ABSTRACT
Defective viral genomes (DVGs) are parasitic viral sequences containing point mutations, deletions, or duplications that might interfere with replication. DVGs are often associated with viral passage at high multiplicities of infection in culture systems but have been increasingly reported in clinical specimens. To date however, only RNA viruses have been shown to contain DVGs in clinical specimens. Here, using direct deep sequencing with multiple library preparation strategies and confirmatory digital droplet PCR (ddPCR) of urine samples taken from immunosuppressed individuals, we show that clinical BK polyomavirus (BKPyV) and JC polyomavirus (JCPyV) strains contain widespread genomic rearrangements across multiple loci that likely interfere with viral replication. BKPyV DVGs were derived from BKPyV genotypes Ia, Ib-1, and Ic. The presence of DVGs was associated with specimens containing higher viral loads but never reached clonality, consistent with a model of parasitized replication. These DVGs persisted during clinical infection as evidenced in two separate pairs of samples containing BK virus collected from the same individual up to 302 days apart. In a separate individual, we observed the generation of DVGs after a 57.5-fold increase in viral load. In summary, by extending the presence of DVGs in clinical specimens to DNA viruses, we demonstrate the ubiquity of DVGs in clinical virology.
IMPORTANCE Defective viral genomes (DVGs) can have a significant impact on the production of infectious virus particles. DVGs have only been identified in cultured viruses passaged at high multiplicities of infection and RNA viruses collected from clinical specimens; no DNA virus in the wild has been shown to contain DVGs. Here, we identified BK and JC polyomavirus DVGs in clinical urine specimens and demonstrated that these DVGs are more frequently identified in samples with higher viral loads. The strains containing DVGs had rearrangements throughout their genomes, with the majority affecting genes required for viral replication. Longitudinal analysis showed that these DVGs can persist during an infection but do not reach clonality within the chronically infected host. Our identification of polyomavirus DVGs suggests that these parasitic sequences exist across the many classes of viruses capable of causing human disease.
KEYWORDS: BK virus, DNA virus, JC virus, ddPCR, defective, defective interfering particle, defective interfering genome, defective viral genome, polyomavirus, rearrangement
INTRODUCTION
Defective viral genomes (DVGs) constitute a peculiar group of viral mutants incapable of autonomous replication. These DVGs contain point mutations, deletions, or duplications in their genomes (1). DVGs can either hinder or assist the growth of the virus. The plant pathogen Turnip crinkle virus produces DVGs that assist viral growth by interfering with the plant’s antiviral mechanisms (2, 3). Many DVGs of human-pathogenic viruses produce defective interfering particles that parasitize and interfere with viral replication. These DVGs are thought to contribute to chronic viral infections by promoting survival of the infected host cells or reducing the infectious viral load by suppressing the number of replication-competent virions produced during active infection (4). Increasingly, DVGs are being investigated for their potential as antiviral strategies (1, 5).
Two major types of DVGs have been described for RNA viruses (1). Deletion-type DVGs are truncated versions of the nondefective viral genome that occur when the polymerase skips part of the genome during replication. Deletion DVGs tend to possess identical terminal sequences while missing several or all essential genes required for self-propagation (6, 7). The second type of DVG is the copy-back DVGs. Copy-back and the related snap-back DVGs are rearranged genomes consisting of an authentic terminus followed immediately by an inverted repeat of some or all of that sequence (8). Copy-back DVGs have been reported in many negative-sense RNA viruses, mainly in the Paramyxoviridae family, and are predicted to be the products from the reattachment of RNA-dependent RNA polymerase (RdRp) to the nascent strand, thus copying back the end of the genome (9).
DVGs have frequently been identified in cell culture systems, especially when viral stocks are passaged at high multiplicities of infection, and are increasingly being identified in clinical specimens (4, 10–14). These DVGs contain deletions of variable lengths and across various regions in the genome but generally retain the origin of replication (15). Cultured BK and JC polyomavirus have both been shown to contain deletions spanning the large T antigen (16, 17). Deletions spanning multiple genomic regions, including VP1 and VP2, have been observed when the related simian virus 40 is passaged in cell culture (18–21). To date, only single-stranded RNA viruses have been demonstrated to form DVGs in clinical specimens (4, 22–24). However, the significance and role of DVG during clinical infection are unclear.
Polyomaviruses are nonenveloped, double-stranded DNA viruses with a small, circular genome of approximately 5,000 bp (25). Among the 102 identified species, BK polyomavirus (BKPyV) and JC polyomavirus (JCPyV) are the two most commonly known to infect humans (26). BKPyV and JCPyV are highly prevalent in the population, with most individuals initially getting infected in early childhood and maintaining a lifelong infection thereafter (27, 28). The infections of BKPyV, also known as Human polyomavirus 1, are not associated with disease in immunocompetent individuals but can cause nephropathy and allograft failure in individuals receiving a renal transplant (29–32). In individuals with hemorrhagic cystitis, BKPyV can be found at very high titers in urine (>108 copies/ml of urine) (10). JCPyV, also known as Human polyomavirus 2, is most well known for its association with progressive multifocal leukoencephalopathy (PML) in immunosuppressed individuals and can cause many novel neurological disorders, such as JC virus granule cell neuropathy and JC virus encephalopathy (11). JCPyV can be found in cerebrospinal fluid and urine of individuals with PML at values ranging from 102 to 108 copies/ml (12).
While performing genome recovery of polyomaviruses for urine specimens sent to our clinical laboratory, we noticed many large-scale genomic rearrangements in BKPyV and JCPyV. These rearrangements were recovered independently of the sequencing strategy used and were found across different viral lineages. Polyomavirus DVGs were found more frequently in those specimens that were specifically associated with high viral load specimens but never reached clonality, consistent with a model of defective interfering replication.
RESULTS
Defective polyomavirus genomes in clinical urine samples.
We performed metagenomic shotgun sequencing on DNA extracted from 43 BKPyV-positive clinical urine samples collected from 37 individuals with a median viral load of 1.50 × 108 copies/ml (range, 6.3 × 104 to 4.6 × 1010 copies/ml) and 2 JCPyV-positive clinical urine specimens (see Table S1 in the supplemental material). Of the 45 samples, we recovered 38 complete BKPyV genomes and 11 complete JCPyV genomes and found BKPyV and JCPyV coinfections in 11 of these samples (Fig. 1). We identified cooccurrence of common uropathogenic bacteria present at a frequency greater than 10 reads per million (RPM) in 30 of the 45 polyomavirus-positive samples (Table S2). Reads corresponding to Candida spp. at a frequency greater than 10 RPM were identified in 9 of the 45 samples.
FIG 1.
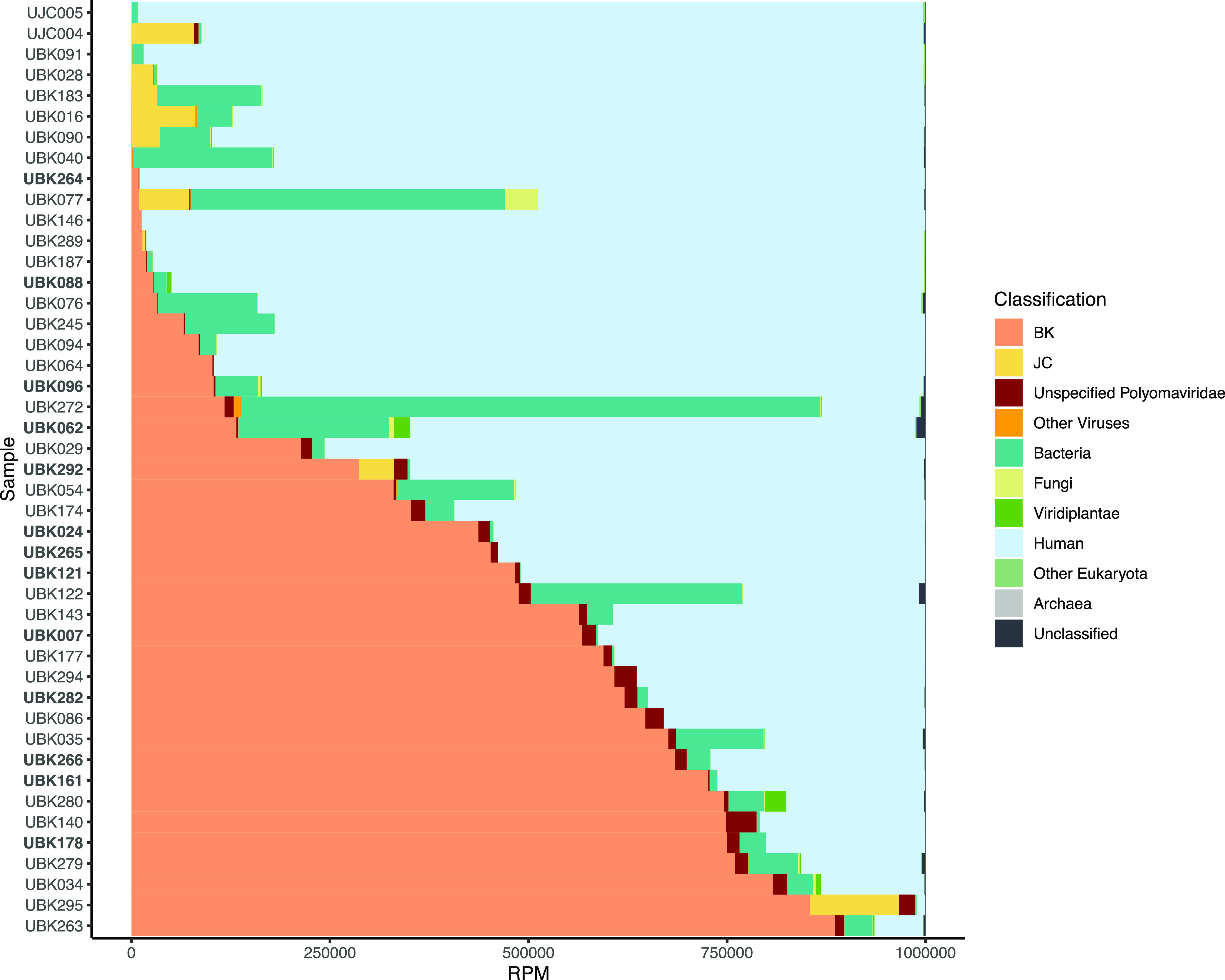
Taxonomic classifications identified by metagenomic analysis, color-coded and normalized by reads per million (RPM). Samples are sorted in ascending order of RPM assigned to BKPyV. The samples with bolded labels contain DVGs.
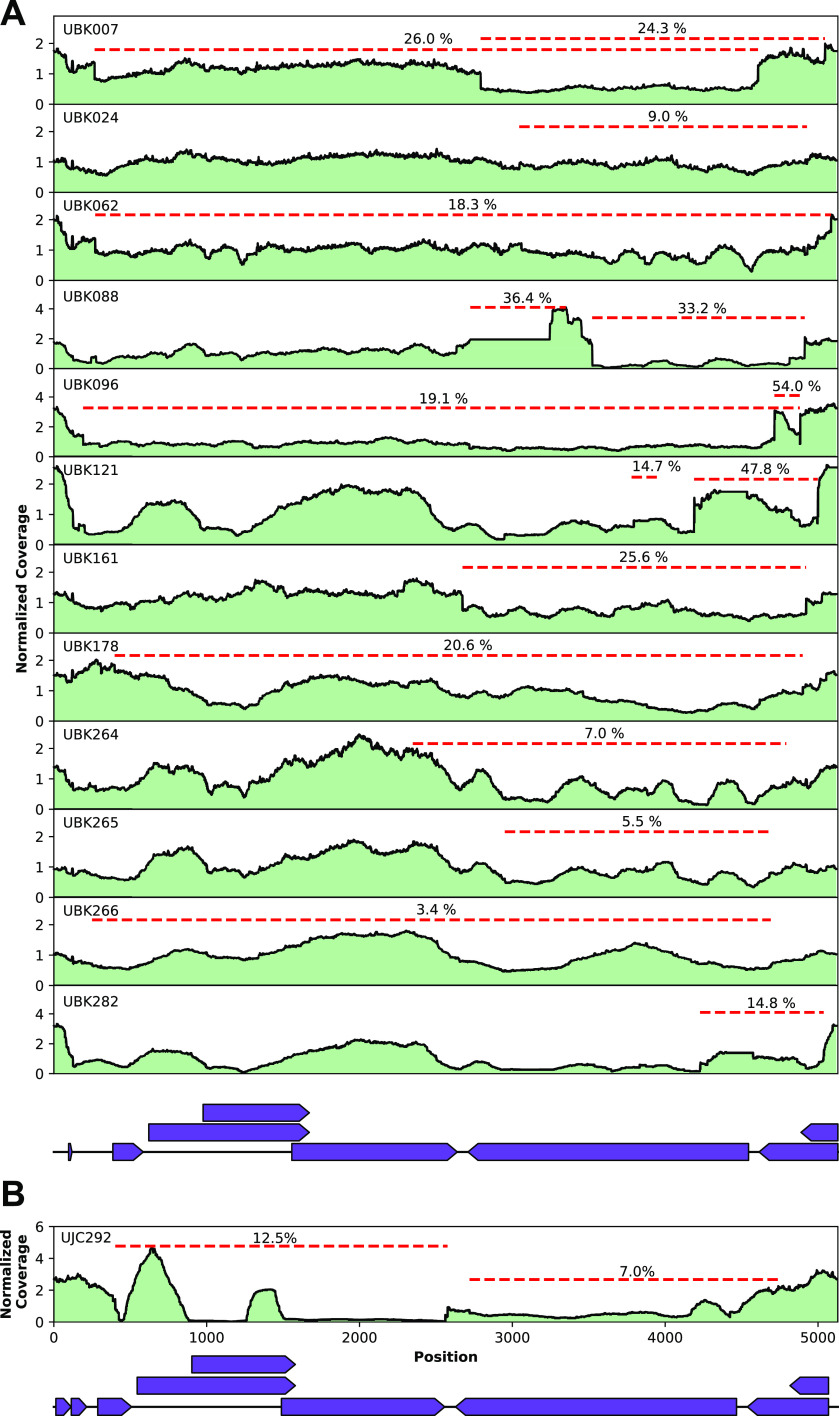
In 13 of the BKPyV-positive samples, we identified large rearrangements or deletions constituting DVGs in BKPyV or JCPyV that were supported by 10 or more sequencing reads and included samples with a sum total frequency of 10% or greater of these rearrangements (Fig. 2, Table 1). Twelve of the thirteen samples with rearrangements were identified in BKPyV genomes collected from eleven different individuals, while the remaining sample was identified in a JCPyV genome.
FIG 2.
(A and B) Coverage maps of defective BKPyV genomes (A) and a JCPyV genome (B) observed in shotgun sequencing data of 13 polyomavirus-positive specimens. The normalized sequencing read coverage across each sample’s consensus viral genome sequence is plotted in green. Junctions present in individual sequencing reads that are representative of the genomic rearrangements present in the DVGs are depicted by red dashes. For these rearrangements, the percentage of reads with the particular junction was calculated relative to the maximum genomic coverage in the analyzed sample.
TABLE 1.
Rearrangement and deletion junction positions and counts for BKPyV DVGs as detected by Geneious Prime using a threshold of at least 10 reads and frequency >10% compared to maximum depth
| Sample | Junction type | Nucleotide start position | Nucleotide end position | Length (bp) | No. of Junction reads | Reads with junction (%) | Genes affected |
|---|---|---|---|---|---|---|---|
| UBK007 | Rearrangement | 268 | 4628 | 763 | 958 | 26.0 | LargeT, SmallT |
| Rearrangement | 2805 | 5068 | 2,262 | 898 | 24.3 | LargeT, SmallT | |
| Deletion | 236 | 346 | 110 | 312 | 8.5 | None | |
| Deletion | 260 | 346 | 86 | 94 | 2.6 | None | |
| Deletion | 275 | 428 | 153 | 67 | 1.8 | Agnoprotein | |
| Deletion | 317 | 324 | 7 | 45 | 1.2 | None | |
| Rearrangement | 262 | 4794 | 591 | 26 | 0.7 | Agnoprotein, LargeT, SmallT | |
| Deletion | 2914 | 3826 | 912 | 25 | 0.7 | LargeT | |
| UBK024 | Deletion | 1739 | 1746 | 7 | 546 | 25.0 | VP1 |
| Deletion | 4841 | 4971 | 130 | 320 | 14.6 | LargeT, SmallT | |
| Rearrangement (inversion) | 3059 | 4950 | 1,891 | 196 | 9.0 | LargeT, SmallT | |
| Rearrangement (inversion) | 61 | 3155 | 2,045 | 148 | 6.8 | Agnoprotein, LargeT, VP1, VP2, VP3 | |
| Deletion | 4647 | 4768 | 121 | 137 | 6.3 | SmallT | |
| Rearrangement (inversion) | 243 | 3354 | 2,028 | 97 | 4.4 | Agnoprotein, LargeT, VP1, VP2, VP3 | |
| Rearrangement | 76 | 4567 | 649 | 95 | 4.3 | Agnoprotein, LargeT, VP1, VP2, VP3 | |
| Rearrangement (inversion) | 47 | 306 | 259 | 78 | 3.6 | None | |
| Rearrangement (inversion) | 3060 | 4974 | 1,914 | 77 | 3.5 | LargeT, SmallT | |
| Deletion | 5119 | 218 | 510 | 72 | 3.3 | LargeT, SmallT | |
| Rearrangement | 611 | 405 | 207 | 51 | 2.3 | All | |
| Rearrangement | 5011 | 4981 | 31 | 43 | 2.0 | All | |
| Rearrangement | 165 | 2215 | 2,049 | 27 | 1.2 | Agnoprotein, VP1, VP2, VP3 | |
| UBK062 | Rearrangement | 272 | 5108 | 292 | 2,696 | 18.3 | All |
| Rearrangement | 49 | 2896 | 2,281 | 1,800 | 12.2 | Agnoprotein, LargeT, VP1, VP2, VP3 | |
| Rearrangement | 3074 | 4953 | 1,878 | 1,123 | 7.6 | LargeT, SmallT | |
| Deletion | 2894 | 3272 | 378 | 1,009 | 6.8 | LargeT | |
| Rearrangement | 2920 | 4168 | 1,247 | 952 | 6.5 | LargeT | |
| Rearrangement | 3272 | 49 | 1,903 | 811 | 5.5 | LargeT, SmallT | |
| Deletion | 3461 | 4191 | 730 | 603 | 4.1 | LargeT | |
| Rearrangement | 220 | 4847 | 501 | 578 | 3.9 | All | |
| Rearrangement | 327 | 5019 | 436 | 423 | 2.9 | All | |
| Rearrangement | 3325 | 4981 | 1,655 | 239 | 1.6 | LargeT, SmallT | |
| Rearrangement | 2779 | 4051 | 1,271 | 200 | 1.4 | LargeT | |
| Rearrangement | 290 | 4951 | 467 | 152 | 1.0 | All | |
| Rearrangement | 288 | 2267 | 1,978 | 116 | 0.8 | Agnoprotein, VP1, VP2, VP3 | |
| Rearrangement | 255 | 4925 | 458 | 111 | 0.8 | All | |
| Rearrangement | 303 | 4943 | 488 | 109 | 0.7 | All | |
| Deletion | 4183 | 4682 | 499 | 102 | 0.7 | LargeT, SmallT | |
| Rearrangement | 2784 | 4872 | 2,087 | 101 | 0.7 | LargeT, SmallT | |
| Deletion | 2904 | 3490 | 586 | 99 | 0.7 | LargeT | |
| Rearrangement | 480 | 2805 | 2,324 | 89 | 0.6 | Agnoprotein, LargeT, VP1, VP2, VP3 | |
| Rearrangement | 3118 | 4872 | 1,753 | 87 | 0.6 | LargeT, SmallT | |
| Rearrangement | 3634 | 4741 | 1,106 | 67 | 0.5 | LargeT, SmallT | |
| Rearrangement | 273 | 4678 | 723 | 66 | 0.5 | All | |
| Rearrangement | 285 | 4856 | 557 | 64 | 0.4 | All | |
| Rearrangement | 330 | 4962 | 496 | 47 | 0.3 | All | |
| Deletion | 3382 | 4314 | 932 | 45 | 0.3 | LargeT | |
| Deletion | 3752 | 4673 | 921 | 40 | 0.3 | LargeT, SmallT | |
| Deletion | 696 | 700 | 4 | 37 | 0.3 | VP2 | |
| Deletion | 4899 | 5098 | 199 | 36 | 0.2 | LargeT, SmallT | |
| Rearrangement | 360 | 4297 | 1,191 | 34 | 0.2 | Agnoprotein, LargeT, VP1, VP2, VP3 | |
| Rearrangement | 3037 | 2850 | 188 | 18 | 0.1 | All | |
| Rearrangement | 271 | 3065 | 2,334 | 16 | 0.1 | Agnoprotein, LargeT, VP1, VP2, VP3 | |
| Deletion | 307 | 692 | 385 | 15 | 0.1 | Agnoprotein, VP2 | |
| Rearrangement | 231 | 5089 | 270 | 15 | 0.1 | All | |
| Rearrangement | 391 | 4634 | 885 | 13 | 0.1 | Agnoprotein, LargeT, VP1, VP2, VP3 | |
| Rearrangement (inversion) | 1141 | 1179 | 38 | 13 | 0.1 | VP2, VP3 | |
| Deletion | 391 | 1047 | 656 | 12 | 0.1 | Agnoprotein, VP2, VP3 | |
| Deletion | 492 | 705 | 213 | 10 | 0.1 | Agnoprotein, VP2 | |
| UBK088 | Deletion | 2737 | 3365 | 628 | 219 | 36.4 | LargeT |
| Rearrangement | 3538 | 4935 | 1,396 | 200 | 33.2 | LargeT, SmallT | |
| Rearrangement | 277 | 4836 | 551 | 56 | 9.3 | All | |
| Rearrangement | 3379 | 245 | 1,974 | 51 | 8.5 | LargeT, SmallT | |
| Rearrangement | 3338 | 3272 | 67 | 27 | 4.5 | All | |
| UBK096 | Rearrangement (inversion) | 4736 | 4905 | 169 | 525 | 54.0 | SmallT |
| Rearrangement (inversion) | 192 | 4903 | 430 | 186 | 19.1 | All | |
| Rearrangement | 2731 | 4963 | 2,231 | 68 | 7.0 | LargeT, SmallT | |
| Rearrangement | 300 | 4650 | 792 | 48 | 4.9 | All | |
| Deletion | 183 | 322 | 139 | 31 | 3.2 | None | |
| Rearrangement (inversion) | 3780 | 3825 | 45 | 23 | 2.4 | LargeT | |
| Rearrangement | 24 | 1851 | 1,826 | 19 | 2.0 | Agnoprotein, VP1, VP2, VP3 | |
| Rearrangement | 2796 | 5043 | 2,246 | 14 | 1.4 | LargeT, SmallT | |
| Rearrangement | 4740 | 2337 | 2,404 | 12 | 1.2 | All | |
| UBK121 | Deletion | 4382 | 4558 | 176 | 6,030 | 55.7 | LargeT |
| Rearrangement (inversion) | 4208 | 5021 | 813 | 5174 | 47.8 | LargeT, SmallT | |
| Deletion | 1898 | 1902 | 4 | 3,634 | 33.5 | VP1 | |
| Deletion | 3747 | 3756 | 9 | 1,700 | 15.7 | LargeT | |
| Deletion | 3797 | 3984 | 187 | 1,587 | 14.7 | LargeT | |
| Rearrangement | 4847 | 3811 | 1,037 | 881 | 8.1 | LargeT, SmallT | |
| Rearrangement (inversion) | 4210 | 5017 | 807 | 774 | 7.1 | LargeT, SmallT | |
| Deletion | 4682 | 4855 | 173 | 753 | 7.0 | SmallT | |
| Rearrangement (inversion) | 96 | 4749 | 442 | 653 | 6.0 | All | |
| Rearrangement | 4302 | 2962 | 1,341 | 611 | 5.6 | All | |
| Rearrangement (inversion) | 4207 | 5020 | 813 | 583 | 5.4 | LargeT, SmallT | |
| Deletion | 3501 | 3511 | 10 | 486 | 4.5 | LargeT | |
| Rearrangement | 4104 | 3467 | 638 | 435 | 4.0 | All | |
| Deletion | 2843 | 3499 | 656 | 271 | 2.5 | LargeT | |
| Rearrangement | 3624 | 4989 | 1,364 | 246 | 2.3 | LargeT, SmallT | |
| Deletion | 1883 | 1890 | 7 | 196 | 1.8 | VP1 | |
| Rearrangement | 3341 | 32 | 1,785 | 171 | 1.6 | LargeT, SmallT | |
| Deletion | 4587 | 4677 | 90 | 152 | 1.4 | SmallT | |
| Rearrangement | 3461 | 3,455 | 7 | 113 | 1.0 | All | |
| Deletion | 2678 | 2686 | 8 | 99 | 0.9 | None | |
| Rearrangement (inversion) | 3097 | 5,046 | 1,949 | 83 | 0.8 | LargeT, SmallT | |
| Rearrangement (inversion) | 4818 | 5070 | 252 | 82 | 0.8 | LargeT, SmallT | |
| Rearrangement (inversion) | 3635 | 4972 | 1,337 | 69 | 0.6 | LargeT, SmallT | |
| Rearrangement (inversion) | 4313 | 4817 | 504 | 63 | 0.6 | LargeT, SmallT | |
| Rearrangement (inversion) | 2770 | 4570 | 1,800 | 51 | 0.5 | LargeT | |
| Rearrangement (inversion) | 3109 | 4254 | 1,145 | 50 | 0.5 | LargeT | |
| Deletion | 4,641 | 4846 | 205 | 50 | 0.5 | SmallT | |
| Deletion | 4308 | 4778 | 470 | 42 | 0.4 | LargeT, SmallT | |
| Rearrangement | 80 | 4639 | 537 | 32 | 0.3 | All | |
| Deletion | 3612 | 3616 | 4 | 29 | 0.3 | LargeT | |
| Deletion | 3550 | 4185 | 635 | 28 | 0.3 | LargeT | |
| Deletion | 664 | 668 | 4 | 26 | 0.2 | VP2 | |
| Rearrangement (inversion) | 155 | 4625 | 625 | 26 | 0.2 | Agnoprotein, LargeT, VP1, VP2, VP3 | |
| Rearrangement (inversion) | 4694 | 4727 | 33 | 26 | 0.2 | SmallT | |
| Rearrangement (inversion) | 3153 | 3984 | 831 | 24 | 0.2 | LargeT | |
| Deletion | 4788 | 4794 | 6 | 22 | 0.2 | SmallT | |
| Rearrangement (inversion) | 3594 | 4299 | 705 | 21 | 0.2 | LargeT | |
| Deletion | 4,694 | 4704 | 10 | 19 | 0.2 | SmallT | |
| Deletion | 4888 | 5094 | 206 | 18 | 0.2 | LargeT, SmallT | |
| Rearrangement (inversion) | 24 | 4273 | 846 | 18 | 0.2 | Agnoprotein, LargeT, VP1, VP2, VP3 | |
| UBK161 | Rearrangement | 2686 | 4944 | 2,257 | 1,369 | 25.6 | LargeT, SmallT |
| Rearrangement | 501 | 2259 | 1,757 | 426 | 8.0 | All | |
| Rearrangement | 2246 | 5041 | 2,318 | 82 | 1.5 | LargeT, SmallT, VP1 | |
| Deletion | 3476 | 3509 | 33 | 29 | 0.5 | LargeT | |
| Rearrangement | 472 | 1707 | 1,234 | 28 | 0.5 | Agnoprotein, VP1, VP2, VP3 | |
| Rearrangement (inversion) | 320 | 340 | 20 | 20 | 0.4 | None | |
| Deletion | 681 | 685 | 4 | 14 | 0.3 | VP2 | |
| Rearrangement | 5094 | 1970 | 1,987 | 14 | 0.3 | All | |
| UBK178 | Rearrangement | 400 | 4922 | 620 | 1,099 | 20.6 | All |
| Deletion | 2931 | 3865 | 934 | 952 | 9.5 | LargeT | |
| Rearrangement (inversion) | 288 | 3478 | 1,951 | 601 | 6.0 | Agnoprotein, LargeT, VP1, VP2, VP3 | |
| Rearrangement (inversion) | 4638 | 5037 | 399 | 590 | 5.9 | LargeT, SmallT | |
| Deletion | 5079 | 365 | 530 | 34 | 0.3 | LargeT, SmallT | |
| Deletion | 710 | 714 | 4 | 32 | 0.3 | VP2 | |
| Rearrangement (inversion) | 2633 | 2711 | 78 | 31 | 0.3 | VP1 | |
| Rearrangement | 353 | 293 | 61 | 27 | 0.3 | All | |
| Rearrangement | 2413 | 4899 | 2,485 | 22 | 0.2 | LargeT, SmallT, VP1 | |
| Rearrangement (inversion) | 3199 | 3356 | 157 | 22 | 0.2 | LargeT | |
| Rearrangement | 478 | 1,534 | 1,055 | 19 | 0.2 | Agnoprotein, VP2, VP3 | |
| Deletion | 2488 | 2,502 | 14 | 18 | 0.2 | VP1 | |
| Deletion | 158 | 301 | 143 | 16 | 0.2 | None | |
| Rearrangement (inversion) | 1838 | 2150 | 312 | 16 | 0.2 | VP1 | |
| Rearrangement | 66 | 4748 | 460 | 15 | 0.2 | All | |
| Rearrangement (inversion) | 1453 | 1463 | 10 | 14 | 0.1 | VP2, VP3 | |
| Rearrangement (inversion) | 137 | 4957 | 321 | 14 | 0.1 | All | |
| Rearrangement (inversion) | 2808 | 3360 | 552 | 13 | 0.1 | LargeT | |
| Deletion | 3446 | 3458 | 12 | 11 | 0.1 | LargeT | |
| Deletion | 477 | 583 | 106 | 10 | 0.1 | Agnoprotein | |
| UBK264 | Rearrangement | 2359 | 4814 | 2,454 | 14 | 7.0 | LargeT, SmallT, VP1 |
| Rearrangement | 276 | 1892 | 1,615 | 12 | 6.0 | Agnoprotein, VP1, VP2, VP3 | |
| UBK265 | Rearrangement | 2964 | 4702 | 1,737 | 553 | 5.5 | LargeT, SmallT |
| Rearrangement | 203 | 2891 | 2,454 | 305 | 3.0 | Agnoprotein, LargeT, VP1, VP2, VP3 | |
| Rearrangement | 304 | 2360 | 2,055 | 215 | 2.1 | Agnoprotein, VP1, VP2, VP3 | |
| Rearrangement | 2622 | 4703 | 2,080 | 104 | 1.0 | LargeT, SmallT, VP1 | |
| Deletion | 270 | 311 | 41 | 87 | 0.9 | None | |
| Rearrangement | 3827 | 4873 | 1,045 | 65 | 0.6 | LargeT, SmallT | |
| Rearrangement | 3429 | 4755 | 1,325 | 52 | 0.5 | LargeT, SmallT | |
| Rearrangement (inversion) | 3894 | 4519 | 625 | 50 | 0.5 | LargeT | |
| Rearrangement (inversion) | 275 | 4495 | 921 | 49 | 0.5 | Agnoprotein, LargeT, VP1, VP2, VP3 | |
| Deletion | 710 | 714 | 4 | 49 | 0.5 | VP2 | |
| Deletion | 180 | 354 | 174 | 49 | 0.5 | None | |
| Rearrangement | 282 | 2741 | 2,458 | 42 | 0.4 | Agnoprotein, LargeT, VP1, VP2, VP3 | |
| Rearrangement | 230 | 3539 | 1,833 | 39 | 0.4 | Agnoprotein, LargeT, VP1, VP2, VP3 | |
| Rearrangement | 3813 | 5001 | 1,187 | 37 | 0.4 | LargeT, SmallT | |
| Rearrangement | 218 | 3599 | 1,761 | 32 | 0.3 | Agnoprotein, LargeT, VP1, VP2, VP3 | |
| Deletion | 3867 | 4649 | 782 | 32 | 0.3 | LargeT, SmallT | |
| Deletion | 3019 | 3925 | 906 | 30 | 0.3 | LargeT | |
| Rearrangement | 3733 | 5104 | 1,370 | 27 | 0.3 | LargeT, SmallT | |
| Rearrangement | 282 | 3837 | 1,587 | 26 | 0.3 | Agnoprotein, LargeT, VP1, VP2, VP3 | |
| Rearrangement | 263 | 2914 | 2,491 | 26 | 0.3 | Agnoprotein, LargeT, VP1, VP2, VP3 | |
| Rearrangement | 281 | 4554 | 869 | 26 | 0.3 | Agnoprotein, LargeT, VP1, VP2, VP3 | |
| Deletion | 498 | 517 | 19 | 25 | 0.3 | Agnoprotein | |
| Rearrangement | 261 | 4826 | 577 | 25 | 0.3 | All | |
| Deletion | 252 | 349 | 97 | 25 | 0.3 | None | |
| Rearrangement | 263 | 4455 | 950 | 24 | 0.2 | Agnoprotein, LargeT, VP1, VP2, VP3 | |
| Rearrangement | 227 | 4703 | 666 | 24 | 0.2 | All | |
| Rearrangement | 266 | 4703 | 705 | 24 | 0.2 | All | |
| Rearrangement (inversion) | 31 | 390 | 359 | 21 | 0.2 | Agnoprotein | |
| Rearrangement | 280 | 4771 | 651 | 19 | 0.2 | All | |
| Rearrangement | 3100 | 4491 | 1,390 | 15 | 0.2 | LargeT | |
| Deletion | 252 | 357 | 105 | 14 | 0.1 | None | |
| UBK266 | Rearrangement | 251 | 4712 | 681 | 83 | 3.4 | All |
| Rearrangement (inversion) | 13 | 307 | 294 | 69 | 2.8 | None | |
| Rearrangement | 3643 | 4721 | 1,077 | 48 | 2.0 | LargeT, SmallT | |
| Rearrangement | 313 | 4610 | 845 | 42 | 1.7 | Agnoprotein, LargeT, VP1, VP2, VP3 | |
| Rearrangement | 260 | 4703 | 699 | 38 | 1.6 | All | |
| Rearrangement | 312 | 4931 | 523 | 37 | 1.5 | All | |
| Rearrangement | 301 | 4778 | 665 | 32 | 1.3 | All | |
| Rearrangement (inversion) | 1857 | 5048 | 1,950 | 28 | 1.2 | LargeT, SmallT, VP1 | |
| Rearrangement | 2862 | 5129 | 2,266 | 27 | 1.1 | LargeT, SmallT | |
| Rearrangement | 324 | 4443 | 1,023 | 26 | 1.1 | Agnoprotein, LargeT, VP1, VP2, VP3 | |
| Deletion | 4851 | 5007 | 156 | 23 | 0.9 | LargeT, SmallT | |
| Rearrangement | 337 | 2519 | 2,181 | 23 | 0.9 | Agnoprotein, VP1, VP2, VP3 | |
| Deletion | 1976 | 2481 | 505 | 22 | 0.9 | VP1 | |
| Rearrangement | 1837 | 4991 | 1,988 | 20 | 0.8 | LargeT, SmallT, VP1 | |
| Deletion | 3937 | 4869 | 932 | 17 | 0.7 | LargeT, SmallT | |
| Rearrangement | 232 | 1615 | 1,382 | 12 | 0.5 | Agnoprotein, VP1, VP2, VP3 | |
| UBK282 | Deletion | 4421 | 4597 | 176 | 2,175 | 30.0 | LargeT |
| Deletion | 1937 | 1941 | 4 | 1,290 | 17.8 | VP1 | |
| Rearrangement (inversion) | 4247 | 5060 | 813 | 1,076 | 14.8 | LargeT, SmallT | |
| Deletion | 3781 | 3790 | 9 | 545 | 7.5 | LargeT | |
| Deletion | 3831 | 4018 | 187 | 503 | 6.9 | LargeT | |
| Rearrangement (inversion) | 4250 | 5057 | 807 | 475 | 6.6 | LargeT, SmallT | |
| Rearrangement | 4886 | 3850 | 1,037 | 422 | 5.8 | All | |
| Rearrangement (inversion) | 95 | 4789 | 440 | 381 | 5.3 | All | |
| Deletion | 2882 | 3538 | 656 | 294 | 4.1 | LargeT | |
| Deletion | 762 | 766 | 4 | 290 | 4.0 | VP2 | |
| Rearrangement | 3663 | 5028 | 1,364 | 247 | 3.4 | LargeT, SmallT | |
| Deletion | 3540 | 3550 | 10 | 220 | 3.0 | LargeT | |
| Deletion | 1922 | 1929 | 7 | 191 | 2.6 | VP1 | |
| Rearrangement | 4143 | 3506 | 638 | 188 | 2.6 | All | |
| Rearrangement (inversion) | 4246 | 5059 | 813 | 156 | 2.2 | LargeT, SmallT | |
| Rearrangement (inversion) | 4249 | 5056 | 807 | 135 | 1.9 | LargeT, SmallT | |
| Rearrangement | 4341 | 3001 | 1,341 | 134 | 1.9 | All | |
| Deletion | 4721 | 4894 | 173 | 130 | 1.8 | SmallT | |
| Rearrangement (inversion) | 2785 | 2859 | 74 | 79 | 1.1 | LargeT | |
| Deletion | 4626 | 4716 | 90 | 58 | 0.8 | SmallT | |
| Rearrangement | 3500 | 3494 | 7 | 52 | 0.7 | All | |
| Rearrangement (inversion) | 2764 | 5058 | 2,294 | 41 | 0.6 | LargeT, SmallT | |
| Deletion | 4364 | 4788 | 424 | 26 | 0.4 | LargeT, SmallT | |
| Rearrangement (inversion) | 4857 | 5109 | 252 | 23 | 0.3 | LargeT, SmallT | |
| Deletion | 4420 | 4610 | 190 | 20 | 0.3 | LargeT | |
| Deletion | 540 | 553 | 13 | 19 | 0.3 | Agnoprotein | |
| Rearrangement | 4665 | 3510 | 1,156 | 18 | 0.3 | All | |
| Rearrangement | 23 | 5122 | 36 | 15 | 0.2 | All | |
| UBK007—using Kapa HyperPrep Plus kit | Rearrangement | 2805 | 5068 | 2,262 | 98 | 17.92 | LargeT, SmallT |
| Rearrangement | 268 | 4628 | 763 | 90 | 16.45 | All | |
| Deletion | 236 | 346 | 110 | 43 | 7.86 | None | |
| Deletion | 275 | 428 | 153 | 20 | 3.66 | Agnoprotein | |
| UBK096—using Kapa HyperPrep Plus kit | Rearrangement (inversion) | 192 | 4903 | 430 | 44 | 41.9 | All |
| Rearrangement (inversion) | 4736 | 4905 | 169 | 35 | 33.33 | LargeT, SmallT |
The nucleotides present in the junctions in the BKPyV strains were separated by a range of 4 to 2,491 nucleotides (nt) (median, 628 nt). Notably, multiple unique junctions (median, 15; range, 2 to 40) were identified in each DVG-containing sample. All 12 BKPyV strains had junctions including the large T antigen, and in 10 of the strains, the most abundant deletion or rearrangement involved the large T antigen (Table 1). The most abundant junction in 11 of the BKPyV strains were internal deletions and, correspondingly, these DVGs were classified as deletion type. In the final strain, UBK096, the most abundant junction was an inversion rearrangement. In the 1 JCPyV strain with detectable junctions, 15 unique junctions, ranging from 285 to 2,302 nucleotides in length (median, 1,917 nt) were identified (Table 2). The most abundant junction was a rearrangement that spanned all three capsid proteins, VP1, VP2, and VP3. Other DVGs in the JCPyV present in specimen UBK292 were classified as deletion type.
TABLE 2.
Rearragement and deletion junction positions and counts for JCPyV DVGs as detected by Geneious Prime using a threshold of at least 10 reads and frequency >10% compared to maximum depth
| Sample | Junction type | Nucleotide start position | Nucleotide end position | Length (bp) | No. of junction reads | Reads with junction (%) | Genes affected |
|---|---|---|---|---|---|---|---|
| JCPyV in UBK292 | Rearrangement | 402 | 2576 | 2,173 | 508 | 12.48 | Agnoprotein, VP1, VP2, VP3 |
| Rearrangement | 425 | 2561 | 2,135 | 356 | 8.75 | Agnoprotein, VP1, VP2, VP3 | |
| Deletion | 103 | 5121 | 285 | 351 | 8.63 | All | |
| Rearrangement | 386 | 4423 | 1,089 | 284 | 6.98 | Agnoprotein, LargeT, VP1, VP2, VP3 | |
| Rearrangement | 2720 | 4739 | 2,018 | 162 | 3.98 | LargeT, SmallT | |
| Rearrangement | 2626 | 4582 | 1,955 | 46 | 1.13 | LargeT, VP1 | |
| Rearrangement | 2640 | 4641 | 2,000 | 44 | 1.08 | LargeT, SmallT, VP1 | |
| Rearrangement | 2794 | 4200 | 1,405 | 38 | 0.93 | LargeT | |
| Rearrangement | 430 | 4721 | 835 | 34 | 0.84 | All | |
| Rearrangement | 2186 | 4159 | 1,972 | 19 | 0.47 | LargeT, VP1 | |
| Rearrangement | 334 | 4502 | 958 | 18 | 0.44 | Agnoprotein, LargeT, VP1, VP2, VP3 | |
| Rearrangement | 2345 | 4648 | 2,302 | 17 | 0.42 | LargeT, SmallT, VP1 | |
| Deletion | 4488 | 5034 | 546 | 14 | 0.34 | LargeT, SmallT | |
| Rearrangement | 287 | 4429 | 984 | 13 | 0.32 | Agnoprotein, LargeT, VP1, VP2, VP3 | |
| Rearrangement | 1043 | 2961 | 1,917 | 11 | 0.27 | LargeT, VP1, VP2, VP3 |
We confirmed that the observed junctions were not an artifact of our library preparation protocol by performing a second method for generating sequencing libraries on a subset of DVG-containing and DVG-negative samples. Identical junctions were found in the DVG-containing samples, and no new DVGs were found in DVG-negative samples using the different library preparation (Fig. S1). We further confirmed select junctions using specific PCR and Sanger sequencing (NCBI BioProject PRJNA657423).
An elevated VP1-to-large T antigen ratio is observed in BKPyV strains containing defective viral genomes and having high viral loads.
We have previously shown that digital droplet PCR (ddPCR) is a cost-effective method to detect and confirm copy number alterations in cultured BKPyV and JCPyV (17). As many of the BKPyV DVGs had rearrangements and deletions spanning the large T antigen at a high frequency and comparatively fewer rearrangements and deletions spanning VP1 (Table 1), we speculated that DVG-containing samples should have a high number of VP1 copies relative to large T antigen copies. Thus, a VP1-to-large T antigen copy number ratio in DVG-containing samples should be greater than 1, reflecting the abundancy of large T antigen defective genomes, while samples without DVGs should have a ratio of 1, indicative of a lack of rearrangements and deletions in these genomes. To confirm our hypothesis, we performed ddPCR targeting the VP1 and large T antigen on the 12 BKPyV strains containing DVGs by deep sequencing and 21 BKPyV strains without DVGs by deep sequencing.
Quantitative analysis with a ddPCR assay showed that the VP1-to-large T antigen copy number ratio was significantly higher in those strains with DVGs (mean, 1.69; range, 0.99 to 3.57) than in those with intact genomes (mean, 1.02; range, 0.91 to 1.31) (P = 0.0002, Kruskal-Wallis test; Fig. 3A). Of note, two strains with DVGs (UBK265/UBK266) had a comparably low level of rearrangements (3 to 5%) by sequencing at the exact loci interrogated by the ddPCR and had a ddPCR VP1/large T antigen ratio equivalent to 1. A VP1/large T antigen ratio cutoff of 1.25 separated the remaining 10 DVG-containing strains confirmed by sequencing from all strains without DVGs except UBK292.
FIG 3.
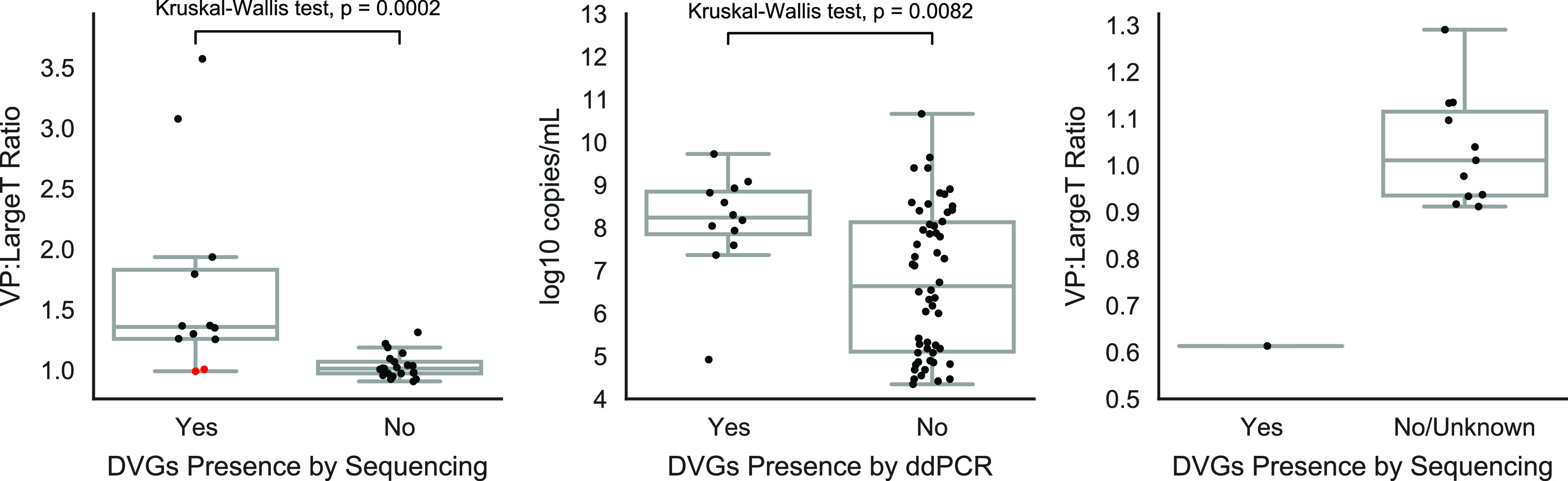
Confirmation of defective viral genomes using droplet digital PCR. (A) The copy number ratio for VP1 and large T antigen is plotted for 33 BKPyV isolates with and without DVGs determined by sequencing. Quartiles for each group are plotted in a box-and-whiskers plot, and error bars are 1.5-fold the interquartile range. Red dots indicate two strains (UBK265/UBK266) with DVGs that had a comparably low level of rearrangements (3 to 5%) at the target loci as determined by sequencing, consistent with their equivalent copy number measured by ddPCR. Statistical comparison was performed via Kruskal-Wallis test. (B) Log10 copies/ml viral load values in 66 BKPyV-positive specimens are displayed. The presence of DVGs was determined by ddPCR using a VP1/large T antigen ratio cutoff of 1.25. Statistical comparison was performed via Kruskal-Wallis test. (C) ddPCR VP1/large T antigen copy number ratios for 12 JCPyV-positive specimens are depicted. One JCPyV specimen contained a sequence-confirmed DVG, while 7 of the 11 other specimens lacked DVGs by sequencing, and the remaining had unknown DVG status.
We next used this ddPCR and established a ratio cutoff to screen 33 additional BKPyV-positive specimens for which high coverage genomes could not be recovered by shotgun sequencing due to low viral load. Notably, just one of the 33 low-viral-load samples had a VP1/large T antigen ratio greater than 1.25, consistent with low viral load infrequently containing DVGs. Furthermore, this analysis showed that specimens that tested positive for copy number alterations by ddPCR (VP1/large T antigen ratio of > 1.25) had a greater than 33-fold higher median viral load than those specimens that had a normal copy number ratio (median, 1.75 × 108 copies/ml versus 5.30 × 106 copies/ml; P = 0.0082, Kruskal-Wallis test) (Fig. 3B).
We next performed ddPCR targeting the VP1 and large T antigen on the JCPyV strain containing DVGs and 11 JCPyV-positive urine specimens, 7 of which did not contain DVGs by deep sequencing. The 11 JCPyV-positive strains had a mean VP1/large T antigen copy number ratio of 1.03 (range, 0.91 to 1.29), while the strain with the DVGs had a ratio of 0.61 (Fig. 3C), consistent with a copy number ratio of 0.58 by sequencing at the targeted loci.
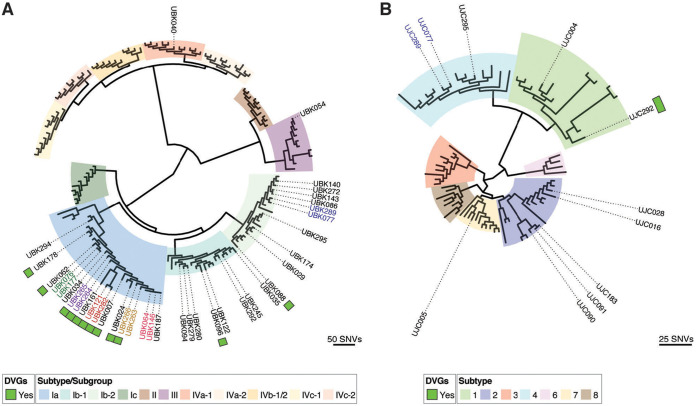
Phylogenetic analysis of BKPyV and JCPyV genomes reveals that strains containing defective viral genomes belong to multiple subgroups and are stable across time.
We next assessed the genetic relatedness of the BKPyV strains that produced DVGs by performing a phylogenetic analysis with 106 representative BKPyV genomes and the 38 total BKPyV genomes recovered in this study (Fig. 4A). Of the 38 strains, 36 belonged to subtype I, 1 belonged to subtype III, and 1 belonged to subtype IV. These observations are consistent with subtype I being the most prevalent subtype in the United States and subtypes II to IV being less frequently detected in North America (33). Of the 36 subtype I strains, 18 belonged to subgroup Ia, 8 belonged to subgroup Ib-1, and 10 belonged to subgroup Ib-2. The 12 strains containing DVGs all belonged to subtype I. Ten of these strains belonged to subgroup Ia, 1 belonged to subgroup Ib-1, and 1 belonged to subgroup Ib-2.
FIG 4.
(A) Phylogenetic tree of 38 BKPyV consensus genomes recovered in this study along with 106 representative BKPyV genomes covering all 11 subtypes. BKPyV samples containing DVGs are indicated by a green square, and those samples longitudinally collected from the same individual are highlighted through coloring of the sample labels. (B) Phylogenetic tree of the 11 JCPyV genomes recovered in this study with 106 JCPyV covering 7 JCPyV subtypes. The samples containing DVGs are indicated with a green square.
Of the 38 BKPyV genomes recovered in this study, 6 pairs of isolates were collected from the same individuals (UBK064/UBK146, UBK077/UBK289, UBK076/UBK177, UBK121/UBK282, UBK264/UBK265, UBK263/UBK266), allowing us to look at longitudinal evolution of BKPyV DVGs. The first three of these pairs (UBK064/UBK146, UBK077/UBK289, UBK076/UBK177) did not contain any DVGs and were genetically identical across a longitudinal sampling time of 7, 487, and 21 days, respectively. Interestingly, in one pair collected 159 days apart, the first isolate, UBK263, only contained intact genomes, while the second isolate, UBK266, contained DVGs. Only 1 consensus single nucleotide variant (SNV) was recovered between these samples, which resulted in a coding change (Glu43Lys) in the agnoprotein. Notably, the BKPyV viral load increased by 57.5× between UBK263 and UBK266, suggesting that the increase in viral load may have contributed to the formation of DVGs. In the final two pairs (UBK121/UBK282 and UBK264/UBK265), both strains contained DVGs. UBK121 and UBK282 were collected 302 days apart and differed by 2 SNVs as well as a 39-bp insertion at the origin. One of the SNVs resulted in a coding change, Lys556Arg, in the large T antigen. UBK264 and UBK265 were collected 41 days apart and differed by 2 SNVs, both of which resulted in coding changes in the agnoprotein (Gln8Arg and Glu43Lys).
We then assessed the genetic relatedness of the 11 JCPyV strains sequenced in this study by performing a phylogenetic analysis with 64 representative JCPyV genomes (Fig. 4B). Two of the strains belonged to subtype 1, 5 belonged to subtype 2, 3 belonged to subtype 4, and 1 belonged to subtype 7. The one JCPyV strain containing DVGs belonged to subtype 1.
BKPyV DVG-containing strains are more likely to contain unfixed mutations in the BC loop of VP1 than non-DVG containing strains.
The host DNA cytosine deaminase APOBEC3B has been shown to shape the intrahost evolution of BKPyV within the kidney (34). APOBEC3B targets cytosines contained within specific trinucleotide motifs (YCD) (35–37). In BKPyV, APOBEC3B recognition sites are present more often on the antisense DNA strand, and APOBEC3B-associated mutagenesis is speculated to contribute to the emergence of antibody escape mutations (36). The antisense DNA strand that encodes the BC loop, which is often a target of neutralizing antibodies, of the VP1 viral capsid protein, has a number of APOBEC3B recognition sites (36, 38, 39). We examined all 38 BKPyV strains for mutations that did not reach fixation (allelic frequency between 10 and 90%) within the BC loop of VP1 that could be attributed to APOBEC3B damage. Of the 38 BKPyV strains, 10 had an unfixed mutation potentially associated with APOBEC3B mutagenesis (Table 3). Four of these strains had a nephropathy-associated mutation, Glu73Gln, attributed to APOBEC3B activity. Interestingly, 8 of the 10 strains with APOBEC3B-associated mutations in the BC loop of VP1 contained DVGs. We then examined all 38 BKPyV strains for unfixed mutations in the BC loop that were unlikely to be associated with APOBEC3B mutagenesis. Three DVG-containing BKPyV strains had additional mutations in the BC loop of VP1, while two of the non-DVG containing strains had additional BC loop mutations. In total, 8 of the 12 (66.7%) DVG-containing BKPyV strains had unfixed BC loop mutations, while just 3 of the 26 (11.5%) DVG-containing strains had such mutations.
TABLE 3.
Unfixed, nonsynonymous mutations located in the BC loop of VP1 identified in the BKPyV strains sequenced in this studya
| Sample | Contains DVGs? | Amino acid mutation | Frequency | APOBEC3B-associated? | APOBEC3B trinucleotide mutation | Strand containing APOBEC3B recognition site |
|---|---|---|---|---|---|---|
| UBK024 | Yes | Glu60Gln | 15.9 | Yes | TCT → TGT | Antisense |
| 66insSerLeu | 31.4 | No | NA | NA | ||
| Lys71Asn | 15.3 | No | NA | NA | ||
| Asn79Ser | 17.1 | No | NA | NA | ||
| Ser80Thr | 15.6 | No | NA | NA | ||
| UBK029 | No | Lys74Asn | 35.3 | No | NA | NA |
| UBK062 | Yes | Leu68Val | 89.7 | Yes | TCT → TGT | Sense |
| Glu82Gln | 40.5 | Yes | TCT → TGT | Antisense | ||
| UBK096 | Yes | Glu73Gln | 38.2 | Yes | TCA → TGA | Antisense |
| Glu73Lys | 25.9 | Yes | TCA → TTA | Antisense | ||
| Glu82Gln | 11.9 | Yes | TCT → TGT | Antisense | ||
| UBK121 | Yes | Asp58Asn | 12.9 | No | NA | NA |
| Glu60Gln | 36.2 | Yes | TCT → TGT | Antisense | ||
| Lys69Gln | 36.8 | No | NA | NA | ||
| Val72Ile | 35.5 | No | NA | NA | ||
| Glu82Gln | 85 | Yes | TCT → TGT | Antisense | ||
| Gln82His | 14.9 | Yes | TCT → TGT | Antisense | ||
| UBK122 | No | Ala72Val | 17.7 | No | NA | NA |
| Glu73Gln | 46.3 | Yes | TCA → TGA | Antisense | ||
| UBK161 | Yes | Gln82His | 22.5 | Yes | TCT → TGT | Antisense |
| UBK177 | No | Glu73Gln | 15.1 | Yes | TCA → TGA | Antisense |
| UBK178 | Yes | Glu73Gln | 13.6 | Yes | TCA → TGA | Antisense |
| Glu82Gln | 88.6 | Yes | TCT → TGT | Antisense | ||
| UBK266 | Yes | Asp60Asn | 31.6 | Yes | TCT → TTT | Antisense |
| UBK282 | Yes | Asp58Asn | 18.9 | No | NA | NA |
| Glu60Gln | 44.1 | Yes | TCT → TGT | Antisense | ||
| Lys69Gln | 45.2 | No | NA | NA | ||
| Val72Ile | 43.1 | No | NA | NA | ||
| Ser78Asn | 10 | No | NA | NA | ||
| Gln82His | 32.9 | Yes | TCT → TGT | Antisense |
NA, not applicable.
Persons with DVG-containing viruses are more likely to have changes to immunosuppressive regimens.
Clinical information was available for 31 individuals with BK viruria, including 22 individuals without any samples containing DVGs and 9 individuals with DVGs identified in at least one clinical sample (Tables 4 and 5). Both the DVG-negative and DVG-containing groups were similar in age (mean, 55 ± 16.3 versus 56 ± 17.7 years) and proportion of females (54.5% and 55.6%), respectively. The clinical setting under which all individuals developed BK viruria was related to immunosuppression secondary to organ transplant. Renal transplant was the most common indication for both groups, with peripheral blood stem cell and multiorgan (kidney and pancreas or kidney and liver) transplants representing a smaller percentage. Comparison of immunosuppressive regimens between DVG groups demonstrated that DVG-containing individuals more often had their dose of mycophenolate stopped or changed to leflunomide. Changes in therapy for the DVG-containing group are perhaps related to their higher viral loads.
TABLE 4.
Clinical characteristics of the individuals in this study
| Characteristic | DVG (–) | DVG (+) |
|---|---|---|
| No. of patients | 22 | 9 |
| Age (yrs), mean (SD) | 55 (16.3) | 56 (17.7) |
| Sex | ||
| Female (n) | 12 | 5 |
| Male (n) | 10 | 4 |
| Diagnosis | ||
| End-stage renal disease (n) | 19 | 8 |
| Hematological malignancy (n) | 3 | 1 |
| Organ transplant | ||
| Kidney (n) | 15 | 8 |
| Peripheral blood stem cell (n) | 3 | 1 |
| Kidney and pancreas (n) | 3 | 0 |
| Kidney and liver (n) | 1 | 0 |
| Change in management | ||
| No change (n) | 10 | 2 |
| Reduce mycophenolate (n) | 4 | 0 |
| Stop mycophenolate (n) | 6 | 3 |
| Change to leflunomide (n) | 2 | 4 |
| Received intravenous immunoglobulin (n) | 3 | 2 |
TABLE 5.
Laboratory and pathology information by DVG status
| Biopsy | DVG (–) |
DVG (+) |
||||
|---|---|---|---|---|---|---|
| Pos | Neg | NRa | Pos | Neg | NRa | |
| Evidence of virus (SV40 IHC), n | 2 | 3 | 17 | 1 | 4 | 4 |
| Evidence of rejection, n | 2 | 6 | 14 | 1 | 6 | 2 |
| Serology | ||||||
| Recipient CMV, n | 13 | 8 | 1 | 7 | 2 | 0 |
| Donor CMV, n | 11 | 9 | 2 | 1 | 8 | 0 |
| Recipient EBV, n | 17 | 3 | 2 | 8 | 0 | 1 |
Pos, positive; neg, negative; NR, not reported;
DVG-containing individuals were more likely to undergo renal biopsy (7 of 9 versus 8 of 22). However, histologic evidence of kidney injury or immunohistochemical staining for the large T antigen was rarely seen (1 and 4 individuals, respectively). Pretransplant serologies demonstrated the majority of individuals in both groups to be cytomegalovirus (CMV) and Epstein-Barr virus (EBV) positive. In the DVG-containing group, individuals more often received organs from donors who were CMV-negative.
DISCUSSION
Here, we report the presence of polyomavirus defective genome segments in clinical specimens and observe the diversity of DVG populations naturally generated across individuals and within a host over the course of infection. Specifically, by deep sequencing, we identified multiple BKPyV strains and one JCPyV strain that contained large deletions or rearrangement junctions in their genomes. Although these junctions have been observed in all six gene segments, most were present in the large T antigen, which is consistent with reports from other viruses in which DVGs most commonly occur in replication-related genes (13). These observations also are consistent with the characteristic features of the polyomavirus DVGs found in culture—major internal deletions with retention of regions that are essential for genome packaging (4, 13, 24).
DVG formation is thought to be a form of viral parasitism, which occurs when multiple viruses infect the same cell (4, 5, 13, 24). Our finding that DVGs were more likely to be present when polyomavirus was present at high copy numbers is consistent with this model. A dramatic increase in viral load was temporally also associated with the only longitudinal specimens in which DVGs newly arose. Alternatively, DVGs may enhance the pathogenicity of the virus by assisting the virus to evade the host’s immune response, as is observed with the Turnip crinkle virus (2, 3). Consistent with this hypothesis, we observed a higher frequency of unfixed mutations in the BC loop of the capsid protein VP1 in DVG-containing BKPyV strains than in those strains without DVGs. Interestingly, many of these BC loop mutations could have arisen through APOBEC3B-associated mutagenesis, which has been speculated to produce antibody escape mutations in BKPyV (36, 38, 39). These unfixed mutations in DVGs may provide the virus with unique and mosaic capsids that are not targeted by the host’s existing set of polyomavirus-directed neutralizing antibodies. In addition, the higher frequency of potential APOBEC3B-associated mutations in DVG-containing strains may suggest that APOBEC3B activity could be driving the formation of DVGs. Alternatively, the bidirectional replication mechanism of polyomaviruses combined with decatenation of linked circular genomes would provide several opportunities for recombination and generation of deletions and other large-scale rearrangements (14, 40, 41).
We also examined the evolution of BKPyV in a small subset of individuals that had longitudinal specimens available. The recovery of 1 to 2 SNVs in specimens taken from persons with longitudinal samples spaced between 1 month and 1 year apart is consistent with the relatively high intrahost evolutionary rate of polyomaviruses, previously measured at 4.9 × 10−4 to 1.2 × 10−3 substitutions per site per year (42). Given that we only detected one newly generated DVG in this study, more work is required to uncover the determinants and rate of DVG generation in polyomaviruses.
One main limitation of our sequencing approach was the use of short-read sequencing, which may have affected our measurements of DVGs in each specimen and did not allow us to link rearrangements across the multiple populations of virus present within the viral population. Recent work in polyomaviruses has demonstrated the promise of long-read sequencing to resolve complex populations of polyomaviruses (43, 44). However, the limited total DNA contained in these samples complicates the use of long-read approaches given the large-scale rearrangements recovered and requirement for multiple cycles of PCR. We also quantitated rearrangements as a percentage of maximum coverage, which may bias quantitation depending on local variations in copy number. We bulwarked our sequencing approach by using multiple library preparations of the same sample to illustrate that deletions and rearrangements recovered were not due to library generation.
Of note, it has yet to be established whether these DVGs identified from the clinical specimens interfere with the replication of the full-genome virus in vitro, and the role of DVGs in natural polyomavirus infections has yet to be addressed. The deletions and rearrangements recovered in our study were generally quite large and affected proteins and domains required for viral replication, consistent with their defective nomenclature. Further studies focusing on the function of specific rearrangements found in polyomavirus DVGs through reverse genetics and cell culture-based approaches are required to help understand their role in replication interference.
In conclusion, by demonstrating the existence of polyomavirus DVGs in human specimens, we extend this broad property of viral evolution to DNA viruses as they exist in human hosts. The polyomaviruses containing DVGs recovered in this study generally had significantly higher copy number of capsid genes to replication genes. Further work is required to determine whether these genetic copy alterations result in differences in protein levels in human samples that could potentially affect pharmacodynamic properties and efficacy profiles of capsid-directed therapies for polyomaviruses.
MATERIALS AND METHODS
Clinical testing and next-generation sequencing.
This work was approved by the University of Washington Institutional Review Board (STUDY00000408). Urine samples were collected from individuals suspected to have a BKPyV or JCPyV infection, and clinical testing was performed at the University of Washington Virology Laboratory using the previously described quantitative PCR assays (16, 45).
BKPyV- or JCPyV-positive urine samples were first filtered using a 0.22-μM filter, and DNA was extracted from these samples using the Quick-DNA viral kit (Zymo). Sequencing libraries were constructed from 2 μl of DNA using the Nextera XT kit (Illumina) and cleaned with 0.6× volumes of AMPure XP beads (Beckman Coulter). For four samples, a second sequencing library was created using the KAPA HyperPrep Plus kit (Roche) and purified with 0.8× volumes of AMPure XP beads. The resulting libraries were then sequenced on 1 × 192-bp or 2 × 300-bp Illumina MiSeq runs.
Identification of defective viral genomes.
Sequencing reads were adapter- and quality-trimmed using Trimmomatic v0.38 (46) and mapped to the BKPyV (NC_001538.1) or JCPyV (NC_001699.1) reference genome. Consensus genomes were manually called, and sequencing reads were then mapped to the respective consensus genome in Geneious Prime (47), with the structural variant, insertion, and deletion detection setting enabled. Because of the high proportion of small deletions contained around the origin and upstream of the agnoprotein, those junctions entirely contained between nucleotides 0 to 300 were discarded and not included in downstream further analysis. Each of the junctions reported here was supported by a minimum of 10 sequencing reads. In addition, we confirmed the presence of the highest-abundance junction identified in each sample using DI-tector (48).
Next, we confirmed the presence of these junctions in a subset of these samples through PCR and Sanger sequencing. PCR amplification was performed using the CloneAmp HiFi Premix (TaKaRa) and the primers listed in Table 6 under the following conditions: hold at 98°C for 2 min, followed by 35 cycles of 98°C for 10 s, 61°C for 15 s and 72°C for 30 s, followed by a final extension at 72°C for 5 min. PCR products were run on a 1.5% agarose gel and extracted using the QIAquick gel extraction kit (Qiagen). Sanger sequencing was performed with the primers listed in Table 6 by Genewiz, Inc.
TABLE 6.
Primer and probe sequences used in this study
| Purpose | Name | Sequence (5′ → 3′) |
|---|---|---|
| For Sanger sequencing | BK-DIP-F2 | GATGGGCAGCCTATGTATGG |
| BK-DIP-R2 | GCAATGGTGGGTCCAAATAATTG | |
| BK62-DIP-1 | GGCTGAAGTATCTGAGACTTGG | |
| BK62-DIP-2 | CTTGCCTGCTTTGCTGTGTAT | |
| For ddPCR | BKV_VP_Fwd | GCCCCAGGAGGTGCTAATC |
| BKV_VP_Rev | CAGGCCTAGAAGTAAAGGCAACA | |
| BKV_VP_Probe | Fam_AGAACTGCTCCTCAATG-MGB | |
| BKV_largeT_Fwd | TTATCTCAGAATCCAGCCTTTCCT | |
| BKV_largeT_Rev | GGCCTGTAGCTGATTTTGCAA | |
| BKV_largeT_Probe | Vic_CCATTCAACAATTCTAG-MGB | |
| JCV_VP_Fwd | CACAGAGCACAAGGCGTACC | |
| JCV_VP_Rev | AAGCAACACTGTTGTGGCAG | |
| JCV_VP_Probe | Fam_TTCCTGATCCCACC_FQ | |
| JCV_LargeT_Fwd | CCAGTGCCTTTTACATCCTC | |
| JCV_LargeT_Rev | GGCCAATAGACAGTGGCAA | |
| JCV_LargeT_Probe | Hex_ATCAAGTAAAGCTGCAGCT_FQ |
Droplet digital PCR detection of defective viral genomes.
ddPCR was performed using the Bio-Rad QX100 system (Bio-Rad, Hercules, CA, USA) and QuantaSoft for data analysis. There are two sets of primers and probes targeting the VP1 and large T antigen regions, respectively (Table 6). Each reaction was performed with Bio-Rad ddPCR supermix for probes with the final concentration of primers at 900 nM and probes at 250 nM and 25 units of HindIII (New England Biolabs). Plasmid BK Dunlop and JC Mad-1 were gifts from Peter Howley (Addgene plasmids no. 25466 and no. 25626) and were used as positive controls for 1:1 VP1/large T-antigen copy number. After droplet generation, droplets were transferred to a 96-well PCR plate and amplified on a 2720 thermal cycler (Applied Biosystems) with the following thermocycling parameters: 94°C for 10 min, followed by 40 cycles of 94°C for 30 s and 60°C for 1 min, and 98°C hold for 10 min. After the thermal cycling, the plate was transferred to a droplet reader. The QuantaSoft software was used for data analysis.
Phylogenetic, metagenomic, and unfixed variant analysis.
We downloaded all 510 complete BKPyV genomes from NCBI GenBank (accessed 16 August 2020) and removed any duplicate genomes from these data set to obtain 402 unique BKPyV genomes. These 402 BKPyV genomes were then classified into the 11 VP1 sequence subtypes and subgroups with BKTyper 1.0 (49). We next randomly selected 10 genomes from each of these subtypes or subgroups for inclusion in our phylogenetic analysis. The 106 representative BKPyV genomes and the 38 complete BKPyV genomes obtained in this study were aligned with MAFFT v7.429 (50). A phylogenetic tree was generated from this alignment using RaxML v8.2.11 (51) and visualized with ggtree (52).
To perform the JCPyV phylogenetic tree, we downloaded all 696 complete JCPyV genomes from NCBI GenBank (accessed 16 January 2021) and removed any duplicate genomes to obtain 565 unique JCPyV genomes. These genomes were then classified into the previously defined JCPyV subtypes (53) based on their VP1 sequence using a custom Python script. We then randomly selected genomes from each subtype. An alignment and a phylogenetic tree were generated as described above with these 64 representative JCPyV genomes and the 11 JCPyV genomes recovered in this study.
Metagenomic analysis was performed as previously described (54), using the metagenomic classifier CLOMP (https://github.com/rcs333/CLOMP). Counts were normalized between samples, and classifications were expressed as reads per million (RPM). Taxonomic classifications of each read were assigned to the most specific NCBI taxonomy ID possible. As such, reads aligning to two or more reference genomes within a taxonomical classification were assigned to the next lowest taxonomical category. Any reads assigned to environmental or artificial sequences were discarded. Reads that matched equally well to two or more different domains were categorized as “unclassified.”
Unfixed variants in the BC loop of the VP1 (amino acids 57 to 89 in the BKPyV reference genome [NC_001538.1]) were detected by mapping sequencing reads to the consensus BKPyV genome obtained for each sample in Geneious Prime (38, 47). A minimum sequencing depth of 10× and allele frequency of greater than 10% were used for calling variants. We considered unfixed variants to have an allele frequency less than 90%.
Data availability.
Sequencing reads and consensus genomes are available under NCBI BioProject PRJNA657423; GenBank accession numbers MW587957 to MW587964, MW587966 to MW587994, and MW588006; and Sequence Read Archive accession numbers SRR13239807 to SRR13239811, SRR13239816 to SRR13239829, SRR13239831 to SRR13239851, SRR13239853 to SRR13239855, SRR13680575, and SRR13680576 (see Table 7).
TABLE 7.
Accession numbers for samples sequenced in this studya
| Sample ID | BKPyV Genome ID | BKPyV GenBank accession no. | JCPyV genome ID | JCPyV GenBank accession no. | Library prepn method 1 | SRA accession no. 1 | Library prepn method 2 | SRA accession no. 2 |
|---|---|---|---|---|---|---|---|---|
| UBK007 | UBK007 | MW587957 | NA | NA | Nextera XT | SRR13239855 | KAPA HyperPrep Plus | SRR13239815 |
| UBK016 | NA | NA | UJC016 | MW587997 | Nextera XT | SRR13239854 | ||
| UBK024 | UBK024 | MW587958 | NA | NA | Nextera XT | SRR13239843 | ||
| UBK028 | NA | NA | UJC028 | MW587998 | Nextera XT | SRR13239832 | ||
| UBK029 | UBK029 | MW587959 | NA | NA | Nextera XT | SRR13239821 | ||
| UBK034 | UBK034 | MW587960 | NA | NA | Nextera XT | SRR13239811 | ||
| UBK035 | UBK035 | MW588006 | NA | NA | Nextera XT | SRR13239810 | ||
| UBK040 | UBK040 | MW587961 | NA | NA | Nextera XT | SRR13239809 | ||
| UBK054 | UBK054 | MW587962 | NA | NA | Nextera XT | SRR13239808 | ||
| UBK062 | UBK062 | MW587963 | NA | NA | Nextera XT | SRR13239807 | ||
| UBK064 | UBK064 | MW587964 | NA | NA | Nextera XT | SRR13239853 | ||
| UBK076 | UBK076 | MW587966 | NA | NA | Nextera XT | SRR13239851 | ||
| UBK077 | UBK077 | MW587967 | UJC077 | MW587999 | Nextera XT | SRR13239850 | ||
| UBK086 | UBK086 | MW587968 | NA | NA | Nextera XT | SRR13239849 | KAPA HyperPrep Plus | SRR13239814 |
| UBK088 | UBK088 | MW587969 | NA | NA | Nextera XT | SRR13239848 | ||
| UBK090 | NA | NA | UJC090 | MW588000 | Nextera XT | SRR13239847 | ||
| UBK091 | NA | NA | UJC091 | MW588001 | Nextera XT | SRR13239846 | ||
| UBK094 | UBK094 | MW587970 | NA | NA | Nextera XT | SRR13239845 | KAPA HyperPrep Plus | SRR13239813 |
| UBK096 | UBK096 | MW587971 | NA | NA | Nextera XT | SRR13239844 | KAPA HyperPrep Plus | SRR13239812 |
| UBK121 | UBK121 | MW587972 | NA | NA | Nextera XT | SRR13239842 | ||
| UBK122 | UBK122 | MW587973 | NA | NA | Nextera XT | SRR13239841 | ||
| UBK140 | UBK140 | MW587974 | NA | NA | Nextera XT | SRR13239840 | ||
| UBK143 | UBK143 | MW587975 | NA | NA | Nextera XT | SRR13239839 | ||
| UBK146 | UBK146 | MW587976 | NA | NA | Nextera XT | SRR13239838 | ||
| UBK161 | UBK161 | MW587977 | NA | NA | Nextera XT | SRR13239837 | ||
| UBK174 | UBK174 | MW587978 | NA | NA | Nextera XT | SRR13239836 | ||
| UBK177 | UBK177 | MW587979 | NA | NA | Nextera XT | SRR13239835 | ||
| UBK178 | UBK178 | MW587980 | NA | NA | Nextera XT | SRR13239834 | ||
| UBK183 | NA | NA | UJC183 | MW588002 | Nextera XT | SRR13239833 | ||
| UBK187 | UBK187 | MW587981 | NA | NA | Nextera XT | SRR13239831 | ||
| UBK245 | UBK245 | MW587982 | NA | NA | Nextera XT | SRR13239829 | ||
| UBK263 | UBK263 | MW587983 | NA | NA | Nextera XT | SRR13239828 | ||
| UBK264 | UBK264 | MW587984 | NA | NA | Nextera XT | SRR13239827 | ||
| UBK265 | UBK265 | MW587985 | NA | NA | Nextera XT | SRR13239826 | ||
| UBK266 | UBK266 | MW587986 | NA | NA | Nextera XT | SRR13239825 | ||
| UBK272 | UBK272 | MW587987 | NA | NA | Nextera XT | SRR13239824 | ||
| UBK279 | UBK279 | MW587988 | NA | NA | Nextera XT | SRR13239823 | ||
| UBK280 | UBK280 | MW587989 | NA | NA | Nextera XT | SRR13239822 | ||
| UBK282 | UBK282 | MW587990 | NA | NA | Nextera XT | SRR13239820 | ||
| UBK289 | UBK289 | MW587991 | UJC289 | MW588003 | Nextera XT | SRR13239819 | ||
| UBK292 | UBK292 | MW587992 | UJC292 | MW588004 | Nextera XT | SRR13239818 | ||
| UBK294 | UBK294 | MW587993 | NA | NA | Nextera XT | SRR13239817 | ||
| UBK295 | UBK295 | MW587994 | UJC295 | MW588005 | Nextera XT | SRR13239816 | ||
| UJC004 | NA | NA | UJC004 | MW587995 | Nextera XT | SRR13680576 | ||
| UJC005 | NA | NA | UJC005 | MW587996 | Nextera XT | SRR13680575 |
NA, not applicable.
Footnotes
Supplemental material is available online only.
jvi.00250-21-s0001.pdf (983.4KB, pdf)
Contributor Information
Alexander L. Greninger, Email: agrening@uw.edu.
Lawrence Banks, International Centre for Genetic Engineering and Biotechnology.
REFERENCES
- 1.Vignuzzi M, López CB. 2019. Defective viral genomes are key drivers of the virus-host interaction. Nat Microbiol 4:1075–1087. 10.1038/s41564-019-0465-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Simon AE, Roossinck MJ, Havelda Z. 2004. Plant virus satellite and defective interfering RNAs: new paradigms for a new century. Annu Rev Phytopathol 42:415–437. 10.1146/annurev.phyto.42.040803.140402. [DOI] [PubMed] [Google Scholar]
- 3.Roux L, Simon AE, Holland JJ. 1991. Effects of defective interfering viruses on virus replication and pathogenesis in vitro and in vivo, p 181–211. In Maramorosch K, Murphy FA, Shatkin AJ (ed), Advances in virus research. Academic Press, San Diego, CA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Genoyer E, López CB. 2019. The impact of defective viruses on infection and immunity. Annu Rev Virol 6:547–566. 10.1146/annurev-virology-092818-015652. [DOI] [PubMed] [Google Scholar]
- 5.López CB. 2019. Unexpected lessons from the neglected: how defective viral genomes became important again. PLoS Pathog 15:e1007450. 10.1371/journal.ppat.1007450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davis AR, Hiti AL, Nayak DP. 1980. Influenza defective interfering viral RNA is formed by internal deletion of genomic RNA. Proc Natl Acad Sci U S A 77:215–219. 10.1073/pnas.77.1.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Perrault J, Semler BL. 1979. Internal genome deletions in two distinct classes of defective interfering particles of vesicular stomatitis virus. Proc Natl Acad Sci U S A 76:6191–6195. 10.1073/pnas.76.12.6191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dimmock NJ, Easton AJ. 2014. Defective interfering influenza virus RNAs: time to reevaluate their clinical potential as broad-spectrum antivirals? J Virol 88:5217–5227. 10.1128/JVI.03193-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nichol ST, O’Hara PJ, Holland JJ, Perrault J. 1984. Structure and origin of a novel class of defective interfering particle of vesicular stomatitis virus. Nucleic Acids Res 12:2775–2790. 10.1093/nar/12.6.2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leung AY, Suen CK, Lie AK, Liang RH, Yuen KY, Kwong YL. 2001. Quantification of polyoma BK viruria in hemorrhagic cystitis complicating bone marrow transplantation. Blood 98:1971–1978. 10.1182/blood.V98.6.1971. [DOI] [PubMed] [Google Scholar]
- 11.Tan CS, Koralnik IJ. 2010. Progressive multifocal leukoencephalopathy and other disorders caused by JC virus: clinical features and pathogenesis. Lancet Neurol 9:425–437. 10.1016/S1474-4422(10)70040-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berger JR, Danaher RJ, Dobbins J, Do D, Miller CS. 2020. Dynamic expression of JC virus in urine and its relationship to serostatus. Mult Scler Relat Disord 41:101972. 10.1016/j.msard.2020.101972. [DOI] [PubMed] [Google Scholar]
- 13.Genoyer E, López CB. 2019. Defective viral genomes alter how Sendai virus interacts with cellular trafficking machinery, leading to heterogeneity in the production of viral particles among infected cells. J Virol 93:e01579-18. 10.1128/JVI.01579-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sowd GA, Li NY, Fanning E. 2013. ATM and ATR activities maintain replication fork integrity during SV40 chromatin replication. PLoS Pathog 9:e1003283. 10.1371/journal.ppat.1003283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O’Neill FJ, Maryon EB, Carroll D. 1982. Isolation and characterization of defective simian virus 40 genomes which complement for infectivity. J Virol 43:18–25. 10.1128/JVI.43.1.18-25.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Greninger AL, Bateman AC, Atienza EE, Wendt S, Makhsous N, Jerome KR, Cook L. 2017. Copy number heterogeneity of JC virus standards. J Clin Microbiol 55:824–831. 10.1128/JCM.02337-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bateman AC, Greninger AL, Atienza EE, Limaye AP, Jerome KR, Cook L. 2017. Quantification of BK virus standards by quantitative real-time PCR and droplet digital PCR is confounded by multiple virus populations in the WHO BKV international standard. Clin Chem 63:761–769. 10.1373/clinchem.2016.265512. [DOI] [PubMed] [Google Scholar]
- 18.Shenk TE, Carbon J, Berg P. 1976. Construction and analysis of viable deletion mutants of simian virus 40. J Virol 18:664–671. 10.1128/JVI.18.2.664-671.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mertz JE, Berg P. 1974. Viable deletion mutants of simian virus 40: selective isolation by means of a restriction endonuclease from Hemophilus parainfluenzae. Proc Natl Acad Sci U S A 71:4879–4883. 10.1073/pnas.71.12.4879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mertz JE, Carbon J, Herzberg M, Davis RW, Berg P. 1975. Isolation and characterization of individual clones of simian virus 40 mutants containing deletions duplications and insertions in their DNA. Cold Spring Harbor Symp Quant Biol 39 Pt 1:69–84. 10.1101/sqb.1974.039.01.012. [DOI] [PubMed] [Google Scholar]
- 21.Tai HT, Smith CA, Sharp PA, Vinograd J. 1972. Sequence heterogeneity in closed simian virus 40 deoxyribonucleic acid. J Virol 9:317–325. 10.1128/JVI.9.2.317-325.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ziegler CM, Botten JW. 2020. Defective interfering particles of negative-strand RNA viruses. Trends Microbiol 28:554–565. 10.1016/j.tim.2020.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alnaji FG, Brooke CB. 2020. Influenza virus DI particles: defective interfering or delightfully interesting? PLoS Pathog 16:e1008436. 10.1371/journal.ppat.1008436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Manzoni TB, López CB. 2018. Defective (interfering) viral genomes re-explored: impact on antiviral immunity and virus persistence. Future Virol 13:493–503. 10.2217/fvl-2018-0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boothpur R, Brennan DC. 2010. Human polyoma viruses and disease with emphasis on clinical BK and JC. J Clin Virol 47:306–312. 10.1016/j.jcv.2009.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pinto M, Dobson S. 2014. BK and JC virus: a review. J Infect 68(Suppl 1):S2–S8. 10.1016/j.jinf.2013.09.009. [DOI] [PubMed] [Google Scholar]
- 27.Taguchi F, Kajioka J, Miyamura T. 1982. Prevalence rate and age of acquisition of antibodies against JC virus and BK virus in human sera. Microbiol Immunol 26:1057–1064. 10.1111/j.1348-0421.1982.tb00254.x. [DOI] [PubMed] [Google Scholar]
- 28.Brown P, Tsai T, Gajdusek DC. 1975. Seroepidemiology of human papovaviruses. Discovery of virgin populations and some unusual patterns of antibody prevalence among remote peoples of the world. Am J Epidemiol 102:331–340. 10.1093/oxfordjournals.aje.a112169. [DOI] [PubMed] [Google Scholar]
- 29.Hirsch HH. 2005. BK virus: opportunity makes a pathogen. Clin Infect Dis 41:354–360. 10.1086/431488. [DOI] [PubMed] [Google Scholar]
- 30.Reploeg MD, Storch GA, Clifford DB. 2001. BK virus: a clinical review. Clin Infect Dis 33:191–202. 10.1086/321813. [DOI] [PubMed] [Google Scholar]
- 31.Starrett GJ, Buck CB. 2019. The case for BK polyomavirus as a cause of bladder cancer. Curr Opin Virol 39:8–15. 10.1016/j.coviro.2019.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Robles MTS, Cantalupo PG, Duray AM, Freeland M, Murkowski M, van Bokhoven A, Stephens-Shields AJ, Pipas JM, Imperiale MJ. 2020. Analysis of viruses present in urine from patients with interstitial cystitis. Virus Genes 56:430–438. 10.1007/s11262-020-01767-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhong S, Randhawa PS, Ikegaya H, Chen Q, Zheng H-Y, Suzuki M, Takeuchi T, Shibuya A, Kitamura T, Yogo Y. 2009. Distribution patterns of BK polyomavirus (BKV) subtypes and subgroups in American, European and Asian populations suggest co-migration of BKV and the human race. J Gen Virol 90:144–152. 10.1099/vir.0.83611-0. [DOI] [PubMed] [Google Scholar]
- 34.Verhalen B, Starrett GJ, Harris RS, Jiang M. 2016. Functional upregulation of the DNA cytosine deaminase APOBEC3B by polyomaviruses. J Virol 90:6379–6386. 10.1128/JVI.00771-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zou J, Wang C, Ma X, Wang E, Peng G. 2017. APOBEC3B, a molecular driver of mutagenesis in human cancers. Cell Biosci 7:29. 10.1186/s13578-017-0156-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peretti A, Geoghegan EM, Pastrana DV, Smola S, Feld P, Sauter M, Lohse S, Ramesh M, Lim ES, Wang D, Borgogna C, FitzGerald PC, Bliskovsky V, Starrett GJ, Law EK, Harris RS, Killian JK, Zhu J, Pineda M, Meltzer PS, Boldorini R, Gariglio M, Buck CB. 2018. Characterization of BK polyomaviruses from kidney transplant recipients suggests a role for APOBEC3 in driving in-host virus evolution. Cell Host Microbe 23:628–635.e7. 10.1016/j.chom.2018.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Burns MB, Temiz NA, Harris RS. 2013. Evidence for APOBEC3B mutagenesis in multiple human cancers. Nat Genet 45:977–983. 10.1038/ng.2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dugan AS, Gasparovic ML, Tsomaia N, Mierke DF, O’Hara BA, Manley K, Atwood WJ. 2007. Identification of amino acid residues in BK virus VP1 that are critical for viability and growth. J Virol 81:11798–11808. 10.1128/JVI.01316-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pastrana DV, Ray U, Magaldi TG, Schowalter RM, Çuburu N, Buck CB. 2013. BK polyomavirus genotypes represent distinct serotypes with distinct entry tropism. J Virol 87:10105–10113. 10.1128/JVI.01189-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fareed GC, Garon GF, Salzman NP. 1972. Origin and direction of simian virus 40 deoxyribonucleic acid replication. J Virol 10:484–491. 10.1128/JVI.10.3.484-491.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Danna KJ, Nathans D. 1972. Bidirectional replication of Simian virus 40 DNA. Proc Natl Acad Sci U S A 69:3097–3100. 10.1073/pnas.69.11.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Domingo-Calap P, Schubert B, Joly M, Solis M, Untrau M, Carapito R, Georgel P, Caillard S, Fafi-Kremer S, Paul N, Kohlbacher O, González-Candelas F, Bahram S. 2018. An unusually high substitution rate in transplant-associated BK polyomavirus in vivo is further concentrated in HLA-C-bound viral peptides. PLoS Pathog 14:e1007368. 10.1371/journal.ppat.1007368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Seppälä H, Virtanen E, Saarela M, Laine P, Paulín L, Mannonen L, Auvinen P, Auvinen E. 2017. Single-molecule sequencing revealing the presence of distinct JC polyomavirus populations in patients with progressive multifocal leukoencephalopathy. J Infect Dis 215:889–895. 10.1093/infdis/jiw399. [DOI] [PubMed] [Google Scholar]
- 44.Czech-Sioli M, Günther T, Therre M, Spohn M, Indenbirken D, Theiss J, Riethdorf S, Qi M, Alawi M, Wülbeck C, Fernandez-Cuesta I, Esmek F, Becker JC, Grundhoff A, Fischer N. 2020. High-resolution analysis of Merkel cell polyomavirus in Merkel cell carcinoma reveals distinct integration patterns and suggests NHEJ and MMBIR as underlying mechanisms. PLoS Pathog 16:e1008562. 10.1371/journal.ppat.1008562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hoffman NG, Cook L, Atienza EE, Limaye AP, Jerome KR. 2008. Marked variability of BK virus load measurement using quantitative real-time PCR among commonly used assays. J Clin Microbiol 46:2671–2680. 10.1128/JCM.00258-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Beauclair G, Mura M, Combredet C, Tangy F, Jouvenet N, Komarova AV. 2018. DI-tector: defective interfering viral genomes’ detector for next-generation sequencing data. RNA 24:1285–1296. 10.1261/rna.066910.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martí-Carreras J, Mineeva-Sangwo O, Topalis D, Snoeck R, Andrei G, Maes P. 2020. BKTyper: free online tool for polyoma BK virus VP1 and NCCR typing. Viruses 12:837. 10.3390/v12080837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Katoh K, Kuma K, Toh H, Miyata T. 2005. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res 33:511–518. 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yu G. 2020. Using ggtree to visualize data on tree-like structures. Curr Protoc Bioinformatics 69:e96. 10.1002/cpbi.96. [DOI] [PubMed] [Google Scholar]
- 53.Cubitt CL, Cui X, Agostini HT, Nerurkar VR, Scheirich I, Yanagihara R, Ryschkewitsch CF, Stoner GL. 2001. Predicted amino acid sequences for 100 JCV strains. J Neurovirol 7:339–344. 10.1080/13550280152537201. [DOI] [PubMed] [Google Scholar]
- 54.Peddu V, Shean RC, Xie H, Shrestha L, Perchetti GA, Minot SS, Roychoudhury P, Huang M-L, Nalla A, Reddy SB, Phung Q, Reinhardt A, Jerome KR, Greninger AL. 2020. Metagenomic analysis reveals clinical SARS-CoV-2 infection and bacterial or viral superinfection and colonization. Clin Chem 66:966–972. 10.1093/clinchem/hvaa106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Sequencing reads and consensus genomes are available under NCBI BioProject PRJNA657423; GenBank accession numbers MW587957 to MW587964, MW587966 to MW587994, and MW588006; and Sequence Read Archive accession numbers SRR13239807 to SRR13239811, SRR13239816 to SRR13239829, SRR13239831 to SRR13239851, SRR13239853 to SRR13239855, SRR13680575, and SRR13680576 (see Table 7).
TABLE 7.
Accession numbers for samples sequenced in this studya
| Sample ID | BKPyV Genome ID | BKPyV GenBank accession no. | JCPyV genome ID | JCPyV GenBank accession no. | Library prepn method 1 | SRA accession no. 1 | Library prepn method 2 | SRA accession no. 2 |
|---|---|---|---|---|---|---|---|---|
| UBK007 | UBK007 | MW587957 | NA | NA | Nextera XT | SRR13239855 | KAPA HyperPrep Plus | SRR13239815 |
| UBK016 | NA | NA | UJC016 | MW587997 | Nextera XT | SRR13239854 | ||
| UBK024 | UBK024 | MW587958 | NA | NA | Nextera XT | SRR13239843 | ||
| UBK028 | NA | NA | UJC028 | MW587998 | Nextera XT | SRR13239832 | ||
| UBK029 | UBK029 | MW587959 | NA | NA | Nextera XT | SRR13239821 | ||
| UBK034 | UBK034 | MW587960 | NA | NA | Nextera XT | SRR13239811 | ||
| UBK035 | UBK035 | MW588006 | NA | NA | Nextera XT | SRR13239810 | ||
| UBK040 | UBK040 | MW587961 | NA | NA | Nextera XT | SRR13239809 | ||
| UBK054 | UBK054 | MW587962 | NA | NA | Nextera XT | SRR13239808 | ||
| UBK062 | UBK062 | MW587963 | NA | NA | Nextera XT | SRR13239807 | ||
| UBK064 | UBK064 | MW587964 | NA | NA | Nextera XT | SRR13239853 | ||
| UBK076 | UBK076 | MW587966 | NA | NA | Nextera XT | SRR13239851 | ||
| UBK077 | UBK077 | MW587967 | UJC077 | MW587999 | Nextera XT | SRR13239850 | ||
| UBK086 | UBK086 | MW587968 | NA | NA | Nextera XT | SRR13239849 | KAPA HyperPrep Plus | SRR13239814 |
| UBK088 | UBK088 | MW587969 | NA | NA | Nextera XT | SRR13239848 | ||
| UBK090 | NA | NA | UJC090 | MW588000 | Nextera XT | SRR13239847 | ||
| UBK091 | NA | NA | UJC091 | MW588001 | Nextera XT | SRR13239846 | ||
| UBK094 | UBK094 | MW587970 | NA | NA | Nextera XT | SRR13239845 | KAPA HyperPrep Plus | SRR13239813 |
| UBK096 | UBK096 | MW587971 | NA | NA | Nextera XT | SRR13239844 | KAPA HyperPrep Plus | SRR13239812 |
| UBK121 | UBK121 | MW587972 | NA | NA | Nextera XT | SRR13239842 | ||
| UBK122 | UBK122 | MW587973 | NA | NA | Nextera XT | SRR13239841 | ||
| UBK140 | UBK140 | MW587974 | NA | NA | Nextera XT | SRR13239840 | ||
| UBK143 | UBK143 | MW587975 | NA | NA | Nextera XT | SRR13239839 | ||
| UBK146 | UBK146 | MW587976 | NA | NA | Nextera XT | SRR13239838 | ||
| UBK161 | UBK161 | MW587977 | NA | NA | Nextera XT | SRR13239837 | ||
| UBK174 | UBK174 | MW587978 | NA | NA | Nextera XT | SRR13239836 | ||
| UBK177 | UBK177 | MW587979 | NA | NA | Nextera XT | SRR13239835 | ||
| UBK178 | UBK178 | MW587980 | NA | NA | Nextera XT | SRR13239834 | ||
| UBK183 | NA | NA | UJC183 | MW588002 | Nextera XT | SRR13239833 | ||
| UBK187 | UBK187 | MW587981 | NA | NA | Nextera XT | SRR13239831 | ||
| UBK245 | UBK245 | MW587982 | NA | NA | Nextera XT | SRR13239829 | ||
| UBK263 | UBK263 | MW587983 | NA | NA | Nextera XT | SRR13239828 | ||
| UBK264 | UBK264 | MW587984 | NA | NA | Nextera XT | SRR13239827 | ||
| UBK265 | UBK265 | MW587985 | NA | NA | Nextera XT | SRR13239826 | ||
| UBK266 | UBK266 | MW587986 | NA | NA | Nextera XT | SRR13239825 | ||
| UBK272 | UBK272 | MW587987 | NA | NA | Nextera XT | SRR13239824 | ||
| UBK279 | UBK279 | MW587988 | NA | NA | Nextera XT | SRR13239823 | ||
| UBK280 | UBK280 | MW587989 | NA | NA | Nextera XT | SRR13239822 | ||
| UBK282 | UBK282 | MW587990 | NA | NA | Nextera XT | SRR13239820 | ||
| UBK289 | UBK289 | MW587991 | UJC289 | MW588003 | Nextera XT | SRR13239819 | ||
| UBK292 | UBK292 | MW587992 | UJC292 | MW588004 | Nextera XT | SRR13239818 | ||
| UBK294 | UBK294 | MW587993 | NA | NA | Nextera XT | SRR13239817 | ||
| UBK295 | UBK295 | MW587994 | UJC295 | MW588005 | Nextera XT | SRR13239816 | ||
| UJC004 | NA | NA | UJC004 | MW587995 | Nextera XT | SRR13680576 | ||
| UJC005 | NA | NA | UJC005 | MW587996 | Nextera XT | SRR13680575 |
NA, not applicable.