ABSTRACT
Vibrio parahaemolyticus cells transit from free-swimming to surface adapted lifestyles, such as swarming colonies and three-dimensional biofilms. These transitions are regulated by sensory modules and regulatory networks that involve the second messenger cyclic diguanylate monophosphate (c-di-GMP). In this work, we show that a previously uncharacterized c-di-GMP phosphodiesterase (VP1881) from V. parahaemolyticus plays an important role in modulating the c-di-GMP pool. We found that the product of VP1881 promotes its own expression when the levels of c-di-GMP are low or when the phosphodiesterase (PDE) is catalytically inactive. This behavior has been observed in a class of c-di-GMP receptors called trigger phosphodiesterases, and hence we named the product of VP1881 TpdA, for trigger phosphodiesterase A. The absence of tpdA showed a negative effect on swimming motility while, its overexpression from an isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible promoter showed a positive effect on both swimming and swarming motility and a negative effect on biofilm formation. Changes in TpdA abundance altered the expression of representative polar and lateral flagellar genes, as well as that of the biofilm-related gene cpsA. Our results also revealed that autoactivation of the native PtpdA promoter is sufficient to alter c-di-GMP signaling responses such as swarming and biofilm formation in V. parahaemolyticus, an observation that could have important implications in the dynamics of these social behaviors.
IMPORTANCE c-di-GMP trigger phosphodiesterases (PDEs) could play a key role in controlling the heterogeneity of biofilm matrix composition, a property that endows characteristics that are potentially relevant for sustaining integrity and functionality of biofilms in a variety of natural environments. Trigger PDEs are not always easy to identify based on their sequence, and hence not many examples of these type of signaling proteins have been reported in the literature. Here, we report on the identification of a novel trigger PDE in V. parahaemolyticus and provide evidence suggesting that its autoactivation could play an important role in the progression of swarming motility and biofilm formation, multicellular behaviors that are important for the survival and dissemination of this environmental pathogen.
KEYWORDS: Vibrio parahaemolyticus, bacterial genetics, biofilms, c-di-GMP, flagellar motility, second messenger, signal transduction, swarming
INTRODUCTION
Bacteria have developed multiple strategies to tackle life-threatening situations such as attack by predators and bacterial competitors and the limitation of nutrients in oligotrophic environments. One of the most frequently recurring strategies for survival involves the formation of biofilms, a self-made enclosure where a bacterial population is protected by an extracellular matrix that is typically composed of exopolysaccharides, proteins, and DNA (1). This communal life provides such an important fitness advantage that it has been recently estimated that most bacteria and archaea in our planet are found to form biofilms (2). Although biofilms offer multiple benefits, their making impose an important energetic burden that needs to be overlooked by cells experiencing nutritional limitations and looking for qualitative rather than quantitative strategies of survival. Thus, the transition from a planktonic to a communal multicellular lifestyle is regulated by sophisticated regulatory networks in which the second messenger cyclic diguanylate monophosphate (c-di-GMP) plays a central role (3, 4).
c-di-GMP is synthesized by proteins with diguanylate cyclase (DGC) activity and degraded by c-di-GMP-specific phosphodiesterases (PDEs). These enzymes have conserved catalytic domains; the GGDEF domain is present in DGCs, and the EAL or HD-GYP domains are present in PDEs (3–5). The signaling module is completed by c-di-GMP receptors with a diversity of activities, including transcriptional regulators, ATPases, flagellar motor switch regulators, and deacetylases (6–14). Several bacterial genomes encode tens of c-di-GMP synthesizers and degraders (https://www.ncbi.nlm.nih.gov/Complete_Genomes/c-di-GMP.html) (3, 6). In most cases, it is not clear how much individual c-di-GMP metabolic enzymes contribute to redundancy of a particular signaling network or what specific role they play in signal transduction pathways. Such is the case of the c-di-GMP signaling network of the environmental pathogen Vibrio parahaemolyticus. This bacterium uses two social behaviors that are oppositely regulated by c-di-GMP, biofilm formation and a type of coordinated motility known as swarming motility. These two social strategies could potentially drive the colonization success of this pathogen, but it is not clear how this bacterium integrates information to transition from a planktonic lifestyle to a multicellular mode of life on surfaces.
Biofilm formation in V. parahaemolyticus requires the production of the capsular exopolysaccharide CPSA, made by biosynthetic proteins encoded in the cpsA to cpsK locus (15, 16). c-di-GMP levels positively regulate the expression of the cpsA to cpsK locus through the activation of the c-di-GMP receptors CpsR, CpsQ, and ScrO (15, 17). The main drivers of the activation of biofilm matrix production are CpsQ and ScrO (17). Motility, on the other hand, requires the production of either a polar flagellum, in the case of swimming motility, or lateral flagella that are used for swarming over solid surfaces (18–20). While the influence of c-di-GMP on the expression of polar flagellar genes in V. parahaemolyticus has not been fully addressed, it has been shown that the expression of lateral flagellar genes (laf) is negatively influenced by an increase in c-di-GMP levels (21). The c-di-GMP signaling module composed of the bifunctional DGC-PDE ScrC and the modulator proteins ScrA and ScrB controls swarming motility and biofilm formation in response to a self-made signaling molecule named the S signal (15, 21–24). When the periplasmic protein ScrB binds the S signal made by ScrA, it switches the activity of ScrC from a DGC to a PDE. This activity switch favors swarming motility and blocks biofilm formation (23). Besides ScrC, the PDE ScrG has been shown to affect swarming motility and biofilm formation in opposite ways (25). More recently, two GGDEF domain-containing proteins (ScrJ and ScrL) and a degenerate EAL domain-containing protein (LafV) were found to be negative modulators of the expression of laf genes (26). Although our understanding of the c-di-GMP signaling modules present in V. parahaemolyticus keeps growing, we are still at the early stages in their characterization and in defining their precise role in controlling multicellularity and social dynamics.
In this report, we characterized the role of a c-di-GMP phosphodiesterase encoded by the VP1881 gene. The expression of this gene was found to be repressed under surface adaptation conditions (27), and hence we speculated that its activity could be important for bacteria to transition from motile to sessile lifestyles. Our results revealed that this PDE is a key contributor to the maintenance of c-di-GMP levels in V. parahaemolyticus. We also found that expression of VP1881 is induced at low c-di-GMP levels in a mechanism that depends on its own product. Strikingly, the magnitude of positive autoregulation was augmented in the presence of the catalytically inactive variant VP1881AAA. This type of behavior is typical of trigger phosphodiesterases; these are PDEs that degrade c-di-GMP when available but exert an additional regulatory action in the absence of the second messenger (28). Since the product of VP1881 behaved like previously described trigger PDEs, we named it TpdA, for trigger phosphodiesterase A.
In summary, our results uncover the role of TpdA as a novel modulator of swimming motility in V. parahaemolyticus and provide evidence suggesting that induction of the native PtpdA promoter can positively regulate swarming motility and negatively regulate biofilm formation. This regulatory scheme could play a key role in controlling surface colonization and social dynamics in this environmental pathogen.
RESULTS
The gene VP1881 encodes an active phosphodiesterase that modulates the global c-di-GMP pool.
The gene VP1881 encodes a putative phosphodiesterase with a conserved EAL domain. It has been reported that its expression is downregulated in cells grown over a solid surface (27). This could suggest that VP1881 may be involved in the transition from motile to sessile lifestyles. To evaluate if the PDE encoded by the gene VP1881 is capable of modulating c-di-GMP levels, we used a genetic reporter that allows the determination of relative abundance of this second messenger in living cells (29). c-di-GMP positively controls the production of the fluorescent reporter TurboRFP by acting on two c-di-GMP-riboswitches arrayed in tandem upstream of the structural gene. The expression of the reporter is normalized using the fluorescence intensity of Amcyan, a fluorescence reporter that is coexpressed with TurboRFP but independently of c-di-GMP levels. We compared c-di-GMP abundance between the wild-type (WT) strain and genetic backgrounds that lack or overproduce VP1881 (Fig. 1). As an internal control, we analyzed c-di-GMP abundance in a strain overproducing the previously characterized PDE ScrG (25).
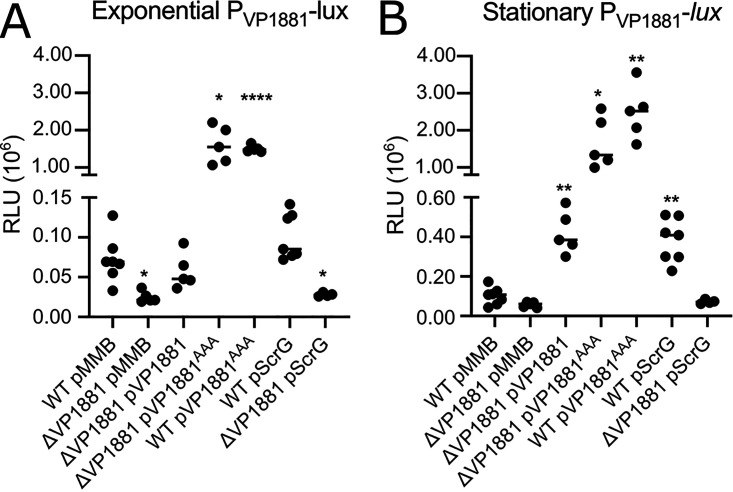
FIG 1.
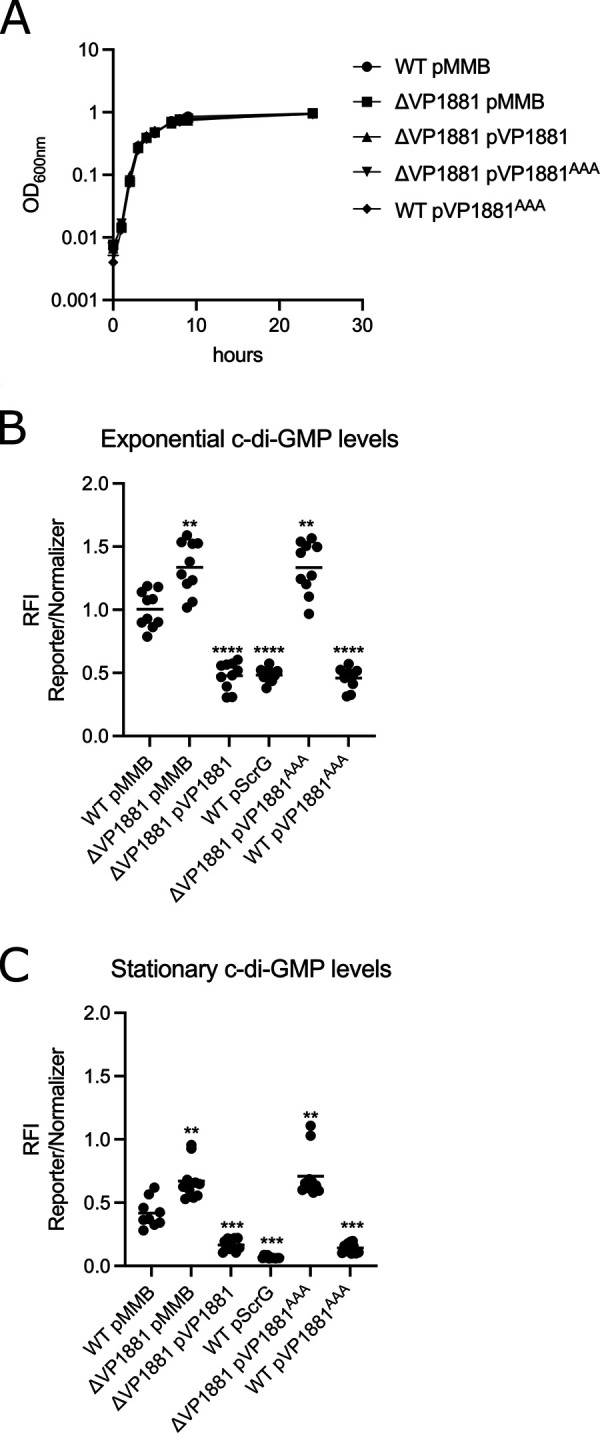
The product of the gene VP1881 modulates c-di-GMP abundance. (A) Growth curve plots from three biological replicates from strains harboring either the empty plasmid pMMB67EH-Gm (pMMB) or overexpression constructs containing VP1881 or VP1881AAA. (B and C) Scatterplots of relative fluorescence intensity (RFI) values from at least 10 biological replicates from each strain of interest harboring a c-di-GMP biosensor, grown to the exponential phase (B) or the stationary phase (C) in the presence of 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). RFI refers to the ratio of arbitrary fluorescence units from the c-di-GMP reporter TurboRFP and the calibrator Amcyan. The horizontal line represents the mean RFI. Overexpression of VP1881 and its variants in the presence of IPTG was achieved using expression constructs that contain an IPTG-inducible promoter. The empty plasmid pMMB67EH-Gm (pMMB) was used as a negative control. Means were compared to the wild type (WT) using Brown-Forsythe and Welch analysis of variance (ANOVA) tests followed by Dunnett’s T3 multiple-comparison test. Adjusted P values of ≤0.05 were deemed statistically significant. **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001.
First, we analyzed the effect on growth of either the absence or overexpression of VP1881. Our results showed that neither the absence nor the overexpression of VP1881 from an isopropyl-β-d-thiogalactopyranoside (IPTG) inducible promoter had a noticeable effect on the growth of these strains compared to the WT strain harboring the empty plasmid pMMB67EH-Gm (pMMB) (Fig. 1A).
The absence of VP1881 resulted in 33% and 61% increases in c-di-GMP levels compared to those of the WT strain in cells grown to the exponential and stationary phases, respectively (Fig. 1B and C). Overexpression of VP1881 in the ΔVP1881 mutant strain from an IPTG-inducible promoter (pVP1881) resulted in 52% and 60% decreases in c-di-GMP levels compared to those of the WT strain in cells grown to the exponential and stationary phases, respectively (Fig. 1B and C). A strain overproducing the previously characterized PDE ScrG from an IPTG-inducible promoter (pScrG) showed 52% and 84% decreases in c-di-GMP levels compared to those of the WT strain in cells grown to the exponential and stationary phases, respectively (Fig. 1B and C). These results strongly suggest that the product of VP1881 is capable of modulating the c-di-GMP pool inside living cells. We also overproduced a VP1881 variant in which the EAL amino acids of the active site were changed to AAA (VP1881AAA); these amino acid changes typically result in an inactive PDE (30–33). Overexpression of VP1881AAA from an IPTG-inducible promoter (pVP1881AAA) did not complement the phenotype of increased c-di-GMP levels in the ΔVP1881 mutant strain compared to the WT strain (Fig. 1B and C). This result strongly suggests that the ability of VP1881 to modulate the abundance of c-di-GMP depends on the presence of a conserved EAL motif. Interestingly, although overproduction of VP1881AAA did not alter c-di-GMP levels in the ΔVP1881 mutant strain, it significantly reduced c-di-GMP accumulation in a WT genetic background (Fig. 1B and C). A possible explanation for this counterintuitive result could be that overproduction of VP1881AAA in the WT background induces the expression of the endogenous copy of VP1881, resulting in reduced c-di-GMP accumulation. Hence, we hypothesized that overproduction of VP1881 and/or VP1881AAA would result in an upregulation of VP1881 gene expression.
The gene VP1881 encodes a trigger phosphodiesterase capable of promoting its own expression.
To analyze the expression of VP1881, we used a transcriptional fusion (PVP1881-luxCDABE) composed of the regulatory region of VP1881 and a bacterial luciferase reporter encoded by the luxCDABE cluster. During exponential growth, the expression of the transcriptional fusion decreased by 66% in the ΔVP1881 mutant strain compared to that in the WT strain (Fig. 2A). We did not observe significant differences in expression between the ΔVP1881 mutant and the WT strains in cells grown to the stationary phase (Fig. 2B). Overproduction of VP1881 from an IPTG-inducible promoter complemented the expression of the fusion to WT levels during the exponential growth phase and resulted in a 318% increase in expression compared to the WT strain in cells grown to the stationary phase (Fig. 2). These results suggest that the product of VP1881 promotes its own expression. This autoregulation could be an indirect effect of lowering the global c-di-GMP pool. To address this, we analyzed the expression of the fusion in the ΔVP1881 pVP1881AAA strain that does not have reduced levels of c-di-GMP compared to the WT strain. In this genetic background, the expression of the fusion increased 2,110% and 1,551% compared to the WT strain in cells grown to the exponential or stationary phases, respectively (Fig. 2), strongly suggesting that changes in c-di-GMP are not fully required to promote activation of the fusion and that the inactive variant VP1881AAA is better than the active VP1881 protein at inducing expression of the PVP1881 promoter. A similar level of induction of the PVP1881 promoter was observed in the WT pVP1881AAA strain (Fig. 2), suggesting that reduced c-di-GMP levels do not have a significant additive effect on the expression of the PVP1881 promoter when the variant VP1881AAA is being overproduced.
FIG 2.
VP1881 regulates its own expression. (A and B) Scatterplots of relative light units (RLU) from at least 5 biological replicates from each strain of interest harboring the transcriptional fusion pBBR-PVP1881-lux, grown to the exponential phase (A) or the stationary phase (B) in the presence of 0.1 mM IPTG. RLU are arbitrary light units per milliliter divided by optical density at 600 nm. The RLU value is directly proportional to the activity of the PVP1881 promoter. The horizontal line represents the mean RLU value. Overexpression of VP1881 and its variants was achieved using expression constructs that contain an IPTG-inducible promoter. The empty plasmid pMMB67EH-Gm (pMMB) was used as a negative control. Means were compared to the WT using Brown-Forsythe and Welch ANOVA tests followed by Dunnett’s T3 multiple-comparison test. Adjusted P values of ≤0.05 were deemed statistically significant. *, P ≤ 0.05; **, P ≤ 0.01; ****, P ≤ 0.0001.
To further evaluate at what level c-di-GMP regulates the expression of VP1881, we overproduced the PDE ScrG in the WT and the ΔVP1881 strains. We analyzed the expression of the PVP1881-luxCDABE fusion in these genetic backgrounds with decreased c-di-GMP levels. Expression of this fusion was not significantly affected by the overproduction of ScrG in exponentially grown cells (Fig. 2A). In contrast, in cells grown to stationary phase overproduction of ScrG resulted in a 279% increase in the expression of the fusion (Fig. 2B). However, the ScrG-dependent increase in expression of the PVP1881-luxCDABE fusion was not observed in the ΔVP1881 background (Fig. 2B). Hence, it appears that c-di-GMP acts upstream of the product of VP1881 and limits its ability to promote its own expression.
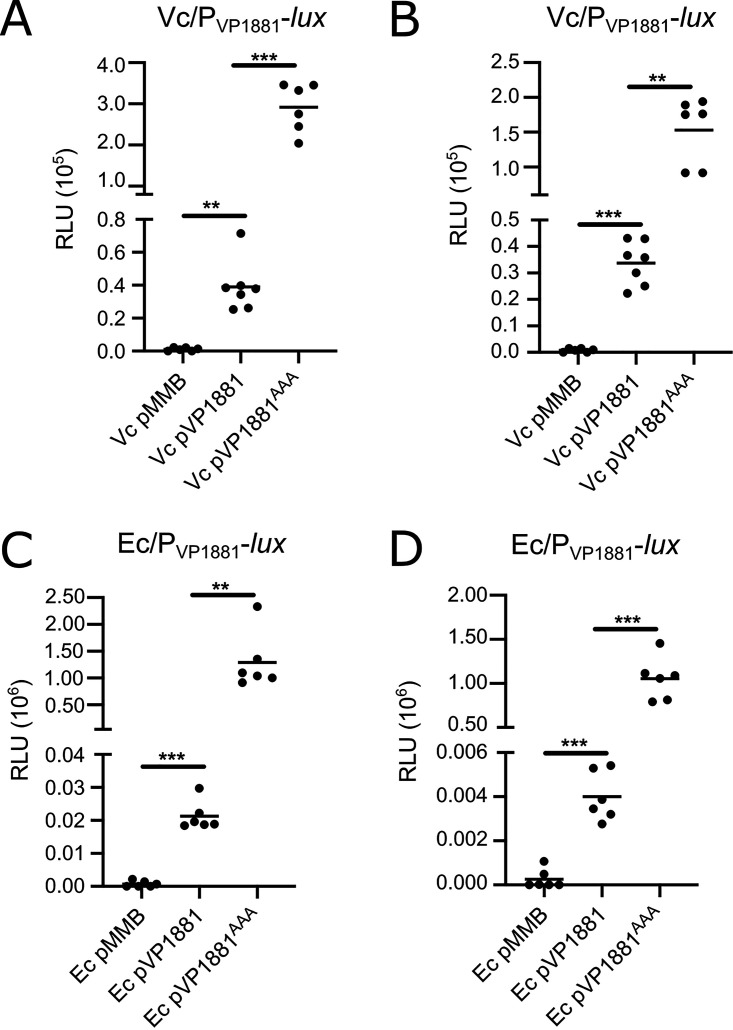
We also wanted to evaluate if the autoregulation of VP1881 could be reconstituted in heterologous hosts that lack an orthologue of VP1881. We did not find potential orthologues of VP1881 in the genomes of Vibrio cholerae O1 El Tor N16961 and Escherichia coli K-12 MG1655 using a bidirectional best-hit analysis, and hence we chose these two genetic backgrounds to study autoregulation of VP1881. In both V. cholerae and E. coli, we observed expression of the PVP1881-luxCDABE fusion only when we overproduced VP1881 or VP1881AAA from an IPTG-inducible promoter (Fig. 3). The expression of the fusion was several orders of magnitude higher in cells overproducing the inactive variant VP1881AAA. These results showed that autoregulation of VP1881 can occur in these two heterologous hosts. In a similar fashion to what occurs in V. parahaemolyticus, the VP1881AAA variant was more effective in activating the transcription of VP1881 in both V. cholerae and E. coli. It remains to be shown what other factors, if any, participate in the activation of this gene’s expression.
FIG 3.
VP1881 regulates its own expression in heterologous hosts. Scatterplots of relative light units (RLU) from at least 6 biological replicates from each strain of interest harboring the transcriptional fusion pBBR-PVP1881-lux are shown. Expression of the pBBR-PVP1881-lux fusion was analyzed in Vibrio cholerae (Vc) cells grown to the exponential phase (A) or the stationary phase (B) and in Escherichia coli (Ec) cells grown to the exponential phase (C) or the stationary phase (D) in the presence of 0.1 mM IPTG. RLU are arbitrary light units per milliliter divided by optical density at 600 nm (OD600). The RLU value is directly proportional to the activity of the PVP1881 promoter. The horizontal line represents the mean RLU value. Overexpression of VP1881 and its variants was achieved using expression constructs that contain an IPTG-inducible promoter. The empty plasmid pMMB67EH-Gm (pMMB) was used as a negative control. Means were compared using Brown-Forsythe and Welch ANOVA tests followed by Dunnett’s T3 multiple-comparison test. Adjusted P values of ≤0.05 were deemed statistically significant. **, P ≤ 0.01; ***, P ≤ 0.001.
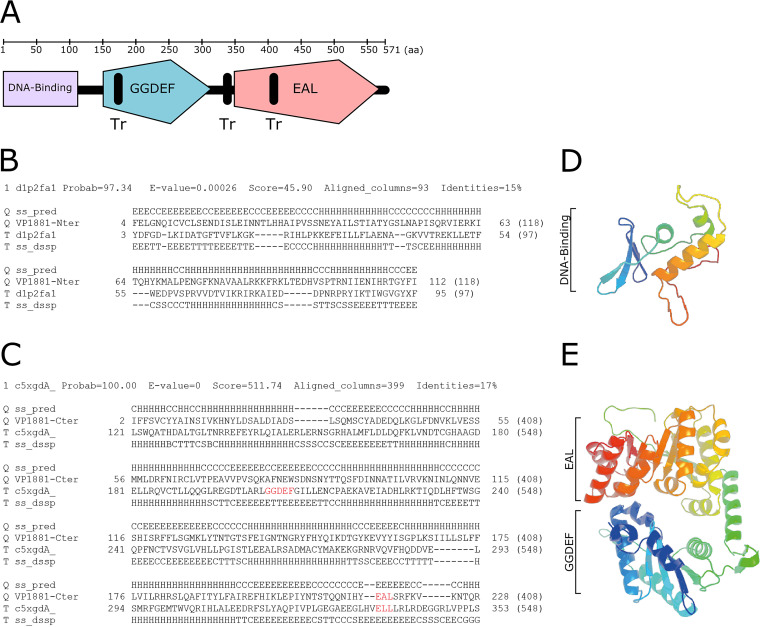
The analysis of the domain architecture of VP1881 using InterProScan (https://www.ebi.ac.uk/interpro/search/sequence/) revealed the presence of potential transmembrane regions and a conserved EAL domain located at the carboxyl terminus (C terminus) of the protein (Fig. 4). No other conserved domains were found in the middle region or in the amino terminus (N terminus) of the protein by using this tool, which strongly relies on protein primary sequence conservation. We next used Phyre2 to predict the domain topology of VP1881; this complementary tool uses secondary and tertiary structure prediction and a template-based modeling approach to define domain boundaries and their potential function (Fig. 4) (34). The first 118 amino acid residues of VP1881 were modeled with 97.3% confidence as a DNA/RNA-binding 3-helical bundle fold. The template used to generate the model was the crystal structure d1p2fa1, which corresponds to the DNA-binding domain of the response regulator DrrB from Thermotoga maritima (Fig. 4B and D) (35). We next analyzed the region between the amino acids 164 and 571 of VP1881. This region was modeled with 100% confidence as a tandem GGDEF-EAL domain, using as a template the crystal structure c5xgdA, which corresponds to the protein PA0861 from Pseudomonas aeruginosa (Fig. 4C and E) (36). These observations suggest that VP1881, in addition to the conserved EAL domain, contains a degenerated GGDEF-like domain and a putative DNA-binding domain. Validation of these domains requires further experimentation and could provide important clues regarding the mechanism involved in the autoregulation of VP1881. In particular, the presence of a DNA-binding domain could support the hypothesis that VP1881 is capable of directly activating its own transcription.
FIG 4.
VP1881 has a putative DNA-binding domain, a putative degenerated GGDEF domain, and a conserved EAL domain. (A) Schematic representation of the domain architecture of VP1881 based on predictions from InterProScan and Phyre2. Tr, transmembrane region. (B and C) Sequence alignment and secondary structure prediction of the N terminus (B) and middle region (C) of VP1881, with templates used for generating three-dimensional (3D) model structures. In the secondary structure prediction (ss_pred), the letter E symbolizes beta strands, the letter H symbolizes alpha helixes, the letter C symbolizes coils, the letter T symbolizes a hydrogen-bonded turn, and the letter S a bend. The GGDEF and EAL motifs are colored red in the alignment. (D and E) Ribbon diagrams representing modeled 3D structures of the putative DNA-binding domain of VP1881 (D) and the GGDEF-EAL domains (E) of VP1881, generated with Phyre2 using templates d1p2fa1 and c5xgdA, respectively. Secondary structures located closer to the N terminus are colored with colder colors, while structures located closer to the C terminus are colored with warmer colors.
The evidence presented so far strongly suggests that VP1881 has a dual function, namely, c-di-GMP degradation and modulation of its own expression. These two functions appear to be negatively correlated, which is a characteristic of a type of c-di-GMP effectors known as trigger phosphodiesterases (28). From here on we will refer to VP1881 as tpdA, which stands for trigger phosphodiesterase A.
The PDE TpdA modulates swimming motility and the expression of flaA.
Based on the most recent census of c-di-GMP related proteins, the genome of V. parahaemolyticus RIMD 2210633 encodes 16 proteins with tandem GGDEF-EAL domains, 13 proteins containing the EAL domain, and 5 with the HD-GYP domain (https://www.ncbi.nlm.nih.gov/Complete_Genomes/c-di-GMP.html) (3, 6, 37). It is yet unknown how many of these proteins are involved in controlling the levels of c-di-GMP during growth under standard laboratory conditions and how many participate in the regulation of motile to sessile transitions. Higher levels of c-di-GMP favor biofilm formation, while lower levels of c-di-GMP favor motility of V. parahaemolyticus cells (22, 23, 25). The results presented above suggest that TpdA is an important contributor in the regulation of the global c-di-GMP pool during growth in rich medium. To determine if phenotypes associated with altered c-di-GMP levels are modified in the absence of tpdA, we first compared the abilities of the ΔtpdA mutant strain and the WT strain to swim in soft agar (lysogeny broth [LB] plus 0.3% agar). The absence of tpdA resulted in a 48% decrease in swimming motility compared to that of the WT strain (Fig. 5). Complementation of the ΔtpdA strain with the pTpdA plasmid resulted in a 64% increase in swimming motility compared to that of the WT strain (Fig. 5). On the other hand, overproduction of TpdAAAA did not complement the motility phenotype of the ΔtpdA strain (Fig. 5). These results suggest that the absence and the overproduction of TpdA affect swimming motility in opposite manners, and this is likely due to changes in c-di-GMP levels. We have shown that overproduction of TpdAAAA in the WT background increases the activity of the tpdA promoter and alters c-di-GMP levels (Fig. 1 and 2). However, we did not observe a clear difference between the motility of the WT and WT pTpdAAAA strains (Fig. 5). This could suggest that, under these conditions, overexpression of tpdA from its own promoter is not sufficient to affect swimming motility.
FIG 5.
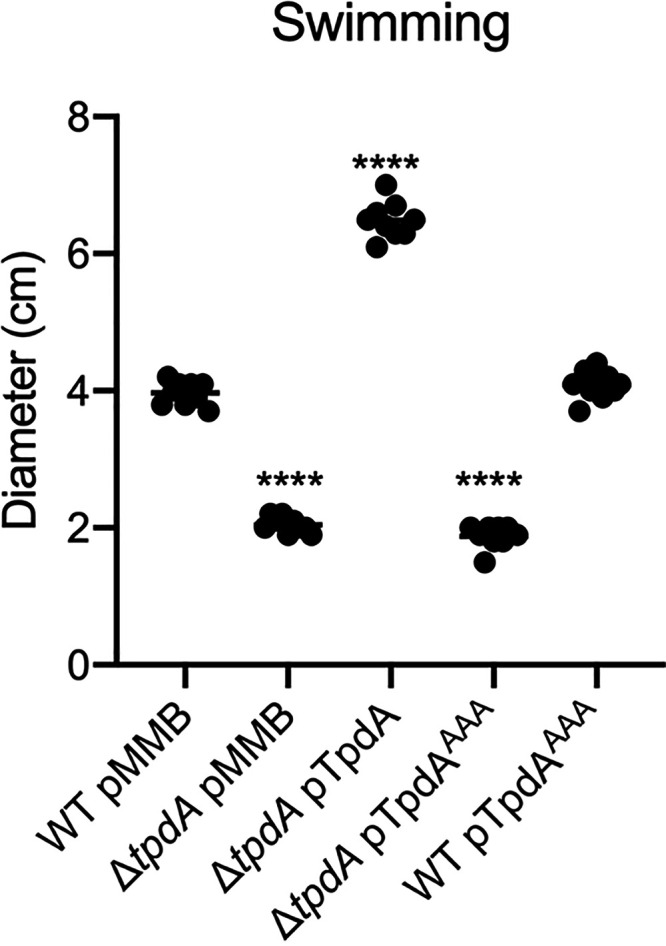
TpdA (VP1881) modulates swimming motility in soft agar. Scatterplots representing the diameter of the migration halo in soft agar (0.3%) plates containing 0.1 mM IPTG, of at least 9 biological replicates from each strain of interest are shown. The horizontal line represents the mean diameter of migration. Overexpression of TpdA and its variants was achieved using expression constructs that contain an IPTG-inducible promoter. The empty plasmid pMMB67EH-Gm (pMMB) was used as a negative control. Means were compared to the WT strain harboring the pMMB plasmid, using an ordinary one-way ANOVA test followed by a Dunnett’s multiple-comparison test. Adjusted P values of ≤0.05 were deemed statistically significant. ****, P ≤ 0.0001.
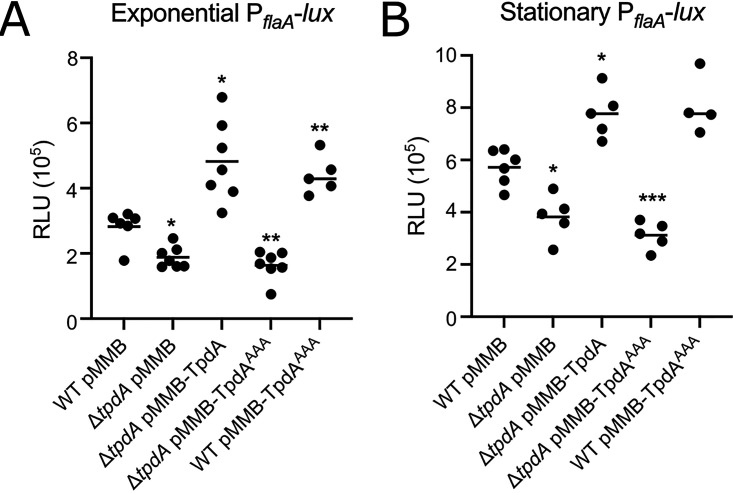
Changes in c-di-GMP levels can alter the expression of flagellar genes in different vibrios (21, 27, 38). To analyze if the absence or overproduction of TpdA could alter the expression of a polar flagellar gene in V. parahaemolyticus, we generated the transcriptional fusion PflaA-luxCDABE with the promoter of flaA, one of the flagellins that compose the polar propeller (18, 39, 40). Similar to the effect observed in swimming motility, during exponential growth, the expression of the fusion decreased 33% in the ΔtpdA mutant strain compared to that of the WT strain. Expression of the fusion in the ΔtpdA pTpdA complemented strain increased by 71% compared to that of the WT strain (Fig. 6A). The overproduction of TpdAAAA was unable to complement the decreased expression of the fusion in the ΔtpdA mutant strain compared to that of the WT strain (Fig. 6A). On the other hand, expression of the fusion in the WT pTpdAAAA strain showed a 56% increase compared to that of the WT strain (Fig. 6A).
FIG 6.
TpdA modulates the expression of the polar flagellar gene flaA. Shown are scatterplots of relative light units (RLU) from at least 5 biological replicates from each strain of interest harboring the transcriptional fusion pBBR-PflaA-lux, grown to the exponential phase (A) or the stationary phase (B) in the presence of 0.1 mM IPTG. RLU are arbitrary light units per ml divided by the optical density at 600 nm. The RLU value is directly proportional to the activity of the PflaA promoter. The horizontal line represents the mean RLU value. Overexpression of TpdA and its variants was achieved using expression constructs that contain an IPTG-inducible promoter. The empty plasmid pMMB67EH-Gm (pMMB) was used as a negative control. Means were compared to the WT using Brown-Forsythe and Welch ANOVA tests followed by Dunnett’s T3 multiple-comparison test. Adjusted P values of ≤0.05 were deemed statistically significant. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001.
In all of the strains where we observed increased expression of flaA, we also observed decreased accumulation of c-di-GMP (Fig. 1). A similar pattern of flaA expression was observed in cells grown to the stationary phase, but under these conditions, the expression of flaA was higher than that in exponentially grown cells (Fig. 6B). These results strongly suggest that c-di-GMP acts downstream of TpdA in regulating flaA expression. The fact that the overproduction of TpdAAAA promotes the expression of flaA in the WT genetic background suggests that induction of endogenous tpdA expression is sufficient not only to alter c-di-GMP levels but perhaps also polar flagellar gene expression. Although we observed an increase in the expression of flaA in the WT pTpdAAAA strain, we did not notice changes in motility compared to that of the WT strain (Fig. 5). It is possible that the TpdA positive regulatory feedback loop is affected by growth conditions (e.g., soft agar versus shaking cultures), and this is something that will be evaluated in future studies.
Overproduction of the PDE TpdA promotes swarming motility and expression of the swarming gene lafA.
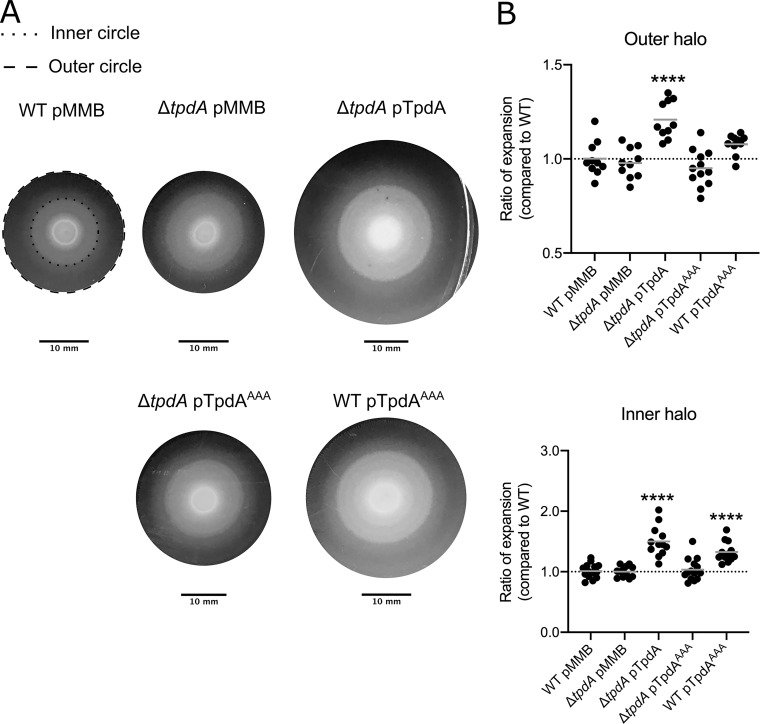
V. parahaemolyticus is capable of swarming over solid surfaces by using lateral flagella that are structurally different from the polar flagellum and are regulated by specific signal transduction circuits (19, 20, 41). Similarly to swimming motility, swarming motility is negatively regulated by c-di-GMP (22, 23, 25). We evaluated if the absence or overproduction of TpdA and the TpdAAAA variant alters swarming motility compared to that of the WT strain. Swarming colonies of each strain formed several distinct concentric halos on the surface of the swarming agar plates (Fig. 7A). We measured the diameters of the outer and inner halos and compared these to the corresponding halos of the WT strain growing in the same swarm plate. Swarming motility of the ΔtpdA mutant strain was not different from that of the WT strain (Fig. 7B). However, the swarming motility of the ΔtpdA pTpdA strain was significantly increased compared to that of the WT strain, with the differences between these strains being more evident for the inner swarming halo (Fig. 7B). The ΔtpdA pTpdAAAA strain swarmed like the WT strain (Fig. 7B). On the other hand, the WT pTpdAAAA strain showed a significant increase in swarming motility compared to that of the WT strain (Fig. 7B). These results indicate that overproduction of TpdA from a synthetic promoter or from its native promoter can promote swarming motility.
FIG 7.
Overproduction of TpdA modulates swarming motility. (A) Representative images of the swarming halos of the strains of interest on swarming plates containing 0.1 mM IPTG. The outer and inner halos of the swarming colony used for measurement are depicted as dashed and dotted lines, respectively. (B) Scatterplots showing ratios of the diameters of the migration halos of the strain of interest and the WT strain grown over the same swarming plate. The migration of at least 10 biological replicates of each strain of interest was analyzed. The horizontal line represents the mean ratio of migration. Overexpression of TpdA and its variants was achieved using expression constructs that contain an IPTG-inducible promoter. The empty plasmid pMMB67EH-Gm (pMMB) was used as a negative control. Means were compared to the WT strain harboring the pMMB plasmid using an ordinary one-way ANOVA test followed by a Dunnett’s multiple-comparison test. Adjusted P values of ≤0.05 were deemed statistically significant. ****, P ≤ 0.0001.
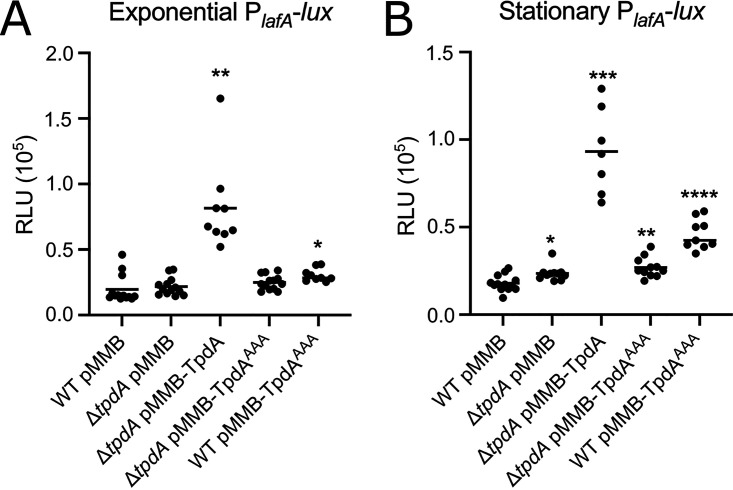
To further evaluate if the absence or overproduction of TpdA affects the expression of a lateral flagellar gene, we generated a transcriptional fusion with the regulatory region of the lateral flagellin A gene lafA (19). In cells grown to the exponential phase, the absence of TpdA did not significantly affect the expression of the PlafA-luxCDABE fusion, while the overexpression of TpdA, but not its inactive variant TpdAAAA, promoted a marked increase in the expression of the fusion in a ΔtpdA genetic background compared to that of the WT strain (Fig. 8A). Similar to what was observed for flaA, the expression of lafA was induced as a consequence of overproducing TpdAAAA in a WT genetic background (Fig. 8A). Based on the results shown above (Fig. 1 and 2), this is likely due to the overexpression of the endogenous copy of tpdA and decreased c-di-GMP accumulation in this genetic background (WT pTpdAAAA). These results suggest that overexpression of tpdA, either from an IPTG-inducible promoter or its own promoter, is capable of affecting the expression of the lateral flagellar gene lafA.
FIG 8.
Overproduction of TpdA induces the expression of the lateral flagellar gene lafA. Shown are scatterplots of relative light units (RLU) from at least 7 biological replicates from each strain of interest harboring the transcriptional fusion pBBR-PlafA-lux, grown to the exponential phase (A) or the stationary phase (B) in the presence of 0.1 mM IPTG. RLU are arbitrary light units per milliliter divided by optical density at 600 nm. The RLU value is directly proportional to the activity of the PlafA promoter. The horizontal line represents the mean RLU value. Overexpression of TpdA and its variants was achieved using expression constructs that contain an IPTG-inducible promoter. The empty plasmid pMMB67EH-Gm (pMMB) was used as negative control. Means were compared to the WT using Brown-Forsythe and Welch ANOVA tests followed by Dunnett’s T3 multiple-comparison test. Adjusted P values of ≤0.05 were deemed statistically significant. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001;
Overproduction of the PDE TpdA negatively controls biofilm formation and cpsA expression.
Biofilm formation is tightly controlled by the levels of the c-di-GMP pool. High and low levels of c-di-GMP promote or impede biofilm formation, respectively (3). We analyzed if the ΔtpdA, ΔtpdA pTpdA, ΔtpdA pTpdAAAA, and the WT pTpdAAAA strains had altered biofilm formation at the liquid-solid interface, compared to the WT strain. The ΔtpdA mutant strain made similar levels of biofilms compared to the WT strain under our experimental conditions, while the ΔtpdA pTpdA strain showed reduced biofilm formation (Fig. 9). The overproduction of the inactive variant TpdAAAA did not alter biofilm formation in the ΔtpdA genetic background but did so in the WT background (Fig. 9). Since we demonstrated that TpdAAAA can promote the expression of the PtpdA promoter, we speculate that the biofilm deficiency of the WT pTpdAAAA strain is mainly due to increased production of TpdA (WT) from its endogenous promoter. These results suggest that overproduction of TpdA, either from an IPTG-inducible promoter or from its own promoter, can severely impair biofilm formation under our experimental conditions.
FIG 9.
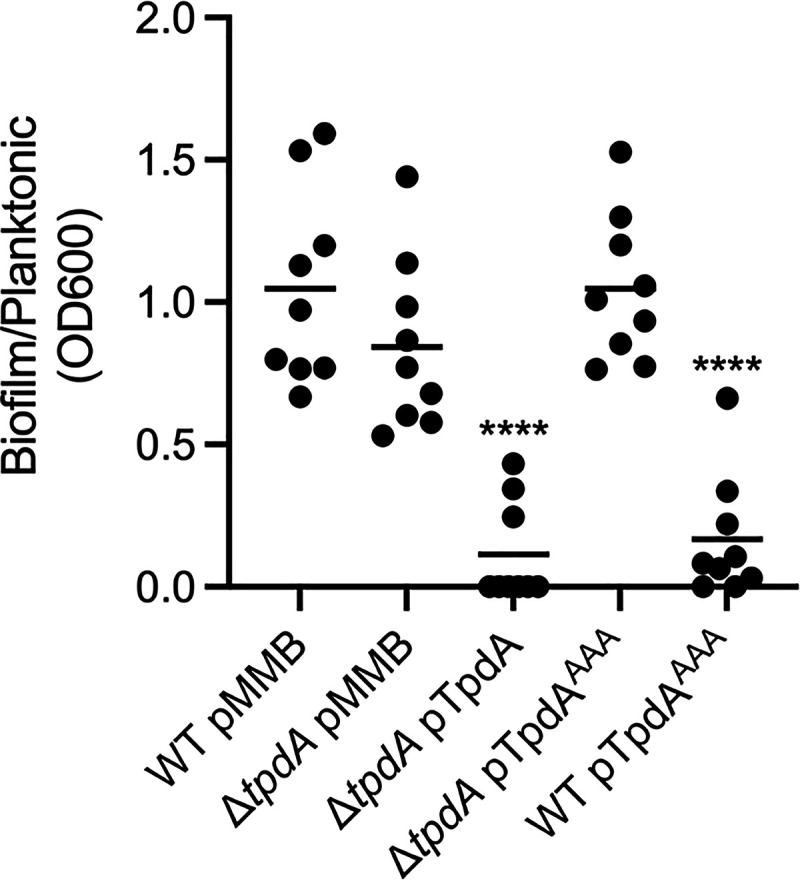
Overproduction of TpdA inhibits biofilm formation at the liquid-solid interface. Shown is a scatterplot showing the OD600 ratio of stained biofilms to planktonic cells grown statically in glass tubes at 30°C. The horizontal line represents the mean. Overexpression of TpdA and its variants was achieved using expression constructs that contain an IPTG-inducible promoter. The empty plasmid pMMB67EH-Gm (pMMB) was used as a negative control. Means were compared to the WT strain harboring the pMMB plasmid using an ordinary one-way ANOVA test followed by a Dunnett’s multiple-comparison test. Adjusted P values of ≤0.05 were deemed statistically significant. ****, P ≤ 0.0001.
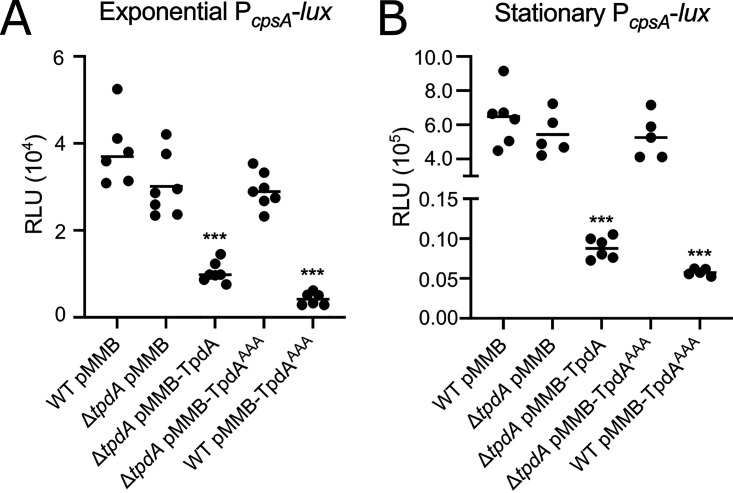
The cpsA to cpsK gene cluster encodes proteins involved in the biosynthesis of the capsular exopolysaccharide CPSA that forms part of the biofilm extracellular matrix (15, 21). To further evaluate if TpdA regulates the transcription of biofilm-related genes, we measured the activity of the promoter of cpsA (PcpsA-luxCDABE) in the same genetic backgrounds used to evaluate biofilm formation. We found a decreased expression of the PcpsA-luxCDABE fusion in the same strains that had reduced levels of biofilm formation, that is, the ones with increased production of TpdA (Fig. 10). This effect was more pronounced during the stationary phase (Fig. 10B). The effect of the overproduction of TpdAAAA on cpsA expression was fully dependent on the presence of a WT copy of tpdA in the genetic background (Fig. 10). Together, these results suggest that the overexpression of tpdA negatively affect the expression of cpsA. The effect of the overexpression of tpdA on biofilm formation and cpsA expression is likely due to a reduction in c-di-GMP levels.
FIG 10.
Overproduction of TpdA negatively influences expression of the biofilm gene cpsA. Shown are scatterplots of relative light units (RLU) from at least 5 biological replicates from each strain of interest harboring the transcriptional fusion pBBR-PcpsA-lux, grown to the exponential phase (A) or the stationary phase (B) in the presence of 0.1 mM IPTG. RLU are arbitrary light units per milliliter divided by optical density at 600 nm (OD600). The RLU value is directly proportional to the activity of the PcpsA promoter. The horizontal line represents the mean RLU value. Overproduction of TpdA and its variants was achieved using expression constructs that contain an IPTG-inducible promoter. The empty plasmid pMMB67EH-Gm (pMMB) was used as a negative control. Means were compared to the WT using Brown-Forsythe and Welch ANOVA tests followed by Dunnett’s T3 multiple-comparison test. Adjusted P values of ≤0.05 were deemed statistically significant. ***, P ≤ 0.001.
DISCUSSION
The content of the V. parahaemolyticus genome suggests that this bacterium has an important signaling potential, especially for forming c-di-GMP signaling modules. The few modules that have been explored to this date have a role in controlling biofilm formation and swarming motility (17, 22, 23, 25, 26). In this report, we unveiled a new c-di-GMP signaling component involved in the regulation of planktonic motility and surface behaviors, the PDE TpdA. This PDE promotes its own expression when c-di-GMP levels are low and also when it is catalytically inactive. Its ability to alter the c-di-GMP pool had an effect on swimming motility, swarming motility, and biofilm formation. We found that overproduction of the catalytically inactive TpdAAAA variant positively regulates swarming motility and negatively regulates biofilm formation, but only when a WT endogenous copy of tpdA is present in the genetic background. Since we also observed that overproduction of TpdAAAA strongly induces the expression of the endogenous PtpdA promoter, we propose that upregulation of tpdA could be a trigger mechanism to turn off biofilm matrix production and/or stimulate swarming motility.
TpdA has some of the characteristics of a group of c-di-GMP effectors named trigger phosphodiesterases. Trigger phosphodiesterases have a regulatory or signaling function that anticorrelates with their ability to degrade c-di-GMP (28). The first trigger phosphodiesterase described in the literature was PdeR (formerly YciR) from E. coli (42, 43). The primary function of PdeR is the negative regulation exerted on the expression of the biofilm regulator csgD, and its secondary activity is as a PDE. PdeR regulates the expression of csgD by interfering with the interaction between the DGC DgcM and the transcriptional regulator MlrA, which is required to activate the transcription of csgD (28, 43–45). Conditions that favor biofilm formation favor the production of a local c-di-GMP pool generated by the DGC DgcE (45). PdeR detects this local c-di-GMP pool and, in response, releases both DgcM and MlrA, enabling activation of csgD and biofilm matrix production (28, 43). This sophisticated regulatory network enables heterogeneity of biofilm matrix production among cells residing inside colony biofilms, a property that has been suggested to confer structural advantages to these three-dimensional edifices in E. coli (44). E. coli has an additional trigger phosphodiesterase named PdeL (formerly YahA) (46). In contrast to PdeR, PdeL has a DNA binding domain of the helix-turn-helix class in addition to the active EAL domain (46–48). PdeL positively regulates its own expression directly, under conditions where the concentration of c-di-GMP is very low (28, 47). The physiological implications of the potential positive feedback loop involving PdeL are still not clear; however, it has been recently reported that this trigger PDE can regulate the expression of flagellar genes (49).
The bioinformatic analysis of the domain architecture of TpdA revealed that this protein may have a DNA-binding domain at its N terminus. Although no strong conclusions can be drawn from this prediction, the genetic evidence presented in this work supports a model where TpdA is able to activate its own transcription under conditions where c-di-GMP is low or TpdA′s enzymatic activity is inhibited. This mode of action resembles that of PdeL from E. coli (28, 47, 49). Our results showed that the TpdAAAA variant cannot alter the expression of flaA, lafA, or cpsA in a ΔtpdA genetic background, which could suggest that TpdA is not a direct regulator of these genes. However, we cannot rule out the possibility that TpdA regulates other genes associated with c-di-GMP-controlled phenotypes. The analysis of the amino acid sequence of TpdA also revealed putative transmembrane regions that may anchor this protein to the cell membrane. Some of these transmembrane regions overlapped with predicted domains (GGDEF-like and EAL), and hence it is unclear if they are functional. However, it would not be unprecedented for a membrane-anchored protein to act as a transcriptional regulator. One of the most well-known regulators of virulence in V. cholerae, ToxR, is a membrane-bound protein (50, 51). ToxR has a winged helix that is located at the N terminus (50, 51). The DNA-binding domain that was predicted at the N terminus of TpdA was modeled based on the structure of the winged helix of the response regulator DrrB, which belongs to the OmpR/PhoB family (35). These are intriguing observations that require further analysis in order to clarify the regulatory mechanism involved in the autoactivation of tpdA.
In previous reports, it was shown that the expression of tpdA was downregulated in surface-grown cells compared to that in planktonic cells (27). Based on our observations, we speculate that the differences in expression of tpdA in these two conditions could originate from changes in the abundance of c-di-GMP sensed by TpdA. However, we cannot discard the possibility of the existence of other regulators that control the expression of tpdA in response to surface adaptation or other unknown signals. For instance, we observed activity of the PtpdA promoter in the absence of TpdA in V. parahaemolyticus, but almost none in V. cholerae or E. coli. These differences in TpdA dependence could be due to the presence of additional regulators in V. parahaemolyticus that are absent in the heterologous hosts analyzed.
The absence of TpdA resulted in reduced swimming motility. We still do not know if TpdA is a specific regulator of swimming motility or if the motility phenotype of the ΔtpdA mutant strain is the consequence of a small increase in the levels of the global c-di-GMP pool. Multiple c-di-GMP receptors have been shown to influence motility in V. cholerae, such as the Plz proteins and the MshE ATPase from the type IV pili MSHA (14, 30, 52–55). These are also conserved in V. parahaemolyticus and could be the effector responsible for the swimming phenotype in the absence of TpdA (https://www.ncbi.nlm.nih.gov/Complete_Genomes/c-di-GMP.html) (3, 6, 37). It is interesting that the absence of TpdA affected swimming motility but not swarming motility, given that both types of motility are negatively regulated by c-di-GMP. However, these behaviors not only are controlled by different types of flagella but also happen on different types of surfaces. Future studies should evaluate if TpdA is differently involved in controlling the c-di-GMP pool under conditions that are relevant for different types of planktonic and social behaviors.
The abundance of TpdA affected the expression of flaA, lafA, and cpsA, likely due to its ability to degrade c-di-GMP. The expression of polar flagellar genes from V. cholerae is regulated negatively by c-di-GMP through the effector FlrA (56). The orthologue of FlrA in V. parahaemolyticus is FlaK; these two proteins are 78.9% identical, and hence FlaK might also be capable of detecting c-di-GMP and this molecule could interfere with its ability to activate flagellar gene expression. In fact, the expression of lateral flagellar genes is negatively regulated by c-di-GMP in V. parahaemolyticus (21), but the identity of the effector is unknown. LafK, the master regulator of lateral flagellar gene expression, is a σ54-dependent transcriptional regulator that is phylogenetically related to FlrA and FlrC (19, 27). Both FlrA and FlrC have been shown to be c-di-GMP effectors (56, 57), so it would be interesting to evaluate if the activity of their relative LafK is also influenced by c-di-GMP levels. The expression of cpsA, on the other hand, is known to be regulated by the c-di-GMP effectors CpsQ and ScrO (17, 21). It is not known if these effectors interact with specialized c-di-GMP signaling modules or if they respond mainly to changes in the global c-di-GMP pool. The small increase in c-di-GMP abundance in the absence of TpdA was not enough to promote cpsA expression and biofilm formation. We do not know if similar increases in c-di-GMP generated by other c-di-GMP signaling modules can exert a response that promotes biofilm formation, in which case an argument for specificity could be made.
Our results show that the upregulation of tpdA can be triggered in conditions where c-di-GMP levels are low or when TpdA cannot degrade c-di-GMP. It is yet unclear if a particular set of DGCs and PDEs control the abundance of c-di-GMP during the process of biofilm formation or surface colonization and what type of signals may intervene in the dynamic control of this second messenger in V. parahaemolyticus. The PDEs ScrC and ScrG control biofilm formation and swarming motility; however, although ScrC appears to be at the top of the hierarchy of regulators of swarming, the contribution of either ScrC or ScrG to the global c-di-GMP pool still needs further analysis (22, 23, 25). The absence of TpdA in the V. parahaemolyticus strain RIMD2210633 results in a noticeable increase in c-di-GMP accumulation, but we do not know how this increase compares to conditions where other PDEs, such as ScrC or ScrG, are absent. A systematic analysis of the contribution of each of the c-di-GMP metabolizing enzymes from V. parahaemolyticus would be key in order to have a better understanding of c-di-GMP signaling in this human pathogen.
TpdA has the potential to be a key player in controlling the kinetics of biofilm development and swarming motility; however, several questions remain regarding its regulation and activity during the development of these surface behaviors. Addressing these important unknown aspects of TpdA function and other c-di-GMP-related enzymes and receptors will further our understanding of the computing network that informs the cell whether to move or not to move and whether to engage in social interactions.
MATERIALS AND METHODS
Strains and growth conditions.
Strains and plasmids used in this work are listed and described in Table 1. All strains were grown in lysogeny broth (LB) (1% tryptone, 0.5% yeast extract, and 1% NaCl [pH 7.5]). E. coli was grown at 37°C, while V. parahaemolyticus and V. cholerae were grown at 30°C under shaking conditions at 200 rpm. Solid medium was prepared using LB and 1.5% agar. The following antibiotics were added at the following concentrations to select for specific genotypes: streptomycin at 200 μg/ml, gentamicin at 15 μg/ml, chloramphenicol at 20 μg/ml for E. coli and 5 μg/ml for vibrios, and rifampin at 100 μg/ml for V. cholerae.
TABLE 1.
List of strains and plasmids used in this study
| Strain or plasmid | Relevant genotype | Source and/or reference |
|---|---|---|
| E. coli strains | ||
| SY327 (λpir) | Δ(lac pro) argE(Am) rif nalA recA56 λpir | |
| BW25113 | F- Δ(araB-araD)567 Δ(rhaD-rhaB)568 ΔlacZ4787(::rrnB-3) hsdR514 rph-1 | Keio Collection |
| SM10 (λpir) | thi thr leu tonA lacY supE recA RP4-2-Tc::Mu λpir Kmr | 59 |
| V. cholerae strains | ||
| DZ_Vp_42 | RIMD2210633 Str | 60, 61 |
| DZ_Vp_113 | DZ_Vp_42 ΔtpdA Str | This work |
| Plasmids | ||
| pDZ_63 (pRE118) | oriT oriV sacB aphA Kmr | 62 |
| pDZ_118 | pRE118-tpdAdel Kmr | This work |
| pRK2073 | Derivative of the mobilization helper plasmid pRK2013; Spcr | Lourdes Girard (63) |
| pFY_1554 (pMMB67EH-Gm) | Expression vector harboring lacI and containing a Ptac promoter; Gmr | Samuel Miller and Fitnat Yildiz |
| pDZ_93 | VP1881 cloned in pFY_1554 | This work |
| pDZ_156 | VP1881-AAA cloned in pFY_1554 | This work |
| pDZ_117 | scrG cloned in pFY_1554 | This work |
| pFY_4535 | c-di-GMP biosensor containing the hok-sok region from pXB300; Gmr | 29 |
| pDZ_119 | c-di-GMP biosensor cloned in a pBBRMCS backbone; Cmr | This work |
| pFY_691 (pBBRlux) | luxCDABE-based promoter fusion vector; Cmr | 64 |
| pDZ_92 | pBBRlux-PVP1881 Cmr | This work |
| pDZ_91 | pBBRlux-PflaA Cmr | This work |
| pDZ_90 | pBBRlux-PlafA Cmr | This work |
| pDZ_46 | pBBRlux-PcpsA Cmr | This work |
Generation of plasmids and conjugation.
Standard procedures for DNA amplification and DNA assembly were used. The high-fidelity DNA polymerase Q5 (New England BioLabs) was used in all of the PCRs. Primers used for PCR amplification are listed in Table 2.
TABLE 2.
List of primers used in this study
| Primer | Sequence (5′–3′) | Construct |
|---|---|---|
| DZ_64 | CGACGGATCCCAAGCTTCTTTAAAAGCGATCAACTCAAC | pDZ_118 |
| DZ_65 | CTGCGGTTTCAACGCAGATTTGGTTTCC | pDZ_118 |
| DZ_66 | AATCTGCGTTGAAACCGCAGATAGTGCTC | pDZ_118 |
| DZ_67 | CATGAATTCCCGGGAGAGCTATAATTAAGGCCCAACTCATG | pDZ_118 |
| DZ_51 | CGAGCTCATGATTAGGTTTGAACTTGG | pDZ_93 |
| DZ_52 | CGGGATCCCTAGAAATGCAGAGGATAG | pDZ_93 |
| DZ_107 | CAGGAAACAGAATTCGAGCTATGATTAGGTTTGAACTTGG | pDZ_156 |
| DZ_108 | CGAGAGGCGGCGGCGTAGTGAATATTTTGCTGC | pDZ_156 |
| DZ_109 | TATTCACTACGCCGCCGCCTCTCGATTCAAAGTGAAG | pDZ_156 |
| DZ_110 | CCTGCAGGTCGACTCTAGAGCTAGAAATGCAGAGGATAG | pDZ_156 |
| DZ_53 | CCTGCAGGTCGACTCTAGAGCTATGAATTCAATGAATATCGG | pDZ_117 |
| DZ_54 | CAGGAAACAGAATTCGAGCTTTGTATCTTAATAATGCGCATG | pDZ_117 |
| DZ_225 | GCAACTAGAGGCATCAAATAAAAC | pDZ_119 |
| DZ_226 | ATTTGCCATCCATTTTGCG | pDZ_119 |
| DZ_227 | CGGTGGCGGCCGCTCTAGAACTAGTCCCAAGCTTGGCAAAACAAC | pDZ_119 |
| DZ_228 | CCATTTTGCGGCCGCAACTAGAGGATCCAGAGTTTGTAGAAACGCAAAAAG | pDZ_119 |
| DZ_49 | CGAGCTCCGAGTGAATACTCGGGCTTT | pDZ_92 |
| DZ_50 | GACTAGTACGCAGATTTGGTTTCCAAG | pDZ_92 |
| DZ_47 | CGAGCTCTTTTAAACCTAAGACGAGAACCTCA | pDZ_91 |
| DZ_48 | GACTAGTCGTTAATCGCCATAATTGATCTC | pDZ_91 |
| DZ_45 | CGAGCTCCCAACCCTCATGGCTAAACT | pDZ_90 |
| DZ_46 | GACTAGTTTGATAAAGCCATCTTAGTCTCCTT | pDZ_90 |
| DZ_7 | CTATAGGGCGAATTGGAGCTCCCATCAAGAAATTAAACCTTAG | pDZ_46 |
| DZ_8 | ATTTGCGGCCGCAACTAGAGGATCCCATAAAGGTGACAATGACAG | pDZ_46 |
The structural gene VP1881 was PCR amplified and cloned using the CloneJet PCR cloning kit (Thermo Fisher Scientific) following the manufacturer’s instructions. To subclone into pMMB67EH-Gm (pMMB67EH-Gentamicin), the destination plasmid and pJET-VP1881 were independently digested with the restriction enzymes SacI-HF and BamHI-HF (New England BioLabs). The digested fragments were purified using the DNA Clean & Concentrator-5 kit and the Zymoclean gel DNA recovery kit (Zymo Research), respectively. The DNA fragments were ligated using a T4 DNA ligase (New England BioLabs). The point mutations in VP1881 to generate the VP1881AAA variant were introduced by splicing by overlap extension PCR (SOEing PCR). Two PCR fragments encompassing the whole sequence of VP1881 were fused by SOEing PCR; the overlapping sequence that enabled assembly contained the point mutations that result in an AAA motif. The PCR product and the destination plasmid (pMMB67EH-Gm) were digested with the restriction enzymes EcoRI-HF and XbaI (New England BioLabs) and ligated with T4 DNA ligase. The gene scrG (VP1377) was PCR amplified using primers that enable an isothermal assembly into pMMB67EH-Gm linearized with SacI and BamHI. The assembly was achieved using the NEBuilder HiFi DNA assembly master mix (New England BioLabs), using a miniaturized reaction mix that included 50 ng of each DNA fragment and 2 μl of the master mix in a 6-μl reaction mixture. The assembly reaction mixture was incubated at 50°C for 1 h before being used for transformation.
The upstream and downstream homology arms used for the deletion of VP1881 were PCR amplified and assembled into the suicide plasmid pRE118, linearized with SacI-HF and XbaI. DNA assembly was achieved using the NEBuilder HiFi DNA assembly master mix using a similar protocol as described above.
The c-di-GMP biosensor is based on one reported and validated previously (29). We amplified the biosensor cassette from pFY_4535 and cloned it into the backbone of pBBRlux lacking the luxCDABE locus. The destination plasmid was generated by inverse PCR, and the c-di-GMP biosensor cassette was cloned by isothermal assembly. We also introduced the PtetRA cassette from plasmid pXB300 in this plasmid; this promoter was not used for the experiments reported in this study.
The fragments used to generate the transcriptional fusions (except for PcpsA) were initially cloned using the CloneJet PCR cloning kit and then subcloned into the plasmid pBBRlux, linearized with SacI and SpeI. The fragment corresponding to PcpsA was cloned into the plasmid pBBRlux linearized with SacI and BamHI using the NEBuilder HiFi DNA assembly master mix.
The sequence fidelity of all DNA assemblies was confirmed by DNA sequencing at the DNA Sequencing and Synthesis Unit of the Instituto de Biotecnología–UNAM.
Plasmids were moved into the desired genetic background through biparental or triparental mating using the mobilizing strain SM10λpir or the helper plasmid pRK2073.
Generation of a mutant strain with the deletion of VP1881.
The plasmid pDZ_118 was used to delete VP1881 by double recombination. This is a suicide plasmid that has an R6K origin of replication that requires the presence of the π protein to be functional. It also contains a kanamycin resistance cassette that enables selection of single recombinants and a sacB gene that enables the isolation of double recombinants by counterselection. The recombination substrate regions corresponded to fragments of approximately 500 bp located upstream and downstream of the coding region of VP1881. The deletion construct also contains an in-frame truncation of the structural gene with a length of 117 bp. The plasmid pDZ_118 was transferred from the E. coli strain SY327λpir to the recipient strain V. parahaemolyticus RIMD2210633 through triparental mating. Single recombinants were selected on LB agar plates containing streptomycin (200 μg/ml) and kanamycin (30 μg/ml). Single colonies were grown with agitation (200 rpm) in MLB broth (LB broth with 3% NaCl) at 30°C overnight. The cultures were serially diluted and spread over MLB-sucrose solid medium (MLB, 10% sucrose, and 1.5% agar) and incubated at room temperature overnight. Sucrose-resistant colonies were replica plated on LB agar plates, with or without 30 μg/ml kanamycin. Double recombinants that had lost the VP1881 gene were identified through colony PCR.
Analysis of growth curves.
Overnight cultures were prepared in 3 ml of LB in the presence of 15 μg/ml gentamicin and incubated at 30°C with agitation (200 rpm). The overnight cultures were diluted 1:200 in fresh LB medium (3 ml) containing 15 μg/ml gentamicin and 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) and incubated at 30°C with agitation (200 rpm). The optical density of the culture at 600 nm (OD600), was measured using the Epoch 2 microplate spectrophotometer at different time points using 100-μl aliquots of culture transferred to clear 96-well plates. Three independent biological replicates were analyzed, and the data were plotted using GraphPad Prism 9.
Determination of relative abundance of c-di-GMP using a c-di-GMP biosensor.
The levels of c-di-GMP were determined using a c-di-GMP biosensor as previously reported (29), with some modifications. Overnight cultures of the strains of interest harboring the c-di-GMP biosensor and a pMMB67EH-Gm-derived plasmid were diluted 1:200 in 20 ml of LB broth containing 15 μg/ml gentamicin, 5 μg/ml chloramphenicol, and 0.1 mM IPTG, and incubated at 30°C with agitation (200 rpm). Cultures were grown to either the exponential (0.4 to 0.6 OD600) or the stationary phase (overnight, approximately 3.0 OD600). Samples of 1 ml, obtained from exponentially grown cultures were centrifuged at 10,000 × g for 1 min, and the cell pellet was resuspended in 100 μl of triple-distilled water. Samples of 100 μl, obtained from cultures grown to the stationary phase, were centrifuged at 10,000 × g for 1 min, and the cell pellet was resuspended in 100 μl of triple-distilled water. The resuspended samples were transferred to black 96-well plates, and the fluorescence was measured using the plate reader Synergy H1 (BioTek). The excitation and emission settings for the detection of Amcyan and TurboRFP were 420/520 nm and 550/580 nm, respectively. The relative fluorescence intensity (RFI) was calculated by dividing the arbitrary fluorescent intensity units of TurboRFP by those of Amcyan. Experiments were repeated at least three times.
Luminescence assay.
Overnight cultures of the strains of interest harboring a transcriptional fusion in the pBBRlux plasmid and a pMMB67EH-Gm-derived plasmid were diluted 1:200 in 20 ml of LB broth containing 15 μg/ml gentamicin and 5 μg/ml chloramphenicol and incubated at 30°C with agitation (200 rpm). Cultures were grown to either the exponential (0.4 to 0.6 OD600) or the stationary phase (approximately 1.5 to 2.0 OD600). Samples of 200 μl, obtained from the grown cultures, were transferred to white 96-well plates with clear bottoms, and the optical density (OD600) and the luminescence of the samples were measured using a Synergy H1 plate reader (BioTek). The integration time for luminescence detection was 5 s. The OD600 and arbitrary light units of a clean LB sample were used as a blank. Samples with arbitrary light units below the blank were considered nondetectable and were treated as zero values. The relative light units (RLU) were calculated as arbitrary light units per ml divided by the OD600. Experiments were repeated at least three times.
Bioinformatic analysis.
The bidirectional best hit analysis using the primary sequence of VP1881 from V. parahaemolyticus RIMD 2210633 and proteins from V. cholerae O1 El Tor N16961 and E. coli K-12 MG1655 was performed using blastp (https://blast.ncbi.nlm.nih.gov/Blast.cgi).
The domain architecture of the full sequence of VP1881 was analyzed using InterProScan (https://www.ebi.ac.uk/interpro/search/sequence/). Analysis of the secondary and tertiary structure predictions of the N terminus, the middle region, and the C terminus of VP1881 was carried out using Phyre2 (http://www.sbg.bio.ic.ac.uk/∼phyre2/html/page.cgi?id=index) (34).
Swimming motility assays.
Swimming motility plates were prepared with LB broth and 0.3% agar. Gentamicin (15 μg/ml) was added to the medium to select for the presence of pMMB67EH-Gm-derived plasmids, and 0.1 mM IPTG was added to promote induction of the Ptac promoter. Soft agar (30 ml) was poured into individual plastic petri dishes (100 mm × 15 mm) and left to dry from 3 to 4 h at room temperature. Three single colonies of each strain of interest were individually inoculated into the center of separate swimming motility plates using sterile wooden toothpicks. The plates were incubated at 25°C for 17 h, and the migration diameter of the swimming colonies was measured manually. At least three independent experiments were done with each of the strains of interest.
Swarming motility assays.
The induction of swarmer cell differentiation and the motility assays were performed as described before with some modifications (58). Swarming plates were prepared with 2.5% heart infusion broth (Bacto) and 1.5% agar. Calcium chloride (CaCl2), 2,2′-bipyridyl, and IPTG were added to a final concentration of 4 mM, 0.05 mM, and 0.1 mM, respectively. Gentamicin was added to a final concentration of 2.4 μg/ml, since higher concentrations inhibited swarming under our experimental conditions. Swarming agar (15 ml) was poured into 100-mm × 15-mm plastic petri dishes, and all bubbles were removed carefully with heat. The swarm plates were left to dry for 3 h at room temperature with the lid on and then for an additional 45 to 50 min without the lid at 37°C.
Overnight cultures of the strain of interest were diluted 1:100 in 1 ml of LB broth containing 15 μg/ml gentamicin and 0.1 mM IPTG and grown with agitation at 30°C until they reached an OD600 between 0.8 and 1.0. Each plate was inoculated with 2 μl of the WT pMMB strain and 2 μl of a strain of interest and sealed carefully with parafilm. Biologically independent duplicates from each strain were assayed in at least three independent experiments. The inoculated swarming plates were incubated at 24°C for approximately 20 h. At the end of the experiment, the swarming halos were photographed and the digital images were used to determine the diameter of the migration halos using Fiji software (NIH). The photographs were converted to 8-bit images and two circles, corresponding to an inner and an outer migrating halo that were consistently identified in each experiment (Fig. 6A), were traced using the oval selection tool and saved as regions of interest (ROIs). The Feret’s diameters of each experimental strain were normalized by dividing by the values of the control strain from the same plate.
Liquid-solid interface biofilm assays.
Overnight cultures of the strains of interest were diluted 1:100 in 1 ml of LB broth with 15 μg/ml gentamicin and 0.1 mM IPTG and incubated in borosilicate glass tubes with a volume capacity of 10 ml under static conditions at 30°C for 6 h. Biological duplicates of each strain were analyzed in three independent experiments. Uninoculated LB broth with the same concentrations of gentamicin and IPTG was incubated under the same conditions and used as a blank. Samples (200 μl) of the planktonic static cultures were transfer to a clear 96-well plate, and the OD600 was measured using an Epoch 2 microplate reader. The remaining culture was carefully discarded by decanting in a 10% bleach solution and washed twice with tap water. The tubes containing the biofilms were left to dry upside-down overnight at room temperature. The biofilm formed at the liquid-solid interface was stained with a 0.1% solution of crystal violet for approximately 1 min. The residual stain and three subsequent washes with tap water were carefully discarded in a 10% bleach solution. The stained biofilms were left to dry for 10 min. The stain from the biofilm was solubilized in 1.2 ml of absolute ethanol for 30 min under static conditions. Afterwards, 200-μl samples were transferred to a clear 96-well plate, and the OD600 was measured using an Epoch 2 microplate reader. The OD600 of the crystal violet stain from the biofilms was divided by the OD600 of the corresponding planktonic culture to account for differences in growth under static conditions. Samples where the crystal violet staining showed OD600 values below those of the blank were considered nondetectable and were treated as zero values.
ACKNOWLEDGMENTS
This work was financially supported by DGAPA-PAPIIT (UNAM) under project IA200519, awarded to David Zamorano-Sánchez.
We are thankful to Eugenio Lopez-Bustos, Paul Gaytan, Santiago Becerra, and Jorge A. Yañez, from Unidad de Síntesis y Secuenciación, Instituto de Biotecnología-UNAM, for their technical assistance in oligonucleotide synthesis and DNA sequencing. We are thankful to Jose Arcadio Farías-Rico for his guidance and discussion on the structure prediction analysis conducted in this work.
Contributor Information
David Zamorano-Sánchez, Email: zamorano@ccg.unam.mx.
George O’Toole, Geisel School of Medicine at Dartmouth.
REFERENCES
- 1.Flemming H-C, Wingender J, Szewzyk U, Steinberg P, Rice SA, Kjelleberg S. 2016. Biofilms: an emergent form of bacterial life. Nat Rev Microbiol 14:563–575. 10.1038/nrmicro.2016.94. [DOI] [PubMed] [Google Scholar]
- 2.Flemming HC, Wuertz S. 2019. Bacteria and archaea on Earth and their abundance in biofilms. Nat Rev Microbiol 17:247–260. 10.1038/s41579-019-0158-9. [DOI] [PubMed] [Google Scholar]
- 3.Römling U, Galperin MY, Gomelsky M. 2013. Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol Mol Biol Rev 77:1–52. 10.1128/MMBR.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jenal U, Reinders A, Lori C. 2017. Cyclic di-GMP: second messenger extraordinaire. Nat Rev Microbiol 15:271–284. 10.1038/nrmicro.2016.190. [DOI] [PubMed] [Google Scholar]
- 5.Hengge R. 2009. Principles of c-di-GMP signalling in bacteria. Nat Rev Microbiol 7:263–273. 10.1038/nrmicro2109. [DOI] [PubMed] [Google Scholar]
- 6.Chou S, Galperin MY. 2016. Diversity of cyclic Di-GMP-binding proteins and mechanisms. J Bacteriol 198:32–46. 10.1128/JB.00333-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Trampari E, Stevenson CEM, Little RH, Wilhelm T, Lawson DM, Malone JG. 2015. Bacterial rotary export ATPases are allosterically regulated by the nucleotide second messenger cyclic-di-GMP. J Biol Chem 290:24470–24483. 10.1074/jbc.M115.661439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu Z, Zhang H, Zhang X, Jiang H, Liu C, Wu F, Qian L, Hao B, Czajkowsky DM, Guo S, Xu Z, Bi L, Wang S, Li H, Tan M, Yan W, Feng L, Hou J, Tao S. 2019. Interplay between the bacterial protein deacetylase CobB and the second messenger c‐di‐ GMP. EMBO J 38. 10.15252/embj.2018100948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fang X, Gomelsky M. 2010. A post-translational, c-di-GMP-dependent mechanism regulating flagellar motility. Mol Microbiol 76:1295–1305. 10.1111/j.1365-2958.2010.07179.x. [DOI] [PubMed] [Google Scholar]
- 10.Boehm A, Kaiser M, Li H, Spangler C, Kasper CA, Ackermann M, Kaever V, Sourjik V, Roth V, Jenal U. 2010. Second messenger-mediated adjustment of bacterial swimming velocity. Cell 141:107–116. 10.1016/j.cell.2010.01.018. [DOI] [PubMed] [Google Scholar]
- 11.Subramanian S, Gao X, Dann CE, Kearns DB. 2017. MotI (DgrA) acts as a molecular clutch on the flagellar stator protein MotA in Bacillus subtilis. Proc Natl Acad Sci U S A 114:13537–13542. 10.1073/pnas.1716231114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paul K, Nieto V, Carlquist WC, Blair DF, Harshey RM. 2010. The c-di-GMP binding protein YcgR controls flagellar motor direction and speed to affect chemotaxis by a “backstop brake” mechanism. Mol Cell 38:128–139. 10.1016/j.molcel.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roelofs KG, Jones CJ, Helman SR, Shang X, Orr MW, Goodson JR, Galperin MY, Yildiz FH, Lee VT. 2015. Systematic identification of cyclic-di-GMP binding proteins in Vibrio cholerae reveals a novel class of cyclic-di-GMP-binding ATPases associated with type II secretion systems. PLoS Pathog 11:e1005232. 10.1371/journal.ppat.1005232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jones CJ, Utada A, Davis KR, Thongsomboon W, Zamorano Sanchez D, Banakar V, Cegelski L, Wong GCL, Yildiz FH. 2015. C-di-GMP regulates motile to sessile transition by modulating MshA pili biogenesis and near-surface motility behavior in Vibrio cholerae. PLoS Pathog 11:e1005068-27. 10.1371/journal.ppat.1005068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Güvener ZT, McCarter LL. 2003. Multiple regulators control capsular polysaccharide production in Vibrio parahaemolyticus. J Bacteriol 185:5431–5441. 10.1128/jb.185.18.5431-5441.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yildiz FH, Visick KL. 2009. Vibrio biofilms: so much the same yet so different. Trends Microbiol 17:109–118. 10.1016/j.tim.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kimbrough JH, Thomas Cribbs J, McCarter LL. 2020. Homologous c-di-GMP-binding Scr transcription factors orchestrate biofilm development in Vibrio parahaemolyticus. J Bacteriol 202. 10.1128/JB.00723-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim YK, McCarter LL. 2000. Analysis of the polar flagellar gene system of Vibrio parahaemolyticus. J Bacteriol 182:3693–3704. 10.1128/jb.182.13.3693-3704.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stewart BJ, McCarter LL. 2003. Lateral flagellar gene system of Vibrio parahaemolyticus. J Bacteriol 185:4508–4518. 10.1128/jb.185.15.4508-4518.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Merino S, Shaw JG, Tomás JM. 2006. Bacterial lateral flagella: an inducible flagella system. FEMS Microbiol Lett 263:127–135. 10.1111/j.1574-6968.2006.00403.x. [DOI] [PubMed] [Google Scholar]
- 21.Ferreira RBR, Chodur DM, Antunes LCM, Trimble MJ, McCarter LL. 2012. Output targets and transcriptional regulation by a cyclic dimeric GMP-responsive circuit in the Vibrio parahaemolyticus scr network. J Bacteriol 194:914–924. 10.1128/JB.05807-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ferreira RBR, Antunes LCM, Greenberg EP, McCarter LL. 2008. Vibrio parahaemolyticus ScrC modulates cyclic dimeric GMP regulation of gene expression relevant to growth on surfaces. J Bacteriol 190:851–860. 10.1128/JB.01462-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trimble MJ, McCarter LL. 2011. Bis-(3′-5′)-cyclic dimeric GMP-linked quorum sensing controls swarming in Vibrio parahaemolyticus. Proc Natl Acad Sci U S A 108:18079–18084. 10.1073/pnas.1113790108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Enos-Berlage JL, Guvener ZT, Keenan CE, McCarter LL. 2005. Genetic determinants of biofilm development of opaque and translucent Vibrio parahaemolyticus. Mol Microbiol 55:1160–1182. 10.1111/j.1365-2958.2004.04453.x. [DOI] [PubMed] [Google Scholar]
- 25.Kim Y-K, McCarter LL. 2007. ScrG, a GGDEF-EAL protein, participates in regulating swarming and sticking in Vibrio parahaemolyticus. J Bacteriol 189:4094–4107. 10.1128/JB.01510-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kimbrough JH, McCarter LL. 2020. Identification of three new GGDEF and EAL domain-containing proteins participating in the Scr surface colonization regulatory network in Vibrio parahaemolyticus. J Bacteriol 203:e00409-20. 10.1128/JB.00409-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gode-Potratz CJ, Kustusch RJ, Breheny PJ, Weiss DS, McCarter LL. 2011. Surface sensing in Vibrio parahaemolyticus triggers a programme of gene expression that promotes colonization and virulence. Mol Microbiol 79:240–263. 10.1111/j.1365-2958.2010.07445.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hengge R. 2016. Trigger phosphodiesterases as a novel class of c-di-GMP effector proteins. Philos Trans R Soc Lond B Biol Sci 371:20150498. 10.1098/rstb.2015.0498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zamorano-Sánchez D, Xian W, Lee CK, Salinas M, Thongsomboon W, Cegelski L, Wong GCL, Yildiz FH. 2019. Functional specialization in Vibrio cholerae diguanylate cyclases: distinct modes of motility suppression and c-di-GMP production. mBio 10:e00670-19. 10.1128/mBio.00670-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu X, Beyhan S, Lim B, Linington RG, Yildiz FH. 2010. Identification and characterization of a phosphodiesterase that inversely regulates motility and biofilm formation in Vibrio cholerae. J Bacteriol 192:4541–4552. 10.1128/JB.00209-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ahumada-Manuel CL, Martínez-Ortíz IC, Hsueh BY, Guzmán J, Waters CM, Zamorano-Sánchez D, Espín G, Núñez C. 2020. Increased c-di-GMP levels lead to the production of alginates of high molecular mass in Azotobacter vinelandii. J Bacteriol 202:e00134-20. 10.1128/JB.00134-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Petersen E, Mills E, Miller SI. 2019. Cyclic-di-GMP regulation promotes survival of a slow-replicating subpopulation of intracellular Salmonella Typhimurium. Proc Natl Acad Sci U S A 116:6335–6340. 10.1073/pnas.1901051116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xin L, Zeng Y, Sheng S, Chea RA, Liu Q, Li HY, Yang L, Xu L, Chiam KH, Liang ZX. 2019. Regulation of flagellar motor switching by c-di-GMP phosphodiesterases in Pseudomonas aeruginosa. J Biol Chem 294:13789–13799. 10.1074/jbc.RA119.009009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE. 2015. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10:845–858. 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robinson VL, Wu T, Stock AM. 2003. Structural analysis of the domain interface in DrrB, a response regulator of the OmpR/PhoB subfamily. J Bacteriol 185:4186–4194. 10.1128/jb.185.14.4186-4194.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu C, Liew CW, Wong YH, Tan ST, Poh WH, Manimekalai MSS, Rajan S, Xin L, Liang ZX, Grüber G, Rice SA, Lescar J. 2017. Insights into biofilm dispersal regulation from the crystal structure of the PAS-GGDEF-EAL region of RbdA from Pseudomonas aeruginosa. J Bacteriol 200. 10.1128/JB.00515-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Amikam D, Galperin MY. 2006. PilZ domain is part of the bacterial c-di-GMP binding protein. Bioinformatics 22:3–6. 10.1093/bioinformatics/bti739. [DOI] [PubMed] [Google Scholar]
- 38.Beyhan S, Tischler AD, Camilli A, Yildiz FH. 2006. Transcriptome and phenotypic responses of Vibrio cholerae to increased cyclic di-GMP level. J Bacteriol 188:3600–3613. 10.1128/JB.188.10.3600-3613.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McCarter LL. 1995. Genetic and molecular characterization of the polar flagellum of Vibrio parahaemolyticus. J Bacteriol 177:1595–1609. 10.1128/jb.177.6.1595-1609.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stewart BJ, McCarter LL. 1996. Vibrio parahaemolyticus FlaJ, a homologue of FliS, is required for production of a flagellin. Mol Microbiol 20:137–149. 10.1111/j.1365-2958.1996.tb02496.x. [DOI] [PubMed] [Google Scholar]
- 41.Belas R, Simon M, Silverman M. 1986. Regulation of lateral flagella gene transcription in Vibrio parahaemolyticus. J Bacteriol 167:210–218. 10.1128/jb.167.1.210-218.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weber H, Pesavento C, Possling A, Tischendorf G, Hengge R. 2006. Cyclic-di-GMP-mediated signalling within the sigma network of Escherichia coli. Mol Microbiol 62:1014–1034. 10.1111/j.1365-2958.2006.05440.x. [DOI] [PubMed] [Google Scholar]
- 43.Lindenberg S, Klauck G, Pesavento C, Klauck E, Hengge R. 2013. The EAL domain protein YciR acts as a trigger enzyme in a c-di-GMP signalling cascade in E. coli biofilm control. EMBO J 32:2001–2014. 10.1038/emboj.2013.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Serra DO, Hengge R. 2019. A c-di-GMP-based switch controls local heterogeneity of extracellular matrix synthesis which is crucial for integrity and morphogenesis of Escherichia coli macrocolony biofilms. J Mol Biol 431:4775–4793. 10.1016/j.jmb.2019.04.001. [DOI] [PubMed] [Google Scholar]
- 45.Hengge R. 2020. Linking bacterial growth, survival, and multicellularity – small signaling molecules as triggers and drivers. Curr Opin Microbiol Elsevier Ltd [DOI] [PubMed] [Google Scholar]
- 46.Schmidt AJ, Ryjenkov DA, Gomelsky M. 2005. The ubiquitous protein domain EAL is a cyclic diguanylate-specific phosphodiesterase: enzymatically active and inactive EAL domains. J Bacteriol 187:4774–4781. 10.1128/JB.187.14.4774-4781.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reinders A, Hee CS, Ozaki S, Mazur A, Boehm A, Schirmer T, Jenal U. 2016. Expression and genetic activation of cyclic di-GMP-specific phosphodiesterases in Escherichia coli. J Bacteriol 198:448–462. 10.1128/JB.00604-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sundriyal A, Massa C, Samoray D, Zehender F, Sharpe T, Jenal U, Schirmer T. 2014. Inherent regulation of EAL domain-catalyzed hydrolysis of second messenger cyclic di-GMP. J Biol Chem 289:6978–6990. 10.1074/jbc.M113.516195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yilmaz C, Rangarajan AA, Schnetz K. 2020. The transcription regulator and c-di-GMP phosphodiesterase PdeL represses motility in Escherichia coli. J Bacteriol 203:e00427-20. 10.1128/JB.00427-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Miller VL, Taylor RK, Mekalanos JJ. 1987. Cholera toxin transcriptional activator ToxR is a transmembrane DNA binding protein. Cell 48:271–279. 10.1016/0092-8674(87)90430-2. [DOI] [PubMed] [Google Scholar]
- 51.Crawford JA, Krukonis ES, DiRita VJ. 2003. Membrane localization of the ToxR winged-helix domain is required for TcpP-mediated virulence gene activation in Vibrio cholerae. Mol Microbiol 47:1459–1473. 10.1046/j.1365-2958.2003.03398.x. [DOI] [PubMed] [Google Scholar]
- 52.Beyhan S, Odell LS, Yildiz FH. 2008. Identification and characterization of cyclic diguanylate signaling systems controlling rugosity in Vibrio cholerae. J Bacteriol 190:7392–7405. 10.1128/JB.00564-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cotter PA, Stibitz S. 2007. c-di-GMP-mediated regulation of virulence and biofilm formation. Curr Opin Microbiol 10:17–23. 10.1016/j.mib.2006.12.006. [DOI] [PubMed] [Google Scholar]
- 54.Pratt JT, Tamayo R, Tischler AD, Camilli A. 2008. PilZ domain proteins bind cyclic diguanylate and regulate diverse processes in Vibrio cholerae. J Biol Chem 148:825–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Floyd KA, Lee CK, Xian W, Nametalla M, Valentine A, Crair B, Zhu S, Hughes HQ, Chlebek JL, Wu DC, Hwan Park J, Farhat AM, Lomba CJ, Ellison CK, Brun YV, Campos-Gomez J, Dalia AB, Liu J, Biais N, Wong GCL, Yildiz FH. 2020. c-di-GMP modulates type IV MSHA pilus retraction and surface attachment in Vibrio cholerae. Nat Commun 11. 10.1038/s41467-020-15331-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Srivastava D, Hsieh M-L, Khataokar A, Neiditch MB, Waters CM. 2013. Cyclic di-GMP inhibits Vibrio cholerae motility by repressing induction of transcription and inducing extracellular polysaccharide production. Mol Microbiol 90:1262–1276. 10.1111/mmi.12432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chakraborty S, Biswas M, Dey S, Agarwal S, Chakrabortty T, Ghosh B, Dasgupta J. 2020. The heptameric structure of the flagellar regulatory protein FlrC is indispensable for ATPase activity and disassembled by cyclic-di-GMP. J Biol Chem 295:16960–16974. 10.1074/jbc.RA120.014083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Heering J, Alvarado A, Ringgaard S. 2017. Induction of cellular differentiation and single cell imaging of Vibrio parahaemolyticus swimmer and swarmer cells. J Vis Exp (123):55842. 10.3791/55842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Simon R, Priefer U, Pühler A. 1983. A broad host range mobilization system for In Vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Bio/Technology 1:784–791. 10.1038/nbt1183-784. [DOI] [Google Scholar]
- 60.Makino K, Oshima K, Kurokawa K, Yokoyama K, Uda T, Tagomori K, Iijima Y, Najima M, Nakano M, Yamashita A, Kubota Y, Kimura S, Yasunaga T, Honda T, Shinagawa H, Hattori M, Iida T. 2003. Genome sequence of Vibrio parahaemolyticus: a pathogenic mechanism distinct from that of V. cholerae. Lancet 361:743–749. 10.1016/S0140-6736(03)12659-1. [DOI] [PubMed] [Google Scholar]
- 61.Whitaker WB, Parent MA, Naughton LM, Richards GP, Blumerman SL, Boyd EF. 2010. Modulation of responses of Vibrio parahaemolyticus O3:K6 to pH and temperature stresses by growth at different salt concentrations. Appl Environ Microbiol 76:4720–4729. 10.1128/AEM.00474-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Edwards RA, Keller LH, Schifferli DM. 1998. Improved allelic exchange vectors and their use to analyze 987P fimbria gene expression. Gene 207:149–157. 10.1016/s0378-1119(97)00619-7. [DOI] [PubMed] [Google Scholar]
- 63.Leong SA, Ditta GS, Helinski DR. 1982. Heme biosynthesis in Rhizobium: identification of a cloned gene coding for delta-aminolevulinic acid synthetase from Rhizobium meliloti. J Biol Chem 257:8724–8730. [PubMed] [Google Scholar]
- 64.Lenz DH, Mok KC, Lilley BN, Kulkarni RV, Wingreen NS, Bassler BL. 2004. The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae. Cell 118:69–82. 10.1016/j.cell.2004.06.009. [DOI] [PubMed] [Google Scholar]