ABSTRACT
Accurate and early susceptibility results could reduce overuse of broad-spectrum antibiotics for empirical treatment of bacteremia. Direct disk diffusion testing (dDD) using nonstandardized inocula directly from blood cultures could facilitate earlier narrowing of antibiotics. To determine the predictive value of dDD compared with standardized antimicrobial susceptibility testing (AST), we performed a retrospective cohort study of 582 blood cultures from 495 pediatric patients with bacteremia. Positive and negative predictive value (PPV: number of isolates susceptible by both dDD and AST divided by the total number of isolates susceptible by dDD; NPV: number of isolates not susceptible [either intermediate or resistant] by both dDD and AST divided by the total number of isolates not susceptible by dDD), sensitivity, specificity, and 95% confidence interval were calculated for each bacterium-antibiotic combination. We evaluated the Antibiotic Spectrum Index of prescribed antibiotics to assess change in antibiotic prescribing after availability of Gram stain, dDD, and AST results. dDD results were available a median of 21 h before AST results. dDD had PPVs of ≥96% for most organism-antibiotic pairs, including 100% (CI 96 to 100%) for Staphylococcus aureus with oxacillin and 99% (CI 93 to 100%) for Enterobacterales with ceftriaxone. NPVs of dDD were variable and frequently lower than the PPV. Very major errors and major errors occurred in 31/5,454 (0.6%) and 231/5,454 (4.2%) organism-antibiotic combinations, respectively. Antibiotics were narrowed in 30% of cases after a dDD result and a further 25% of cases after AST result. dDD is highly predictive of susceptibility for many common organism-antibiotic combinations and provides actionable information one day earlier than standard susceptibility approaches. dDD has the potential to facilitate earlier deescalation to narrow-spectrum antibiotic treatment.
KEYWORDS: antimicrobial stewardship, antimicrobial susceptibility, bacteremia, pediatrics, diagnostic stewardship
INTRODUCTION
Proliferation of multidrug-resistant bacteria through repeated exposure to broad-spectrum antibiotics is an alarming and growing public health threat. The burden of multidrug-resistant pathogens is already substantial, with 2.8 million infections in the United States and 700,000 deaths globally each year (1, 2). Hospitalized patients with bacteremia are typically treated empirically with broad-spectrum antibiotics while organism identification and antibiotic susceptibility results are pending. Limiting the use of broad-spectrum antibiotics in this context has the potential to reduce the development of multidrug-resistant organisms and adverse drug events (3).
Rapid diagnostic tests are important elements of the antimicrobial stewardship toolkit and have been shown to decrease the use of broad-spectrum antibiotics in the treatment of invasive bacterial infections (4). Increasingly, rapid molecular diagnostic tools can provide actionable information earlier than conventional technologies (5–7). Implementation of these tools has had a mixed impact on antibiotic prescribing, with some leading to more rapid deescalation of antibiotics and others leading to antibiotic escalation (8–10). These strategies have been shown to be generally cost-effective, especially when paired with antimicrobial stewardship interventions (11).
Direct disk diffusion testing (dDD) is a phenotypic approach to antimicrobial susceptibility testing that utilizes nonstandardized disk diffusion to provide rapid preliminary susceptibility results (12–14). Specifically, the concentration of bacteria in the inoculum is not adjusted to the 0.5 McFarland standard. There has been limited evaluation of the accuracy of these dDD results in comparison to standardized final antimicrobial susceptibility testing (AST), particularly in bacteria from children (13, 15). If the rapid dDD results were shown to accurately predict final susceptible results, decisions based on dDD could decrease the use of broad-spectrum antibiotics by facilitating earlier targeted therapy.
The antibiotic spectrum index (ASI) is a metric that captures changes in the breadth of antibiotic prescribing. The ASI assigns a score to each antibiotic corresponding to the breadth of its coverage and, using a large cohort of pediatric patients with pneumonia, has been demonstrated to provide an additional dimension of variability in capturing prescribing patterns beyond the traditional metric of days of therapy (DOT) per 1,000 patient days (16).
We sought to evaluate the accuracy of dDD in predicting final susceptibility results for bacterial isolates in blood cultures from pediatric patients, and to use the ASI to evaluate whether the spectrum of prescribed antibiotics was changed by clinicians after dDD results became available.
MATERIALS AND METHODS
Subject selection.
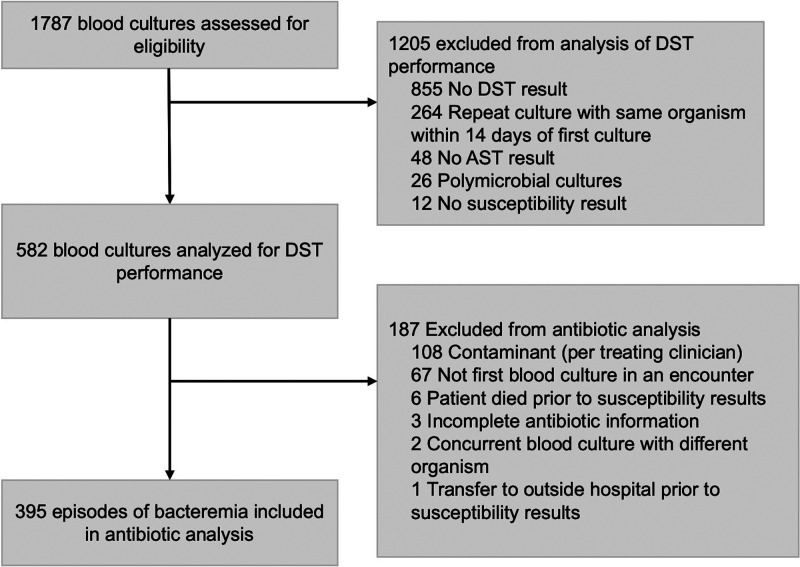
The study was approved by the Boston Children’s Hospital Institutional Review Board. We performed a retrospective cohort study. We collected data from the electronic medical record using an enterprise data warehouse to screen for all positive blood cultures collected at Boston Children’s Hospital between 1 January 2017 and 20 October 2019. Isolates were excluded if a dDD or AST result was missing or if the blood culture was polymicrobial. If a patient had repeat blood cultures positive for the same organism within 14 days of the first positive culture, only the first positive culture was included (Fig. 1). For secondary analysis of antibiotic administration in relation to the timing of susceptibility results, we excluded blood cultures if medication information was unavailable, if the organism was assessed by the treating clinicians to be a contaminant, or if the blood culture was not the first with that organism in a given patient encounter (Fig. 1). For this antibiotic analysis, we recorded patient characteristics and laboratory and clinical parameters within 24 h of blood culture collection.
FIG 1.
Cohort assembly.
Susceptibility testing.
Blood cultures were incubated using the BD Bactec FX system (BD, Franklin Lakes, NJ) and when bacterial growth was identified, a Gram stain was performed. If the Gram stain identified Gram-positive cocci in clusters or Gram-negative bacilli, dDD was performed. dDD is not performed on streptococci and enterococci in our institution, as prior data suggest this approach is not accurate for these bacteria (15). Blood culture bottles were processed within an hour of the time they were detected as positive. A transfer device (Covidien Monoject Angel Wing Safety Female) with a 3-ml syringe (BD) was used to access the blood culture bottle and draw broth into the syringe. Three drops of blood culture broth were added to a Mueller-Hinton agar plate and spread evenly over the surface of the plate in a basket-weave pattern, and antibiotic disks (BD BBL Sensi-Disc) placed within 15 min (12). A single size Covidien venting needle was used throughout. Quality control was performed weekly and with each new shipment or lot for all dDD panels. After inoculation, plates were incubated overnight (18 to 24 h) at 37°C prior to interpretation. For Gram-positive bacteria, dDD was performed and reported using antibiotic disks containing cefoxitin (30 μg), clindamycin (2 μg), erythromycin (15 μg), tetracycline (30 μg), and trimethoprim-sulfamethoxazole (1.25/23.75 μg). Cefoxitin is used as a proxy for oxacillin in susceptibility testing. For Gram-negative organisms, dDD was performed and reported using amikacin (30 μg), ampicillin (10 μg), ampicillin/sulbactam (20 μg), cefepime (30 μg), ceftazidime (30 μg), ceftriaxone (30 μg), ciprofloxacin (5 μg), gentamicin (10 μg), meropenem (10 μg), piperacillin-tazobactam (110 μg), and trimethoprim-sulfamethoxazole (1.25/23.75 μg). Ampicillin, ampicillin/sulbactam, ceftazidime, ceftriaxone, and piperacillin-tazobactam are routinely suppressed for species that can develop antibiotic resistance by constitutive expression of a chromosomal ampC gene. Susceptible, intermediate, and resistant interpretations were based upon CLSI guidelines (17). dDD results were posted to the patient’s chart as a “preliminary report” with the accompanying sentence “Preliminary susceptibility results are based on nonstandardized testing, final susceptibility results may differ.” Treating clinicians were not notified of this result by either the microbiology lab or the antimicrobial stewardship team. Once standardized antimicrobial susceptibility results were available, they were posted to the patient’s chart as “Final” and dDD results were removed from the chart. The following sentence accompanied the posting of final results: “All displayed susceptibility results are definitive, preliminary results are not displayed.” Standardized antimicrobial susceptibility testing was performed after subculturing via a standardized inoculum using automated broth microdilution with the Vitek 2 (bioMérieux, France) using the Vitek cards GP67, GN72, and GN73. Results from the Vitek-2 were used as the reference method to evaluate most results of dDD. Because we have noted errors in results for piperacillin-tazobactam and cefepime with the Vitek-2 for Gram-negative bacilli, disk diffusion susceptibility testing was performed for these two antibiotics with a standardized inoculum following CLSI guidelines. Routine identification was performed by matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) (Bruker MALDI Biotyper CA System) with additional phenotypic identification by Vitek-2 if needed. Genetic resistance marker testing is not performed at our institution.
Antibiotic use.
All antibiotic orders in the study period were identified and examined for start date and time and end date and time to determine which antibiotics were active after blood culture collection and at the time of Gram stain results (empirical antibiotics), after Gram stain result and at the time of dDD result (post-Gram stain), after dDD result and at the time of AST result (post-dDD), and 24 h after AST result (post-AST). The spectra of antibiotics were represented using the antibiotic spectrum index (ASI) (16). The ASI is a tool designed and validated to measure and compare the relative breadth of antibiotic spectrum used in hospitals. Each antibiotic is assigned a score, with one point given for each clinically relevant pathogen treated by the antibiotic, with a scale ranging from 0 points to 14 points. A higher ASI score indicates that an antibiotic has a broad spectrum, while a lower score indicates that an antibiotic has a narrow spectrum. Examples of clinically relevant pathogens that each contribute a point to the ASI of an individual antibiotic include methicillin-susceptible Staphylococcus aureus (MSSA), methicillin-resistant S. aureus (MRSA), vancomycin-resistant Enterococcus faecalis, Pseudomonas aeruginosa, AmpC beta-lactamase producers, and extended-spectrum beta-lactamase (ESBL)-producing Escherichia coli and Klebsiella species. Examples of ASI scores for specific antibiotics include oxacillin (ASI = 1), ampicillin (ASI = 2), cefazolin (ASI = 3), clindamycin (ASI = 4), ceftazidime (ASI = 4), ceftriaxone (ASI = 5), vancomycin (ASI = 5), linezolid (ASI = 6), cefepime (ASI = 6), ciprofloxacin (ASI = 6), piperacillin-tazobactam (ASI = 8), and meropenem (ASI = 10).
Statistical analysis.
We summarized patient characteristics using frequencies and proportions for categorical variables and medians and interquartile ranges for continuous variables. We calculated positive and negative predictive value (PPV: number of isolates susceptible by both dDD and AST divided by the total number of isolates susceptible by dDD; NPV: number of isolates not susceptible [either intermediate or resistant] by both dDD and AST divided by the total number of isolates not susceptible by dDD) and 95% confidence interval for each bacterium-antibiotic combination. We also calculated sensitivity (proportion of all susceptible isolates by AST that were identified as susceptible by dDD) and specificity (proportion of all nonsusceptible isolates by AST that were identified as nonsusceptible by dDD) for each bacterium-antibiotic combination. Agreement of susceptible, intermediate, and resistant results (categorical agreement) between dDD and AST was calculated. Discrepancies were identified and classified as very major errors (a susceptible result by dDD and resistant result by AST), major errors (resistant by dDD and susceptible by AST), and minor errors (any discrepancy between dDD and AST involving an intermediate result). Analyses were performed using R version 3.6.3 (R Core Team, Vienna, Austria).
RESULTS
Study population.
A total of 1,787 blood cultures were collected during the study period, of which 1,205 (67%) were excluded from analysis, most frequently as a consequence of missing susceptibility data (either dDD or AST). The resulting 582 blood cultures and 5,454 organism-antibiotic pairs were assessed to determine dDD performance (Fig. 1). Also, 67 positive cultures were excluded from antibiotic analysis to limit this analysis to the first episode of bacteremia per patient encounter. Of the remaining 515 positive blood cultures, 108 (21%) were considered contaminants by the treating clinician. Of these, 106/108 blood cultures identified as contaminants grew coagulase-negative staphylococci. After all exclusion criteria were applied, 395 episodes of bacteremia remained and underwent evaluation of antibiotic administration (Fig. 1).
Age of included subjects ranged from 0 months to 39 years (Table 1). Data on race and ethnicity were missing for 19% and 28% of patients, respectively. A minority of patients (37%) were female. A total of 18% of patients were neutropenic at the time of blood culture collection (Table 1), and 28 different bacterial species were included in analysis of dDD performance (Table 2).
TABLE 1.
Characteristics and laboratory results of patients treated for bacteremia (n = 395)
| Parameter | Value |
|---|---|
| Age (n [%]) | |
| 0 to <2 mo | 52 (13) |
| 2 mo to <1 yr | 64 (16) |
| 1 yr to 5 yrs | 102 (26) |
| 6 yrs to 10 yrs | 49 (12) |
| 11 yrs to 18 yrs | 83 (21) |
| Older than 18 yrs | 45 (11) |
| Race (n [%]) | |
| Asian | 22 (4.4) |
| Black or African American | 40 (8.1) |
| White | 233 (47) |
| Other | 107 (21.6) |
| Unknown | 94 (18.9) |
| Ethnicity (n [%]) | |
| Hispanic or Latinx | 29 (7) |
| Not Hispanic or Latinx | 231 (58) |
| Unknown | 135 (34) |
| Sex (n [%]) | |
| Male | 250 (63) |
| Female | 145 (37) |
| Primary service (n [%]) | |
| Medical | 145 (37) |
| Oncology/transplant | 77 (19) |
| Surgical | 116 (29) |
| Intensive care unit | 57 (14) |
| White blood cell count, cells/μla (median [IQR]) | 8.1 (4.4,14.2) |
| Severe neutropeniab (n [%]) | 58 (18) |
| C-reactive protein, mg/dlc (median [IQR]) | 6.8 (2.3,15.3) |
n = 330. IQR, interquartile range.
Absolute neutrophil count (ANC) < 500.
n = 193.
TABLE 2.
Organisms included in evaluation of direct disk diffusion testing performance (n = 582)
| Organism | n (%) |
|---|---|
| Acinetobacter speciesa | 6 (1.0) |
| Citrobacter speciesb | 6 (1.0) |
| Enterobacter cloacae | 21 (3.6) |
| Escherichia coli | 46 (7.9) |
| Escherichia vulneris | 1 (0.2) |
| Klebsiella speciesc | 57 (9.8) |
| Pantoea species | 1 (0.2) |
| Proteus mirabilis | 2 (0.3) |
| Pseudomonas aeruginosa | 17 (2.9) |
| Pseudomonas fluorescens group | 1 (0.2) |
| Salmonella species | 7 (1.2) |
| Salmonella enterica serovar Typhimurium | 4 (0.7) |
| Serratia liquefaciens | 1 (0.2) |
| Serratia marcescens | 16 (2.7) |
| Staphylococcus aureus | 137 (23.5) |
| Coagulase-negative staphylococcid | 259 (44.5) |
Includes Acinetobacter calcoaceticus-baumannii complex (n = 2), Acinetobacter radioresistens (n = 2), Acinetobacter ursingii (n = 2), and Acinetobacter species (n = 1).
Includes Citrobacter freundii complex (n = 5) and Citrobacter koseri (n = 1).
Includes Klebsiella aerogenes (n = 4), Klebsiella oxytoca/Raoutella ornithinolytica (n = 10), Klebsiella pneumoniae (n = 41), and Klebsiella variicola (n = 2).
Includes Staphylococcus capitis (n = 20), Staphylococcus epidermidis (n = 176), Staphylococcus haemolyticus (n = 7), Staphylococcus hominis (n = 44), Staphylococcus pettenkoferi (n = 4), and Staphylococcus warneri (n = 8).
Rates of antibiotic susceptibility in the studied cohort, as determined by standardized final susceptibility results, are shown in Table 3 and Table 4. Of note, 70% of E. coli and 77% of Klebsiella spp. were susceptible to ceftriaxone, and 78% of Staphylococcus aureus and 32% of coagulase-negative staphylococci were susceptible to oxacillin.
TABLE 3.
Antibiotic susceptibility (% susceptible) of Gram-negative organisms isolated from blood culture
| Organism (no. tested)a | % susceptible to:b |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Amikacin | Ampicillin | Ampicillin/sulbactam | Cefepime | Ceftazidime | Ceftriaxone | Ciprofloxacin | Gentamicin | Meropenem | Piperacillin/tazobactam | SXT | |
| Acinetobacter species (6) | 100 | 17 | 33 | 83 | 17 | - | 100 | 100 | 100 | 17 | 100 |
| Citrobacter species (6) | 100 | - | - | 100 | 17 | 17 | 100 | 83 | 100 | 0 | 83 |
| Enterobacter cloacae complex (21) | 100 | - | - | 90 | - | - | 100 | 95 | 100 | 0 | 95 |
| Escherichia coli (46) | 100 | 35 | 46 | 78 | 85 | 70 | 70 | 80 | 100 | 93 | 54 |
| Klebsiella species (57) | 100 | - | 58 | 88 | 79 | 77 | 81 | 96 | 95 | 86 | 74 |
| Pseudomonas aeruginosa (17) | 100 | - | - | 88 | 76 | - | 88 | 94 | 71 | 65 | - |
| Salmonella species (11) | - | 64 | - | - | - | 82 | - | - | - | - | 82 |
| Serratia marcescens (16) | 100 | - | - | 100 | - | - | 94 | 100 | 100 | - | 100 |
Definitive antibiotic susceptibility among Gram-negative organisms isolated from blood cultures included in initial cohort. Organisms excluded due to low numbers include Escherichia vulneris (n = 1), Pantoea species (n = 1), Proteus mirabilis (n = 2), Pseudomonas fluorescens group (n = 1), and Serratia liquefaciens (n = 1).
SXT, trimethoprim-sulfamethoxazole; the symbol “-” indicates not tested.
TABLE 4.
Antibiotic susceptibility of Gram-positive organisms isolated from blood culture
| Organism (no. tested) | % susceptible to:a |
||||
|---|---|---|---|---|---|
| Clindamycin | Oxacillin | Tetracycline | SXT | Vancomycin | |
| Staphylococcus aureus (137) | 80 | 78 | 99 | 99 | 100 |
| Coagulase-negative staphylococci (259) | 44 | 32 | 85 | 59 | 100 |
| Staphylococcus capitis (20) | 80 | 85 | 95 | 100 | 100 |
| Staphylococcus epidermidis (176) | 40 | 24 | 88 | 55 | 100 |
| Staphylococcus haemolyticus (7) | 0 | 0 | 100 | 0 | 100 |
| Staphylococcus hominis (44) | 45 | 32 | 68 | 64 | 100 |
| Staphylococcus pettenkoferi (4) | 75 | 25 | 75 | 50 | 100 |
| Staphylococcus warneri (8) | 63 | 88 | 75 | 100 | 100 |
SXT, trimethoprim-sulfamethoxazole.
Direct disk diffusion testing performance.
We evaluated the performance of dDD for each organism-antibiotic pair. Organism-antibiotic pairs with at least 20 isolates tested are shown in Table 5. Although there were fewer than 20 isolates of Pseudomonas aeruginosa, this species was included because of its clinical relevance. Susceptibility to oxacillin by dDD for S. aureus and coagulase-negative staphylococci was highly predictive of susceptibility by AST (PPV 100%, CI: 96 to 100% and PPV 97%, CI: 89 to 100%, respectively). However, resistance to oxacillin by dDD for S. aureus did not predict resistance by AST well (NPV 60%, CI: 45 to 74%). The predictive value of dDD to identify the susceptibility of Staphylococcus aureus to clindamycin was lower than that of oxacillin, at 86% (CI: 75 to 93%). The predictive value of dDD to identify the susceptibility of Enterobacterales to commonly used antibiotics was greater than 99%, with the lower bound of all 95% confidence intervals at ≥93%. The predictive value of susceptibility by dDD for Pseudomonas aeruginosa was similarly high, albeit with fewer isolates tested and wider confidence intervals. The accuracy of dDD in predicting nonsusceptibility for Enterobacterales varied substantially by antibiotic, ranging from meropenem (NPV = 5%, CI: 1 to 16%) to gentamicin (NPV = 100%, CI: 74 to 100%). In general, dDD was highly specific, with values at or near 100% for most organism-antibiotic combinations; exceptions included Staphylococcus aureus and clindamycin, Pseudomonas aeruginosa and ciprofloxacin, and Enterobacterales and piperacillin-tazobactam. Overall, sensitivity tended to be lower than specificity.
TABLE 5.
Predictive values of direct disk diffusion testing (dDD) of clinical isolates from patients with bacteremia to identify organism susceptibility to specific antibiotics
| Organism and antibiotic (no. of isolates tested)e | Positive predictive valuec (% [95% CI])e | Negative predictivec value (% [95% CI]) | Sensitivityd (% [95% CI]) | Specificityd (% [95% CI]) |
|---|---|---|---|---|
| Staphylococcus aureus | ||||
| Clindamycin (137) | 86 (75, 93) | 27 (17, 39) | 54 (45, 63) | 64 (47, 82) |
| Oxacillin (137) | 100 (96, 100) | 60 (45, 74) | 81 (74, 89) | 100 (100, 100) |
| SXT (137) | 100 (97, 100) | 25 (1, 81) | 98 (95, 100) | 100 (100, 100) |
| Coagulase-negative staphylococcia | ||||
| Clindamycin (254) | 88 (80, 94) | 81 (75, 87) | 73 (65, 86) | 92 (88, 97) |
| Oxacillin (251) | 97 (89, 100) | 89 (83, 93) | 73 (63, 82) | 99 (97, 100) |
| SXT (254) | 100 (97, 100) | 76 (71, 86) | 81 (75, 87) | 100 (100, 100) |
| Pseudomonas aeruginosa | ||||
| Cefepime (17) | 100 (78, 100) | 100 (16, 100) | 100 (100, 100) | 100 (100, 100) |
| Ceftazidime (17) | 100 (74, 100) | 80 (28, 100) | 92 (78, 100) | 100 (100, 100) |
| Ciprofloxacin (17) | 93 (68, 100) | 50 (1, 99) | 93 (81, 100) | 50 (0, 100) |
| Gentamicin (17) | 100 (78, 100) | 50 (1, 99) | 94 (82, 100) | 100 (100, 100) |
| Meropenem (17) | 100 (72, 100) | 83 (36, 100) | 92 (76, 100) | 100 (100, 100) |
| TZP (17) | 100 (72, 100) | 100 (48, 100) | 100 (100, 100) | 100 (100, 100) |
| Enterobacterales b | ||||
| Cefepime (135) | 100 (97, 100) | 77 (55, 92) | 96 (92, 99) | 100 (100, 100) |
| Ceftriaxone (110) | 99 (93, 100) | 64 (46, 79) | 85 (77, 92) | 96 (88, 100) |
| Ciprofloxacin (161) | 99 (95, 100) | 62 (48, 75) | 83 (77, 90) | 97 (92, 100) |
| Gentamicin (149) | 99 (97, 100) | 100 (74, 100) | 100 (100, 100) | 100 (100, 100) |
| Meropenem (149) | 100 (97, 100) | 5 (1, 16) | 72 (65, 79) | 100 (100, 100) |
| TZP (84) | 99 (93, 100) | 75 (35, 97) | 97 (94, 100) | 86 (60, 100) |
| Enterobacter cloacae complex | ||||
| Cefepime (20) | 100 (82, 100) | 100 (48, 100) | 100 (100, 100) | 100 (100, 100) |
| Gentamicin (20) | 100 (82, 100) | 100 (22, 100) | 100 (100, 100) | 100 (100, 100) |
| Meropenem (20) | 100 (78, 100) | 0 (0, 52) | 75 (56, 94) | NA |
| Escherichia coli | ||||
| Ampicillin (45) | 100 (66, 100) | 81 (64, 92) | 56 (32, 81) | 100 (100, 100) |
| Cefepime (37) | 100 (87, 100) | 70 (35, 93) | 90 (79, 100) | 100 (100, 100) |
| Ceftazidime (44) | 100 (90, 100) | 55 (23, 83) | 87 (76, 98) | 100 (100, 100) |
| Ceftriaxone (43) | 97 (82, 100) | 79 (49, 95) | 90 (80, 100) | 92 (76, 100) |
| Ciprofloxacin (45) | 100 (88, 100) | 81 (54, 96) | 91 (81, 100) | 100 (100, 100) |
| Gentamicin (44) | 100 (90, 100) | 100 (63, 100) | 100 (100, 100) | 100 (100, 100) |
| Klebsiella pneumoniae | ||||
| Cefepime (35) | 100 (87, 100) | 75 (35, 97) | 93 (84, 100) | 100 (100, 100) |
| Ceftazidime (41) | 100 (86, 100) | 44 (20, 70) | 74 (59, 88) | 100 (100, 100) |
| Ceftriaxone (40) | 100 (86, 100) | 53 (27, 79) | 78 (64, 92) | 100 (100, 100) |
| Ciprofloxacin (41) | 100 (85, 100) | 53 (29, 76) | 71 (55, 87) | 100 (100, 100) |
| Gentamicin (41) | 100 (91, 100) | 100 (16, 100) | 100 (100, 100) | 100 (100, 100) |
| Meropenem (40) | 100 (86, 100) | 13 (2, 38) | 63 (48, 78) | 100 (100, 100) |
| TZP (34) | 96 (82, 100) | 67 (22, 96) | 93 (84, 100) | 80 (45, 100) |
Includes Staphylococcus capitis, Staphylococcus epidermidis, Staphylococcus haemolyticus, Staphylococcus hominis, Staphylococcus pettenkoferi, and Staphylococcus warneri.
Includes Citrobacter freundii complex, Citrobacter koseri, Enterobacter cloacae complex, Escherichia coli, Escherichia vulneris, Klebsiella aerogenes, Klebsiella oxytoca, Klebsiella pneumoniae, Klebsiella variicola, Pantoea species, Proteus mirabilis, Salmonella species, Salmonella enterica serovar Typhimurium, Serratia liquefaciens, and Serratia marcescens.
Positive predictive value = number of isolates susceptible by both dDD and AST divided by total number of isolates susceptible by dDD. Negative predictive value = number of isolates not susceptible (either intermediate or resistant) by both dDD and AST divided by the total number of isolates not susceptible by dDD.
Sensitivity = ability of dDD to identify susceptible AST results (i.e., among susceptible [by AST] isolates, the percentage identified as susceptible by dDD). Specificity = ability of dDD to identify not susceptible AST results (i.e., among not susceptible [by AST] isolates, the percentage identified as not susceptible by dDD).
SXT, trimethoprim-sulfamethoxazole; TZP, piperacillin-tazobactam; CI, confidence interval.
There was categorical agreement in 4,778/5,454 (87.6%) of organism-antibiotic combinations. Very major errors occurred in 31/5,454 (0.6%), major errors occurred in 231/5,454 (4.2%), and minor errors occurred in 4,14/5,454 (7.6%) of organism-antibiotic combinations. Of the 31 very major errors, 19 involved clindamycin (5% of 382 clindamycin-organism combinations), 5 involved ampicillin-sulbactam (0.8% of 571 ampicillin-sulbactam-organism combinations), 2 involved ceftriaxone (1% of 193 ceftriaxone-organism combinations), 2 involved oxacillin (1% of 379 oxacillin-organism combinations), and 1 each involved cefazolin, linezolid, and rifampin (Table 6). Of the 31 very major errors, none resulted in use of ineffective therapy and thus none were associated with harm to the patients.
TABLE 6.
Agreement of direct disk diffusion testing (dDD) with standardized antimicrobial susceptibility testing (AST) using CLSI breakpoints
| Organism and antibiotic (no. of isolates tested)c | Susceptible, intermediate, and resistant agreement (no. [%]) | Very major errorsd (no. [%]) | Major errorsd (no. [%]) | Minor errorsd (no. [%]) |
|---|---|---|---|---|
| Staphylococcus aureus | ||||
| Clindamycin (137) | 68 (49.6) | 10 (7.3) | 0 (0) | 59 (43.1) |
| Oxacillin (137) | 117 (85.4) | 0 (0) | 20 (14.6) | 0 (0) |
| SXT (137) | 133 (97.1) | 0 (0) | 3 (2.2) | 1 (0.7) |
| Coagulase-negative staphylococcia | ||||
| Clindamycin (245) | 195 (79.6) | 9 (3.7) | 3 (1.2) | 38 (15.5) |
| Oxacillin (242) | 221 (91.3) | 2 (0.8) | 19 (7.9) | 0 (0) |
| SXT (245) | 217 (88.6) | 0 (0) | 22 (9) | 6 (2.4) |
| Pseudomonas aeruginosa | ||||
| Cefepime (17) | 16 (94.1) | 0 (0) | 0 (0) | 1 (5.9) |
| Ceftazidime (17) | 16 (94.1) | 0 (0) | 0 (0) | 1 (5.9) |
| Ciprofloxacin (17) | 15 (88.2) | 0 (0) | 0 (0) | 2 (11.8) |
| Gentamicin (17) | 16 (94.1) | 0 (0) | 0 (0) | 1 (5.9) |
| Meropenem (17) | 15 (88.2) | 0 (0) | 1 (5.9) | 1 (5.9) |
| TZP (16) | 16 (100) | 0 (0) | 0 (0) | 0 (0) |
| Enterobacterales b | ||||
| Cefepime (135) | 130 (96.3) | 0 (0) | 5 (3.7) | 0 (0) |
| Ceftriaxone (110) | 96 (87.3) | 1 (0.9) | 2 (1.8) | 11 (10) |
| Ciprofloxacin (161) | 135 (83.9) | 0 (0) | 5 (3.1) | 21 (13) |
| Gentamicin (149) | 149 (100) | 0 (0) | 0 (0) | 0 (0) |
| Meropenem (149) | 107 (71.8) | 0 (0) | 5 (3.4) | 37 (24.8) |
| TZP (84) | 81 (96.4) | 0 (0) | 0 (0) | 3 (3.6) |
| Enterobacter cloacae complex | ||||
| Cefepime (20) | 20 (100) | 0 (0) | 0 (0) | 0 (0) |
| Gentamicin (20) | 20 (100) | 0 (0) | 0 (0) | 0 (0) |
| Meropenem (20) | 15 (75) | 0 (0) | 0 (0) | 5 (25) |
| Escherichia coli | ||||
| Ampicillin (45) | 38 (84.4) | 0 (0) | 1 (2.2) | 6 (13.3) |
| Cefepime (37) | 34 (91.9) | 0 (0) | 3 (8.1) | 0 (0) |
| Ceftazidime (44) | 38 (84.4) | 0 (0) | 3 (6.7) | 4 (8.9) |
| Ceftriaxone (43) | 39 (90.7) | 1 (2.3) | 0 (0) | 3 (7) |
| Ciprofloxacin (45) | 42 (93.3) | 0 (0) | 0 (0) | 3 (6.7) |
| Gentamicin (44) | 44 (100) | 0 (0) | 0 (0) | 0 (0) |
| Klebsiella pneumoniae | ||||
| Cefepime (35) | 33 (94.3) | 0 (0) | 2 (5.7) | 0 (0) |
| Ceftazidime (41) | 32 (78) | 0 (0) | 3 (7.3) | 6 (14.6) |
| Ceftriaxone (40) | 33 (82.5) | 0 (0) | 1 (2.5) | 6 (15) |
| Ciprofloxacin (41) | 30 (73.2) | 0 (0) | 3 (7.3) | 8 (19.5) |
| Gentamicin (41) | 41 (100) | 0 (0) | 0 (0) | 0 (0) |
| Meropenem (40) | 25 (62.5) | 0 (0) | 3 (7.5) | 12 (30) |
| TZP (34) | 32 (94.1) | 0 (0) | 0 (0) | 2 (5.9) |
Includes Staphylococcus capitis, Staphylococcus epidermidis, Staphylococcus haemolyticus, Staphylococcus hominis, Staphylococcus pettenkoferi, and Staphylococcus warneri.
Includes Citrobacter freundii complex, Citrobacter koseri, Enterobacter cloacae complex, Escherichia coli, Escherichia vulneris, Klebsiella aerogenes, Klebsiella oxytoca, Klebsiella pneumoniae, Klebsiella variicola, Pantoea species, Proteus mirabilis, Salmonella species, Salmonella enterica serovar Typhimurium, Serratia liquefaciens, and Serratia marcescens.
SXT, trimethoprim-sulfamethoxazole; TZP, piperacillin-tazobactam.
Very major errors: a susceptible result by dDD and a resistant result by AST. Major error: a resistant result by dDD and a susceptible result by AST. Minor error: a discrepancy between dDD and AST results involving an intermediate result.
Antibiotic use.
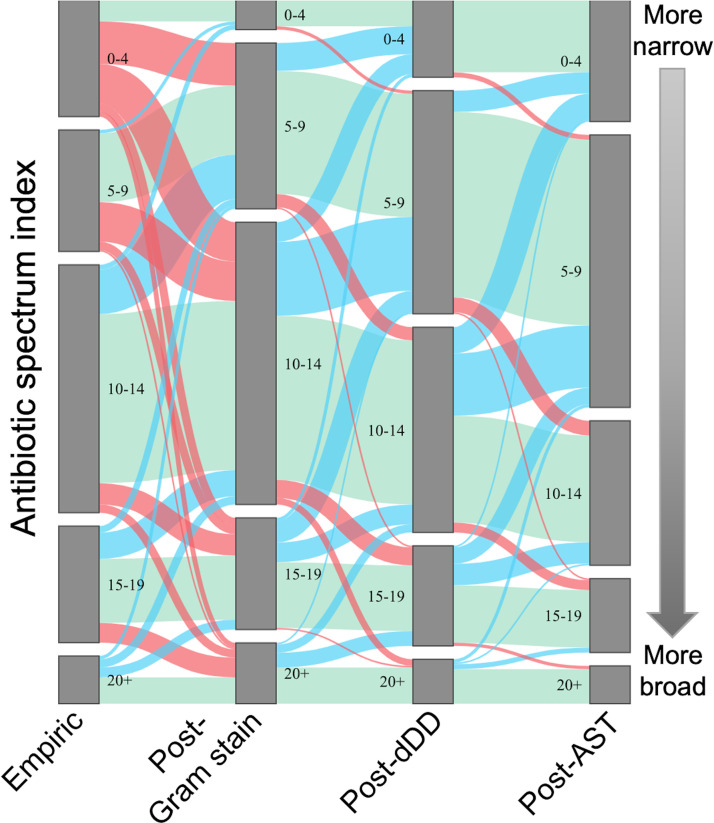
The dDD results were available at a median of 24 h (interquartile range [IQR] 18 to 28 h) after the Gram stain result, and AST results were available at a median of 45 h (IQR 38 to 49 h) after Gram stain result. Antibiotic spectra of prescribed empirical treatment of bacteremia, treatment after Gram stain result, treatment after dDD result, and treatment after AST result are shown in Fig. 2. Antibiotic spectrum indices were grouped to visualize the distribution of breadth of prescribed antibiotics, with 0 to 4 indicating the narrowest therapy, 5 to 9 narrow-moderate therapy, 10 to 14 moderate spectrum therapy, 15 to 19 broad, and 20+ very broad therapy (Fig. 2). The proportion of patients who were treated with antibiotics within a given ASI range is represented by the height of the gray columns. The green bands represent the patients whose ASI remained in the same category from one time point to the next, the blue bands represent the patients whose ASI decreased, and the red bands represent the patients whose ASI increased. The most frequent aggregate ASI range of antibiotics chosen as empirical treatment of bacteremia was 10 to 14, occurring in 38% (151/396) of cases, followed by 5 to 9, 0 to 4, 15 to 19, and 20+ (19%, 18%, 18%, and 7%, respectively). After Gram stain results, 30% of patients (118/396) had antibiotics broadened, 21% (81/396) had antibiotics narrowed, and 50% (197/396) maintained the same aggregate ASI score. After the dDD result, 29% (116/396) of patients had antibiotics narrowed, 64% (253/396) kept the same ASI score, and 7% (27/396) had their antibiotics broadened. This resulted in 34% of patients having an aggregate ASI of 5 to 9, 32% with an aggregate ASI of 10 to 14, and 15%, 12%, and 7% having an aggregate ASI of 15 to 19, 0 to 4, and 20+, respectively. After final standardized susceptibility results were available, a further 25% (97/396) of patients had antibiotics narrowed, 70% (278/396) maintained the same spectrum, and 5% (21/396) were broadened.
FIG 2.
Distribution of spectrum of antibiotics ordered to treat bacteremia in pediatric patients. Antibiotic spectrum, as measured by the antibiotic spectrum index (ASI), is represented in this Sankey diagram on the y axis. The height of each group indicates the proportion of patients with an aggregate antibiotic spectrum falling in that category at four time points. The ASI of empirical antibiotics was measured at the time of the Gram stain result to provide time for a treating physician to settle on an empirical regimen (labeled as Empiric). The post-Gram stain ASI was measured at the time of the dDD result (labeled as post-Gram stain). The ASI with the dDD result was measured at the time of the AST result (labeled as post-dDD). The ASI with the AST result was measured 24 h after the AST result was available (labeled as post-AST). Blue bands between the columns indicate the proportion of patients whose ASI decreased, green bands the proportion of patients whose ASI remained the same, and red bands the proportion whose ASI increased between two time points.
DISCUSSION
In this study, of bacterial isolates from children with positive blood cultures, we found that dDD had high specificity and high positive predictive value to identify antibiotic susceptibility for many clinically relevant bacteria, equivalent to or exceeding that of available molecular diagnostic approaches used in bacteremia (18). Notably, dDD had high PPVs to identify oxacillin susceptibility for Staphylococcus aureus, as well as ceftriaxone and cefepime susceptibilities for Enterobacterales. By providing earlier and accurate oxacillin susceptibility information, dDD has the potential to reduce empirical vancomycin use in the treatment of bacteremia, a longstanding antimicrobial stewardship priority with the potential to result in reduced vancomycin resistance (19, 20). Cefepime and carbapenems are frequently used in the empirical treatment of bacteremia, particularly in immunocompromised patients (21, 22). Reliably predictive cefepime susceptibility could facilitate discontinuation of empirical carbapenem therapy up to 1 day earlier than with conventional susceptibility testing. This intervention could potentially help slow the development of carbapenem-resistant Enterobacterales, identified as an urgent threat by the Centers for Disease Control and Prevention (1, 23). dDD had low rates of very major errors, occurring in only 0.6% of all organism-antibiotic combinations. The rate of major errors was higher at 4.2% and, in this context, could lead to excessively broad antibiotic use until final susceptibility is available. Of the 31 very major errors in our study, 19 involved clindamycin. All 19 of these errors were a consequence of erythromycin-inducible clindamycin resistance, which is not detected in our dDD but is detected by our AST. Placing the erythromycin and clindamycin disks adjacent to each other during dDD might allow detection of erythromycin-inducible clindamycin resistance. This approach was not addressed in the current study but could be investigated in the future.
dDD could serve as an antimicrobial stewardship tool to identify a susceptible result early, before a definitive standardized susceptibility result is available, and facilitate antibiotic narrowing in patients who are stable or improving on empirical therapy. In contrast, if a patient is failing to improve on empirical therapy, coverage can be broadened while awaiting definitive susceptibility results. As dDD has variable and often low negative predictive value and sensitivity, using a resistant dDD result to broaden therapy in a clinically stable or improving patient could lead to unnecessary broadening of antibiotics, as has been seen with other diagnostic tools incorporated into stewardship programs (10). While descriptive, our analysis of antibiotic use suggests that in our institution, clinicians may be using dDD results more frequently to narrow antibiotics rather than to broaden antibiotics. With receipt of a resistant result by dDD, some clinicians may broaden therapy regardless of clinical context, whereas others may continue therapy if the patient is improving. Prospective studies are needed to further evaluate the impact of dDD on antibiotic prescribing, and microbiology labs might consider selectively reporting only dDD results demonstrated to have high predictive value.
Improved diagnostic tests for resistant Gram-negative organisms rank as the greatest unmet diagnostic need among infectious disease physicians (24). There are numerous genes that confer antimicrobial resistance in these bacteria and molecular diagnostic techniques, while rapid, can be limited by the ability to capture all of the clinically relevant resistance mechanisms (25). The phenotypic approach of dDD for susceptibility testing can capture the consequences of resistance mechanisms regardless of the genetic underpinnings, with the exception of inducible mechanisms of resistance, and thus has particularly notable potential as an antimicrobial stewardship tool for Gram-negative bacteria. dDD also has the potential to be a valuable antimicrobial stewardship tool for Gram-positive bacteria. While genetic resistance marker testing is a more rapid option than dDD for these bacteria, it is also more expensive than dDD (10).
We observed similar rates of agreement and error between dDD and AST compared to those reported by Edelmann et al. and Menon et al. (13, 15). To our knowledge, our study is the largest evaluation of dDD in terms of organism and antibiotic combinations, the largest evaluation of dDD and Staphylococcus aureus, and the first to evaluate the sensitivity, specificity, positive predictive value, and negative predictive value of dDD to detect susceptibility (13, 15). Our study is also the first to describe antibiotic prescribing in relation to dDD result availability in pediatric patients. Compared with other commercially available phenotypic susceptibility tests, dDD tests were not as rapid. dDD results were available a median of 24 h (IQR 18 to 28 h) after the Gram stain result, compared with 7 h for automated fluorescence in situ hybridization technology with morphokinetic cellular analysis (26).
We did not perform a cost-effectiveness analysis as part of this study, but with a materials cost of $1.45 per sample at our institution, dDD has the potential to be a useful antimicrobial stewardship tool that could be readily adopted. Future data about cost effectiveness would provide further insight about the value of this approach.
This study had several limitations. The study was observational with no control group. Although we were able to describe changes in prescribed antibiotics in relation to the timing of Gram stain, dDD, and AST results, we were not able to confirm the reason for selection of a specific antibiotic, which could be based on other factors such as allergies or medication interactions. More antibiotics are reported via AST than dDD (vancomycin for Gram-positive cocci and cefazolin and levofloxacin for Gram-negative rods) and it is possible that this information influenced the clinician’s choice of antibiotic. In addition, we could not separate the influence of organism identification (which appears slightly earlier than the dDD result) on decisions to narrow or broaden from empirical therapy. The number of blood cultures with Gram-negative species, including Pseudomonas aeruginosa or Enterobacter cloacae complex, was small, limiting the precision of the estimate of accuracy of dDD in predicting final AST for these organisms. Additionally, the positive and negative predictive values of dDD in predicting susceptibility are influenced by rates of resistance, which were fairly low in our cohort, so local antibiograms should be considered when generalizing the findings of this study.
Direct disk diffusion testing is highly predictive of antibiotic susceptibility for many important bacteria and is available 1 day earlier than standardized final susceptibility results. dDD has the potential to facilitate earlier deescalation to narrow-spectrum antibiotic treatment, and its low cost could make it an accessible antimicrobial stewardship tool in low-resource settings. Prospective studies using direct disk diffusion testing with integrated decision support and/or antimicrobial stewardship program interventions to guide antibiotic therapy are needed to assess the impact of dDD on antibiotic prescribing practices.
ACKNOWLEDGMENTS
We thank Erica Ullo for technical assistance with direct disk diffusion testing.
We report no conflicts of interest.
Footnotes
Supplemental material is available online only.
Contributor Information
Timothy J. Savage, Email: timothy.savage@childrens.harvard.edu.
Patricia J. Simner, Johns Hopkins
REFERENCES
- 1.Centers for Disease Control. 2019. Antibiotic resistance threats in the United States. Centers for Disease Control, Atlanta, GA. 10.15620/cdc:82532. [DOI] [Google Scholar]
- 2.Interagency Coordination Group on Antimicrobial Resistance (IACG). 2019. No time to wait: securing the future from drug-resistant infections. World Health Organization, Geneva, Switzerland. https://www.who.int/antimicrobial-resistance/interagency-coordination-group/final-report/en/.
- 3.Tamma PD, Avdic E, Li DX, Dzintars K, Cosgrove SE. 2017. Association of adverse events with antibiotic use in hospitalized patients. JAMA Intern Med 177:1308–1315. 10.1001/jamainternmed.2017.1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Banerjee R, Teng CB, Cunningham SA, Ihde SM, Steckelberg JM, Moriarty JP, Shah ND, Mandrekar JN, Patel R. 2015. Randomized trial of rapid multiplex polymerase chain reaction-based blood culture identification and susceptibility testing. Clin Infect Dis 61:1071–1080. 10.1093/cid/civ447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Messacar K, Parker SK, Todd JK, Dominguez SR. 2017. Implementation of rapid molecular infectious disease diagnostics: the role of diagnostic and antimicrobial stewardship. J Clin Microbiol 55:715–723. 10.1128/JCM.02264-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.She RC, Bender JM. 2019. Advances in rapid molecular blood culture diagnostics: healthcare impact, laboratory implications, and multiplex technologies. J Appl Lab Med 3:617–630. 10.1373/jalm.2018.027409. [DOI] [PubMed] [Google Scholar]
- 7.van Belkum A, Burnham CAD, Rossen JWA, Mallard F, Rochas O, Dunne WM. 2020. Innovative and rapid antimicrobial susceptibility testing systems. Nat Rev Microbiol 18:299–311. 10.1038/s41579-020-0327-x. [DOI] [PubMed] [Google Scholar]
- 8.Hogan CA, Ebunji B, Watz N, Kapphahn K, Rigdon J, Mui E, Meng L, Alegria W, Holubar M, Deresinski S, Banaei N. 2020. Impact of rapid antimicrobial susceptibility testing in Gram-negative rod bacteremia: a quasi-experimental study. J Clin Microbiol 58:e00360-20. 10.1128/JCM.00360-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tseng AS, Kasule SN, Rice F, Mi L, Chan L, Seville MT, Grys TE. 2018. Is it actionable? An evaluation of the rapid PCR-based blood culture identification panel on the management of Gram-positive and Gram-negative blood stream infections. Open Forum Infect Dis 5:ofy308. 10.1093/ofid/ofy308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Juttukonda LJ, Katz S, Gillon J, Schmitz J, Banerjee R. 2019. Impact of a rapid blood culture diagnostic test in a children’s hospital depends on gram-positive versus gram-negative organism and day versus night shift. J Clin Microbiol 58:e01400-19. 10.1128/JCM.01400-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pliakos EE, Andreatos N, Shehadeh F, Ziakas PD, Mylonakis E. 2018. The cost-effectiveness of rapid diagnostic testing for the diagnosis of bloodstream infections with or without antimicrobial stewardship. Clin Microbiol Rev 31:e00095-17. 10.1128/CMR.00095-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Esser VM, Elefson DONE. 1969. Microbiology laboratory, Department of Pathology, Mercy Hospital, Des Moines, Iowa. [Google Scholar]
- 13.Edelmann A, Pietzcker T, Wellinghausen N. 2007. Comparison of direct disk diffusion and standard microtitre broth dilution susceptibility testing of blood culture isolates. J Med Microbiol 56:202–207. 10.1099/jmm.0.46937-0. [DOI] [PubMed] [Google Scholar]
- 14.Humphries RM, Kircher S, Ferrell A, Krause KM, Malherbe R, Hsiung A, Burnham CAD. 2018. The continued value of disk diffusion for assessing antimicrobial susceptibility in clinical laboratories: report from the clinical and laboratory standards institute methods development and standardization working group. J Clin Microbiol 56:e00437-18. 10.1128/JCM.00437-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Menon V, Lahanas S, Janto C, Lee A. 2016. Utility of direct susceptibility testing on blood cultures: is it still worthwhile? J Med Microbiol 65:501–509. 10.1099/jmm.0.000259. [DOI] [PubMed] [Google Scholar]
- 16.Gerber JS, Hersh AL, Kronman MP, Newland JG, Ross RK, Metjian TA. 2017. Development and application of an antibiotic spectrum index for benchmarking antibiotic selection patterns across hospitals. Infect Control Hosp Epidemiol 38:993–997. 10.1017/ice.2017.94. [DOI] [PubMed] [Google Scholar]
- 17.Clinical and Laboratory Standards Institute (CLSI). 2017. Performance standards for antimicrobial susceptibility testing, supplement M100, 27th edition. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 18.Peker N, Couto N, Sinha B, Rossen JW. 2018. Diagnosis of bloodstream infections from positive blood cultures and directly from blood samples: recent developments in molecular approaches. Clin Microbiol Infect 24:944–955. 10.1016/j.cmi.2018.05.007. [DOI] [PubMed] [Google Scholar]
- 19.Larson EL, Mayhall CG, Nichols RL, Russell BJ, Adams AB, Fleming DW. 1995. Recommendations for preventing the spread of vancomycin resistance. Recommendations of the Hospital Infection Control Practices Advisory Committee (HICPAC). MMWR Recomm Rep 44:1–13. [PubMed] [Google Scholar]
- 20.Karandikar MV, Milliren CE, Zaboulian R, Peiris P, Sharma T, Place AE, Sandora TJ. 2019. Limiting vancomycin exposure in pediatric oncology patients with febrile neutropenia may be associated with decreased vancomycin-resistant Enterococcus incidence. J Pediatric Infect Dis Soc 9:428–436. 10.1093/jpids/piz064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Freifeld AG, Bow EJ, Sepkowitz KA, Boeckh MJ, Ito JI, Mullen CA, Raad II, Rolston KV, Young JAH, Wingard JR, Infectious Diseases Society of America . 2011. Clinical practice guideline for the use of antimicrobial agents in neutropenic patients with cancer: 2010 Update by the Infectious Diseases Society of America. Clin Infect Dis 52:e56–e93. 10.1093/cid/cir073. [DOI] [PubMed] [Google Scholar]
- 22.Children’s Oncology Group. 2015. Guideline for the management of fever and neutropenia in children with cancer and/or undergoing hematopoietic stem-cell transplantation. Children’s Oncology Group, Monrovia, CA. http://childrensoncologygroup.org/downloads/COG_SC_FN_Guideline_Document.pdf. [Google Scholar]
- 23.Hussein K, Sprecher H, Mashiach T, Oren I, Kassis I, Finkelstein R. 2009. Carbapenem resistance among Klebsiella pneumoniae isolates risk factors, molecular characteristics, and susceptibility patterns. Infect Control Hosp Epidemiol 30:666–671. 10.1086/598244. [DOI] [PubMed] [Google Scholar]
- 24.Blaschke AJ, Hersh AL, Beekmann SE, Ince D, Polgreen PM, Hanson KE. 2015. Unmet diagnostic needs in infectious disease. Diagn Microbiol Infect Dis 81:57–59. 10.1016/j.diagmicrobio.2014.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cox G, Wright GD. 2013. Intrinsic antibiotic resistance: mechanisms, origins, challenges and solutions. Int J Med Microbiol 303:287–292. 10.1016/j.ijmm.2013.02.009. [DOI] [PubMed] [Google Scholar]
- 26.Charnot-Katsikas A, Tesic V, Love N, Hill B, Bethel C, Boonlayangoor S, Beavis KG. 2017. Use of the Accelerate Pheno system for identification and antimicrobial susceptibility testing of pathogens in positive blood cultures and impact on time to results and workflow. J Clin Microbiol 56:e01166-17. 10.1128/JCM.01166-17. [DOI] [PMC free article] [PubMed] [Google Scholar]