SUMMARY
Mitochondria and chloroplasts are organelles with high iron demand that are particularly susceptible to iron-induced oxidative stress. Despite the necessity of strict iron regulation in these organelles, much remains unknown about mitochondrial and chloroplast iron transport in plants. Here, we propose that Arabidopsis Ferroportin 3 (FPN3) is an iron exporter dual-targeted to mitochondria and chloroplasts. FPN3 is expressed in shoots regardless of iron conditions, but its transcripts accumulate under iron deficiency in roots. fpn3 mutants cannot grow as well as wild type under iron-deficient conditions and their shoot iron levels are lower compared to wild type. Analyses of iron homeostasis gene expression in fpn3 mutants and ICP-MS measurements show that iron levels in the mitochondria and chloroplasts are increased relative to wild type, consistent with the proposed role of FPN3 as a mitochondrial/plastid iron exporter. In iron deficient fpn3 mutants, abnormal mitochondrial ultrastructure was observed, whereas chloroplast ultrastructure was not affected, implying that FPN3 plays a critical role in the mitochondria. Overall, our study suggests that FPN3 is essential for optimal iron homeostasis.
Keywords: Iron, Ferroportin, Mitochondria, Chloroplast, Arabidopsis, Transport
INTRODUCTION
Iron serves as a critical redox cofactor in vital cellular processes. Nevertheless, excess or improperly regulated iron can cause deleterious effects by generating hydroxyl radicals via the Fenton reaction (Halliwell and Gutteridge, 1992). Therefore, iron homeostasis must be tightly maintained in all organisms, including plants. As photosynthetic organisms, plants use iron as an essential cofactor in both respiration and photosynthesis. At the same time, plant cells must regulate iron to ensure adequate supply while avoiding oxidative stress (Shcolnick and Keren, 2006). Despite being an abundant element in the soil, iron is one of the most limiting nutrients for plant growth – it has extremely low bioavailability under aerobic conditions at neutral or alkaline pH (Marschner, 2012; Colombo et al., 2014). Iron homeostasis in plants is of particular interest, because understanding its mechanisms will provide insights to improving agriculture and human health (Vasconcelos et al., 2017).
Dicots acquire iron by a reduction-based mechanism that is induced under iron deficiency (Jeong et al., 2017; Connorton et al., 2017; Kobayashi et al., 2018; Brumbarova et al., 2015). Ferric chelates in the rhizosphere are solubilized by the release of protons (Santi and Schmidt, 2009) and reduced to ferrous iron by FERRIC REDUCTASE OXIDASE 2 (FRO2) (Robinson et al., 1999). Coumarins released from iron-deficient roots are considered to aid this process as well (Clemens and Weber, 2016; Schmidt et al., 2014; Fourcroy et al., 2014). Ferrous iron is then transported into the root via IRON-REGULATED TRANSPORTER 1 (IRT1) (Connolly et al., 2002; Vert et al., 2002; Varotto et al., 2002). Once iron reaches the root vasculature, it is loaded into the xylem by FERROPORTIN 1 (FPN1) (Morrissey et al., 2009) and chelated with citrate, which is transported into the xylem by the FERRIC REDUCTASE DEFECTIVE 3 (FRD3) transporter (Durrett et al., 2007). The iron-citrate complex is then translocated to shoots. For lateral translocation in shoots, iron-nicotianamine (NA) complexes are formed and translocated from leaves to seeds via the phloem by YELLOW STRIPE-LIKE (YSL) family members (DiDonato et al., 2004; Schaaf et al., 2005; Waters et al., 2006). Two YSL members, YSL1 and YSL3, were shown to regulate long-distance iron-deficiency signals from shoots (Kumar et al., 2017) and are responsible for loading iron into the seeds (Le Jean et al., 2005; Waters et al., 2006). Oligopeptide transporter 3 (OPT3) also plays a role in transmitting shoot-to-root iron signals and regulates the redistribution of iron from source to sink tissues, such as from old leaves to seeds or to developing tissues (Mendoza-Cózatl et al., 2014; Zhai et al., 2014; Stacey et al., 2008).
In addition to iron acquisition and its distribution between tissues, iron transport across subcellular compartments is crucial for proper iron homeostasis. In particular, chloroplasts and mitochondria need a substantial amount of iron; many components of the photosynthetic and respiratory electron transport chains use iron as a cofactor. Fe-S cluster assembly occurs in these organelles and heme biosynthesis occurs in plastids (Masuda et al., 2003; Tanaka et al., 2011). Proteins involved in the last stage of heme biosynthesis are also present in mitochondria (Balk and Schaedler, 2014). Chloroplasts are the most iron-rich organelle in plant cells and accounts for 60-80% of iron found in a leaf cell (Terry and Low, 1982; Shikanai et al., 2003). Meanwhile, in the mitochondria, iron is the major micronutrient present with a molar ratio of 26:8:6:1 for Fe:Zn:Cu:Mn (Tan et al., 2010). Mitochondria and chloroplasts are also highly susceptible to oxidative stress due to reactive oxygen species (ROS) generated by the electron transport chain. The iron sequestering protein ferritin is present in mitochondria and plastids (Briat et al., 2010; Zancani et al., 2004) and is involved in preventing iron-induced oxidative stress (Ravet et al., 2009). In the mitochondria, frataxin, which is involved in Fe-S cluster and heme biogenesis, also plays a role in protection against oxidative stress (Gomez-Casati et al., 2018).
Iron transport in mitochondria and chloroplasts is not as well understood as the mechanisms of iron acquisition in the roots, but molecular and physiological studies have been gradually contributing to understanding iron regulation in chloroplasts and mitochondria. In chloroplasts, physiological studies suggest that ferric chelates move across the outer membrane (Bughio et al., 1997; Solti et al., 2012; Müller et al., 2019), but ferrous iron is imported into chloroplasts across the inner membrane (Bughio et al., 1997; Shingles et al., 2002). The ferric chelate reductase, FRO7, reduces iron for chloroplast iron acquisition (Jeong et al., 2008), and PERMEASE IN CHLOROPLAST 1 (PIC1) mediates iron transport into chloroplasts via interaction with NiCo (Duy et al., 2007; Duy et al., 2011). A recent study in Brassica napus proposes that NiCo may also be involved in iron sensing or iron release from chloroplasts (Pham et al., 2020). The Arabidopsis MitoFerrinLike1 (Mfl1) has also been reported to import iron into chloroplasts (Tarantino et al., 2011). In addition, ATP-binding cassette transporters, ABCI10, NAP14/ABCI11, and ABCI12, may also contribute to iron uptake into chloroplasts (Shimoni-Shor et al., 2010; Voith von Voithenberg et al., 2019). YSL4 and YSL6 have been reported to efflux iron-NA complexes from the chloroplasts (Divol et al., 2013). However, both transporters were also identified in the tonoplast proteome (Jaquinod et al., 2007) and were targeted to the vacuolar and intracellular membranes (Conte et al., 2013). ZmFRD4 was reported as a potential thylakoid iron importer in maize, although direct evidence for iron transport remains to be found (Zhang et al., 2017).
In the mitochondria, a mitoferrin ortholog from rice, MITOCHONDRIAL IRON TRANSPORTER (MIT), was identified as a mitochondrial iron importer (Bashir et al., 2011). Two Arabidopsis mitoferrin orthologs, MIT1 and MIT2, have recently been reported as mitochondrial iron importers that mediate cellular iron homeostasis and are essential for embryogenesis (Jain et al., 2019). A reduction-based mechanism might be involved in moving iron in and out of mitochondria because ferric chelate reductases, FRO3 and FRO8, are localized to the mitochondria in Arabidopsis (Jeong and Connolly, 2009; Heazlewood et al., 2004). The two mitochondrial FROs are likely to play non-overlapping roles, as FRO3 is induced by iron deficiency, whereas FRO8 is not iron-regulated (Mukherjee et al., 2006).
In this study, we investigated an Arabidopsis FPN family member, FPN3/IREG3. FPNs, also known as solute carrier (SCL) group 40A1 and IRON REGULATED (IREG) transporters, efflux iron from the cytoplasm (Drakesmith et al., 2015). FPNs have been extensively studied in vertebrate species, in which the only FPN is localized to the plasma-membrane and exports iron from the cell into the plasma (Drakesmith et al., 2015). In many plant species, multiple FPN/IREG paralogs are present and localize to different sub-cellular compartments. IREG1 of the nickel hyperaccumulator Psychotria gabriellae (Merlot et al., 2014), and buckwheat IREG (Yokosho et al., 2016) were identified on the tonoplast. A nodule-specific FPN2 in Medicago truncatula that is targeted to the symbiosome membrane has also been identified (Escudero et al., 2020). In Arabidopsis, three FPN/IREG family members were identified according to phylogenetic analysis (Schaaf et al., 2006). FPN1/IREG1 (At2g38460) and FPN2/IREG2 (At5g03570) share 77 % identity, whereas FPN3/IREG3 (At5g26820) shares about 20% identity with FPN1 and FPN2 (Figure 1). FPN1/IREG1 localizes to the plasma membrane of cells in the stele (Morrissey et al., 2009), and FPN2/IREG2 is localized on the vacuolar membrane (Morrissey et al., 2009; Schaaf et al., 2006) and its expression is primarily detected in the cortex (Morrissey et al., 2009). Previously, FPN3/IREG3 was reported as Multiple Antibiotic Resistance 1 (MAR1), a chloroplast protein that allows antibiotics to opportunistically enter chloroplasts (Conte et al., 2009). While it was speculated that MAR1 may be involved in iron homeostasis, its potential role in iron regulation and its physiological function were not previously studied. Here, we report that FPN3 is dual-targeted to mitochondria and chloroplasts, and provide results suggesting that FPN3 exports iron from these organelles. Furthermore, our study provides key evidence indicating that FPN3 plays a critical role in iron homeostasis and its function is important in the mitochondria as evidenced by the drastic morphological changes in fpn3 mitochondria under iron deficient conditions.
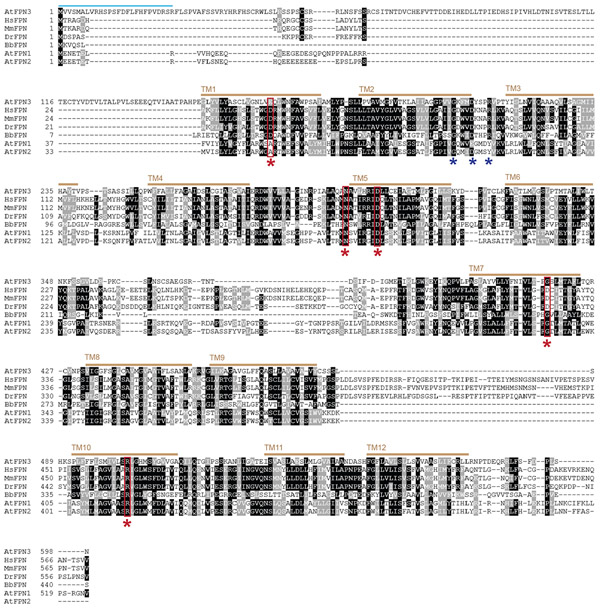
Figure 1. Sequence alignment of FPNs.
Multiple sequence alignment was performed using T-Coffee (Notredame et al., 2000) with FPN amino acid sequences of Arabidopsis thaliana (AtFPN1, AtFPN2, AtFPN3), Homo sapiens (HsFPN), Mus musculus (MmFPN), Danio reio (DrFPN), and Bdellovibrio bacteriovorus (BbFPN). Identical residues are shaded in black, whereas similar residues are shaded in grey. Red stars indicate residues involved in iron binding and transport in human and bacterial FPNs (Bonaccorsi di Patti et al., 2015; Taniguchi et al., 2015). The blue stars indicate motif A, GX3DX3R, which is conserved in MFS members. The predicted transit peptide, denoted as a green bar, suggests a chloroplast or mitochondrial localization (Schwacke et al., 2003). The brown bars indicate predicted transmembrane domains.
RESULTS
Iron binding and transport amino acid residues of human FPN are mostly conserved in Arabidopsis FPN3
To investigate the potential role of FPN3 in iron homeostasis, we first examined if conserved iron binding and transport residues of other FPN members are present in FPN3. Structural modeling and in vitro assays with mutated variants of human FPN identified amino acids that are critical for iron binding and transport (Bonaccorsi di Patti et al., 2014; Taniguchi et al., 2015). These residues were mostly conserved across FPN orthologs in human, mouse, zebrafish, frog, bacteria, and Arabidopsis FPN paralogs (Taniguchi et al., 2015). For example, Asp39 and Asp181 of HsFPN were essential for iron transport (Bonaccorsi di Patti et al., 2014; Taniguchi et al., 2015). The Aps181 of HsFPN was highly conserved across FPNs examined, and the presence of Glu, which is synonymous to Asp, in FPN3 at the position corresponding to Asp39 of HsFPN, was consistent with its potential role in iron binding/transport (Figure 1). Asp39 of HsFPN was not preserved in AtFPN1 and AtFPN2, which are closely related paralogs but distantly related to FPN3 (Schaaf et al., 2006), as Ala was found in lieu of Glu or Asp at the corresponding position (Figure 1). As FPN1 and FPN2 are also known to transport iron (Morrissey et al., 2009), this Asp residue might not be essential for iron transport by Arabidopsis FPNs. In HsFPN, Asn174, Asp 325, and Arg466, were also found to affect iron efflux although not directly as an iron binding site (Bonaccorsi di Patti et al., 2014; Taniguchi et al., 2015). Among these residues, Asn174 and Arg466 of HsFPN were highly preserved in FPN3 and other FPNs examined (Figure 1). The HsFPN Asp325 was conserved among vertebrate FPNs, but Gly was found in the FPNs of Arabidopsis and bacteria (Figure 1). Overall, the conservation of multiple residues that are known to be directly or indirectly involved in iron binding/transport in FPN3 imply that FPN3 is likely to transport ferrous iron, which is a major substrate of FPN orthologs (Drakesmith et al., 2015; Taniguchi et al., 2015).
Heterologously expressed FPN3 exports iron from mitochondria in yeast
Multiple chloroplast proteins have been targeted to the mitochondria when expressed in fungi (Versaw and Harrison, 2002; Jeong et al., 2008; Hurt et al., 1986; Pfaller et al., 1989; Brink et al., 1994). Based on the prior report on MAR1 that indicated chloroplast localization (Conte et al., 2009) and the predicted transit peptides that suggested targeting to chloroplasts or mitochondria (Schwacke et al., 2003), we tested if FPN3 would be able to complement mitochondrial iron transporter mutants of Saccharomyces cerevisiae. After verifying that FPN3 is localized to mitochondria using Western blots with mitochondrial fractions of yeast cells expressing FPN3-FLAG (Figure S1), we expressed FPN3 in Δmmt1/2, which lacks mitochondrial iron exporters. Previous studies showed that Δmmt1/2 did not have a strong phenotype, but the deletion of MMT1/2 in a Δccc1 background, which lacks a vacuolar iron importer, resulted in a slightly decreased sensitivity to high iron compared to Δccc1 (Li et al., 2014). Additionally, Δccc1Δmmt1/2 cells expressing MMT1/2 are highly sensitive to high iron due to increased cytosolic iron (Li et al., 2014). In the present study, we expressed FPN3 in Δccc1Δmmt1/2 under high iron conditions to test if phenotypes similar to Δccc1Δmmt1/2 cells expressing MMT1/2 would arise. As previously observed by Li et al. (2014), the expression of MMT1/2 resulted in reduced growth in high iron (Figure 2A, 2B). The growth of Δccc1Δmmt1/2 expressing FPN3 was decreased in high iron and approximated that of Δccc1Δmmt1/2 expressing MMT1/2 (Figure 2A, 2B). Thus, our high iron growth assay results from both plates and liquid cultures suggest that FPN3 exports iron from the mitochondria (Figure 2A, 2B). We also expressed FPN3 in the Δmrs3Δmrs4 yeast mutant, which lacks the mitochondrial iron importers, Mrs3 and Mrs4, to test if FPN3 is likely to import iron into mitochondria. The Δmrs3Δmrs4 mutant does not have a strong iron phenotype, but exhibits increased sensitivity to oxidative stress compared to wild type cells (Mühlenhoff et al., 2003; Foury and Roganti, 2002). Expression of FPN3 in Δmrs3Δmrs4 did not rescue the oxidative stress sensitivity of Δmrs3Δmrs4 (Figure S2), which would be expected if FPN3 were a mitochondrial iron importer. By contrast, FPN3 expression exacerbated the phenotype, as observed in Δmrs3Δmrs4 cells overexpressing MMT1/2 (Figure S2), indicating that FPN3 is not likely to be importing iron into the mitochondria.
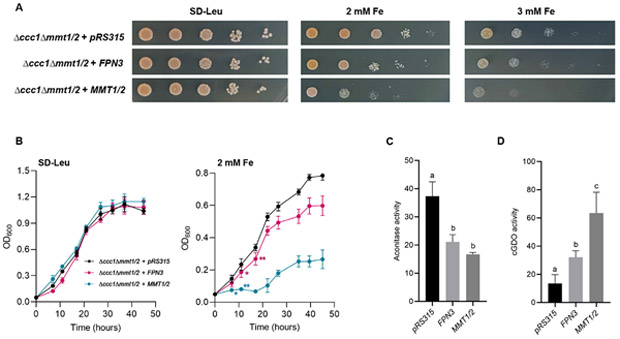
Figure 2. Heterologous expression of FPN3 in yeast.
(A) Spot assays in high iron media (SD-Leu with 2 mM and 3 mM Fe) or control media (SD-Leu) with Δccct1Δmmt1/2 cells expressing FPN3 or MMT1/2, or transformed with an empty vector, pRS315. (B) Growth assays with liquid culture with Δccct1Δmmt1/2 cells expressing FPN3 or MMT1/2, or transformed with an empty vector, pRS315. Significant differences compared to the negative control were determined by two-way ANOVA followed by Tukey’s multiple comparison test (*: p<0.05; **: p<0.001; n=6; Error bars = SD). From the third time point and beyond, cells expressing FPN3 or MMT1/2 were significantly different from the negative control (p<0.001) but labels were omitted for simplicity of the graph. (C) Aconitase activity (nmol/mg protein/min) of wild type (DY150) cells expressing FPN3 or MMT1/2 or transformed with an empty vector, pRS426. Significant differences were determined by one-way ANOVA followed by Tukey’s test and denoted with different letters (p<0.01; n=6; Error bars = SD). (D) cGDO activity (mmol/mg protein/min) of wild type (DY150) cells expressing FPN3, MMT1/2, or the empty vector, pRS315. Significant differences were determined by one-way ANOVA followed by Tukey’s test and denoted with different letters (p<0.01; n=6; Error bars = SD).
Based on the results from Δccc1Δmmt1/2 and Δmrs3Δmrs4 cells expressing FPN3 or MMT1/2, we hypothesized that FPN3 is a mitochondrial iron exporter. If FPN3 is exporting iron from the mitochondria, mitochondrial iron levels should be lower in cells overexpressing FPN3. To test this idea, we measured the activity of aconitase in mitochondria to indirectly assess mitochondrial iron levels. Aconitase is a Fe-S enzyme involved in the tricarboxylic acid cycle, and its activity correlates with the availability of mitochondrial iron. For example, decreased aconitase activity has been detected in the yeast Δmrs3/4 cells and rice plants defective in mitochondrial iron import (Foury and Roganti, 2002; Bashir et al., 2011). Although aconitase is present in the mitochondria and the cytosol, mitochondrial aconitase accounts for most of the cellular aconitase activity in yeast, and yeast cytosolic aconitase is not involved in regulating iron metabolism unlike in mammalian cells (Regev-Rudzki et al., 2005). Aconitase activity of wild type cells expressing FPN3 was similar to cells expressing MMT1/2 and exhibited about 30% lower activity than that of the negative control with an empty vector (Figure 2C), suggesting that FPN3 is exporting iron from the mitochondria.
As an alternative approach to testing mitochondrial iron export by FPN3, we examined if cytosolic iron levels increased in cells expressing FPN3. We co-transformed FPN3 or control plasmids along with a plasmid that expresses the bacterial gentisate 1,2-dioxygenase (GDO) in the yeast cytoplasm (Li et al., 2012) and measured GDO activity. GDO uses iron as a cofactor and the activity of cytosolic GDO (c-GDO), which has been confirmed to localize to the cytoplasm in yeast, correlates with the amount of cytosolic iron. Thus, c-GDO can serve as an indicator of cytosolic iron levels in yeast (Li et al., 2012; Li et al., 2014). We detected significantly higher c-GDO activity in cells expressing FPN3 or MMT1/2, whereas only background level activity was observed in cells with the empty vector control (Figure 2D). This result is in agreement with our findings from yeast growth tests and aconitase activity assays (Figure 2A-C). Overall, our yeast results consistently supported the hypothesis that FPN3 is a mitochondrial iron exporter and is functionally analogous to Mmt1/2.
FPN3 is expressed in the shoots, roots, flowers, and siliques, and is iron-regulated in roots
To detect the expression of FPN3 at the tissue level, we fused the promoter region of FPN3 to the β-glucuronidase (GUS) reporter gene (FPN3p-GUS), transformed the construct into wild type Arabidopsis, and conducted GUS histochemical staining. FPN3p-GUS was expressed in shoots and in the root cap of seedlings from an early stage, and in floral organs and siliques (Figure 3A-F). These results are in agreement with FPN3 expression data reported in multiple transcriptomics studies (Schmid et al., 2005; Zimmermann et al., 2004; Dinneny et al., 2008; Winter et al., 2007).
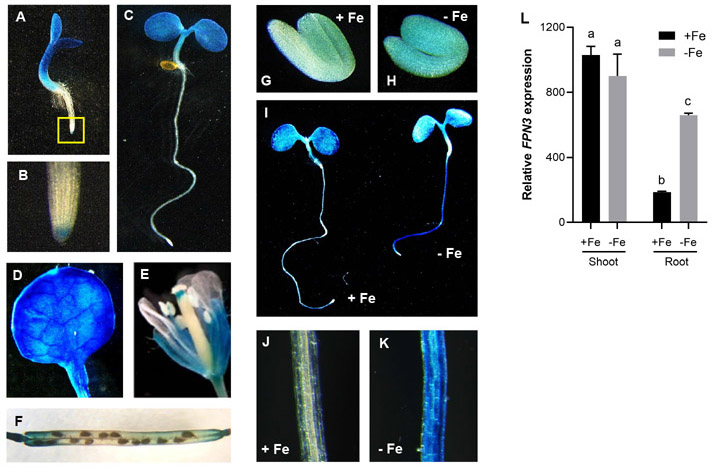
Figure 3. FPN3 expression detected by FPN3p-GUS and RT-qPCR.
(A-C) Histochemical staining of FPN3p-GUS seedlings germinated under iron sufficient conditions on day 3 (A) and day 5 (C), and a close-up image of the root tip of a day 3 seedling (B; boxed region in A). (D) FPN3p-GUS expression in the first true leaf of a 3-week old plant. (E, F) FPN3p-GUS staining in floral organs (E) and the silique (F). (G, H) FPN3p-GUS staining in 1-day-old imbibing seedlings germinated on iron deficient (G) or sufficient (H) media. (I) FPN3p-GUS staining in 3-day-old seedlings germinated on iron deficient (−Fe) or sufficient (+Fe) media. (J, K) Close-up view of iron sufficient root (J) and iron deficient root (K). Representative images of seedlings or organs from at least 6 individuals are shown. (L) Steady state transcript level of FPN3 relative to the internal control, ACT2, in shoots and roots of wild type plants treated under iron deficient or sufficient conditions are shown. Transcript levels were detected by RT-qPCR. Statistically significant groups are denoted with different letters (ANOVA; Tukey’s test; *: p<0.05; n=3; error bars = SD).
To determine whether FPN3 expression is regulated by iron, we performed GUS staining with FPN3p-GUS plants grown under iron-deficient or iron-sufficient conditions. In 1-day old seedlings, FPN3p-GUS expression was more intense in the roots of plants grown under iron-deficient conditions compared to plants from iron-sufficient conditions (Figure 3G, H). We detected stronger staining in roots of 3-day old seedlings grown in iron-deficient medium than in roots from iron-sufficient medium (Figure 3I-K), which was corroborated by our RT-qPCR results that showed approximately 4.5-fold higher level of FPN3 transcript level in iron-deficient roots (Figure 3L). Likewise, higher steady state levels of FPN3 transcripts have been detected in iron-deficient roots in multiple transcriptomic datasets (Mai et al., 2016; Dinneny et al., 2008; Yang et al., 2010; Park et al., 2019; Khan et al., 2018; Buckhout et al., 2009). Close up images of GUS stained roots indicated that FPN3 was broadly expressed throughout multiple cell layers (Figure 3K). According to the cell-specific transcriptomics dataset by Dinneny et al. (2008), FPN3 expression increased under iron deficiency, and was detected in the epidermis, columella root cap, cortex, and steel, with the particularly high expression in the cortex. In the shoots, FPN3p-GUS staining was prominent under both iron sufficient and deficient conditions (Figure 3I-K). Constitutive expression of FPN3 in the shoots regardless of the iron status of the plant was also detected by RT-qPCR (Figure 3I, L) and transcriptomic datasets from multiple studies (Khan et al., 2018; Rodríguez-Celma et al., 2013; Park et al., 2020).
FPN3 is iron-regulated primarily by local signals in roots
Studies with multiple plant species have revealed that two types of iron-deficiency signals regulate the iron-deficiency response in the roots: the local signal determined by the iron level in the rhizosphere, and the systemic, long distance signal dictated by the iron status of the shoots (Vert et al., 2003; Grusak and Pezeshgi, 1996; García et al., 2013; Bienfait et al., 1987).
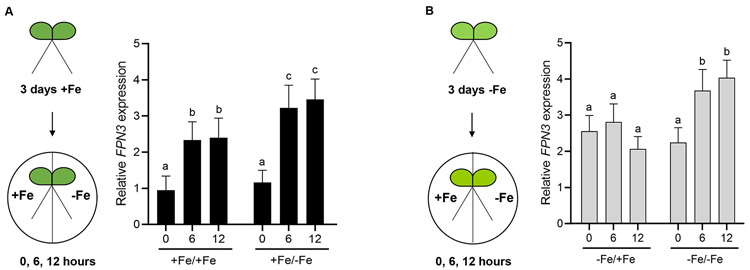
The differential level of FPN3 transcripts in roots and shoots prompted us to test if the iron-dependent FPN3 accumulation in roots is regulated by a local or long-distance signal. Thus, we generated two lateral roots, or split roots, and each root was subjected to different iron growth conditions. The iron growth conditions for our split-root experiments were optimized following the method by Kumar et al. (2017). Plants were grown under iron-deficient or iron-sufficient conditions for 3 days prior to the split media experiment and RNA was extracted from each side of the split root after 0, 6, or 12 hours after transfer for RT-qPCR (Figure 4). If FPN3 level in the roots is regulated by a long-distance signal from the shoots, then FPN3 expression is expected to be higher in both sides of the split roots of plants that were initially grown in iron deficient medium, regardless of the iron conditions for each side of the split roots. Our data suggested that the increased level of FPN3 transcripts in iron deficient roots is primarily driven by the local iron conditions (Figure 4). In the split root transferred from iron sufficient conditions to iron deficient conditions, we observed the greatest, approximately 3-fold, increase in FPN3 transcript levels (Figure 4A). Between the split roots pre-grown under iron deficiency, only the split root subjected to an additional duration of iron deficiency exhibited higher levels of FPN3 over the time course tested, which indicated that a systemic signal based on the shoot iron status did not dictate FPN3 expression in roots (Figure 4B). FPN3 expression was not affected when both split roots from the same seedling were transferred from iron sufficient to sufficient conditions (Figure S3). The steady state level of FPN3 transcripts in split-root plants with both roots transferred to the same condition verified that cutting the roots did not affect FPN3 expression (Figure S3). Overall, our split-root results suggest that FPN3 expression in roots is primarily regulated by the local iron status in the roots.
Figure 4. Steady state FPN3 transcript levels in split-roots.
(A, B) Schematic overview of iron growth conditions prior to RNA extraction and comparison of FPN3 expression in split roots. Seedlings were grown under iron-sufficient (+Fe; A) or deficient (−Fe; B) conditions, then each root was transferred to iron-deficient or iron-sufficient medium for 0, 6, or 12 hours. The iron growth conditions of each split root are denoted as: +Fe/+Fe for growth in iron sufficient conditions followed by iron sufficiency treatment; +Fe/−Fe for growth in iron sufficient conditions followed by iron deficiency treatment; −Fe/+Fe for growth in iron deficient conditions followed by iron sufficiency treatment; −Fe/−Fe for growth in iron deficient conditions followed by iron deficiency treatment. Steady state FPN3 transcript level detected by RT-qPCR with roots from +Fe/+Fe condition was set at 1 for comparison and expression ratios were plotted. Significant differences compared to 0 hour within each condition were determined by one-way ANOVA followed by Dunnett’s test and denoted with different letters (p<0.05; **: p<0.01; n=5; error bars = SD).
Growth of fpn3 mutants is reduced under iron deficiency
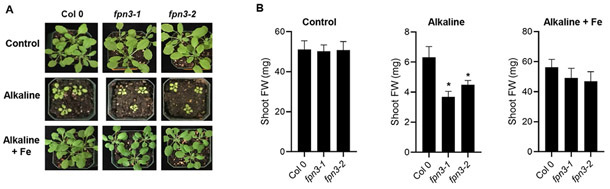
We then conducted phenotypic analyses with fpn3 plants to understand the function of FPN3 in iron homeostasis. Based on the increased FPN3 expression in roots under iron deficiency (Figure 3I-L), we hypothesized that fpn3 mutants might have less iron available for growth or development. To test this idea, we germinated two T-DNA insertion lines, fpn3-1 and fpn3-2, which had significantly reduced levels of FPN3 transcripts (Figure S4A, B), in alkaline soil, where iron availability is drastically reduced. We observed that fpn3 single mutants were smaller than wild type in alkaline soil, but this phenotype was recovered when watered with soluble iron (Figure 5A, 5B). As an alternative approach to verify the low iron growth phenotype, we quantified the root lengths and shoot fresh weights of fpn3 and wild type seedlings germinated in iron sufficient medium (Figure S4C) or in medium without iron (Figure S4D). Under iron deficient conditions, fpn3 mutant seedlings exhibited decreased shoot fresh weights (Figure S4D). The growth defect of fpn3 on iron-deficient medium and alkaline soil consistently showed that FPN3 is necessary for optimal growth under iron limiting conditions.
Figure 5. Growth of fpn3 under iron deficient conditions.
(A) Wild type (Col 0) and fpn3 single mutants germinated and grown in control soil or alkaline soil (~pH 8) with or without iron irrigation. (B) Quantified shoot fresh weights of wild type (Col 0), fpn3 single mutants, germinated and grown in control soil or alkaline soil. Mean values of 36 to 40 individuals are shown. Statistically significant differences compared to Col 0 are denoted (One-way ANOVA; Dunnett’s test; *: p <0.05; error bars = SD).
Iron content is reduced in fpn3 mutant leaves
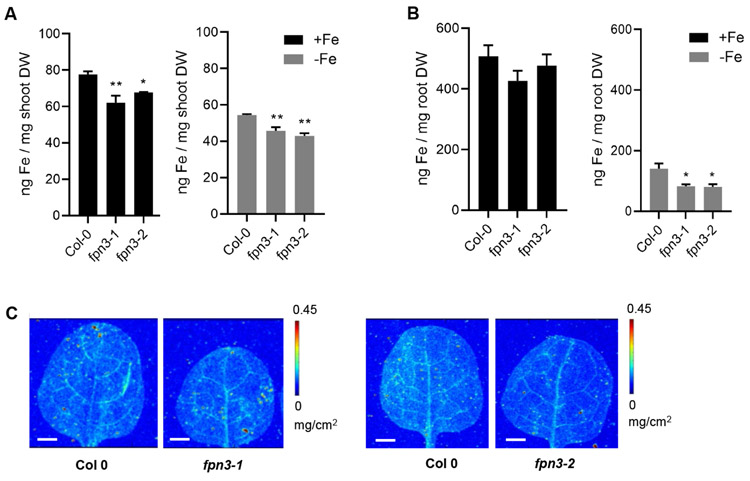
To test if iron content of fpn3 mutant leaves is affected, we conducted bulk elemental analysis of shoot and root tissues from plants grown under iron deficient and sufficient conditions using inductively coupled plasma mass spectrometry (ICP-MS). ICP-MS results revealed that iron content of fpn3 mutant shoot was significantly lower than that of wild type under both iron sufficient and deficient conditions (Figure 6A). In roots, iron content was not statistically different between wild type and fpn3 mutants under iron sufficient conditions while iron content was significantly lower in fpn3 mutant roots than in wild type roots under iron deficiency (Figure 6B). We then performed synchrotron x-ray fluorescence imaging (SXRF) to examine the amount and distribution of iron in leaves of fpn3 and wild type. The overall distribution of iron was not different among genotypes (Figure 6C). These results suggested that the function of FPN3 is necessary to properly maintain iron levels in the leaves regardless of iron conditions, and in the roots under iron deficiency.
Figure 6. Elemental analysis of fpn3 shoots and roots.
(A) Iron content of shoot tissue from plants grown under iron sufficient (+Fe) and deficient (−Fe) conditions measured by ICP-MS. (B) Iron content of root tissue from plants grown under iron sufficient (+Fe) and deficient (−Fe) conditions measured by ICP-MS. Mean values of pooled quadruplicate ICP-MS samples are shown with error bars (SE). Significant differences compared to Col 0 are denoted (One-way ANOVA; Dunnett’s test; *: p<0.05; **: p<0.01). (C) Synchrotron x-ray fluorescence microscopy images of iron distribution in leaves of wild type (Col 0) and fpn3 leaves. The first true leaves of 22-day old plants grown in iron sufficient conditions were imaged at a resolution of 30 x 30 μm with 0.2 sec dwell time. Representative images of leaves from three individuals are shown. Scale bars = 1 mm.
FPN3-GFP is dual-targeted to the mitochondria and plastids in Arabidopsis
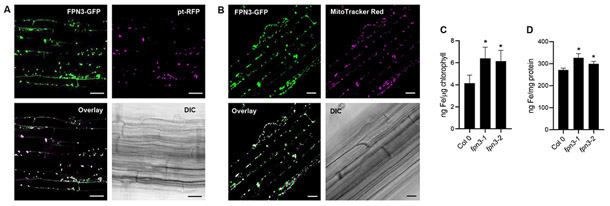
To determine the subcellular localization of FPN3, we generated transgenic Arabidopsis plants stably expressing both 35Sp-FPN3-GFP and 35Sp-pt-RFP, a plastid marker with the transit peptide of rubisco small subunit from tobacco fused to RFP (Nelson et al., 2007). We observed 72% co-localization of FPN3-GFP with the plastid marker (Figure 7A). The FPN3-GFP signal that did not overlap with the plastid marker (Figure 7A) suggested that FPN3-GFP might be dual-targeted. To test the localization of FPN3-GFP in mitochondria, we stained transgenic Arabidopsis plants stably expressing 35Sp-FPN3-GFP with MitoTracker Red, a mitochondrial dye. We verified that 83% of the GFP signal co-localized with the mitochondrial staining (Figure 7B). The total percentage of FPN3-GFP signal that co-localized with mitochondrial or plastid signal exceeds 100% indicating that a portion of the co-localized signal overlaps. Previous reports have revealed frequent co-localization of mitochondria and chloroplasts/plastids (Zhang et al., 2020; Oikawa et al., 2015; Nelson et al., 2007; Sparkes, 2018), which most likely explains the partially overlapping co-localization signal detected in our results. Overall, our data confirm the plastid localization of FPN3-GFP previously reported by Conte et al. (2009) and provide further evidence that FPN3 is dual-targeted to the mitochondria as well.
Figure 7. Subcellular localization of FPN3-GFP and elemental analysis of fpn3 mitochondria and chloroplasts.
(A) Confocal images of an optical section of Arabidopsis root stably co-expressing FPN3-GFP and pt-RFP, and its bright field image. Overlay image of FPN3-GFP with pt-RFP shows co-localization. Scale bars = 25 μm. (B) Confocal images of an optical section of Arabidopsis root stably expressing FPN3-GFP and stained with MitoTracker Red with its bright field image. Overlay image of FPN3-GFP with MitoTracker Red shows co-localization. Scale bars = 25 μm. (C, D) Elemental analysis of chloroplasts and mitochondria isolated from fpn3 shoots. (C) Iron content of chloroplasts measured by ICP-MS and normalized with chlorophyll content. (D) Iron content of mitochondria normalized with mitochondrial protein. Mean values of quadruplicate chloroplast samples and triplicate mitochondrial samples are shown with error bars (SD). Significant differences compared to the wild type, Col 0, are denoted (One-way ANOVA followed by Dunnett’s test; *: p<0.05).
Mitochondria and chloroplasts of fpn3 accumulate iron
Based on the subcellular localization of FPN3 in the mitochondria and chloroplasts (Figure 7A, 7B) and our yeast results that suggest FPN3 is exporting iron from these organelles (Figure 2), we predicted that iron would accumulate in the mitochondria and chloroplasts of fpn3. Thus, we isolated these organelles from fpn3 and wild type shoots. To examine the purity of our samples, Western blots were done with the chloroplasts or mitochondrial samples using antibodies against cytosolic or organellar markers (Figure S7, S8). We then quantified metal accumulation in these organelles using ICP-MS. As predicted, chloroplasts and mitochondria from both fpn3-1 and fpn3-2 contained significantly more iron compared to their counterparts in wild type (Figure 7C, 7D). The accumulation of iron in fpn3 mutant chloroplasts and mitochondria provides strong evidence that supports the role of FPN3 in exporting iron from the mitochondria and chloroplast.
The expression of organellar iron homeostasis genes is altered in the fpn3 mutants
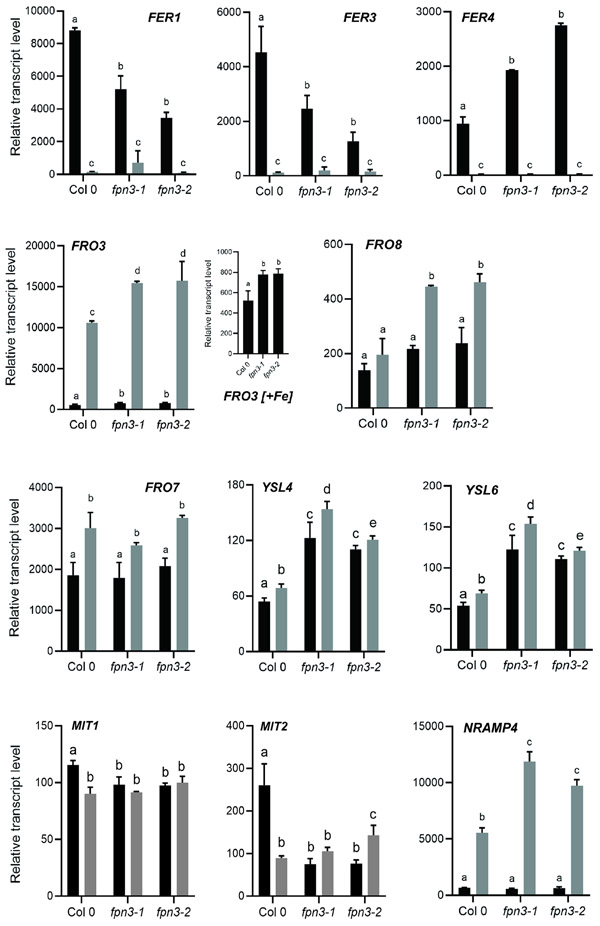
Trafficking of iron between subcellular compartments is crucial for cellular iron homeostasis. Mitochondria and chloroplasts prepared from leaves of fpn3 mutants accumulated more iron (Figure 7C, 7D), whereas leaves of the mutant accumulated less iron compared to wild type (Figure 6). Thus, we predicted that transcript levels of organellar iron responsive genes would be affected in fpn3 mutant lines. We first analyzed the expression of vegetative ferritin genes, FERRITIN1 (FER1), FER3, and FER4, whose transcripts accumulate under iron sufficiency and decrease when iron is low (Petit et al., 2001; Arnaud et al., 2006). We found that transcript levels of FER1 and FER3, which encode plastid-localized ferritins, were significantly lower in iron-sufficient shoots of fpn3 compared to wild type (Figure 8), which is consistent with our results showing less iron accumulation in fpn3 shoots (Figure 6A). The lower steady state level of FER1/3 in fpn3 shoots with more iron in their chloroplasts (Figures 7C and 8) was analogous to the increased FER1 level in pic1, which is unable to import iron into chloroplasts (Duy et al., 2007). In contrast, the expression of FER4, a dual-targeted ferritin that localizes to mitochondria and plastids, was approximately 2 and 2.9-fold higher in fpn3-1 and fpn3-2, respectively, compared to wild type (Figure 8).
Figure 8. Expression of organellar iron homeostasis genes in fpn3 shoots.
Steady-state level transcripts relative to the internal control, ACT2, under iron sufficient (black bars) and iron deficient (grey bars) conditions detected by NanoString. Seedlings were grown in B5 without sucrose for 2 weeks and transferred to iron-deficient or iron-sufficient media for 3 days. Mean values with SD are shown. Significant differences compared to Col 0 samples from the same treatment are denoted with different letters (One-way ANOVA; Dunnett’s test; p<0.05; n=4).
Steady-state expression of two mitochondrial iron importers, MIT1 and MIT2, is higher in iron sufficient conditions than in iron deficient conditions (Jain et al., 2019). In fpn3 mutant shoots, MIT1 and MIT2 transcript levels were significantly lower than in wild type, and their expression was not higher in iron sufficient conditions than in iron deficient conditions (Figure 8). FRO3, which encodes a mitochondrial ferric chelate reductase and has been used as a marker for iron deficiency (Mukherjee et al., 2006), exhibited about 46% and 31% higher expression in iron-deficient shoots of fpn3-1 and fpn3-2, respectively, compared to wild type (Figure 8). Additionally, FRO3 expression was significantly higher in fpn3 shoots than in wild type under iron-sufficient conditions (Figure 8). Along with the expression profiles of FER1 and FER3, elevated FRO3 expression levels in fpn3 shoots is consistent with less cytosolic iron (Figure 6). Expression of another mitochondrial ferric chelate reductase gene, FRO8, was ~30% higher in iron-deficient fpn3 shoots than in iron-sufficient fpn3 shoots and was increased 2-fold in fpn3 shoots compared to wild type shoots under iron-deficient conditions (Figure 8). In contrast to the mitochondrial ferric chelate reductase genes, expression of FRO7, which encodes the chloroplast ferric chelate reductase, was not significantly different between the fpn3 mutants and wild type; FRO7 transcript levels were increased in iron deficiency, but similar across all three genotypes (Figure 8). We noted that the expression of YSL4 and YSL6, which encode transporters that efflux iron-NA from the chloroplasts (Divol et al., 2013) or vacuoles (Jaquinod et al., 2007; Conte et al., 2013), was significantly increased in fpn3 mutant shoots as compared to wild type (Figure 8). NATURAL RESISTANCE-ASSOCIATED MACROPHAGE PROTEIN 4 (NRAMP4) encodes a vacuolar iron effluxer and is induced by iron deficiency (Lanquar et al., 2005), and its expression was significantly higher in iron-deficient shoots of fpn3 compared to wild type (Figure 8). This observation is in accordance with the expression profiles of FER1, FER3 and FRO3 in shoots (Figure 8), which indicated that fpn3 shoots have less iron than wild type shoots, and our elemental analysis data, which revealed less iron accumulation in fpn3 shoots compared to wild type shoots (Figure 6).
fpn3 mitochondria exhibit abnormal morphology under iron deficient conditions
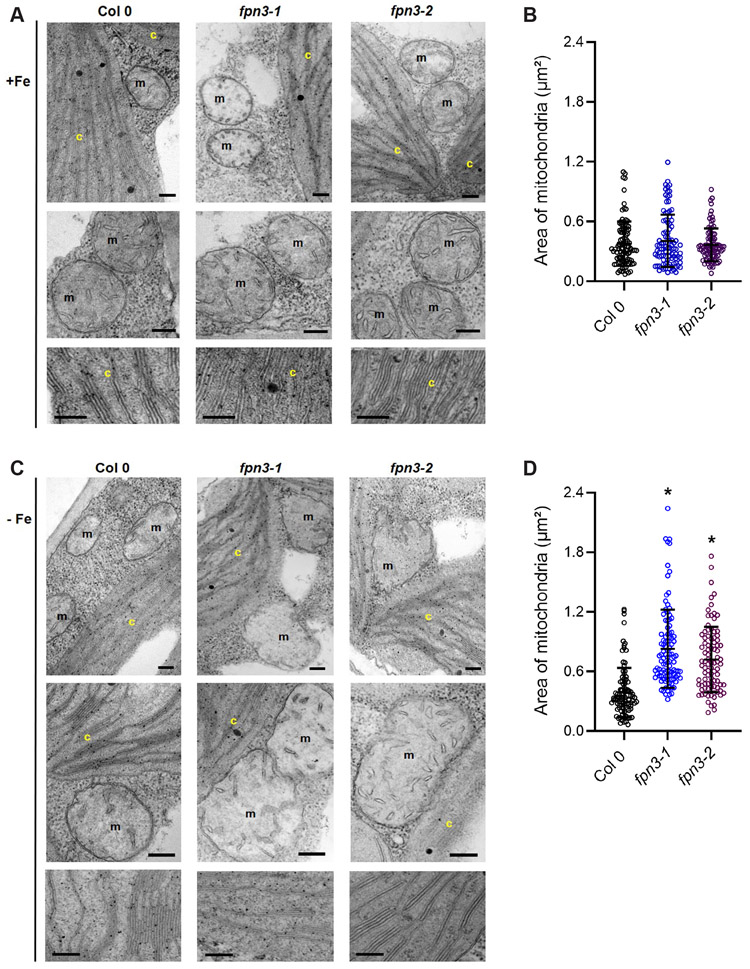
Abnormal chloroplast ultrastructure has been observed in mutants defective in iron metabolism of multiple plants species grown under iron deficiency (Bogorad et al., 1959; Duy et al., 2007; Platt-Aloia et al., 1983; Stocking, 1975; Vigani et al., 2015). In addition to chloroplasts, the ultrastructure of mitochondria has also been shown to be affected by iron deficiency in plants (Pascal and Douce, 1993; Vigani et al., 2015). Therefore, we examined the ultrastructure of chloroplasts and mitochondria in fpn3 and wild type leaves using transmission electron microscopy (TEM). Our TEM images revealed that mitochondria of iron-deficient fpn3 mutants were enlarged and the outer and inner membranes appeared deformed (Figure 9C), but such phenotypes were not detected under iron sufficient conditions (Figure 9A). By comparing the quantified area of mitochondria, we determined that fpn3 mitochondria were significantly larger than wild type mitochondria under iron deficient conditions (Figure 9D), but not under iron sufficient conditions (Figure 9B). In contrast, no discernable differences were observed in the chloroplasts of fpn3 and wild type; chloroplast morphology and thylakoid ultrastructure were similar in both fpn3 lines and the wild type under both iron sufficient and deficient conditions (Figure 9A, 9C). Overall, the iron-dependent mitochondrial ultrastructure phenotype (Figure 9) strongly suggests that FPN3 function is critical for mitochondria under iron deficient conditions.
Figure 9. Mitochondria and chloroplast ultrastructure of fpn3.
(A) TEM images of the wild type, Col 0, fpn3-1, and fpn3-2 leaf sections with mesophyll cells from iron sufficient conditions. Close-up images of mitochondria (middle row) and thylakoids (bottom row) are shown. (B) Area of mitochondria from iron sufficient conditions. data point represents the quantified area from each mitochondrion section, and the lines represent mean values with SD. Ninety-four to 105 mitochondria from 38 to 41 cells were quantified. (C) TEM images of iron deficient wild type, fpn3-1, and fpn3-2 leaf sections with mesophyll cells. Chloroplasts (c) and mitochondria (m) are labeled in the images. Scale bars = 200 nm. (D) Area of mitochondria from iron deficient conditions. Each data point represents the quantified area from each mitochondrion section, and the lines represent mean values with SD. Ninety-six to 113 mitochondria from 40 to 57 cells were quantified. (one-way ANOVA; Tukey’s multiple comparison test’ *: p<0.01).
DISCUSSION
Unraveling the physiological role of FPN3
FPN3 was previously reported as MAR1 and proposed to opportunistically transport aminoglycoside class antibiotics (Conte et al., 2009; Conte and Lloyd, 2010), but this study adds multiple pieces of information that is critical to understanding the physiological role of FPN3/MAR1.
FPN3 is dual-targeted to mitochondria and chloroplasts:
Our results show that FPN3 is dual-targeted to the mitochondria and chloroplasts, and plays a crucial role in the mitochondria. Conte et al (2009) reported that MAR1 was targeted to chloroplasts based on observations in Arabidopsis protoplasts transiently expressing YFP fusions of MAR1 or its putative transit peptide. Here, we observed that FPN3-GFP is co-localized with mitochondrial and plastid markers in stable transgenic lines (Figure 7A, 7B). As mitochondria and chloroplasts/plastids are frequently observed to colocalize (Zhang et al., 2020; Oikawa et al., 2015; Nelson et al., 2007; Sparkes, 2018), mitochondrial localization may not have been previously detected due to the high density of chloroplasts in the protoplasts imaged by Conte et al (2009). Moreover, our study reveals that iron accumulates in both mitochondria and chloroplasts of fpn3 (Figure 7C, 7D). The abnormal mitochondrial morphology detected in iron deficient fpn3 mutant leaves (Figure 9) indicate that FPN3 plays an essential role in maintaining iron homeostasis in the mitochondria. It is also possible that the chloroplasts may be better equipped with mechanisms that compensate for defective FPN3 function under iron deficient conditions. For example, given the proposed function of YSL4/6 in chloroplasts (Divol et al., 2013), we speculate that YSL4/6 might be compensating for the loss of FPN3 in plastids. Alternatively, based on their proposed role in the vacuole (Conte et al., 2013), the increased YSL4/6 transcript levels in fpn3 is consistent with the higher level of NRAMP4 expression observed in fpn3 (Figure 8). The presence of three ferritins in chloroplasts (FER1, FER3 and FER4) and only one in the mitochondria (FER4) also implies that fpn3 mitochondria might be more susceptible to iron accumulation than fpn3 chloroplasts.
FPN3 is an iron exporter of mitochondria/chloroplasts:
Our results show that FPN3 exports iron from mitochondria and chloroplasts and most likely transports ferrous iron ions. Conte et al (2009) speculated that MAR1 may be importing the polyamine iron chelator, nicotianamine (NA), based on its structural similarity to aminoglycoside class antibiotics. However, the presence of the highly conserved iron ion binding residues in FPN3/IREG3 (Figure 1) and the iron phenotypes of yeast cells expressing FPN3 that are analogous to yeast cells expressing MMT1/2 (Figure 2) strongly support the idea that ferrous iron ions are FPN3/IREG3 substrates. Compared to yeast cells expressing MMT1/2, the growth phenotype and enzymatic activities of yeast cells expressing FPN3 were not as dramatically different from the negative control (Figure 2). This is most likely because the MMT1/2 plasmid we used expresses both MMT1 and MMT2 (Li et al., 2014), whereas FPN3 alone was being expressed in a heterologous system in our assays. Moreover, it is not unusual to observe partial complementation with a heterologous gene (Morrissey et al., 2009; Zhai et al., 2014). In addition to the yeast data (Figure 2), iron accumulation in isolated chloroplasts and mitochondria quantified by ICP-MS (Figure 7C, 7D) supports the role of FPN3/IREG3 as an iron exporter of mitochondria and chloroplasts. A recent study speculated that BnMAR1, the FPN3/IREG3 ortholog in Brassica napus, might be involved in iron release from the plastids based on the correlation between its gene expression profile and chloroplast iron content (Pham et al., 2020).
FPN family members identified to date transport iron from the cytoplasm. As metal transporters of the same family generally retain the direction of transport in respect to the cytoplasm, an organellar FPN is likely to transport iron into the organellar lumen. According to the endosymbiotic origin of mitochondria and chloroplasts (Roger et al., 2017; Zimorski et al., 2014; Yoon et al., 2004), it is possible that proteins of these organelles could retain their ancestral directionality as seen in respiratory proteins (Nirody et al., 2020). Under the hypothesis that FPN3 is of bacterial origin, its directionality is consistent with iron export from its ancestral cytoplasm, i.e., from the organellar lumen into the eukaryotic cytoplasm. Based on the subcellular localization of FPN3 (Figure 7A, 7B) and the endosymbiotic origin of mitochondria and chloroplasts (Roger et al., 2017; Zimorski et al., 2014; Raven and Allen, 2003; Rockwell et al., 2014), it is reasonable to speculate that FPN3 diverged from a bacterial ancestor. Prokaryotic ferroportins have not been as extensively studied as their vertebrate orthologs, but a bacterial FPN from Bdellovibrio bacteriovorus has been reported (Bonaccorsi di Patti et al., 2015; Taniguchi et al., 2015).
Although the mechanism of transport via FPN has not been fully understood, structural and biochemical studies indicated that the Bdellovibrio bacteriovorus FPN3 ortholog, BdFPN, transports iron and other divalent cations along their concentration gradient in a uniporter-like manner (Taniguchi et al., 2015). FPN2/IREG2 transports iron as shown by complementation tests with the yeast Δccc1 mutant (Morrissey et al., 2009), but has also been shown to transport nickle and cobalt based on experiments in yeast and plants (Schaaf et al., 2006; Morrissey et al., 2009). We noted that the content of other essential metals, such as manganese, zinc, and copper, was also affected in fpn3 chloroplasts and mitochondria (Figure S9), which suggests that FPN3 might transport other metals in addition to iron. In particular, zinc accumulation was significantly higher in both mitochondria and chloroplasts of fpn3 than in wild type (Figure S9). While manganese and copper accumulation in fpn3 mitochondria and chloroplasts was also significantly different from that of wild type, this may reflect secondary effects because the pattern of changes was not consistent between the two organelles (Figure S9). At the tissue level, we did not observe significant differences in the distribution or content of zinc, manganese, or copper. SXRF indicated that zinc and manganese distribution in the leaves was similar in fpn3 and wild type (Figure S5), and accumulation of zinc, manganese, and copper in the roots and shoots of fpn3 was comparable to that of wild type (Figure S6). Further studies will be necessary to test if other metals could also be transported by FPN3.
Differential regulation of FPN3 in roots and shoots:
We show that FPN3 is constitutively expressed regardless of the iron conditions in the shoots, whereas its transcript levels are increased by iron deficiency in the roots. We note that our data contradict the results of Conte et al (2009), who reported the down-regulation of MAR1 expression in iron deficient seedlings by RT-qPCR. However, multiple transcriptomics datasets corroborate our results and show that FPN3/IREG3 transcript level is higher in wild type roots under iron deficiency (Mai et al., 2016; Dinneny et al., 2008; Yang et al., 2010; Park et al., 2019; Khan et al., 2018; Buckhout et al., 2009) but is not iron-regulated in shoots (Khan et al., 2018; Rodríguez-Celma et al., 2013; Park et al., 2020). The differential expression of FPN3 in roots and shoots under iron-deficient or iron-sufficient conditions (Figure 3G-L) implies that FPN3 might play different physiological roles in roots and shoots via differentially regulating mitochondrial iron trafficking in those tissues. A previous study has provided evidence that mitochondrial iron status could distinctively impact iron regulation in roots and shoots by revealing that transcript and metabolic profiles of roots and shoots were differentially affected by the induction of local iron deficiency in the mitochondria (Vigani et al., 2016).
Proposed role of FPN3 in the regulation of iron in roots and shoots
We speculate that FPN3 may be releasing iron into the root cytoplasm so that it can be reallocated within the cell to help cope with iron deficiency and to adjust metabolic needs of the plant under iron deficiency. As a response to iron deficiency, metabolic pathways in plants, including those that occur in chloroplasts and mitochondria, are coordinately remodeled in both roots and shoots (López-Millán et al., 2013; Thimm et al., 2001; Rodríguez-Celma et al., 2013). Our split root data revealed that the transcript level of FPN3 in roots is primarily regulated in response to the local availability of iron (Figure 4). However, in the split-root transferred from iron sufficient to sufficient conditions, a 2-fold increase in FPN3 transcript level was detected (Figure 4A) and FPN3 expression was not significantly reduced 12 hours after split roots grown in iron deficient medium was transferred to iron sufficient medium (Figure 4B). This observation implied that FPN3 expression in split roots exposed to iron sufficient local environments may be partially regulated by iron deficiency perceived in the other half of the split root via a systemic response.
In the shoots, FPN3 is constitutively expressed regardless of the plant iron status (Figure 3G-L) and throughout development according to transcriptomic datasets (Winter et al., 2007). FPN3 might be releasing iron to protect from iron-induced oxidative stress in the shoots. It has been proposed that plant ferritins play a major role in preventing iron-induced oxidative stress (Ravet et al., 2009). FER4 encodes a dual-targeted ferritin that localizes to mitochondria and plastids. FER4 is the only mitochondrial ferritin, as mitochondria of fer4 cells are devoid of ferritins (Tarantino, Santo, et al., 2010). The pronounced increase in FER4 transcripts in fpn3-1 and fpn3-2 (Figure 8) is likely caused the higher level of iron accumulating in the mitochondria and chloroplasts of these mutants (Figure 7C, D).
Although FPN3 does not appear to be iron-regulated at the transcriptional level in the shoots, the abnormal mitochondria morphology detected in iron deficient fpn3 leaves suggest a critical role for FPN3 in the mitochondria under iron deficiency (Figure 9). Because FRO8 is not regulated by the iron status in wild type (Figure 8; Mukherjee et al., 2006), the higher level of FRO8 in iron-deficient fpn3 shoots (Figure 8) may also imply a dysregulation of iron-dependent mitochondrial function. While the mechanism behind the swollen mitochondria of iron deficient fpn3 shoots remains to be understood, enlarged mitochondria have been reported in an early study with iron-deficient rice seedlings (Mori et al., 1991), and impaired ion homeostasis is known to affect mitochondrial volume (Szabo and Zoratti, 2014; Teardo et al., 2015). Swollen mitochondria have also been observed upon anaerobic treatment, decreased metabolism, or oxidative stress (Vartapetian et al., 2003; Yoshinaga et al., 2005; Lee et al., 2002). We postulate that oxidative stress or metabolic constraints caused by dysregulated iron homeostasis in iron deficient fpn3 mitochondria might have resulted in the compromised mitochondrial ultrastructure. We further speculate that the reduced copper level in fpn3 mitochondria (Figure S9B) may indicate that fpn3 mitochondria are more susceptible to oxidative stress than fpn3 chloroplasts, which did not exhibit morphological differences (Figure 9A, 9C) and contained more copper than wild type chloroplasts (Figure S9A). A recent study has shown that oxidative stress in copper-deficient plants was associated with the accumulation of reactive oxygen species in the mitochondria due to impaired cytochrome c function (Ishka and Vatamaniuk, 2020).
Steady state transcript levels of FPN1 in fpn3 shoots under both iron sufficient and deficient conditions were increased, while FPN1 expression is decreased in iron deficient fpn3 roots (Suppl Fig 10). These results imply that FPN1 might contribute to reducing shoot iron content by limiting iron loading into the root xylem and increasing iron efflux from the plasma membrane in the shoots. The expression of FPN2, the other FPN paralog, was not affected in fpn3 (Suppl Fig 10). Further studies will be necessary to explain how iron accumulation in fpn3 mitochondria and chloroplasts leads to less iron at the tissue level. Nevertheless, the observations from rice mit-2, a viable mutant with reduced expression of the rice mitochondrial iron importer, OsMIT, are in line with the idea that mitochondrial iron might affect iron regulation at the tissue level; this mutant contained more iron in its shoots despite accumulating less iron in their mitochondria (Bashir et al., 2011).
Notably, steady state transcript levels of the Arabidopsis mitochondrial iron importer genes, MIT1 and MIT2, exhibit expression profiles opposite to that of FPN3. In the roots, MIT1 and MIT2 expression is not iron-regulated but is higher than in the shoots (Jain et al., 2019), whereas moderate increase of MIT1/2 expression was detected in iron sufficient shoots (Figure 8; Jain; et al. 2019). Expression of the mitochondrial iron importer, OsMIT, was also decreased in iron deficient shoots (Bashir et al., 2011). The contrasting expression profiles of MITs and FPN3 led us to postulate that shoot mitochondria might prioritize to export iron regardless of the iron status and only import iron when it is sufficiently available, whereas root mitochondria steadily import iron and only export it when the plant is iron deficient. We further speculate that the intricate balance of mitochondrial iron trafficking, particularly iron export by FPN3, is necessary for optimal iron regulation at the tissue level. This idea agrees with our observation that iron content was reduced in fpn3 shoots and in iron-deficient fpn3 roots where FPN3 is highly expressed, whereas iron accumulation was not affected in iron sufficient roots where FPN3 is expression is low (Figure 3L, Figure 6B).
Implications of a mitochondrial iron exporter
Mitochondrial metabolism was found to be less affected than chloroplast metabolism under mild iron deficiency (Hantzis et al., 2017). However, it is evident that improper FPN3 function appears to have a greater impact on the mitochondria than chloroplasts (Figure 9). The mitochondrial phenotype of fpn3 signifies the importance of iron export from the mitochondria under iron deficient conditions. It has been well-established that chloroplasts are the most iron-rich organelle in plant cells and vacuoles serve as the major iron storage compartment, and several studies have provided evidence that mitochondria can contribute as an additional intracellular iron reservoir. In yeast Δccc1 mutant cells that lack the vacuolar iron importer Ccc1, overexpressing MRS3 and MRS4, which encode mitochondrial iron importers of the mitoferrin family, suppressed iron toxicity (Lin et al., 2011). Furthermore, MMT1 and MMT2, which belong to the cation diffusion facilitator (CDF) family, export mitochondrial iron and overexpression of MMT1/2 results in multiple phenotypes that support the idea that mitochondria could function as an iron storage organelle (Li et al., 2014). It is noteworthy that mitochondrial iron import mediated by mitoferrins is common across multiple organisms, such as in yeast, mammals, worm, fruit fly, rice, and Arabidopsis (Paradkar et al., 2009; Shaw et al., 2006; Metzendorf et al., 2009; Ren et al., 2012; Bashir et al., 2011; Mühlenhoff et al., 2003; Jain et al., 2019). Unlike mitochondrial iron import, we speculate that mechanisms to export mitochondrial iron are more diverse. FPNs do not exist in yeast and mitochondrial FPNs have not been found in vertebrates or invertebrates, to our knowledge. In mammalian mitochondria, an ATP-binding cassette (ABC) transporter, ABCB8, is involved in exporting iron (Ichikawa et al., 2012). The presence of a mitochondrial iron exporter is necessary, but not sufficient to suggest that mitochondria might function as a subcellular iron store. Nevertheless, our work shows that FPN3 is regulating mitochondrial iron and implies a potential role of mitochondria as an iron reservoir. It is possible that fpn3 mutants might be increasing FER4 to protect from iron toxicity due to the lack of an iron exporter. The higher expression of FER4 in fpn3 than in wild type (Figure 8) is also consistent with the view that mitochondria could store iron. We postulate that mitochondrial ferritin may play a more dynamic function to assist iron storage in addition to protecting mitochondria from iron-induced oxidative damage.
In conclusion, our work has advanced our understanding of the physiological roles of FPN3 by providing evidence that FPN3 is an iron exporter dual-targeted to mitochondria and chloroplasts. Moreover, our TEM results indicate that FPN3 function is more crucial for mitochondria. Further studies are necessary to elucidate the mechanisms underlying the phenotypes observed and the potential effect of FPN3 on mitochondrial and chloroplast metabolism and oxidative stress. Future work should also address potential crosstalk among chloroplasts, mitochondria, and vacuoles to comprehensively understand organellar iron trafficking and homeostasis in plant cells.
MATERIALS AND METHODS
Yeast high iron growth assays
For growth tests in high iron media, pRS315-FPN3, pRS315-MMT1/2, or pRS315 vector alone was transformed into Δccc1Δmmt1/2 (Li et al., 2014). The Δccc1Δmmt1/2 strain, the MMT1/2 plasmid, and yeast vectors were kind gifts from Jerry Kaplan. For spot assays, serial dilutions of overnight yeast cultures were spotted onto SD-Leu plates with appropriate auxotrophic supplements containing 2 mM iron (2 mM ferrous ammonium sulfate hexahydrate). The negative control plates contained media with no added iron. Cells growth at 30°C was monitored over 2-5 days. For growth assays in liquid culture, overnight yeast cultures were inoculated to a starting OD600 of 0.05 into SD-Leu with or without 2 mM iron supplementation and grown in a shaker incubator at 30°C, and cell density was measured at OD600 at the indicated time points.
Aconitase assay
Wild type DY150 cells were transformed with FPN3, MMT1/2, or pRS315 vector alone. These transformants were inoculated in appropriate selective media (SD-Leu), grown overnight at 30°C, and sub-cultured in SD-Leu supplemented with 250 μM FeSO4 until the cells reached mid-log phase. The subcultures were harvested and washed with cold phosphate-buffered saline (PBS), and protein lysate was prepared using PBS with protease inhibitor (Pierce) via homogenization with glass beads for 10 minutes at 4°C. Samples used in the aconitase assay were normalized with total protein concentration. Aconitase activity was assayed and quantified using the BioAssay Systems EnzyChrom Aconitase Assay Kit, following manufacturer’s instructions.
c-GDO assay
Wild type DY150 cells were co-transformed with FPN3, MMT1/2, or pRS315 and the c-GDO plasmid, and grown as described above for the aconitase assay. Cell lysates were prepared with these transformants by glass bead homogenization, and GDO activity was assayed as described by Li et al (2014). The assay reaction consisted of 20 mM Tris-HCl (pH 8.0) and 0.1 mM 2,3-dihydroxy-benzoic acid (gentisic acid) as the substrate. The enzyme activity was determined based on the absorbance measured at 340 nm using an extinction coefficient of 10.2 cm−1mm−1.
Plant materials and growth conditions
Two independent alleles of fpn3 T-DNA insertion lines, fpn3-1 (SALK_034189) and fpn3-2 (SALK_009286), were obtained from the Salk collection (Alonso et al., 2003). RT-qPCR verified that FPN3 expression was drastically reduced in these lines (Figure S4). Arabidopsis plants were grown at 22 °C under a 16/8-hour light/dark cycle in the Conviron A1000 growth chamber.
Plant growth test under iron-deficient conditions
For alkaline soil tests, seedlings were germinated and grown in normal soil (Pro-Mix; pH ~5.8), alkaline soil (pH ~8), or alkaline soil watered with soluble iron (0.5 mM Fe-EDDHA). Alkaline soil was prepared by adding approximately 1.3 g of calcium oxide per liter of soil solution (Kim et al., 2006). For growth tests on plates, seedlings were germinated vertically for 12 days on either iron-deficient media (Murashige and Skoog (MS) without iron (Caisson MSP33) supplemented with 300 μM ferrozine) or iron-sufficient media (MS with 100 μM FeNa-EDTA (Caisson MSP34). Seedlings were photographed, and ImageJ was used to quantify root lengths. Shoots were separated and weighed on the MT5 Analytical Microbalance (Mettler Toledo).
Generation of FPN3p-GUS transgenic lines and histochemical staining
A fragment containing 890 bp of upstream sequence and the FPN3 start codon was amplified from a BAC clone, F2P16 (GenBank AF007270), obtained from the Arabidopsis Biological Resource Center (ABRC) using primers 5’-cacccactttctcttttgttagattctagttg-3’ and 5’-cattctataaat-tgattctcctcttctcc-3’ and cloned into pENTR/D-TOPO (Invitrogen). The construct was cloned into pGWB533 (Nakagawa et al., 2007) using Gateway LR Clonase II (Invitrogen). The resulting FPN3p-GUS construct was moved into Agrobacterium tumefaciens strain LBA4404 and transformed into wild type Col 0 plants using the floral dip method (Clough and Bent, 1998). pGWB533 was from Tsuyoshi Nakagawa (Addgene plasmid #74872; RRID: Addgene_74872).
For histochemical staining, seedlings or tissues of FPN3p-GUS plants were fixed and incubated with 5-bromo-4-chloro-3-indolyl β–D-glucuronide as described by (Jefferson et al., 1987). At least five seedlings from three independent T2 or T3 lines were examined, and images were obtained using the LeicaM165 FC dissecting scope and Leica LAS EZ 3.4.0 software.
Split root assays
For split-root samples for RT-qPCR, plants were prepared following the protocol described by Kumar et al. (2017) with slight modifications. Wild type plants were grown for 5 days on iron-sufficient medium, cut at the primary roots and grown for 7 days to induce split roots, and transferred to iron-deficient or iron-sufficient medium for 3 days. The plants were then washed with 0.1M citrate buffer and moved to split plates, in which one of the split roots was placed on iron-deficient medium and the other root on iron-sufficient medium. Plants were kept on split plates for 0, 6 and 12 hours before harvesting roots for RNA preparation.
Generation of FPN3-GFP transgenic lines
The coding region of FPN3 was cloned from a BAC clone, F2P16 (GenBank AF007270), obtained from the Arabidopsis Biological Resource Center (ABRC) using primers 5’-caccatggttgtttcaatggctttgg-3’ and 5’-atttgagagagggtcgaaggag-3’ and cloned into pENTR/D-TOPO (Invitrogen). The final construct, 35Sp-FPN3-GFP, was cloned into pGWB505 (Nakagawa et al., 2007) using Gateway LR Clonase II (Invitrogen). 35Sp-FPN3-GFP and 35Sp-pt-RFP were co-transformed into Agrobacterium tumefaciens strain LBA4404, and then into wild type Col 0 using the floral dip method (Clough and Bent, 1998). pGWB505 was a gift from Tsuyoshi Nakagawa (Addgene plasmid #74847; RRID: Addgene_74847) and 35Sp-pt-rb (Nelson et al., 2007) was obtained from ABRC (stock #CD3-1000).
Confocal microscopy
Arabidopsis roots stably expressing 35Sp-FPN3-GFP were imaged with the Nikon A1 Spectral Detector Confocal with FLIM Module at the Light Microscopy Core, Institute for Applied Life Science at University of Massachusetts, Amherst. For mitochondrial co-localization roots were stained with MitoTracker Red FM (Invitrogen) to label the mitochondria prior to imaging, by incubating roots in a final concentration of 1 μM MitoTracker Red FM for 30 minutes. Z-stack images were taken with FITC and TRITC channels. Images were collected and processed with NIS-Elements software.
Chloroplast isolation
Chloroplast isolation was conducted following (Smith et al., 2003). Arabidopsis plants were grown for 4 weeks on soil. Roughly 10 g of shoot tissue was harvested, rinsed in digestion buffer (20 mM MES-KOH, pH 5.2, 400 mM sorbitol, 0.5 mM CaCl2), and incubated in digestion enzyme solution (10 ml digestion buffer, 0.04 g Macerozyme R-10 (bioWORLD), 0.2 g Cellulase (bioWORLD) for 3.5 h under light. During digestion, a 40%:85% AT Percoll step gradient was prepared on ice. Upon completion of digestion, the solution was filtered through cheesecloth to harvest protoplasts. To isolate protoplasts, the filtered solution was suspended in digestion buffer and centrifuged at 100 g for 5 min at 4°C, the supernatant was discarded, then the isolated protoplasts were resuspended in protoplast resuspension buffer (20 mM MES-KOH, pH 6.0, 400 mM sorbitol, 0.5 mM CaCl2) before centrifuging at 100 g for 2 min at 4°C and removing supernatant. Next, protoplasts were resuspended in protoplast breakage buffer (20 mM tricine-KOH, pH 8.4, 300 mM sorbitol, 5 mM EDTA, 5 mM EGTA, 10 mM NaHCO3, 0.05 g BSA added per 50 mL of solution before use) and immediately filtered through a protoplast rupturing device (a syringe tube with the end cut off, with a 20 μm and 10 μm mesh, respectively, attached via electrical tape). Next, the broken chloroplasts were layered onto the 40%:85% AT Percoll step gradient and centrifuged at 2500 g for 10 min at 4°C, with the brake off. A green band was visualized at the 40%85% AT Percoll interface and harvested. The resulting solution was diluted in HEPES-sorbitol buffer, pH 8.0 (50 mM HEPES-KOH, pH 8.0, 330 mM sorbitol) and centrifuged at 700 g for 5 min at 4°C. After removing the supernatant, resulting chloroplasts were resuspended in HEPES-sorbitol buffer, pH 8.0.
Chlorophyll content (chlorophyll a and b) was quantified with the NanoDrop One Spectrophotometer (ThermoFisher) following manufacturer’s instructions. The following equations were used to quantify chlorophyll a and chlorophyll b content, respectively: Ca = 14.85A666 – 5.14A650; Cb = 25.48A650 – 7.36A666 (Porra et al., 1989; Barnes et al., 1992). Purity was confirmed via Western blots with organellar markers.
Mitochondrial isolation
Mitochondria were isolated from four-week-old Arabidopsis leaves using a protocol adapted from (Keech et al., 2005). Prior to the isolation, 30mL of continuous 50% Percoll gradient (50% (v/v) Percoll, 0.3M sucrose, 10 mM TES, 1 mM EDTA, 10 mM KH2PO4, 1 mM glycine, pH adjusted to 7.5 with KOH) was centrifuged at 39000 g for 40 min and kept at 4°C. Roughly 40-50g of tissue was homogenized in pre-chilled grinding buffer B (0.3M sucrose, 60 m TES, 2 mM EDTA, 10mM KH2PO4, 25 mM tetrasodium pyrophosphate, 1 mM glycine, 1% (w/v) polyvinyl-pyrrolidone-40, 1% (w/v) defatted bovine serum albumin (BSA), with 50 mM sodium ascorbate and 20 mM cysteine added and readjustment of the pH to 8.0 with KOH just prior to grinding) using a hand blender, filtered through a 20-μm nylon mesh, and centrifuged at 2500 g for 5 minutes to remove intact chloroplasts and thylakoid membranes. The supernatant was centrifuged at 15000 g for 15 minutes. The pellet obtained was resuspended in wash buffer B (0.3M sucrose, 10mM TES, 10mM KH2PO4, pH adjusted to 8.0 with KOH) and gently homogenized in a chilled glass homogenizer. The resuspended pellet was then layered on top of the 50% Percoll gradient and centrifuged at 15000 g for 20 minutes; each 30 mL density gradient has a maximum load of mitochondria from 30g of fresh leaf tissue. After centrifugation, the mitochondria formed a whitish band near the bottom of the tube. This band was aspirated and resuspended in 0.2-0.3 mL of wash buffer B to obtain a roughly 20-fold dilution. All procedures were conducted in the cold room or at 4°C. Mitochondrial protein was quantified using DC Protein Assay (Bio-Rad).
SXRF Imaging
The spatial distribution of iron, manganese, and zinc in hydrated leaf tissues was imaged via SXRF at the F3 station, a bending magnet beamline with multilayer monochromator, at the Cornell High Energy Synchrotron Source. Seeds were directly germinated on soil and grown at 22°C, 14 h light/10h dark photoperiod at a photo flux density of 110 μmol/m2/sec. The nutrient solution contained the following components: 1.25 mM KNO3, 0.625 mM KH2PO4, 0.5 mM MgSO4, 0.5 mM Ca(NO3)2, 10 μM Fe(III)-N,N’-Bis(2-hydroxybenxyl)ethylene-diamine-N, N’-diacetic acid (HBED), a water soluble iron chelate, 17.5 μM H3BO3, 3.5 μM MnCl2, 0.125 μM CuSO4, 0.25 μM ZnSO4, 0.05 μM Na2MoO4, 2.5 μM NaCl, and 0.0025 μM CoCl2. The hydroponic solution was changed every week. Data were analyzed and processed as described in Yan et al 2017. Briefly, the first true leaves from 22 day-old plants were detached immediately prior to imaging, placed in the wet chamber made between two layers of metal-free Kapton™ film and mounted onto 35 mm slide mounts. The 2D raster maps were acquired using a focused, monochromatic incident x-ray beam at 12.8 keV and photon flux of approximately 3x1010 photons/sec. Samples were scanned at the resolution of 30 x 30 μm and acquisition time of 0.2 sec per data point. These settings did not cause damage to plant tissues within 6-8h scans required for analysis of the full set of genotypes. Element-specific x-ray fluorescence was detected using a Vortex ME-4 Silicon Drift Detector (SDD). Calibration of XRF equipment were done using a uniform thin metal film standard during each experiment. Data were processed with Praxes, a software that employs PyMca libraries in batch mode (Solé et al., 2007).
ICP-MS analysis
Shoot iron content of Col 0, fpn3-1, and fpn3-2 was measured by ICP-MS and normalized with dry weight. Plants were grown following the same procedure as the growth test on low iron plates. Dried shoot, root, chloroplast, and mitochondrial samples of Col 0, fpn3-1, and fpn3-2 were digested with nitric acid and analyzed with the Perkin-Elmer NexION 350D ICP-MS at the Mass Spectrometry Core, Institute for Applied Life Science at University of Massachusetts, Amherst. Metal content was determined by normalizing the sample metal concentration (parts per billion; ppb) with the dry mass (mg; shoot and root tissue), chlorophyll content (μg; isolated chloroplast) or total protein (μg; isolated mitochondria) of each sample.
Gene expression analyses
Total RNA isolated with the Agilent Plant RNA Minikit from root or shoot tissue of plants grown under iron-deficient or iron-sufficient conditions were used for gene expression analyses. The quantity and purity of RNA were examined using NanoDrop One (Thermo Scientific), and the integrity of RNA was assessed by electrophoresis using a bleach gel (Aranda et al., 2012). For RT-qPCR, first strand cDNA was synthesized from total RNA using Quanta qScript, and qPCR was conducted using Bio-Rad SYBR Green Supermix in CFX-Connect Real-Time PCR Detection System (Bio-Rad). The relative transcript level of genes was calculated following the ΔCt method (Schmittgen and Livak, 2008), using ACTIN2 (ACT2) as an internal control. Primers were designed using QuantPrime (Arvidsson et al., 2008) and the sequences are listed in Table S1. For gene expression analysis with Nanostring (Geiss et al., 2008), total RNA was hybridized with a custom CodeSet that included probes specific to iron homeostasis genes of interest and internal control genes, ACT2 and UBC, which was designed by NanoString Technologies. Gene expression analysis was conducted with the NanoString MAX/FLEX nCounter (NanoString Technologies) at Dartmouth College and nCounter SPRINT Profiler System (NanoString Technologies) at the Molecular Genetics Core Facility at Boston Children’s Hospital following the manufacturer’s instructions and processed using nSolver 4.0. The custom CodeSet sequences are listed in Table S 2.
Transmission electron microscopy (TEM)
Excised cotyledons of Arabidopsis seedlings treated under iron sufficient or deficient conditions were fixed by immersing them in 2.5% glutaraldehyde/ 2% paraformaldehyde in 0.1 M Na Cacodylate buffer (pH 7.2) and incubating them under vacuum for 30 min. After this primary fixation, the samples were rinsed three times in fresh fixation buffer for 10 min. each time and were secondarily fixed with 1.0% osmium tetroxide in ddH2O (w/v) for 1hr at room temperature. The samples were then rinsed again three times in ddH2O and then placed in 1% aqueous uranyl acetate (w/v) in the refrigerator overnight (tertiary fixation). After three more rinses in ddH2O, the samples were then dehydrated through a graded series of ethanol (10%, 30%, 50%, 75%, 85%, 95%, and 100% for 3 changes). Infiltration was accomplished by running the samples through ethanol 100%: Spurr’s resin-hard formulation (75:25 / V:V) for 1 h at room temperature, then to ethanol 100%: Spurr’s resin-hard formulation (50:50 / V:V) and finally to ethanol 100%: Spurr’s resin-hard formulation (25:75 / V:V) overnight. The following day the samples were transferred through 5 changes of 100% Spurr’s epoxy resin each 1 hr long, and then placed in molds and polymerized for two days at 68° C. Ultrathin sections (approx. 70 nm thick) were collected onto 200 mesh copper support grids and contrasted with Uranyl Acetate and Lead Citrate and then imaged using a CM 10 transmission electron microscope, under 100Kv accelerating voltage. Images were recorded with a Gatan Erlangshen CCD Digital camera.
Supplementary Material
Figure S1. Mitochondrial localization of FPN3-FLAG in yeast
Figure S2. Heterologous expression of FPN3 in Δmrs3/4
Figure S3. FPN3 expression in split root control seedlings transferred to the same conditions.
Figure S4. FPN3 T-DNA insertion lines and quantification of growth phenotypes
Figure S5. Elemental map of manganese and zinc in fpn3 leaves.
Figure S6. Manganese, zinc, and copper content in roots and shoots
Figure S7. Western blots with isolated chloroplasts.
Figure S8. Western blots with isolated mitochondria.
Figure S9. Manganese, zinc, and copper content in chloroplasts and mitochondria
Figure S10. FPN1 and FPN2 expression in wild type and fpn3
Methods S1. Western blots
Table S1. RT-qPCR primer sequence
Table S2. NanoString custom codeset target sequence
ACKNOWLEDGEMENTS
We are grateful to Mary Lou Guerinot and Jerry Kaplan for feedback on this manuscript. We also thank Jerry Kaplan for the yeast strains and plasmids used in this study, Liangtao Li for the yeast Western blot and help with aconitase assays, Elsbeth Walker for advice on the split root experiments, Inhwan Hwang for the pCamV3-GFP and F1-ATPase-RFP plasmids, James Chambers for assistance with confocal microscopy, Britney Privett for assistance with NanoString, Steve Eyles for help with ICP-MS, and Lara Strittmatter and UMass Medical School Electron Microscopy Facility for assistance with TEM. We also thank Veronica Voronina, Joye Yang, and Aleks Merkovich for technical assistance at various stages of this project.
This work was supported by the National Science Foundation (NSF) grant to JJ (IOS-1754969) and OKV (IOS-1656321 and IOS-1754966), the Gregory S. Call Undergraduate Research Program (LK, KT, EYP, FH, JGI, MC, AK, DC, CD, EMP), Doelling Undergraduate Research Fund (LK, KT, EYP, JGI, AK, DC, CD, EMP), Schupf Scholars Program (JZ), and the Sarles Fellowship (MC). The transmission electron microscopy was supported by the National Center for Research Resources award (SI0OD021580) to the UMass Medical School Electron Microscopy Facility. The authors are solely responsible for the content of this paper and do not necessarily represent the official views of the National Center for Research Resources, National Institutes of Health, or NSF. CHESS is supported by the NSF and NIH/NIGMS via NSF Award DMR-1332208.
Footnotes
CONFLICT OF INTEREST STATEMENT
The authors declare no conflict of interest.
SUPPORTING INFORMATION
Additional Supporting Information may be found in the online version of this article.
DATA AVAILABILITY STATEMENT
All relevant data can be found within the manuscript and its supporting materials.
REFERENCES
- Alonso JM, Stepanova AN, Leisse TJ, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science, 301, 653–657. [DOI] [PubMed] [Google Scholar]
- Aranda PS, LaJoie DM and Jorcyk CL (2012) Bleach gel: a simple agarose gel for analyzing RNA quality. Electrophoresis, 33, 366–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnaud N, Murgia I, Boucherez J, Briat J-F, Cellier F and Gaymard F (2006) An iron-induced nitric oxide burst precedes ubiquitin-dependent protein degradation for Arabidopsis AtFer1 ferritin gene expression. J. Biol. Chem, 281, 23579–23588. [DOI] [PubMed] [Google Scholar]
- Balk J and Schaedler TA (2014) Iron cofactor assembly in plants. Annu. Rev. Plant Biol, 65, 125–153. [DOI] [PubMed] [Google Scholar]
- Barnes JD, Balaguer L, Manrique E, Elvira S and Davison AW (1992) A reappraisal of the use of DMSO for the extraction and determination of chlorophylls a and b in lichens and higher plants. Environ Exp Bot, 32, 85–100. [Google Scholar]
- Bashir K, Ishimaru Y, Shimo H, et al. (2011) The rice mitochondrial iron transporter is essential for plant growth. Nat. Commun, 2, 322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienfait HF, de Weger LA and Kramer D (1987) Control of the development of iron-efficiency reactions in potato as a response to iron deficiency is located in the roots. Plant Physiol., 83, 244–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogorad L, Pires G and Swift H (1959) The structure of chloroplasts in leaf tissue of iron deficient Xanthium. Brookhaven Symp. Biol [Google Scholar]
- Bonaccorsi di Patti MC, Polticelli F, Cece G, Cutone A, Felici F, Persichini T and Musci G (2014) A structural model of human ferroportin and of its iron binding site. FEBS J., 281, 2851–2860. [DOI] [PubMed] [Google Scholar]
- Bonaccorsi di Patti MC, Polticelli F, Tortosa V, Furbetta PA and Musci G (2015) A bacterial homologue of the human iron exporter ferroportin. FEBS Lett., 589, 3829–3835. [DOI] [PubMed] [Google Scholar]
- Briat J-F, Duc C, Ravet K and Gaymard F (2010) Ferritins and iron storage in plants. Biochim. Biophys. Acta, 1800, 806–814. [DOI] [PubMed] [Google Scholar]
- Brink S, Flügge UI, Chaumont F, Boutry M, Emmermann M, Schmitz U, Becker K and Pfanner N (1994) Preproteins of chloroplast envelope inner membrane contain targeting information for receptor-dependent import into fungal mitochondria. J. Biol. Chem, 269, 16478–16485. [PubMed] [Google Scholar]
- Brumbarova T, Bauer P and Ivanov R (2015) Molecular mechanisms governing Arabidopsis iron uptake. Trends Plant Sci., 20, 124–133. [DOI] [PubMed] [Google Scholar]
- Buckhout TJ, Yang TJW and Schmidt W (2009) Early iron-deficiency-induced transcriptional changes in Arabidopsis roots as revealed by microarray analyses. BMC Genomics, 10, 147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bughio N, Takahashi M, Yoshimura E, Nishizawa NK and Mori S (1997) Light-Dependent Iron Transport into Isolated Barley Chloroplasts. Plant and Cell Physiology, 38, 101–105. [Google Scholar]
- Clemens S and Weber M (2016) The essential role of coumarin secretion for Fe acquisition from alkaline soil. Plant Signal. Behav, 11, e1114197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ and Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J., 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Colombo C, Palumbo G, He J-Z, Pinton R and Cesco S (2014) Review on iron availability in soil: interaction of Fe minerals, plants, and microbes. J. Soils Sediments, 14, 538–548. [Google Scholar]
- Connolly EL, Fett JP and Guerinot ML (2002) Expression of the IRT1 metal transporter is controlled by metals at the levels of transcript and protein accumulation. Plant Cell, 14, 1347–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connorton JM, Balk J and Rodríguez-Celma J (2017) Iron homeostasis in plants - a brief overview. Metallomics, 9, 813–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conte S, Stevenson D, Furner I and Lloyd A (2009) Multiple antibiotic resistance in Arabidopsis is conferred by mutations in a chloroplast-localized transport protein. Plant Physiol., 151, 559–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conte SS, Chu HH, Rodriguez DC, Punshon T, Vasques KA, Salt DE and Walker EL (2013) Arabidopsis thaliana Yellow Stripe1-Like4 and Yellow Stripe1-Like6 localize to internal cellular membranes and are involved in metal ion homeostasis. Front. Plant Sci, 4, 283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conte SS and Lloyd AM (2010) The MAR1 transporter is an opportunistic entry point for antibiotics. Plant Signal. Behav, 5, 49–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiDonato RJ, Roberts LA, Sanderson T, Eisley RB and Walker EL (2004) Arabidopsis Yellow Stripe-Like2 (YSL2): a metal-regulated gene encoding a plasma membrane transporter of nicotianamine-metal complexes. Plant J., 39, 403–414. [DOI] [PubMed] [Google Scholar]
- Dinneny JR, Long TA, Wang JY, et al. (2008) Cell identity mediates the response of Arabidopsis roots to abiotic stress. Science, 320, 942–945. [DOI] [PubMed] [Google Scholar]
- Divol F, Couch D, Conéjéro G, Roschzttardtz H, Mari S and Curie C (2013) The Arabidopsis YELLOW STRIPE LIKE4 and 6 transporters control iron release from the chloroplast. Plant Cell, 25, 1040–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drakesmith H, Nemeth E and Ganz T (2015) Ironing out Ferroportin. Cell Metab., 22, 777–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrett TP, Gassmann W and Rogers EE (2007) The FRD3-mediated efflux of citrate into the root vasculature is necessary for efficient iron translocation. Plant Physiol., 144, 197–205.17351051 [Google Scholar]
- Duy D, Stübe R, Wanner G and Philippar K (2011) The chloroplast permease PIC1 regulates plant growth and development by directing homeostasis and transport of iron. Plant Physiol., 155, 1709–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duy D, Wanner G, Meda AR, von Wirén N, Soll J and Philippar K (2007) PIC1, an ancient permease in Arabidopsis chloroplasts, mediates iron transport. Plant Cell, 19, 986–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escudero V, Abreu I, Tejada-Jiménez M, et al. (2020) Medicago truncatula Ferroportin2 mediates iron import into nodule symbiosomes. New Phytol. [DOI] [PubMed] [Google Scholar]
- Fourcroy P, Sisó-Terraza P, Sudre D, et al. (2014) Involvement of the ABCG37 transporter in secretion of scopoletin and derivatives by Arabidopsis roots in response to iron deficiency. New Phytol., 201, 155–167. [DOI] [PubMed] [Google Scholar]
- Foury F and Roganti T (2002) Deletion of the mitochondrial carrier genes MRS3 and MRS4 suppresses mitochondrial iron accumulation in a yeast frataxin-deficient strain. J. Biol. Chem, 277, 24475–24483. [DOI] [PubMed] [Google Scholar]
- García MJ, Romera FJ, Stacey MG, Stacey G, Villar E, Alcántara E and Pérez-Vicente R (2013) Shoot to root communication is necessary to control the expression of iron-acquisition genes in Strategy I plants. Planta, 237, 65–75. [DOI] [PubMed] [Google Scholar]
- Geiss GK, Bumgarner RE, Birditt B, et al. (2008) Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat. Biotechnol, 26, 317–325. [DOI] [PubMed] [Google Scholar]
- Gomez-Casati DF, Busi MV and Pagani MA (2018) Plant frataxin in metal metabolism. Front. Plant Sci, 9, 1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grusak MA and Pezeshgi S (1996) Shoot-to-Root Signal Transmission Regulates Root Fe(III) Reductase Activity in the dgl Mutant of Pea. Plant Physiol., 110, 329–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliwell B and Gutteridge JM (1992) Biologically relevant metal ion-dependent hydroxyl radical generation. An update. FEBS Lett., 307, 108–112. [DOI] [PubMed] [Google Scholar]
- Hantzis LJ, Kroh GE, Jahn CE, Cantrell M, Peers G, Pilon M and Ravet K (2017) A program for iron economy during deficiency targets specific Fe proteins. Plant Physiol., 176, 596–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heazlewood JL, Tonti-Filippini JS, Gout AM, Day DA, Whelan J and Millar AH (2004) Experimental analysis of the Arabidopsis mitochondrial proteome highlights signaling and regulatory components, provides assessment of targeting prediction programs, and indicates plant-specific mitochondrial proteins. Plant Cell, 16, 241–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurt EC, Soltanifar N, Goldschmidt-Clermont M, Rochaix JD and Schatz G (1986) The cleavable pre-sequence of an imported chloroplast protein directs attached polypeptides into yeast mitochondria. EMBO J., 5, 1343–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichikawa Y, Bayeva M, Ghanefar M, et al. (2012) Disruption of ATP-binding cassette B8 in mice leads to cardiomyopathy through a decrease in mitochondrial iron export. Proc. Natl. Acad. Sci. USA, 109, 4152–4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishka MR and Vatamaniuk OK (2020) Copper deficiency alters shoot architecture and reduces fertility of both gynoecium and androecium in Arabidopsis thaliana. BioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain A, Dashner ZS and Connolly EL (2019) Mitochondrial Iron Transporters (MIT1 and MIT2) Are Essential for Iron Homeostasis and Embryogenesis in Arabidopsis thaliana. Front. Plant Sci, 10, 1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaquinod M, Villiers F, Kieffer-Jaquinod S, Hugouvieux V, Bruley C, Garin J and Bourguignon J (2007) A Proteomics Approach Highlights a Myriad of Transporters in the Arabidopsis thaliana Vacuolar Membrane. Plant Signal. Behav, 2, 413–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Jean M, Schikora A, Mari S, Briat J-F and Curie C (2005) A loss-of-function mutation in AtYSL1 reveals its role in iron and nicotianamine seed loading. Plant J., 44, 769–782. [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA and Bevan MW (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J., 6, 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong J, Cohu C, Kerkeb L, Pilon M, Connolly EL and Guerinot ML (2008) Chloroplast Fe(III) chelate reductase activity is essential for seedling viability under iron limiting conditions. Proc. Natl. Acad. Sci. USA, 105, 10619–10624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong J and Connolly EL (2009) Iron uptake mechanisms in plants: Functions of the FRO family of ferric reductases. Plant Sci., 176, 709–714. [Google Scholar]
- Jeong J, Merkovich A, Clyne M and Connolly EL (2017) Directing iron transport in dicots: regulation of iron acquisition and translocation. Curr. Opin. Plant Biol, 39, 106–113. [DOI] [PubMed] [Google Scholar]
- Keech O, Dizengremel P and Gardeström P (2005) Preparation of leaf mitochondria fromArabidopsis thaliana. Physiol. Plant, 124, 403–409. [Google Scholar]
- Khan MA, Castro-Guerrero NA, McInturf SA, et al. (2018) Changes in iron availability in Arabidopsis are rapidly sensed in the leaf vasculature and impaired sensing leads to opposite transcriptional programs in leaves and roots. Plant Cell Environ., 41, 2263–2276. [DOI] [PubMed] [Google Scholar]
- Kim SA, Punshon T, Lanzirotti A, Li L, Alonso JM, Ecker JR, Kaplan J and Guerinot ML (2006) Localization of iron in Arabidopsis seed requires the vacuolar membrane transporter VIT1. Science, 314, 1295–1298. [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Nozoye T and Nishizawa NK (2018) Iron transport and its regulation in plants. Free Radic. Biol. Med, 133, 11–20. [DOI] [PubMed] [Google Scholar]
- Kumar RK, Chu H-H, Abundis C, Vasques K, Rodriguez DC, Chia J-C, Huang R, Vatamaniuk OK and Walker EL (2017) Iron-Nicotianamine Transporters Are Required for Proper Long Distance Iron Signaling. Plant Physiol., 175, 1254–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanquar V, Lelièvre F, Bolte S, et al. (2005) Mobilization of vacuolar iron by AtNRAMP3 and AtNRAMP4 is essential for seed germination on low iron. EMBO J., 24, 4041–4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Singh AP, Chung GC, Kim YS and Kong IB (2002) Chilling root temperature causes rapid ultrastructural changes in cortical cells of cucumber (Cucumis sativus L.) root tips. J. Exp. Bot, 53, 2225–2237. [DOI] [PubMed] [Google Scholar]
- Li L, Miao R, Bertram S, Jia X, Ward DM and Kaplan J (2012) A role for iron-sulfur clusters in the regulation of transcription factor Yap5-dependent high iron transcriptional responses in yeast. J. Biol. Chem, 287, 35709–35721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Miao R, Jia X, Ward DM and Kaplan J (2014) Expression of the yeast cation diffusion facilitators Mmt1 and Mmt2 affects mitochondrial and cellular iron homeostasis: evidence for mitochondrial iron export. J. Biol. Chem, 289, 17132–17141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Li L, Jia X, Ward DM and Kaplan J (2011) Genetic and biochemical analysis of high iron toxicity in yeast: iron toxicity is due to the accumulation of cytosolic iron and occurs under both aerobic and anaerobic conditions. J. Biol. Chem, 286, 3851–3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Millán A-F, Grusak MA, Abadía A and Abadía J (2013) Iron deficiency in plants: an insight from proteomic approaches. Front. Plant Sci, 4, 254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mai H-J, Pateyron S and Bauer P (2016) Iron homeostasis in Arabidopsis thaliana: transcriptomic analyses reveal novel FIT-regulated genes, iron deficiency marker genes and functional gene networks. BMC Plant Biol., 16, 211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marschner P ed. (2012) Marschner’s Mineral Nutrition of Higher Plants 3rd ed.,. [Google Scholar]
- Masuda T, Suzuki T, Shimada H, Ohta H and Takamiya K (2003) Subcellular localization of two types of ferrochelatase in cucumber. Planta. [DOI] [PubMed] [Google Scholar]
- Mendoza-Cózatl DG, Xie Q, Akmakjian GZ, et al. (2014) OPT3 is a component of the iron-signaling network between leaves and roots and misregulation of OPT3 leads to an over-accumulation of cadmium in seeds. Mol. Plant, 7, 1455–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlot S, Hannibal L, Martins S, Martinelli L, Amir H, Lebrun M and Thomine S (2014) The metal transporter PgIREG1 from the hyperaccumulator Psychotria gabriellae is a candidate gene for nickel tolerance and accumulation. J. Exp. Bot, 65, 1551–1564. [DOI] [PubMed] [Google Scholar]
- Metzendorf C, Wu W and Lind MI (2009) Overexpression of Drosophila mitoferrin in l(2)mbn cells results in dysregulation of Fer1HCH expression. Biochem. J, 421, 463–471. [DOI] [PubMed] [Google Scholar]
- Mori S, Nishizawa N, Hayashi H and Chino M (1991) Why are young rice plants highly susceptible to iron deficiency? … interactions in plants. [Google Scholar]
- Morrissey J, Baxter IR, Lee J, Li L, Lahner B, Grotz N, Kaplan J, Salt DE and Guerinot ML (2009) The ferroportin metal efflux proteins function in iron and cobalt homeostasis in Arabidopsis. Plant Cell, 21, 3326–3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mühlenhoff U, Stadler JA, Richhardt N, Seubert A, Eickhorst T, Schweyen RJ, Lill R and Wiesenberger G (2003) A specific role of the yeast mitochondrial carriers MRS3/4p in mitochondrial iron acquisition under iron-limiting conditions. J. Biol. Chem, 278, 40612–40620. [DOI] [PubMed] [Google Scholar]
- Mukherjee I, Campbell NH, Ash JS and Connolly EL (2006) Expression profiling of the Arabidopsis ferric chelate reductase (FRO) gene family reveals differential regulation by iron and copper. Planta, 223, 1178–1190. [DOI] [PubMed] [Google Scholar]
- Müller B, Kovács K, Pham H-D, et al. (2019) Chloroplasts preferentially take up ferric-citrate over iron-nicotianamine complexes in Brassica napus. Planta, 249, 751–763. [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Suzuki T, Murata S, et al. (2007) Improved Gateway binary vectors: high-performance vectors for creation of fusion constructs in transgenic analysis of plants. Biosci. Biotechnol. Biochem, 71, 2095–2100. [DOI] [PubMed] [Google Scholar]
- Nelson BK, Cai X and Nebenführ A (2007) A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J., 51, 1126–1136. [DOI] [PubMed] [Google Scholar]
- Nirody JA, Budin I and Rangamani P (2020) ATP synthase: Evolution, energetics, and membrane interactions. J. Gen. Physiol, 152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notredame C, Higgins DG and Heringa J (2000) T-Coffee: A novel method for fast and accurate multiple sequence alignment. J. Mol. Biol, 302, 205–217. [DOI] [PubMed] [Google Scholar]
- Oikawa K, Matsunaga S, Mano S, et al. (2015) Physical interaction between peroxisomes and chloroplasts elucidated by in situ laser analysis. Nat. Plants, 1, 15035. [DOI] [PubMed] [Google Scholar]
- Paradkar PN, Zumbrennen KB, Paw BH, Ward DM and Kaplan J (2009) Regulation of mitochondrial iron import through differential turnover of mitoferrin 1 and mitoferrin 2. Mol. Cell. Biol, 29, 1007–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park EY, Tsuyuki KM, Hu F, Lee J and Jeong J (2019) PRC2-Mediated H3K27me3 Contributes to Transcriptional Regulation of FIT-Dependent Iron Deficiency Response. Front. Plant Sci, 10, 627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park EY, Tsuyuki KM, Parsons EM and Jeong J (2020) PRC2-mediated H3K27me3 modulates shoot iron homeostasis in Arabidopsis thaliana. Plant Signal. Behav, 1784549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascal N and Douce R (1993) Effect of Iron Deficiency on the Respiration of Sycamore (Acer pseudoplatanus L.) Cells. Plant Physiol., 103, 1329–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit JM, Briat JF and Lobréaux S (2001) Structure and differential expression of the four members of the Arabidopsis thaliana ferritin gene family. Biochem. J, 359, 575–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaller R, Pfanner N and Neupert W (1989) Mitochondrial protein import. Bypass of proteinaceous surface receptors can occur with low specificity and efficiency. J. Biol. Chem, 264, 34–39. [PubMed] [Google Scholar]
- Pham HD, Pólya S, Müller B, et al. (2020) The developmental and iron nutritional pattern of PIC1 and NiCo does not support their interdependent and exclusive collaboration in chloroplast iron transport in Brassica napus. Planta, 251, 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt-Aloia KA, Thomson WW and Terry N (1983) Changes in plastid ultrastructure during iron nutrition-mediated chloroplast development. Protoplasma, 114-114, 85–92. [Google Scholar]
- Porra RJ, Thompson WA and Kriedemann PE (1989) Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochimica et Biophysica Acta (BBA) - Bioenergetics, 975, 384–394. [Google Scholar]
- Raven JA and Allen JF (2003) Genomics and chloroplast evolution: what did cyanobacteria do for plants? Genome Biol., 4, 209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravet K, Touraine B, Boucherez J, Briat J-F, Gaymard F and Cellier F (2009) Ferritins control interaction between iron homeostasis and oxidative stress in Arabidopsis. Plant J., 57, 400–412. [DOI] [PubMed] [Google Scholar]
- Regev-Rudzki N, Karniely S, Ben-Haim NN and Pines O (2005) Yeast aconitase in two locations and two metabolic pathways: seeing small amounts is believing. Mol. Biol. Cell, 16, 4163–4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Y, Yang S, Tan G, et al. (2012) Reduction of mitoferrin results in abnormal development and extended lifespan in Caenorhabditis elegans. PLoS One, 7, e29666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson NJ, Procter CM, Connolly EL and Guerinot ML (1999) A ferric-chelate reductase for iron uptake from soils. Nature, 397, 694–697. [DOI] [PubMed] [Google Scholar]
- Rockwell NC, Lagarias JC and Bhattacharya D (2014) Primary endosymbiosis and the evolution of light and oxygen sensing in photosynthetic eukaryotes. Front. Ecol. Evol, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Celma J, Pan IC, Li W, Lan P, Buckhout TJ and Schmidt W (2013) The transcriptional response of Arabidopsis leaves to Fe deficiency. Front. Plant Sci, 4, 276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roger AJ, Muñoz-Gómez SA and Kamikawa R (2017) The origin and diversification of mitochondria. Curr. Biol, 27, R1177–R1192. [DOI] [PubMed] [Google Scholar]
- Santi S and Schmidt W (2009) Dissecting iron deficiency-induced proton extrusion in Arabidopsis roots. New Phytol., 183, 1072–1084. [DOI] [PubMed] [Google Scholar]
- Schaaf G, Honsbein A, Meda AR, Kirchner S, Wipf D and von Wirén N (2006) AtIREG2 encodes a tonoplast transport protein involved in iron-dependent nickel detoxification in Arabidopsis thaliana roots. J. Biol. Chem, 281, 25532–25540. [DOI] [PubMed] [Google Scholar]
- Schaaf G, Schikora A, Häberle J, Vert G, Ludewig U, Briat J-F, Curie C and von Wirén N (2005) A putative function for the arabidopsis Fe-Phytosiderophore transporter homolog AtYSL2 in Fe and Zn homeostasis. Plant Cell Physiol., 46, 762–774. [DOI] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Schölkopf B, Weigel D and Lohmann JU (2005) A gene expression map of Arabidopsis thaliana development. Nat. Genet, 37, 501–506. [DOI] [PubMed] [Google Scholar]
- Schmidt H, Günther C, Weber M, Spörlein C, Loscher S, Böttcher C, Schobert R and Clemens S (2014) Metabolome analysis of Arabidopsis thaliana roots identifies a key metabolic pathway for iron acquisition. PLoS One, 9, e102444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen TD and Livak KJ (2008) Analyzing real-time PCR data by the comparative CT method. Nat. Protoc, 3, 1101–1108. [DOI] [PubMed] [Google Scholar]
- Schwacke R, Schneider A, van der Graaff E, Fischer K, Catoni E, Desimone M, Frommer WB, Flügge U-I and Kunze R (2003) ARAMEMNON, a novel database for Arabidopsis integral membrane proteins. Plant Physiol., 131, 16–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw GC, Cope JJ, Li L, et al. (2006) Mitoferrin is essential for erythroid iron assimilation. Nature, 440, 96–100. [DOI] [PubMed] [Google Scholar]
- Shcolnick S and Keren N (2006) Metal homeostasis in cyanobacteria and chloroplasts. Balancing benefits and risks to the photosynthetic apparatus. Plant Physiol., 141, 805–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikanai T, Müller-Moulé P, Munekage Y, Niyogi KK and Pilon M (2003) PAA1, a P-type ATPase of Arabidopsis, functions in copper transport in chloroplasts. Plant Cell, 15, 1333–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimoni-Shor E, Hassidim M, Yuval-Naeh N and Keren N (2010) Disruption of Nap14, a plastid-localized non-intrinsic ABC protein in Arabidopsis thaliana results in the over-accumulation of transition metals and in aberrant chloroplast structures. Plant Cell Environ., 33, 1029–1038. [DOI] [PubMed] [Google Scholar]
- Shingles R, North M and McCarty RE (2002) Ferrous ion transport across chloroplast inner envelope membranes. Plant Physiol., 128, 1022–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MD, Schnell DJ, Fitzpatrick L and Keegstra K (2003) In vitro analysis of chloroplast protein import. Curr. Protoc. Cell Biol, Chapter 11, Unit11.16. [DOI] [PubMed] [Google Scholar]
- Solé VA, Papillon E, Cotte M, Walter P and Susini J (2007) A multiplatform code for the analysis of energy-dispersive X-ray fluorescence spectra. Spectrochimica Acta Part B At. Spectrosc, 62, 63–68. [Google Scholar]
- Solti A, Kovács K, Basa B, Vértes A, Sárvári E and Fodor F (2012) Uptake and incorporation of iron in sugar beet chloroplasts. Plant Physiol. Biochem, 52, 91–97. [DOI] [PubMed] [Google Scholar]
- Sparkes I (2018) Lessons from optical tweezers: quantifying organelle interactions, dynamics and modelling subcellular events. Curr. Opin. Plant Biol, 46, 55–61. [DOI] [PubMed] [Google Scholar]
- Stacey MG, Patel A, McClain WE, Mathieu M, Remley M, Rogers EE, Gassmann W, Blevins DG and Stacey G (2008) The Arabidopsis AtOPT3 protein functions in metal homeostasis and movement of iron to developing seeds. Plant Physiol., 146, 589–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stocking CR (1975) Iron Deficiency and the Structure and Physiology of Maize Chloroplasts. Plant Physiol., 55, 626–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo I and Zoratti M (2014) Mitochondrial channels: ion fluxes and more. Physiol. Rev, 94, 519–608. [DOI] [PubMed] [Google Scholar]
- Tan Y-F, O’Toole N, Taylor NL and Millar AH (2010) Divalent metal ions in plant mitochondria and their role in interactions with proteins and oxidative stress-induced damage to respiratory function. Plant Physiol., 152, 747–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka R, Kobayashi K and Masuda T (2011) Tetrapyrrole Metabolism in Arabidopsis thaliana. Arabidopsis Book, 9, e0145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi R, Kato HE, Font J, Deshpande CN, Wada M, Ito K, Ishitani R, Jormakka M and Nureki O (2015) Outward- and inward-facing structures of a putative bacterial transition-metal transporter with homology to ferroportin. Nat. Commun, 6, 8545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarantino D, Morandini P, Ramirez L, Soave C and Murgia I (2011) Identification of an Arabidopsis mitoferrinlike carrier protein involved in Fe metabolism. Plant Physiol. Biochem, 49, 520–529. [DOI] [PubMed] [Google Scholar]
- Tarantino D, Santo N, Morandini P, Casagrande F, Braun H-P, Heinemeyer J, Vigani G, Soave C and Murgia I (2010) AtFer4 ferritin is a determinant of iron homeostasis in Arabidopsis thaliana heterotrophic cells. J. Plant Physiol, 167, 1598–1605. [DOI] [PubMed] [Google Scholar]
- Teardo E, Carraretto L, De Bortoli S, Costa A, Behera S, Wagner R, Lo Schiavo F, Formentin E and Szabo I (2015) Alternative splicing-mediated targeting of the Arabidopsis GLUTAMATE RECEPTOR3.5 to mitochondria affects organelle morphology. Plant Physiol., 167, 216–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry N and Low G (1982) Leaf chlorophyll content and its relation to the intracellular localization of iron. J. Plant Nutr, 5, 301–310. [Google Scholar]
- Thimm O, Essigmann B, Kloska S, Altmann T and Buckhout TJ (2001) Response of Arabidopsis to iron deficiency stress as revealed by microarray analysis. Plant Physiol., 127, 1030–1043. [PMC free article] [PubMed] [Google Scholar]
- Varotto C, Maiwald D, Pesaresi P, Jahns P, Salamini F and Leister D (2002) The metal ion transporter IRT1 is necessary for iron homeostasis and efficient photosynthesis in Arabidopsis thaliana. Plant J., 31, 589–599. [DOI] [PubMed] [Google Scholar]
- Vartapetian BB, Andreeva I and Generozova IP (2003) Functional electron microscopy in studies of plant response and adaptation to anaerobic stress. Annals of …. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasconcelos MW, Gruissem W and Bhullar NK (2017) Iron biofortification in the 21st century: setting realistic targets, overcoming obstacles, and new strategies for healthy nutrition. Curr. Opin. Biotechnol, 44, 8–15. [DOI] [PubMed] [Google Scholar]
- Versaw WK and Harrison MJ (2002) A chloroplast phosphate transporter, PHT2;1, influences allocation of phosphate within the plant and phosphate-starvation responses. Plant Cell, 14, 1751–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vert G, Grotz N, Dédaldéchamp F, Gaymard F, Guerinot ML, Briat J-F and Curie C (2002) IRT1, an Arabidopsis transporter essential for iron uptake from the soil and for plant growth. Plant Cell, 14, 1223–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vert GA, Briat J-F and Curie C (2003) Dual regulation of the Arabidopsis high-affinity root iron uptake system by local and long-distance signals. Plant Physiol., 132, 796–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigani G, Bashir K, Ishimaru Y, et al. (2016) Knocking down mitochondrial iron transporter (MIT) reprograms primary and secondary metabolism in rice plants. J. Exp. Bot, 67, 1357–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigani G, Faoro F, Ferretti AM, Cantele F, Maffi D, Marelli M, Maver M, Murgia I and Zocchi G (2015) Three-Dimensional Reconstruction, by TEM Tomography, of the Ultrastructural Modifications Occurring in Cucumis sativus L. Mitochondria under Fe Deficiency. PLoS One, 10, e0129141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voith von Voithenberg L, Park J, Stübe R, Lux C, Lee Y and Philippar K (2019) A Novel Prokaryote-Type ECF/ABC Transporter Module in Chloroplast Metal Homeostasis. Front. Plant Sci, 10, 1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters BM, Chu H-H, Didonato RJ, Roberts LA, Eisley RB, Lahner B, Salt DE and Walker EL (2006) Mutations in Arabidopsis yellow stripe-like1 and yellow stripe-like3 reveal their roles in metal ion homeostasis and loading of metal ions in seeds. Plant Physiol., 141, 1446–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV and Provart NJ (2007) An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS One, 2, e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang TJW, Lin W-D and Schmidt W (2010) Transcriptional profiling of the Arabidopsis iron deficiency response reveals conserved transition metal homeostasis networks. Plant Physiol., 152, 2130–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokosho K, Yamaji N, Mitani-Ueno N, Shen RF and Ma JF (2016) An Aluminum-Inducible IREG Gene is Required for Internal Detoxification of Aluminum in Buckwheat. Plant Cell Physiol., 57, 1169–1178. [DOI] [PubMed] [Google Scholar]
- Yoon HS, Hackett JD, Ciniglia C, Pinto G and Bhattacharya D (2004) A molecular timeline for the origin of photosynthetic eukaryotes. Mol. Biol. Evol, 21, 809–818. [DOI] [PubMed] [Google Scholar]
- Yoshinaga K, Arimura SI, Niwa Y and Tsutsumi N (2005) Mitochondrial Behaviour in the Early Stages of ROS Stress Leading to Cell Death in Arabidopsis thaliana. Annals of …. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zancani M, Peresson C, Biroccio A, et al. (2004) Evidence for the presence of ferritin in plant mitochondria. Eur. J. Biochem, 271, 3657–3664. [DOI] [PubMed] [Google Scholar]
- Zhai Z, Gayomba SR, Jung H-I, et al. (2014) OPT3 Is a Phloem-Specific Iron Transporter That Is Essential for Systemic Iron Signaling and Redistribution of Iron and Cadmium in Arabidopsis. Plant Cell, 26, 2249–2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X-Y, Zhang X, Zhang Q, Pan X-X, Yan L-C, Ma X-J, Zhao W-Z, Qi X-T and Yin L-P (2017) Zea mays Fe deficiency-related 4 (ZmFDR4) functions as an iron transporter in the plastids of monocots. Plant J., 90, 147–163. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Sampathkumar A, Kerber SM-L, Swart C, Hille C, Seerangan K, Graf A, Sweetlove L and Fernie AR (2020) A moonlighting role for enzymes of glycolysis in the co-localization of mitochondria and chloroplasts. Nat. Commun, 11, 4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L and Gruissem W (2004) GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol., 136, 2621–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimorski V, Ku C, Martin WF and Gould SB (2014) Endosymbiotic theory for organelle origins. Curr. Opin. Microbiol, 22, 38–48. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Mitochondrial localization of FPN3-FLAG in yeast
Figure S2. Heterologous expression of FPN3 in Δmrs3/4
Figure S3. FPN3 expression in split root control seedlings transferred to the same conditions.
Figure S4. FPN3 T-DNA insertion lines and quantification of growth phenotypes
Figure S5. Elemental map of manganese and zinc in fpn3 leaves.
Figure S6. Manganese, zinc, and copper content in roots and shoots
Figure S7. Western blots with isolated chloroplasts.
Figure S8. Western blots with isolated mitochondria.
Figure S9. Manganese, zinc, and copper content in chloroplasts and mitochondria
Figure S10. FPN1 and FPN2 expression in wild type and fpn3
Methods S1. Western blots
Table S1. RT-qPCR primer sequence
Table S2. NanoString custom codeset target sequence
Data Availability Statement
All relevant data can be found within the manuscript and its supporting materials.