ABSTRACT
Ehrlichia chaffeensis causes human monocytic ehrlichiosis by replicating within phagosomes of monocytes/macrophages. A function disruption mutation within the pathogen’s ECH_0660 gene, which encodes a phage head-to-tail connector protein, resulted in the rapid clearance of the pathogen in vivo, while aiding in induction of sufficient immunity in a host to protect against wild-type infection challenge. In this study, we describe the characterization of a cluster of seven genes spanning from ECH_0659 to ECH_0665, which contained four genes encoding bacterial phage proteins, including the ECH_0660 gene. Assessment of the promoter region upstream of the first gene of the seven genes (ECH_0659) in Escherichia coli demonstrated transcriptional enhancement under zinc and iron starvation conditions. Furthermore, transcription of the seven genes was significantly higher under zinc and iron starvation conditions for E. chaffeensis carrying a mutation in the ECH_0660 gene compared to the wild-type pathogen. In contrast, for the ECH_0665 gene mutant with the function disruption, transcription from the genes was mostly similar to that of the wild type or was moderately downregulated. Recently, we reported that this mutation caused a minimal impact on the pathogen’s in vivo growth, as it persisted similarly to the wild type. The current study is the first to describe how zinc and iron contribute to E. chaffeensis biology. Specifically, we demonstrated that the functional disruption in the gene encoding the phage head-to-tail connector protein in E. chaffeensis results in the enhanced transcription of seven genes, including those encoding phage proteins, under zinc and iron limitation.
IMPORTANCE Ehrlichia chaffeensis, a tick-transmitted bacterium, causes human monocytic ehrlichiosis by replicating within phagosomes of monocytes/macrophages. A function disruption mutation within the pathogen’s gene encoding a phage head-to-tail connector protein resulted in the rapid clearance of the pathogen in vivo, while aiding in induction of sufficient immunity in a host to protect against wild-type infection challenge. In the current study, we investigated if the functional disruption in the phage head-to-tail connector protein gene caused transcriptional changes resulting from metal ion limitations. This is the first study describing how zinc and iron may contribute to E. chaffeensis replication.
KEYWORDS: Rickettsiales, Anaplasmataceae pathogens, tick-borne diseases, metal ion deficiency, Ehrlichia chaffeensis, iron, metal ions, zinc
INTRODUCTION
The obligate intracellular rickettsial pathogen Ehrlichia chaffeensis grows within the phagosomes of monocytes/macrophages and causes human monocytic ehrlichiosis (HME), a potentially life-threatening zoonotic disease (1, 2). This pathogen is transmitted primarily from an infected Amblyomma americanum tick (3, 4). Common signs and symptoms of HME range from influenza-like symptoms to a life-threatening disease (5). E. chaffeensis also causes a similar disease in dogs, in addition to infecting white-tailed deer and coyotes (6). Several other obligate tick-transmitted Anaplasmataceae family pathogens, including Ehrlichia ewingii, Ehrlichia canis, Ehrlichia muris subspecies eauclairensis, and Anaplasma phagocytophilium, are also known to cause diseases in people (7–9). These pathogens have reduced genomes and are also evolved for intracellular survival within arthropod hosts and cause persistent infections in vertebrate hosts (6). E. chaffeensis replication within phagosomes inhibits phagosome-lysosome fusion, and the pathogen evades the host immune responses by several mechanisms (10–18). Ehrlichia species and other related Anaplasmataceae pathogens depend on the host for nutritional sources such as metabolites and metal ions for survival (6, 19).
While iron and zinc are two essential metal ions, neither of them is readily accessible within a host for an intracellular pathogen (20). Zinc and iron are also known to play essential roles in host innate and adaptive immune functions (21, 22). During pathogen infection, nutritional immunity can be triggered that involves the host-imposed metal ion deprivation or poisoning to counter infection progression (23, 24). Iron can catalyze the formation of reactive oxygen species (ROS) during macrophage-based bacterial killing by damaging bacterial membranes, proteins, and DNA (25). Bacterial pathogens also employ proficient ways to regulate the acquisition of the metal ions to both support nutritional requirements and to evade metal ion-mediated host response (24). Previous studies reported that E. chaffeensis-containing phagosomes and, similarly, Mycobacterium tuberculosis-containing phagosomes in human monocytes accumulate transferrin receptors (26, 27). E. chaffeensis inclusions are early endosomes, which selectively accumulate transferrin receptors, possibly facilitating the acquisition of iron by the pathogen from the host cell cytoplasm. A recent study described upregulation of iron-regulated genes under iron starvation conditions in a related bacterium, Ehrlichia ruminantium, by a predicted master regulatory protein, ErxR (28).
Simplification and assimilation of bacteriophage genomes into bacterial chromosomes is well documented (29). Such a process, leading to residual phage genomes being integrated into bacterial chromosomes, serves as a secretion system to support bacterial growth or provide enzymes that benefit the bacteria by helping them to adapt to adverse environments (30–32). The bacteriophage-derived secretion systems in bacteria are used for multiple functions, including the translocation of effector proteins and metal ion transporters (30, 33). Rickettsial pathogens, including E. chaffeensis, contain only two known secretion systems, the type I secretion system (T1SS) and the type IV secretion system (T4SS) (6). We recently discovered that a function disruption mutation within a E. chaffeensis gene (ECH_0660) encoding a phage head-to-tail connector protein causes the bacterial attenuation and leading to its rapid clearance from vertebrate hosts (34, 35).
In the current study, we assessed how the ECH_0660 mutation contributes to E. chaffeensis attenuation by performing molecular characterization of the seven genes spanning from ECH_0659 to ECH_0665, as four genes from this cluster encode phage-related proteins, including ECH_0660. We described how deprivation of zinc and iron impacts gene expression from the gene cluster in wild-type E. chaffeensis and mutants of the pathogen with gene function disruptions in two of the seven genes, ECH_0660 and ECH_0665.
RESULTS
Analysis of E. chaffeensis DNA sequence surrounding the ECH_0660 gene.
A genomic segment spanning E. chaffeensis genes ECH_0659 to ECH_0665 (seven genes) was evaluated for its relationship with the ECH_0660 gene encoding the phage head-to-tail connector protein in which we reported a disruption mutation impacting in vivo growth (34) (see Fig. S1 in the supplemental material). Intergenic spacer sequences are absent or ranged in size from 43 to 141 bp long in some of the seven genes assessed. The intergenic spacer regions are absent between the coding regions of ECH_0660, ECH_0661, and ECH_0662. Furthermore, the protein coding sequences for these three genes are overlapped by a few nucleotides, namely, four nucleotides between ECH_0660 and ECH_0661 and 23 nucleotides between ECH_0661 and ECH_0662. ECH_0659 encodes a hypothetical protein, while ECH0661, ECH_0662, and ECH_0665 are similar to ECH_0660 in encoding predicted bacteriophage proteins, phage major tail protein, phage stopper protein, and phage terminase large subunit protein, respectively. Structural prediction analysis revealed a greater degree of structural homology for the four predicted phage proteins of E. chaffeensis with previously documented structural phage proteins and a protein required for phage DNA packaging (see Fig. S2 in the supplemental material) (36). ECH_0663 and ECH_0664 genes are paralogs encoding hypothetical proteins sharing 34.5% homology. To determine if the seven genes are conserved in other related Anaplasmataceae family bacteria, genomes of seven organisms belong to the genera Anaplasma, Ehrlichia, Neorickettsia, and Wolbachia were compared (see Fig. S3 in the supplemental material). Homologs for all seven genes were identified in other Ehrlichia species. Anaplasma phagocytophilum lacks only an ECH_0663 homolog (one of the two paralogs), while both ECH_0663 and ECH_0664 homologs are absent in Anaplasma marginale and Wolbachia. Neorickettsia sennetsu contains only one of the seven genes.
Promoter segment upstream of the first gene of the seven genes responds to the metal ion restriction.
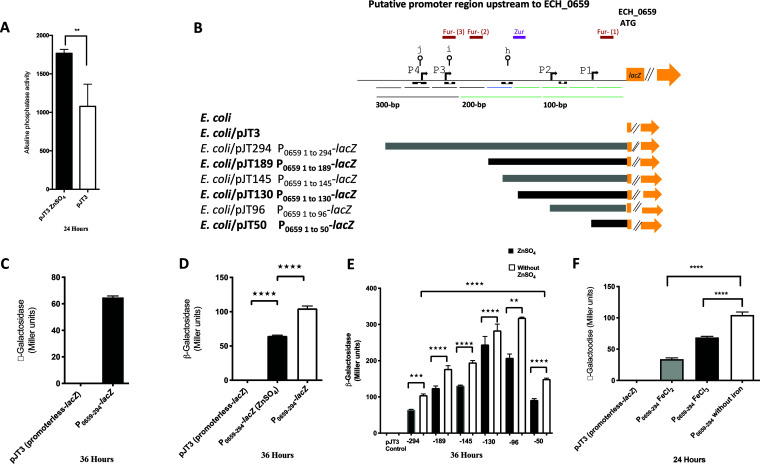
Bioinformatic analysis of the putative 294-bp-long promoter segment that is located upstream of the coding sequence of the first of the seven genes revealed the presence of several inverted repeat (IR) sequences with the potential to form intrastrand secondary structures (Fig. 1). Furthermore, the sequence included putative binding sites that likely interact with zinc- and iron-responsive proteins (37). To test whether transcription from the seven genes could be influenced by these metal ions, the promoter segment was cloned in front of the lacZ reporter gene in a plasmid, and β-galactosidase activity was tested in E. coli XL1-Blue MRF′ under conditions of chemically defined medium adjusted by the availability of metal ions. To assess the zinc restriction, E. coli transformed with a plasmid vector (pJT3) was first grown under phosphate starvation conditions in the culture medium with or without 15 μM ZnSO4 for 24 h. The endogenous alkaline phosphatase (PhoA) activity expressed from the E. coli chromosome served as an ideal positive control for our study to measure the impact of zinc starvation assessed from the E. coli lysates, as it requires zinc for its optimal activity (38). A reduction of 40% of PhoA activity was observed without zinc, compared to when it was present (Fig. 2A). We then analyzed the activity for the E. coli lysate containing the recombinant plasmid with the full-length promoter (pJT294) or segments of it with 5′ end deletions (Fig. 2B). The β-galactosidase (β-Gal) activity was observed only for the recombinant plasmid containing the full-length promoter (Fig. 2C). Further, the β-Gal activity increased by 30% when zinc was absent in the culture medium compared to that when it was present (Fig. 2D). The 5′ end deletion segments of the promoter caused enhanced β-Gal expression when zinc was absent (Fig. 2E), with minimal β-Gal activity observed for the shortest segment of 50 bp. Similarly, the full-length recombinant construct expressed detectible β-Gal activity with or without 100 μM FeCl2 or FeCl3, and the enzyme activity was significantly greater in the absence of iron compared to that in its presence (Fig. 2F).
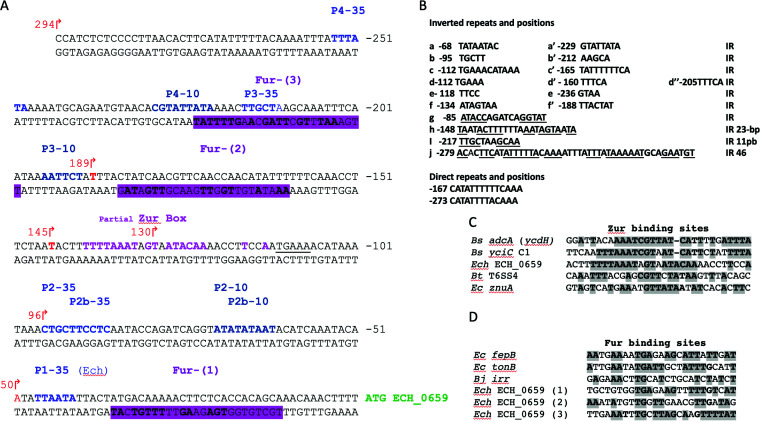
FIG 1.
The putative promoter region upstream of the ECH_0659 coding region contains DNA binding motifs that likely contribute to metal ion regulation and transcription. (A) Putative promoter region upstream of the ECH_0659 coding region. Regions that resemble the consensus for the Fur-binding site are indicated by magenta shading and in boldface type and are labeled Fur-(1), Fur-(2), and Fur-(3). The region that resembles the consensus for the Zur-binding site is indicated in boldface type in magenta color and labeled “partial Zur box.” Predicted −35 and −10 sequences are shown in blue and cyan-blue text, respectively, and are labeled P1-35, P2-35, P2b-35, P3-35, P4-35, P2-10, P2b-10, P3-10, and P4-10. (B) Inverted repeat sequences that are separated are a-a′, b, b′, c-c′, d-d′-d″, e-e′ and f-f′; inverted repeats that are located at close proximity are identified with underlines and listed as h, i, and j. Direct repeats are shown after the inverted repeats. (C and D) Zur (C) and Fur (D) partial consensus binding site sequences from other Gram-positive and Gram-negative bacteria are indicated by gray shading. Bt, Burkholderia thailandensis; Bs, Bacillus subtilis; Bj, Bradyrhizobium japonicum; Ec, E. coli; Ech, E. chaffeensis.
FIG 2.
A promoter region upstream of ECH_0659 assessed in an Escherichia coli surrogate system using zinc- and iron-sufficient or depletion conditions. (A) Endogenous alkaline phosphatase activity (PhoA) of the E. coli XL1-Blue MRF′ strain harboring the plasmid vector pJT3. Bacterial cells were grown in chemically defined medium (Tris-glucose) with zinc or phosphate restriction or supplemented with 15 μM ZnSO4 or 64 mM buffer phosphate. For iron response, bacterial cells were grown in Tris-glucose medium, iron depleted or supplemented with 100 μM FeCl2 or 100 μM FeCl3, as described in Materials and Methods. (B) Schematic diagram of the ECH_0659-promoter region and transcriptional fusion constructed pJT294, pJT189, pJT145, pJT130, pJT96, pJT50, and the control pJT3 plasmid, showing the putative binding regions for Zur and Fur (colored boxes), the −10 and −35 promoter elements (bent arrows), and the secondary structures (lines with open circles [j, i, and h]). Inverted repeat sequences are indicated with black boxes. The numbering of the nucleotides is relative to the ECH_0659 translational start codon. (C, D, E, and F) β-Gal activity for each construct is expressed as Miller units and was measured in Escherichia coli as described in Materials and Methods. The values are results from three independent experiments. Statistical significance was determined by one-way analysis of variance (ANOVA) followed by Tukey’s multiple-comparison test (**, P < 0.01; ***, P < 0.001; ****, P < 0.0001).
Transcripts from the seven genes assessed for the E. chaffeensis wild type and ECH_0660 and ECH_0665 mutants.
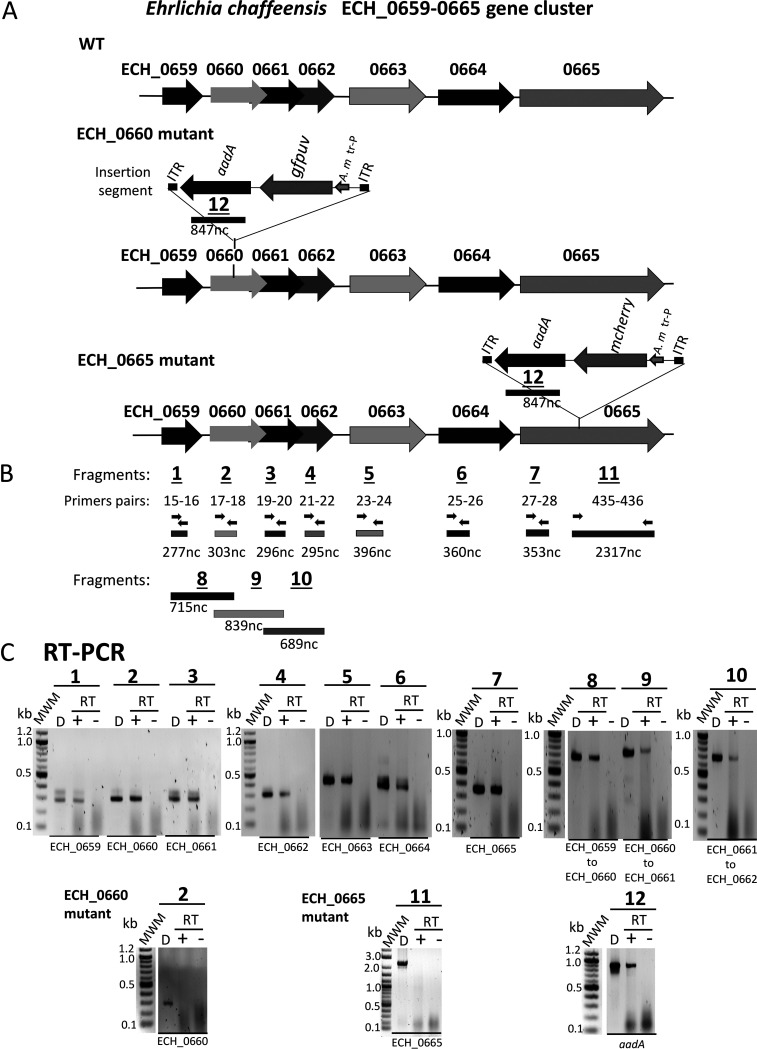
Transcripts for all seven genes were detected in the RNA recovered from the E. chaffeensis wild type cultured in the canine macrophage cell line DH82 (Fig. 3A to C). As intergenic spacer regions are absent or too small and range in size from 43 to 141 bp between the genes of all seven genes (Fig. S1), and given that the promoter prediction analysis identified several putative binding sites for transcriptional regulators in the sequence upstream from the ECH_0659 coding region (Fig. 1), we assessed if a polycistronic message(s) is transcribed from these genes. Reverse transcription-PCR (RT-PCR) assays targeting the coding regions overlapping from ECH_0659 to ECH_0662 yielded predicted amplicons (Fig. 3B and C), while such products were absent when assessed for the presence of overlapping transcripts between the genes from ECH_0662 to ECH_0665 (data not shown). These results suggest that the genes ECH_0659 to ECH_0662 are cotranscribed as a single transcript, while the remaining three genes, spanning from ECH_0663 to ECH_0665, are not part of the polycistronic message. To determine the impact of the ECH_0660 gene disruption mutation (34), RT-PCR analysis was carried out similarly for this mutant. Transcription was not detected for ECH_0660 (Fig. 3C), although the mutant tested positive for the remaining six genes (data not shown). We recently generated another mutant with functional disruption in ECH_0665, the last gene of the gene cluster (39). Unlike the ECH_0660 mutation, this mutation has no impact on the pathogen’s in vivo growth, as it progressed very similarly to that of the wild type when assessed in a vertebrate host (39). For this mutant, transcription was absent for ECH_0665 (Fig. 3C), while the remaining gene transcripts were detected (data not shown). In both ECH_0660 and ECH_0665 mutants, the RT-PCR products targeting the insertion-specific aadA gene were also observed (Fig. 3C).
FIG 3.
Transcripts from the seven genes assessed for the E. chaffeensis wild type and the ECH_0660 and ECH_0665 mutants. (A) Schematic representation of the seven genes on E. chaffeensis wild-type chromosome, and ECH_0660 and ECH_0665 mutants. (B) Reverse transcription-PCR (RT-PCR) targets identified. The primer pairs and their estimated products are represented with arrows and bars, respectively. (C) RT-PCR data presented for the amplicons generated targeting the 7 genes. “D” refers to a positive control with genomic DNA as the template; + and − refer to the RT-PCR assays performed with or without reverse transcriptase, respectively. Molecular weight markers (MWM) were included when resolving the PCR products to help locating specific amplicons. Expected amplicons for the internal coding regions of all seven genes were detected for the wild type when reverse transcriptase was added but were absent for the mutation insertion regions of ECH_0660 and ECH_0665 mutants (PCRs 2 and 11, respectively). Also, amplicons from overlapping coding regions of genes were detected as shown. For both ECH_0660 and ECH_0665 mutants, the transcription of the aadA gene from the Himar1 transposon (Tn) insertion was also observed (PCR 12).
Effects of zinc and iron starvation on the E. chaffeensis wild type and ECH_0660 and ECH_0665 mutant strains.
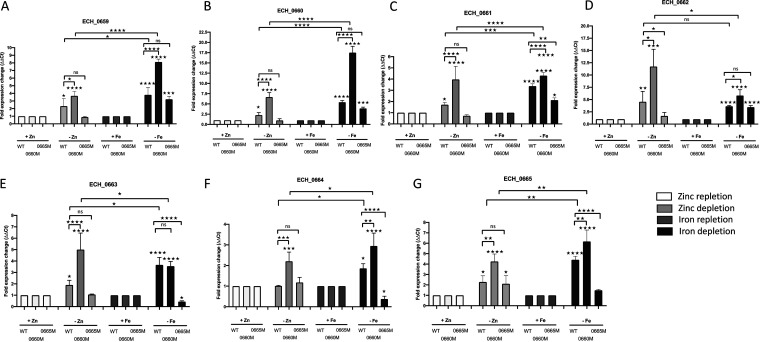
As we found only the predicted promoter region upstream of the seven genes and because this promoter responded to zinc and iron depletion when assessed in the heterologous E. coli surrogate system, we evaluated how E. chaffeensis responds to zinc and iron starvation in culture. The E. chaffeensis wild type and two mutants with ECH_0660 or ECH_0665 gene function disruptions were assessed by reverse transcription-quantitative PCR (qRT-PCR) for transcriptional changes from the cluster of seven genes, following their growth in medium containing zinc and with or without the zinc chelator N,N,N′,N′-tetrakis(2-pyridylmethyl)ethylenediamine (TPEN) (Fig. 4). We similarly assessed the iron starvation in medium containing FeSO4 and with or without the inclusion of the iron chelator 2,2′-bipyridyl (BIP) (Fig. 4). The relative gene expression in the presence of the zinc chelator for ECH_0659 was increased 2.3-fold for the wild type and 3.7-fold for the ECH_0660 mutant but did not show significant alteration in ECH_0665 mutant (Fig. 4A). The iron chelation is more pronounced for this gene, where we observed a 3.8-fold increase for the wild type. Similarly, for the ECH_0665 mutant, the transcripts increased by 3.2-fold, whereas an 8.1-fold increase was observed for the ECH_0660 mutant (Fig. 4A). Similarly, ECH_0660 transcripts for zinc chelation resulted in a 2.3-fold increase for the wild type and a 6.7-fold increase for the ECH_0660 mutant for gene coding region upstream of the insertion mutation site, while 1-fold downregulation was noted for the ECH_0665 mutant (Fig. 4B). The iron chelation was also more enhanced for these gene transcripts, which increased by 5.4-fold for the wild type and 17.5-fold for the ECH_0660 mutant, whereas it declined 1.5-fold for the ECH_0665 mutant compared to that of the wild type (Fig. 4B). For ECH_0661, the relative transcription without zinc was increased 1.7-fold for the wild type, while expression increased 4-fold for the ECH_0660 mutant and decreased to 0.7-fold for the ECH_0665 mutant (Fig. 4C). As in ECH_0659 and ECH_0660 gene transcripts, iron chelation resulted in considerably more transcripts for ECH_0661; these gene transcripts increased to 3.4-fold, 4.3-fold, and 2.1-fold for the wild type, ECH_0660 mutant, and ECH_0665 mutants, respectively (Fig. 4C). The removal of zinc from the medium caused a much more pronounced enhancement of the ECH_0662 transcription, as 4.6-fold and 11.7-fold transcript levels were observed in the wild type and the ECH_0660 mutant, respectively, and with no significant difference for the ECH_0665 mutant (Fig. 4D). While iron chelation also triggered ECH_0662 transcription, its effect is less compared to that of zinc chelation, with similar values for the wild type and ECH_0665 mutants (3.6- and 3.5-fold, respectively), whereas for the ECH_0660 mutant, it was another 2-fold more (5.8-fold) (Fig. 4D). ECH_0663 transcript levels increased during zinc chelation 1.9-fold for the wild type and 5-fold for the ECH_0660 mutant, with no significant change for the ECH_0665 mutant, whereas for the iron chelation, the wild type and the ECH_0660 mutant had similar increases of transcripts (3.7- and 3.6-fold, respectively), and transcription declined 0.5-fold for the ECH_0665 mutant (Fig. 4E). ECH_0664 relative gene expression was moderately increased during zinc chelation only for the ECH_0660 mutant bacteria, while no differences in the transcript levels were observed for both the wild type and the ECH_0665 mutant (Fig. 4F). Similarly, a moderate increase in transcript levels was observed in bacteria cultured under conditions of iron chelation for the wild type (1.9-fold) and the ECH_0660 mutant (3-fold) but was downregulated for the ECH_0665 mutant (0.4-fold) (Fig. 4F). Gene expression changes for the last gene (ECH_0665) also increased moderately, about 2.3-fold for the wild type and 2.1-fold for the ECH_0665 mutant (for the gene coding region upstream of the mutation site), and 4.3-fold for the ECH_0660 mutant. With the iron chelation, the transcription enhancement was more noticeable; 4.4-fold and 6.2-fold, respectively, for the wild type and ECH_0660 mutant bacteria and no significant difference for the ECH_0665 mutant (Fig. 4G). Together, transcripts of all seven genes were significantly upregulated for the wild type under both the zinc and iron depletion conditions, and the transcription levels were significantly enhanced for the ECH_0660 mutant, while essentially similar enhancements were observed in the wild type and downregulation was observed in the ECH_0665 mutant.
FIG 4.
Effects of zinc and iron starvation on the E. chaffeensis wild type and ECH_0660 and ECH_0665 mutants. The genes ECH_0659 to ECH_0665 from E. chaffeensis are upregulated under zinc and iron depletion conditions. The expression of ECH_0659 (A), ECH_0660 (B), ECH_0661 (C), ECH_0662 (D), ECH_0663 (E), ECH_0664 (F), and ECH_0665 (G) from wild-type, ECH_0660 mutant, and ECH_0665 mutant organisms was measured during the stationary phase of infection under zinc repletion or depletion conditions, and similarly under iron repletion or depletion conditions, by quantitative real-time RT-PCR. The data represent the mean ± standard deviation (SD) from 2 biological replicas, each of which comprises 3 technical replicas. Fold expression changes are normalized using gyrB as an endogenous reference gene (ΔCT). Statistical significance was determined by one-way analysis of variance (ANOVA) followed by Tukey’s multiple-comparison test (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001).
DISCUSSION
In earlier studies, we reported that a mutation in the E. chaffeensis ECH_0660 gene encoding the phage head-to-tail connector protein results in the rapid clearance of the pathogen from vertebrate hosts (white-tailed deer and dogs). In contrast, persistent infection occurs with wild-type E. chaffeensis and the ECH_0665 mutant (34, 35, 39, 40). Pathogenic intracellular bacteria are often challenged by the restricted availability of metal ions such as iron and zinc in the host cells (20). Starvation of the metal ions triggers enhances expression of certain metal ion response genes in bacteria. In this study, we characterized functional disruption mutations in the ECH_0660 and ECH_0665 genes of E. chaffeensis encoding two predicted phage proteins, the head-to-tail-connector protein and the phage terminase enzyme, respectively. We assessed how the wild type and the two mutants respond to depletion of two important metal ions, zinc and iron. The wild type and the mutant bacteria had distinct gene expression profiles in response to depletion of the two metal ions. Specifically, chelation of the metal ions triggered significant transcriptional enhancement for most of the genes from the seven genes for the wild type. Transcription enhancement was significantly higher for all seven genes under zinc starvation conditions for the ECH_0660 mutant, while iron starvation similarly caused enhanced gene expression from six genes. Contrary to these observations, the transcripts for the ECH_0665 gene mutant, previously shown to have a minimal impact on the bacterial in vivo growth (39), were more or less similar to those of wild-type E. chaffeensis. We used TPEN, a chelator known to chelate other divalent metal ions such as cadmium, cobalt, nickel, and copper (41), to chelate zinc from E. chaffeensis cultures. Thus, the gene expression changes observed may also be the result of depleting other metal ions. As the E. coli surrogate system selectively demonstrated the impact of zinc, we reasoned that the TPEN chelation effects primarily resulted from zinc starvation.
Zinc and iron play essential roles in bacterial cells to serve as cofactors for multiple enzymes, to support the structural integrity of proteins, and to regulate protein expression from several bacterial genes (42). These metal ions are not readily accessible for intracellular pathogens (20). Pathogenic bacteria employ unique strategies in acquiring metal ions from a host cell during intracellular replication (24). We observed that the predicted promoter sequence upstream of the seven genes activates reporter gene expression, and it was significantly higher in medium with limited zinc or iron when assessed in the E. coli surrogate system. Although the E. coli-based analysis may not be ideal for assessing an E. chaffeensis promoter function, this heterologous system is currently the best means of investigating bacterial gene regulation, considering the lack of desirable genetic tools for the pathogen (11, 43–49). Assessment of the effect of promoter deletion analysis suggested that one or more of the putative DNA binding motifs within the sequence responds to zinc or iron depletion. The E. chaffeensis genome includes genes encoding a zinc transporter protein (ECH_0067) and an iron transport system substrate-binding protein (ECH_0189). However, it is yet to be determined if these proteins are involved in metal ion transport. A recent study in a related Ehrlichia species (Ehrlichia ruminantium) suggested that iron restriction causes upregulation of a T4SS protein (VirB), an outer membrane-associated protein (MAP1), and two predicted transcriptional regulators (Tr1 and ErxR) (28). In the current study, we demonstrated significant transcriptional enhancement for the seven genes when depleting zinc and iron during E. chaffeensis growth for the wild-type bacteria. The transcriptional enhancement was significantly greater for the ECH_0660 mutant, a mutation known to exert an in vivo growth defect (34, 35, 50). In contrast, transcript levels were similar to those of the wild type, or slightly downregulated for the ECH_0665 mutant. We recently reported that this mutation has a minimal impact on its in vivo growth (39). Together, the data presented in the current study suggest that the growth defect mutation in the ECH_0660 gene encoding the phage head-to-tail connector protein triggers significant upregulation of transcription from the seven genes assessed when metal ions are chelated, while the mutation in the ECH_0665 gene encoding the phage terminase protein leads to the pathogen maintaining transcription levels more or less similar to those of wild-type E. chaffeensis.
We reported earlier that the ECH_0660, ECH_0663, and ECH_0665 gene products are membrane-associated proteins, as judged by mass spectrometry analysis of purified total and immunogenic E. chaffeensis membrane fractions (51). We recently reported that the ECH_0660 gene mutant caused minimal variation in the bacterial global transcriptome and proteomes compared to the wild-type E. chaffeensis (52, 53) when assessed in standard culture medium (54). This mutant having the growth defect allowed the vertebrate host to initiate a potent immune response to confer protection against wild-type pathogen infection challenge (34, 35). The data presented in the current study suggest that the functional disruption in the predicted phage head-to-tail connector protein gene is likely detrimental in altering the pathogen’s ability to obtain metal ions to support its growth within a phagosome of infected host macrophages. Our previously described mutation in the ECH_0660 gene causing in vivo attenuation may have caused a metabolic burden for E. chaffeensis resulting in the upregulation of gene expression from several genes likely involved in the metal ion uptake.
MATERIALS AND METHODS
Bioinformatics.
The E. chaffeensis genomic region (GenBank accession number CP000236.1) spanning from the ECH_0659 to ECH_0665 genes was subjected to data searching as described below. Inverted repeat and direct repeat sequences in DNA were identified by visual examination and by using the palindromic sequence finder REPFIND (55). Promoter −35 and −10 element sequences for Ehrlichia genes were predicted with the BPROM program (56). Genomic DNA sequences of Ehrlichia canis (Jake), Ehrlichia muris (AS145), Ehrlichia ruminantium (Welgevonden), Anaplasma phagocytophilum (HZ), Anaplasma marginale (St. Maries), Wolbachia (wHa), and Neorickettsia sennetsu were obtained from the NCBI website. The genome data were compared with the gene cluster from ECH_0659 to ECH_0665 to identify the gene homologs. DNA and protein alignments were performed with ClustalW and ClustalX, respectively. Database searches were performed using NCBI BLAST and CDD databases (57).
Bacterial strains, plasmids, and media.
The bacterial strains and plasmids used in this study are listed in Table S1 in the supplemental material. Escherichia coli strains were routinely grown at 37°C in Luria–Bertani (LB) medium. When required, the medium was supplemented with an appropriate antibiotic. Tris-glucose (TG) medium (58) was used for zinc response assessment, with or without 15 μM ZnSO4. Similarly, the iron response was assessed in TG medium with or without 100 μM FeCl2 or 100 μM FeCl3. For determining β-galactosidase (β-Gal) or alkaline phosphatase (PhoA) activities, 100-μl aliquots taken at 24 h or 72 h were used as described below.
Construction of ECH_0659 promoter-lacZ fusion plasmids.
DNA manipulations to prepare recombinant plasmids and E. coli transformations were carried out by standard molecular cloning protocols as in (59). All primers used in this study were synthesized by Integrated DNA Technology (Coralville, IA) and are listed in Table S2 in the supplemental material. DNA fragments containing the entire 294-bp putative promoter region upstream of the ECH_0659 gene coding sequence were amplified by PCR using E. chaffeensis genomic DNA as the template and with the primer sets specific for the segment (Table S2). PCR products were cloned using primers designed for directional cloning into KpnI-BamHI sites of the pJT3 plasmid (60) to create the recombinant plasmid pJT294. Deletion constructs were prepared similarly with 5′-end sequential deletions.
Enzymatic assays.
β-Galactosidase (β-Gal) activity was assessed on the cultures supplemented with 50 μg 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) · ml−1. Quantitative evaluation of β-Gal was carried out using permeabilized cells obtained from stationary-phase cultures grown with or without 15 μM ZnSO4, or with or without 100 μM FeCl2/FeCl3, and incubated with o-nitrophenyl-β-d-galactopyranoside (ONPG) substrate (Sigma-Aldrich) as previously described (61). Quantitative evaluation of alkaline phosphatase (PhoA) was carried out similarly using cultures grown under phosphate starvation conditions with or without 15 μM ZnSO4 and incubated with 4-nitrophenyl phosphate (4 mg · ml−1) substrate (Sigma-Aldrich) as described previously (58). Average values (±standard deviations) for activity units were calculated from three independent assays, each carried out in triplicate.
Treatment of E. chaffeensis-infected macrophages (DH82 cells) with metal chelators and subsequent RNA analysis.
E. chaffeensis (Arkansas isolate) wild-type, ECH_0660 mutant (34), and ECH_0665 mutant (39) cells were grown in the canine macrophage cell line DH82 (ATCC, Manassas, VA) in minimal essential medium (MEM) as previously described (54). For the zinc response assay, culture medium was supplemented with 15 μM ZnSO4, with or without 5 μM N,N,N′,N′-tetrakis(2-pyridylmethyl)ethylenediamine (TPEN, a zinc chelator; Sigma-Aldrich, St. Louis, MO) (63). To define the growth characteristics under iron-supplemented or -depleted conditions, E. chaffeensis cultures were propagated in medium supplemented with 100 μM FeSO4 and with or without 0.15 mM 2,2′-bipyridyl (BIP, an iron chelator; Sigma-Aldrich) (62). TPEN or BIP was added independently to the cultures at 72 h postinoculation when infectivity was about 80 to 90%. After 24 h, cultures were harvested by centrifugation at 14,000 × g for 10 min. The pellet was resuspended with a half volume of the medium and mixed with 2.5 ml of RNAlater stabilization solution (Thermo Fisher) and stored at −4°C until RNA extraction. Total RNA was recovered, DNase treated, and then reverse transcribed using SuperScript III reverse transcriptase (Invitrogen). The cDNA was used in PCR analysis using Platinum Taq DNA polymerase (Invitrogen) using primers specific for each gene. The resulting PCR products were separated by electrophoresis in 1.5% agarose gel. Real-time PCR amplification of the cDNA was performed with 2× PowerUp SYBR green master mix (Applied Biosystems, Life Technologies) as specified by the manufacturer, with 1 μl cDNA (corresponding to 0.1 to 0.2 μg of total RNA) and using a gene-specific primer set. Two independent experiments were performed for every gene transcript with samples examined in triplicate for each experiment in a StepOnePlus real-time PCR system (Applied Biosystems by Life Technologies). Relative expression levels of the specific transcripts between different biological conditions for wild-type and mutant E. chaffeensis were calculated using the relative abundance of gyrB as an internal reference standard for normalization and expressed as fold differences by calculating 2−(ΔΔCT) (CT, threshold cycle) (64). The final qRT-PCR data were presented as the means of two experiments.
Statistics.
The statistical differences between experimental groups were assessed by one-way analysis of variance (ANOVA) followed by Tukey’s multiple-comparison test or Student’s t test. Differences with a P value of <0.05 were considered significant.
ACKNOWLEDGMENTS
This work was supported by the PHS grants AI070908 and AI152418 from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, USA.
The funders had no role in the study design, data collection, and analysis, the decision to publish, or the preparation of the manuscript.
We thank Donald Harbidge at the KSU CVM molecular biology core facility for his help in performing qRT-PCR analysis.
Footnotes
Supplemental material is available online only.
Contributor Information
Roman R. Ganta, Email: rganta@vet.k-state.edu.
Laurie E. Comstock, Brigham and Women's Hospital/Harvard Medical School
REFERENCES
- 1.Anderson BE, Dawson JE, Jones DC, Wilson KH. 1991. Ehrlichia chaffeensis, a new species associated with human ehrlichiosis. J Clin Microbiol 29:2838–2842. 10.1128/JCM.29.12.2838-2842.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dawson JE, Anderson BE, Fishbein DB, Sanchez JL, Goldsmith CS, Wilson KH, Duntley CW. 1991. Isolation and characterization of an Ehrlichia sp. from a patient diagnosed with human ehrlichiosis. J Clin Microbiol 29:2741–2745. 10.1128/JCM.29.12.2741-2745.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ewing SA, Dawson JE, Kocan AA, Barker RW, Warner CK, Panciera RJ, Fox JC, Kocan KM, Blouin EF. 1995. Experimental transmission of Ehrlichia chaffeensis (Rickettsiales: Ehrlichieae) among white-tailed deer by Amblyomma americanum (Acari: Ixodidae). J Med Entomol 32:368–374. 10.1093/jmedent/32.3.368. [DOI] [PubMed] [Google Scholar]
- 4.Dumler JS, Barbet AF, Bekker CP, Dasch GA, Palmer GH, Ray SC, Rikihisa Y, Rurangirwa FR. 2001. Reorganization of genera in the families Rickettsiaceae and Anaplasmataceae in the order Rickettsiales: unification of some species of Ehrlichia with Anaplasma, Cowdria with Ehrlichia and Ehrlichia with Neorickettsia, descriptions of six new species combinations and designation of Ehrlichia equi and ‘HGE agent’ as subjective synonyms of Ehrlichia phagocytophila. Int J Syst Evol Microbiol 51:2145–2165. 10.1099/00207713-51-6-2145. [DOI] [PubMed] [Google Scholar]
- 5.Paddock CD, Childs JE. 2003. Ehrlichia chaffeensis: a prototypical emerging pathogen. Clin Microbiol Rev 16:37–64. 10.1128/cmr.16.1.37-64.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dunning Hotopp JC, Lin M, Madupu R, Crabtree J, Angiuoli SV, Eisen JA, Eisen J, Seshadri R, Ren Q, Wu M, Utterback TR, Smith S, Lewis M, Khouri H, Zhang C, Niu H, Lin Q, Ohashi N, Zhi N, Nelson W, Brinkac LM, Dodson RJ, Rosovitz MJ, Sundaram J, Daugherty SC, Davidsen T, Durkin AS, Gwinn M, Haft DH, Selengut JD, Sullivan SA, Zafar N, Zhou L, Benahmed F, Forberger H, Halpin R, Mulligan S, Robinson J, White O, Rikihisa Y, Tettelin H. 2006. Comparative genomics of emerging human ehrlichiosis agents. PLoS Genet 2:e21. 10.1371/journal.pgen.0020021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buller RS, Arens M, Hmiel SP, Paddock CD, Sumner JW, Rikhisa Y, Unver A, Gaudreault-Keener M, Manian FA, Liddell AM, Schmulewitz N, Storch GA. 1999. Ehrlichia ewingii, a newly recognized agent of human ehrlichiosis. N Engl J Med 341:148–155. 10.1056/NEJM199907153410303. [DOI] [PubMed] [Google Scholar]
- 8.Andersson M, Råberg L. 2011. Wild rodents and novel human pathogen Candidatus Neoehrlichia mikurensis, Southern Sweden. Emerg Infect Dis 17:1716–1718. 10.3201/eid1709.101058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen SM, Dumler JS, Bakken JS, Walker DH. 1994. Identification of a granulocytotropic Ehrlichia species as the etiologic agent of human disease. J Clin Microbiol 32:589–595. 10.1128/JCM.32.3.589-595.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Popov VL, Yu XJ, Walker DH. 2000. The 120 kDa outer membrane protein of Ehrlichia chaffeensis: preferential expression on dense-core cells and gene expression in Escherichia coli associated with attachment and entry. Microb Pathog 28:71–80. 10.1006/mpat.1999.0327. [DOI] [PubMed] [Google Scholar]
- 11.Wakeel A, den Dulk-Ras A, Hooykaas PJ, McBride JW. 2011. Ehrlichia chaffeensis tandem repeat proteins and Ank200 are type 1 secretion system substrates related to the repeats-in-toxin exoprotein family. Front Cell Infect Microbiol 1:22. 10.3389/fcimb.2011.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dunphy PS, Luo T, McBride JW. 2014. Ehrlichia chaffeensis exploits host SUMOylation pathways to mediate effector-host interactions and promote intracellular survival. Infect Immun 82:4154–4168. 10.1128/IAI.01984-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mohan Kumar D, Yamaguchi M, Miura K, Lin M, Los M, Coy JF, Rikihisa Y. 2013. Ehrlichia chaffeensis uses its surface protein EtpE to bind GPI-anchored protein DNase X and trigger entry into mammalian cells. PLoS Pathog 9:e1003666. 10.1371/journal.ppat.1003666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mohan Kumar D, Lin M, Xiong Q, Webber MJ, Kural C, Rikihisa Y. 2015. EtpE binding to DNase X induces ehrlichial entry via CD147 and hnRNP-K recruitment, followed by mobilization of N-WASP and actin. mBio 6:e01541-15–e01515. 10.1128/mBio.01541-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rikihisa Y. 2015. Molecular pathogenesis of Ehrlichia chaffeensis infection. Annu Rev Microbiol 69:283–304. 10.1146/annurev-micro-091014-104411. [DOI] [PubMed] [Google Scholar]
- 16.Kader M, Alaoui-El-Azher M, Vorhauer J, Kode BB, Wells JZ, Stolz D, Michalopoulos G, Wells A, Scott M, Ismail N. 2017. MyD88-dependent inflammasome activation and autophagy inhibition contributes to Ehrlichia-induced liver injury and toxic shock. PLoS Pathog 13:e1006644. 10.1371/journal.ppat.1006644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lina TT, Luo T, Velayutham TS, Das S, McBride JW. 2017. Ehrlichia activation of Wnt-PI3K-mTOR signaling inhibits autolysosome generation and autophagic destruction by the mononuclear phagocyte. Infect Immun 85:e00690-17. 10.1128/IAI.00690-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Green RS, Izac JR, Naimi WA, O'Bier N, Breitschwerdt EB, Marconi RT, Carlyon JA. 2020. Ehrlichia chaffeensis EplA interaction with host cell protein disulfide isomerase promotes infection. Front Cell Infect Microbiol 10:500. 10.3389/fcimb.2020.00500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin M, Liu H, Xiong Q, Niu H, Cheng Z, Yamamoto A, Rikihisa Y. 2016. Ehrlichia secretes Etf-1 to induce autophagy and capture nutrients for its growth through RAB5 and class III phosphatidylinositol 3-kinase. Autophagy 12:2145–2166. 10.1080/15548627.2016.1217369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma L, Terwilliger A, Maresso W. 2015. Iron and zinc exploitation during bacterial pathogenesis. Metallomics 7:1541–1554. 10.1039/c5mt00170f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nairz M, Haschka D, Demetz E, Weiss G. 2014. Iron at the interface of immunity and infection. Front Pharmacol 5:152. 10.3389/fphar.2014.00152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gammoh NZ, Rink L. 2017. Zinc in infection and inflammation. Nutrients 9:624. 10.3390/nu9060624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Becker KW, Skaar EP. 2014. Metal limitation and toxicity at the interface between host and pathogen. FEMS Microbiol Rev 38:1235–1249. 10.1111/1574-6976.12087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lopez CA, Skaar EP. 2018. The impact of dietary transition metals on host-bacterial interactions. Cell Host Microbe 23:737–748. 10.1016/j.chom.2018.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang J, Pantopoulos K. 2011. Regulation of cellular iron metabolism. Biochem J 434:365–381. 10.1042/BJ20101825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barnewall RE, Rikihisa Y, Lee EH. 1997. Ehrlichia chaffeensis inclusions are early endosomes which selectively accumulate transferrin receptor. Infect Immun 65:1455–1461. 10.1128/IAI.65.4.1455-1461.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clemens DL, Horwitz MA. 1996. The Mycobacterium tuberculosis phagosome interacts with early endosomes and is accessible to exogenously administered transferrin. J Exp Med 184:1349–1355. 10.1084/jem.184.4.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moumène A, Gonzalez-Rizzo S, Lefrançois T, Vachiéry N, Meyer DF. 2017. Iron starvation conditions upregulate Ehrlichia ruminantium type IV secretion system, tr1 transcription factor and map1 genes family through the master regulatory protein ErxR. Front Cell Infect Microbiol 7:535. 10.3389/fcimb.2017.00535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lorenz N, Reiger M, Toro-Nahuelpan M, Brachmann A, Poettinger L, Plener L, Lassak J, Jung K. 2016. Identification and initial characterization of prophages in Vibrio campbellii. PLoS One 11:e0156010. 10.1371/journal.pone.0156010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Costa TR, Felisberto-Rodrigues C, Meir A, Prevost MS, Redzej A, Trokter M, Waksman G. 2015. Secretion systems in Gram-negative bacteria: structural and mechanistic insights. Nat Rev Microbiol 13:343–359. 10.1038/nrmicro3456. [DOI] [PubMed] [Google Scholar]
- 31.Figueroa-Bossi N, Uzzau S, Maloriol D, Bossi L. 2001. Variable assortment of prophages provides a transferable repertoire of pathogenic determinants in Salmonella. Mol Microbiol 39:260–271. 10.1046/j.1365-2958.2001.02234.x. [DOI] [PubMed] [Google Scholar]
- 32.Marrs B. 1974. Genetic recombination in Rhodopseudomonas capsulata. Proc Natl Acad Sci U S A 71:971–973. 10.1073/pnas.71.3.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peñil-Celis A, Garcillán-Barcia MP. 2019. Crosstalk between type VI secretion system and mobile genetic elements. Front Mol Biosci 6:126. 10.3389/fmolb.2019.00126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cheng C, Nair AD, Indukuri VV, Gong S, Felsheim RF, Jaworski D, Munderloh UG, Ganta RR. 2013. Targeted and random mutagenesis of Ehrlichia chaffeensis for the identification of genes required for in vivo infection. PLoS Pathog 9:e1003171. 10.1371/journal.ppat.1003171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nair AD, Cheng C, Jaworski DC, Ganta S, Sanderson MW, Ganta RR. 2015. Attenuated mutants of Ehrlichia chaffeensis induce protection against wild-type infection challenge in the reservoir host and in an incidental host. Infect Immun 83:2827–2835. 10.1128/IAI.00487-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chaban Y, Lurz R, Brasilès S, Cornilleau C, Karreman M, Zinn-Justin S, Tavares P, Orlova EV. 2015. Structural rearrangements in the phage head-to-tail interface during assembly and infection. Proc Natl Acad Sci U S A 112:7009–7014. 10.1073/pnas.1504039112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fillat MF. 2014. The FUR (ferric uptake regulator) superfamily: diversity and versatility of key transcriptional regulators. Arch Biochem Biophys 546:41–52. 10.1016/j.abb.2014.01.029. [DOI] [PubMed] [Google Scholar]
- 38.Kim EE, Wyckoff HW. 1991. Reaction mechanism of alkaline phosphatase based on crystal structures: two-metal ion catalysis. J Mol Biol 218:449–464. 10.1016/0022-2836(91)90724-K. [DOI] [PubMed] [Google Scholar]
- 39.Wang Y, Nair ADS, Alhassan A, Jaworski DC, Liu H, Trinkl K, Hove P, Ganta CK, Burkhardt N, Munderloh UG, Ganta RR. 2020. Multiple Ehrlichia chaffeensis genes critical for its persistent infection in a vertebrate host are identified by random mutagenesis coupled with in vivo infection assessment. Infect Immun 88:e00316-20. 10.1128/IAI.00316-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McGill JL, Nair AD, Cheng C, Rusk RA, Jaworski DC, Ganta RR. 2016. Vaccination with an attenuated mutant of Ehrlichia chaffeensis induces pathogen-specific CD4+ T cell immunity and protection from tick-transmitted wild-type challenge in the canine host. PLoS One 11:e0148229. 10.1371/journal.pone.0148229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sigdel TK, Easton JA, Crowder MW. 2006. Transcriptional response of Escherichia coli to TPEN. J Bacteriol 188:6709–6713. 10.1128/JB.00680-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Palmer LD, Skaar EP. 2016. Transition metals and virulence in bacteria. Annu Rev Genet 50:67–91. 10.1146/annurev-genet-120215-035146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peddireddi L, Cheng C, Ganta RR. 2009. Promoter analysis of macrophage- and tick cell-specific differentially expressed Ehrlichia chaffeensis p28-Omp genes. BMC Microbiol 9:99. 10.1186/1471-2180-9-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cheng Z, Miura K, Popov VL, Kumaga Y, Rikihisa Y. 2011. Insights into the CtrA regulon in development of stress resistance in obligatory intracellular pathogen Ehrlichia chaffeensis. Mol Microbiol 82:1217–1234. 10.1111/j.1365-2958.2011.07885.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cheng Z, Lin M, Rikihisa Y. 2014. Ehrlichia chaffeensis proliferation begins with NtrY/NtrX and PutA/GlnA upregulation and CtrA degradation induced by proline and glutamine uptake. mBio 5:e02141. 10.1128/mBio.02141-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu H, Jakkula L, Von Ohlen T, Ganta RR. 2016. Sequence determinants spanning −35 motif and AT-rich spacer region impacting Ehrlichia chaffeensis sigma 70-dependent promoter activity of two differentially expressed p28 outer membrane protein genes. DNA Res 23:495–505. 10.1093/dnares/dsw034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McClure EE, Chávez A, Shaw DK, Carlyon JA, Ganta RR, Noh SM, Wood DO, Bavoil PM, Brayton KA, Martinez JJ, McBride JW, Valdivia RH, Munderloh UG, Pedra J. 2017. Engineering of obligate intracellular bacteria: progress, challenges and paradigms. Nat Rev Microbiol 15:544–558. 10.1038/nrmicro.2017.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu H, Ganta RR. 2019. Sequence determinants spanning −10 motif and spacer region implicated in unique Ehrlichia chaffeensis sigma 32-dependent promoter activity of dnaK gene. Front Microbiol 10:1772. 10.3389/fmicb.2019.01772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Duan N, Ma X, Cui H, Wang Z, Chai Z, Yan J, Li X, Feng Y, Cao Y, Jin Y, Bai F, Wu W, Rikihisa Y, Cheng Z. 2021. Insights into the mechanism regulating the differential expression of the P28-OMP outer membrane proteins in obligatory intracellular pathogen Ehrlichia chaffeensis. Emerg Microbes Infect 10:461–471. 10.1080/22221751.2021.1899054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cheng C, Nair AD, Jaworski DC, Ganta RR. 2015. Mutations in Ehrlichia chaffeensis causing polar effects in gene expression and differential host specificities. PLoS One 10:e0132657. 10.1371/journal.pone.0132657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Seo GM, Cheng C, Tomich J, Ganta RR. 2008. Total, membrane, and immunogenic proteomes of macrophage- and tick cell-derived Ehrlichia chaffeensis evaluated by liquid chromatography-tandem mass spectrometry and MALDI-TOF methods. Infect Immun 76:4823–4832. 10.1128/IAI.00484-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kondethimmanahalli C, Ganta R. 2018. Impact of three different mutations in Ehrlichia chaffeensis in altering the global gene expression patterns. Sci Rep 8:6162. 10.1038/s41598-018-24471-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kondethimmanahalli C, Liu H, Ganta RR. 2019. Proteome analysis revealed changes in protein expression patterns caused by mutations in Ehrlichia chaffeensis. Front Cell Infect Microbiol 9:58. 10.3389/fcimb.2019.00058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cheng C, Ganta RR. 2008. Laboratory maintenance of Ehrlichia chaffeensis and Ehrlichia canis and recovery of organisms for molecular biology and proteomics studies. Curr Prot Microbiol 9. 10.1002/9780471729259.mc03a01s9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Betley JN, Frith MC, Graber JH, Choo S, Deshler JO. 2002. A ubiquitous and conserved signal for RNA localization in chordates. Curr Biol 12:1756–1761. 10.1016/S0960-9822(02)01220-4. [DOI] [PubMed] [Google Scholar]
- 56.Solovyev V, Salamov A. 2011. Automatic annotation of microbial genomes and metagenomic sequences, p 61–78. In Li RW (ed) Metagenomics and its applications in agriculture, biomedicine and environmental studies. Nova Science Publishers, Hauppauge, NY. [Google Scholar]
- 57.Marchler-Bauer A, Anderson JB, Cherukuri PF, DeWeese-Scott C, Geer LY, Gwadz M, He S, Hurwitz DI, Jackson JD, Ke Z, Lanczycki CJ, Liebert CA, Liu C, Lu F, Marchler GH, Mullokandov M, Shoemaker BA, Simonyan V, Song JS, Thiessen PA, Yamashita RA, Yin JJ, Zhang D, Bryant SH. 2005. CDD: a Conserved Domain Database for protein classification. Nucleic Acids Res 33:D192–D196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Torres A, Juárez MD, Cervantes R, Espitia C. 2001. Molecular analysis of Mycobacterium tuberculosis phosphate specific transport system in Mycobacterium smegmatis: characterization of recombinant 38 kDa (PstS-1). Microb Pathog 30:289–297. 10.1006/mpat.2001.0434. [DOI] [PubMed] [Google Scholar]
- 59.Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- 60.Juárez-Rodríguez MD, Torres-Escobar A, Demuth DR. 2013. Construction of new cloning, lacZ reporter and scarless-markerless suicide vectors for genetic studies in Aggregatibacter actinomycetemcomitans. Plasmid 69:211–222. 10.1016/j.plasmid.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- 62.Kaes C, Katz A, Hosseini MW. 2000. Bipyridine: the most widely used ligand. A review of molecules comprising at least two 2,2′-bipyridine units. Chem Rev 100:3553–3590. 10.1021/cr990376z. [DOI] [PubMed] [Google Scholar]
- 63.Cho YE, Lomeda RA, Ryu SH, Lee JH, Beattie JH, Kwun IS. 2007. Cellular Zn depletion by metal ion chelators (TPEN, DTPA, and chelex resin) and its application to osteoblastic MC3T3-E1 cells. Nutr Res Pract 1:29–35. 10.4162/nrp.2007.1.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Winer J, Jung CK, Shackel I, Williams PM. 1999. Development and validation of real-time quantitative reverse transcriptase-polymerase chain reaction for monitoring gene expression in cardiac myocytes in vitro. Anal Biochem 270:41–49. 10.1006/abio.1999.4085. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1 and S2 and Fig. S1 to S3. Download JB.00027-21-s0001.pdf, PDF file, 3.41 MB (3.4MB, pdf)