LETTER
The global spread of extended-spectrum β-lactamase (ESBL) is a matter of great concern given the high morbidity and mortality associated with invasive infections. The most frequent ESBLs are of the CTX-M type, followed by the TEM and SHV variants, being reported in all Enterobacterales species and mostly among Klebsiella pneumoniae and Escherichia coli species (1, 2).
Several techniques have been developed to rapidly identify ESBL producers, such as molecular techniques based on PCR amplification and/or sequencing, biochemical identification of ESBL activity, and immunological detection of ESBLs (3–7). Among them, the NG-Test CTX-M MULTI assay (NG Biotech, Guipry, France) can rapidly detect CTX-M-type ESBLs from bacterial culture and requires minimal hands-on time and no specific equipment. This test was evaluated against 172 CTX-M producers and showed excellent sensitivity and specificity for detection of CTX-M-like enzymes (6, 8). Of note, it was previously evaluated using a collection of 23 Klebsiella oxytoca strains, among which four produced an acquired CTX-M-type ESBL, and the test showed 100% specificity and 100% sensitivity (6).
By evaluating the performance of rapid tests for screening carriers of acquired ESBL producers (6), we identified a single K. oxytoca isolate (strain N1061) that was positive with the NG-Test CTX-M MULTI but remained negative for blaCTX-M and other ESBL genes upon screening by PCR. Therefore, we aimed to determine the exact mechanisms leading to this discrepant result.
Whole-genome sequencing (WGS) of K. oxytoca N1061 was performed, and data analysis identified two β-lactamase genes, encoding the broad-spectrum β-lactamases TEM-1 and OXY-1-2. The naturally produced OXY-type β-lactamases from K. oxytoca mostly belonging to two main types (OXY-1 and OXY-2 with 88.66% amino acid identity) that display a peculiar ESBL hydrolysis spectrum when overexpressed, usually evidenced by an aztreonam-resistance phenotype (9).
With the purpose to decipher whether the OXY β-lactamase production might indeed lead to the false-positive result of the NG-Test CTX-M MULTI, this test was evaluated using 20 non-CTX-M-producing K. oxytoca clinical strains (Table 1). The absence of blaCTX-M genes was assessed by negative multiplex PCRs (10). The NG-Test CTX-M MULTI assay gave positive results for 6 out of those 22 strains (Table 1).
TABLE 1.
Evaluation of NG-Test CTX-M MULTI and MIC results of clinical K. oxytoca isolates and OXY-producing E. coli recombinant strains
| Strain | OXY variant | NG-Test CTX-M MULTIc | Minimal inhibitory concn (MIC) to:d |
||||||
|---|---|---|---|---|---|---|---|---|---|
| PIP | PTZ | CEF | CTX | CAZ | ATM | IMP | |||
| N85 | OXY-1-2 | + | >256 | >256 | >256 | 4 | 1 | >256 | 0.25 |
| N988 | OXY-1-3 | + | >256 | >256 | 256 | 0.5 | 0.25 | 16 | 0.25 |
| N1061 | OXY-1-3 | + | >256 | >256 | >256 | 1 | 1 | 32 | 0.25 |
| S175 | OXY-1-3 | + | >256 | >256 | 256 | 0.5 | 0.25 | 64 | 0.25 |
| N629 | OXY-1-4 | + | >256 | >256 | 128 | 0.25 | 0.25 | 32 | 0.25 |
| S148 | OXY-1-9 | + | >256 | >256 | 128 | 0.25 | 0.25 | 16 | 0.125 |
| N109 | OXY-2-1 | − | >256 | >256 | >256 | 1 | 0.25 | 64 | 0.125 |
| N828 | OXY-2-2 | − | >256 | >256 | >256 | 1 | 0.25 | 32 | 0.25 |
| N29 | OXY-2-3 | − | >256 | >256 | >256 | 4 | 1 | >256 | 0.25 |
| N496 | OXY-2-4 | − | >256 | >256 | >256 | >32 | 2 | >256 | 0.25 |
| R1057a | OXY-2-5 | − | >256 | >256 | >256 | >32 | >256 | 128 | 0.25 |
| N288 | OXY-2-6 | − | >256 | >256 | >256 | 1 | 0.5 | 64 | 0.25 |
| N683 | OXY-2-6 | − | >256 | >256 | >256 | 1 | 0.5 | 64 | 0.25 |
| N713 | OXY-2-6 | − | >256 | >256 | >256 | >32 | 1 | >256 | 0.5 |
| N794 | OXY-2-6 | − | >256 | >256 | >256 | 0.5 | 0.25 | 16 | 0.125 |
| R225b | OXY-2-6 | − | >256 | >256 | >256 | >32 | 64 | 64 | 0.5 |
| N94 | OXY-2-8 | − | >256 | >256 | >256 | 4 | 1 | >256 | 0.125 |
| N202 | OXY-2-8 | − | >256 | >256 | 256 | 0.5 | 0.25 | 16 | 0.125 |
| N630 | OXY-2-8 | − | >256 | >256 | >256 | 1 | 0.25 | 32 | 0.25 |
| S10 | OXY-2-8 | − | 128 | 16 | 64 | 0.125 | 0.5 | 1 | 0.125 |
| N761 | OXY-2-12 | − | >256 | >256 | >256 | 1 | 0.25 | 32 | 0.125 |
| R1056a | OXY-2-14 | − | >256 | >256 | >256 | >32 | 4 | >256 | 0.5 |
| OXY-1 T10 | OXY-1-2 | + | >256 | >256 | 256 | 2 | 0.5 | >256 | 0.25 |
| OXY-2 T10 | OXY-2-1 | − | >256 | 32 | >256 | 0.25 | 0.25 | 16 | 0.125 |
| OXY-1-mut T10 (KKS → NAA) | OXY-mut | − | >256 | >256 | >256 | 0.5 | 0.5 | 256 | 0.25 |
ESBL phenotype.
AmpC coproducer.
(+), positive result; (−), negative result.
PIP, piperacillin; PTZ, piperacillin-tazobactam; CEF, cephalotin; CTX, cefotaxime; CAZ, ceftazidime; ATM, aztreonam; IMP, imipenem.
Then, PCR amplification followed by sequencing was performed to amplify the two main OXY-type β-lactamase genes, using two primer pairs (Table S1 in the supplemental material). Hence, OXY-1-like-variants were identified among six strains giving a positive NG-Test CTX-M MULTI result, and OXY-2-like variants were identified in 16 strains that gave a negative NG-CTX-M MULTI result (Table 1). All strains producing an OXY-1-like β-lactamase, differing from one to four amino acids, gave a positive result with the NG-Test, whereas all OXY-2-like-producing strains remained negative (Table 1).
To further investigate this phenomenon, cloning of representative blaOXY-1 and blaOXY-2-type genes using corresponding amplicons was performed using the pCR-Blunt II-TOPO recipient vector (Fisher Scientific AG, Reinarch, Switzerland), followed by the transformation of recombinant plasmids in the same Escherichia coli TOP10 background. OXY-1-2- and OXY-2-1-producing E. coli recombinant strains showed positivity and negativity using the NG-Test CTX-M MULTI assay, respectively (Fig. S1). These data suggest that OXY-1-type amino acid sequences cross-react with the monoclonal antibodies included in the test.
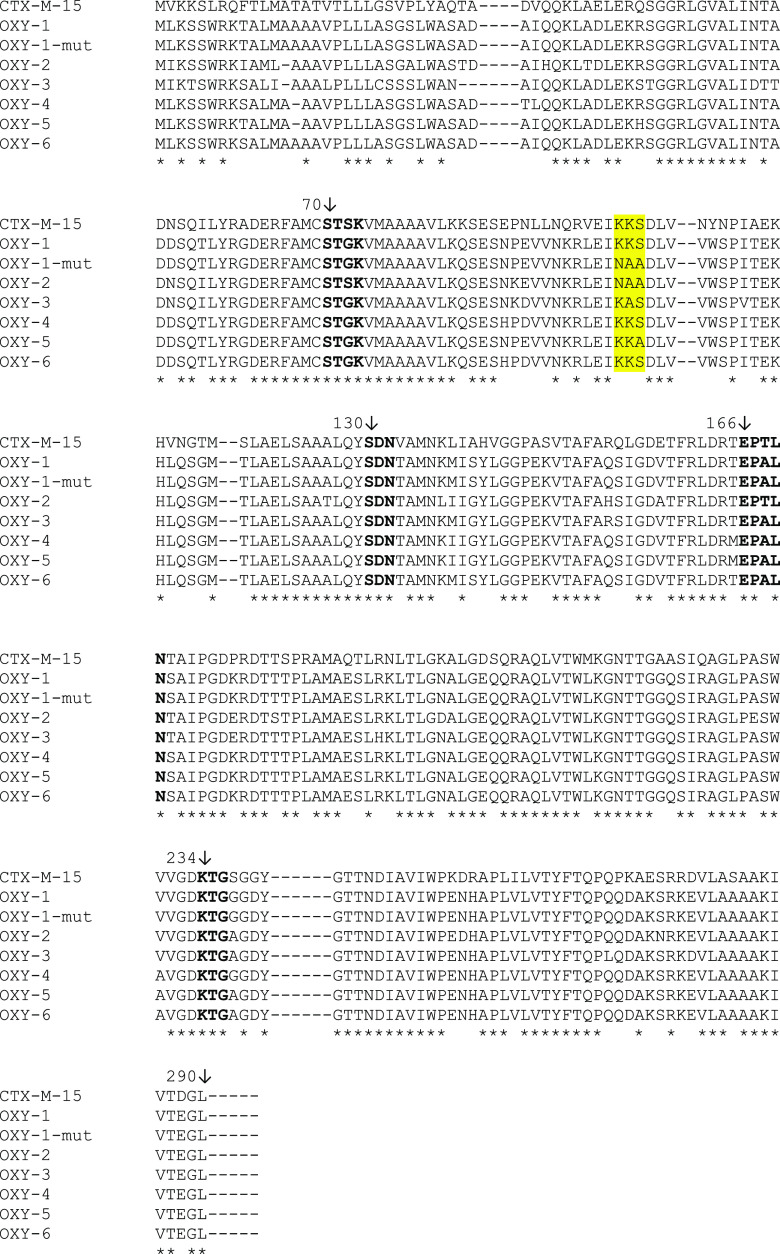
A comparative analysis of the OXY-1, OXY-2, and CTX-M amino acid sequences was performed in silico by Clusta1W (http://www.ebi.ac.uk/clustalw) (Fig. 1). Three consecutive residues (Lys-Lys-Ser [KKS], positions Ambler 101 to 103) of the OXY-1 protein sequence were also identified in the CTX-M-15 sequence, but not in that of OXY-2 (Asn-Ala-Ala [NAA]). Therefore, we hypothesized that residues of the KKS triplet might be targets of at least one of the monoclonal antibodies of the NG-Test CTX-M MULTI assay, since it was located at the external part of the folded protein (Fig. S2).
FIG 1.
Alignment of CTX-M-15, OXY-variants, and OXY-1-mut amino acid sequences according to Ambler’s numbering scheme. The selected residues of OXY protein sequences that might be a potential target of the monoclonal antibodies are indicated in yellow. Asterisks indicate the identical residues in the β-lactamases. Boldface residues are the highly conserved residues involved in the catalytic site of the class A β-lactamases.
In order to further explore this hypothesis, site-direct mutagenesis was performed to modify the KKS residues of OXY-1-2 β-lactamase using the Q5 site-directed mutagenesis kit (New England BioLabs) with primers (Table S1) generated by the NEBaseChanger online tool (http://nebasechanger.neb.com/) and pTOPO-OXY-1-2 recombinant plasmid as the template. These residues were replaced with the NAA residues present in the OXY-2-1 protein sequence (Fig. 1). A negative NG-Test CTX-M MULTI result was obtained with the mutated OXY-1-producing E. coli recombinant strain, confirming our initial hypothesis (Table 1 and Fig. S1).
Six OXY-type enzymes have been reported in K. oxytoca since the discovery of OXY-1 (11, 12) (Fig. 1). The OXY-1, OXY-4, and OXY-6 β-lactamases contain the KKS residues at positions 101 to 103. In contrast, OXY-3 and OXY-5 harbor Lys-Ala-Ser (KAS) and Lys-Lys-Ala (KKA) residues, respectively. Therefore, it is likely that all K. oxytoca strains, except those producing OXY-2-type β-lactamases, might also lead to a positive NG-Test (Fig. 1).
Our results show that OXY-1-type β-lactamases cross-react with monoclonal antibodies contained in the NG-Test CTX-M MULTI assay, giving rise to false-positive results. It remains surprising that none of the 19 K. oxytoca strains that did not produce a CTX-M-type β-lactamase and were tested in a previous study did not provide a false-positive result (6). It might be possible that all those K. oxytoca produced only an OXY-2-type β-lactamase, since sequences of the corresponding genes were not provided.
By investigating an international collection of K. oxytoca isolates, a higher incidence of OXY-2-type β-lactamases (42 isolates, 61.8% of the total) was observed compared to OXY-1-type β-lactamases (17 isolates, 25% of the total) (11). Results from our study, together with the higher prevalence of OXY-2-like versus OXY-1-like enzymes, suggest an overall predominance of OXY-2 variants over OXY-1 variants (9).
Considering that immunological detection of CTX-M-type ESBLs is now increasingly used in routine laboratories (at least in Europe, where those tests are available), and that the NG-Test CTX-M MULTI assay is actually claimed to exhibit an excellent sensitivity and specificity for detection of CTX-M-like ESBLs, it is important to highlight that false-positive results may also be obtained. Such false positivity may occur with K. oxytoca, leading to inappropriate diagnostic results and consequently inappropriate antibiotic escalation for treating K. oxytoca infections. Furthemore, false-positive results might lead to unnecessary isolation of patients, with natural-gut carriers of K. oxytoca being falsely flagged as ESBL-producer carriers.
ACKNOWLEDGMENTS
This work has been funded by the by the Swiss National Reference center for Emerging Antibiotic Resistance (NARA), by the Immunocolitest Project (ANAES, Ecobio2, France), and by the SNF grant (Emerging Antibiotic Resistance 2020–2023, 310030_188801, Switzerland).
Footnotes
Supplemental material is available online only.
Contributor Information
Patrice Nordmann, Email: patrice.nordmann@unifr.ch.
Patricia J. Simner, Johns Hopkins
REFERENCES
- 1.Bevan ER, Jones AM, Hawkey PM. 2017. Global epidemiology of CTX-M β-lactamases: temporal and geographical shifts in genotype. J Antimicrob Chemother 72:2145–2155. 10.1093/jac/dkx146. [DOI] [PubMed] [Google Scholar]
- 2.Rossolini GM, D'Andrea MM, Mugnaioli C. 2008. The spread of CTX-M-type extended-spectrum β-lactamases. Clin Microbiol Infect 14:33–41. 10.1111/j.1469-0691.2007.01867.x. [DOI] [PubMed] [Google Scholar]
- 3.Naas T, Cuzon G, Truong H, Bernabeu S, Nordmann P. 2010. Evaluation of a DNA microarray, the check-points ESBL/KPC array, for rapid detection of TEM, SHV, and CTX-M extended-spectrum β-lactamases and KPC carbapenemases. Antimicrob Agents Chemother 54:3086–3092. 10.1128/AAC.01298-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nordmann P, Dortet L, Poirel L. 2012. Rapid detection of extended-spectrum-β-lactamase-producing Enterobacteriaceae. J Clin Microbiol 50:3016–3022. 10.1128/JCM.00859-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Demord A, Poirel L, D'Emidio F, Pomponio S, Nordmann P. 2020. Rapid ESBL NP Test for rapid detection of expanded-spectrum β lactamase producers in Enterobacterales. Microb Drug Resist 10.1089/mdr.2020.0391. [DOI] [PubMed] [Google Scholar]
- 6.Blanc DS, Poncet F, Grandbastien B, Greub G, Senn L, Nordmann P. 2021. Evaluation of the performance of rapid tests for screening carriers of acquired ESBL-producing Enterobacterales and their impact on the turnaround time. J Hosp Infect 108:19–24. 10.1016/j.jhin.2020.10.013. [DOI] [PubMed] [Google Scholar]
- 7.Bernabeu S, Ratnam KC, Boutal H, Gonzalez C, Vogel A, Devilliers K, Plaisance M, Oueslati S, Malhotra-Kumar S, Dortet L, Fortineau N, Simon S, Volland H, Naas T. 2020. A lateral flow immunoassay for the rapid identification of CTX-M-producing Enterobacterales from culture plates and positive blood cultures. Diagnostics (Basel) 10:764. 10.3390/diagnostics10100764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bianco G, Boattini M, Iannaccone M, Cavallo R, Costa C. 2020. Evaluation of the NG-Test CTX-M MULTI immunochromatographic assay for the rapid detection of CTX-M extended-spectrum-β-lactamase producers from positive blood cultures. J Hosp Infect 105:341–343. 10.1016/j.jhin.2020.02.009. [DOI] [PubMed] [Google Scholar]
- 9.Naas T, Oueslati S, Bonnin RA, Dabos ML, Zavala A, Dortet L, Retailleau P, Iorga BI. 2017. Beta-lactamase database (BLDB)—structure and function. J Enzyme Inhib Med Chem 32:917–919. 10.1080/14756366.2017.1344235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Woodford N, Fagan EJ, Ellington MJ. 2006. Multiplex PCR for rapid detection of genes encoding CTX-M extended-spectrum β-lactamases. J Antimicrob Chemother 57:154–155. 10.1093/jac/dki412. [DOI] [PubMed] [Google Scholar]
- 11.Fevre C, Jbel M, Passet V, Weill FX, Grimont PA, Brisse S. 2005. Six groups of the OXY β-Lactamase evolved over millions of years in Klebsiella oxytoca. Antimicrob Agents Chemother 49:3453–3462. 10.1128/AAC.49.8.3453-3462.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Izdebski R, Fiett J, Urbanowicz P, Baraniak A, Derde LP, Bonten MJ, Carmeli Y, Goossens H, Hryniewicz W, Brun-Buisson C, Brisse S, Gniadkowski M,. MOSAR WP2, WP3 and WP5 Study Groups. 2015. Phylogenetic lineages, clones and β-lactamases in an international collection of Klebsiella oxytoca isolates non-susceptible to expanded-spectrum cephalosporins. J Antimicrob Chemother 70:3230–3237. 10.1093/jac/dkv273. [DOI] [PubMed] [Google Scholar]