ABSTRACT
Aspergillus niger is an opportunistic pathogen commonly found in a variety of indoor and outdoor environments. An environmental isolate of A. niger from a pig farm was resistant to itraconazole, and in-depth investigations were conducted to better understand cellular responses that occur during growth when this pathogen is exposed to an antifungal. Using a combination of cultivation techniques, antibiotic stress testing, and label-free proteomics, this study investigated the physiological and metabolic responses of A. niger to sublethal levels of antifungal stress. Challenging A. niger with itraconazole inhibited growth, and the MIC was estimated to be > 16 mg · liter−1. Through the proteome analysis, 1,305 unique proteins were identified. During growth with 2 and 8 mg · liter−1 itraconazole, a total of 91 and 50 proteins, respectively, were significantly differentially expressed. When challenged with itraconazole, A. niger exhibited decreased expression of peroxidative enzymes, increased expression of an ATP-binding cassette (ABC) transporter most likely involved as an azole efflux pump, and inhibited ergosterol synthesis; however, several ergosterol biosynthesis proteins increased in abundance. Furthermore, reduced expression of proteins involved in the production of ATP and reducing power from both the tricarboxylic acid (TCA) and glyoxylate cycles was observed. The mode of action of triazoles in A. niger therefore appears more complex than previously anticipated, and these observations may help highlight future targets for antifungal treatment.
KEYWORDS: resistance, resistance mechanisms, antifungal, fungi, physiology, triazole, ergosterol synthesis
INTRODUCTION
Aspergillus niger is an opportunistic pathogen commonly found in a variety of indoor and outdoor environments (1). The spores of A. niger can be easily aerosolized (2) and have the potential to be deposited in bronchioles of the human respiratory tract (3). Infections caused by A. niger can lead to the development of allergic bronchopulmonary aspergillosis (ABPA) or invasive aspergillosis, both of which can be lethal in susceptible patient groups (4, 5).
Patients suspected of having fungal infection typically undergo a variety of different diagnostic tests, such as chest radiographs, culturing of the suspected infectious agent(s) from bronchoalveolar lavage fluid (BALF) and biopsy specimens, sputum screening, and/or blood antigen testing (6). Upon diagnosis, patients often receive long-term antifungal treatment with or without surgical debridement (6, 7).
While fungal resistance to triazoles is known to develop in patients treated with triazoles, patients who are azole naive have also been noted to develop triazole-resistant fungal infections (8). The triazole resistance may have derived from overuse of homologous triazoles in agricultural settings, which has conferred cross-resistance to medical triazoles (9).
Recent research has shown that multiple environmental isolates of Aspergillus spp. were resistant to different antifungals, such as itraconazole, voriconazole, and caspofungin, which are used to treat invasive aspergillosis in human patients (6). This is of concern, as antimicrobial resistance is associated with worse patient outcomes (10). Only a few studies have investigated A. niger’s susceptibility in general, and the MIC of itraconazole for different isolates of A. niger has previously been estimated to be in the range of 0.03 to 32 mg · liter−1 (11–15). In diverse occupational settings, workers have been observed to be exposed to airborne A. niger, e.g., cleaning workers (16). Waste collection workers can inhale up to 2.3 × 104 CFU of this fungus during a work day (17). Ear infections have been diagnosed among biowaste workers, and A. niger has been isolated from the ear discharge (18). Compost workers from Germany exposed to fungal bioaerosols developed ABPA, which was treated with corticosteroids and itraconazole (19). Therefore, the presence of antifungal-resistant A. niger in occupational settings is of concern.
The mechanisms which confer resistance to triazoles are typically ascribed to mutations in the 14-α-sterol demethylase (cyp51a) gene (20), the overexpression of efflux pumps (21), or single-base substitutions in the transcription factor subunits, such as HapE, initiating changes altering cyp51a expression (22). The effect of these resistance mechanisms can be seen by the increased tolerance of Aspergillus spp. to concentrations of triazoles which would normally inhibit growth, thereby conferring resistance to triazoles.
Although the aforementioned mechanisms have been proposed for the acquisition of resistance to triazoles, these have mainly focused on the most common etiological agent of aspergillosis, Aspergillus fumigatus (23). However, the metabolic pathways which are changed upon challenge with triazoles in A. fumigatus may not necessarily be the same ones as in A. niger.
Itraconazole’s mode of action is to inhibit ergosterol synthesis, which causes stalled cell growth and promotes cell death. The inhibition of the ergosterol synthesis is believed to be caused by formation of a complex between itraconazole and the heme iron of the fungal cytochrome P450 (24, 25), which leads to inhibition of 14α-demethylase. This breakdown in the production of new ergosterol causes an accumulation of toxic sterol intermediates (14-α-methyl-3,6-diol) and formation of a defective cell membrane whose permeability and function are altered (26, 27). Some other postulated mechanisms of itraconazole are inhibition of the peroxidative enzymes, resulting in accumulation of intracellular hydrogen peroxide, and the triggering by antifungal triazoles of the formation of mitochondrial reactive oxygen species (ROS) by the mitochondrial respiratory complex I (24, 28–30).
The aim of this study was to elucidate the acute antibiotic impact and cellular response when A. niger was challenged with itraconazole. The approach was to study the differential proteome to diagnose physiological states and identify the processes affected by the antifungal compound. The study compared the effect at sublethal doses (2 and 8 mg · liter−1), which are well below the isolate’s MIC, thereby excluding a concentration-dependent toxic effect.
RESULTS
Antifungal susceptibility testing of A. niger isolate 351.
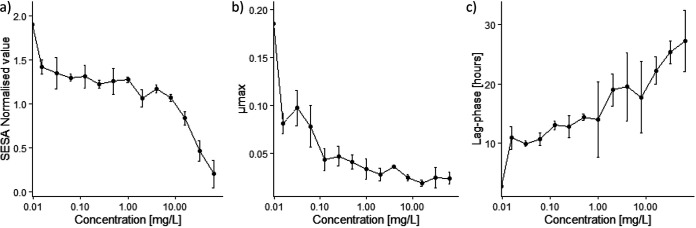
A total of 15 A. niger isolates were sampled from straw found in the pig farm. They displayed different levels of resistance to itraconazole (MICs between 0.5 and >16 mg · liter−1), voriconazole (MICs between 0.5 and >16 mg · liter−1), and caspofungin (MICs between 0.0625 and 16 mg · liter−1) (our unpublished work). Initially, an assessment of the effect of itraconazole against A. niger isolate 351 was made, and even the lowest concentration of itraconazole appeared to have an inhibitory effect on the growth of the isolate (Fig. 1a; also, see Fig. S1 in the supplemental material). Almost complete inhibition of the cell growth was seen at concentrations of itraconazole above 16 mg · liter−1, and therefore, the MIC was estimated to be >16 mg · liter−1. An antifungal concentration of 0.5 mg · liter−1 resulted in a 4-fold decrease in the maximum growth rate (Fig. 1b) and a 5-fold increase in the lag phase (Fig. 1c).
FIG 1.
To measure the effect of itraconazole on A. niger isolate 351, concentrations ranging from 0 to 64 mg · liter−1 were tested. (a) Segmentation extraction surface area (SESA) normalized values measured after 48 h showing the biomass produced by a single isolate of Aspergillus niger challenged with different concentrations of itraconazole. (b) Maximum growth rate (μmax) of Aspergillus niger isolate 351 at different concentrations of itraconazole. (c) Lag phase for different concentrations of itraconazole.
Strain verification and genome quality.
Matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) was used to identify isolate 351 as A. niger. To confirm this result and to generate a genomic reference database of the isolate, the genome of A. niger isolate 351 was sequenced with a total of 513,820 reads, and a length of 1.79 Gb was generated. After trimming the read length, N50 was estimated at 13,487 and the number of reads was 103,449, resulting in ∼41× coverage. Based on BUSCO (benchmarking universal single-copy orthologs) analysis, it was estimated that the genome had a completeness of 98.2%.
To verify that the isolate was indeed A. niger, a nucleotide BLAST search against NCBI was conducted (for the BLASTn results, see Table S1). The search showed a clear affiliation with A. niger, with near 100% similarity to the A. niger genomic contig An01c0390 (AM269986.1). The genome was investigated for well-known Cyp51A point mutations (M217I, L98H, G54W, P216L, F219I, M220V/K/T, and G448S) as well as potential tandem repeats in the promoter region of cyp51A. No tandem repeats (TR34 or TR46) in isolate 351 were observed, and the M217I point mutation was found in the genomic data.
Proteomic analysis of Aspergillus niger challenged with itraconazole.
The differential proteomic analysis resulted in detection of 1,305 proteins. When A. niger was treated with 2 and 8 mg · liter−1 itraconazole, a total of 91 and 50 proteins were found to have significantly changed in abundance relative to the control (P < 0.05; log2 < −0.5 and log2 > 0.5) with 41 and 23 proteins showing an increase and 50 and 27 proteins showing a decrease in abundance, respectively (Table 1 and Fig. S2; for the full list of significantly expressed proteins, see Table S1). Analysis of differential A. niger proteomes upon treatment with itraconazole at two concentrations below the MIC was conducted in biological quadruplicates, in order to identify the physiological response and molecular targets of itraconazole. This led to the identification of several significantly altered biochemical pathways, including the glyoxylate cycle, the tricarboxylic acid (TCA) cycle, peroxidase activity, and ergosterol synthesis.
TABLE 1.
Top 10 proteins which were found to have significant changes in abundance at 2 and 8 mg · liter−1 itraconazole
| Itraconazole concn (mg liter−1) | Upregulated |
Downregulated |
||||||
|---|---|---|---|---|---|---|---|---|
| P value | Difference | Protein ID | Protein name or description | P value | Difference | Protein IDs | Protein name or description | |
| 2 | 3.8E−05 | 6.68 | A0A117E0T6 | Sterol 24-C-methyltransferase | 0.0025 | −5.19 | XP_001389347.1 | Isocitrate lyase |
| 5.9E−07 | 5.41 | A0A117DW86 | ABC transporter | 0.0011 | −3.94 | A0A100IR47 | Oxaloacetate acetylhydrolase | |
| 1.8E−05 | 4.02 | XP_025478936.1 | Glutathione peroxidase Hyr1 | 0.0004 | −3.52 | A0A100IPP9 | Geranylgeranyl pyrophosphate synthase | |
| 1.7E−03 | 3.16 | A0A117E2H8 | Choline oxidase | 0.0000 | −3.19 | A0A100IHE1 | Glycerol-3-phosphate dehydrogenase | |
| 2.1E−02 | 2.85 | A0A100IEW1 | F5/8 type C domain protein | 0.0010 | −3.05 | A0A124BYU7 | Allergen | |
| 4.4E−03 | 2.72 | G3Y367 | Protein disulfide-isomerase | 0.0087 | −3.03 | XP_025476110.1 | Hypothetical protein BO87DRAFT_379724 | |
| 4.0E−04 | 2.03 | A0A100IRZ3 | Protein disulfide-isomerase | 0.0067 | −2.90 | A0A124BY46 | Pathogenesis-associated protein Cap20 | |
| 1.1E−03 | 2.02 | XP_001391010.1 | Hit family protein 1 | 0.0089 | −2.54 | A0A117DVZ9 | Catalase | |
| 1.1E−03 | 1.79 | G3XSV7 | Uncharacterized protein | 0.0202 | −2.38 | A0A100I3Y8 | Aldehyde dehydrogenase | |
| 1.6E−03 | 1.68 | A0A100I439 | Translationally controlled tumor protein homolog | 0.0077 | −2.30 | A0A100IQA9 | Alcohol dehydrogenase (adhA) | |
| 8 | 4.2E−05 | 6.40 | A0A117E0T6 | Sterol 24-C-methyltransferase | 0.0019 | −5.92 | XP_001389347.1 | Isocitrate lyase |
| 3.1E−06 | 5.04 | A0A117DW86 | ABC transporter | 0.0058 | −4.81 | XP_025485096.1 | Superoxide dismutase [Mn], mitochondrial | |
| 1.1E−02 | 3.70 | A0A100IIZ2 | Cyp51A | 0.0006 | −4.41 | A0A100IR47 | Oxaloacetate acetylhydrolase | |
| 1.0E−04 | 3.61 | XP_025478936.1 | Glutathione peroxidase Hyr1 | 0.0029 | −3.36 | A0A100IPP9 | Geranylgeranyl pyrophosphate synthase | |
| 9.0E−03 | 2.81 | A0A100I6D1 | FAD-binding PCMH-type domain-containing protein | 0.0015 | −2.36 | A0A124BYU7 | Allergen | |
| 1.5E−04 | 1.86 | A0A100IA04 | Aspartic protease PEPAd | 0.0100 | −2.31 | A0A117DVZ9 | Catalase | |
| 7.0E−03 | 1.74 | XP_001391010.1 | Hit family protein 1 | 0.0001 | −2.30 | A0A100IHE1 | Glycerol-3-phosphate dehydrogenase | |
| 6.9E−03 | 1.40 | XP_001395889.1 | Actin-related protein 2/3 complex subunit ARC19 | 0.0001 | −2.23 | A0A100IKP3 | Peroxisomal multifunctional beta-oxidation protein | |
| 5.2E−03 | 1.38 | G3XS64 | ATP synthase subunit delta, mitochondrial | 0.0060 | −2.18 | XP_025476110.1 | Hypothetical protein BO87DRAFT_379724 | |
| 1.5E−03 | 1.37 | A0A100II93 | N,N-dimethylglycine oxidase | 0.0052 | −2.10 | G3XT80 | Thioredoxin reductase | |
Significantly expressed proteins were categorized according to their gene ontologies (GO) (Table S1). Enrichment analysis was carried out to identify functional classes that are significantly overrepresented in the set of differentially expressed proteins.
Many proteins which were significantly more abundant upon itraconazole treatment belonged to the categories of intracellular proteins (protein disulfide-isomerase [G3Y367], translationally controlled tumor protein [TCTP] homologue [A0A100I439] [31], and cyclophilin type CYPA peptidyl-prolyl cis-trans isomerase [XP_025478857.1] [P < 0.006 and log2 > 0.84]), ATP synthesis (ATP phosphoribosyltransferase [G3XPV6] and ATP synthase subunit delta [G3XS64] [P = 0.005 and log2 > 1.17]), and cell redox homeostasis (thioredoxin domain-containing protein [G3Y722] and protein disulfide-isomerase [G3Y367] [P < 0.0044 and log2 > 1.28]). The protein with the highest increase in abundance during treatment with 2 and 8 mg · liter−1 itraconazole was sterol 24-C-methyltransferase (A0A117E0T6) (P < 0.00004 and log2 > 6.4), while the protein with the second highest increase in expression when treated with both 2 and 8 mg · liter−1 itraconazole was an ABC transporter (A0A117DW86) (P < 0.000003 and log2 > 5.04) (Table 1).
The proteins with decreased abundance upon itraconazole treatment belonged to the categories of peroxidase activity (catalase [A0A117DVZ9], catalase-peroxidase [A0A117DWC4], and superoxide dismutase [XP_025485096.1] [P value < 0.009 and log2 < −1.46]), catalytic activity (pyruvate carboxylase [A0A100IA53], aspartate aminotransferase [XP_001397865.2], glutamate decarboxylase [A0A100IU53], and pyruvate kinase [XP_001391973.1] [P < 0.01 and log2 <−0.92]), tricarboxylic acid cycle (isocitrate dehydrogenase [A0A100IUT3] and citrate synthase [XP_001393983.1] [P < 0.001 and log2 < −1.09]), and ATP binding (plasma membrane ATPase [A0A100IRV8], diphosphomevalonate decarboxylase [A0A100IAV5], and homoserine kinase [XP_001398437.1] [P < 0.003 and log2 < −0.76]). Diphosphomevalonate decarboxylase is involved in isoprenoide biosynthesis and thus also in the biosynthesis of ergosterol. The protein with the greatest decrease in abundance due to treatment of both 2 and 8 mg · liter−1 itraconazole was isocitrate lyase (XP_001389347.1) (P < 0.0025 and log2 < −5.2).
When treatment with 2 mg · liter−1 itraconazole was compared to treatment with 8 mg · liter−1, no significantly differently expressed proteins were detected using a criterion with a P value of <0.05 and log2 values of <−0.5 and >0.5. Therefore, the responses of A. niger to the two different concentrations of itraconazole were not significantly different from each other.
DISCUSSION
In this study, we investigated susceptibility and proteomic profiles of A. niger isolate 351 treated with itraconazole at two concentrations below its MIC. Complementary to target-centric approaches, the approach used in this study was chosen to provide an insight into the physiological responses and thereby to better understand the antibiotic mechanism. Previously studies have examined A. niger’s response to antifungals (voriconazole and itraconazole) when exposed to one concentration and used a two-dimensional gel electrophoretic separation method coupled with mass-spectrometric analysis to identify the proteins expressed upon treatment with an antifungal compound (32, 33), whereas we examined A. niger’s response to two different concentrations of itraconazole and used a shotgun proteomic approach for identification of the abundant proteins.
Genome quality and strain verification.
The fungus was isolated from straw in a pig farm and identified as A. niger by both MALDI-TOF MS and whole-genome sequencing. The purpose of the genome sequencing, besides identifying the isolate, was to function as a reference for the proteome data obtained. Therefore, a completeness of 98.2% and a coverage of ∼41 gave a sufficient depth to be used as a reference genome for the proteomic data.
Antifungal susceptibility of Aspergillus niger isolate 351.
To determine whether the isolate of A. niger was susceptible or resistant to azole compounds, we confirmed the updated EUCAST clinical breakpoints (v.10.0) (34), stating a breakpoint value of 4 mg · liter−1. A. niger isolate 351 was able to grow at concentrations up to 16 mg · liter−1 and is therefore categorizable as being highly resistant to itraconazole (8, 11–14). The large discrepancy in the determined MICs could be related to the fact that a relatively small number of Aspergillus spp. have acquired resistance mechanisms (12, 15).
Differentially expressed proteins.
Overall, the proteins with significant differential expression during growth in the presence of itraconazole are either part of the energy metabolism or responsible for cell maintenance and therefore indicate vital responses to maintain cell vitality during antifungal stress.
The proteins found to be more significant abundant upon treatment at both 2 and 8 mg · liter−1 itraconazole was the sterol 24-C-methyltransferase, which is an important protein in ergosterol biosynthesis (35). Since itraconazole inhibits Cyp51A, an enzyme responsible for an intermediate step in ergosterol biosynthesis, the physiological response by A. niger appears to increase the synthesis of sterol 24-C-methyltransferase in an attempt to minimize shortage of ergosterol by the inhibition from itraconazole (36). The target enzyme of itraconazole was found to be the third most upregulated protein at 8 mg · liter−1 and is known to contribute to clinical triazole resistance in the closely related species A. fumigatus when overexpressed (37). Thus, higher expression of Cyp51A appears to be a compensation strategy by the isolate during increased exposure to itraconazole. The three most commonly identified resistance mechanisms of triazole resistance in the closely related fungus are mutations in the sterol-demethylase gene cyp51A, overexpression of cyp51A, and overexpression of drug efflux pumps (37). The identification of Cyp51A as the third most upregulated protein at 8 mg · liter−1 and as not detectable at 2 mg · liter−1 indicates that A. niger needs to activate another resistance/compensation mechanism to maintain ergosterol biosynthesis. However, other research has suggested that the increase of sterol 24-C-methyltransferase results in the accumulation of toxic sterol intermediates and disruption of the ergosterol homeostasis (27, 28). The genomic data for this isolate were inspected for well-known Cyp51A point mutations (M217I, L98H, G54W, P216L, F219I, M220V/K/T, and G448S) (38, 39) as well as tandem repeats in the promoter region of cyp51A. The results from this confirm that no tandem repeats (TR34 or TR46) were present in isolate 351, suggesting that some other mechanism led to the overexpression of cyp51A. Only the M217I point mutation was found in the genomic data. This mutation has previously been associated with higher levels of itraconazole resistance in Aspergillus terreus (40).
Multidrug-resistant fungal cells frequently obtain their resistance through modifications in the quality or quantity of target enzyme, reduced access to the target, or a combination of these mechanisms (38, 41). At the time of writing, there are no reports of Aspergillus spp. modifying triazole antifungals as a mechanism of resistance.
Active efflux mechanisms through ATP-binding cassette (ABC) transporters have previously been associated with high-level triazole resistance (39, 42, 43). In this study, ABC transporters were found to be significantly more abundant when A. niger was challenged with itraconazole, and this is most likely the reason for this particular isolate’s high level of resistance. The ABC transporter (A0A117DW86) has an orthologous gene in A. fumigatus (cdr1B) with the transcription factor AtrR and has been described as an azole transporter (39). Notably, the transcription factor coregulates the ABC transporter (Cdr1B) and the drug target (Cyp51A) (44). However, AtrR most likely cooperates with a sterol-regulatory element binding protein (SrbA) to drive the transcription of Cyp51A (45–47). ABC transporters, in general, have a broad specificity for hydrophobic compounds, including antifungals (42), and the data presented in this study therefore suggest that the efflux pumps are of importance as potential resistance mechanisms to triazoles in A. niger.
Among the group of proteins with decreased abundance upon treatment with itraconazole were proteins related to peroxidase activity, including catalase-peroxidase, catalase, and superoxide dismutase (SOD). These proteins are responsible for detoxifying ROS and converting them to water and oxygen to avoid uncontrolled radical formation in the cell (48). Differentially expressed proteins in response to antibiotic compounds have previously been described with fungal cells challenged with miconazole, an imidazole with a mechanism similar to that of itraconazole (29). An increased production of intracellular ROS would be expected to cause damages in the fungal hyphae, which could impair protein homeostasis and reduce growth. In this study, we found that itraconazole inhibits the expression of peroxidases, which is in agreement with previously postulated effects of itraconazole (24) and a universal induction of oxidative stress caused by all antibiotics (49). When exposed to itraconazole, A. niger exhibits both increased production of ROS and a decrease in expressed peroxidative enzymes. This increase of ROS production and decrease in ability to respond to ROS stress are in agreement with previous reports which propose a clear connection between the production of ROS and drug-induced disruption of the ergosterol homeostasis (29). Similar induction of ROS production and inhibition of peroxidative enzymes have been observed in different fungal genera, including Aspergillus, Candida, and Cryptococcus (28, 29), and therefore, this appears to be a more general response to azole antifungals. However, previous studies have also found that fungi often respond to antifungal drugs by increasing their antioxidant stress response (33, 50), which contradicts the findings in this study. The general downregulation of antioxidative enzymes, such as catalase, may be yet another resistance response upon treatment with itraconazole, since catalase activity has been shown to potentiate antifungal azoles (50). One antioxidative enzyme was found to be upregulated upon treatment with itraconazole, the glutathione peroxidase Hyr1, which plays a crucial role in the defense against ROS (51).
Isocitrate lyase, a central enzyme in the glyoxylate cycle (52), was the protein showing the most significant decrease in expression after treatment with itraconazole at both 2 and 8 mg · liter−1. The proteomic data showed that itraconazole inhibits both the TCA cycle and the glyoxylate cycle. Such inhibition results in an energy-poor state and decreases cell proliferation and viability. Whether this is a direct primary effect caused by itraconazole or a secondary derived effect is not yet understood.
Overall, this study demonstrates the cellular responses of the environmental Aspergillus niger isolate 351 to the antifungal triazole itraconazole. As demonstrated both in this study and elsewhere, even the lowest concentrations reduced cellular growth (Fig. 1), and this is most likely due to the inhibition of the ergosterol synthesis and to the decreased expression of peroxidative enzymes along with the resulting accumulation of ROS. However, the fungus is able to survive, albeit at lower activity. This is consistent with the increased abundance of the ABC transporter, an efflux pump with a broad specificity toward hydrophobic compounds such as itraconazole. The effect of itraconazole on the physiology of A. niger appears to be more complex than expected, with several modes of actions occurring simultaneously. The proteomic analysis also revealed a decrease in the energy synthesis and production of reducing powers from both TCA and glyoxylate cycle. Moreover, some significantly expressed proteins identified in this study may be possible candidates for further studies in relation to their role in drug resistance and their possible future use as targets for antifungal treatment.
In summary, the present study explored the physiological and metabolic responses of A. niger upon treatment with itraconazole. Two concentrations of itraconazole below the isolate’s estimated MIC with a breakpoint around 16 mg · liter−1 showed very similar changes in protein expression. This observation suggests that A. niger responds to itraconazole independently at all concentrations below the MIC. The mode of action appears to be complicated, with multiple mechanisms occurring simultaneously. These cover inhibition of the ergosterol synthesis, increase in the expression of efflux transporters, and reduced expression of peroxidative enzymes, which support the mutation in cyp51A (M217I) found in the genome. Furthermore, these inhibitions appear to be accompanied by a decreased production of energy and reducing power. The investigated isolate uses three of the five commonly described resistance mechanisms for A. fumigatus: reduced interaction affinity (point mutation), overexpression of the target protein, and overexpression of efflux pumps. Knowledge of the physiological changes caused by exposure to antifungals provides a better platform for development of optimized antifungal treatments.
MATERIALS AND METHODS
Sample collection.
Isolates of Aspergillus niger were obtained from straw collected on a Danish pig farm, and their antifungal resistance profiles were investigated in this study. In brief, isolates of A. niger were extracted from straw in a pig farm, cultured on dichloran glycerol agar supplemented with 100 mg · liter−1 chloramphenicol (DG18) (Oxoid), identified using matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) on a Microflex Biotyper system (Bruker Daltonics), and tested for their resistance to itraconazole, voriconazole, amphotericin B, and caspofungin. Isolates of A. niger were found to be resistant to multiple antifungals (itraconazole, voriconazole, and caspofungin). We chose one isolate which exhibited high resistance to itraconazole to examine its physiological and metabolic responses when challenged with itraconazole at different concentrations.
Antifungal susceptibility testing.
Antifungal susceptibility testing was performed using the oCelloScope system (BioSense Solutions ApS). Different concentrations of itraconazole (Sigma-Aldrich) were used, ranging from 0 to 64 mg · liter−1, dissolved in dimethyl sulfoxide (DMSO), and stored at −80°C until used. The fungus was grown in RPMI medium (with 2% glucose and MOPS [morpholinepropanesulfonic acid] buffer [Sigma-Aldrich] at a pH of 7.0), starting with a conidial concentration of 5 × 105 conidia per ml, and all concentrations of itraconazole were tested in quadruplicate. The camera focus and illumination level were set to 2,790 μm and 300, respectively. Image acquisition was performed in one scan area in the center of each well, which was scanned every 30 min for 48 h. The growth kinetics were determined by image stack processing using the segmentation extracted surface area (SESA) algorithm, included in the UniExplorer software (BioSense Solutions ApS).
High-molecular-weight-DNA extraction.
A. niger was grown in RPMI medium (with 2% glucose and MOPS buffer [Sigma-Aldrich] at a pH of 7.0) for 5 days at 37°C and 150 rpm. Mycelium was harvested by filtering the liquid using Miracloth (Popolini), followed by a rinsing step of the mycelium with 20 ml sterile double-distilled water (ddH2O). Mycelium was freeze-dried for 1 day and ground in a mortar to a powder. DNA from the freeze-dried and ground mycelium was extracted using a phenol-chloroform procedure (53) with an RNase A treatment step (Qiagen), and high-molecular-weight (HMW) genomic DNA was extracted using the Qiagen Genomic-tip 20/G (Qiagen). Procedures were performed according to the manufacturers’ suggested protocols.
The quality of the HMW DNA was assessed by measuring the DNA concentration using a NanoDrop 1000 instrument and a Qubit dsDNA HS assay kit following the manufacturer’s recommendations. The lengths of the HMW DNA fragments were determined using Agilent genomic DNA ScreenTape following the manufacturer’s recommendations. Small DNA fragments were removed using Short Read Eliminator XS (Circulomics) according to the manufacturer’s recommendations.
Whole-genome sequencing and bioinformatics.
DNA repair, end preparation, adapter ligation, cleanup, and priming were done following the manufacturer’s protocol (MinION; Oxford Nanopore Technologies). In brief, DNA ends were repaired and dA tailed using the NEBNext end repair/dA-tailing module. Sequencing adapters were ligated onto the prepared ends following DNA end repair. The resulting library was loaded onto a single MinION R9.4.1 (106) flow cell and sequenced for 72 h.
Base calling and sequence quality analysis were performed using Guppy v3.2.10 (Guppy is available only to ONT customers via their community site, https://community.nanoporetech.com) and NanoPlot v1.24.0 (55), respectively. Reads were filtered for quality (>Q8) and length (>5,000 bp) using FiltLong v0.2.0 (56). Assembly of the reads were done using minimap2 v2.17 and miniasm v0.3 (57), and the following polish was done using Racon v1.3.3 (58) and medaka v1.0.1 (59). Assessment of genome assembly and annotation completeness was done using BUSCO v3.0.2 (60).
To verify the strain of A. niger used in this study, a nucleotide BLAST search (61) against NCBI was conducted for the assembled genome, and a protein BLAST search against NCBI was also conducted for the predicted proteins from the BUSCO analysis.
Protein extraction.
A. niger was grown in RPMI medium (with 2% glucose and MOPS buffer [Sigma-Aldrich] at a pH of 7.0) with 0, 2, and 8 mg · liter−1 itraconazole for 2 days at 37°C and harvested by centrifugation at 4,000 × g for 8 min. The three conditions were tested with four biological replicates, and A. niger grown without antifungal was the control. The pelleted mycelium was washed in ddH2O, centrifuged at 4,000 × g for 8 min, washed a second time in ddH2O with cOmplete protease inhibitor cocktail (Roche), mixed according to the manufacturer’s recommendations, and centrifuged at 4,000 × g for 8 min. The supernatant was discarded, and the pellet was resuspended in 385 μl triethylammonium bicarbonate (TEAB) resuspension buffer (0.05 M TEAB buffer stock, 1.0 mg · liter−1 sodium deoxycholate [NaDOC] [pH ≤ 8]) and 385 μl B-PER buffer (bacterial protein extraction reagent; Thermo Scientific). The suspension was transferred to an adaptive focused acoustic (AFA) Millitube (1.0 ml; Covaris) and treated with adaptive focused acoustic using a Covaris focused ultrasonicator (M220 M-series) in a total of four cycles (peak incident power of 75 W, a duty factor of 10%, 200 cycles per burst, and 180 s per cycle at 6°C). To collect cell debris as a pellet, the samples were subsequently centrifuged at 16,000 × g for 10 min. The supernatant was transferred to new Eppendorf tubes and stored at 4°C until further use.
Quantitative spectrometry.
For the preparation of the differential proteome, 12 A. niger isolate 351 cultures were used: 4 biological replicates with either 0, 2, or 8 mg · liter−1 itraconazole. Samples for liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis were prepared as previously described (62), with minor modifications: a NanoDrop One instrument (Thermo Scientific) was used to roughly estimate the protein concentration, measured at 280 nm, and in-solution digestion was performed on approximately 15 μg of protein from each sample. Peptides were desalted using a modified StageTip protocol (63, 64), with one modification: the concentration of R3 and R2 was doubled.
Tryptic peptides were analyzed by automated LC–electrospray ionization (ESI)–MS/MS, as previously described (65), with some modifications. A top 20 method was used to acquire the MS data, with an MS1 with an injection time of 50 ms and resolution of 60,000, an MS2 with an injection time of 45 ms, and resolution of 15,000, using an isolation window of 1.2 m/z, and the normalized collision energy was set to 28 eV.
Bioinformatic processing.
The raw data were analyzed with MaxQuant (v1.6.10.43) (66) using the search engine Andromeda (67), with carbamidomethylation set as a fixed modification and methionine oxidation as a variable modification and with a false discovery rate (FDR) of 1%, using label-free quantification (LFQ) as implemented in MaxQuant. Data were cross-referenced with three databases: an in-house-constructed proteome from Aspergillus niger isolate 351 (PRJEB41506 and Table S1) and two reference proteomes of Aspergillus niger, An76 (UP000068243) and ATCC 1015 (UP000009038), downloaded from UniProt (accessed 20 October 2020).
The generated output file was imported into Perseus (v1.6.10.0) (68). Student’s t tests were used to investigate differential protein expression and were performed on log2-transformed LFQ values using a significance level of a P value of ≤0.05 and permutation-based FDR at 5%. Fold change was expressed as the ratio of averaged LFQ values of a protein across at least 3 of 4 replications of A. niger isolate 351 challenged with itraconazole divided by the averaged LFQ value of the same protein observed in the respective control experiment.
Gene ontology enrichment analysis.
Proteins that were statistically differentially expressed were uploaded to FungiFun2 (69) and functionally annotated using standard settings.
Data availability.
The genome of A. niger has been uploaded to the European Nucleotide Archive (ENA) with the accession number PRJEB41506. The mass spectrometry proteomics data have been deposited in the ProteomeXchange Consortium via the PRIDE (70) partner repository with the data set identifiers PXD022738 and https://doi.org/10.6019/PXD022738.
ACKNOWLEDGMENTS
This work was supported by the Novo Nordisk Foundation (NNF16OC0021818).
We thank Trine Sørensen and Celine Petersen for guidance in whole-genome sequencing on MinION and processing of the raw reads.
We declare no conflict of interest.
Footnotes
Supplemental material is available online only.
aac.02549-20-s000s1.pdf (685.1KB, pdf)
aac.02549-20-s000s2.xlsx (4.7MB, xlsx)
REFERENCES
- 1.Rintala H, Pitkäranta M, Täubel M. 2012. Microbial communities associated with house dust. Adv Appl Microbiol 78:75–120. doi: 10.1016/B978-0-12-394805-2.00004-X. [DOI] [PubMed] [Google Scholar]
- 2.Li X, Zhang T, Wang S. 2019. Aerosolization of Aspergillus niger spores from colonies on different positions of a circular tube. E3S Web Conf 111:e02030. doi: 10.1051/e3sconf/201911102030. [DOI] [Google Scholar]
- 3.White JK, Nielsen JL, Madsen AM. 2020. Potential respiratory deposition and species composition of airborne culturable, viable, and non-viable fungi during occupancy in a pig farm. Atmosphere (Basel) 11:639. doi: 10.3390/atmos11060639. [DOI] [Google Scholar]
- 4.Barac A, Ong DSY, Jovancevic L, Peric A, Surda P, Spiric VT, Rubino S. 2018. Fungi-induced upper and lower respiratory tract allergic diseases: one entity. Front Microbiol 9:583. doi: 10.3389/fmicb.2018.00583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Person AK, Chudgar SM, Norton BL, Tong BC, Stout JE. 2010. Aspergillus niger: an unusual cause of invasive pulmonary aspergillosis. J Med Microbiol 59:834–838. doi: 10.1099/jmm.0.018309-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jenks J, Hoenigl M. 2018. Treatment of aspergillosis. J Fungi 4:98. doi: 10.3390/jof4030098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reischies F, Hoenigl M. 2014. The role of surgical debridement in different clinical manifestations of invasive aspergillosis. Mycoses 57:1–14. doi: 10.1111/myc.12224. [DOI] [PubMed] [Google Scholar]
- 8.Snelders E, Huis in 't Veld RAG, Rijs AJMM, Kema GHJ, Melchers WJG, Verweij PE. 2009. Possible environmental origin of resistance of Aspergillus fumigatus to medical triazoles. Appl Environ Microbiol 75:4053–4057. doi: 10.1128/AEM.00231-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Azevedo M-M, Faria-Ramos I, Cruz LC, Pina-Vaz C, Gonçalves Rodrigues A. 2015. Genesis of azole antifungal resistance from agriculture to clinical settings. J Agric Food Chem 63:7463–7468. doi: 10.1021/acs.jafc.5b02728. [DOI] [PubMed] [Google Scholar]
- 10.Cosgrove SE. 2006. The relationship between antimicrobial resistance and patient outcomes: mortality, length of hospital stay, and health care costs. Clin Infect Dis 42:S82–S89. doi: 10.1086/499406. [DOI] [PubMed] [Google Scholar]
- 11.Dannaoui E, Persat F, Monier MF, Borel E, Piens MA, Picot S. 1999. In-vitro susceptibility of Aspergillus spp. isolates to amphotericin B and itraconazole. J Antimicrob Chemother 44:553–555. doi: 10.1093/jac/44.4.553. [DOI] [PubMed] [Google Scholar]
- 12.Mello E, Posteraro B, Vella A, De Carolis E, Torelli R, D'Inzeo T, Verweij PE, Sanguinetti M. 2017. Susceptibility testing of common and uncommon Aspergillus species against posaconazole and other mold-active. Antimicrob Agents Chemother 61:e00168-17. doi: 10.1128/AAC.00168-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tokarzewski S, Ziółkowska G, Nowakiewicz A. 2012. Susceptibility testing of Aspergillus niger strains isolated from poultry to antifungal drugs—a comparative study of the disk diffusion, broth microdilution (M 38-A) and Etest methods. Pol J Vet Sci 15:125–133. doi: 10.2478/v10181-011-0123-7. [DOI] [PubMed] [Google Scholar]
- 14.Borman AM, Fraser M, Palmer MD, Szekely A, Houldsworth M, Patterson Z, Johnson EM. 2017. MIC distributions and evaluation of fungicidal activity for amphotericin B, itraconazole, voriconazole, posaconazole and caspofungin and 20 species of pathogenic filamentous fungi determined using the CLSI broth microdilution method. J Fungi 3:27. doi: 10.3390/jof3020027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arendrup MC, Cuenca-Estrella M, Lass-Flörl C, Hope WW. 2013. Breakpoints for antifungal agents: an update from EUCAST focussing on echinocandins against Candida spp. and triazoles against Aspergillus spp. Drug Resist Updat 16:81–95. doi: 10.1016/j.drup.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 16.Lu R, Tendal K, Frederiksen MW, Uhrbrand K, Li Y, Madsen AM. 2020. Strong variance in the inflammatory and cytotoxic potentials of Penicillium and Aspergillus species from cleaning workers’ exposure in nursing homes. Sci Total Environ 724:138231. doi: 10.1016/j.scitotenv.2020.138231. [DOI] [PubMed] [Google Scholar]
- 17.Madsen AM, Frederiksen MW, Jacobsen MH, Tendal K. 2020. Towards a risk evaluation of workers’ exposure to handborne and airborne microbial species as exemplified with waste collection workers. Environ Res 183:109177. doi: 10.1016/j.envres.2020.109177. [DOI] [PubMed] [Google Scholar]
- 18.Bünger J, Antlauf-Lammers M, Schulz TG, Westphal GA, Müller MM, Ruhnau P, Hallier E. 2000. Health complaints and immunological markers of exposure to bioaerosols among biowaste collectors and compost workers. Occup Environ Med 57:458–464. doi: 10.1136/oem.57.7.458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Poole CJM, Wong M. 2013. Allergic bronchopulmonary aspergillosis in garden waste (compost) collectors-occupational implications. Occup Med (Lond) 63:517–519. doi: 10.1093/occmed/kqt097. [DOI] [PubMed] [Google Scholar]
- 20.Montesinos I, Argudín MA, Hites M, Ahajjam F, Dodémont M, Dagyaran C, Bakkali M, Etienne I, Jacobs F, Knoop C, Patteet S, Lagrou K. 2017. Culture-based ethods and molecular tools for azole-resistant Aspergillus fumigatus detection in a Belgian university hospital. J Clin Microbiol 55:2391–2399. doi: 10.1128/JCM.00520-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Slaven JW, Anderson MJ, Sanglard D, Dixon GK, Bille J, Roberts IS, Denning DW. 2002. Increased expression of a novel Aspergillus fumigatus ABC transporter gene, atrF, in the presence of itraconazole in an itraconazole resistant clinical isolate. Fungal Genet Biol 36:199–206. doi: 10.1016/S1087-1845(02)00016-6. [DOI] [PubMed] [Google Scholar]
- 22.Camps SMT, Dutilh BE, Arendrup MC, Rijs AJMM, Snelders E, Huynen MA, Verweij PE, Melchers WJG. 2012. Discovery of a hapE mutation that causes azole resistance in Aspergillus fumigatus through whole genome sequencing and sexual crossing. PLoS One 7:e50034. doi: 10.1371/journal.pone.0050034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kousha M, Tadi R, Soubani AO. 2011. Pulmonary aspergillosis: a clinical review. Eur Respir Rev 20:156–174. doi: 10.1183/09059180.00001011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Piérard G, Arrese J, Piérard-Franchimont C. 2000. Itraconazole. Expert Opin Pharmacother 1:287–304. doi: 10.1517/14656566.1.2.287. [DOI] [PubMed] [Google Scholar]
- 25.Monk BC, Tomasiak TM, Keniya MV, Huschmann FU, Tyndall JDA, O'Connell JD, Cannon RD, McDonald JG, Rodriguez A, Finer-Moore JS, Stroud RM. 2014. Architecture of a single membrane spanning cytochrome P450 suggests constraints that orient the catalytic domain relative to a bilayer. Proc Natl Acad Sci U S A 111:3865–3870. doi: 10.1073/pnas.1324245111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zuckerman JM, Tunkel AR. 1994. Itraconazole: a new triazole antifungal agent. Infect Control Hosp Epidemiol 15:397–410. doi: 10.1086/646938. [DOI] [PubMed] [Google Scholar]
- 27.Shapiro RS, Robbins N, Cowen LE. 2011. Regulatory circuitry governing fungal development, drug resistance, and disease. Microbiol Mol Biol Rev 75:213–267. doi: 10.1128/MMBR.00045-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ferreira GF, Baltazar L de M, Alves Santos JR, Monteiro AS, Fraga LA, de O, Resende-Stoianoff MA, Santos DA. 2013. The role of oxidative and nitrosative bursts caused by azoles and amphotericin B against the fungal pathogen Cryptococcus gattii. J Antimicrob Chemother 68:1801–1811. doi: 10.1093/jac/dkt114. [DOI] [PubMed] [Google Scholar]
- 29.Kobayashi D, Kondo K, Uehara N, Otokozawa S, Tsuji N, Yagihashi A, Watanabe N. 2002. Endogenous reactive oxygen species is an important mediator of miconazole antifungal effect. Antimicrob Agents Chemother 46:3113–3117. doi: 10.1128/AAC.46.10.3113-3117.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shekhova E, Kniemeyer O, Brakhage AA. 2017. Induction of mitochondrial reactive oxygen species production by itraconazole, terbinafine, and amphotericin B as a mode of action against Aspergillus fumigatus. Antimicrob Agents Chemother 61:e00978-17. doi: 10.1128/AAC.00978-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bommer UA, Thiele BJ. 2004. The translationally controlled tumour protein (TCTP). Int J Biochem Cell Biol 36:379–385. doi: 10.1016/s1357-2725(03)00213-9. [DOI] [PubMed] [Google Scholar]
- 32.Amarsaikhan N, Albrecht-Eckardt D, Sasse C, Braus GH, Ogel ZB, Kniemeyer O. 2017. Proteomic profiling of the antifungal drug response of Aspergillus fumigatus to voriconazole. Int J Med Microbiol 307:398–408. doi: 10.1016/j.ijmm.2017.07.011. [DOI] [PubMed] [Google Scholar]
- 33.Gautam P, Mushahary D, Hassan W, Upadhyay SK, Madan T, Sirdeshmukh R, Sundaram CS, Sarma PU. 2016. In-depth 2-DE reference map of Aspergillus fumigatus and its proteomic profiling on exposure to itraconazole. Med Mycol 54:524–536. doi: 10.1093/mmy/myv122. [DOI] [PubMed] [Google Scholar]
- 34.Arendrup MC, Friberg N, Mares M, Kahlmeter G, Meletiadis J, Guinea J, Andersen CT, Arikan-Akdagli S, Barchiesi F, Chryssanthou E, Hamal P, Järv H, Klimko N, Kurzai O, Lagrou K, Lass-Flörl C, Matos T, Muehlethaler K, Rogers TR, Velegraki A, Arikan S,. Subcommittee on Antifungal Susceptibility Testing (AFST) of the ESCMID European Committee for Antimicrobial Susceptibility Testing (EUCAST). 2020. How to interpret MICs of antifungal compounds according to the revised clinical breakpoints v. 10.0 European Committee on Antimicrobial Susceptibility Testing (EUCAST). Clin Microbiol Infect 26:1464–1472. doi: 10.1016/j.cmi.2020.06.007. [DOI] [PubMed] [Google Scholar]
- 35.Nes WD, Zhou W, Ganapathy K, Liu JL, Vatsyayan R, Chamala S, Hernandez K, Miranda M. 2009. Sterol 24-C-methyltransferase: aenzymatic target for the disruption of ergosterol biosynthesis and homeostasis in Cryptococcus neoformans. Arch Biochem Biophys 481:210–218. doi: 10.1016/j.abb.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 36.Alcazar-Fuoli L, Mellado E. 2013. Ergosterol biosynthesis in Aspergillus fumigatus: its relevance as an antifungal target and role in antifungal drug resistance. Front Microbiol 3:439. doi: 10.3389/fmicb.2012.00439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rybak JM, Fortwendel JR, Rogers PD. 2019. Emerging threat of triazole-resistant Aspergillus fumigatus. J Antimicrob Chemother 74:835–842. doi: 10.1093/jac/dky517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shishodia SK, Tiwari S, Shankar J. 2019. Resistance mechanism and proteins in Aspergillus species against antifungal agents. Mycology 10:151–165. doi: 10.1080/21501203.2019.1574927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hagiwara D, Watanabe A, Kamei K, Goldman GH. 2016. Epidemiological and genomic landscape of azole resistance mechanisms in Aspergillus fungi. Front Microbiol 7:1382. doi: 10.3389/fmicb.2016.01382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arendrup MC, Jensen RH, Grif K, Skov M, Pressler T, Johansen HK, Lass-Flörl C. 2012. In vivo emergence of Aspergillus terreus with reduced azole susceptibility and a Cyp51A M217I alteration. J Infect Dis 206:981–985. doi: 10.1093/infdis/jis442. [DOI] [PubMed] [Google Scholar]
- 41.Ghannoum MA, Rice LB. 1999. Antifungal agents: mode of action, mechanisms of resistance, and correlation of these mechanisms with bacterial resistance. Clin Microbiol Rev 12:501–517. doi: 10.1128/CMR.12.4.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cannon RD, Lamping E, Holmes AR, Niimi K, Baret PV, Keniya MV, Tanabe K, Niimi M, Goffeau A, Monk BC. 2009. Efflux-mediated antifungal drug resistance. Clin Microbiol Rev 22:291–321. doi: 10.1128/CMR.00051-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rajendran R, Mowat E, McCulloch E, Lappin DF, Jones B, Lang S, Majithiya JB, Warn P, Williams C, Ramage G. 2011. Azole resistance of Aspergillus fumigatus biofilms is partly associated with efflux pump activity. Antimicrob Agents Chemother 55:2092–2097. doi: 10.1128/AAC.01189-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hagiwara D, Miura D, Shimizu K, Paul S, Ohba A, Gonoi T, Watanabe A, Kamei K, Shintani T, Moye-Rowley WS, Kawamoto S, Gomi K. 2017. A novel Zn2-Cys6 transcription factor AtrR plays a key role in an azole resistance mechanism of Aspergillus fumigatus by co-regulating cyp51A and cdr1B expressions. PLoS Pathog 13:e1006096. doi: 10.1371/journal.ppat.1006096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Paul S, Stamnes M, Thomas GH, Liu H, Hagiwara D, Gomi K, Filler SG, Moye-Rowley WS. 2019. AtrR is an essential determinant of azole resistance in Aspergillus fumigatus. mBio 10:e02563-18. doi: 10.1128/mBio.02563-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dhingra S, Cramer RA. 2017. Regulation of sterol biosynthesis in the human fungal pathogen Aspergillus fumigatus: opportunities for therapeutic development. Front Microbiol 8:92. doi: 10.3389/fmicb.2017.00092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Willger SD, Puttikamonkul S, Kim KH, Burritt JB, Grahl N, Metzler LJ, Barbuch R, Bard M, Lawrence CB, Cramer RA. 2008. A sterol-regulatory element binding protein is required for cell polarity, hypoxia adaptation, azole drug resistance, and virulence in Aspergillus fumigatus. PLoS Pathog 4:e1000200. doi: 10.1371/journal.ppat.1000200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nakonieczna J, Michta E, Rybicka M, Grinholc M, Gwizdek-Wiśniewska A, Bielawski KP. 2010. Superoxide dismutase is upregulated in Staphylococcus aureus following protoporphyrin-mediated photodynamic inactivation and does not directly influence the response to photodynamic treatment. BMC Microbiol 10:323. doi: 10.1186/1471-2180-10-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dwyer DJ, Kohanski MA, Collins JJ. 2009. Role of reactive oxygen species in antibiotic action and resistance. Curr Opin Microbiol 12:482–489. doi: 10.1016/j.mib.2009.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Martins D, Nguyen D, English AM. 2019. Ctt1 catalase activity potentiates antifungal azoles in the emerging opportunistic pathogen Saccharomyces cerevisiae. Sci Rep 9:9185. doi: 10.1038/s41598-019-45070-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Inoue Y, Matsuda T, Sugiyama KI, Izawa S, Kimura A. 1999. Genetic analysis of glutathione peroxidase in oxidative stress response of Saccharomyces cerevisiae. J Biol Chem 274:27002–27009. doi: 10.1074/jbc.274.38.27002. [DOI] [PubMed] [Google Scholar]
- 52.Lee YV, Wahab HA, Choong YS. 2015. Potential inhibitors for isocitrate lyase of Mycobacterium tuberculosis and non-M. tuberculosis: a summary. Biomed Res Int 2015:895453. doi: 10.1155/2015/895453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cassago A, Panepucci RA, Baião AMT, Henrique-Silva F. 2002. Cellophane based mini-prep method for DNA filamentous fungus Trichoderma reesei. BMC Microbiol 2:14. doi: 10.1186/1471-2180-2-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Reference deleted.
- 55.De Coster W, D'Hert S, Schultz DT, Cruts M, Van Broeckhoven C. 2018. NanoPack: visualizing and processing long-read sequencing data. Bioinformatics 34:2666–2669. doi: 10.1093/bioinformatics/bty149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wick R. 2019. Filtlong. GitHub. [Google Scholar]
- 57.Li H. 2016. Minimap and miniasm: fast mapping and de novo assembly for noisy long sequences. Bioinformatics 32:2103–2110. doi: 10.1093/bioinformatics/btw152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vaser R, Sović I, Nagarajan N, Šikić M. 2017. Fast and accurate de novo genome assembly from long uncorrected reads. Genome Res 27:737–746. doi: 10.1101/gr.214270.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Oxford Nanopore Technologies Ltd. 2018. medaka: sequence correction provided by ONT Research. GitHub. [Google Scholar]
- 60.Waterhouse RM, Seppey M, Simao FA, Manni M, Ioannidis P, Klioutchnikov G, Kriventseva EV, Zdobnov EM. 2018. BUSCO applications from quality assessments to gene prediction and phylogenomics. Mol Biol Evol 35:543–548. doi: 10.1093/molbev/msx319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 62.Peydaei A, Bagheri H, Gurevich L, de Jonge N, Nielsen JL. 2020. Impact of polyethylene on salivary glands proteome in Galleria melonella. Comp Biochem Physiol Part D Genomics Proteomics 34:100678. doi: 10.1016/j.cbd.2020.100678. [DOI] [PubMed] [Google Scholar]
- 63.Rappsilber J, Mann M, Ishihama Y. 2007. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat Protoc 2:1896–1906. doi: 10.1038/nprot.2007.261. [DOI] [PubMed] [Google Scholar]
- 64.Yu Y, Smith M, Pieper R. 2014. A spinnable and automatable StageTip for high throughput peptide desalting and proteomics. Protoc Exch: 2–5. [Google Scholar]
- 65.García-Moreno PJ, Gregersen S, Nedamani ER, Olsen TH, Marcatili P, Overgaard MT, Andersen ML, Hansen EB, Jacobsen C. 2020. Identification of emulsifier potato peptides by bioinformatics: application to omega-3 delivery emulsions and release from potato industry side streams. Sci Rep 10:690. doi: 10.1038/s41598-019-57229-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cox J, Mann M. 2008. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 67.Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, Mann M. 2011. Andromeda: a peptide search engine integrated into the MaxQuant environment. J Proteome Res 10:1794–1805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- 68.Tyanova S, Temu T, Cox J. 2016. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat Protoc 11:2301–2319. doi: 10.1038/nprot.2016.136. [DOI] [PubMed] [Google Scholar]
- 69.Priebe S, Kreisel C, Horn F, Guthke R, Linde J. 2015. FungiFun2: a comprehensive online resource for systematic analysis of gene lists from fungal species. Bioinformatics 31:445–446. doi: 10.1093/bioinformatics/btu627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Perez-Riverol Y, Csordas A, Bai J, Bernal-Llinares M, Hewapathirana S, Kundu DJ, Inuganti A, Griss J, Mayer G, Eisenacher M, Pérez E, Uszkoreit J, Pfeuffer J, Sachsenberg T, Yilmaz Ş, Tiwary S, Cox J, Audain E, Walzer M, Jarnuczak AF, Ternent T, Brazma A, Vizcaíno JA. 2019. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res 47:D442–D450. doi: 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The genome of A. niger has been uploaded to the European Nucleotide Archive (ENA) with the accession number PRJEB41506. The mass spectrometry proteomics data have been deposited in the ProteomeXchange Consortium via the PRIDE (70) partner repository with the data set identifiers PXD022738 and https://doi.org/10.6019/PXD022738.