ABSTRACT
APOBEC3 proteins play pivotal roles in defenses against retroviruses, including HIV-1, as well as retrotransposons. Presumably due to the evolutionary arms race between the hosts and retroelements, APOBEC3 genes have rapidly evolved in primate lineages through sequence diversification, gene amplification and loss, and gene fusion. Consequently, modern primates possess a unique set or “repertoire” of APOBEC3 genes. The APOBEC3 gene repertoire of humans has been well investigated. There are three types of catalytic domains (Z domain; A3Z1, A3Z2, and A3Z3), 11 Z domains, and 7 independent genes, including 4 genes encoding double Z domains. However, the APOBEC3 gene repertoires of nonhuman primates remain largely unclear. Here, we characterize APOBEC3 gene repertoires among primates and investigated the evolutionary scenario of primate APOBEC3 genes using phylogenetic and comparative genomics approaches. In the 21 primate species investigated, we identified 145 APOBEC3 genes, including 69 double-domain type APOBEC3 genes. We further estimated the ages of the respective APOBEC3 genes and revealed that APOBEC3B, APOBEC3D, and APOBEC3F are the youngest in humans and were generated in the common ancestor of Catarrhini. Notably, invasion of the LINE1 retrotransposon peaked during the same period as the generation of these youngest APOBEC3 genes, implying that LINE1 invasion was one of the driving forces of the generation of these genes. Moreover, we found evidence suggesting that sequence diversification by gene conversions among APOBEC3 paralogs occurred in multiple primate lineages. Together, our analyses reveal the hidden diversity and the complicated evolutionary scenario of APOBEC3 genes in primates.
IMPORTANCE In terms of virus-host interactions and coevolution, the APOBEC3 gene family is one of the most important subjects in the field of retrovirology. APOBEC3 genes are composed of a repertoire of subclasses based on sequence similarity, and a paper by LaRue et al. provides the standard guideline for the nomenclature and genomic architecture of APOBEC3 genes. However, it has been more than 10 years since this publication, and new information, including RefSeq, which we used in this study, is accumulating. Based on accumulating knowledge, APOBEC3 genes, particularly those of primates, should be refined and reannotated. This study updates knowledge of primate APOBEC3 genes and their genomic architectures. We further inferred the evolutionary scenario of primate APOBEC3 genes and the potential driving forces of APOBEC3 gene evolution. This study will be a landmark for the elucidation of the multiple aspects of APOBEC3 family genes in the future.
KEYWORDS: APOBEC3, evolution, primates
INTRODUCTION
It is evident that some viral infections are pathogenic and lethal to hosts. However, hosts are not always vulnerable to viral infections but possess a repertoire of antiviral genes to regulate pathogenic viral infections. The long-lasting coexistence and coevolution of hosts and viruses led to the evolution of antiviral genes in hosts, and the virus-driven evolution of antiviral genes is considered the “Red Queen effect” (reviewed in references 1 to 4). As a hallmark of the antiviral genes that have undergone an evolutionary arms race with pathogenic viruses, some antiviral genes, such as ZC3HAV1 (also known as zinc-finger antiviral protein [ZAP]) (5), bone marrow stromal antigen 2 (BST2; also known as tetherin) (6–8), and radical SAM domain-containing 2 (RSAD2; also known as viperin) (9, 10), are under strong diversifying selection. In particular, the RSAD2 gene is carried by eukaryotes to prokaryotes and functions as an antiviral gene (11). In addition, some family genes such as interferon-induced transmembrane (IFITM) genes (12–14), tripartite motif-containing (TRIM) genes (15–18), and apolipoprotein B mRNA-editing enzyme catalytic polypeptide-like 3 (APOBEC3; A3) genes (1, 19) are antiviral and under diversifying selection. In addition to diversifying selection, IFITM family genes (14), TRIM family genes (18), and A3 family genes (4, 19, 20) have undergone amplification by tandem gene duplication.
A3 family proteins are cellular cytidine deaminases and members of AID/APOBEC superfamily (reviewed in references 21 and 22). AID/APOBEC superfamily proteins, including A3 proteins, commonly possess a zinc-dependent catalytic domain (Z domain) with a conserved HxE/PCxxC motif (reviewed in references 21 and 22). There are seven A3 members (A3A, A3B, A3C, A3D, A3F, A3G, and A3H) in humans, clustered in the locus sandwiched by the CBX6 and CBX7 genes on chromosome 22 (herein we call this genomic region “canonical A3 locus”) (20, 23). Some A3 proteins, such as A3D, A3F, A3G, and A3H, are potent inhibitors of infection by human immunodeficiency virus type 1 (HIV-1) (24–29). To counteract antiviral A3 proteins, an accessory protein of HIV-1, viral infectivity factor (Vif), degrades the A3 proteins expressed in HIV-1-infected cells via the ubiquitin-proteasome pathway (reviewed in references 4 and 30). Vif is well conserved in a variety of lentiviruses, including HIV-1 and simian immunodeficiency viruses (SIVs), and neutralizes antiviral A3 proteins expressed in hosts (reviewed in references 4 and 31).
The A3 Z domains are classified into three classes based on sequence similarity: A3Z1, A3Z2, and A3Z3 domains (reviewed in references 4 and 20). Human A3A, A3C, and A3H possess single A3Z1, A3Z2, and A3Z3 domains, respectively, while the other four A3s harbor double Z domains: A3Z2-A3Z1 in A3B and A3G and A3Z2-A3Z2 in A3D and A3F (reviewed in references 4 and 20). Previous studies have estimated that the origins of A3Z1, A3Z2, and A3Z3 are the common ancestor of Eutheria, and the common ancestor of primates possessed the three genes containing the respective single Z domains (20, 32). Since the human genome encodes seven A3 genes and four of the seven genes harbor double Z domains, these observations suggest that A3 genes were amplified by tandem gene duplication and that some of them were concatenated during primate evolution to generate novel A3 genes harboring two Z domains (4, 20). In addition, Yang et al. (33) and we (19) have recently revealed that some primate A3 genes are encoded in different regions of the canonical A3 locus. Because these A3 genes located in the noncanonical A3 loci lack introns, it is suggested that these A3 genes were amplified by retrotransposition (here, we refer to retrotransposition-mediated amplification of intronless A3 genes as “retrocopying”) during primate evolution.
To elucidate the evolutionary scenario of primate A3 genes in depth, which includes gene amplification (by tandem gene duplication and retrocopying), gene loss, and the formation of double-domain A3 genes, a repertoire of A3 genes in diverse primates should be determined at a high resolution. Although the gene structure of human A3 genes is well studied, the A3 gene repertoires of nonhuman primates, including nonhuman Hominidae, Old World monkeys (OWMs), New World monkeys (NWMs), and Prosimians, have not been characterized deeply. Some previous studies have addressed this issue using information on primate genome sequences (1, 19, 32). In fact, we recently revealed the numbers of A3 Z domains encoded by the genomes of 160 mammalian species, including 30 primate species (19). Nevertheless, the details of the gene structures of A3 gene repertoires in nonhuman primates (e.g., what kinds of double-domain type A3 genes are present in each primate species) remain largely unknown.
In this study, we systematically reannotated primate A3 genes using the coding sequence (CDS) data set provided by the RefSeq database (34). Since the CDS annotation in RefSeq is based on transcriptomic information (i.e., mRNA sequences), as well as genomic information (34), we can investigate whether respective A3 Z domains are expressed as single- or double-domain-type genes by splicing. Moreover, we examined the prevalence, age, and origin of each A3 gene in primates in depth. Here, we further describe the complicated evolutionary scenario of primate A3 genes and address factors that potentially drive the evolution of primate A3 genes.
RESULTS
Reannotation of primate AID/APOBEC genes.
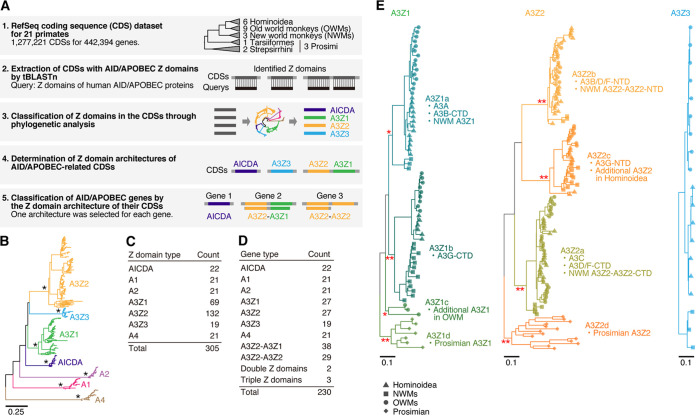
To elucidate the evolutionary history of AID/APOBEC genes in primates at a high resolution, we undertook systematic reannotation of primate AID/APOBEC genes (Fig. 1A). We surveyed the CDS database provided by RefSeq (release 101) (34) for 21 primate species (see Data Set S1 in the supplemental material) and extracted 454 CDSs (consisting of 230 genes) displaying sequence similarity with the Z domains of human AID/APOBECs. We classified the identified Z domains into seven categories (AICDA, A1, A2, A3Z1, A3Z2, A3Z3, and A4) through phylogenetic analysis. Finally, we resolved the Z-domain architectures of the respective AID/APOBEC-related CDSs and subsequently classified the AID/APOBEC-related genes according to the Z-domain architectures of their related CDSs.
FIG 1.
Reannotation of AID/APOBEC genes in primates. (A) Scheme for the reannotation of AID/APOBEC-related genes in primates from the RefSeq database. (B) Phylogenetic tree of the Z domains of primate AID/APOBEC genes. One representative CDS was selected for each gene, and only Z domains in the CDS were used for the analysis. The tree was reconstructed by the NJ method according to the nucleotide sequences of the Z domains. *, bootstrap value = 100. (C) Number of AID/APOBEC Z domains identified in this study. (D) Number of respective types of AID/APOBEC genes. (E) Phylogenetic trees of primates A3Z1, A3Z2, and A3Z3. The subclassifications of A3Z1 and A3Z2 (A3Z1a–1d and A3Z2a–2d, respectively) are indicated. Information on primate lineages is indicated at the tip of the tree. *, bootstrap value > 70; **, bootstrap value > 90.
In the 21 primate species, we identified a total of 305 Z domains in 230 AID/APOBEC genes (Fig. 1B to D; see also Data Set S2). As described previously (19), A3Z1 and A3Z2 have been highly amplified in primates (Fig. 1B and C). Genes with double Z domains were detected only in A3 genes, and half of the A3 genes (69/145) encode double Z domains (Fig. 1D; see also Data Set S2). The majority of the double domain-type A3s in primates show A3Z2-A3Z1 (e.g., A3B and A3G) or A3Z2-A3Z2 (e.g., A3D and A3F) architectures (Fig. 1D).
Origin of A3 genes encoding double Z domains in primates.
Compared with the other AID/APOBEC Z domains, including A3Z3s, A3Z1s, and A3Z2s, have been amplified and diversified, with further complexity, in primates (Fig. 1C) (19). We investigated the codon sites under diverse selection in primate A3Z1, A3Z2, and A3Z3 sequences using the mixed effects model of evolution algorithm (MEME) (35) (see Data Set S3). Consistent with a previous study (19), a larger number of codon sites under diversifying selection were detected in A3Z1 and A3Z2 than in A3Z3, suggesting that A3Z1 and A3Z2 have become more diversified during primate evolution than A3Z3 (see Data Set S3).
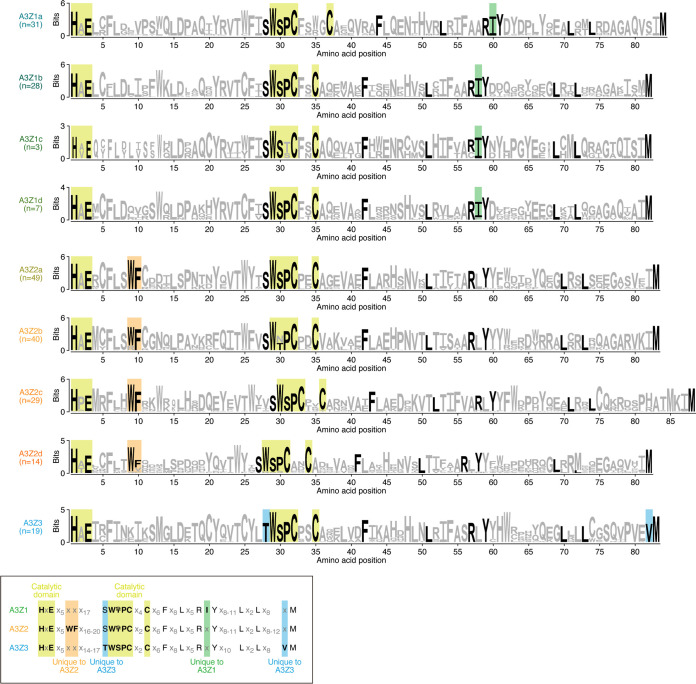
To trace the evolutionary history of primate A3Z1s and A3Z2s, we classified primate A3Z1s and A3Z2s into more detailed phylogenetic clusters (A3Z1a–1d and A3Z2a–2d, respectively) (Fig. 1E). Clusters A3Z1a–1b and A3Z2a–2c contain Z domains identified from Hominoidea, OWMs, and NWMs, suggesting that these clusters formed in the common ancestor of Simiiformes, which includes Hominoidea, OWMs, and NWMs. On the other hand, A3Z1d- and A3Z2d-type Z domains were detected specifically in Prosimians, and A3Z1c was detected only in OWMs. The known motifs of the catalytic domain of cytidine deaminase (i.e., HxE and WS/TPCx2-4C) (20) were well conserved across the A3 domains we identified (Fig. 2). As an exception, the catalytic motif (WS/TPCx2-4C) was degraded in A3Z1c, an OWM-specific A3 Z domain, suggesting that the deaminase activity of A3Z1c has been lost during evolution (Fig. 2). We additionally confirmed that the amino acid residues that have been reported as unique in A3Z1, A3Z2, and A3Z3 (20) are highly conserved in the corresponding classes of the A3 Z domains in this study (Fig. 2).
FIG 2.
Sequence motifs conserved in A3 Z domains. Logo plots of amino acid sequences for respective clusters of A3 Z domains are shown. The yellow square indicates the amino acid residues comprising the catalytic domain of A3 proteins. Also, amino acid residues that have been reported as specific to A3Z1, A3Z2, and A3Z3 (20) are annotated. The logo plots were created using WebLogo 3 (59) (http://weblogo.threeplusone.com/create.cgi).
The human genome encodes four double-domain A3 genes: A3B, A3D, A3F, and A3G (4, 20). According to the cluster defined above, we next investigated the Z-domain architectures of primate double-domain A3s in depth (Fig. 1E). A3D and A3F commonly possess a A3Z2-A3Z2 architecture (4, 20). In our classification, the architectures of both A3D and A3F were A3Z2b-A3Z2a (Fig. 1E), suggesting that these two genes arose via tandem gene duplication of an ancestral A3Z2b-A3Z2a gene.
Similar to A3D and A3F, A3B and A3G are double-domain type A3 genes, and these two genes have an A3Z2-A3Z1 architecture. However, our classification showed that these are composed of the different types of Z domains: A3Z2b-A3Z1a for A3B and A3Z2c-A3Z1b for A3G (Fig. 1E). These observations suggest that A3B and A3G were generated independently. In particular, the C-terminal domain (CTD) of A3B clusters together with A3As (A3Z1a), whereas its N-terminal domain (NTD) clusters with the NTDs of A3Ds and A3Fs (A3Z2b). In the case of A3G, both its NTD (A3Z2c) and its CTD (A3Z1b) formed unique clusters (Fig. 1E). Consistent with a previous report (32), these results suggest that A3B arose via fusion of an A3D/F-NTD-like Z domain (A3Z2b here) and an A3A-like Z domain (A3Z1a here).
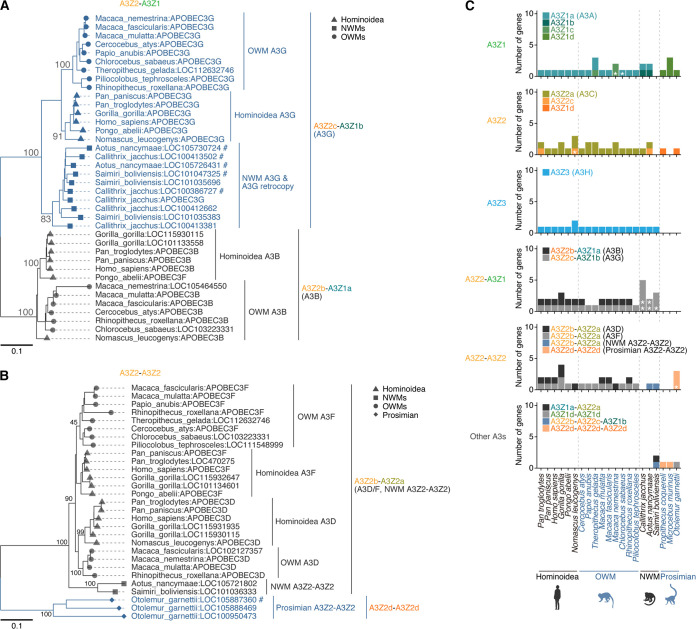
To estimate the age of the formation of the respective types of double-domain A3 genes, we next examined the prevalence of these A3 genes among primates (Fig. 3). A3G-like (A3Z2c-A3Z1b) genes are present in Hominoidea, OWMs, and NWMs but absent in Prosimians (Fig. 3A and C). Consistent with our previous findings (19), these observations suggest that the A3G-like (A3Z2c-A3Z1b) gene was formed in the common ancestor of Simiiformes. On the other hand, A3B-like (A3Z2b-A3Z1a) genes are present only in Catarrhini (i.e., Hominoidea and OWMs) (Fig. 3A and C), suggesting that the A3B-like (A3Z2b-A3Z1a) gene was formed in the common ancestor of Catarrhini. Although A3D and A3F are present only in Catarrhini, a single copy of the gene encoding the same Z-domain architecture (A3Z2b-A3Z2a) is present in NWMs (Fig. 3B and C). This NWM-specific A3Z2b-A3Z2a gene is encoded in the A3 gene cluster, which is present between CBX6 and CBX7 (4), supporting that this gene is an authentic member of the A3 gene family (Fig. 4A). These results suggest that the A3D/F-like gene (A3Z2b-A3Z2a) formed in the common ancestor of Simiiformes and subsequently was duplicated in the common ancestor of Catarrhini, resulting in the formation of A3D and A3F genes.
FIG 3.
Prevalence of double-domain A3 genes in primates. (A and B) Phylogenetic trees for A3Z2-A3Z1 (A) and A3Z2-A3Z2 (B) genes in primates. Species names and official gene symbols assigned by RefSeq are indicated at the tips of the tree. #, A3 genes generated by retrocopying (i.e., intronless A3 genes). (C) Number of respective types of A3 genes in primate species. *, A3 genes generated by retrocopying (i.e., intronless A3 genes). The detailed information is summarized in Data Set S2.
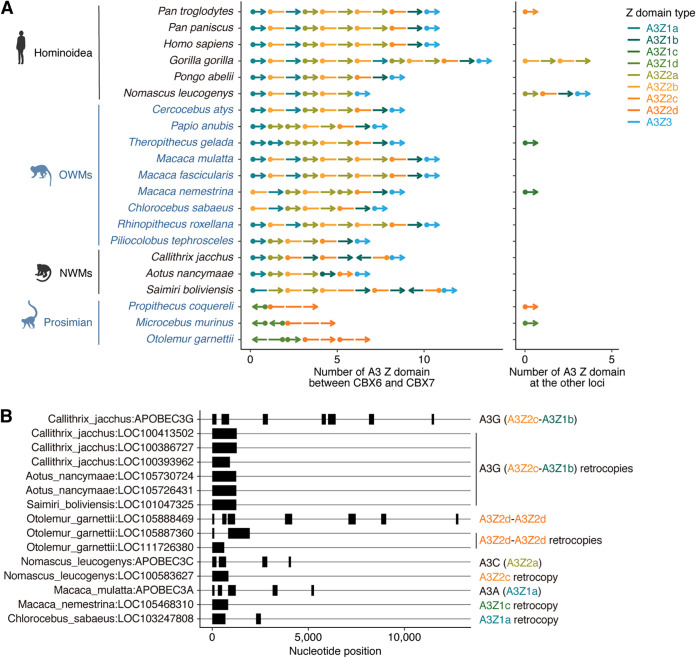
FIG 4.
Genomic location of primate A3 genes. (A) Genomic synteny of primate A3 genes. A segment represents a Z domain. A dot and arrowhead denote the start and end of the gene, respectively. Only nonretrocopy A3 genes are shown. (Left) A3 genes in the canonical A3 locus, which is situated between CBX6 and CBX7. (Right) A3 genes located in genomic loci other than the canonical A3 locus (e.g., unplaced scaffold sequences). (B) A3 retrocopy genes identified in primates. Genomic CDS structures of respective genes are shown. In addition to retrocopy genes, several A3 genes in the canonical A3 locus are shown. The detailed information is summarized in Data Set S2.
Consistent with a previous report (19), prosimians (including Strepsirrhini and Tarsiiformes) did not possess the A3Z3 gene (Fig. 3C), suggesting loss of this gene in the prosimian lineage. Instead, another type of A3Z2-A3Z2 (A3Z2d-A3Z2d) gene was present. Because this prosimian-specific A3Z2d domain is phylogenetically different from other A3Z2 domains (Fig. 1E), the A3Z2-A3Z2 formations in Simiiformes and Prosimians likely occurred in parallel during primate evolution. Moreover, this type of prosimian-specific A3Z2d-A3Z2d gene was amplified not only via tandem gene duplication but also via gene retrocopying, as with A3G in NWMs (19, 33) (Fig. 4B). These data support that this prosimin-specific A3Z2d-A3Z2d authentically exists and is expressed in germ line cells, where heritable retrocopying events occur. In addition, we identified other types of A3 retrocopies in Nomascus leucogens (A3Z2c), Macaca nemestrina (A3Z1c), and Chlorocebus sabaeus (A3Z1a), suggesting that the formation of novel A3 genes via retrocopy has occurred in multiple primate lineages (Fig. 4B).
Gene conversions on the multiple A3 Z domains.
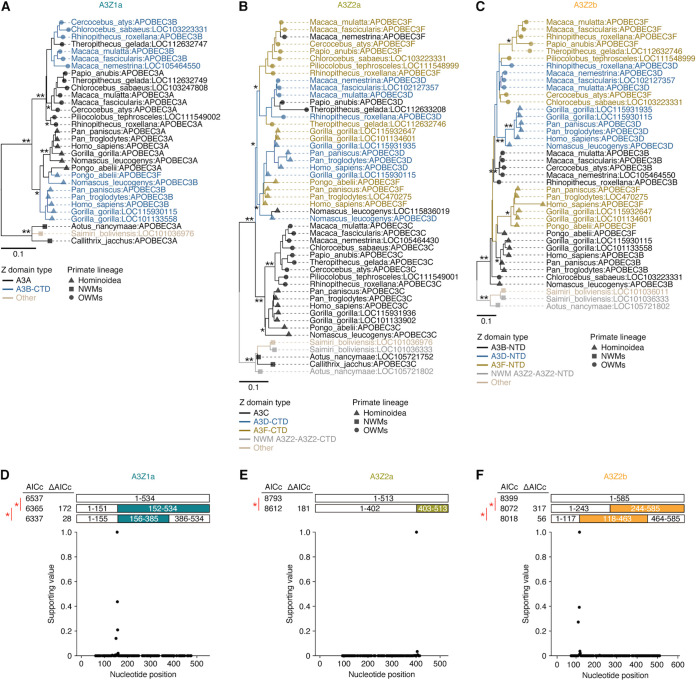
To further investigate the evolutionary trajectory of the youngest double-domain A3 genes in primates (i.e., A3B, A3D, and A3F), which were generated in the common ancestor of Catarrhini, we reconstructed the phylogenetic trees of three types of Z domains (i.e., A3Z1a, A3Z2a, and A3Z2b) related to the A3 genes above (Fig. 5A to C).
FIG 5.
Gene conversions of multiple A3 Z domains. (A to C) Phylogenetic trees of A3Z1a (A), A3Z2a (B), and A3Z2b (C). The trees were reconstructed by the NJ method according to the nucleotide sequences of Z domains. The bootstrap value is denoted on the corresponding node. (D to F) Gene conversion events detected by GARD (36) in A3Z1a (D), A3Z2a (E), and A3Z2b (F). (Top) Statistical comparison of the evolutionary models with and without gene conversion event(s). The Akaike information criterion correction (AICc) value of each model and the ΔAICc value between models are indicated. An asterisk denotes a P value of <0.01 in the model comparison. (Bottom) Relative support values of the recombination breakpoints detected by GARD. The highest breakpoint signal was normalized as one. *, bootstrap value > 70; **, bootstrap value > 90.
We first examined the topology of the trees for A3Z1a (including A3A and A3B-CTD) (Fig. 5A). Z domains corresponding to A3A and A3B-CTD were mingled together and clustered according to the primate lineages (i.e., Hominoidea and OWMs) instead of the gene types (A3A and A3B-CTD). This tree topology suggests that the A3A and A3B genes did not independently evolve after the speciation of Hominoidea and OWMs (i.e., after the formation of A3B [Fig. 3A and C]). This implies the possibility that gene conversion occurred between A3A and A3B-CTD. Therefore, we evaluated the possibility of gene conversion events between A3A and A3B-CTD using a genetic algorithm for recombination detection (GARD) (36). The GARD analysis showed that the model with one or two recombination events outperformed the model with no recombination in explaining the evolution of A3Z1a (Fig. 5D), supporting the occurrence of gene conversion(s) between A3A and A3B-CTD.
Similarly, we examined the topology of the trees for A3Z2a (including A3C, A3D-CTD and A3F-CTD) and A3Z2b (including A3B-NTD, A3D-NTD, and A3F-NTD) (Fig. 5B and C). In the case of the A3Z2a domain, the A3C genes in primates formed a unique cluster and showed a topology concordant with the primate phylogeny (Fig. 5B). On the other hand, similar to the A3Z1a domain above, A3D-CTD and A3F-CTD were mingled together and clustered according to primate lineage, Hominoidea and OWMs, instead of gene type (Fig. 5B). In the case of the A3Z2b domain, A3B-NTD and A3F-NTD in Hominoidea clustered together, though the bootstrap support value was not high (Fig. 5C; bootstrap value = 14). These results suggest that gene conversion also occurred for A3Z2a and A3Z2b. Therefore, we evaluated the possibility of gene conversion events in A3Z2a and A3Z2b using GARD (Fig. 5E and F). In both cases, recombination events with statistical significance were detected, supporting the occurrence of gene conversions for these genes.
LINE1 is a possible driving force for the generation of the A3B gene.
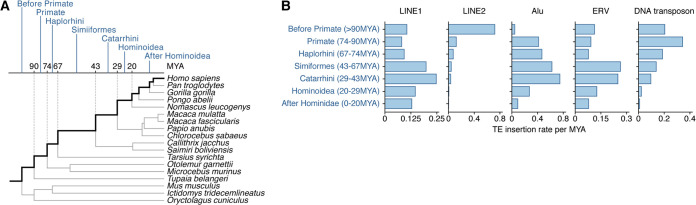
It has been considered that one of the major roles of primate A3s is to restrict the invasion of retroviruses, including endogenous retroviruses (ERVs) or other types of transposable elements (TEs) (e.g., LINEs and SINEs) (19, 37). In addition, our data suggest that the generation of A3B and the duplication of A3D/F occurred in the common ancestor of Catarrhini (Fig. 3). To investigate the evolutionary event(s) driving the generation of these A3 genes, we examined whether intensive invasion of retroviruses or TEs occurred in the common ancestor of Catarrhini. We estimated the insertion date of the respective loci of five TE categories (LINE1, LINE2, Alu, DNA transposon, and ERV) in the human genome using a comparative genomic approach (19) and subsequently quantified the amount of TE insertions in each evolutionary period (Fig. 6A). As illustrated in Fig. 6B, the invasion of LINE1 peaked around the age of the common ancestor of Catarrhini (i.e., 29 to 43 million years ago [MYA]). On the other hand, invasions of other TEs, such as LINE2, DNA transposons, and ERV, peaked at different periods. As an exception, the period of Alu invasion peak overlapped with that of LINE1 (Fig. 6B), consistent with the knowledge that Alu is a nonautonomous retroelement and is transposed by LINE1 (38). Altogether, these findings suggest the possibility that the origin of the A3B (A3Z2b-A3Z1a) gene and/or the duplication of A3D/F (A3Z2b-A3Z2a) genes in the common ancestor of Catarrhini were driven by the invasion of LINE1 (Fig. 7).
FIG 6.
Estimation of the number of TE insertions in each evolutionary period. (A) Definition of evolutionary periods. (B) Estimated insertion amount of TEs per MYA in respective periods. The results for five TE categories (LINE1, LINE2, Alu, ERV, and DNA transposon) are shown.
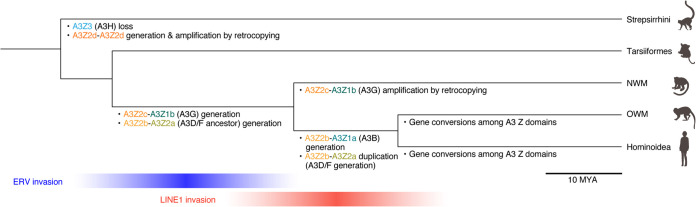
FIG 7.
Complex evolutionary history of A3 genes in primates. (Top) The evolution of A3 genes in primates. In the Strepsirrhini lineage, A3Z3 loss and A3Z2d-A3Z2d generation, followed by retrocopying occurred. In the common ancestor of Simiiformes (Hominoidea, OWMs, and NWMs), A3Z2c-A3Z1b (A3G) (19) and A3Z2b-A3Z2a (the ancestor of A3D and A3F) were generated. In the NWM lineage, A3Z2c-A3Z1b (A3G) was amplified by retrocopying (19, 33). In the common ancestor of Hominoidea and OWMs, A3Z2b-A3Z1a (A3B) was generated. In addition, A3Z2b-A3Z2a was duplicated (i.e., A3D and A3F were generated). In the lineages of Hominoidea and OWMs, gene conversions among A3 Z domains occurred multiple times. (Bottom) Association of invasions of retroelements with primate A3 evolution. The invasion of ERVs (blue) in the human genome peaked at the age of the common ancestor of Simiiformes, when A3Z2c-A3Z1b (A3G) was generated (19). On the other hand, LINE1 invasion peaked at the age of the common ancestor of Hominoidea and OWMs, when A3B, A3D, and A3F were generated.
DISCUSSION
In this study, we conducted molecular phylogenetic and evolutionary analyses by using the CDSs provided by RefSeq (34) and uncovered the evolutionary scenario of the evolution of primate A3 genes. The results of this study and previous works (4, 19, 20, 32, 33) show that the evolution of primate A3 genes is more complicated than expected (Fig. 7). In particular, we classified the A3Z1 and A3Z2 genes into A3Z1a–1d and A3Z2a–2d and revealed when and how double domain A3 genes were generated during primate evolution. Moreover, we addressed evolutionary events that might have contributed to driving the evolution of A3 family genes.
Previous studies have suggested that three core (i.e., single-domain) A3 genes, A3Z1, A3Z2 and A3Z3, were generated in the common ancestor of Eutheria (20, 32). On the other hand, Homininae, including humans, possess four double-domain A3 genes: A3B (A3Z2b-A3Z1a), A3D (A3Z2b-A3Z2a), A3F (A3Z2b-A3Z2a), and A3G (A3Z2c-A3Z1b). Our recent study showed that A3G (A3Z2c-A3Z1b) was generated in the common ancestor of Simiiformes (19). In contrast to A3G, we revealed that A3B (A3Z2b-A3Z1a) was generated independently of A3G, and that it is younger than A3G: the A3B gene was generated in the common ancestor of Hominidae and OWMs (Fig. 7). Regarding A3D and A3F, NWMs carry a single copy of the A3D/F-like gene (A3Z2b-A3Z2a) in their genome, suggesting that an A3D/F-like gene (A3Z2b-A3Z2a) was generated in the common ancestor of Simiiformes and then duplicated into A3D and A3F in the common ancestor of Hominidae and OWMs via tandem gene duplication (Fig. 7).
It is assumed that the physiological role of A3 genes is the regulation of exogenous retroviruses, including HIV-1, and endogenous retroelements, including ERVs, LINEs, and SINEs (reviewed in reference 37). We recently showed that the rapid evolution and amplification of mammalian A3 genes would be driven by the invasion of retroelements (19). In the present study, we deeply assessed the timing of the generation of double-domain A3 genes and the invasion of a variety of retroelements during primate evolution (Fig. 6 and 7). Consistent with our recent study (19), invasion of ERVs peaked in the common ancestor of Hominidae, OWMs, and NWMs (Fig. 6), which is consistent with the generation of the A3G gene and an ancestral A3D/F-like gene (A3Z2b-A3Z2a) (Fig. 7). On the other hand, the invasions of LINE1 and Alu peaked in the common ancestor of Catarrhini (Fig. 6). This period overlaps with the generation of the A3B gene and the tandem duplication of A3D and A3F (Fig. 7). A3B potently suppresses the growth of LINE1 (39–41), whereas A3F inhibits the replication of vif-deleted HIV-1 (42), HERV-K (43), and LINE1 (40). Altogether, these findings suggest that LINE1 invasion in the common ancestor of Catarrhini is a driving force of the evolution of primate A3 genes.
The genome of a prosimian (Otolemur garnettii) encodes a unique double-domain A3 gene, the A3Z2d-A3Z2d gene (Fig. 3B and C). Because the sequence of this gene is different from that of A3D/F (A3Z2b-A3Z2a) in Catarrhini, it is suggested that this bushbaby-specific A3Z2d-A3Z2d gene was generated independently of A3D/F. Interestingly, this bushbaby-specific A3Z2d-A3Z2d gene was amplified by retrocopying (Fig. 4B). As reported previously (19, 33), amplification of A3G genes by retrocopying is prominent in the NWM lineage. As retrocopying-mediated A3 gene amplification was observed in multiple lineages of primates, retrocopy-mediated A3 gene amplification frequently occurred during primate evolution. Although the biological significance of the frequent retrocopying of A3 genes during primate evolution and its evolutionary driving force remain unclear, as described above, certain A3 proteins potentially suppress LINE1 retrotransposition (39–41). Therefore, one might assume that A3 gene amplification via retroelement machinery hijacking is a self-regulatory system for modulating excessive retrotransposition of retroelements.
In addition to gene amplification mediated by tandem duplication and retrocopying, we show evidence suggesting gene conversion between the paralogs of primate A3 genes. In particular, although the generation of novel A3 genes and A3 gene amplification were not observed after the divergence of Hominidae and OWMs, we found evidence suggesting gene conversion between A3A and A3B-CTD (A3Z1a domain), between A3D-CTD and A3F-CTD (A3Z2a domain), and between A3D-NTD and A3F-NTD (Fig. 5). Gene conversions that occur between paralogs reduce sequence diversity between paralogs (i.e., reduction in intraspecies sequence diversity) while increasing sequence diversity between orthologs (i.e., increase in interspecies sequence diversity), which may result in the difference in antiviral activity of orthologs of some primate A3 genes. For instance, it is known that the anti-HIV-1 activity of A3D and A3F differs among primates: human A3F is stronger than human A3D (28, 44), but chimpanzee A3F is weaker than chimpanzee A3D (45). The difference in the antiviral activity of great ape A3D and A3F may be attributed to gene conversion between these genes. Moreover, it is known that human A3D and A3F bind to HIV-1 Vif via their CTDs (46–52); in addition, human and chimpanzee A3D/F can be degraded by the Vif proteins of HIV-1 (25, 53) and an SIV infecting chimpanzee (SIVcpz) (45). However, it is of interest that chimpanzee A3D is resistant to degradation mediated by the Vif proteins of certain SIVs (e.g., SIVrcm and SIVmus), the putative ancestors of SIVcpz, but that chimpanzee A3F is degraded by them (45). The difference in the sensitivity of chimpanzee A3D and A3F to the Vif proteins of SIVrcm and SIVmus may also be due to gene conversion between these paralogs. Thus, one may assume that gene conversion between A3 paralogs promoted escape of degradation by ancestral Vif-like factor(s).
By using the CDSs from RefSeq, we elucidated multiple aspects of primate A3 evolution. However, some issues remain unclear because of technical limitations. First, because primate A3 genes are highly similar and repetitive, we were unable to define whether some sequences are authentic. For instance, although two gorilla A3B sequences were obtained from RefSeq (LOC115930115 and LOC101133558), we could not determine whether gorilla carries two A3B genes or two haplotypes of gorilla A3B genes are present or whether the results was due to sequence errors. Second, although it is evident that gene conversion has occurred between some A3 paralogs, the exact breakpoints for gene conversions in these genes remain unknown. Third, this study is mainly based on genomic sequence data, and the expression of primate A3 genes at the mRNA or protein level was not addressed. Nevertheless, we show the evolutionary scenario of primate A3 genes and in particular reveal the timing of the generation of double-domain A3 genes. To deeply elucidate the evolution of primate A3 genes, further investigations, including deep and accurate sequencing of the genomic regions encoding A3 genes, expression profiles of these genes and experimental verification of the antiviral activity of these genes, will be needed.
In summary, we reveal the evolution of primate A3 genes, which is more complicated than expected. Primate A3 genes have been diversified and amplified by tandem gene duplication, retrocopying, and gene conversion. Diversification and functional differentiation of antiviral genes can lead to the establishment of species-specific antiviral defenses, which play pivotal roles in regulating cross-species viral transmission. Our findings suggest that the roles of primate A3 genes as “species barriers” (25–29) are attributable to the rapid and complicated evolution of A3 genes driven by retroelements.
MATERIALS AND METHODS
Data downloads.
The sequence data and metadata of RefSeq CDSs (release 101) for the genomes of 21 primates (summarized in Data Set S1) were downloaded using the command “ncbi-genome-download” (version 0.3.0 [https://github.com/kblin/ncbi-genome-download]; download date, 28 October 2020). LiftOver chain files (summarized in Data Set S4) used for estimating TE insertion dates were downloaded from the UCSC genome browser website (http://hgdownload.soe.ucsc.edu/goldenPath/hg19/liftOver/).
Reannotation of AID/APOBEC genes in primates.
To extract CDSs displaying sequence similarity to the Z domains of AID/APOBECs, the sequences of RefSeq CDSs for 21 primates were scanned using tBLASTn (v2.6.0) with an E value threshold of 1.0E–3 (54). The amino acid sequences of the Z domains of human AID/APOBECs (summarized in Data Set S5) were used as query sequences. Hit regions that overlapped each other for >30 bp were clustered using bedtools cluster (v2.27.0) (55). Subsequently, one top-hit region in each cluster was selected according to the alignment coverage (i.e., the alignment length divided by query length). The hit region with <0.5 alignment coverage was discarded. The hit region matching the antisense strand of the CDS was also discarded. The sequences of the hit regions were extracted from CDSs using bedtools getfasta (55). In this study, extracted sequences were defined as sequences displaying similarity to AID/APOBEC Z domains, and related CDSs were defined as CDSs encoding the Z domains.
Phylogenetic analysis was performed to classify the nucleotide sequences of the extracted AID/APOBEC Z domains. Multiple sequence alignment was carried out using MAFFT (version 7.407) with the FFT-NS-i algorithm (56), with an alignment site with <0.75 site coverage (i.e., the number of aligned sequences divided by the total number of sequences) eliminated. The phylogenetic tree was reconstructed by the neighbor-joining (NJ) method using MEGA X (57) (model, maximum composite likelihood with site heterogeneity in substitution rate [+G]; d, transitions + transversions; gap treatment, pairwise deletion). The robustness of the tree topology was evaluated by 100 bootstrap analyses. The class of the respective Z domain was defined according to this phylogenetic tree.
The Z domain architectures of the respective AID/APOBEC-related CDSs were determined according to the Z-domain class defined above. Since multiple CDSs are sometimes recorded for one gene, one “representative CDS” per gene was (basically) selected to assign one Z-domain architecture to each gene. First, the longest CDS was selected for each type of Z-domain architecture of one gene. If CDSs with distinct Z-domain architectures were present for one gene, representative CDS(s) were selected as follows: if the number of Z domains was different among CDSs, the CDS with the larger number of Z domains was selected; if equal, both CDSs were retained. Finally, the Z-domain architecture of a gene was assigned according to that of the representative CDS. If two representative CDSs were present for a gene, both Z-domain architectures were retained for the gene.
Detailed information on the identified AID/APOBEC Z domains, CDSs, and genes is summarized in Data Set S2.
Genomic synteny analysis of A3 genes.
CBX6 and CBX7 in respective species were extracted as orthologous genes of human CBX6 and CBX7, as defined by the National Center for Biotechnology Information (https://ftp.ncbi.nih.gov/gene/DATA/gene_orthologs.gz; download date, 29 October 2020). Genomic locations of CBX6 and CBX7 in respective species were extracted from the metadata files of RefSeq CDSs (the file “_feature_table.txt”). AID/APOBEC genes located between CBX6 and CBX7 were annotated using bedtools intersect (55).
Identification of AID/APOBEC-related retrocopy genes.
Regarding the representative CDS (described above under “Reannotation of AID/APOBEC genes in primates”), the number of introns sandwiched by exons overlapping with the CDS was counted. Intron information was extracted from the header of CDSs in the fasta file provided by RefSeq (the file “_cds_from_genomic.fna”). A2 and A4 genes were excluded from the analysis because these genes have no or few introns (19). Genes with representative CDSs having a “partial” tag (i.e., CDSs lacking definite start or stop codons) were also excluded. Genes in which the representative CDS has ≤1 intron were regarded as retrocopy genes. We confirmed that all of the identified A3 retrocopy genes are located outside the canonical A3 gene cluster (see Data Set S2).
Reconstruction of phylogenetic trees.
The phylogenetic trees shown in Fig. 1E, 3A, and 3B were reconstructed as follows. The multiple sequence alignment was generated using MAFFT with the E-INS-i (56); the alignment site with <0.95 site coverage (i.e., the number of aligned sequences divided by the total number of sequences) was eliminated. The phylogenetic tree was reconstructed by the NJ method using MEGA X (57) (model, maximum composite likelihood with site heterogeneity in substitution rate [+G]; d, transitions + transversions; gap treatment, pairwise deletion). The robustness of the tree topology was evaluated by 100 bootstrap analyses.
The phylogenetic trees shown in Fig. 5A to C were reconstructed as follows. The multiple sequence alignment was constructed using MAFFT with the auto option. In the multiple sequence alignment, the alignment site with <0.95 site coverage (i.e., the number of aligned sequences divided by the total number of sequences) was eliminated. The phylogenetic tree was reconstructed by the NJ method using MEGA X (57). The robustness of tree topology was evaluated by 100 bootstrap analyses.
Detection of codon sites under diversifying selection.
We first constructed codon-based multiple sequence alignments (MSAs) of primate A3Z domains (A3Z1, A3Z2, and A3Z3) using MUSCLE (58) implemented in MEGA X (57). In the MSA, alignment sites with <10% site coverage were eliminated using the in-house script “select_alignment_site.Py.” which is available in the GitHub repository (https://github.com/TheSatoLab/primate_A3_repertoire_and_evolution). Subsequently, sequences with gaps in >10% alignment sites were eliminated using the script above.
To identify codon sites under diversifying selection in the MSA, we performed dN/dS analysis with the branch-site model using MEME (35) implemented at the Datamonkey website (http://datamonkey.org).
Detection of gene conversion events.
Gene conversion events were detected by GARD (36) using a web server (http://www.datamonkey.org/gard) with default options.
Estimation of the amount of TE insertion in each time period.
The amount of TE insertion in each time period was estimated using ortholog distribution-based methods (19). The genomic positions of respective TE loci in the human genome (hg19) determined by RepeatMasker (http://www.repeatmasker.org) in a previous study (19) were used. The LiftOver program (http://genome.ucsc.edu/cgi-bin/hgLiftOver) and chain files were used to convert the genomic coordinates of TEs in the human genome to those in other species using the option “Minmatch = 0.5.” If conversion was successful, we inferred that the orthologous copy of the TEs was likely present in the corresponding genome. The insertion dates of the respective TE loci were estimated from ortholog distributions according to the scheme described in a previous study (19). The relative amount of TE insertion in each period was calculated as the total sequence length of TEs inserted in that period divided by that in all periods. Subsequently, the insertion amount of TEs per MYA was calculated in the respective period.
Data visualization.
Phylogenetic trees were visualized with ggtree (http://bioconductor.org/packages/release/bioc/html/ggtree.html). Other data were visualized with ggplot2 (https://ggplot2.tidyverse.org/).
Data availability.
The raw data and computational codes are available from the GitHub repository (https://github.com/TheSatoLab/primate_A3_repertoire_and_evolution).
ACKNOWLEDGMENTS
We thank Yoshio Koyanagi and Naoko Misawa (Kyoto University, Kyoto, Japan) and Mai Suganami, Akiko Oide, Mai Fujimi, and Miyabishara Yokoyama (The University of Tokyo, Tokyo, Japan) for generous support.
This research was funded in part by AMED Research Program on HIV/AIDS grants 19fk0410019 (to K.S.) and 19fk0410014 (to K.S.); AMED Research Program on Emerging and Re-emerging Infectious Diseases grants 20fk0108146 (to K.S.), 19fk010817 (to K.S.), and 20fk0108270 (to K.S.); JST J-RAPID grant JPMJJR2007 (to K.S.); JST SICORP (e-ASIA) grant JPMJSC20U1 (to K.S.); JST CREST grant JPMJCR20H4 (to K.S.); JSPS KAKENHI Grant-in-Aid for Scientific Research B grant 18H02662 (to K.S.); JSPS KAKENHI Grant-in-Aid for Scientific Research on Innovative Areas grants 16H06429 (to K.S.), 16K21723 (to K.S.), 17H05813 (to K.S.), and 19H04826 (to K.S.); JSPS KAKENHI Early-Career Scientists grant 20K15767 (to J.I.); JSPS Fund for the Promotion of Joint International Research (Fostering Joint International Research) grant 18KK0447 (to K.S.); JSPS Research Fellow PD grant 19J01713 (to J.I.); the ONO Medical Research Foundation (to K.S.); the Ichiro Kanehara Foundation (to K.S.); the Lotte Foundation (to K.S.); the Mochida Memorial Foundation for Medical and Pharmaceutical Research (to K.S.); the Daiichi Sankyo Foundation of Life Science (to K.S.); the Sumitomo Foundation (to K.S.); the Uehara Foundation (to K.S.); and the Takeda Science Foundation (to K.S.).
We have no conflicts of interest to declare.
Footnotes
Supplemental material is available online only.
jvi.00144-21-s0001.xlsx (10.7KB, xlsx)
jvi.00144-21-s0002.xlsx (133.6KB, xlsx)
jvi.00144-21-s0003.xlsx (11.2KB, xlsx)
jvi.00144-21-s0004.xlsx (10.1KB, xlsx)
jvi.00144-21-s0005.xlsx (10.5KB, xlsx)
jvi.00144-21-s0006.pdf (16.3KB, pdf)
Contributor Information
Kei Sato, Email: KeiSato@g.ecc.u-tokyo.ac.jp.
Frank Kirchhoff, Ulm University Medical Center.
REFERENCES
- 1.Sawyer SL, Emerman M, Malik HS. 2004. Ancient adaptive evolution of the primate antiviral DNA-editing enzyme APOBEC3G. PLoS Biol 2:E275. 10.1371/journal.pbio.0020275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sato K, Gee P, Koyanagi Y. 2012. Vpu and BST2: still not there yet? Front Microbiol 3:131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Duggal NK, Emerman M. 2012. Evolutionary conflicts between viruses and restriction factors shape immunity. Nat Rev Immunol 12:687–695. 10.1038/nri3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nakano Y, Aso H, Soper A, Yamada E, Moriwaki M, Juarez-Fernandez G, Koyanagi Y, Sato K. 2017. A conflict of interest: the evolutionary arms race between mammalian APOBEC3 and lentiviral Vif. Retrovirology 14:31. 10.1186/s12977-017-0355-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kerns JA, Emerman M, Malik HS. 2008. Positive selection and increased antiviral activity associated with the PARP-containing isoform of human zinc-finger antiviral protein. PLoS Genet 4:e21. 10.1371/journal.pgen.0040021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu J, Chen K, Wang JH, Zhang C. 2010. Molecular evolution of the primate antiviral restriction factor tetherin. PLoS One 5:e11904. 10.1371/journal.pone.0011904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gupta RK, Hué S, Schaller T, Verschoor E, Pillay D, Towers GJ. 2009. Mutation of a single residue renders human tetherin resistant to HIV-1 Vpu-mediated depletion. PLoS Pathog 5:e1000443. 10.1371/journal.ppat.1000443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takeuchi JS, Ren F, Yoshikawa R, Yamada E, Nakano Y, Kobayashi T, Matsuda K, Izumi T, Misawa N, Shintaku Y, Wetzel KS, Collman RG, Tanaka H, Hirsch VM, Koyanagi Y, Sato K. 2015. Coevolutionary dynamics between tribe Cercopithecini tetherins and their lentiviruses. Sci Rep 5:16021. 10.1038/srep16021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lim ES, Wu LI, Malik HS, Emerman M. 2012. The function and evolution of the restriction factor Viperin in primates was not driven by lentiviruses. Retrovirology 9:55. 10.1186/1742-4690-9-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seo JY, Yaneva R, Cresswell P. 2011. Viperin: a multifunctional, interferon-inducible protein that regulates virus replication. Cell Host Microbe 10:534–539. 10.1016/j.chom.2011.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bernheim A, Millman A, Ofir G, Meitav G, Avraham C, Shomar H, Rosenberg MM, Tal N, Melamed S, Amitai G, Sorek R. 2021. Prokaryotic viperins produce diverse antiviral molecules. Nature 589:120–124. 10.1038/s41586-020-2762-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao X, Li J, Winkler CA, An P, Guo JT. 2018. IFITM genes, variants, and their roles in the control and pathogenesis of viral infections. Front Microbiol 9:3228. 10.3389/fmicb.2018.03228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hickford D, Frankenberg S, Shaw G, Renfree MB. 2012. Evolution of vertebrate interferon inducible transmembrane proteins. BMC Genomics 13:155. 10.1186/1471-2164-13-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Z, Liu J, Li M, Yang H, Zhang C. 2012. Evolutionary dynamics of the interferon-induced transmembrane gene family in vertebrates. PLoS One 7:e49265. 10.1371/journal.pone.0049265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sawyer SL, Wu LI, Emerman M, Malik HS. 2005. Positive selection of primate TRIM5alpha identifies a critical species-specific retroviral restriction domain. Proc Natl Acad Sci U S A 102:2832–2837. 10.1073/pnas.0409853102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson WE, Sawyer SL. 2009. Molecular evolution of the antiretroviral TRIM5 gene. Immunogenetics 61:163–176. 10.1007/s00251-009-0358-y. [DOI] [PubMed] [Google Scholar]
- 17.Tareen SU, Sawyer SL, Malik HS, Emerman M. 2009. An expanded clade of rodent Trim5 genes. Virology 385:473–483. 10.1016/j.virol.2008.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sawyer SL, Emerman M, Malik HS. 2007. Discordant evolution of the adjacent antiretroviral genes TRIM22 and TRIM5 in mammals. PLoS Pathog 3:e197. 10.1371/journal.ppat.0030197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ito J, Gifford RJ, Sato K. 2020. Retroviruses drive the rapid evolution of mammalian APOBEC3 genes. Proc Natl Acad Sci U S A 117:610–618. 10.1073/pnas.1914183116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.LaRue RS, Andresdottir V, Blanchard Y, Conticello SG, Derse D, Emerman M, Greene WC, Jonsson SR, Landau NR, Lochelt M, Malik HS, Malim MH, Munk C, O’Brien SJ, Pathak VK, Strebel K, Wain-Hobson S, Yu XF, Yuhki N, Harris RS. 2009. Guidelines for naming nonprimate APOBEC3 genes and proteins. J Virol 83:494–497. 10.1128/JVI.01976-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Conticello SG. 2008. The AID/APOBEC family of nucleic acid mutators. Genome Biol 9:229. 10.1186/gb-2008-9-6-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Conticello SG, Langlois MA, Yang Z, Neuberger MS. 2007. DNA deamination in immunity: AID in the context of its APOBEC relatives. Adv Immunol 94:37–73. 10.1016/S0065-2776(06)94002-4. [DOI] [PubMed] [Google Scholar]
- 23.Jarmuz A, Chester A, Bayliss J, Gisbourne J, Dunham I, Scott J, Navaratnam N. 2002. An anthropoid-specific locus of orphan C to U RNA-editing enzymes on chromosome 22. Genomics 79:285–296. 10.1006/geno.2002.6718. [DOI] [PubMed] [Google Scholar]
- 24.Sheehy AM, Gaddis NC, Choi JD, Malim MH. 2002. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418:646–650. 10.1038/nature00939. [DOI] [PubMed] [Google Scholar]
- 25.Wiegand HL, Doehle BP, Bogerd HP, Cullen BR. 2004. A second human antiretroviral factor, APOBEC3F, is suppressed by the HIV-1 and HIV-2 Vif proteins. EMBO J 23:2451–2458. 10.1038/sj.emboj.7600246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zheng YH, Irwin D, Kurosu T, Tokunaga K, Sata T, Peterlin BM. 2004. Human APOBEC3F is another host factor that blocks human immunodeficiency virus type 1 replication. J Virol 78:6073–6076. 10.1128/JVI.78.11.6073-6076.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Simon V, Zennou V, Murray D, Huang Y, Ho DD, Bieniasz PD. 2005. Natural variation in Vif: differential impact on APOBEC3G/3F and a potential role in HIV-1 diversification. PLoS Pathog 1:e6. 10.1371/journal.ppat.0010006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hultquist JF, Lengyel JA, Refsland EW, LaRue RS, Lackey L, Brown WL, Harris RS. 2011. Human and rhesus APOBEC3D, APOBEC3F, APOBEC3G, and APOBEC3H demonstrate a conserved capacity to restrict Vif-deficient HIV-1. J Virol 85:11220–11234. 10.1128/JVI.05238-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chaurasiya KR, McCauley MJ, Wang W, Qualley DF, Wu T, Kitamura S, Geertsema H, Chan DS, Hertz A, Iwatani Y, Levin JG, Musier-Forsyth K, Rouzina I, Williams MC. 2014. Oligomerization transforms human APOBEC3G from an efficient enzyme to a slowly dissociating nucleic acid-binding protein. Nat Chem 6:28–33. 10.1038/nchem.1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harris RS, Dudley JP. 2015. APOBECs and virus restriction. Virology 479–480:131–145. 10.1016/j.virol.2015.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harris RS, Anderson BD. 2016. Evolutionary paradigms from ancient andongoing conflicts between the lentiviral Vif protein and mammalian APOBEC3 enzymes. PLoS Pathog 12:e1005958. 10.1371/journal.ppat.1005958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Münk C, Willemsen A, Bravo IG. 2012. An ancient history of gene duplications, fusions and losses in the evolution of APOBEC3 mutators in mammals. BMC Evol Biol 12:71. 10.1186/1471-2148-12-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang L, Emerman M, Malik HS, McLaughlin RNJ. 2020. Retrocopying expands the functional repertoire of APOBEC3 antiviral proteins in primates. Elife 9. 10.7554/eLife.58436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O’Leary NA, Wright MW, Brister JR, Ciufo S, Haddad D, McVeigh R, Rajput B, Robbertse B, Smith-White B, Ako-Adjei D, Astashyn A, Badretdin A, Bao Y, Blinkova O, Brover V, Chetvernin V, Choi J, Cox E, Ermolaeva O, Farrell CM, Goldfarb T, Gupta T, Haft D, Hatcher E, et al. 2016. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res 44:D733–D475. 10.1093/nar/gkv1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murrell B, Wertheim JO, Moola S, Weighill T, Scheffler K, Kosakovsky Pond SL. 2012. Detecting individual sites subject to episodic diversifying selection. PLoS Genet 8:e1002764. 10.1371/journal.pgen.1002764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kosakovsky Pond SL, Posada D, Gravenor MB, Woelk CH, Frost SD. 2006. GARD: a genetic algorithm for recombination detection. Bioinformatics 22:3096–3098. 10.1093/bioinformatics/btl474. [DOI] [PubMed] [Google Scholar]
- 37.Refsland EW, Harris RS. 2013. The APOBEC3 family of retroelement restriction factors. Curr Top Microbiol Immunol 371:1–27. 10.1007/978-3-642-37765-5_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dewannieux M, Esnault C, Heidmann T. 2003. LINE-mediated retrotransposition of marked Alu sequences. Nat Genet 35:41–48. 10.1038/ng1223. [DOI] [PubMed] [Google Scholar]
- 39.Wissing S, Montano M, Garcia-Perez JL, Moran JV, Greene WC. 2011. Endogenous APOBEC3B restricts LINE-1 retrotransposition in transformed cells and human embryonic stem cells. J Biol Chem 286:36427–36437. 10.1074/jbc.M111.251058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stenglein MD, Harris RS. 2006. APOBEC3B and APOBEC3F inhibit L1 retrotransposition by a DNA deamination-independent mechanism. J Biol Chem 281:16837–16841. 10.1074/jbc.M602367200. [DOI] [PubMed] [Google Scholar]
- 41.Marchetto MCN, Narvaiza I, Denli AM, Benner C, Lazzarini TA, Nathanson JL, Paquola ACM, Desai KN, Herai RH, Weitzman MD, Yeo GW, Muotri AR, Gage FH. 2013. Differential L1 regulation in pluripotent stem cells of humans and apes. Nature 503:525–529. 10.1038/nature12686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liddament MT, Brown WL, Schumacher AJ, Harris RS. 2004. APOBEC3F properties and hypermutation preferences indicate activity against HIV-1 in vivo. Curr Biol 14:1385–1391. 10.1016/j.cub.2004.06.050. [DOI] [PubMed] [Google Scholar]
- 43.Lee YN, Bieniasz PD. 2007. Reconstitution of an infectious human endogenous retrovirus. PLoS Pathog 3:e10. 10.1371/journal.ppat.0030010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nakano Y, Misawa N, Juarez-Fernandez G, Moriwaki M, Nakaoka S, Funo T, Yamada E, Soper A, Yoshikawa R, Ebrahimi D, Tachiki Y, Iwami S, Harris RS, Koyanagi Y, Sato K. 2017. HIV-1 competition experiments in humanized mice show that APOBEC3H imposes selective pressure and promotes virus adaptation. PLoS Pathog 13:e1006348. 10.1371/journal.ppat.1006348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Etienne L, Bibollet-Ruche F, Sudmant PH, Wu LI, Hahn BH, Emerman M. 2015. The role of the antiviral APOBEC3 gene family in protecting chimpanzees against lentiviruses from monkeys. PLoS Pathog 11:e1005149. 10.1371/journal.ppat.1005149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nakashima M, Ode H, Kawamura T, Kitamura S, Naganawa Y, Awazu H, Tsuzuki S, Matsuoka K, Nemoto M, Hachiya A, Sugiura W, Yokomaku Y, Watanabe N, Iwatani Y. 2016. Structural insights into HIV-1 Vif-APOBEC3F interaction. J Virol 90:1034–1047. 10.1128/JVI.02369-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Siu KK, Sultana A, Azimi FC, Lee JE. 2013. Structural determinants of HIV-1 Vif susceptibility and DNA binding in APOBEC3F. Nat Commun 4:2593. 10.1038/ncomms3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Richards C, Albin JS, Demir Ö, Shaban NM, Luengas EM, Land AM, Anderson BD, Holten JR, Anderson JS, Harki DA, Amaro RE, Harris RS. 2015. The binding interface between human APOBEC3F and HIV-1 Vif elucidated by genetic and computational approaches. Cell Rep 13:1781–1788. 10.1016/j.celrep.2015.10.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Albin JS, LaRue RS, Weaver JA, Brown WL, Shindo K, Harjes E, Matsuo H, Harris RS. 2010. A single amino acid in human APOBEC3F alters susceptibility to HIV-1 Vif. J Biol Chem 285:40785–40792. 10.1074/jbc.M110.173161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bohn MF, Shandilya SM, Albin JS, Kouno T, Anderson BD, McDougle RM, Carpenter MA, Rathore A, Evans L, Davis AN, Zhang J, Lu Y, Somasundaran M, Matsuo H, Harris RS, Schiffer CA. 2013. Crystal structure of the DNA cytosine deaminase APOBEC3F: the catalytically active and HIV-1 Vif-binding domain. Structure 21:1042–1050. 10.1016/j.str.2013.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kitamura S, Ode H, Nakashima M, Imahashi M, Naganawa Y, Kurosawa T, Yokomaku Y, Yamane T, Watanabe N, Suzuki A, Sugiura W, Iwatani Y. 2012. The APOBEC3C crystal structure and the interface for HIV-1 Vif binding. Nat Struct Mol Biol 19:1005–1010. 10.1038/nsmb.2378. [DOI] [PubMed] [Google Scholar]
- 52.Land AM, Shaban NM, Evans L, Hultquist JF, Albin JS, Harris RS. 2014. APOBEC3F determinants of HIV-1 Vif sensitivity. J Virol 88:12923–12927. 10.1128/JVI.02362-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dang Y, Wang X, Esselman WJ, Zheng YH. 2006. Identification of APOBEC3DE as another antiretroviral factor from the human APOBEC family. J Virol 80:10522–10533. 10.1128/JVI.01123-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10:421. 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26:841–842. 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549. 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Crooks GE, Hon G, Chandonia JM, Brenner SE. 2004. WebLogo: a sequence logo generator. Genome Res 14:1188–1190. 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The raw data and computational codes are available from the GitHub repository (https://github.com/TheSatoLab/primate_A3_repertoire_and_evolution).