ABSTRACT
The aim of current study was to explore the mechanism of microRNA 142-5p (miR-142-5p) in cervical cancer through mediating the phosphoinositol-3-kinase adaptor protein 1 (PIK3AP1)/PI3K/AKT axis. To this end, reverse transcription-quantitative PCR (RT-qPCR) and Western blot analysis results revealed that miR-142-5p was poorly expressed, whereas PIK3AP1 was highly expressed, in cervical cancer tissues and cells. Furthermore, miR-142-5p was hypermethylated in cervical cancer, as reflected by methylation-specific PCR (MS-PCR) and chromatin immunoprecipitation (ChIP) assessment of enrichment of DNMT1/DNMT3a/DNMT3b in the promoter region of miR-142-5p. A target binding relationship between miR-142-5p and PIK3AP1 was established, showing that miR-142-5p targeted and inhibited the expression of PIK3AP1. Loss- and gain-of-function assays were conducted to determine the roles of miR-142-5p and PIK3AP1 in cervical cancer cells. CCK-8, flow cytometry, and Transwell assay results revealed that overexpression of miR-142-5p in cervical cancer cells downregulated PIK3AP1 and inhibited the PI3K/AKT signaling pathway, leading to reduced proliferation, migration, and invasion capacity of cervical cancer cells but enhanced apoptosis. Collectively, epigenetic regulation of miR-142-5p targeted PIK3AP1 to inactivate the PI3K/AKT signaling pathway, thus suppressing development of cervical cancer, which presents new targets for the treatment of cervical cancer.
KEYWORDS: cervical cancer, microRNA 142-5p, phosphoinositol-3-kinase adaptor protein 1, migration, phosphatidylinositol 3-kinase adaptor protein 1/AKT
INTRODUCTION
Cervical cancer remains a major cause of cancer-related morbidity around the globe (1). More than 500,000 women are diagnosed with cervical cancer annually, which brings more than 300,000 deaths worldwide, despite recognition that the high-risk subtype of human papillomavirus (HPV) is the main cause of the disease (2). Researches show that population-wide vaccination in adult women against the HPV associated with cervical may significantly reduce the incidence of cervical cancer (3). Furthermore, cervical cancer screening through detection and treatment of high cervical intraepithelial neoplasia has been highly successful in preventing this cancer (4). Standard therapeutic methods include external irradiation and brachytherapy with cisplatin chemotherapy (5). However, resistance to chemotherapy and radiotherapy is a major obstacle to the effective treatment of advanced cervical cancer (6).
A prior report has shown that microRNAs (miRNAs) are associated with the recurrence and metastasis of cervical cancer (7). miRNAs are small noncoding RNAs that regulate a variety of target genes and therefore participate in the regulation of a variety of biological and pathological processes, including the onset and development of cancer (8). The expression of miR-142-5p is reportedly downregulated in cervical cancer (9), and miR-142-5p expression is silenced in HPV-positive cells (10). Moreover, miRNA-142-3p is poorly expressed in cervical cancer, and its low expression is linked to poor overall survival and progression-free survival (11). We applied the Starbase database in the present study to predict binding sites between miR-142-5p and the 3′ untranslated region (UTR) region of phosphoinositol-3-kinase adaptor protein 1 (PIK3AP1) mRNA. PIK3AP1 is an adaptor protein originally isolated from B cells, which can promote cancer proliferation and chemosensitivity (12). A previous report has suggested that PIK3AP1 expression is elevated in gastric cancer and that patients with high PIK3AP1 expression in tumors have dismal survival (13). Additionally, PIK3AP1 has been reported to activate the PI3K/AKT signaling pathway (13), which is involved in a variety of biological processes and is often abnormally activated in human cancers (14). Furthermore, PIK3AP1 and AKT gene expression are positively correlated with cancer (12). Investigation of the relation between PI3K/AKT and cervical cancer shows that ARHGAP17 inhibits tumor progression by inhibiting the PI3K/AKT signaling pathway (15), whereas activation of mitogen-activated protein kinase (MAPK), PI3K/AKT, and hTERT signaling promotes tumor growth and metastasis and regulates radiosensitivity of cervical cancer (16). Therefore, this study aimed to explore how miR-142-5p affects cervical cancer through mediating the PIK3AP1/PI3K/AKT axis.
RESULTS
miR-142-5p is poorly expressed in cervical cancer tissues and cells, and overexpression of miR-142-5p inhibits the proliferation, migration, and invasion of cancer cells and promotes apoptosis.
The expression of miR-142-5p is downregulated in cervical cancer (9). To determine the influence of miR-142-5p on the occurrence and development of cervical cancer, we measured the expression of miR-142-5p in cervical cancer tissues and cells by reverse transcription-quantitative PCR (RT-qPCR), which revealed significantly lower miR-142-5p expression in cervical cancer tissues and cells than that in normal samples (Fig. 1A). The association between miR-142 and survival of patients with cervical cancer included in The Cancer Genome Atlas (TCGA) showed a better survival rate of patients with relatively high expression of miR-142 (Fig. 1B), indicating a prognostic role of miR-142.
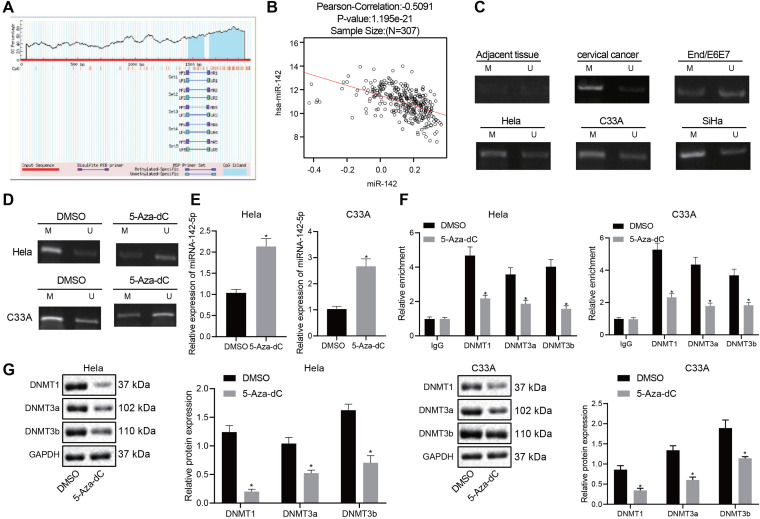
FIG 1.
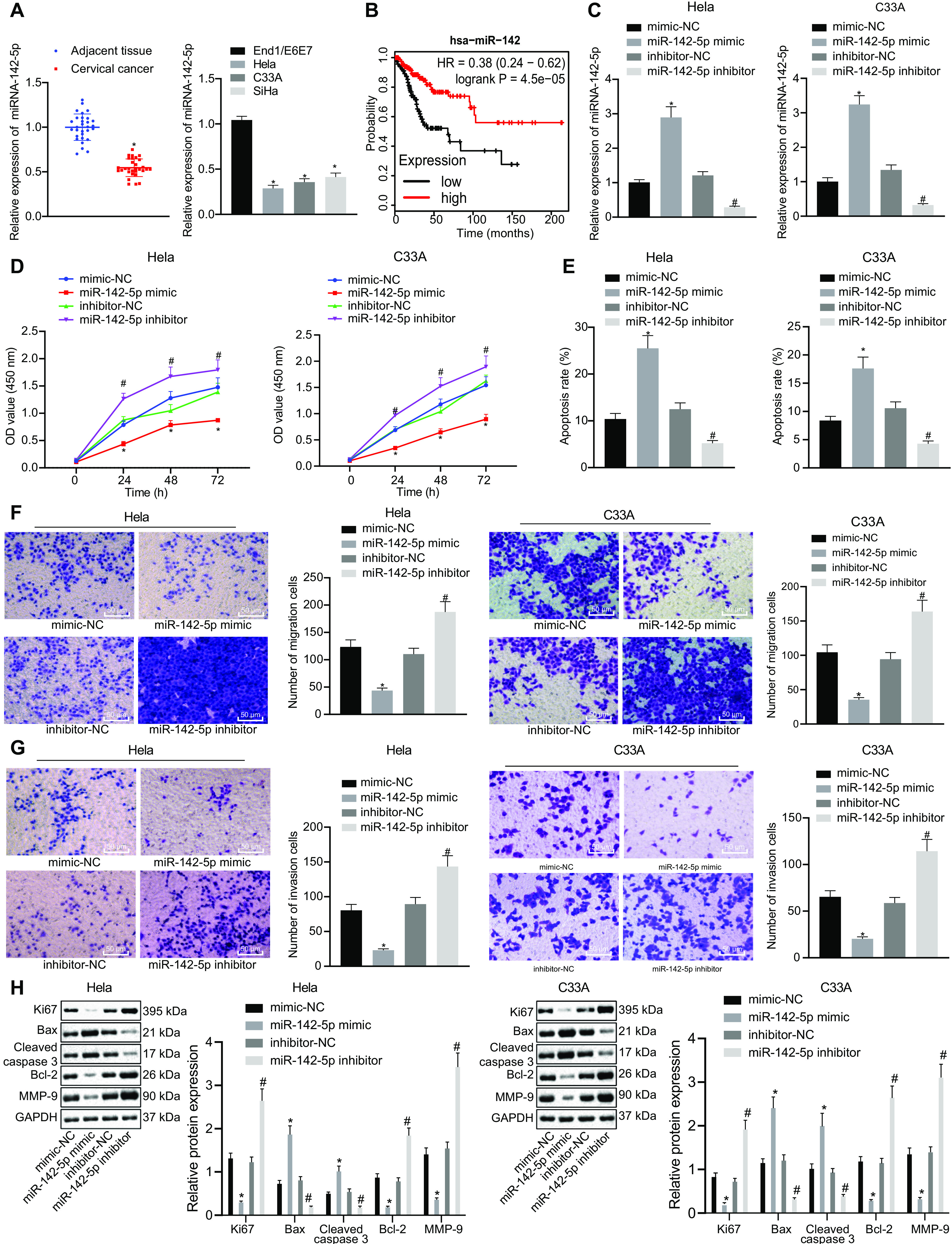
miR-142-5p is poorly expressed in cervical cancer tissues and cells, and overexpression of miR-142-5p inhibits the proliferation, migration, and invasion of cancer cells and promotes apoptosis. HeLa and C33A were transfected with miR-142-5p mimic or miR-142-5p inhibitor. (A) The expression of miR-142-5p in the clinical tissues and cells of cervical cancer was detected by RT-qPCR (n = 30). (B) Survival analysis of miR-142 in the clinical treatment of TCGA cervical cancer. The x axis represents the survival time, and the y axis represents the survival rate, with red indicating the survival status of patients with high expression and black indicating the survival status of patients with low expression. (C) miR-142-5p expression in the transfected HeLa and C33A cells was measured using RT-qPCR. (D) Proliferation was detected using CCK-8. (E) Apoptosis was examined by flow cytometry. (F and G) Migration and invasion were tested using the Transwell assay (×200). (H) The expression of related genes was monitored by Western blot analysis. The measurement data are expressed as means ± standard deviations. The paired t test was used to compare the carcinoma and adjacent tissues. The data comparison between multiple groups was performed using ANOVA, followed by Tukey’s post hoc test. For data comparison between groups at different time points, repeated-measurement ANOVA was used, followed by Tukey’s post hoc test. *, P < 0.05 versus adjacent tissues or mimic-NC-transfected HeLa and C33A cells; #, P < 0.05 versus inhibitor-NC-transfected HeLa and C33A cells.
To further study the effects of miR-142-5p on the biological characteristics of cervical cancer cells, cervical cancer cells (HeLa and C33A) were transfected with miR-142-5p mimic or miR-142-5p inhibitor. The results of RT-qPCR analysis showed that miR-142-5p expression was notably elevated in miR-142-5p mimic-transfected HeLa and C33A cervical cancer cells but was markedly downregulated upon miR-142-5p inhibitor transfection, thus validating the successful transfection efficiency (Fig. 1C). The cell counting kit 8 (CCK-8) assay showed that the proliferation of HeLa and C33A cells was dramatically reduced after the cells were transfected with miR-142-5p mimic (Fig. 1D). Flow cytometry showed that the apoptosis rate in HeLa and C33A cells transfected with miR-142-5p mimic was strikingly increased (Fig. 1E). Results obtained using the Transwell assay illustrated that migration and invasion ability were significantly reduced in HeLa and C33A cells upon treatment with miR-142-5p mimic (Fig. 1F and G). Western blot analysis to determine the expression of related genes showed remarkably decreased expression of Ki67, B-cell lymphoma 2 (Bcl-2), and matrix metalloproteinase 9 (MMP-9), whereas Bcl-2-associated X (Bax) and C-caspase 3 expression was significantly boosted in miR-142-5p mimic-transfected HeLa and C33A cells (Fig. 1H). However, treatment of the cells with miR-142-5p inhibitor provoked opposite results (Fig. 1E to H). Taken together, our results show that the expression of miR-142-5p was downregulated in cervical cancer cells, whereas the upregulation of miR-142-5p induced inhibited proliferation, migration, and invasion as well as accelerated apoptosis.
The promoter of miR-142-5p is methylated in cervical cancer and its expression is inhibited.
A hypermethylated CpG island exists in the promoter region of miR-142-5p (17). The MethPrimer (http://www.urogene.org/cgi-bin/methprimer/methprimer.cgi) website also confirmed the presence of a CpG island in the promoter region of miR-142-5p (Fig. 2A). Correlation analysis of miR-142 expression data and methylation data in TCGA cervical cancer data showed a significantly negative correlation between the degree of methylation and the expression of miR-142 (Fig. 2B). Analysis using methylation-specific PCR (MS-PCR) showed that the methylation level of miR-142-5p in cervical cancer tissues and cells was significantly increased compared to that in adjacent tissues and normal cells (Fig. 2C). Subsequently, HeLa and C33A cells were treated with the DNA methylation inhibitor 5-Aza-dC, which provoked a significant reduction in the methylation level of miR-142-5p and elevated expression of miR-142-5p (Fig. 2D and E). Since the catalytic action of methyltransferase enzyme is required for DNA methylation, we adopted chromatin immunoprecipitation (ChIP) for enrichment analysis of DNMT1, DNMT3a, and DNMT3b in the promoter region of the miR-142-5p gene. Results revealed that the enrichment of DNMT1, DNMT3a, and DNMT3b in the promoter region of miR-142-5p gene was significantly reduced after treatment with 5-Aza-dC (Fig. 2F). Next, Western blot analysis was used to determine the expression of DNA methylases DNMT1, DNMT3a, and DNMT3b after 5-Aza-dC treatment (Fig. 2G); the results demonstrated that 5-Aza-dC treatment markedly reduced the expression of DNMT1, DNMT3a, and DNMT3b (Fig. 2G). The above results indicated that in cervical cancer, the promoter of miR-142-5p was methylated and its expression was inhibited.
FIG 2.
The promoter of miR-142-5p is methylated in cervical cancer and its expression is inhibited. (A) Enrichment analysis of CPG island in the miR-142 promoter region. (B) The correlation between miR-142 methylation level and expression in TCGA cervical cancer was analyzed. The x axis represents the methylated beta value of miR-142, the y axis represents the expression value of miR-142, and the upper part represents Pearson’s correlation coefficient and the corresponding P value. (C) The methylation level of miR-142-5p in cervical cancer tissues and cells was measured using MS-PCR. M, methylated band; U, unmethylated band. (D) After treatment of HeLa and C33A cells with the DNA methylation inhibitor 5-Aza-dc, the methylation level of miR-142-5p was detected by MS-PCR. (E) The expression of miR-142-5p was measured with RT-qPCR after HeLa and C33A cells were treated with 5-Aza-dc. (F) DNA-methylating enzymes DNMT1, DNMT3a, and DNMT3b were enriched in the promoter region of the miR-142-5p gene as measured by ChIP assay. (G) The protein expression of DNMT1, DNMT3a, and DNMT3b was examined after HeLa and C33A cells were treated with 5-Aza-dc as determined by Western blot analysis. The measurement data are expressed as means ± standard deviations. The two groups of data conforming to the normal distribution were compared using the unpaired t test. The data comparison between multiple groups was performed using ANOVA, followed by Tukey’s post hoc test. The experiment was repeated three times. *, P < 0.05 versus DMSO-treated HeLa and C33A cells.
PIK3AP1 is a downstream target gene of miR-142-5p and is highly expressed in cervical cancer tissues and cells.
In search of miR-142-5p downstream regulatory mechanism, we downloaded a cervical cancer expression data set GSE63514 through the GEO database and obtained a large number of differentially expressed genes through differential analysis of gene expression in normal samples and tumor samples in the database (Fig. 3A), which are provided in the supplementary table at https://doi.org/10.6084/m9.figshare.13264832.v1. Next, we predicted the target genes of miR-142-5p from the Starbase database (http://starbase.sysu.edu.cn/starbase2/) and intersected these target genes with the top 200 differentially upregulated genes in GSE63514 (Fig. 3B), which yielded seven differentially expressed genes (CEP55, PIK3AP1, KIF14, NRIP3, DTL, MCM10, and DEPDC1 genes). As depicted by Fig. 4, RT-qPCR analysis of these seven genes showed a significant increase in the expression of PIK3AP1 in cervical cancer tissues compared with adjacent tissues. We analyzed the Starbase database and then predicted that miR-142-5p bound to the PIK3AP1 mRNA 3′ UTR (Fig. 3C). Meanwhile, TCGA cervical cancer data also showed increased expression of PIK3AP1 in tumors (Fig. 3D). Western blot analysis displayed significantly increased expression of PIK3AP1 in cervical cancer tissues and cells compared with that in adjacent tissues and normal cells (Fig. 3E).
FIG 3.
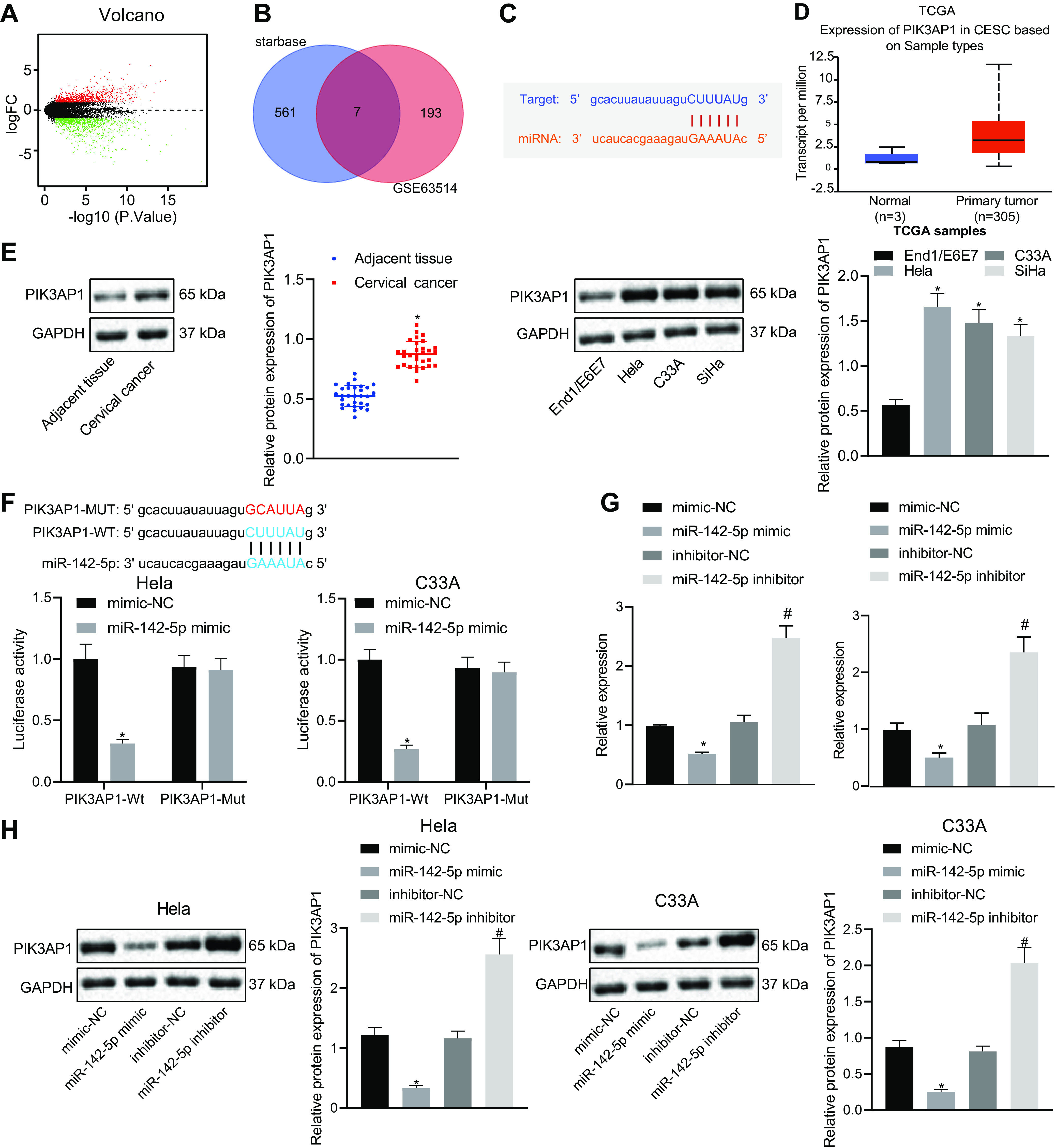
PIK3AP1 expression is elevated in cervical cancer and miR-142-5p targets and inhibits the expression of PIK3AP1. (A) The differentially expressed genes were obtained through differential analysis of gene expression in normal samples and tumor samples from the GSE63514 data set. The x axis represents logFC, and the y axis indicates −log10 (P value); the green dots stand for the genes with significantly low expression in the disease, and the red dots represent the genes with significantly high expression in the disease sample. (B) Venn diagram of the intersection of predicted target genes and the top 200 differentially upregulated genes obtained from GSE63514. (C) The binding site of miR-142-5p to the PIK3AP1 mRNA 3′ UTR was predicted using the Starbase database. (D) Expression of PIK3AP1 in TCGA cervical cancer was observed. The x axis represents the sample type, the y axis represents the expression value, the blue boxes represent the expression of PIK3AP1 in normal samples, and the red boxes represent the expression of PIK3AP1 in tumor samples. (E) The protein expression of PIK3AP1 in cervical cancer tissues (n = 30) and cells was measured using Western blot analysis. (F) The target binding of miR-142-5p to PIK3AP1 was verified using the dual-luciferase reporter gene assay. (G) The mRNA expression of PIK3AP1 in cotransfected cells was detected by RT-qPCR. (H) The protein expression of PIK3AP1 in cotransfected cells was detected by Western blot analysis. The measurement data are expressed as means ± standard deviations. The paired t test was used for the comparison between the cancerous tissues and the adjacent tissues, and the unpaired t test was used for the data comparison between two groups. The data comparison between multiple groups was performed using ANOVA, followed by Tukey’s post hoc test. The experiment was repeated three times. *, P < 0.05 versus adjacent tissues or mimic-NC-transfected cells; #, P < 0.05 versus inhibitor-NC transfected cells.
FIG 4.
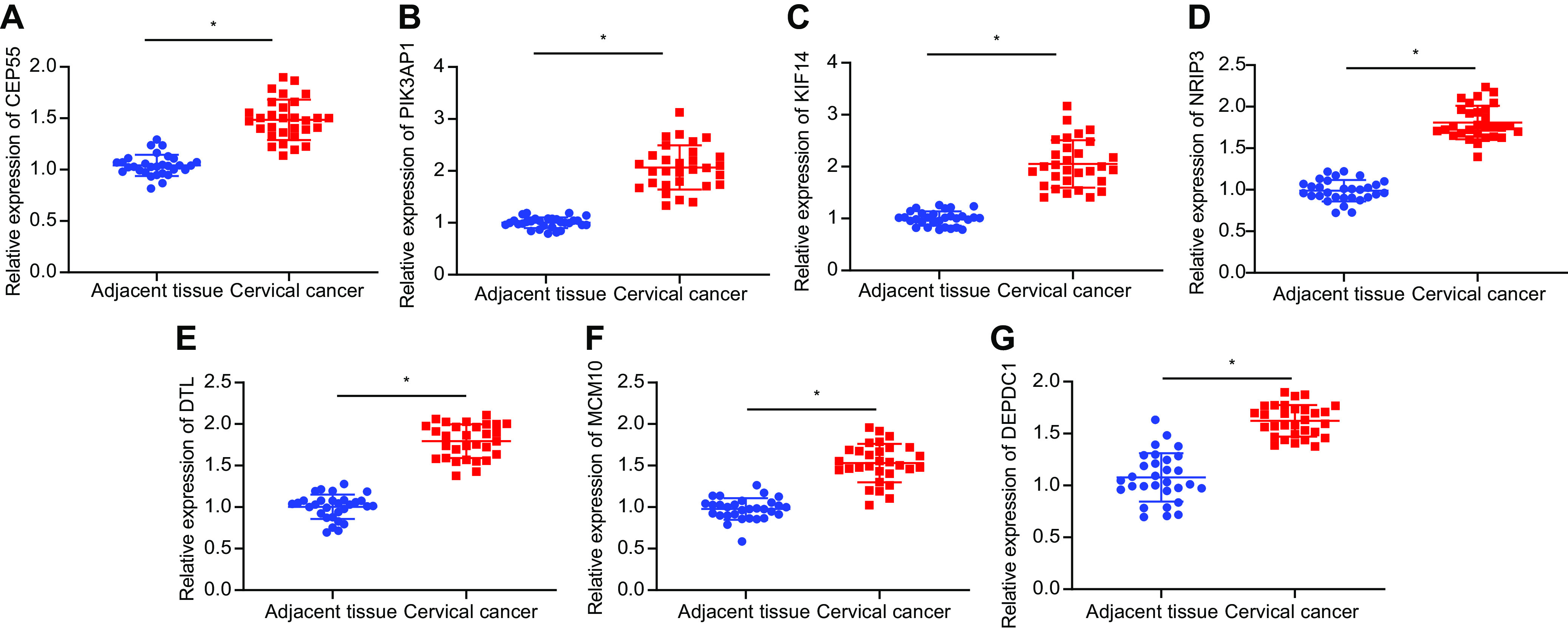
The mRNA expression of CEP55, PIK3AP1, KIF14, NRIP3, DTL, MCM10, and DEPDC1 in cervical cancer and adjacent tissues determined with RT-qPCR (n = 30). The measurement data are expressed as means ± standard deviations. ANOVA was used for the comparison among multiple groups, followed by Tukey’s post hoc test. *, P < 0.05 versus adjacent tissues.
The target binding of miR-142-5p to PIK3AP1 was verified by dual-luciferase reporter gene assay, which showed that HeLa and C33A cells cotransfected with the PIK3AP1 wild type (PIK3AP1-Wt) and miR-142-5p mimic showed a weaker luciferase activity in comparison with that of cells cotransfected with PIK3AP1-Wt and negative (NC) mimic. However, there was no significant difference in luciferase activity between cells cotransfected with PIK3AP1 mutant (PIK3AP1-Mut) and miR-142-5p mimic and cells cotransfected with PIK3AP1-Mut and NC mimic (Fig. 3F), thus suggesting that miR-142-5p targeted and bound to PIK3AP1. To further investigate the regulation of miR-142-5p on PIK3AP1 expression, HeLa and C33A cells were treated with miR-142-5p mimic or miR-142-5p inhibitor. The results of RT-qPCR and Western blot analysis exhibited that PIK3AP1 expression was notably downregulated after HeLa and C33A cells were treated with miR-142-5p mimic but upregulated by treatment with miR-142-5p inhibitor (Fig. 3G and H). The above studies showed that PIK3AP1 was highly expressed in cervical cancer and that miR-142-5p targeted and suppressed the expression of PIK3AP1.
Overexpression of miR-142-5p suppresses the proliferation, migration, and invasion of cervical cancer cells and promotes apoptosis through targeting the PIK3AP1/PI3K/AKT axis.
We next tested the effect of miR-142-5p on the biological characteristics of cervical cancer cells through targeting and inhibiting PIK3AP1 in HeLa and C33A cells. The results of Western blot analysis exhibited that PIK3AP1 expression was higher in HeLa and C33A cells cotransfected with mimic-NC and overexpressed PIK3AP1 (oe-PIK3AP1) than that in cells cotransfected with mimic-NC and oe-NC. In addition, PIK3AP1 expression was significantly decreased in HeLa and C33A cells cotransfected with miR-142-5p mimic and oe-NC in comparison with that in cells cotransfected with mimic-NC and oe-NC. Moreover, the expression of PIK3AP1 was strikingly boosted in HeLa and C33A cells cotransfected with miR-142-5p mimic and oe-PIK3AP1 compared to that in cells cotransfected with miR-142-5p mimic and oe-NC (Fig. 5A).
FIG 5.
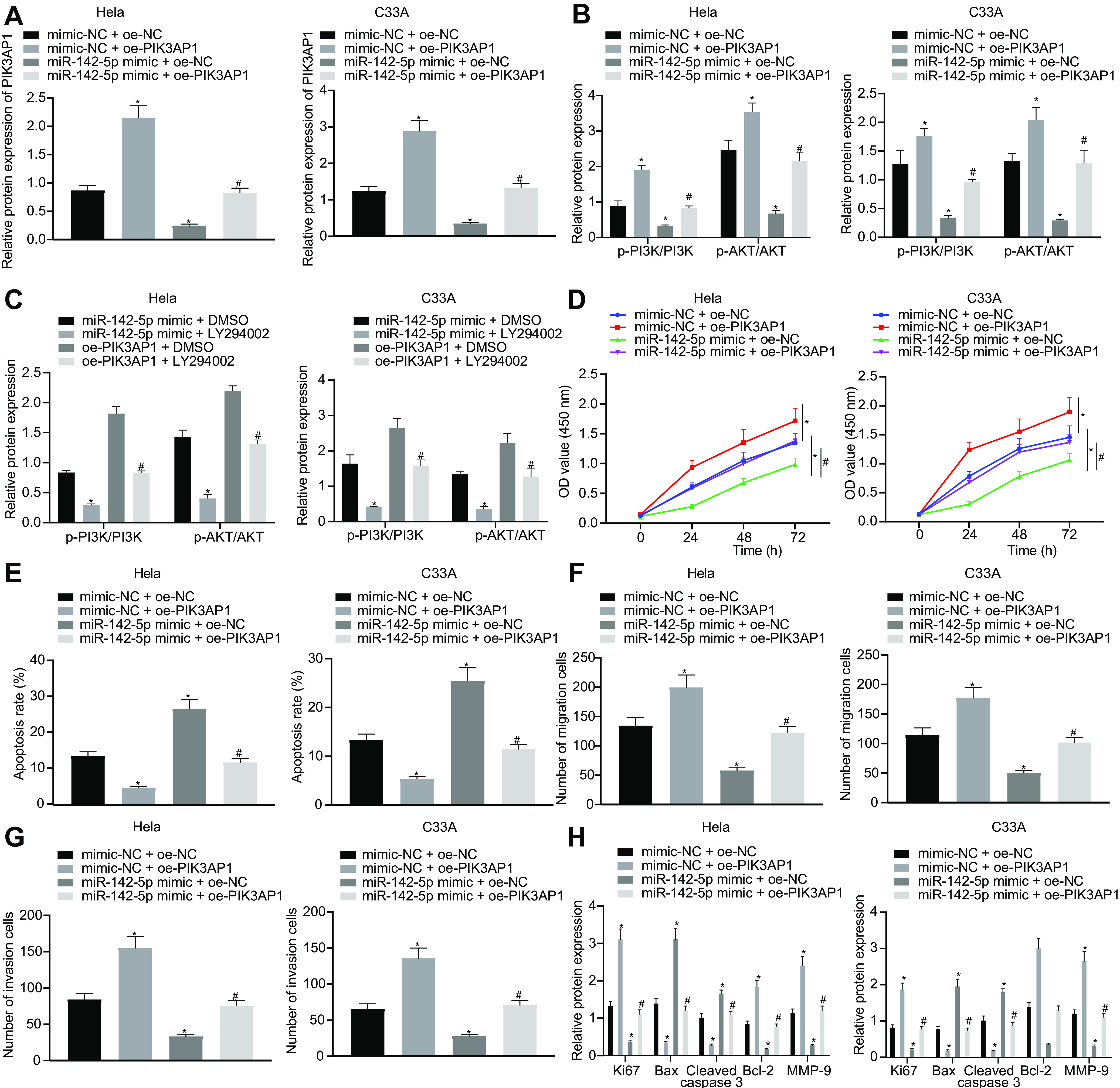
Overexpression of miR-142-5p suppresses the proliferation, migration, and invasion of cervical cancer cells and promotes their apoptosis through inhibiting PIK3AP1 expression. (A) The expression of PIK3AP1 was detected using Western blot analysis. (B) The expression of PI3K/AKT signaling pathway-related proteins was detected using Western blot analysis. (C) The expression of PI3K/AKT signaling pathway-related proteins after LY294002 treatment was detected using Western blot analysis. (D) The proliferation ability was measured using CCK-8. (E) Apoptosis was tested by flow cytometry. (F) The migration ability was evaluated by the means of the Transwell assay. (G) The invasion ability was evaluated by the Transwell assay. (H) The expression of related genes was detected using Western blot analysis. The measurement data are expressed as means ± standard deviations. The data comparison between multiple groups was performed using ANOVA, followed by Tukey’s post hoc test. *, P < 0.05 versus HeLa and C33A cells cotransfected with mimic-NC and oe-NC; #, P < 0.05 versus HeLa and C33A cells cotransfected with miRNA-142-5p mimic and oe-NC. The experiment was repeated three times.
A prior report has demonstrated that PIK3AP1 can activate the PI3K/AKT signaling pathway (13). Therefore, we performed Western blot analysis to determine the expression of related proteins in the PI3K/AKT signaling pathway, which showed that HeLa and C33A cells cotransfected with mimic-NC and oe-PIK3AP1 had significantly increased phosphorylation levels of PI3K and AKT compared to those of HeLa and C33A cells cotransfected with mimic-NC and oe-NC, while cells cotransfected with miR-142-5p mimic and oe-NC showed opposite results. In the HeLa and C33A cells transfected with miR-142-5p mimic, the further treatment with oe-PIK3AP1 induced notably higher phosphorylation levels of PI3K and AKT than those in cells treated with oe-NC (Fig. 5B), thus showing that miR-142-5p could inhibit the activation of the PI3K/AKT signaling pathway through targeting and suppressing the expression of PIK3AP1. Next, the effect of miR-142-5p on the biological characteristics of cervical cancer cells by regulating the PIK3AP1/PI3K/AKT axis was studied by the further treatment with LY294002, a PI3K/AKT signaling pathway inhibitor. The results of Western blot analysis showed that the phosphorylation levels of PI3K and AKT in HeLa and C33A cells cotreated withmiR-142-5p mimic and LY294002 were remarkably downregulated compared to those in cells cotreated with miR-142-5p mimic and dimethyl sulfoxide (DMSO). Meanwhile, the cotreatment of HeLa and C33A cells with oe-PIK3AP1 and LY294002 induced significantly decreased levels of PI3K and AKT compared to those in cells cotreated with oe-PIK3AP1 and DMSO (Fig. 5C).
To explore the effects of the miR-142-5p-dependent PIK3AP1/PI3K/AKT axis on cell proliferation, migration, invasion, and apoptosis abilities, we adopted CCK-8 (Fig. 5D), flow cytometry (Fig. 5E), and the Transwell assay (Fig. 5F and G), which showed that the proliferation, migration, and invasion abilities in HeLa and C33A cells cotransfected with mimic-NC and oe-PIK3AP1 were notably enhanced, while apoptosis ability was reduced in comparison with that in HeLa and C33A cells cotransfected with mimic-NC and oe-NC. However, we found opposite results in HeLa and C33A cells cotransfected with miR-142-5p mimic and oe-NC and cells cotransfected with mimic-NC and oe-NC. Additionally, the proliferation, migration, and invasion abilities in cells cotransfected with miR-142-5p mimic and oe-PIK3AP1 were significantly increased, but apoptosis was reduced, compared to those of cells cotransfected with miR-142-5p mimic and oe-NC. Further Western blot analysis displayed that, compared to that in cells cotransfected with mimic-NC and oe-NC, the expression of Ki67, Bcl-2, and MMP-9 was notably upregulated, whereas Bax expression was downregulated in HeLa and C33A cells cotransfected with mimic-NC and oe-PIK3AP1; we saw opposite effects in cells transfected with miR-142-5p mimic and oe-NC. Moreover, the expression of Ki67, Bcl-2, and MMP-9 was significantly increased, while Bax expression was decreased, in HeLa and C33A cells cotransfected with miR-142-5p mimic and oe-PIK3AP1 compared with findings in cells cotransfected with miR-142-5p mimic and oe-NC (Fig. 5H). The proliferation, migration, and invasion abilities in HeLa and C33A cells cotreated with miR-142-5p mimic and LY294002 were strikingly reduced while cell apoptosis was enhanced relative to those in cells cotreated with miR-142-5p mimic and DMSO. Similarly, proliferation, migration, and invasion abilities in cells cotreated with oe-PIK3AP1 and LY294002 were also notably reduced while cell apoptosis was enhanced in comparison with those in cells cotreated with oe-PIK3AP1 and DMSO. Western blot analysis of related proteins showed that HeLa and C33A cells cotreated with miR-142-5p mimic and LY294002 had downregulated protein expression of Ki67, Bcl-2, and MMP-9 but upregulated Bax protein expression compared to those in cells cotreated with miR-142-5p mimic and DMSO. The cotreatment of HeLa and C33A cells with oe-PIK3AP1 and LY294002 also resulted in elevated expression of Ki67, Bcl-2, and MMP-9 but decreased Bax expression compared to those in cells cotreated with oe-PIK3AP1 and DMSO (Fig. 6). Taken together, the results show that miR-142-5p restrained cell proliferation, migration, and invasion as well as promoted apoptosis of cervical cancer cells by regulating the PIK3AP1/PI3K/AKT axis.
FIG 6.
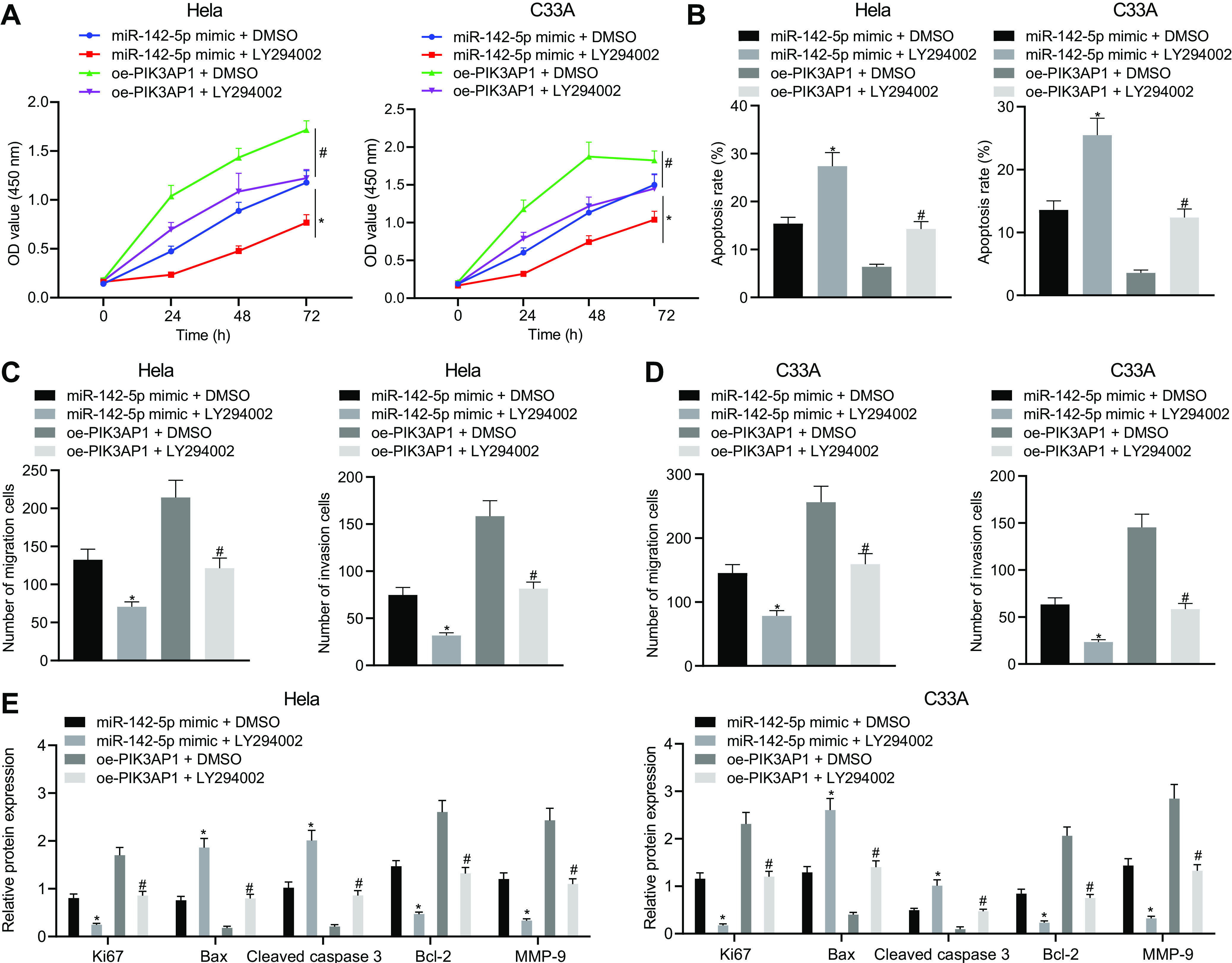
The upregulation of miR-142-5p inhibits the proliferation, migration, and invasion of cervical cancer cells and promotes their apoptosis by regulating the PIK3AP1/PI3K/AKT axis. (A) The proliferation ability was measured using CCK-8. (B) Apoptosis was tested by flow cytometry. (C) The migration ability was evaluated by the means of the Transwell assay (×200). (D) The invasion ability was evaluated by the means of the Transwell assay (×200). (E) The protein expression of related genes was detected using Western blot analysis. The measurement data are expressed as means ± standard deviations. ANOVA was used for the comparison among multiple groups, followed by Tukey’s post hoc test. For data comparison between groups at different time points, repeated-measurement ANOVA was used, with Tukey’s posttest. The experiment was repeated three times. *, P < 0.05 versus HeLa and C33A cells cotransfected with mimic-NC and oe-NC or cotreated with miR-142-5p mimic and DMSO; #, P < 0.05 versus HeLa and C33A cells cotransfected with miR-142-5p mimic and oe-NC or cotreated with oe-PIK3AP1 and DMSO.
Upregulation of miR-142-5p inhibits cervical cancer tumor growth and metastasis through mediating the PIK3AP1/PI3K/AKT axis.
Finally, we studied the effect of miR-142-5p on tumorigenesis in nude mice by regulating the axis of PIK3AP1/PI3K/AKT in vivo. We introduced miR-142-5p antagomir or LY294002 (PI3K/AKT signaling pathway inhibitor) into the nude mice xenografting with cervical cancer cells. The results of RT-qPCR showed that the expression of miR-142-5p in nude mice cotreated with miR-142-5p antagomir and DMSO was notably downregulated compared with that obtained with antagomir-NC and DMSO treatment. However, the expression of miR-142-5p in mice cotreated with miR-142-5p antagomir and LY294002 showed no significant difference compared with that in mice cotreated with miR-142-5p antagomir and DMSO. Compared with the mice treated with agomir-NC and DMSO, the nude mice cotreated with miR-142-5p agomir and DMSO exhibited markedly upregulated expression of miR-142-5p (Fig. 7A). The results of Western blot analysis exhibited that PIK3AP1 expression and phosphorylation levels of PI3K and AKT in nude mice cotreated with miR-142-5p antagomir and DMSO were all significantly boosted in comparison with findings in mice cotreated with antagomir-NC and DMSO. However, the expression of PIK3AP1 was unchanged, while phosphorylation levels of PI3K and AKT were decreased, in mice cotreated with miR-142-5p antagomir and LY294002 compared with those in mice cotreated with the miR-142-5p antagomir and DMSO. Compared with the mice cotreated with agomir-NC and DMSO, the nude mice cotreated with miR-142-5p agomir and DMSO exhibited a striking reduction in the expression of PIK3AP1 and phosphorylation levels of PI3K and AKT (Fig. 7B).
FIG 7.
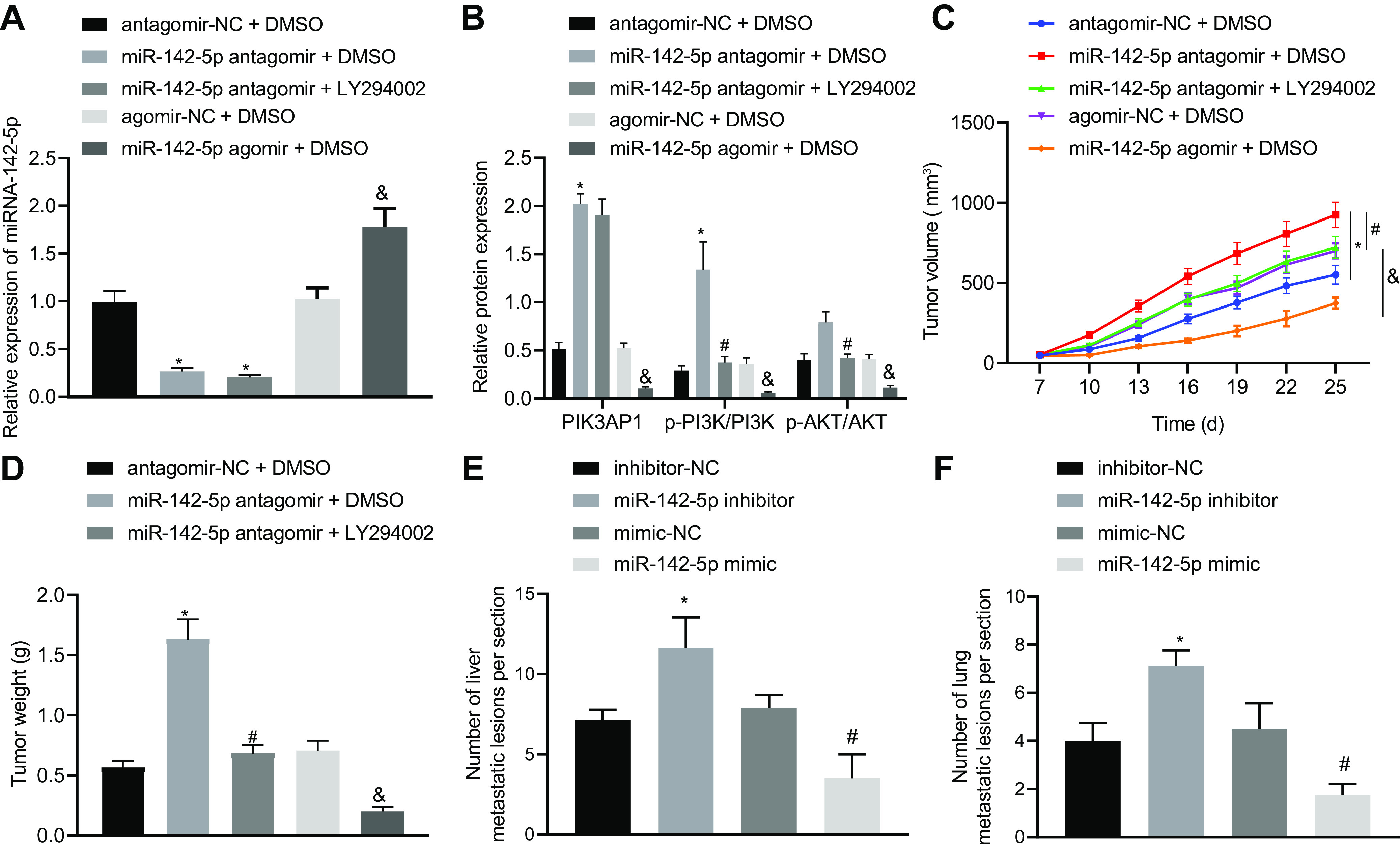
Restoration of miR-142-5p inhibits cervical cancer tumor growth and metastasis through mediating the PIK3AP1/PI3K/AKT axis. (A) miR-142-5p expression in cervical cancer xenograft tissues of mice was measured using RT-qPCR. (B) The related protein expression of PIK3AP1 and the PI3K/AKT signaling pathway in cervical cancer tissues of mice was detected using Western blot analysis. (C) The volume of transplanted tumor in nude mice was examined. (D) The weight of transplanted tumors in nude mice was evaluated (n = 8). *, P < 0.05 versus mice cotreated with antagomir-NC and DMSO; #, P < 0.05 versus mice cotreated with miR-142-5p antagomir and DMSO; &, P < 0.05 versus mice cotreated with agomir-NC and DMSO. (E) The number of liver metastases of mice injected with stably transfected HeLa cervical cancer cells treated with inhibitor-NC, miR-142-5p inhibitor, mimic-NC, or miR-142-5p mimic. (F) The number of lung metastases of mice injected with stably transfected HeLa cervical cancer cells treated with inhibitor-NC, miR-142-5p inhibitor, mimic-NC, or miR-142-5p mimic (n = 8). *, P < 0.05 versus mice injected with stably transfected cervical cancer cells expressing inhibitor-NC; #, P < 0.05 versus mice injected with stably transfected cervical cancer cells expressing mimic-NC. Measurement data are expressed as means ± standard deviations. ANOVA was conducted for comparison among multiple groups, followed by Tukey’s post hoc test. For data comparison between groups at different time points, repeated-measurement ANOVA was used, with Tukey’s posttest.
The tumor volume and weight of xenografts in mice cotreated with miR-142-5p antagomir and DMSO were significantly increased compared to those in mice cotreated with antagomir-NC and DMSO. Moreover, the tumor volume and weight of xenografts in mice cotreated with miR-142-5p antagomir and LY294002 were notably decreased in comparison with those in mice cotreated with miR-142-5p antagomir and DMSO. Compared with the mice cotreated with agomir-NC and DMSO, the nude mice cotreated with miR-142-5p agomir and DMSO exhibited notably decreased tumor volume and weight (Fig. 7C and D).
The stably transfected HeLa cervical cancer cells treated with inhibitor-NC, miR-142-5p inhibitor, mimic-NC, or miR-142-5p mimic were injected into mice via a tail vein. As revealed in Fig. 7E and F, nude mice injected with HeLa cells expressing miR-142-5p inhibitor exhibited an increased number of metastases in the liver and lungs compared with inhibitor-NC treatment, while opposite effects were observed in response to miR-142-5p mimic relative to mimic-NC. Taken together, these results show that miR-142-5p inhibited the growth and metastasis of cervical cancer tumors in vivo by regulating the PIK3AP1/PI3K/AKT signal axis.
DISCUSSION
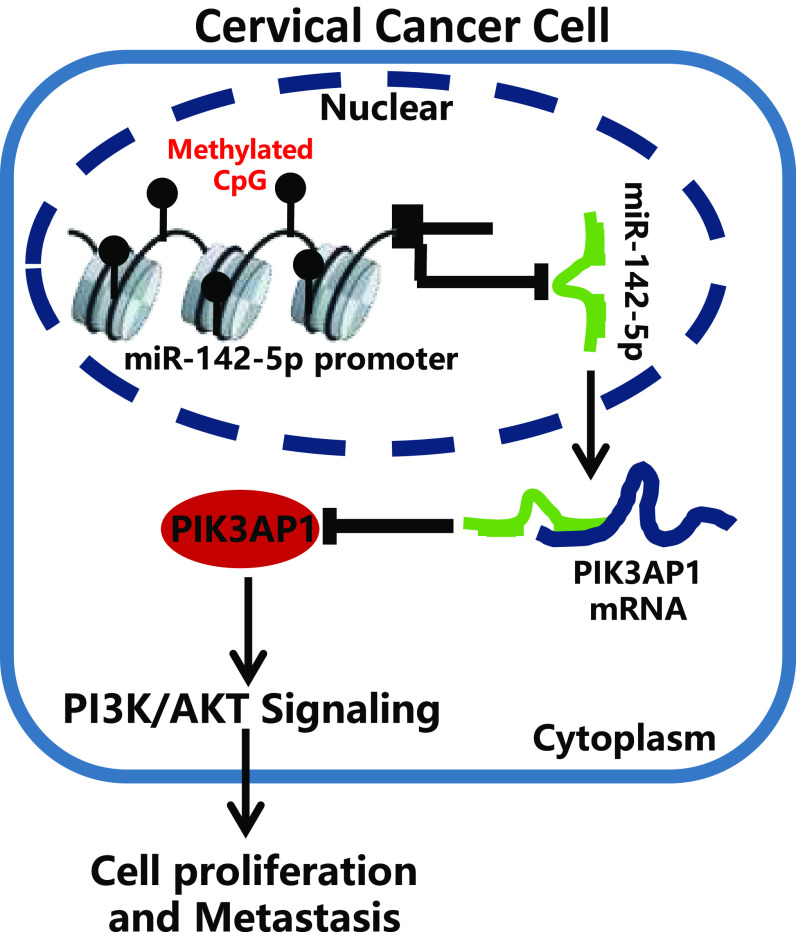
Cervical cancer ranks as the fourth most common malignancy diagnosed in women worldwide, despite knowledge of the link with HPV infection, and there is a serious problem with access to adequate treatment, especially in developing countries (18). Cervical cancer screening is essential for early detection and treatment of precancerous cells and cervical cancer (19), although many women continue to die of this largely preventable disease (20). The major issues for the limited treatment of cervical cancer lie in inadequate screening, in conjunction with the inherent invasiveness (21). Meanwhile, radiation-resistant cervical cancer is likely to cause local recurrence, distant metastasis, and reduced survival (22). New evidence suggests that disorders of miRNA regulation contribute to the development and progression of cervical cancer (23). The objective of this study was to investigate the effect of miR-142-5p on the PIK3AP1/PI3K/AKT axis in cervical cancer. These results revealed that overexpression of miR-142-5p could suppress the progression of cervical cancer through targeting and inhibiting PIK3AP1 expression via the PI3K/AKT signaling pathway (Fig. 8).
FIG 8.
Possible molecular mechanism of miR-142-5p in the development of cervical cancer through mediating the PIK3AP1/PI3K/AKT axis.
Initially, the study revealed that miR-142-5p was poorly expressed in cervical cancer and that overexpression of miR-142-5p was conducive to inhibited proliferation, migration, and invasion of cervical cancer cells and promoted apoptosis. Consistent with our findings, the expression of miR-142-5p is also reported to be downregulated in cervical cancer (9). A significant decrease in the level of miR-142-3p has been observed previously in cervical cancer tissues, and patients with low expression of miR-142-3 have poor overall survival and progression-free survival (24). Moreover, in human cervical cancer tissues and a group of cell lines, the expression of miR-142 is significantly downregulated, and the overexpression of miR-142 reduces the proliferation and invasiveness of representative SiHa and HeLa cells and enhances apoptosis (25). Our next results showed that the promoter of miR-142-5p was methylated and its expression was inhibited in cervical cancer. The relations between miR-142-5p and promoter methylation level have been investigated previously in hepatic biliary atresia, where the overexpression of miR-142-5p and elevated DNMT1 mRNA level were negatively correlated with DNMT3a/DNMT3b and where upregulation of miR-142-5p strikingly decreased the overexpression of DNMTs (26). Triclosan-induced miR-142-5p expression inhibits the P450c17 enzyme by suppressing the JAK1/STAT1 pathway and its downstream Sp1/DNMT1/DAX1 cascade (27). Furthermore, the higher the degree of HPV methylation, the higher the severity of the disease (28), and the gene shows a higher level of methylation in cervical cancer than that in healthy persons (29).
Next, we found that PIK3AP1 was the downstream target gene of miR-142-5p and that its expression was elevated in cervical cancer. Concordant with the present results, PIK3AP1 is known to be involved in a key oncogenic pathway and is upregulated in the peripheral blood of breast cancer patients (30). PIK3AP1 expression is also elevated in gastric cancer, and such patients with high PIK3AP1 expression in tumors have a trend toward poorer survival (13). Our subsequent experiments showed that restoration of miR-142-5p targeted and inhibited the expression of PIK3AP1 to restrain proliferation, migration, and invasion of cervical cancer cells and enhance apoptosis. Conversely, a previous study showed that depletion of miR-142-5p could directly regulate CYR61 expression to promote tumor metastasis in gastric cancer (31). We now report that miR-142-5p inhibited the proliferation, migration, and invasion of cervical cancer cells and promoted their apoptosis by regulating the PIK3AP1/PI3K/AKT axis. Consistent with our results, PIK3AP1 and AKT gene expression are positively correlated during the immune inflammatory response of GES-1 cells induced by Helicobacter pylori (12). Apart from that, ARHGAP17 inhibits tumor progression by inhibiting the PI3K/AKT signaling pathway in cervical cancer (15). Activation of MAPK, PI3K/AKT, and hTERT signaling promotes tumor growth and metastasis and regulates radiosensitivity to cervical cancer (16). In our final experiments, we confirmed in vivo that miR-142-5p elevation repressed the growth of xenograft via suppressing PIK3AP1/PI3K/AKT expression. To sum up, overexpression of miR-142-5p could suppress the progression of cervical cancer through inhibiting the PIK3AP1/PI3K/AKT axis. This study offers a new perspective on potential targeted therapy of cervical cancer and sets the stage for further elucidation and targeted prevention.
MATERIALS AND METHODS
Ethical statement.
The study protocol was approved by the Ethics Committee of West China Second University Hospital and conducted in compliance with the Declaration of Helsinki. The experiments involving animals were performed in accordance with the principles for laboratory animals of the National Institutes of Health.
Collection of tissue samples.
In this study, we recruited 30 patients with cervical cancer who underwent surgery in Sichuan Integrative Medicine Hospital from August 2013 to September 2015. Tissue specimens, including paired cervical cancer tissues and normal adjacent tissues, were collected. All patients had diagnosis confirmed by the pathology department. Tissue specimens were placed at –80°C immediately after surgery for subsequent studies.
Cell culture and grouping.
Normal cervical immortalized epithelial cell line END1/E6E7 and cervical cancer cell lines HeLa, C33A, and SiHa were purchased from the American Type Culture Collection (ATCC; Manassas, VA). These cells were cultured in Dulbecco’s modified Eagle medium (DMEM) containing 1% penicillin and streptomycin and 10% fetal bovine serum (FBS), in a constant-temperature incubator at 37°C with 5% CO2, and passaged every 2 to 3 days until they reached the logarithmic growth stage.
Cervical cancer cells were transfected using a Lipofectamine 2000 (Invitrogen, Carlsbad, CA) kit (11668019; Thermo Fisher Scientific, Madison, WI) with NC mimic (negative-control plasmids of miR-142-5p mimic), miR-142-5p mimic (miR-142-5p mimic plasmids), NC inhibitor (negative-control plasmids of miR-142-5p inhibitor), miR-142-5p inhibitor (miR-142-5p inhibitor plasmids), oe-NC (negative-control plasmids of overexpressed PIK3AP1), or oe-PIK3AP1 (overexpressed PIK3AP1 plasmids). Then, the uninfected cervical cancer cells or stably transfected cervical cancer cells were cotreated with 7.5 μM 5-Aza-dC, DMSO, or 100 ng/ml of LY294002 for inhibition of the PI3K/AKT signaling pathway. All the mimics and inhibitors were synthesized, plasmid vectors were constructed, and the sequencing was identified and tested by Guangzhou Ribobio Biotechnology Co., Ltd. (Guangzhou, China). In brief, 4 μg of the target plasmid and 10 μl of Lipofectamine 2000 were diluted with 250 μl of serum-free Opti-MEM medium (Gibco, Carlsbad, CA) and mixed. The mixture was left at room temperature for 5 min, mixed evenly for 20 min, and added into the culture well after 20 min. The culture well was then placed in a 37°C, 5% CO2 incubator for 6 h. The medium was replaced with the complete medium, and culture continued for 48 h prior to collection of the cells to measure the transfection efficiency for subsequent experiments.
RT-qPCR.
TRIzol (Invitrogen) was used to extract total RNA from tissues and cells, and the total RNA concentration and purity were detected by a NanoDrop 2000 trace UV spectrophotometer (1011U; Nanodrop Technologies, Wilmington, DE). According to the instructions of the TaqMan microRNA assay reverse transcription primer kit (4427975; Applied Biosystems, Waltham, MA), RNA was reverse transcribed to generate cDNA, and primers of miR-142-5p were designed. The sequence was synthesized by TaKaRa Bio (Table 1). A Fast SYBR green PCR kit (Applied Biosystems) was used to prepare the reaction system, and real-time fluorescent quantitative PCR detection was performed using an ABI7500 quantitative PCR instrument (Applied Biosystems). U6 was considered the internal reference. The threshold cycle (2−ΔΔCT) method was adopted to calculate relative transcription levels of target genes, which were expressed as ΔΔCT = ΔCT experimental group − ΔCT control group, ΔCT = CT (target gene) − CT (internal reference). The relative transcription level of target gene mRNA = 2−ΔΔCT. Three parallel wells were set for each sample, and each experiment was repeated three times.
TABLE 1.
RT-qPCR primer sequencesa
| Target | Primer sequence (5′–3′) |
|---|---|
| miR-142-5p (human) | F: 5′-AACTCCAGCTGGTCCTTAG-3′ |
| R: 5′-TCTTGAACCCTCATCCTGT-3′ | |
| U6 (human) | F: 5′-GCTTCGGCAGCACATATACTAAAAT-3′ |
| R: 5′-CGCTTCACGAATTTGCGT-3′ | |
| miR-142-5p (mouse) | F: 5′-CGCGCGCATAAAGTAGAAAGCAC-3′ |
| R: 5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTC-3′ | |
| U6 (mouse) | F: 5′-CTCGCTTCGGCAGCACA-3′ |
| R: 5′-AACGCTTCACGAATTTGCGT-3′ |
miR-142-5p, microRNA-142-5p; U6, U6 snRNA; F, forward; R, reverse.
Western blot analysis.
Radioimmunoprecipitation assay lysate containing phenylmethanesulfonyl fluoride (P0013C; Beyotime Biotechnology, Shanghai, China) was used to extract the total protein in the tissues or cells. Then, the proteins were incubated on ice for 30 min at 4°C and centrifuged at 8,000 × g for 10 min, and the supernatant was removed. The total protein concentration was detected with the bicinchoninic acid disodium kit. Next, 50-μg portions of protein were dissolved in 2× sodium dodecyl sulfate (SDS) loading buffer. After boiling for 5 min, the samples were subjected to SDS-polyacrylamide gel electrophoresis, transferred to a polyvinylidene fluoride (PVDF) membrane, and blocked with 5% skimmed milk powder at room temperature for 1 h. Then, the PVDF membrane was coincubated at 4°C overnight with the following diluted primary antibodies: rabbit anti-PIK3AP1 (ab237629, 1:1,000), PI3K (number 4257, 1:1,000; CST, Beverly, MA), p-PI3K (no. 4228, 1:1,000; CST), AKT (ab8805, 1:500), p-AKT (ab38449, 1:800), Ki67 (ab92742, 1:1,500), Bax (ab32503, 1:1,000), cleaved caspase 3 (ab32042, 1:500), Bcl-2 (ab182858, 1:2,000), MMP-9 (ab38898, 1:1,000), DNMT1 (ab188453, 1:10,000), DNMT3a (ab188470, 1:2,000), DNMT3b (ab79822, 1:1,000), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH; ab9485, 1:2,500). Antibodies were from Abcam (Cambridge, UK) except where indicated. The membrane was washed 3 times with Tris-buffered saline with Tween 20 (TBST) for 10 min each time and then incubated with horseradish peroxidase (HRP)-labeled secondary antibody goat anti-rabbit immunoglobulin G (IgG) H&L (ab97051, 1:2,000; Abcam) for 1 h. Afterwards, the membrane was rinsed with TBST, developed using an ECL fluorescence detection kit (BB-3501; Amersham, Little Chalfont, UK), exposed using a Bio-Rad image analysis system (Bio-Rad, Hercules, CA), and analyzed by Quantity One v4.6.2 software. The relative protein content was expressed by the gray value of the corresponding protein band/the gray value of the GAPDH protein band. The experiment was repeated three times and averaged.
CCK-8.
The proliferation of HeLa and C33A cells was detected with a CCK-8 kit (CK04; Dojindo, Kumamoto, Japan). Cervical cancer cells in the logarithmic growth phase were selected and seeded at a density of 2 × 103/well in 96-well plates for preculture for 24 h, whereupon the cells were transfected. After transfection for 48 h, 10 µl of CCK-8 reagent was added at 0, 24, 48, and 72 h and incubated at 37°C for 4 h. The absorbance values of the wells at the wavelength of 450 nm were measured using a microplate reader. The values were proportional to the cell proliferation in the medium, and the cell growth curve was accordingly plotted.
Transwell assay.
Cervical cancer cells to be tested were put into the upper chamber of Transwell culture chamber (3413; Beijing Unique Biotechnology Co., Ltd., Beijing, China). Then, the cells were incubated at 37°C for 4 to 5 h. The transfected cells were diluted with 100 μl of serum-free medium to form cell suspension (1 × 106 cells/ml). After seeding, 600 μl of DMEM containing 20% FBS was added into the lower chamber. Three parallel wells were set for each group. After incubation for 24 h at 37°C under 5% CO2, the upper surface cells of the membrane were wiped with cotton balls, and the remaining cells were fixed with 5% glutaraldehyde, stained with 0.1% crystal violet for 5 min at 4°C, and observed under an inverted fluorescence microscope (TE2000; Nikon, China), with counting in 4 or 5 visual fields. Matrigel gel (YB356234; Shanghai Yu Bo Biotech Co., Ltd., Shanghai, China) was added to the Transwell plate chamber for the invasion experiments, with other operations unchanged.
Flow cytometry.
After transfection with cervical cancer cells in each group for 48 h, trypsin without EDTA was used to detach the cells. Then, the cells were collected in flow tube and centrifuged, and the supernatant was discarded. The cells were washed 3 times with cold phosphate-buffered saline (PBS), and the supernatant was separated by centrifugation. Annexin V-fluorescein isothiocyanate (FITC)-polyimide (PI) staining solution was prepared with annexin V-FITC, PI, and HEPES buffer solution at a volume ratio of 1:2:50, in line with instructions in the annexin V-FITC apoptosis assay kit (C1065; Beyotime Biotechnology). Next, cells were resuspended with the staining solution (1 × 106 cells/100 μl) supplemented with 1 ml of HEPES buffer and incubated at room temperature with oscillation for 15 min. The excitation wavelength was recorded at 488 nm with a flow cytometer, and the fluorescence of FITC and PI was detected by 525-nm and 620-nm band-pass filters, respectively, for detection of apoptosis. The experiment was repeated three times.
MS-PCR.
The genomic DNA of cervical cancer tissues and cells was extracted by using the genomic DNA extraction kit of TIANGEN (Beijing, China) according to the instructions of the kit. The DNA concentration and purity were determined by UV spectrophotometry, and the DNA was stored at –80°C for later use. An EZ DNA methylation kit (Zymo Research, Irvine, CA) was used to treat the DNA with sodium sulfite, followed by desulfurization and purification by a reaction column. The purified DNA was used for subsequent PCRs. Methylated and nonmethylated primers were designed for the enrichment area of the miR-142-5p gene promoter CPG island (Table 2). The reaction products were analyzed by agarose gel electrophoresis, gel electrophoresis imaging, and an image analysis system. Each experiment was repeated three times.
TABLE 2.
Primer sequences
| Gene | Primer sequence (5′–3′) |
|---|---|
| miR-142-5p-M | F: AGTTATCGTTTATAAGGTTTAGGGC |
| R: CACTATCTATCTATCCGTCGACGTA | |
| miR-142-5p-U | F: TTATTGTTTATAAGGTTTAGGGTGG |
| R: CACTATCTATCTATCCATCAACATA |
ChIP.
When confluence reached 70 to 80%, the cervical cancer cells were fixed with 1% formaldehyde at room temperature for 10 min to cross-link the DNA and protein. After cross-linking, the cells were randomly broken by ultrasonic treatment, with 15 cycles at intervals of 10 s to break into fragments of appropriate size. After centrifugation at 13,000 rpm and 4°C, the supernatant was collected and divided into two tubes, to which were added negative-control antibody rabbit anti-IgG (ab172730, 1:100; Abcam) and target protein-specific antibodies DNMT1 (ab13537, 1:100; Abcam), DNMT3a (ab2850, 1:100; Abcam), and DNMT3b (ab2851, 1:100; Abcam), and incubated overnight at 4°C. Protein agarose-Sepharose chromatography was used to precipitate endogenous DNA-protein complexes. After a brief centrifugation, the supernatant was absorbed and the nonspecific complexes were washed, de-cross-linked at 65°C overnight, extracted in phenol-chloroform, and purified to recover DNA fragments, followed by detection of the enrichment of DNMT1, DNMT3a, and DNMT3b in the promoter region of the miR-142-5p gene.
Dual-luciferase reporter gene assay.
A website (Starbase2 [http://starbase.sysu.edu.cn/starbase2/]) was used to predict that PIK3AP1 may be the downstream regulatory gene of miR-142-5p. Human embryonic kidney HEK293T cells were cultured in DMEM containing 10% FBS at 37°C with 5% CO2. The cDNA fragment of the PIK3AP1 3′ UTR (PIK3AP1-Wt) containing the binding site of miR-142-5p was inserted into the pmirGLO vector. The PIK3AP1 3′ UTR cDNA fragment (PIK3AP1-Mut) was synthesized by point mutation and inserted into the pmirGLO vector, with verification of the inserted sequences by sequencing. The recombinant vector pmirGLO-PIK3AP1-Wt or pmirGLO-PIK3AP1-Mut was cotransfected into HEK293T cells with miR-142-5p mimic (miR-142-5p overexpression sequence) or NC mimic (negative-control sequence) by the liposome transfection method, and the cells were incubated for 48 h before being collected and lysed. Next, 100 μl of lysate supernatant was taken and added with 100 μl of Renilla luciferase detection working solution to detect Renilla luciferase activity. In addition, 100 μl of lysate supernatant was taken and added with 100 μl of firefly luciferase detection reagent, followed by mixing and detection of the firefly luciferase activity. A multifunctional SpectraMax M5 microplate reader (Molecular Devices, Shanghai, China) was used for this purpose, with the interval set to 2 s and the measurement time at 10 s to detect Renilla luciferase activity and firefly luciferin enzyme activity.
Construction of mouse model.
Forty specific-pathogen-free (SPF)-grade female BALB/c nude mice (aged 6 weeks, weighing 15 to 18 g) were purchased from Shanghai SLAC Laboratory Animal Co., Ltd. (Shanghai, China). HeLa cervical cancer cells in the logarithmic growth phase were prepared into a cell suspension with a concentration of about 1 × 107 cells/ml. The prepared cell suspension was injected into the skin of the left underarm of nude mice with a 1-ml syringe to establish the subcutaneous xenograft model of nude mice. When the tumor tissue volume reached 50 mm3, the nude mice were randomly divided into 5 groups with 8 mice in each group. NC antagomir plus DMSO, miR-142-5p antagomir plus DMSO, or miR-142-5p antagomir plus LY294002 was injected into subcutaneous xenografts in nude mice. The mice were injected with 10 nmol of NC antagomir, 10 mg/kg (of body weight) of DMSO, 10 nmol of miR-142-5p antagomir, or 10 mg/kg of LY294002. All groups of mice were subjected to the injections once a week for 5 weeks, whereupon they were euthanized. The subcutaneous transplanted tumor was removed and weighed, and the tumor tissues were extracted for Western blot analysis.
Thirty-two SPF female BALB/c nude mice (6 weeks, 15 to 18 g) were purchased from Shanghai SLAC Laboratory Animal Co., Ltd., and raised in an SPF environment. The nude mice were randomly divided into 4 groups with 8 mice in each group. Next, the stably transfected HeLa cervical cancer cells treated with inhibitor-NC, miR-142-5p inhibitor, mimic-NC, or miR-142-5p mimic were constructed. Stably transfected cells (1 × 106 cells/ml) were resuspended in 100 μl of normal saline and injected into mice via a tail vein. Seven weeks later, the BALB/c nude mice were euthanized, and the number of lung and liver metastases was counted.
Bioinformatic analysis.
Correlation between miR-142 expression and methylation level in cervical cancer was analyzed through the LinkOmics database (http://www.linkedomics.org/admin.php). The correlation between miR-142-5p and cervical cancer survival was analyzed using the miRpower database (http://kmplot.com/analysis/index.php?p=service&cancer=pancancer_mirna). The cervical cancer expression database GSE63514 was obtained through the GEO database (https://www.ncbi.nlm.nih.gov/geo/), which included 24 normal samples and 28 tumor samples, in which the normal samples were taken as NC. The R language limma package was used for differential expression analysis, and the false-discovery rate (FDR) method was used to correct P values, with absolute log fold change (|logFC|) of >1 and adjusted P value (adj.P) of <0.05 as the screening criteria for differentially expressed genes. The expression of PIK3AP1 in TCGA cervical cancer was retrieved using the UALCAN database (http://ualcan.path.uab.edu/analysis.html), and the binding sites of miR-142-5p and PIK3AP1 were predicted using Starbase2 (http://starbase.sysu.edu.cn/starbase2/).
Statistical analysis.
Statistical analysis was conducted by SPSS 21.0 (IBM, Armonk, NY). The measurement data were expressed as means ± standard deviations. The paired t test was used to compare the carcinoma and adjacent tissues. The data comparison between multiple groups was performed using one-way analysis of variance (ANOVA), followed by Tukey’s post hoc test. For data comparison between groups at different time points, repeated measurement ANOVA was used, followed by Tukey’s post hoc test. A P value of <0.05 indicated that the difference was statistically significant.
Data availability.
The data sets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
ACKNOWLEDGMENTS
We express gratitude to the reviewers for their valuable comments.
We have no funding to report.
Junliang Guo, Tian Tang, and Jinhong Li conceived and designed research. Jinhong Li and Yihong Yang performed the experiments. Junliang Guo, Tian Tang, and Yi Quan analyzed the data. Long Zhang and Wei Huang interpreted the results of the experiments. Muchuan Zhou, Wei Huang, and Yi Quan prepared the figures. Junliang Guo, Tian Tang, and Jinhong Li drafted the manuscript. Long Zhang and Muchuan Zhou edited and revised the manuscript. Junliang Guo, Tian Tang, Jinhong Li, Yihong Yang, Yi Quan, Long Zhang, Wei Huang, and Muchuan Zhou approved the final version of the manuscript.
We have no competing interests to report.
REFERENCES
- 1.Cancer Genome Atlas Research Network, et al. 2017. Integrated genomic and molecular characterization of cervical cancer. Nature 543:378–384. doi: 10.1038/nature21386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cohen PA, Jhingran A, Oaknin A, Denny L. 2019. Cervical cancer. Lancet 393:169–182. doi: 10.1016/S0140-6736(18)32470-X. [DOI] [PubMed] [Google Scholar]
- 3.Denny L, de Sanjose S, Mutebi M, Anderson BO, Kim J, Jeronimo J, Herrero R, Yeates K, Ginsburg O, Sankaranarayanan R. 2017. Interventions to close the divide for women with breast and cervical cancer between low-income and middle-income countries and high-income countries. Lancet 389:861–870. doi: 10.1016/S0140-6736(16)31795-0. [DOI] [PubMed] [Google Scholar]
- 4.Zheng M, Hou L, Ma Y, Zhou L, Wang F, Cheng B, Wang W, Lu B, Liu P, Lu W, Lu Y. 2019. Exosomal let-7d-3p and miR-30d-5p as diagnostic biomarkers for non-invasive screening of cervical cancer and its precursors. Mol Cancer 18:76. doi: 10.1186/s12943-019-0999-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dryden-Peterson S, Bvochora-Nsingo M, Suneja G, Efstathiou JA, Grover S, Chiyapo S, Ramogola-Masire D, Kebabonye-Pusoentsi M, Clayman R, Mapes AC, Tapela N, Asmelash A, Medhin H, Viswanathan AN, Russell AH, Lin LL, Kayembe MKA, Mmalane M, Randall TC, Chabner B, Lockman S. 2016. HIV infection and survival among women with cervical cancer. J Clin Oncol 34:3749–3757. doi: 10.1200/JCO.2016.67.9613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choi CH, Chung JY, Kang JH, Paik ES, Lee YY, Park W, Byeon SJ, Chung EJ, Kim BG, Hewitt SM, Bae DS. 2020. Chemoradiotherapy response prediction model by proteomic expressional profiling in patients with locally advanced cervical cancer. Gynecol Oncol 157:437–443. doi: 10.1016/j.ygyno.2020.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wei WF, Zhou CF, Wu XG, He LN, Wu LF, Chen XJ, Yan RM, Zhong M, Yu YH, Liang L, Wang W. 2017. MicroRNA-221-3p, a TWIST2 target, promotes cervical cancer metastasis by directly targeting THBS2. Cell Death Dis 8:3220. doi: 10.1038/s41419-017-0077-5. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 8.Si W, Shen J, Zheng H, Fan W. 2019. The role and mechanisms of action of microRNAs in cancer drug resistance. Clin Epigenetics 11:25. doi: 10.1186/s13148-018-0587-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tang T, Wong HK, Gu W, Yu MY, To KF, Wang CC, Wong YF, Cheung TH, Chung TK, Choy KW. 2013. MicroRNA-182 plays an onco-miRNA role in cervical cancer. Gynecol Oncol 129:199–208. doi: 10.1016/j.ygyno.2012.12.043. [DOI] [PubMed] [Google Scholar]
- 10.Wald AI, Hoskins EE, Wells SI, Ferris RL, Khan SA. 2011. Alteration of microRNA profiles in squamous cell carcinoma of the head and neck cell lines by human papillomavirus. Head Neck 33:504–512. doi: 10.1002/hed.21475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma J, Yu J, Liu J, Yang X, Lou M, Liu J, Feng F, Ji P, Wang L. 2017. MicroRNA-302a targets GAB2 to suppress cell proliferation, migration and invasion of glioma. Oncol Rep 37:1159–1167. doi: 10.3892/or.2016.5320. [DOI] [PubMed] [Google Scholar]
- 12.Zhang A, Yan S, Cao M, Wu D, Zhou J, Yu Z, Wu M, Liu Y, Lu S, Hu G, Zhao J. 2020. Abnormal methylation of PIK3AP1 was involved in regulating the immune inflammatory response of GES-1 cells induced by Helicobacter pylori. Biochem Biophys Res Commun 524:36–42. doi: 10.1016/j.bbrc.2020.01.007. [DOI] [PubMed] [Google Scholar]
- 13.Zhang F, Li K, Yao X, Wang H, Li W, Wu J, Li M, Zhou R, Xu L, Zhao L. 2019. A miR-567-PIK3AP1-PI3K/AKT-c-Myc feedback loop regulates tumour growth and chemoresistance in gastric cancer. EBioMedicine 44:311–321. doi: 10.1016/j.ebiom.2019.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang Q, Jiang W, Hou P. 2019. Emerging role of PI3K/AKT in tumor-related epigenetic regulation. Semin Cancer Biol 59:112–124. doi: 10.1016/j.semcancer.2019.04.001. [DOI] [PubMed] [Google Scholar]
- 15.Guo Q, Xiong Y, Song Y, Hua K, Gao S. 2019. ARHGAP17 suppresses tumor progression and up-regulates P21 and P27 expression via inhibiting PI3K/AKT signaling pathway in cervical cancer. Gene 692:9–16. doi: 10.1016/j.gene.2019.01.004. [DOI] [PubMed] [Google Scholar]
- 16.Che Y, Li Y, Zheng F, Zou K, Li Z, Chen M, Hu S, Tian C, Yu W, Guo W, Luo M, Deng W, Zou L. 2019. TRIP4 promotes tumor growth and metastasis and regulates radiosensitivity of cervical cancer by activating MAPK, PI3K/AKT, and hTERT signaling. Cancer Lett 452:1–13. doi: 10.1016/j.canlet.2019.03.017. [DOI] [PubMed] [Google Scholar]
- 17.Teinturier P, Dechambre H. 1968. Study of anteversion of the hip in children. Rev Chir Orthop Reparatrice Appar Mot 54:545–551. (In French.) [PubMed] [Google Scholar]
- 18.Small W, Jr, Bacon MA, Bajaj A, Chuang LT, Fisher BJ, Harkenrider MM, Jhingran A, Kitchener HC, Mileshkin LR, Viswanathan AN, Gaffney DK. 2017. Cervical cancer: a global health crisis. Cancer 123:2404–2412. doi: 10.1002/cncr.30667. [DOI] [PubMed] [Google Scholar]
- 19.Fowler CI, Saraiya M, Moskosky SB, Miller JW, Gable J, Mautone-Smith N. 2017. Trends in cervical cancer screening in Title X-funded health centers—United States, 2005–2015. MMWR Morb Mortal Wkly Rep 66:981–985. doi: 10.15585/mmwr.mm6637a4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Benard VB, Thomas CC, King J, Massetti GM, Doria-Rose VP, Saraiya M. 2014. Vital signs: cervical cancer incidence, mortality, and screening—United States, 2007–2012. MMWR Morb Mortal Wkly Rep 63:1004–1009. [PMC free article] [PubMed] [Google Scholar]
- 21.Olusola P, Banerjee HN, Philley JV, Dasgupta S. 2019. Human papilloma virus-associated cervical cancer and health disparities. Cells 8:622. doi: 10.3390/cells8060622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang T, Xue X, Peng H. 2019. Therapeutic delivery of miR-29b enhances radiosensitivity in cervical cancer. Mol Ther 27:1183–1194. doi: 10.1016/j.ymthe.2019.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu Y, Wu F, Liu Y, Zhao Q, Tang H. 2019. DNMT1 recruited by EZH2-mediated silencing of miR-484 contributes to the malignancy of cervical cancer cells through MMP14 and HNF1A. Clin Epigenetics 11:186. doi: 10.1186/s13148-019-0786-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li M, Li BY, Xia H, Jiang LL. 2017. Expression of microRNA-142-3p in cervical cancer and its correlation with prognosis. Eur Rev Med Pharmacol Sci 21:2346–2350. [PubMed] [Google Scholar]
- 25.Zhou D, Chen L, Yang K, Jiang H, Xu W, Luan J. 2017. SOCS molecules: the growing players in macrophage polarization and function. Oncotarget 8:60710–60722. doi: 10.18632/oncotarget.19940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang Y, Jin Z, Dong R, Zheng C, Huang Y, Zheng Y, Shen Z, Chen G, Luo X, Zheng S. 2018. MicroRNA-29b/142-5p contribute to the pathogenesis of biliary atresia by regulating the IFN-gamma gene. Cell Death Dis 9:545. doi: 10.1038/s41419-018-0605-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Duan P, Huang X, Ha M, Li L, Liu C. 2020. miR-142-5p/DAX1-dependent regulation of P450c17 contributes to triclosan-mediated testosterone suppression. Sci Total Environ 717:137280. doi: 10.1016/j.scitotenv.2020.137280. [DOI] [PubMed] [Google Scholar]
- 28.Bowden SJ, Kalliala I, Veroniki AA, Arbyn M, Mitra A, Lathouras K, Mirabello L, Chadeau-Hyam M, Paraskevaidis E, Flanagan JM, Kyrgiou M. 2019. The use of human papillomavirus DNA methylation in cervical intraepithelial neoplasia: a systematic review and meta-analysis. EBioMedicine 50:246–259. doi: 10.1016/j.ebiom.2019.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu NY, Zhang X, Chu T, Zhu S, Deng Y, Zhou Y, Wang Y, Zhao X, Liu L, Fang C, Wang Y, Liou YL, Cai J, Wang J. 2019. High methylation of ZNF582 in cervical adenocarcinoma affects radiosensitivity and prognosis. Ann Transl Med 7:328. doi: 10.21037/atm.2019.06.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Politopoulos I, Gibson J, Tapper W, Ennis S, Eccles D, Collins A. 2011. Composite likelihood-based meta-analysis of breast cancer association studies. J Hum Genet 56:377–382. doi: 10.1038/jhg.2011.23. [DOI] [PubMed] [Google Scholar]
- 31.Yan J, Yang B, Lin S, Xing R, Lu Y. 2019. Downregulation of miR-142-5p promotes tumor metastasis through directly regulating CYR61 expression in gastric cancer. Gastric Cancer 22:302–313. doi: 10.1007/s10120-018-0872-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data sets used and/or analyzed during the current study are available from the corresponding author on reasonable request.