ABSTRACT
Lung adenocarcinoma (LUAD) is a common type of malignancy of lung cancers. Long intergenic noncoding RNAs (lincRNAs) have emerged as crucial regulators of various cancers, including LUAD. LINC01006 is a newly discovered long noncoding RNA (lncRNA) whose function in LUAD remains to be explored. This study is to explore the role of LINC01006 in LUAD. Quantitative real-time PCR (RT-qPCR) analysis and Western blotting were used to determine the expression levels and protein levels, respectively. Functional assays and animal experiments investigated the role of LINC01006 both in vivo and in vitro. Moreover, TOP/FOP assay was performed to detect the activation of the Wnt/β-catenin signaling pathway. The interaction between LINC01006 and microRNA 29-2-3-p (miR-29-2-3-p)/catenin beta 1 (CTNNB1) was explored by RNA binding protein immunoprecipitation (RIP), RNA pulldown, luciferase reporter assays, and rescue experiments. According to the results, LINC01006 was highly expressed in LUAD tissues and cell lines. LINC01006 knockdown significantly suppressed cell proliferative, migratory, and epithelial-mesenchymal transition (EMT) capacities and tumor development. Moreover, LINC01006 enhanced CTNNB1 via sequestering miR-129-2-3p and activated the Wnt/β-catenin pathway in LUAD. Overall, LINC01006 promotes LUAD development via activating the Wnt/β-catenin pathway, implying that LINC01006 might be a promising biomarker for LUAD treatment.
KEYWORDS: LINC01006, miR-129-2-3p, CTNNB1, LUAD, Wnt/β-catenin pathway
INTRODUCTION
As the most leading cause of cancer death all around the world, lung cancer is characterized with fastest-growing occurrence and mortality (1). Non-small cell lung cancer (NSCLC) constitutes approximately 80% of lung cancers, among which lung adenocarcinoma (LUAD) is the most prevalent (2). Despite the fact that early diagnosis and treatment of LUAD have improved, the 5-year overall survival rate for LUAD patients remains low (3, 4). Regulation of the Wnt/β-catenin pathway has been reported to exert crucial functions in cancers, including LUAD via impacting tumorigenesis, chemotherapy resistance, radiotherapy resistance, and so on (5–7). Therefore, exploring mechanisms involved in LUAD development and identifying more effective molecular targets for LUAD are urgent.
Long noncoding RNAs (lncRNAs) are a set of transcripts more than 200 nucleotides in length with no protein-coding abilities (8). They have been reported as crucial regulators in multiple cellular processes, such as cell differentiation, growth, and apoptosis, thereby affecting cancer progression (9–11). For instance, lncRNA TUG1 enhances the expression of Runx2 through sponging microRNA 204-5p (miR-204-5p), promoting osteoblast differentiation in calcific aortic valve disease (CAVD) (12). LncRNA HOTAIR propels cell growth, metastasis, and apoptosis via the miR-20a-5p/HMGA2 axis in breast cancer (13). Additionally, lncRNAs have exerted functions in lung cancer (14, 15). LncRNA H19 depresses miR-196b to elevate LIN28B, facilitating cell proliferation in lung cancer (16). HOTAIR expression is elevated in lung cancers and correlates with metastasis and poor prognosis (17).
As a kind of lncRNA, long intergenic noncoding RNA (lincRNA) was also recorded as an important regulator in cancer. Among the upregulated lincRNAs found on the GEPIA database (http://gepia2.cancer-pku.cn/#index), LINC01006 is a novel lincRNA with 758 bp and has seldom been reported in cancers, not to mention in LUAD. Furthermore, according to the GTEx database, LINC01006 was found to be notably downregulated in healthy lung tissues. Therefore, we chose LINC01006 for further study. LINC01006 functions as a modular scaffold to induce other active or repressive regulator, playing an important role in imprinting control, immune responses, as well as cancer development. For example, LINC01006 facilitates cell proliferation and metastasis in pancreatic cancer by sequestering miR-2682-5p and regulating HOXB8 expression (18). Besides, LINC01006 has also been reported to be involved in the process of gastric cancer (19). However, the mechanism by which LINC01006 functions in LUAD development remains to be elucidated. The goal of this paper is to investigate the role of LINC01006 in LUAD in more detail and probe into its potential regulatory mechanism.
RESULTS
LINC01006 promotes cell proliferation, migration, and EMT in LUAD.
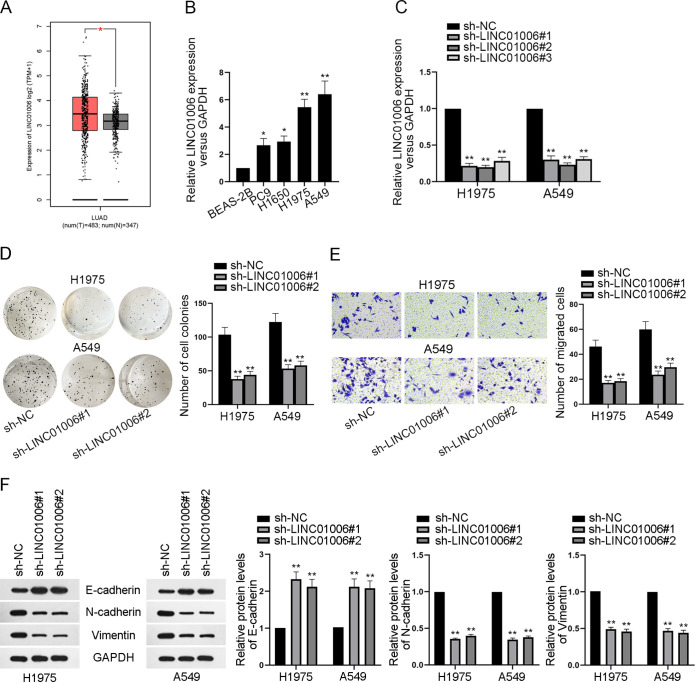
To investigate the relevance of LINC01006 to LUAD, we first explored the expression of LINC01006 in LUAD tissues. According to the GEPIA database (http://gepia2.cancer-pku.cn/#index), LINC01006 was significantly elevated in LUAD tissues in than in healthy tissues (Fig. 1A). Besides, quantitative real-time PCR (RT-qPCR) analysis revealed that LINC01006 was overexpressed in LUAD cell lines (PC9, H1650, H1975, and A549) compared with the nontumor cell line (BEAS-2B) (Fig. 1B). Hence, we conjectured that LINC01006 was an oncogene in LUAD cells. In order to further disclose the role of LINC01006 in LUAD, a series of functional assays were carried out. Before that, we first knocked down LINC01006 in H1975 and A549 cell lines via the transfection of sh-NC and sh-LINC01006#1/2/3, respectively, to verify the knockdown efficiency of these plasmids (Fig. 1C). Considering that LINC01006#1 and LINC01006#2 functioned better than LINC01006#3 at silencing the expression of LINC01006 in LUAD cells, LINC01006#1 and LINC01006#2 were selected for the knockdown in following assays. Colony formation assay was implemented to detect the proliferation of H1975 and A549 cell lines. Results suggested that depletion of LINC01006 efficiently attenuated cell proliferation (Fig. 1D). Moreover, transwell assay demonstrated that the number of cells migrating into the lower chambers declined after the silence of LINC01006, indicating that LINC01006 downregulation inhibited the migratory ability of LUAD cells (Fig. 1E). Besides, Western blotting was applied to evaluate the protein levels of EMT markers after LINC01006 was silenced. It turned out that knockdown of LINC01006 overtly enhanced the protein levels of epithelial marker (E-cadherin). In contrast, protein levels of mesenchymal markers (N-cadherin and vimentin) were terribly decreased, suggesting the reduction in cell epithelial-mesenchymal transition (EMT) (Fig. 1F). Taken together, LINC01006 exerts the carcinogenic function in LUAD cells.
FIG 1.
LINC01006 promotes cell proliferation, migration, and EMT in LUAD. (A) The expression of LINC01006 in healthy lung tissues and LUAD tumor tissues was analyzed by examining the GEPIA database (http://gepia2.cancer-pku.cn/#index). (B) LINC01006 expression in human healthy lung epithelial cell line (BEAS-2B) and LUAD cell lines (PC9, H1650, H1975, and A549) was evaluated by RT-qPCR. (C) RT-qPCR was applied to assess the knockdown efficiency of sh-LINC01006#1, sh-LINC01006#2, and sh-LINC01006#3. (D) Colony formation assay was implemented to investigate the proliferative capacity of LUAD cells. (E) Cell migration before and after LINC01006 knockdown was examined by transwell assay. (F) The protein levels of epithelial marker (E-cadherin) and mesenchymal markers (N-cadherin and vimentin) were analyzed by Western blotting. *, P < 0.05; **, P < 0.01.
Depletion of LINC01006 inhibits LUAD tumor growth in vivo.
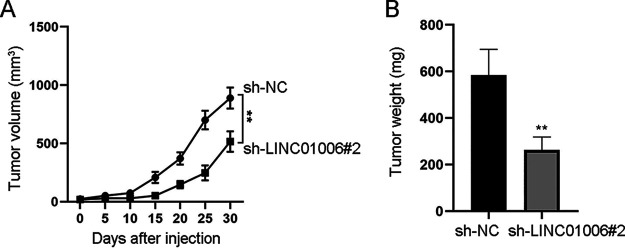
To further illustrate the function of LINC01006 on LUAD tumor growth, we performed animal experiments. As shown in Fig. 2A, tumor growth was much slower, and tumor volume was much smaller in the sh-LINC01006#2 group compared with the control group. As expected, the weight of tumors excised from mice injected with the sh-LINC01006#2-transfected cells was obviously less than that of the control group (Fig. 2B). Overall, LINC01006 promotes LUAD tumor growth in vivo.
FIG 2.
Depletion of LINC01006 inhibits LUAD tumor growth in vivo. (A) Tumor growth curve displayed the tumor volume and tumor growth rate. (B) Tumor weight in sh-NC and sh-LINC01006#2 groups was assessed. **, P < 0.01.
Downregulation of LINC01006 inactivates the Wnt/β-catenin signaling pathway in LUAD cells.
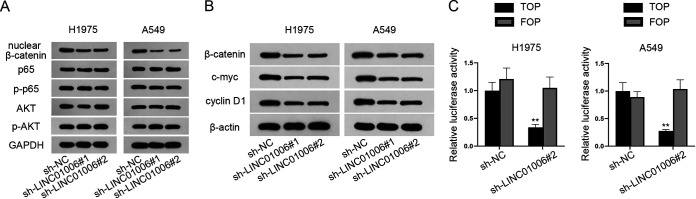
It has been widely reported that various signaling pathways are involved in lncRNA-mediated cancer progression (20, 21). We speculated there might be one signaling pathway participating in LUAD development. Through Western blot analysis, protein levels of key factors (β-catenin, p65, phosphorylated p65 [p-p65], AKT, and phosphorylated AKT [p-AKT]) in several common signaling pathways were evaluated after LINC01006 was knocked down. The analysis indicated that the protein level of β-catenin (the marker of Wnt/β-catenin signaling pathway) was decreased dramatically, whereas that of p65/p-p65 or AKT/p-AKT (markers of NF-κB or phosphatidylinositol 3-kinase [PI3K]/AKT pathways, respectively) was not significantly affected in H1975 and A549 cells (Fig. 3A). It was preliminarily postulated that it was the Wnt signaling pathway, rather than the NF-κB and PI3K/AKT pathway that participates in LUAD development. Later, this assumption was further verified through Western blot analysis. The result unveiled that protein levels of β-catenin, c-myc, and cyclin D1 (Wnt signaling pathway-related proteins) in H1975 and A549 cells were both markedly decreased after the knockdown of LINC01006 (Fig. 3B). Then, the TOP/FOP flash assay was applied to evaluate the activity of the Wnt/β-catenin pathway. The outcome showed that the luciferase activity of the TOP vector declined significantly due to LINC01006 suppression, while no significant change of FOP luciferase activity was displayed (Fig. 3C). Therefore, it was convincing that LINC01006 activates the Wnt/β-catenin signaling pathway to function in LUAD cells.
FIG 3.
Downregulation of LINC01006 inactivates the Wnt/β-catenin signaling in LUAD cells. (A) Key proteins (β-catenin, p65, p-p65, AKT, and p-AKT) in common signaling pathways were measured by Western blot assay when LINC01006 was knocked down; GAPDH served as the internal reference. (B) Western blot assay was performed to assess the expression of protein markers (β-catenin, c-myc, and cyclin D1) in the Wnt/β-catenin signaling pathway. β-Actin was used as the internal reference. (C) The TOP/FOP flash assay was conducted to measure the activity of Wnt/β-catenin signaling pathway in H1975 and A549 cells transfected with sh-NC or sh-LINC01006#2. **, P < 0.01.
LINC01006 acts as sponges for miR-129-2-3p to upregulate CTNNB1.
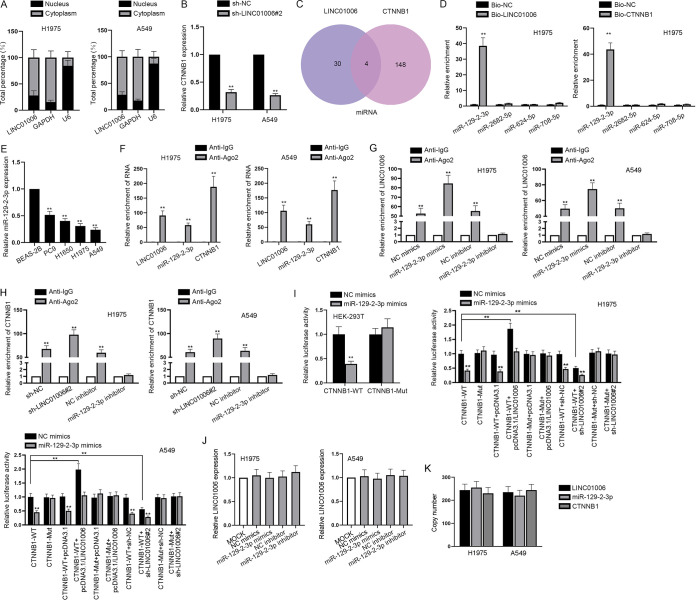
Since the signaling pathway regulated via LINC01006 has been determined, the regulatory mechanism of LINC01006 in the activation of Wnt/β-catenin signaling pathway activity was investigated in LUAD cells. Through a subcellular fractionation assay, we found that LINC01006 was chiefly abundant in cytoplasm both in H1975 and A549 cells (Fig. 4A). For this reason, we hypothesized that LINC01006 could activate the Wnt signaling pathway at the posttranscriptional level. Emerging evidence has demonstrated that competing endogenous RNA (ceRNA) was an important posttranscriptional mechanism to regulate gene expression and biological behaviors of cells in cancers (22, 23). In view of the fact that CTNNB1 was the mRNA of β-catenin protein, we conjectured that LINC01006 might activate the Wnt/β-catenin pathway in LUAD cells by function as a ceRNA via acting as sponges for miRNAs and enhancing CTNNB1 expression. As expected, we observed that knockdown of LINC01006 markedly reduced CTNNB1 levels in LUAD cells based on the result of RT-qPCR (Fig. 4B). With the implementation of the starBase database (http://starbase.sysu.edu.cn/), four miRNAs (miR-129-2-3p, miR-2682-5p, miR-624-5p, and miR-7085p) were predicted to bind to both LINC01006 and CTNNB1 (Fig. 4C). To determine the actual miRNA sponged by LINC01006, RNA pulldown assay was conducted. The result showed that miR-129-2-3p was conspicuously more enriched in Bio-LINC01006 and Bio-CTNNB1 groups than the other three miRNAs, while no miRNA was significantly enriched in negative-control (NC) groups (Fig. 4D). In this way, the interaction between miR-129-2-3p and LINC01006 or CTNNB1 was verified. Besides, the observable downregulation of miR-129-2-3p in LUAD cells further validated the reasonability of regarding miR-129-2-3p as downstream of LINC01006 (Fig. 4E). To further investigate the interaction among LINC01006, miR-129-2-3p, and CTNNB1, mechanism assays were implemented. RNA binding protein immunoprecipitation (RIP) assays showed that LINC01006, miR-129-2-3p, and CTNNB1 were all enriched in Ago2 antibody precipitates (Fig. 4F). In addition, the enrichment of LINC01006 in Ago2 antibody precipitates was increased or decreased significantly compared with the control group when miR-129-2-3p was upregulated or downregulated (Fig. 4G). As expected, when LINC01006 was silenced, the level of CTNNB1 enriched in Ago2 antibody precipitates was much higher than that in the control group (Fig. 4H). Moreover, the enrichment of CTNNB1 in Ago2 antibody was observed to be dramatically decreased because of the miR-129-2-3p knockdown (Fig. 4H). Therefore, the coexistence of these three RNAs in RNA-induced silencing complexes (RISCs) and the sponge role of LINC01006 targeting miR-129-2-3p to upregulate CTNNB1 were proved validly. Then, the results of luciferase reporter assays revealed that miR-129-2-3p mimics effectively decreased the luciferase activity of CTNNB1-WT (WT stands for wild type). In contrast, the luciferase activity of CTNNB1-Mut (Mut stands for mutant) was not significantly affected when being cotransfected with neither NC mimics nor miR-129-3p mimics (Fig. 4I). Further, the luciferase activity of CTNNB1-WT was enhanced significantly and exceeded the original state when LINC01006 was overexpressed, while it was weakened a lot when LINC01006 was knocked down in the appearance of NC mimics or miR129-2-3p mimics. However, the luciferase activity of CTNNB1-Mut was little affected by upregulation or downregulation of LINC01006 in H1975 and A549 cells (Fig. 4I). Besides, neither overexpression nor knockdown of miR-129-2-3p led to distinct change of LINC01006 expression in LUAD cells (Fig. 4J). So far, it was verified that miR-129-2-3p binds to both CTNNB1 and LINC01006, forming the ceRNA relationship. Furthermore, copy numbers of LINC01006, CTNNB1, and miR-129-2-3p in LUAD cells were determined through RT-qPCR. Results showed that LINC01006 and CTNNB1 were both at levels similar to that of miR-129-2-3p (Fig. 4K). Considering that there is only one binding site between miR-129-2-3p and LINC01006 or CTNNB1, the competition between LINC01006 and CTNNB1 for binding to miR-129-2-3p in LUAD cells was further exhibited. In a word, LINC01006 acts as a sponge for miR-129-2-3p to upregulate CTNNB1.
FIG 4.
LINC01006 acts as a sponge for miR-129-2-3p to upregulate CTNNB1. (A) Subcellular fractionation was performed to locate LINC01006 in LUAD cells. (B) The effect of LINC01006 downregulation on the relative expression level of CTNNB1 was evaluated by RT-qPCR analysis. (C) Four potential miRNAs binding to LINC01006 and CTNNB1 were discovered with the application of bioinformatics. (D) miR-129-2-3p was screened out as the miRNA that could bind to both LINC01006 and CTNNB1 through RNA pulldown assay. (E) The expression of miR-129-2-3p in healthy lung epithelial cell line (BEAS-2B) and LUAD cell lines (PC9, H1650, H1975, and A549) was examined. (F) RIP assay was implemented to demonstrate the coexistence of LINC01006, miR-129-2-3p, and CTNNB1 in RNA-induced silencing complexes (RISCs). (G) RIP assays researching the enrichment of LINC01006 were carried out when miR-129-2-3p was overexpressed or downregulated in LUAD cells. (H) RIP assays researching the enrichment of CTNNB1 were done when the expression of LINC01006 or miR-129-2-3p was inhibited in LUAD cells. (I) Luciferase reporter assay was carried out to verify the interaction between miR-129-2-3p and LINC01006 or CTNNB1 in HEK-293T or LUAD cells. (J) RT-qPCR was performed to detect the relative expression level of LINC01006 after either the overexpression or downregulation of miR-129-2-3p. (K) Copy numbers of LINC01006, miR-129-2-3p, and CTNNB1 in H1975 and A549 cells were measured by RT-qPCR. **, P < 0.01.
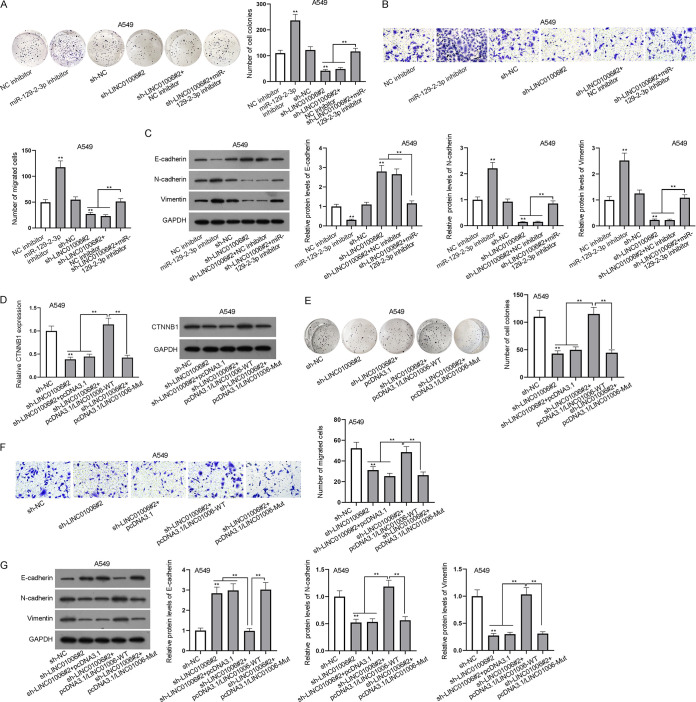
LINC01006 promotes proliferation, migration, and EMT of LUAD cells via sponging miR-129-2-3p.
To attest the effectiveness of LINC01006/miR-129-2-3p/CTNNB1 axis in LUAD cells, a series of functional assays were conducted. As shown in Fig. 5A, compared with negative controls, the proliferation of LUAD cells was enhanced after the downregulation of miR-129-2-3p while being weakened by the depletion of LINC01006. Interestingly, the inhibitory effect on cell proliferation caused by silenced LINC01006 was reversed by miR-129-2-3p inhibitor (Fig. 5A). Additionally, a transwell assay was applied to detect the migratory capacity of LUAD cells. The results showed that the knockdown of miR-129-2-3p facilitated LUAD cell migration and miR-129-2-3p inhibitor counteracted the suppressive function on cell migration resulting from the knockdown of LINC01006 (Fig. 5B). Moreover, Western blotting was employed to analyze the EMT-associated proteins in LUAD cells. The results showed that miR-129-2-3p silence obviously reduced the protein level of E-cadherin while elevating that of N-cadherin or vimentin. As expected, the enhanced E-cadherin protein levels induced by low expression of LINC01006 were reversed by miR-129-2-3p inhibitor. Similarly, the reduced N-cadherin and vimentin protein levels caused by downregulation of LINC01006 were recovered by miR-129-2-3p inhibitor (Fig. 5C). To further verify whether the promoting effect of LINC01006 on LUAD cell progression relies on its binding to miR-129-2-3p and modulation of CTNNB1, we evaluated the mRNA and protein levels of CTNNB1 via RT-qPCR and Western blotting after the knockdown or overexpression of LINC01006. The result exhibited that the impeded CTNNB1 expression caused by LINC01006 silence was rescued by the overexpression of LINC01006-WT while being seldom influenced by the upregulation of LINC01006-Mut at neither the mRNA nor protein level (Fig. 5D). Afterwards, we carried out the other set of rescue assays to determine the influence of LINC01006-WT or LINC01006-Mut on the biological behaviors of LUAD cells transfected with sh-LINC01006#2. The colony formation assay showed that the overexpression of LINC01006-WT reversed the decreased cell proliferation caused by ablation of LINC01006, whereas LINC01006-Mut upregulation had no similar effect (Fig. 5E). Likewise, transwell migration assays suggested that upregulated expression of LINC01006-WT reversed the inhibitory effect induced by LINC01006 knockdown on cell migration, while LINC01006-Mut overexpression made no obvious difference in cell migration (Fig. 5F). Furthermore, the increased protein level of E-cadherin due to silenced LINC01006 was notably lowered by overexpressed LINC01006-WT instead of LINC01006-Mut (Fig. 5G). Consistently, the LINC01006-WT overexpression rescued protein levels of N-cadherin and vimentin which were inhibited by the knockdown of LINC01006 but mutant-type LINC01006 failed to countervail the influence of sh-LINC01006#2 (Fig. 5G). According to the above results, LINC01006 plays a promotive role in LUAD cell progression via competitively binding to miR-129-2-3p to upregulate CTNNB1.
FIG 5.
LINC01006 promotes proliferation, migration, and EMT of LUAD cells by acting as a sponge for miR-129-2-3p. (A) Colony formation assay was used to evaluate A549 cell proliferation. (B) Cell migration was appraised by transwell assay. (C) Western blotting was conducted to analyze EMT-related protein levels in A549 cells. (D) The mRNA and protein levels of CTNNB1 were evaluated by RT-qPCR and Western blot analyses. (E) Colony formation assay was used to evaluate A549 cell proliferation in the presence of wild-type and mutant LINC01006 after the transfection of sh-LINC01006#2. (F) The migratory ability of A549 cells was appraised by transwell assay in the presence of wild-type and mutant LINC01006 after the transfection of sh-LINC01006#2. (G) Western blotting was performed to analyze EMT-related protein (E-cadherin, N-cadherin, and vimentin) levels in different groups. **, P < 0.01.
LINC01006 facilitates proliferation, migration, and EMT of LUAD cells via enhancing CTNNB1 to activate the Wnt/β-catenin pathway.
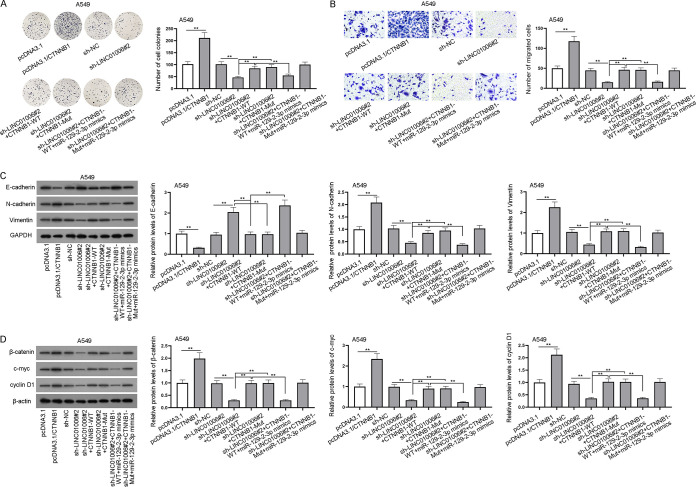
Previous assays have proved that the regulation of LINC01006 in LUAD cells was achieved by sponging miR-129-2-3p to modulate CTNNB1. Hence, assays were conducted to further reveal the interaction among LINC01006, miR-129-2-3p, and CTNNB1 in LUAD cells and the effect of their interaction on cell proliferation, migration, and EMT, as well as activation of the Wnt/β-catenin pathway. From colony formation and transwell assays, we found that the proliferation and migration of A549 cells was enhanced or inhibited in pcDNA3.1/CTNNB1 or sh-LINC01006#2 groups compared with the control groups, and the suppressive effect of LINC01006 downregulation on cell proliferation and migration was offset by CTNNB1-WT or CTNNB1-Mut (Fig. 6A and B). Intriguingly, the overexpression of miR-129-2-3p offset only the accelerative influence of CTNNB1-WT on cell proliferation and migration rather than that of CTNNB1-Mut (Fig. 6A and B). Meanwhile, CTNNB1-WT/Mut upregulation could reverse the hindered EMT induced by the depletion of LINC01006 (Fig. 6C). As expected, the upregulation of miR-129-2-3p made no significant difference to cell EMT in sh-LINC01006#2 plus CTNNB1-Mut groups while impairing that in sh-LINC01006#2 plus CTNNB1-WT groups (Fig. 6C). Finally, through Western blot analysis, we found that protein levels of markers in the Wnt/β-catenin signaling pathway (β-catenin, c-myc, and cyclin D1) were decreased by LINC01006 knockdown but augmented by CTNNB1-WT/Mut overexpression (Fig. 6D). In accordance with the supposition, the overexpression of miR-129-2-3p decreased the levels of β-catenin, c-myc, or cyclin D1 in groups transfected with CTNNB1-WT while exerting no obvious effect on those in sh-LINC01006#2 plus CTNNB1-Mut groups (Fig. 6D). In summary, LINC01006 upregulates CTNNB1 through binding to miR-129-2-3p, thus activating the Wnt/β-catenin signaling pathway and facilitating LUAD cell proliferation, migration, and EMT.
FIG 6.
LINC01006 facilitates proliferation, migration, and EMT of LUAD cells via enhancing CTNNB1 to activate the Wnt/β-catenin pathway. (A) Colony formation assay was carried out to evaluate the proliferative ability of A549 cells in different groups: pcDNA3.1, pcDNA3.1/CTNNB1, sh-NC, sh-LINC01006#2, sh-LINC01006#2 plus CTNNB1-WT, sh-LINC01006#2 plus CTNNB1-Mut, sh-LINC01006#2 plus CTNNB1-WT plus miR-129-2-3p-mimics and sh-LINC01006#2 plus CTNNB1-Mut plus miR-129-2-3p-mimics. (B) Transwell assay was performed to assess cell migration in the above different groups. (C) E-cadherin, N-cadherin, and vimentin protein levels in the above different groups were measured via Western blotting. (D) The protein levels of β-catenin, c-myc, and cyclin D1 in Wnt/β-catenin signaling pathways in the above different groups were measured by Western blotting. **, P < 0.01.
DISCUSSION
LUAD has been regarded as a malignancy difficult to cure all the time (4). Meanwhile, increasing research has proved that lncRNAs are closely associated with LUAD (15, 24). For instance, LINC00857 was discerned to promote lung cancer progression (25). LINC00312 induced the migration of LUAD by regulating YBX1 (26). Therefore, the critical value of studying the role of lincRNA in LUAD is apparent. Located in the intergenic region, LINC01006 was found to be highly expressed in LUAD cells. Functional studies and animal experiments also revealed that downregulation of LINC01006 could significantly hinder LUAD cell proliferation, migration, and EMT as well as the tumor weight and volume in vivo. Afterwards, by using Western blot assays, we evaluated the proteins of several common signaling pathways after LINC01006 was silenced. The results indicated that LINC01006 played an important role in the activation of the Wnt signaling pathway in LUAD cells.
It is well-known that β-catenin is connected with the Wnt pathway, and CTNNB1 is the mRNA which can be translated into β-catenin. Here, we found that knockdown of LINC01006 could downregulate CTNNB1 expression dramatically. Therefore, we hypothesized and verified that LINC01006 regulates LUAD cell progression by regulating CTNNB1 to participate in the Wnt/β-catenin pathway.
Mounting evidence has indicated that lncRNAs regulate mRNAs via acting as sponges for miRNAs (27, 28). In this paper, we found four applicable miRNAs, among which miR-129-2-3p was selected out as the bona fide target gene of LINC01006 by RNA pulldown assay. Subsequently, RIP and luciferase reporter assays validated the ceRNA connection among LINC01006, miR-129-2-3, and CTNNB1. Furthermore, rescue experiments declared that miR-129-2-3p inhibitor or CTNNB1 overexpression could recover the inhibitory effect generated by LINC01006 silencing on LUAD cell progression. In addition, upregulation of CTNNB1 also rescued the inactivation of Wnt signaling pathway caused by LINC01006 depletion. Moreover, the overexpression of miR-129-2-3p was found to be effective in counteracting the effect of upregulation of CTNNB1-WT while little affecting biological behaviors of LUAD cells transfected with CTNNB1-Mut containing the mutant binding sites with miR-129-2-3p. In summary, our study unveiled the carcinogenic role of LINC01006 in LUAD progression by acting as sponges for miR-129-2-3p, regulating CTNNB1 and activating Wnt/β-catenin pathway, providing a novel insight into LUAD.
Considering that the relationship between LINC01006 and LUAD was unclear before this current study, our study is an essential supplement for the study on both LINC01006 and LUAD. RNAs related to LUAD and relevant ceRNA networks will be further studied in the future.
MATERIALS AND METHODS
Cell culture.
Human LUAD cell lines (PC9, H1650, H1975, and A549) and a human healthy lung epithelial cell line (BEAS-2B) were selected for cell culture. H1650, H1975, A549, and BEAS-2B cell lines were all obtained from American Type Culture Collection (ATCC) (Manassas, VA, USA), while PC9 cell lines were purchased from Huatuo Biotechnology Co., Ltd. (Shenzhen, China). PC9, H1650, and H1975 cell lines were cultured in RPMI 1640 medium (Thermo Fisher Scientific) with 10% fetal bovine serum (FBS) (Solarbio). A549 cell lines were cultured in F-12K medium (2 mM l-glutamine, 1,500 mg/liter sodium bicarbonate, and 10% FBS). BEAS-2B cell lines were cultured in BEGM (5 ng/ml EGF, 70 ng/ml phosphoethanolamine, and 10% FBS). All cells were cultured at 37°C with 5% CO2.
Cell transfection.
The short hairpin RNAs (shRNAs) specifically targeting LINC01006, as well as their corresponding negative-control shRNAs (sh-NCs), were all chemically synthesized by Genechem (Shanghai, China). In addition, pcDNA3.1 vectors were subcloned with LINC01006-WT/Mut or CTNNB1-WT/Mut for overexpression, and empty pcDNA3.1 vector was used as a negative control. MiR-129-2-3p mimics/inhibitor and their corresponding NC mimics or NC inhibitor were procured from RiboBio (Shanghai, China). All the cell transfections were conducted with Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA).
Quantitative real-time PCR (RT-qPCR) analysis.
Total RNAs were isolated from cultured cell lines through TRIzol reagent (Sigma, St. Louis, MO, USA). Then, reverse transcription was conducted in RevertAid First Strand cDNA Synthesis kit (Thermo Fisher, IL, USA). With the application of SYBR green PCR master mix (Applied Biosystems, Foster City, CA, USA), RT-qPCR was implemented in accordance with the manufacturer’s instructions. Based on the 2−ΔΔCt method, relative expression was normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Assays were conducted independently three times.
Colony formation assay.
H1975 and A549 cells were plated into six-well plates (200 cells/well) and incubated for 2 weeks. Then, cells were fixed before being stained with 0.1% crystal violet. After that, cell colonies were counted manually. Assays were conducted independently three times.
Transwell migration assay.
Cells were first seeded into the upper chambers added with serum-free medium. Meanwhile, the lower chambers were filled with culture medium containing 10% FBS. After 24 h, the migrated cells in the lower chambers were fixed and were observed using a microscope after staining with crystal violet. Assays were conducted independently three times.
Western blot analysis.
Cells were lysed by using radioimmunoprecipitation assay (RIPA) buffer (Thermo Fisher, IL, USA). Then, extracted proteins were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and subsequently transferred to polyvinylidene difluoride (PVDF) membranes (Millipore, Billerica, MA, USA). After the membranes were blocked with nonfat milk, they were successively incubated with primary antibodies at 4°C overnight and secondary antibodies for 1 h in the dark room. Primary antibodies against β-catenin, p65, p-p65, AKT, p-AKT, c-myc, cyclin D1, GAPDH, β-actin, E-cadherin, N-cadherin, and vimentin were all bought from Abcam (Cambridge, USA). The quantification of proteins was accomplished with the application of Immobilon Western chemiluminescent horseradish peroxidase (HRP) substrate (Bio-Rad, CA, USA). Assays were conducted independently three times.
RIP.
RIP assay was conducted in RNA binding protein immunoprecipitation kit (Millipore, Burlington, MA, USA). In brief, cell lysates were incubated in RIP buffer supplemented with Argonaute 2 (Ago2) antibodies or control immunoglobulin G (IgG) antibodies. Finally, RNAs were extracted from the precipitates and subjected to RT-qPCR analysis. Assays were conducted independently three times.
RNA pulldown assay.
H1975 or A549 cells were lysed and cultured with biotinylated (Bio-) control sequences, LINC01006 or CTNNB1. Magnetic beads were then added into the complexes comprising cell lysate and Bio-NC, Bio-LINC01006, or Bio-CTNNB1. When the precipitation was fully finished, RNAs were extracted and purified from the precipitates pulled down by magnetic beads for RT-qPCR analysis. Assays were conducted independently three times.
Subcellular fractionation.
Via cytoplasmic and Nuclear RNA purification kit (Norgen, Thorold, ON, Canada), cytoplasmic and nuclear fragments were isolated from each other. Then, expression levels of LINC01006 were analyzed by RT-qPCR. GAPDH or U6 was regarded as cytoplasmic or nuclear control, respectively. Assays were conducted independently three times.
Luciferase reporter assay and TOP/FOP flash assay.
For luciferase reporter assay, wild-type (WT) and mutant (Mut) sequences of CTNNB1-3′ untranslated region (UTR) containing wild-type or mutant binding sites with miR-129-2-3p were amplified and subcloned into pmirGLO dual-luciferase vectors (Promega) to generate pmirGLO-CTNNB1-WT/Mut. Afterwards, NC mimics or miR-129-2-3p mimics with or without pcDNA3.1, pcDNA3.1/LINC01006, sh-NC, or sh-LINC01006#2 were cotransfected with the aforementioned plasmids into LUAD or HEK-293T cells. For TOP/FOP flash assay, Wnt/β-catenin reporter vector TOP/FOP was cotransfected into H1975 and A549 cells with sh‐NC or sh‐LINC01006#2. Luciferase activity was measured by the Dual Luciferase Reporter assay kit (Promega, Madison, WI, USA). Assays were conducted independently three times.
Animal experiments.
Nude mice (at the age of 4 to 5 weeks) were acquired from SLRC Laboratory Animal Company (Shanghai, China) and housed under specific-pathogen-free (SPF) conditions. LUAD cells were transfected with sh-NC or sh-LINC01006#2 and suspended in PBS. After that, cells were subcutaneously injected into the mice. Tumor growth and tumor volume were detected every 5 days. After 1 month, the mice were euthanized. Then the mouse tumors were excised and weighed. All animal studies were approved by the ethics committee of the Tianjin Medical University Cancer Institute & Hospital.
Statistical analysis.
Statistical analysis was performed with GraphPad Prism 6.0 (GraphPad, San Diego, CA, USA). All the data were expressed as means ± standard deviations (SD). Statistical differences between groups were analyzed by Student’s t test or one-way or two-way analysis of variance (ANOVA). All the experiments were carried out separately in triplicate. Differences were regarded to be statistically significant when the P value was less than 0.05.
ACKNOWLEDGMENTS
We express sincere gratitude to all lab personnel in this study.
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
We declare that we have no conflicts of interest.
REFERENCES
- 1.Nasim F, Sabath BF, Eapen GA. 2019. Lung cancer. Med Clin North Am 103:463–473. 10.1016/j.mcna.2018.12.006. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R, Naishadham D, Jemal A. 2013. Cancer statistics, 2013. CA Cancer J Clin 63:11–30. 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 3.Denisenko TV, Budkevich IN, Zhivotovsky B. 2018. Cell death-based treatment of lung adenocarcinoma. Cell Death Dis 9:117. 10.1038/s41419-017-0063-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hutchinson BD, Shroff GS, Truong MT, Ko JP. 2019. Spectrum of lung adenocarcinoma. Semin Ultrasound CT MR 40:255–264. 10.1053/j.sult.2018.11.009. [DOI] [PubMed] [Google Scholar]
- 5.Stewart DJ. 2014. Wnt signaling pathway in non-small cell lung cancer. J Natl Cancer Inst 106:djt356. 10.1093/jnci/djt356. [DOI] [PubMed] [Google Scholar]
- 6.Rapp J, Jaromi L, Kvell K, Miskei G, Pongracz JE. 2017. WNT signaling − lung cancer is no exception. Respir Res 18:167. 10.1186/s12931-017-0650-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen J, Hu L, Chen J, Pan Q, Ding H, Xu G, Zhu P, Wen X, Huang K, Wang Y. 2016. Detection and analysis of Wnt pathway related lncRNAs expression profile in lung adenocarcinoma. Pathol Oncol Res 22:609–615. 10.1007/s12253-016-0046-9. [DOI] [PubMed] [Google Scholar]
- 8.Wahlestedt C. 2013. Targeting long non-coding RNA to therapeutically upregulate gene expression. Nat Rev Drug Discov 12:433–446. 10.1038/nrd4018. [DOI] [PubMed] [Google Scholar]
- 9.Tang J, Jiang R, Deng L, Zhang X, Wang K, Sun B. 2015. Circulation long non-coding RNAs act as biomarkers for predicting tumorigenesis and metastasis in hepatocellular carcinoma. Oncotarget 6:4505–4515. 10.18632/oncotarget.2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ballantyne RL, Zhang X, Nuñez S, Xue C, Zhao W, Reed E, Salaheen D, Foulkes AS, Li M, Reilly MP. 2016. Genome-wide interrogation reveals hundreds of long intergenic noncoding RNAs that associate with cardiometabolic traits. Hum Mol Genet 25:3125–3141. 10.1093/hmg/ddw154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luo G, Wang M, Wu X, Tao D, Xiao X, Wang L, Min F, Zeng F, Jiang G. 2015. Long non-coding RNA MEG3 inhibits cell proliferation and induces apoptosis in prostate cancer. Cell Physiol Biochem 37:2209–2220. 10.1159/000438577. [DOI] [PubMed] [Google Scholar]
- 12.Yu C, Li L, Xie F, Guo S, Liu F, Dong N, Wang Y. 2018. LncRNA TUG1 sponges miR-204-5p to promote osteoblast differentiation through upregulating Runx2 in aortic valve calcification. Cardiovasc Res 114:168–179. 10.1093/cvr/cvx180. [DOI] [PubMed] [Google Scholar]
- 13.Zhao W, Geng D, Li S, Chen Z, Sun M. 2018. LncRNA HOTAIR influences cell growth, migration, invasion, and apoptosis via the miR-20a-5p/HMGA2 axis in breast cancer. Cancer Med 7:842–855. 10.1002/cam4.1353. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14.Chen R, Li WX, Sun Y, Duan Y, Li Q, Zhang AX, Hu JL, Wang YM, Gao YD. 2017. Comprehensive analysis of lncRNA and mRNA expression profiles in lung cancer. Clin Lab 63:313–320. 10.7754/Clin.Lab.2016.160812. [DOI] [PubMed] [Google Scholar]
- 15.Li L, Peng M, Xue W, Fan Z, Wang T, Lian J, Zhai Y, Lian W, Qin D, Zhao J. 2018. Integrated analysis of dysregulated long non-coding RNAs/microRNAs/mRNAs in metastasis of lung adenocarcinoma. J Transl Med 16:372. 10.1186/s12967-018-1732-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ren J, Fu J, Ma T, Yan B, Gao R, An Z, Wang D. 2018. LncRNA H19-elevated LIN28B promotes lung cancer progression through sequestering miR-196b. Cell Cycle 17:1372–1380. 10.1080/15384101.2018.1482137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Loewen G, Jayawickramarajah J, Zhuo Y, Shan B. 2014. Functions of lncRNA HOTAIR in lung cancer. J Hematol Oncol 7:90. 10.1186/s13045-014-0090-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang L, Wang Y, Zhang L, You G, Li C, Meng B, Zhou M, Zhang M. 2019. LINC01006 promotes cell proliferation and metastasis in pancreatic cancer via miR-2682-5p/HOXB8 axis. Cancer Cell Int 19:320. 10.1186/s12935-019-1036-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu X, Chen F, Shao Y, Xu D, Guo J. 2017. Long intergenic non-protein coding RNA 1006 used as a potential novel biomarker of gastric cancer. Cancer Biomark 21:73–80. 10.3233/CBM-170273. [DOI] [PubMed] [Google Scholar]
- 20.Peng WX, Koirala P, Mo YY. 2017. LncRNA-mediated regulation of cell signaling in cancer. Oncogene 36:5661–5667. 10.1038/onc.2017.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu J, Markowitz GJ, Wang X. 2016. Noncoding RNAs regulating cancer signaling network. Adv Exp Med Biol 927:297–315. 10.1007/978-981-10-1498-7_11. [DOI] [PubMed] [Google Scholar]
- 22.Qi X, Zhang DH, Wu N, Xiao JH, Wang X, Ma W. 2015. ceRNA in cancer: possible functions and clinical implications. J Med Genet 52:710–718. 10.1136/jmedgenet-2015-103334. [DOI] [PubMed] [Google Scholar]
- 23.Wang Y, Hou J, He D, Sun M, Zhang P, Yu Y, Chen Y. 2016. The emerging function and mechanism of ceRNAs in cancer. Trends Genet 32:211–224. 10.1016/j.tig.2016.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang Z, Li H, Wang Z, Yang Y, Niu J, Liu Y, Sun Z, Yin C. 2018. Microarray expression profile of long non-coding RNAs in human lung adenocarcinoma. Thorac Cancer 9:1312–1322. 10.1111/1759-7714.12845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang L, He Y, Liu W, Bai S, Xiao L, Zhang J, Dhanasekaran SM, Wang Z, Kalyana-Sundaram S, Balbin OA, Shukla S, Lu Y, Lin J, Reddy RM, Carrott PW, Jr, Lynch WR, Chang AC, Chinnaiyan AM, Beer DG, Zhang J, Chen G. 2016. Non-coding RNA LINC00857 is predictive of poor patient survival and promotes tumor progression via cell cycle regulation in lung cancer. Oncotarget 7:11487–11499. 10.18632/oncotarget.7203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peng Z, Wang J, Shan B, Li B, Peng W, Dong Y, Shi W, Zhao W, He D, Duan M, Cheng Y, Zhang C, Duan C. 2018. The long noncoding RNA LINC00312 induces lung adenocarcinoma migration and vasculogenic mimicry through directly binding YBX1. Mol Cancer 17:167. . 10.1186/s12943-018-0920-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feng C, Zhao Y, Li Y, Zhang T, Ma Y, Liu Y. 2019. LncRNA MALAT1 promotes lung cancer proliferation and gefitinib resistance by acting as a miR-200a sponge. Arch Bronconeumol 55:627–633. 10.1016/j.arbres.2019.03.026. [DOI] [PubMed] [Google Scholar]
- 28.Chen J, Lin Y, Jia Y, Xu T, Wu F, Jin Y. 2019. LncRNA HAND2-AS1 exerts anti-oncogenic effects on ovarian cancer via restoration of BCL2L11 as a sponge of microRNA-340-5p. J Cell Physiol 234:23421–23436. 10.1002/jcp.28911. [DOI] [PubMed] [Google Scholar]