Abstract
Machine learning and linear regression models using CGM and participant data reduced HbA1c estimation error by up to 26% compared to the GMI formula, and exhibit superior performance in estimating the median of HbA1c at the cohort level, potentially of value for remote clinical trials interrupted by COVID-19.
Keywords: Continuous glucose monitoring, HbA1c estimation
1. Introduction
A better understanding of how data from continuous glucose monitors (CGMs) correspond to hemoglobin A1c (HbA1c) is necessary to link CGM data to long-term health outcomes in people with diabetes.1 , 7 Clinical studies in which patient HbA1c measurements have been interrupted by COVID-19 rely on estimates of HbA1c from CGM data. The Glucose Management Indicator (GMI) is the accepted method for using CGM-derived mean glucose to estimate lab-tested HbA1c. However, GMI often produces large estimation errors in patients with HbA1c > 8%.3 Improving the accuracy of HbA1c estimation could reduce the need for in-person HbA1c testing and allow for the continuation of research studies during which HbA1c testing has been interrupted by COVID-19. We hypothesize that the accuracy of HbA1c estimation can be improved by accounting for additional patient information and using machine learning (ML) methods.
2. Materials and methods
CGM, HbA1c, and demographic data were aggregated from four cohorts described in studies listed by the Type 1 Diabetes (T1D) Exchange, along with an additional cohort described in a study on a lifestyle intervention for teenagers with T1D, for a total of five cohorts.4., 5., 6, 7, 8, 9. Lab-tested HbA1c values were accompanied by at least five days (and up to 90 days) of CGM recordings (Table 1 ). Multiple HbA1c values could be included from each participant so long as they were preceded by sufficient CGM data. Demographic data included age, race, gender, and ethnicity. Due to data sparsity, HbA1c values <5.5% or >11.5% were excluded, as were participants whose self-identified race was neither white nor Black.
Table 1.
For each cohort, the minimum, first quartile, median, third quartile, and maximum number of days of CGM data available for each participant. Most participants in cohorts 1 and 2 have approximately one week of CGM data, whereas most participants in cohorts 3, 4, and 5 have two or more weeks of CGM data.
| Number of days of CGM data available |
|||||||
|---|---|---|---|---|---|---|---|
| Cohort # | N | Median HbA1c | Min | Q1 | Median | Q3 | Max |
| 1 | 495 | 9.3 | 5 | 7 | 7 | 7 | 8 |
| 2 | 238 | 8.6 | 5 | 6 | 6 | 7 | 15 |
| 3 | 782 | 8.4 | 5 | 11 | 35 | 56 | 84 |
| 4 | 1886 | 7.2 | 5 | 12 | 40 | 77 | 90 |
| 5 | 811 | 7.1 | 5 | 66 | 83 | 88 | 90 |
The statistics calculated for the CGM data associated with each HbA1c were mean, standard deviation, coefficient of variation, and percent of time in: hypoglycemia [<54 mg/dL], clinical hypoglycemia [54–69], target range [70–180], conservative target range [70–140], above target range [181–250], and far above target range [>250].
Using HbA1c as the response variable and all available CGM glucose statistics and demographics as the features, the following models were trained: L1-regularized regression (LASSO),10 LASSO containing two-way interactions between all features, and random forest (RF) regression.11 For comparison, ordinary least squares (OLS) regression of HbA1c on mean glucose and race (OLSmgr) was fit.7
After restricting HbA1c measurements to those for which an additional HbA1c had been measured for the same participant at least 70 days prior, all of the models were re-trained with prior HbA1c as an additional feature. For the full and restricted sets of HbA1c values, 5-fold cross-validation (CV) was used to tune the parameters of each model and to identify the top performing model. The performance of each model was compared to GMI at the level of each HbA1c value along with the median and interquartile range (IQR) of HbA1c for each participant cohort. The paired Wilcoxon signed-rank test was applied to the estimation errors measured using out-of-sample root-mean-squared error (RMSE) averaged across folds, each of which contained distinct participant data. For a practical comparison of model performance, the proportions of participants for whom the model estimate was within 0.5 and 1 percentage point of true HbA1c were calculated.
3. Results and discussion
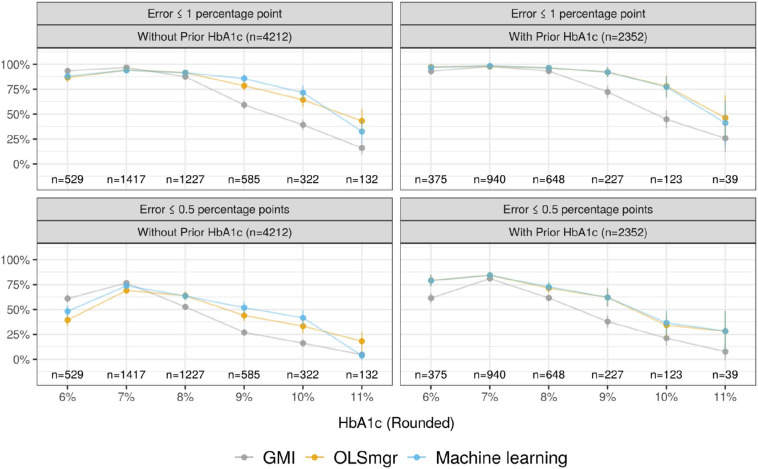
4212 HbA1c measurements from 1182 participants were collected. The top performing ML model that did not use prior HbA1c was the RF model, which produced an average error of 0.67 percentage points (SD = 0.03), 19% lower than the GMI average of 0.83 percentage points (SD = 0.02) (p < 0.001). OLSmgr had an average error 8% higher than the RF model. Respectively, the HbA1c estimates of the RF model and GMI were within 1 percentage point of true HbA1c for 87% and 81% of participants, and within 0.5 percentage points for 60% and 54% of participants. The stronger performance of the RF model was especially pronounced at HbA1c values of 9% and 10% (Fig. 1 ). In cohorts with median HbA1c > 8%, the RF and OLSmgr model estimated the median HbA1c of participants more accurately than GMI, with similar accuracy in the remaining cohorts (Table 2 ). Across cohorts, the models estimated the IQR of HbA1c with similar accuracy.
Fig. 1.
For each HbA1c value rounded to the nearest whole number, the plots show the proportion of model estimates that were within 1 percentage point (top) and 0.5 percentage points (bottom) of true HbA1c, with 95% confidence intervals for each proportion. Non-overlapping confidence intervals indicate statistical significance at the 0.05 level. The left plots compare GMI to an OLS model accounting for race and mean glucose (OLSmgr) and a random forest (RF) model accounting for many CGM metrics and demographics (Machine learning). The right plots compare GMI to OLSmgr fit with prior HbA1c and a LASSO model accounting for prior HbA1c along with the same covariates as the RF model (Machine learning). The sample sizes indicate the number of HbA1c values that fall in each rounded HbA1c bucket.
Table 2.
For each cohort, the true median and IQR of participant HbA1c values, along with the raw error observed when estimating these two quantities using GMI, OLSmgr, and the random forest (RF) model without accounting for prior HbA1c values. Compared to GMI, the OLSmgr and RF models more accurately estimate median HbA1c in cohorts with median HbA1c > 8%, with approximately similar performance in the remaining cohorts. Across cohorts, the OLSmgr and RF models estimate the IQR of participant HbA1c values approximately as accurately as GMI.
| Median HbA1c |
IQR HbA1c |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Cohort # | N | True value | GMI error | OLSmgr error | RF error | True value | GMI error | OLSmgr error | RF error |
| 1 | 495 | 9.3 | +0.8 | +0.4 | +0.2 | 1.5 | +0.2 | +0.1 | +0.4 |
| 2 | 238 | 8.6 | +0.4 | +0.1 | −0.1 | 1.1 | +0.1 | 0.0 | +0.2 |
| 3 | 782 | 8.4 | +0.9 | +0.3 | 0.0 | 1.9 | +0.6 | +0.4 | +0.5 |
| 4 | 1886 | 7.2 | 0.0 | −0.2 | −0.1 | 1.1 | +0.2 | +0.2 | +0.1 |
| 5 | 811 | 7.0 | −0.1 | −0.3 | −0.2 | 0.9 | +0.2 | +0.2 | +0.3 |
2352 HbA1c measurements from 872 participants were paired with an additional HbA1c measured at least 70 days prior. The best performing ML model that accounted for prior HbA1c was LASSO, with an average error of 0.49 percentage points (SD = 0.01), 26% lower than the GMI average of 0.67 percentage points (SD = 0.01) (p < 0.001). OLSmgr fit with prior HbA1c had an average error 0.4% higher than the LASSO model. Respectively, the HbA1c estimates of the LASSO model and GMI were within 1 percentage point of true HbA1c for 95% and 89% of participants, and within 0.5 percentage points for 74% and 64% of participants. The stronger performance of the LASSO model was especially pronounced at HbA1c values of 9% and 10% when measuring error within 1 percentage point, and at 9% when measuring error within 0.5 percentage points (Fig. 1). Performance at the cohort level was similar to that of the models excluding prior HbA1c (Table 3 ).
Table 3.
For each cohort, the true median and IQR of participant HbA1c values, along with the raw error observed when estimating these two quantities using GMI, OLSmgr, and the LASSO model when accounting for a prior HbA1c value. Cohort sizes are smaller than in Table 2 because not all participants had a prior HbA1c value. Cohort 3 is excluded from the table since none of its participants had an additional HbA1c value. Compared to GMI, the OLSmgr and LASSO models more accurately estimate median HbA1c in cohorts with median HbA1c > 8%, with approximately similar performance in the remaining cohorts. For cohorts 2, 4, and 5, the OLSmgr and LASSO models estimate the IQR of participant HbA1c values approximately as well as GMI, with slightly worse accuracy in cohort 1 that is likely attributable to random variation.
| Median HbA1c |
IQR HbA1c |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Cohort # | N | True value | GMI error | OLSmgr error | LASSO error | True value | GMI error | OLSmgr error | LASSO error |
| 1 | 277 | 9.2 | +0.6 | +0.1 | +0.1 | 1.4 | +0.1 | +0.3 | +0.4 |
| 2 | 119 | 8.5 | +0.3 | −0.1 | −0.1 | 1.0 | +0.1 | +0.2 | +0.2 |
| 4 | 1465 | 7.2 | +0.1 | 0.0 | 0.0 | 1.0 | +0.2 | 0.0 | +0.1 |
| 5 | 490 | 7.0 | −0.2 | −0.1 | −0.1 | 0.9 | +0.3 | +0.2 | +0.2 |
The simple OLS model estimating HbA1c from mean glucose, race, and prior HbA1c performs nearly as well as the LASSO model with access to many additional features, suggesting that the performance of GMI could be dramatically improved by accounting for only two additional features and without sacrificing interpretability. That ML models cannot substantially outperform linear regression suggests that HbA1c cannot be perfectly estimated from standard demographic features or summary BG metrics. Without access to more granular CGM data12 or patients' red blood cell characteristics,13 we suspect that no ML model can perform substantially better than a simple OLS model accounting for mean glucose, race, and prior HbA1c.
In order to increase the sample size and make the models applicable in clinical settings with imperfect patient adherence, the present analysis allowed the inclusion of participants with as few as 5 days of data or HbA1c measurements as recently as 70 days before evaluation. We expect that application to patients with more days of data would only improve performance. Furthermore, we note that our data was restricted to patients who identified as Black or white (Hispanic or non-Hispanic), so additional data from patients who identify with other race groups would make our models applicable to a wider patient population. Finally, we note that all participants had type 1 diabetes, so the results may not generalize to patients with type 2 diabetes.
To mitigate the challenges of applying this work in a clinical setting, we have made the code available online.14 The linear models presented are no more difficult to implement than GMI and still improve performance. A potential use of the model, in an ongoing investigation of the effect of CGM use on glucose management, is to interpolate HbA1c measurements missed due to disruption caused by COVID-19. Using the pre-COVID HbA1c measurements will allow the investigators to determine whether the algorithm proposed here or GMI provides more accurate estimates.
4. Conclusions
ML models using CGM and participant data reduced HbA1c estimation error by up to 26% compared to the GMI formula. The performance gains of the models were pronounced at higher HbA1c values; for example, for participants with HbA1c values close to 9%, the ML models were up to 24% more likely than GMI to estimate HbA1c within 0.5 percentage points.
In clinical trials, while the accuracy of the HbA1c is important, one major concern is the change in HbA1c. Our model's improved accuracy at high HbA1c values offers an advantage over GMI, which underestimates high values of HbA1c and may thus systematically underestimate reductions in HbA1c. The performance limitations of GMI for high values of HbA1c are likely due to the fundamental limitations of modeling a non-linear phenomenon with a single variable linear model. Our non-linear and multi-variable linear models overcome this limitation. Future investigation should determine whether HbA1c estimation error can be further reduced by accounting for longitudinal data on the association of CGM data and HbA1c, and whether similar ML models perform substantially better than GMI for patients with type 2 diabetes. Further, the ML models could be trained to predict other measures of long-term glucose control, such as glycated albumin.15 , 16
Acknowledgements
All authors made substantial contributions to the design, execution, and interpretation of the analyses and contributed to the development of the manuscript by drafting, reviewing, and critically commenting on the text at each stage of development. JG and DS take responsibility for the contents of the letter. DMM has had research support from the NIH, JDRF, NSF, and the Helmsley Charitable Trust and his institution has had research support from Medtronic, Dexcom, Insulet, Bigfoot Biomedical, Tandem, and Roche. DMM was supported by 5P30DK11607403. DMM has consulted for Abbott, the Helmsley Charitable Trust, Sanofi, Novo Nordisk, Eli Lilly, Medtronic, and Insulet. JG is supported by the National Science Foundation through a National Science Foundation Graduate Research Fellowship. AW is supported by the Department of Defense through a National Defense Science and Engineering Graduate Fellowship. The funders had no role in the design of the study, data acquisition, nor analysis. The other authors report no relevant disclosures.
Footnotes
Declaration of competing interest: DMM has had research support from the NIH, JDRF, NSF, and the Helmsley Charitable Trust and his institution has had research support from Medtronic, Dexcom, Insulet, Bigfoot Biomedical, Tandem, and Roche. DMM was supported by 5P30DK11607403. DMM has consulted for Abbott, the Helmsley Charitable Trust, Sanofi, Novo Nordisk, Eli Lilly, Medtronic, and Insulet. JG is supported by the National Science Foundation through a National Science Foundation Graduate Research Fellowship. AW was supported by the Department of Defense through a National Defense Science and Engineering Graduate Fellowship. The funders had no role in the design of the study, data acquisition, nor analysis. The other authors report no relevant funding or disclosures.
References
- 1.Danne T., Nimri R., Battelino T., Bergenstal R.M., Close K.L., DeVries J.H., et al. International consensus on use of continuous glucose monitoring. Diabetes Care. 2017;40:1631–1640. doi: 10.2337/dc17-1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bergenstal R.M., Beck R.W., Close K.L., Grunberger G., Sacks D.B., Kowalski A., et al. Glucose management indicator (GMI): a new term for estimating A1C from continuous glucose monitoring. Diabetes Care. 2018;41:2275–2280. doi: 10.2337/dc18-1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Type 1 Diabetes Exchange - Public Site [online]. Available from https://public.jaeb.org/t1dx/stdy.
- 5.JDRF CGM Study Group JDRF randomized clinical trial to assess the efficacy of real-time continuous glucose monitoring in the management of type 1 diabetes: research design and methods. Diabetes Technol Ther. 2008;10(4):310–321. doi: 10.1089/dia.2007.0302. [DOI] [PubMed] [Google Scholar]
- 6.Aleppo G., Ruedy K.J., Riddlesworth T.D., Kruger D.F., Peters A.L., Hirsch I., et al. REPLACE-BG: a randomized trial comparing continuous glucose monitoring with and without routine blood glucose monitoring in adults with well-controlled type 1 diabetes. Diabetes Care. 2017;40:538–545. doi: 10.2337/dc16-2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bergenstal R.M., Gal R.L., Connor C.G., Gubitosi-Klug R., Kruger D., Olson B.A., et al. Racial differences in the relationship of glucose concentrations and hemoglobin A1c levels. Ann Intern Med. 2017;167:95–102. doi: 10.7326/M16-2596. [DOI] [PubMed] [Google Scholar]
- 8.Nwosu B.U., Maranda L., Cullen K., Greenman L., Fleshman J., McShea N., et al. A randomized, double-blind, placebo-controlled trial of adjunctive metformin therapy in overweight/obese youth with type 1 diabetes. PLoS One. 2015;10 doi: 10.1371/journal.pone.0137525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mayer-Davis EJ, Maahs DM, Seid M, Crandell J, Bishop FK, Driscoll KA, Hunter CM, Kichler JC, Standiford D, Thomas JM, FLEX Study Group. Efficacy of the Flexible Lifestyles Empowering Change intervention on metabolic and psychosocial outcomes in adolescents with type 1 diabetes (FLEX): a randomised controlled trial. The Lancet Child & Adolescent Health 2018;9:635–646 PubMed. [DOI] [PMC free article] [PubMed]
- 10.Lasso (statistics). https://en.wikipedia.org/wiki/Lasso_(statistics).
- 11.Random forest. https://en.wikipedia.org/wiki/Random_forest.
- 12.Fabris C., Heinemann L., Beck R., Cobelli C., Kovatchev B. Estimation of hemoglobin A1c from continuous glucose monitoring data in individuals with type 1 diabetes: is time in range all we need? Diabetes Technol Ther. 2020;22:501–508. doi: 10.1089/dia.2020.0236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cohen R.M., Franco R.S., Khera P.K., Smith E.P., Lindsell C.J., Ciraolo P.J., et al. Red cell life span heterogeneity in hematologically normal people is sufficient to alter HbA1c. Blood. 2008;112:4284–4291. doi: 10.1182/blood-2008-04-154112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.https://github.com/joshuagrossman/a1c
- 15.Desouza C.V., Rosenstock J., Zhou R., Holcomb R.G., Fonseca V.A. Glycated albumin at 4 weeks correlates with a1c levels at 12 weeks and reflects short-term glucose fluctuations. Endocr Pract. 2015;21:1195–1203. doi: 10.4158/EP14570.OR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Desouza C.V., Holcomb R.G., Rosenstock J., Frias J.P., Hsia S.H., Klein E.J., et al. Results of a study comparing glycated albumin to other glycemic indices. J Clin Endocrinol Metab. 2020;105:677–687. doi: 10.1210/clinem/dgz087. [DOI] [PMC free article] [PubMed] [Google Scholar]