Abstract
Post-prostatectomy radiotherapy requires accurate annotation of the prostate bed (PB), i.e., the residual tissue after the operative removal of the prostate gland, to minimize side effects on surrounding organs-at-risk (OARs). However, PB segmentation in computed tomography (CT) images is a challenging task, even for experienced physicians. This is because PB is almost a “virtual” target with non-contrast boundaries and highly variable shapes depending on neighboring OARs. In this work, we propose an asymmetric multi-task attention network (AMTA-Net) for the concurrent segmentation of PB and surrounding OARs. Our AMTA-Net mimics experts in delineating the non-contrast PB by explicitly leveraging its critical dependency on the neighboring OARs (i.e., the bladder and rectum), which are relatively easy to distinguish in CT images. Specifically, we first adopt a U-Net as the backbone network for the low-level (or prerequisite) task of the OAR segmentation. Then, we build an attention sub-network upon the backbone U-Net with a series of cascaded attention modules, which can hierarchically transfer the OAR features and adaptively learn discriminative representations for the high-level (or primary) task of the PB segmentation. We comprehensively evaluate the proposed AMTA-Net on a clinical dataset composed of 186 CT images. According to the experimental results, our AMTA-Net significantly outperforms current clinical state-of-the-arts (i.e., atlas-based segmentation methods), indicating the value of our method in reducing time and labor in clinical workflow. Our AMTA-Net also presents better performance than the technical state-of-the-arts (i.e., the deep learning-based segmentation methods), especially for the most indistinguishable and clinically critical part of the PB boundaries.
Keywords: Segmentation, prostate bed, computed tomography, deep learning, multi-task, attention mechanism
1. Introduction
Prostate cancer is a common type of men cancer. According to the latest cancer statistics (Siegel et al. (2020)), there would be more than 190, 000 newly diagnosed cases and 33, 000 deaths associated with the prostate cancer in the United States this year, accounting for more than 1 in 5 new diagnoses and the second cancer mortality (after lung cancer) in men. Radical prostatectomy, i.e., a surgical operation to resect the prostate gland, is one of the most effective treatments when the cancer is believed to be confined to the prostate. However, after the radical prostatectomy, a few cancerous tissues may yet remain in the residual part of the prostate gland, as well as the surgical bed and some adjacent tissues. This region is clinically defined as prostate bed (PB) or prostatic fossa, which would develop a recurrence even metastasis without additional treatment. To eliminate the residual cancerous tissues, postoperative radiotherapy on the PB is commonly carried out as a standard adjuvant or salvage setting for the radical prostatectomy. Precisely delineating the target volume of PB in planning computed tomography (CT) images is a prerequisite for the efficacy of postoperative radiotherapy.
However, accurate PB segmentation in CT images is a unique task of great difficulty. As the example shown in Fig. 1, the PB is an anatomical region situated between the bladder and rectum in the male pelvis. It mainly consists of the residual prostatic tissues and some adjacent volumes of the bladder, bordering on the anterior surface of the rectum. In contrast to the prostate gland, whose boundary can be distinguished by the intensity change, the PB is not an intact structure with practical boundaries. This makes it often referred to as a “virtual” volume or “invisible” target in the literature (Hwee et al. (2011); Delpon et al. (2016); Latorzeff et al. (2017)). In clinical practice, physicians typically follow a set of complicated consensus guidelines (Poortmans et al. (2007); Wiltshire et al. (2007); Sidhom et al. (2008); Michalski et al. (2010)) to delineate the contour of PB. The recommended protocols are normally based on the PB’s spatial dependency on the surrounding organs-at-risk (OARs), i.e., the bladder and rectum. In other words, it is almost impossible to annotate PB in CT images merely relying on the intensity contrast. Furthermore, since the PB mainly consists of soft tissues, its size and shape could be significantly different across patients, highly affected by the status (full or empty) of the neighboring bladder and rectum. These reasons make PB segmentation fundamentally different from and much harder than the segmentation of other pelvic organs.
Fig. 1:
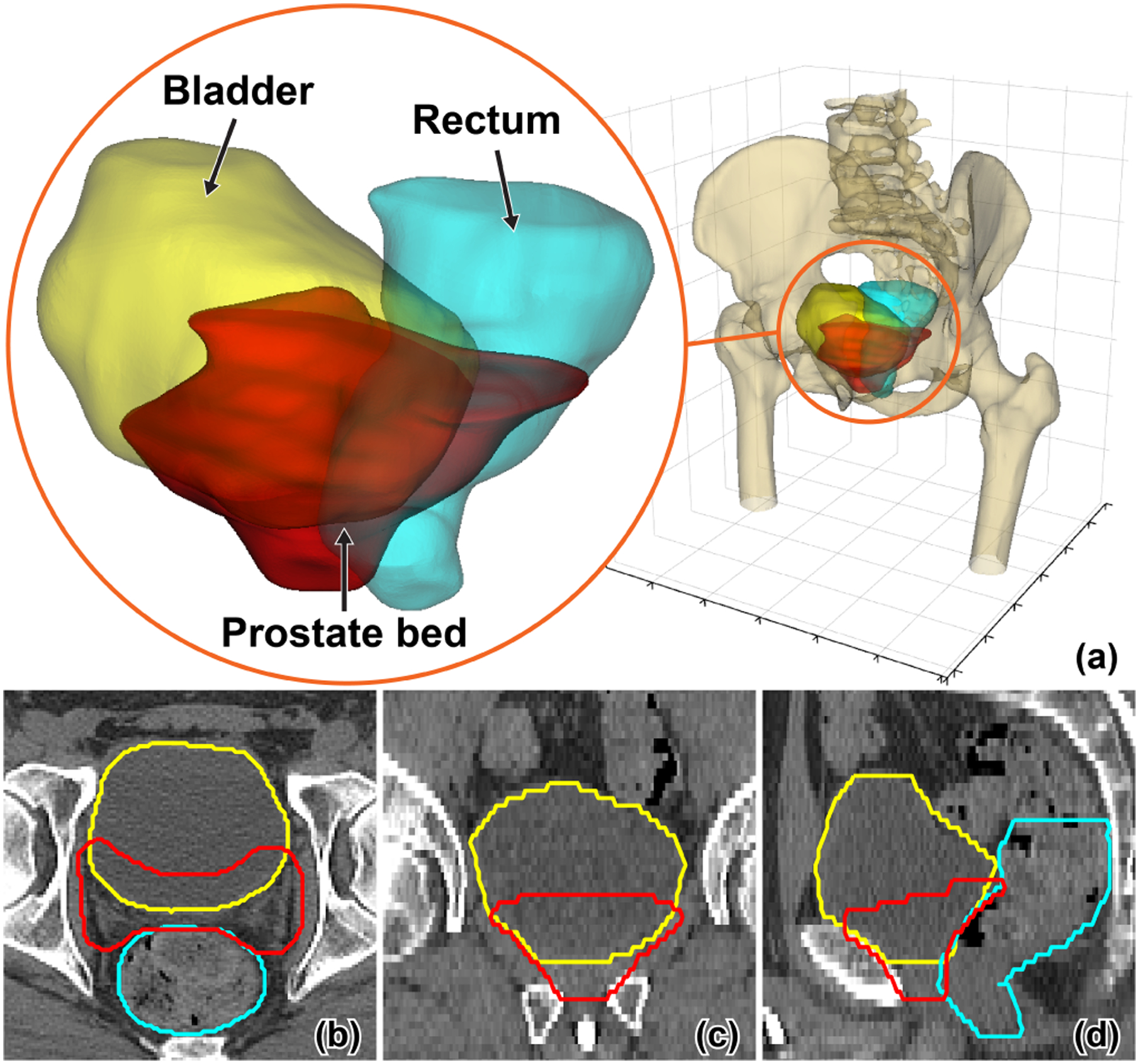
A representative sample of the post-prostatectomy case visualized in 3D (a) and 2D (b,c,d) views. The prostate bed, bladder, and rectum are displayed in red, yellow, and cyan color, respectively.
Although automated workflows are highly desired in the clinic, there are only a few such kind of methods (Hwee et al. (2011); Delpon et al. (2016)) proposed for PB segmentation. All these studies involve the atlas-based segmentation (ABS) method, which is a general methodology widely used in medical image analysis (Mohamed et al. (2006); Zhan et al. (2007); Wu et al. (2011); Jia et al. (2012); Iglesias and Sabuncu (2015)). Capitalizing on image registration techniques, these methods typically align the input image to the labeled atlas images, by which the segmentation contours of the atlases are further mapped to the unlabeled input image. According to the experimental results reported in the literature Hwee et al. (2011) and Delpon et al. (2016), the ABS methods are efficient for the delineation of high-contrast organs (e.g., the femoral heads), while cannot work well for the non-contrast PB. In recent years, benefiting from the task-oriented representation learning and end-to-end combination of local-to-global information, the fully convolutional network (FCN) (Long et al. (2015)) and its variants (Ronneberger et al. (2015)) have witnessed enormous progress in semantic segmentation. Although there is no yet research on deep learning-based method for PB segmentation, we hypothesize that FCN can be a good candidate solution for our problem, considering its compelling performance on some related applications such as the pelvic organ segmentation (Xu et al. (2018); He et al. (2019); Nie et al. (2019); Wang et al. (2019, 2020b,a)). However, the challenge is that general network architectures without explicit modeling of the dependency of PB to the adjacent OARs may fail to delineate this “virtual” target with non-contrast boundaries.
In this paper, we propose an asymmetric multi-task attention network (AMTA-Net) for the segmentation of PB in CT images. Mimicking the clinical workflow for manual PB delineation, our AMTA-Net is designed as an asymmetric multi-task model, in which the segmentation of OARs (i.e., bladder and rectum) serves as a low-level (or prerequisite) task providing guidance to the high-level (or primary) task of PB segmentation. Specifically, we first exploit a U-Net as the backbone network for the low-level task of OAR segmentation. Upon the backbone U-Net, we then build an attention sub-network for the high-level task of PB segmentation. The attention sub-network consists of a series of cascaded attention modules, which can hierarchically transfer the relevant OAR features from the backbone U-Net and adaptively learn discriminative representations for accurate PB segmentation. In addition, the output of the backbone is skip-connected as the initial input of the attention sub-network, which provides contextual information to refine the segmentation of the non-contrast PB.
In summary, the main contribution of this work is four-fold:
We leverage the power of deep learning to handle the challenging problem of PB segmentation in CT images, in which the target suffers from non-contrast boundaries and highly irregular shapes. To the best of our knowledge, this is the first exploration using a deep learning-based method to deal with this unique problem.
Inspired by the clinical workflow for manual PB delineation, we explicitly formulate the PB segmentation as a high-level task depending on the low-level task of OAR segmentation. Accordingly, a novel asymmetric multi-task network architecture, i.e., the AMTA-Net, is proposed to infer the PB mask from the structural information of the bladder and rectum.
We design a series of attention modules in the proposed AMTA-Net, which can learn task-oriented feature representations for accurate PB segmentation by transferring the relevant OAR features from the backbone network.
As a by-product of PB segmentation, our AMTA-Net can simultaneously segment the bladder and rectum, whose contour is also required in the post-prostatectomy radiotherapy to protect the OARs.
To evaluate the performance of the proposed AMTA-Net for PB segmentation, we conduct extensive experiments on a clinical dataset consisting of 186 CT images acquired from different patients. According to the experimental results, our AMTA-Net not only outperforms the clinical state-of-the-arts (i.e., the ABS methods) by a significant margin, but also achieves better performance in comparison to the technical state-of-the-arts (i.e., the deep learning-based methods for general image segmentation), especially for the most indistinguishable and clinically critical part of the PB boundaries.
It is worth noting that, this work is an extension of a preliminary conference publication (Xu et al. (2020)). In addition to a more detailed literature review, other major extensions in this journal paper include 1) more state-of-the-art deep learning-based methods are included in comparison with the proposed AMTA-Net, 2) more systematic evaluations are performed to verify the significance of the improvement achieved by the proposed AMTA-Net when compared with the state-of-the-arts, 3) a set of ablation studies are conducted to justify the effectiveness of the essential designs in the proposed AMTA-Net, 4) comprehensive discussions are presented to analyze network design and some other factors that contribute to accurate PB segmentation, and 5) further investigations on the issues related to inter-observer variability and radiotherapy dose distribution.
The rest of the paper is organized as follows. Section 2 gives an overview of previous works related to the PB segmentation and multi-task deep learning for medical image analysis. Section 3 presents the detailed designs of the proposed AMTA-Net. In Section 4, we conduct extensive experiments on a clinical dataset to evaluate the performance of our AMTA-Net and verify the efficacy of our designs. Some specific issues are discussed in Section 5. Finally, we conclude this work in Section 6.
2. Related Works
In this section, we briefly review previous works on automatic PB segmentation and multi-task deep learning for medical image analysis.
2.1. Prostate bed segmentation
In clinical workflow, the segmentation of PB in CT images is commonly carried out by the physicians using manual contouring tools. A set of complicated consensus guidelines have been proposed as professional support (Poortmans et al. (2007); Wiltshire et al. (2007); Sidhom et al. (2008); Michalski et al. (2010)). However, due to the divergence in physicians’ experience and knowledge, the manually delineated contours may present significant inter-observer variabilities, despite the use of rigorous contouring protocols and guidelines (Latorzeff et al. (2017)). Although automatic methods could be more robust in delineating PB with relatively higher efficiency, there are only a few ABS methods proposed for this challenging problem. Leveraging image registration techniques, the ABS methods typically align an input image to the labeled atlas images and then map the atlas segmentations to the unlabeled input image. Hwee et al. (2011) proposed to use a commercial ABS software with 75 atlas images to segment the PB. According to their experimental results, the ABS method is significantly faster than the manual contouring procedure, while the segmentation accuracy on PB is far from the requirement for clinical use (with the mean dice similarity coefficient around 0.47). A similar conclusion was drawn in a later research by Delpon et al. (2016). They compared five commercial ABS systems for the segmentation of PB and surrounding OARs. The results showed that these ABS methods consistently perform well in the segmentation of high-contrast targets such as the femoral heads but cannot reliably delineate the non-contrast PB. Overall, the limited performance of the conventional ABS method is mainly caused by two reasons: (1) they typically rely on intensity information for image registration, while the PB is a “virtual” object with non-contrast boundary; (2) they cannot effectively model the geometric correlation between the PB and surrounding OARs, which is critical for the segmentation of the non-contrast PB.
2.2. Multi-task deep learning for medical image analysis
As a popular machine learning strategy, multi-task learning (MTL) aims to improve the performance of multiple tasks by jointly learning a unified model, under the assumption that these tasks can be complementary to each other (Zhang and Yang (2017)). The strategy of MTL has been successfully applied to developing deep convolutional neural networks (CNNs) for various medical image analyzing tasks. For example, Moeskops et al. (2016) used a single CNN to perform the simultaneous segmentation of different tissues from different imaging modalities, achieving equivalent performance to that of multiple CNNs individually trained for each task. Xue et al. (2018) proposed a multi-task relationship learning method for full left ventricle quantification in cardiac MR images. This method can automatically learn the relationship between different tasks in an end-to-end fashion to improve the generalization capacity of the entire deep network. Bragman et al. (2018) combined uncertainty model with multi-task learning to rebalance the weights of CT synthesis task and OAR segmentation task for MR-only radiotherapy treatment planning. Lian et al. (2020) proposed a hierarchical FCN for the joint localization of brain atrophy and diagnosis of Alzheimer’s disease with the whole-brain MR images. Most of these existing multi-task CNNs for medical image analysis were designed to perform symmetric knowledge transformation between any two coupled tasks, i.e., different tasks are placed at the same level and learned equally. However, this is not always the real case in practice, e.g., the segmentation of PB relies on the surrounding OARs, while non-contrast PB is hard to bring additional information for the OARs, which are much easier to annotate. This challenge features that our task of automatic PB segmentation desires an asymmetric multi-task learning model (Lee et al. (2016)), which can leverage the inter-task dependency to perform better in some specific tasks.
3. Method
The schematic diagram of our AMTA-Net is shown in Fig. 2. It mainly consists of two sub-networks: (1) a backbone with the U-Net architecture for the low-level task of OARs (i.e., bladder and rectum) segmentation, and (2) an attention sub-network built upon the backbone for the high-level task of PB segmentation. The attention sub-network consists of a series of Attention Modules (AMs), with the inner structure specified in Fig. 3.
Fig. 2:
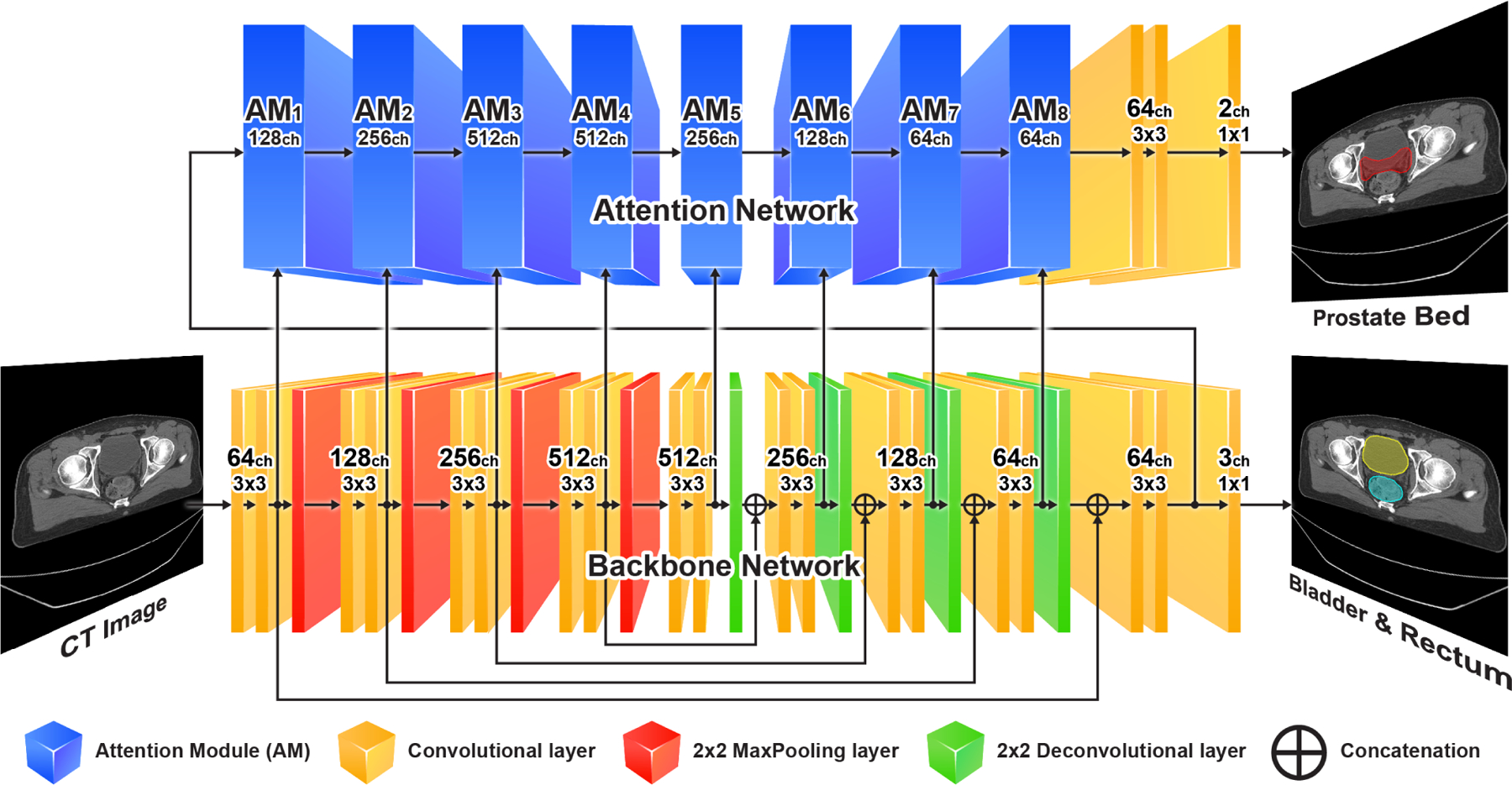
Schematic diagram of the AMTA-Net. It consists of a backbone U-Net (bottom) and an attention sub-network (top). The attention sub-network consists of a series of cascaded attention modules (AM). The inner structure of the AM is illustrated in Fig. 3.
Fig. 3:
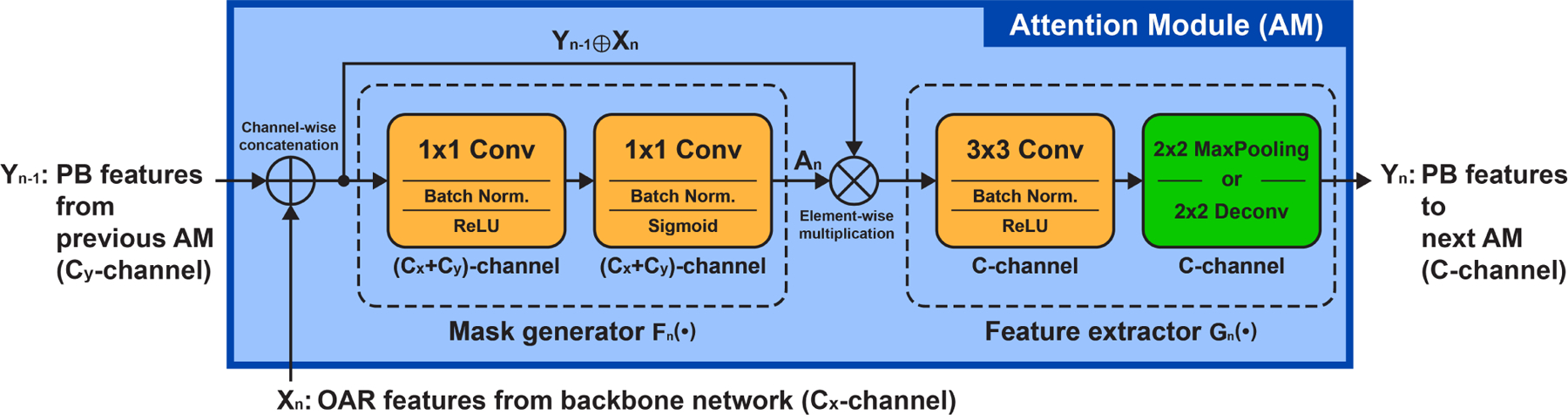
Inner structure of the n-th attention module (AM) in the attention sub-network.
3.1. Backbone network for OAR segmentation
In post-prostatectomy radiotherapy, accurate contouring of the bladder and rectum is not only a prerequisite for the OAR definition to minimize side effects on normal tissues, but also provides an essential reference for the physicians to delineate the clinical target volume (CTV) of PB. Based on this observation, in our design, we consider OAR segmentation as a foundation for the PB segmentation. To achieve accurate OAR segmentation in CT images, our AMTA-Net adopts a U-Net (Ronneberger et al. (2015)) as the backbone network to predict pixelwise OAR masks, considering that this classical FCN architecture and its variants have shown promising results in the task of CT pelvic organs segmentation. As shown in the bottom of Fig. 2, the backbone network mainly consists of an encoding path, a decoding path, and the skip connections between them. Both the encoding path and the decoding path are composed of four cascaded convolutional blocks. The numbers labeled on each convolutional block denote its output channel and kernel size (e.g., “512ch 3×3” denotes a convolutional layer with 3×3 kernels and 512 output channels). All the convolutional layers are followed by a batch normalization layer (Ioffe and Szegedy (2015)) and a rectified linear unit (ReLU) (Nair and Hinton (2010)) except for the last 1×1 convolutional layer, which is followed by a softmax layer. Notably, the backbone U-Net not only outputs the probability maps for the OAR segmentation, but also shares its intermediate feature maps with the subsequent attention sub-network. These shared intermediate feature maps can be seen as a feature pool containing both image spatial details and OAR structural information. From this feature pool, the attention sub-network can learn discriminative representations for the PB segmentation with the guidance of the OAR structures.
3.2. Attention sub-network for PB segmentation
Since the PB boundary largely depends on the shape of the bladder and rectum, it is intuitive to assume that the feature representations for the PB segmentation are highly correlated with those for the OAR segmentation, although they are not exactly the same. Therefore, inspired by the attention mechanism proposed in symmetric multi-task learning (Liu et al. (2019)), we build an attention sub-network upon the backbone U-Net to adaptively learn the PB features from the OAR features. As shown in the top of Fig. 2, the attention sub-network mainly consists of a series of cascaded attention modules (AMs). Besides the serial connection with the preceding AM, each AM laterally connects to a convolutional block in the backbone network. As mentioned before, the intermediate feature maps shared by each convolutional block of the backbone form a feature pool, from which the AMs can select the most relevant features to learn task-oriented representations for accurate PB segmentation.
The inner structure of the AM is illustrated in Fig. 3, by which the PB feature representations in a stage of the attention sub-network are jointly determined by the backbone and the preceding stage of the attention sub-network. Specifically, for the n-th AM (i.e., AMn) in the attention sub-network, there are two inputs: 1) the features generated by the preceding AMn−1 (the cascaded input denoted as Yn−1), and 2) the features from the n-th convolutional block of the backbone (the lateral input denoted as Xn). The AM first adopts a light-weight block (namely mask generator) consisting of two cascaded 1×1 convolutional layers to learn an adaptive attention mask from the input Yn−1 and Xn. Let the mask generator be a function Fn(·), the adaptive attention mask An (with the value of each element·between [0, 1]) can be denoted as:
| (1) |
where the symbol ⊕ denotes the concatenation of two tensors by channel. The attention map An has the same size as the input Yn−1 ⊕ Xn. It is then used to tailor the input Yn−1 ⊕ Xn by element-wise multiplication. After that, a 3×3 convolutional layer followed by a 2×2 pooling/deconvolutional layer is employed as a feature extractor Gn(·) to further generate discriminative feature representation Yn for the PB segmentation:
| (2) |
where the symbol ⊗ denotes the element-wise multiplication. The final pooling/deconvolutional layer is used in the encoding/decoding path of the attention sub-network to down-/upsample the output feature maps, ensuring the spatial consistency with the corresponding convolutional layers in the backbone network. For the first attention module AM1, its cascaded input Y0 is connected to the output of the second last convolutional layer of the backbone network, while the lateral input X1 comes from the first convolutional block of the backbone. Similar to the auto-context strategy, the combination of these two inputs provides both strong semantic information and rich image details to assist the construction of the attention sub-network.
3.3. Loss function
In the training stage, the model parameters are optimized by minimizing the following multi-task loss function:
| (3) |
where Lpb and Loar are the PB segmentation loss from the attention sub-network and the OAR segmentation loss from the backbone network, respectively. λpb and λoar are the corresponding loss weights. In our experiments, we set λpb = 1 and λoar = 1 to get optimal results on PB segmentation. We will show experimental results in Section 5.2 to justify this setting. Considering the class imbalance between the foreground and background pixels, we define Lpb and Loar as the dice loss function (Milletari et al. (2016)):
| (4) |
where P and denote the C-channel predicted mask and the ground-truth mask, respectively. pci and are the values of the i-th pixel in the c-th channel of the predicted mask P and the ground-truth mask , respectively.
3.4. Implementation details
The proposed AMTA-Net takes 2D CT slices as the input. We combine three adjacent slices to compose a 3-channel input image, which provides more spatial context to infer the segmentation mask of the center slice. For the effect of the input adjacent slice number, we will investigate it in Section 5.1. All the CT slices are center-cropped and resampled to a uniform size of 128×128 with a spatial resolution of 2×2 mm2 through bi-linear interpolation. Pixel intensities are rescaled from [−200, 800] Hounsfield Unit (HU) to [0, 1] by linear mapping. The intensities below −200 HU (beyond 800 HU) are clipped to 0 (1). To mitigate overfitting, in the training stage, we randomly translate and rotate the input CT slices in a range of [−5.00, 5.00] mm and [−0.05, 0.05] rad, respectively.
We train the model for 100 epochs with a base learning rate of 10−3 and a batch size of 144 (a mini-batch size of 24 on six graphic cards). The model achieving the highest accuracy (i.e., a weighted average dice similarity coefficient on the PB, bladder, and rectum with the weights of 0.5, 0.25, and 0.25, respectively) on the validation set is stored as the final model, which is then used to perform the inference on the testing set. We implement the model using the PyTorch framework on Ubuntu 16.04 (x64) operating system. All the training procedures are conducted on a server computer equipped with two Intel(R) Xeon(R) E5–2650 CPUs working at 2.20GHz and six NVIDIA TITAN Xp graphic cards with 12 GBytes of memory each. Model parameters are initialized using the Xavier algorithm (Glorot and Bengio (2010)) and optimized using the back-propagation algorithm (LeCun et al. (1998)) and Adam optimizer (Kingma and Ba (2014)). The implementation of the proposed method is publicly available on GitHub1.
4. Experiments
4.1. Dataset
In this study, all the experiments are conducted on a clinical dataset composed of 186 post-prostatectomy patients collected in one hospital (i.e., the Radiation Oncology Department, UNC-Chapel Hill, U.S.) from year 2009 to 2019. Each case has one planning CT image and the corresponding segmentation masks of PB, bladder, and rectum. All these masks are manually delineated by one of three expert physicians and modified/verified by the rest two physicians to mitigate the inter-observer variability. We use these masks as ground-truth for training and evaluation. The slice number of each CT image varies from 98 to 270, resulting in a total of 26, 133 slices. All these slices have a uniform size of 512×512. The slice in-plane spacing ranges from 0.81 mm to 1.37 mm, and the slice thickness varies in two values of 3.00 mm (184 cases) and 1.50 mm (2 cases). We randomly divide the dataset into five folds in terms of patients and conduct 5-fold cross validation to evaluate the performance of different models (three folds for training, one fold for validation, and one fold for testing in each iteration). Five cases with severe metal artifact caused by artificial femoral heads are excluded when used for testing.
4.2. Metrics
We compute the mean value and standard deviation of Dice Similarity Coefficient (DSC) and Average Symmetric Surface Distance (ASD) on all cases for PB, bladder, and rectum as the metrics to quantitatively evaluate the model performance. Specifically, we use the DSC as the primary metric to quantify the overlap ratio between the predicted mask and the ground-truth mask:
| (5) |
where Vp and Vg denote the volume of the predicted mask and the ground-truth mask, respectively. The ASD is used as a secondary metric to measure the shape conformity between the predicted mask and the ground-truth mask:
| (6) |
where Sp and Sg denote the surfaces of the predicted mask and the ground-truth mask, respectively. d(a, S) is the minimum distance between point a to surface S.
4.3. Competing methods
The competing methods evaluated in the experiments can be categorized into two classes: 1) the ABS methods which represent the clinical state-of-the-art for PB segmentation, and 2) the deep learning-based segmentation methods which represent the technical state-of-the-art for general image segmentation.
4.3.1. Atlas-based segmentation methods
As we introduced in Section 1, automatic PB segmentation has been rarely studied before except for a few explorations that use commercial software integrating the atlas-based segmentation (ABS) methods. Therefore, to demonstrate the clinical value of our AMTA-Net, we conduct a comparison with the commercial ABS software. In this experiment, we not only list the results of five commercial ABS software reported in literature to perform a qualitative comparison, but also apply one representative ABS software (i.e., the MIM Maestro® with a recently released version of 6.9.6) on our dataset to conduct a quantitative comparison. We apply the same 5-fold cross validation to evaluate the ABS software as our method. In each iteration of the cross validation, four folds of cases are used to build the atlas database and one fold of cases are used for testing. The major hyper-parameter of the ABS software is the number of the matched case used to aggregate the output segmentation. We determine this parameter by grid-search strategy and finally set it to an optimal value of eight. In the result of the ABS software, the segmentation fails in one case, whose DSC and ASD dramatically deviate from the average. We find this failed case has an exceptionally full bladder and rectum, which is not common for radiotherapy. Therefore, to avoid introducing bias, we exclude this failed case from the result of the ABS software but still count it in the result of other competing methods (including ours).
4.3.2. Deep learning-based segmentation methods
To demonstrate the technical novelties in our design, we compare our method to the state-of-the-art deep networks for general image segmentation and the variants of them, including:
U-Net (Ronneberger et al. (2015)), V-Net (Milletari et al. (2016)), and VoxResNet (Chen et al. (2018)): Three single-task FCN models for medical image segmentation, which typically consist of an encoding (downsample) path and a decoding (upsample) path.
Multi-task U-Net (MT-U-Net): A symmetric multi-task model derived from U-Net. It consists of one encoding path shared by two sibling decoding paths, which are used for PB segmentation and OAR segmentation, respectively.
Multi-task Attention U-Net (MTA-U-Net) (Liu et al. (2019)): A symmetric multi-task model integrating attention mechanisms. A backbone U-Net extracts task-shared features while two sibling attention sub-networks built upon the backbone U-Net extract task-specific features for PB segmentation and OAR segmentation, respectively.
Both MT-U-Net and MTA-U-Net have symmetric network architectures where the PB segmentation task and OAR segmentation task are equally learned. It means that the learning of the PB segmentation task takes no explicit reference from the neighboring OAR structures but implicit reference by feature sharing with the OAR segmentation task. All the competing networks are implemented in 2D manners and trained using the same configurations as our method introduced in Section 3.4. We also apply the same 5-fold cross validation as ours to evaluate the competing networks. Although this experimental setting ensures that the network architecture is the sole variable in the comparison between different competing methods, it may not guarantee all of these methods to reach the full potential since they might need different hyper-parameters to further optimize their performance.
4.4. Comparison with other methods
4.4.1. Comparison with ABS methods
The experimental results of the commercial ABS software are summarized in the top part of Table 1. According to the reported data in literature (the top six rows in Table 1), all the ABS software show relatively low accuracy on PB (with a mean DSC lower than 70%), indicating the inherent difficulty in PB segmentation. Among these ABS software, MIM has relatively better performance and wider application in the clinic. Thus, we adopt a recently released version of the MIM ABS software on our dataset to conduct a direct comparison with our method. According to the experimental results shown in the last row of the top part of Table 1, the proposed AMTA-Net significantly outperforms the MIM ABS software on both PB segmentation and OAR segmentation. Our mean DSC on PB is 8.55% (75.67% v.s. 67.12%) higher than that of the MIM ABS software. This superior performance demonstrates that the proposed AMTA-Net can largely reduce the labor and time costs in clinical workflow, justifying the application value of our method.
Table 1:
Quantitative comparison between different methods
| Methods | DSC [mean(std) %] | ASD [mean(std) mm] | ||||
|---|---|---|---|---|---|---|
| PB | Bladder | Rectum | PB | Bladder | Rectum | |
| ABS(MIM*,Hwee et al. (2011)) | 47.00(16.00)† | 67.00(18.00)† | 58.00(9.00)† | - | - | - |
| ABS(WFB*,Delpon et al. (2016)) | 56.00(10.00)† | 76.00(12.00)† | 73.00(7.00)† | - | - | - |
| ABS(MIM*,Delpon et al. (2016)) | 61.00(9.00)† | 80.00(14.00)† | 75.00(7.00)† | - | - | - |
| ABS(ABAS*,Delpon et al. (2016)) | 67.00(13.00)† | 81.00(13.00)† | 75.00(9.00)† | - | - | - |
| ABS(SPICE*,Delpon et al. (2016)) | 37.00(9.00)† | 76.00(26.00)† | 68.00(12.00)† | - | - | - |
| ABS(RS*,Delpon et al. (2016)) | 51.00(17.00)† | 59.00(15.00)† | 49.00(12.00)† | - | - | - |
| ABS(MIM*) | 67.12(10.47) | 67.28(15.97) | 62.97(11.75) | 3.51(1.59) | 5.63(3.33) | 4.85(3.04) |
| U-Net(Ronneberger et al. (2015)) | 73.29(7.43) | - | - | 2.79(1.27) | - | - |
| V-Net(Milletari et al. (2016)) | 73.36(7.89) | - | - | 2.67(1.22) | - | - |
| VoxResNet(Chen et al. (2018)) | 74.58(7.45) | - | - | 2.55(1.11) | - | - |
| MT-U-Net | 74.41(7.23) | 87.84(10.04) | 80.22(7.82) | 2.58(1.21) | 1.62(2.12) | 2.50(1.85) |
| MTA-U-Net(Liu et al. (2019)) | 75.03(7.11) | 88.46(8.08) | 80.31(9.23) | 2.51(1.13) | 1.44(1.30) | 2.56(2.91) |
| Ours | 75.67(6.56) | 88.40(8.72) | 80.35(9.36) | 2.42(1.03) | 1.47(1.22) | 2.60(3.00) |
MIM: MIM Maestro (MIM Software); WFB: WorkFlow Box (Mirada Medical); ABAS: Atlas-Based Autosegmentation (Elekta); SPICE: Pinnacle (Philips); RS: RayStation (RaySearch Laboratories);
indicates a reported result conducted from other datasets.
4.4.2. Comparison with deep learning-based methods
The experimental results of the deep learning-based methods are summarized in the bottom part of Table 1. According to the results, we can have the following observations:
For all the competing methods, the deep learning-based methods outperform the conventional ABS methods by a significant margin in both PB and OAR segmentation tasks. This result demonstrates the superiority of the deep learning methodology in dealing with the pelvic organs that have variable shapes and low-/non-contrast boundaries.
For the deep learning-based methods, the multi-task networks (MT-U-Net, MTA-U-Net, and ours) generally outperform the single-task networks (U-Net, V-Net, and VoxResNet). This result indicates that, by jointly learning the PB segmentation task and the OAR segmentation task, the multi-task networks can leverage the knowledge used for the OAR segmentation to facilitate the PB segmentation, thus achieve higher accuracy than the single-task networks that learn the PB segmentation task individually.
For the multi-task networks (MT-U-Net, MTA-U-Net, and ours), our method achieves the highest accuracy on PB and comparable performance on OARs when compared with the other two competitors. This result indicates the superior performance of our method for the specific problem of PB segmentation.
According to the experimental results shown in Table 1, the improvement margin of our method over other deep learning-based methods is smaller than that over the ABS methods. Therefore, we conduct paired t-test to check whether our improvement margin is statistically significant or not. Specifically, we calculate two-sided p-value between the results of our method and the deep learning-based competitors. The significance level is defined as a thresholding p-value of 0.05. In Table 2, we list the p-values along with the corresponding DSC and ASD of all the deep learning-based methods. We can see our result is significantly different with most of the competing results except for the ASD against the MTA-U-Net (see the top part of Table 2, Whole PB volume). To investigate which part of the PB volume our method gain the most, we separate the CT slices of the PB volume into two parts, according to whether the ground-truth PB contour is overlapped or non-overlapped with the OAR contour. As a result, we get 2,190 OAR-overlapped slices and 1,108 non-OAR-overlapped slices in the entire dataset, respectively. For the OAR-overlapped slices, our result is significantly better than all the competing results with p-value < 0.05 (see the middle part of Table 2, PB contours overlapped with OARs). Meanwhile, for the non-OAR-overlapped slices, our result is also better than most of the competitors. But the difference within the multi-task deep learning-based methods is not statistically significant (see the bottom part of Table 2, PB contours non-overlapped with OARs). This result indicates that the superior performance of our method mainly comes from the OAR-overlapped slices where the PB boundaries are usually non-contrast and highly correlated with the OAR shape. This improvement is especially meaningful to the post-prostatectomy radiotherapy, where the PB contour accuracy in the OAR-overlapped slices has more impact than that in the non-OAR-overlapped slices. This is because the therapeutic dose has high chance to hurt the surrounding healthy tissues in the region where the PB is overlapped with the OARs. Therefore, the physicians often need extra efforts to carefully define the PB boundary in the OAR-overlapped slices.
Table 2:
Statistical significance testing on the results of deep learning-based methods
| U-Net | V-Net | VoxResNet | MT-U-Net | MTA-U-Net | Ours | |
|---|---|---|---|---|---|---|
| Whole PB volume | ||||||
| DSC [mean(std) %] | 73.29(7.43) | 73.36(7.89) | 74.58(7.45) | 74.41(7.23) | 75.03(7.11) | 75.67(6.56) |
| p-value | <0.0001 | <0.0001 | 0.0066 | 0.0001 | 0.0194 | - |
| ASD [mean(std) mm] | 2.79(1.27) | 2.67(1.22) | 2.55(1.11) | 2.58(1.21) | 2.51(1.13) | 2.42(1.03) |
| p-value | <0.0001 | <0.0001 | 0.0238 | 0.0006 | 0.0663 | - |
| PB contour overlapped with OARs (counted on 2,190 CT slices) | ||||||
| DSC [mean(std) %] | 79.75(14.49) | 78.80(16.12) | 79.20(17.19) | 80.50(14.57) | 80.78(14.80) | 81.82(13.40) |
| p-value | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | - |
| ASD [mean(std) mm] | 3.58(3.16) | 3.71(3.56) | 3.75(4.24) | 3.39(3.24) | 3.34(3.31) | 3.14(2.76) |
| p-value | <0.0001 | <0.0001 | <0.0001 | <0.0001 | 0.0074 | - |
| PB contour non-overlapped with OARs (counted on 1,108 CT slices) | ||||||
| DSC [mean(std) %] | 76.54(16.78) | 75.42(17.40) | 75.39(17.58) | 77.67(14.82) | 77.83(15.89) | 78.01(15.41) |
| p-value | 0.0002 | <0.0001 | 0.0003 | 0.3887 | 0.0897 | - |
| ASD [mean(std) mm] | 3.28(3.45) | 3.34(3.02) | 3.34(3.42) | 2.97(2.35) | 3.04(3.02) | 3.00(2.96) |
| p-value | 0.0061 | <0.0001 | 0.0403 | 0.8203 | 0.1578 | - |
The underline indicates a result that has no statistically significant difference with our result regarding its p-value > 0.05. Best result is highlighted in bold.
Fig. 4 visualizes some results on the OAR-overlapped slices. In these slices, the PB shows non-contrast boundary. Less intensity information can be leveraged to infer the contour. We can see the proposed method performs better than the competing methods, demonstrateing the effectiveness of our design towards the non-contrst PB boundary.
Fig. 4:
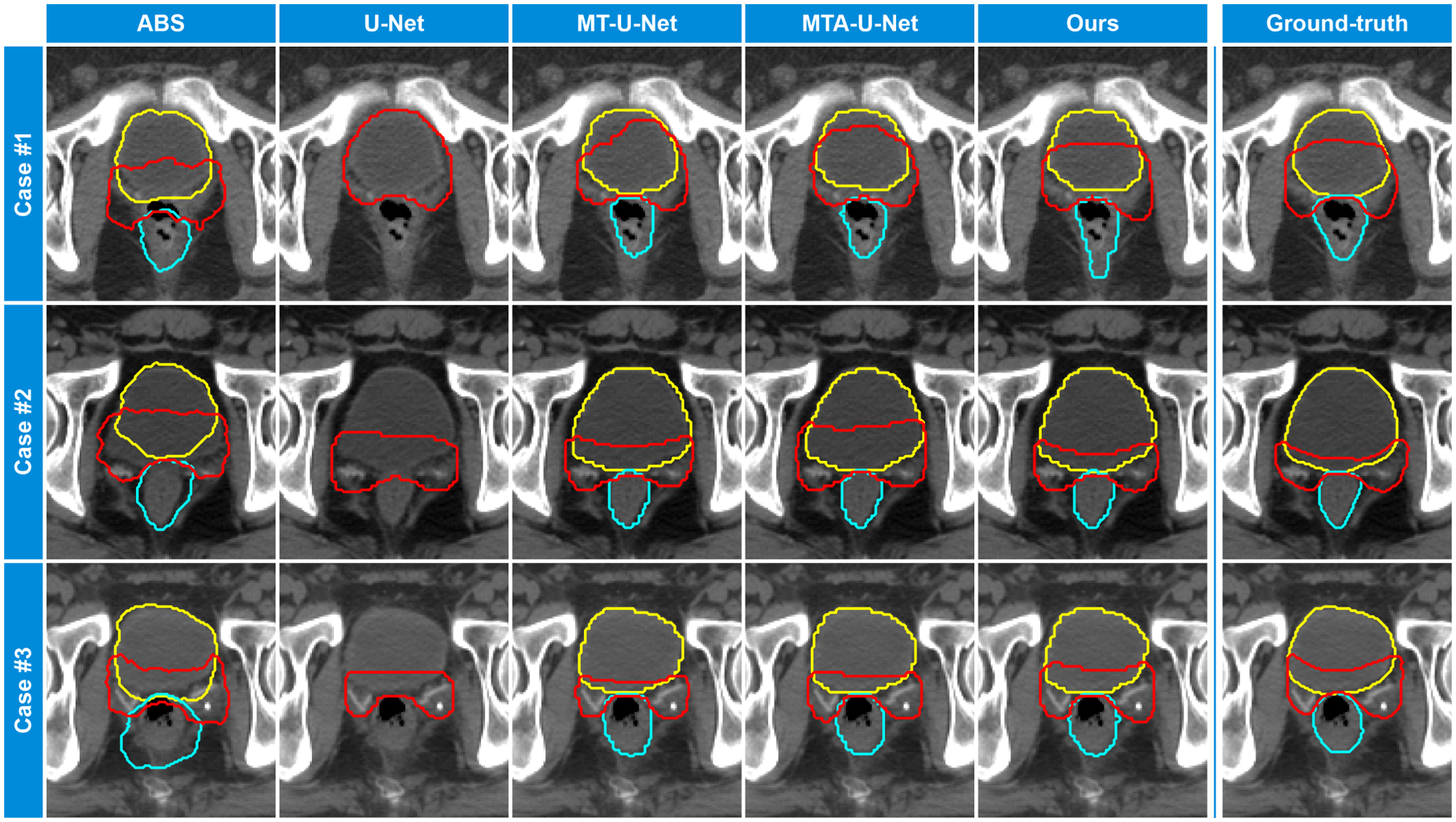
Results visualization. From left to right columns: contours generated by ABS method, U-Net, MT-U-Net, MTA-U-Net, and the proposed AMTA-Net. The ground-truth contours are shown in the rightmost column. The prostate bed, bladder, and rectum are displayed in red, yellow, and cyan color, respectively.
4.5. Ablation experiments
Overall, there are three key designs contributing to the superior performance of the proposed AMTA-Net: (i) the multi-task learning strategy jointly considering PB segmentation and OAR segmentation, (ii) the asymmetric network architecture explicitly modeling the one-sided structural dependency between PB and OARs, and (iii) the attention mechanisms flexibly selecting and transferring discriminative features for PB segmentation. In this section, we will conduct a set of ablation experiments on our AMTA-Net to justify the efficacy of these three designs.
4.5.1. Effectiveness of multi-task learning strategy
In our AMTA-Net, the multi-task learning strategy is realized by simultaneously training the backbone network for OAR segmentation and the attention sub-network for PB segmentation. When we mute the output of the backbone network by removing its last convolutional layer, the AMTA-Net degenerates to a single-task model focusing on PB segmentation. As the experimental results shown in Table 3, this ablation single-task model (Table 3, Ours w/o OAR prediction) achieves lower accuracy than the proposed AMTA-Net, demonstrating the effectiveness of the multi-task learning strategy in facilitating accurate PB segmentation.
Table 3:
Quantitative results of ablation experiments.
| Models | DSC of PB [mean(std) %] | ASD of PB [mean(std) mm] |
|---|---|---|
| Ours w/o OAR prediction | 74.20(7.40) | 2.65(1.25) |
| Ours w/o asym-architecture | 74.93(7.21) | 2.51(1.18) |
| Ours w/o attention | 75.15(6.62) | 2.46(0.99) |
| Ours | 75.67(6.56) | 2.42(1.03) |
4.5.2. Effectiveness of asymmetric network architecture
According to the clinical experience of the physicians, the delineation of PB highly relies on the neighboring OARs’ contour, while the non-contrast PB hardly bring additional information for the OAR segmentation. Inspired by this experience, our AMTA-Net is designed in an asymmetric architecture to explicitly leverage the one-sided dependency between PB and OARs. To justify the efficacy of this design, we re-organize our AMTA-Net to a symmetric architecture and compare this symmetric counterpart with our AMTA-Net. Specifically, we clone a sibling attention sub-network in our AMTA-Net to specially segment the OARs, and mute the original output of the backbone network. The number of feature maps in the backbone network is also cut in half to keep the network capacity (the number of trainable parameters) equivalent to our AMTA-Net. According to the experimental results shown in Table 3, this symmetric variant (Table 3, Ours w/o asym-architecture) achieves lower accuracy than our AMTA-Net, demonstrating the effectiveness of the asymmetric network architecture for PB segmentation.
4.5.3. Effectiveness of attention mechanisms
In our AMTA-Net, the attention mechanisms are implemented through the element-wise multiplication between the input feature maps and the self-learned soft attention masks. If we fix all these attention masks to an identity map, the attention mechanisms in our AMTA-Net will become invalid, and all the original features input to the attention sub-network will be directly used for the PB segmentation. According to our experimental results (Table 3, Ours w/o attention), disabling the attention mechanisms in our AMTA-Net leads to a decrease in final accuracy. This result indicates that, benefiting from the attention mechanisms, the proposed attention sub-network can focus on the discriminative parts of the input features, thus producing higher accuracy in PB segmentation.
5. Discussion
5.1. Effects of input adjacent slice number
The proposed AMTA-Net is fully implemented using 2D convolutional neural networks. To take more spatial context into consideration, we combine the adjacent slices with the center slice to compose a multi-channel image as the input. The number of the input slices is a hyper-parameter, which could affect the results of our method. To investigate this effect, we conduct an experiment on this hyper-parameter and list the experimental results in Table 4. It can be seen that the model using multi-slice input can achieve higher segmentation accuracy than the model using single-slice input. However, when we increase the number of the input adjacent slices further, the model performance does not get more improvement. We attribute this result to the highly variable shapes of the target organs along the axial direction. It is ideal for implementing the proposed method fully in a 3D manner. However, the large memory footprints of the 3D networks make it infeasible to directly expand our model to 3D structures without any compromise in image size and resolution, which would affect the segmentation accuracy. The training set in terms of 3D CT volumes is also much smaller than that in terms of 2D CT slices. This may increase the risk of over-fitting. To this end, we choose to implement our AMTA-Net in 2D manners.
Table 4:
Quantitative results of the proposed AMTA-Net inputting different number of adjacent slices.
| Number of input slices | DSC [mean(std) %] | ASD [mean(std) mm] | ||||
|---|---|---|---|---|---|---|
| PB | Bladder | Rectum | PB | Bladder | Rectum | |
| 1 | 75.14(7.19) | 87.60(9.03) | 80.29(6.95) | 2.47(1.15) | 1.65(1.58) | 2.48(1.64) |
| 3 | 75.67(6.56) | 88.40(8.72) | 80.35(9.36) | 2.42(1.03) | 1.47(1.22) | 2.60(3.00) |
| 5 | 75.32(6.87) | 88.46(8.72) | 79.74(9.59) | 2.47(1.07) | 1.47(1.33) | 2.76(3.08) |
| 7 | 75.56(6.87) | 88.25(9.30) | 80.22(7.67) | 2.49(1.10) | 1.56(1.65) | 2.54(1.89) |
5.2. Task balance between PB and OAR segmentation
As mentioned in Section 3.3, the contribution from the PB segmentation task and the OAR segmentation task to the final training objective is weighted by two hyper-parameters, i.e.,λpb and λoar. To determine the optimal balance between these two tasks, we separately train our AMTA-Net using different value combinations of λpb and λoar. The experimental results are listed in Table 5 and plotted in Fig. 5. It can be seen that the PB segmentation task reaches the highest accuracy when it is learned with equal importance to the OAR segmentation task (λpb : λoar = 1 : 1). This can be explained that, since the PB segmentation highly relies on the OAR segmentation, a larger ratio of λpb : λoar would bring worse results on OAR segmentation thus degrade the PB segmentation, while decreasing the ratio of λpb : λoar would directly degrade the PB segmentation. Therefore, we set λpb = 1 and λoar = 1 in our training stage.
Table 5:
Quantitative results of the proposed AMTA-Net trained with different weights of PB and OAR segmentation tasks.
| λpb : λoar | DSC [mean(std) %] | ASD [mean(std) mm] | ||||
|---|---|---|---|---|---|---|
| PB | Bladder | Rectum | PB | Bladder | Rectum | |
| 1 : 2 | 75.16(6.93) | 88.20(10.33) | 80.19(7.82) | 2.51(1.05) | 1.56(2.12) | 2.59(2.06) |
| 1 : 1 | 75.67(6.56) | 88.40(8.72) | 80.35(9.36) | 2.42(1.03) | 1.47(1.22) | 2.60(3.00) |
| 2 : 1 | 75.63(7.04) | 87.17(11.02) | 79.67(7.80) | 2.46(1.19) | 1.69(1.78) | 2.64(2.01) |
| 4 : 1 | 74.93(7.46) | 87.23(9.63) | 78.54(9.67) | 2.53(1.20) | 1.67(1.57) | 2.89(3.09) |
| 8 : 1 | 74.27(7.31) | 85.80(11.85) | 77.72(9.86) | 2.59(1.15) | 1.97(2.26) | 3.06(2.53) |
Fig. 5:
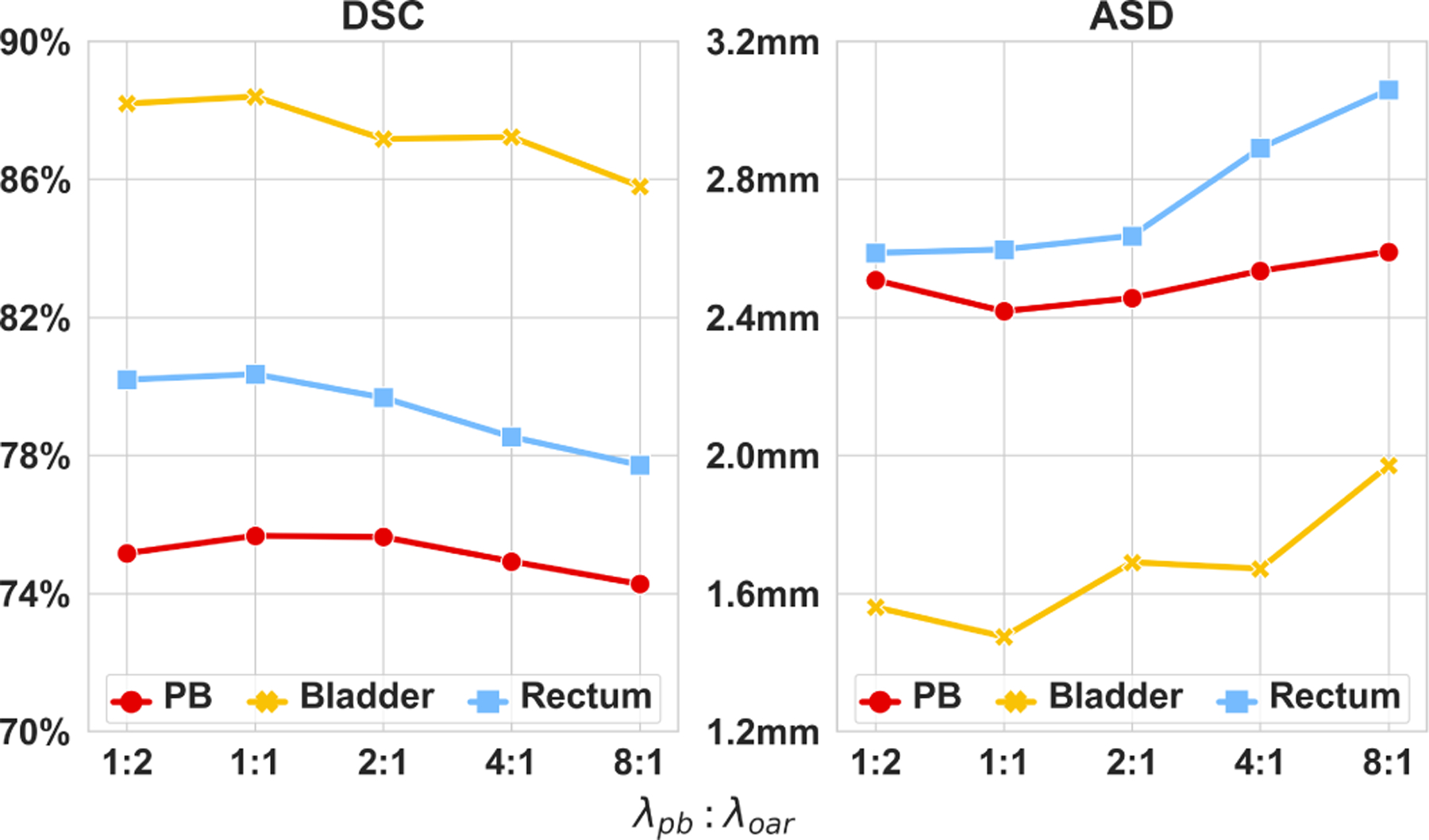
Mean DSC and ASD of the proposed AMTA-Net trained with different weights of PB segmentation task and OAR segmentation task.
5.3. Differences to multi-task attention U-Net
It is worth mentioning that, although our AMTA-Net and the competing method MTA-U-Net (Liu et al. (2019)) both leverage attention mechanisms to construct multi-task networks, the methodological designs of these two methods are fundamentally different. First, our network uses an asymmetric architecture to explicitly leverage the one-sided dependency between PB and OARs, while MTA-U-Net is a general symmetric multi-task network that treats all tasks equally. Second, the attention mechanisms in these two methods are implemented through different attention modules (AM). The AM used in our method pays attention to both the PB features extracted by the preceding AM and the OAR features from the backbone, while the AM in MTA-U-Net only considers the shared features in the backbone network. Third, our AMTA-Net has fewer trainable parameters than MTA-U-Net (about 75% of MTA-U-Net), implying higher parameter efficiency and better generalization capacity of our network.
5.4. Results discussion in clinical context
The absolute DSC of PB (75.67%) is not as high as that of the bladder (88.40%) and rectum (80.35%), indicating the inherent difficulty in PB segmentation compared with other organs. We attribute this to the fact that the PB has a large part of non-contrast boundaries which highly rely on the shape of surrounding organs and expert opinion rather than local intensities.
The proposed AMTA-Net outperforms the conventional ABS method by a significant margin. The superior performance demonstrates its clinical value since it can largely reduce the labor and time costs in the clinical workflow. The proposed method also outperforms the technical state-of-the-arts (i.e., the deep learning-based methods for general image segmentation), verifying the application-oriented methodological novelties in our design. It is worth noting that, our AMTA-Net achieves a statistically significant improvement in the most clinically critical part of the PB volume (i.e., the region overlapped with the neighboring OARs), where the physicians often need extra efforts to fine-tune the contours. This improvement has clinical benefits in the context of radiotherapy since the dose distribution in these regions needs to be made very sharp (about 10% of prescription dose per mm) to protect the normal tissues.
5.5. Effects of inter-observer variability
As discussed in the previous literature, the inter-observer variability in target contouring may be the most significant contributor to the uncertainty in radiation treatment (RT) planning (Hwee et al. (2011)). However, it is an inherent difficulty that always exists in the manual contouring procedure for PB, a “virtual” target, even with the use of consensus guidelines (Latorzeff et al. (2017)). To mitigate the effect of the inter-observer variability, in our clinical practice, the planning PB contour is defined by three experienced physicians following a two-step workflow: Step 1) One of the three physicians will manually delineate an initial contour following some pre-defined protocols. Step 2) The other two physicians will make further modification and verification on the initial contour to reach a consensus. Therefore, the resulting PB annotation in our study can be seen as a consensus opinion from the expert group. Since we use this consensus annotation as our training target (ground-truth), the trained deep network is expected to be able to behave as similar as the expert group, whose opinion suffers less inter-observer variability.
To demonstrate the significant inter-observer variability in the PB segmentation, we conduct an experiment to compare the contouring result of the individual observers (i.e., the contour delineated by one physician in Step 1 or by some other resident physicians) with that of the expert group (i.e., the consensus contour in Step 2, also the ground-truth annotation we used for training and evaluation). We conduct this comparison on a subset of our dataset, which contains 24 cases2. The comparison result is shown in Table 6. We also provide the result of other methods yielded from the same sub dataset.
Table 6:
Comparison with the individual human observer.
| Segmentation performed by | DSC of PB [mean(std) %] | ASD of PB [mean(std) mm] |
|---|---|---|
| Individual human observer | 72.22(12.76) | 3.03(1.86) |
| ABS | 65.88(9.52) | 3.84(1.54) |
| U-Net | 71.06(9.76) | 3.32(1.82) |
| V-Net | 71.46(10.72) | 3.08(1.69) |
| VoxResNet | 73.24(9.23) | 2.88(1.50) |
| MT-U-Net | 72.56(9.57) | 2.98(1.74) |
| MTA-U-Net | 73.12(10.06) | 3.03(1.89) |
| Ours | 74.46(8.86) | 2.67(1.37) |
It can be seen that, even for the human observer, the manual segmentation by individuals only achieves a mean dice of 72.22% when compared with the expert group opinion, demonstrating the severe inter-observer variability and the inherent difficulty in the PB segmentation. On the other hand, our method achieves a mean dice of 74.46%. This result proves the clinical value of our study, especially given the fact that the proposed method is fully automatic, which can significantly save the time and labor cost in manual delineation. Furthermore, this experiment also demonstrates the effectiveness of our method in alleviating the inter-observer variability, which is meaningful to mitigating the uncertainty in RT planning (Hwee et al. (2011)).
5.6. Impact on radiotherapy dose distribution
In the workflow of RT planning, PB and OAR segmentation act as a prerequisite for the subsequent radiation dose optimization. The accuracy of PB and OAR contour could directly affect the quality of the dose distribution, which is correlative to the treatment efficacy. As we have demonstrated that our method achieves higher PB segmentation accuracy than other competitors, it would be desired to know how much gain could this improvement bring to the RT dose distribution. Therefore, we investigate the RT dose of 10 cases in our dataset, where our method achieves higher PB contour accuracy than other methods. We calculate the dose-volume histograms (DVHs) using the PB and OAR contours generated by different segmentation methods. Fig. 6 shows the DVHs of two example cases. For brevity, we only evaluate the DVH associated with the multi-organ segmentation methods (including the ABS method, MT-U-Net, MTA-U-Net, and our method) in this experiment since the multi-organ segmentation methods generally present better performance than the single-organ segmentation methods.
Fig. 6:
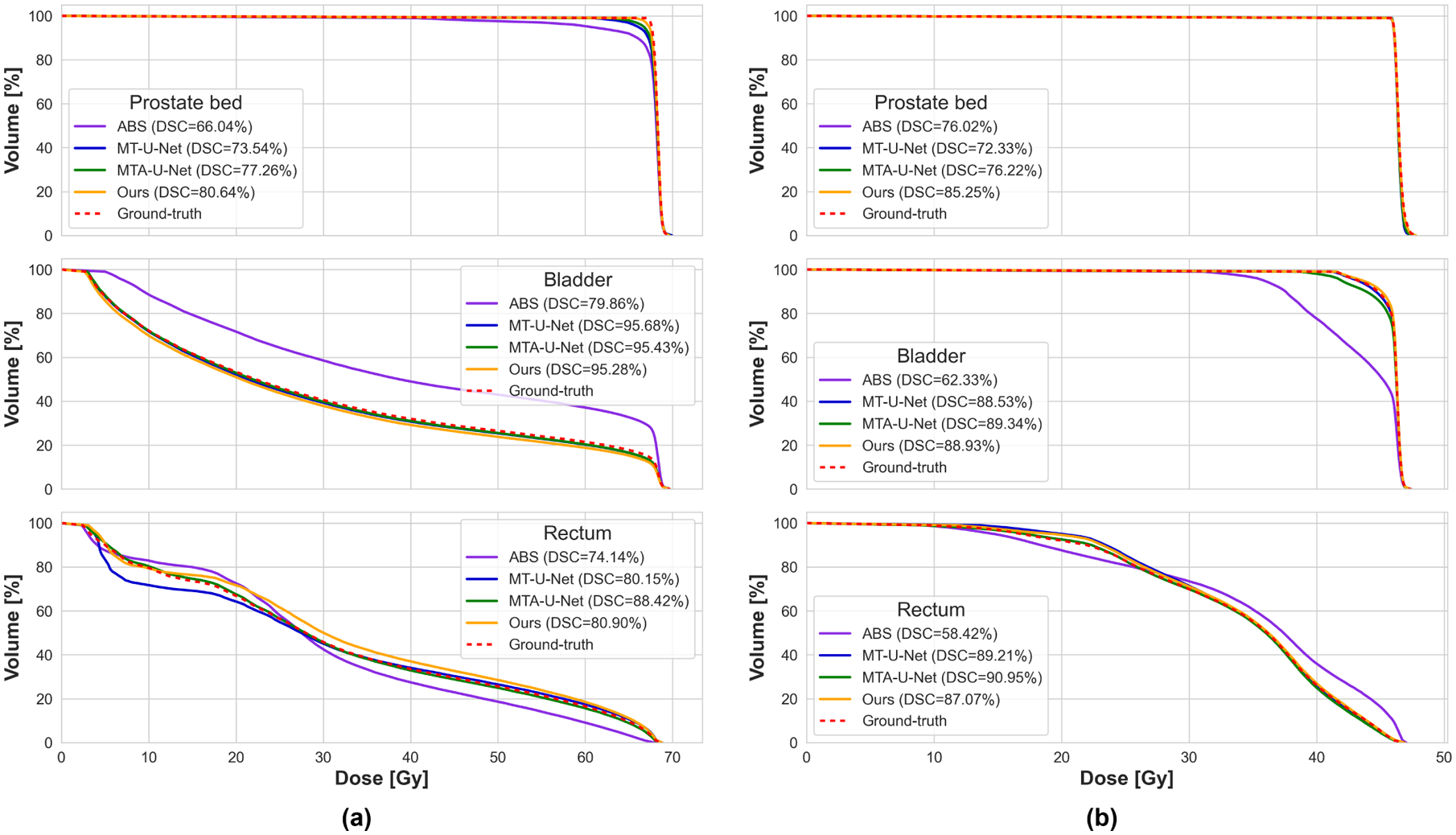
Two example cases (a) and (b) showing dose-volume histograms (DVHs) calculated on PB (top), bladder (middle), and rectum (bottom) contours generated by different methods. The DSC of corresponding segmentation is indicated in the legend.
According to our observation, we generally find that higher segmentation accuracy can contribute to a better shaped DVH, which is closer to the ground-truth. To quantify the quality of the dose distribution, we calculate the prescription dose coverage on the PB volume and list the corresponding PB segmentation DSC in Table 7. The prescription dose coverage is a criterion widely used in radiotherapy to measure the planning dose quality, which is defined as the percentage volume of the target getting the dose higher than the prescription dose. Ideally, we want 100% volume of the PB to get covered by the prescription dose.
Table 7:
Correlation between segmentation accuracy and prescription dose coverage on PB.
| PB contour from | DSC [mean(std) %] | Coverage [mean(std) %] |
|---|---|---|
| ABS | 61.78(10.77) | 83.83(13.10) |
| MT-U-Net | 71.96(12.49) | 88.68(14.04) |
| MTA-U-Net | 73.28(11.55) | 91.90(14.77) |
| Ours | 77.37(10.01) | 93.51(11.09) |
| Ground-truth | 100.00 | 99.50(0.92) |
From Table 7 we can see the segmentation accuracy shows a positive correlation with the prescription dose coverage on PB, which is consistent with our observation on the DVH. On the other hand, it seems the coverage is less sensitive to the PB segmentation accuracy since the PB contour with a mean DSC of 77.37% can still achieve a mean coverage of 93.51%. Similar phenomenons also exist in some DVHs (see the top DVH in Fig. 6(b)), where the DSC of PB contours generated by different methods varies in a large range, but the corresponding DVHs are almost the same as the ground-truth. We attribute this insensitivity to the fact that both the DVH and the coverage criterion only care about the dose distribution inside the evaluated contour. If the PB segmentation is smaller than the ground-truth (under-segmented), although the segmentation accuracy is low, the DVH and coverage could still look good.
Since the uncertainties in PB segmentation of radiotherapy plan will directly impact on bladder irradiation, it will be highly clinically relevant to study the correlation between urinary toxicity and accuracy of PB segmentation. However, this topic is out of our current research design, and we leave it for the readers who work in related domains.
6. Conclusion
In this work, we propose an Asymmetric Multi-Task Attention Network (AMTA-Net) to address the challenging problem of PB segmentation in CT images. The proposed AMTA-Net mainly consists of two parts: The first part is a backbone network with U-Net architecture used to conduct the low-level (or prerequisite) task of OAR segmentation. Based on the backbone network, the second part is an attention sub-network used to perform the high-level (or primary) task of PB segmentation. The attention sub-network consists of a series of cascaded attention modules, which hierarchically select and transfer the most relevant OAR features in the backbone network to generate discriminative feature representations for accurate PB segmentation. Three key properties of the proposed AMTA-Net (i.e., the multi-task learning strategy, the asymmetric network architecture, and the attention mechanisms) contribute to the PB segmentation. We comprehensively evaluate the proposed AMTA-Net on a clinical dataset composed of 186 CT images. The experimental results show that the proposed AMTA-Net significantly outperforms the clinical state-of-the-art methods (i.e., the ABS methods) and also presents better performance in comparison to the technical state-of-the-art methods (i.e., the deep learning-based methods for general image segmentation), especially for the most indistinguishable and clinically critical part of PB boundaries, demonstrating the clinical value and technical novelty of our method.
Highlights.
The first exploration using deep learning methods to segment prostate bed in CT image
Asymmetric multi-task network taking reference from OAR to facilitate PB segmentation
Attention mechanisms selecting discriminative features for accurate PB segmentation
Concurrent segmentation of bladder and rectum for OAR definition in radiotherapy
Acknowledgments
This work was supported in part by NIH grant CA206100.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Because the contour delineated by an individual physician is a temporary intermediate product of the final planning contours, it is not necessarily archived in all cases. We only recalled this kind of contour in 24 cases in our dataset.
References
- Bragman FJ, et al. , 2018. Uncertainty in multitask learning: joint representations for probabilistic mr-only radiotherapy planning, in: International Conference on Medical Image Computing and Computer-Assisted Intervention, Springer. pp. 3–11. [Google Scholar]
- Chen H, et al. , 2018. Voxresnet: Deep voxelwise residual networks for brain segmentation from 3d mr images. NeuroImage 170, 446–455. [DOI] [PubMed] [Google Scholar]
- Delpon G, et al. , 2016. Comparison of automated atlas-based segmentation software for postoperative prostate cancer radiotherapy. Frontiers in oncology 6, 178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glorot X, Bengio Y, 2010. Understanding the difficulty of training deep feedforward neural networks, in: Proceedings of the thirteenth international conference on artificial intelligence and statistics, pp. 249–256. [Google Scholar]
- He K, et al. , 2019. Pelvic organ segmentation using distinctive curve guided fully convolutional networks. IEEE transactions on medical imaging 38, 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwee J, et al. , 2011. Technology assessment of automated atlas based segmentation in prostate bed contouring. Radiation Oncology 6, 110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias JE, Sabuncu MR, 2015. Multi-atlas segmentation of biomedical images: a survey. Medical image analysis 24, 205–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioffe S, Szegedy C, 2015. Batch normalization: Accelerating deep network training by reducing internal covariate shift. arXiv preprint arXiv:1502.03167. [Google Scholar]
- Jia H, Yap PT, Shen D, 2012. Iterative multi-atlas-based multi-image segmentation with tree-based registration. NeuroImage 59, 422–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingma DP, Ba J, 2014. Adam: A method for stochastic optimization. arXiv preprint arXiv:1412.6980. [Google Scholar]
- Latorzeff I, et al. , 2017. Delineation of the prostate bed: the invisible target is still an issue? Frontiers in oncology 7, 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeCun Y, et al. , 1998. Gradient-based learning applied to document recognition. Proceedings of the IEEE 86, 2278–2324. [Google Scholar]
- Lee G, et al. , 2016. Asymmetric multi-task learning based on task relatedness and loss, in: International Conference on Machine Learning, pp. 230–238. [Google Scholar]
- Lian C, et al. , 2020. Hierarchical fully convolutional network for joint atrophy localization and Alzheimer’s disease diagnosis using structural MRI. IEEE Transactions on Pattern Analysis and Machine Intelligence 42, 880–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, et al. , 2019. End-to-end multi-task learning with attention, in: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 1871–1880. [Google Scholar]
- Long J, et al. , 2015. Fully convolutional networks for semantic segmentation, in: Proceedings of the IEEE conference on computer vision and pattern recognition, pp. 3431–3440. [Google Scholar]
- Michalski JM, et al. , 2010. Development of rtog consensus guidelines for the definition of the clinical target volume for postoperative conformal radiation therapy for prostate cancer. International Journal of Radiation Oncology*Biology*Physics 76, 361–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milletari F, et al. , 2016. V-net: Fully convolutional neural networks for volumetric medical image segmentation, in: 2016 Fourth International Conference on 3D Vision (3DV), IEEE. pp. 565–571. [Google Scholar]
- Moeskops P, et al. , 2016. Deep learning for multi-task medical image segmentation in multiple modalities, in: International Conference on Medical Image Computing and Computer-Assisted Intervention, Springer. pp. 478–486. [Google Scholar]
- Mohamed A, Zacharaki EI, Shen D, Davatzikos C, 2006. Deformable registration of brain tumor images via a statistical model of tumor-induced deformation. Medical image analysis 10, 752–763. [DOI] [PubMed] [Google Scholar]
- Nair V, Hinton GE, 2010. Rectified linear units improve restricted boltzmann machines, in: Proceedings of the 27th international conference on machine learning (ICML-10), pp. 807–814. [Google Scholar]
- Nie D, et al. , 2019. Strainet: Spatially varying stochastic residual adversarial networks for mri pelvic organ segmentation. IEEE transactions on neural networks and learning systems 30, 1552–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poortmans P, et al. , 2007. Guidelines for target volume definition in postoperative radiotherapy for prostate cancer, on behalf of the eortc radiation oncology group. Radiotherapy and Oncology 84, 121–127. [DOI] [PubMed] [Google Scholar]
- Ronneberger O, et al. , 2015. U-net: Convolutional networks for biomedical image segmentation, in: International Conference on Medical image computing and computer-assisted intervention, Springer. pp. 234–241. [Google Scholar]
- Sidhom MA, et al. , 2008. Post-prostatectomy radiation therapy: consensus guidelines of the australian and new zealand radiation oncology genitourinary group. Radiotherapy and Oncology 88, 10–19. [DOI] [PubMed] [Google Scholar]
- Siegel RL, et al. , 2020. Cancer statistics, 2020. CA: A Cancer Journal for Clinicians; 70, 7–30. [DOI] [PubMed] [Google Scholar]
- Wang S, et al. , 2019. Ct male pelvic organ segmentation using fully convolutional networks with boundary sensitive representation. Medical image analysis 54, 168–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, et al. , 2020a. Ct male pelvic organ segmentation via hybrid loss network with incomplete annotation. IEEE Transactions on Medical Imaging. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, et al. , 2020b. Iterative label denoising network: Segmenting male pelvic organs in ct from 3d bounding box annotations. IEEE Transactions on Biomedical Engineering. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiltshire KL, et al. , 2007. Anatomic boundaries of the clinical target volume (prostate bed) after radical prostatectomy. International Journal of Radiation Oncology*Biology*Physics 69, 1090–1099. [DOI] [PubMed] [Google Scholar]
- Wu G, Jia H, Wang Q, Shen D, 2011. Sharpmean: groupwise registration guided by sharp mean image and tree-based registration. NeuroImage 56, 1968–1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, et al. , 2018. Automatic bladder segmentation from ct images using deep cnn and 3d fully connected crf-rnn. International Journal of Computer Assisted Radiology and Surgery 13, 967–975. [DOI] [PubMed] [Google Scholar]
- Xu X, et al. , 2020. Asymmetrical multi-task attention u-net for the segmentation of prostate bed in ct image, in: International Conference on Medical Image Computing and Computer-Assisted Intervention, Springer. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue W, et al. , 2018. Full left ventricle quantification via deep multitask relationships learning. Medical image analysis 43, 54–65. [DOI] [PubMed] [Google Scholar]
- Zhan Y, Ou Y, Feldman M, Tomaszeweski J, Davatzikos C, Shen D, 2007. Registering histologic and mr images of prostate for image-based cancer detection. Academic radiology 14, 1367–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Yang Q, 2017. A survey on multi-task learning. arXiv preprint arXiv:1707.08114. [Google Scholar]


