Abstract
The SARS-CoV-2 pandemic is a worldwide public health emergency. Despite the beginning of a vaccination campaign, the search for new drugs to appropriately treat COVID-19 patients remains a priority. Drug repurposing represents a faster and cheaper method than de novo drug discovery. In this study, we examined three different network-based approaches to identify potentially repurposable drugs to treat COVID-19. We analyzed transcriptomic data from whole blood cells of patients with COVID-19 and 21 other related conditions, as compared with those of healthy subjects. In addition to conventionally used drugs (e.g., anticoagulants, antihistaminics, anti-TNFα antibodies, corticosteroids), unconventional candidate compounds, such as SCN5A inhibitors and drugs active in the central nervous system, were identified. Clinical judgment and validation through clinical trials are always mandatory before use of the identified drugs in a clinical setting.
Keywords: COVID-19, Drug repurposing, Corticosteroid, Heparin, Inflammatory bowel disease, Septic shock
Graphical Abstract
1. Introduction
Drug repurposing consists of the use of an existing active pharmaceutical ingredient already on the market for a different indication [1]. This approach offers several advantages compared with the development of a new drug, including a faster and cheaper process due to consolidated knowledge regarding the drug’s safety and toxicity and higher success rates in introducing the drug to the market since it has already been tested in clinical trials [1]. The pharmacological base of drug repurposing relies on the fact that some diseases share common biological targets and that one drug may have several targets and thus may be able to treat different diseases [1]. In this framework, computational approaches could offer valuable help in identifying potential candidates for systematic drug repurposing. Within this context, network medicine, which applies tools and concepts from network theory to elucidate the relation between molecular level perturbations and phenotypic disease manifestations, represents a highly promising approach [2], [3], [4], [5], [6], [7]. Network medicine builds on the hypothesis that the human interactome (i.e., the network of all physical and functional molecular interactions within the cell) can be interpreted as a map and that diseases are local perturbations [8]. However, the molecular determinants of a given disease are not randomly scattered, but tend to be co-localized and agglomerated in specific regions of the interactome, generating a disease module [3], [9]. Network-based approaches allow the identification of the specific interactome neighborhood that is perturbed in a specific disease, thus unveiling disease-disease relationships and guiding the search for therapeutic targets [10], [11], [12], [13], [14], [15], [16], [17], [18], [19]. This approach could also be used to identify repurposable drugs for human diseases, including COVID-19 [20], [21], [22], [23], [24], [25].
COVID-19 has quickly been recognized as a worldwide public health emergency. To date, the World Health Organization (WHO) reports more than 170,000,000 confirmed cases of COVID-19 globally and more than three million deaths (World Health Organization, https://covid19.who.int/).
Although a huge vaccination campaign is in progress worldwide, novel emerging variants limit the efficacy of the campaign and indicate that patient management still remains a priority from a clinical perspective. As such, in addition to developing novel therapies targeting viral infection, drug repurposing represents a successful strategy to improve the identification of effective approaches.
Many computational tools for drug repurposing in COVID-19 patients have been developed and most are based on three-dimensional analysis of the drug structure in relation to the viral and/or host targets and their binding affinities and interactions [26]. Among identified repurposable drugs, some target viral proteins, including antiviral drugs that inhibit viral RNA polymerase (e.g., favipiravir, remdesivir) or viral protease (e.g., lopinavir, prulifloxacin, tegobuvir, bictegravir, nelfinavir, and darunavir) [26]. Other drugs act on human cells and can block virus entry by several mechanisms, including inhibiting TMPRSS2 and other cell-surface proteases involved in SARS‑CoV-2 activation (e.g., camostat mesylate and bromhexine), blocking clathrin-mediated endocytosis (e.g. chlorpromazine, baricitinib, and ruxolitinib), or preventing endocytosis by increasing endosomal pH (e.g., chloroquine and hydroxychloroquine). Although in vitro studies showed controversial results, these drugs have advanced to clinical trials either alone or in combination [27].
In parallel with targeting virus replication and cell entry, it is becoming evident that the host immune response plays a pivotal role in disease evolution. It has been reported that patients with severe COVID-19 disease present, in the early phases, hyperactivation of the innate immune response with cytokine storm resulting in a massive inflammatory response that later turns toward massive chronic basal inflammation characterized by a refractory immune state [28]. However, inappropriate adaptive immune response seems to play a crucial role in the late phase of the disease [29], [30], which is probably linked to immune checkpoint activation and immune system exhaustion [29]. This massive immune response has paved the way for testing several immunomodulatory agents in parallel with antiviral drugs [28], [31].
Several immunomodulatory and anti-inflammatory agents have been tested to control cytokine storm. Tocilizumab, a monoclonal antibody against IL-6 receptors normally used for the treatment of diseases such as rheumatoid arthritis, was promising at first, though subsequent clinical trials did not provide unequivocal results on the benefit of tocilizumab in COVID-19 patients [32]. Corticosteroids appear to be effective in the treatment of COVID-19 patients, and many trials have confirmed that dexamethasone may be used for hospitalized subjects with severe SARS-CoV-2 infection. Other drugs able to control inflammation, such as baricitinib, ruxolitinib, and eculizumab, are currently under clinical evaluation. Some anticoagulant and antiplatelet drugs have also been suggested to be effective in the treatment of COVID-19 patients. In particular, heparin was found to limit hypercoagulability in COVID-19 patients, exert anti-inflammatory effects, and reduce mortality in hospitalized patients. Several clinical trials are currently evaluating heparin treatment efficacy in hospitalized patients with COVID-19 [33].
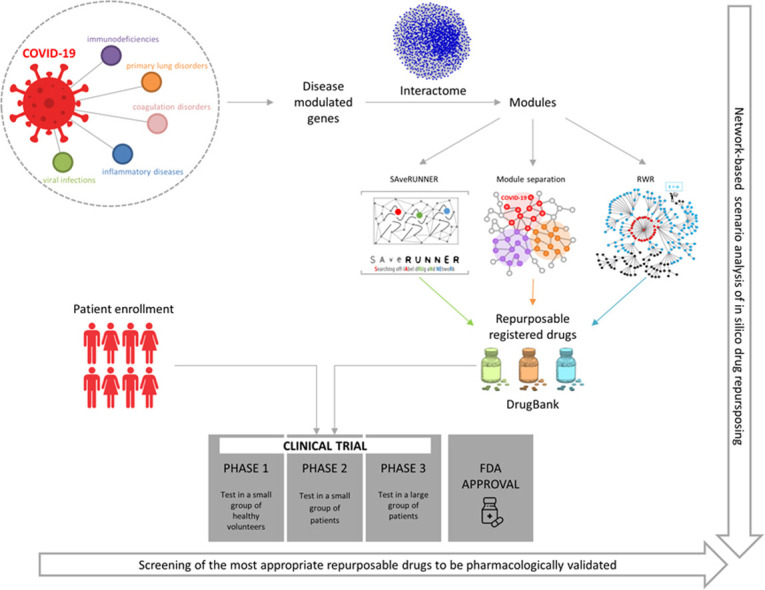
Despite these findings, robust clinical evidence is currently only available for a very limited number of drugs. Therefore, the aim of this paper was to identify drugs that could be repurposed for the treatment of patients with COVID-19 through a network medicine approach. We exploited the computational power of network medicine to explore the different scenarios provided by three different in silico analyses. In fact, a single study might not be enough to cover the multiform clinical frame of the disease. We used transcriptomic data from whole blood cells, including all innate and adaptive immune system cells, of patients with COVID-19 and other inflammatory conditions, infections, or conditions with some clinical features in common with COVID-19. For each disease, we identified the genes that were most deregulated compared with healthy subjects. We then selected functionally related genes and verified that they were co-localized in the human interactome, thus generating a functional coherent disease module. This allowed us to identify drugs targeting proteins that were within or in proximity to the COVID-19 module. Moreover, we also identified drugs that could be potentially repositioned for COVID-19 among those with an original medical indication for a disease whose module was in the COVID-19 neighborhood. Our in-silico analysis provided new pharmacological hypotheses to be explored and experimentally validated.
2. Methods
2.1. Data retrieval
Whole blood transcriptomic data for COVID-19 and 21 other diseases, including bacterial and viral infections, inflammatory diseases, immunodeficiency, primary lung, and coagulation disorders [34], were selected from the Gene Expression Omnibus (GEO) database. All datasets also included transcriptomic data of healthy controls.
Notably, the COVID-19 dataset was only recently deposited and is the first available concerning whole-genome gene expression data on whole blood cells. All patients were hospitalized for community-acquired lower respiratory tract infection with SARS-CoV-2 within the first 24 h of hospital admission. Details for each dataset are reported in Supplementary Table 1. The human interactome was downloaded from Cheng et al. [20]. This version of the interactome is composed of 217,160 protein–protein interactions connecting 15,970 unique proteins. Drug-target interactions were downloaded from DrugBank [35], which contains 13,563 drug entries, including 2627 approved small molecule drugs, 1373 approved biologics, 131 nutraceuticals, and over 6370 experimental drugs (released 22–04–2020) [35]. The target Uniprot IDs provided by DrugBank were mapped to Entrez gene IDs using the BioMart – Ensembl tool [36]. For our analysis, we selected a total of 1873 Food and Drug Administration (FDA)-approved drugs with at least one annotated target.
2.2. Disease-modulated genes and their localization in the human interactome
In order to identify genes that were most modulated by the disease, we computed differentially expressed genes between pathological and healthy conditions for every dataset using the following R packages: limma [37] , to analyze microarray data, and Deseq2 [38], to analyze RNA-seq data. The thresholds for p-value and Log2FC for each disease are reported in Supplementary Table 1. We mapped a list of the disease’s modulated genes on the human interactome to identify the ones in the same connected subnetwork (i.e., the largest connected component), and thus functionally related. In order to test whether this subnetwork forms a statistically significant disease module, for each analyzed disease we randomly selected groups of proteins in the human interactome with the same size and degree distribution as the original list of disease deregulated genes and the following three metrics were computed: 1) the size of the largest connected component (LCC); 2) the number of interactions in the LCC; and 3) the total number of interactions. The three metrics were then z-score normalized by applying a degree-preserving randomization procedure, expecting a p value ≤ 0.05 for genes forming a statistically significant disease module [39]. Log2FC thresholds were chosen to guarantee the topological organization of disease deregulated genes in statistically significant modules.
2.3. SAveRUNNER
To predict and prioritize off-label drug indications for COVID-19, we used a novel network-based algorithm for drug repurposing called SAveRUNNER (Searching off-lAbel dRUg aNd NEtwoRk) [40], [41]. Specifically, SAveRUNNER predicts drug-disease associations by quantifying the interplay between the drug targets and disease-associated proteins in the human interactome via a novel network-based similarity measure that rewards associations between drugs and diseases located in the same network neighborhoods. SAveRUNNER requires a list of disease genes, a list of drug targets, and a reference interactome. As disease genes, we used the genes falling in the COVID-19 module; as drug targets, we assembled target information of the 1873 FDA-approved drugs obtained from DrugBank [35]; and as a reference interactome we used the version provided by Cheng et al. [20]. A comprehensive description of SAveRUNNER algorithm can be found in [40], [41].
2.4. Network-based disease similarity
2.4.1. Network module separation
To measure the vicinity between the COVID-19 module and the other disease modules in the human interactome network, we used the non-Euclidean separation distance [9] defined in Eq. (1):
| (1) |
where is the network proximity defined in Eq. (2):
| (2) |
and is the shortest distance between the element of module and the element of module . A negative value for the separation measure indicates that two disease modules are in the same neighborhood of the human interactome, and thus they overlap; whereas a positive value for the separation measure indicates that two disease modules are topologically well separated. To evaluate the significance of module separation across two disease-specific modules (A, B), we built a reference distance distribution corresponding to the expected distance between two randomly selected groups of proteins with the same size and degree distribution as the original two disease-specific modules (A, B). The random selection was repeated 1000 times to build the reference distance distribution. The module separation measure was z-score normalized by using the mean and standard deviation of the reference distribution. Subsequently, the p value for the given z statistic was calculated. A p value ≤ 0.05 indicated that the separation between two disease-specific modules in the human interactome was more (or less) than that expected by chance.
2.4.2. Random Walk with Restart
The Random Walk with Restart (RWR) algorithm is another network-based approach to measure the closeness between the COVID-19 module and the other 19 disease modules in the human interactome network. RWR is an algorithm based on an intuitive concept that revolves around random walks. Given a random walker starting from a given node x, there are two different options at each iteration: either moving to one of its neighboring nodes or returning to x with a certain probability. Formally, the RWR algorithm can be described by Eq. (3):
| (3) |
where is the network adjacency matrix, representing the matrix of transitions between nodes, whose element denotes the transition probability of going from node j to node i; E is the starting point vector, whose element is equal to 1 if i is a starting node, 0 otherwise; is a probabilities vector, whose element denotes the probability of being at node i at iteration ; is a number ranging in (0,1), and expresses the probability of “restarting” from the starting point node at each iteration. At iteration t = 0, the value of is equal to E. The probabilities vector will be iteratively calculated until the point of converge is reached (i.e., , or the difference between the probability to stay and the probability to move is lower than a given threshold). Eventually, the RWR returns the vector R of the steady-state probabilities for each node in the network as output.
We ran RWR by considering the adjacency matrix , built from the human interactome as a transition matrix, and the genes in the COVID-19 module as elements of the vector E. For each disease module, we averaged RWR steady-state probabilities corresponding with each module element and obtained a mean probability for each disease, i.e., the probability to reach it starting from COVID-19. This disease probability was then normalized by using the modified z-score defined in Eq. (4):
| (4) |
where is the disease probability, is the median value of distribution of all disease probabilities, MAD is the median absolute deviation defined as the median of the absolute difference of the observation from the sample median (i.e., median(), and c is a scale factor equal to 0.6745, such that for normal distribution, is equal to the standard z-score [42]. We termed this normalized disease probability ‘COVID-19 closeness’. Values of COVID-19 closeness that were outside the overall distribution pattern of the normalized disease probabilities were defined as outliers. A commonly used rule is to define a data point as an outlier if it is more than above the third quartile or below the first quartile. This means that low outliers are below (i.e., the farthest diseases from COVID-19) and high outliers are above (i.e., the closest diseases to COVID-19). Values of COVID-19 closeness that are outside the upper and lower quartiles are usually indicated as upper and lower whiskers, respectively. Diseases corresponding to high outliers as well as upper whiskers are more likely to be reached by the random walker starting from COVID-19.
3. Results
3.1. Functionally-related and co-localized disease-related genes in the human interactome
In this study, we first compared the gene expression profile of COVID-19 and 21 diseases in which inflammatory and immune processes are involved with the profile of healthy controls in order to identify the highest modulated genes under pathological conditions (Supplementary Table 1). We mapped these genes on the human interactome, which is a network of proteins (nodes) in which the edges are the physical and functional interactions occurring between them, to evaluate whether they had the propensity to aggregate in local, disease-specific neighborhoods of the human interactome, thus making them functionally-related genes. For each disease, we extracted the largest connected component from the subnetwork composed of genes that were modulated in that specific disease condition and verified whether these genes presented a statistically significant ability to generate a disease module. For subsequent analyses, we selected only the diseases that satisfied this module hypothesis, in accordance with the organizing principles of network medicine [3], [9], [39]. Notably, we found that for all diseases analyzed (with the exception of ankylosing spondylitis and chronic spontaneous urticaria), the deregulated genes formed statistically significant modules ( Table 1). Thus, we considered 20 diseases, including COVID-19, for subsequent analyses.
Table 1.
Module search results for the analyzed datasets. LCC = largest connected component.
| Disease name | Number of LCC nodes |
Number of LCC edges |
Number of total interactions |
|||
|---|---|---|---|---|---|---|
| observation | p-value | observation | p-value | observation | p-value | |
| Ankylosing spondylitis | 216 | 0.6 | 825 | 0.005 | 827 | 0.008 |
| Crohn’s disease | 399 | 2E-13 | 1120 | 3E-39 | 1147 | 2E-43 |
| Chronic obstructive pulmonary disease (COPD) | 163 | 0.0002 | 409 | 0.0002 | 418 | 0.0003 |
| Chronic spontaneous urticaria | 183 | 0.5 | 444 | 0.1 | 476 | 0.02 |
| Community-acquired pneumonia | 421 | 0.01 | 1347 | 1E-11 | 1359 | 2E-11 |
| Common variable immunodeficiency | 25 | 0.02 | 24 | 0.03 | 30 | 0.01 |
| COVID-19 | 314 | 0.00001 | 441 | 3E-9 | 514 | 5E-17 |
| Dermatomyositis | 11 | 2E-11 | 23 | 2E-25 | 24 | 5E-17 |
| H3N2 flu | 170 | 0.03 | 270 | 0.001 | 282 | 6E-4 |
| H1N1 flu | 278 | 0.01 | 1309 | 2E-25 | 1331 | 5E-27 |
| Inflammatory bowel disease | 412 | 5E-16 | 1139 | 9E-38 | 1161 | 7E-42 |
| Inclusion body myositis | 3 | 0.01 | 4 | 0.01 | 5 | 0.007 |
| Infective endocarditis | 202 | 0.05 | 359 | 0.003 | 373 | 0.001 |
| Primary lung cancer | 333 | 0.0006 | 1185 | 2E-24 | 1192 | 2E-24 |
| Polymyositis | 12 | 0.004 | 14 | 0.00004 | 16 | 0.0002 |
| Rheumatoid arthritis | 178 | 0.03 | 321 | 1E-5 | 343 | 2E-7 |
| Sarcoidosis | 476 | 0.04 | 1731 | 4E-22 | 1745 | 3E-22 |
| SARS | 33 | 0.04 | 62 | 2E-6 | 65 | 3E-6 |
| Septic shock | 363 | 0.001 | 1202 | 1E-13 | 1223 | 6E-15 |
| Tuberculosis | 469 | 0.006 | 1829 | 8E-22 | 1843 | 7E-22 |
| Ulcerative colitis | 329 | 0.05 | 488 | 0.04 | 508 | 0.05 |
| Venous thromboembolism | 513 | 0.04 | 2975 | 0.00002 | 2977 | 0.00003 |
3.2. Drug-disease module vicinity
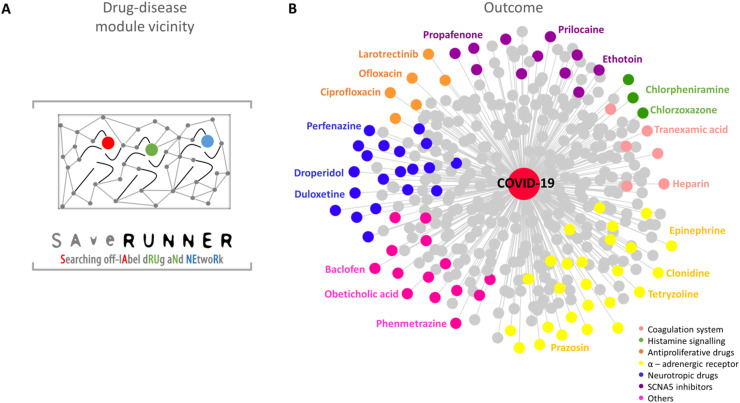
To discover novel repurposable drugs and evaluate the magnitude to which a given drug can be repositioned for COVID-19, we exploited the recently developed SAveRUNNER algorithm [40]. The rationale behind SAveRUNNER builds on the hypothesis that for a drug to be effective against a specific disease, its associated targets (drug module) and the disease-specific associated genes (disease module) should be located nearby in the human interactome [20] ( Fig. 1 A).
Fig. 1.
SAveRUNNER. A) The network-based algorithm used to identify off-label drug indications against COVID-19 [40]. B) The SAveRUNNER outcome network showing the high-confidence predicted drug-disease associations (p-value ≤ 0.05) connecting COVID-19 with 399 FDA-approved non-COVID-19 drugs. Drugs are colored according to the targeted pathways reported in the legend.
Using SAveRUNNER, we computed the similarity between each drug module and the COVID-19 module together with the corresponding statistical significance obtained through a degree-preserving randomization procedure. We obtained a weighted bipartite drug-disease network, where the link between a drug and a disease is appreciated if the corresponding drug targets and disease genes are located nearby in the interactome to a greater extent than what would be expected by chance (Fig. 1 B). The weight of their interaction corresponds with the similarity measure between the corresponding drug and disease module. In our study, SAveRUNNER identified 399 repurposable drugs for COVID-19 (Supplementary Table 2). Focusing on the top-ranked predicted drugs (similarity greater than 0.8), we observed molecules involved in the modulation of the coagulation system (e.g., heparin and tranexamic acid), antihistaminic drugs, mast cell stabilizers (e.g., chlorzoxazone and chlorpheniramine), anti-proliferative drugs including tyrosine kinase (TRK) inhibitors and antibiotics (e.g., larotrectinib and ciprofloxacin), alpha-adrenergic receptor agents (e.g., clonidine and prazosin); drugs affecting the central nervous system (e.g., perfenazine and droperidol), and inhibitors of the sodium voltage-gated channel alpha subunit 5 (SCN5A), which is involved in cardiac rhythm control (e.g., propafenone and prilocaine), among others ( Table 2, Supplementary Table 3, and Fig. 1 B).
Table 2.
Potential Drug Repurposing for COVID-19. SAveRUNNER-identified repurposable drugs for SARS-CoV-2 showing a similarity greater than 0.8. * indicates drugs under investigation in clinical trials.
| Approved Therapeutic Use | Drugs | Potential application in COVID-19 | |||
|---|---|---|---|---|---|
| Antiplatelet/anticoagulant | Heparin* | Reduce the risk of thrombosis | |||
| Fibrinolytics | Streptokinase | ||||
| Pro-coagulants | Tranexamic acid* | Aminocaproic Acid | Limit viral entry | ||
| Polysulphates | Pentosan polysulphate | Chondroitin sulphate | |||
| Histamine H1 - receptors antagonists | Diphenylpyraline | Chlorpheniramine | Limit cytokine storm | ||
| Mast cells stabilizers | Chlorzoxazone | ||||
| Tropomyosin receptor kinase B inhibitors | Entrectinib | Larotrectinib | Reduce immune cells proliferation | ||
| Other Antiproliferatives | d-Serine | Podofillotoxin | |||
| Fluoroquinolones | Ofloxacin | Ciprofloxacin | |||
| α1-adrenergic receptors blockers | Nicergoline | Dapiprazole | Moxisylyte | Prazosin* | Limit cytokine storm |
| Silodosin | Tamsulosin | Alfuzosin | Phenoxybenzamine | ||
| Phentolamine | |||||
| α2-adrenergic receptors agonists | Apraclonidine | Guanabenz | Guanfacine | Levonordefrin | |
| Brimonidine | Clonidine* | ||||
| α1-agonists | Ergometrine | Metaraminol | Tetryzoline | Methoxamine | Sustain blood pressure in case of septic shock |
| Midodrine | Oxymetazoline | Phenylephrine | Phenylpropanolamine | ||
| Xylometazoline | Epinephrine | Naphazoline | |||
| Phenothiazines and Antipsychotics | Perphenazine | Thioridazine | Thiothixene | Periciazine | Limit cytokine storm through modulation of Dopaminergic, Adrenergic and/or Serotoninergic receptors |
| Pipotiazine | Prochlorperazine | Flupentixol | |||
| Serotonin-norepinephrine reuptake Inhibitors (SNRI) | Duloxetine | Sibutramine | Venlafaxine | ||
| Serotonin antagonist and reuptake inhibitors (SARI) | Nefazodone | Lorpiprazole | |||
| Dopaminergics | Armodafinil | Diethylpropion | Modafinil | Solriamfetol | |
| Benzphetamine | Fenoldopam | ||||
| Antidopaminergics | Droperidol | Methylergometrine | Acetophenazine | Lumateperone | |
| Anti-epileptics | Fosphenytoin | Ethotoin | Mephenytoin | Limit macrophages acrivation | |
| Antiarrhythmic agents | Ajmaline | Encainide | Indecainide | Moricizine | |
| Tocainide | Propafenone | Vernakalant | |||
| Local anaesthetics | Benzonatate | Prilocaine | |||
3.3. Disease-disease module vicinity
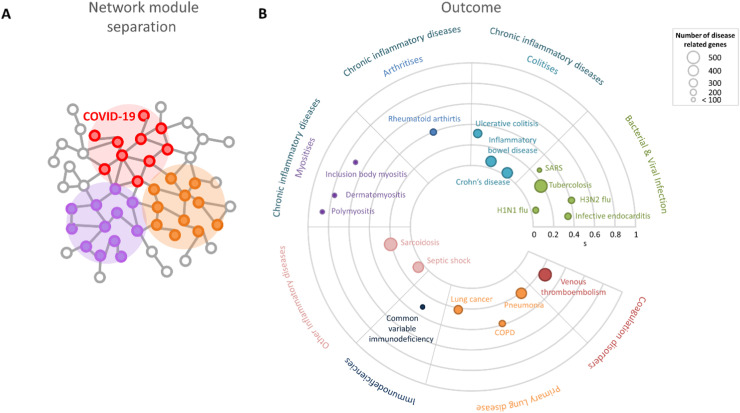
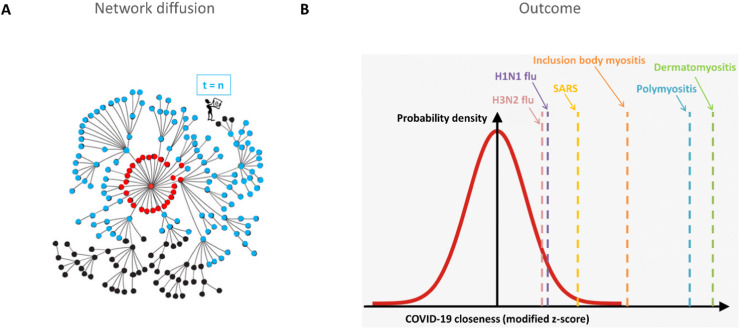
To find similarities between COVID-19 and the other 19 considered diseases, we implemented two network-based approaches: (1) network module separation, which quantified the topological distance between a disease module and the COVID-19 module in the human interactome network ( Fig. 2 A); (2) the RWR algorithm, which calculated the probability of a random walker reaching a disease module starting from the COVID-19 module ( Fig. 3 A).
Fig. 2.
Network module separation. A) Schematic representation of disease-related gene modules and their topological distances in the human interactome. B) Radial plot reporting the network module separation measure(s) between COVID-19 and the other 19 diseases. Each disease is represented as a circle whose radius reflects the number of disease-related genes associated with it. The farther a disease is from the center, the more distant its module is from the COVID-19 module in the human interactome.
Fig. 3.
Random Walk with Restart (RWR). A) Sketch of the RWR algorithm applied on the human interactome. Red nodes represent the starting point nodes, light blue nodes represent all visited nodes at the end of the algorithm run, and black nodes represent the nodes of the human interactome that were not visited. B) Distribution of modified z-score-normalized probabilities (COVID-19 closeness) of nodes that were visited by the RWR algorithm starting from nodes belonging to the COVID-19 module. Diseases that are high outliers and upper whiskers are highlighted in the figure.
We observed that every separation value was statistically significant (see Methods), and we considered diseases whose separation values were less than the 15th percentile of the distribution of all separation values. We found H1N1 flu, Crohn’s disease, inflammatory bowel disease, and septic shock to be the closest diseases to COVID-19 (Supplementary Table 2 and Fig. 2 B).
From the RWR algorithm, we selected only those diseases that ranked within the outliers or upper whiskers, i.e., diseases that were more likely to be reached by the random walker starting from COVID-19 (see Methods). These diseases included dermatomyositis, polymyositis, and inclusion body myositis in the outlier category, whereas SARS, H1N1, and H3N2 flu appeared as upper whiskers (Supplementary Table 2 and Fig. 3 B).
Interestingly, the RWR approach confirmed the results obtained with network module separation, in which H1N1 flu resulted as one of the closest diseases to COVID-19 in the interactome, with respect to the other analyzed diseases.
4. Discussion
SARS-CoV-2 has become a pandemic infection with a global health priority. There is an urgent need to find more effective drugs to reduce disease burden and mortality. In this context, drug repurposing represents a strategy to be exploited. Since it is known that host immune response plays a pivotal role in disease evolution, we used a network medicine approach to analyze transcriptomic data from the blood cells of patients affected by COVID-19 or other inflammatory conditions, infections, or diseases sharing some clinical features with COVID-19 in order to find the most important pathways associated with immune system activation under these conditions.
In particular, we first identified the most modulated genes in each disease condition and then verified whether they might be functionally related and co-localized in the human interactome, forming functional coherent disease modules that could include genes with a critical role in immune response regulation. Even though the cause-effect relationship cannot be directly inferred by expression data, it is reasonable to assume that disease co-modulated genes are functionally coordinated in response to an external stimulus, implying that they might be part of the same pathways or biological functions, and may influence each other or be influenced by the same underlying mechanism(s). Inspired by the organizing principles of the network medicine paradigm, we evaluated whether disease deregulated genes had the propensity to aggregate in local disease-specific neighborhoods of the human interactome, thus being functionally-related genes displaying a statistically significant tendency to form dense disease modules [3], [9], [39]. The accurate identification and localization of these disease modules represents the first step toward a systematic understanding of molecular-level pathological mechanisms, together with the prediction of novel disease-disease relationships. Indeed, if two disease modules overlap or are in the immediate vicinity within the interactome, local perturbations causing one disease can disrupt pathways of the other disease module as well, resulting in common clinical and pathobiological characteristics [9].
In order to analyze the multiform clinical frame of the disease from different scenarios, we used three different network medicine approaches to select drugs commonly used for the treatment of other conditions that could be repurposed for use in COVID-19 patients.
It should be bear in mind that computational approaches are useful to generate new pharmacological hypotheses that need to be tested and validated experimentally. The potential use of the identified drugs for COVID-19 treatment will have to be carefully evaluated taking into account their possible side effects that can be found at DrugBank database website (https://go.drugbank.com/)[35]. Being aware of this important limitation we discussed the different identified drug classes considering the available information from literature focusing on their possible effects on the immune response modulation and infection natural history in COVID-19 patients.
SAveRUNNER algorithm was used to identify repurposable drugs from DrugBank that could target the COVID-19 module or its neighborhood. Results show that drugs involved in the modulation of the coagulation system, histamine receptors, mast cell stability, immune cell proliferation, adrenergic receptors, serotonin receptors, or sodium channel SCN5A (sodium voltage-gated channel alpha subunit 5) function may have a great impact on immune system response in COVID-19 patients.
SAveRUNNER identified drugs acting on COVID-19-related genes, regardless of their specific effects. As a consequence, some drugs identified may be beneficial while others might be detrimental, and a critical and clinical evaluation that also considers the stage of SARS-CoV-2 infection is always essential [29]. Below we briefly discuss drug classes with a similarity greater than 0.8 (Table 2).
4.1. Drugs active on the coagulation system
SAveRUNNER analysis found different compounds with a mechanism of action involving the modulation of the coagulation system that could be repurposed for COVID-19. This observation positively correlates with SARS-CoV-2 infection, where a severe impairment in the coagulation system leading to thrombosis is frequently observed [43]. Indeed, heparin has been tested as a prophylactic treatment and was demonstrated to improve disease-specific mortality [44]. Heparin clearly emerged in our analysis, supporting the potential and accuracy of SAveRUNNER software in identifying repurposable drugs. Several clinical trials testing the effect of different heparin formulations in COVID-19 are ongoing, including NEBUHEPA (NCT04530578), which is evaluating the effect of nebulized heparin in patients with COVID-19-related acute respiratory distress syndrome (ARDS). The same analysis identified chondroitin sulphate and pentosan polysulphate, which showed a lower activity compared to heparin in inhibiting platelet aggregation inhibition and was also shown to interact with spike proteins, thus reducing virion internalization and blocking inflammation and the cytokine storm associated with antigenic epitope exposition [45]. Another drug that has emerged is streptokinase, a fibrinolytic drug used in severe acute thrombosis. A case series report showed that streptokinase is effective in COVID-19 patients [46]. Intriguingly, the analysis also identified tranexamic acid, which is normally used as a pro-coagulant agent during bleeding due to its ability to inhibit circulating plasminogen and other proteases, leading to thrombus stabilization [47]. Since plasminogen is one of the proteases necessary for spike-protein cleavage, thus allowing virion interaction with angiotensin-converting enzyme 2 (ACE2)-expressing cells [47], the possibility of using tranexamic acid in COVID-19 patients is currently being tested in clinical trials (NCT04338126).
4.2. H1-inhibitors and mast cell stabilizers
Histamine is a proinflammatory molecule produced by mast cells that mediates type I hypersensitivity reactions. Mast cell abundance in human airways supports the potential relevance of this mediator in SARS-CoV-2 infection. Short-term effects of mast cell degranulation and histamine release include increased vascular permeability, vasodilation, immune cell recruitment, and platelet activation [48], [49]. Moreover, histamine release induces interleukin 6 (IL-6), leukotrienes, and the production of other inflammatory prostaglandins, thus triggering the activation of innate response [49]. Mast cell stabilization and blocking histamine signaling might be fundamental in controlling the cytokine storm, which is typical of the early stages of SARS-CoV-2 infection [49]. Indeed, our analysis highlighted a potential repurposing of chlorzoxazone, a mast cell stabilizer that blocks calcium channels and inhibits degranulation as well as leukotriene and cytokine production [50]. Similarly, diphenylpyraline and chlorpheniramine, which are commonly used antihistaminic drugs, could potentially block the early phase of cytokine storm during SARS-CoV-2 infection [49]. Of note, emerging evidence currently supports a direct antiviral effect of targeting the histamine pathway in SARS-CoV-2 in vitro [51].
4.3. Antiproliferative drugs and antibiotics with antiproliferative activity
This category includes both tyrosine kinase inhibitors (TRK inhibitors) [Entrectinib, Larotrectinib] and drugs inhibiting the activity of human topoisomerase II, such as fluoroquinolones (ofloxacin, ciprofloxacin) [52], [53]. Due to their ability to act as antiproliferative molecules, these drugs have emerged as promising antitumor repurposed compounds. We can reasonably expect that their inhibition of cell proliferation might be useful to limit also the immune cell proliferation and consequent cytokine storm during SARS-CoV-2 infection [29], [54], [55].
Of note, the potential relevance of TRK inhibitor repurposing has been confirmed by other drug repurposing studies [56]. In addition, fluoroquinolones have been found to have antiviral activity in vitro, thus supporting a potential benefit in COVID-19 patients in limiting bacterial superinfection [57].
4.4. α-adrenergic receptor agents
SAveRUNNER analysis found that α1-antagonists and α2-agonists could be repurposed for COVID-19. Indeed, data suggest that α1 adrenergic receptor activation may induce pro-inflammatory cytokine secretion in innate cells, thus suggesting the possibility that blocking α1 adrenergic receptors might limit the cytokine storm that characterizes severe COVID-19 patients [58]. Rose et al. found that men with a confirmed or suspected COVID-19 diagnosis who were on treatment with α1 adrenergic receptor antagonists prior to hospitalization had reduced in-hospital mortality (OR: −36%) compared to those who were not taking α1 adrenergic receptor antagonist medications [59]. As such, blocking alpha-adrenergic signaling in the immune system might be successful, particularly in early-stage infection, and indeed prazosin (α1-antagonist) is now being tested in a clinical trial (NCT04365257). Similarly, α2-agonists such as clonidine could be repurposed during COVID-19 to limit ARDS and inflammatory response [60]. Intriguingly, α1-agonism could stimulate immune system response and could be considered in COVID-19 patients in case of septic shock [61].
4.5. Drugs active in the central nervous system
Several drugs active in the central nervous system have been identified as repurposable for COVID-19 following SAveRUNNER analysis, particularly tricyclic compounds, drugs active in serotonin signaling (i.e. SNRI and SARI), and dopaminergics and dopamine antagonists. Interestingly, Hoertel et al. [62] suggested a possible role of both SSRI and non-SSRI antidepressants in reducing the risk of death and intubation in patients hospitalized for SARS-CoV-2 infection. Recent evidence highlighted that these drugs may have an effect on both innate and adaptive immunity:
-
-
Phenotyazine and antipsychotic drugs have known effects on α-adrenergic and histaminergic receptors, and could therefore act as possible immune system modulators [49], [63]. Moreover, different drugs belonging to this class were shown to possess antiviral properties, suggesting potential repurposing for COVID-19 [64].
-
-
SNRI and SARI might also have a modulatory effect on the immune system, particularly on lymphocytes, which express serotonin receptor 5-hydroxytriptamine 2 (5-HT2). In rat and mouse models, fluoxetine treatment produced a significant reduction in TNFα and IFNγ production. In SARS-CoV-2 infection, drugs modulating the serotonin signaling might be repurposed as cytokine storm regulators [65].
-
-
Dopamine receptors are expressed in different immune cell subtypes and their effect on immune response modulation is still debated. Dopaminergic stimulation reduces TNF-α and ROS production in neutrophils, though it stimulates mast cell degranulation and monocyte chemotaxis. In addition, dopamine stimulation appeared to be protective in a mouse model of peritonitis [66]. In this scenario, targeting the dopaminergic pathway emerged as a potential strategy to limit cytokine storm during COVID-19. However, a clear role of dopaminergic system activation in the context of immune response is debated, and more research is necessary to better define the role of dopamine in immune system modulation [66], [67].
4.6. Drugs acting on SCN5A sodium channels
SCN5A sodium channels are commonly expressed in excitable tissue, particularly neurons and myocytes. Most identified SCN5A inhibitors are anti-arrhythmic drugs, local anesthetics, or anti-epileptics. Recent evidence highlighted that the SCN5A channel is involved in macrophage activation and plays a pivotal role in host antiviral response by inducing the phosphorylation and nuclear translocation of the transcription factor ATF2 [68]. Moreover, in LPS-activated macrophages, SCN5A regulates endosomal acidification and stimulates phagocytosis. Although this process protects the host during acute infections, it may also promote tissue injury [69]. Interestingly, endosomal and lysosomal acidification allow viral cellular entrance [29]. In this context, SCN5A inhibitors may contribute to controlling both systemic inflammation and viral infection. Other studies reported that macrophages present an anti-inflammatory phenotype in mice expressing human SCN5A [70].
Consistent with these observations, propafenone, a SCN5A inhibitor used for its anti-arrhythmic properties, was suggested as a possible inhibitor of spike protein cleavage and SARS-CoV-2 cellular penetration [71]. However, SCN5A inhibitors present a series of limitations that need to be taken into account, including arrhythmia.
Among other drugs (Supplementary Table 3), SAveRUNNER also identified baclofen, a GABA-B agonist commonly used in neurodegenerative diseases as an antispastic. A recent computational analysis identified it as a TNFα inhibitor [72]. Since TNFα is one of the main inflammatory signals of innate immunity, baclofen might be repurposed as a mitigator of cytokine storm in SARS-CoV-2 infection [29]. Studies in mouse models found that FXR activation reduces the levels of circulating NF-KB and other proinflammatory cytokines, such as MCP-1. Obeticholic acid, an FXR agonist mainly indicated for the treatment of biliostasis, was shown to exert anti-inflammatory activities observed in the reduction of liver inflammation [73] and was identified by a computational study and proven in vitro to inhibit SARS-CoV-2 ligation to human ACE2 [74].
Network module separation was used to find diseases with a module close to that of COVID-19. Our hypothesis is that drugs used to treat these diseases may also be beneficial in COVID-19 patients. We found that septic shock, Crohn’s disease, inflammatory bowel disease (IBD), and H1N1 flu modules were very close to the COVID-19 module. Consistently, it was observed that COVID-19 patients with increased immune system activation present an elevated incidence of sepsis [29]. Moreover, the significative proximity between the COVID-19, IBD, and Crohn’s disease modules is not surprising since literature data support that COVID-19 and IBD immune system activation share several similarities and that some drugs used for IBD appear to also be effective for COVID-19 patients [75]. Of the drugs reported in the DrugBank database for septic shock treatment, both epinephrine and norepinephrine were also identified by the SAveRUNNER algorithm, suggesting that SARS-CoV-2 infection and septic shock share common epinephrine or norepinephrine targets. Naloxone is another drug used to treat septic shock and is currently under investigation in COVID-19 patients. Mesalazine and sulfasalazine, two anti-inflammatory drugs, are used for Crohn’s disease treatment and have also been identified by the SAveRUNNER algorithm.
Two monoclonal anti-tumor necrosis factor alpha antibodies (adalimumab and infliximab) and many corticosteroid drugs (budesonide, methotrexate, prednisolone, prednisone, and hydrocortisone) used for Crohn’s disease are currently being tested in COVID-19 in several clinical trials. This is not surprising since the role of corticosteroids as anti-inflammatory drugs is well known. Of note, the RECOVERY randomized clinical trial showed that the use of dexamethasone resulted in lower 28-day mortality in those receiving either invasive mechanical ventilation or oxygen alone [76].
In addition, an increase in TNFα, a strong pro-inflammatory cytokine, has been observed in patients affected by COVID-19 [31]. Some evidence suggests that TNFα inhibition may downregulate ACE2 expression and shedding, thus reducing viral entry into cells [31]. Several drugs, including an antiviral drug (oseltamivir), anti-inflammatory drugs (naproxen and acetylsalicylic acid), a beta-2 adrenergic receptor agonist (salbutamol), and an analgesic/antipyretic drug (acetaminophen), are used for flu treatment and are currently in clinical trials. However, several antiviral drugs, including oseltamivir, do not seem to exert a robust effect against the SARS-CoV-2 virus [27].
The immune system and the sympathetic nervous system are highly connected through post-ganglionic sympathetic nerve fibers, which secrete norepinephrine that innervates both primary and secondary lymphoid tissues. Both innate and adaptive immune system cells express adrenergic receptors, mainly β2. There is evidence that glucocorticoids and other β2-receptor agonists suppress macrophage secretion of TNFα and other inflammatory cytokines in response to lipopolysaccharide, reducing inflammatory damage. It has been shown that norepinephrine drives alternative M2 macrophage development, characterized by an anti-inflammatory phenotype [58], and that β2 adrenergic receptors modulate the activation of several innate immune cells and consequently modulate T and B cell response. However, the role of β2 receptors on the immune system is still debated and some authors have reported a pro-inflammatory role [77].
The RWR algorithm was used to search for diseases whose drugs may also perturb the COVID-19 module. We identified H1N1 and H3N2 flu, SARS-CoV-1 infection, dermatomyositis, polymyositis, and inclusion body myositis. These findings support the documented similarity between SARS-CoV-2 and SARS-CoV-1 infections [29]. Interestingly, both the network module separation approach and the RWR highlighted the disease modules of H1N1 flu and SARS-CoV-2 infection. Several corticosteroids used for dermatomyositis and polymyositis (prednisolone, prednisone, hydrocortisone, methylprednisolone, betamethasone, and methylprednisolone hemisuccinate) and corticotropin are currently in clinical trials for COVID-19. Notably, triamcinolone is a corticosteroid drug that is used for diseases identified by both the network module separation approach and RWR ( Table 3).
Table 3.
Drugs used for the diseases identified by the network module separation approach and RWR. * indicates drugs identified by SAveRUNNER. Drugs currently under investigation in clinical trials, as reported in DrugBank, are highlighted in bold .
| Network module separation |
Random Walk with Restart |
|||
|---|---|---|---|---|
| Septic shock | Crohn’s disease | H1N1 flu | Dermatomyositis | Polymyositis |
| Epinephrine* | Mesalazine* | Acetaminophen* | Corticotropin | |
| Norepinephrine* | Sulfasalazine* | Cetirizine* | Methylprednisolone | |
| Naloxone | Adalimumab | Chlorpheniramine* | Betamethasone | |
| Budesonide | Phenylephrine* | Bupivacaine | ||
| Infliximab | Pseudoephedrine* | Methylprednisolone hemisuccinate | ||
| Naproxen | ||||
| Oseltamivir | ||||
| Salbutamol | ||||
| Acetylsalicylic acid | ||||
| Ascorbic acid | ||||
| Methotrexate | Methotrexate | |||
| Triamcinolone | Triamcinolone | |||
| Prednisolone | Prednisolone | |||
| Prednisone | Prednisone | |||
| Hydrocortisone | Hydrocortisone | |||
In conclusion, we used a network medicine approach to generate new pharmacological hypotheses for the COVID-19 treatment. While some of the in-silico identified drugs are already under evaluation in clinical trials, others have been proposed by expert opinion or other computational studies to be potentially effective in COVID-19 patients. SAveRUNNER analysis also identified novel drug categories, including drugs known to be active in the central nervous system and sodium channel blockers, that could be repurposed in COVID-19 patients.
The in-silico methodology has many limitations, including the need to test and validate the identified drugs. Indeed, the potential benefits as well as the risks of possible adverse reactions, mainly due to the multi-target action of many compounds, must be carefully evaluated and proved. Moreover, an efficient translation should also take into account pharmacokinetic aspects that could impact the clinical applicability of repurposed drugs.
CRediT authorship contribution statement
Pasquale Sibilio: Formal analysis, Validation, Writing – original draft. Simone Bini: Data curation, Visualization, Writing – original draft. Giulia Fiscon: Formal analysis, Validation. Marialuisa Sponziello: Writing – review & editing, Visualization. Federica Conte: Formal analysis, Validation. Valeria Pecce: Writing – review & editing, Visualization. Cosimo Durante: Data curation, Writing – review & editing. Paola Paci: Methodology, Conceptualization, Supervision, Writing – original draft. Rosa Falcone: Conceptualization, Data curation. Giuseppe Danilo Norata: Data curation, Writing – review & editing. Lorenzo Farina: Methodology, Project administration, Writing – original draft. Antonella Verrienti: Conceptualization, Supervision, Writing – original draft.
Acknowledgements
P.S, S.B, and R.F. contributed to this paper as part of their Ph.D. studies in Biotechnologies and Clinical Medicine at Sapienza University of Rome. L.F. was financially supported by a Sapienza University of Rome grant entitled “Network medicine-based machine learning and graph theory algorithms for precision oncology” - n. RM1181642AFA34C2. The manuscript has been edited by Melissa Kerr.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.biopha.2021.111954.
Appendix A. Supplementary material
Supplementary material
.
Supplementary material
.
Supplementary material
.
Availability and requirements
SAveRUNNER source code is freely available at https://github.com/sportingCode/SAveRUNNER.git, along with a comprehensive user guide.
References
- 1.Jourdan J.-P., Bureau R., Rochais C., Dallemagne P. Drug repositioning: a brief overview. J. Pharm. Pharmacol. 2020;72(9):1145–1151. doi: 10.1111/jphp.13273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sonawane AR, Weiss ST, Glass K., Sharma A. Network Medicine in the Age of Biomedical Big Data. Front Genet [Internet]. 2019 Apr 11 [cited 2020 Apr 7];10. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6470635/. [DOI] [PMC free article] [PubMed]
- 3.Barabási A.-L., Gulbahce N., Loscalzo J. Network medicine: a network-based approach to human disease. Nat. Rev. Genet. 2011;12(1):56–68. doi: 10.1038/nrg2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Silverman E.K., Schmidt H., Anastasiadou E., Altucci L., Angelini M., Badimon L., Balligand J.L., Benincasa G., Capasso G., Conte F., Di Costanzo A., Farina L., Fiscon G., Gatto L., Gentili M., Loscalzo J., Marchese C., Napoli C., Paci P., Petti M., Quackenbush J., Tieri P., Viggiano D., Vilahur G., Glass K., Baumbach J. Molecular networks in network medicine: development and applications. Wiley Interdiscip. Rev. Syst. Biol. Med. 2020;12(6) doi: 10.1002/wsbm.1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tieri P., Farina L., Petti M., Astolfi L., Paci P., Castiglione F. In: Encyclopedia of Bioinformatics and Computational Biology. Ranganathan S., Gribskov M., Nakai K., Schönbach C., editors. Academic Press; Oxford: 2019. Network Inference and Reconstruction in Bioinformatics; pp. 805–813.https://www.sciencedirect.com/science/article/pii/B9780128096338202902 [Google Scholar]
- 6.Conte F., Fiscon G., Licursi V., Bizzarri D., D’Antò T., Farina L., Paci P. A paradigm shift in medicine: a comprehensive review of network-based approaches. Biochim. Biophys. Acta (BBA) Gene Regul. Mech. 2020;1863(6) doi: 10.1016/j.bbagrm.2019.194416. [DOI] [PubMed] [Google Scholar]
- 7.Fiscon G., Conte F., Farina L., Paci P. Network-based approaches to explore complex biological systems towards network medicine. Genes. 2018;9(9) doi: 10.3390/genes9090437. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6162385/ [cited 2021 May 4] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caldera M., Buphamalai P., Müller F., Menche J. Interactome-based approaches to human disease. Curr. Opin. Syst. Biol. 2017;3:88–94. [Google Scholar]
- 9.Menche J., Sharma A., Kitsak M., Ghiassian S.D., Vidal M., Loscalzo J., et al. Uncovering disease-disease relationships through the incomplete interactome. Science. 2015;347(6224) doi: 10.1126/science.1257601. https://science.sciencemag.org/content/347/6224/1257601 [cited 2019 Nov 22] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Falcone R., Conte F., Fiscon G., Pecce V., Sponziello M., Durante C., Farina L., Filetti S., Paci P., Verrienti A. BRAFV600E-mutant cancers display a variety of networks by SWIM analysis: prediction of vemurafenib clinical response. Endocrine. 2019;64(2):406–413. doi: 10.1007/s12020-019-01890-4. [DOI] [PubMed] [Google Scholar]
- 11.Pecce V., Verrienti A., Fiscon G., Sponziello M., Conte F., Abballe L., Durante C., Farina L., Filetti S., Paci P. The role of FOSL1 in stem-like cell reprogramming processes. Sci. Rep. 2021;11:14677. doi: 10.1038/s41598-021-94072-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paci P., Colombo T., Fiscon G., Gurtner A., Pavesi G., Farina L. SWIM: a computational tool to unveiling crucial nodes in complex biological networks. Sci. Rep. 2017;7 doi: 10.1038/srep44797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Paci P., Fiscon G., Conte F., Licursi V., Morrow J., Hersh C., Cho M., Castaldi P., Glass K., Silverman E.K., Farina L. Integrated transcriptomic correlation network analysis identifies COPD molecular determinants. Sci. Rep. 2020;10(1):1–18. doi: 10.1038/s41598-020-60228-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fiscon G., Conte F., Paci P. SWIM tool application to expression data of glioblastoma stem-like cell lines, corresponding primary tumors and conventional glioma cell lines. BMC Bioinforma. 2018;19(Suppl 15):436. doi: 10.1186/s12859-018-2421-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fiscon G., Conte F., Licursi V., Nasi S., Paci P. Computational identification of specific genes for glioblastoma stem-like cells identity. Sci. Rep. 2018;8(1):7769. doi: 10.1038/s41598-018-26081-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fiscon G., Pegoraro S., Conte F., Manfioletti G., Paci P. Gene network analysis using SWIM reveals interplay between the transcription factor-encoding genes HMGA1, FOXM1, and MYBL2 in triple-negative breast cancer. FEBS Lett. 2021;595:1569–1586. doi: 10.1002/1873-3468.14085. https://febs.onlinelibrary.wiley.com/doi/abs/10.1002/1873-3468.14085 [cited 2021 May 3] [DOI] [PubMed] [Google Scholar]
- 17.Grimaldi A.M., Conte F., Pane K., Fiscon G., Mirabelli P., Baselice S., Giannatiempo R., Messina F., Franzese M., Salvatore M., Paci P., Incoronato M. The new paradigm of network medicine to analyze breast cancer phenotypes. Int. J. Mol. Sci. 2020;21(18):6690. doi: 10.3390/ijms21186690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Panebianco V., Pecoraro M., Fiscon G., Paci P., Farina L., Catalano C. Prostate cancer screening research can benefit from network medicine: an emerging awareness. Npj Syst. Biol. Appl. 2020;6(1):1–6. doi: 10.1038/s41540-020-0133-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fiscon G., Conte F., Farina L., Pellegrini M., Russo F., Paci P. In: MicroRNA Target Identification: Methods and Protocols [Internet] Laganà A., editor. Springer; New York, NY: 2019. Identification of disease–miRNA networks across different cancer types using SWIM; pp. 169–181. [cited 2021 May 7] [DOI] [PubMed] [Google Scholar]
- 20.Cheng F., Desai R.J., Handy D.E., Wang R., Schneeweiss S., Barabási A.-L., Loscalzo J. Network-based approach to prediction and population-based validation of in silico drug repurposing. Nat. Commun. 2018;9(1):2691. doi: 10.1038/s41467-018-05116-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheng F., Liu C., Jiang J., Lu W., Li W., Liu G., Zhou W., Huang J., Tang Y. Prediction of drug-target interactions and drug repositioning via network-based inference. PLOS Comput. Biol. 2012;8(5) doi: 10.1371/journal.pcbi.1002503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou Y., Hou Y., Shen J., Huang Y., Martin W., Cheng F. Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Disco. 2020;6(1):1–18. doi: 10.1038/s41421-020-0153-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fang J., Zhang P., Wang Q., Zhou Y., Chiang C.-W., Chen R., et al. Network-based Translation of GWAS Findings to Pathobiology and Drug Repurposing for Alzheimer’s Disease. medRxiv. 2020 Jan 18;2020.01.15.20017160.
- 24.Gysi DM, Valle ÍD, Zitnik M., Ameli A., Gan X., Varol O., et al. Network Medicine Framework for Identifying Drug Repurposing Opportunities for COVID-19. ArXiv200407229 Cs Q-Bio Stat [Internet]. 2020 Apr 15 [cited 2020 May 17]; Available from: http://arxiv.org/abs/2004.07229. [DOI] [PMC free article] [PubMed]
- 25.Fiscon G., Conte F., Amadio S., Volonté C., Paci P. Drug repurposing: a network-based approach to Amyotrophic Lateral Sclerosis. Neurotherapeutics. 2021 doi: 10.1007/s13311-021-01064-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sahoo B.M., Ravi Kumar B.V.V., Sruti J., Mahapatra M.K., Banik B.K., Borah P. Drug repurposing strategy (DRS): emerging approach to identify potential therapeutics for treatment of novel coronavirus infection. Front. Mol. Biosci. 2021;8 doi: 10.3389/fmolb.2021.628144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nitulescu G.M., Paunescu H., Moschos S.A., Petrakis D., Nitulescu G., Ion G.N.D., Spandidos D.A., Nikolouzakis T.K., Drakoulis N., Tsatsakis A. Comprehensive analysis of drugs to treat SARS‑CoV‑2 infection: mechanistic insights into current COVID‑19 therapies (Review) Int. J. Mol. Med. 2020;46(2):467–488. doi: 10.3892/ijmm.2020.4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim J.S., Lee J.Y., Yang J.W., Lee K.H., Effenberger M., Szpirt W., Kronbichler A., Shin J.I. Immunopathogenesis and treatment of cytokine storm in COVID-19. Theranostics. 2021;11(1):316–329. doi: 10.7150/thno.49713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.López-Collazo E., Avendaño-Ortiz J., Martín-Quirós A., Aguirre L.A. Immune response and COVID-19: a mirror image of sepsis. Int. J. Biol. Sci. 2020;16(14):2479–2489. doi: 10.7150/ijbs.48400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pum A., Ennemoser M., Adage T., Kungl A.J. Cytokines and chemokines in SARS-CoV-2 infections—therapeutic strategies targeting cytokine storm. Biomolecules. 2021;11(1):91. doi: 10.3390/biom11010091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rizk J.G., Kalantar-Zadeh K., Mehra M.R., Lavie C.J., Rizk Y., Forthal D.N. Pharmaco-immunomodulatory therapy in COVID-19. Drugs. 2020;80(13):1267–1292. doi: 10.1007/s40265-020-01367-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kulanthaivel S., Kaliberdenko V.B., Balasundaram K., Shterenshis M.V., Scarpellini E., Abenavoli L. Tocilizumab in SARS-CoV-2 patients with the syndrome of cytokine storm: a narrative review. Rev. Recent Clin. Trials. 2021;16(2):138–145. doi: 10.2174/1574887115666200917110954. [DOI] [PubMed] [Google Scholar]
- 33.Scavone C., Mascolo A., Rafaniello C., Sportiello L., Trama U., Zoccoli A., et al. Therapeutic strategies to fight COVID-19: Which is the status artis? Br. J. Pharmacol. 2021 doi: 10.1111/bph.15452. https://bpspubs.onlinelibrary.wiley.com/doi/abs/10.1111/bph.15452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barrett T., Wilhite S.E., Ledoux P., Evangelista C., Kim I.F., Tomashevsky M., Marshall K.A., Phillippy K.H., Sherman P.M., Holko M., Yefanov A., Lee H., Zhang N., Robertson C.L., Serova N., Davis S., Soboleva A. NCBI GEO: archive for functional genomics data sets—update. Nucleic Acids Res. 2013;41(D1):D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wishart D.S., Feunang Y.D., Guo A.C., Lo E.J., Marcu A., Grant J.R., Sajed T., Johnson D., Li C., Sayeeda Z., Assempour N., Iynkkaran I., Liu Y., Maciejewski A., Gale N., Wilson A., Chin L., Cummings R., Le D., Pon A., Knox C., Wilson M. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018;46:D1074–D1082. doi: 10.1093/nar/gkx1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kinsella R.J., Kähäri A., Haider S., Zamora J., Proctor G., Spudich G., Almeida-King J., Staines D., Derwent P., Kerhornou A., Kersey P., Flicek P. Ensembl BioMarts: a hub for data retrieval across taxonomic space. Database J. Biol. Database. Curation. 2011;2011 doi: 10.1093/database/bar030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7) doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paci P., Fiscon G., Conte F., Wang R.-S., Farina L., Loscalzo J. Gene co-expression in the interactome: moving from correlation toward causation via an integrated approach to disease module discovery. Npj Syst. Biol. Appl. 2021;7(1):1–11. doi: 10.1038/s41540-020-00168-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fiscon G., Conte F., Farina L., Paci P. SAveRUNNER: a network-based algorithm for drug repurposing and its application to COVID-19. PLOS Comput. Biol. 2021;17(2) doi: 10.1371/journal.pcbi.1008686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fiscon G., Paci P. SAveRUNNER: an R-based tool for drug repurposing. BMC Bioinform. 2021;22(1):150. doi: 10.1186/s12859-021-04076-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Iglewicz B., Hoaglin D. How to detect and handle outliers. ASQC Basic Ref. Qual. Control Stat. Tech. 1993:16. [Google Scholar]
- 43.Iba T., Warkentin T.E., Thachil J., Levi M., Levy J.H. Proposal of the definition for COVID-19-associated coagulopathy. J. Clin. Med. 2021;10(2):191. doi: 10.3390/jcm10020191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Price L.C., Garfield B., Bleakley C., Keeling A.G.M., Mcfadyen C., McCabe C., Ridge C.A., Wort S.J., Price S., Arachchillage D.J. Rescue therapy with thrombolysis in patients with severe COVID-19-associated acute respiratory distress syndrome. Pulm. Circ. 2020;10(4) doi: 10.1177/2045894020973906. 2045894020973906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu J., Li J., Arnold K., Pawlinski R., Key NS. Using heparin molecules to manage COVID‐2019. Res Pract Thromb Haemost [Internet]. 2020 Apr 23 [cited 2021 Jan 27]; Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7264589/. [DOI] [PMC free article] [PubMed]
- 46.Caballero López A., Herrera Cartaya C., Chávez González E., González Reinoso D., Caballero Font JA, Sánchez Vera N., et al. Pulmonary Thrombosis in COVID-19 Treated by Thrombolysis: A Small Case Series Using Streptokinase. Semin Thromb Hemost. 2020 Dec 30. [DOI] [PubMed]
- 47.Ogawa H., Asakura H. Consideration of tranexamic acid administration to COVID-19 patients. Physiol. Rev. 2020;100(4):1595–1596. doi: 10.1152/physrev.00023.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Demopoulos C., Antonopoulou S., Theoharides T.C. COVID-19, microthromboses, inflammation, and platelet activating factor. BioFactors Oxf. Engl. 2020;46(6):927–933. doi: 10.1002/biof.1696. [DOI] [PubMed] [Google Scholar]
- 49.Eldanasory O.A., Eljaaly K., Memish Z.A., Al-Tawfiq J.A. Histamine release theory and roles of antihistamine in the treatment of cytokines storm of COVID-19. Travel Med. Infect. Dis. 2020;37 doi: 10.1016/j.tmaid.2020.101874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sestili P., Stocchi V. Repositioning Chromones for Early Anti-inflammatory Treatment of COVID-19. Front Pharmacol [Internet]. 2020 Jun 5 [cited 2021 Jan 27];11. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7289983/. [DOI] [PMC free article] [PubMed]
- 51.Reznikov LR, Norris MH, Vashisht R., Bluhm AP, Li D., Liao Y.-SJ, et al. Identification of antiviral antihistamines for COVID-19 repurposing. Biochem Biophys Res Commun [Internet]. 2020 Dec 3 [cited 2021 Jan 27]; Available from: http://www.sciencedirect.com/science/article/pii/S0006291X20321409. [DOI] [PMC free article] [PubMed]
- 52.Yadav V., Talwar P. Repositioning of fluoroquinolones from antibiotic to anti-cancer agents: an underestimated truth. Biomed. Pharm. 2019;111:934–946. doi: 10.1016/j.biopha.2018.12.119. [DOI] [PubMed] [Google Scholar]
- 53.Abdel-Aal M.A.A., Abdel-Aziz S.A., Shaykoon M.S.A., Abuo-Rahma G.E.-D.A. Towards anticancer fluoroquinolones: a review article. Arch. Pharm. 2019;352(7) doi: 10.1002/ardp.201800376. [DOI] [PubMed] [Google Scholar]
- 54.Rasaeifar B., Gomez-Gutierrez P., Perez J.J. Molecular features of non-selective small molecule antagonists of the bradykinin receptors. Pharmaceuticals. 2020;13(9):259. doi: 10.3390/ph13090259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Assar S., Nosratabadi R., Azad H.K., Masoumi J., Mohamadi M., Hassanshahi G. A review of immunomodulatory effects of fluoroquinolones. Immunol. Investig. 2020;0(0):1–20. doi: 10.1080/08820139.2020.1797778. [DOI] [PubMed] [Google Scholar]
- 56.Abhithaj J., Dileep F., Sharanya C.S., Arun K.G., Sadasivan C., E Jayadevi V. Repurposing simeprevir, calpain inhibitor IV and a cathepsin F inhibitor against SARS-CoV-2 and insights into their interactions with Mpro. J. Biomol. Struct. Dyn. 2020:1–12. doi: 10.1080/07391102.2020.1813200. [DOI] [PubMed] [Google Scholar]
- 57.Scroggs S.L.P., Offerdahl D.K., Flather D.P., Morris C.N., Kendall B.L., Broeckel R.M., Beare P.A., Bloom M.E. Fluoroquinolone antibiotics exhibit low antiviral activity against SARS-CoV-2 and MERS-CoV. Viruses. 2020;13(1):8. doi: 10.3390/v13010008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sharma D., Kunamneni A. Recent progress in the repurposing of drugs/molecules for the management of COVID-19. Expert Rev. Anti Infect. Ther. 2020;0(0):1–9. doi: 10.1080/14787210.2021.1860020. [DOI] [PubMed] [Google Scholar]
- 59.Rose L., Graham L., Koenecke A., Powell M., Xiong R., Shen Z., et al. The Association Between Alpha-1 Adrenergic Receptor Antagonists and In-Hospital Mortality from COVID-19. MedRxiv Prepr Serv Health Sci. 2020 Dec 22; [DOI] [PMC free article] [PubMed]
- 60.Hyoju S.K., Zaborina O., van Goor H. SARS-CoV-2 and the sympathetic immune response: dampening inflammation with antihypertensive drugs (Clonidine and Propranolol) Med. Hypotheses. 2020;144 doi: 10.1016/j.mehy.2020.110039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Luo P., Liu D., Li J. Epinephrine use in COVID-19: friend or foe? Eur. J. Hosp. Pharm. 2021;28(1):1. doi: 10.1136/ejhpharm-2020-002295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hoertel N., Sánchez-Rico M., Vernet R., Beeker N., Jannot A.-S., Neuraz A., Salamanca E., Paris N., Daniel C., Gramfort A., Lemaitre G., Bernaux M., Bellamine A., Lemogne C., Airagnes G., Burgun A., Limosin F., AP-HP / Universities / INSERM COVID- Research Collaboration and AP-HP COVID CDR I. Association between antidepressant use and reduced risk of intubation or death in hospitalized patients with COVID-19: results from an observational study. Mol. Psychiatry. 2021:1–14. doi: 10.1038/s41380-021-01021-4. [DOI] [PubMed] [Google Scholar]
- 63.Grisanti L.A., Perez D.M., Porter J.E. Modulation of immune cell function by α(1)-adrenergic receptor activation. Curr. Top. Membr. 2011;67:113–138. doi: 10.1016/B978-0-12-384921-2.00006-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Otręba M., Kośmider L., Rzepecka-Stojko A. Antiviral activity of chlorpromazine, fluphenazine, perphenazine, prochlorperazine, and thioridazine towards RNA-viruses. A review. Eur. J. Pharmacol. 2020;887 doi: 10.1016/j.ejphar.2020.173553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Di Rosso M.E., Palumbo M.L., Genaro A.M. Immunomodulatory effects of fluoxetine: a new potential pharmacological action for a classic antidepressant drug? Pharmacol. Res. 2016;109:101–107. doi: 10.1016/j.phrs.2015.11.021. [DOI] [PubMed] [Google Scholar]
- 66.Pinoli M., Marino F., Cosentino M. Dopaminergic regulation of innate immunity: a review. J. NeuroImmune Pharmacol. J. Soc. NeuroImmune Pharmacol. 2017;12(4):602–623. doi: 10.1007/s11481-017-9749-2. [DOI] [PubMed] [Google Scholar]
- 67.Thomas Broome S., Louangaphay K., Keay K.A., Leggio G.M., Musumeci G., Castorina A. Dopamine: an immune transmitter. Neural Regen. Res. 2020;15(12):2173–2185. doi: 10.4103/1673-5374.284976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jones A., Kainz D., Khan F., Lee C., Carrithers M.D. Human macrophage SCN5A activates an innate immune signaling pathway for antiviral host defense*. J. Biol. Chem. 2014;289(51):35326–35340. doi: 10.1074/jbc.M114.611962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Carrithers M.D., Dib-Hajj S., Carrithers L.M., Tokmoulina G., Pypaert M., Jonas E.A., Waxman S.G. Expression of the voltage-gated sodium channel NaV1.5 in the macrophage late endosome regulates endosomal acidification. J. Immunol. Balt. Md 1950. 2007;178(12):7822–7832. doi: 10.4049/jimmunol.178.12.7822. [DOI] [PubMed] [Google Scholar]
- 70.Rahgozar K., Wright E., Carrithers L.M., Carrithers M.D. Mediation of protection and recovery from experimental autoimmune encephalomyelitis by macrophages expressing the human voltage-gated sodium channel NaV1.5. J. Neuropathol. Exp. Neurol. 2013;72(6):489–504. doi: 10.1097/NEN.0b013e318293eb08. [DOI] [PubMed] [Google Scholar]
- 71.Singh S., Florez H. Bioinformatic study to discover natural molecules with activity against COVID-19. F1000Research. 2020;9:1203. [DOI] [PMC free article] [PubMed]
- 72.Li G., Ruan S., Zhao X., Liu Q., Dou Y., Mao F. Transcriptomic signatures and repurposing drugs for COVID-19 patients: findings of bioinformatics analyses. Comput. Struct. Biotechnol. J. 2021;19:1–15. doi: 10.1016/j.csbj.2020.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Verbeke L., Mannaerts I., Schierwagen R., Govaere O., Klein S., Vander Elst I., Windmolders P., Farre R., Wenes M., Mazzone M., Nevens F., van Grunsven L.A., Trebicka J., Laleman W. FXR agonist obeticholic acid reduces hepatic inflammation and fibrosis in a rat model of toxic cirrhosis. Sci. Rep. 2016;6:33453. doi: 10.1038/srep33453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Carino A., Moraca F., Fiorillo B., Marchianò S., Sepe V., Biagioli M., Finamore C., Bozza S., Francisci D., Distrutti E., Catalanotti B., Zampella A., Fiorucci S. Hijacking SARS-CoV-2/ACE2 receptor interaction by natural and semi-synthetic steroidal agents acting on functional pockets on the receptor binding domain. Front. Chem. 2020;8 doi: 10.3389/fchem.2020.572885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yang C., Xiao S.-Y. COVID-19 and inflammatory bowel disease: a pathophysiological assessment. Biomed. Pharmacother. 2021;135 doi: 10.1016/j.biopha.2021.111233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.RECOVERY Collaborative Group, Horby P., Lim W.S., Emberson J.R., Mafham M., Bell J.L., et al. Dexamethasone in Hospitalized Patients with Covid-19. N. Engl. J. Med. 2021;384(8):693–704. doi: 10.1056/NEJMoa2021436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Barbieri A., Robinson N., Palma G., Maurea N., Desiderio V., Botti G. Can beta-2-adrenergic pathway be a new target to Combat SARS-CoV-2 hyperinflammatory syndrome?—lessons learned from cancer. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.588724. https://www.frontiersin.org/articles/10.3389/fimmu.2020.588724/full cited 2021 Jan 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material
Supplementary material
Data Availability Statement
SAveRUNNER source code is freely available at https://github.com/sportingCode/SAveRUNNER.git, along with a comprehensive user guide.