LETTER
Recent data published in the literature reported on the performance of the quantitative Roche Elecsys anti-SARS-CoV-2 (severe acute respiratory syndrome coronavirus 2) S assay (1–3). The specificity of the test was between 99.8 and 100% (1–3). However, for sensitivity calculations, Higgins et al. included 167 samples from 167 reverse transcription-PCR (RT-PCR)-positive patients and reported sensitivities of 67.1% and 82.5% at 0 to 14 and at >14 days after RT-PCR positivity, respectively (1). We noticed that the follow-up period was short, 73 days. The absence of samples with a longer-term follow-up might have mitigated the true sensitivity of the test, and this is particularly important as this pandemic advances.
In order to complement the current published data on the Roche Elecsys anti-SARS-CoV-2 S and nucleocapsid protein (NCP) assays, we report here a series of 229 samples from 105 symptomatic patients (mean age, 56 years; range, 25 to 95 years) with a confirmed SARS-CoV-2 RT-PCR (LightMix Modular SARS-CoV E-gene set, Roche Diagnostics, Basel, Switzerland). Among these patients, 57 were females and 48 were males. The majority of patients (n = 88; 83.8%) developed a nonsevere form of coronavirus disease 2019 (COVID-19), while the others (n = 17; 16.2%) required hospitalization.
Samples were retrospectively collected from 26 March 2020 to 18 February 2021 at the Clinique Saint-Luc Bouge (Namur, Belgium). Residual frozen samples from routine diagnostic testing were used. The median time between the symptom onset and the RT-PCR was 3 days (interquartile range [IQR], 1 to 7 days).
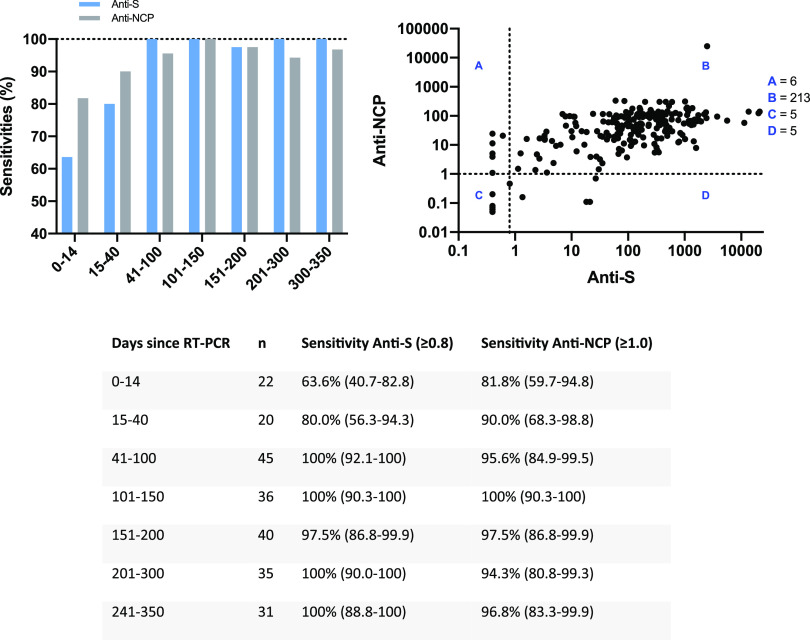
Using the same time intervals as those used by Higgins et al. (1), we found similar sensitivities for the 0- to 14-day and 15- to 40-day time frames. Although the confidence intervals were overlapping, higher sensitivities were observed using the NCP assay (Fig. 1). It is noteworthy that the sensitivities increased for samples collected >40 days and remained elevated up to 350 days after RT-PCR positivity. Similar sensitivities at median days of 48 and 139 days since diagnosis (97.6% versus 98.2%) were found in the study of Schaffner et al. (2) using the Roche Elecsys anti-SARS-CoV-2 S assay.
FIG 1.
Anti-SARS-CoV-2 antibody response (anti-S and anti-NCP) by days after PCR positivity, as measured by the quantitative Roche Elecsys anti-SARS-CoV-2 S assay and by the qualitative Roche Elecsys anti-SARS-CoV-2 NCP assay. In the top left bar graph, the x axis shows the number of days after PCR positivity.
Out of our 229 samples tested, 5 samples were positive for anti-S antibodies but negative for anti-NCP antibodies and 6 samples were positive for anti-NCP antibodies but negative for anti-S antibodies. The global agreement between the two assays was 94.8%, a score similar to that found with multiplex assays (4, 5).
Our results support that the maximal sensitivity of the anti-S assay of Roche Diagnostics may be better than reported previously (1) and that the absence of seroconversion observed in this previous study may be due to insufficient follow-up of patients. The data of Schaffner et al. (2) are in line with our findings. Other studies that evaluated the anti-S assay from Roche Diagnostics included samples only up to 44 days after RT-PCR (3, 6). Other independent long-term studies are needed to confirm our data. Our results suggest that negative samples before 40 days after possible contact should be retested after this period to increase detection of seropositive subjects.
As the pandemic advances, subjects may have been in contact with SARS-CoV-2 for many months, and the capacity of serological assays to detect the seropositivity in these subjects is of utmost importance. A recent report suggests that a change in vaccine policy to give previously infected individuals only one dose of the vaccine would not negatively impact their antibody response and may consequently free up millions of vaccine doses (7). Therefore, it is reassuring to observe a sensitivity close to 100% in samples collected ≥40 and up to 350 days after RT-PCR positivity, since this could be an important parameter in future vaccine administration strategies.
Contributor Information
Julien Favresse, Email: j.favresse@labstluc.be.
Alexander J. McAdam, Boston Children’s Hospital
REFERENCES
- 1.Higgins V, Fabros A, Kulasingam V. 2021. Quantitative measurement of anti-SARS-CoV-2 antibodies: analytical and clinical evaluation. J Clin Microbiol 59:e03149-20. 10.1128/JCM.03149-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schaffner A, Risch L, Aeschbacher S, Risch C, Weber MC, Thiel SL, Jungert K, Pichler M, Grossmann K, Wohlwend N, Lung T, Hillmann D, Bigler S, Bodmer T, Imperiali M, Renz H, Kohler P, Vernazza P, Kahlert CR, Twerenbold R, Paprotny M, Conen D, Risch M. 2020. Characterization of a pan-immunoglobulin assay quantifying antibodies directed against the receptor binding domain of the SARS-CoV-2 S1-subunit of the spike protein: a population-based study. J Clin Med 9:3989. 10.3390/jcm9123989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Muench P, Jochum S, Wenderoth V, Ofenloch-Haehnle B, Hombach M, Strobl M, Sadlowski H, Sachse C, Torriani G, Eckerle I, Riedel A. 2020. Development and validation of the Elecsys anti-SARS-CoV-2 immunoassay as a highly specific tool for determining past exposure to SARS-CoV-2. J Clin Microbiol 58:e01694-20. 10.1128/JCM.01694-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gillot C, Douxfils J, Cadrobbi J, Laffineur K, Dogne JM, Elsen M, Eucher C, Melchionda S, Modaffarri E, Tre-Hardy M, Favresse J. 2020. An original ELISA-based multiplex method for the simultaneous detection of 5 SARS-CoV-2 IgG antibodies directed against different antigens. J Clin Med 9:3752. 10.3390/jcm9113752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Favresse J, Brauner J, Bodart N, Vigneron A, Roisin S, Melchionda S, Douxfils J, Ocmant A. 2020. An original multiplex method to assess five different SARS-CoV-2 antibodies. Clin Chem Lab Med 10.1515/cclm-2020-1652. [DOI] [PubMed] [Google Scholar]
- 6.Merrill AE, Jackson JB, Ehlers A, Voss D, Krasowski MD. 2020. Head-to-head comparison of two SARS-CoV-2 serology assays. J Appl Lab Med 5:1351–1357. 10.1093/jalm/jfaa125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krammer F, Srivastava K, Simon V. 2021. Robust spike antibody responses and increased reactogenicity in seropositive individuals after a single dose of SARS-CoV-2 mRNA vaccine. medRxiv 10.1101/2021.01.29.21250653. [DOI]