ABSTRACT
During enteric salmonellosis, neutrophil-generated reactive oxygen species alter the gut microenvironment, favoring survival of Salmonella Typhimurium. While type 3 secretion system 1 (T3SS-1) and flagellar motility are potent Salmonella Typhimurium agonists of the neutrophil respiratory burst in vitro, neither of these pathways alone is responsible for stimulation of a maximal respiratory burst. To identify Salmonella Typhimurium genes that impact the magnitude of the neutrophil respiratory burst, we performed a two-step screen of defined mutant libraries in coculture with human neutrophils. We first screened Salmonella Typhimurium mutants lacking defined genomic regions and then tested single-gene deletion mutants representing particular regions under selection. A subset of single-gene deletion mutants was selected for further investigation. Mutants in four genes, STM1696 (sapF), STM2201 (yeiE), STM2112 (wcaD), and STM2441 (cysA), induced an attenuated respiratory burst. We linked the altered respiratory burst to reduced T3SS-1 expression and/or altered flagellar motility for two mutants (ΔSTM1696 and ΔSTM2201). The ΔSTM2441 mutant, defective for sulfate transport, formed aggregates in minimal medium and adhered to surfaces in rich medium, suggesting a role for sulfur homeostasis in the regulation of aggregation/adherence. We linked the aggregation/adherence phenotype of the ΔSTM2441 mutant to biofilm-associated protein A and flagellins and hypothesize that aggregation caused the observed reduction in the magnitude of the neutrophil respiratory burst. Our data demonstrate that Salmonella Typhimurium has numerous mechanisms to limit the magnitude of the neutrophil respiratory burst. These data further inform our understanding of how Salmonella may alter human neutrophil antimicrobial defenses.
KEYWORDS: Salmonella, cysteine metabolism, flagellar motility, neutrophils, respiratory burst
INTRODUCTION
Nontyphoidal Salmonella (NTS) infections account for the largest disease burden of foodborne illnesses globally (1). Acute enteric disease caused by NTS is characterized by marked neutrophilic enteritis and inflammatory diarrhea (2). Neutrophil influx into the gut lumen is induced by effector proteins secreted by the type 3 secretion system 1 (T3SS-1) encoded within the horizontally acquired locus, Salmonella pathogenicity island 1 (3). Neutrophilic influx benefits NTS by eliminating competing microbes and generating new nutrients to support NTS metabolism in the gut lumen (4–6). However, neutrophils also limit the systemic spread of NTS in the host (7). Given this, it is apparent that there is a complicated interplay between NTS and neutrophils that impacts NTS fitness during infection.
A hallmark of the neutrophil antimicrobial response is the production of reactive oxygen species (ROS) through the respiratory burst (8, 9). Generation of ROS occurs through assembly of the multicomponent NADPH oxidase enzyme complex at the neutrophil phagolysosome and/or cell membrane, with ROS released intracellularly, extracellularly, or both (10, 11). The magnitude of the neutrophil respiratory burst is influenced by interactions with the invading pathogen. Lipopolysaccharide, a pathogen-associated molecular pattern located in the bacterial outer membrane, is one factor that primes the neutrophil for a robust respiratory burst (12, 13). In addition, both the Salmonella Typhimurium T3SS-1 and flagellar motility are important agonists of the neutrophil respiratory burst in vitro (14). However, biofilm-associated NTS, encased in an extracellular matrix composed of proteins, carbohydrates, and extracellular DNA (reviewed in reference 15), elicits a diminished neutrophil respiratory burst compared to that in planktonic cells (16). Taken together, these findings suggest that NTS has numerous mechanisms to alter the magnitude of the neutrophil respiratory burst, which likely help to facilitate its survival in different host compartments.
The mechanisms by which NTS modulates the respiratory burst are unknown. Although the T3SS-1 and flagellar motility are key Salmonella Typhimurium agonists of the neutrophil respiratory burst, deletion of both elements does not completely abrogate the neutrophil respiratory burst in vitro (14). Therefore, we hypothesized that additional S. Typhimurium factors would serve as agonists of the neutrophil respiratory burst. Using a two-step genetic screen, we identified genes that were associated with significant changes in the neutrophil respiratory burst and characterized the genes for effects on T3SS-1 expression and swimming motility. We found that cellular aggregation mediated by a defect in sulfate import reduced the magnitude of the neutrophil respiratory burst. Together, this work provides mechanistic insight into how S. Typhimurium modulates the magnitude of the human neutrophil respiratory burst, a phenomenon that likely facilitates its survival during enteric infection.
(This article was submitted to an online preprint archive [17].)
RESULTS
Library screen for S. Typhimurium factors that alter the neutrophil respiratory burst.
Flagellar motility and the T3SS-1 are potent S. Typhimurium agonists of the neutrophil respiratory burst (14). To identify additional mechanisms by which S. Typhimurium influences the neutrophil respiratory burst, we performed a two-step genetic screen using defined libraries of S. Typhimurium mutants (18). The multigene deletion (MGD) library consisted of 151 defined mutants, each containing deletions of ∼4 to 40 contiguous genes, with total library coverage of nearly half of the S. Typhimurium genome. Individual mutants from the MGD library were cocultured with neutrophils to establish mutants that alter the magnitude of the intracellular neutrophil respiratory burst. A mutant was considered to cause an altered neutrophil respiratory burst if it was significantly different from the wild type (WT) at any time after coculture (see Table S1 in the supplemental material). We identified 45 MGD mutants that elicited an altered the respiratory burst, with 2 mutants covering overlapping genomic regions. Three mutants deleted for the T3SS-1 and its effectors and 3 mutants deleted for flagellar components elicited a reduced respiratory burst in our screen, confirming our ability to identify known agonists of the neutrophil respiratory burst with this screening methodology. Four MGD mutants covered 25 genes or more and were excluded from further analysis due to the likelihood of additive effects of multiple genes affecting the respiratory burst. We chose 21 of the remaining 33 MGD mutants for further testing. We confirmed the mutant identity for 19 of 21 mutants by PCR and prioritized the genes from 19 confirmed genomic regions for further analysis (see Table S2). Twelve of the 19 MGD mutants elicited a higher respiratory burst, while 7 MGD mutants elicited a lower respiratory burst than the WT organism (Table S2). A minilibrary of 134 single-gene deletion (SGD) mutants from the prioritized genomic regions was assembled.
In the second step of our screen, we interrogated the effects of our mutant minilibrary on the neutrophil respiratory burst, as measured by rhodamine fluorescence. We found 42 mutants from 16 genomic regions that elicited an altered respiratory burst compared to that of the WT organism (see Tables S3 and S4). Of the 42 SGD mutants, 20 elicited an increased respiratory burst and 22 elicited a decreased respiratory burst compared to that of the WT organism. We selected seven SGD mutants for further evaluation in neutrophil (PMN)-Salmonella coculture, 3 of which elicited a reduced respiratory burst (ΔSTM0018, ΔSTM2112, and ΔSTM2441) and 4 of which elicited an increased respiratory burst (ΔSTM1213, ΔSTM1690, ΔSTM1696, and ΔSTM2201). The seven mutations were moved into a clean genetic background and retested in coculture with neutrophils. Four mutants (ΔSTM1696, ΔSTM2112, ΔSTM2201, and ΔSTM2441) elicited a significantly attenuated neutrophil respiratory burst compared to that of the WT organism when grown to both stationary and exponential phase, while three mutations had no effect on the neutrophil respiratory burst (Fig. 1). The phenotypes of two of our mutants, in ΔSTM1696 and ΔSTM2201 strains, were reversed when tested in the clean genetic background (Table S4). Overall, our screening strategy identified 4 mutants that stimulated an altered neutrophil respiratory burst compared to that of the WT organism.
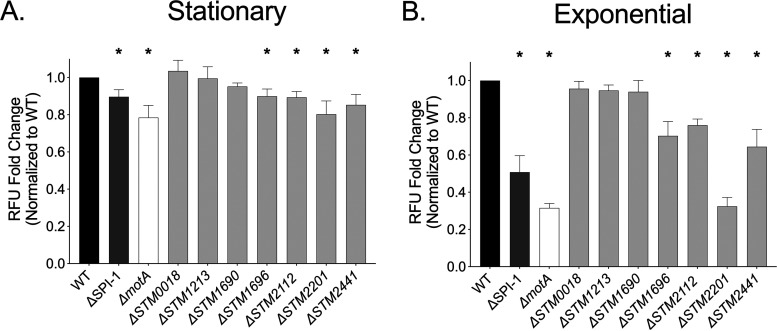
FIG 1.
Neutrophil respiratory burst is blunted in response to four single gene deletion mutants. The neutrophil respiratory burst as measured by rhodamine fluorescence elicited by the WT (HA420) and the ΔSPI-1 (JE598), ΔmotA (JE1202), ΔSTM0018 (JE963), ΔSTM1213 (JE965), ΔSTM1690 (JE967), ΔSTM1696 (JE969), ΔSTM2112 (JE971), ΔSTM2201 (JE973), and ΔSTM2441 (JE975) mutants at an MOI of 50:1. Neutrophils were exposed for 2 h to bacteria grown to stationary phase (A) or 1 h to bacteria grown to late-exponential phase (B). The relative fluorescence elicited by each mutant was normalized to that of the WT organism for each donor. Bars indicate means ± standard errors of the means (SEMs) from triplicate samples using blood from 4 different donors. *, P < 0.05 between the WT and indicated mutant by Student’s t test.
Effects of mutations on swimming motility and T3SS-1 expression.
We investigated possible mechanisms for the alteration in the neutrophil respiratory burst elicited by the four identified mutants. First, we performed swimming assays to determine the contribution of each gene to flagellar motility. We found a significant defect in swimming motility for the ΔSTM1696, ΔSTM2201, and ΔSTM2441 mutants (Fig. 2A and B). The ΔSTM2201 mutant was nonmotile. Since the magnitude of the respiratory burst stimulated by the ΔSTM2201 mutant was comparable to that of the nonmotile ΔmotA mutant, it is likely that the severe motility defect of the ΔSTM2201 mutant explains the observed alteration in the neutrophil respiratory burst.
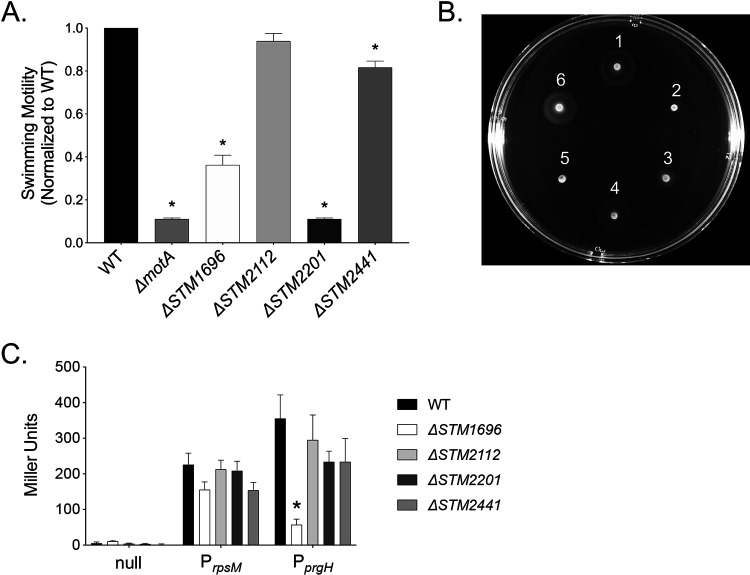
FIG 2.
Swimming motility and T3SS-1 expression in mutants eliciting an altered neutrophil respiratory burst. (A) Normalized overnight cultures of the WT and the ΔmotA (JE1202), ΔSTM1696 (JE969), ΔSTM2112 (JE971), ΔSTM2201 (JE973), and ΔSTM2441 (JE975) mutants were spotted onto swimming plates. Cell spread was measured 4 h postinoculation. Each assay was performed in 4 to 5 replicates on 3 separate occasions. (B) Representative photograph of a swimming plate (1, WT; 2, ΔmotA mutant; 3, ΔSTM1696 mutant; 4, ΔSTM2112 mutant; 5, ΔSTM2201 mutant; 6, ΔSTM2441 mutant). (C) Activation of a T3SS-1 apparatus promoter (PprgH) in late-exponential-phase growth as determined by β-galactosidase activity (from 5 independent experiments). Plasmid constructs with the indicated promoter driving lacZY expression included: null (promoter-less lacZY), PrpsM (positive control), and PprgH (T3SS-1 promoter). Bars represent the means ± SEMs. *, P < 0.05 between the WT and mutant by Student’ t test.
Next, we evaluated the effects of our mutations on expression of the T3SS-1 using a plasmid containing a T3SS-1 apparatus gene promoter (PprgH) driving the expression of lacZY. We found reduced T3SS-1 expression for the ΔSTM1696 mutant only (Fig. 2C). The combination of decreased motility and altered T3SS-1 expression for the ΔSTM1696 mutant suggests that one or both of the observed defects contribute to the weakened neutrophil respiratory burst. Through these experiments evaluating both swimming motility and T3SS-1 expression, we were able to assign likely causes for the observed defects in neutrophil respiratory burst for both the ΔSTM1696 and ΔSTM2201 mutants.
Colanic acid and the neutrophil respiratory burst.
Colanic acid is a negatively charged polysaccharide capsule that helps to maintain transmembrane potential and the proton motive force during envelope stress (19). Colanic acid is produced by enzymes encoded within the colanic acid capsule biosynthetic gene cluster (STM2118-STM2099), and its production is stimulated by low temperatures (<30°C) and biofilm-forming conditions (20–22). Since colanic acid is not typically produced in rich media at body temperature, we found it surprising that two mutants in the biosynthetic gene cluster, ΔSTM2112 (ΔwcaD) and ΔSTM2114 (ΔwcaB) were identified in the screen.
To establish whether colanic acid acts as an agonist of the neutrophil respiratory burst under our assay conditions, we investigated the neutrophil respiratory burst in response to a complete colanic acid capsule cluster mutant (Δwza) deleted for wza through wcaM (STM2118-STM2099) (19). We observed no alterations to the neutrophil respiratory burst for the Δwza mutant in comparison to the that of the isogenic WT (Fig. 3). Since mutants within the colanic acid biosynthesis pathway located downstream of the WcaJ glycosyl transferase could accumulate toxic intermediates that affect viability, we evaluated the growth of the ΔSTM2112 mutant in different media (23). We found no growth defects in either rich or minimal media (see Fig. S1A and B). Neutrophil-Salmonella coculture medium includes 10% normal human serum and therefore contains complement. Colanic acid protects Escherichia coli against complement-mediated killing (24); therefore, we hypothesized the ΔSTM2112 mutant would be sensitive to the serum in our assay. We found no growth defect for the ΔSTM2112 mutant after 2 h, the duration of time used for our neutrophil-Salmonella coculture (Fig. S1C). Our data suggest that a defective colanic acid capsule was not the likely cause for the reduced neutrophil respiratory burst elicited by the ΔSTM2112 mutant under the conditions tested.
FIG 3.
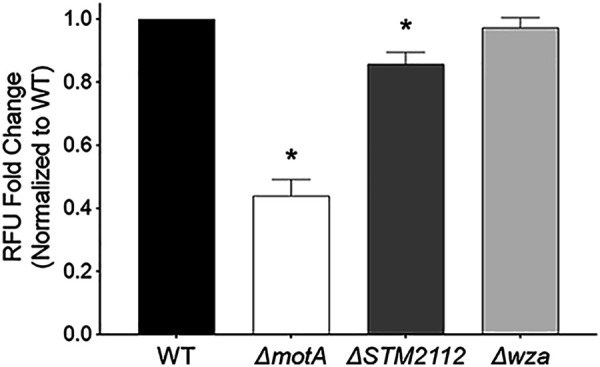
Deletion of the complete colanic acid biosynthetic cluster has no effect on the neutrophil respiratory burst. Neutrophils were exposed to the WT (WN150) or the ΔmotA (JE1762), ΔSPI-1 (JE1760), ΔSTM2112 (JE1800), and Δwza (JP245) mutants grown to late-exponential phase, and the neutrophil respiratory burst was measured by rhodamine fluorescence after a 1-h coculture. Data analysis as for Fig. 1. Bars indicate means ± SEMs from triplicate samples using blood from 3 different donors. *, P < 0.05 between the WT and mutant by Student’s t test.
Sulfate import stimulates the neutrophil respiratory burst.
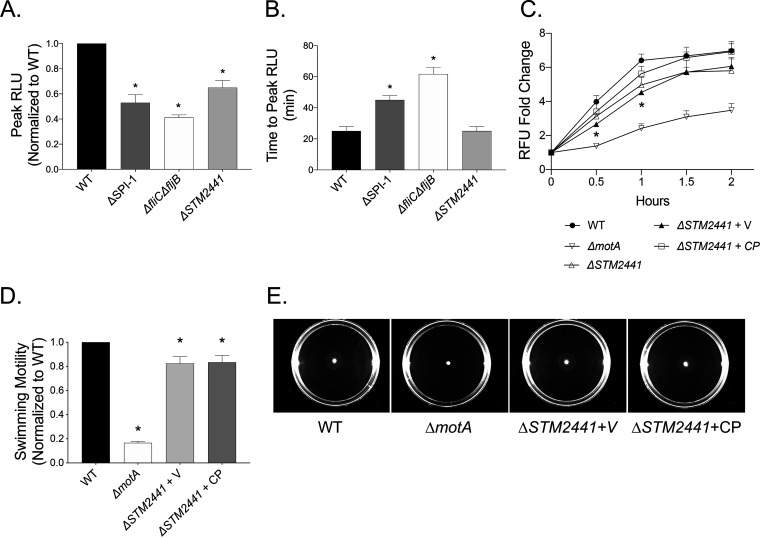
STM2441 (cysA) encodes the ATPase subunit of the SulT sulfate permease responsible for importing sulfate and thiosulfate into the cell (25). Our data demonstrate that the ΔSTM2441 mutant elicited a lower magnitude neutrophil respiratory burst than the WT (Fig. 1). To verify our observations, we tested the effect of the ΔSTM2441 mutant on the neutrophil respiratory burst using luminol, which measures both intracellular and extracellular reactive oxygen species (26). We found that the peak magnitude of the neutrophil respiratory burst elicited by the ΔSTM2441 mutant was decreased compared to that of the WT organism (Fig. 4A and Fig. S2), but there was no effect of the mutation on the time to peak respiratory burst (Fig. 4B and Fig. S2). Next, we constructed a low-copy-number plasmid containing an intact copy of STM2441 with its native promoter to evaluate the phenotypes of the mutant complemented in trans. Complementation of the ΔSTM2441 mutant in trans completely restored the intracellular neutrophil respiratory burst to WT levels, definitively linking the altered respiratory burst to deletion of STM2441 (Fig. 4C). Finally, we found no reversal of the swimming defect for the complemented ΔSTM2441 mutant (Fig. 4D and E). Together, our data demonstrate that neutrophils produce a muted respiratory burst in response to the ΔSTM2441 mutant, a phenotype that is not likely related to the small reduction in swimming motility.
FIG 4.
Deletion of ΔSTM2441 is linked to altered total and intracellular neutrophil respiratory burst. The peak neutrophil respiratory burst normalized to the WT for the same donor (A) and time to peak neutrophil respiratory burst (B) as measured by luminol-enhanced chemiluminescence upon exposure to the WT or the ΔmotA (JE1202), ΔSTM2441 (JE975), or ΔfliC ΔfljB (JE524) mutants grown to late-exponential phase. (C) Neutrophils were exposed to the same mutants as in panels A and B and ΔSTM2441+V (JE975/pWSK29), and ΔSTM2441+CP (JE975/pWSK29::STM2441) with respiratory burst measured by rhodamine fluorescence as for Fig. 1. Bars and data points indicate means ± SEMs from triplicate samples using blood from 3 different donors. (D) Normalized overnight cultures of the WT and the ΔmotA, ΔSTM2441+V, and ΔSTM2441+CP mutants were spotted onto swimming plates. Analysis as for Fig. 3. (E) Representative photographs of swimming plates. *, P < 0.05 between the indicated mutant and the WT by Student’s t test.
Sulfate import defect induces cellular aggregation.
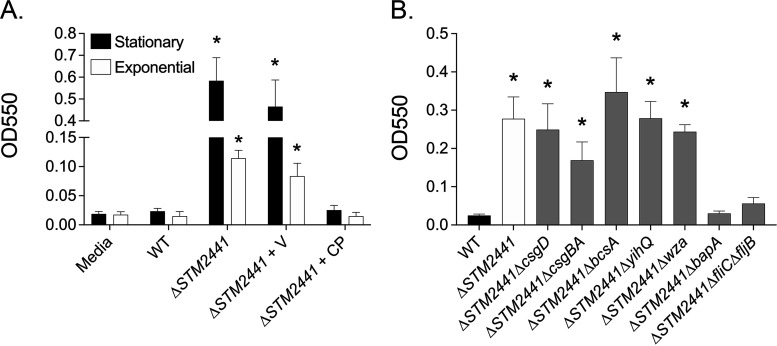
The ΔcysA mutant is a cysteine auxotroph, because sulfate import is essential for cysteine biosynthesis (27, 28). We investigated the growth of the ΔSTM2441 mutant in both rich and minimal media. As anticipated, we found that the ΔSTM2441 mutant has a growth defect, reversed by complementation in trans, in M9 minimal medium which contains MgSO4 as the sole sulfur source (Fig. 5A). The addition of l-cysteine to M9 minimal medium completely reversed the growth defect, confirming that the ΔSTM2441 mutant is a cysteine auxotroph (Fig. 5B). We found no growth abnormalities in rich medium (LB) or medium used for Salmonella-neutrophil coculture, which contains 0.2 mM l-cystine and 0.4 mM MgSO4 (see Fig. S3A and B).
FIG 5.
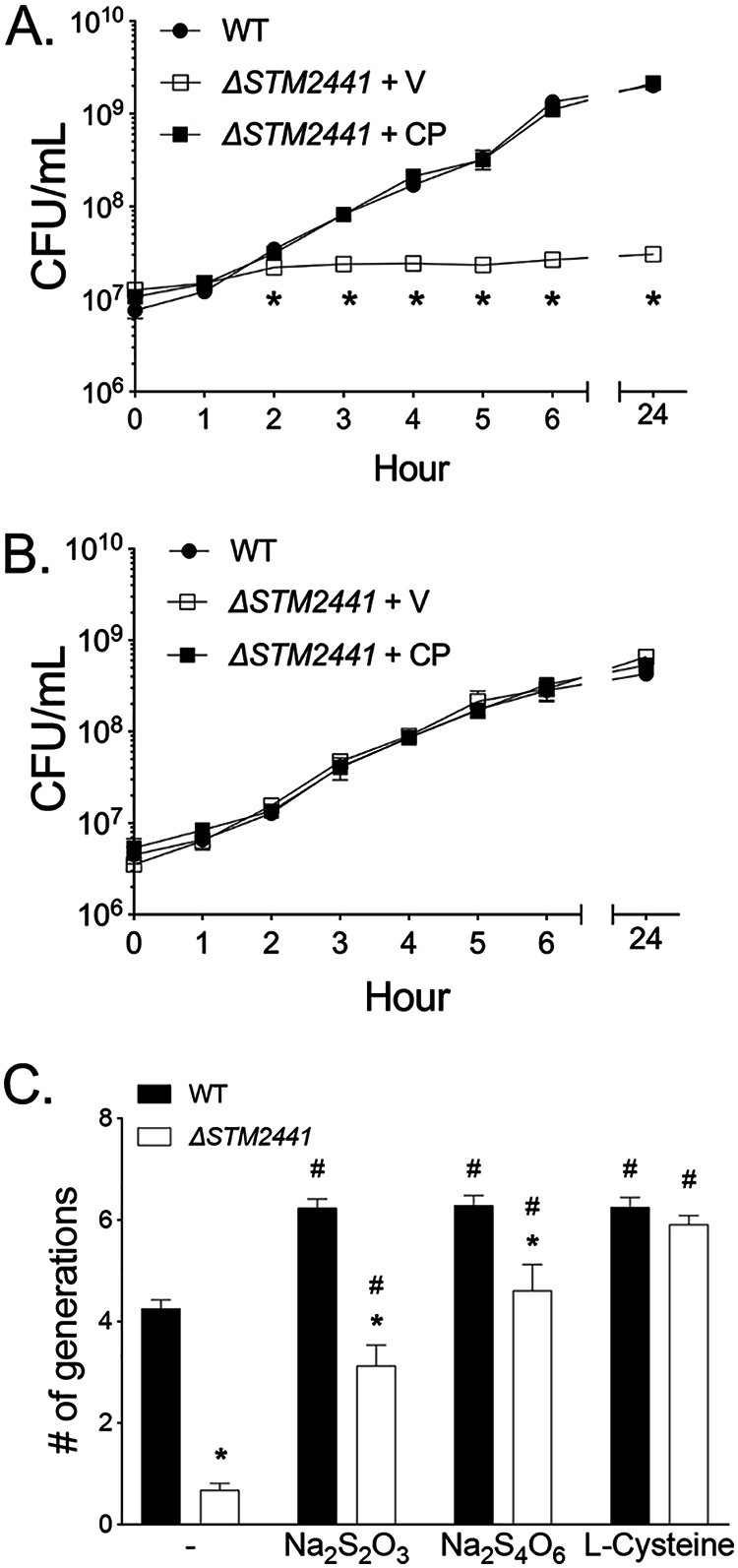
Growth defect of the ΔSTM2441 mutant cysteine auxotroph is partially reversed by thiosulfate and tetrathionate. Growth of the WT, ΔSTM2441+V (JE975/pWSK29), and ΔSTM2441+CP (JE975/pWSK29::STM2441) in M9 minimal broth (A) or M9 with l-cysteine (1 mM) (B). (A and B) Overnight cultures were diluted 1:100 into indicated media and samples taken hourly to determine CFU per milliliter. Data points represent means ± SEMs. CFU data were log transformed and statistical significance determined by two-way ANOVA. *, P < 0.05 between WT and ΔSTM2441+V. (C) Growth of the WT and ΔSTM2441 (JE975) mutant in SF broth with no sulfur source (−) or supplemented with sodium thiosulfate (1 mM Na2S2O3), sodium tetrathionate (0.5 mM Na2S4O6), or l-cysteine (1 mM). Overnight cultures were diluted 1:100 into indicated media, and the number of generations was determined after 24 h of growth. Statistical significance was determined by Student’s t test. *, P < 0.05 between WT and ΔSTM2441 in a given medium. #, P < 0.05 between the indicated medium and nonsupplemented medium for a given strain. Experiments were performed on three independent occasions.
Since sulfate and thiosulfate can be used in sulfate assimilation pathways, we hypothesized that other sulfur sources known to be present in the host could rescue the growth defect of the ΔSTM2441 mutant (4, 25, 29). We evaluated growth in media with thiosulfate or tetrathionate as sole sulfur sources. We found that both thiosulfate and tetrathionate partially restored growth of the ΔSTM2441 mutant (Fig. 5C). The partial restoration in growth suggests that Salmonella has one or more other import mechanisms that allow selective import of thiosulfate and/or tetrathionate. While performing experiments in minimal medium, we identified macroscopic cellular aggregates within the liquid medium that also adhered to the flask walls after 24 h of growth (see Fig. S4A). The adherent aggregates were positive for crystal violet staining and were most abundant in media supplemented with tetrathionate and thiosulfate (Fig. S4B). These observations led us to investigate whether cellular aggregates could form under the bacterial preparation conditions used for the neutrophil respiratory burst assay.
Bacteria prepared for neutrophil respiratory burst assays were grown in rich medium to either stationary or exponential growth. Using crystal violet staining, we detected bacterial adherence to the surface of conical tubes at the liquid-air interface of bacteria grown to both stationary and exponential phases for the ΔSTM2441 mutant (Fig. 6A), with no apparent adherence of the WT organism, as expected. The surface adhesion was reversed in the complemented ΔSTM2441 mutant (Fig. 6A). These data suggest that defective import of sulfate induces inappropriate bacterial aggregation, a phenotype that may explain the altered neutrophil respiratory burst stimulated by the ΔSTM2441 mutant.
FIG 6.
Adherence of ΔSTM2441 mutant to surfaces is linked to biofilm-associated protein and flagellins. (A) Crystal violet staining was used to quantify surface-adherent bacteria from the WT and the ΔSTM2441 mutant grown at 37°C with agitation to stationary and exponential phases. (B) Surface-adherent bacteria from the WT, ΔSTM2441 mutant (JE1391), and double mutants grown to stationary phase as determined by crystal violet staining. Crystal violet was quantified by optical density (550 nm). Experiments were performed on 4 (A) or 6 (B) independent occasions. Bars represent means ± SEMs. *, P < 0.05 between the WT and mutant as determined by Student’s t test.
We hypothesized that cellular aggregation in the ΔSTM2441 mutant was due to induction of extracellular matrix components of biofilms. Using a genetic approach, we tested whether the inappropriate aggregation observed in the ΔSTM2441 mutant aggregation could be relieved by deletion of genes involved in biofilm, including csgD (biofilm master regulator), csgBA (curli fimbriae), bcsA (cellulose), bapA (biofilm associated protein A), yihQ (O-antigen capsule), and wza-wcaM (colanic acid) (reviewed in reference 30). We also tested a fliC and fljB (flagellins) double mutant, since flagella can be incorporated into biofilm extracellular matrix and are important for initial surface attachment (31). The deletion of either ΔbapA or ΔfliC ΔfljB in the ΔSTM2441 mutant background (ΔSTM2441 ΔbapA and ΔSTM2441 ΔfliC ΔfljB double mutants) eliminated the adherence of the ΔSTM2441 mutant (Fig. 6B). None of the single biofilm extracellular matrix component mutants adhered to plastic under the assay conditions used (see Fig. S5). Together, these data indicate that the aggregation stimulated by defective sulfate import in S. Typhimurium is due to both the biofilm-associated protein A (BapA) and flagellins (FliC and/or FljB).
DISCUSSION
In this study, we used a two-step screening approach to discover new Salmonella Typhimurium agonists of the neutrophil respiratory burst. We first tested the effects of deletion of large genomic regions (multigene deletion [MGD] mutants) on the magnitude of the neutrophil respiratory burst and followed with testing mutants in individual genes (single-gene deletion mutants) corresponding to disrupted genomic regions identified in the first step. We identified four mutants, in ΔSTM1696, ΔSTM2112, ΔSTM2201, and ΔSTM2441, that elicited a consistently diminished neutrophil respiratory burst. We were unable to determine a mechanism for the reduction in neutrophil respiratory burst for one mutant (ΔSTM2112). In the other three mutants, the attenuated respiratory burst response of neutrophils was linked to abnormal swimming motility (ΔSTM1696 and ΔSTM2201), reduced T3SS-1 expression (ΔSTM1696), and cellular aggregation (ΔSTM2441). The cellular aggregation observed in the ΔSTM2441 mutant was linked to biofilm-associated protein A and flagellins.
Our two-step genetic screen allowed us to interrogate nearly half of the S. Typhimurium genome (∼1,959 genes) using only 151 mutants. The purpose of the screening steps was preliminary identification of potential gene targets that influenced the neutrophil respiratory burst. This screening approach has been used to identify mechanisms of cytosolic replication in epithelial cells (32), poultry colonization (33), and resistance mechanisms to an antibiofilm drug (34). Our prior work demonstrated that expression of the T3SS-1 and flagellar motility are agonists of the neutrophil respiratory burst (14). Mutants with deleted genes encoding flagella and T3SS-1 apparatus and effector proteins contained within the MGD library elicited a lower neutrophil respiratory burst in the MGD screen. Identification of known neutrophil respiratory burst agonists during the screening stage demonstrated that the methodology used could identify new genomic regions that influence the neutrophil respiratory burst. One shortcoming of our approach is the possibility that additive effects of multiple gene deletions generated false positives. Three of the 19 genomic regions that were prioritized for study in the second step did not contain single genes that altered the neutrophil respiratory burst and are considered false-positive results. Another potential shortcoming of the library screening strategy was that bacteria were grown to stationary phase prior to exposure to neutrophils. The expression of many genes is altered during stationary phase compared to that during exponential growth; therefore, some mutants may not have been identified because the neutrophil response might only be altered when expression of a given gene is induced (35, 36). Finally, our use of two individual blood donors for library screening resulted in limited statistical power. Although the sample size was low, this strategy permitted preliminary identification of mutants that influenced the neutrophil respiratory burst which was followed by confirmatory experiments with enhanced statistical power. Despite these shortcomings, the identification of the internal controls in this assay indicated the methodology was able to identify new agonists of the neutrophil respiratory burst.
STM1696 (sapF) is part of the sapABCDF operon, which confers antimicrobial peptide (AMP) resistance in Salmonella by exporting AMPs extracellularly (37, 38). We hypothesized that loss of this AMP resistance may have been associated with the altered neutrophil respiratory burst by reducing the viability of the ΔSTM1696 mutant after phagocytosis. We found that the ΔSTM1696 mutant has decreased flagellar motility and T3SS-1 expression, which are known agonists of the respiratory burst. Although we did not rule out that defective export of AMPs by the ΔSTM1696 mutant could have contributed to the altered neutrophil respiratory burst, our data suggest that the altered expression of two important agonists of the neutrophil respiratory burst are a likely cause(s) of the observed phenotype. Which of these processes is more important for the neutrophil response and why they are altered in the ΔSTM1696 mutant require further study.
The putative LysR type transcriptional regulator, STM2201 (yeiE), is uncharacterized in Salmonella Typhimurium (39). We correlated the diminished neutrophil respiratory burst for the ΔSTM2201 mutant to a lack of swimming motility, as neutrophils responded to the ΔSTM2201 mutant from a clean genetic background in a similar way as to the nonmotile ΔmotA mutant. This phenotype is in contrast to the phenotype observed for the ΔSTM2201 mutant from the minilibrary, which elicited a greater magnitude respiratory burst than the WT. The contrasting respiratory burst responses elicited by the mutant during our screen versus in a clean genetic background demonstrates the need for rigorous confirmation of mutant phenotypes, as there can be potential confounding factors associated with mutants arrayed in a library. Although the YeiE regulon has been described in E. coli, it does not include any genes responsible for motility (40). To our knowledge, this is the first report of STM2201 (yeiE) playing a role in swimming motility. Further work is needed to characterize the mechanism by which STM2201 contributes to altered swimming motility.
STM2112 (wcaD) encodes a colanic acid polymerase within the colanic acid biosynthetic cluster. As an exopolysaccharide, colanic acid could act as an agonist of the neutrophil respiratory burst through stimulation of neutrophil pattern recognition receptors. However, our method of bacterial preparation was not performed under conditions known to induce colanic acid expression (20–22). Furthermore, the lack of an altered neutrophil respiratory burst in response to the colanic acid operon deletion mutant (Δwza) suggests that colanic acid deletion is not the likely cause for the altered neutrophil respiratory burst in response to the ΔSTM2112 mutant in our work. Consistent with our findings, a mutant in colanic acid biosynthesis (ΔwcaM) elicited no change in neutrophil respiratory burst when in planktonic growth (16). Since mutations in enzymes downstream of the initial glycosyl transferase in the colanic acid biosynthesis pathway (WcaJ) lead to accumulation of toxic intermediates (23), we hypothesize that the ΔSTM2112 mutant has reduced viability in the presence of neutrophils, leading to the observed reduction in neutrophil respiratory burst.
We found a blunted neutrophil respiratory burst in response to the ΔSTM2441 mutant and correlated this finding to the development of cellular aggregates. STM2441 (cysA) encodes the ATPase subunit of the sulfate permease (cysPTWA and sbp) to import sulfate and thiosulfate into the cell for cysteine and methionine biosynthesis (25, 41). We confirmed that the ΔSTM2441 mutant is a cysteine auxotroph and found that it forms aggregates both under sulfur-limited conditions and in rich media. Disruption of sulfate reduction, cysteine biosynthesis, and cysteine catabolism is associated with biofilm development in E. coli in the absence of canonical environmental cues (42–44). Biofilm-associated Salmonella Typhimurium elicit a blunted neutrophil respiratory burst compared to that of planktonic cells, and the blunted respiratory burst is linked to curli fimbriae and O-antigen capsule (16). Curli fimbriae are a major component of the Salmonella biofilm extracellular matrix and are recognized by innate immune receptors Toll-like receptor 2 (TLR2) and TLR9 and the nod-like receptor protein 3 inflammasome (45–48). In spite of the importance of curli fimbriae in biofilm extracellular matrix, we observed no change to cellular aggregation for the ΔSTM2441 mutant when also deleted for ΔcsgBA (curli fimbriae) or ΔcsgD (regulator of biofilm formation). We hypothesize that the observed aggregates formed by the ΔSTM2441 mutant may have biofilm-like properties but do not represent mature biofilms; therefore, the mechanisms by which they reduce the neutrophil respiratory burst may differ.
We linked both BapA and flagellins to the inappropriate aggregation of the ΔSTM2441 mutant. BapA is a large 386-kDa surface protein that is involved in initial surface attachment and promotes cell-to-cell interactions leading to biofilm formation at the air-liquid interface (49). Flagellar motility also contributes to the initial bacterial attachment to surfaces during biofilm formation (31, 50, 51). One possibility for the reduced neutrophil respiratory burst is that production of the large surface-associated protein BapA could shield other Salmonella agonists from recognition by neutrophils. Another possibility is that BapA, which is composed of a 27-tandem repeat of bacterial immunoglobulin-like domains, may alter the immune response (52). For example, Staphylococcus aureus possesses two surface-associated proteins with immunoglobulin-binding repeat domains, Sbi and SpA, that block neutrophil Fc receptors in order to evade the immune response (53). Finally, the formation of cellular aggregates by the ΔSTM2441 mutant may also effectively serve to reduce the number of individual bacteria detected by neutrophils, thereby reducing the magnitude of the respiratory burst (14). Further investigation is needed to elucidate the mechanism(s) by which defects in sulfate acquisition lead to the formation of bacterial aggregates in S. Typhimurium and how this phenotype impacts neutrophil antimicrobial responses.
Using a two-step screening approach to identify Salmonella mutants that influence the human neutrophil respiratory burst, we confirmed four genes as important for stimulation of a maximal neutrophil respiratory burst. Two mutants had intriguing phenotypes. We found that STM2201, a putative Lys-R transcriptional regulator, appears to act as a regulator of flagellar motility. We also found that disruption of sulfate acquisition leads to inappropriate cellular aggregation and a resultant reduction in neutrophil respiratory burst. Further investigation into both of these genes is warranted to gain new insight into the myriad of mechanisms by which S. Typhimurium can influence the magnitude of the host immune response.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
All bacterial strains are derivatives of Salmonella enterica serovar Typhimurium ATCC 14028s (Table 1). The multigene deletion (MGD) and single-gene deletion (SGD) mutant library collections were previously described (18). Mutations were moved into a clean genetic background by P22 transduction, and antibiotic cassettes were removed as described previously (54, 55). Unless otherwise indicated, bacteria were grown in Luria-Bertani (LB-Miller) broth or LB agar supplemented with nalidixic acid (50 mg/liter), kanamycin (50 mg/liter), chloramphenicol (20 mg/liter), carbenicillin (100 mg/liter), and tetracycline (20 mg/liter). For assays using bacteria from stationary-phase growth, bacteria were grown in LB broth at 37°C with agitation (225 rpm) overnight. For assays using bacteria from late-exponential-phase growth, bacteria were prepared by diluting overnight cultures 1:100 in LB broth followed by incubation at 37°C with agitation for 3.5 h.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Genotype or descriptiona | Reference or source |
|---|---|---|
| Strains | ||
| HA420 | ATCC 14028.s (spontaneous Nalr) | 61 |
| WN150 | ATCC 14028.s | 62 |
| JE598 | HA420 ΔSPI-1::cm (Nalr, Cmr) | 14 |
| JE183 | HA420 ΔssaK::kan (Nalr, Kanr) | This study |
| JE524 | 14028.s ΔfliC::kan ΔfljB::cm (Kanr, Cmr) | 14 |
| JE1202 | HA420 ΔmotA::kan (Nalr, Kanr) | 14 |
| JE963 | HA420 ΔSTM0018::kan (Nalr, Kanr) | This study |
| JE965 | HA420 ΔSTM1213::kan (Nalr, Kanr) | This study |
| JE967 | HA420 ΔSTM1690::kan (Nalr, Kanr) | This study |
| JE969 | HA420 ΔSTM1696::kan (Nalr, Kanr) | This study |
| JE971 | HA420 ΔSTM2112::kan (Nalr, Kanr) | This study |
| JE973 | HA420 ΔSTM2201::kan (Nalr, Kanr) | This study |
| JE975 | HA420 ΔSTM2441::kan (Nalr, Kanr) | This study |
| JE239 | HA420 carrying pNN387 (Nalr, Kanr, Cmr) | 63 |
| JE240 | HA420 carrying pNN387::rpsMp (Nalr, Kanr, Cmr) | 63 |
| JE241 | HA420 carrying pNN387::prgHp (Nalr, Kanr, Cmr) | 63 |
| JE1318 | JE969 carrying pNN387 (Nalr, Kanr, Cmr) | This study |
| JE1320 | JE969 carrying pNN387::rpsMp (Nalr, Kanr, Cmr) | This study |
| JE1322 | JE969 carrying pNN387::prgHp (Nalr, Kanr, Cmr) | This study |
| JE1324 | JE971 carrying pNN387 (Nalr, Kanr, Cmr) | This study |
| JE1326 | JE971 carrying pNN387::rpsMp (Nalr, Kanr, Cmr) | This study |
| JE1328 | JE971 carrying pNN387::prgHp (Nalr, Kanr, Cmr) | This study |
| JE1330 | JE973 carrying pNN387 (Nalr, Kanr, Cmr) | This study |
| JE1332 | JE973 carrying pNN387::rpsMp (Nalr, Kanr, Cmr) | This study |
| JE1333 | JE973 carrying pNN387::prgHp (Nalr, Kanr, Cmr) | This study |
| JE1335 | JE975 carrying pNN387 (Nalr, Kanr, Cmr) | This study |
| JE1337 | JE975 carrying pNN387::rpsMp (Nalr, Kanr, Cmr) | This study |
| JE1339 | JE975 carrying pNN387::prgHp (Nalr, Kanr, Cmr) | This study |
| JP245 | WN150 Δwza::tetRA (Tetr) | 19 |
| JE1760 | WN150 ΔSPI-1::cm (Cmr) | This study |
| JE1762 | WN150 ΔmotA::kan (Kanr) | This study |
| JE1800 | WN150 ΔSTM2112::kan (Kanr) | This study |
| JE1673 | JE975 carrying pWSK29 (Nalr, Kanr, Ampr) | This study |
| JE1675 | JE975 carrying pWSK29::STM2441 (Nalr, Kanr, Ampr) | This study |
| JE1391 | HA420 ΔSTM2441::frt (Nalr) | This study |
| SF15 | 14028s ΔcsgBA::kan (Kanr) | 65 |
| JE758 | HA420 ΔcsgD::kan (Nalr, Kanr) | 34 |
| JE1939 | HA420 ΔcsgBA::kan (Nalr, Kanr) | This study |
| JE1953 | HA420 ΔbcsA::kan (Nalr, Kanr) | This study |
| JE1963 | HA420 ΔbapA::kan (Nalr, Kanr) | This study |
| JE2018 | HA420 ΔyihQ::cm (Nalr, Cmr) | This study |
| JE1978 | HA420 Δwza::tetRA (Nalr, Tetr) | This study |
| JE1919 | HA420 ΔfliC::frt ΔfljB::frt (Nalr) | This study |
| JE1872 | JE1391 ΔcsgD::kan (Nalr, Kanr) | This study |
| JE2020 | JE1391 ΔcsgBA::kan (Nalr, Kanr) | This study |
| JE2022 | JE1391 ΔbcsA::kan (Nalr, Kanr) | This study |
| JE2024 | JE1391 ΔbapA::kan (Nalr, Kanr) | This study |
| JE2026 | JE1391 ΔyihQ::cm (Nalr, Kanr) | This study |
| JE2031 | JE1391 Δwza-wcaM::tetRA (Nalr, Tetr) | This study |
| JE2033 | JE1919 ΔSTM2441::kan (Nalr, Kanr) | This study |
| Plasmids | ||
| pCP20 | Flp recombinase; Ampr | 55 |
| pWSK29 | Cloning vector; Ampr | 59 |
| pWSK29::STM2441 | pWSK29::STM2441; Ampr | This study |
| pNN387 | Single-copy vector containing promoterless lacZY; Cmr | 66 |
| pNN387::rpsMp | pNN387::rpsMp; Cmr | 63 |
| pNN387::prgHp | pNN387::prgHp; Cmr | 63 |
Nal, nalidixic acid; Cm, chloramphenicol; Kan, kanamycin; Amp, ampicillin; Tet, tetracycline.
For growth curves, overnight cultures were diluted 1:100 into 50 ml LB or M9 minimal medium and grown at 37°C with agitation (225 rpm) for 24 h. M9 minimal medium (48 mM Na2HPO4, 22 mM KH2PO4, 9 mM NaCl, 19 mM NH4Cl, 0.1 mM CaCl2, and 2 mM MgSO4) was supplemented with 0.2% (wt/vol) dextrose as a carbon source and 1 mM l-cysteine, where indicated (56). Samples were taken hourly for 6 h and once at 24 h, diluted, and plated to enumerate CFU. Each experiment was performed on three independent occasions.
For growth in neutrophil-Salmonella coculture medium, bacteria were washed and diluted in phosphate-buffered saline (PBS) and added to 100 μl RPMI 1640 medium (with l-glutamine, without phenol red; Gibco) supplemented with 1 mM Ca2+, 1 mM Mg2+, and 10% normal human serum from male AB donors (Corning) to achieve a final concentration of 5 × 106 CFU. Cultures were incubated standing at 37°C with 5% CO2 for 2 h and then serially diluted and plated to enumerate CFU. Each experiment was performed on three independent occasions.
For analysis of the effects of sulfur on bacterial viability, a sulfur-free (SF) minimal medium was used. SF medium was a modification of M9 minimal medium using 2 mM MgCl2 as a magnesium source. SF medium was supplemented with 0.5 mM sodium tetrathionate (Na2S4O6), 1 mM sodium thiosulfate (Na2S2O3), or 1 mM l-cysteine where indicated. Overnight cultures were diluted 1:100 into SF media and grown at 37°C with agitation for 24 h. Bacterial viability was assessed at 24 h by serial dilution and plating to enumerate CFU. The number of generations was established as (log10[final CFU] − log10[start CFU])/0.301. Each experiment was performed on three independent occasions.
Human subjects.
Neutrophils were isolated from peripheral blood of healthy adult volunteers. All participants provided written informed consent. The study was approved by the Institutional Research Ethics Committee of North Carolina State University (IRB approval number 616).
Neutrophil respiratory burst in coculture with Salmonella.
Neutrophils (PMN) were isolated from whole blood by a Ficoll gradient centrifugation technique as previously described (14). Isolated PMNs exhibited greater than 98% viability and 95% purity as determined by trypan blue exclusion. Purified PMNs were suspended in RPMI 1640 medium (with l-glutamine, without phenol red; Gibco) supplemented with 1 mM Ca2+, 1 mM Mg2+, and 10% normal human serum from male AB donors (Corning) to a final concentration of 1.15 × 106 PMN/ml. Neutrophils were primed with human recombinant granulocyte-macrophage colony-stimulating factor (GMCSF; 30 ng/ml) for 30 min at 37°C with 5% CO2.
For library screening, bacteria from stationary-phase growth were diluted 1:1 in phosphate-buffered saline (PBS) prior to addition to the neutrophil culture. For individual mutant strain evaluation, bacteria from stationary- and late-exponential-growth phases were used where indicated to inoculate neutrophil cultures. Bacteria were washed and diluted in PBS to achieve a multiplicity of infection (MOI) of 50:1. Bacteria were serially diluted and plated on LB agar to establish the final MOI.
Intracellular ROS was measured by fluorescence of rhodamine generated by oxidation of dihydrorhodamine-123 (DHR-123) as previously described (14). Neutrophils were allowed to settle for 10 min on black polystyrene 96-well plates coated with 5% fetal calf serum (FCS) prior to addition of 10 μM DHR-123 and either bacteria or controls. Negative controls in each plate included neutrophils with assay medium (RPMI medium plus 10% human serum) and bacterial growth medium (LB) to ensure neutrophils were not inappropriately activated. Neutrophils stimulated with phorbol 12-myristate 13-acetate (PMA) (50 ng/ml; Sigma-Aldrich) were included on each plate as a positive control. A mutant we previously determined to have an altered respiratory burst (ΔSPI-1) was also included on each plate as a control for the assay (14). Plates were incubated at 37°C with 5% CO2. Relative fluorescence units (RFU; 485 nm excitation, 528 nm emission) were determined prior to incubation and then hourly for 3 h for library screening and every 30 min for 2 h for individual strain evaluation (Synergy HTX; BioTek). Neutrophils from 2 different blood donors were used for library screens, and neutrophils from 3 to 4 different blood donors were used for individual strain evaluation.
Total intracellular and extracellular ROS were measured by luminol (5-amino-2,3-dihydro-1,4-phthalazinedione; 1 mM; Sigma-Aldrich) enhanced chemiluminescence as previously described (14). Briefly, neutrophils were aliquoted onto a white polystyrene 96-well plate coated with 5% FCS. Neutrophils were allowed to settle for 10 min prior to addition of luminol, and a baseline luminescence measurement was obtained (integration time, 1s) (Synergy HTX; BioTek). Neutrophils were then stimulated with bacteria or PMA as a positive control. The plate was incubated at 37°C and luminescence measured every 5 min for 90 min. The peak and time to peak luminescence were determined for each strain and normalized to the WT where indicated.
Bacterial motility assays.
Swimming motility was assayed on plates containing 0.3% Difco Bacto agar (LB Miller base, 25 g/liter) as described (57). Overnight cultures were grown at 37°C with agitation, and cell concentration was normalized by the optical density at 600 nm (OD600). Bacteria were spotted onto plates and incubated at 37°C for 4 h. The diameter of each colony was measured and compared to that for the wild-type organism on the same plate. Each assay was performed on three separate occasions in 4 to 5 replicates. Swimming assays were repeated on individual plates for photographs (ChemiDoc MP).
β-Galactosidase assays.
Bacteria bearing plasmid constructs were grown overnight at 37°C with agitation in LB broth supplemented with the appropriate antibiotic. For induction of T3SS-1 expression, overnight cultures were diluted 1:100 into LB with antibiotic and incubated at 37°C with agitation for 3.5 h. β-Galactosidase activity was determined from cell pellets using standard methodology and Miller units determined using the following equation: 1,000 × [OD420 − (1.75 × OD550)]/[time × volume × OD600] (58). Experiments were performed on five separate occasions.
Complementing plasmid construction.
Genomic DNA was isolated from S. Typhimurium using the GenElute bacterial genomic DNA kit (Sigma-Aldrich). Restriction endonuclease sequences were incorporated into the following primer sequences to facilitate cloning: STM2441BamH1Fwd, 5′-GTAGGA TCCGCGATTTTACTGGGCGCATC-3′, and STM2441Kpn1Rev, 5′-GTAGGTACCTGTAATTTGACCAGCGGCGT-3′. A 1.5-kb product for STM2441 including the native promoter was generated from template genomic DNA using Q5 DNA polymerase (New England BioLabs) with an annealing temperature of 72°C and an extension time of 40 s for 35 cycles. The expected size of the PCR product was confirmed by agarose gel electrophoresis. The PCR product was digested with the restriction endonucleases BamHI and KpnI (New England BioLabs) and purified (QIAquick PCR purification kit; Qiagen). The low-copy-number vector, pWSK29 (59), was sequentially digested with BamHI and KpnI followed by dephosphorylation with shrimp alkaline phosphatase (New England BioLabs). Ligation of the product and linearized vector was performed overnight at 14°C with T4 DNA ligase (New England BioLabs). The resulting construct was transformed into DH5α Escherichia coli by heat shock, and transformants were obtained by selection on LB agar with carbenicillin and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal; 40 μg/ml). Transformants were purified twice and the plasmids were isolated (QIAprep miniprep kit; Qiagen). The insert size length was determined by restriction digestion of the plasmid followed by agarose gel electrophoresis, and the expected sequence was confirmed by Sanger sequencing (Eton Bioscience). The complementing plasmid was transformed into restriction-deficient modification-positive S. Typhimurium LB5000 by electroporation, and transformants were isolated by selection on LB agar with carbenicillin and purified twice (60). Plasmids were then transformed into the mutant by electroporation. The resulting strains were purified twice and then stored in glycerol stocks at −80°C.
Crystal violet assays.
Bacteria were grown to both stationary and late-exponential phases in 50-ml conical tubes in 5 ml volume. Cell density was determined by OD600. Nonadherent cells were removed by decanting, and tubes were washed twice with double-distilled water (ddH2O). Adherent cells were stained with 0.1% crystal violet for 15 min followed by 4 washes with ddH2O. Tubes were inverted and allowed to dry overnight. The crystal violet was solubilized in 30% acetic acid by vortexing and quantified by measurement of OD550.
Statistical analyses.
For MGD library screening, an altered respiratory burst was determined using the raw rhodamine RFU data, comparing each mutant to the WT control on the same 96-well plate by a two-way analysis of variance (ANOVA) with a Dunnett’s test for multiple comparisons. For screening the SGD minilibrary, we normalized the RFU elicited by SGD mutants to the RFU elicited by the WT on the same 96-well plate at a given time to reduce bias between plates. Statistical significance was determined by two-way ANOVA with a Dunnett’s test for multiple comparisons. For all other assays, statistical significance was determined using Student’s t test or two-way ANOVA with Dunnett’s correction for multiple comparisons where indicated. Significance was set at a P value of <0.05. Analyses were performed using GraphPad Prism version 8.0.
ACKNOWLEDGMENTS
A portion of this work was supported by startup funds from the University of Wisconsin—Madison (to J.R.E.). T.L.W. was supported in part by 5T32OD01113. J.R.E. was supported in part by USDA-NIFA 2018-67017-27632 and NIAID K08AI108794. M.K.S. was supported in part by NIH Office of the Director K01OD015136.
The funders played no role in the experimental design or execution of the work.
Footnotes
Supplemental material is available online only.
iai.00701-20-s0001.pdf (398.1KB, pdf)
iai.00701-20-s0002.xlsx (25.2KB, xlsx)
iai.00701-20-s0003.xlsx (19.4KB, xlsx)
iai.00701-20-s0004.xlsx (31.7KB, xlsx)
iai.00701-20-s0001.xlsx (26KB, xlsx)
Contributor Information
J. R. Elfenbein, Email: jelfenbein@wisc.edu.
Manuela Raffatellu, University of California San Diego School of Medicine.
REFERENCES
- 1.Kirk MD, Pires SM, Black RE, Caipo M, Crump JA, Devleesschauwer B, Dopfer D, Fazil A, Fischer-Walker CL, Hald T, Hall AJ, Keddy KH, Lake RJ, Lanata CF, Torgerson PR, Havelaar AH, Angulo FJ. 2015. World Health Organization estimates of the global and regional disease burden of 22 foodborne bacterial, protozoal, and viral diseases, 2010: a data synthesis. PLoS Med 12:e1001921. doi: 10.1371/journal.pmed.1001921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Santos RL, Zhang S, Tsolis RM, Baumler AJ, Adams LG. 2002. Morphologic and molecular characterization of Salmonella Typhimurium infection in neonatal calves. Vet Pathol 39:200–215. doi: 10.1354/vp.39-2-200. [DOI] [PubMed] [Google Scholar]
- 3.Zhang S, Santos RL, Tsolis RM, Stender S, Hardt W-D, Bäumler AJ, Adams LG. 2002. The Salmonella enterica serotype Typhimurium effector proteins SipA, SopA, SopB, SopD, and SopE2 act in concert to induce diarrhea in calves. Infect Immun 70:3843–3855. doi: 10.1128/iai.70.7.3843-3855.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Winter SE, Thiennimitr P, Winter MG, Butler BP, Huseby DL, Crawford RW, Russell JM, Bevins CL, Adams LG, Tsolis RM, Roth JR, Baumler AJ. 2010. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature 467:426–429. doi: 10.1038/nature09415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu JZ, Jellbauer S, Poe AJ, Ton V, Pesciaroli M, Kehl-Fie TE, Restrepo NA, Hosking MP, Edwards RA, Battistoni A, Pasquali P, Lane TE, Chazin WJ, Vogl T, Roth J, Skaar EP, Raffatellu M. 2012. Zinc sequestration by the neutrophil protein calprotectin enhances Salmonella growth in the inflamed gut. Cell Host Microbe 11:227–239. doi: 10.1016/j.chom.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thiennimitr P, Winter SE, Winter MG, Xavier MN, Tolstikov V, Huseby DL, Sterzenbach T, Tsolis RM, Roth JR, Bäumler AJ. 2011. Intestinal inflammation allows Salmonella to use ethanolamine to compete with the microbiota. Proc Natl Acad Sci U S A 108:17480–17485. doi: 10.1073/pnas.1107857108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Behnsen J, Perez-Lopez A, Nuccio SP, Raffatellu M. 2015. Exploiting host immunity: the Salmonella paradigm. Trends Immunol 36:112–120. doi: 10.1016/j.it.2014.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eckstein MR, Baehner RL, Nathan DG. 1971. Amino acid oxidase of leukocytes in relation to H2O2-mediated bacterial killing. J Clin Invest 50:1985–1991. doi: 10.1172/JCI106690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clifford DP, Repine JE. 1982. Hydrogen peroxide mediated killing of bacteria. Mol Cell Biochem 49:143–149. doi: 10.1007/BF00231175. [DOI] [PubMed] [Google Scholar]
- 10.DeLeo FR, Allen LA, Apicella M, Nauseef WM. 1999. NADPH oxidase activation and assembly during phagocytosis. J Immunol 163:6732–6740. [PubMed] [Google Scholar]
- 11.Babior BM, Lambeth JD, Nauseef W. 2002. The neutrophil NADPH oxidase. Arch Biochem Biophys 397:342–344. doi: 10.1006/abbi.2001.2642. [DOI] [PubMed] [Google Scholar]
- 12.DeLeo FR, Renee J, McCormick S, Nakamura M, Apicella M, Weiss JP, Nauseef WM. 1998. Neutrophils exposed to bacterial lipopolysaccharide upregulate NADPH oxidase assembly. J Clin Invest 101:455–463. doi: 10.1172/JCI949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zagryazhskaya A, Galkina S, Grishina Z, Viryasova G, Romanova J, Lazarenko M, Steinhilber D, Sud'ina G. 2012. Neutrophil cellular responses to various Salmonella Typhimurium LPS chemotypes. Salmonella - a diversified superbug. IntechOpen, London, United Kingdom. doi: 10.5772/29087. [DOI] [Google Scholar]
- 14.Westerman TL, Bogomolnaya L, Andrews-Polymenis HL, Sheats MK, Elfenbein JR. 2018. The Salmonella type-3 secretion system-1 and flagellar motility influence the neutrophil respiratory burst. PLoS One 13:e0203698. doi: 10.1371/journal.pone.0203698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tursi SA, Tükel Ç. 2018. Curli-containing enteric biofilms inside and out: matrix composition, immune recognition, and disease implications. Microbiol Mol Biol Rev 82:e00028-18. doi: 10.1128/MMBR.00028-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hahn MM, Gunn JS. 2020. Salmonella extracellular polymeric substances modulate innate phagocyte activity and enhance tolerance of biofilm-associated bacteria to oxidative stress. Microorganisms 8:253. doi: 10.3390/microorganisms8020253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Westerman TL, Sheats MK, Elfenbein JR. 8 November 2020. Sulfate import in Salmonella Typhimurium impacts bacterial aggregation and the neutrophil respiratory burst. BioRxiv doi: 10.1101/2020.11.06.37243. [DOI] [PMC free article] [PubMed]
- 18.Porwollik S, Santiviago CA, Cheng P, Long F, Desai P, Fredlund J, Srikumar S, Silva CA, Chu W, Chen X, Canals R, Reynolds MM, Bogomolnaya L, Shields C, Cui P, Guo J, Zheng Y, Endicott-Yazdani T, Yang HJ, Maple A, Ragoza Y, Blondel CJ, Valenzuela C, Andrews-Polymenis H, McClelland M. 2014. Defined single-gene and multi-gene deletion mutant collections in Salmonella enterica sv. Typhimurium. PLoS One 9:e99820. doi: 10.1371/journal.pone.0099820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pando JM, Karlinsey JE, Lara JC, Libby SJ, Fang FC. 2017. The Rcs-regulated colanic acid capsule maintains membrane potential in Salmonella enterica serovar Typhimurium. mBio 8:e00808-17. doi: 10.1128/mBio.00808-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Danese PN, Pratt LA, Kolter R. 2000. Exopolysaccharide production is required for development of Escherichia coli K-12 biofilm architecture. J Bacteriol 182:3593–3596. doi: 10.1128/jb.182.12.3593-3596.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prigent-Combaret C, Vidal O, Dorel C, Lejeune P. 1999. Abiotic surface sensing and biofilm-dependent regulation of gene expression in Escherichia coli. J Bacteriol 181:5993–6002. doi: 10.1128/JB.181.19.5993-6002.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Prigent-Combaret C, Prensier G, Le Thi TT, Vidal O, Lejeune P, Dorel C. 2000. Developmental pathway for biofilm formation in curli-producing Escherichia coli strains: role of flagella, curli and colanic acid. Environ Microbiol 2:450–464. doi: 10.1046/j.1462-2920.2000.00128.x. [DOI] [PubMed] [Google Scholar]
- 23.Ranjit DK, Young KD. 2016. Colanic acid intermediates prevent de novo shape recovery of Escherichia coli spheroplasts, calling into question biological roles previously attributed to colanic acid. J Bacteriol 198:1230–1240. doi: 10.1128/JB.01034-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miajlovic H, Cooke NM, Moran GP, Rogers TRF, Smith SG. 2014. Response of extraintestinal pathogenic Escherichia coli to human serum reveals a protective role for Rcs-regulated exopolysaccharide colanic acid. Infect Immun 82:298–305. doi: 10.1128/IAI.00800-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aguilar-Barajas E, Diaz-Perez C, Ramirez-Diaz MI, Riveros-Rosas H, Cervantes C. 2011. Bacterial transport of sulfate, molybdate, and related oxyanions. Biometals 24:687–707. doi: 10.1007/s10534-011-9421-x. [DOI] [PubMed] [Google Scholar]
- 26.Bedouhene S, Moulti-Mati F, Hurtado-Nedelec M, Dang PM, El-Benna J. 2017. Luminol-amplified chemiluminescence detects mainly superoxide anion produced by human neutrophils. Am J Blood Res 7:41–48. [PMC free article] [PubMed] [Google Scholar]
- 27.Dreyfuss J. 1964. Characterization of a sulfate- and thiosulfate-transporting system in Salmonella Typhimurium. J Biol Chem 239:2292–2297. doi: 10.1016/S0021-9258(20)82233-9. [DOI] [PubMed] [Google Scholar]
- 28.Barrett EL, Chang GW. 1979. Cysteine auxotrophs of Salmonella Typhimurium which grow without cysteine in a hydrogen/carbon dioxide atmosphere. J Gen Microbiol 115:513–516. doi: 10.1099/00221287-115-2-513. [DOI] [PubMed] [Google Scholar]
- 29.Furne J, Springfield J, Koenig T, DeMaster E, Levitt MD. 2001. Oxidation of hydrogen sulfide and methanethiol to thiosulfate by rat tissues: a specialized function of the colonic mucosa. Biochem Pharmacol 62:255–259. doi: 10.1016/s0006-2952(01)00657-8. [DOI] [PubMed] [Google Scholar]
- 30.Gunn JS, Bakaletz LO, Wozniak DJ. 2016. What's on the outside matters: the role of the extracellular polymeric substance of Gram-negative biofilms in evading host immunity and as a target for therapeutic intervention. J Biol Chem 291:12538–12546. doi: 10.1074/jbc.R115.707547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pratt LA, Kolter R. 1998. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol Microbiol 30:285–293. doi: 10.1046/j.1365-2958.1998.01061.x. [DOI] [PubMed] [Google Scholar]
- 32.Wrande M, Andrews-Polymenis H, Twedt DJ, Steele-Mortimer O, Porwollik S, McClelland M, Knodler LA. 2016. Genetic determinants of Salmonella enterica serovar Typhimurium proliferation in the cytosol of epithelial cells. Infect Immun 84:3517–3526. doi: 10.1128/IAI.00734-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang HJ, Bogomolnaya LM, Elfenbein JR, Endicott-Yazdani T, Reynolds MM, Porwollik S, Cheng P, Xia XQ, McClelland M, Andrews-Polymenis H. 2016. Novel two-step hierarchical screening of mutant pools reveals mutants under selection in chicks. Infect Immun 84:1226–1238. doi: 10.1128/IAI.01525-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Griewisch KF, Pierce JG, Elfenbein JR. 2020. Genetic determinants of Salmonella resistance to the biofilm inhibitory effects of a synthetic 4-oxazolidinone small molecule. Appl Environ Microbiol 86:e01120-20. doi: 10.1128/AEM.01120-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Proshkin S, Rahmouni AR, Mironov A, Nudler E. 2010. Cooperation between translating ribosomes and RNA polymerase in transcription elongation. Science 328:504–508. doi: 10.1126/science.1184939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kroger C, Colgan A, Srikumar S, Handler K, Sivasankaran SK, Hammarlof DL, Canals R, Grissom JE, Conway T, Hokamp K, Hinton JC. 2013. An infection-relevant transcriptomic compendium for Salmonella enterica serovar Typhimurium. Cell Host Microbe 14:683–695. doi: 10.1016/j.chom.2013.11.010. [DOI] [PubMed] [Google Scholar]
- 37.Parra-Lopez C, Baer MT, Groisman EA. 1993. Molecular genetic analysis of a locus required for resistance to antimicrobial peptides in Salmonella Typhimurium. EMBO J 12:4053–4062. doi: 10.1002/j.1460-2075.1993.tb06089.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rodas PI, Contreras I, Mora GC. 2010. Salmonella enterica serovar Typhi has a 4.1 kb genetic island inserted within the sapABCDF operon that causes loss of resistance to the antimicrobial peptide protamine. J Antimicrob Chemother 65:1624–1630. doi: 10.1093/jac/dkq197. [DOI] [PubMed] [Google Scholar]
- 39.McClelland M, Sanderson KE, Spieth J, Clifton SW, Latreille P, Courtney L, Porwollik S, Ali J, Dante M, Du F, Hou S, Layman D, Leonard S, Nguyen C, Scott K, Holmes A, Grewal N, Mulvaney E, Ryan E, Sun H, Florea L, Miller W, Stoneking T, Nhan M, Waterston R, Wilson RK. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852–856. doi: 10.1038/35101614. [DOI] [PubMed] [Google Scholar]
- 40.Gao Y, Yurkovich JT, Seo SW, Kabimoldayev I, Dräger A, Chen K, Sastry AV, Fang X, Mih N, Yang L, Eichner J, Cho B-K, Kim D, Palsson BO. 2018. Systematic discovery of uncharacterized transcription factors in Escherichia coli K-12 MG1655. Nucleic Acids Res 46:10682–10696. doi: 10.1093/nar/gky752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sirko A, Hryniewicz M, Hulanicka D, Böck A. 1990. Sulfate and thiosulfate transport in Escherichia coli K-12: nucleotide sequence and expression of the cysTWAM gene cluster. J Bacteriol 172:3351–3357. doi: 10.1128/jb.172.6.3351-3357.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sturgill G, Toutain CM, Komperda J, O'Toole GA, Rather PN. 2004. Role of CysE in production of an extracellular signaling molecule in Providencia stuartii and Escherichia coli: loss of cysE enhances biofilm formation in Escherichia coli. J Bacteriol 186:7610–7617. doi: 10.1128/JB.186.22.7610-7617.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rossi E, Motta S, Mauri P, Landini P. 2014. Sulfate assimilation pathway intermediate phosphoadenosine 59-phosphosulfate acts as a signal molecule affecting production of curli fibres in Escherichia coli. Microbiology (Reading) 160:1832–1844. doi: 10.1099/mic.0.079699-0. [DOI] [PubMed] [Google Scholar]
- 44.Hufnagel DA, Price JE, Stephenson RE, Kelley J, Benoit MF, Chapman MR. 2017. Thiol starvation induces redox-mediated dysregulation of Escherichia coli biofilm components. J Bacteriol 200:e00389-17. doi: 10.1128/JB.00389-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tükel Ç, Wilson RP, Nishimori JH, Pezeshki M, Chromy BA, Bäumler AJ. 2009. Responses to amyloids of microbial and host origin are mediated through Toll-like receptor 2. Cell Host Microbe 6:45–53. doi: 10.1016/j.chom.2009.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tükel C, Nishimori JH, Wilson RP, Winter MG, Keestra AM, van Putten JPM, Bäumler AJ. 2010. Toll-like receptors 1 and 2 cooperatively mediate immune responses to curli, a common amyloid from enterobacterial biofilms. Cell Microbiol 12:1495–1505. doi: 10.1111/j.1462-5822.2010.01485.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tursi SA, Lee EY, Medeiros NJ, Lee MH, Nicastro LK, Buttaro B, Gallucci S, Wilson RP, Wong GC, Tükel Ç. 2017. Bacterial amyloid curli acts as a carrier for DNA to elicit an autoimmune response via TLR2 and TLR9. PLoS Pathog 13:e1006315. doi: 10.1371/journal.ppat.1006315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rapsinski GJ, Wynosky-Dolfi MA, Oppong GO, Tursi SA, Wilson RP, Brodsky IE, Tükel Ç. 2015. Toll-like receptor 2 and NLRP3 cooperate to recognize a functional bacterial amyloid, curli. Infect Immun 83:693–701. doi: 10.1128/IAI.02370-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Latasa C, Roux A, Toledo-Arana A, Ghigo JM, Gamazo C, Penades JR, Lasa I. 2005. BapA, a large secreted protein required for biofilm formation and host colonization of Salmonella enterica serovar Enteritidis. Mol Microbiol 58:1322–1339. doi: 10.1111/j.1365-2958.2005.04907.x. [DOI] [PubMed] [Google Scholar]
- 50.Wood TK, González Barrios AF, Herzberg M, Lee J. 2006. Motility influences biofilm architecture in Escherichia coli. Appl Microbiol Biotechnol 72:361–367. doi: 10.1007/s00253-005-0263-8. [DOI] [PubMed] [Google Scholar]
- 51.Friedlander RS, Vogel N, Aizenberg J. 2015. Role of flagella in adhesion of Escherichia coli to abiotic surfaces. Langmuir 31:6137–6144. doi: 10.1021/acs.langmuir.5b00815. [DOI] [PubMed] [Google Scholar]
- 52.Guttula D, Yao M, Baker K, Yang L, Goult BT, Doyle PS, Yan J. 2019. Calcium-mediated protein folding and stabilization of Salmonella biofilm-associated protein A. J Mol Biol 431:433–443. doi: 10.1016/j.jmb.2018.11.014. [DOI] [PubMed] [Google Scholar]
- 53.Smith EJ, Visai L, Kerrigan SW, Speziale P, Foster TJ. 2011. The Sbi protein is a multifunctional immune evasion factor of Staphylococcus aureus. Infect Immun 79:3801–3809. doi: 10.1128/IAI.05075-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sternberg NL, Maurer R. 1991. Bacteriophage-mediated generalized transduction in Escherichia coli and Salmonella Typhimurium. Methods Enzymol 204:18–43. doi: 10.1016/0076-6879(91)04004-8. [DOI] [PubMed] [Google Scholar]
- 55.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sambrook J, Fritsch E, Maniatis T. 1989. Molecular cloning: a laboratory Manual. vol 3. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 57.Bogomolnaya LM, Aldrich L, Ragoza Y, Talamantes M, Andrews KD, McClelland M, Andrews-Polymenis HL. 2014. Identification of novel factors involved in modulating motility of Salmonella enterica serotype typhimurium. PLoS One 9:e111513. doi: 10.1371/journal.pone.0111513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 59.Wang RF, Kushner SR. 1991. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene 100:195–199. doi: 10.1016/0378-1119(91)90366-J. [DOI] [PubMed] [Google Scholar]
- 60.Bullas LR, Ryu JI. 1983. Salmonella Typhimurium LT2 strains which are r− m+ for all three chromosomally located systems of DNA restriction and modification. J Bacteriol 156:471–474. doi: 10.1128/JB.156.1.471-474.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bogomolnaya LM, Santiviago CA, Yang H-J, Baumler AJ, Andrews-Polymenis HL. 2008. 'Form variation' of the O12 antigen is critical for persistence of Salmonella Typhimurium in the murine intestine. Mol Microbiol 70:1105–1119. doi: 10.1111/j.1365-2958.2008.06461.x. [DOI] [PubMed] [Google Scholar]
- 62.Navarre WW, Porwollik S, Wang Y, McClelland M, Rosen H, Libby SJ, Fang FC. 2006. Selective silencing of foreign DNA with low GC content by the H-NS protein in Salmonella. Science 313:236–238. doi: 10.1126/science.1128794. [DOI] [PubMed] [Google Scholar]
- 63.Zheng Y, Sambou T, Bogomolnaya LM, Cirillo JD, McClelland M, Andrews-Polymenis H. 2013. The EAL domain containing protein STM2215 (rtn) is needed during Salmonella infection and has cyclic di-GMP phosphodiesterase activity. Mol Microbiol 89:403–419. [DOI] [PubMed] [Google Scholar]
- 64.Reference deleted.
- 65.Weening EH, Barker JD, Laarakker MC, Humphries AD, Tsolis RM, Bäumler AJ. 2005. The Salmonella enterica serotype Typhimurium lpf, bcf, stb, stc, std, and sth fimbrial operons are required for intestinal persistence in mice. Infect Immun 73:3358–3366. doi: 10.1128/IAI.73.6.3358-3366.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Elledge SJ, Sugiono P, Guarente L, Davis RW. 1989. Genetic selection for genes encoding sequence-specific DNA-binding proteins. Proc Natl Acad Sci U S A 86:3689–3693. doi: 10.1073/pnas.86.10.3689. [DOI] [PMC free article] [PubMed] [Google Scholar]