ABSTRACT
A major goal of HIV vaccine design is to elicit broadly neutralizing antibodies (bNAbs). Such bNAbs target HIV’s trimeric, membrane-embedded envelope glycoprotein spikes (mEnv). Soluble Env (sEnv) trimers have been used as vaccines, but engineering sEnvs for stability, multivalency, and desired antigenicity is problematic and deletes key neutralizing epitopes on glycoprotein 41 (gp41) while creating neoepitopes that elicit unwanted antibodies. Meanwhile, multivalent mEnv vaccines are challenging to develop due to trimer instability and low mEnv copy number amid other extraneous proteins on virus-like particles. Here, we describe a multivalent mEnv vaccine platform that does not require protein engineering or extraneous proteins. mEnv trimers were fixed, purified, and combined with naked liposomes in mild detergent. On removal of detergent, mEnv spikes were observed embedded in liposome particles (mean diameter, 133 nm) in correct orientation. These particles were recognized by HIV bNAbs and not non-NAbs and are designated mEnv liposomes (MELs). Following a sequential immunization scheme in rabbits, MELs elicited antibodies that neutralized tier 2 HIV isolates. Analysis of serum antibody specificities, including those to epitopes involving a missing conserved N-glycosylation site at position 197 near the CD4 binding site on two of the immunogens, provides clues on how NAb responses can be improved with modified immunogens. In sum, MELs are a biochemically defined platform that enables rational immunization strategies to elicit HIV bNAbs using multimerized mEnv.
IMPORTANCE A vaccine that induced broadly neutralizing antibodies against HIV would likely end the AIDS pandemic. Such antibodies target membrane-embedded envelope glycoprotein spikes (mEnv) that HIV uses to enter cells. Due to HIV Env’s low expression and instability, soluble stabilized Env trimers have been used as vaccine candidates, but these have an altered base that disrupts targets of HIV broadly neutralizing antibodies that bind near the membrane and are not available for all HIV isolates. Here, we describe membrane Env liposomes (MELs) that display a multivalent array of stable mEnvs on liposome particles. MELs showed the expected antibody recognition properties, including targeting parts of mEnv missing on soluble Envs. Immunization with MELs elicited antibodies that neutralized diverse HIV isolates. The MEL platform facilitates vaccine development with potentially any HIV Env at high valency, and a similar approach may be useful for eliciting antibodies to membrane-embedded targets of therapeutic interest.
KEYWORDS: HIV, envelope, gp120, gp41, immunogen, liposome, vaccine
INTRODUCTION
HIV/AIDS afflicts more than 38 million people worldwide and is currently without a practical cure (1). Modest efficacy of a vaccine against HIV infection has been observed and was correlated with the activity of certain nonneutralizing antibodies in humans (2, 3). However, to effectively curb the pandemic, which involves a high diversity of circulating viral isolates, a vaccine most likely will need to reproducibly elicit HIV broadly neutralizing antibodies (bNAbs), something that described vaccines have failed to do (4, 5).
HIV bNAbs must recognize the envelope glycoprotein trimeric spike in its membrane-embedded state (mEnv). Many early vaccines had consisted of soluble glycoprotein (gp) subunits of Env, e.g., gp120 outer subunit, gp160 precursor, or artificially truncated gp140s, which lacked proper processing by furin and omitted the transmembrane (TM) domain and C-terminal tail (CTT) of subunit gp41 (6). These subunit vaccines elicited antibodies with limited capacity to neutralize HIV and targeted mainly variable epitopes (7–10); they also lacked well-ordered trimeric structures and were missing important bNAb epitopes (11).
More recently, soluble Env (sEnv) gp140s have been engineered with trimer stabilizing mutations, e.g., SOSIP (12, 13), UFO (14), and NFL (15), which recapitulate quaternary epitopes of HIV bNAbs (e.g., PGT145, PGT151, 35O22, and 3BC176) while simultaneously occluding several non-NAb epitopes (e.g., b6, F105, and 7B2) (16–18). Immunization with these sEnvs has elicited NAbs against relatively resistant (tier 2) primary viruses, often to variable loops or gaps in Env’s glycan shield (19–22). NAb responses have been enhanced in some cases in which sEnv immunogens were chemically cross-linked (23–25), multimerized on nanoparticles or liposomes (26–28), and/or engineered with N-linked glycosylation site mutations. A sequential prime-boost regimen using such approaches has recently elicited sporadic titers of bNAbs (25).
Immunization with sEnvs often elicits disproportionately high titers of non-NAbs to the truncated base (28, 29), while key epitopes of bNAbs 2F5, 4E10, and 10E8 against the membrane-proximal external region (MPER) are disrupted or missing (30, 31). Differences in conformational dynamics between sEnv and native mEnv have also been noted (32). Virus-like particles (VLPs) display mEnv but may elicit off-target, autoreactive, or distracting antibody responses due to Env debris or extraneous proteins from both the virus and the host cell (23, 33–35). Genetic vaccines such as mRNA vaccines have gained recent attention as a promising platform (36) but do not allow control over the heterogeneity, quality, and multimerization of the translated protein and do not permit cross-linking for added stability, which are features that have been linked to favorable antibody responses to HIV Env (37). Hence, an mEnv vaccine alternative that addresses these concerns is desirable to diversify approaches and enable new hypotheses.
Here, we developed a vaccine platform that displays multivalent, well-ordered mEnv spikes on liposomes, termed MELs. mEnv was purified readily from cell lines (38) and assembled into liposomes in the correct orientation with MPER exposed and CTT buried. Notably, MELs were devoid of extraneous proteins and displayed stable, cross-linked mEnv trimers in a multivalent array. In an immunization experiment, MELs elicited antibodies that neutralized select tier 2 isolates of HIV. Hence, MELs should be a useful platform for rational vaccine design involving HIV mEnv and other membrane proteins for which membrane display influences important epitopes.
RESULTS
Creation and characterization of fixed mEnv spikes.
We previously described an HIV Env nanoparticle immunogen in which mEnv was captured on antibody-coated nanobeads (23). In that approach, the capture antibody was an undesirable vaccine component, and mEnv yields were low from the transient transfection of cells. Here, we took advantage of a recently described stable cell line strategy that increased mEnv yield by more than 10-fold (38). The mEnv of this cell line is a variant of the clade B isolate ADA, termed Comb-mut (ADA.CM), which was selected previously for high trimer stability (39). We also generated two new mEnv cell lines, JRFL.TD15 and CH505.N197D, to produce mutant Envs of a clade B isolate and a clade C transmitted/founder (T/F) isolate, respectively, as will be described further below.
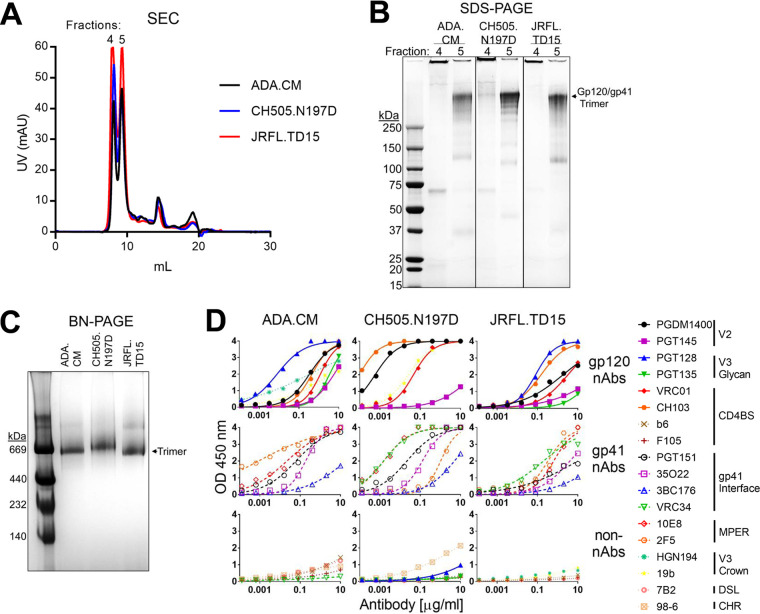
mEnv-expressing cells were fixed in glutaraldehyde (GA), a clinically approved chemical cross-linker shown previously to have helped in eliciting HIV NAbs (24, 40). While cross-linking can diminish binding to Env by certain NAbs, for example, when a NAb binds to GA reactive lysines or a conformation other than that which is fixed, cross-linking can also stabilize or enhance specific quaternary bNAb epitopes (23). Fixed cells were solubilized in the detergent n-dodecyl-β-d-maltoside (DDM), and mEnv was affinity purified using the trimer-specific antibody PGT151. Size exclusion chromatography of purified mEnv revealed two major peaks, the first being aggregated mEnv while the second was trimeric mEnv that was used for subsequent biophysical studies (Fig. 1A and B). Purified mEnvs ADA.CM, CH505.N197D, and JRFL.TD15 were verified to be trimeric using blue native (BN)-PAGE and denaturing SDS-PAGE (Fig. 1B and C). The trimers were recognized by several bNAbs to gp120 and gp41, including those to quaternary epitopes, and showed minimal binding by non-NAbs (Fig. 1D). These data show the mEnvs are stable and trimeric and have native-like antigenicity. Of note, mEnvs were produced in yields sufficient for immunizations, i.e., ∼0.5 mg of pure mEnv per liter of culture medium.
FIG 1.
Purified mEnvs are trimeric and retain native antigenicity. (A) mEnv, which was GA cross-linked and PGT151 affinity purified, was analyzed using size exclusion chromatography (SEC). (B) Two mEnv-containing fractions from SEC were analyzed by denaturing Coomassie SDS-PAGE. (C) MEnv (5 μg; from fraction 5 in panel B) was analyzed using BN-PAGE stained with Coomassie blue. (D) Purified mEnv was captured using lectin from Galanthus nivalis (GNL) and probed in an ELISA using a panel of antibodies. Antibodies are classified as NAbs if they neutralized cognate virus with an IC50 of <50 μg/ml. NAbs against all isolates include PGDM1400, PGT145, PGT128, PGT135, VRC01, CH103, PGT151, 35O22, 3BC176, and 10E8. Non-NAbs against all isolates include b6, F105, 7B2, and 98-6. VRC34 neutralizes CH505.N197D and JRFL.TD15 but not ADA.CM. PGT128, 2F5, and PGT135 neutralize ADA.CM and JRFL.TD15 but not CH505.N197D. HGN194 neutralizes ADA.CM but not CH505.N197D or JRFL.TD15. 19b neutralizes ADA.CM and CH505.N197D but not JRFL.TD15.
Incorporation of mEnv into liposomes.
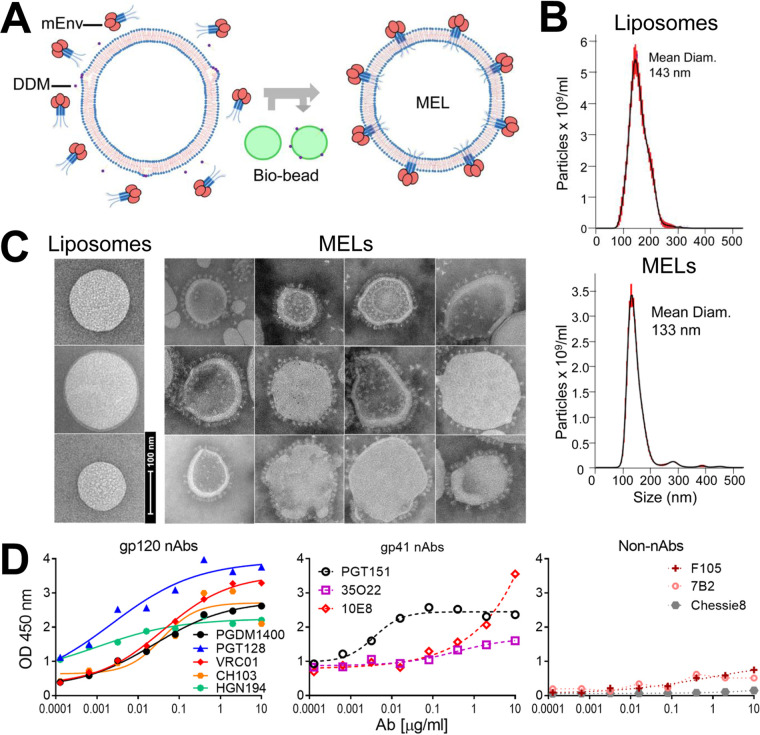
To see if mEnv could incorporate into liposomes, we adopted an approach described for making proteoliposomes (Fig. 2A) (41, 42). Thus, naked liposomes (70% 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine [POPC], 30% cholesterol), formed by extrusion through a membrane with 100-nm pores, were treated with DDM at a concentration predetermined to saturate, but not fully solubilize, the lipid bilayer. Purified mEnv was added, and then the detergent was removed by adsorption using polystyrene Bio-beads (43). Mean particle size of both naked liposomes and the mEnv-liposome mixture was ∼140 nm, as determined by nanoparticle tracking analysis (NTA; Fig. 2B). Notably, negative-stain electron microscopy (EM) revealed prominent, outward-protruding spikes on the liposomes, which we termed membrane-Env liposomes, or MELs (Fig. 2C).
FIG 2.
Production, properties, and antigenicity of mEnv liposomes (MELs). (A, left) Naked liposomes were treated with the mild detergent DDM, which destabilizes the membrane bilayer, and then combined with purified mEnv trimers. (Right) Detergent was depleted from the liposome-mEnv mixture by repeated incubations with Bio-beads, which yields MELs decorated with an array of spikes. Data shown in this and subsequent panels are from JRFL.TD15 MELs, but similar results were observed with ADA.CM and CH505.N197D. (B) Nanoparticle tracking analysis (NTA) reveals liposomes are monodispersed with a mean diameter of 143 nm; MELs containing mEnv remain monodispersed with a mean diameter of 133 nm. (C) Negative-stain EM shows MELs are embedded with mEnv trimeric spikes. (D) ELISA data show that MELs are bound by bNAbs but not by non-NAbs and the anti-CTT antibody Chessie8. MELs were labeled with a 2% molar ratio of biotin-DOPE and captured on streptavidin-coated microwells.
We probed the antigenicity of the MELs using a panel of antibodies in an enzyme-linked immunosorbent assay (ELISA). As expected, the MELs were bound efficiently by most bNAbs, while binding by non-NAbs was minimal (Fig. 2D). We also probed MELs using the anti-CTT antibody, Chessie8 (44), and found that it did not bind the MELs but did bind to detergent-solubilized mEnvs, which is in agreement with the EM images showing that MELs were decorated with embedded spikes pointing outwards. We conclude that mEnv incorporated into the bilayer of MELs in the correct orientation with the ectodomain and MPER accessible to bNAbs and the CTT buried within the liposome.
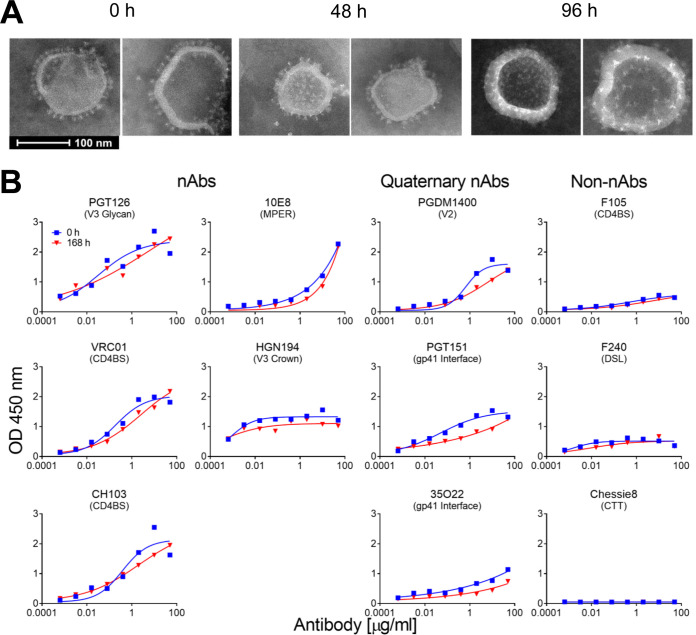
The stability of MELs was analyzed over time at physiological temperature using negative-stain EM and a capture ELISA. MELs appeared to retain the embedded spikes and overall appearance for at least 96 h, as visualized by negative-stain EM (Fig. 3A). Additionally, after 7 days at 37°C, MELs were recognized efficiently by NAbs to the CD4 binding site (CD4BS), N332 glycan supersite, V3, and MPER (Fig. 3B), while binding by three quaternary bNAbs, PGDM1400, PGT151, and 35O22, modestly decreased. Importantly, binding by non-NAbs or anti-CTT antibody to MELs did not increase after the incubation. We surmised trimeric mEnv and MELs were stable enough to elicit relevant humoral responses in animals.
FIG 3.
Stability of MELs in solution. (A) Negative-stain EM images of MELs following 0, 48, and 96 h of incubation periods at 37°C. (B) Antibody binding properties of MELs before and after incubation for 168 h at 37°C, as analyzed by streptavidin-capture ELISA as described for Fig. 2D.
MELs elicit HIV neutralizing antibodies.
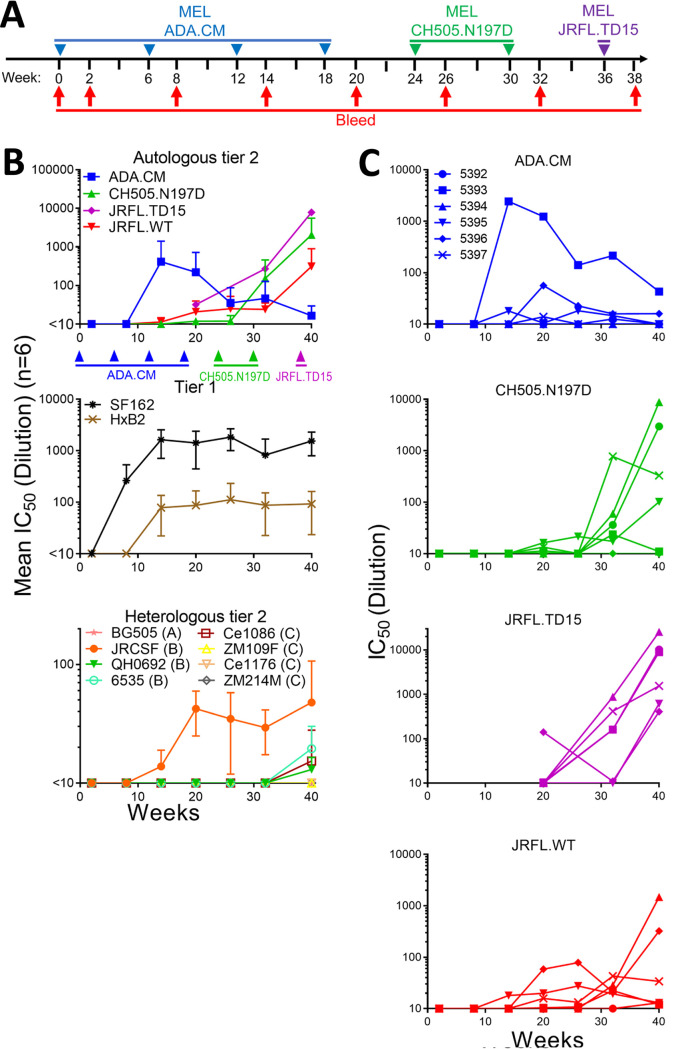
Next, we explored the immunogenicity of MELs. We chose to immunize with MELs in alum plus CpG using New Zealand White (NZW) rabbits, as we had used these adjuvants for a prior rabbit study in which sporadic tier 2 autologous NAb responses were elicited (23). We note that although CpG ODN 1826 reportedly blocked binding of V2 bNAbs to SOSIP gp140 (45), we saw no effect of CpG ODN 2007 on the reactivity of any bNAbs to mEnvs in an ELISA (data not shown). Rabbits were inoculated with ADA.CM MELs every 6 weeks for four immunizations (Fig. 4A).
FIG 4.
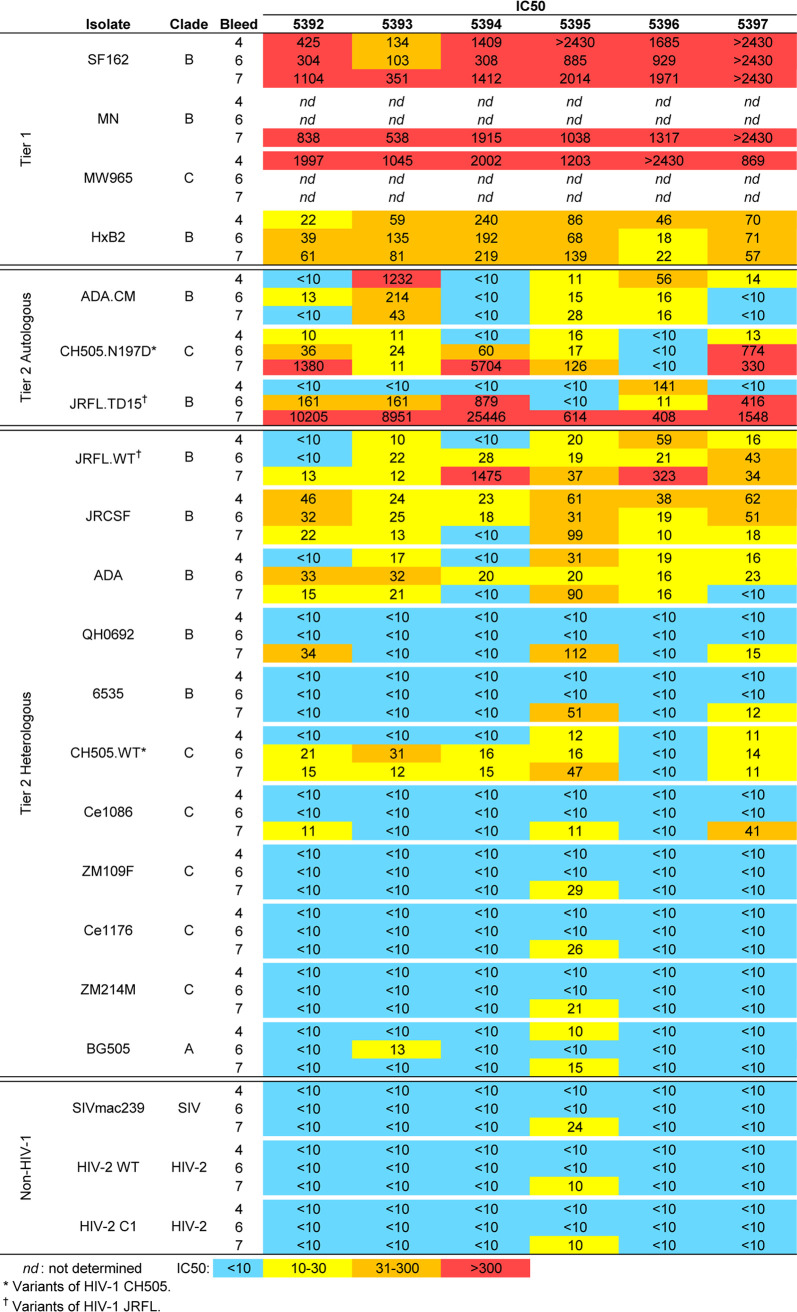
Immunization with MELs in rabbits elicited HIV NAbs. (A) Schedule of immunization of NZW rabbits with MELs. Rabbits were immunized with MELs seven times, i.e., ADA.CM (4 times), CH505.N197D (2 times), and JRFL.TD15 (1 time), 6 weeks apart. (B) Kinetics of neutralization of autologous and heterologous tier 1 and tier 2 isolates by MEL immune sera. Mean IC50s of six rabbit sera from each bleed against the three immunizing or autologous isolates and JRFL.WT (top), heterologous tier 1 isolates (middle), and a panel of heterologous tier 2 isolates (bottom). The lowest serum dilution tested was 1:10, so any serum that did not reach an IC50 at a 1:10 dilution has been defined as <10. (C) Kinetics of neutralization of immunizing virus strains by individual rabbit sera.
Sera from ADA.CM immunized rabbits were tested for HIV neutralizing activity in a standardized assay using TZM-bl target cells. Tier 1a strains SF162 and MW965 were neutralized with mean 50% inhibitory concentrations (IC50s) of >1:1,000, while neutralization of tier 1b strain HxB2 was modest but consistent with a mean IC50 of about 1:100 (Fig. 4B and 5). Sera from four of six rabbits showed autologous neutralization against ADA.CM, with rabbit serum 5393 reaching an IC50 of >1:1,200 (Fig. 4B and C). Heterologous primary isolates were also neutralized by the sera, including the clade B isolates JRCSF (mean IC50 of 1:42 for six responders) and JRFL (mean IC50 of 1:31 for four responders), while a clade C mutant virus CH505.N197D was also neutralized (mean IC50 of 1:14 for four responders).
Encouraged by the MEL ADA.CM immunogenicity results, we chose to boost the animals using a phylogenetically distant Env, anticipating that such a strategy might broaden serum neutralization if sera could already weakly neutralize the boosting mEnv. For the first boost, we chose CH505.N197D, since 4 out of 6 rabbit sera showed detectable neutralization of this mEnv and a stable cell line of it had already been prepared. CH0505 is a transmitted/founder (TF) virus from a donor that developed bNAbs to the CD4BS (46), and CH0505.N197D is an N-glycosylation site mutant that showed increased sensitivity to CD4BS bNAbs without increased binding by nonneutralizing CD4BS antibodies b6 and F105 (Fig. 1D; see also Fig. 10B). Following two boosts with CH505.N197D MELs, NAb titers increased against CH505.N197D virus (Fig. 5). Weak neutralization also increased against CH505.WT and was observed in 5 of 6 animals (average IC50, 1:20), whereas previously it was only detectable in two animals (average IC50, 1:12). Notably, rabbit 5397 neutralized CH505.N197D with an IC50 of ∼1:1,000. Meanwhile, although serum neutralization of ADA.CM decreased, neutralization against ADA and JRCSF was unchanged, and NAb titers against the heterologous isolate JRFL became more consistent (Fig. 4B and C and 5).
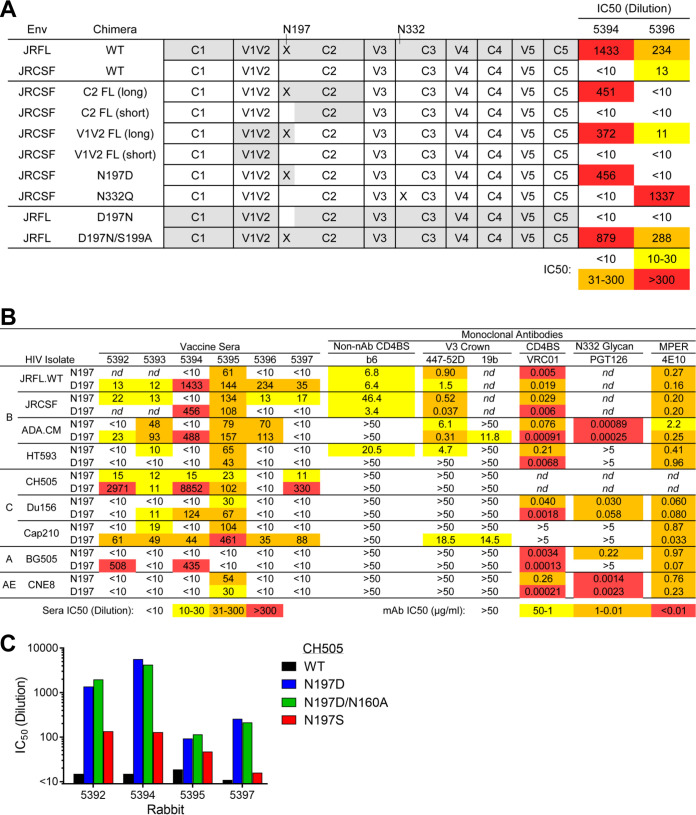
FIG 10.
MELs elicited potent NAbs to a glycan hole at position 197. (A) Neutralization by sera 5394 and 5396 against a panel of domain-swapped HIV Env chimeras between JRFL and the more resistant JRCSF as well as against N-glycosylation site mutants N197 and N332 of JRCSF. (B) N197D mutants of diverse HIV isolates were tested for neutralization by sera from the final bleed postsequential MEL immunization (left) as well as by a panel of bNAbs and non-NAbs (right). JRFL and JRCSF and their N197 mutants were tested for neutralization by V3 antibody F425-B4e8 and MPER antibody 2F5 rather than 447-52D and 4E10, respectively. (C) Rabbit sera from the final bleed were tested for neutralization against CH505.WT, CH505.N197D, CH505.N197D.N160A (knocks out V2 bNAb neutralization), and CH505.N197S.
FIG 5.
HIV neutralization breadth and potency of MEL immunized rabbit sera. Sera taken from rabbits were tested in neutralization assays against a cross-clade panel of autologous and heterologous HIV isolates. Data shown are reciprocal serum dilutions at the IC50 from bleeds 4 (after ADA.CM), 6 (after CH505.N197D), and 7 (after JRFL.TD15) for each isolate.
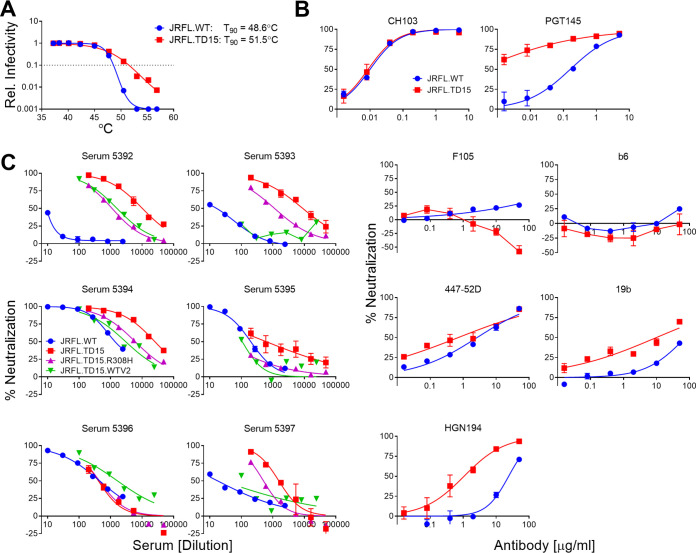
Because boosting with CH505.N197D improved heterologous neutralization, we chose to boost the animals once more. We had generated an mEnv cell line previously, JRFL.TD15, which incorporates TD15 mutations shown to stabilize soluble JRFL gp140 trimers (47). The functional stability of JRFL.TD15 mEnv, i.e., the temperature at which an Env pseudovirus loses 90% infectivity in an hour (T90) (48), was indeed found to be 3°C higher than that of the JRFL wild type (WT) (Fig. 6A). Of note, JRFL.TD15 showed increased sensitivity to V2 and V3 antibodies compared to JRFL.WT (Fig. 6B). JRFL.TD15 virus was also more sensitive than JRFL.WT to 4 of 6 rabbit sera following boosting with CH505.N197D MELs (Fig. 5). We note that the JRFL.TD15 cell line expresses Env with a truncation in the CTT as a result of a spontaneous mutation in the incorporated env transgene, similar to that which occurred in the ADA.CM cell line (38). The boost with JRFL.TD15 MELs strongly increased NAb titers against JRFL.TD15 virus in all animals, up to >1:25,000 (Fig. 5 and 6C). Neutralization titers against JRFL.WT were lower but had increased in 4 of 6 animals, with 50-fold and 15-fold increases in animals 5394 and 5396, respectively. Neutralization also increased against CH505.N197D with several sera, up to 1:5,700 (Fig. 5). Heterologous neutralization, albeit sporadic and weak, increased against JRCSF, and, notably, could now be observed against the clade B isolates QH0692 and 6535 and the clade C isolate Ce1086.
FIG 6.
Stability and neutralization sensitivity of HIV JRFL.TD15. (A) Stability of function of JRFL.TD15 and JRFL.WT mEnvs was studied by determining the relative infectivity of cognate pseudovirions incubated for an hour at different temperatures; the temperature at which infectivity is reduced by 90% (T90) is indicated to the left. (B) Neutralization of JRFL.TD15 by narrow neutralizing antibodies against V3 (447-52D, 19b, and HGN194) and CD4BS (F105 and b6), as well as by bNAbs to CD4BS (CH103) and to V2 (PGT145). (C) MEL-immunized rabbit sera were assayed for neutralization against JRFL.WT, JRFL.TD15, and mutants JRFL.TD15.R308H and JRFL.TD15.WTV2, which bear JRFL.WT V3 and V2 domains, respectively.
Antibody binding specificities in MEL immune sera.
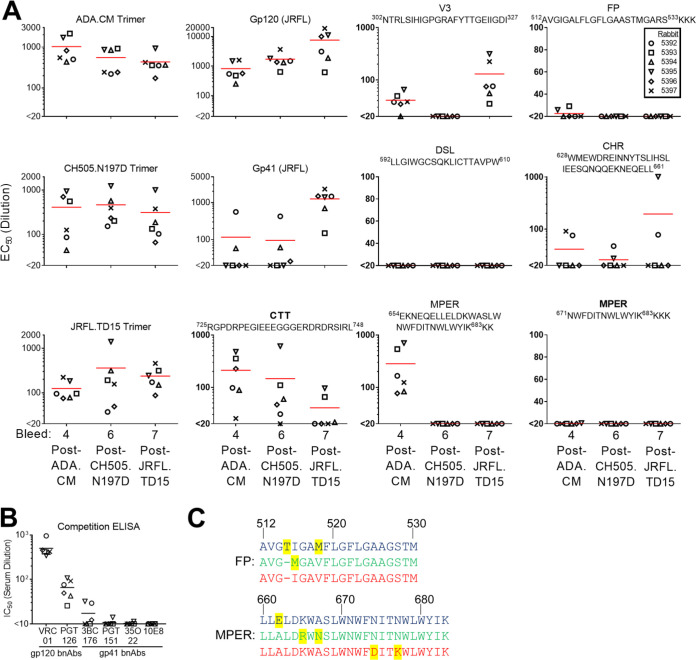
MEL ADA.CM immune sera were probed in an ELISA and found to contain high antibody binding titers against trimeric ADA.CM and monomeric gp120, with lower titers to recombinant gp41 (Fig. 7A). Serum antibodies also bound to the full MPER peptide (amino acids [aa] 654 to 683) containing the 2F5 and 4E10/10E8 epitopes but not to the C-terminal MPER peptide (aa 670 to 683), suggesting that elicited MPER antibodies targeted mostly N-terminal epitopes in the MPER. Serum antibodies bound to V3 peptides of gp120 and the immunodominant Kennedy epitope region in the gp41 CTT. The latter result indicates the CTT was exposed to B cells, perhaps from disrupted MELs. Antibody titers to the FP were detectable but low, and no binding was observed to the gp41 disulfide loop that is occluded in mEnv trimer structures (13, 16, 38).
FIG 7.
Antibody binding specificities in MEL antisera revealed by ELISA. (A) Sera from bleeds 4 (post-ADA.CM), 6 (post-CH505.N197D), and 7 (post-JRFL.TD15) were tested for binding to each of the three immunizing mEnvs as well as recombinant gp120 (JRCSF) and gp41 (JRFL) and various antigens and peptides of HIV Env. (B) Sera from the final bleed were tested for the ability to block biotinylated bNAb binding to JRFL.TD15 trimer. The biotinylated bNAbs include those against CD4BS (VRC01), N332 glycan supersite (PGT126), gp120-gp41 interface (3BC176, PGT151, and 35O22), and the MPER (10E8). (C) Alignment of the FP (aa 512 to 530) and MPER (aa 660 to 683) of mEnvs used in the sequential immunization. ADA.CM sequence is in blue, CH505.N197D sequence is in green, and JRFL.TD15 sequence is in red. Residues that differ in one isolate relative to the other two are highlighted in yellow.
The immune sera following boosting with CH505.N197D and JRFL.TD15 MELs showed diminished titers in ELISAs against gp41 epitopes MPER, FP, and CTT, while titers to monomeric gp120 rose about 10-fold (Fig. 7A). Of note, some residues of the N-terminal MPER, FP, and CTT are mismatched between ADA.CM, CH505.N197D, and JRFL.TD15 (Fig. 7C). Titers against V3 peptide decreased to undetectable levels from boosting with CH505.N197D, perhaps due to limited exposure of the V3 crown (Fig. 1D) and sequence differences in V3 between CH505.N197D and ADA.CM. V3 antibody titers rebounded following the boost with JRFL.TD15 that is modestly sensitive to V3 crown antibodies (Fig. 7A).
Immune sera from the final bleed were also tested in a competition ELISA for the ability to block bNAbs from binding to JRFL.TD15 mEnv. All six sera robustly blocked binding by VRC01 and PGT126 to the CD4BS and N332 glycan supersite, respectively (Fig. 7B). Two sera, which were shown to recognize a CHR peptide (Fig. 7A), weakly blocked binding of bNAb 3BC176 to the gp120-gp41 interface (Fig. 7B). Altogether, serum specificities to Env seem to be diverse and often overlap the CD4BS or N332 supersite, less often with the gp41 epitopes.
Neutralization specificities in MEL immune sera.
Next, we determined the HIV NAb specificities elicited by MEL immunization. We found the most potent neutralizing serum against ADA.CM, 5393, did not neutralize the parental isolate, ADA. Of the seven mutations that differentiate ADA.CM from ADA, we found the V1 alteration (N139/I140 deletion, N142S) was sufficient to make ADA as sensitive to 5393 as ADA.CM and, hence, is the likely target of NAbs in this serum (Fig. 8A).
FIG 8.
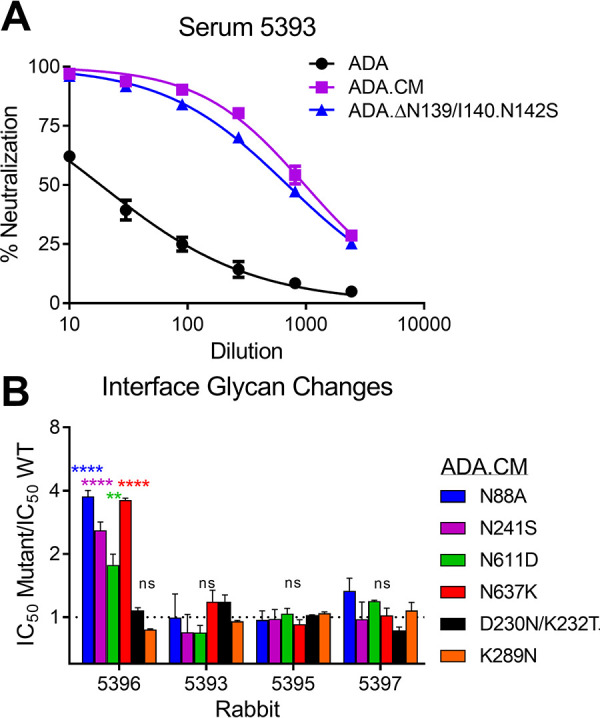
ADA.CM neutralization activities in sera 5393 and 5396 target V1 and the gp120-gp41 interface. (A) Serum 5393 from bleed 3, following immunization with ADA.CM, was tested for neutralization of ADA.CM, parental ADA, and an ADA mutant containing an N139/I140 deletion and N142S substitution in V1 of gp120. (B) Four rabbit sera from bleed 4 that neutralized ADA.CM with an IC50 of >10 were tested in neutralization assays against ADA.CM with glycan knockout mutations (N88A, N241S, N611D, and N637K) or glycan hole-filling mutations (D230N/K232T and K289N) near the gp120-gp41 interface.
Removal of consensus N-linked glycosylation sites on Env (i.e., glycan holes) naturally exposes the underlying protein surface and has been associated with eliciting NAbs against autologous HIV (19, 21, 49, 50). ADA.CM lacks two relatively conserved N-linked glycosylation sites at N289 and N230, so we asked whether the sera could neutralize ADA.CM mutants with N-glycosylation sites restored at these sites (K289N and D230N/K232T) as well as with other N-glycosylation site knockouts near this gp120-gp41 interface (N88A, N241S, N611D, and N637K). The N-glycosylation site mutations did not affect neutralization by most sera, but the four knockout mutations did significantly increase neutralization by serum 5396, suggesting that some NAbs in this serum target the gp120-gp41 interface (Fig. 8B).
We used V3 crown and MPER linear peptides as “dump-in” reagents to see if they could block HIV serum neutralization. As anticipated, V3 peptide blocked neutralization by anti-V3 antibody F425-B4e8 and blocked most serum neutralization of the tier 1a SF162 strain (Fig. 9A and B). Neutralization of heterologous tier 2 isolate Ce1086 was also abrogated by addition of V3 peptide, as was neutralization of JRCSF, but only in two of three animals. V3 peptide competition did not affect the potent neutralization of JRFL.WT and CH505.N197D and only decreased neutralization of JRFL.TD15 and heterologous tier 2 isolate QH0692 about 2-fold. An MPER peptide did not block neutralization with any serum or isolate tested (data not shown).
FIG 9.
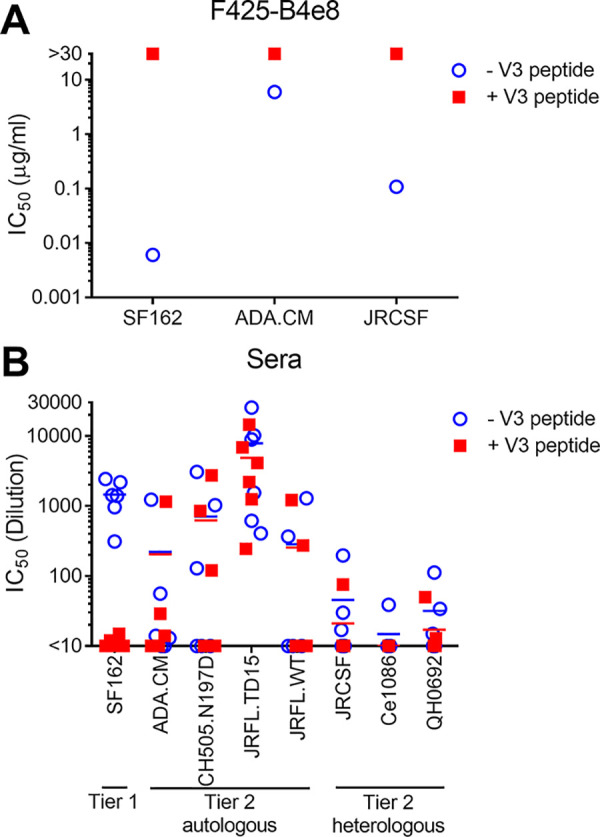
Serum neutralization of autologous isolates and some heterologous tier 2 isolates cannot be blocked by V3 peptide. Neutralization by monoclonal antibody F425-B4e8 that binds to the V3 crown (A) and sera from the final bleed (B) was tested in a neutralization assay against HIV isolates in the presence or absence of V3 peptide (JRFL sequence, NTRLSIHIGPGRAFYTTGEIIGDI).
To further assess the neutralization discrepancy between JRFL.TD15 and JRFL.WT, we generated R308H and WT-V2 mutants of JRFL.TD15 that revert V3 and V2, respectively, to that of JRFL.WT. On average, R308H decreased the IC50 of the sera 8-fold, ranging from no change (serum 5396) to a 24-fold decrease (serum 5395), so it accounted for some, but not all, of the difference in sensitivity between JRFL.TD15 and JRFL.WT (Fig. 6C). JRFL.TD15.WTV2, on the other hand, decreased the neutralization of JRFL.TD15 back to JRFL.WT levels for 4 of the 6 sera, suggesting that most JRFL.TD15 NAbs that do not neutralize JRFL.WT target V2.
V2 NAbs reportedly have been elicited by soluble CH505 immunogens (51). Hence, we generated an N160A mutant of CH505.N197D that knocks out neutralization by V2 bNAbs (52). Removing the N160 glycosylation site did not affect serum neutralization of CH505.N197D, suggesting the elicited NAbs do not target this conserved region of V2 (Fig. 10C).
Envs CH505.N197D and JRFL.TD15 both lack N-linked glycosylation sites at position 197 near the CD4BS, so we anticipated some NAbs to these viruses target the common glycan hole. We made use of the fact that sera from two animals, 5394 and 5396, neutralized JRFL but not the tier 2 isolate JRCSF that has the N197 glycosylation site. Therefore, domain-substituted chimeras of JRFL and JRCSF were tested for neutralization by these sera. Serum 5394 neutralized JRCSF.N197D and JRCSF chimeras engrafted with JRFL V2 or C2 domains that also introduced D197, while 5396 did not (Fig. 10A). However, an N-linked glycosylation site knock-in mutation, D197N, in JRFL abrogated neutralization by both sera. Of note, serum 5396 potently neutralized JRCSF N332Q, but serum 5394 did not. Overall, each serum has distinct NAb specificities against the 197 glycan hole on JRFL.
To understand antibody specificities to the N197 glycan hole in more detail, we made N197D mutants of isolates ADA.CM, HT593, Du156, CAP210, BG505, and CNE8, in addition to mutants of CH505, JRCSF, and JRFL, described above. Sera 5396 and 5397 only neutralized JRFL and CH505.N197D, respectively (Fig. 10B). However, serum 5394 could neutralize Du156, BG505, ADA.CM, Cap210, CH505, JRCSF, and JRFL isolates with D197, for a total of 7 out of 10 N197 glycan-deleted isolates. Serum 5392 neutralized BG505.N197D (IC50, 1:500) as well as CH505.N197D, revealing modest breadth against N197D mutants.
Serum neutralization was also compared using CH505.N197D and CH505.N197S. The N197S mutant of CH505 was less sensitive than N197D to all tested sera but was more sensitive than CH505.WT (Fig. 10C). These data suggest some NAbs elicited to the CH505 N197 glycan hole depend in part on the Asp side chain at position 197.
The V5 loop of gp120 plays a role in the epitopes of many CD4BS bNAbs (53–55). Therefore, we generated V5 mutants of JRFL and CH505 to test whether serum NAbs targeting the N197D glycan hole have features in common with described CD4BS bNAbs. Replacing V5 of JRFL with that of JRCSF had little effect on neutralization by sera 5394 and 5396 (Table 1). Likewise, a DT insertion in V5 of CH505.N197D, an insertion found in evolutionary variants of CH505 that reduces sensitivity to CD4BS bNAb CH103 (46), had a limited effect on most serum neutralization. However, the DT insertion did decrease neutralization of CH505.N197D by sera 5392 and 5397 by 2.4-fold and 5-fold, respectively, suggesting some NAbs elicited against the N197 glycan hole also interact with V5.
TABLE 1.
Effect of V5 substitutions on serum neutralization
| Serum | IC50 (dilution) |
Fold decrease (WT/mutant) | |||
|---|---|---|---|---|---|
| CH505.N197D | CH505.N197D.V5-DTa | JRFL | JRFL.V5-CSFb | ||
| 5392 | 1,424 | 600 | 2.4 | ||
| 5394 | 4,002 | 2,467 | 1.6 | ||
| 5395 | 46 | 52 | 0.9 | ||
| 5397 | 138 | 27 | 5.1 | ||
| 5394 | 803 | 542 | 1.5 | ||
| 5396 | 180 | 385 | 0.5 | ||
Contains a DT insertion in the V5 domain of gp120.
JRFL V5 domain was replaced with the V5 domain of JRCSF.
DISCUSSION
Membrane Env is the sole target of HIV NAbs and, hence, is a reasonable platform for a vaccine, but its development has lagged behind that of sEnv. Here, we describe MELs, which contain purified mEnv spikes embedded and arrayed on the membrane of liposomes. Whereas VLP vaccines have been described previously, these typically have a low copy number of mEnv (34). High Env VLPs (hVLPs) have been developed more recently that display more than 100 spikes (38), but, like most VLPs, also carry Env debris, membrane proteins from the host cell, and viral proteins like capsid and matrix protein, which may divert the immune response. While we did not compare MELs to cognate sEnv and hVLPs in this immunization study, studies to do so are underway. With MELs, any Env can, in principle, be embedded, and GA can be used to stabilize mEnvs in a native trimeric state. In contrast, the majority of Envs fail to form well-ordered sEnv trimers, including reasonably stable mEnvs, such as ADA.CM and CH0505 (39, 51), yet the latter were used here in MELs to elicit tier 2 NAbs.
Virosomes (56), nanodiscs (57), bicelles (58), and capture nanoparticles (23, 28, 59) are possible alternatives to MELs for displaying mEnv but either lack multivalent display or contain extraneous proteins that may divert the immune response. With MELs, T helper and B cell epitopes are exclusively from mEnv and may activate B cells efficiently via B cell receptor cross-linking (60). Size and composition of nanoparticles can affect trafficking in lymphatics, as can antigen presentation and processing by dendritic cells (61); hence, future studies are warranted to determine whether changing MEL properties can be used to improve immune responses. Mixed MELs may also be prepared using many different Envs to potentially favor activation of naive B cells against conserved epitopes (62).
MELs were shown to elicit autologous NAbs in most animals but heterologous neutralization was limited, and it is also unclear to what extent cross-neutralization is mediated by a single NAb or multiple NAbs with narrow specificity. However, the sequential Env regimen used was likely suboptimal. A prior study that sporadically elicited NAbs with greater breadth involved priming with Envs containing a glycan hole at the CD4BS and then boosted with Envs that had N-glycosylation sites restored (25). In contrast, we hyperimmunized using an mEnv without such a glycan hole likely too many times before boosting with heterologous mEnvs that were also highly unrelated. Indeed, the animal that developed the most potent NAbs against ADA.CM, 5393, showed weaker cross-neutralizing responses with heterologous Env boosting, suggesting that overpriming subverted the development of breadth. The second mEnv, CH505.N197D, contains a CD4BS-proximal glycan hole at position 197 and is sensitive to the bNAb CH103 UCA (D. P. Leaman and M. B. Zwick, unpublished data). Neutralization titers to this mEnv rose rapidly on boosting and did so again on boosting with JRFL.TD15, which also lacks the N197 glycosylation site. Thus, it would be of interest to immunize again in rabbits and in human Ig knock-in mice (63), using CH505.N197D and JRFL.TD15 in the prime and then boosting with glycan-restored Envs to see if NAbs develop against more conserved elements of the CD4BS more effectively.
With respect to the MPER and FP, antibody binding titers in the MEL immune sera were low relative to the glycan-deficient CD4BS and variable loops that have been shown to be highly immunogenic (21, 64, 65). However, elicitation of significant NAb responses to the FP has reportedly required priming with multivalent epitope-focused FP immunogens, and similar approaches using MPER focused immunogens have shown some, albeit more limited, enhancement in on-target antibody responses (66). Priming with FP- or MPER-specific peptides or scaffold proteins that engage bNAb UCAs, followed by boosts with natively presented FP and MPER on MELs, may be warranted. Significant sequence differences in the MPER and FP exist between the Envs we used as well, which reportedly affect antibody recognition, sometimes profoundly (67, 68). In the case of the FP, the priming ADA.CM mEnv contains a very rare insertion of a Thr residue, and, indeed, weak initial binding titers to FP were found to disappear on boosting with CH505.N197D that lacks the Thr insertion. Antibody responses might improve using Envs that have similar sequences in these regions, as might higher accessibility and/or affinity for bNAb UCAs (31). Stabilization of mEnv without chemical cross-linking and the use of specific lipids might further enhance the immunogenicity of MELs. Finally, an MEL approach may be useful for eliciting antibodies against difficult membrane proteins more generally on pathogens or cancerous or diseased cells, especially when membrane display influences proximal key epitopes. Future work will focus on developing MEL vaccination in animals to reproducibly elicit NAbs against the CD4BS by the N197 glycan as well as to the gp41 interface and MPER at the base of mEnv (69, 70).
MATERIALS AND METHODS
Reagents. (i) Cells.
TZM-bl cells were obtained from the NIH ARRRP. HEK 293T cells were purchased from the ATCC. High Env cell lines are discussed in more detail below.
(ii) Antibodies.
Anti-HIV monoclonal antibodies b12, HGN194, 7B2, PGT128, PGT126, PGDM1400, VRC01, CH103, CH103 UCA, PGT151, 35O22, 10E8, F105, and 3BC176 were produced in-house as described previously (71). 2G12 and 2F5 were purchased from Polymun. Mouse antibody Chessie 8 was a gift from George Lewis (University of Maryland).
(iii) Proteins and peptides.
Monomeric JRFL gp120 was purchased from Progenics, and JRFL gp41 MBP-fusion protein M41xt (aa 535 to 681) was produced in Escherichia coli in-house (72). The following peptides were synthesized by GenScript: JRV3 (302NTRLSIHIGPGRAFYTTGEIIGDI327), C34 (CH3O-628WMEWDREINNYTSLIHSLIEESQNQQEKNEQELL663-NH2), FP-bio (512AVGIGALFLGFLGAAGSTMGARS534-biotin), PID-bio (592LLGIWGCSQKLICTTAVPW610-biotin), MPER peptides PDT-081 (654EKNEQELLELDKWASLWNWFDITNWLWYIKKK683) and 94-1 (671NWFDITNWLWYIKKKK683), and CTT peptides CTT-2 (725RGPDRPEGIEEEGK737) and CTT-3 (738GERDRDRSIRL748).
mEnv cell lines.
The cell line ADA.CM (V4) was described previously (38). The cell line CH505.N197D was prepared similarly using the CH505 Env sequence (46), in which mutation N197D was introduced by lentiviral transduction followed by iterative rounds using fluorescence-activated cell sorting to select for a PGT145high b6low phenotype. Likewise, the stable cell line expressing JRFL.TD15 mEnv was prepared using the sEnv sequence, described elsewhere (47), which was connected to the remaining MPER-TM-CTT sequence of wild-type JRFL. Of note, sequencing of DNA prepared from this cell line revealed a frameshift in the CTT at nucleotide position 2134, resulting in the following altered CTT sequence and early truncation: 710QGYSPCPSRPCCPPPAAPTAPRASRRRAASATATAPAAW*749. The cell lines were prepared fresh from freezer stocks, were passaged fewer than 20 times in Dulbecco’s modified Eagle medium (DMEM) containing 10% fetal bovine serum (FBS), 2 mM l-glutamine, 100 U/ml penicillin, 100 μg/ml streptomycin, and 2.5 μg/ml puromycin, and were kept from becoming overgrown by splitting every 3 to 4 days.
Virus production.
HIV was produced as a pseudotyped virus by transient transfection of HEK293T cells using Env plasmid DNA, pSG3ΔEnv backbone plasmid, and 25-kDa polyethyleneimine as the transfection reagent, as previously described (48).
Production and purification of mEnv.
Cell lines overexpressing mEnv were grown in shaker flasks at 37°C in an 8% CO2 atmosphere to a density of 4 × 106 cells/ml and then pelleted by centrifugation at 300 × g for 5 min. The cell pellet was resuspended in phosphate-buffered saline (PBS) and treated with 15 mM glutaraldehyde (GA; Sigma) at room temperature for 10 min. Unreacted GA was quenched for 15 min in 50 mM Tris-HCl, after which treated cells were solubilized for 20 min in 0.5% DDM (Sigma). Cell debris was removed by centrifugation at 3,000 × g for 15 min at 4°C, and then the supernatant was clarified by spinning at 80,000 × g in an Optima ultracentrifuge (Beckman) for 1 h at 4°C. mEnv was affinity purified using the trimer-specific bNAb PGT151 as previously described (13, 73). Briefly, PGT151-coupled protein A Sepharose beads (GE Healthcare) were added to the supernatant and rotated overnight at 4°C. The PGT151 beads were washed with TN75 (20 mM Tris, pH 8, 500 mM NaCl, 0.1% DDM, 0.003% sodium deoxycholate), and bound mEnv was eluted in 3 M MgCl2. The trimer fraction was purified by size exclusion chromatography on a Superdex 200 10/300 GL column in TBS plus 0.1% DDM plus 0.003% sodium deoxycholate using an ÄKTA Pure 25L high-performance liquid chromatography instrument (GE Healthcare) at a flow rate of 0.75 ml/min.
MEL production.
Naked liposomes were prepared by dissolving POPC and cholesterol (Avanti) in chloroform at a 70:30 molar ratio. For liposomes to be analyzed by ELISA, 18:1 Biotinyl Cap PE (Avanti) was added to the lipids at a 2% molar ratio. Lipids were dried in a vacuum overnight, hydrated in PBS (20 mg/ml total lipid) with constant shaking for 2 h at 37°C, and then sonicated for 30 s. The resulting liposomes were extruded 14 times through 1-μm, 0.8-μm, 0.4-μm, 0.2-μm, and 0.1-μm filters using a mini-extrusion device (Avanti) at room temperature (RT) and then diluted to 4 mg/ml in PBS. DDM was added to a final concentration of 0.1% (1.9 mM, detergent to lipid molar ratio of 0.3:1) to destabilize the lipid bilayer (74). Purified mEnv trimers were added at a 1:10 trimer/lipid ratio (wt/wt; with final concentrations of 1.9 mM DDM and 7.2 μM sodium deoxycholate), and the mixture was incubated at RT for 30 min. To remove detergent, polystyrene Bio-beads SM2 (Bio-Rad), prewashed once with methanol and five times with water, were added at 40 mg/ml, and the sample was rotated at RT for 30 min. Liposomes were treated thrice more with fresh Bio-beads at 4°C for 1 h, overnight, and 2 h, respectively. MELs in the supernatant were drawn off the polystyrene beads using a pipette and stored at 4°C.
BN-PAGE.
BN-PAGE was performed using the NativePAGE gel system (ThermoFisher) as previously described (75), except that samples were run on 3 to 12% gels. Coomassie staining was performed using Simply Blue safe stain (ThermoFisher) according to the manufacturer’s instructions.
SDS-PAGE.
Purified mEnv trimers (5 μg) were incubated in Laemmli buffer (Bio-Rad) containing 50 mM dithiothreitol (DTT) for 5 min at 100°C before loading on 8 to 16% Tris-glycine gels (ThermoFisher). Gels were electrophoresed at RT in running buffer (25 mM Tris, 192 mM glycine, 0.1% SDS, pH 8.3) at 150 V for 1 h and then Coomassie stained as described above.
ELISAs. (i) Capture ELISA.
Galanthus nivalis lectin (GNL) capture ELISAs were performed as described previously (23). Briefly, microtiter wells were coated with GNL (5 μg/μl) in PBS overnight at 4°C, and then purified mEnv trimers (2 μg/μl) in PBS plus 0.1% DDM were captured for 2 h at 37°C. Plates were blocked with 4% nonfat dry milk (NFDM) in PBS for 1 h at 37°C. Primary and horseradish peroxidase (HRP)-conjugated secondary antibody incubations were performed for 1 h and 45 min, respectively, in PBS plus 0.05% Tween 20 plus 0.4% NFDM at 37°C.
(ii) Direct ELISA.
Antigens (5 μg/μl) in PBS were coated onto microtiter wells overnight at 4°C, and ELISAs were performed as described above without the antigen capture step.
(iii) MEL ELISA.
MEL ELISAs were performed as described above, except microtiter wells were coated with streptavidin (5 μg/μl) and then blocked with 4% nonfat dry milk. MELs that incorporated biotinylated DOPE were captured on wells at 37°C for 2 h. Subsequent steps were done as described above but without detergent.
(iv) Competition ELISA.
A capture ELISA was performed as described above, except serial dilutions of sera were added to wells at twice the final concentration. After 5 min, biotinylated bNAb was added at a concentration previously determined to produce 50% maximum signal, and the mixture was incubated for 1 h at 37°C. bNAb binding was detected using streptavidin-HRP (Jackson).
NTA.
A NanoSight N300 instrument (Malvern) was used to determine the dispersity and size of liposomes and MELs by following a previously described method (38).
Negative-stain EM.
Liposomes were applied for 2 min onto glow-discharged, carbon-coated 400-Cu mesh grids (Electron Microscopy Sciences). Excess sample was removed by gentle contact with tissue paper, and the grids were placed on a droplet of 2% phosphotungstic acid (PTA) solution (pH 6.9) for 2 min. Excess stain was removed and grids were examined on a Philips CM100 electron microscope (FEI) at 80 kV. Images were acquired using a Megaview III charge-coupled device (CCD) camera (Olympus Soft Imaging Solutions).
Rabbit immunization.
Rabbit studies were approved and carried out in accordance with protocols provided to the Institutional Animal Care and Use Committee (IACUC) at Scripps Research (La Jolla, CA) under approval number 07-0026-5. Six New Zealand White (NZW) rabbits were immunized with a MEL formulation containing a total of 250 μg mEnv added to 100 μg CpG ODN 2007 (Invivogen) and alum (Pierce) adjuvant, with a MEL/alum ratio of 1:2 by volume. Injections were made bilaterally with 0.6 ml per injection via the subcutaneous route within 1 h of the formulation. MELs were used within 3 days of preparation. Blood was drawn from the marginal ear vein 4 and 10 days postinjection for the preparation of peripheral blood mononuclear cells and serum, respectively.
Neutralization assays.
Pseudotyped HIV-1 neutralization assays were performed using TZM-bl target cells, as previously described (23). Briefly, TZM-bl cells were seeded onto a 96-well plate in 100 μl of growth media and incubated overnight at 37°C before the addition of virus. Virus was coincubated with antibody or serum (previously heat inactivated at 56°C for 30 min) at 37°C for 1 h and then added to TZM-bl cells. Infectivity was determined 72 h later by adding Bright-Glo (Promega) and measuring luciferase activity using a Synergy H1 plate reader (BioTek).
ACKNOWLEDGMENTS
We thank Daniel Sands, Mark Ochoa, and Trevor Biddle for technical support. We thank Jeong Hyun Lee, Jidnyasa Ingale, Shridhar Bale, and John Elder for helpful discussions on mEnv purification and MEL production. The Scripps Research Microscopy Core performed electron microscopy, and Scripps Research Animal Resources carried out the animal procedures.
Funding support was from National Institutes of Health (NIAID) grants AI114401 and AI098602 (M.B.Z.) and P01 AI104722 (R.T. Wyatt), and the James B. Pendleton Charitable Trust (M.B.Z.) enabled the purchase of an AKTA Pure instrument.
Contributor Information
Michael B. Zwick, Email: zwick@scripps.edu.
Viviana Simon, Icahn School of Medicine at Mount Sinai.
REFERENCES
- 1.UNAIDS. 2020. UNAIDS data 2020. https://www.unaids.org/sites/default/files/media_asset/2020_aids-data-book_en.pdf. Accessed 5 Nov 2020.
- 2.Tomaras GD, Plotkin SA. 2017. Complex immune correlates of protection in HIV-1 vaccine efficacy trials. Immunol Rev 275:245–261. 10.1111/imr.12514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rerks-Ngarm S, Pitisuttithum P, Nitayaphan S, Kaewkungwal J, Chiu J, Paris R, Premsri N, Namwat C, de Souza M, Adams E, Benenson M, Gurunathan S, Tartaglia J, McNeil JG, Francis DP, Stablein D, Birx DL, Chunsuttiwat S, Khamboonruang C, Thongcharoen P, Robb ML, Michael NL, Kunasol P, Kim JH. 2009. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N Engl J Med 361:2209–2220. 10.1056/NEJMoa0908492. [DOI] [PubMed] [Google Scholar]
- 4.Haynes BF, Burton DR, Mascola JR. 2019. Multiple roles for HIV broadly neutralizing antibodies. Sci Transl Med 11:eaaz2686. 10.1126/scitranslmed.aaz2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bjorkman PJ. 2020. Can we use structural knowledge to design a protective vaccine against HIV-1? HLA 95:95–103. 10.1111/tan.13759. [DOI] [PubMed] [Google Scholar]
- 6.Spearman P. 2006. Current progress in the development of HIV vaccines. Curr Pharm Des 12:1147–1167. 10.2174/138161206776055859. [DOI] [PubMed] [Google Scholar]
- 7.Gilbert P, Wang M, Wrin T, Petropoulos C, Gurwith M, Sinangil F, D’Souza P, Rodriguez‐Chavez IR, DeCamp A, Giganti M, Berman PW, Self SG, Montefiori DC. 2010. Magnitude and breadth of a nonprotective neutralizing antibody response in an efficacy trial of a candidate HIV-1 gp120 vaccine. J Infect Dis 202:595–605. 10.1086/654816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Spearman P, Lally MA, Elizaga M, Montefiori D, Tomaras GD, McElrath MJ, Hural J, De Rosa SC, Sato A, Huang Y, Frey SE, Sato P, Donnelly J, Barnett S, Corey LJ. 2011. A trimeric, V2-deleted HIV-1 envelope glycoprotein vaccine elicits potent neutralizing antibodies but limited breadth of neutralization in human volunteers. J Infect Dis 203:1165–1173. 10.1093/infdis/jiq175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang X, Wyatt R, Sodroski J. 2001. Improved elicitation of neutralizing antibodies against primary human immunodeficiency viruses by soluble stabilized envelope glycoprotein trimers. J Virol 75:1165–1171. 10.1128/JVI.75.3.1165-1171.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mascola JR, Snyder SW, Weislow OS, Belay SM, Belshe RB, Schwartz DH, Clements ML, Dolin R, Graham BS, Gorse GJ, Keefer MC, McElrath MJ, Walker MC, Wagner KF, McNeil JG, McCutchan FE, Burke DS, National Institute of Allergy and Infectious Diseases . 1996. Immunization with envelope subunit vaccine products elicits neutralizing antibodies against laboratory-adapted but not primary isolates of human immunodeficiency virus type 1. J Infect Dis 173:340–348. 10.1093/infdis/173.2.340. [DOI] [PubMed] [Google Scholar]
- 11.Ringe RP, Sanders RW, Yasmeen A, Kim HJ, Lee JH, Cupo A, Korzun J, Derking R, van Montfort T, Julien JP, Wilson IA, Klasse PJ, Ward AB, Moore JP. 2013. Cleavage strongly influences whether soluble HIV-1 envelope glycoprotein trimers adopt a native-like conformation. Proc Natl Acad Sci U S A 110:18256–18261. 10.1073/pnas.1314351110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sanders RW, Vesanen M, Schuelke N, Master A, Schiffner L, Kalyanaraman R, Paluch M, Berkhout B, Maddon PJ, Olson WC, Lu M, Moore JP. 2002. Stabilization of the soluble, cleaved, trimeric form of the envelope glycoprotein complex of human immunodeficiency virus type 1. J Virol 76:8875–8889. 10.1128/jvi.76.17.8875-8889.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sanders RW, Derking R, Cupo A, Julien JP, Yasmeen A, de Val N, Kim HJ, Blattner C, de la Pena AT, Korzun J, Golabek M, de Los Reyes K, Ketas TJ, van Gils MJ, King CR, Wilson IA, Ward AB, Klasse PJ, Moore JP. 2013. A next-generation cleaved, soluble HIV-1 Env trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS Pathog 9:e1003618. 10.1371/journal.ppat.1003618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kong L, He L, de Val N, Vora N, Morris CD, Azadnia P, Sok D, Zhou B, Burton DR, Ward AB, Wilson IA, Zhu J. 2016. Uncleaved prefusion-optimized gp140 trimers derived from analysis of HIV-1 envelope metastability. Nat Commun 7:12040. 10.1038/ncomms12040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sharma SK, de Val N, Bale S, Guenaga J, Tran K, Feng Y, Dubrovskaya V, Ward AB, Wyatt RT. 2015. Cleavage-independent HIV-1 Env trimers engineered as soluble native spike mimetics for vaccine design. Cell Rep 11:539–550. 10.1016/j.celrep.2015.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Julien JP, Cupo A, Sok D, Stanfield RL, Lyumkis D, Deller MC, Klasse PJ, Burton DR, Sanders RW, Moore JP, Ward AB, Wilson IA. 2013. Crystal structure of a soluble cleaved HIV-1 envelope trimer. Science 342:1477–1483. 10.1126/science.1245625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee JH, Ozorowski G, Ward AB. 2016. Cryo-EM structure of a native, fully glycosylated, cleaved HIV-1 envelope trimer. Science 351:1043–1048. 10.1126/science.aad2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stadtmueller BM, Bridges MD, Dam KM, Lerch MT, Huey-Tubman KE, Hubbell WL, Bjorkman PJ. 2018. DEER spectroscopy measurements reveal multiple conformations of HIV-1 SOSIP envelopes that show similarities with envelopes on native virions. Immunity 49:235–246. 10.1016/j.immuni.2018.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McCoy LE, van Gils MJ, Ozorowski G, Messmer T, Briney B, Voss JE, Kulp DW, Macauley MS, Sok D, Pauthner M, Menis S, Cottrell CA, Torres JL, Hsueh J, Schief WR, Wilson IA, Ward AB, Sanders RW, Burton DR. 2016. Holes in the glycan shield of the native HIV envelope are a target of trimer-elicited neutralizing antibodies. Cell Rep 16:2327–2338. 10.1016/j.celrep.2016.07.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sanders RW, van Gils MJ, Derking R, Sok D, Ketas TJ, Burger JA, Ozorowski G, Cupo A, Simonich C, Goo L, Arendt H, Kim HJ, Lee JH, Pugach P, Williams M, Debnath G, Moldt B, van Breemen MJ, Isik G, Medina-Ramirez M, Back JW, Koff WC, Julien JP, Rakasz EG, Seaman MS, Guttman M, Lee KK, Klasse PJ, LaBranche C, Schief WR, Wilson IA, Overbaugh J, Burton DR, Ward AB, Montefiori DC, Dean H, Moore JP. 2015. HIV-1 neutralizing antibodies induced by native-like envelope trimers. Science 349:aac4223. 10.1126/science.aac4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Crooks ET, Tong T, Chakrabarti B, Narayan K, Georgiev IS, Menis S, Huang X, Kulp D, Osawa K, Muranaka J, Stewart-Jones G, Destefano J, O'Dell S, LaBranche C, Robinson JE, Montefiori DC, McKee K, Du SX, Doria-Rose N, Kwong PD, Mascola JR, Zhu P, Schief WR, Wyatt RT, Whalen RG, Binley JM. 2015. Vaccine-elicited tier 2 HIV-1 neutralizing antibodies bind to quaternary epitopes involving glycan-deficient patches proximal to the CD4 binding site. PLoS Pathog 11:e1004932. 10.1371/journal.ppat.1004932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hessell AJ, Malherbe DC, Pissani F, McBurney S, Krebs SJ, Gomes M, Pandey S, Sutton WF, Burwitz BJ, Gray M, Robins H, Park BS, Sacha JB, LaBranche CC, Fuller DH, Montefiori DC, Stamatatos L, Sather DN, Haigwood NL. 2016. Achieving potent autologous neutralizing antibody responses against tier 2 HIV-1 viruses by strategic selection of envelope immunogens. J Immunol 196:3064–3078. 10.4049/jimmunol.1500527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leaman DP, Lee JH, Ward AB, Zwick MB. 2015. Immunogenic display of purified chemically cross-linked HIV-1 spikes. J Virol 89:6725–6745. 10.1128/JVI.03738-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schiffner T, Pallesen J, Russell RA, Dodd J, de Val N, LaBranche CC, Montefiori D, Tomaras GD, Shen X, Harris SL, Moghaddam AE, Kalyuzhniy O, Sanders RW, McCoy LE, Moore JP, Ward AB, Sattentau QJ. 2018. Structural and immunologic correlates of chemically stabilized HIV-1 envelope glycoproteins. PLoS Pathog 14:e1006986. 10.1371/journal.ppat.1006986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dubrovskaya V, Tran K, Ozorowski G, Guenaga J, Wilson R, Bale S, Cottrell CA, Turner HL, Seabright G, O'Dell S, Torres JL, Yang L, Feng Y, Leaman DP, Vázquez Bernat N, Liban T, Louder M, McKee K, Bailer RT, Movsesyan A, Doria-Rose NA, Pancera M, Karlsson Hedestam GB, Zwick MB, Crispin M, Mascola JR, Ward AB, Wyatt RT. 2019. Vaccination with glycan-modified HIV NFL envelope trimer-liposomes elicits broadly neutralizing antibodies to multiple sites of vulnerability. Immunity 51:915–929. 10.1016/j.immuni.2019.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McGuire AT, Gray MD, Dosenovic P, Gitlin AD, Freund NT, Petersen J, Correnti C, Johnsen W, Kegel R, Stuart AB, Glenn J, Seaman MS, Schief WR, Strong RK, Nussenzweig MC, Stamatatos L. 2016. Specifically modified Env immunogens activate B-cell precursors of broadly neutralizing HIV-1 antibodies in transgenic mice. Nat Commun 7:10618. 10.1038/ncomms10618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morris CD, Azadnia P, de Val N, Vora N, Honda A, Giang E, Saye-Francisco K, Cheng Y, Lin X, Mann CJ, Tang J, Sok D, Burton DR, Law M, Ward AB, He L, Zhu J. 2017. Differential antibody responses to conserved HIV-1 neutralizing epitopes in the context of multivalent scaffolds and native-like gp140 trimers. mBio 8:e00036-17. 10.1128/mBio.00036-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bale S, Goebrecht G, Stano A, Wilson R, Ota T, Tran K, Ingale J, Zwick MB, Wyatt RT. 2017. Covalent linkage of HIV-1 trimers to synthetic liposomes elicits improved B cell and antibody responses. J Virol 91:e00443-17. 10.1128/JVI.00443-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu JK, Crampton JC, Cupo A, Ketas T, van Gils MJ, Sliepen K, de Taeye SW, Sok D, Ozorowski G, Deresa I, Stanfield R, Ward AB, Burton DR, Klasse PJ, Sanders RW, Moore JP, Crotty S. 2015. Murine antibody responses to cleaved soluble HIV-1 envelope trimers are highly restricted in specificity. J Virol 89:10383–10398. 10.1128/JVI.01653-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krebs SJ, Kwon YD, Schramm CA, Law WH, Donofrio G, Zhou KH, Gift S, Dussupt V, Georgiev IS, Schatzle S, McDaniel JR, Lai YT, Sastry M, Zhang B, Jarosinski MC, Ransier A, Chenine AL, Asokan M, Bailer RT, Bose M, Cagigi A, Cale EM, Chuang GY, Darko S, Driscoll JI, Druz A, Gorman J, Laboune F, Louder MK, McKee K, Mendez L, Moody MA, O'Sullivan AM, Owen C, Peng D, Rawi R, Sanders-Buell E, Shen CH, Shiakolas AR, Stephens T, Tsybovsky Y, Tucker C, Verardi R, Wang K, Zhou J, Zhou T, Georgiou G, Alam SM, Haynes BF, Rolland M. 2019. Longitudinal analysis reveals early development of three MPER-directed neutralizing antibody lineages from an HIV-1-infected individual. Immunity 50:677–691. 10.1016/j.immuni.2019.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang L, Irimia A, He L, Landais E, Rantalainen K, Leaman DP, Vollbrecht T, Stano A, Sands DI, Kim AS, Poignard P, Burton DR, Murrell B, Ward AB, Zhu J, Wilson IA, Zwick MB. 2019. An MPER antibody neutralizes HIV-1 using germline features shared among donors. Nat Commun 10:5389. 10.1038/s41467-019-12973-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu M, Ma X, Castillo-Menendez LR, Gorman J, Alsahafi N, Ermel U, Terry DS, Chambers M, Peng D, Zhang B, Zhou T, Reichard N, Wang K, Grover JR, Carman BP, Gardner MR, Nikic-Spiegel I, Sugawara A, Arthos J, Lemke EA, Smith AB, III, Farzan M, Abrams C, Munro JB, McDermott AB, Finzi A, Kwong PD, Blanchard SC, Sodroski JG, Mothes W. 2019. Associating HIV-1 envelope glycoprotein structures with states on the virus observed by smFRET. Nature 568:415–419. 10.1038/s41586-019-1101-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Crooks ET, Moore PL, Franti M, Cayanan CS, Zhu P, Jiang P, de Vries RP, Wiley C, Zharkikh I, Schulke N, Roux KH, Montefiori DC, Burton DR, Binley JM. 2007. A comparative immunogenicity study of HIV-1 virus-like particles bearing various forms of envelope proteins, particles bearing no envelope and soluble monomeric gp120. Virology 366:245–262. 10.1016/j.virol.2007.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cantin R, Methot S, Tremblay MJ. 2005. Plunder and stowaways: incorporation of cellular proteins by enveloped viruses. J Virol 79:6577–6587. 10.1128/JVI.79.11.6577-6587.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Poon B, Hsu JF, Gudeman V, Chen IS, Grovit-Ferbas K. 2005. Formaldehyde-treated, heat-inactivated virions with increased human immunodeficiency virus type 1 env can be used to induce high-titer neutralizing antibody responses. J Virol 79:10210–10217. 10.1128/JVI.79.16.10210-10217.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pardi N, Hogan MJ, Porter FW, Weissman D. 2018. mRNA vaccines–a new era in vaccinology. Nat Rev Drug Discov 17:261–279. 10.1038/nrd.2017.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Medina-Ramírez M, Sanders RW, Sattentau QJ. 2017. Stabilized HIV-1 envelope glycoprotein trimers for vaccine use. Curr Opin HIV AIDS 12:241–249. 10.1097/COH.0000000000000363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stano A, Leaman DP, Kim AS, Zhang L, Autin L, Ingale J, Gift SK, Truong J, Wyatt RT, Olson AJ, Zwick MB. 2017. Dense array of spikes on HIV-1 virion particles. J Virol 91:415–417. 10.1128/JVI.00415-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leaman DP, Zwick MB. 2013. Increased functional stability and homogeneity of viral envelope spikes through directed evolution. PLoS Pathog 9:e1003184. 10.1371/journal.ppat.1003184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Soldemo M, Adori M, Stark JM, Feng Y, Tran K, Wilson R, Yang L, Guenaga J, Wyatt RT, Karlsson Hedestam GB. 2017. Glutaraldehyde cross-linking of HIV-1 Env trimers skews the antibody subclass response in mice. Front Immunol 8:1654. 10.3389/fimmu.2017.01654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Seddon AM, Curnow P, Booth PJ. 2004. Membrane proteins, lipids and detergents: not just a soap opera. Biochim Biophys Acta 1666:105–117. 10.1016/j.bbamem.2004.04.011. [DOI] [PubMed] [Google Scholar]
- 42.Geertsma ER, Nik Mahmood NA, Schuurman-Wolters GK, Poolman B. 2008. Membrane reconstitution of ABC transporters and assays of translocator function. Nat Protoc 3:256–266. 10.1038/nprot.2007.519. [DOI] [PubMed] [Google Scholar]
- 43.Rigaud JL, Mosser G, Lacapere JJ, Olofsson A, Levy D, Ranck JL. 1997. Bio-Beads: an efficient strategy for two-dimensional crystallization of membrane proteins. J Struct Biol 118:226–235. 10.1006/jsbi.1997.3848. [DOI] [PubMed] [Google Scholar]
- 44.Steckbeck JD, Sun C, Sturgeon TJ, Montelaro RC. 2013. Detailed topology mapping reveals substantial exposure of the “cytoplasmic” C-terminal tail (CTT) sequences in HIV-1 Env proteins at the cell surface. PLoS One 8:e65220. 10.1371/journal.pone.0065220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ozorowski G, Cupo A, Golabek M, LoPiccolo M, Ketas TA, Cavallary M, Cottrell CA, Klasse PJ, Ward AB, Moore JP. 2018. Effects of adjuvants on HIV-1 envelope glycoprotein SOSIP trimers in vitro. J Virol 92:e00381-18. 10.1128/JVI.00381-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liao HX, Lynch R, Zhou T, Gao F, Alam SM, Boyd SD, Fire AZ, Roskin KM, Schramm CA, Zhang Z, Zhu J, Shapiro L, Mullikin JC, Gnanakaran S, Hraber P, Wiehe K, Kelsoe G, Yang G, Xia SM, Montefiori DC, Parks R, Lloyd KE, Scearce RM, Soderberg KA, Cohen M, Kamanga G, Louder MK, Tran LM, Chen Y, Cai F, Chen S, Moquin S, Du X, Joyce MG, Srivatsan S, Zhang B, Zheng A, Shaw GM, Hahn BH, Kepler TB, Korber BT, Kwong PD, Mascola JR, Haynes BF. 2013. Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus. Nature 496:469–476. 10.1038/nature12053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guenaga J, Dubrovskaya V, de Val N, Sharma SK, Carrette B, Ward AB, Wyatt RT. 2015. Structure-guided redesign increases the propensity of HIV Env to generate highly stable soluble trimers. J Virol 90:2806–2817. 10.1128/JVI.02652-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Agrawal N, Leaman DP, Rowcliffe E, Kinkead H, Nohria R, Akagi J, Bauer K, Du SX, Whalen RG, Burton DR, Zwick MB. 2011. Functional stability of unliganded envelope glycoprotein spikes among isolates of human immunodeficiency virus type 1 (HIV-1). PLoS One 6:e21339. 10.1371/journal.pone.0021339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou T, Doria-Rose NA, Cheng C, Stewart-Jones GBE, Chuang GY, Chambers M, Druz A, Geng H, McKee K, Kwon YD, O'Dell S, Sastry M, Schmidt SD, Xu K, Chen L, Chen RE, Louder MK, Pancera M, Wanninger TG, Zhang B, Zheng A, Farney SK, Foulds KE, Georgiev IS, Joyce MG, Lemmin T, Narpala S, Rawi R, Soto C, Todd JP, Shen CH, Tsybovsky Y, Yang Y, Zhao P, Haynes BF, Stamatatos L, Tiemeyer M, Wells L, Scorpio DG, Shapiro L, McDermott AB, Mascola JR, Kwong PD. 2017. Quantification of the impact of the HIV-1-glycan shield on antibody elicitation. Cell Rep 19:719–732. 10.1016/j.celrep.2017.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Voss JE, Andrabi R, McCoy LE, de Val N, Fuller RP, Messmer T, Su CY, Sok D, Khan SN, Garces F, Pritchard LK, Wyatt RT, Ward AB, Crispin M, Wilson IA, Burton DR. 2017. Elicitation of neutralizing antibodies targeting the V2 apex of the HIV envelope trimer in a wild-type animal model. Cell Rep 21:222–235. 10.1016/j.celrep.2017.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Saunders KO, Verkoczy LK, Jiang C, Zhang J, Parks R, Chen H, Housman M, Bouton-Verville H, Shen X, Trama AM, Scearce R, Sutherland L, Santra S, Newman A, Eaton A, Xu K, Georgiev IS, Joyce MG, Tomaras GD, Bonsignori M, Reed SG, Salazar A, Mascola JR, Moody MA, Cain DW, Centlivre M, Zurawski S, Zurawski G, Erickson HP, Kwong PD, Alam SM, Levy Y, Montefiori DC, Haynes BF. 2017. Vaccine induction of heterologous tier 2 HIV-1 neutralizing antibodies in animal models. Cell Rep 21:3681–3690. 10.1016/j.celrep.2017.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Walker LM, Phogat SK, Chan-Hui PY, Wagner D, Phung P, Goss JL, Wrin T, Simek MD, Fling S, Mitcham JL, Lehrman JK, Priddy FH, Olsen OA, Frey SM, Hammond PW, Kaminsky S, Zamb T, Moyle M, Koff WC, Poignard P, Burton DR. 2009. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science 326:285–289. 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fera D, Schmidt AG, Haynes BF, Gao F, Liao HX, Kepler TB, Harrison SC. 2014. Affinity maturation in an HIV broadly neutralizing B-cell lineage through reorientation of variable domains. Proc Natl Acad Sci U S A 111:10275–10280. 10.1073/pnas.1409954111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schommers P, Gruell H, Abernathy ME, Tran MK, Dingens AS, Gristick HB, Barnes CO, Schoofs T, Schlotz M, Vanshylla K, Kreer C, Weiland D, Holtick U, Scheid C, Valter MM, van Gils MJ, Sanders RW, Vehreschild JJ, Cornely OA, Lehmann C, Fätkenheuer G, Seaman MS, Bloom JD, Bjorkman PJ, Klein F. 2020. Restriction of HIV-1 escape by a highly broad and potent neutralizing antibody. Cell 180:471–489. 10.1016/j.cell.2020.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Umotoy J, Bagaya BS, Joyce C, Schiffner T, Menis S, Saye-Francisco KL, Biddle T, Mohan S, Vollbrecht T, Kalyuzhniy O, Madzorera S, Kitchin D, Lambson B, Nonyane M, Kilembe W, Poignard P, Schief WR, Burton DR, Murrell B, Moore PL, Briney B, Sok D, Landais E. 2019. Rapid and focused maturation of a VRC01-class HIV broadly neutralizing antibody lineage involves both binding and accommodation of the N276-glycan. Immunity 51:141–154. 10.1016/j.immuni.2019.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Moser C, Amacker M, Kammer AR, Rasi S, Westerfeld N, Zurbriggen R. 2007. Influenza virosomes as a combined vaccine carrier and adjuvant system for prophylactic and therapeutic immunizations. Expert Rev Vaccines 6:711–721. 10.1586/14760584.6.5.711. [DOI] [PubMed] [Google Scholar]
- 57.Witt KC, Castillo-Menendez L, Ding H, Espy N, Zhang S, Kappes JC, Sodroski J. 2017. Antigenic characterization of the human immunodeficiency virus (HIV-1) envelope glycoprotein precursor incorporated into nanodiscs. PLoS One 12:e0170672. 10.1371/journal.pone.0170672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rantalainen K, Berndsen ZT, Antanasijevic A, Schiffner T, Zhang X, Lee WH, Torres JL, Zhang L, Irimia A, Copps J, Zhou KH, Kwon YD, Law WH, Schramm CA, Verardi R, Krebs SJ, Kwong PD, Doria-Rose NA, Wilson IA, Zwick MB, Yates JR, III, Schief WR, Ward AB. 2020. HIV-1 envelope and MPER antibody structures in lipid assemblies. Cell Rep 31:107583. 10.1016/j.celrep.2020.107583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Khairil Anuar INA, Banerjee A, Keeble AH, Carella A, Nikov GI, Howarth M. 2019. Spy&Go purification of SpyTag-proteins using pseudo-SpyCatcher to access an oligomerization toolbox. Nat Commun 10:1734. 10.1038/s41467-019-09678-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chackerian B, Peabody DS. 2020. Factors that govern the induction of long-lived antibody responses. Viruses 12:74. 10.3390/v12010074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brisse M, Vrba SM, Kirk N, Liang Y, Ly H. 2020. Emerging concepts and technologies in vaccine development. Front Immunol 11:583077. 10.3389/fimmu.2020.583077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kanekiyo M, Joyce MG, Gillespie RA, Gallagher JR, Andrews SF, Yassine HM, Wheatley AK, Fisher BE, Ambrozak DR, Creanga A, Leung K, Yang ES, Boyoglu-Barnum S, Georgiev IS, Tsybovsky Y, Prabhakaran MS, Andersen H, Kong WP, Baxa U, Zephir KL, Ledgerwood JE, Koup RA, Kwong PD, Harris AK, McDermott AB, Mascola JR, Graham BS. 2019. Mosaic nanoparticle display of diverse influenza virus hemagglutinins elicits broad B cell responses. Nat Immunol 20:362–372. 10.1038/s41590-018-0305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Williams WB, Zhang J, Jiang C, Nicely NI, Fera D, Luo K, Moody MA, Liao HX, Alam SM, Kepler TB, Ramesh A, Wiehe K, Holland JA, Bradley T, Vandergrift N, Saunders KO, Parks R, Foulger A, Xia SM, Bonsignori M, Montefiori DC, Louder M, Eaton A, Santra S, Scearce R, Sutherland L, Newman A, Bouton-Verville H, Bowman C, Bomze H, Gao F, Marshall DJ, Whitesides JF, Nie X, Kelsoe G, Reed SG, Fox CB, Clary K, Koutsoukos M, Franco D, Mascola JR, Harrison SC, Haynes BF, Verkoczy L. 2017. Initiation of HIV neutralizing B cell lineages with sequential envelope immunizations. Nat Commun 8:1732. 10.1038/s41467-017-01336-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Townsley S, Li Y, Kozyrev Y, Cleveland B, Hu SL. 2016. Conserved role of an N-linked glycan on the surface antigen of human immunodeficiency virus type 1 modulating virus sensitivity to broadly neutralizing antibodies against the receptor and coreceptor binding sites. J Virol 90:829–841. 10.1128/JVI.02321-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gray ES, Madiga MC, Hermanus T, Moore PL, Wibmer CK, Tumba NL, Werner L, Mlisana K, Sibeko S, Williamson C, Abdool Karim SS, Morris L. 2011. The neutralization breadth of HIV-1 develops incrementally over four years and is associated with CD4+ T cell decline and high viral load during acute infection. J Virol 85:4828–4840. 10.1128/JVI.00198-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dennison SM, Sutherland LL, Jaeger FH, Anasti KM, Parks R, Stewart S, Bowman C, Xia SM, Zhang R, Shen X, Scearce RM, Ofek G, Yang Y, Kwong PD, Santra S, Liao HX, Tomaras G, Letvin NL, Chen B, Alam SM, Haynes BF. 2011. Induction of antibodies in rhesus macaques that recognize a fusion-intermediate conformation of HIV-1 gp41. PLoS One 6:e27824. 10.1371/journal.pone.0027824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cheng C, Duan H, Xu K, Chuang G-Y, Corrigan AR, Geng H, O’Dell S, Ou L, Chambers M, Changela A, Chen X, Foulds KE, Sarfo EK, Jafari AJ, Hill KR, Kong R, Liu K, Todd JP, Tsybovsky Y, Verardi R, Wang S, Wang Y, Wu W, Zhou T, Arnold FJ, Doria-Rose NA, Koup RA, McDermott AB, Scorpio DG, Worobey M, Shapiro L, Mascola JR, Kwong PD. 2020. Immune monitoring reveals fusion peptide priming to imprint cross-clade HIV-neutralizing responses with a characteristic early B cell signature. Cell Rep 32:107981. 10.1016/j.celrep.2020.107981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nelson JD, Kinkead H, Brunel FM, Leaman D, Jensen R, Louis JM, Maruyama T, Bewley CA, Bowdish K, Clore GM, Dawson PE, Frederickson S, Mage RG, Richman DD, Burton DR, Zwick MB. 2008. Antibody elicited against the gp41 N-heptad repeat (NHR) coiled-coil can neutralize HIV-1 with modest potency but non-neutralizing antibodies also bind to NHR mimetics. Virology 377:170–183. 10.1016/j.virol.2008.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Verkoczy L, Chen Y, Zhang J, Bouton-Verville H, Newman A, Lockwood B, Scearce RM, Montefiori DC, Dennison SM, Xia SM, Hwang KK, Liao HX, Alam SM, Haynes BF. 2013. Induction of HIV-1 broad neutralizing antibodies in 2F5 knock-in mice: selection against membrane proximal external region-associated autoreactivity limits T-dependent responses. J Immunol 191:2538–2550. 10.4049/jimmunol.1300971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xu K, Acharya P, Kong R, Cheng C, Chuang GY, Liu K, Louder MK, O'Dell S, Rawi R, Sastry M, Shen CH, Zhang B, Zhou T, Asokan M, Bailer RT, Chambers M, Chen X, Choi CW, Dandey VP, Doria-Rose NA, Druz A, Eng ET, Farney SK, Foulds KE, Geng H, Georgiev IS, Gorman J, Hill KR, Jafari AJ, Kwon YD, Lai YT, Lemmin T, McKee K, Ohr TY, Ou L, Peng D, Rowshan AP, Sheng Z, Todd JP, Tsybovsky Y, Viox EG, Wang Y, Wei H, Yang Y, Zhou AF, Chen R, Yang L, Scorpio DG, McDermott AB, Shapiro L. 2018. Epitope-based vaccine design yields fusion peptide-directed antibodies that neutralize diverse strains of HIV-1. Nat Med 24:857–867. 10.1038/s41591-018-0042-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gift SK, Leaman DP, Zhang L, Kim AS, Zwick MB. 2017. Functional stability of HIV-1 envelope trimer affects accessibility to broadly neutralizing antibodies at its apex. J Virol 91:e01216-17. 10.1128/JVI.01216-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nelson JD, Brunel FM, Jensen R, Crooks ET, Cardoso RM, Wang M, Hessell A, Wilson IA, Binley JM, Dawson PE, Burton DR, Zwick MB. 2007. An affinity-enhanced neutralizing antibody against the membrane-proximal external region of human immunodeficiency virus type 1 gp41 recognizes an epitope between those of 2F5 and 4E10. J Virol 81:4033–4043. 10.1128/JVI.02588-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ringe RP, Yasmeen A, Ozorowski G, Go EP, Pritchard LK, Guttman M, Ketas TA, Cottrell CA, Wilson IA, Sanders RW, Cupo A, Crispin M, Lee KK, Desaire H, Ward AB, Klasse PJ, Moore JP. 2015. Influences on the design and purification of soluble, recombinant native-like HIV-1 envelope glycoprotein trimers. J Virol 89:12189–12210. 10.1128/JVI.01768-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lambert O, Levy D, Ranck JL, Leblanc G, Rigaud JL. 1998. A new “gel-like” phase in dodecyl maltoside-lipid mixtures: implications in solubilization and reconstitution studies. Biophys J 74:918–930. 10.1016/S0006-3495(98)74015-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Leaman DP, Kinkead H, Zwick MB. 2010. In-solution virus capture assay helps deconstruct heterogeneous antibody recognition of human immunodeficiency virus type 1. J Virol 84:3382–3395. 10.1128/JVI.02363-09. [DOI] [PMC free article] [PubMed] [Google Scholar]