ABSTRACT
Hepatitis B virus (HBV) capsid or core protein (HBc) consists of an N-terminal domain (NTD) and a C-terminal domain (CTD) connected by a short linker peptide. Dynamic phosphorylation and dephosphorylation of HBc regulate its multiple functions in capsid assembly and viral replication. The cellular cyclin-dependent kinase 2 (CDK2) plays a major role in HBc phosphorylation and, furthermore, is incorporated into the viral capsid, accounting for most of the “endogenous kinase” activity associated with the capsid. The packaged CDK2 is thought to play a role in phosphorylating HBc to trigger nucleocapsid disassembly (uncoating), an essential step during viral infection. However, little is currently known on how CDK2 is recruited and packaged into the capsid. We have now identified three RXL motifs in the HBc NTD known as cyclin docking motifs (CDMs), which mediate the interactions of various CDK substrates/regulators with CDK/cyclin complexes. Mutations of the CDMs in the HBc NTD reduced CTD phosphorylation and diminished CDK2 packaging into the capsid. Also, the CDM mutations showed little effects on capsid assembly and pregenomic RNA (pgRNA) packaging but impaired the integrity of mature nucleocapsids. Furthermore, the CDM mutations blocked covalently closed circular DNA (CCC DNA) formation during infection while having no effect on or enhancing CCC DNA formation via intracellular amplification. These results indicate that the HBc NTD CDMs play a role in CDK2 recruitment and packaging, which, in turn, is important for productive infection.
IMPORTANCE Hepatitis B virus (HBV) is an important global human pathogen and persistently infects hundreds of millions of people, who are at high risk of cirrhosis and liver cancer. HBV capsid packages a host cell protein kinase, the cyclin-dependent kinase 2 (CDK2), which is thought to be required to trigger disassembly of the viral nucleocapsid during infection by phosphorylating the capsid protein, a prerequisite for successful infection. We have identified docking sites on the capsid protein for recruiting CDK2, in complex with its cyclin partner, to facilitate capsid protein phosphorylation and CDK2 packaging. Mutations of these docking sites reduced capsid protein phosphorylation, impaired CDK2 packaging into HBV capsids, and blocked HBV infection. These results provide novel insights regarding CDK2 packaging into HBV capsids and the role of CDK2 in HBV infection and should facilitate the development of antiviral drugs that target the HBV capsid protein.
KEYWORDS: capsid, core protein, covalently closed circular DNA, cyclin, cyclin docking motif, cyclin-dependent kinase 2, endogenous kinase, hepadnavirus, hepatitis B virus, infection
INTRODUCTION
Chronic hepatitis B virus (HBV) infection affects approximately 292 million people worldwide, which results in 887,000 deaths annually primarily from complications of cirrhosis and hepatocellular carcinoma (1, 2). Current therapies for chronic HBV infection can reduce viral replication and alleviate diseases but rarely cure the infection (1, 3). HBV belongs to the family Hepadnaviridae, a family of hepatotropic DNA viruses with a ca. 3.2-kb partially double-stranded, relaxed circular (RC) DNA genome in an icosahedral capsid, which is in turn enclosed in an envelope studded with three viral envelope (surface) proteins (4, 5). During infection, the RC DNA from the incoming virion is delivered to the hepatocyte nucleus and converted to the covalently closed circular (CCC) DNA, which is a definitive sign of successful infection. CCC DNA then serves as the transcriptional template for the production of all viral RNAs, including the pregenomic RNA (pgRNA). pgRNA serves as the mRNA for the translation of the HBV capsid (core) protein (HBc) and the reverse transcriptase (RT) protein. HBc, together with the RT protein, packages the pgRNA into an immature nucleocapsid (NC) during capsid assembly in which the pgRNA is converted to a single-stranded (SS) DNA and then to the RC DNA by the RT in a process termed NC maturation. The resulting mature NC, containing the RC DNA, then interacts with the envelope proteins to form virions, which are secreted out of the cell. Interestingly, the mature NC, instead of being enveloped, can also deliver its RC DNA content back to the nucleus to form more CCC DNA in a process called intracellular CCC DNA amplification or recycling (6–8).
HBc contains 183 (or 185, depending on genotypes) residues and can be divided into two domains. The N-terminal domain (NTD) consists of residues 1 to 140, which are linked by a flexible linker to the arginine-rich C-terminal domain (CTD; residues 150 to 183) (9, 10). The NTD is known to self-assemble into capsids at sufficiently high concentrations, but assembly is dependent on, and regulated by, the CTD under physiological conditions in vitro or in vivo (11, 12). The linker was traditionally thought to be merely a spacer between the NTD and CTD but was recently shown to play a major role in multiple stages of viral replication (13, 14). The CTD contains seven conserved phosphor acceptor sites, the dynamic (de)phosphorylation of which plays multiple roles in regulating HBV capsid assembly, pgRNA packaging, and DNA synthesis (15–21). The three S-P sites at positions 155, 162, and 170 are highly conserved major phosphorylation sites (16, 22, 23), whereas T160, S168, S176, and S178 are potential minor phosphorylation sites (15, 21, 24, 25). Our recent work suggests that two S-P sites (S44 and S49) in the NTD may also undergo transient phosphorylation to facilitate disassembly (uncoating) (see below) (25). HBc phosphorylation state does not appear to play an essential role in directing virion formation (24–26). Instead, a region on the capsid surface (the matrix binding domain [MBD]) interacts with a linear, ca. 22-amino-acid-long domain (the matrix domain [MD]) of the L surface protein for virion secretion (26–29), which is further regulated by the HBc linker (14, 21).
HBc can regulate CCC DNA formation, a critical step in the establishment and persistence of HBV infection, through at least three potential mechanisms. First, through its interactions with the envelope proteins for virion secretion, HBc can negatively regulate CCC DNA formation via a negative-feedback mechanism, which can be impaired by mutations in the L envelope protein as well as HBc, resulting in enhanced CCC DNA amplification (14, 30–34). Second, NC disassembly, which is subject to regulation by dynamic HBc phosphorylation and dephosphorylation (25, 35, 36), controls the release of the RC DNA genome and, thus, its subsequent conversion to CCC DNA (14, 25, 37). In fact, NC disassembly may be a critical determinant of HBV host species tropism (38). Third, HBc controls CCC DNA formation via the regulation of RC DNA nuclear import. During NC maturation, the increased negative charge from RC DNA synthesis may induce exposure of the HBc CTD, located inside immature NCs, to the surface of mature NCs, allowing the nuclear localization signals (NLSs) in the CTD to function in delivering the RC DNA into the nucleus for CCC DNA formation (39–41). The HBc function in nuclear import is also subject to regulation by the CTD phosphorylation state (39, 42).
Several host cell protein kinases have been proposed to phosphorylate one or more phosphor acceptor sites in the HBc CTD, including the cyclin-dependent kinase 2 (CDK2), serine-arginine protein kinase (SRPK), protein kinase C (PKC), and the polo-like kinase 1 (PLK1) (22, 23, 43–45). For HBc dephosphorylation, the protein phosphatase 1 (PP1) is reported to be involved in the initial dephosphorylation during pgRNA packaging (46), whereas protein phosphatase 2A (PP2A) is thought to mediate the subsequent dephosphorylation during RC DNA synthesis (14). Furthermore, a so-called “endogenous kinase,” packaged into the HBV capsid that can phosphorylate HBc at S/T residues in the capsid, was discovered decades ago (47, 48). Our previous work identified CDK2 as the major endogenous kinase, which is packaged into capsids independent of other viral proteins or pgRNA (23). Consistent with the known specificity of CDK as a proline-directed kinase (with a preference for S/T-P-X-K/R motifs) (49), CDK2 mainly phosphorylates the three major S-P sites at S155, S162, and S170 in the HBc CTD in vitro and in vivo (23). In addition, CDK2 phosphorylation at these S-P sites may promote subsequent phosphorylation at the other CTD sites by other kinases (15, 43, 50). The packaged CDK2 is thought to be responsible for phosphorylating HBc at its NTD S-P sites (S44 and S49), transiently during infection, to facilitate NC uncoating and thus CCC DNA formation (25).
How CDK2 is recruited to HBc or packaged into capsids remains unknown. In the cell cycle, CDKs are activated by complexing with a particular cyclin partner, which also plays an important role in targeting the partnered CDK to appropriate substrates. CDK2 can be complexed with either cyclin A or E depending on the cell cycle stage and is partnered with cyclin E during the late G1 phase (51, 52). HBV replication is thought to require the host hepatocytes to exit from G0 but then stall in the G1 phase of the cell cycle (53–55). Cyclin proteins contain a conserved domain called cyclin box that is responsible for the recognition of the cyclin docking motifs (CDMs) on the substrates and regulators of CDK/cyclin complexes. One such CDM, with the sequence RXL, mediates association with multiple cyclins, including cyclin E or A (56–59), and together with the phosphor acceptor site sequence helps to recruit the CDK/cyclin complexes to the substrates.
The HBc CTD alone, containing the CDK consensus phosphorylation sites, shows measurable interactions with CDK2 in vitro (21). However, as with other CDK substrates, the phosphor acceptor sites alone are unlikely to be sufficient to recruit CDK2 to HBc and particularly for CDK2 packaging into the capsid. Inspection of the HBc sequences allowed us to identify three highly conserved RXL sequences in its NTD as putative CDMs, potentially helping recruit the CDK2/cyclin complex to HBc and package the kinase into the capsid. Our mutagenesis analysis of these putative CDMs in the HBc NTD provides evidence that these sites indeed play a role in regulating HBc phosphorylation and packaging of CDK2 into the capsid, which, in turn, is important for HBV infection. Our results also reveal that the CDM residues play a key role in regulating the stability/integrity of mature NCs and in virion formation, which in turn have a major effect on CCC DNA formation.
RESULTS
Identification of three putative CDMs in the HBc NTD.
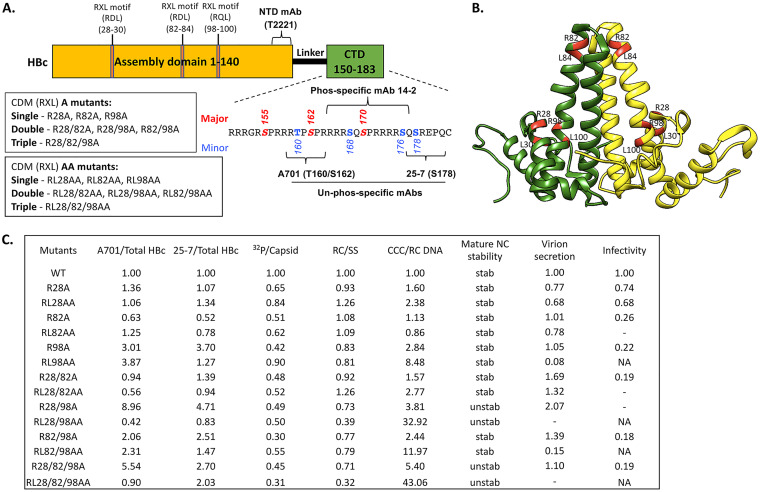
We have reported previously that CDK2 can phosphorylate the three major phosphorylation sites in the HBc CTD and is the major kinase packaged inside HBV capsid (endogenous kinase) (23). CDK2, in complex with cyclins (mainly cyclin E or A), is known to be recruited to specific substrates, in part through cyclin interaction with CDMs in the substrates with the sequence RXL, where X is any amino acid residue (56). Inspection of the HBc sequence indeed showed three such CDMs (RXL) at positions 28 to 30, 82 to 84, and 98 to 100, all in the NTD, separate from the major CDK2 sites of phosphorylation located at the HBc CTD (Fig. 1A and B). To ascertain the role of these three motifs in HBV replication, including the regulation of HBc phosphorylation, we generated two panels of mutants, the A series and the AA series, replacing the conserved R residue at position 28, 82, or 98 alone or in combination with the conserved L residue at position 30, 84, or 100, respectively, either individually or in various combinations, with an A residue (Fig. 1A). If one or more of the CDMs are involved in recruiting CDK2-cyclin to phosphorylate HBc and facilitate CDK2 packaging into capsids, their mutations, by impairing cyclin binding to HBc, diminish CDK2 recruitment to HBc and thus negatively affect HBc phosphorylation by CDK2 and CDK2 packaging to capsids. Both the R and L residues in the RXL motif are important for cyclin docking, with the R side chain involved in ionic interactions with cyclins while the L side chain makes hydrogen bond or van der Waals interactions with cyclins (57). Thus, the AA mutations were anticipated to more severely disrupt CDK2/cyclin docking than the A mutations.
FIG 1.
Summary of the HBc CDM mutants and their phenotypes. (A) Schematic of HBc protein structure highlighting key domains, residues, and MAb epitopes. The NTD, linker, and CTD are indicated. The multiple RXL motifs in the assembly domain (NTD) of HBc at positions 28 to 30, 82 to 84, and 98 to 100 are indicated. The CDM A and CDM AA mutants are described, as is the CTD sequence (positions 150 to 183), with the three major S/T-P phosphorylation motifs (S155, S162, S170) and four minor sites of CTD phosphorylation highlighted. The epitopes/regions recognized by the indicated MAbs are also shown. (B) Localization of the three CDM sites (with the conserved R and L residues in each motif) highlighted in red on the HBc dimer structure (NCBI Protein ID 6HTX) (71). (C) Summary of the effects of the HBc CDM (RXL) mutations on HBc CTD phosphorylation, endogenous kinase activity, and HBV replication in HepG2 cells. The levels of HBc protein that were nonphosphorylated at T160/S162 (within the A701 epitope) or S178 (within the 25-7 epitope) were normalized to total HBc detected by the NTD MAb T2221. The endogenous kinase activity detected by 32P labeling was normalized to the total capsid amount detected by the NTD MAb T2221. RC DNA was normalized to SS DNA as a measure for RC DNA synthesis/stability. The CCC DNA amount normalized to RC DNA represents CCC DNA synthesis efficiency. Mature NC stability was tested by DNase digestion of RC DNA. The DNA virion level was normalized to the intracellular RC DNA amount as a measure of virion secretion efficiency. Viral infectivity was defined by the CCC DNA amount produced during infection by using the same MOI. The normalized level for each parameter from the WT HBc is set at 1.00. unstab, unstable NC; stab, stable NC (similar to WT); NA, not applicable (not enough virion available to assess infectivity); −, undetectable.
The HBc CDM mutations affected CTD phosphorylation.
To determine if the RXL mutants could affect HBc unphosphorylation, we took advantage of two different monoclonal antibodies (MAbs) that selectively recognize the unphosphorylated HBc CTD at two different locations, as we reported before (24). MAb A701 is selective for unphosphorylated S162, which is one of the major CTD sites of phosphorylation, and it may also recognize the unphosphorylated T160, one of the minor phosphorylation sites (Fig. 1A). Another MAb, 25-7, is selective for an unphosphorylated S178 (14), another minor CTD phosphorylation site. An increase in the signals detected by these MAbs could indicate decreased phosphorylation at these CTD sites, although it could also, in principle, reflect increased dephosphorylation at these same sites (see Discussion). We also monitored CTD phosphorylation levels by using MAb 14-2, selective for a phosphorylated epitope between positions 164 and 182 in the HBc CTD, specifically phosphorylated S170. A decrease in the signal detected by Mab 14-2 could indicate decreased phosphorylation at S170, although it could also, in principle, reflect increased dephosphorylation at this site. The total HBc protein levels were measured by using the NTD-specific MAb T2221, recognizing an HBc epitope approximately between positions 130 and 140 and thus detecting HBc independent of its phosphorylation state (Fig. 1A).
We expressed both the A and AA series of the HBc CDM mutants in the human hepatoma cell lines HepG2 and Huh7. Although only results from HepG2 cells are shown here, similar results were also obtained in Huh7 cells regarding HBc phosphorylation state (as well as all the other parameters measured; see below). Following resolution of the wild-type (WT) or mutant HBc proteins (monomers) by SDS-PAGE (Fig. 2A, top four panels) or capsids by native agarose gel electrophoresis (NAGE) (Fig. 2A, bottom four panels), immunoblot analysis was conducted using the above MAbs (Fig. 1A). The levels of the unphosphorylated HBc or capsid at T160/S162 and S178, respectively, detected by MAb A701 and MAb 25-7, and phosphorylated HBc or capsid at S170, detected by 14-2, were all normalized to total HBc or capsid levels detected by T2221 (Fig. 2B). Both the total HBc protein (monomers) and capsid levels from all the A and AA mutants were similar to those of the WT, indicating that the mutations did not significantly affect HBc expression or assembly. Interestingly, all mutants containing the R82A substitution migrated slower on an SDS-PAGE gel than the WT HBc, possibly due to increased phosphorylation or conformational change (Fig. 2A, lanes 3, 5, 7, 8, 11, 13, 15, and 16; see below). Although some differences were observed between the SDS-PAGE and NAGE Western blots for some mutants (e.g., the A701 signals from R28/98A and R28/82/98A), the HBc phosphorylation state was largely consistent between the two assays for all mutants (Fig. 2). Although the minor differences between the two assays for some mutants could be significant (e.g., in terms of capsid assembly) and warrant further studies in the future, we decided to describe below the results from these two assays together as similar results.
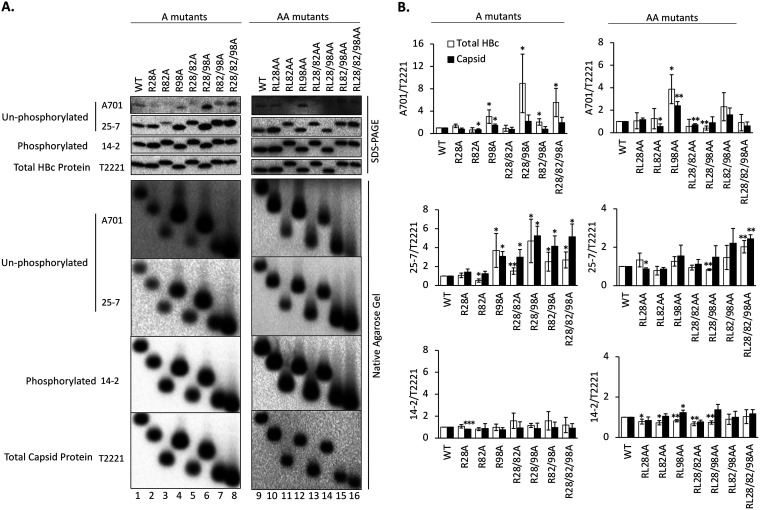
FIG 2.
Effects of the HBc CDM mutations on HBc CTD phosphorylation state. HepG2 cells were transfected with the indicated HBc expression constructs. Five days later, the cytoplasmic lysates were collected. (A) Levels of HBc (top) or assembled capsid (bottom) were measured following SDS-PAGE and transfer to a PVDF membrane or agarose gel electrophoresis and transfer to a nitrocellulose membrane, respectively, using the indicated MAbs. MAb A701 or 25-7 specifically recognizes HBc CTD unphosphorylated at T160 (possibly also S162) or S178, respectively. MAb 14-2 recognizes CTD phosphorylated at S170. Total HBc or capsid levels were determined by the NTD-specific MAb T2221. (B) The A701, 25-7, or 14-2 signals were normalized to the T2221 signals for each CDM mutant or WT HBc, and the normalized values are expressed relative to that of the WT, which was set at 1.00. Total HBc levels (i.e., assembled and nonassembled), either phosphorylated as detected by MAb 14-2 or dephosphorylated as detected by MAb A701 or 25-7, after normalization to all HBc as detected by MAb T2221 were derived from SDS-PAGE Western blots. Similarly, capsid levels (assembled capsids) were derived from native agarose gel electrophoresis/Western blots. Data are presented as the mean and SD of results from at least three independent experiments. Asterisks indicate significant differences in comparison with the WT (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
The levels of nonphosphorylated HBc were significantly increased in the R98-containing R98A, R28/98A, R82/98A and R28/82/98A mutants at both the A701 (T160/S162) and 25-7 (S178) epitopes (Fig. 2A, lanes 4, 6, 7, and 8). The R28/82A double mutant also showed increased unphosphorylation, as detected by 25-7 (Fig. 2A, lane 5), while no obvious differences were detected in the R28A and R82A single mutants. Interestingly, the AA panel of mutants showed less effect on the HBc phosphorylation state at the A701 and 25-7 epitopes than did the corresponding A mutants. However, the mutation involving site 98, RL98AA, still showed the strongest effect on increasing the A701 signal (Fig. 2A, lane 12; Fig. 2B). On the other hand, no major differences could be detected by the phosphor-specific (S170) MAb 14-2 in any of the A or AA mutants (Fig. 2). Therefore, the substitutions at the three potential CDMs (positions 28, 82, and 98) seemed to affect CTD phosphorylation differently and in a phosphorylation site-specific manner, with the RXL motif at positions 98 to 100 playing a dominant role among the three CDMs in influencing the HBc phosphorylation state at the CTD sites monitored. However, in contrast to our anticipation that the AA series mutants would have stronger effects than the A series of mutants due to substitutions at both of the conserved residues (R and L) instead of only the R residue, RL98AA showed less effect in reducing phosphorylation at S178 (but similar or greater effect at S162 [T160]) than R98A (Fig. 2B) (see more on this in Discussion).
Phos-tag SDS-PAGE revealed multiple hypophosphorylated HBc species caused by the CDM mutations.
To determine the effects of the CDM mutations on the HBc phosphorylation state in more detail, we performed Phos-tag SDS-PAGE analysis of HBc proteins from HepG2 cells transfected with the HBV replicon plasmids harvesting the A mutants, as described recently (14, 25). We used HBc expressed and purified from Escherichia coli, which is completely nonphosphorylated, as a nonphosphorylated HBc control. As expected, the E. coli-derived HBc migrated as a single band to the bottom of the gel and was strongly detected by MAbs A701 and 25-7, selective for the unphosphorylated HBc, as well as by MAb T2221 (used to detect total HBc) (Fig. 3A), but was not detected at all by MAb 14-2, selective for the phosphorylated CTD (Fig. 3A). We also used the HBc 3A mutant, in which the three major CTD phosphorylation sites (S155, S162, S170) (Fig. 1A) are replaced with Ala, expressed in the HEK293T cells, as a hypophosphorylated control (25). For total HBc, MAb T2221 detected a series of 5 closely spaced bands in this mutant near the bottom of the gel (Fig. 3B), with band 1 comigrating with E. coli-derived HBc. MAb A701, selective for unphosphorylated HBc at S162/T160, detected 4 bands comigrating with bands 1 to 4 detected by T2221. On the other hand, MAb 25-7, selective for unphosphorylated HBc at S178, detected predominantly the fastest-migrating (i.e., completely nonphosphorylated) species. In contrast, no band was detected by MAb 14-2, consistent with the complete loss of phosphorylation at S170 due to the three Ala substitutions. These control HBc proteins thus indicated that the Phos-tag gel can resolve different HBc phosphor species at high resolution, consistent with our recent results (14, 25), and combined with antibodies selective for different phospho-isoforms may allow identification of HBc species phosphorylated at different sites.
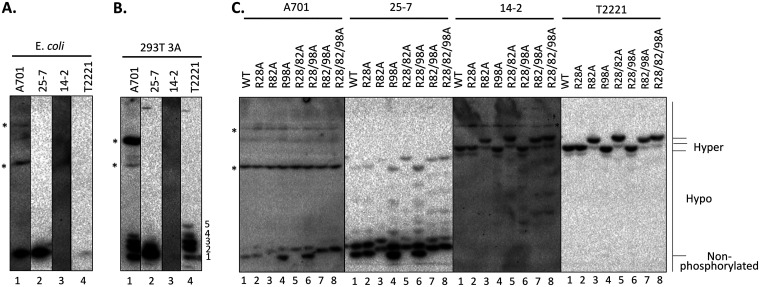
FIG 3.
Analysis of the phosphorylation state of the HBc CDM mutants by Phos-tag SDS-PAGE. HepG2 cells were transfected with HBV replicon constructs expressing the indicated CDM mutants or WT HBc. (C) The transfected cells were harvested at 5 days posttransfection, cell lysates were subjected to Phos-tag SDS-PAGE, and HBc proteins were detected by immunoblot analysis using the indicated anti-HBc MAbs. (A and B) HBc protein expressed and purified from E. coli control (A) or cytoplasmic lysate from pCI-HBc-3A-transfected HEK293T cells (B) was also used as a nonphosphorylated or hypophosphorylated HBc control, respectively. Hyper, hyperphosphorylated HBc; Hypo, hypophosphorylated HBc. *, nonspecific background bands. The numerals 1 to 5 denote the hypophosphorylated HBc-3A species, with species 1 comigrating with the unphosphorylated HBc.
As reported by us and others (14, 25, 45), the hyperphosphorylated WT HBc expressed in human cells could be resolved into two major species migrating near the top of the gel, consistent with their severely retarded mobility in the gel (Fig. 3C, lane 1), which can be detected by MAb T2221. In contrast, neither of the these hyperphosphorylated species was detected by MAb A701 or 25-7, indicating that these HBc species were highly phosphorylated at the A701 and 25-7 epitopes (T160/S162 and S178). The phosphor-specific MAb 14-2 detected the same two bands as those detected by T2221 in the WT HBc, confirming that these hyperphosphorylated HBc species were indeed phosphorylated at S170. In contrast, MAb A701 detected two fast-migrating, hypophosphorylated species (Fig. 3C, lane 1), including the fastest-migrating species that comigrated with the nonphosphorylated HBc from E. coli. MAb 25-7 detected, besides the fastest-migrating species that comigrated with the nonphosphorylated HBc from E. coli, a series of hypophosphorylated species migrating between the nonphosphorylated HBc and the hyperphosphorylated WT HBc expressed in HepG2 cells. The hypophosphorylated (and potentially nonphosphorylated) HBc species from HepG2 cells were not detected by MAb T2221 (Fig. 3C, lane 1), indicating that these hypophosphorylated (nonphosphorylated at S178) HBc species were present at only very low levels, with the majority WT HBc being hyperphosphorylated.
As with the regular SDS-PAGE analysis, any mutant containing the R82A substitution, including R82A, R28/82A, R82/98A and R28/82/98A, migrated significantly slower than the WT HBc on the Phos-tag gel, irrespective of the MAbs used for detection (Fig. 3C, lanes 3, 5, 7, and 8 versus lane 1). Although it remains possible that the slower mobility of these mutants was due to a change caused by the R82A substitution in the HBc state of phosphorylation at some sites that could not be monitored here, the fact that all four MAbs that were nonselective or were selective for phosphorylated or nonphosphorylated HBc at three (four) different CTD sites all detected this upshift suggested instead that this substitution likely caused a conformational change leading to the slower mobility of mutants harboring the substitution. Compared with the total HBc signals detected by MAb T2221, MAbs A701 and 25-7 detected stronger un- or hypophosphorylated (fast-migrating) HBc species, which were unphosphorylated at T160/S162 or S178, respectively, in all mutants containing the R98A substitution, i.e., R98A, R28/98A, R82/98A, R28/82/98A, than in the WT (Fig. 3C, lanes 4, 6, 7, and 8 versus lane 1). In addition, MAb 25-7 detected a series of hypophosphorylated HBc species in these same four mutants, which were absent or weaker in the WT, in the middle of the gel, indicating that these mutants contained increased levels of multiple hypophosphorylated species (nonphosphorylated at S178), consistent with the increased nonphosphorylation revealed by the regular SDS-PAGE described above (Fig. 2A).
Although no major differences were found between any of the A mutants and WT HBc by the phosphor-specific MAb 14-2 in regular SDS-PAGE, our Phos-tag gel analysis using this MAb revealed clear differences in HBc phosphorylation for some A mutants. As expected, the major HBc species in the WT and all A mutants detected by 14-2 were the same two hyperphosphorylated species at the top of the gel as those detected by T2221, indicating that these predominant hyperphosphorylated species were both phosphorylated at S170, which was not dramatically affected in any of the A mutants. However, all the R98A-containing mutants, as well as the R28/82A mutant, showed multiple heterogeneous bands in the middle of the gel representing hypophosphorylated species (Fig. 3C, lanes 4 to 8), which were absent in the WT. These results indicated that the R98A substitution by itself, and R28A and R82A substitutions in combination, did decrease HBc phosphorylation significantly to generate the multiple hypophosphorylated species detectable by MAb 14-2 on the Phos-tag gel (i.e., remaining phosphorylated at S170 but unphosphorylated at other sites), in accordance with the increased levels of unphosphorylated HBc detected by A701 and 25-7.
In contrast to the R98A mutant, the single-substitution CDM mutants R28A and R82A showed little to no effects on HBc phosphorylation in comparison with the WT, based on the Phos-tag gel analysis and detection with either the non-phosphor-specific MAbs A701 and 25-7 or the phosphor-specific MAb 14-2 (Fig. 3C, lanes 2 and 3), consistent with the regular SDS-PAGE analysis described above (Fig. 2B). Thus, both the regular and Phos-tag SDS-PAGE and Western blot analyses indicated that among the three CDMs in the HBc NTD, the RXL motif at positions 98 to 100 played a dominant role in enhancing HBc phosphorylation (in particular, at T160/S162 and S178), possibly by recruiting the CDK2-cyclin complex. We also attempted Phos-tag gel analysis of the AA mutants but did not observe any significant differences between the mutants and the WT HBc, consistent with their weaker effects on CTD phosphorylation, in comparison with the A mutants, revealed by the regular SDS-PAGE and Western blot analyses described above.
The CDM mutant capsids showed decreased levels of endogenous kinase activity in vitro.
To test whether the CDM (RXL) motifs could serve to recruit CDK2 into HBV capsids, we isolated WT and mutant capsids from HepG2 cells and conducted the endogenous kinase reaction in vitro to measure the levels of endogenous kinase activity in the different capsids. In addition, we attempted to block the endogenous kinase activity by inhibitor CDK2iii, an ATP-competitive inhibitor highly specific to CDK2 (25). Consistent with a role for the CDMs in facilitating the packaging of the endogenous kinase (mainly CDK2), all CDM mutants showed various levels of decrease in the endogenous kinase activity compared with that of the WT, after normalization of the in vitro phosphorylated capsid amount to the total capsid amount (Fig. 4). Among the single-A mutants, the R98A mutant appeared to show the most decrease in endogenous kinase activity in comparison with the WT, consistent with the predominant role of this CDM in recruiting the CDK2-cyclin complex to HBc as revealed above. Surprisingly, the double substitution at this same site showed only a weak effect on endogenous kinase activity (see Discussion). In general, the decrease in endogenous kinase activity was more pronounced in the double- and triple-A or AA mutants (Fig. 4B), suggesting that the CDMs may work cooperatively to facilitate kinase packaging into the capsid.
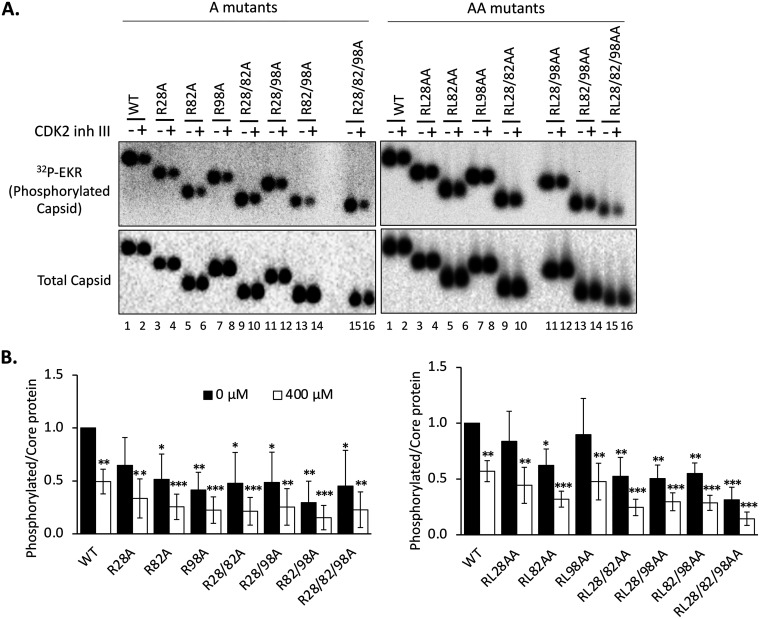
FIG 4.
Effect of the HBc CDM mutations on the endogenous kinase activity in capsids. (A) HepG2 cells were transfected and harvested as described in the legend to Fig. 2. An in vitro endogenous kinase reaction (EKR) assay was conducted with the indicated HBV capsids without or with 400 μM CDK2 inhibitor III (inh III). The reaction products were resolved by agarose gel electrophoresis, phosphorylated capsids labeled by the endogenous kinase were detected by autoradiography (top panel), and total capsid levels were measured by Western blot analysis by using the HBc MAb T2221 (bottom). (B) The effects of the CDM mutations on the EKR efficiency were calculated by normalizing the phosphorylation (32P) signal of the WT HBc or CDM mutants, with or without the inhibitor, to the respective capsid level and are expressed relative to the normalized signal of the WT capsid in the absence of the inhibitor, which was set at 1.0. Data are presented as the mean and SD of results from three independent experiments. Asterisks indicate significant differences in comparison with the WT (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
The CDK2-specific inhibitor reduced the endogenous kinase activity in the WT capsid as anticipated (Fig. 4A, lane 2). However, as we reported before (23), residual endogenous kinase activity remained, suggesting that other kinases may also be packaged and could replace CDK2 to phosphorylate the capsid during the endogenous kinase reaction. Furthermore, the CDK2 inhibitor also reduced the endogenous kinase activity in all CDM mutant capsids, suggesting that other than the three putative CDM sites in the HBc NTD, additional HBc sequences may also mediate low levels of CDK2 packaging into capsids, consistent with the association between CDK2 and the HBc CTD that we detected before (21).
The HBc CDM mutants showed no significant defect on pgRNA packaging.
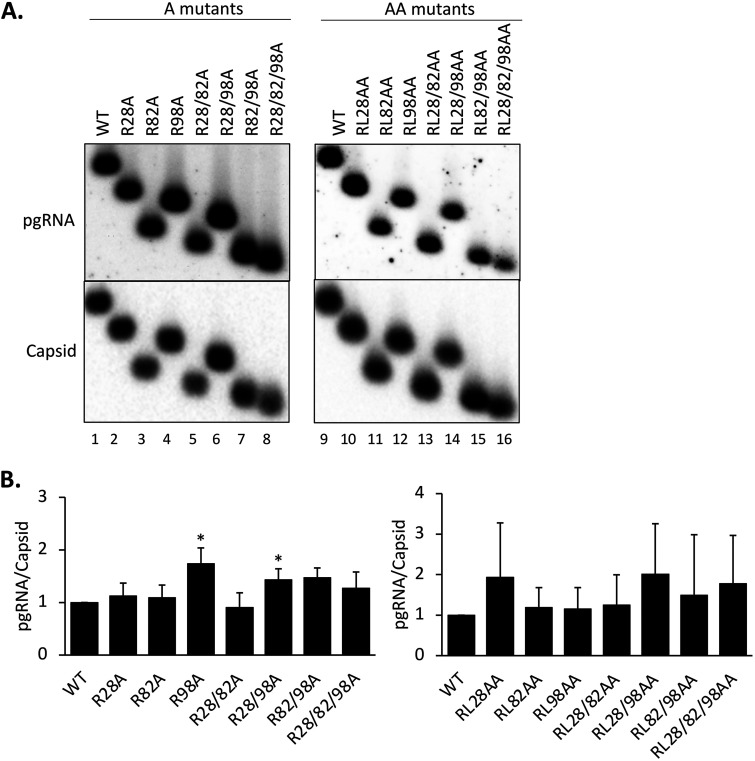
We were next interested in the effects of the RXL mutations on the ability of HBc to package the pgRNA. Levels of WT and mutant capsids from transfected HepG2 cells were measured by the NAGE assay. Levels of HBV pgRNA packaging was normalized to the capsid levels for the WT HBc and each mutant. The results showed that none of the CDM mutants had any defect in pgRNA packaging (Fig. 5). The R98A and R28/98A mutants showed a modestly increased (less than 2-fold) level of pgRNA packaging.
FIG 5.
Effects of the HBc CDM mutations on pgRNA packaging. HepG2 cells were transfected with the HBV replicon constructs harboring the CDM A mutants and harvested as described in the legend to Fig. 3. For the CDM AA mutants, HepG2 cells were cotransfected with the indicated HBc expression constructs together with the HBV genomic construct defective in HBc expression and harvested at 5 days posttransfection. (A) Cytoplasmic lysate was analyzed for pgRNA packaging, following agarose gel electrophoresis and transfer to nitrocellulose membrane, using the antisense HBV RNA probe and MAb T2221 to detect the packaged pgRNA and assembled capsids, respectively. (B) RNA packaging efficiency was determined by normalizing the levels of RNA packaging to those of capsids, with the efficiency from the WT set at 1.0. Data are presented as the mean and SD of results from three independent experiments. Asterisks indicate significant differences in comparison with the WT (*, P < 0.05).
Combined CDM mutations at both sites 28 and 98 impaired the stability/integrity of mature NCs.
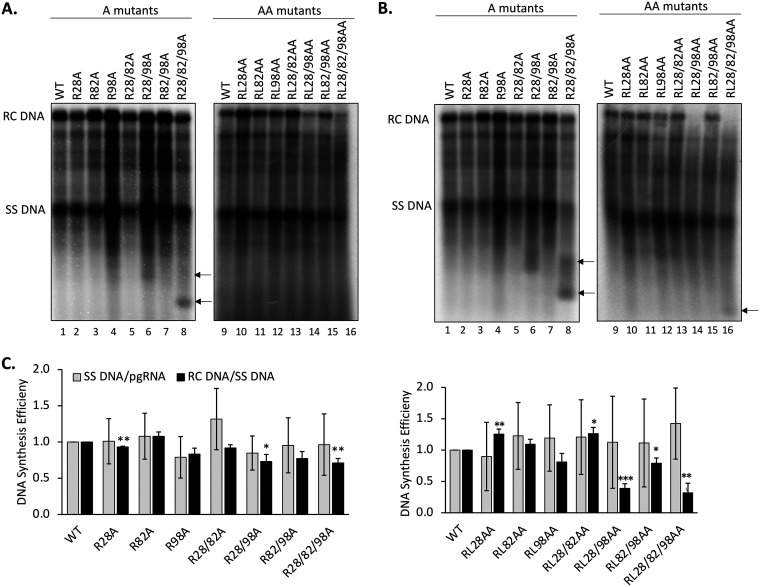
To determine the effects of the CDM mutations on HBV DNA synthesis, viral DNA was released from NCs by SDS/proteinase K treatment and detected by Southern blotting analysis, either with no pretreatment (Fig. 6A) or with pretreatment with DNase (Fig. 6B), which is intended to degrade plasmid DNA but can also degrade viral NC (core) DNA if NCs are unable to protect their DNA content (14, 38). By comparing the levels of the various DNA species with and without DNase pretreatment, we could evaluate the overall levels of viral DNA synthesis as well as the integrity or stability of viral NCs harboring different viral DNA species. We found that most of the A series of mutants showed no significant defect in DNA synthesis as all DNA species were detectable, with or without DNase treatment (Fig. 6). The core DNA levels in R98A, R28/98A and R82/98A showed a slight increase over that of the WT (Fig. 6A and B, lanes 4, 6, and 7), but this could be accounted for by the slightly increased pgRNA packaging in these mutants (Fig. 5). Interestingly, R28/98A and especially R28/82/98A produced small DNA fragments running faster than the SS DNA (with apparent sizes in the 600- to 800-bp range) (Fig. 6A, lanes 6 and 8), which were further increased following DNase treatment (Fig. 6B, lanes 6 and 8). In addition, after DNase treatment, the RC DNA levels in these mutants were reduced (Fig. 6B, lanes 6 and 8), suggesting that both the RC DNA-containing NCs from these mutants were unstable, leading to the digestion of their DNA content to the small fragments observed. R28/82/98A produced DNA fragments even smaller than those in R28/98A, suggesting that the R-to-A substitutions at all three sites may have contributed to further NC disruption than the substitutions at CDM sites 28 and 98.
FIG 6.
Effects of the HBc CDM mutations on core DNA levels. HepG2 cells were transfected and harvested as described in the legend to Fig. 5. HBV NC-associated DNA (core DNA) was released from cytoplasmic lysate of transfected cells, without (A) or with (B) Turbo DNase pretreatment, by SDS-proteinase K digestion and analyzed by Southern blotting using an HBV DNA probe. Arrows indicate the short DNA fragments in lanes 6 and 8 in panel A and lanes 6, 8, and 16 in panel B, likely derived from incomplete DNA degradation. (C) The minus-strand (SS) DNA synthesis efficiency is represented by the relative levels of SS DNA normalized to those of the packaged pgRNA, and the SS DNA/pgRNA ratios are compared with that of the WT, which was set at 1.0. The plus-DNA strand synthesis efficiency was calculated by normalizing the relative levels of RC DNA to those of the SS DNA, and the RC DNA/SS RNA ratios were compared with that of the WT, which was set at 1.0. Data are presented as the mean and SD of results from three independent experiments. Asterisks indicate significant differences in comparison with the WT (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
For the AA series of mutants, it was notable that RL28/98AA and RL28/82/98AA showed significantly reduced levels of RC DNA compared with the WT even without prior DNase treatment (Fig. 6A, lanes 14 and 16). After DNase pretreatment, these two mutants showed no detectable RC DNA, indicating that the low levels of RC DNA in these two mutants could not be protected by their mature NCs (Fig. 6B, lanes 14 and 16). Also, a small DNA fragment appeared in the RL28/82/98AA mutants after DNase treatment, which migrated a bit faster than those in R28/82/98A, suggesting more complete DNA degradation in RL28/82/98AA than R28/82/98A but still partial protection of the small DNA fragment. Therefore, whereas the single R-to-A substitutions in R28/98A and R28/82/98A already destabilized the mature NCs, the additional L-to-A substitutions in RL28/98AA and RL28/82/98AA led to their further destabilization/disruption. The other AA mutants showed little to no effect on viral DNA synthesis.
CDM mutations at sites 28 and 98 increased PF-RC and CCC DNA levels during intracellular amplification.
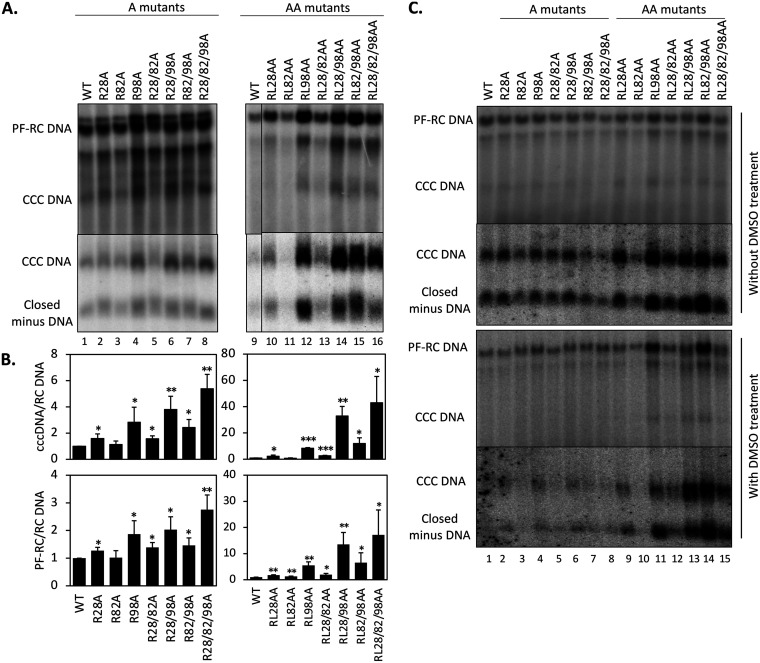
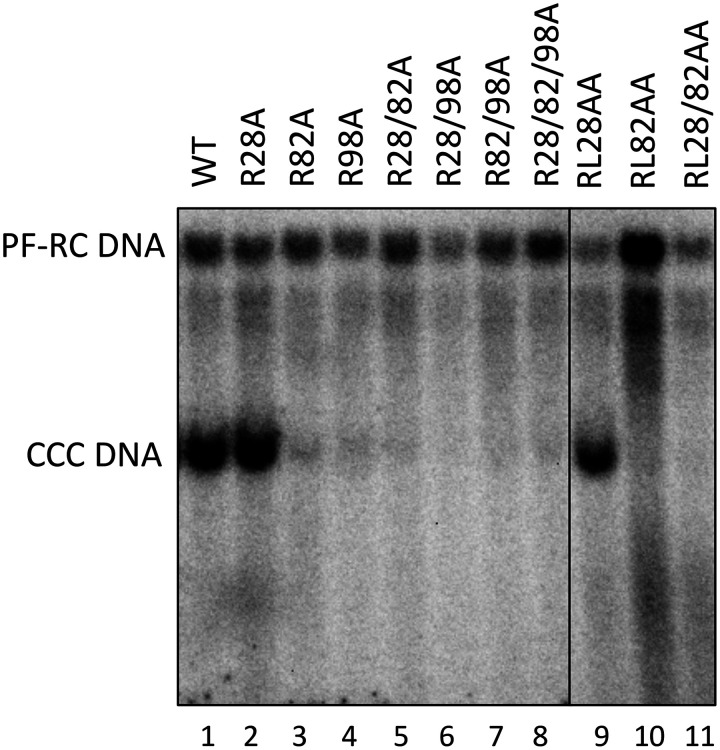
To ascertain the potential effects of the CDM mutations on HBV CCC DNA formation, protein-free (PF) DNAs, including PF-RC DNA and CCC DNA, isolated from transfected HepG2 cells were analyzed by a Southern blot assay. In order to detect the CCC DNA more robustly and accurately, we digested the PF DNAs by exonuclease I (Exo I) and III, which remove noncovalently closed DNA but preserve CCC DNA and covalently closed minus-strand DNA intermediate (60). HBV PF-RC DNA and CCC DNA levels were then normalized to those of the core RC DNA, the ultimate precursor to both PF-RC DNA and CCC DNA. The results showed that whereas the single or double substitutions at site 82 (i.e., R82A and RL82AA) showed little effect, the A and AA mutants containing substitutions at site 28 or 98 increased the levels of PF-RC DNA and CCC DNA compared with the WT (Fig. 7A). The increase ranged from 2- to 40-fold. The substitutions at site 98 showed a stronger effect than those at site 28, with the AA mutants showing stronger effects than the A mutants, and both A and AA mutants with mutations at sites 28 and 98 showed additive/synergistic effects (Fig. 7B).
FIG 7.
Effects of the HBc CDM mutations on CCC DNA formation. HepG2 cells were transfected and harvested as described in the legend to Fig. 5. (A) PF DNA, including PF-RC and CCC DNA, was purified and digested by DpnI as described in Materials and Methods to remove plasmid DNA copurifying with the HBV PF DNA (top panel). HBV PF DNA was further treated with Exo I and III to remove noncovalently closed DNA for specific detection of CCC DNA (bottom panel). (B) CCC DNA levels (top panel) and PF-RC DNA levels (bottom panel) from the WT and mutants were normalized to those of core RC DNA. Normalized CCC DNA or PF-RC DNA level from the mutants was compared to that of the WT, which was set at 1.0. Data are presented as the mean and SD of results from three independent experiments. Asterisks indicate significant differences in comparison with the WT (*, P < 0.05; **, P < 0.01; ***, P < 0.001). (C) HepG2-NTCP cells were transfected and harvested as described in the legend to Fig. 5. PF DNA was extracted from transfected HepG2-NTCP cells cultured without (top two panels) or with (bottom two panels) supplementation with 2% DMSO after transfection. DNA was analyzed by Southern blot analysis. Closed minus-strand DNA was derived from PF-RC DNA in which the minus strand is covalently closed but the plus strand remains open (60).
To determine if the differences in CCC DNA formation by the HBc mutants in HepG2-NTCP during infection versus the parental HepG2 cells during transfection might be due to potential clonal differences among HepG2 cells or cell culture conditions (HepG2-NTCP cells were cultured with diethyl sulfoxide [DMSO] after infection whereas HepG2 cells were not), we transfected the HBc mutants into HepG2-NTCP cells and treated the transfected HepG2-NTCP cells with or without DMSO. We obtained essentially the same results regarding HBV replication, including CCC DNA formation, in transfected HepG2-NTCP cells with or without DMSO treatment as in transfected HepG2 cells (Fig. 7C; and data not shown), which indicated that the differential effects of the CDM mutations on CCC DNA levels in infected HepG2-NTCP cells versus transfected HepG2 cells were not due to clonal variation of HepG2 cells or cell culture conditions.
The L100A substitution severely impaired virion secretion.
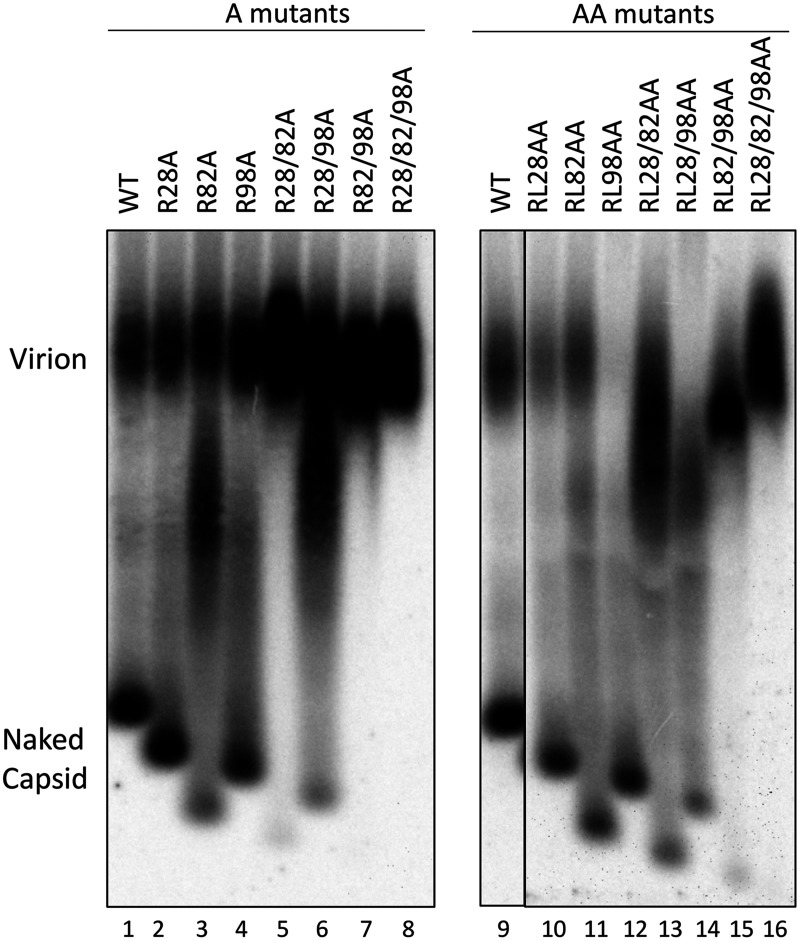
To determine the effects of the CDM mutations on virion secretion, HBV particles released from cell culture supernatants of transfected HepG2 cells were resolved by NAGE, followed by Southern blotting, to detect virion-associated DNA. For most single-site A or AA mutants, the virion DNA levels were similar to those of the WT (Fig. 8, lanes 1 to 4 and 9 to 11). However, RL98AA showed almost no virion DNA signal (Fig. 8, lane 12). On the other hand, all double- and triple-site A and AA mutants showed apparently less naked capsid release and increased virion signals than the WT (Fig. 8, lanes 5 to 8 and 13 to 16). However, these results could be explained by the aggregation of naked capsids, leading to their comigration with virions for these mutants. Therefore, to determine the virion secretion levels for these mutants, the concentrated supernatant from these mutants, along with the WT as well as the virion secretion-defective RL98AA mutant, was fractionated using CsCl density gradient ultracentrifugation to separate virions from naked capsids based on the higher density of naked capsids than virions (24, 61, 62).
FIG 8.
Effects of the HBc CDM mutations on virion secretion. HepG2 cells were transfected as described in the legend to Fig. 5. Cell culture supernatant was collected on day 5 posttransfection. HBV particles in concentrated culture supernatant were resolved by agarose gel electrophoresis and transferred to a nitrocellulose membrane. HBV DNA in virions and naked capsids was detected by using the HBV DNA probe.
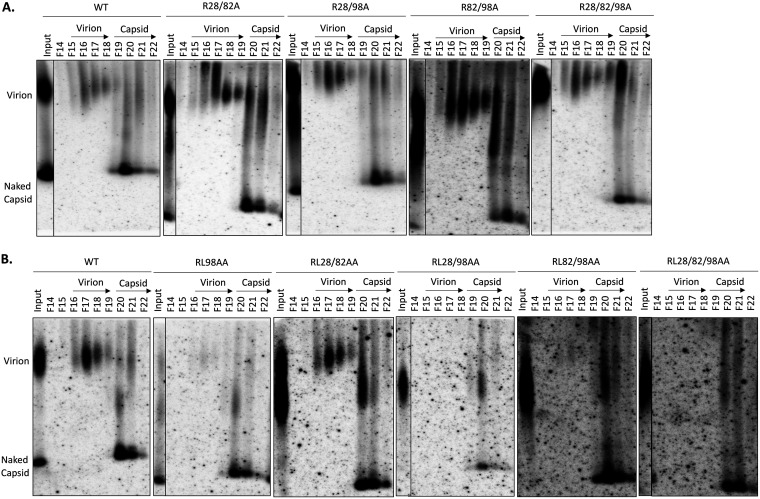
The CsCl gradient fraction results showed that all double- and triple-site A mutants secreted virions at levels comparable to that of the WT (Fig. 9A). Together with the results shown in Fig. 8, these results showed that none of the A mutants adversely affected secretion of complete virions. As expected from the secretion block detected with unfractionated supernatant above (Fig. 8), RL98AA showed very few virions (Fig. 9B). Moreover, all RL98AA-containing mutants showed dramatically reduced virion levels compared with the WT, with RL28/98AA and RL28/82/98AA showing no detectable virions (Fig. 9B). In contrast, the RL28/82AA mutant, with AA substitutions at both positions 28 and 82 but not at position 98, showed WT-like virion secretion. These results thus indicated that it was the AA substitutions at position 98 alone that blocked virion secretion.
FIG 9.
CsCl density gradient analysis of virion secretion by HBc CDM mutants. HepG2 (A) and Huh7 cells (B) were transfected with the indicated constructs. Cell culture supernatant was collected on day 5 posttransfection. Concentrated culture supernatant was fractionated on isopycnic CsCl density gradients. Selected fractions (F14 to F22) were resolved by native agarose gel electrophoresis. Following transfer to a nitrocellulose membrane, HBV DNA in virions and naked capsid released into the culture supernatant were detected by using the HBV-specific DNA probe. The virion and capsid fractions are indicated.
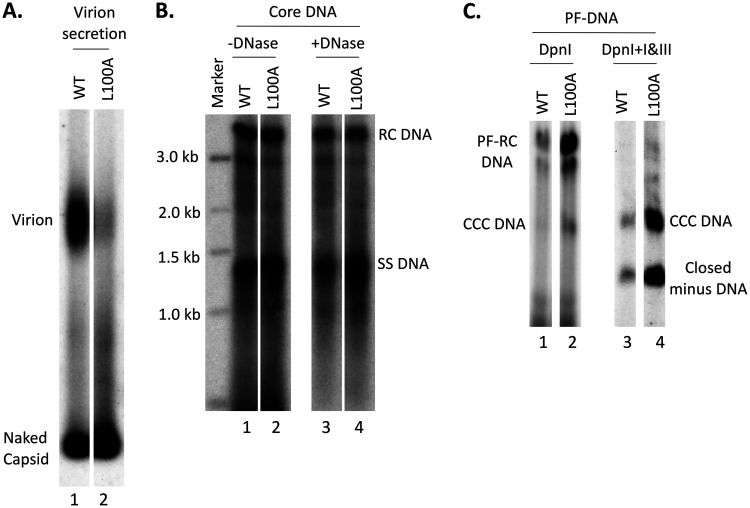
Since R98A had little effect on virion secretion, the secretion defect of RL98AA was likely caused by the L100A substitution. To test if the L100A substitution alone could block virion secretion, we constructed the L100A single substitution mutant (i.e., without the R98A mutation). We found that this single substitution, alone, was indeed sufficient to block virion secretion, with little effects on intracellular DNA synthesis or NC stability (Fig. 10A and B). Due to the severe block in virion secretion, L100A also dramatically increased PF-RC DNA and CCC DNA levels (by ca. 6- and 8-fold, respectively) (Fig. 10C). Indeed, this effect of L100A probably accounted for most of the increase in PF-RC and CCC DNA levels by all L100A-containing mutants, i.e., RL98AA, RL28/98AA, RL82/98AA, and RL28/82/98AA (Fig. 7A, lanes 12 and 14 to 16).
FIG 10.
Effects of the HBc L100A mutation on virion secretion, core DNA, and PF DNAs. HepG2 cells were transfected as described in the legend to Fig. 5. (A) Cell culture supernatant was collected on day 5 posttransfection. HBV particles in concentrated culture supernatant were resolved by agarose gel electrophoresis and transferred to a nitrocellulose membrane. HBV DNA in virions and naked capsids was detected by Southern blot analysis using the HBV DNA probe. (B) Core DNA released from cytoplasmic lysate of L100A-transfected HepG2 cells, without (lanes 1 and 2) or with (lanes 3 and 4) Turbo DNase pretreatment, was analyzed by Southern blot analysis as described in the legend to Fig. 6. (C) PF DNA from L100A-transfected HepG2 cells was analyzed by Southern blot analysis as described in the legend to Fig. 7.
CDM mutants at position 82 or 98 blocked HBV infection in HepG2-NTCP cells.
To test if the CDM mutants affected viral infection, we measured HBV CCC DNA formation, the definitive sign of productive HBV infection, in infected HepG2-NTCP cells at 3 days postinfection, when only CCC DNA formation from the input virus occurs and intracellular CCC DNA amplification has not started (25, 60, 63). We prepared WT and CDM mutant inocula using concentrated culture supernatant from transfected HepG2 cells, except for the four mutants containing the L100A substitution (RL98AA, RL28/98AA, RL82/98AA, and RL28/82/98AA) that secreted little to no DNA virions (Fig. 9B). HepG2-NTCP cells were infected with the WT and mutant viruses at equal multiplicities of infection (MOI) (ca. 400 genome equivalents [GE]/cell), and PF DNAs including CCC DNA were extracted from infected cells 3 days after infection and analyzed by Southern blotting. The results showed that among all the mutants tested, R28A showed infectivity similar to that of the WT and RL28AA showed slightly reduced infectivity (Fig. 1C and (11), indicating that this CDM site does not play an essential role in infection. The A (R82A, R98A, R28/82A, R28/98A, R82/98A, and R28/82/98A) and AA (RL28AA, RL82AA, and RL28/82AA) mutations at sites 82 and 98 strongly diminished infectivity (Fig. 1C and 11). To test further whether these mutants were indeed defective in infection, we increased the mutant inocula by 2 to 5 times during infection and still observed little infection by the mutants (data not shown). Since all these mutants could form CCC DNA at levels similar to or higher than the WT during intracellular amplification (Fig. 7), these results clearly demonstrated that the CDM mutations could differentially affect CCC DNA formation during infection versus intracellular amplification (Fig. 1C).
FIG 11.
Effects of the HBc CDM mutations on infection. HBV inocula were prepared from HepG2 cells cotransfected with the indicated WT or mutant HBc expression construct plus the HBc-defective replicon. HBV PF DNA was extracted from HepG2-NTCP cells 3 days after infection and measured by Southern blot analysis using a 32P-labeled HBV DNA probe.
DISCUSSION
Our ongoing attempts to understand how CDK2 is packaged into HBV capsids led us to the identification of three conserved CDMs (RXL) in the HBc NTD at positions 28 to 30, 82 to 84, and 98 to 100. Our mutagenesis analysis of these CDMs in general supports a role for these CDMs in facilitating the recruitment to HBc, and packaging into capsids, of CDK2. In particular, substitutions of the CDM at positions 98 to 100, and to a lesser degree at the other two CDMs as well, decreased the levels of HBc phosphorylation in transfected cells at two different CTD sites (S162, S178, and possibly also T160 and other sites). At least some of the combined substitutions at two or three of the CDMs together showed stronger reduction in CTD phosphorylation, consistent with the notion that the CDMs act together to recruit the kinase for CTD phosphorylation. Furthermore, most of the CDM mutants showed decreased endogenous kinases activity in the capsid (less packaging of CDK2). Whereas none of the CDM mutants showed strong defects in pgRNA packaging, mature NCs containing mutations at both CDM sites 28 and 98 were destabilized, which was associated with significantly enhanced CCC DNA formation. While mutants containing the L100A substitution were defective in virion secretion, implicating L100 as an important residue in envelope interaction, mutations at either CDM site 82 or 98 caused a severe defect in infection, supporting a role of the CDMs and the packaged CDK2 in the infection process (Fig. 12).
FIG 12.
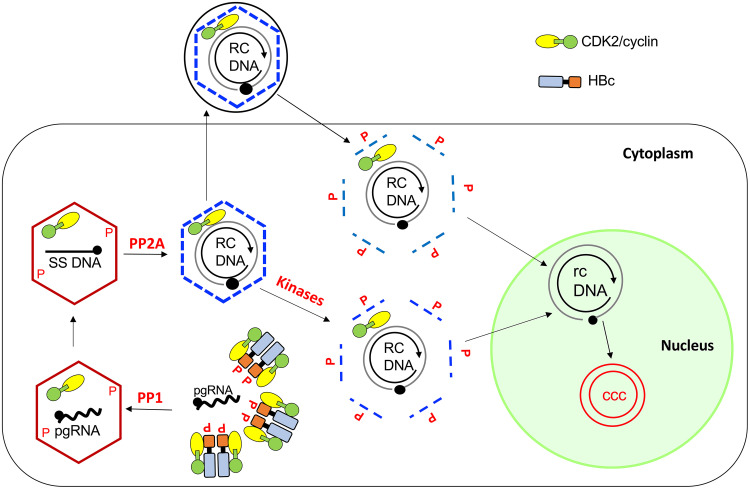
Model for CDK2/cyclin regulation of HBV NC assembly and disassembly during intracellular CCC DNA amplification and de novo infection. HBc subunits are heavily phosphorylated at the CTD in the cytoplasm by host protein kinases, including the CDK2/cyclin complex and others, and are partially dephosphorylated during pgRNA packaging by PP1. CDK2/cyclin is packaged into immature NCs during NC assembly. HBc is subsequently further dephosphorylated by PP2A during NC maturation. Rephosphorylation of the mature NC in the virion during infection triggers NC disassembly (uncoating) to facilitate release of the RC DNA to the nucleus for CCC DNA formation. During intracellular CCC DNA amplification, cellular kinases from the cytoplasm may phosphorylate HBc in the mature NC to initiate its disassembly such that the packaged kinase may not be essential for uncoating in this pathway of CCC DNA formation. See the text for details.
Among the three CDMs at the HBc NTD, mutations at site 98 had the strongest effect, in comparison to the WT, in reducing CTD phosphorylation, as reflected by the increase in nonphosphorylated HBc detected by both MAbs A701 and 25-7, selective for nonphosphorylated S162 (and possibly T160) and S178, respectively, suggesting that site 98 played a dominant role in CDK2/cyclin recruitment. Phos-tag gel analysis also further revealed that the R98A substitution, alone or in combination with other mutations, generated multiple hypophosphorylated HBc species that were absent or present only at very low levels in the WT, indicative of decreased and more heterogeneous phosphorylation than with the WT. In addition, double- and triple-A mutations involving site 98, such as R28/98A and R28/82/98A, decreased HBc phosphorylation even more than substitutions at site 98 alone, particularly at S162 (T160), suggesting that site 28 cooperates with site 98 in recruiting CDK2 to phosphorylate these HBc CTD sites.
However, the double substitutions (i.e., both the R and L residues in the CDM) at site 98 (RL98AA) actually had less effect in reducing phosphorylation at S178 than the single R-to-A substitution at the same site (R98A), even though the double substitution may be expected to disrupt cyclin/CDK2 binding more severely than the single substitution. Furthermore, the effects of CDM mutations could vary depending on the CTD phosphorylation sites (S162 [T160] versus S178) (especially in the case of RL98AA) (see Fig. 1C for a summary). These results are likely a reflection of the complex regulation of HBc phosphorylation dynamics. For example, kinases other than CDK2 that can also mediate HBc phosphorylation may be affected by the CDM mutations in ways that are difficult to predict. Moreover, the phosphorylation state at one CTD site can affect that at the other sites. For example, the S-P site phosphorylation at the HBc CTD was reported to prime phosphorylation at the other CTD sites; however, we recently found that S-to-A substitutions at the three CTD S-P sites, which prevent their phosphorylation, could also enhance phosphorylation at one or both of the S-Q sites in the CTD (11, 14, 25, 43). This interdependent nature of the CTD phosphorylation dynamics can further confound the interpretation of the direct versus indirect effects of the CDM mutations. Finally, as non- or hypophosphorylated HBc may be less stable than (hyper)phosphorylated HBc (11), the steady-state levels of HBc phosphorylation may also be affected by the stability of the different phosphor isoforms, which can further be affected by the CDM mutations. In particular, the A and AA substitutions combined at sites 28 and 98 (R28/98A, R28/82/98A, RL28/98AA, and RL28/82/98AA) led to capsid destabilization, which may be related to the surprising results that the AA mutations at these two or three sites together led to less nonphosphorylated HBc species than the A mutations (Fig. 1C). For example, it is possible that AA mutations at these sites actually led to a more pronounced reduction in HBc phosphorylation than the corresponding A mutations but that the rapid degradation (loss) of these severely underphosphorylated or nonphosphorylated species would lead to their apparent decrease. The close proximity of CDM sites 28 and 98 on the capsid structure (Fig. 1B) increases the likelihood that combined mutations at these sites could alter some aspects of the capsid structure and dynamics.
Our mutational analysis of the HBc CDMs in general supports a role for these sites, in a collaborative manner, in facilitating CDK2 packaging into HBV capsids. The endogenous kinase activity decreased significantly in almost all CDM mutants, with the mutations at site 82 and especially at site 98 reducing it more strongly than those at site 28 (Fig. 1C), in comparison with the WT, suggesting that the RXL motifs at positions 82 to 84 and especially at positions 98 to 100 functioned as the major cyclin docking sites, which is consistent with the strongest effect in reducing CTD phosphorylation in cells by site 98 mutation. Surprisingly, the reduction in endogenous kinase activity in RL98AA was less severe than that in R98A, in comparison to the WT. One possible explanation is that the RL98AA capsid contained more non- and hypophosphorylated HBc species than R98A, which meant that more HBc sites could be available for phosphorylation during the endogenous kinase reaction in vitro, thus generating the stronger signals observed. In any case, low levels of CDK2 were apparently still packaged into all the mutant capsids, possibly due to the interaction of the HBc CTD with CDK2 (21). On the other hand, the reduction in endogenous kinase activity that we could measure could be an underestimate of the reduction in CDK2 packaging, as other cellular kinases may be packaged at increased levels when CDK2 packaging is reduced due to the CDM mutations. The above-mentioned interdependence of phosphorylation at the different CTD sites can further complicate the interpretation of the endogenous kinase reaction results. Also, the exact cyclin partner(s) for CDK2 that is packaged remains unknown. Given the cell cycle preference of HBV replication in the G1 phase of the cell cycle (52–54), it is possible that the HBc CDMs interact with cyclin E for packaging into the capsid. Since only a single copy of CDK2 is packaged (23), HBc-CDK2 (cyclin) interactions are likely transient and of low affinity, so it is difficult to identify the cyclin component(s) directly or to determine the reduction in CDK2/cyclin packaging caused by the CDM mutations more accurately, e.g., by immunoblotting or mass spectrometry methods. Our attempts to directly detect the HBc-cyclin E interaction through immunoprecipitation have been unsuccessful so far (H. Liu and J. Hu, unpublished results). However, we found that overexpression of CDK2 and cyclin E2 could increase HBc phosphorylation expressed in transfected human cells, which could be abrogated by at least some of the CDM mutations (H. Liu and J. Hu, unpublished results), further supporting a role of CDK2/cyclin E in phosphorylating HBc in vivo as well as the potential packaging of cyclin E into the HBV capsid.
We have recently suggested that the packaged CDK2 in HBV capsids facilitates HBV CCC DNA synthesis during infection by possibly phosphorylating the virion capsid from inside to trigger disassembly (uncoating) by increasing the electrostatic repulsion between the RC DNA and the phosphorylated capsid (25). Also, the host hepatocytes are normally stationary and may thus lack the kinases needed to phosphorylate the incoming virion capsid at the start of infection. Here, by reducing kinase packaging, we provide additional evidence further confirming an important role of the packaged CDK2 in supporting efficient HBV infection. The more severely impaired endogenous kinase activity in site 82 and 98 mutations was generally correlated with more impaired CCC DNA formation during infection, whereas the mutations at site 28 showed little impairment in the endogenous kinase activity or CCC DNA formation during infection. As discussed above, it is plausible that the three CDMs work cooperatively to recruit and package CDK2/cyclin into the capsid, as suggested by the results that mutations at two or three CDMs showed more severe reduction in endogenous kinase activity and blocked infection more strongly.
The reduction in HBc phosphorylation levels at S162 (and T160) and S178 caused by mutations at CDM sites 28, 82, and 98, individually or in some combination, did not significantly impair pgRNA packaging or RC DNA synthesis, consistent with previous reports (28, 64). On the other hand, multiple reports indicate that both HBV pgRNA packaging and DNA synthesis depend on dynamic HBc phosphorylation and dephosphorylation (24, 35, 36, 65). The stability of RC DNA-containing mature NCs is also thought to be dependent on HBc dephosphorylation based on the charge balance hypothesis (14, 35, 36, 66). To interpret the apparent lack of effects of the CDM mutations on these processes, it is important to keep in mind that the HBc phosphorylation state at CTD sites other than S162 (and T162) and S178 cannot yet be monitored directly due to the lack of antibodies selective for their state of phosphorylation. Potential changes at these other sites might have compensated for any deleterious effects of reduced HBc phosphorylation at S162 (and T160) and S178 on pgRNA packaging or DNA synthesis. Moreover, as ca. 90% of HBV capsids formed in human cells are empty and ca. only 10% of capsids contain pgRNA or HBV DNA (5, 62), the mutations might not have affected the HBc state of phosphorylation in the minor population of pgRNA or DNA-containing NCs, which is difficult to ascertain without separating these NCs from empty capsids, a difficult task at present.
Interestingly, all the A and AA substitution mutants at CDM sites 28 and 98, but not at site 82, enhanced CCC DNA formation during intracellular amplification. Other than the impairment of virion formation by the L100A substitution, which could have accounted for the effect of this mutation on CCC DNA formation (see below), combined substitutions at these two CDM sites with or without mutations at site 82 (R28/98A, RL28/98AA, R28/82/98A, and RL28/82/98AA) appreciably affected the stability/integrity of mature NCs, which could also have increased CCC DNA formation by enhancing NC uncoating and RC DNA release. Interestingly, degradation of the NC DNA, which was further increased by adding exogenous DNase, in R28/98A and R28/82/98A and in RL28/82/98AA after DNase digestion, led to the accumulation of small DNA fragments with apparent sizes of hundreds of base pairs. Future studies to determine how the NC DNA is degraded to these fragments, which were then apparently protected from further digestion, in these mutants, may provide insights into how the DNA genome is released from the capsid (i.e., uncoating), a process essential for HBV replication (and for many other viruses) and yet poorly understood. It remains possible that even mutations at CDM site 28 or 98 alone have also affected NC stability, albeit less drastically than the combined mutations at both of these sites, which escaped detection by our DNase protection assay but could, nevertheless, have accounted for the increased CCC DNA levels by these mutants.
How the CDM mutations at CDM sites 28 and 98 destabilized the mature NCs remains to be determined. At the levels that we could discern, there did not appear to be any clear correlation between the HBc phosphorylation state and the NC stability of the CDM mutants. We have reported that mutations in the HBc NTD (e.g., L60A and I126A) (67) and linker (e.g., S141A and L143I) (14) similarly impair mature NC stability and increase CCC DNA formation. Structural effects on the capsid, in addition to or instead of alterations in HBc phosphorylation state, may account for the effects of these mutations on NC stability/integrity. These two CDM sites, at positions 28 to 30 and 98 to 100, are located in close proximity on the capsid structure (Fig. 1B) and may function together to regulate maturation-associated NC structural changes, in order to accommodate the increasing negative charge and internal pressure as a result of RC DNA synthesis (37, 66, 68). These structural dynamics during NC maturation may be impaired in the mutants that selectively destabilize mature NCs. Previously, it was reported that substitutions at the HBc dimer interface (e.g., V124A) also selectively reduced RC DNA levels, which was thought to be due to alterations of capsid structural dynamics impairing RC DNA synthesis (69). In light of our results here and reported recently (14), those dimer interface mutants may also destabilize mature NCs.
All mutants containing the L100A substitution also had increased CCC DNA, likely due to the block in virion secretion leading to the overamplification of CCC DNA, analogous to the effects of the envelope mutations (30, 31, 33). When this substitution was combined with the NC destabilization mutations at CDM sites 28 and 98 (in RL28/98AA and RL28/82/98AA), synergistic effects on CCC DNA formation were observed (Fig. 1C), as expected. L100, an important residue newly discovered here for virion secretion, lies close to L95 and K96 on the capsid structure, two residues previously shown to be also critical for mature NC envelopment and virion formation, and thus may be part of the MBD on the capsid interacting with the MD of the L envelope protein (26, 27, 70, 71).
As a third potential mechanism to increase CCC DNA formation, some of the CDM mutations might result in increased nuclear import of RC DNA in mature NCs. The HBc CTD, which harbors multiple NLSs, can be localized to both the interior and exterior of the capsid (41, 72, 73), which is also subject to regulation by the HBc phosphorylation state as well as the nucleic acid content of the capsid. This possibility has been difficult to test experimentally due to the difficulty in specifically tracking the nuclear import of mature NCs versus immature NCs or empty capsids.
As discussed above, the effects on CDK2/cyclin binding and CDK2 packaging by the AA series of mutants, compared to the A series of mutants, might have been confounded by the potential effects of the additional L-to-A substitutions, present in the AA but not A mutants, on HBc stability and structure. In the case of the effect of L100A on virion secretion, it is unlikely due to any effects on CDK2/cyclin E binding, given that the CTD state of phosphorylation per se does not affect virion secretion significantly (24). Similarly, others have recently reported that HBc residues 27, 31, 98, and 101, which are near two of the CDMs studied here, are part of a nucleolar retention signal (74). Although it seems unlikely that any effects we observed here were related to nucleolar localization, mutations at the CDM sites could certainly have pleiotropic effects beyond affecting cyclin binding, which warrants future studies.
The striking differential effects of the CDM mutations on CCC DNA formation during intracellular amplification versus de novo infection highlights the different role of the packaged CDK2 and the capsid structure in general in regulating CCC DNA formation through these two pathways. We have excluded here any potential effects of clonal variation of the HepG2 cells or cell culture conditions (i.e., DMSO treatment). The packaged CDK2 in the capsid, while critical for efficient capsid uncoating during infection, appears to be less critical during intracellular CCC DNA amplification, as none of the CDM mutations here impaired this pathway (Fig. 12). In addition, NC destabilization due to some of the CDM mutations, while enhancing CCC DNA formation during intracellular amplification, may lead to premature NC disruption during the virion entry process and thus block CCC DNA formation during infection. We have also recently identified other HBc mutations, in the linker region, that similarly block CCC DNA formation during infection but not during intracellular CCC DNA amplification (14), suggesting that multiple regions of HBc are involved in the infection process. Future studies to elucidate the mechanism underlying this differential effect of the HBc mutations should bring new insights into the requirements of the HBV infection process, which remains poorly understood at present.
MATERIALS AND METHODS
Plasmid constructs.
pCI-HBc-WT and pCI-HBc-3A for expression of the WT or 3A mutant (containing S-to-A substitutions at positions 155, 162, and 170) HBc proteins have been previously described (11). The panel of cyclin docking motif (CDM) mutants of HBc, in the same plasmid backbone pCI as in pCI-HBc-WT, including the CDM A mutants R28A, R82A, R98A, R28/82A, R28/98A, R82/98A, and R28/82/98A, were constructed by changing the first conserved residue in the CDM motif (RXL), R28, R82, or R98, individually or in various combinations to A via PCR-mediated mutagenesis (In-Fusion cloning). Similarly, the CDM AA mutant panel, including RL28AA, RL82AA, RL98AA, RL28/82AA, RL28/98AA, RL82/98AA, and RL28/82/98AA, were constructed by changing both the conserved R (position 28, 82, or 98) and L (position 30, 84, or 100) residues to A individually or in various combinations, as with the CDM A mutants. Moreover, pCI-HBc-L100A was constructed by changing L100 to A. The HBV replicon plasmid pCIΔA-HBV-HBc-WT has been described before (25, 67, 75, 76). The entire panel of CDM A mutants were subcloned from the pCI-HBc background into pCIΔA-HBV-HBc-WT and express the mutated HBV pgRNA carrying the same HBc CDM A mutations as in the pCI-HBc CDM A mutant panel. pCIΔA-HBV-HBc-C− was constructed by replacement of the 4th codon (GAC) in the HBc open reading frame (ORF) with a stop codon (TAG), which is capable of supporting viral replication upon complementation with HBc (14).
Cell culture and transient transfection.
The human hepatoma cell lines HepG2 and Huh7, as well as HepG2-NTCP cells expressing the HBV receptor NTCP (25, 60), were cultured in Dulbecco’s modified Eagle’s medium (DMEM)-F12 supplemented with 10% fetal bovine serum (FBS) (HyClone) and 50 μg/ml of penicillin-streptomycin, as previously described (24, 25). HepG2 cells were seeded in 60-mm dishes and transfected at 70% confluence with 4 μg (total) of plasmid using X-tremeGENE HP DNA transfection reagent (Roche). Huh7 cells were transfected with the FuGENE 6 transfection reagent (Promega). A 1:1 mass ratio of each plasmid was used when two plasmids were used for cotransfections (24).
SDS-PAGE and Western blot analysis.
Cells were lysed with NP40 lysis buffer, and HBc proteins in the cytoplasmic lysate were resolved by 12.5% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), transferred to a polyvinylidene difluoride (PVDF) membrane, and detected by the indicated antibodies. The mouse MAb clone T2221 (catalog no. 2AHC24) (13), specific for the HBc NTD, was purchased from Tokyo Future Style. The HBc CTD-specific MAbs A701 and 25-7, which specifically recognize unphosphorylated T160/S162 and S178, respectively, have been described before (11, 13, 14, 24). The HBc CTD-specific rabbit MAb 14-2 was also custom-made (Abcam) and recognizes a phosphorylated CTD epitope between positions 164 and 183, specifically the phosphorylated S170 (61, 77).
Phos-tag gel electrophoresis.
Phos-tag SDS-PAGE was performed by adding Phos-tag acrylamide AAL-107 (Wako Pure Chemical Corporation) plus MnCl2 to the acrylamide solutions for conventional SDS-PAGE resolving gels, as described previously (14, 25). Briefly, cytoplasmic lysate containing HBc proteins was boiled in SDS sample buffer containing 1 mM MnCl2 and 10% β-mercaptoethanol (β-ME) for 10 min and then resolved on an SDS-polyacrylamide gel (10%, acrylamide/bisacrylamide ratio of 37.5:1) containing 100 μM Phos-tag acrylamide and 200 μM MnCl2. The gel was soaked in transfer buffer (39 mM glycine, 48 mM Tris, 20% [vol/vol] methanol [MeOH]) containing 10 mM EDTA with gentle shaking for 30 min for three times to chelate the Mn2+ left in the gel that could impact the transfer efficiency. Subsequently, the gel was soaked in the transfer buffer without EDTA with gentle shaking for 10 min before transfer to a PVDF membrane.
Nucleocapsid assembly and virion secretion assay.
To detect NC or virion particles, cytoplasmic lysate or DNase I-digested polyethylene glycol 8000 (PEG 8000)-precipitated cell culture supernatant was resolved on a 1% native agarose gel (native agarose gel electrophoresis [NAGE]) by using previously reported procedures (62, 78). Briefly, after NAGE and transfer of the viral particles to a nitrocellulose membrane, the pgRNA packaged into NCs was detected using a 32P-labeled minus-sense riboprobe, and the viral DNA associated with the particles was detected using a 32P-labeled HBV DNA probe. The 32P signals were quantified using a phosphor imaging system (Azura). The same membrane was subsequently incubated with the indicated HBc-specific antibodies to detect HBc.
Isolation and analysis of viral DNA.
HBV core DNA (NC-associated DNA) and protein-free (PF) DNAs were isolated 5 days posttransfection as previously described (30, 60). Briefly, for isolation of HBV core DNA, cells were lysed in the NP40 lysis buffer and the cytoplasmic lysates were digested with 0.6 mg/ml of proteinase K (PK) and 0.5% SDS at 37°C for 1 h. To test the NC integrity, cytoplasmic lysate was incubated with Turbo DNase (0.2 U/μl) at 37°C for 30 min, and then the digestion was terminated by adding EDTA (30 mM final concentration). The DNase-pretreated lysate was subsequently subjected to PK/SDS digestion as described above. The released core DNA was resolved on a 1.5% agarose gel as described previously (13). PF DNA (including PF-RC DNA and CCC DNA) was isolated using a modified Hirt extraction method as previously described (30). PF DNA was further digested by DpnI (NEB) to remove the transfected plasmids. To detect CCC DNA more robustly and more accurately, PF DNA was digested with exonuclease I and III as described previously (60), which removes single-stranded DNA and double-stranded DNA, respectively, from 3′ ends. The PF DNA was resolved on a 1.2% agarose gel. HBV core and PF DNA were detected by standard Southern blot analysis using a 32P-labeled HBV DNA probe. Hybridization signals were visualized by phosphor imaging as described above.
EKR assay.
An endogenous kinase reaction (EKR) assay to measure the packaging of CDK2 into HBV capsids was performed as described before (23, 25). Briefly, 15-μl volumes of NP40 lysates from transfected HepG2 or Huh7 cells were digested at 37°C with proteinase K for 30 min (final concentration, 0.5 mg/ml). The reaction was then stopped by adding the proteinase K inhibitor (Calbiochem) (final concentration, 1 mM) and incubating the mixture for 10 min at room temperature. EKR assay was carried out by adding the kinase reaction buffer (10 mM sodium phosphate, pH 7.0, 10 mM MgCl2, 0.01% Brij) supplemented with fresh EDTA-free protease inhibitor cocktail (Roche), 1 mM dithiothreitol (DTT), and 5 μCi [γ-32P]ATP (3,000 Ci/mmol, 10 mCi/ml; Perkin Elmer) to the PK-digested lysates and incubating the mixture at 37°C for 1 h (23, 25). In reaction mixtures that included the CDK2 inhibitor III [2(bis-(hydroxyethyl)amino)-6-(4-methoxybenzylamino)-9-isopropyl-purine] (Millipore Sigma; catalog no. 238803), the inhibitor was added before the start of the reaction at a final concentration of 400 μM. CDK2 inhibitor III is a potent, selective, reversible, and ATP-competitive inhibitor of Cdk2 (50% inhibitory concentrations [IC50s], 0.5 μM for Cdk2/A and Cdk2/E, 4.2 μM for Cdk1/B, and 215 μM for Cdk4/D1) (23). It inhibits other kinases only at much higher concentrations (IC50 > 1.25 mM for MAPK, PKA, and PKC). Therefore, at 400 μM, it should remain specific for CDK. We found that this fairly high concentration was needed to obtain consistent inhibitory effects in our EKR assay, presumably due to the fact that CDK2 packaged in capsids, rather than the purified enzyme, was used in the EKR assay. The IC50 values may be increased due to issues with accessibility to the packaged kinase by the inhibitor. The reaction mixture was loaded onto a 1% agarose gel. After electrophoresis, the gel was washed in distilled water for several hours to remove the free isotope from the gel. Upon transfer to a nitrocellulose membrane, the radiolabeled capsids were visualized by phosphor imaging (Azura). The same membrane was subsequently incubated with MAb T2221 to detect the capsid.
Analysis of HBV virion secretion by CsCl density gradient fractionation.
Cell culture supernatant containing HBV particles (virions and naked NCs) was concentrated by PEG precipitation, digested by DNase I at a final concentration of 1 mg/ml at 37°C for 1 h to eliminate residual plasmid DNA, and then fractionated by isopycnic CsCl gradient ultracentrifugation (62). The purified virion fractions or concentrated and DNase-digested supernatant samples were analyzed by NAGE, and the viral DNA associated with the particles was detected using a 32P-labeled HBV DNA probe.
HBV infection of HepG2-NTCP cells.
HepG2-NTCP cells were seeded in collagen I-coated 35-mm tissue culture dishes to reach 100% confluence the next day. HBV inoculum harvested from transiently transfected HepG2 or Huh7 cells was added to HepG2-NTCP cells at a multiplicity of infection (MOI) of 400 genome equivalents (GE)/cell in the presence of 4% PEG 8000 (25, 63). After 16 h of incubation, the inoculum was removed and the cells were maintained in complete cell culture medium supplemented with 2% DMSO. The cells were harvested 3 days postinfection for extraction of PF DNA, which was detected by Southern blot analysis as described above.
Quantification and statistical analysis.
All experiments were repeated at least three times. DNA or RNA signals from Southern blot analysis were detected and quantified by phosphor imaging using the Sapphire biomolecular imager (Azure Biosystems). Protein signals from Western blot analysis were detected and quantified using the Image Lab system 6.0.1 (Bio-Rad) as described before (14, 25, 61). Subtraction of background signals was done separately for each individual lane. All quantifications were done within the linear range of the instruments in the absence of signal saturation. The data are shown as mean ± standard deviation (mean ± SD), with statistical analysis performed using the two-tailed, unpaired Student's t test.
ACKNOWLEDGMENTS
We thank Laurie Luckenbaugh for excellent technical assistance, Joseph Wang for advice on structural analysis, and Jason Liao at the Penn State Cancer Institute for statistical assistance.
This work was supported by NIH grants R37AI043453 and R01Al127670 to J.H.
Contributor Information
Jianming Hu, Email: juh13@psu.edu.
J.-H. James Ou, University of Southern California
REFERENCES
- 1.Hu J, Protzer U, Siddiqui A. 2019. Revisiting hepatitis B virus: challenges of curative therapies. J Virol 93:e01032-19. 10.1128/JVI.01032-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.WHO. 2017. Global hepatitis report 2017. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 3.Revill PA, Chisari FV, Block JM, Dandri M, Gehring AJ, Guo H, Hu J, Kramvis A, Lampertico P, Janssen HLA, Levrero M, Li W, Liang TJ, Lim S-G, Lu F, Penicaud MC, Tavis JE, Thimme R, Zoulim F, ICE-HBV Senior Advisors . 2019. A global scientific strategy to cure hepatitis B. Lancet Gastroenterol Hepatol 4:545–558. 10.1016/S2468-1253(19)30119-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hu J. 2016. Hepatitis B virus virology and replication, p 1–34. In Liaw Y-F, Zoulim F (ed), Hepatitis B virus in human diseases. Humana Press, Totowa, NJ. [Google Scholar]
- 5.Hu J, Liu K. 2017. Complete and incomplete hepatitis B virus particles: formation, function, and application. Viruses 9:56. 10.3390/v9030056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tuttleman JS, Pourcel C, Summers J. 1986. Formation of the pool of covalently closed circular viral DNA in hepadnavirus-infected cells. Cell 47:451–460. 10.1016/0092-8674(86)90602-1. [DOI] [PubMed] [Google Scholar]
- 7.Hu J, Seeger C. 2015. Hepadnavirus genome replication and persistence. Cold Spring Harb Perspect Med 5:a021386. 10.1101/cshperspect.a021386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Summers J, Mason WS. 1982. Replication of the genome of a hepatitis B-like virus by reverse transcription of an RNA intermediate. Cell 29:403–415. 10.1016/0092-8674(82)90157-x. [DOI] [PubMed] [Google Scholar]
- 9.Zlotnick A, Venkatakrishnan B, Tan Z, Lewellyn E, Turner W, Francis S. 2015. Core protein: a pleiotropic keystone in the HBV lifecycle. Antiviral Res 121:82–93. 10.1016/j.antiviral.2015.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Viswanathan U, Mani N, Hu Z, Ban H, Du Y, Hu J, Chang J, Guo JT. 2020. Targeting the multifunctional HBV core protein as a potential cure for chronic hepatitis B. Antiviral Res 182:104917. 10.1016/j.antiviral.2020.104917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ludgate L, Liu K, Luckenbaugh L, Streck N, Eng S, Voitenleitner C, Delaney WE, Hu J. 2016. Cell-free hepatitis B virus capsid assembly dependent on the core protein C-terminal domain and regulated by phosphorylation. J Virol 90:5830–5844. 10.1128/JVI.00394-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rat V, Pinson X, Seigneuret F, Durand S, Herrscher C, Lemoine R, Burlaud-Gaillard J, Raynal PY, Hourioux C, Roingeard P, Tramier M, de Rocquigny H. 2020. Hepatitis B virus core protein domains essential for viral capsid assembly in a cellular context. J Mol Biol 432:3802–3819. 10.1016/j.jmb.2020.04.026. [DOI] [PubMed] [Google Scholar]
- 13.Liu K, Luckenbaugh L, Ning X, Xi J, Hu J. 2018. Multiple roles of core protein linker in hepatitis B virus replication. PLoS Pathog 14:e1007085. 10.1371/journal.ppat.1007085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xi J, Luckenbaugh L, Hu J. 2021. Multiple roles of PP2A binding motif in hepatitis B virus core linker and PP2A in regulating core phosphorylation state and viral replication. PLoS Pathog 17:e1009230. 10.1371/journal.ppat.1009230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jung J, Hwang SG, Chwae Y-J, Park S, Shin H-J, Kim K. 2014. Phosphoacceptors threonine 162 and serines 170 and 178 within the carboxyl-terminal RRRS/T motif of the hepatitis B virus core protein make multiple contributions to hepatitis B virus replication. J Virol 88:8754–8767. 10.1128/JVI.01343-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lan YT, Li J, Liao W, Ou J. 1999. Roles of the three major phosphorylation sites of hepatitis B virus core protein in viral replication. Virology 259:342–348. 10.1006/viro.1999.9798. [DOI] [PubMed] [Google Scholar]
- 17.Schlicht HJ, Bartenschlager R, Schaller H. 1989. The duck hepatitis B virus core protein contains a highly phosphorylated C terminus that is essential for replication but not for RNA packaging. J Virol 63:2995–3000. 10.1128/JVI.63.7.2995-3000.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu M, Summers J. 1994. Multiple functions of capsid protein phosphorylation in duck hepatitis B virus replication. J Virol 68:4341–4348. 10.1128/JVI.68.7.4341-4348.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nassal M. 1992. The arginine-rich domain of the hepatitis B virus core protein is required for pregenome encapsidation and productive viral positive-strand DNA synthesis but not for virus assembly. J Virol 66:4107–4116. 10.1128/JVI.66.7.4107-4116.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lewellyn EB, Loeb DD. 2011. Serine phosphoacceptor sites within the core protein of hepatitis B virus contribute to genome replication pleiotropically. PLoS One 6:e17202. 10.1371/journal.pone.0017202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu K, Ludgate L, Yuan Z, Hu J. 2015. Regulation of multiple stages of hepadnavirus replication by the carboxyl-terminal domain of viral core protein in trans. J Virol 89:2918–2930. 10.1128/JVI.03116-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Daub H, Blencke S, Habenberger P, Kurtenbach A, Dennenmoser J, Wissing J, Ullrich A, Cotten M. 2002. Identification of SRPK1 and SRPK2 as the major cellular protein kinases phosphorylating hepatitis B virus core protein. J Virol 76:8124–8137. 10.1128/jvi.76.16.8124-8137.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ludgate L, Ning X, Nguyen DH, Adams C, Mentzer L, Hu J. 2012. Cyclin-dependent kinase 2 phosphorylates S/T-P sites in the hepadnavirus core protein C-terminal domain and is incorporated into viral capsids. J Virol 86:12237–12250. 10.1128/JVI.01218-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ning X, Basagoudanavar SH, Liu K, Luckenbaugh L, Wei D, Wang C, Wei B, Zhao Y, Yan T, Delaney W, Hu J. 2017. Capsid phosphorylation state and hepadnavirus virion secretion. J Virol 91:e00092-17. 10.1128/JVI.00092-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luo J, Xi J, Gao L, Hu J. 2020. Role of hepatitis B virus capsid phosphorylation in nucleocapsid disassembly and covalently closed circular DNA formation. PLoS Pathog 16:e1008459. 10.1371/journal.ppat.1008459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu K, Hu J. 2019. Secretion of empty or complete hepatitis B virions: envelopment of empty capsids versus mature nucleocapsids. Future Virology 14:95–105. 10.2217/fvl-2018-0128. [DOI] [Google Scholar]
- 27.Pairan A, Bruss V. 2009. Functional surfaces of the hepatitis B virus capsid. J Virol 83:11616–11623. 10.1128/JVI.01178-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ponsel D, Bruss V. 2003. Mapping of amino acid side chains on the surface of hepatitis B virus capsids required for envelopment and virion formation. J Virol 77:416–422. 10.1128/jvi.77.1.416-422.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ning X, Luckenbaugh L, Liu K, Bruss V, Sureau C, Hu J. 2018. Common and distinct capsid and surface protein requirements for secretion of complete and genome-free hepatitis B virions. J Virol 92:e00272-18. 10.1128/JVI.00272-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gao W, Hu J. 2007. Formation of hepatitis B virus covalently closed circular DNA: removal of genome-linked protein. J Virol 81:6164–6174. 10.1128/JVI.02721-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lentz TB, Loeb DD. 2011. Roles of the envelope proteins in the amplification of covalently closed circular DNA and completion of synthesis of the plus-strand DNA in hepatitis B virus. J Virol 85:11916–11927. 10.1128/JVI.05373-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Summers J, Smith PM, Huang MJ, Yu MS. 1991. Morphogenetic and regulatory effects of mutations in the envelope proteins of an avian hepadnavirus. J Virol 65:1310–1317. 10.1128/JVI.65.3.1310-1317.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lenhoff RJ, Summers J. 1994. Coordinate regulation of replication and virus assembly by the large envelope protein of an avian hepadnavirus. J Virol 68:4565–4571. 10.1128/JVI.68.7.4565-4571.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guo H, Jiang D, Zhou T, Cuconati A, Block TM, Guo JT. 2007. Characterization of the intracellular deproteinized relaxed circular DNA of hepatitis B virus: an intermediate of covalently closed circular DNA formation. J Virol 81:12472–12484. 10.1128/JVI.01123-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Basagoudanavar SH, Perlman DH, Hu J. 2007. Regulation of hepadnavirus reverse transcription by dynamic nucleocapsid phosphorylation. J Virol 81:1641–1649. 10.1128/JVI.01671-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perlman DH, Berg EA, O'Connor PB, Costello CE, Hu J. 2005. Reverse transcription-associated dephosphorylation of hepadnavirus nucleocapsids. Proc Natl Acad Sci U S A 102:9020–9025. 10.1073/pnas.0502138102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cui X, Ludgate L, Ning X, Hu J. 2013. Maturation-associated destabilization of hepatitis B virus nucleocapsid. J Virol 87:11494–11503. 10.1128/JVI.01912-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cui X, Guo J-T, Hu J. 2015. Hepatitis B virus covalently closed circular DNA formation in immortalized mouse hepatocytes associated with nucleocapsid destabilization. J Virol 89:9021–9028. 10.1128/JVI.01261-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liao W, Ou J-h. 1995. Phosphorylation and nuclear localization of the hepatitis B virus core protein: significance of serine in the three repeated SPRRR motifs. J Virol 69:1025–1029. 10.1128/JVI.69.2.1025-1029.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rabe B, Vlachou A, Panté N, Helenius A, Kann M. 2003. Nuclear import of hepatitis B virus capsids and release of the viral genome. Proc Natl Acad Sci U S A 100:9849–9854. 10.1073/pnas.1730940100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen C, Wang JC-Y, Zlotnick A. 2011. A kinase chaperones hepatitis B virus capsid assembly and captures capsid dynamics in vitro. PLoS Pathog 7:e1002388. 10.1371/journal.ppat.1002388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gallucci L, Kann M. 2017. Nuclear import of hepatitis B virus capsids and genome. Viruses 9:21. 10.3390/v9010021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Diab A, Foca A, Fusil F, Lahlali T, Jalaguier P, Amirache F, N'Guyen L, Isorce N, Cosset F-L, Zoulim F, Andrisani O, Durantel D. 2017. Polo‐like‐kinase 1 is a proviral host factor for hepatitis B virus replication. Hepatology 66:1750–1765. 10.1002/hep.29236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kann M, Gerlich WH. 1994. Effect of core protein phosphorylation by protein kinase C on encapsidation of RNA within core particles of hepatitis B virus. J Virol 68:7993–8000. 10.1128/JVI.68.12.7993-8000.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Heger-Stevic J, Zimmermann P, Lecoq L, Böttcher B, Nassal M. 2018. Hepatitis B virus core protein phosphorylation: identification of the SRPK1 target sites and impact of their occupancy on RNA binding and capsid structure. PLoS Pathog 14:e1007488. 10.1371/journal.ppat.1007488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hu Z, Ban H, Zheng H, Liu M, Chang J, Guo JT. 2020. Protein phosphatase 1 catalyzes HBV core protein dephosphorylation and is co-packaged with viral pregenomic RNA into nucleocapsids. PLoS Pathog 16:e1008669. 10.1371/journal.ppat.1008669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Albin C, Robinson W. 1980. Protein kinase activity in hepatitis B virus. J Virol 34:297–302. 10.1128/JVI.34.1.297-302.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Feitelson MA, Marion PL, Robinson WS. 1982. Core particles of hepatitis B virus and ground squirrel hepatitis virus. II. Characterization of the protein kinase reaction associated with ground squirrel hepatitis virus and hepatitis B virus. J Virol 43:741–748. 10.1128/JVI.43.2.741-748.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Malumbres M. 2014. Cyclin-dependent kinases. Genome Biol 15:122. 10.1186/gb4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Elia AE, Rellos P, Haire LF, Chao JW, Ivins FJ, Hoepker K, Mohammad D, Cantley LC, Smerdon SJ, Yaffe MB. 2003. The molecular basis for phosphodependent substrate targeting and regulation of Plks by the Polo-box domain. Cell 115:83–95. 10.1016/s0092-8674(03)00725-6. [DOI] [PubMed] [Google Scholar]
- 51.Morgan DO. 1997. Cyclin-dependent kinases: engines, clocks, and microprocessors. Annu Rev Cell Dev Biol 13:261–291. 10.1146/annurev.cellbio.13.1.261. [DOI] [PubMed] [Google Scholar]
- 52.Morgan DO. 2007. The cell cycle: principles of control. New Science Press, London, United Kingdom. [Google Scholar]
- 53.Bouchard M, Giannakopoulos S, Wang EH, Tanese N, Schneider RJ. 2001. Hepatitis B virus HBx protein activation of cyclin A–cyclin-dependent kinase 2 complexes and G1 transit via a Src kinase pathway. J Virol 75:4247–4257. 10.1128/JVI.75.9.4247-4257.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gearhart TL, Bouchard MJ. 2010. Replication of the hepatitis B virus requires a calcium-dependent HBx-induced G1 phase arrest of hepatocytes. Virology 407:14–25. 10.1016/j.virol.2010.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yeh C-T, Wong SW, Fung Y-K, Ou J-H. 1993. Cell cycle regulation of nuclear localization of hepatitis B virus core protein. Proc Natl Acad Sci U S A 90:6459–6463. 10.1073/pnas.90.14.6459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wood DJ, Endicott JA. 2018. Structural insights into the functional diversity of the CDK-cyclin family. Open Biol 8:180112. 10.1098/rsob.180112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brown NR, Noble ME, Endicott JA, Johnson LN. 1999. The structural basis for specificity of substrate and recruitment peptides for cyclin-dependent kinases. Nat Cell Biol 1:438–443. 10.1038/15674. [DOI] [PubMed] [Google Scholar]
- 58.Adams PD, Sellers WR, Sharma SK, Wu AD, Nalin CM, Kaelin WG. Jr, 1996. Identification of a cyclin-cdk2 recognition motif present in substrates and p21-like cyclin-dependent kinase inhibitors. Mol Cell Biol 16:6623–6633. 10.1128/mcb.16.12.6623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Takeda DY, Wohlschlegel JA, Dutta A. 2001. A bipartite substrate recognition motif for cyclin-dependent kinases. J Biol Chem 276:1993–1997. 10.1074/jbc.M005719200. [DOI] [PubMed] [Google Scholar]
- 60.Luo J, Cui X, Gao L, Hu J. 2017. Identification of intermediate in hepatitis B virus covalently closed circular (CCC) DNA formation and sensitive and selective CCC DNA detection. J Virol 91:e00539-17. 10.1128/JVI.00539-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hong X, Luckenbaugh L, Mendenhall M, Walsh R, Cabuang L, Soppe S, Revill P, Burdette D, Feierbach B, Delaney W, Hu J. 2020. Characterization of hepatitis B precore/core-related antigens. J Virol 95:e01695-20. 10.1128/JVI.01695-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ning X, Nguyen D, Mentzer L, Adams C, Lee H, Ashley R, Hafenstein S, Hu J. 2011. Secretion of genome-free hepatitis B virus—single strand blocking model for virion morphogenesis of para-retrovirus. PLoS Pathog 7:e1002255. 10.1371/journal.ppat.1002255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Luo J, Luckenbaugh L, Hu H, Yan Z, Gao L, Hu J. 2020. Involvement of host ATR-CHK1 pathway in hepatitis B virus covalently closed circular DNA formation. mBio 11:e03423-19. 10.1128/mBio.03423-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wu S, Luo Y, Viswanathan U, Kulp J, Cheng J, Hu Z, Xu Q, Zhou Y, Gong G-Z, Chang J, Li Y, Guo J-T. 2018. CpAMs induce assembly of HBV capsids with altered electrophoresis mobility: implications for mechanism of inhibiting pgRNA packaging. Antiviral Res 159:1–12. 10.1016/j.antiviral.2018.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhao Q, Hu Z, Cheng J, Wu S, Luo Y, Chang J, Hu J, Guo J-T. 2018. Hepatitis B virus core protein dephosphorylation occurs during pregenomic RNA encapsidation. J Virol 92:e02139-17. 10.1128/JVI.02139-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chua PK, Tang F-M, Huang J-Y, Suen C-S, Shih C. 2010. Testing the balanced electrostatic interaction hypothesis of hepatitis B virus DNA synthesis by using an in vivo charge rebalance approach. J Virol 84:2340–2351. 10.1128/JVI.01666-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cui X, Luckenbaugh L, Bruss V, Hu J. 2015. Alteration of mature nucleocapsid and enhancement of covalently closed circular DNA formation by hepatitis B virus core mutants defective in complete virion formation. J Virol 89:10064–10072. 10.1128/JVI.01481-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dhason MS, Wang JC-Y, Hagan MF, Zlotnick A. 2012. Differential assembly of hepatitis B virus core protein on single-and double-stranded nucleic acid suggest the dsDNA-filled core is spring-loaded. Virology 430:20–29. 10.1016/j.virol.2012.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tan Z, Pionek K, Unchwaniwala N, Maguire ML, Loeb DD, Zlotnick A. 2015. The interface between HBV capsid proteins affects self-assembly, pgRNA packaging, and reverse transcription. J Virol 89:3275–3284. 10.1128/JVI.03545-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pastor F, Herrscher C, Patient R, Eymieux S, Moreau A, Burlaud-Gaillard J, Seigneuret F, de Rocquigny H, Roingeard P, Hourioux C. 2019. Direct interaction between the hepatitis B virus core and envelope proteins analyzed in a cellular context. Sci Rep 9:16178. 10.1038/s41598-019-52824-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bottcher B, Nassal M. 2018. Structure of mutant hepatitis B core protein capsids with premature secretion phenotype. J Mol Biol 430:4941–4954. 10.1016/j.jmb.2018.10.018. [DOI] [PubMed] [Google Scholar]
- 72.Yu X, Jin L, Jih J, Shih C, Zhou ZH. 2013. 3.5 Å cryoEM structure of hepatitis B virus core assembled from full-length core protein. PLoS One 8:e69729. 10.1371/journal.pone.0069729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Selzer L, Kant R, Wang JC-Y, Bothner B, Zlotnick A. 2015. Hepatitis B virus core protein phosphorylation sites affect capsid stability and transient exposure of the C-terminal domain. J Biol Chem 290:28584–28593. 10.1074/jbc.M115.678441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nair S, Zlotnick A. 2021. HBV core protein is in flux between cytoplasmic, nuclear, and nucleolar compartments. mBio 12:e03514-20. 10.1128/mBio.03514-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fallows DA, Goff SP. 1995. Mutations in the epsilon sequences of human hepatitis B virus affect both RNA encapsidation and reverse transcription. J Virol 69:3067–3073. 10.1128/JVI.69.5.3067-3073.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hu J, Flores D, Toft D, Wang X, Nguyen D. 2004. Requirement of heat shock protein 90 for human hepatitis B virus reverse transcriptase function. J Virol 78:13122–13131. 10.1128/JVI.78.23.13122-13131.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hong X, Luckenbaugh L, Perlman DH, Revill PA, Wieland SF, Menne S, Hu J. 18 January 2021. Characterization and application of precore/core-related antigens in animal models of hepatitis B virus infection. Hepatology 10.1002/hep.31720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Luckenbaugh L, Kitrinos K, Delaney IW, Hu J. 2015. Genome‐free hepatitis B virion levels in patient sera as a potential marker to monitor response to antiviral therapy. J Viral Hepat 22:561–570. 10.1111/jvh.12361. [DOI] [PMC free article] [PubMed] [Google Scholar]