ABSTRACT
Uropathogenic Escherichia coli (UPEC) is a major pathogen that causes urinary tract infection (UTI). This bacterium adheres to and internalizes within urinary tract cells, where it aggregates and subsequently forms biofilm-like multicellular colonies that protect UPEC from antimicrobial agents and the host’s immune system. Here, we show that OmpX, an outer membrane protein, plays a role in the pathogenesis of UPEC in renal cells. Deletion of ompX decreased bacterial internalization and aggregation within kidney epithelial cells and also impaired the colonization of mouse urinary tracts, but the ompX mutant still adhered to the epithelial cells at a level similar to that of the parent strain. FlhD, the master regulator of flagellum-related genes, had a low expression level in the ompX mutant compared to the parent strain, and the ompX mutant exhibited defective motility due to lower flagellar production than the parent strain. The fliC mutant, which lacks flagella, exhibited lower levels of bacterial internalization and aggregation than the parent strain. Additional deletion of ompX in the fliC mutant did not further decrease bacterial internalization. These combined results suggest that OmpX contributes to flagellar production in UPEC and then sustains UPEC virulence associated with bacterial internalization and aggregation within urinary tract cells and colonization in the urinary tract.
KEYWORDS: virulence, pathogenesis, biofilm, motility, urinary tract infection, pyelonephritis, flagella, gene regulation
INTRODUCTION
Urinary tract infection (UTI) is one of the most common infectious diseases. Uropathogenic Escherichia coli (UPEC) is a major pathogen that is estimated to cause over 80% of uncomplicated UTIs (1, 2). UPEC is categorized in a pathogenic subgroup termed extraintestinal pathogenic E. coli (ExPEC), which causes infectious disease outside the intestines. When UPEC enters the urinary tract, the bacteria adhere to and internalize within bladder epithelial cells, where they aggregate and form biofilm-like microbial colonies, termed intracellular bacterial communities (IBCs) (3). IBCs protect UPEC from antimicrobial agents, which may be associated with the failure of antimicrobial chemotherapy and the recurrence of infection (4, 5). After the bacteria ascend the ureters and reach the kidneys, if chemotherapy fails, this may result in irreversible kidney failure and/or septicemia.
Fimbriae and flagella are major protein structures responsible for the pathogenicity of UPEC. Type 1 fimbriae are required for bacterial adhesion to and internalization within bladder epithelial cells, while P-type fimbriae are proposed to play a role in the pathogenesis of ascending UTIs and pyelonephritis in humans (6–8). Flagella are required for bacterial migration to infection sites as the bacteria colonize the bladder, and flagellum-mediated motility contributes to bacterial fitness and aggregation within bladder epithelial cells (9–11). Flagella also contribute to bacterial entry into renal collecting duct cells, and it has been proposed that they allow the bacteria to ascend from the bladder and initiate kidney infections (12). It has been demonstrated that antibodies against flagella prevent the dissemination of UPEC into the kidneys (13).
OmpX is an outer membrane protein composed of an eight-stranded β-barrel structure with membrane-spanning regions (14). Although this protein was originally described in Enterobacter cloacae, its homolog and paralog proteins have been identified in other Gram-negative pathogens, including E. coli, Salmonella enterica, Yersinia pestis, and Klebsiella pneumoniae (15–19). There are some reports that these proteins are implicated in bacterial virulence. In a study on E. coli, deletion of ompX in an ExPEC strain isolated from the lung of a diseased pig decreased the virulence in systemically infected mice and alveolar basal epithelial cells (20).
We are interested in characterizing proteins that contribute to the pathogenicity of UPEC in the urinary tract. In this study, we show that a UPEC ompX mutant colonized the kidneys of UTI mice and internalized and aggregated within human kidney epithelial cells with lower efficiency than the parent strain but still adhered to the cells at a level similar to that of the parent strain. The ompX mutant exhibited defective motility due to a low level of flagellin expression compared to the parent strain. We also show that the decreased ability of the UPEC ompX mutant to internalize within epithelial cells of the kidney is associated with defective flagellum production and motility.
RESULTS
Deletion of the ompX gene reduces UPEC colonization in the kidneys of mice.
To characterize the role of OmpX in UPEC pathogenesis, we constructed an in-frame deletion mutant of the ompX gene and estimated the abilities of the parent CFT073 strain and its ompX mutant to colonize in the mouse urinary tract. C3H/HeN female mice were transurethrally infected with 1 × 108 CFU of either the ompX mutant or the parent strain, and the titers of bacteria in the bladder and kidneys were determined at 48 h postinfection. Although no statistical difference was observed in the numbers of bacteria in the bladder between the ompX mutant and the parent strain, mice infected with the ompX mutant exhibited lower UPEC burdens in the kidneys than those infected with the parent strain (median numbers of CFU for bladder, 1.8 × 104 for the parent strain and 4.9 × 103 for the ompX mutant; median numbers for kidneys, 4.7 × 104 for the parent strain and 3.4 × 103 for the ompX mutant) (Fig. 1). We confirmed that the reduction of CFU in the mouse kidneys for the ompX mutant was not due to a growth defect because the growth rates of the ompX mutant were essentially the same as those of the parent strain when cultured in LB medium, artificial urine medium (AUM), and RPMI 1640 media (Fig. S1).
FIG 1.
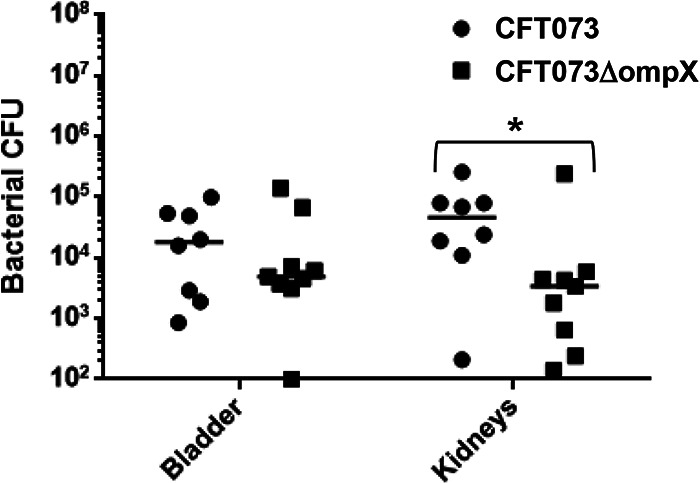
Colonization by the parent strain (CFT073) and ompX mutant in the bladders and kidneys of mice with UTIs. The female mice (n = 3 for each group) were infected with the parent strain or ompX mutant. At 48 h postinfection, cell numbers of bacteria isolated from the bladder and kidneys were determined as CFU. We repeated experiments independently three times, but one mouse infected with the parent strain died within 48 h of infection. Each data point represents a sample from an individual mouse (n = 8 for the parent strain and n = 9 for the ompX mutant). Horizontal bars show median values. *, P < 0.05 relative to the value for the parent strain.
Deletion of the ompX gene reduces bacterial internalization in and aggregation within renal epithelial cells.
From the result of the mouse experiment, we hypothesized that the ompX gene might also contribute to bacterial adhesion to and/or internalization within the kidney cells, and we compared the abilities of the parent strain and the ompX mutant to adhere to and internalize within HTB-44 kidney epithelial cells using a gentamicin protection assay. No significant difference was seen in the total adherence/internalization values between the parent strain and the ompX mutant (2.96 ± 0.38% for the parent strain and 2.80 ± 0.74% for the ompX mutant) (Fig. 2A). However, the ompX mutant exhibited an approximately 3-fold-lower degree of internalization alone than the parent strain (0.020 ± 0.003% for the parent strain and 0.0073 ± 0.0016% for the ompX mutant) (Fig. 2B). We confirmed that the internalization of the ompX mutant was elevated by the introduction of pTrc99KompX, the complementation plasmid for the ompX gene (Fig. 2C). We also characterized UPEC colonies within the kidney epithelial cells and then inoculated the green fluorescent protein (GFP)-expressing parent strain or ompX mutant into HTB-44 cells. The ompX mutant formed smaller and fewer colonies than the parent strain. When the exogenous ompX expression plasmid pTH18krompX was introduced, the mutant formed colonies similar to those of the parent strain (Fig. 3A to C). These observations suggest that the ompX gene is responsible for optimal internalization and aggregation within the kidney epithelial cells.
FIG 2.
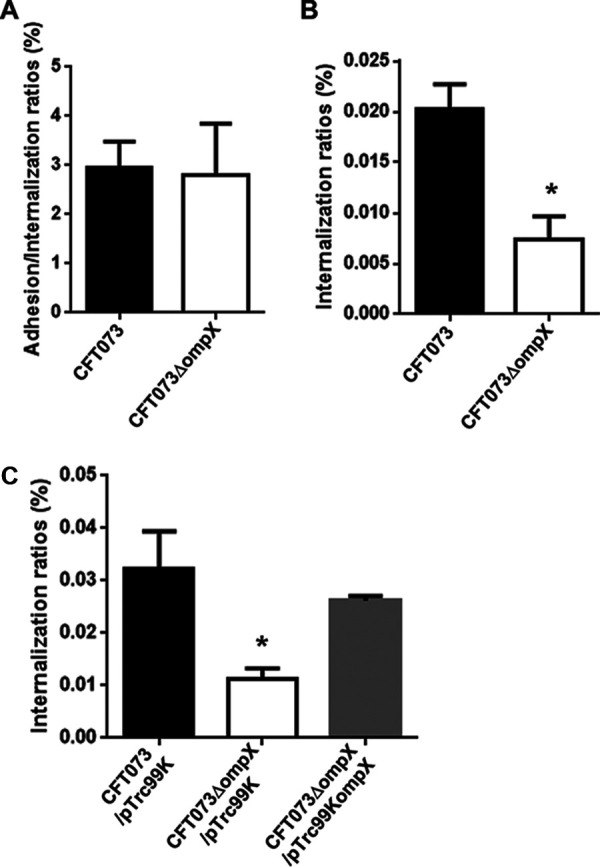
Adhesion to and internalization in kidney epithelial cells (HTB-44) of the parent strain (CFT073) and the ompX mutant (A and B) or the parent strain and the ompX mutant carrying pTrc99K (empty vector) or pTrc99KompX (ompX expression plasmid) (C). Values are percent CFU of adhered/internalized (A) and internalized (B and C) bacteria relative to total bacterial cell numbers. Data are means from three independent experiments; error bars indicate standard deviations. *, P < 0.05 relative to values for CFT073 (A and B) or CFT073/pTrc99K (C).
FIG 3.
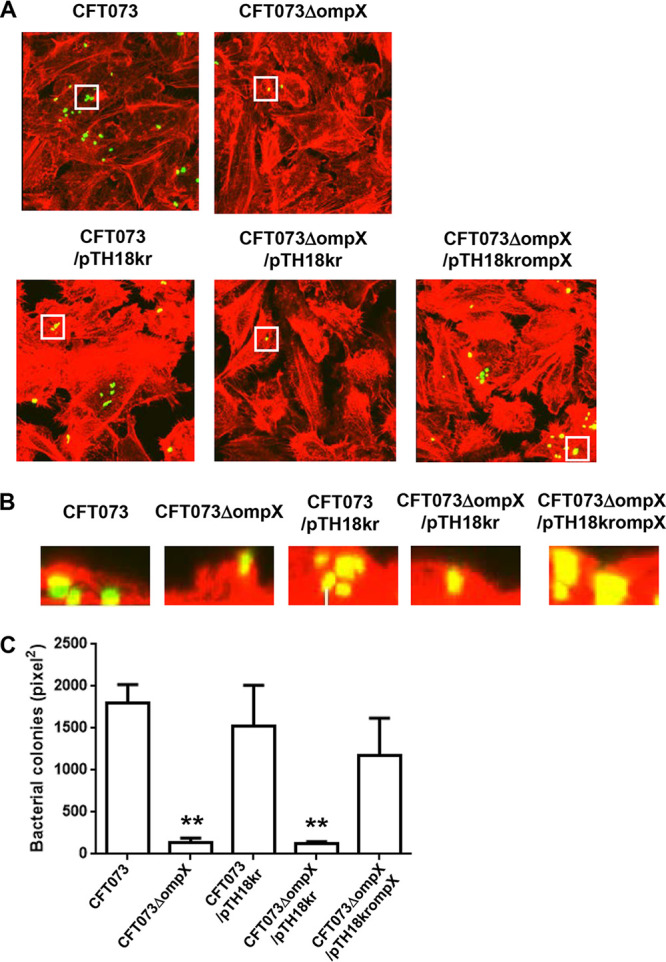
Aggregation within kidney epithelial cells (HTB-44) for the parent strain and the ompX mutant or the parent strain and the ompX mutant carrying pTH18kr (empty vector) or pTH18krompX (ompX expression plasmid). Bacteria carrying a green fluorescence protein (GFP) expression plasmid, pTurboGFP-B, and HTB-44 cells stained with rhodamine-phalloidin were imaged with green and red fluorescence, respectively, using a 100× objective. Images were taken from above (A), and cross-sectional images (B) correspond to the white boxes in panel A. The experiment was repeated twice, and similar results were obtained. (C) Aggregated bacteria within HTB-44 cells were quantified by representing levels of colonized bacteria as areas (in square pixels) of GFP. Microscopy data are means from three fields of view, and error bars indicate standard deviations. **, P < 0.01 relative to the value for the parent strain CFT073.
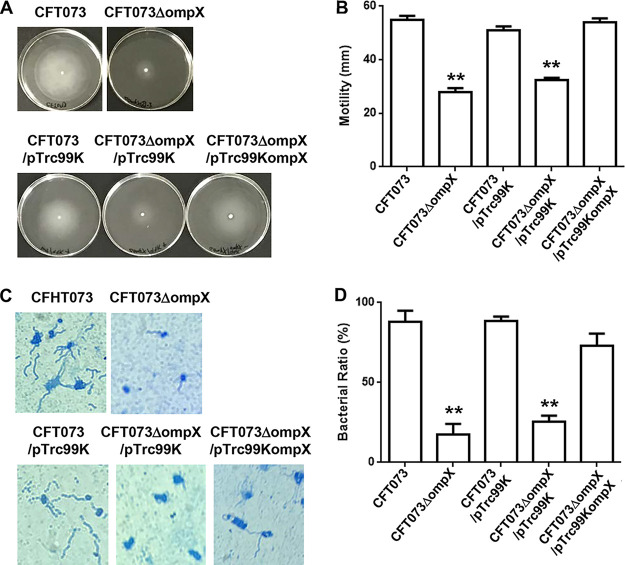
The ompX mutant exhibits lower motility, due to lower flagellar production, than the parent strain but produces P-type and type 1 fimbriae at the same level as the parent strain.
Flagella and flagellum-associated motility contribute to the internalization of UPEC in renal cells, leading to kidney infections (12, 13). Therefore, the reduced bacterial internalization and aggregation within the kidney epithelial cells by ompX deletion may be associated with decreased flagellar production and motility. To test this hypothesis, we compared the flagellar production and motility of the ompX mutant with those of the parent strain. We found that the ompX mutant exhibited lower motility than the parent strain, and decreased motility in the ompX mutant was restored to the parent level by the introduction of pTrc99KompX (Fig. 4A and B). Flagellar staining showed that the ompX mutant produced fewer flagella than the parent strain (Fig. 4C and D).
FIG 4.
Motilities and flagellar production for the parent strain (CFT073) and the ompX mutant or the parent strain and the ompX mutant carrying pTrc99K (empty vector) or pTrc99KompX (ompX expression plasmid). (A) Bacterial migration on LB medium containing 0.3% agar. (B) Diameters reflecting bacterial migration on the agar. Data are means from three independent experiments; error bars indicate standard deviations. (C) Flagella and bacterial cells were stained with Victoria blue/tannic acid were pictured using a 100× objective. (D) Ratios of bacteria observed with flagella to ∼120 to 150 randomly selected bacteria on microscopy, presented as percentages. Data are means, and error bars indicate standard deviations. **, P < 0.01 relative to the value for CFT073.
Flagellin, encoded by fliC, is a major component of flagella (21). We assessed the level of flagellin by Western blotting. At the beginning of the experiment, we used a commercial fliC antibody, but we were unable to detect FliC in the parent strain. For this reason, we constructed strains carrying pTH18krfliC-VSVG, which produce the recombinant vesicular stomatitis virus G protein (VSVG)-tagged FliC protein under an innate fliC promoter on the pTH18kr vector for the parent strain and the ompX mutant. We then detected FliC-VSVG in UPEC cell lysates and secreted fractions by Western blotting with the VSVG antibody. The ompX mutant produced less FliC-VSVG than the parent strain, and the FliC-VSVG level was elevated when pTrc99AompX, the ompX expression plasmid, was introduced (Fig. 5A). We confirmed that the fliC mutant internalized within the kidney epithelial cells with low efficiency compared to the parent strain, and deletion of ompX in the fliC mutant did not further decrease the bacterial internalization rate within kidney epithelial cells (Fig. 5B and C). We also observed that introduction of the fliC expression plasmid elevated the level of internalization to the parent level (Fig. 5D). Like the ompX mutant, the fliC mutant formed small colonies with a low frequency within kidney epithelial cells compared to the parent strain (Fig. 6A to C).
FIG 5.
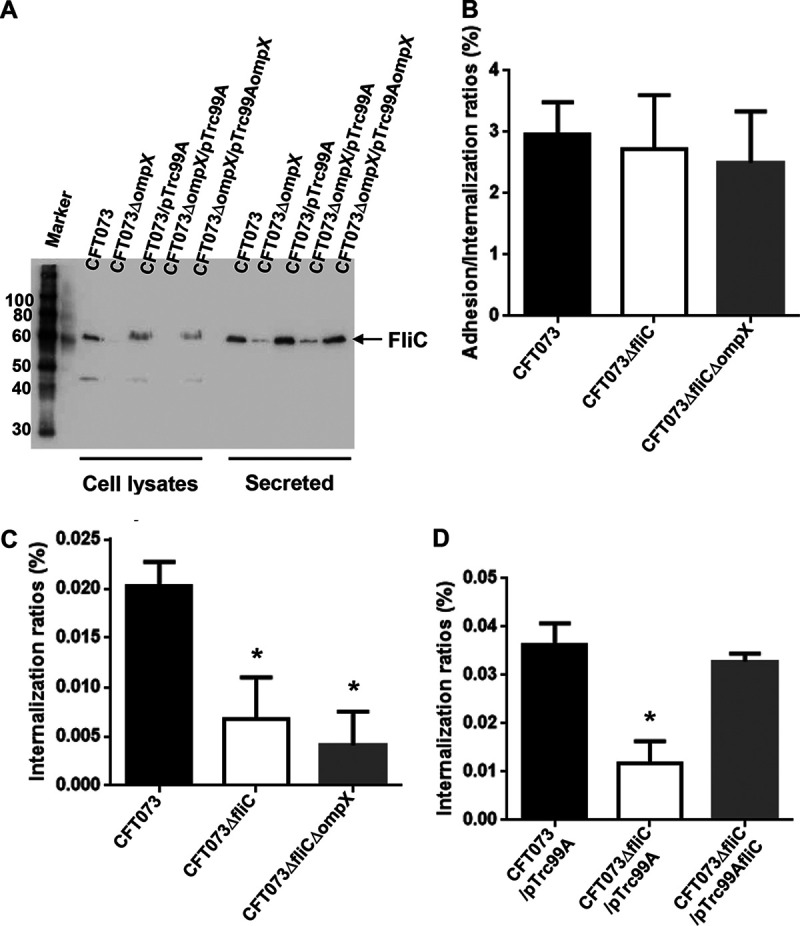
FliC expression in the ompX mutant and contribution of fliC to bacterial adhesion to and internalization within the kidney epithelial cells. (A) Western blots of cell lysates and secreted proteins from the parent strain (CFT073) and the ompX mutant containing a VSVG-tagged FliC expression plasmid (pTH18krfliC-VSVG) and pTrc99A (empty vector) or pTrc99AompX (ompX expression plasmid). Locations of molecular mass standards (in kilodaltons) are shown on the left. VSVG-tagged FliC was visualized by probing with a VSVG antibody. Adhesion to and internalization in kidney epithelial cells (HTB-44) of the parent strain, fliC mutant, and fliC/ompX double mutant (B and C) or the parent strain and the fliC mutant carrying pTrc99A (empty vector) or pTrc99AfliC (fliC expression plasmid) (D). Values are percent CFU of adhered/internalized (B) and internalized (C and D) bacteria relative to total bacterial cell numbers. Data are means from three independent experiments; error bars indicate standard deviations. *, P < 0.05 relative to CFT073 (B and C) or CFT073/pTrc99A (D).
FIG 6.
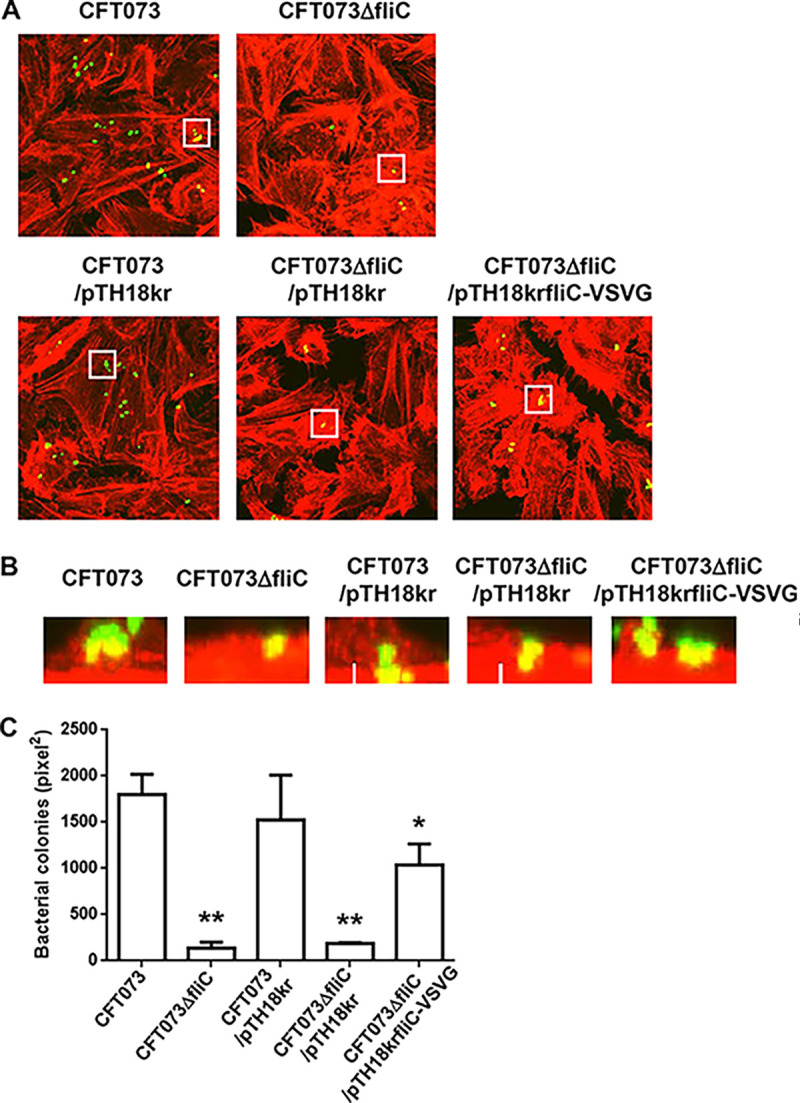
Aggregation within kidney epithelial cells (HTB-44) for the parent strain and the fliC mutant or the parent strain and the fliC mutant carrying pTH18kr (empty vector) or pTH18krfliC-VSVG (fliC expression plasmid). Bacteria carrying a GFP expression plasmid, pTurboGFP-B, and HTB-44 cells stained with rhodamine-phalloidin were imaged with green and red fluorescence, respectively, using a 100× objective. Images were taken from above (A), and cross-sectional images (B) correspond to the white boxes in panel A. The experiment was repeated twice, and similar results were obtained. (C) Aggregated bacteria within HTB-44 cells were quantified by determining levels of colonized bacteria as areas (in square pixels) of GFP. Microscopy data are means from three fields of view, and error bars indicate standard deviations. *, P < 0.05, and **, P < 0.01, relative to CFT073.
The P-type fimbria is the second most common fimbria produced by UPEC CFT073 and is thought to be closely associated with pyelonephritis in humans (7, 8). We measured the transcription levels of papA, which encodes a P-type fimbrial component, and found no apparent difference in papA levels between the parent strain and the ompX mutant (Fig. 7A). We also measured the transcription levels of fimA, which encodes a major component of the type 1 fimbria, the most common fimbria, and fimbrial activity by determining agglutination titers of guinea pig erythrocytes. However, no apparent difference was observed between the parent strain and the ompX mutant in fimA transcript levels or agglutination titers (64 for both the parent strain and the ompX mutant) (Fig. 7A).
FIG 7.
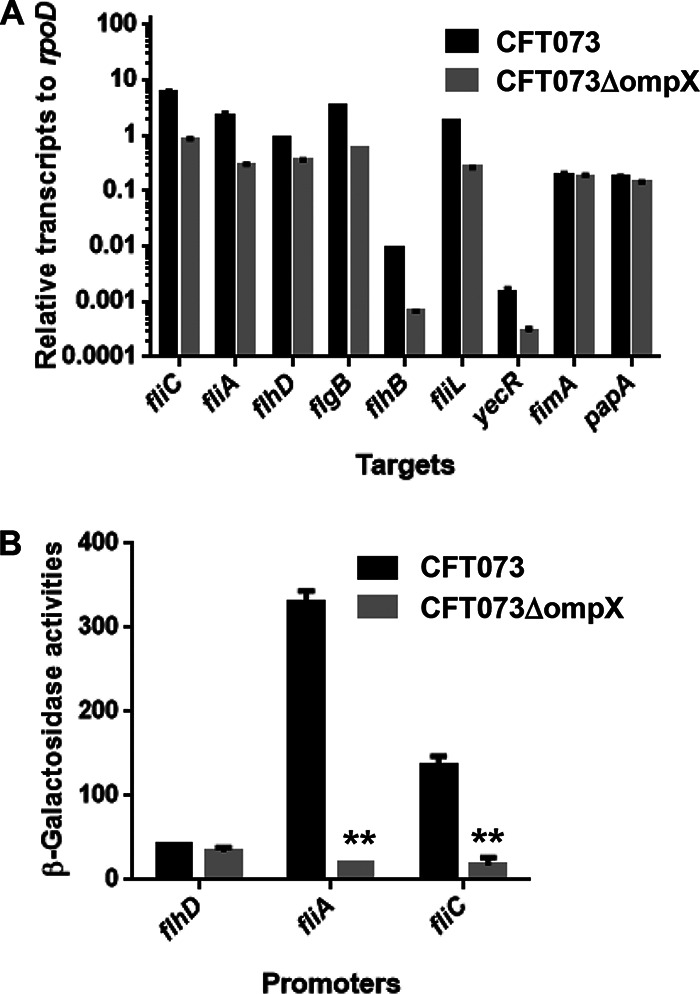
Transcript levels and promoter activities of flagellum-related and fimbrial genes in the parent strain (CFT073) and the ompX mutant. (A) Transcript levels were determined relative to that of rpoD. Data are means for two biological replicates; error bars indicate the ranges. (B) β-Galactosidase activities corresponding to flhD, fliA, and fliC promoter activities in the parent strain and the ompX mutant containing pNNflhD-P, pNNfliA-P, or pNNfliC-P, the lacZ reporter plasmid. Data are means from three independent experiments; error bars indicate standard deviations. **, P < 0.01 relative to CFT073.
These observations suggest that defective internalization and aggregation in the ompX mutant involve lower levels of flagellar production than those in the parent strain, as the ompX mutant produces P-type and type 1 fimbriae at the same level as the parent strain.
Deletion of ompX decreases transcription of flhD, which encodes a master regulator for flagellar expression, and leads to a reduction in flagellar production.
To address how the deletion of ompX decreases flagellar production, we investigated the mechanism of flagellin expression. fliC gene expression is activated by FliA, a flagellar biosynthesis sigma factor, and the fliA gene is activated by FlhD, a master regulator of flagellar expression (22). We measured the transcription levels of fliA and flhD in addition to fliC using quantitative PCR (qPCR) and found that fliA and flhD levels in the ompX mutant were lower than those in the parent strain (Fig. 7A). FlhD also activates the transcription of genes that encode the flagellar components FlgB, FlhB, and FliL and the gene yecR, which is not related to flagellar production (23, 24). As with fliA, the transcription levels of these genes in the ompX mutant were lower than those in the parent strain (Fig. 7A). This observation suggests that the ability of FlhD to activate transcription of fliA, followed by induction of FliC expression, is decreased by ompX deletion. We also compared the promoter activities of these genes via lacZ expression from the reporter plasmids pNNfliC-P, pNNfliA-P, and pNNflhD-P between the ompX mutant and the parent strain. The promoter activities of fliC and fliA corresponding to LacZ expression from pNNfliC-P and pNNfliA-P, respectively, in the ompX mutant were lower than those in the parent strain; however, no significant difference in flhD levels was observed between the two strains (Fig. 7B).
These combined results suggest that the ompX gene contributes to maintaining transcription of flhD but does not affect promoter activity.
Deletion of ompX does not impair the outer membrane.
Since OmpX is an outer membrane protein, its defect may compromise outer membrane integrity, possibly contributing to the attenuated virulence of the ompX mutant. We investigated the susceptibility of the ompX mutant and the parent strain to sodium deoxycholate and SDS by MIC assays to test this hypothesis. There was no significant difference in susceptibility to these reagents between the parent strain and the ompX mutant (MICs of sodium deoxycholate were 25,600 μg/ml and MICs of SDS were 32,800 μg/ml for both the parent strain and the ompX mutant). Therefore, the outer membrane is unlikely to be impaired by deletion of the ompX gene.
DISCUSSION
The roles of OmpX in bacterial pathogenesis have been characterized in some pathogens, such as S. enterica, Y. pestis, K. pneumoniae, and E. coli including ExPEC. Deletion of ompX in these bacterial species attenuated virulence (15–20). One E. coli study suggested that the ompX gene contributes to the virulence of pig lung disease-related ExPEC in the respiratory system and during systemic infection, because deletion of ompX decreased mouse mortality and bacterial distribution in systemically infected mice and decreased adhesion to and invasion of alveolar basal epithelial cells (20).
In this study, we investigated the role of OmpX in the pathogenesis of UPEC. A previous study, which used the UPEC strain J96, reported that the ompX gene was not required for bacterial adhesion to host laryngeal epithelial cells (15). Our study, which used the UPEC strain CFT073 and urinary tract cells, suggests that the ompX gene is unlikely to contribute to bacterial adhesion. However, we found evidence that an ompX gene is required for optimal internalization and aggregation within kidney epithelial cells. In addition, we found that the ompX mutant exhibited defective motility due to a low level of flagellar production, which is associated with decreased levels of internalization and aggregation, because a nonflagellar fliC mutant also exhibited low levels of internalization and aggregation, and deletion of ompX did not further reduce these levels. Thus, we suggest that OmpX is implicated in the flagellum-associated pathogenesis of UPEC in the urinary tract. In another study, a fliC mutant was shown to exhibit a growth disadvantage in the urinary tracts of mice when it was inoculated with the parent strain (in which fliC is intact); however, the mutant still colonized at a rate similar to that of the parent strain in solo infection experiments (9, 10). P-type fimbriae are thought to contribute to virulence in the kidney (7, 8). However, our qPCR analyses indicated that OmpX was unlikely to be associated with the expression of the papA gene. Therefore, the decreased rate of kidney colonization observed in the mice infected with the ompX mutant does not seem to be associated with defects in either flagella or P-type fimbriae, implying that there are still unknown factors that participate in OmpX pathogenicity.
Flagellum expression is activated by the transcriptional regulator FlhD (22). The ompX mutant exhibited a low level of flhD transcription compared to the parent strain; therefore, we concluded that the decrease in flagellum production in the ompX mutant was caused by reduced FlhD expression. However, the promoter activity of flhD in the ompX mutant was similar to that of the parent strain. It remains unclear how the deletion of ompX reduces the transcription level of flhD. The ompX gene may contribute to the stability of flhD mRNA, perhaps by protecting flhD mRNA from RNase activity, either directly or via RNA binding proteins.
Distinctive differences have been observed between some E. coli strains regarding the effects of ompX deletion on flagellum-associated motility and type 1 fimbrial production (20, 25). The deletion of ompX in a pig lung disease-related ExPEC strain attenuated virulence in systemically infected mice and promoted motility (20). We do not know why the effect of ompX deletion on motility in this ExPEC strain was inconsistent with results for UPEC. Expression of flagellum-related genes is regulated by many regulatory proteins, including CRP, LrhA, H-NS, QseB, RcsB, OmpR, and CytR, in addition to FlhD (26–32). These proteins activate or repress flagellar gene expression, either directly or via FlhD. Several outer membrane proteins, including OmpX, affect the activity of other proteins, such as RpoE (15, 33). Therefore, the activity of repressor proteins produced by this ExPEC strain, but not UPEC, may be reduced by ompX deletion; this reduction may then be overcome by FlhD expression in ExPEC. One study demonstrated that deletion of ompX in a nonpathogenic K-12 strain, producing type 1 fimbriae, decreased motility, while it did not decrease motility in a strain that lacked type 1 fimbriae (25). This implies that type 1 fimbria production may affect flagellar expression and motility by deletion of ompX. Another study found that type 1 fimbria production was promoted in the ompX mutant of the nonpathogenic K-12 strain, while we observed no significant difference in type 1 fimbria production in UPEC (25). Similar to the case of motility, the inconsistent effects of ompX deletion on type 1 fimbria production may be due to the different regulatory mechanisms of the fim genes, which encode type 1 fimbria proteins, in UPEC and nonpathogenic K-12 strains. OmpX has been proposed as a potential target for the treatment of Yersinia infections (34). To expand this idea to E. coli infections, extensive information related to the regulatory mechanism of type 1 fimbrial and flagellar expression, together with proteins affected by OmpX, would be necessary.
OmpX is a member of the outer membrane protein family having an eight-stranded β-barrel structure, which is closely related to the structure of OmpA (14). However, the roles of OmpA in UPEC pathogenicity differ from those of OmpX. Deleting ompA impaired colony formation in UTI in mice; however, the ompA mutant retained motility and the ability to aggregate within bladder epithelial cells at a level similar to that of the strain with intact ompA (11, 35). Although OmpX and OmpA share highly conserved transmembrane domain structures, OmpX lacks a periplasmic domain, while OmpA contains one. Therefore, this periplasmic domain may participate in determining the roles of these proteins in pathogenicity.
In conclusion, we have characterized the roles of OmpX in E. coli pathogenesis and flagellum expression associated with UPEC virulence. This study provides an insight into the molecular mechanisms involved in the virulence of UPEC.
MATERIALS AND METHODS
Bacterial strains, host cells, and culture conditions.
The bacterial strains and plasmids used in this study are listed in Table 1.Bacteria were grown in Luria-Bertani (LB) medium. Optical density at 600 nm (OD600) was measured as an indicator of cell growth. Antibiotics were added to the growth medium for marker selection and maintenance of plasmids at the following concentrations: 45 μg/ml chloramphenicol, 50 μg/ml kanamycin, and 150 μg/ml ampicillin. HTB-44 kidney epithelial cells were cultured as previously described (36).
TABLE 1.
Strains and plasmids used in this studya
| Strain or plasmid | Relevant genotype/phenotype | Reference |
|---|---|---|
| Strains | ||
| CFT073 | Parent strain (ATCC 700928) | ATCC 700928 |
| CFT073ΔompX | ompX mutant of CFT073 | This work |
| CFT073ΔfliC | fliC mutant of CFT073 | 11 |
| CFT073ΔfliCΔompX | fliC and ompX double mutant of CFT073 | This work |
| Plasmids | ||
| pKO3 | Temperature-sensitive vector for gene targeting, sacB, Cmr | 37 |
| pTrc99K | Vector for IPTG-inducible expression; Kmr | 38 |
| pTrc99KompX | ompX expression plasmid; Kmr | This work |
| pTrc99A | Vector for IPTG-inducible expression; Apr | 38 |
| pTrc99AompX | ompX expression plasmid; Apr | This work |
| pTrc99AfliC | fliC expression plasmid; Apr | This work |
| pTH18kr | Low-copy-no. plasmid; Kmr | 39 |
| pTH18krfliC-VSVG | C-terminally VSVG-tagged FliC expression plasmid; Kmr | This work |
| pTH18krompX | ompX expression plasmid; Kmr | This work |
| pNN387 | Single-copy plasmid with promoterless lacZ; Cmr | 40 |
| pNNflhD-P | flhD promoter reporter; Cmr | This work |
| pNNfliA-P | fliA promoter reporter; Cmr | This work |
| pNNfliC-P | fliC promoter reporter; Cmr | This work |
| pTurboGFP-B | GFP expression plasmid; Apr | Evrogen |
Cmr, chloramphenicol resistance; Kmr, kanamycin resistance; Apr, ampicillin resistance.
Cloning and mutant construction.
The ompX gene deletion was produced by sequence overlap extension PCR, as previously described (37), using the primer pairs ompX-delta1/ompX-delta2 and ompX-delta3/ompX-delta4, as shown in Table 2. The upstream flanking DNA comprised 450 bp and the first four amino acid codons. The downstream flanking DNA comprised the last two amino acid codons, the stop codon, and 450 bp of DNA. This deletion construct was ligated into the temperature-sensitive plasmid pKO3 (37) and introduced into UPEC strains. We selected sucrose-resistant chloramphenicol-sensitive colonies at 30°C. To construct the OmpX expression plasmids pTrc99KompX and pTrc99AompX, we amplified the ompX gene by PCR using primer pairs (Table 2) and ligated the product into the pTrc99K and pTrc99A vectors (38). A FliC expression plasmid, pTrc99AfliC, was constructed by PCR amplification of the fliC gene and ligation of its product into the pTrc99A vector (38).
TABLE 2.
Primers used in this study
| Primer | DNA sequence (5′-3′) | Use |
|---|---|---|
| ompX-delta1 | GCGGGATCCGACTTAGCTAACGAGGCTCC | ompX mutant construction |
| ompX-delta2 | ATATCACCGAAGTGATTAGAAGCGAATTTTTTTCATAACCACCTC | ompX mutant construction |
| ompX-delta3 | TTTGAGGTGGTTATGAAAAAAATTCGCTTCTAATCACTTCGGTG | ompX mutant construction |
| ompX-delta4 | GCGGTCGACAAACAGACGATTTACTGCGC | ompX mutant construction |
| pTrcompX-F | GCGCCATGGAAAAAATTGCATGTCTTTC | pTrc99K/pTrc99AompX constructions |
| pTrcompX-R | GCGGGATCCTTAGAAGCGGTAACCAACACC | pTrc99K/pTrc99AompX constructions |
| pTrcfliC-F | GCGCCATGGCACAAGTCATTAATACC | pTrc99AfliC construction |
| pTrcfliC-R | GCGGTCGACTTAACCCTGCAGCAGAGAC | pTrc99AfliC construction |
| pTHfliC-F | GCGAAGCTTCCTGACCCGACTCCCAGCG | pTH18krfliC-VSVG construction |
| pTHfliC-VSVG-R | GCGGGATCCTTATTTTCCTAATCTATTCATTTCAATATCTGTATAACCCTGCAGCAGAGACAG | pTH18krfliC-VSVG construction |
| pTHompX-F | GCGGGATCCCCCGTTTTGACTAAAATGCG | pTH18krompX construction |
| pTHompX-R | GCGAAGCTTTTAGAAGCGGTAACCAACACC | pTH18krompX construction |
| flhD-PF | GCGGCGGCCGCTGCGGTTAATCTCCCGTAAG | pNNflhD-P construction |
| flhD-PR | GCGAGTACTTATTCCCACCCAGAATAACC | pNNflhD-P construction |
| fliA-PF | GCGGCGGCCGCCTGAGACTGACGGCAACGCC | pNNfliA-P construction |
| fliA-PR | GCGAAGCTTCACGATAAACAGCCCTGCG | pNNfliA-P construction |
| fliC-PF | GCGGCGGCCGCCCTGACCCGACTCCCAGCG | pNNfliC-P construction |
| fliC-PR | GCGAAGCTTGATTCGTTATCCTATATTGC | pNNfliC-P construction |
| rrsA-qPCR-F | CGGTGGAGCATGTGGTTTAA | Quantitative real-time PCR |
| rrsA-qPCR-R | GAAAACTTCCGTGGATGTCAAGA | Quantitative real-time PCR |
| rpoD-qPCR-F | CAAGCCGTGGTCGGAAAA | Quantitative real-time PCR |
| rpoD-qPCR-R | GGGCGCGATGCACTTCT | Quantitative real-time PCR |
| fimA-qPCR-F | TGCGGGTAGCGCAACAA | Quantitative real-time PCR |
| fimA-qPCR-R | ACGCAGTCCCTGTTTTATCCA | Quantitative real-time PCR |
| papA-qPCR-F | TTTTTCGGGTGTCCCAAGTG | Quantitative real-time PCR |
| papA-qPCR-R | TGTTGCACCGACGGTCTGT | Quantitative real-time PCR |
| flhD-qPCR-F | GACAACGTTAGCGGCACTGA | Quantitative real-time PCR |
| flhD-qPCR-R | TTGATTGGTTTCTGCCAGCTT | Quantitative real-time PCR |
| fliA-qPCR-F | CGAGCGTGGAACTTGACGAT | Quantitative real-time PCR |
| fliA-qPCR-R | CGACGGCATTAAGTAACCCAAT | Quantitative real-time PCR |
| fliC-qPCR-F | TCCACTGAAAGCTCTGGATGAA | Quantitative real-time PCR |
| fliC-qPCR-R | CCCAGGGATGAACGGAATT | Quantitative real-time PCR |
| flgB-qPCR-F | TCAGGCTCGCGATATCGATT | Quantitative real-time PCR |
| flgB-qPCR-R | CCGTCCACGTTGCATGACT | Quantitative real-time PCR |
| flhB-qPCR-F | TCGCGCTGCGCATTC | Quantitative real-time PCR |
| flhB-qPCR-R | TTCAAGCGTCGGGACGTTA | Quantitative real-time PCR |
| fliL-qPCR-F | ACTGGCATTCGCATCAGGTT | Quantitative real-time PCR |
| fliL-qPCR-R | GGCACGACGCGTTGCT | Quantitative real-time PCR |
| yecR-qPCR-F | GACGCGGCAACAGGTATTGT | Quantitative real-time PCR |
| yecR-qPCR-R | CGGGCATGCTGCAAAAA | Quantitative real-time PCR |
To construct a C-terminal vesicular stomatitis virus glycoprotein (VSVG)-tagged FliC expression plasmid, pTH18krfliC-VSVG, the DNA containing the fliC coding region and its 300-bp upstream region was amplified with pTHfliC-F and pTHfliC-VSVG-R primers and ligated into the HindIII and BamHI sites in pTH18kr (39), the low-copy-number plasmid. This DNA fragment contains a cis-regulatory element for fliC gene expression. In addition, although the pTH18kr vector has a lac promoter sequence upstream of the HindIII and BamHI sites, the introduced fliC gene is oriented in a reverse direction relative to the lac promoter. Therefore, we expected that the resulting bacterial construct would produce FliC as a C-terminal VSVG-tagged protein from its native promoter.
The pHT18krompX plasmid was constructed by amplifying ompX and the 263-bp upstream region including the promoter and ligating the product into pTH18kr. Since this plasmid is compatible with pTurboGFP-B, these plasmids were introduced together into the ompX mutant to characterize bacterial colonies within kidney epithelial cells using fluorescent images. We also constructed pNNflhD-P, pNNfliA-P, and pNNfliC-P, lacZ reporter plasmids to measure flhD, fliA, and fliC promoter activities. We PCR amplified the 1,200-bp upstream region of flhD and the 300-bp upstream regions of fliA and fliC, respectively, and ligated these products into pNN387 (40) with a promoterless lacZ. All constructs were confirmed by DNA sequencing.
Urinary tract infections in mice.
We estimated UPEC virulence using a UTI mouse model as previously described (11). Bacterial suspensions in phosphate-buffered saline (PBS) (1 × 108 CFU) were administered to 8-week-old C3H/HeN female mice via transurethral catheterization. The numbers of CFU in the bladder and kidneys 48 h postinfection were determined by counting colonies grown on XM-G agar. All animal studies were approved by the Animal Research Committee of Gunma University (approval number 19-094).
Infection of kidney epithelial cells.
UPEC cell adhesion and internalization within HTB-44 cells were assessed using a gentamicin protection assay as previously described (41). The numbers of adherent and/or internalized bacterial cells were determined as ratios of CFU (as percentages) to total cell CFU. We also imaged the bacteria in HTB-44 cells using confocal microscopy, as previously described (41). A UPEC strain carrying a green fluorescence protein (GFP) expression plasmid, pTurboGFP-B (Evrogen, Moscow, Russia), was inoculated into cultured HTB-44 cells and incubated for 2 h. Noninternalized bacteria were washed out with gentamicin and PBS+ (PBS containing 0.5 mM MgCl2 and 1 mM CaCl2). The HTB-44 cells were stained with rhodamine-phalloidin (Life Technologies, Carlsbad, CA, USA). Fluorescent images were acquired using an Olympus FV1200 IX81 microscope and processed using FV10-ASW software (Olympus Corp., Tokyo, Japan).
Hemagglutination assays.
To estimate the activity of type 1 fimbriae, we tested the hemagglutination of guinea pig red blood cells as previously described (41).
Motility assays.
To evaluate bacterial motility, the overnight cultures were spotted onto LB medium containing 0.3% agar and grown for 8 h at 37°C under an atmosphere of 5% CO2.
Flagellum staining.
Bacteria were cultured for 24 h at 30°C in heart Infusion medium containing 1.5% agar. Flagella were stained with Victoria blue/tannic acid solution as previously described (11).
RNA extraction and quantitative real-time PCR.
We grew bacteria to late logarithmic growth phase (OD600, ∼0.5). Total-RNA extraction, cDNA synthesis, and real-time PCR were carried out as previously described (11). The constitutively expressed rrsA and rpoD genes were used as internal controls.
Western blotting.
To detect VSVG-tagged FliC from UPEC, bacteria were grown to early stationary phase and separated by centrifugation. The cell pellets were resuspended in 50 mM phosphate buffer containing 8 M urea and then lysed by sonication. Secreted proteins were precipitated from the supernatants with 10% trichloroacetic acid (TCA) and dissolved in Laemmli sample buffer (Bio-Rad Laboratories, Hercules, CA). Cell lysates (7.5 μg) and secreted proteins were separated on duplicate 10% acrylamide Tris-glycine SDS-PAGE gels. One gel was stained with Coomassie brilliant blue stain (CBB), and the other was electroblotted onto a polyvinylidene fluoride (PVDF) membrane (Bio-Rad Laboratories, Hercules, CA). VSVG-tagged FliC was detected with VSVG antibody (Sigma Chemical) and an anti-rabbit horseradish peroxidase-conjugated immunoglobulin G (IgG) secondary antibody (Sigma-Aldrich Co. LLC., St. Louis, MO) using a SuperSignal West Pico kit (Thermo Fisher Scientific, Waltham, MA). VSVG-tagged FliC protein bands were visualized using a LAS-4010 luminescent image analyzer (GE Healthcare Japan, Tokyo).
Promoter assay.
UPEC strains carrying pNNflhD-P, pNNfliA-P, or pNNfliC-P, the LacZ reporter plasmid, were grown at 37°C in LB medium. β-Galactosidase activities from lacZ expression in cell lysates were determined by Miller’s method (42).
Deoxycholate and SDS susceptibility assays.
Susceptibility of bacteria to deoxycholate and SDS was estimated by the MIC assay using a serial agar dilution method. Five microliters of 100-fold-diluted overnight cultures (∼50,000 cells) was inoculated onto a LB agar plate containing sodium deoxycholate and SDS and incubated for 16 h at 37°C. The MICs were determined as the lowest concentrations at which growth was inhibited.
Statistical analysis.
The P value in each assay except UTI mouse experiments was determined by the unpaired t test with GraphPad Prism version 6.00. To determine P value in the UTI mouse experiment, we used the Mann-Whitney test data with GraphPad Prism version 6.00.
ACKNOWLEDGMENTS
This study was kindly supported by JSPS KAKENHI “Grant-in-Aid for Scientific Research (C)” grant number 19K07533 and Research Program on Emerging and Re-emerging Infectious Diseases from Japan Agency Research and Development, AMED grant number 21fk0108604h0901.
Footnotes
Supplemental material is available online only.
iai.00721-20-s0001.pdf (90.3KB, pdf)
Contributor Information
Hidetada Hirakawa, Email: hirakawa@gunma-u.ac.jp.
Igor E. Brodsky, University of Pennsylvania
REFERENCES
- 1.Hayami H, Takahashi S, Ishikawa K, Yasuda M, Yamamoto S, Wada K, Kobayashi K, Hamasuna R, Minamitani S, Matsumoto T, Kiyota H, Tateda K, Sato J, Hanaki H, Masumori N, Nishiyama H, Miyazaki J, Fujimoto K, Tanaka K, Uehara S, Matsubara A, Ito K, Hayashi K, Kurimura Y, Ito S, Takeuchi T, Narita H, Izumitani M, Nishimura H, Kawahara M, Hara M, Hosobe T, Takashima K, Chokyu H, Matsumura M, Ihara H, Uno S, Monden K, Sumii T, Kawai S, Kariya S, Sato T, Yoshioka M, Kadena H, Matsushita S, Nishi S, Hosokawa Y, Shirane T, Yoh M, Watanabe S, et al. 2019. Second nationwide surveillance of bacterial pathogens in patients with acute uncomplicated cystitis conducted by Japanese Surveillance Committee from 2015 to 2016: antimicrobial susceptibility of Escherichia coli, Klebsiella pneumoniae, and Staphylococcus saprophyticus. J Infect Chemother 25:413–422. 10.1016/j.jiac.2019.02.021. [DOI] [PubMed] [Google Scholar]
- 2.Zhang L, Foxman B. 2003. Molecular epidemiology of Escherichia coli mediated urinary tract infections. Front Biosci 8:e235–e244. 10.2741/1007. [DOI] [PubMed] [Google Scholar]
- 3.Anderson GG, Palermo JJ, Schilling JD, Roth R, Heuser J, Hultgren SJ. 2003. Intracellular bacterial biofilm-like pods in urinary tract infections. Science 301:105–107. 10.1126/science.1084550. [DOI] [PubMed] [Google Scholar]
- 4.Mulvey MA, Lopez-Boado YS, Wilson CL, Roth R, Parks WC, Heuser J, Hultgren SJ. 1998. Induction and evasion of host defenses by type 1-piliated uropathogenic Escherichia coli. Science 282:1494–1497. 10.1126/science.282.5393.1494. [DOI] [PubMed] [Google Scholar]
- 5.Mulvey MA, Schilling JD, Hultgren SJ. 2001. Establishment of a persistent Escherichia coli reservoir during the acute phase of a bladder infection. Infect Immun 69:4572–4579. 10.1128/IAI.69.7.4572-4579.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martinez JJ, Mulvey MA, Schilling JD, Pinkner JS, Hultgren SJ. 2000. Type 1 pilus-mediated bacterial invasion of bladder epithelial cells. EMBO J 19:2803–2812. 10.1093/emboj/19.12.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lane MC, Mobley HL. 2007. Role of P-fimbrial-mediated adherence in pyelonephritis and persistence of uropathogenic Escherichia coli (UPEC) in the mammalian kidney. Kidney Int 72:19–25. 10.1038/sj.ki.5002230. [DOI] [PubMed] [Google Scholar]
- 8.Lillington J, Geibel S, Waksman G. 2014. Biogenesis and adhesion of type 1 and P pili. Biochim Biophys Acta 1840:2783–2793. 10.1016/j.bbagen.2014.04.021. [DOI] [PubMed] [Google Scholar]
- 9.Wright KJ, Seed PC, Hultgren SJ. 2005. Uropathogenic Escherichia coli flagella aid in efficient urinary tract colonization. Infect Immun 73:7657–7668. 10.1128/IAI.73.11.7657-7668.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lane MC, Lockatell V, Monterosso G, Lamphier D, Weinert J, Hebel JR, Johnson DE, Mobley HL. 2005. Role of motility in the colonization of uropathogenic Escherichia coli in the urinary tract. Infect Immun 73:7644–7656. 10.1128/IAI.73.11.7644-7656.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hirakawa H, Suzue K, Kurabayashi K, Tomita H. 2019. The Tol-Pal system of uropathogenic Escherichia coli is responsible for optimal internalization into and aggregation within bladder epithelial cells, colonization of the urinary tract of mice, and bacterial motility. Front Microbiol 10:1827. 10.3389/fmicb.2019.01827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pichon C, Hechard C, Du Merle L, Chaudray C, Bonne I, Guadagnini S, Vandewalle A, Le Bouguenec C. 2009. Uropathogenic Escherichia coli AL511 requires flagellum to enter renal collecting duct cells. Cell Microbiol 11:616–628. 10.1111/j.1462-5822.2008.01278.x. [DOI] [PubMed] [Google Scholar]
- 13.Schwan WR. 2008. Flagella allow uropathogenic Escherichia coli ascension into murine kidneys. Int J Med Microbiol 298:441–447. 10.1016/j.ijmm.2007.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vogt J, Schulz GE. 1999. The structure of the outer membrane protein OmpX from Escherichia coli reveals possible mechanisms of virulence. Structure 7:1301–1309. 10.1016/s0969-2126(00)80063-5. [DOI] [PubMed] [Google Scholar]
- 15.Mecsas J, Welch R, Erickson JW, Gross CA. 1995. Identification and characterization of an outer membrane protein, OmpX, in Escherichia coli that is homologous to a family of outer membrane proteins including Ail of Yersinia enterocolitica. J Bacteriol 177:799–804. 10.1128/jb.177.3.799-804.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heffernan EJ, Harwood J, Fierer J, Guiney D. 1992. The Salmonella typhimurium virulence plasmid complement resistance gene rck is homologous to a family of virulence-related outer membrane protein genes, including pagC and ail. J Bacteriol 174:84–91. 10.1128/jb.174.1.84-91.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kolodziejek AM, Hovde CJ, Minnich SA. 2012. Yersinia pestis Ail: multiple roles of a single protein. Front Cell Infect Microbiol 2:103. 10.3389/fcimb.2012.00103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kolodziejek AM, Schnider DR, Rohde HN, Wojtowicz AJ, Bohach GA, Minnich SA, Hovde CJ. 2010. Outer membrane protein X (Ail) contributes to Yersinia pestis virulence in pneumonic plague and its activity is dependent on the lipopolysaccharide core length. Infect Immun 78:5233–5243. 10.1128/IAI.00783-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Climent N, Ferrer S, Rubires X, Merino S, Tomas JM, Regue M. 1997. Molecular characterization of a 17-kDa outer-membrane protein from Klebsiella pneumoniae. Res Microbiol 148:133–143. 10.1016/S0923-2508(97)87644-9. [DOI] [PubMed] [Google Scholar]
- 20.Meng X, Liu X, Zhang L, Hou B, Li B, Tan C, Li Z, Zhou R, Li S. 2016. Virulence characteristics of extraintestinal pathogenic Escherichia coli deletion of gene encoding the outer membrane protein X. J Vet Med Sci 78:1261–1267. 10.1292/jvms.16-0071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Minamino T, Imada K. 2015. The bacterial flagellar motor and its structural diversity. Trends Microbiol 23:267–274. 10.1016/j.tim.2014.12.011. [DOI] [PubMed] [Google Scholar]
- 22.Soutourina OA, Bertin PN. 2003. Regulation cascade of flagellar expression in Gram-negative bacteria. FEMS Microbiol Rev 27:505–523. 10.1016/S0168-6445(03)00064-0. [DOI] [PubMed] [Google Scholar]
- 23.Fitzgerald DM, Bonocora RP, Wade JT. 2014. Comprehensive mapping of the Escherichia coli flagellar regulatory network. PLoS Genet 10:e1004649. 10.1371/journal.pgen.1004649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Macnab RM. 1992. Genetics and biogenesis of bacterial flagella. Annu Rev Genet 26:131–158. 10.1146/annurev.ge.26.120192.001023. [DOI] [PubMed] [Google Scholar]
- 25.Otto K, Hermansson M. 2004. Inactivation of ompX causes increased interactions of type 1 fimbriated Escherichia coli with abiotic surfaces. J Bacteriol 186:226–234. 10.1128/jb.186.1.226-234.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Soutourina O, Kolb A, Krin E, Laurent-Winter C, Rimsky S, Danchin A, Bertin P. 1999. Multiple control of flagellum biosynthesis in Escherichia coli: role of H-NS protein and the cyclic AMP-catabolite activator protein complex in transcription of the flhDC master operon. J Bacteriol 181:7500–7508. 10.1128/JB.181.24.7500-7508.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lehnen D, Blumer C, Polen T, Wackwitz B, Wendisch VF, Unden G. 2002. LrhA as a new transcriptional key regulator of flagella, motility and chemotaxis genes in Escherichia coli. Mol Microbiol 45:521–532. 10.1046/j.1365-2958.2002.03032.x. [DOI] [PubMed] [Google Scholar]
- 28.Bertin P, Terao E, Lee EH, Lejeune P, Colson C, Danchin A, Collatz E. 1994. The H-NS protein is involved in the biogenesis of flagella in Escherichia coli. J Bacteriol 176:5537–5540. 10.1128/jb.176.17.5537-5540.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clarke MB, Sperandio V. 2005. Transcriptional regulation of flhDC by QseBC and sigma (FliA) in enterohaemorrhagic Escherichia coli. Mol Microbiol 57:1734–1749. 10.1111/j.1365-2958.2005.04792.x. [DOI] [PubMed] [Google Scholar]
- 30.Francez-Charlot A, Laugel B, Van Gemert A, Dubarry N, Wiorowski F, Castanie-Cornet MP, Gutierrez C, Cam K. 2003. RcsCDB His-Asp phosphorelay system negatively regulates the flhDC operon in Escherichia coli. Mol Microbiol 49:823–832. 10.1046/j.1365-2958.2003.03601.x. [DOI] [PubMed] [Google Scholar]
- 31.Shin S, Park C. 1995. Modulation of flagellar expression in Escherichia coli by acetyl phosphate and the osmoregulator OmpR. J Bacteriol 177:4696–4702. 10.1128/jb.177.16.4696-4702.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hirakawa H, Takita A, Kato M, Mizumoto H, Tomita H. 2020. Roles of CytR, an anti-activator of cyclic-AMP receptor protein (CRP) on flagellar expression and virulence in uropathogenic Escherichia coli. Biochem Biophys Res Commun 521:555–561. 10.1016/j.bbrc.2019.10.165. [DOI] [PubMed] [Google Scholar]
- 33.Mecsas J, Rouviere PE, Erickson JW, Donohue TJ, Gross CA. 1993. The activity of sigma E, an Escherichia coli heat-inducible sigma-factor, is modulated by expression of outer membrane proteins. Genes Dev 7:2618–2628. 10.1101/gad.7.12b.2618. [DOI] [PubMed] [Google Scholar]
- 34.Erova TE, Rosenzweig JA, Sha J, Suarez G, Sierra JC, Kirtley ML, van Lier CJ, Telepnev MV, Motin VL, Chopra AK. 2013. Evaluation of protective potential of Yersinia pestis outer membrane protein antigens as possible candidates for a new-generation recombinant plague vaccine. Clin Vaccine Immunol 20:227–238. 10.1128/CVI.00597-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nicholson TF, Watts KM, Hunstad DA. 2009. OmpA of uropathogenic Escherichia coli promotes postinvasion pathogenesis of cystitis. Infect Immun 77:5245–5251. 10.1128/IAI.00670-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fogh J. 1978. Cultivation, characterization, and identification of human tumor cells with emphasis on kidney, testis, and bladder tumors. Natl Cancer Inst Monogr 1978:5–9. [PubMed] [Google Scholar]
- 37.Link AJ, Phillips D, Church GM. 1997. Methods for generating precise deletions and insertions in the genome of wild-type Escherichia coli: application to open reading frame characterization. J Bacteriol 179:6228–6237. 10.1128/jb.179.20.6228-6237.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hirakawa H, Nishino K, Yamada J, Hirata T, Yamaguchi A. 2003. Beta-lactam resistance modulated by the overexpression of response regulators of two-component signal transduction systems in Escherichia coli. J Antimicrob Chemother 52:576–582. 10.1093/jac/dkg406. [DOI] [PubMed] [Google Scholar]
- 39.Hashimoto-Gotoh T, Yamaguchi M, Yasojima K, Tsujimura A, Wakabayashi Y, Watanabe Y. 2000. A set of temperature sensitive-replication/-segregation and temperature resistant plasmid vectors with different copy numbers and in an isogenic background (chloramphenicol, kanamycin, lacZ, repA, par, polA). Gene 241:185–191. 10.1016/S0378-1119(99)00434-5. [DOI] [PubMed] [Google Scholar]
- 40.Elledge SJ, Davis RW. 1989. Position and density effects on repression by stationary and mobile DNA-binding proteins. Genes Dev 3:185–197. 10.1101/gad.3.2.185. [DOI] [PubMed] [Google Scholar]
- 41.Kurabayashi K, Agata T, Asano H, Tomita H, Hirakawa H. 2016. Fur represses adhesion to, invasion of, and intracellular bacterial community formation within bladder epithelial cells and motility in uropathogenic Escherichia coli. Infect Immun 84:3220–3231. 10.1128/IAI.00369-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miller JH. 1992. A short course in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]