ABSTRACT
Limited information is available on whether blaKPC-containing plasmids from isolates in a hospital outbreak can be differentiated from epidemiologically unrelated blaKPC-containing plasmids based on sequence data. This study aimed to evaluate the performance of three approaches to distinguish epidemiologically related from unrelated blaKPC-containing pKpQiL-like IncFII(k2)-IncFIB(pQiL) plasmids. Epidemiologically related isolates were subjected to short- and long-read whole-genome sequencing. A hybrid assembly was performed, and plasmid sequences were extracted from the assembly graph. Epidemiologically unrelated plasmid sequences were extracted from GenBank. Pairwise comparisons of epidemiologically related and unrelated plasmids based on SNPs using snippy and of phylogenetic distance using Roary and using a similarity index that penalizes size differences between plasmids (Stoesser index) were performed. The percentage of pairwise comparisons misclassified as genetically related or as clonally unrelated was determined using different genetic thresholds for genetic relatedness. The ranges of number of SNPs, Roary phylogenetic distance, and Stoesser index overlapped between the epidemiologically related and unrelated plasmids. When a genetic similarity threshold that classified 100% of epidemiologically related plasmid pairs as genetically related was used, the percentages of plasmids misclassified as epidemiologically related ranged from 6.7% (Roary) to 20.8% (Stoesser index). Although epidemiologically related plasmids can be distinguished from unrelated plasmids based on genetic differences, blaKPC-containing pKpQiL-like IncFII(k2)-IncFIB(pQiL) plasmids show a high degree of sequence similarity. The phylogenetic distance as determined using Roary showed the highest degree of discriminatory power between the epidemiologically related and unrelated plasmids.
KEYWORDS: KPC, plasmids, transmission
INTRODUCTION
Infections with carbapenem-resistant Enterobacteriaceae are associated with increased mortality and have emerged as an urgent public health threat (1–3). Worldwide, one of the most frequent mechanisms for carbapenem resistance in Enterobacteriaceae is the production of Klebsiella pneumoniae carbapenemase (KPC) (4). Nosocomial transmission plays an important role in spreading these KPC-producing Enterobacteriaceae, and several hospital outbreaks have been described (5–8). Molecular typing is often used to detect the source and route of transmission in these hospital outbreak settings (9). Whole-genome sequencing of bacterial isolates is currently the ultimate tool for molecular typing, enabling comparison of the entire bacterial chromosome to identify bacterial clones with great precision using either a gene-by-gene or single-nucleotide-polymorphism (SNP) approach (10). However, the gene encoding KPC production, blaKPC, is located not on the bacterial chromosome but on large conjugative resistance plasmids. These plasmids can transfer between isolates, and nosocomial transmission of these plasmids can go undetected when only molecular typing of the bacterial chromosome is performed (11).
One of the most predominant plasmid types containing blaKPC are the pKpQiL-like IncFII(k2)-IncFIB(pQiL) plasmids, which have spread across the globe (12–14). Several reports have already described plasmid spread between different isolates and patients during hospital outbreaks (15–19). Analyzing these resistance plasmids is typically performed by generating a combination of short- and long-read sequence data, enabling hybrid assembly algorithms to perform complete plasmid assemblies (17, 20–22). Accurately distinguishing epidemiologically related from unrelated plasmids (e.g., based on the number of SNPs) is essential to detect or exclude nosocomial plasmid transmission in outbreaks. However, limited information is available on whether blaKPC-containing plasmids from isolates in a hospital outbreak can be differentiated from epidemiologically unrelated blaKPC-containing plasmids based on sequence data. This study aimed to evaluate the performance of three approaches, based on determining SNPs, phylogenetic distance, and a similarity index, to distinguish epidemiologically related from unrelated blaKPC-containing pKpQiL-like IncFII(k2)-IncFIB(pQiL) plasmids.
RESULTS
The percentage of whole-genome multilocus sequence typing (wgMLST) allele differences between the 3 K. pneumoniae isolates ranged from 0.002% (Pk1 versus Pk2) to 0.770% (Pk1 [and Pk2] versus Pk4). The percentage of wgMLST allele differences between the 2 Klebsiella aerogenes isolates was 0.979%. In all epidemiologically related isolates, a circular plasmid contig containing a blaKPC-2 gene (located within a Tn4401a transposon) and an IncFII(k2) and IncFIB plasmid replicon was detected and extracted from the assembly graph (Table 1). The plasmid contigs were either 113,638 or 113,639 bp, had a GC content of 53.9%, and contained the acquired beta-lactam resistance genes blaTEM1A and blaOXA-9. Fifteen epidemiologically unrelated plasmids were detected in and extracted from GenBank. The plasmids were isolated from patients in 5 different countries (Greece, Italy, the United States, Australia, and the United Kingdom) between 2007 and 2017 and ranged in size from 99,142 to 117,916 bp (Table 1) (13, 14, 23). Global sequence alignment revealed 13 plasmids to be entirely homologous (see Fig. S1 in the supplemental material). In the other 8 plasmids, only a limited number of structural rearrangements, deletions, or/and insertions were detected (Fig. S1).
TABLE 1.
Plasmids included from the outbreak and GenBanka
| Patient | Host species | Host MLST | Plasmid name | Epidemiologically related | Accession no. | Yr isolated | Location | Plasmid size (bp) | No. of contigs/plasmid |
|---|---|---|---|---|---|---|---|---|---|
| P1 (index) | K. pneumoniae | 258 | Pk1 | Yes | SAMEA7502064 | 2013 | Breda, Netherlands | 113,639 | 1 |
| P2 | K. pneumoniae | 258 | Pk2 | Yes | SAMEA7502065 | 2013 | Breda, Netherlands | 113,638 | 1 |
| P3 | K. aerogenes | NA | Pk3 | Yes | SAMEA7502066 | 2013 | Breda, Netherlands | 113,639 | 1 |
| P3 | E. coli | 2598 | Pe1 | Yes | SAMEA7502069 | 2013 | Breda, Netherlands | 113,639 | 1 |
| P3 | K. pneumoniae | 309 | Pk4 | Yes | SAMEA7502067 | 2014 | Breda, Netherlands | 113,639 | 1 |
| P5 | K. aerogenes | NA | Pk5 | Yes | SAMEA7502068 | 2013 | Breda, Netherlands | 113,639 | 1 |
| NA | K. pneumoniae | 258 | pKP1504-kpc | No | NC_023903.1 | 2008 | Athens, Greece | 113,640 | NA |
| NA | K. pneumoniae | 147 | pKP1780-kpc | No | NC_023904.1 | 2009 | Heraklion, Greece | 113,622 | NA |
| NA | K. pneumoniae | 234 | pKpQiL-234 | No | NC_025187.1 | 2009 | Central New Jersey, USA | 114,464 | NA |
| NA | K. pneumoniae | 10 | pKpQiL-10 | No | NC_025166.1 | 2010 | New York City, NY, USA | 113,639 | NA |
| NA | K. pneumoniae | 35 | pKP3913-kpc | No | NC_023906.1 | 2011 | Athens, Greece | 113,640 | NA |
| NA | K. pneumoniae | 11 | pKP1870-kpc | No | NC_023905.1 | 2009 | Agios Nikolaos, Greece | 116,047 | NA |
| NA | E. coli | 131 | pKpQIL-Ec | No | NC_025167.1 | 2010 | Northern New Jersey, USA | 99,142 | NA |
| NA | K. pneumoniae | 258 | pKpQIL-531 | No | NZ_CP008833.1 | 2013 | Bethesda, MD, USA | 113,639 | NA |
| NA | K. pneumoniae | 258 | pUHKPC07 | No | NZ_CP011986.1 | 2007 | Cleveland, OH, USA | 113,639 | NA |
| NA | K. pneumoniae | 258 | pUHKPC33 | No | NZ_CP011991.1 | 2008 | Cleveland, OH, USA | 113,638 | NA |
| NA | K. pneumoniae | 258 | p500_1420 | No | NZ_CP011981.1 | 2012 | Northeastern Ohio, USA | 130,552 | NA |
| NA | K. pneumoniae | 258 | CR14_p3 | No | NZ_CP015395.1 | 2012 | Valhalla, NY, USA | 116,419 | NA |
| NA | K. pneumoniae | NA | KPN207_p2 | No | NZ_LT216438.1 | 2016b | United Kingdom | 117,916 | NA |
| NA | K. pneumoniae | 258 | pAUSMDU8079-2 | No | NZ_CP022693.1 | 2017b | Australia | 113,639 | NA |
| NA | K. pneumoniae | 258 | pIT-01C03 | No | HG969995 | 2011 | Milan, Italy | 113,642 | NA |
NA, not available.
Year and location of GenBank submission are given; no further information is available.
The number of pairwise comparisons was 15 for the epidemiologically related plasmids and 120 for the epidemiologically unrelated plasmids (including the plasmid from the index patient) (Table 2). The number of aligned bases used for the SNP analysis between each plasmid and the reference ranged from 80,591 to 111,332 (see Table S1). The median number of SNPs varied significantly between the two groups (P < 0.001) (Table 2). However, the range of SNPs overlapped, ranging from 0 to 1 for the epidemiologically related plasmids and from 0 to 674 for the epidemiologically unrelated plasmids (Table 2; Table S2). A total of 192 genes were detected, of which 56 were present in all plasmids. The phylogenetic distance as determined using Roary ranged from 0.00 to 0.06 between the epidemiologically related plasmids and from 0.00 to 1.69 between the epidemiologically unrelated plasmids (P < 0.001) (Table 2; Table S3). The Stoesser similarity index ranged from 0.99 to 1 between the epidemiologically related plasmids and from 0.51 to 1 between the epidemiologically unrelated plasmids (P < 0.001) (Table 2; Table S4). Between the three comparison methods, the number of SNPs and the Roary phylogenetic distance showed the highest degree of correlation, with a Spearman’s rank correlation coefficient of 0.820 (P < 0.001). The Spearman’s rank correlation coefficients between the Stoesser similarity index and the number of SNPs or Roary phylogenetic distance were −0.65 (P < 0.001) and −0.77 (P < 0.001), respectively.
TABLE 2.
Number of pairwise comparisons, SNPs, genes variably present or present, and combined number of SNPs and variable gene presence between the plasmidsa
| Parameter | SNPs |
Phylogenetic distance using Roary |
Stoesser similarity index |
|||
|---|---|---|---|---|---|---|
| Related | Unrelated | Related | Unrelated | Related | Unrelated | |
| Median | 0 | 29 | 0.00 | 0.37 | 1.00 | 0.90 |
| Range | 0–1 | 0–674 | 0.00–0.06 | 0.00–1.69 | 1.00–0.99 | 1–0.51 |
The number of pairwise comparisons for related plasmids was 15, and the number for unrelated plasmids was 120.
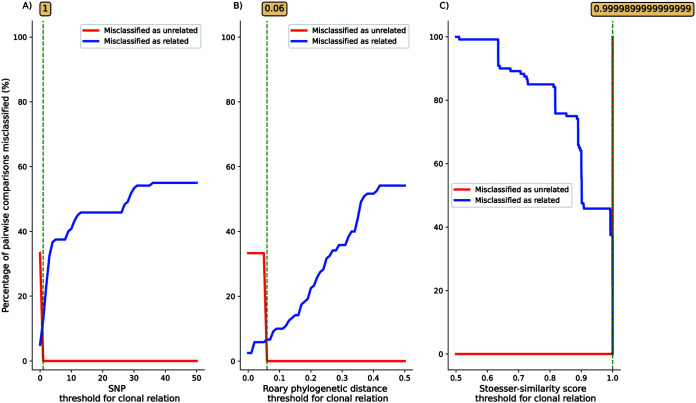
When the threshold was set at the minimal value (or maximal value for the Stoesser similarity index) that classified 100% of epidemiologically related plasmid pairs as genetically related, the percentage of presumed epidemiologically unrelated plasmid pairs that were classified as epidemiologically related was 12.5% with the SNPs approach (Fig. 1A), 6.7% with the Roary phylogenetic distance approach (Fig. 1B), and 20.8% for the Stoesser similarity index approach (Fig. 1C).
FIG 1.
Percentage of pairwise comparisons of epidemiologically related and unrelated plasmids misclassified as either genetically related (blue) or genetically unrelated (red) using different thresholds of (A) number of SNPs, (B) Roary phylogenetic distance, and (C) Stoesser similarity index. Green dotted lines with numbers in boxes show minimal threshold values (or maximal value for the Stoesser similarity index) that classified 100% of epidemiologically related plasmid pairs as genetically related.
DISCUSSION
In this study on the genetic diversity of blaKPC-containing IncF plasmids, epidemiologically related plasmids were on average genetically more related than epidemiologically unrelated plasmids. However, the genetic diversity of plasmids was limited, which precluded 100% correct classification of epidemiologically related and unrelated isolates. When a genetic similarity threshold that classified 100% of epidemiologically related plasmid pairs as genetically related was used, the percentages of plasmids misclassified as epidemiologically related based on the sequence data ranged from 6.7% to 20.8% depending on the comparison method. The phylogenetic distance determined using Roary showed the highest discriminatory power between the epidemiologically related and unrelated plasmids. Since Roary relies primarily on differences in gene presence to determine the phylogenetic distance between sequences, solely using Roary to differentiate epidemiologically related from unrelated plasmid sequences could lead to falsely dismissing plasmid transmission when large rearrangements occur between different plasmids present in an outbreak. These within-outbreak plasmid rearrangements have been described in previous studies (18, 24). Within-outbreak rearrangements might have less effect on a SNP-based analysis, which uses only blocks of shared sequences for its analysis. Therefore, it might be necessary to tailor the method used, e.g., SNPs or Roary (or both), to differentiate between epidemiologically related and unrelated plasmids to each specific plasmid outbreak setting. The relatively poor performance of the Stoesser index in this analysis could be due to the presence of 9 plasmids of equal sequence length (5 epidemiologically related; 4 epidemiologically unrelated), which is an important parameter in determining the phylogenetic distance in this method.
Molecular plasmid typing has been used to demonstrate the transmission of blaKPC-containing plasmids between bacterial isolates and patients (16, 17). However, previous studies did not include similar plasmids of unrelated patients in the analysis. Our findings on the degree of sequence similarity between blaKPC-containing plasmids isolated from epidemiologically related and unrelated patients can be used to reveal a possible horizontal transfer of blaKPC-containing IncFII(k2)-IncFIB(pQiL) plasmids in hospital outbreaks. Despite this, when the lowest (or highest for the Stoesser-similarity index) threshold for clonal relatedness that classified 100% of epidemiologically related plasmid pairs as genetically related was used, the percentage of presumed epidemiologically unrelated plasmid pairs that were classified as genetically related was higher in blaKPC-containing IncFII(k2)-IncFIB(pQiL) plasmids than the percentages described when the bacterial chromosome was typed in a previous study (25). This relatively high percentage of misclassifications compared to bacterial chromosome typing was present in all three comparison methods, suggesting a relatively stable plasmid content. Several other studies have also described a high degree of sequence similarity between blaKPC-containing plasmids isolated from epidemiologically unrelated patients (13, 23). However, to the best of our knowledge, this is the first study comparing the sequence similarity between epidemiologically related and unrelated blaKPC-containing IncFII(k2)-IncFIB(pQiL) plasmids.
The limited number of differences between plasmids isolated from epidemiologically unrelated patients observed in this study is also seen for other resistance plasmids (26, 27). OXA-48-containing plasmids belonging to the IncL/M replicon type have a far wider host range than the blaKPC-containing IncFII(k2)-IncFIB(pQiL) plasmids included in our study. Despite this, a recent study has stated that even these OXA-48 containing plasmids from outbreak-related patients could not be distinguished from similar plasmids of non-outbreak-related patients based on SNPs (26). Interestingly, other studies found blaKPC-containing plasmids to be highly variable in their nucleotide sequence within the same outbreak and even within patients (18, 28). This suggests that the plasmid content could also be highly unstable in vivo during hospital outbreaks. Therefore, it could well be that transmission of blaKPC-containing IncF plasmids within hospital outbreaks cannot be dismissed based on sequence dissimilarity between the different plasmids investigated.
This study included bacterial isolates encompassing three different species isolated from an extensively described outbreak with clear epidemiological links between the different outbreak patients. Moreover, the epidemiologically related blaKPC-containing IncFII(k2)-IncFIB(pQiL) plasmids were compared to a set of similar unrelated plasmids using both a SNP and gene-by-gene approach in a setting where blaKPC-containing Enterobacteriaceae are nonendemic. This limits the possibility of falsely classifying blaKPC-containing plasmids as epidemiologically related. Furthermore, all epidemiologically related blaKPC-containing IncFII(k2)-IncFIB(pQiL) plasmid sequences in this study were single circular contigs in the assembly graph.
This study has several limitations. Only blaKPC-containing IncFII(k2)-IncFIB(pQiL) plasmid sequences of six isolates in one outbreak were included. Therefore, it remains unknown whether plasmids belonging to other incompatibility groups and containing other resistance genes also show a high degree of sequence similarity to epidemiologically related plasmids and whether Roary also shows the highest discriminatory power between the epidemiologically related and unrelated plasmids in other plasmid types. Moreover, studies that include more epidemiologically related and unrelated blaKPC-containing IncFII(k2)-IncFIB(pQiL) plasmids are needed to confirm our findings. Additionally, the relatively high occurrence of horizontal gene transfer of blaKPC-containing IncFII(k2)-IncFIB(pQiL) plasmids detected in the outbreak included here could be facilitated by the use of antibiotics in 3 of the 6 patients included in our outbreak. Therefore, future studies should be performed, including epidemiologically related blaKPC-containing IncFII(k2)-IncFIB(pQiL) plasmids isolated in other outbreaks, to confirm our findings.
To conclude, although epidemiologically related blaKPC-containing IncF plasmids can be distinguished from unrelated blaKPC-containing IncF plasmids based on genetic differences, epidemiologically related and unrelated blaKPC-containing IncF plasmids show a high degree of sequence similarity. The phylogenetic distance as determined using Roary showed the highest degree of discriminatory power between the epidemiologically related and unrelated blaKPC-containing IncF plasmids.
MATERIALS AND METHODS
Outbreak and plasmid selection.
From June to December 2013, an outbreak occurred in an 800-bed teaching hospital (approximately 40,000 admissions/year) and a 150-bed nursing home in Breda, the Netherlands. In total, six patients were colonized or infected with KPC-producing Enterobacteriaceae belonging to three different species (K. pneumoniae, K. aerogenes, and Escherichia coli). The outbreak was recognized on 13 July 2013, when a KPC-producing K. pneumoniae isolate was detected in the pleural fluid of patient 2 in the pulmonary ward. This event followed the earlier repatriation of patient 1 from an intensive care unit in Greece on June 2013, who was found to be colonized with a KPC-producing K. pneumoniae isolate 2 days after admission. Patient 1 was transferred to a nursing home in August 2013, where further transmission occurred between November and December 2013 from patient 1 to 4 other patients in the nursing home. The outbreak was comprehensively described by Weterings et al. (6).
Epidemiologically related isolates (i.e., belonging to the outbreak) were selected in such a way that at least every unique blaKPC-containing isolate, based on species identification, antimicrobial susceptibility testing, and molecular typing (6), was included. Moreover, one additional KPC-producing K. pneumoniae isolate was included that was cultured only 2 months after the outbreak period in February 2014 from a rectal swab taken from patient 3. The antimicrobial susceptibility testing results for this isolate differed from those of the outbreak KPC-producing K. pneumoniae isolates, as it was susceptible to ciprofloxacin, tobramycin, and trimethoprim-sulfamethoxazole according to Vitek-2 (bioMérieux, Marcy-l’Étoile, France) automated susceptibility testing using EUCAST breakpoints (v10.0). Epidemiologically unrelated plasmid sequences containing a blaKPC-2 gene and the replicon IncFIIK2-FIB were extracted from GenBank on 11 September 2017 (see the supplemental materials and methods). To ensure epidemiological unrelatedness, only the first uploaded plasmid sequence per geographic location per year was included, and no sequences with a Dutch origin and isolated in 2013/2014 were included in the analysis.
Whole-genome sequencing and analysis.
Epidemiologically related isolates were subjected to short-read sequencing on an Illumina MiSeq platform using Nextera XT chemistry generating 250-bp paired-end reads (Illumina, San Diego, CA, USA) and long-read sequencing on a MinION sequencer using the FLO-MIN106D flow cell and the SQK RBK004 rapid barcoding sequencing kit according to the standard protocol provided by the manufacturer (Oxford Nanopore Technologies, Oxford, United Kingdom). A hybrid assembly of long-read and short-read sequence data was performed using Unicycler v.0.8.4 (29). Whole-genome MLST (wgMLST) (core and accessory genome) was performed for all sequenced isolates using Ridom SeqSphere+, version 4.1.9 (Ridom, Münster, Germany). Species-specific typing schemes were used as described by Kluytmans-van den Bergh et al. (25). The pairwise genetic difference between isolates of the same species was calculated by dividing the number of allele differences by the total number of alleles shared between the two sequences in the wgMLST typing scheme. The sequenced isolates’ genomes were uploaded to the online bioinformatic tools ResFinder v.3.1 and PlasmidFinder v.2.0 (Center for Genomic Epidemiology, Technical University of Denmark, Lyngby, Denmark) (30, 31). Acquired resistance genes were called when at least 60% of the length of the best matching gene in the ResFinder database was covered with a sequence identity of at least 90%. Plasmid replicon genes were called when at least 60% of the sequence length of the replicon gene in the PlasmidFinder database was covered with a sequence identity of at least 80%.
Plasmid analysis.
Circular components created by the hybrid assembly that were smaller than 1,000 kb and contained a blaKPC gene were extracted from the assembly graph using BANDAGE v0.8.1 (32). All extracted plasmid components and the plasmid sequences extracted from the GenBank were annotated using Prokka v1.13.3 (33). A global sequence alignment of the extracted plasmids was performed using progressiveMAUVE v2.4.0. The number of SNPs between the plasmids were calculated using snippy v4.4.5 (reference sequence: GenBank accession number NC_014016.1). A pangenome was constructed, and phylogenetic distance based on gene presence/absence between the different plasmids was determined using Roary v1.13.2 (34). All plasmids were pairwise aligned using dnadiff, and a similarity index was calculated between the different plasmids as described by Stoesser et al. (28) (Stoesser similarity index). This similarity index can vary from 0 (completely unsimilar plasmids) to 1 (identical plasmids) and, contrary to both Roary phylogenetic distance and number of SNPs, penalizes size differences between plasmids. Pairwise comparisons of SNPs, Roary phylogenetic distance, and Stoesser similarity were performed between (i) epidemiologically related plasmids and (ii) the first plasmid isolated from the index patient in the outbreak and the epidemiologically unrelated plasmid sequences extracted from GenBank. The percentage of pairwise comparisons misclassified as genetically related or as clonally unrelated was determined using different genetic thresholds for genetic relatedness for all three comparison methods: SNP thresholds tested ranged from 0 to 50 with steps of 1, Roary thresholds from 0.0 to 0.5 with steps of 0.01, and Stoesser index thresholds from 0.5 to 1 with steps of 0.00001.
Statistical analysis.
The median number of SNPs, the median Roary phylogenetic distance, and the median Stoesser similarity index were calculated for epidemiologically related and unrelated plasmids. Medians were compared using Mann-Whitney U tests (SciPy v1.5.0). Spearman’s rank correlation coefficients between the number of SNP differences, Roary phylogenetic distance, and the Stoesser similarity index were calculated using SciPy v1.5.0.
Data availability.
Short- and long-read sequence data included in this study are available from the European Nucleotide Archive of the European Bioinformatics Institute under study accession number PRJEB41009.
ACKNOWLEDGMENT
We declare no competing interests.
Footnotes
Supplemental material is available online only.
aac.00147-21-s0005.pdf (1.8MB, pdf)
aac.00147-21-s0001.xlsx (10.8KB, xlsx)
aac.00147-21-s0002.xlsx (11.1KB, xlsx)
aac.00147-21-s0003.xlsx (12.2KB, xlsx)
aac.00147-21-s0004.xlsx (12.3KB, xlsx)
REFERENCES
- 1.Tascini C, Lipsky BA, Iacopi E, Ripoli A, Sbrana F, Coppelli A, Goretti C, Piaggesi A, Menichetti F. 2015. KPC-producing Klebsiella pneumoniae rectal colonization is a risk factor for mortality in patients with diabetic foot infections. Clin Microbiol Infect 21:790.e1–790.e3. doi: 10.1016/j.cmi.2015.04.010. [DOI] [PubMed] [Google Scholar]
- 2.Xu L, Sun X, Ma X. 2017. Systematic review and meta-analysis of mortality of patients infected with carbapenem-resistant Klebsiella pneumoniae. Ann Clin Microbiol Antimicrob 16:18. doi: 10.1186/s12941-017-0191-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Munoz-Price LS, Poirel L, Bonomo RA, Schwaber MJ, Daikos GL, Cormican M, Cornaglia G, Garau J, Gniadkowski M, Hayden MK, Kumarasamy K, Livermore DM, Maya JJ, Nordmann P, Patel JB, Paterson DL, Pitout J, Villegas MV, Wang H, Woodford N, Quinn JP. 2013. Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect Dis 13:785–796. doi: 10.1016/S1473-3099(13)70190-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Duin D, Doi Y. 2017. The global epidemiology of carbapenemase-producing Enterobacteriaceae. Virulence 8:460–469. doi: 10.1080/21505594.2016.1222343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ruppé E, Olearo F, Pires D, Baud D, Renzi G, Cherkaoui A, Goldenberger D, Huttner A, François P, Harbarth S, Schrenzel J. 2017. Clonal or not clonal? Investigating hospital outbreaks of KPC-producing Klebsiella pneumoniae with whole-genome sequencing. Clin Microbiol Infect 23:470–475. doi: 10.1016/j.cmi.2017.01.015. [DOI] [PubMed] [Google Scholar]
- 6.Weterings V, Zhou K, Rossen JW, van Stenis D, Thewessen E, Kluytmans J, Veenemans J. 2015. An outbreak of colistin-resistant Klebsiella pneumoniae carbapenemase-producing Klebsiella pneumoniae in the Netherlands (July to December 2013), with inter-institutional spread. Eur J Clin Microbiol Infect Dis 34:1647–1655. doi: 10.1007/s10096-015-2401-2. [DOI] [PubMed] [Google Scholar]
- 7.Carbonne A, Thiolet JM, Fournier S, Fortineau N, Kassis-Chikhani N, Boytchev I, Aggoune M, Séguier JC, Sénéchal H, Tavolacci MP, Coignard B, Astagneau P, Jarlier V. 2010. Control of a multi-hospital outbreak of KPC-producing Klebsiella pneumoniae type 2 in France, September to October 2009. Eurosurveillance 15:4–9. doi: 10.2807/ese.15.48.19734-en. [DOI] [PubMed] [Google Scholar]
- 8.David S, Reuter S, Harris SR, Glasner C, Feltwell T, Argimon S, Abudahab K, Goater R, Giani T, Errico G, Aspbury M, Sjunnebo S, Feil EJ, Rossolini GM, Aanensen DM, Grundmann H, the EuSCAPE Working Group. 2019. Epidemic of carbapenem-resistant Klebsiella pneumoniae in Europe is driven by nosocomial spread. Nat Microbiol 4:1919–1929. doi: 10.1038/s41564-019-0492-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Quainoo S, Coolen JPM, van Hijum SAFT, Huynen MA, Melchers WJG, van Schaik W, Wertheim HFL. 2017. Whole-genome sequencing of bacterial pathogens: the future of nosocomial outbreak analysis. Clin Microbiol Rev 30:1015–1063. doi: 10.1128/CMR.00016-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schürch AC, Arredondo-Alonso S, Willems RJL, Goering RV. 2018. Whole genome sequencing options for bacterial strain typing and epidemiologic analysis based on single nucleotide polymorphism versus gene-by-gene–based approaches. Clin Microbiol Infect 24:350–354. doi: 10.1016/j.cmi.2017.12.016. [DOI] [PubMed] [Google Scholar]
- 11.Willemsen I, van Esser J, Kluytmans-van den Bergh M, Zhou K, Rossen JW, Verhulst C, Verduin K, Kluytmans J. 2016. Retrospective identification of a previously undetected clinical case of OXA-48-producing K. pneumoniae and E. coli: the importance of adequate detection guidelines. Infection 44:107–110. doi: 10.1007/s15010-015-0805-7. [DOI] [PubMed] [Google Scholar]
- 12.Doumith M, Findlay J, Hirani H, Hopkins KL, Livermore DM, Dodgson A, Woodford N. 2017. Major role of pKpQIL-like plasmids in the early dissemination of KPC-type carbapenemases in the UK. J Antimicrob Chemother 72:2241–2248. doi: 10.1093/jac/dkx141. [DOI] [PubMed] [Google Scholar]
- 13.Chen L, Chavda KD, Melano RG, Jacobs MR, Koll B, Hong T, Rojtman AD, Levi MH, Bonomo RA, Kreiswirth BN. 2014. Comparative genomic analysis of kpc-encoding pkpqil-like plasmids and their distribution in New Jersey and New York hospitals. Antimicrob Agents Chemother 58:2871–2877. doi: 10.1128/AAC.00120-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leavitt A, Chmelnitsky I, Carmeli Y, Navon-Venezia S. 2010. Complete nucleotide sequence of KPC-3-encoding plasmid pKpQIL in the epidemic Klebsiella pneumoniae sequence type 258. Antimicrob Agents Chemother 54:4493–4496. doi: 10.1128/AAC.00175-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tofteland S, Naseer U, Lislevand JH, Sundsfjord A, Samuelsen Ø. 2013. A long-term low-frequency hospital outbreak of KPC-producing Klebsiella pneumoniae involving intergenus plasmid diffusion and a persisting environmental reservoir. PLoS One 8:e59015. doi: 10.1371/journal.pone.0059015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mathers AJ, Cox HL, Kitchel B, Bonatti H, Brassinga AKC, Carroll J, Scheld WM, Hazen KC, Sifri CD. 2011. Molecular dissection of an outbreak of carbapenem-resistant Enterobacteriaceae reveals intergenus KPC carbapenemase transmission through a promiscuous plasmid. mBio 2:e00204-11. doi: 10.1128/mBio.00204-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Conlan S, Thomas PJ, Deming C, Park M, Lau AF, Dekker JP, Snitkin ES, Clark TA, Luong K, Song Y, Tsai Y-C, Boitano M, Dayal J, Brooks SY, Schmidt B, Young AC, Thomas JW, Bouffard GG, Blakesley RW, Mullikin JC, Korlach J, Henderson DK, Frank KM, Palmore TN, Segre JA, NISC Comparative Sequencing Program. 2014. Single-molecule sequencing to track plasmid diversity of hospital-associated carbapenemase-producing Enterobacteriaceae. Sci Transl Med 6:254ra126. doi: 10.1126/scitranslmed.3009845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sheppard AE, Stoesser N, Wilson DJ, Sebra R, Kasarskis A, Anson LW, Giess A, Pankhurst LJ, Vaughan A, Grim CJ, Cox HL, Yeh AJ, Sifri CD, Walker AS, Peto TE, Crook DW, Mathers AJ, Modernising Medical Microbiology (MMM) Informatics Group. 2016. Nested Russian doll-like genetic mobility drives rapid dissemination of the carbapenem resistance gene blaKPC. Antimicrob Agents Chemother 60:3767–3778. doi: 10.1128/AAC.00464-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martin J, Phan HTT, Findlay J, Stoesser N, Pankhurst L, Navickaite I, De Maio N, Eyre DW, Toogood G, Orsi NM, Kirby A, Young N, Turton JF, Hill RLR, Hopkins KL, Woodford N, Peto TEA, Walker AS, Crook DW, Wilcox MH. 2017. Covert dissemination of carbapenemase-producing Klebsiella pneumoniae (KPC) in a successfully controlled outbreak: long- and short-read whole-genome sequencing demonstrate multiple genetic modes of transmission. J Antimicrob Chemother 72:3025–3034. doi: 10.1093/jac/dkx264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wick RR, Judd LM, Gorrie CL, Holt KE. 2017. Completing bacterial genome assemblies with multiplex MinION sequencing. Microb Genom 3:000132. doi: 10.1099/mgen.0.000132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.George S, Pankhurst L, Hubbard A, Votintseva A, Stoesser N, Sheppard AE, Mathers A, Norris R, Navickaite I, Eaton C, Iqbal Z, Crook DW, Phan HTT. 2017. Resolving plasmid structures in Enterobacteriaceae using the MinION nanopore sequencer: assessment of MinION and MinION/Illumina hybrid data assembly approaches. Microb Genom 3:000118. doi: 10.1099/mgen.0.000118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arredondo-Alonso S, Willems RJ, van Schaik W, Schürch AC. 2017. On the (im)possibility of reconstructing plasmids from whole-genome short-read sequencing data. Microb Genom 3:000128. doi: 10.1099/mgen.0.000128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Papagiannitsis CC, Di Pilato V, Giani T, Giakkoupi P, Riccobono E, Landini G, Miriagou V, Vatopoulos AC, Rossolini GM. 2016. Characterization of KPC-encoding plasmids from two endemic settings, Greece and Italy. J Antimicrob Chemother 71:2824–2830. doi: 10.1093/jac/dkw227. [DOI] [PubMed] [Google Scholar]
- 24.Stohr JJJM, Verweij JJ, Buiting AGM, Rossen JWA, Kluytmans JAJW. 2020. Within-patient plasmid dynamics in Klebsiella pneumoniae during an outbreak of a carbapenemase-producing Klebsiella pneumoniae. PLoS One 15:e0233313. doi: 10.1371/journal.pone.0233313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kluytmans-van den Bergh MFQ, Rossen JWA, Bruijning-Verhagen PCJ, Bonten MJM, Friedrich AW, Vandenbroucke-Grauls CMJE, Willems RJL, Kluytmans JAJW. 2016. Whole-genome multilocus sequence typing of extended-spectrum-beta-lactamase-producing Enterobacteriaceae. J Clin Microbiol 54:2919–2927. doi: 10.1128/JCM.01648-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hidalgo L, de Been M, Rogers MRC, Schürch AC, Scharringa J, van der Zee A, Bonten MJM, Fluit AC. 2019. Sequence-based epidemiology of an OXA-48 plasmid during a hospital outbreak. Antimicrob Agents Chemother 63:e01204-19. doi: 10.1128/AAC.01204-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roer L, Overballe-Petersen S, Hansen F, Johannesen TB, Stegger M, Bortolaia V, Leekitcharoenphon P, Korsgaard HB, Seyfarth AM, Mossong J, Wattiau P, Boland C, Hansen DS, Hasman H, Hammerum AM, Hendriksen RS. 2019. ST131 fimH22 Escherichia coli isolate with a blaCMY-2/IncI1/ST12 plasmid obtained from a patient with bloodstream infection: highly similar to E. coli isolates of broiler origin. J Antimicrob Chemother 74:557–560. doi: 10.1093/jac/dky484. [DOI] [PubMed] [Google Scholar]
- 28.Stoesser N, Phan HTT, Seale AC, Aiken Z, Thomas S, Smith M, Wyllie D, George R, Sebra R, Mathers AJ, Vaughan A, Peto TEA, Ellington MJ, Hopkins KL, Crook DW, Orlek A, Welfare W, Cawthorne J, Lenney C, Dodgson A, Woodford N, Walker AS, Aiken Z, Akinremi O, Ali A, Cawthorne J, Cleary P, Crook DW, Decraene V, Dodgson A, Doumith M, Ellington MJ, George R, Grimshaw J, Guiver M, Hill R, Hopkins KL, Jones R, Lenney C, Mathers AJ, McEwan A, Moore G, Neilson M, Neilson S, Peto TEA, Phan HTT, Regan M, Seale AC, Stoesser N, Turner-Gardner J, et al. 2020. Genomic epidemiology of complex, multispecies, plasmid-borne blaKPC carbapenemase in Enterobacterales in the United Kingdom from 2009 to 2014. Antimicrob Agents Chemother 64:1–18. doi: 10.1128/AAC.02244-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wick RR, Judd LM, Gorrie CL, Holt KE. 2017. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol 13:e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carattoli A, Zankari E, García-Fernández A, Voldby Larsen M, Lund O, Villa L, Møller Aarestrup F, Hasman H. 2014. In silico detection and typing of plasmids using plasmidfinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV. 2012. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wick RR, Schultz MB, Zobel J, Holt KE. 2015. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics 31:3350–3352. doi: 10.1093/bioinformatics/btv383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 34.Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, Holden MTG, Fookes M, Falush D, Keane JA, Parkhill J. 2015. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31:3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Short- and long-read sequence data included in this study are available from the European Nucleotide Archive of the European Bioinformatics Institute under study accession number PRJEB41009.