcus infantarius, Streptococcus gallolyticus, Streptococcus pneumoniae, Aggregatibacter aphrophilus, Aggregatibacter paraphrophilus, Aggregatibacter actinomycetemcomitans, Abiotrophia defectiva, Gemella, Staphylococcus aureus, coagulase-negative staphylococci, and Aspergillus. The reason for enriching this control group with E. faecalis is a recent publication suggesting a cross-seroreactivity, based on immunoblotting, between E. faecalis and Bartonella (28). (ii) The second group consisted of patients suspected to have chronic Q fever (n = 34), with anti-phase I IgG titers of 1:1,600 to 1:51,200, determined using IFA performed at the national reference laboratory for Q fever, the Israel Institute for Biological Research, Ness-Ziona, Israel. (iii) The third group consisted of patients seropositive for Chlamydia pneumoniae (either IgG ≥ 1:64, IgA ≥ 1:32, or both) (n = 20) and seronegative for Chlamydia trachomatis and Chlamydia psittaci, using IFA performed at the National Chlamydia and Mycoplasma Center, Rabin Medical Center, Petah Tikva, Israel. (iv) The last group consisted of patients with brucellosis and serum agglutination test value of ≥1:160 (n = 10).
DNA extraction and PCR.
Genomic DNA was extracted from tissue specimens and bacteria using the QIAamp DNA minikit (Qiagen, Valencia, CA) or E.Z.N.A. bacterial DNA kit (Omega Bio-Tek) and plasmid DNA was extracted using the QIAprep spin miniprep kit (Qiagen), all according to the manufacturers’ instructions, and kept at −20°C until further analysis. Conventional PCR was performed using primers targeting various genes (Tables 2 and 3), followed by DNA sequencing of 1 to 4 PCR products per specimen (DNA Sequencing Unit at the G. S. Wise Faculty of Life Sciences, Tel Aviv University, and Hy Laboratories Ltd., Rehovot, Israel) and analysis using Chromas (version 2.6; Technelysium, South Brisbane, Australia) and BLAST search in the GenBank database (https://www.ncbi.nlm.nih.gov/GenBank/).
TABLE 3.
Sequences of oligonucleotides relevant to this study
| Oligonucleotide use and name | Oligonucleotide sequence (5′→3′) | Target organism | Target gene | Product size (bp) | Reference |
|---|---|---|---|---|---|
| Conventional PCR | |||||
| BhCS.781p | GGGGACCAGCTCATGGTGG | Bartonella sp. | gltA | 379 | 37 |
| BhCS.1137 | AATGCAAAAAGAACAGTAAACA | Bartonella sp. | |||
| gltA-F | GCTATGTCTGCATTCTATCA | Bartonella sp. | gltA | 790 | 38 |
| gltA-R | GATCYTCAATCATTTCTTTCCA | Bartonella sp. | |||
| Barton-1 | TAACCGATATTGGTTGTGTTGAAG | Bartonella sp. | ribC | 588 | 39 |
| Barton-2 | TAAAGCTAGAAAGTCTGGCAACATAACG | Bartonella sp. | |||
| PAPn1 | TTCTAGGAGTTGAAACCGAT | B. henselae | pap31 | 277 | 40 |
| PAPn2 | GAAACACCACCAGCAACATA | B. henselae | |||
| ftsZ-F | ATTAATCTGCAYCGGCCAGA | Bartonella sp. | ftsZ | 935 | 38 |
| ftsZ-R | ACVGADACACGAATAACACC | Bartonella sp. | |||
| groEL-F | GAACTNGAAGATAAGTTNGAA | Bartonella sp. | groEL | 1,459 | 38 |
| groEL-R | AATCCATTCCGCCCATTC | Bartonella sp. | |||
| rpoB-F | CGCATTGGCTTACTTCGTATG | Bartonella sp. | rpoB | 893 | 38 |
| rpoB-R | GTAGACTGATTAGAACGCTG | Bartonella sp. | |||
| 16S-27f | AGAGTTTGATCMTGGCTCAG | Eubacteria | 16S rRNA | 862-3 | 41 |
| 16S-907r | CCGTCAATTCMTTTRAGTTT | Eubacteria | 42 | ||
| Real-time PCR | |||||
| ribC-bart-F1 | GGTTTGGCTGAGATYATTGA | Bartonella sp. | ribC | 185 | Present study |
| ribC-bart-R1 | TCAAGTGTATGRCGAATAAT | Bartonella sp. | Present study | ||
| Bartonella genus SimpleProbe | GAAGGTGAXITGCAGTTCGTT-Ph | Bartonella sp. | ribC | Present study | |
| B. henselae SimpleProbe | TTTCCTXIACAAGTACCAAA-Ph | B. henselae | ribC | Present study | |
| B. quintana SimpleProbe | TCCTGCAAGTXITCCAACAAAATTCACGCCCTT-Ph | B. quintana | ribC | Present study | |
| β-globin-F | TACACATATTGACCAAATCAGGGTA | Human | β-globin | 165 | Present study |
| β-globin-R | TTTAGAATGGTGCAAAGAGGC | Human | β-globin | Present study | |
| β-globin-probe | Cy5-TACTTTCCCTAATCTCTTTCTTTCAGGGCAAT-BBQ | Human | β-globin | Present study |
Real-time PCR was developed in our research laboratory and has been used by the clinical microbiology laboratory since 2017. This assay was designed to specifically identify B. henselae and B. quintana, which account for >95% of all Bartonella endocarditis cases, and also identify other Bartonella spp. implicated in endocarditis at the genus level. Bartonella genus-specific degenerate primers (Sigma-Aldrich, Rehovot, Israel) were used to amplify a 185-bp fragment of the ribC gene. The resulting PCR product was detected postamplification using 3 SimpleProbe probes for the identification of Bartonella genus, B. henselae, and B. quintana DNA (TIB MOLBIOL, Berlin, Germany, and Dyn Diagnostics, Migdal HaEmeq, Israel) (Table 3). Once hybridized to its target sequence, the SimpleProbe probe emits more fluorescence than the free probe, which is self-quenched. Each probe has a distinctive melting temperature (Tm) at which the probe no longer binds the amplified target, resulting in an instantaneous drop in the fluorescent signal. As a result, changes in fluorescence are based solely on the hybridization status of the probe. Even minor changes in the target DNA sequence, such as single nucleotide polymorphisms, might result in a detectable shift of the melting curve, thus facilitating pathogen identification, without sequencing the amplicon (technical note no. LC 18/2004, Roche Applied Science). PCR conditions were as follows: 5 min at 95°C followed by 50 cycles of 10 s at 95°C, 20 s at 55°C, and 10 s at 72°C. The melting-curve analysis step was performed postamplification and consisted of 60 s at 95°C and 120 s at 37°C, with gradual increase to 80°C and cooling for 30 s. Real-time PCR and analysis were performed using the LightCycler 96 instrument and software version 1.1 (Roche Diagnostics, Basel, Switzerland). For the construction of positive-control plasmids, we cloned the ribC PCR amplicons of B. henselae and B. quintana into high-copy-number plasmids designated pMR-ribC185-Bh and pMR-ribC185-Bq, respectively (PCR cloning kit; Qiagen, Valencia, CA). Thirty, 300, and 3000 copies of both were used as positive controls along with the clinical specimen DNA in a multiplex real-time PCR assay. The analytical sensitivity was determined to be 3 to 30 plasmid copies per reaction (M. Giladi, unpublished data). To identify Bartonella, melting peaks of unknown specimens were compared to those of known standards (positive-control plasmids). Each run of clinical specimens also included a negative no-template control and an internal control gene (human β-globin) to verify template DNA integrity and absence of PCR inhibitors in the specimens. For the purpose of this study, all clinical specimens were subjected to both real-time PCR and conventional PCR except for four patients for whom tissue or DNA samples were not available for real-time PCR (Table 2). All PCR products of both real-time PCR and conventional PCR assays were sequenced to corroborate the identification of the Bartonella sp. To determine its specificity, the real-time PCR assay was performed on DNA extracted from non-Bartonella bacteria known to be associated with culture-negative endocarditis as presented in Table 1.
RESULTS
Serology.
Of 17 patients with B. henselae endocarditis who were tested with EIA using B. henselae-derived antigen, 16 (94%) had anti-B. henselae IgG titers of ≥1:800 and 1 patient had an IgG titer of 1:400 (range, 1:400 to 1:12,800). Of 17 patients with B. quintana endocarditis who were tested using B. quintana-derived antigen, 16 (94%) had anti-B. quintana IgG titers of >1:800 and 1 patient had an IgG titer of 1:200 (range, 1:200 to 1:26,600). One patient with B. koehlerae endocarditis had an anti-B. koehlerae IgG titer of 1:1,600 when tested using the corresponding antigen. Significant cross-reactivity between B. henselae, B. quintana, and B. koehlerae was found in all sera tested (Table 2). Two or more serum specimens taken from the same patient at least 1 month apart were available for 12 patients, 10 with B. henselae and 2 with B. quintana endocarditis (Fig. 1). The median duration to achieve IgG titer decrease of two dilutions, observed in 11 patients, was 5 months (range, 2 to 41 months).
FIG 1.
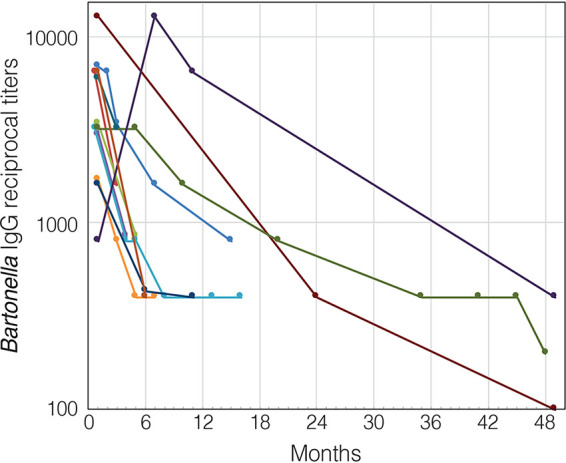
Kinetics of anti-Bartonella IgG antibodies expressed as reciprocal titers in a logarithmic scale in patients with Bartonella endocarditis (n = 12). Only results for patients with 2 or more serum specimens taken at least 1 month apart are presented. Each circle represents the mean enzyme immunoassay optical density of triplicate determinations for 1 patient. Each patient is marked with a different color.
The anti-Bartonella IgG titer distribution among patients with Bartonella endocarditis and patients in the control groups tested to evaluate specificity is presented in Fig. 2. Of 62 patients with blood culture-positive endocarditis, 58 (94%) were considered seronegative, with an anti-Bartonella IgG titer of <1:100, while 4 patients (6%), 3 of them with Enterococcus faecalis endocarditis, had an IgG titer of 1:100. Of 34 patients with serological profile consistent with chronic Q fever, 20 (59%) were seronegative, while 5 patients were seropositive for Bartonella henselae and/or B. quintana with IgG titers of 1:100, 4 patients with 1:200, 4 patients with 1:400, and 1 patient with an anti-Bartonella IgG titer of 1:800. Of note, among the 14 patients with chronic Q fever and cross-reactivity with Bartonella, anti-Bartonella IgG titers were 3 to 7 dilutions lower than the matching anti-Coxiella burnetii phase I IgG titers. Of the patients with Bartonella endocarditis, only one patient (3%), with an anti-Bartonella IgG titer of 1:3,200, tested positive for Q fever, with a phase I IgG titer of 1:200. Of 20 patients with Chlamydia pneumoniae infection, 18 (90%) were seronegative for Bartonella (IgG < 1:100) and 2 were seropositive, with IgG titers of 1:100 and 1:200. Of 10 patients with brucellosis, 9 (90%) were seronegative for Bartonella and 1 had an IgG titer of 1:100.
FIG 2.
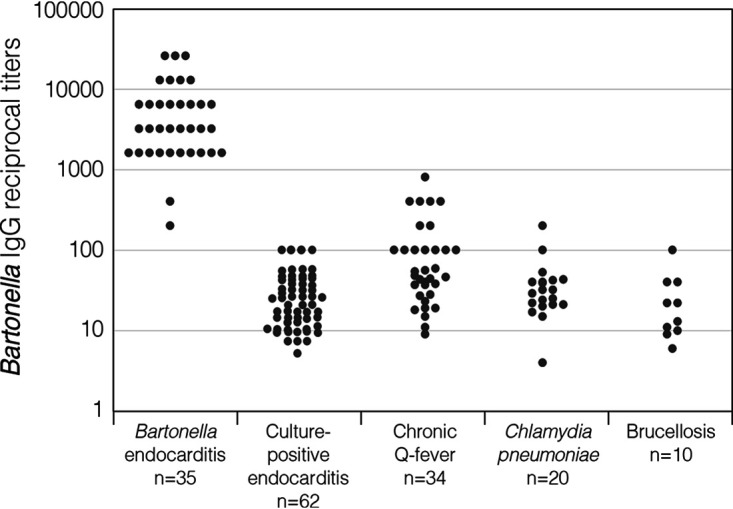
Distribution of anti-Bartonella IgG reciprocal titers, expressed in a logarithmic scale, in patients with Bartonella endocarditis and 4 control groups: patients with culture-positive endocarditis, patients with anti-Coxiella burnetii IgG phase I antibody titers of ≥1:1,600, Chlamydia pneumoniae-seropositive patients, and patients with brucellosis and serum agglutination test values of ≥1:160. Each circle represents the mean enzyme immunoassay optical density of triplicate determinations for 1 patient.
Molecular diagnosis.
We first evaluated the performance of the Bartonella genus-specific probe to identify Bartonella spp. known to cause endocarditis. All species tested were identified, each with a single fluorescent peak, some with a unique Tm (e.g., B. elizabethae, Tm, 43°C, and B. washoensis, Tm, 50°C) and others with overlapping Tms which do not allow interspecies separation (B. henselae, B. quintana, B. koehlerae, and B. vinsonii, Tm, 57 to 58°C) (Fig. 3 and Table 1). DNA sequencing confirmed the species identity.
FIG 3.
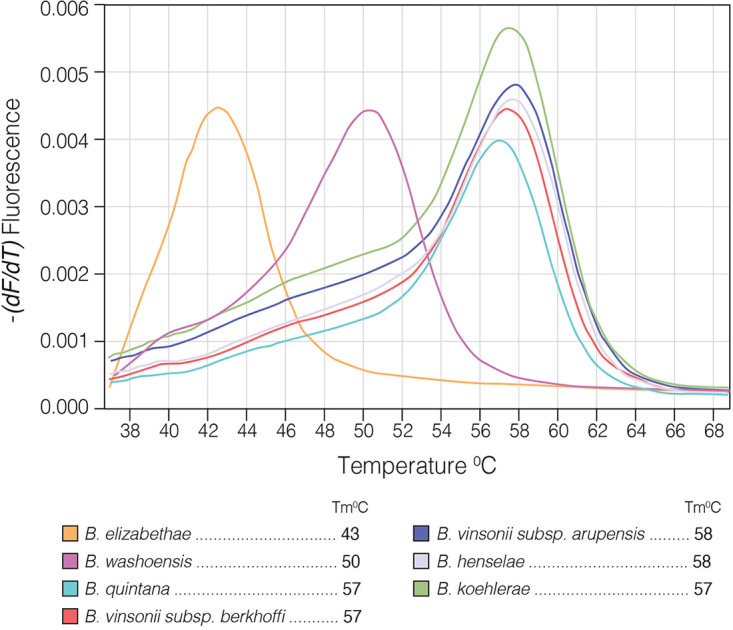
Multiplex real-time PCR performed on 7 Bartonella spp. reported to cause endocarditis in humans, utilizing a Bartonella genus-specific probe. The melting temperature (Tm) was calculated by plotting the negative derivative of fluorescence over temperature (−dF/dT). The Tm value was defined as the peak of the curve. Values were rounded to the nearest integer.
The real-time PCR assay performed on infected cardiac specimens was positive for all 33 tested specimens and was in complete agreement with results of the sequence-based diagnosis of the conventional PCR. The assay specifically identified all cases of B. henselae and B. quintana endocarditis based on the fluorescent pattern and Tm values. Tm-1 (range, 49°C to 51°C) and Tm-3 (68°C to 71°C) were generated by the specific B. henselae and B. quintana probes, respectively. Tm-2 (57°C to 61°C), representing the genus-specific probe, was common to B. henselae, B. quintana, and B. koehlerae (Table 2). Figures 4 and 5 present the identification of B. quintana DNA in infected native aortic valve and B. henselae DNA in infected native mitral valve, respectively, of two patients with culture-negative endocarditis. Specificity of the real-time PCR assay was established when the multiplex assay, including Bartonella genus, B. henselae, and B. quintana probes, was performed using DNA of non-Bartonella culture negative-associated bacteria and resulted in negative fluorescent signals (Table 1).
FIG 4.
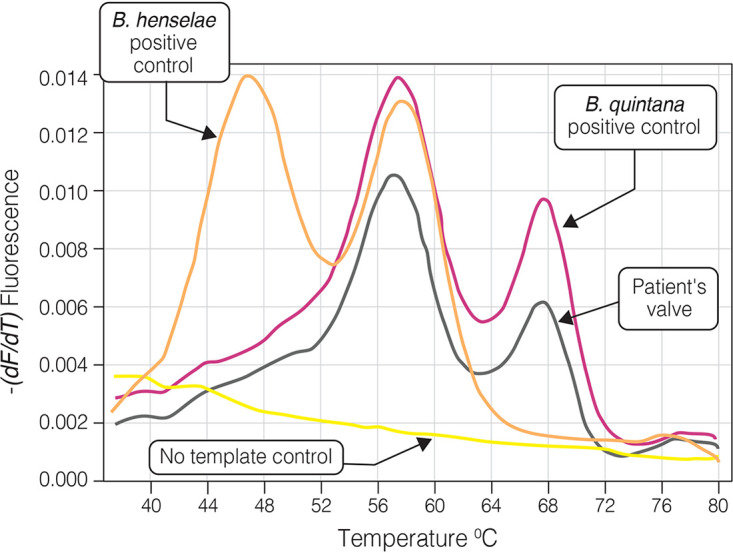
Multiplex real-time PCR using 3 SimpleProbe probes: Bartonella genus-specific, B. henselae-specific, and B. quintana-specific probes. PCR was performed on valvular tissue of a 65-year-old patient with native aortic valve Bartonella quintana endocarditis (patient number 34 [Table 2]). Two fluorescent peaks corresponding to melting temperatures (Tms) of 57.2°C and 67.6°C are the result of detachment of the genus-specific and B. quintana-specific probes from the PCR product, respectively. Plasmids pMR-ribC185-Bq and pMR-ribC185-Bh were used as positive controls for B. quintana and B. henselae, respectively.
FIG 5.
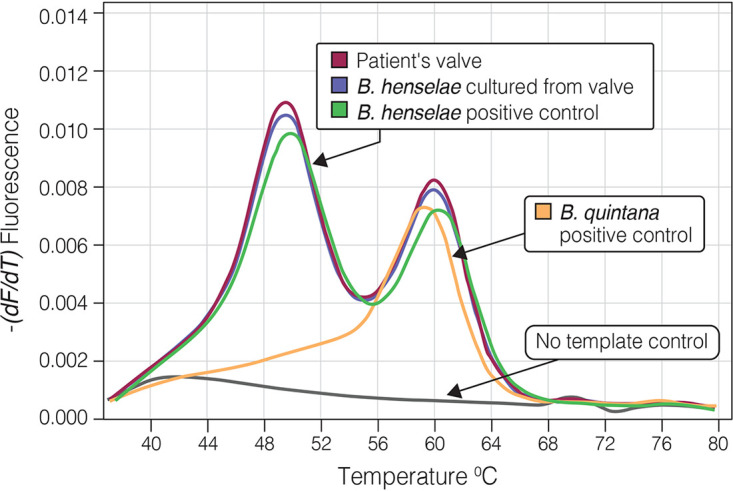
Multiplex real-time PCR using 2 SimpleProbe probes, B. henselae-specific and genus-specific probes. PCR was performed on valvular tissue of a 56-year-old patient with native mitral valve Bartonella henselae endocarditis (patient number 13 [Table 2]). Two fluorescent peaks corresponding to Tms of 49.3°C and 59.8°C are the result of detachment of the B. henselae-specific and genus-specific probes from the PCR product, respectively. Plasmids pMR-ribC185-Bq and pMR-ribC185-Bh were used as positive controls for B. quintana and B. henselae, respectively. The peak corresponding to the B. quintana-specific probe is not seen since the B. quintana-specific probe was not used in this specific assay.
Cultures.
Of the 28 clinical cardiac specimens available for culture, 1 (4%) was positive, growing B. henselae. Two blood cultures were positive, one growing B. henselae and the other B. quintana.
DISCUSSION
This report describes the modalities used in a national reference center for the diagnosis of Bartonella endocarditis. The fact that all Bartonella endocarditis cases in Israel are referred for diagnoses to a single laboratory permitted analysis of the performance of these methods for 37 patients diagnosed with Bartonella endocarditis during a 20-year period. To date, the mainstay of the literature on Bartonella endocarditis, and in particular description of diagnostic methods, stems from series originating in France and one series from the United Kingdom, with limited data reported elsewhere (1, 18, 29–31). In fact, a recent review published in 2019 identified only 13 cases of B. quintana endocarditis reported from North America, although Bartonella spp. are believed to be the most common cause of culture-negative endocarditis in the United States (32, 33).
IFA, either as a “homemade” or a commercial product, is the most commonly used serological method for the diagnosis of Bartonella infections, including Bartonella endocarditis, utilized among others by the national reference center for Q fever and Bartonella infection in France and the Centers for Disease Control and Prevention (CDC) in the United States (1, 34). In comparison with EIA, IFA has several limitations. Preparation of antigen requires cultivation of Bartonella spp. in Vero cells or other cell lines. Attempts to use agar-derived antigens have resulted in autoagglutination of the Bartonella bacilli, which interferes with the IFA (34). In addition, IFAs in general suffer from interobserver variation and are also not suitable for automation or for screening large numbers of specimens. We found that anti-Bartonella IgG titers of ≥1:800 were recorded for 94% of patients with Bartonella endocarditis, while the remaining patients, though PCR positive on cardiac infected tissue, demonstrated lower titers. A previous study from France utilizing an IFA on 106 patients with Bartonella endocarditis also demonstrated that IgG titers of ≥1:800 correlate with endocarditis in 94% of cases (1). Clinicians should be aware, however, that relatively low IgG titers, obtained by either IFA or EIA, may represent rare cases of B. henselae or B. quintana endocarditis but may also be attributable to non-quintana, non-henselae Bartonella spp., as was demonstrated in our B. koehlerae endocarditis patient who had IgG titer of 1:400 when tested using B. henselae antigen, compared with a titer of 1:1,600 when B. koehlerae antigen was incorporated in the EIA. Similarly, an IgG titer of <1:800 was reported for patients with B. alsatica endocarditis who were tested with IFA using either B. henselae or B. quintana antigens (1).
Although, similarly to IFA, our EIA cannot distinguish between endocarditis due to B. henselae and B. quintana as a result of cross-seroreactivity between these two species, the clinical significance of this finding is limited since the medical approaches to B. henselae and B. quintana endocarditis are quite similar (17, 31). More concerning is the cross-reactivity between Bartonella spp. and Coxiella burnetii, since they are important etiologic agents of culture-negative endocarditis with very similar clinical characteristics yet with dissimilar practices for antimicrobial treatment and serological follow-up (17, 18). Of 34 patients with serology consistent with chronic Q fever, we found that 13 patients (38%) showed cross-seroreactivity with Bartonella spp.; however, all had IgG titers of <1:800, except 1 patient (3%) with anti-Coxiella burnetii phase I IgG titer of 1:6,400 who tested positive for Bartonella with an IgG titer of 1:800. Cross-reaction between C. burnetii antibodies and Bartonella antigen have been reported before (17, 35). La Scola and Raoult (17) have found that using IFA, more than 50% of chronic Q fever patients had sera which reacted to a significant level with B. henselae and/or B. quintana. Although cross-adsorption studies and Western blot analysis have been shown to assist identification of the true culprit in cases with cross-reactivity while also increasing the diagnostic sensitivity (1), these assays are not currently available in most clinical microbiology laboratories (1, 17). However, Bartonella-Coxiella cross-seroreactivity infrequently poses a clinical quandary since in the majority of cases, IgG titers against the true pathogen are several dilutions higher than those of the pathogen causing the false-positive serology and thus allowing for the correct diagnosis. To further explore the specificity of our EIA, we tested sera from several control groups. We have also demonstrated low-level cross-reactivity in cases of Chlamydia pneumoniae infections similarly to a previous study using an IFA (19), in cases infected with Brucella melitensis, a pathogen phylogenetically close to Bartonella, and in cases with culture-positive bacterial endocarditis. Cross-reactivity, however, was uncommon in these cases and always in low titers, supporting an anti-Bartonella IgG titer of ≥1:800 as a cutoff for Bartonella endocarditis diagnosis even in the presence of cross-reactivity. Such low-level cross-reactivity does not necessarily indicate antigenic similarity but may also be the result of polyclonal B cell activation, known to occur in endocarditis (27, 36). Of particular interest is a subgroup of patients with E. faecalis endocarditis, since a recent publication demonstrated cross-seroreactivity with this pathogen in Bartonella-infected endocarditis patients tested by Western immunoblotting (28). We have tested serum samples from 31 patients with E. faecalis endocarditis, and although 10% were Bartonella seropositive, none demonstrated high-level seropositivity (>1:100) that might interfere with the interpretation of Bartonella EIA. Further studies are needed to evaluate the added value of Western blotting, which may increase diagnostic sensitivity (at the expense of lower specificity). The potential benefit, however, must be weighed against the increased burden placed on the microbiology laboratory that does not perform Western immunoblotting or cross-adsorption studies on a routine basis.
We also showed that the multiplex real-time PCR assay performed on cardiac tissue accurately identified B. quintana and B. henselae, which are responsible for almost all Bartonella endocarditis cases in humans, without the need for DNA sequencing. The unique melting curves generated by the interaction of the genus-specific and species-specific probes with the PCR product results in 2 distinct Tms, 9 to 11°C apart, which are easily recognized. Melting peaks of clinical specimens were compared to those of known standards (plasmids pMR-ribC185-Bh and pMR-ribC185-Bq). This comparison to standards is essential since Tm is not a constant value and a significant Tm shift may occur due to multiple factors, such as oligonucleotide concentration, cation concentration, particularly free magnesium (which by itself is inversely related to the amount of deoxynucleoside triphosphates [dNTPs], number of probes and primers, or amount and length of the target DNA), and other components of the reaction. We have observed a Tm shift of 1 to 4°C (Tables 1 and 2); however, all probe-generated peaks of a tested specimen and positive controls are shifted in the same direction and magnitude, resulting in a fixed interval between peaks. We demonstrated that the genus-specific probe also identified 5 non-henselae, non-quintana Bartonella spp. known to cause endocarditis. B. alsatica and “Candidatus Bartonella mayotimonensis,” also reported to rarely cause endocarditis, were not tested with our real-time PCR assay; however, due to the use of degenerate primers designated to encompass a large number of Bartonella spp. and the characteristics of the genus-specific probe, we predict that they will be recognized by this probe. To further support this prediction, we tested 21 additional Bartonella spp. isolated from various hosts, including ruminants, rodents, felines, squirrels, a kangaroo, and a bat, from different continents, including North America, Africa, Europe, Asia, and Australia, and all were recognized by the genus-specific probe, resulting in a single peak with variable Tms (Giladi, unpublished). Along these lines, we expect that if a new Bartonella sp. is implicated in endocarditis, it will be recognized by the real-time PCR assay. Contrary to the case with B. quintana and B. henselae, which are accurately identified by specific probes, the identity of other species recognized by the genus probe alone needs to be corroborated by either DNA sequencing or a second PCR to distinguish between members of the same melting group. As non-henselae, non-quintana Bartonella spp. are extremely rare causes of endocarditis, a decision to add additional specific probes for the identification of these species is unlikely to be supported by cost-benefit analysis. However, if future epidemiological studies demonstrate that the incidence of another endocarditis-associated Bartonella sp. is increased, new species-specific probes can be incorporated into this assay. The high specificity of the real-time PCR system is achieved by using Bartonella-specific degenerate primers to amplify part of the ribC gene and 3 Bartonella-specific probes. This was demonstrated by testing a large number of bacterial pathogens known to be associated with culture-negative endocarditis, including members of the HACEK group (a group of Gram-negative bacilli consisting of Haemophilus spp., A. actinomycetemcomitans, Cardiobacterium hominis, Eikenella corrodens, and Kingella spp.), nutritionally variant streptococci (e.g., Abiotrophia spp.), Coxiella burnetii, Legionella pneumophila, and Tropheryma whipplei, none of which produced false-positive results. The high positivity rate of PCR performed on infected cardiac tissue is in agreement with previous studies reporting 93 to 98% positivity rates of PCR performed on valvular specimens in spite of the fact that in one of these studies, 62% of the specimens were obtained from patients receiving antibiotic therapy. To summarize, the current study highlights the SimpleProbe real-time PCR as a sensitive and specific method which is also rapid and high throughput for the detection and differentiation of Bartonella spp. causing endocarditis.
In conclusion, we have demonstrated that EIA coupled with a novel real-time PCR assay can play an important role in the laboratory diagnosis of Bartonella endocarditis and expand the diagnostic arsenal at the disposal of the clinical microbiologist. Since serology remains the major diagnostic tool in the absence of tissue suitable for PCR, recognizing its many pitfalls is essential to avoid incorrect diagnosis.
ACKNOWLEDGMENT
This study was partially funded by a grant from the Israel Innovation Authority, Israel’s Ministry of Economy.
Contributor Information
Michael Giladi, Email: michael.giladi@gmail.com.
Brad Fenwick, University of Tennessee at Knoxville.
REFERENCES
- 1.Edouard S, Nabet C, Lepidi H, Fournier PE, Raoult D. 2015. Bartonella, a common cause of endocarditis: a report on 106 cases and review. J Clin Microbiol 53:824–829. 10.1128/JCM.02827-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fournier PE, Thuny F, Richet H, Lepidi H, Casalta JP, Arzouni JP, Maurin M, Celard M, Mainardi JL, Caus T, Collart F, Habib G, Raoult D. 2010. Comprehensive diagnostic strategy for blood culture-negative endocarditis: a prospective study of 819 new cases. Clin Infect Dis 51:131–140. 10.1086/653675. [DOI] [PubMed] [Google Scholar]
- 3.Houpikian P, Raoult D. 2005. Blood culture-negative endocarditis in a reference center: etiologic diagnosis of 348 cases. Medicine (Baltimore) 84:162–173. 10.1097/01.md.0000165658.82869.17. [DOI] [PubMed] [Google Scholar]
- 4.Daly JS, Worthington MG, Brenner DJ, Moss CW, Hollis DG, Weyant RS, Steigerwalt AG, Weaver RE, Daneshvar MI, O’Connor SP. 1993. Rochalimaea elizabethae sp. nov. isolated from a patient with endocarditis. J Clin Microbiol 31:872–881. 10.1128/JCM.31.4.872-881.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roux V, Eykyn SJ, Wyllie S, Raoult D. 2000. Bartonella vinsonii subsp. berkhoffii as an agent of afebrile blood culture-negative endocarditis in a human. J Clin Microbiol 38:1698–1700. 10.1128/JCM.38.4.1698-1700.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Olarte L, Ampofo K, Thorell EA, Sanderson S, Doby E, Pavia AT, Rosado H, Raoult D, Socolovschi C, Hersh AL. 2012. Bartonella vinsonii endocarditis in an adolescent with congenital heart disease. Pediatr Infect Dis J 31:531–534. 10.1097/INF.0b013e31824ba95a. [DOI] [PubMed] [Google Scholar]
- 7.Fenollar F, Sire S, Wilhelm N, Raoult D. 2005. Bartonella vinsonii subsp. arupensis as an agent of blood culture-negative endocarditis in a human. J Clin Microbiol 43:945–947. 10.1128/JCM.43.2.945-947.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Avidor B, Graidy M, Efrat G, Leibowitz C, Shapira G, Schattner A, Zimhony O, Giladi M. 2004. Bartonella koehlerae, a new cat-associated agent of culture-negative human endocarditis. J Clin Microbiol 42:3462–3468. 10.1128/JCM.42.8.3462-3468.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Raoult D, Roblot F, Rolain JM, Besnier JM, Loulergue J, Bastides F, Choutet P. 2006. First isolation of Bartonella alsatica from a valve of a patient with endocarditis. J Clin Microbiol 44:278–279. 10.1128/JCM.44.1.278-279.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jeanclaude D, Godmer P, Leveiller D, Pouedras P, Fournier PE, Raoult D, Rolain JM. 2009. Bartonella alsatica endocarditis in a French patient in close contact with rabbits. Clin Microbiol Infect 15(Suppl 2):110–111. 10.1111/j.1469-0691.2008.02187.x. [DOI] [PubMed] [Google Scholar]
- 11.Lin EY, Tsigrelis C, Baddour LM, Lepidi H, Rolain JM, Patel R, Raoult D. 2010. Candidatus Bartonella mayotimonensis and endocarditis. Emerg Infect Dis 16:500–503. 10.3201/eid1603.081673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giladi M, Kletter Y, Avidor B, Metzkor-Cotter E, Varon M, Golan Y, Weinberg M, Riklis I, Ephros M, Slater L. 2001. Enzyme immunoassay for the diagnosis of cat-scratch disease defined by polymerase chain reaction. Clin Infect Dis 33:1852–1858. 10.1086/324162. [DOI] [PubMed] [Google Scholar]
- 13.Tasher D, Raucher-Sternfeld A, Tamir A, Giladi M, Somekh E. 2017. Bartonella quintana, an unrecognized cause of infective endocarditis in children in Ethiopia. Emerg Infect Dis 23:1246–1252. 10.3201/eid2308.161037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Metzkor-Cotter E, Kletter Y, Avidor B, Varon M, Golan Y, Ephros M, Giladi M. 2003. Long-term serological analysis and clinical follow-up of patients with cat scratch disease. Clin Infect Dis 37:1149–1154. 10.1086/378738. [DOI] [PubMed] [Google Scholar]
- 15.Habot-Wilner Z, Trivizki O, Goldstein M, Kesler A, Shulman S, Horowitz J, Amer R, David R, Ben-Arie-Weintrob Y, Bakshi E, Almog Y, Sartani G, Vishnevskia-Dai V, Kramer M, Bar A, Kehat R, Ephros M, Giladi M. 2018. Cat-scratch disease: ocular manifestations and treatment outcome. Acta Ophthalmol 96:e524–e532. 10.1111/aos.13684. [DOI] [PubMed] [Google Scholar]
- 16.Jost M, Latz A, Ballhorn W, Kempf VAJ. 2018. Development of a specific and sensitive enzyme-linked immunosorbent assay as an in vitro diagnostic tool for detection of Bartonella henselae antibodies in human serum. J Clin Microbiol 56:e01329-18. 10.1128/JCM.01329-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.La Scola B, Raoult D. 1996. Serological cross-reactions between Bartonella quintana, Bartonella henselae, and Coxiella burnetii. J Clin Microbiol 34:2270–2274. 10.1128/JCM.34.9.2270-2274.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fournier PE, Mainardi JL, Raoult D. 2002. Value of microimmunofluorescence for diagnosis and follow-up of Bartonella endocarditis. Clin Diagn Lab Immunol 9:795–801. 10.1128/cdli.9.4.795-801.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Drancourt M, Mainardi JL, Brouqui P, Vandenesch F, Carta A, Lehnert F, Etienne J, Goldstein F, Acar J, Raoult D. 1995. Bartonella (Rochalimaea) quintana endocarditis in three homeless men. N Engl J Med 332:419–423. 10.1056/NEJM199502163320702. [DOI] [PubMed] [Google Scholar]
- 20.Avidor B, Kletter Y, Abulafia S, Golan Y, Ephros M, Giladi M. 1997. Molecular diagnosis of cat scratch disease: a two-step approach. J Clin Microbiol 35:1924–1930. 10.1128/JCM.35.8.1924-1930.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Avidor B, Varon M, Marmor S, Lifschitz-Mercer B, Kletter Y, Ephros M, Giladi M. 2001. DNA amplification for the diagnosis of cat-scratch disease in small-quantity clinical specimens. Am J Clin Pathol 115:900–909. 10.1309/Y5WN-8DFD-WLVT-KKAD. [DOI] [PubMed] [Google Scholar]
- 22.Giladi M, Maman E, Paran D, Bickels J, Comaneshter D, Avidor B, Varon-Graidy M, Ephros M, Wientroub S. 2005. Cat-scratch disease-associated arthropathy. Arthritis Rheum 52:3611–3617. 10.1002/art.21411. [DOI] [PubMed] [Google Scholar]
- 23.Landes M, Maor Y, Mercer D, Habot-Wilner Z, Bilavsky E, Chazan B, Cohen R, Glikman D, Strahilevitz J, Katzir M, Litachevsky V, Melamed R, Guri A, Shaked H, Perets O, Wiener-Well Y, Stren A, Paul M, Zimhony O, Srugo I, Rahav G, Bishara J, Kuperman AA, Ben-Ami R, Ephros M, Giladi M. 23 November 2019. Cat scratch disease presenting as fever of unknown origin is a unique clinical syndrome. Clin Infect Dis 10.1093/cid/ciz1137. [DOI] [PubMed] [Google Scholar]
- 24.Maman E, Bickels J, Ephros M, Paran D, Comaneshter D, Metzkor-Cotter E, Avidor B, Varon-Graidy M, Wientroub S, Giladi M. 2007. Musculoskeletal manifestations of cat scratch disease. Clin Infect Dis 45:1535–1540. 10.1086/523587. [DOI] [PubMed] [Google Scholar]
- 25.Li JS, Sexton DJ, Mick N, Nettles R, Fowler VG, Jr, Ryan T, Bashore T, Corey GR. 2000. Proposed modifications to the Duke criteria for the diagnosis of infective endocarditis. Clin Infect Dis 30:633–638. 10.1086/313753. [DOI] [PubMed] [Google Scholar]
- 26.Durack DT, Lukes AS, Bright DK. 1994. New criteria for diagnosis of infective endocarditis: utilization of specific echocardiographic findings. Duke Endocarditis Service. Am J Med 96:200–209. 10.1016/0002-9343(94)90143-0. [DOI] [PubMed] [Google Scholar]
- 27.Holland TL, Bayer AS, Fowler VG. 2020. Endocarditis and intravascular infections, p 1068–1108. In Bennett JE, Dolin R, Blaser MJ (ed), Mandell, Douglas, and Bennett’s principles and practice of infectious diseases, 9th ed, vol 1. Elsevier, Inc, Philadelphia, PA. [Google Scholar]
- 28.Arregle F, Gouriet F, Amphoux B, Edouard S, Chaudet H, Casalta JP, Habib G, Fournier PE, Raoult D. 2019. Western immunoblotting for the diagnosis of Enterococcus faecalis and Streptococcus gallolyticus infective endocarditis. Front Cell Infect Microbiol 9:314. 10.3389/fcimb.2019.00314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fournier PE, Lelievre H, Eykyn SJ, Mainardi JL, Marrie TJ, Bruneel F, Roure C, Nash J, Clave D, James E, Benoit-Lemercier C, Deforges L, Tissot-Dupont H, Raoult D. 2001. Epidemiologic and clinical characteristics of Bartonella quintana and Bartonella henselae endocarditis: a study of 48 patients. Medicine (Baltimore) 80:245–251. 10.1097/00005792-200107000-00003. [DOI] [PubMed] [Google Scholar]
- 30.Raoult D, Fournier PE, Drancourt M, Marrie TJ, Etienne J, Cosserat J, Cacoub P, Poinsignon Y, Leclercq P, Sefton AM. 1996. Diagnosis of 22 new cases of Bartonella endocarditis. Ann Intern Med 125:646–652. 10.7326/0003-4819-125-8-199610150-00004. [DOI] [PubMed] [Google Scholar]
- 31.Chaloner GL, Harrison TG, Birtles RJ. 2013. Bartonella species as a cause of infective endocarditis in the UK. Epidemiol Infect 141:841–846. 10.1017/S0950268812001185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lam JC, Fonseca K, Pabbaraju K, Meatherall BL. 2019. Case report: Bartonella quintana endocarditis outside of the Europe-African gradient: comprehensive review of cases within North America. Am J Trop Med Hyg 100:1125–1129. 10.4269/ajtmh.18-0929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chambers HF, Bayer AS. 2020. Native-valve infective endocarditis. N Engl J Med 383:567–576. 10.1056/NEJMcp2000400. [DOI] [PubMed] [Google Scholar]
- 34.Regnery RL, Olson JG, Perkins BA, Bibb W. 1992. Serological response to “Rochalimaea henselae” antigen in suspected cat-scratch disease. Lancet 339:1443–1445. 10.1016/0140-6736(92)92032-b. [DOI] [PubMed] [Google Scholar]
- 35.Cooper MD, Hollingdale MR, Vinson JW, Costa J. 1976. A passive hemagglutination test for diagnosis of trench fever due to Rochalimaea quintana. J Infect Dis 134:605–609. 10.1093/infdis/134.6.605. [DOI] [PubMed] [Google Scholar]
- 36.Kaell AT, Volkman DJ, Gorevic PD, Dattwyler RJ. 1990. Positive Lyme serology in subacute bacterial endocarditis. A study of four patients. JAMA 264:2916–2918. 10.1001/jama.1990.03450220082027. [DOI] [PubMed] [Google Scholar]
- 37.Norman AF, Regnery R, Jameson P, Greene C, Krause DC. 1995. Differentiation of Bartonella-like isolates at the species level by PCR-restriction fragment length polymorphism in the citrate synthase gene. J Clin Microbiol 33:1797–1803. 10.1128/JCM.33.7.1797-1803.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bai Y, Malania L, Alvarez Castillo D, Moran D, Boonmar S, Chanlun A, Suksawat F, Maruyama S, Knobel D, Kosoy M. 2013. Global distribution of Bartonella infections in domestic bovine and characterization of Bartonella bovis strains using multi-locus sequence typing. PLoS One 8:e80894. 10.1371/journal.pone.0080894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johnson G, Ayers M, McClure SCC, Richardson SE, Tellier R. 2003. Detection and identification of Bartonella species pathogenic for humans by PCR amplification targeting the riboflavin synthase gene (ribC). J Clin Microbiol 41:1069–1072. 10.1128/jcm.41.3.1069-1072.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zeaiter Z, Fournier P-E, Raoult D. 2002. Genomic variation of Bartonella henselae strains detected in lymph nodes of patients with cat scratch disease. J Clin Microbiol 40:1023–1030. 10.1128/jcm.40.3.1023-1030.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harmsen D, Dostal S, Roth A, Niemann S, Rothganger J, Sammeth M, Albert J, Frosch M, Richter E. 2003. RIDOM: comprehensive and public sequence database for identification of Mycobacterium species. BMC Infect Dis 3:26. 10.1186/1471-2334-3-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alsmark CM, Frank AC, Karlberg EO, Legault B-A, Ardell DH, Canbäck B, Eriksson A-S, Näslund AK, Handley SA, Huvet M, La Scola B, Holmberg M, Andersson SGE. 2004. The louse-borne human pathogen Bartonella quintana is a genomic derivative of the zoonotic agent Bartonella henselae. Proc Natl Acad Sci U S A 101:9716–9721. 10.1073/pnas.0305659101. [DOI] [PMC free article] [PubMed] [Google Scholar]


