LETTER
Integrase strand transfer inhibitors (INSTIs), in combination with other antivirals, are widely used for the treatment of HIV-1-infected individuals. HIV-1 resistance to INSTIs is typically associated with mutations in the viral integrase gene (1). However, recent studies suggested that mutations in the HIV-1 3′-polypurine tract (3′-PPT) may also confer viral resistance to INSTIs. Specifically, Malet et al. (2) reported in an in vitro selection experiment that a high concentration of dolutegravir (DTG) selected for mutations in the 3′-PPT that conferred INSTI resistance. Wijting et al. (3) reported on the selection of mutations in the 3′-PPT, but not integrase gene, in the virus from an infected individual who failed DTG maintenance monotherapy. However, no follow-up studies have ascertained whether the 3′-PPT mutations reported by Wijting et al. reduce INSTI sensitivity in phenotypic analyses of drug resistance. The primary objective of this study was to address this critical knowledge gap.
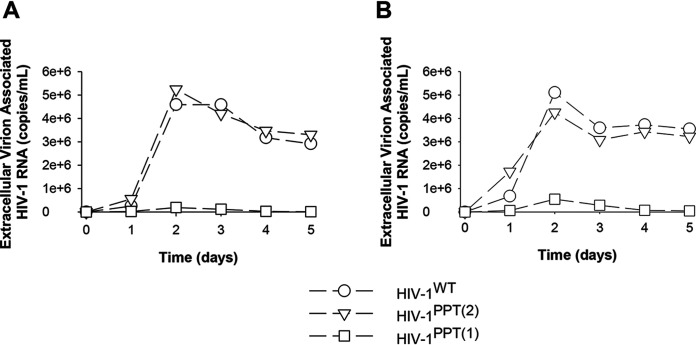
Using site-directed mutagenesis, we constructed subtype B HIV-1LAI infectious viruses in which the 3′-PPT sequence was changed from 5′-TTAAAAGAAAAGGGGGG-3′ (HIV-1WT) to 5′-TTAAAAGAAAAGCAGTΔACTGGAA-3′ (HIV-1PPT(1)) or 5′-TTAAAAGAAAAGGGAGC-3′ (HIV-1PPT(2)), in line with the mutations reported by Malet et al. (2) and Wijting et al. (3), respectively. We first assessed whether the 3′-PPT mutations affected the replication capacity of HIV-1 using a relatively low (27 ng p24) or high (81 ng p24) input of virus (Fig. 1). We found that HIV-1PPT(2) replicated as efficiently as HIV-1WT. In contrast, we could barely detect replication of HIV-1PPT(1), suggesting that the mutations dramatically reduce replication efficiency. In light of this diminished replication efficiency, HIV-1PPT(1) was excluded from further analysis. HIV-1 susceptibility to the INSTIs DTG, cabotegravir, elvitegravir, and raltegravir was assessed in a single-cycle assay, as described previously (4). We found that HIV-1WT and HIV-1PPT(2) exhibited similar susceptibility to each of the INSTIs (Table 1). Previously published studies have suggested that HIV-1 reverse transcriptase (RT)-mediated plus-strand DNA synthesis from the 3′-PPT is exquisitely sensitive to inhibition by nonnucleoside RT inhibitors (NNRTIs) (5–7). Therefore, we also assessed HIV-1WT and HIV-1PPT(2) susceptibility to efavirenz and rilpivirine and found that the 3′-PPT mutations conferred neither resistance nor hypersusceptibility to these drugs (Table 1). Collectively, these data underscore that the 3′-PPT mutations reported by Wijting et al. do not confer INSTI resistance, and thus it is unlikely that their selection can be attributed to the observed DTG failure in the HIV-1-infected individual.
FIG 1.
Replication capacity of HIV-1WT, HIV-1PPT(1), and HIV-1PPT(2). TZM-bl cells were infected with 27 ng p24 (A) or 81 ng p24 (B) of each virus. A single-copy assay for extracellular virion-associated HIV-1 RNA was used to quantify virus production, as described previously (9). Data are reported as the average of two independent experiments.
TABLE 1.
Susceptibility of HIV-1 containing mutations in the 3′-PPT to INSTIs, efavirenz, and rilpivirine
| Inhibitor by drug class | EC50 (nM)a of: |
Fold changeb | |
|---|---|---|---|
| HIV-1WT | HIV-1PPT(2) | ||
| INSTI | |||
| DTG | 0.66 ± 0.23 | 0.85 ± 0.42 | 1.3 |
| Cabotegravir | 0.32 ± 0.06 | 0.37 ± 0.02 | 1.1 |
| Elvitegravir | 0.75 ± 0.51 | 0.76 ± 0.50 | 1.0 |
| Raltegravir | 4.75 ± 0.68 | 5.16 ± 0.82 | 1.1 |
| NNRTI | |||
| Efavirenz | 0.91 ± 0.05 | 1.08 ± 0.50 | 1.2 |
| Rilpivirine | 0.42 ± 0.08 | 0.52 ± 0.18 | 1.2 |
Concentrations of drug required to inhibit viral replication by 50% (EC50). Data reported as a mean ± standard deviation from at least 3 independent experiments.
Mean fold change in EC50 of mutant versus wild-type (WT) virus. EC50 values between HIV-1WT and HIV-1PPT(2) were compared for statistically significant differences (P < 0.05) using a t test. None were found to be statistically significant.
Clinical management of HIV-1 infection relies on genotypic analyses of drug resistance that inform on the utility of including a specific drug class in a combination regimen. In this regard, it is important to know whether mutations outside the HIV-1 integrase gene contribute to INSTI resistance. Malet et al. (8) recently reported that the selection of mutations in the 3′-PPT in individuals failing INSTI resistance was rare, and our study highlights that if they do arise, they do not necessarily contribute to INSTI resistance. As such, genotypic analysis of the 3′-PPT in HIV-1 resistance to INSTIs is not warranted.
ACKNOWLEDGMENTS
This study was supported by research grants R01AI081571 and P50 AI150481 to N.S.-C. from the National Institutes of Health.
Tsinghua scholars (Y.W.) receive their cost-of-living support from the China Scholarship Council (CSC).
REFERENCES
- 1.Anstett K, Brenner B, Mesplede T, Wainberg MA. 2017. HIV drug resistance against strand transfer integrase inhibitors. Retrovirology 14:36. doi: 10.1186/s12977-017-0360-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Malet I, Subra F, Charpentier C, Collin G, Descamps D, Calvez V, Marcelin AG, Delelis O. 2017. Mutations located outside the integrase gene can confer resistance to HIV-1 integrase strand transfer inhibitors. mBio 8:e00922-17. doi: 10.1128/mBio.00922-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wijting I, Lungu C, Rijnders B, van der Ende ME, Pham HT, Mesplede T, Pas SD, Voermans J, Schuurman R, van de Vijver D, Boers P, Gruters RA, Boucher C, van Kampen J. 2018. HIV-1 resistance dynamics in patients with virologic failure to dolutegravir maintenance monotherapy. J Infect Dis 218:688–697. doi: 10.1093/infdis/jiy176. [DOI] [PubMed] [Google Scholar]
- 4.Giacobbi NS, Sluis-Cremer N. 2017. In vitro cross-resistance profiles of rilpivirine, dapivirine, and MIV-150, nonnucleoside reverse transcriptase inhibitor microbicides in clinical development for the prevention of HIV-1 infection. Antimicrob Agents Chemother 61:e00277-17. doi: 10.1128/AAC.00277-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grobler JA, Dornadula G, Rice MR, Simcoe AL, Hazuda DJ, Miller MD. 2007. HIV-1 reverse transcriptase plus-strand initiation exhibits preferential sensitivity to non-nucleoside reverse transcriptase inhibitors in vitro. J Biol Chem 282:8005–8010. doi: 10.1074/jbc.M608274200. [DOI] [PubMed] [Google Scholar]
- 6.Biondi MJ, Beilhartz GL, McCormick S, Götte M. 2010. N348I in HIV-1 reverse transcriptase can counteract the nevirapine-mediated bias toward RNase H cleavage during plus-strand initiation. J Biol Chem 285:26966–26975. doi: 10.1074/jbc.M110.105775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Betancor G, Álvarez M, Marcelli B, Andrés C, Martínez MA, Menéndez-Arias L. 2015. Effects of HIV-1 reverse transcriptase connection subdomain mutations on polypurine tract removal and initiation of (+)-strand DNA synthesis. Nucleic Acids Res 43:2259–2270. doi: 10.1093/nar/gkv077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Malet I, Delelis O, Nguyen T, Leducq V, Abdi B, Morand-Joubert L, Calvez V, Marcelin AG. 2019. Variability of the HIV-1 3′ polypurine tract (3′PPT) region and implication in integrase inhibitor resistance. J Antimicrob Chemother 74:3440–3444. doi: 10.1093/jac/dkz377. [DOI] [PubMed] [Google Scholar]
- 9.Cillo AR, Vagratian D, Bedison MA, Anderson EM, Kearney MF, Fyne E, Koontz D, Coffin JM, Piatak M, Jr, Mellors JW. 2014. Improved single-copy assays for quantification of persistent HIV-1 viremia in patients on suppressive antiretroviral therapy. J Clin Microbiol 52:3944–3951. doi: 10.1128/JCM.02060-14. [DOI] [PMC free article] [PubMed] [Google Scholar]