ABSTRACT
Real-time PCR (RT-PCR) is widely used to diagnose human pathogens. RT-PCR data are traditionally analyzed by estimating the threshold cycle (CT) at which the fluorescence signal produced by emission of a probe crosses a baseline level. Current models used to estimate the CT value are based on approximations that do not adequately account for the stochastic variations of the fluorescence signal that is detected during RT-PCR. Less common deviations become more apparent as the sample size increases, as is the case in the current SARS-CoV-2 pandemic. In this work, we employ a method independent of CT value to interpret RT-PCR data. In this novel approach, we built and trained a deep learning model, qPCRdeepNet, to analyze the fluorescent readings obtained during RT-PCR. We describe how this model can be deployed as a quality assurance tool to monitor result interpretation in real time. The model’s performance with the TaqPath COVID19 Combo Kit assay, widely used for SARS-CoV-2 detection, is described. This model can be applied broadly for the primary interpretation of RT-PCR assays and potentially replace the CT interpretive paradigm.
KEYWORDS: artificial intelligence, deep learning, RT-PCR, COVID-19, TaqPath, SARS-CoV-2, real-time PCR
INTRODUCTION
Clinical laboratories commonly use real-time PCR (RT-PCR) to detect human pathogens, including SARS-CoV-2 (1). RT-PCR combines PCR amplification chemistry with fluorescent probe detection of amplified products to determine the presence of genetic material specific to the pathogen of interest (1). RT-PCR data are traditionally analyzed by estimating the threshold cycle (CT) at which the exponential phase of the fluorescence signal crosses a baseline threshold. Appropriate determination of the threshold is critical to the determination of a positive or negative result. Current models for estimating the CT value are based on approximations of generalized sigmoid functions or by analyzing the first- and second-order derivatives of the PCR time series (2–6). Not all reactions result in a well-defined generalized sigmoid function. Stochastic variations (i.e., noise) in the fluorescence signal exist to various degrees in all RT-PCRs. A baseline fluorescence is determined early in the PCR, and this baseline is used to establish a threshold. Noise early in the PCR can impede correct threshold determination, which can cause errors in the qualitative interpretation of the results.
Here, we apply a novel approach, qPCRdeepNet. The qPCRdeepNet model employs a deep convolutional neural network (ConvNet) (7) to analyze RT-PCR fluorescence data as a global image. The deep convolutional neural network is a type of machine learning model widely used in a range of image analysis tasks (7). The large volume of patient samples and overstretched laboratory personnel during the COVID19 pandemic created a need for autonomous, real-time quality measures to ensure accurate result interpretation (8). The model’s training was performed on a data set collected at our institution using an adapted CDC 2019-nCov assay (9) to learn the various shapes of RT-PCR (fluorescence) signals. The model was then tested on a TaqPath COVID-19 Combo Kit assay, and qPCRdeepNet performance was compared to that of the FDA EUA interpretation software, Applied Biosystems COVID-19 Interpretive Software v1.3 and v1.5, commonly used in clinical laboratories with this assay. To demonstrate the broad applicability of qPCRdeepNet, the model was also tested on other microbiology and oncology RT-PCR assays. We discuss the high performance of qPCRdeepNet and how we use the model as an autonomous, real-time quality assurance step for diagnostic RT-PCR assays.
MATERIALS AND METHODS
The RT-PCR per-cycle data for each reaction represents an array of fluorescent readings (Rn values). The length of the Rn array is defined by the assay manufacturer and is usually ≥40. For the current analysis, the first 40 cycles (Rn values) were used. These values were extracted from ABI 7500 Fast EDS files (https://github.com/nzxzxw/edsbreaker). The Rn array was then z-score normalized and plotted as a two-dimensional (2D) 299- by 299-pixel image. The images were plotted without axis tick marks or values to exclude them from potentially being learned as features. Instead, a dashed line, at the zero z score, was added to all images to provide the convolutional neural network with a scale of the z values on a given image.
Deep convolutional neural network-based software (qPCRdeepNet) was built for this analysis. Written in Python language, the software includes standard deep-learning libraries Tensorflow, Keras, and Scikit-lean (10, 11). The architecture of the network, shown in Fig. S1 in the supplemental material, consists of three sets of feature-map blocks plus a fully connected multilayer perceptron, i.e., a dense neural network block. The composition of each block is shown and includes a stacked 2D convolution, activation, pooling, and batch normalization layers. qPCRdeepNet training was done using class stratification with a split of 9:1 between training/testing data. The training set was further split 8:2 into training/validation sets, where the software was continuously tested on the validation set, with early stopping implemented based on the performance on the validation set, both in terms of accuracy and decay of the loss function. No training was performed on the validation sets.
Images from RT-PCRs, labeled as positive or negative, from 7,763 samples set up with an adapted CDC 2019-nCov assay (9) were used for training and initial testing. Each sample consists of three individual PCRs (N1, N2, and RNP control), each producing its own fluorescence array. These images were classified initially based on CT values, i.e., positive (CT of ≤40) or negative (CT of >40). The set was then reviewed by visual inspection to ensure the classifications were correct. As mentioned above, using a 9 to 1 split, the training set included 20,961 images, and the test set included 2,328 images. Both sets contained approximately 43% positive images.
Further testing, without retraining, was performed on 50,146 samples run on the TaqPath COVID-19 Combo Kit assay (ThermoFisher Scientific, Waltham, MA). A total of 200,584 images were generated from EDS files from each of four channels (or dyes), which represent three SARS-CoV-2 targets (ORF1ab, N, and S genes) and an internal control (MS2 bacteriophage). Each sample was classified as not detected, detected, or inconclusive for the presence of SARS-CoV-2 using Applied Biosystems (ABI) COVID-19 Interpretive Software v1.3 or qPCRdeepNet using the rule set stated in the TaqPath interpretation guidelines (12). As a comparison, we also interpreted a subset of samples using Interpretive Software v1.5.
Further testing, without retraining, was performed on (i) 950 samples with the TaqMan SARS-CoV-2, Flu A, Flu B multiplex assay (ThermoFisher), (ii) 1,306 samples with TaqMan SARS-CoV-2, Flu A/B, RSV multiplex assay (ThermoFisher), and (iii) 87 samples with TaqMan JAK2 V617F cast-PCR mutation detection assay (ThermoFisher). The assays were run per the manufacturer’s protocols. For all assays, the EDS files were used to generate a z-score-normalized plot as described above. These plots were interpreted by qPCRdeepNet to make a positive (detected) or negative (not detected) interpretation for the target analyte. These results were compared to those of the standard CT method (detected, CT of ≤40; not detected, CT of >40) per the manufacturer’s protocols. All RT-PCR experiments described above were run on an Applied Biosystems 7500 Fast PCR system (ThermoFisher), and CT values were generated using Applied Biosystems 7500 software v2.3 (ThermoFisher). For any discrepant cases, the true call was determined based on visual inspection of z-score-normalized amplification plots looking for the presence of sigmoid amplification of fluorescence signal (Fig. 1B).
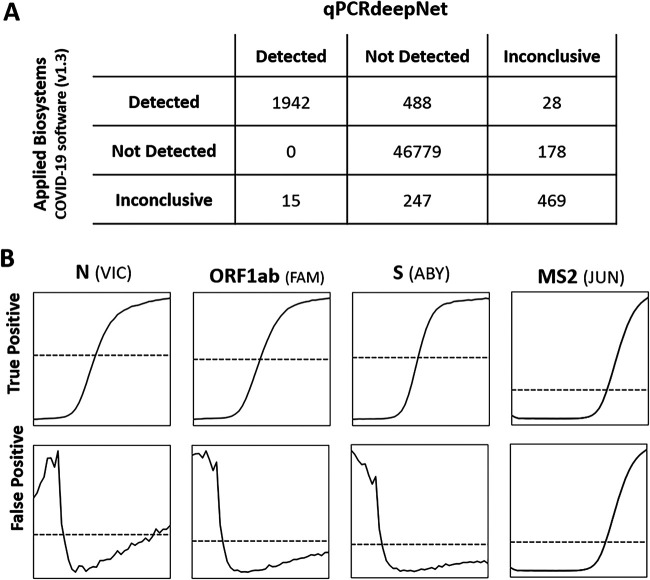
FIG 1.
(A) Performance of qPCRdeepNet classifier versus Applied Biosystems COVID-19 Interpretive Software v1.3, shown as a confusion matrix. (B) Representative z-score-normalized plots of true-positive cases, interpreted as detected by both qPCRdeepNet classifier and ABI Interpretive Software v1.3, and false-negative cases, interpreted as not detected by qPCRdeepNet and detected by ABI Interpretive Software v1.3. The latter case shows three negative curves for SARS-CoV-2 genes N, ORF1ab, and S genes and a positive curve for the internal control, MS2, as is expected for a sample in which SARS-CoV-2 is not detected. The early high level of signal in SARS-CoV-2 genes is a common cause of false positives by ABI Interpretive Software v1.3. Dashed line, z = 0.
Data availability.
A self-contained version of the test software, qPCRdeepNet, was made publicly available at the GitHub public repository (https://www.github.com/davidalouani/qPCRdeepNet). The repository includes data examples and detailed instructions for software installation.
RESULTS
The performance of qPCRdeepNet, trained on the CDC 2019-nCov assay (9), on the training and validation sets as a function of the training time (epoch) for accuracy, loss-function decay, and area under the concentration-time curve (AUC) is shown in Fig. S2 in the supplemental material. The model shows a robust performance as well as fast convergence between the model predictions on training and validation sets. The absence of deviations of the model’s performance, at later epochs, between the training set and validation set indicates the absence of overfitting. The performance of qPCRdeepNet in correctly picking the class prediction (positive or negative) for the CDC assay set is shown in Table S1.
qPCRdeepNet was further tested on 50,146 clinical samples run on the TaqPath COVID-19 Combo Kit assay and analyzed by Applied Biosystems (ABI) COVID-19 Interpretive Software v1.3 to evaluate the performance of the model. There was a 98% agreement in calls between the two methods (Fig. 1A), with a 1% false-positive rate calculate relative to the qPCRdeepNet prediction. Examining closely the false-positives group, i.e., interpreted as detected by ABI Interpretive Software v1.3 and not detected by qPCRdeepNet, there were 488 such cases. Visual inspection of this group showed that out of 488 samples with discrepant interpretation, 486 were indeed misinterpreted by ABI Interpretive Software v1.3. This is consistent with prior observations (8) that ABI Interpretive Software v1.3 produced a relatively high rate of false-positive calls. The ABI Interpretive Software v1.3 false-negative rate was zero relative to qPCRdeepNet prediction and visual inspection.
Before implementation of the qPCRdeepNet as a quality measure, we used an ad hoc surrogate method, the “N gene rule,” to identify potential false-positive interpretations. The N gene rule was a simple mechanism in which any sample interpreted as detected by TaqPath software v1.3 but that had an undermined CT value for the N gene would be flagged for repeat, as it most likely represented a false-positive call. The implementation of the N gene rule as a quality measure with ABI Interpretive Software v1.3 reduced the number of false-positive calls to 272.
While the manuscript was undergoing submission, ABI Interpretive Software v1.5 was released. To determine the performance differences between v1.3 and v1.5, the EDS files from all plates that contained a false-positive call (by v1.3 software) were run with the ABI Interpretive Software v1.5. For these problematic plates (plates containing false positives), there was 86% agreement in calls between v1.3 and v1.5 (Fig. 2A). This was similar to the 85% agreement observed between v1.3 and qPCRdeepNet for these problematic plates. ABI Interpretive Software v1.5 corrected nearly 97% (470 of 486) of false-positive calls made by v1.3 relative to qPCRdeepNet and visual inspection. Further confidence in the performance of qPCRdeepNet came in comparing its performance with ABI Interpretive Software v1.5 for these problematic plates, demonstrating 99% agreement in calls (Fig. 2B). The performance of the various approaches on the plates containing false positives is summarized in Table 1.
FIG 2.
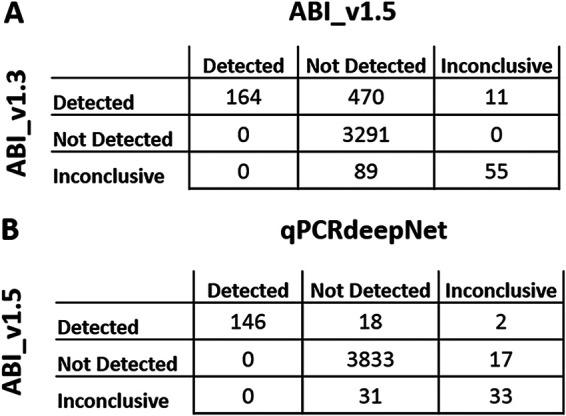
Performance of qPCRdeepNet classifier versus Applied Biosystems COVID-19 Interpretive Software (ABI) v1.5 for the subset of PCR plates that contained false-positive calls by the ABI Interpretive Software v1.3. (A) Confusion matrix of interpretation by ABI v1.3 versus ABI v1.5. (B) Confusion matrix of ABI v1.5 versus qPCRdeepNet.
TABLE 1.
Performance metrics by different interpretative methodsa
| Assay | No. of samples |
PPV | NPV | Inconclusive rate (%) | |||
|---|---|---|---|---|---|---|---|
| True positive | False positive | True negative | False negative | ||||
| ABI v1.3 | 148 | 486 | 3,291 | 0 | 0.23 | 1.00 | 3.8 |
| ABI v1.3 + N gene | 148 | 272 | 3,505 | 0 | 0.35 | 1.00 | 3.8 |
| ABI v1.5 | 148 | 16 | 3,850 | 0 | 0.90 | 1.00 | 1.6 |
| qPCRdeepNet | 146 | 0 | 3,882 | 2 | 1.00 | 1.00 | 1.2 |
Metrics were determined by using the subset of the TaqPath COVID-19 Combo Kit assay RT-PCR plates that contained false-positive calls by ABI Interpretive Software v1.3. These metrics are derived by using visual inspection as a reference. PPV, positive predictive value; NPV, negative predictive value.
To determine the applicability of qPCRdeepNet to other RT-PCR assays, we tested its performance on three other RT-PCR assays: TaqMan SARS-CoV-2, Flu A, Flu B multiplex assay, (2) TaqMan SARS-CoV-2, Flu A/B, RSV multiplex assay, and (3) TaqMan JAK2 V617F Cast-PCR mutation detection assay. qPCRdeepNet was not retrained with data from any of these assays. To assess performance, the qPCRdeepNet binary call (detected or not detected) was compared to the binary call based on CT value as determined by ABI 7500 software v2.3 (CT of ≤40, detected; CT of >40, not detected) for each of the analytes in each sample. For the multiplex assays, three different pathogens (or analytes) were assessed in each sample. Comparisons of calls were made at the pathogen (or analyte) level. There was similar high performance between the qPCRdeepNet and traditional CT methods (Table 2), although the positive predictive value was improved using qPCRdeepNet for the multiplex assays. These results confirm the applicability of this approach as an independent way to analyze RT-PCR data.
TABLE 2.
Performance metrics of qPCRdeepNet- versus CT-based qualitative interpretation for multiplex SARS-CoV2/InfA/InfB, multiplex SARS-CoV2/InfAB/RSV, and Cast-PCR JAK2 assaysa
| Assay | No. of samples |
CT method |
qPCRdeepNet |
||
|---|---|---|---|---|---|
| PPV | NPV | PPV | NPV | ||
| Multiplex SARS-CoV2/InfA/InfB | 950 | 0.96 | 0.99 | 0.99 | 0.99 |
| Multiplex SARS-CoV2/InfAB/RSV | 1,306 | 0.97 | 0.99 | 0.99 | 0.99 |
| CastPCR JAK2 assay | 87 | 1.00 | 1.00 | 1.00 | 1.00 |
These metrics are derived by using visual inspection as a reference. Positive predictive value (PPV) and negative predictive value (NPV) are shown.
DISCUSSION
Current models for analysis of RT-PCR assays based on functional approximations, although very good, are not fully adequate to accommodate the large variations of noise that become apparent in mass testing with multiple operators, as is the case with the current pandemic. A quality tool, agnostic to the noise that may exist in RT-PCR data, is needed to examine the global shape of the fluorescent signal. This visual inspection, or image analysis, that is often done in clinical laboratories is especially suited for deep convolutional network.
RT-PCR-based SARS-CoV-2 assays are subject to errors that include both false-negative and false-positive results. The varied performance of these tests is well documented (13). Although most RT-PCR-based assays are highly specific (13), false-positive interpretations can result from technical factors, such as contamination during sample preparation, or analytical factors, such as errors in how the fluorescent signal is interpreted. The qPCRdeepNet deep learning model can be used to check, autonomously and in real time, the interpretation of the RT-PCR fluorescent signal to improve the accuracy of the interpretation. We use qPCRdeepNet as part of larger laboratory information system (LIS) that our laboratory has created to connect information data flow. The implementation and interface of the LIS workflow is shown in Fig. S4 and S6 in the supplemental material.
The extent of false-positive cases using ABI Interpretive Software v1.3 in this study may be greater than that experienced in other clinical laboratories. This study took place before the FDA and ThermoFisher issued a warning that inadequate results may result due to inadequate vortexing and centrifugation of the RT-PCR plate (8) and overlapped the implementation of the TaqPath COVID-19 assay to a new and large workforce largely unfamiliar with RT-PCR.
This study provides evidence that the version 1.5 upgrade to the Applied Biosystems COVID-19 Interpretive Software greatly reduced false-positive interpretations that occurred in version 1.3. Since the implementation of qPCRdeepNet as a routine quality assurance measure for the TaqPath COVID-19 assay, which spanned both v1.3 and v1.5 implementation, we have stopped 0.2% (13/5591) and 0.1% (3/2,265) of cases, as interpreted by v1.3 and v.15, respectively, from being reported as false positives (Fig. S3).
We have demonstrated that qPCRdeepNet can serve as an alternative to traditional CT-based methods of RT-PCR interpretation for qualitative RT-PCR assays. Currently, we use a concurrent interpretative approach that combines both CT and qPCRdeepNet interpretation models to identify misinterpreted cases. In summary, we describe a novel deep learning method (qPCRdeepNet) for the qualitative interpretation of RT-PCR fluorescent signal. The method is independent of traditional analysis approaches and can be deployed either as an autonomous real-time quality check of traditional CT-based interpretation or as a primary analysis method for RT-PCR assays.
ACKNOWLEDGMENTS
No outside funding was used to support this investigation.
We have no conflicts of interest to disclose.
Footnotes
Supplemental material is available online only.
Contributor Information
Navid Sadri, Email: navid.sadri@uhhospitals.org.
John P. Dekker, National Institute of Allergy and Infectious Diseases
REFERENCES
- 1.Espy MJ, Uhl JR, Sloan LM, Buckwalter SP, Jones MF, Vetter EA, Yao JDC, Wengenack NL, Rosenblatt JE, Cockerill FR, Smith TF. 2006. Real-time PCR in clinical microbiology: applications for routine laboratory testing. Clin Microbiol Rev 19:165–256. doi: 10.1128/CMR.19.1.165-256.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tichopad A, Dilger M, Schwarz G, Pfaffl MW. 2003. Standardized determination of real-time PCR efficiency from a single reaction set-up. Nucleic Acids Res 31:e122. doi: 10.1093/nar/gng122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tichopad A, Didier A, Pfaffl MW. 2004. Inhibition of real-time RT-PCR quantification due to tissue-specific contaminants. Mol Cell Probes 18:45–50. doi: 10.1016/j.mcp.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 4.Gunay M, Goceri E, Balasubramaniyan R. 2016. Machine learning for optimum CT-prediction for qPCR, p 588–592. 2016 15th IEEE Int Conf Machine Learning Appl (ICMLA). IEEE, Anaheim, CA. doi: 10.1109/ICMLA.2016.0103. [DOI] [Google Scholar]
- 5.Tellinghuisen J, Spiess A-N. 2014. Comparing real-time quantitative polymerase chain reaction analysis methods for precision, linearity, and accuracy of estimating amplification efficiency. Anal Biochem 449:76–82. doi: 10.1016/j.ab.2013.12.020. [DOI] [PubMed] [Google Scholar]
- 6.Zhao S, Fernald RD. 2005. Comprehensive algorithm for quantitative real-time polymerase chain reaction. J Comput Biol 12:1047–1064. doi: 10.1089/cmb.2005.12.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.LeCun Y, Bengio Y, Hinton G. 2015. Deep learning. Nature 521:436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 8.US FDA. 2020. Risk of inaccurate results with Thermo Fisher Scientific TaqPath COVID-19 Combo Kit–letter to clinical laboratory staff and health care providers. US FDA, Washington, DC. [Google Scholar]
- 9.Rhoads DD, Cherian SS, Roman K, Stempak LM, Schmotzer CL, Sadri N. 2020. Comparison of Abbott ID Now, DiaSorin Simplexa, and CDC FDA emergency use authorization methods for the detection of SARS-CoV-2 from nasopharyngeal and nasal swabs from individuals diagnosed with COVID-19. J Clin Microbiol 58:e00760-20. doi: 10.1128/JCM.00760-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abadi M, Agarwal A, Barham P, Brevdo E, Chen Z, Citro C, Corrado GS, Davis A, Dean J, Devin M, Ghemawat S, Goodfellow I, Harp A, Irving G, Isard M, Jia Y, Jozefowicz R, Kaiser L, Kudlur M, Levenberg J, Mane D, Monga R, Moore S, Murray D, Olah C, Schuster M, Shlens J, Steiner B, Sutskever I, Talwar K, Tucker P, Vanhoucke V, Vasudevan V, Viegas F, Vinyals O, Warden P, Wattenberg M, Wicke M, Yu Y, Zheng X. 2016. TensorFlow: large-scale machine learning on heterogeneous distributed systems. arXiv 1603.04467 [cs]. https://arxiv.org/abs/1603.04467.
- 11.Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Müller A, Nothman J, Louppe G, Prettenhofer P, Weiss R, Dubourg V, Vanderplas J, Passos A, Cournapeau D, Brucher M, Perrot M, Duchesnay É. 2018. Scikit-learn: machine learning in Python. arXiv 1201.0490 [cs]. https://arxiv.org/abs/1201.0490.
- 12.ThermoFisher Scientific. 2020. TaqPath COVID-19 Combo Kit. ThermoFisher Scientific, Waltham, MA. [Google Scholar]
- 13.Ravi N, Cortade DL, Ng E, Wang SX. 2020. Diagnostics for SARS-CoV-2 detection: a comprehensive review of the FDA-EUA COVID-19 testing landscape. Biosens Bioelectron 165:112454. doi: 10.1016/j.bios.2020.112454. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
A self-contained version of the test software, qPCRdeepNet, was made publicly available at the GitHub public repository (https://www.github.com/davidalouani/qPCRdeepNet). The repository includes data examples and detailed instructions for software installation.