Summary
Astrocytes are a viable source for generating new neurons via direct conversion. However, little is known about the neurogenic cascades triggered in astrocytes from different regions of the CNS. Here, we examine the transcriptome induced by the proneural factors Ascl1 and Neurog2 in spinal cord-derived astrocytes in vitro. Each factor initially elicits different neurogenic programs that later converge to a V2 interneuron-like state. Intriguingly, patch sequencing (patch-seq) shows no overall correlation between functional properties and the transcriptome of the heterogenous induced neurons, except for K-channels. For example, some neurons with fully mature electrophysiological properties still express astrocyte genes, thus calling for careful molecular and functional analysis. Comparing the transcriptomes of spinal cord- and cerebral-cortex-derived astrocytes reveals profound differences, including developmental patterning cues maintained in vitro. These relate to the distinct neuronal identity elicited by Ascl1 and Neurog2 reflecting their developmental functions in subtype specification of the respective CNS region.
Keywords: spinal cord, astrocytes, direct reprogramming, Ascl1, Neurog2, patch-seq, single-cell RNA-seq, patterning genes
Graphical abstract
Highlights
-
•
Ascl1 and Neurog2 induce initially distinct transcriptomes in spinal cord astrocytes
-
•
Neurons induced by Ascl1 or Neurog2 converge to a V2 interneuron-like state
-
•
Patch-seq shows functional and transcriptional heterogeneity with low correlation
-
•
Developmentally established patterning genes are maintained in astrocytes in vitro
Kempf et al. show that Ascl1 and Neurog2 elicit initially divergent transcriptional programs in spinal cord astrocytes converging later to a V2 interneuron-like state, according to their developmental role and patterning cues in astrocytes. Patch-seq analysis reveals the heterogeneity of fate conversion with little correlation between transcriptome and electrophysiological properties.
Introduction
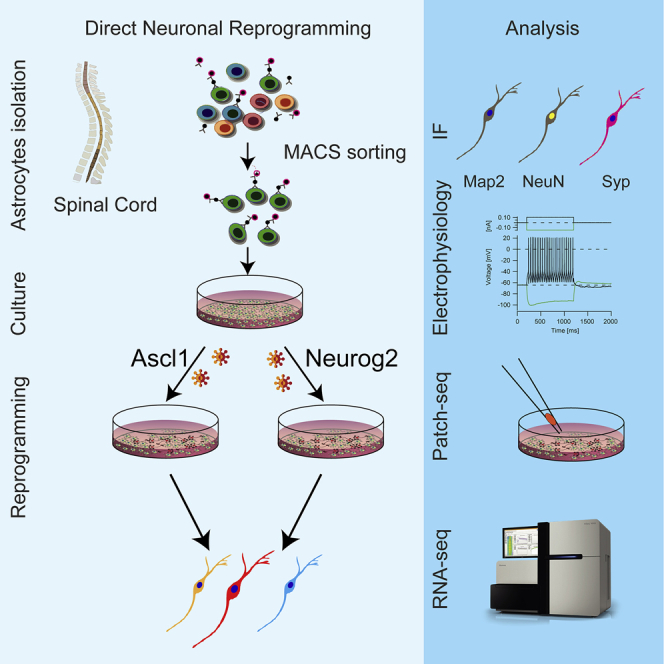
Direct reprogramming of local glial cells into neurons in the central nervous system (CNS) has become a promising approach for neuronal replacement in disease (Barker et al., 2018). Pioneered by in vitro conversion of astrocytes from the cerebral cortex (Berninger et al., 2007; Heinrich et al., 2010; Heins et al., 2002), this approach is often based on the expression of proneural factors (e.g., Ascl1 and Neurog2), master regulators and pioneer factors in the conversion process both in vitro (Berninger et al., 2007; Masserdotti et al., 2015; Smith et al., 2016; Vierbuchen et al., 2010; Wapinski et al., 2013) and in vivo (Guo et al., 2014; Liu et al., 2015; Mattugini et al., 2019; Rivetti di Val Cervo et al., 2017; Torper et al., 2013). Cortical astrocyte cultures allowed identifying major reprogramming hurdles (Gascón et al., 2016; Russo et al., 2020), whose manipulation in vivo led to improving the reprogramming efficiency from 10% to over 90% (Gascón et al., 2016).
However, the use of neuronal reprogramming for therapy requires the generation of adequate neuronal subtypes specific to different CNS regions. For instance, the induction of most of the spinal cord (SC) neuronal diversity has not yet been achieved (Su et al., 2014b), besides motoneuron generation from fibroblasts (Abernathy et al., 2017; Church et al., 2021; Meyer et al., 2014; Son et al., 2011) and neurons with different neurotransmitter identities from NG2 glia in vivo (Tai et al., 2021). Ascl1 and Neurog2 are involved in generating specific interneuron subtypes in the developing SC (Lu et al., 2015; Misra et al., 2014): Ascl1 specifies excitatory neurons (Borromeo et al., 2014; Mizuguchi et al., 2006), and Neurog2 is a downstream effector of Ptf1a, instructing GABAergic interneurons (Henke et al., 2009) or motorneurons (Lee et al., 2020). In the ventral SC, both Ascl1 and Neurog2 specify a V2 interneuron identity (Parras et al., 2002), specifically a GABAergic V2b interneuron fate (Misra et al., 2014). In forebrain development, Ascl1 instructs inhibitory GABAergic neurons and Neurog1/2 glutamatergic neurons (Kovach et al., 2013; Parras et al., 2002), which is reflected in the neuronal subtypes generated by these proneural genes from cortical astrocytes (Berninger et al., 2007; Heinrich et al., 2010; Masserdotti et al., 2015). Recently, differences in the reprogramming efficiency of astrocytes obtained from various CNS regions have been reported (Chouchane et al., 2017; Hu et al., 2019; Rao et al., 2021). However, the mechanisms underlying direct neuronal reprogramming of region-specific astrocytes are largely unknown. Here, we investigated the neurogenic programs induced by Ascl1 and Neurog2 in SC-derived astrocytes, the neuronal identity of reprogrammed neurons, and how the regional specification of SC astrocytes may influence the fate of neurons reprogrammed by Ascl1 or Neurog2.
Results
Direct conversion of SC-derived astrocytes into functional neurons
Protocols used for culturing cortical astrocytes (Heinrich et al., 2011) (Figure S1A) were not successful to enrich for SC astrocytes from postnatal day (P) 2–3 mice, as glial fibrillary acidic protein (Gfap)-positive astrocytes were below 50% (Figures S1B and S1C; mean = 41.7%, confidence interval [CI] = 20.6). To enrich for astrocytes, we isolated ACSA-2+ cells (Kantzer et al., 2017) via magnetic-activated cell sorting (MACS). Indeed, the majority of sorted cells were now astrocytes after 8 days in culture (Gfap+, mean = 79.4%, CI = 9.8; Sox9+: mean = 85.9%, CI = 7.6; Figures S1D and S1E) with a low abundance of oligodendrocyte progenitors (Olig2+, mean = 6%, CI = 11.6; expression data in Figure S1H) or microglia (Iba1+, mean = 1%, CI = 1) and no detectable neuroblasts (Dcx+) or neurons (β3-tubulin+) (Figure S1E). Thus, ACSA2-MACS yielded highly enriched cultures of astrocytes to explore astrocyte-to-neuron reprogramming.
SC-derived astrocytes were transduced with retrovirus expressing either Ascl1 or Neurog2 under the CAG promoter (Heinrich et al., 2010, 2011), and, 8 days post-infection (DPI), the proportion of β3-tubulin immunoreactive cells with neuronal morphology among transduced cells was quantified (Gascón et al., 2016; Masserdotti et al., 2015) (Figure 1A). A relatively low proportion of neuronal cells was observed (Neurog2, mean = 9.21%, CI = 6.23; Ascl1, mean = 18.07%, CI = 21.11; Figures 1B and 1C; Gfap+/DsRed+ in Figure 1D), in line with previous reports (Hu et al., 2019) and considerably lower than the efficiency observed in cerebral cortex gray matter (GM)-derived astrocytes, using the same factors and culture conditions (∼60% for Neurog2, ∼40% for Ascl1) (Heinrich et al., 2010; Hu et al., 2019). Treatment with small molecules (forskolin and dorsomorphin) (Gascón et al., 2016; Liu et al., 2013; Smith et al., 2016) remarkably enhanced the conversion efficiency of SC astrocytes by about 3 times (Neurog2, mean = 31.20%, CI = 19.30; Ascl1, mean = 55.15%, CI = 45.11; Figures 1B and 1C), suggesting the presence of similar reprogramming hurdles as observed in other cells (Gascón et al., 2016; Liu et al., 2013). Notably, few oligodendrocyte progenitors were also detected in these cultures (Figures S1F and S1G).
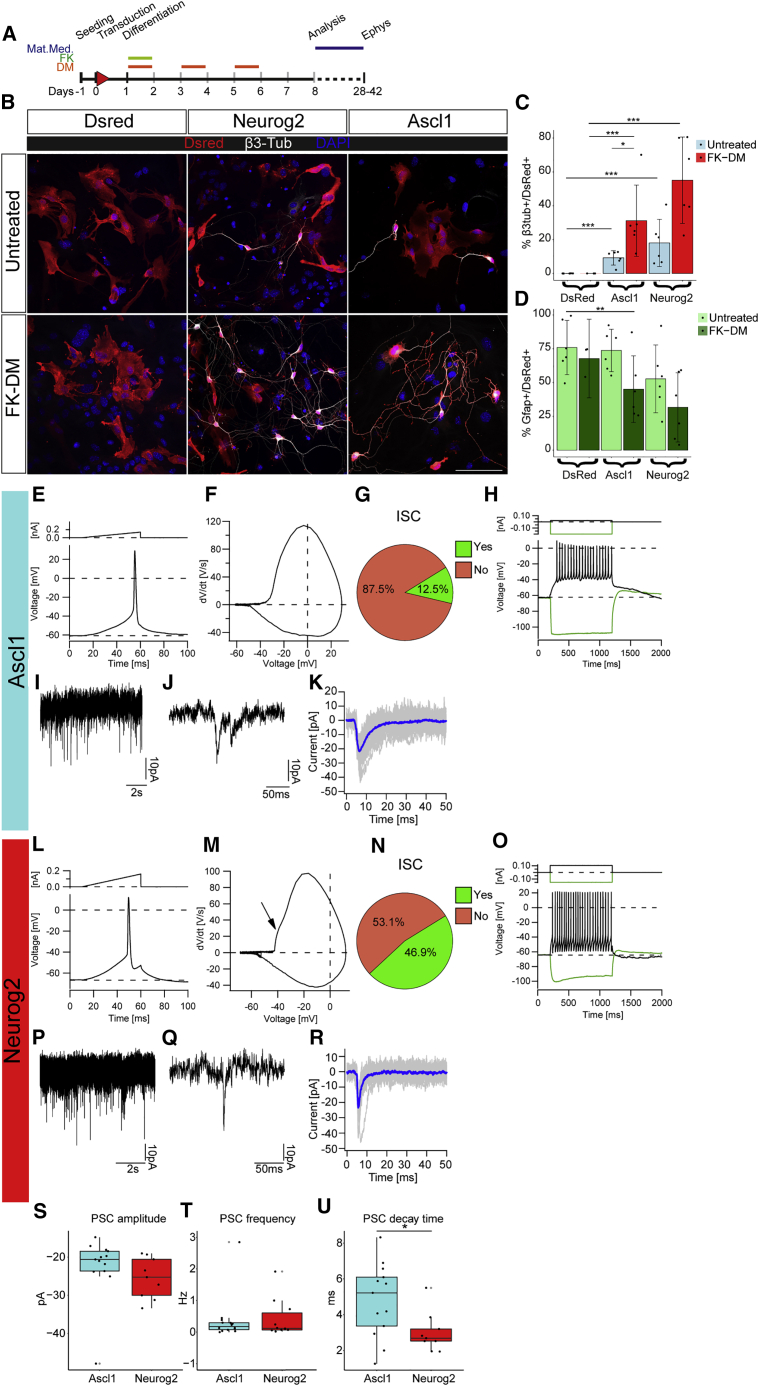
Figure 1.
Direct conversion of SC-derived astrocytes into functional neurons
(A) Scheme of the experimental design.
(B) Micrographs depicting cells transduced with control (DsRed), Ascl1-, and Neurog2-encoding virus and converted into neuronal cells. Forskolin (FK) and dorsomorphin (DM) treatment is shown (lower panels). Scale bar: 100 μm.
(C and D) Barplot of %β3-tub+/DsRed+ cells with neuronal morphology (C) and %Gfap+/DsRed+ (D) at 8DPI with mean and CI (95% confidence for n = 6). Linear regression model performed. n= 6 independent experiments; ∗p < 0.05; ∗∗p < 0.001; ∗∗∗p < 0.0001.
(E, F, L, and M) Examples of a single action potential (E and L) and corresponding phaseplane plots of the action potential in Ascl1- (F) or Neurog2- (M) iNs. Arrow (M) indicates the initial spike component (ISC).
(G and N) Pie charts depicting the percent of cells with ISC among iNs (3/24 and 15/32 cells recorded, respectively). n = 3 independent experiments.
(H and O) Example of an evoked train of action potentials (Ascl1-iNs, H; Neurog2-iNs, O). The green line shows a hyperpolarizing voltage response upon injection of hyperpolarizing current steps (1 s, 150 pA), characterized by a decline of amplitude during ongoing current injection (H) or without such decline (O). Injection of suprathreshold depolarizing current pulses (1 s, 105 pA, upper traces, black) evoked a continuous pattern train of action potentials (H and O), followed by a marked depolarization and long-lasting after-hyperpolarization.
(I, J, P, and Q) Examples of spontaneous synaptic activity in cultures transduced with Ascl1 (I, magnification in J) or Neurog2 (P, magnification in Q).
(K and R) Graphs of the averaged PSC from the cells in (J) and (R), respectively.
(S–U) Boxplots showing PSC amplitude (S), frequency (T), and decay time (U) in Ascl1 and Neurog2 iNs. Each dot represents a cell. ∗p < 0.05.
Electrophysiology performed at 4–6 weeks after infection (28–42 DPI) showed that most reprogrammed neuronal cells generated an action potential (75% and 84.4% of Ascl1- and Neurog2-iNeurons [iNs], respectively; Figures S1I and S1J) and exhibited similar passive and active membrane properties (Figures S1K–S1P); the action potential duration was significantly higher in Ascl1- than in Neurog2-reprogrammed neurons (Figure S1M), suggesting a lower density of Na+-channels in Ascl1-converted neurons. Single evoked action potential (Figures 1E and 1L) showed an initial spike component (ISC, so-called prepotential; Crochet et al., 2004; Golding and Spruston, 1998; Figures 1F, 1G, 1M, and 1N), while suprathreshold depolarizing current pulses induced repetitive spikes in both Ascl1- and Neurog2-converted neurons (Figures 1H, 1O, S1Q, and S1R). Importantly, both Ascl1- and Neurog2-reprogrammed neurons showed post-synaptic currents (PSC; Figures 1I–1K and 1P–1R), with similar amplitude and frequency (Figures 1S and 1T) but different decay times (Figure 1U). Thus, Ascl1 and Neurog2 reprogrammed SC-derived astrocytes into functional iNs (Yang et al., 2011).
As the electrophysiological properties suggested some differences between the Ascl1- and Neurog2-iNs, we explored the reprogramming at early and late time points at the molecular level.
Transcriptional programs induced by Ascl1 and Neurog2 at early stages in SC astrocytes
To analyze the programs induced at early stages, we timed the induction of target genes using our previously established approach with the hydroxytamoxifen (OHT)-inducible forms of Ascl1 and Neurog2 (Masserdotti et al., 2015). After 24 h of OHT treatment, Ascl1ERT2 and Neurog2ERT2 localized to the nucleus (Figures S2A–S2C), and their protein level was similar (Figure S2D). Following 4 days of OHT treatment, both factors converted SC astrocytes into neuronal cells with similar efficiency as the constitutively expressed factors at 8 DPI (Ascl1ERT2, β3-tubulin+/DsRed+ cells, mean = 25.92%, CI = 20.35; Neurog2ERT2, mean = 8.64%, CI = 3.38; no small molecules; Figures 2A–2C). For RNA sequencing (RNA-seq) (Bagnoli et al., 2018), transduced cells were selected via fluorescence-activated cell sorting (FACS; Figure S2E) 24 h after OHT treatment. To control for OHT-related gene expression, we compared proneural-factor-induced programs to DsRed-OHT-treated samples (Figures 2D–2P and S2F–S2L).
Figure 2.
Transcriptional changes upon Ascl1ERT2- and Neurog2ERT2-induced direct reprogramming at early stages
(A) Scheme of the experimental design. Red bars indicate the time of OHT treatment.
(B) Micrographs of cells expressing Neurog2ERT2 and Ascl1ERT2 in the absence (upper panel) or presence (lower panel) of OHT. Scale bar: 100 μm.
(C) Barplot of %β3-tub+/DsRed+ cells with neuronal morphology following Neurog2ERT2 and Ascl1ERT2 activation with OHT at 8DPI. Mean and CI (95%) are shown (n = 3 for Ascl1ERT2 untreated; n = 4 for Neurog2ERT2 untreated; n = 6 for all the others). Linear regression model applied. ∗∗∗p < 0.0001.
(D–P) Gene expression analysis showing bar plots (D and K) and Venn diagrams (E and L) of the number of genes differentially up- (D) or downregulated (K) (padj < 0.01 and log2FC > 0.5 or < −0.5) upon Ascl1ERT2 and Neurog2ERT2 induction after 24 h and the associated top GO terms as indicated (F–I for upregulated; M–P for downregulated) related to biological process (BP; F and M) or molecular function (MF; G and N) from commonly regulated genes (E and L) or specifically by Ascl1ERT2 (H and O) or Neurog2ERT2 (I and P). In (J), a heatmap is shown of most differentially regulated genes as indicated on the left side (padj < 0.01 and absolute log2FC > 1).
(Q) Scatterplot comparing Neurog2ERT2- and Ascl1ERT2-regulated genes (padj < 0.01 and log2FC > 2); transcription factors (TFs) in bold (blue, Ascl1 specific; black, Neurog2 specific).
(R) Network of most relevant TFs and chromatin modifiers induced after 24 h by Neurog2ERT2 (orange), Ascl1ERT2 (blue), and common (yellow). Bigger nodes correspond to the reprogramming factors; only connected nodes are plotted. Line width corresponds to StringDB scores.
(S) Example pathways identified after GSEA in Neurog2ERT2 and Ascl1ERT2 differentially expressed genes (Data S1).
Both factors induced drastic transcriptional changes compared to control (Figures 2D and 2K; Data S1). Consistent with previous data (Masserdotti et al., 2015), the programs differed profoundly, with only 17.6% of the upregulated genes common between the Ascl1ERT2- and Neurog2ERT2-induced cascades (log2FC > 1 and padj < 0.01, Figure 2E). This is 5 times higher than the proportion of genes commonly upregulated by these factors in GM astrocytes (Masserdotti et al., 2015). Gene Ontology (GO) analysis of commonly upregulated genes highlighted genes involved in nervous system development and regulation of neurogenesis (e.g., Insm1, Sox11, Sox4, Dll1; Figure 2F; Data S1) and in transcriptional activity (e.g., Rcor2, Ncor2, Cbfa2t3, and many transcription factors [TFs]; Figure 2G; Data S1). When focusing on genes uniquely induced by Ascl1ERT2 (107) or Neurog2ERT2 (40) (log2FC > 1, padj < 0.01 for 1 TF and log2FC ≤ 0 for the other; Figures 2E, 2J, S2H, and S2I), we found the former to specifically regulate genes related to muscle activity (e.g., Tnn1, Tnnt2; Figure 2H; Data S1), in line with previous observations on Ascl1 regulating alternative fates (Lee et al., 2020; Treutlein et al., 2016). Conversely, Neurog2ERT2 specifically regulated genes related to cytokine signaling, axon guidance/cell adhesion (e.g., Robo1, Cxcr4; Figure 2I), and regulation of GTPase activity (e.g., Chn2, Itga6, Ntrk3; Figure 2I; Data S1).
Both factors triggered the significant downregulation of many genes (Figure 2K), of which 25.8% were common (log2FC < −1, padj < 0.01; Figure 2L). These were connected to vasoconstriction (e.g., Avpr1a, Ednra, Edn1; Figure 2M; Data S1), suggesting the repression of astrocyte-specific functions (Alfaro-Cervello et al., 2012; Filosa et al., 2016), and involved the re-organization of the extracellular matrix (Figure 2N; Data S1). Typical genes expressed in astrocytes were also downregulated (e.g., Aqp4, Slc1a3, Fgfr3; Figure S2J) (Weng et al., 2019), as well as some expressed in glial progenitors (e.g., Pax6, Fabp7, Vimentin; Figure S2K) (Treutlein et al., 2016). Stringent criteria for gene selection revealed a subset of genes uniquely downregulated by Ascl1ERT2 (40 genes; Figure 2L), associated to phosphorylation and actin organization (e.g., Tec, Nrp1, Actr3b, Figure 2O; Data S1); conversely, Neurog2ERT2 specifically downregulated genes (43; Figure 2L) were associated with purine metabolism (e.g., Aldoa, Bcl2l1, Eno1b, Uqcrc1; Figure 2P; Data S1). Taken together, Ascl1ERT2 and Neurog2ERT2 not only induced different neurogenic programs at an early stage, but they also suppressed different aspects of the starter cell identity.
We next explored the expression changes of TFs and chromatin remodeling factors because important in fate specification and being a significant fraction of the regulated genes (13.9% in Ascl1ERT2 and 12.5% in Neurog2ERT2; log2FC > 1, padj < 0.01; Figure S2L; Data S1). Among the common upregulated TFs (log2FC > 1, padj < 0.01; Figure S2L), some have already been implicated in direct neuronal reprogramming (e.g., Hes6, Insm1, Sox11, Sox4, Prox1, Trnp1, Zbtb18, and Ezh2; Liu et al., 2018; Masserdotti et al., 2015; Wapinski et al., 2013). Interestingly, many other TFs were regulated specifically by Ascl1ERT2 (e.g., Id3, Gata3, Klf10, Lhx3; Figure 2Q) or Neurog2ERT2 (e.g., Zbtb16, Sap18, Nhlh1; Figure 2Q; Data S1), including Gata3 and Lhx3, associated to V2 interneurons and motoneurons (Andrzejczuk et al., 2018; Misra et al., 2014). We also noted that both factors induced the expression of their respective endogenous genes (Data S1).
To identify important common TF- and chromatin-remodeler-specific downstream effectors, we constructed a gene expression regulatory network (Su et al., 2014a) by ranking TFs according to their induction, statistical significance and expression level (Figure 2R; see STAR Methods). The sub-network composed of shared genes (in yellow; Figure 2R) comprised many factors already found in cortical astrocyte reprogramming (e.g., Hes6, Sox11, Sox4, Atoh8, Olig1/2, Ncor2; Masserdotti et al., 2015), suggesting that this network may be common in different regions. Interestingly, the Ascl1ERT2-specific subnetwork was mainly composed of TFs (e.g., Sox8, Id1, Id2, Id3, Sox4, Sox11; Figure 2R, left), while the Neurog2ERT2-specific subnetwork was associated with many chromatin modifiers (e.g., Hmgb2, Smarcc1, Smarce1, Arid1a; Figure 2R, right) with Tcf4, the heterodimeric partner of Ascl1 and Neurog2 (Wang and Baker, 2015), linking them (Quevedo et al., 2019). Gene set enrichment analysis (GSEA) (Subramanian et al., 2005) supported this observation, as genes related to “chromatin binding” and “chromatin assembly and disassembly” were more enriched in Neurog2ERT2 than in Ascl1ERT2 differentially expressed genes (Figure 2S).
Together, these data indicated that Ascl1ERT2 and Neurog2ERT2 activity quickly induced very different neurogenic cascades, characterized by pan-neuronal and TF-specific downstream cascades, which contribute to establish the new neuronal identity.
Molecular characterization of Ascl1- and Neurog2-iNs
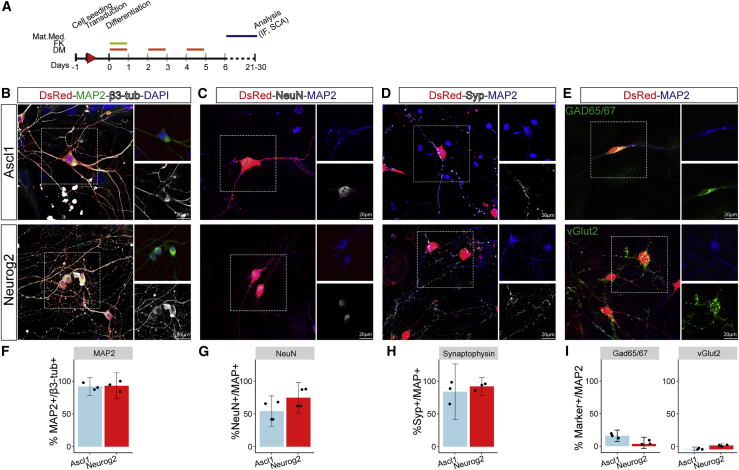
To explore the neuronal identity elicited by these factors in SC astrocytes, we analyzed reprogrammed cells at later stages (21–30 DPI, Figure 3A): most iNs (β3-tubulin+DsRed+) were positive for the mature pan-neuronal markers Map2 (Ascl1, mean = 91.77%, CI = 10.54; Neurog2, mean = 92.93%, CI = 15.74; Figures 3B and 3F), NeuN (Ascl1, mean = 58.43%, CI = 25.70; Neurog2, mean = 79.03%, CI = 25.63; Figures 3C and 3G), and synaptophysin (Ascl1, mean = 84.10%, CI = 33.22; Neurog2, mean = 91.99%, CI = 10.2; Figures 3D and 3H). Some iNs were positive for the GABAergic marker Gad65/67 (Ascl1, mean = 15.93%, CI = 6.12; Neurog2, mean = 5.218%, CI = 4.60; Figures 3E and 3I, upper micrograph), while very few were positive for the glutamatergic presynaptic marker vGlut2, visible as punctate staining along the processes (Ascl1, mean = 0.65%, CI = 1.96; Neurog2, mean = 5.83%, CI = 1.9; Figures 3E and 3I, lower micrograph), similar to previous observations (Hu et al., 2019).
Figure 3.
Expression of neuronal markers in Ascl1 and Neurog2 iNs
(A) Scheme of the experimental design. IF, immunofluorescence; SCA, single-cell analysis. Bars indicate the day of treatment with different small molecules.
(B–E) Micrographs depicting pan-neuronal, GABAergic, and glutamatergic markers as indicated in Ascl1- (top) and Neurog2- (bottom) iNs at 21–24 DPI. Scale bar: 20 μm.
(F–I) Bar charts of percentage of mature neuronal markers (F–H) and GAD65/67 or vGlut2 (I) in Ascl1- and Neurog2-iNs as indicated. n = 3 independent experiments.
To better investigate iNs’ identity, we collected single cells at 21–28 DPI (Figure S3A) and subjected them to RNA-seq (Picelli et al., 2014). Morphometric analysis (Figures S3B–S3E) did not reveal significant differences between Ascl1- and Neurog2-iNs. Single-cell RNA-seq analysis (scRNA-seq) was performed on 33 iNs (48 collected cells), identified by the expression of DsRed, small soma and the presence of two or more long and thin processes (11 Ascl1-iNs, 22 Neurog2-iNs), and six astrocytes (Ascl1-transduced with a flat morphology). Principal component analysis (PCA) clearly separated astrocytes and iNs (Figure 4A), with Ascl1-iNs clustering farther away from non-reprogrammed astrocytes than from Neurog2-iNs (Figure 4A). In line with the criteria for neuronal selection, hundreds of genes were differentially expressed between iNs and non-reprogrammed astrocytes (padj < 0.01; Figures 4B and 4E; Data S2), with many upregulated genes common to Ascl1- and Neurog2-iNs (Figure 4C; GO in Figure 4D; Data S2). Thus, Ascl1- and Neurog2-iNs become more similar after the initial induction of very different neurogenic programs (for specific differences, see Figures S3F and S3G). GSEA (Figures S3H and S3I; Data S2) confirmed the enrichment for genes involved in synapse formation and neuronal activity for both TF-mediated reprogramming cascades. The few genes commonly downregulated (Figure 4F) were involved in detoxification (Figure 4G; Data S2) and β-oxidation (GSEA; Figures S3J and S3K; Data S2), well-known astrocyte hallmarks (Russo et al., 2020).
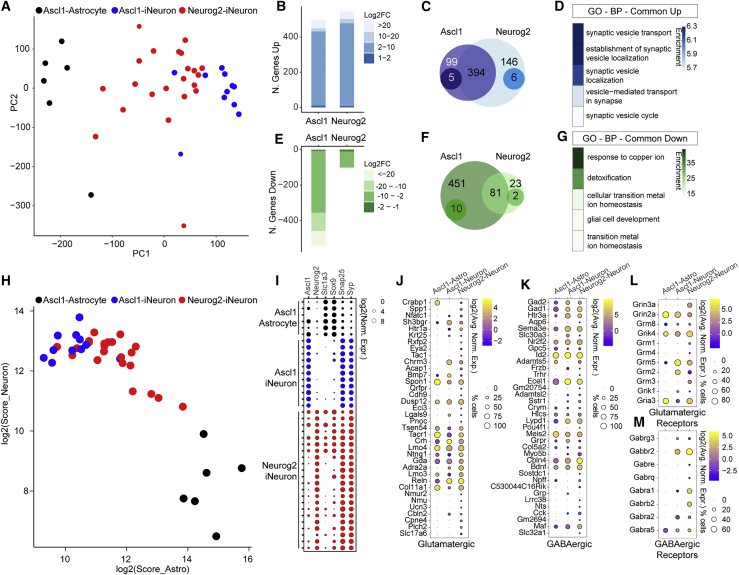
Figure 4.
scRNA-seq of Ascl1- and Neurog2-iNs
(A) PCA of the 39 single cells isolated by patching. Neurog2-iNs (red dots) are localized between Ascl1-astrocytes (black dots) and Ascl1-iNs (blue dots).
(B, C, E, and F) Barplots (B and E) and Venn diagrams (C and F) of the number of genes upregulated (B and C) or downregulated (E and F) (padj < 0.01) between iNs and astrocytes. In (C) and (F), genes were considered if log2FC > |1| and padj < 0.01. Unique genes (darker circles) were considered if, for example, log2(FC-Ascl1) > 1, padj < 0.01, and log2(FC-Neurog2) ≤ 0, or, for example, log2(FC-Neurog2) > = 1, padj < 0.01, and log2(FC-Ascl1) ≤ 0.
(D and G) Top 5 GO terms (BP) from commonly upregulated (D) or downregulated (G) genes. Colors depict the enrichment over expected genes.
(H) Scatterplot depicting each cell based on log2(Astrocyte score) and log2(Neuronal score).
(I–M) Bubble plots showing the log2(expression) of selected genes (I), glutamatergic (J), GABAergic (K), and the respective receptors (L and M) in single cells. Size of the circles reflects the proportion of cells expressing the marker in each category (in J–M); color depicts the log2(average of normalized expression) (in J–M).
Interestingly, Neurog2-iNs were widely dispersed in the PCA plot, with some close to non-reprogrammed astrocytes (Figure 4A). As also the number of significantly downregulated genes was lower in Neurog2- than in Ascl1-iNs (Figure 4E), we examined whether this was due to an incomplete conversion. We generated an “astrocyte score” and a “neuronal score” (Astro-Score and Neuron-Score, respectively) by summing the normalized expression values of 68 markers for astrocytes (merging those used in Tripathy et al. [2018] and those identified in Weng et al. [2019]; averaged expression in Figure S3L) and 21 neuronal markers (including neuronal vesicles coding genes; averaged expression in Figure S3M). Ascl1-astrocytes (black dots, Figure 4H) were distinct from Ascl1-iNs; however, some Neurog2-iNs were close to the Ascl1-astrocytes, thus reflecting a rather incomplete conversion. Of note, Ascl1-iNs also retained the expression of some markers for astrocytes (e.g., Fgfr3, Nfia; Figure S3L), thus indicating the persistence of some aspects of the starter cell identity.
To determine the potential cause of the unsuccessful conversion, we examined the expression of endogenous Ascl1 and Neurog2, as they were upregulated at an early stage by the forced expression of Ascl1 and Neurog2 (Data S1). Endogenous Neurog2 was expressed in all Neurog2-iNs at a similar level (Figure 4I) and, likewise, Ascl1 in Ascl1-iNs. However, a significant proportion of Neurog2-iNs also expressed high levels of Ascl1 (Figure 4I), and these cells were closer to non-reprogrammed astrocytes (Figure S3N); conversely, Neurog2 was barely detected in Ascl1-iNs. This correlated with the high expression of the markers for astrocytes Slc1a3 and Sox9 (Figure 4I) but had no effect on the neuronal program, as Snap25 and Syp were similarly expressed. Thus, the induction of endogenous Ascl1 in Neurog2-iNs might perturb the neuronal conversion and maintain some glial traits, as Ascl1 is also involved in the generation of glial lineages (Kelenis et al., 2018).
To determine the neuronal subtype identity elicited by these TFs, we analyzed the expression of genes characteristic for glutamatergic or GABAergic neurons and their respective receptors (Figures 4L and 4M). This revealed a surprisingly similar transmitter identity between the iNs induced by Ascl1 and Neurog2: both expressed some glutamatergic (Figure 4J) and GABAergic markers (Figure 4K) and their receptors (Figures 4L and 4M) (Häring et al., 2018). However, they differed in the expression of specific neuropeptides, such as galanin (Gal) in the Ascl1-iNs (Figure S3O), or in specific receptors (e.g., Cnr1, Hrh3, Adra2a, and Chrna3 in Neurog2-iNs; Htr5b in Ascl1-iN; Figures S3P–S3R) (Ren et al., 2019).
Taken together, no clear-cut difference in the neuronal identity was detectable in Ascl1- versus Neurog2-iNs at later stages. However, this may be due to the heterogeneity in reprogramming, a surprising finding given the rather homogeneous maturity observed by electrophysiology (Figure 1). We therefore combined these modes of analysis using patch sequencing (patch-seq) to investigate the degree of correlation between electrophysiological and transcriptional maturity of iNs and their neuronal subtype identity.
Molecular analysis of electrophysiologically characterized Ascl1- and Neurog2-iNs using patch-seq
To directly correlate gene expression and electrophysiology, transduced SC-derived astrocytes were subjected to patch-seq (Cadwell et al., 2017; Winterer et al., 2019) from 14 DPI onward. First, we confirmed the high similarity of Ascl1- and Neurog2-iNs over DsRed-control (as in Figure 4C)—irrespective of their electrophysiological properties (Figure S4E; Data S3)— identifying only 72 genes specific for Ascl1 (Figures S4E and S4F; GO terms in Figure S4J; Data S3) and 51 upregulated only in Neurog2-iNs (Figures S4E and S4G; GO terms in Figure S4K; Data S3). Markers of astrocytes (e.g., Aqp4, Nfia, Sox9) were still expressed, though at lower levels than in DsRed-transduced astrocytes (Figure S4H).
Next, we grouped the cells based on their electrophysiological properties resulting in four groups (Figures S4A–S4D; STAR Methods): a control group (DsRed-transduced astrocytes; Figures 5A and 5B) and three classes of iNs. The different classes of iNs could be characterized by either high resting membrane potential and no spike or off-spike occurring after the depolarization (class 1 iNs; Figures 5C and 5D), small overshooting and immature spike (class 2 iNs; Figures 5E and 5F), or overshooting and (repetitive) firing (class 3 iNs; Figures 5G and 5H). Control cells and iNs were separated in the PCA plot (Figure 5I), with no detectable clustering among the diverse iN subgroups, as even class 1 and class 3 were intermingled (Figure 5I). This suggests that the electrophysiological properties of neurons may be determined mostly at the post-transcriptional level and/or by a small part of the transcriptional program. Differential expression analysis compared to control (log2FC > 1; padj < 0.01; Data S4) revealed 747 differentially expressed genes in class 1 iNs, 1,351 in class 2 iNs, and 1,384 in class 3 iNs, with 712 regulated in all and 1,298 co-regulated in the class 2 and class 3 iNs (Figure 5J). Genes differentially regulated in both class 2 and class 3 iNs were associated with synaptic activity (Data S4 for GO terms). Noting the presence of potassium channel (K-channel) subunit coding genes (e.g., Kcnb1, Kcns2; GO in Figure 5N; Data S4) among the molecular signature of class 3 iNs (Figure 5K) and the expression of glutamate receptor subunit coding genes (e.g., Grik3, Grm2; Figure 5L; GO Figure 5M; Data S4) in class 2 iNs signature (Figure 5L), we analyzed their expression in more detail (Figures 5O and S4L). Overall, both K-channels and glutamate receptors were more expressed in class 2 and class 3 iNs than in class 1 iNs and control astrocytes. Likewise, glutamatergic and GABAergic markers were expressed more in class 2 and class 3 iNs (Figures S4M and S4N). Some K-channels were expressed in both class 2 and class 3 iNs (e.g., Kcnc3), while others seemed specific to one of the proneural factors (e.g., Kcnf1 for Neurog2; Kcns2 for Ascl1; Figure 5O); glycinergic markers were barely detected (Figure S4O) (Callahan et al., 2019). Taken together, electrophysiology clearly identifies functional neuronal properties but cannot account for the transcriptional fidelity of conversion. Transcriptome analysis reveals the molecular state of the converted cells but cannot attribute functional features to neurons. Notably, the expression of some K-channels correlates with some functional properties of iNs.
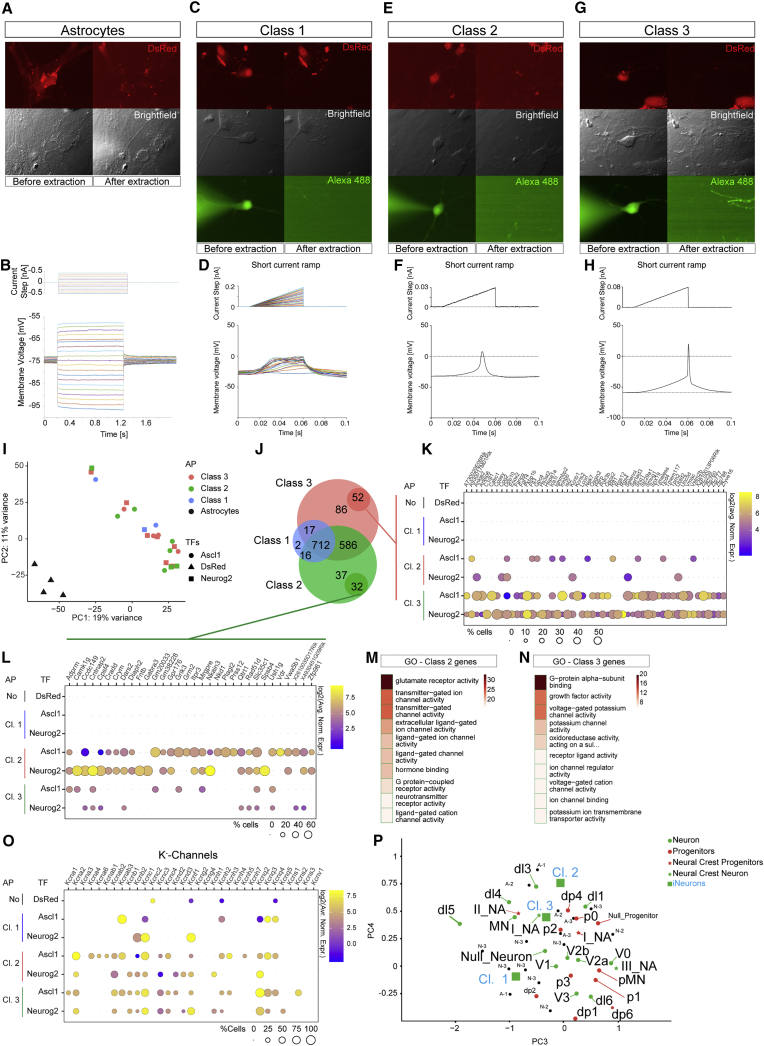
Figure 5.
Patch-seq analysis of Ascl1- and Neurog2-iNs
(A, C, E, and G) Epifluorescent and brightfield pictures of cells selected for electrophysiological and transcriptome analysis. Shown are a control astrocyte (A; n = 4), class 1 non-firing reprogrammed cells (C; Ascl1 n = 2, Neurog2 n = 1), class 2 immature firing iNs (E, Ascl1, n = 5; Neurog2, n = 3), and class 3 firing iNs (G; Ascl1, n = 4; Neurog2, n = 7).
(B, D, F, and H) Examples of current steps (upper plots) and the corresponding membrane voltage responses (lower plots) for the cell types indicated in (A), (C), (E), and (G). For the astrocytes (B), long current step pulses are shown with hyperpolarizing and depolarizing steps. For the reprogrammed cells, short current ramp steps are shown with either all traces (class 1, D) or single traces (class 2, F; class 3, H).
(I) PCA plot of 26 cells analyzed by patch-seq. Control cells (black triangles) are separated from iNs.
(J) Venn diagram of the genes shared among different classes of iNs.
(K, L, and O) Bubble plots showing the expression of genes highly expressed in class 3 (K) or class 2 (L) iNeurons and K-channels (O) in the different subgroups.
(M and N) Top 10 GO terms (MF) from highly expressed genes in class 2 (M) or class 3 iNs (N). Colors depict the enrichment over expected genes.
(P) PCA of iNs (clustered according to classes in C, E, and G) compared to SC progenitors and neurons from Delile et al. (2019). Each black dot represents an iN. A-n, Ascl1-Class n; N-n, Neurog2-Class n.
Next, we compared our dataset with a published dataset of SC neurons (Delile et al., 2019). Control astrocytes were separated from neurons (Figure S4P); a closer look revealed that only the class 3 iNs appeared in the center of the cloud of endogenous SC neurons, while class 1 and class 2 iNs were at the margins (Figure 5P). Class 3 iNs were closer to V2b interneurons and p2, their progenitors (Karunaratne et al., 2002; Li et al., 2005). Consistent with this, Neurog2- and Ascl1-firing iNs were very close together, with some Ascl1 class 3 iNs closer to the p2 progenitors and some Neurog2 class 3 iNs closer to the V2b interneurons. Thus, both TFs elicited a rather similar neuronal subtype identity in SC-derived astrocytes, in pronounced contrast to previous observations following the conversion of astrocytes from other regions (Heinrich et al., 2010; Hu et al., 2019).
Region-specific identity markers are maintained in vitro
The above results clearly showed the generation of different iNs in the SC compared to cortex and midbrain astrocyte reprogramming (Heinrich et al., 2010; Liu et al., 2015). One possible cause for this may be the regionalized gene expression in the starter cells. To determine this, we analyzed the transcriptome of MACS astrocytes from the cortical GM and the SC, immediately after MACS (acute) or cultured for 7 days under identical conditions (culture; Figure 6A). PCA separated the samples according to the region (PC1) and acute isolation versus culture (PC2; Figure S5A). Comparison between SC and GM astrocytes revealed similar differences in gene expression for both acutely isolated and cultured astrocytes (Figures 6B and 6C; Data S5). Known patterning genes from development were among the most differentially expressed genes in each condition analyzed (e.g., Otx1, Emx2, Foxg1 in GM astrocytes; Hox genes in SC astrocytes; Figure S5B) (Hébert and Fishell, 2008; Philippidou and Dasen, 2013; Sagner and Briscoe, 2019). Indeed, GO term analysis supported the forebreain origin of GM astrocytes (Figures 6D and 6F; Data S5) and the SC signature for SC astrocytes (e.g., Hox genes; Figures 6E, 6F, 6H, and 6I; Data S5) from both acutely isolated and in vitro cells.
Figure 6.
Transcriptional differences between astrocytes from SC and cerebral cortex GM
(A) Scheme of sample collection for RNA-seq.
(B and C) Venn diagram of the transcriptome of GM and SC astrocytes acutely isolated (B) or following 7 days in culture (C).
(D–H) Top 5 GO terms (BP) associated with genes differentially expressed in acutely isolated GM (D) and SC (E) and cultured GM (G) and SC (H) astrocytes.
(F and I) Heatmaps of the relative expression of genes comprised in the top GO for each region in acute (F) and cultured condition (I).
(J) Bubble plot depicting the percentage of cells and log2(averaged normalized expression) of patterning genes in different classes of iNs (dataset from Figure 5).
(K and L) Network analysis of TFs, enriched chromatin modifiers, and RNA-binding protein in cultured SC astrocytes (K) or cultured GM astrocytes (L) with main hubs in blue.
(M and O) Examples of pathways identified by GSEA after comparing cultured GM and SC astrocytes transcriptome. SC neural tube patterning genes (M); GM-related forebrain regionalization genes (O).
(N and P) Barplots depicting the log2FC of genes associated with (M) (shown in N) or with (O) (shown in P).
(Q) Overlap between the genes induced by Ascl1ERT2 and Neurog2ERT2 in GM (log2FC > 1.3 and p < 0.05) and SC (log2FC > 2 and padj < 0.01) astrocytes at 24 h.
(R and S) Top 5 GO terms (R; cellular compartment, CC) and sketch of the cellular localization (S) of 20 commonly upregulated genes. Neurod4 is regulated in SC-derived astrocytes by Ascl1ERT2 only with padj = 0.06.
Interestingly, the transcriptional profile of cultured GM astrocytes was enriched for vasculature-related genes (Figures 6G and 6I; Data S5), supporting the notion of astrocytes as important players for blood-brain barrier formation (Langen et al., 2019). Of note, primary cultures of astrocytes from both regions showed high similarity with their acutely isolated counterparts (Figures S5C and S5F), with differences related to angiogenesis (in vivo for both GM and SC; Figures S5D and S5G; Data S6) and wound healing (Figures S5E and S5H; Data S6). Furthermore, acutely isolated cells, either from cortical GM or SC, expressed higher levels of astrocyte markers than did their cultured counterparts (Figures S5I and S5J). Importantly, the expression of region-specific patterning genes was maintained in iNs (Hox genes; Figure 6J), thus supporting the concept that neuronal identity is influenced by the regionalization of the starter astrocyte population.
To investigate the programs in astrocytes cultured from GM and the SC, we identified differentially expressed TFs, chromatin modifiers, and RNA-binding proteins and constructed a gene expression regulatory network (Su et al., 2014a). Three distinct branches emerged in SC astrocytes (Figure 6K): one related to RNA-binding proteins (red circles) (Matera and Wang, 2014), another comprising genes for neural tube development (e.g., Pax6, FoxP1, green circles), and the third including chromatin-related factors (e.g., Ssrp1, Hmgb2, Dnmt1, blue circles). Conversely, network analysis on genes from GM astrocytes revealed one ramified network (Figure 6L), in which FoxG1, Mef2c, and Cebpb seemed to be main hubs. GSEA highlighted that SC astrocytes express “dorso-ventral neural tube patterning” genes (Figures 6M and 6N; Data S5), and GM astrocytes express a higher number of “forebrain regionalization” genes (Figures 6O and 6P). In summary, both acutely isolated and in vitro cultured GM and SC astrocytes exhibited transcriptome differences and retained specific developmental and patterning hallmarks and gene networks (Figures 6 and S5), with high relevance for specifying neuronal subtypes.
Common and region-specific cascades induced by Ascl1 and Neurog2
As patterning genes are retained in iNs (Figure 6J), possibly influencing the reprograming, we compared Ascl1ERT2- and Neurog2ERT2-induced neurogenic cascades in SC- (Figure 2) and GM-derived astrocytes, obtained from published microarray data (Masserdotti et al., 2015) and induced for the same time (24 h) without any small-molecule treatment. This revealed a profound influence of the starter cells on the induced programs: most regulated genes were specific for a specific combination of starter cell and TF (Figure 6Q), with only 27.3% of Ascl1ERT2- and 12% of Neurog2ERT2-upregulated genes common between the astrocytes isolated from different regions (Figure 6Q). Irrespective of the starter cell, a large fraction of genes upregulated by Ascl1ERT2 (176 genes; Figures 6Q, yellow background, and S5K) were associated with neuronal functions (Figure 6R); among them, genes such as Calm1, Pvalb; the transcription factors Lhx3, Lmo1, and Sox8; and the signaling molecules Bmp2, Bmp7, Sema6b, and Wnt9a (Figure S5K). Neurog2ERT2 regulated 22 genes common to both regions (Figure 6Q, green background), among which were several synapse-associated protein coding genes, such as Cnr1, Cplx2, Lrrtm1, and Cadm3 (Figure S5L). Overall, 20 genes (Figures 6Q and 6S, blue background) were commonly induced by both proneural factors from the different types of astrocytes, including many transcription factors (Hes6, Insm1, Neurod4, Prox1, Sox11, and Trnp1) (Masserdotti et al., 2015). Thus, the starter astrocyte regional identity profoundly influences the reprogramming process for each proneural factor with few pan-neuronal core regulators.
Discussion
Direct conversion of SC-derived astrocytes showed that Ascl1 and Neurog2 elicit functionally mature neurons, with a signature reminiscent of ventral SC interneuron identity, despite initially inducing very different transcriptional cascades. The distinct neuronal fate instructed from SC versus cortex astrocytes unraveled the potent influence of the regional identity of astrocytes, shown here to be maintained in vitro. These data further support the concept that astrocytes are well suited as starter cells for neuronal reprogramming: their regional specification and the programs elicited by the proneural factors may allow achieving the correct regional neuronal identity. Finally, our patch-seq data demonstrate the need for both electrophysiology and transcriptomics to ensure adequate conversion and fate acquisition.
Distinct molecular programs elicited by Ascl1 and Neurog2 in SC astrocytes
Using the inducible forms of Ascl1 and Neurog2, we identified the early induction of two largely distinct molecular programs (Figure 2), possibly also influenced by differences in the timing of the reprogramming or in the reprogramming efficiency. These programs comprise many TFs (Figure 2Q): of note, Neurog2ERT2 induced more chromatin remodeling factors than Ascl1ERT2 (Figure 2R), suggesting that Neurog2 may require specific co-factors and complexes to promote and stabilize the neuronal cascade (Figures 2R and 2S). Conversely, Ascl1 can apparently act as on-target pioneer factor (Wapinski et al., 2013) either alone or by recruiting endogenously expressed co-factors. Both TFs repressed astrocyte-specific genes (Figure S2I), while no neural stem cell marker was induced (Figure S2J), supporting the direct conversion of astrocytes without passing through a more stem-cell-like state, as is the case in fibroblast-to-neuron direct conversion (Treutlein et al., 2016), or upon the forced expression of Sox2 in pericytes (Karow et al., 2018).
It was surprising to see no clear specification toward a GABAergic or glutamatergic identity (Figure 3) of iNs, contrary to recent reports (Hu et al., 2019) and different from cortical GM astrocytes reprogramming (Heinrich et al., 2010). Both Ascl1 and Neurog2 iNs from SC astrocytes were close to a ventral SC GABAergic V2b interneuron type (Figure 5P), consistent with the expression of Gad1 and Gad2 in some firing Ascl1 or Neurog2 iNs (Figure S4N). While most iNs did not fully complete the GABAergic neurotransmitter identity (Figure 3I), our results reveal that Ascl1 and Neurog2 do not instruct GABAergic versus glutamatergic neurons, as in the cortex astrocytes (Heinrich et al., 2010) and, to some extent, the midbrain astrocytes (Liu et al., 2015). Rather, the crosstalk between the reprogramming factors and the cellular context shapes the neuronal outcome: the persistence of patterning TFs in astrocytes of the respective regions may influence the developmental role of the proneural factors. During SC development, V2a and V2b interneurons derive from p2 progenitors, which express Ascl1, Neurog1, and Neurog2 in a mosaic pattern. Ascl1 promotes a V2a progenitor fate and Neurog2 inhibits a V2a progenitor fate, thereby promoting V2b neuronal identity (Misra et al., 2014). The acquired fate elicited by reprogramming closely reflects the role of these factors in SC development and may explain why some Neurog2-iNs can progress a bit further toward the V2b neuronal identity. Thus, Neurog2 can also induce GABAergic neuronal subtypes depending on its developmental role and the regional identity of the starter cells.
Our analysis further showed that Ascl1 and Neurog2 can interfere with each other, as shown for V2b interneuron fate acquisition (Misra et al., 2014) and in the telencephalon (Fode et al., 2000; Kovach et al., 2013; Schuurmans et al., 2004). Furthermore, Ascl1 is also involved in gliogenesis throughout the CNS, including the SC (Kelenis et al., 2018; Vue et al., 2014). As such, the co-expression of endogenous Ascl1 and Neurog2 in some Neurog2 iNs correlated with an incomplete repression of the astrocytic signature (Figures 4I, S3L, and S4H). However, Ascl1 and Neurog2 are co-expressed at E12.5 in the dorsal telencephalic ventricular zone (Britz et al., 2006), and double-knockout (KO) embryos showed specific defects in neurogenesis (Dennis et al., 2017; Nieto et al., 2001), further supporting their context-dependent activity; as such, in fibroblasts their co-expression rather improves reprogramming into neurons (Herdy et al., 2019; Ladewig et al., 2012). Thus, the function of these TFs is context dependent despite their common neurogenic role: astrocytes provide a context that allows predictions of the induced fate based on CNS development from radial glial cells.
The contribution of regionalization to the reprogramming process
Developmentally instructed patterning information was present in acutely isolated astrocytes and maintained in vitro, with GM-derived astrocytes expressing TFs characteristic of the dorsal telencephalon (e.g., Emx1, Otx1) and SC-derived astrocytes expressing Hox genes (Figure S5B). Thus, the maintenance of regionalization contributes to the induction of different neurogenic cascades by the same TFs in astrocytes from different regions (Figure 6Q), resulting in the generation of different neurons, as reported for different brain regions including distinct thalamic nuclei in vitro and in vivo (Herrero-Navarro et al., 2021; Mattugini et al., 2019; Qian et al., 2020; Zhou et al., 2020). Of note, GM astrocytes could be more efficiently reprogrammed than SC astrocytes, suggesting a broader effect of regionalization on direct conversion. For example, the transcriptional and proteomic context in different astrocytes could regulate proneural gene activity (e.g., via the expression of specific cofactors or via their phosphorylation; Ali et al., 2014; Hindley et al., 2012), a very relevant aspect for in vivo repair. Only few genes were induced by both TFs in both regions (e.g., NeuroD4, Hes6, Insm1, Prox1, and Sox11; Figures 6Q and 6R) (Masserdotti et al., 2015), thus possibly representing the “pan-neurogenic” core network sufficient to instruct a neuronal fate in different cell types, including astrocytes from other CNS regions.
In summary, these data provide compelling evidence for a major contribution of the starter cell in shaping the ability of Ascl1 and Neurog2 to exploit their reprogramming potential, which calls for the identification of starter-cell-specific cocktails of reprogramming factors. Furthermore, it suggests that many reprogrammed cells retain features of their original identity, prompting the need for molecular characterization of the final neuronal outcome.
Patch-seq reveals low correlation between electrophysiology and transcriptome of single iNs
The gold standard of iNs is electrophysiology, and rightly so, as firing and synaptic connectivity are central to their functional roles. However, thus far, the electrophysiological and transcriptional state of single iNs using patch-seq was not assessed. Here, we show that there is no correlation in the overall clustering of different electrophysiological classes of iNs according to their transcriptome, suggesting that these major functional differences might depend on only few genes (which, hence, would not influence the clustering) or on other aspects (e.g., post-translational modifications). Indeed, channels and receptors have a long half-life as proteins and often do not appear as distinguishing features in scRNA-seq data. Therefore, the transcriptome cannot predict the electrophysiological state of iNs, even though we unraveled the higher expression of some K-channels in the most mature iNs (Figures 5N and 5O). These contribute to maintain the resting membrane potential and repolarize neurons, a distinctive feature of firing iNs. Importantly, electrophysiology cannot be used as a predictor of the overall fate conversion at the transcriptional level. Hence, it is essential to complement electrophysiology with transcriptional analysis as quality control for the identity of iNs, to avoid incomplete phenotypes especially for in vivo neuronal replacement.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse anti-β-III-Tubulin | Sigma-Aldrich | Cat# T8660; RRID: AB_477590 |
| Mouse anti-GFAP | Dako | Cat# Z0334; RRID: AB_100013482 |
| Rabbit anti-GFAP | Sigma-Aldrich | Cat# G3893; RRID: AB_477010 |
| Rat anti-RFP | Chromotek | Cat# 5F8; RRID: AB_2336064 |
| Rat anti-RFP | Rockland | Cat# 600-401-379; RRID:AB_2209751 |
| Anti-MAP2 | Millipore | Cat# MAB378, RRID:AB_94967 |
| Anti-MAP | Millipore | Cat# AB5622, RRID:AB_91939 |
| Anti-vGlut2 | Synaptic Systems | Cat# 135402; RRID:AB_2187539 |
| Anti-Gad65/67 | Sigma-Aldrich | Cat# G5163; RRID:AB_477019 |
| Anti-synaptophysin | Synaptic Systems | Cat# 101 011 RRID:AB_887824) |
| Anti-NeuN | Merck/Millipore | MAB377; RRID:AB_2298772 |
| Anti-Ascl1 | BD PharMingen | RRID:AB_396479 |
| Anti-Neurog2 | Gift from DJ Anderson, Caltech, California | N/A |
| Anti-Sox9 | Sigma-Aldrich | Cat# AB5535; RRID:AB_2239761 |
| Anti-Olig2 | Millipore | Cat# MABN50; RRID:AB_10807410 |
| Anti-Iba1 | Synaptic System | Cat# 234013; RRID:AB_2661873 |
| Anti-Dcx | Millipore | Cat# ab2253; RRID:AB_1586992 |
| Anti-Mouse Alexa Fluor 488 | Molecular Probes | Cat# A-21202; RRID: AB_141607 |
| Anti-Mouse IgG2b 633 | Innovative Research | Cat# A21146; RRID: AB_1500899 |
| Anti-Mouse IgG1 647 | Molecular Probes | Cat# A21240; RRID: AB_141658 |
| Anti-Mouse IgG1 Biotin | Southernbiotech | Cat# 1070-08; RRID: AB_2794413 |
| Anti-Rabbit Alexa Fluor 488 | Molecular Probes | Cat# A21206; RRID: AB_141708 |
| Anti-rabbit-Cy5 | ImmunoResearch | Cat # 111-175-144 RRID: AB_2338013 |
| Anti-rabbit Cy3 | ImmunoResearch | Cat# 711-165-152 RRID: AB_2307443 |
| Anti-Rat Cy3 | ImmunoResearch | Cat# 112-165-167 RRID: AB_2338251 |
| Streptavidin Alex Fluor 405 | Thermo Fisher | Cat# S32351 |
| Chemicals, peptides, and recombinant proteins | ||
| EGF | GIBCO | Cat# PHG0311 |
| bFGF | GIBCO | Cat# 13256029 |
| Poly-D-Lysine | Sigma-Aldrich | Cat# P0899 |
| B27 | GIBCO | Cat# 17504044 |
| HBSS medium | Thermo Fisher | Cat# 24020117 |
| HEPES | Thermo Fisher | Cat# 15630080 |
| DMEM/F12 | Thermo Fisher | Cat# 10565018 |
| trypsin/EDTA 0,25% | Thermo Fisher | Cat# 25200056 |
| Neurobasal Medium | GIBCO | Cat# 21103149 |
| Glucose | GIBCO | Cat# A2494001 |
| GluataMAX | GIBCO | Cat# 35050061 |
| OptiMEM – GlutaMAX | Thermo Fisher | Cat# 51985-026 |
| EGTA | Sigma-Aldrich | Cat# E3889 |
| Lipofectamine 2000 | Thermo Fisher | Cat# 11668019 |
| Triton X-100 | Sigma-Aldrich | Cat# T9284 |
| BDNF | Peprotech | Cat# 450-02 |
| GDNF | Peprotech | Cat# 450-10 |
| cAMP | Sigma-Aldrich | Cat# D0260 |
| NT3 | Peprotech | Cat# 450-03 |
| N2 | Invitrogen | Cat# 17502048 |
| Forskolin | Sigma-Aldrich | Cat# F-6886 |
| Dorsomorphin | Sigma-Aldrich | Cat# P-5499 |
| Hydroxyl-Tamoxifen | Sigma-Aldrich | Cat# H-7904 |
| Bovine Serum Albumine (BSA) | Sigma-Aldrich | Cat# A9418 |
| Critical commercial assays | ||
| Arcturus PicoPure RNA Isolation Kit | Thermo Fisher | Cat# 12204-01 |
| Agencourt AMPure XP | Beckman Coulter | Cat# 10136224 |
| Deposited data | ||
| Bulk RNA-seq | This study | GEO: GSE174238 |
| Patch-Seq – Figure 4 | This study | GEO: GSE173977 |
| Patch-Seq – Figure 5 | This study | GEO: GSE173978 |
| SC_vs_GM Astrocytes | This study | GEO: GSE173979 |
| Experimental models: Organisms/strains | ||
| C57BL/6 | LMU animal Facility | N/A |
| Recombinant DNA | ||
| RV CAG-Neurog2-IRES-DsRedExpress2 | Gascón et al., 2016 | N/A |
| RV CAG-Ascl1-IRES-DsRed | Heinrich et al., 2010 | N/A |
| RV CAG-DsRedExpress2 | Heinrich et al., 2010 | N/A |
| RV CAG-Ascl1ERT2-IRES-DsRed | Masserdotti et al., 2015 | N/A |
| RV CAG-ERT2-Neurog2-IRES-DsRed | This study | N/A |
| Software and algorithms | ||
| ZEN software | Zeiss | https://www.zeiss.com/microscopy/en_us/products/microscope-software RRID:SCR_013672 |
| ImageJ | ImageJ | https://imagej.net RRID: SCR_003070 |
| GraphPad Prism 5.0 | GraphPad Software | http://www.graphpad.com:443/ RRID:SCR_002798 |
| Adobe Photoshop | Adobe Photoshop | https://www.adobe.com RRID: SCR_014199 |
| Adobe Illustrator | Adobe Illustrator | https://www.adobe.com RRID:SCR_010279 |
| Microsoft Excel | Microsoft Excel | https://www.microsoft.com/en-gb/ RRID:SCR_016137 |
| RStudio | RStudio | https://www.rstudio.com/ RRID: SCR_000432 |
| TopGO v.2.34.0 | Alexa and Rahnenfuhrer, 2018 | https://bioconductor.org/packages/release/bioc/html/topGO.html |
| DESeq2 v. 1.22.2 | Love et al., 2014 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| Ggplot2 v.3.2.0 | Wickham, 2016 | https://cran.r-project.org/web/packages/ggplot2/index.html |
| fgsea | Sergushichev, 2016 | https://bioconductor.org/packages/release/bioc/html/fgsea.html |
| Other | ||
| Aqua Poly/Mount | Polysciences | Cat# 18606-20 |
| pluriStrainer Mini | pluriselect | Cat# 43-10040-40 |
| NucBlue | ThermoFisher | Cat# R37605 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Dr. Giacomo Masserdotti (giacomo.masserdotti@helmholtz-muenchen.de).
Materials availability
Plasmids generated in this study are available upon request.
Data and code availability
-
•
RNASeq data have been deposited to GEO (GSE174238; GSE173977; GSE173978; GSE173979) and publicly available. Accession numbers are listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to re-analyze the data reported in this paper is available from the lead contact upon request.
Experimental model and subject details
Wild-type mice (primary cell culture, IHC, RNA-sequencing)
All experimental procedures in this study, done at the LMU Munich, were performed in accordance with German and European Union guidelines and were approved by the government of Upper Bavaria. Primary cultures of astrocytes from spinal cords were obtained from brains of C57BL/6J mice of P2-3 days of age; primary cultures of gray matter cortex were obtained from brains of C57BL/6J mice of P6-7 days of age; no specific gender was considered. Mice were fed ad libitum and housed with 12/12 h light and dark cycle and kept under specific-pathogen-free (SPF) conditions.
Primary cultures of astrocytes
Three to six postnatal (P2-P3) mice were sacrificed and their entire spinal cord isolated from the vertebrae, after carefully ablating the dorsal root ganglia. For cortical gray matter astrocytes, 2 postnatal (P6-P7) mice were sacrificed, brain extracted and only the gray matter of the cerebral cortex was isolated, after paying attention to remove the sub-ventricular zone and the hippocampus. Both spinal cords and gray matter tissue was dissociated to obtain a single cell suspension using the OctoMACs protocol (Miltenyi Biotec), according to manufacturer’s instruction. Astrocytes were subsequently isolated using anti- ACSA-2 MACS-microBead Technology (Miltenyi Biotec) according to manual’s instruction (Kantzer et al., 2017). Primary cultures of gray matter-derived astrocytes were expanded in uncoated plastic flasks, while spinal cord-astrocytes plated on poly-D-lysine (PDL, Sigma-Aldrich)-coated flasks. Cells were grown in T12 (spinal cord) or T25 (gray matter) flasks in medium consisting of DMEM/F12 (1:1) with GlutaMax, 10% fetal bovine serum (FBS), glucose, penicillin /streptomycin, and 1x B27 serum-free-supplement, 10 ng/ml epidermal growth factor (EGF), and 10 ng/ml basic fibroblast growth factor (bFGF) (astro-medium). Primary cultures of astrocytes were maintained in an incubator for 6-8 days at 37°C and 5% CO2. Cells were passaged at 80%–90% confluency using 0.05% trypsin/EDTA and plated on poly-D-lysine coated glass coverslips at a density of 50,000-60,000 cells per coverslip (in 24-well plates) or in PDL-coated 6-well plates (Nunc) at a density of 300,000 cells per dish in fresh and complete astro-medium, supplemented with EGF and bFgf. Cells were either fixed at the time of infection, to evaluate the purity of the cultures, or infected with retroviral particles 12-16 hours after plating. For the experiment in Figure 6, cells were collected 1 day after re-plating.
Methods details
Plasmids and viral production
The plasmid containing Ascl1ERT2 has been previously described (Masserdotti et al., 2015). Neurog2ERT2 was generated by cloning ERT2 at the 5′- of Neurog2 cDNA. The cDNA was cloned downstream of the CAG promoter and followed by an Intra-Ribosome-Entry-Site (IRES) and DsRedExpress2; similarly, Ascl1 and Neurog2 have been cloned in the same retroviral backbone, characterized by the CAG promoter (CMV enhancer, chicken beta-actin promoter and a large sysnthetic intron), and the IRES-DsRedExpress2 sequence; control virus was generated by cloning DsRedExpress2 downstream of the CAG promoter (Gascón et al., 2016; Heinrich et al., 2010, 2011). This allows to identify transduced cells and quantify the reprogramming efficiency over the transduced cells. To produce viral particles, retrovirus-encoding plasmids were transfected in 293GPG (Ory et al., 1996) and collected via ultracentrifugation at 27.000 rpm for 2 hours after 3, 5 and 7 days, as previously described (Heinrich et al., 2011). Pellet was resuspended in 100μl of PBS 1X (added with 5mM MgCl2) and aliquots stored at −80°C until use.
Transduction
Primary cultures of astrocytes were plated in 24-well plates at a density of 50,000-60,000 cells per well; the next day, cells were infected with viral particles, according to the experimental design. Twenty-four to 36 hours later, astro-medium was replaced with fresh medium consisting of DMEM/F12 (1:1), penicillin/streptomycin, supplemented with 1x B27 and GlutaMax, glucose but not FBS, EGF and FGF (differentiation medium) and the cells maintained in culture until 6-7 days post-transfection in 9% CO2 incubator. For FACSorting, astrocytes were plated in 6-well plates pre-coated with PDL at a concentration of 300,000-350,000 cells per well. The following day, cells were transduced with the retroviral particles. One day later, medium was replaced with differentiation medium and cells were harvested 24 hours later (see scheme in Figure 2A). The viruses used are listed in the key resource table and were produced as previously described (Heinrich et al., 2011; Masserdotti et al., 2015).
Long term culture of reprogrammed astrocytes
To improve the survival of the iNeurons for the characterization of the neuronal subtypes, electrophysiology, single-cell experiments (patch-seq), cultures were treated with maturation medium (BDNF 20ng/ml, GDNF 20ng/ml, N2, NT3 20ng/ml, cAMP 100μM) every fourth day, starting from day 8DPI. Catalog numbers are indicated in Key resource table.
Fluorescence-activated cell sorting
Astrocytes were washed twice with 1x PBS, treated with trypsin (0,05% in EDTA) for 5 minutes, before resuspending in pre-warmed DMEM/F12 phenol-red free medium was added. Cells were harvested by centrifugation (1,000 rpm, 5min, 4°C), washed twice with 1x PBS and resuspended in 300μl of DMEM/F12 (1:1), phenol-red-free. Single cell suspension was filtrated using a 40-μm cell strainer and a drop of NucBlue was added to label living cells. Gates were defined using negative control (un-transduced cultures of astrocytes) and positive (cultures of astrocytes transduced with DsRed-encoding virus). The following lasers were used: x axis: 582/15 (DsRed); y axis 530/30 (FITC) (no signal in the channel; detecting autofluoresence). 20000-to-30000 events were collected per sample in a 1.5 mL tube containing 300μl of 0.05% BSA in PBS1X. Samples were kept in ice until the end of sorting, then harvested by centrifugation (1,000 rpm, 5min, 4°C). RNA was extracted with Arcturus PicoPure RNA isolation kit, according to manufacturer’s instruction.
Immunocytochemistry
Cells were fixed in 4% paraformaldehyde (PFA) in 1X PBS for 10 min. at room temperature, washed in 1X PBS twice for 5 minutes, and stored up to a month at 4°C before staining. Specimen were incubated in primary antibodies (see Key Resource table) in PBS1X containing 4% Bovine Serum Albumin (BSA) and 0.5% Triton X-100 for 2 hours at room temperature or overnight at 4°C. After washing three times for 5 minutes with PBS, cells were incubated with the appropriate species- or subclass-specific secondary antibodies, with or without DAPI to label nuclei (blue), diluted 1:10000, for 1 hour in the dark at room temperature. Optionally, after incubating with primary antibodies and washing with PBS, biotin-labeled secondary antibodies were used at a dilution of 1:200 for 1 hour, followed by streptavidin-coupled fluorophores (1:500) for another hour. Coverslips were then mounted with Aqua Poly/Mount. List of primary and secondary antibodies can be found in the Key Resource table.
Bulk RNA sequencing
Transcriptional changes upon Ascl1ERT2 or Neurog2ERT2 activation
Cells from five biological replicates per condition (DsRed+OHT, Ascl1ERT2+OHT, NeurogERT2+OHT) and 2 replicates for DsRed-OHT-untreated condition were sorted and their RNA extracted with PicoPure Kit (Arcturus, Kit0204). One entire biological replicate (DsRed+OHT, Ascl1ERT2+OHT, NeurogERT2+OHT) was removed from the analysis because of the low reads in one of the samples (DsRed+OHT). GEO number GSE174238.
Transcriptome of acutely isolated astrocytes cultured astrocytes
Following MACS procedure, cells were collected for RNA extraction (“acute samples”). Alternatively, MACS-sorted astrocytes were grown in flasks as described. Upon 80% confluency, cells were re-seeded in 6-well plates with fresh astrocyte-medium, as for being transduced. One day later, cells were collected and processed for RNA extraction. Five to six independent biological samples were collected per condition (GM n = 5 both acute and culture; SC, n = 6 both acute and culture). GEO number: GSE173979.
RNA was isolated on column using the PicPure™ RNAextraction kit (Applied Biosystems) and RNA quality and concentration were evaluated with an Agilent BioAnalyzer 2100 (Agilent). All included samples had a RIN > 9. 3μg of RNA from each sample was used to generate the RNA-seq libraries using bulk-adapted mcSCRB-seq protocol (Bagnoli et al., 2018): cDNA was generated by oligo-dT primers containing well-specific (e.g., sample specific) barcodes and unique molecular identifiers (UMIs). Unincorporated barcode primers were digested using Exonuclease I (Thermo Fisher). cDNA was pre-amplified using KAPA HiFi HotStart polymerase (Roche) and pooled before Nextera libraries were constructed from 0.8 ng of pre-amplified cleaned up cDNA using Nextera XT Kit (Illumina). 3′ ends were enriched with a custom P5 primer (P5NEXTPT5, IDT) and libraries were size selected using 2% E6 Gel Agarose EX Gels (Life Technologies), cut out in the range of 300–800 bp, and extracted using the Monarch DNA Gel Extraction Kit (New England Biolabs) according to manufacturer’s recommendations. Libraries were paired end sequenced on an Illumina HiSeq 1500 instrument. Sixteen bases were sequenced within the first read to obtain cellular and molecular barcodes, and 50 bases were sequenced in the second read into the cDNA fragment. An additional eight bases were sequenced to obtain the i7 barcode. On average, we sequence around 20 million read/sample. Gene-based transcripts counts were obtained by running the zUMI pipeline (Parekh et al., 2018) (version 0.0.2) using Ensembl annotation release 81.
RNA-seq analysis
The analysis was performed using R (3.5.3) and RStudio (version 1.2.1335). See Key Resources Table for packages used. As no significant difference was found between OHT-treated and OHT-untreated Ds-Red-transduced cells (unsupervised clustering, Figure S2F, Principal Component analysis, Figure S2G, no genes differentially expressed with padj < 0.01, Data S1), we compared proneural factor induced programs to DsRed-OHT treated samples. Gene Ontology enrichment analysis was performed using the package “TopGO” in RStudio: differentially expressed genes (Log2FC > 1, padj < 0.01 or Log2FC < −1, padj < 0.01) were provided as input, while the list of the genes with a pvalue were used as background. Top 20 GO, ranked on the basis of Exact Fisher score (< 0.01), were selected (see Tables S2–S9; S11; S14; S16–S19; S21–S23; S25–S32 in Data S1,S2, S3, S4, S5, and S6). GO terms were then ranked for the enrichment, obtained by diving the number of detected genes versus the number of expected genes, and top 5 were plotted. Gene Set Enrichment Analysis (Subramanian et al., 2005) was performed using the package “fgsea” in Rstudio (Sergushichev, 2016). “An algorithm for fast pre-ranked gene set enrichment analysis using cumulative statistic calculation”).
To define the Astrocyte- and Neuron-Score, normalized counts per each gene were obtained from the package DSEq2 (Love et al., 2014). Genes associated with astrocytes were obtained from Tripathy et al. (2018) and Weng et al. (2019), summed and used as coordinate to plot in ggplot2. To define the TFs in Figures 6K and 6L, TFs were filtered from all differentially expressed genes SC-Astro versus GM-Astro (padj < 0.01).
Single cell RNA-sequencing patch-seq related to Figures 4 and S3
Three- to four-week-old single cells were collected using an electrophysiology setup following a modified protocol (Cadwell et al., 2017). The coverslips were kept for 1-1.5 hours in the chamber for cell picking with a single cell collected every 5-6 minutes. Cells to be aspirated were selected by random screening of the coverslip. Brightfield and fluorescence pictures were taken before and after aspiration to catalog each cell for morphometric analysis and to confirm the successful aspiration of the soma into the electrode. We used borosilicate glass Warner Capillary Glass Tubing with polished ends and the inside filament (Model No. G150F-4) to make the aspiration electrodes with the input resistance between 2-3 MΩ (tested with the Axon Instruments Axopatch 200B amplifier). The intracellular solution contained (in mM): 4 KCl, 2 NaCl, 0.2 EGTA, 135 K-Gluconate, 10 HEPES (K-salt), 4 ATP (Mg-salt), 0.5 GTP (Na-salt), 10 Phospho-creatin, pH = 7.3, 290 mOsm. This intracellular solution was supplemented with RNase Out (1:40, 40 U/ μl, Invitrogen) and glycogen (1:1000 from 20 mg/ml stock, Roche). Each electrode was backfilled with 1.3 μL of this supplemented intracellular solution just before aspiration. To collect the individual aspirated iNeurons, PCR tubes were filled with the following solution as described for the SmartSeq2 method by Picelli et al. (2014). We modified some volumes and concentrations as suggested in the patch-seq method by Cadwell et al. (2017), because it worked better for our experimental setting. Each PCR tube contained 1 μL of 0.6% Triton X-100 (vol/vol) in RNase-free water (sterile, disk filter, stored at 4°C) with 1:40 of RNase Out (Invitrogen), 1 μL of oligo-dT30VN primers (10 μM stock, stored at −80°C), and 1 μL of dNTP (10 mM stock, stored at −20°C). The PCR tubes were kept on ice until the cell was ready to be collected. The PCR tubes were kept at −80°C until ready for the SmartSeq2 reverse transcription and cDNA amplification. Libraries were prepared with MicroPlex Library Preparation Kit v2 (Diagenode, C05010014). Cells were collected from 2 biologically independent reprogramming experiments. Sequencing reads were mapped to the GRCm38 reference genome using STAR software version 2.6.0a. TPM expression values based on ENSEMBL annotation version GRCm38.92 were calculated with RSEM (1.3.0). Data are deposited with the number GSE173977 in GEO.
Single cell-RNA-seq Patch-seq related to Figures 5 and S4
The culture coverslips were transferred to the recording chamber and were constantly perfused with fresh aCSF heated to 28°C using an in-line temperature controller (SH-27B combined with TC-324C, Warner Instruments Corp., Connecticut, USA). The aCSF consisted of (in mM) 125 NaCl, 2.5 KCl, 26 NaHCO3, 1.25 NaH2PO4, 2 CaCl2, 2 MgCl2, 11 D-glucose (pH at 28°C = 7.4, osmolarity = 305–315 mOsm, perfused with a mixture of 95% O2 and 5% CO2). The coverslips were visualized under an upright microscope (Axioskop FS, Zeiss, Oberkochen, Germany) equipped with differential interference contrast (DIC)-infrared optics and epifluorescence (filter set: Zeiss BP450-490, LP520). The fluorescence and infrared images were acquired with a CCD camera (Orca-ER, Hamamatsu, Shizouka, Japan). Single cells were visualized using a 40 × /0.74 N.A. water immersion objective (Olympus). The electrodes for whole-cell patch-clamp recordings were pulled from borosilicate glass capillaries (OD = 1.5 mm, ID = 0.86 mm, Warner Instruments Corp., Connecticut, USA) using a puller (Zeitz-Instruments, Martinsried, Germany). The intracellular solution consisted of (in mM) 126 K-gluconate, 4 KCl, 10 HEPES [4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid], 10 Na2-phosphocreatine, 4 Mg-ATP, 0.3 Na2-GTP, 0.05 Alexa Fluor 488 (Life Technologies), adjusted to pH 7.4 at 28°C and 295–300 mOsm. The electrodes (resistance ∼4 MΩ) were connected to the amplifier’s headstage via a chlorided silver wire. A silver / silver chloride-pellet immersed into the aCSF solution in the chamber served as reference electrode. Somatic whole-cell recordings were made in the current clamp mode using an ELC 03 XS amplifier (npi electronics, Tamm, Germany). Bias and offset current were zeroed before giga seal formation.
Electrophysiological recordings were performed as described in the section Electrophysiology with the few differences: following membrane rupture, the cells were not clamped to −60 mV and resting membrane potentials higher than −50 mV were taken as one of the parameters showing immature reprogrammed cells. Determination of the membrane resistance was only performed by measurement of the amplitude of a voltage deviation induced by a small hyperpolarizing current pulse (1 s, 5 – 10 pA). The cells’ ability to generate action potentials was tested by depolarizing current ramps (50 ms) from 0 – 100 pA. The AP amplitude was measured as the difference between the spike threshold and the spike maximum, the AP half-width was determined as width in ms at spike half-height. Electrophysiological data analysis was performed using MATLAB 2020a (Mathworks, Massachusetts, USA).
Based on the electrophysiological data, recorded cells were divided into 4 categories: Ds-Red transduced astrocytes (control astrocytes); not firing, immature and firing iNeurons. Reprogrammed cells were considered not firing if the resting membrane potential was higher than −40 mV and/or the action potential not present; immature neurons were designated if the resting membrane potential was between −40 mV and −50 mV and the action potential amplitude lower than 40 mV; firing neurons were defined as having a resting membrane potential lower than −50 mV, a train of action potential, whose amplitude was higher than 40mV.
After the recording, the cell was aspirated using previously published protocols (Földy et al., 2016; Winterer et al., 2019). The content of the cell was aspirated by applying negative pressure to the capillary. The intracellular solution did not contain any ribonuclease inhibitors and very small amount of intracellular solution (> 2 μl) was used in the glass capillary to perform the electrophysiological recordings. After the extraction of the cell, the content of the capillary was expulsed into a single 0.2 mL PCR tube by applying positive pressure and breaking off the capillary tip. The PCR tube contained 3 μl of lysis buffer solution which consisted of 0.1% Triton X-100, 5 mM dNTP Mix, 2.5 μM oligo-dT30VN primers, and 1 U/μl RNase OUT. The sample was briefly spun down using a centrifuge and flash-frozen on dry ice. The subsequent reverse transcription (RT) and cDNA amplification was done according to the SmartSeq2 protocol (Picelli et al., 2014). The resulting cDNA fragments were analyzed using High Sensitivity DNA Kit (Agilent Bioanalyzer, Santa Clara, USA). After sequencing, raw reads were de-multiplexed on in-house high-performance-cluster (HPC) using Je (version 2.0.2). The raw sequencing reads were aligned to Ensembl GRCm38 mouse reference genome using STAR aligner (version 2.7.1) with the GeneCounts parameter on. Further processing of the data was performed in RStudio using the Bioconductor package DESeq2 (Love et al., 2014). Bubble plots showing the expression of the genes in different conditions were generated as follows: normalized gene counts per each gene were divided by the gene length (in Kb, obtained from Ensembl); then, such expression values were averaged within each subgroup, and then log2 transformed. Data are deposited with the number GSE173978 in GEO.
Single-cell morphometry
Pictures of individual single cells were subjected to a morphometric analysis. Using ImageJ software, several parameters were manually analyzed: number of primary processes, angles between the processes, the longitudinal axis of the cell soma, and the cross-section area of the cell soma. As n = 2 independent samples, we used nonparametric test for the statistical analysis. For the distribution of the angles between processes, we separated the measured angles into bins separated by 20 degrees and used non-linear regression of the fourth-order polynomial function to curve fit the frequency distribution. All the collected cells were analyzed, with the exception of 1 cell from Neurog2 (Ascl1 n = 20; Neurog2 = 21).
Ranking of transcription factors
Putative lead F (transcription or chromatin modification factor) (Zhang et al., 2015) was assessed by an approach adopted from Rackham et al. (2016). For each factor we established a sphere of influence of up to three level depth using gene-gene relations based on STRING database version 10.5 (Szklarczyk et al., 2017). We considered only relations with total scores > 300, where less than half of the value was attributed to text mining. Using measured difference in RNA-seq expression (data processed with DESeq2; Love et al., 2014) we calculated scores one for F and one for its underling network using following equations:
where: F – factor, FC – fold change, AdjPval – adjusted p value (both from DSeq2), g – gene, Dist – number of steps between g and F, Pnd – parent node degree, n - length of list of genes associated with F.
Subsequently all factors were ranked based on combined ranking of both scores and filtered for the normalized expression (TFs with norm. expression < 200 after induction were excluded for Figure 2, while for Figure 3 genes with norm. exp < 20 were excluded). To provide context to factors driving cell identity, we plotted a network using Cytoscape 3.6 (Su et al., 2014a) with edges width correlating to STRING interaction score. Only factors connected to other TFs were plotted.
Comparison of scRNA-seq with publicly available data
To find if our dataset holds some resemblance to any cell type, we used available data (Delile et al., 2019). We combined iNeuron single cell RNA-seq and public data to calculate average gene expression values for each cell type and PCs (Principal Component) of resulting dataset. To compare cell type similarity, we plotted 3rd and 4th component.
Electrophysiology
For electrophysiological recordings, coverslips were transferred to an organ bath mounted on the stage of an upright microscope (Axioscope FS, Zeiss, Göttingen, Germany). The cell cultures were perfused with a bathing solution consisting of (in mM): NaCl 150, KCl 3, CaCl2 3, MgCl2 2, 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) 10, and D-glucose 10. The pH of the solution was adjusted to 7.4 (NaOH) and its osmolarity ranged between 309 to 313 mOsmol. The perfusion rate was set to 1.4 mL / min and recordings were performed at room temperature (23 – 24°C). The microscope was equipped with differential interference contrast (DIC) optics and with epifluorescence optics for green and red fluorescence (filter sets: Zeiss BP450-490, LP520, Zeiss BP546/12, lP590). Images were taken and displayed using a software-operated CCD microscope camera (ORCA R, Hamamatsu, Herrsching, Germany).The electrodes for whole cell patch-clamp recordings were fabricated from borosilicate glass capillaries (OD: 1.5 mm, ID: 0.86 mm, Hugo Sachs Elektronik-Harvard Apparatus, March-Hugstetten, Germany) and filled with a solution composed of (in mM): potassium gluconate 135, KCl 4, NaCl 2, ethylene glycol-bis(2-aminoethylether)-N,N,N′,N′-tetraacetic acid (EGTA) 0.2, HEPES (potassium salt) 10, adenosine-triphosphate (magnesium salt, ATP[Mg]) 4, sodium guanosine-triphosphate (NaGTP) 0.5, and phosphocreatine 10 (pH: 7.25 – 7.30, osmolarity: 288 – 291 mOsmol). The series resistance determined after establishment of the whole cell recording mode (9 – 17 MΩ) was compensated by 70 – 85%. The recorded signals were amplified (x10 or x20), filtered at 10 or 20 kHz (current clamp) and 3 kHz (voltage clamp), digitized at a sampling rate of 10 or 20 kHz and stored on a computer for offline analysis. Data acquisition and generation of command pulses was done by means of a CED 1401 Power 3 system in conjunction with Signal6 data acquisition software (Cambridge electronic design, Cambridge, England). Data analysis was performed using IGOR Pro 6 (WaveMetrics, Lake Oswego, USA) together with the NeuroMatic IGOR plugin (Version 2) (www.neuromatic.thinkrandom.com). Microscope images were corrected for contrast and brightness by using Photoshop CS3 (Adobe Software Systems, Ireland). By means of the motorized microscope stage, each coverslip was scanned systematically and iNeurons were identified by red fluorescence. Following membrane rupture, the cells were voltage-clamped to a holding potential of −60 mV and kept under this condition until stabilization of the holding current (3 – 5 min). Determination of the input resistance RN was performed either by measurement of the amplitude of a voltage deviation induced by a small hyperpolarizing current pulse (1 s, 5 – 10 pA) or by determining the slope of the current-voltage-curve (IV-curve) at its origin. The somatic membrane time constant Ω was derived by fitting a dual exponential function to the voltage relaxation following cessation of a small hyperpolarizing current pulse and the total membrane capacity CN was estimated using a method (Zemankovics et al., 2010). The cells’ ability to generate action potentials was tested by injecting depolarizing current pulses (50 ms) with increasing current strengths (ΩI: 5 or 10 pA) or by depolarizing current ramps (50 ms) from 0 – 100 pA. The amplitudes of the action potentials (spikes) were measured as the difference between the resting membrane potential and the spike maximum, the spike duration was determined at half-maximum amplitude and the spike threshold was derived from a phase-plane plot (Bean, 2007). The action potential discharge pattern of the cells was investigated by injections of depolarizing current pulses (1 – 2 s), the amplitudes of which were raised in steps (5 or 10 pA) from 0 – 200 pA at a frequency of 0.1 Hz.
All chemicals and drugs were obtained from Sigma-Aldrich (Munich, Germany) and Biotrend (Cologne, Germany), respectively. Data are given as median ± IQR. Statistical comparison of two samples was performed by using Mann-Whitney unpaired two-tailed t test.
Quantification and statistical analysis
Immunostainings were analyzed with an AxioM2 epifluorescence microscope or LSM710 laser-scanning confocal (Carl Zeiss). Digital images were captured using the ZEN2 software (Carl Zeiss). Retroviral vector-transduced cells were quantified from more than 25 randomly chosen 20x fields in at least 3 independent experiments. Quantification for neuronal cells was based on β-III-tubulin immunoreactivity and morphological parameters, e.g., presence of 2 or more processes longer than 3x the cell soma as in Gascón et al. (2016). Astrocytes were quantified based on morphological features and expression of known astrocytic markers (Gfap or Sox9). Analysis of the culture composition (Figure S1) was performed by quantifying 2 tile images, each composed of 25 images (acquired at 20X) in 3 independent biological experiment. To quantify the intensity of Asc12ERT2 and Neurog2ERT2 (Figure S2B), 1 tile image (25 images at 20X) was imported in Fiji: nuclei were identified via DAPI and used to create a mask (define threshold; Set Measurement; Analyze particles (60 < particles < 250). Then, the mask was applied to the tile image to obtain the average intensity per each channel per each particle (that is, each nucleus present in the tile image). We selected the cells whose intensity value of green channel (corresponding to the transcription factor) was clearly above green-negative nuclei. Each tile image provided at the intensity of at least 170 cells. Data were collected for 3 independent cultures and analyzed in RStudio. Average intensity was calculated per each independent experiment. Boxplots show the median and the interquantile range (IQR) Whiskers are calculated as 1.58∗IQR (Figures 1S–1U, S1H, S1K–S1P, S2D, and S4A–S4D). Data presented as bar plots show mean and confidence interval (CI) (Figures 1C, 1D, 2C, 3F–3I, and S3B–S3D).
Data were analyzed with Microsoft Excel, GraphPad Prism 7.0 software RStudio (version 1.2.1335); statistics was performed with linear regression using “lm” function (R Stats package) in RStudio on log2 transformed reprogramming efficiency. Evaluation of the residuals for fitted linear models was performed with the package “DHARMa” (Hartig, 2021). DHARMa: Residual Diagnostics for Hierarchical (Multi-Level / Mixed) Regression Models. R package version 0.3.2.0. https://cran.r-project.org/web/packages/DHARMa/index) in RStudio. Statistical details of the experiments can be found in the figure legends. Significance is based on the p value indicated on the graphs as ∗ p ≤ 0.05, ∗∗ p ≤ 0.01, ∗∗∗p ≤ 0.001.
Acknowledgments
We are very grateful to the excellent technical assistance of Ines Mühlhahn, Detlef Franzen, Manja Thorwirth, and Andrea Steiner-Mezzadri (BMC, LMU); Judith Fischer for FACS training and troubleshooting; Tobias Straub, head of the Bioinformatic Unit at the BMC, for aligning RNA-seq raw data and advice on analysis; Jovica Ninkovic and Mike Myoga for critical comments; and LaFuga (Gene Center, Munich) and Next Generation Sequencing Core Facility (Helmholtz) for sequencing. This work was funded by the German Research Foundation grants SFB 870, SPP1757 (to M.G.), the advanced ERC ChroNeuroRepair (to M.G.), ERA-Net neuron grant MICRONET (to M.G.), the EU consortium NSC Reconstruct (M.G.), German Research Foundation grant TRR274 (to M.G.), and grant EN 1093/2-1 (to A.J. and W.E.). This work is dedicated to Rinaldo Masserdotti.
Author contributions
G.M. conceived and designed the project. J.K. and K.K. performed the isolation, culturing, and most of the reprogramming experiments. J.K. and K.K. evaluated the reprogramming efficiency at 8DPI; J.K. performed and quantified the experiments at day 21. K.K. prepared the cells for FACS analysis and extracted RNA for bulk RNA-seq analysis. B.A.H. prepared primary cultures of astrocytes from cortex GM, collected single cells, processed the samples with SmartSeq2, and prepared the libraries for sequencing; D.P. collected single iNs for patch-seq, processed them via Smart-Seq2 protocol, and performed morphometric analysis. V.B performed patch-seq experiments, analyzed the electrophysiological data, prepared the samples via Smart-Seq2, and performed initial analysis of RNA-seq data. A.J. prepared bulk-adapted mcSCRB-seq libraries; T.S.-E. contributed to establish MACS protocol for SC, trained J.K. and K.K., and performed immunocytochemistry; T.R. performed whole-cell patch-clamp experiments and analyzed the electrophysiological properties of iNs. W.E. provided reagents; G.M. performed FACS and analyzed data from bulk and single-cell RNA-seq; P.S. compared single-cell RNA with publicly available data and analyzed single-cell RNA-seq with G.M.; M.G. contributed to the conceptual organization of the manuscript and the patch-seq data and financed the work. G.M. and M.G. wrote the manuscript, and all authors contributed corrections and comments.
Declaration of interests
The authors declare no competing interests.
Published: July 20, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2021.109409.
Contributor Information
M. Götz, Email: magdalena.goetz@helmholtz-muenchen.de.
G. Masserdotti, Email: giacomo.masserdotti@helmholtz-muenchen.de.
Supplemental information
References
- Abernathy D.G., Kim W.K., McCoy M.J., Lake A.M., Ouwenga R., Lee S.W., Xing X., Li D., Lee H.J., Heuckeroth R.O. MicroRNAs Induce a Permissive Chromatin Environment that Enables Neuronal Subtype-Specific Reprogramming of Adult Human Fibroblasts. Cell Stem Cell. 2017;21:332–348.e9. doi: 10.1016/j.stem.2017.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfaro-Cervello C., Soriano-Navarro M., Mirzadeh Z., Alvarez-Buylla A., Garcia-Verdugo J.M. Biciliated ependymal cell proliferation contributes to spinal cord growth. J. Comp. Neurol. 2012;520:3528–3552. doi: 10.1002/cne.23104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexa A., Rahnenfuhrer G. R Foundation; 2018. topGO: Enrichment Analysis for Gene Ontology, version 2.34.0. [Google Scholar]
- Ali F.R., Cheng K., Kirwan P., Metcalfe S., Livesey F.J., Barker R.A., Philpott A. The phosphorylation status of Ascl1 is a key determinant of neuronal differentiation and maturation in vivo and in vitro. Development. 2014;141:2216–2224. doi: 10.1242/dev.106377. [DOI] [PubMed] [Google Scholar]
- Andrzejczuk L.A., Banerjee S., England S.J., Voufo C., Kamara K., Lewis K.E. Tal1, Gata2a, and Gata3 Have Distinct Functions in the Development of V2b and Cerebrospinal Fluid-Contacting KA Spinal Neurons. Front. Neurosci. 2018;12:170. doi: 10.3389/fnins.2018.00170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagnoli J.W., Ziegenhain C., Janjic A., Wange L.E., Vieth B., Parekh S., Geuder J., Hellmann I., Enard W. Sensitive and powerful single-cell RNA sequencing using mcSCRB-seq. Nat. Commun. 2018;9:2937. doi: 10.1038/s41467-018-05347-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker R.A., Götz M., Parmar M. New approaches for brain repair-from rescue to reprogramming. Nature. 2018;557:329–334. doi: 10.1038/s41586-018-0087-1. [DOI] [PubMed] [Google Scholar]
- Bean B.P. The action potential in mammalian central neurons. Nat. Rev. Neurosci. 2007;8:451–465. doi: 10.1038/nrn2148. [DOI] [PubMed] [Google Scholar]
- Berninger B., Costa M.R., Koch U., Schroeder T., Sutor B., Grothe B., Götz M. Functional properties of neurons derived from in vitro reprogrammed postnatal astroglia. J. Neurosci. 2007;27:8654–8664. doi: 10.1523/JNEUROSCI.1615-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borromeo M.D., Meredith D.M., Castro D.S., Chang J.C., Tung K.C., Guillemot F., Johnson J.E. A transcription factor network specifying inhibitory versus excitatory neurons in the dorsal spinal cord. Development. 2014;141:2803–2812. doi: 10.1242/dev.105866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britz O., Mattar P., Nguyen L., Langevin L.M., Zimmer C., Alam S., Guillemot F., Schuurmans C. A role for proneural genes in the maturation of cortical progenitor cells. Cereb. Cortex. 2006;16:i138–i151. doi: 10.1093/cercor/bhj168. [DOI] [PubMed] [Google Scholar]
- Cadwell C.R., Scala F., Li S., Livrizzi G., Shen S., Sandberg R., Jiang X., Tolias A.S. Multimodal profiling of single-cell morphology, electrophysiology, and gene expression using Patch-seq. Nat. Protoc. 2017;12:2531–2553. doi: 10.1038/nprot.2017.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan R.A., Roberts R., Sengupta M., Kimura Y., Higashijima S.I., Bagnall M.W. Spinal V2b neurons reveal a role for ipsilateral inhibition in speed control. eLife. 2019;8:e47837. doi: 10.7554/eLife.47837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chouchane M., Melo de Farias A.R., Moura D.M.S., Hilscher M.M., Schroeder T., Leão R.N., Costa M.R. Lineage Reprogramming of Astroglial Cells from Different Origins into Distinct Neuronal Subtypes. Stem Cell Reports. 2017;9:162–176. doi: 10.1016/j.stemcr.2017.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church V.A., Cates K., Capano L., Aryal S., Kim W.K., Yoo A.S. Generation of Human Neurons by microRNA-Mediated Direct Conversion of Dermal Fibroblasts. Methods Mol. Biol. 2021;2239:77–100. doi: 10.1007/978-1-0716-1084-8_6. [DOI] [PubMed] [Google Scholar]
- Crochet S., Fuentealba P., Timofeev I., Steriade M. Selective amplification of neocortical neuronal output by fast prepotentials in vivo. Cereb. Cortex. 2004;14:1110–1121. doi: 10.1093/cercor/bhh071. [DOI] [PubMed] [Google Scholar]
- Delile J., Rayon T., Melchionda M., Edwards A., Briscoe J., Sagner A. Single cell transcriptomics reveals spatial and temporal dynamics of gene expression in the developing mouse spinal cord. Development. 2019;146:dev173807. doi: 10.1242/dev.173807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis D.J., Wilkinson G., Li S., Dixit R., Adnani L., Balakrishnan A., Han S., Kovach C., Gruenig N., Kurrasch D.M. Neurog2 and Ascl1 together regulate a postmitotic derepression circuit to govern laminar fate specification in the murine neocortex. Proc. Natl. Acad. Sci. USA. 2017;114:E4934–E4943. doi: 10.1073/pnas.1701495114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filosa J.A., Morrison H.W., Iddings J.A., Du W., Kim K.J. Beyond neurovascular coupling, role of astrocytes in the regulation of vascular tone. Neuroscience. 2016;323:96–109. doi: 10.1016/j.neuroscience.2015.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fode C., Ma Q., Casarosa S., Ang S.L., Anderson D.J., Guillemot F. A role for neural determination genes in specifying the dorsoventral identity of telencephalic neurons. Genes Dev. 2000;14:67–80. [PMC free article] [PubMed] [Google Scholar]
- Földy C., Darmanis S., Aoto J., Malenka R.C., Quake S.R., Südhof T.C. Single-cell RNAseq reveals cell adhesion molecule profiles in electrophysiologically defined neurons. Proc. Natl. Acad. Sci. USA. 2016;113:E5222–E5231. doi: 10.1073/pnas.1610155113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gascón S., Murenu E., Masserdotti G., Ortega F., Russo G.L., Petrik D., Deshpande A., Heinrich C., Karow M., Robertson S.P. Identification and Successful Negotiation of a Metabolic Checkpoint in Direct Neuronal Reprogramming. Cell Stem Cell. 2016;18:396–409. doi: 10.1016/j.stem.2015.12.003. [DOI] [PubMed] [Google Scholar]
- Golding N.L., Spruston N. Dendritic sodium spikes are variable triggers of axonal action potentials in hippocampal CA1 pyramidal neurons. Neuron. 1998;21:1189–1200. doi: 10.1016/s0896-6273(00)80635-2. [DOI] [PubMed] [Google Scholar]
- Guo Z., Zhang L., Wu Z., Chen Y., Wang F., Chen G. In vivo direct reprogramming of reactive glial cells into functional neurons after brain injury and in an Alzheimer’s disease model. Cell Stem Cell. 2014;14:188–202. doi: 10.1016/j.stem.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartig F. DHARMa: Residual Diagnostics for Hierarchical (Multi-Level/Mixed) Regression Models. R package version 0.4.3. 2021 http://florianhartig.github.io/DHARMa/ [Google Scholar]
- Häring M., Zeisel A., Hochgerner H., Rinwa P., Jakobsson J.E.T., Lönnerberg P., La Manno G., Sharma N., Borgius L., Kiehn O. Neuronal atlas of the dorsal horn defines its architecture and links sensory input to transcriptional cell types. Nat. Neurosci. 2018;21:869–880. doi: 10.1038/s41593-018-0141-1. [DOI] [PubMed] [Google Scholar]
- Hébert J.M., Fishell G. The genetics of early telencephalon patterning: some assembly required. Nat. Rev. Neurosci. 2008;9:678–685. doi: 10.1038/nrn2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich C., Blum R., Gascón S., Masserdotti G., Tripathi P., Sánchez R., Tiedt S., Schroeder T., Götz M., Berninger B. Directing astroglia from the cerebral cortex into subtype specific functional neurons. PLoS Biol. 2010;8:e1000373. doi: 10.1371/journal.pbio.1000373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich C., Gascón S., Masserdotti G., Lepier A., Sanchez R., Simon-Ebert T., Schroeder T., Götz M., Berninger B. Generation of subtype-specific neurons from postnatal astroglia of the mouse cerebral cortex. Nat. Protoc. 2011;6:214–228. doi: 10.1038/nprot.2010.188. [DOI] [PubMed] [Google Scholar]
- Heins N., Malatesta P., Cecconi F., Nakafuku M., Tucker K.L., Hack M.A., Chapouton P., Barde Y.A., Götz M. Glial cells generate neurons: the role of the transcription factor Pax6. Nat. Neurosci. 2002;5:308–315. doi: 10.1038/nn828. [DOI] [PubMed] [Google Scholar]
- Henke R.M., Savage T.K., Meredith D.M., Glasgow S.M., Hori K., Dumas J., MacDonald R.J., Johnson J.E. Neurog2 is a direct downstream target of the Ptf1a-Rbpj transcription complex in dorsal spinal cord. Development. 2009;136:2945–2954. doi: 10.1242/dev.035352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herdy J., Schafer S., Kim Y., Ansari Z., Zangwill D., Ku M., Paquola A., Lee H., Mertens J., Gage F.H. Chemical modulation of transcriptionally enriched signaling pathways to optimize the conversion of fibroblasts into neurons. eLife. 2019;8:e41356. doi: 10.7554/eLife.41356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero-Navarro Á., Puche-Aroca L., Moreno-Juan V., Sempere-Ferràndez A., Espinosa A., Susín R., Torres-Masjoan L., Leyva-Díaz E., Karow M., Figueres-Oñate M. Astrocytes and neurons share region-specific transcriptional signatures that confer regional identity to neuronal reprogramming. Sci. Adv. 2021;7:eabe8978. doi: 10.1126/sciadv.abe8978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindley C., Ali F., McDowell G., Cheng K., Jones A., Guillemot F., Philpott A. Post-translational modification of Ngn2 differentially affects transcription of distinct targets to regulate the balance between progenitor maintenance and differentiation. Development. 2012;139:1718–1723. doi: 10.1242/dev.077552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X., Qin S., Huang X., Yuan Y., Tan Z., Gu Y., Cheng X., Wang D., Lian X.F., He C., Su Z. Region-Restrict Astrocytes Exhibit Heterogeneous Susceptibility to Neuronal Reprogramming. Stem Cell Reports. 2019;12:290–304. doi: 10.1016/j.stemcr.2018.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantzer C.G., Boutin C., Herzig I.D., Wittwer C., Reiß S., Tiveron M.C., Drewes J., Rockel T.D., Ohlig S., Ninkovic J. Anti-ACSA-2 defines a novel monoclonal antibody for prospective isolation of living neonatal and adult astrocytes. Glia. 2017;65:990–1004. doi: 10.1002/glia.23140. [DOI] [PubMed] [Google Scholar]
- Karow M., Camp J.G., Falk S., Gerber T., Pataskar A., Gac-Santel M., Kageyama J., Brazovskaja A., Garding A., Fan W. Direct pericyte-to-neuron reprogramming via unfolding of a neural stem cell-like program. Nat. Neurosci. 2018;21:932–940. doi: 10.1038/s41593-018-0168-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karunaratne A., Hargrave M., Poh A., Yamada T. GATA proteins identify a novel ventral interneuron subclass in the developing chick spinal cord. Dev. Biol. 2002;249:30–43. doi: 10.1006/dbio.2002.0754. [DOI] [PubMed] [Google Scholar]
- Kelenis D.P., Hart E., Edwards-Fligner M., Johnson J.E., Vue T.Y. ASCL1 regulates proliferation of NG2-glia in the embryonic and adult spinal cord. Glia. 2018;66:1862–1880. doi: 10.1002/glia.23344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovach C., Dixit R., Li S., Mattar P., Wilkinson G., Elsen G.E., Kurrasch D.M., Hevner R.F., Schuurmans C. Neurog2 simultaneously activates and represses alternative gene expression programs in the developing neocortex. Cereb. Cortex. 2013;23:1884–1900. doi: 10.1093/cercor/bhs176. [DOI] [PubMed] [Google Scholar]
- Ladewig J., Mertens J., Kesavan J., Doerr J., Poppe D., Glaue F., Herms S., Wernet P., Kögler G., Müller F.J. Small molecules enable highly efficient neuronal conversion of human fibroblasts. Nat. Methods. 2012;9:575–578. doi: 10.1038/nmeth.1972. [DOI] [PubMed] [Google Scholar]
- Langen U.H., Ayloo S., Gu C. Development and Cell Biology of the Blood-Brain Barrier. Annu. Rev. Cell Dev. Biol. 2019;35:591–613. doi: 10.1146/annurev-cellbio-100617-062608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Q.Y., Mall M., Chanda S., Zhou B., Sharma K.S., Schaukowitch K., Adrian-Segarra J.M., Grieder S.D., Kareta M.S., Wapinski O.L. Pro-neuronal activity of Myod1 due to promiscuous binding to neuronal genes. Nat. Cell Biol. 2020;22:401–411. doi: 10.1038/s41556-020-0490-3. [DOI] [PubMed] [Google Scholar]
- Li S., Misra K., Matise M.P., Xiang M. Foxn4 acts synergistically with Mash1 to specify subtype identity of V2 interneurons in the spinal cord. Proc. Natl. Acad. Sci. USA. 2005;102:10688–10693. doi: 10.1073/pnas.0504799102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M.L., Zang T., Zou Y., Chang J.C., Gibson J.R., Huber K.M., Zhang C.L. Small molecules enable neurogenin 2 to efficiently convert human fibroblasts into cholinergic neurons. Nat. Commun. 2013;4:2183. doi: 10.1038/ncomms3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Miao Q., Yuan J., Han S., Zhang P., Li S., Rao Z., Zhao W., Ye Q., Geng J. Ascl1 Converts Dorsal Midbrain Astrocytes into Functional Neurons In Vivo. J. Neurosci. 2015;35:9336–9355. doi: 10.1523/JNEUROSCI.3975-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Yu C., Daley T.P., Wang F., Cao W.S., Bhate S., Lin X., Still C., 2nd, Liu H., Zhao D. CRISPR Activation Screens Systematically Identify Factors that Drive Neuronal Fate and Reprogramming. Cell Stem Cell. 2018;23:758–771.e8. doi: 10.1016/j.stem.2018.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu D.C., Niu T., Alaynick W.A. Molecular and cellular development of spinal cord locomotor circuitry. Front. Mol. Neurosci. 2015;8:25. doi: 10.3389/fnmol.2015.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masserdotti G., Gillotin S., Sutor B., Drechsel D., Irmler M., Jørgensen H.F., Sass S., Theis F.J., Beckers J., Berninger B. Transcriptional Mechanisms of Proneural Factors and REST in Regulating Neuronal Reprogramming of Astrocytes. Cell Stem Cell. 2015;17:74–88. doi: 10.1016/j.stem.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matera A.G., Wang Z. A day in the life of the spliceosome. Nat. Rev. Mol. Cell Biol. 2014;15:108–121. doi: 10.1038/nrm3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattugini N., Bocchi R., Scheuss V., Russo G.L., Torper O., Lao C.L., Götz M. Inducing Different Neuronal Subtypes from Astrocytes in the Injured Mouse Cerebral Cortex. Neuron. 2019;103:1086–1095.e5. doi: 10.1016/j.neuron.2019.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer K., Ferraiuolo L., Miranda C.J., Likhite S., McElroy S., Renusch S., Ditsworth D., Lagier-Tourenne C., Smith R.A., Ravits J. Direct conversion of patient fibroblasts demonstrates non-cell autonomous toxicity of astrocytes to motor neurons in familial and sporadic ALS. Proc. Natl. Acad. Sci. USA. 2014;111:829–832. doi: 10.1073/pnas.1314085111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra K., Luo H., Li S., Matise M., Xiang M. Asymmetric activation of Dll4-Notch signaling by Foxn4 and proneural factors activates BMP/TGFβ signaling to specify V2b interneurons in the spinal cord. Development. 2014;141:187–198. doi: 10.1242/dev.092536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuguchi R., Kriks S., Cordes R., Gossler A., Ma Q., Goulding M. Ascl1 and Gsh1/2 control inhibitory and excitatory cell fate in spinal sensory interneurons. Nat. Neurosci. 2006;9:770–778. doi: 10.1038/nn1706. [DOI] [PubMed] [Google Scholar]
- Nieto M., Schuurmans C., Britz O., Guillemot F. Neural bHLH genes control the neuronal versus glial fate decision in cortical progenitors. Neuron. 2001;29:401–413. doi: 10.1016/s0896-6273(01)00214-8. [DOI] [PubMed] [Google Scholar]
- Ory D.S., Neugeboren B.A., Mulligan R.C. A stable human-derived packaging cell line for production of high titer retrovirus/vesicular stomatitis virus G pseudotypes. Proc. Natl. Acad. Sci. USA. 1996;93:11400–11406. doi: 10.1073/pnas.93.21.11400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parekh S., Ziegenhain C., Vieth B., Enard W., Hellmann I. zUMIs - A fast and flexible pipeline to process RNA sequencing data with UMIs. Gigascience. 2018;7:giy059. doi: 10.1093/gigascience/giy059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parras C.M., Schuurmans C., Scardigli R., Kim J., Anderson D.J., Guillemot F. Divergent functions of the proneural genes Mash1 and Ngn2 in the specification of neuronal subtype identity. Genes Dev. 2002;16:324–338. doi: 10.1101/gad.940902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippidou P., Dasen J.S. Hox genes: choreographers in neural development, architects of circuit organization. Neuron. 2013;80:12–34. doi: 10.1016/j.neuron.2013.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picelli S., Faridani O.R., Björklund A.K., Winberg G., Sagasser S., Sandberg R. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 2014;9:171–181. doi: 10.1038/nprot.2014.006. [DOI] [PubMed] [Google Scholar]
- Qian H., Kang X., Hu J., Zhang D., Liang Z., Meng F., Zhang X., Xue Y., Maimon R., Dowdy S.F. Reversing a model of Parkinson’s disease with in situ converted nigral neurons. Nature. 2020;582:550–556. doi: 10.1038/s41586-020-2388-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quevedo M., Meert L., Dekker M.R., Dekkers D.H.W., Brandsma J.H., van den Berg D.L.C., Ozgür Z., van IJcken W.F.J., Demmers J., Fornerod M., Poot R.A. Mediator complex interaction partners organize the transcriptional network that defines neural stem cells. Nat. Commun. 2019;10:2669. doi: 10.1038/s41467-019-10502-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rackham O.J., Firas J., Fang H., Oates M.E., Holmes M.L., Knaupp A.S., Suzuki H., Nefzger C.M., Daub C.O., Shin J.W., FANTOM Consortium A predictive computational framework for direct reprogramming between human cell types. Nat. Genet. 2016;48:331–335. doi: 10.1038/ng.3487. [DOI] [PubMed] [Google Scholar]
- Rao Z., Wang R., Li S., Shi Y., Mo L., Han S., Yuan J., Jing N., Cheng L. Molecular Mechanisms Underlying Ascl1-Mediated Astrocyte-to-Neuron Conversion. Stem Cell Reports. 2021;16:534–547. doi: 10.1016/j.stemcr.2021.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren J., Isakova A., Friedmann D., Zeng J., Grutzner S.M., Pun A., Zhao G.Q., Kolluru S.S., Wang R., Lin R. Single-cell transcriptomes and whole-brain projections of serotonin neurons in the mouse dorsal and median raphe nuclei. eLife. 2019;8:e49424. doi: 10.7554/eLife.49424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivetti di Val Cervo P., Romanov R.A., Spigolon G., Masini D., Martín-Montañez E., Toledo E.M., La Manno G., Feyder M., Pifl C., Ng Y.H. Induction of functional dopamine neurons from human astrocytes in vitro and mouse astrocytes in a Parkinson’s disease model. Nat. Biotechnol. 2017;35:444–452. doi: 10.1038/nbt.3835. [DOI] [PubMed] [Google Scholar]
- Russo G.L., Sonsalla G., Natarajan P., Breunig C.T., Bulli G., Merl-Pham J., Schmitt S., Giehrl-Schwab J., Giesert F., Jastroch M. CRISPR-Mediated Induction of Neuron-Enriched Mitochondrial Proteins Boosts Direct Glia-to-Neuron Conversion. Cell Stem Cell. 2020;28:524–534.e7. doi: 10.1016/j.stem.2020.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagner A., Briscoe J. Establishing neuronal diversity in the spinal cord: a time and a place. Development. 2019;146:dev182154. doi: 10.1242/dev.182154. [DOI] [PubMed] [Google Scholar]
- Schuurmans C., Armant O., Nieto M., Stenman J.M., Britz O., Klenin N., Brown C., Langevin L.M., Seibt J., Tang H. Sequential phases of cortical specification involve Neurogenin-dependent and -independent pathways. EMBO J. 2004;23:2892–2902. doi: 10.1038/sj.emboj.7600278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D.K., Yang J., Liu M.L., Zhang C.L. Small Molecules Modulate Chromatin Accessibility to Promote NEUROG2-Mediated Fibroblast-to-Neuron Reprogramming. Stem Cell Reports. 2016;7:955–969. doi: 10.1016/j.stemcr.2016.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son E.Y., Ichida J.K., Wainger B.J., Toma J.S., Rafuse V.F., Woolf C.J., Eggan K. Conversion of mouse and human fibroblasts into functional spinal motor neurons. Cell Stem Cell. 2011;9:205–218. doi: 10.1016/j.stem.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su G., Morris J.H., Demchak B., Bader G.D. Biological network exploration with Cytoscape 3. Curr. Protoc. Bioinformatics. 2014;47 doi: 10.1002/0471250953.bi0813s47. 8.13.1-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Z., Niu W., Liu M.L., Zou Y., Zhang C.L. In vivo conversion of astrocytes to neurons in the injured adult spinal cord. Nat. Commun. 2014;5:3338. doi: 10.1038/ncomms4338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Mesirov J.P. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sergushichev A.A. 2016. An algorithm for fast preranked gene set enrichment analysis using cumulative statistic calculation. bioRxiv. [DOI] [Google Scholar]
- Szklarczyk D., Morris J.H., Cook H., Kuhn M., Wyder S., Simonovic M., Santos A., Doncheva N.T., Roth A., Bork P. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45:D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai W., Wu W., Wang L.L., Ni H., Chen C., Yang J., Zang T., Zou Y., Xu X.M., Zhang C.L. In vivo reprogramming of NG2 glia enables adult neurogenesis and functional recovery following spinal cord injury. Cell Stem Cell. 2021;28:923–937.e4. doi: 10.1016/j.stem.2021.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torper O., Pfisterer U., Wolf D.A., Pereira M., Lau S., Jakobsson J., Björklund A., Grealish S., Parmar M. Generation of induced neurons via direct conversion in vivo. Proc. Natl. Acad. Sci. USA. 2013;110:7038–7043. doi: 10.1073/pnas.1303829110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treutlein B., Lee Q.Y., Camp J.G., Mall M., Koh W., Shariati S.A., Sim S., Neff N.F., Skotheim J.M., Wernig M., Quake S.R. Dissecting direct reprogramming from fibroblast to neuron using single-cell RNA-seq. Nature. 2016;534:391–395. doi: 10.1038/nature18323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathy S.J., Toker L., Bomkamp C., Mancarci B.O., Belmadani M., Pavlidis P. Assessing Transcriptome Quality in Patch-Seq Datasets. Front. Mol. Neurosci. 2018;11:363. doi: 10.3389/fnmol.2018.00363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierbuchen T., Ostermeier A., Pang Z.P., Kokubu Y., Südhof T.C., Wernig M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vue T.Y., Kim E.J., Parras C.M., Guillemot F., Johnson J.E. Ascl1 controls the number and distribution of astrocytes and oligodendrocytes in the gray matter and white matter of the spinal cord. Development. 2014;141:3721–3731. doi: 10.1242/dev.105270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L.H., Baker N.E. E Proteins and ID Proteins: Helix-Loop-Helix Partners in Development and Disease. Dev. Cell. 2015;35:269–280. doi: 10.1016/j.devcel.2015.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wapinski O.L., Vierbuchen T., Qu K., Lee Q.Y., Chanda S., Fuentes D.R., Giresi P.G., Ng Y.H., Marro S., Neff N.F. Hierarchical mechanisms for direct reprogramming of fibroblasts to neurons. Cell. 2013;155:621–635. doi: 10.1016/j.cell.2013.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng Q., Wang J., Wang J., He D., Cheng Z., Zhang F., Verma R., Xu L., Dong X., Liao Y. Single-Cell Transcriptomics Uncovers Glial Progenitor Diversity and Cell Fate Determinants during Development and Gliomagenesis. Cell Stem Cell. 2019;24:707–723.e8. doi: 10.1016/j.stem.2019.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. Springer-Verlag; 2016. ggplot2: Elegant Graphics for Data Analysis. [Google Scholar]
- Winterer J., Lukacsovich D., Que L., Sartori A.M., Luo W., Földy C. Single-cell RNA-Seq characterization of anatomically identified OLM interneurons in different transgenic mouse lines. Eur. J. Neurosci. 2019;50:3750–3771. doi: 10.1111/ejn.14549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang N., Ng Y.H., Pang Z.P., Südhof T.C., Wernig M. Induced neuronal cells: how to make and define a neuron. Cell Stem Cell. 2011;9:517–525. doi: 10.1016/j.stem.2011.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemankovics R., Káli S., Paulsen O., Freund T.F., Hájos N. Differences in subthreshold resonance of hippocampal pyramidal cells and interneurons: the role of h-current and passive membrane characteristics. J. Physiol. 2010;588:2109–2132. doi: 10.1113/jphysiol.2009.185975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H.M., Liu T., Liu C.J., Song S., Zhang X., Liu W., Jia H., Xue Y., Guo A.Y. AnimalTFDB 2.0: a resource for expression, prediction and functional study of animal transcription factors. Nucleic Acids Res. 2015;43:D76–D81. doi: 10.1093/nar/gku887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H., Su J., Hu X., Zhou C., Li H., Chen Z., Xiao Q., Wang B., Wu W., Sun Y. Glia-to-Neuron Conversion by CRISPR-CasRx Alleviates Symptoms of Neurological Disease in Mice. Cell. 2020;181:590–603.e16. doi: 10.1016/j.cell.2020.03.024. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
RNASeq data have been deposited to GEO (GSE174238; GSE173977; GSE173978; GSE173979) and publicly available. Accession numbers are listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to re-analyze the data reported in this paper is available from the lead contact upon request.