Abstract
The CRISPR-based genome editing technology has opened extremely useful strategies in biological research and clinical therapeutics, thus attracting great attention with tremendous progress in the past decade. Despite its robust potential in personalized and precision medicine, the CRISPR-based gene editing has been limited by inefficient in vivo delivery to the target cells and by safety concerns of viral vectors for clinical setting. In this review, recent advances in tailored nanoparticles as a means of non-viral delivery vector for CRISPR/Cas systems are thoroughly discussed. Unique characteristics of the nanoparticles including controllable size, surface tunability, and low immune response lead considerable potential of CRISPR-based gene editing as a translational medicine. We will present an overall view on essential elements in CRISPR/Cas systems and the nanoparticle-based delivery carriers including advantages and challenges. Perspectives to advance the current limitations are also discussed toward bench-to-bedside translation in engineering aspects.
Keywords: CRISPR, Nanoparticle, Gene editing, Drug delivery, Gene theray
Introduction
Since the discovery of the clustered regularly-interspaced short palindromic repeats (CRISPR) as a part of adaptive prokaryotic immune system, the CRISPR-associated systems (Cas) have revolutionized the field of molecular biology [1]. In particular, the simplicity and the specificity of CRISPR/Cas that can readily edit DNA sequences in vivo are changing the paradigm in biomedical research including disease target validation and genetic disorder mechanism identification as well as epigenetic studies over the decade [2–8]. The CRISPR/Cas-based gene editing consists of two main components: a guide RNA (gRNA) for target specificity and a non-specific CRISPR-associated nuclease (e.g., Cas9). The gRNA is a short synthetic RNA composed of a scaffold sequence (for Cas9-binding) and a spacer or targeting sequence (typically, ~ 20 nucleotides) intended to edit. The Cas9 nuclease is directed by a gRNA to modify a specific chromosomal DNA sequence by inducing a sequence-specific double-strand break (DSB), which is then resolved by error-prone non-homologous end-joining (NHEJ) repair mechanism [9–11]. Consequently, the CRISPR/Cas system can be employed for gene knockout, activation, repression, and epigenetic editing (methylation or demethylation). The CRISPR/Cas is also clinically transforming the therapeutic treatment of diseases from monogenic rare disease to immunotherapy because it potentially corrects disease-causing mutations as a tool of precision medicine. Moreover, high recurrence rate and chemoresistance for most types of cancers highlight the need for new therapeutic modality such as gene therapy. For example, most anticancer drugs require repeated administration, which increases side effects, toxicity, and medical cost, thus severely reduces patients’ quality of life. However, the CRISPR/Cas gene editing has the potential to permanently disrupt tumor survival genes, which enables to overcome the repeated dosing limitations of traditional cancer chemotherapy [12]. Despite the remarkable advances of CRISPR-based gene therapy, safe and efficient delivery of the CRISPR components to the target cell or organ is the main challenge for clinical translation, thus its medical applications still remain limited. In this regard, considering clinical translation of CRISPR/Cas system, the development of safe and efficient platforms to deliver the CRISPR components for accurate therapeutic purpose are one of the crucial factors, which will lead exclusive breakthrough in translational medicine.
Inherent ability of virus to introduce exogenous genetic materials into host cells ensures high transduction efficiency, thus the viral vectors have been commonly adapted to deliver CRISPR components in vivo [13]. Although high delivery efficiency is obviously the main advantage of viral vectors over non-viral delivery platforms, safety concerns including immunogenicity and insertional mutagenesis are major obstacles of viral vectors to be addressed with the development of safe and advanced delivering vehicles [14]. Adeno-associated viruses (AAVs) is one of the most commonly used viral vector due to long-term stability and relatively lower immunogenicity and genomic integration rate, thus several clinical trials using AAVs have been progressed [15]. Nevertheless, recent studies have reported incremental risk of AAV integration in CRISPR for clinical uses including severe insertional mutagenesis and carcinogenesis [16, 17]. For instance, a study of dog with hemophilia treated with AAV up to 10 years showed that the vector can readily insert in many spots across the host’s genome, sometimes near genes affecting cell growth [7].
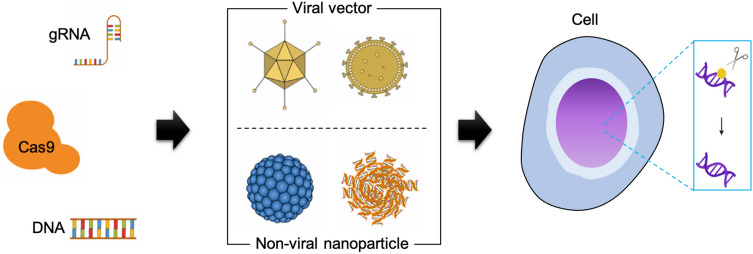
Non-viral delivery systems offer fascinating characteristics such as reduced off-target effects and cytotoxicity over the viral vectors. In addition, non-viral vehicles utilize Cas9 mRNA or proteins with synthetic sgRNA (e.g., ribonucleoprotein (RNP)) to directly deliver the CRISPR components, rather than requiring transcription and translation steps as needed for viral CRISPR delivery. However, the large size of both Cas9 protein (~ 160 kDa) and sgRNA (~ 31 kDa) is one of the major obstacles for non-viral delivery systems. Nanoparticle-based delivery systems as a non-viral vector have been reported to transport the CRISPR components to the target cells and tissues, resulting in promising gene editing and therapeutic efficacy [18–22]. Nanoparticles are attractive because of large loading capacity, scale-up capability, controllable surface modification for specific cell targeting, and minimal immunogenicity. However, lower delivery efficiency to non-liver tissues remains an important challenge for therapeutic translation for the nanoparticle-mediated CRISPR/Cas gene editing systems. Here, we review the recent advances of gene editing technology with particular focus on nanoparticle-mediated CRISPR/Cas delivery (Fig. 1), and provide in-depth overlook on the targeted delivery strategy of the CRISPR/Cas-based translational medicine. As the nanoparticle-mediated delivery systems are the promising non-viral vector for clinically reliable CRISPR-based genome editing, the engineering aspects of material characteristics are thoroughly discussed with potential and challenges in delivering CRISPR/Cas system for translational medicine.
Fig. 1.
Schematic illustration depicting genome-editing with CRISPR-Cas9 system. Viral or non-viral delivery methods have been widely adopted to transport the CRISPR-based genomic editing components including gRNA, Cas9 and donor DNA
Gene editing: past and present
The advent of gene editing tools
The field of molecular biology has been vastly developed over the past years, with gene editing being the most promising one due to its potential applications in gene therapy to treat incurable disease with recent molecular drugs. Efforts on the gene editing technology have also attracted great attention in the field of translational medicine, which provides the game-changer therapeutics in a wide range of cancers and genetic disorders. The core technique of the genome editing is based on the use of engineered nucleases composed of sequence-specific DNA-binding domains fused to a nonspecific DNA cleavage module. Therefore, the gene editing process requires the presence of highly specific nucleases which cleave the desired sequence in the genome, resulting in the creation of site-specific double strand breaks (DSB) and the subsequent DNA modification through the DSB repair pathways [23, 24]. DSB can be repaired by either error prone NHEJ or template assisted error free homology directed repair (HDR). In NHEJ, the DSB is repaired with some random nucleotides inserted or deleted (indels) at the DSB site since there is no template sequence used. As a result, the NHEJ pathway disrupts the gene sequence and is usually used for gene knockout research [25, 26]. The NHEJ is simple and efficient but there is a risk of mutagenesis. On the other hand, the HDR pathway enables use of a homologous donor template, either in the forms of single-stranded oligodeoxynucleotides (ssODNs) or double-stranded DNA (dsDNA), resulting in targeted gene editing including gene knock-in research [27]. Despite being less efficient in editing, the HDR pathway is more accurate (less error-prone) compared to NHEJ pathway. Consequently, the process of genome modification via HDR may pave the way to treating a number of multifactorial genetic disorder diseases since it can modify and correct the mutated genes back to their intended wild-type version [28, 29].
To induce precise modification of specific DNA sequences, researchers have designed different classes of nucleases: meganucleases, zinc-finger nucleases (ZFN), and transcription activator-like effector nucleases (TALEN) [30]. Meganucleases are the first endonuclease discovered; they are very specific endonucleases that can recognize the dsDNA base pairs ranging from 14 to 40, thus acting as scissors to modify the sequences in a certain genome. Their compatibility with broad types of delivery strategies to the target cell has made the meganucleases become widely utilized as a gene editing tool [31, 32]. ZFN and TALEN are chimeric nucleases composed of programmable, sequence-specific DNA-binding module zinc finger and DNA binding domain of TLA, respectively, linked to a nonspecific DNA restriction endonuclease FokI [33–35]. Consequently, both ZFNs and TALENs enable a broad range of genetic modifications by inducing DNA double-strand breaks that stimulate error-prone NHEJ or HDR at specific genomic locations [30]. The simplicity and flexibility of these nucleases are powerful to redefine the boundary of molecular biology, thus being catapulted to the forefront of genetic engineering.
Although the ZFN and TALEN have been proven to be useful in gene therapy and consequently have opened up a wide range of biomedical applications, several challenging aspects still hinder their clinical translations [36]. For example, constructing meganucleases has been an arduous task for researchers since the enzyme exhibits a non-modular configuration, whereby both DNA recognition and cleavage site become intertwined at some points in its domain [37]. TALENs also prove to be challenging because of their large size, and this impedes researchers to have only few options for their delivery [38]. And last but not the least, incremental off-target possibility is one of the crucial concerns once exposed to the host cells [39–41]. However, the utilization of CRISPR-Cas9 system has significantly advanced the current standard to engineer genome and has led the vast breakthrough on molecular biology, biomedical engineering, and translational medicine [42].
CRISPR-Cas as an inherent defensive system of bacteria
The CRISPR system is originally involved in the defense system of bacteria and related species [43]. In fact, a short sequence of the viral DNA is inserted between the bacterial genome’s CRISPR sequences when a virus (e.g., bacteriophage) infects a bacterial cell, which is the first stage (adaptation) of bacterial defense process. In the second stage (expression), the bacterial cell transcribes the CRISPR sequences into pre-crRNA, which is processed afterwards into becoming a mature crRNA. Finally, the complex of crRNA and Cas protein recognize and attack the recurring invading virus’ nucleic acid sequences (the last stage, interference). This is achieved through base complementarity between the target strand and crRNA [44–46]. The insertion of viral DNA sequences onto the bacterial genome’s CRISPR sequences is essential because the memory guides the Cas proteins to attack the same virus and disable its infectious machinery by introducing double-strand breaks.
Over the past years, there has been an extensive research about the diversity of CRISPR-Cas system. The CRISPR-Cas systems are categorized into two main classes, with each class diverging into many types and divisions [47]. The categorized classes are based on the effector molecules used by the CRISPR-Cas systems: class 1 contains multi-subunit effector complexes while class 2 consists of a single multi-domain effector molecule. Class 1 system, which is subdivided into types I, III, and IV, is widely utilized by bacteria and archaea, covering almost ~ 90% of CRISPR-Cas systems. On the other hand, class 2 system, which includes types II, V, and VI, is used exclusively by bacteria, comprising ~ 10% of CRISPR-Cas systems. Each type has its own unique characteristics; for example, presence of protospacer adjacent motif (PAM) is required in type I and type II systems to invade DNA sequences for recognition, while type III system does not need the PAM sequence [48, 49]. Despite having these subtle differences from one another, the CRISPR/Cas systems serve as an adaptive immune response for all the microbial species since the mechanism is specific to the invading virus’ genetic sequences, and this effectively blocks the viral infection when the same virus happens to attack again.
CRISPR as a programmable genome editing tool
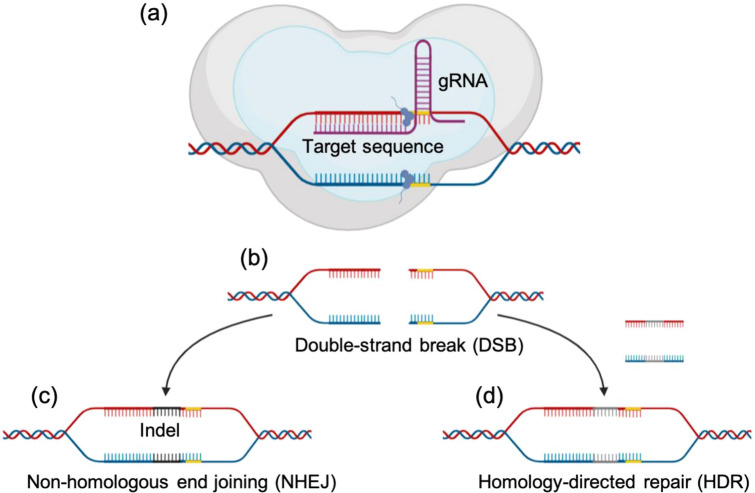
Originally used by bacteria to render the invading virus harmless, the CRISPR has been widely adopted as a new way to alter specific genetic information contained in a certain organism. As a result, the CRISPR has gained tremendous attention and has been demonstrated as a means of potential therapeutics on many kinds of diseases, genetic disorders, and even cancers [50–57]. Generally, the CRISPR-based genome editing system is composed of two main components: either two-part or single guide RNA and Cas protein. The two-part guide RNA composed of crRNA and tracrRNA required for target sequence specificity and complex formation with Cas protein and crRNA as a scaffold, respectively. The Cas9 nuclease is directed by the gRNA to modify a specific chromosomal DNA sequence by inducing a sequence-specific DSB, which is then resolved by error-prone NHEJ repair mechanism or template assisted error free homology directed repair mechanism (Fig. 2).
Fig. 2.
Mechanism of CRISPR-Cas9 as a genome editing tool. The Cas9-gRNA complex makes its way to the target sequence: a the Cas9 nuclease cuts the desired specific sequences. b This process leads to the formation of DSB, which will be consequently repaired by the cell’s DSB repair pathways. c In NHEJ, the breaks are repaired by creating insertions or deletions (indels). d HDR, on the other hand, makes use of a donor template which leads to a precise modification
In addition to the classical CRISPR/Cas9 system, there are many engineered versions of CRISPR system for genome editing such as base editor and prime editor. The base editor (BE) directly converts cytidine to uridine without DNA double strand breaks by fusing cytidine deaminase to endonuclease deficient dCas9. The cytidine deaminase is delivered to target sequence by dCas9/gRNA and convert nearby cytidine to uridine resulting in C to T or G to A substitution. This can be used to convert several disease risk factor genes to its normal sequence without the risk of DSB and subsequent random indel mutation [58]. The primer editor (PE) is a genome editing system without inducing DNA double strand break. This system composed of catalytically impaired Cas9 nickase fused to engineered reverse transcriptase (the prime editor), and prime editing guide RNA(pegRNA) which has crRNA sequence for target specificity, spacerRNA sequence as a scaffold and extended sequence at 3’ as a template for gene editing. At the target region, single strand DNA nick is generated by Cas9, and this nicked strand is replaced with DNA which is synthesized by reverse transcriptase domain of PE using pegRNA as a template [59].
Recently, a number of engineered CRISPR systems has been suggested to improve gene editing efficiency, reduce off-target editing, bypass double strand break and be only activated by external stimuli. The advances in CRISPR system for genome editing emphasize the importance and potential of development of safe and efficient universal CRISPR delivery system as a platform for delivery of not only classical CRISPR/Cas9 system, but also their engineered derivatives for application in translational medicine.
Intracellular delivery of CRISPR components
Source of Cas9 protein
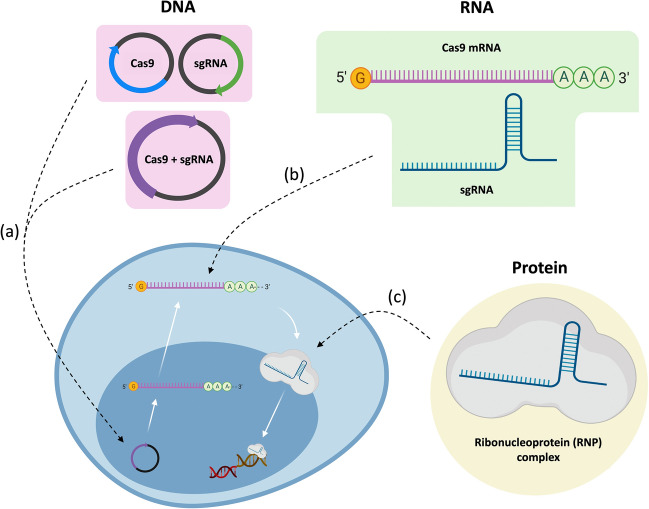
The delivery of CRISPR-based system, in terms of the source and/or format of the Cas9 nuclease, is subdivided into three categories: DNA, mRNA, and protein (Fig. 3) [60, 61]. DNA plasmid as a source of Cas9 nuclease has been most widely used to induce CRISPR-based genome editing, because they are relatively stable, cheap, and easy to manipulate [62–65]. Once the DNA plasmid is delivered into a cell, it should enter the nucleus for transcription of Cas9 mRNA and gRNA. Cas9 protein is then translated from the processed mRNA, and finally Cas9/gRNA complex cuts chromosomal DNA after nuclear translocation. This process is, however, inefficient due to the many steps required for mature Cas9/gRNA ribonucleoprotein expression and proper localization. Furthermore, there is a risk of off-target mutation due to sustained expression of CRISPR system even after target gene editing is finished [66].
Fig. 3.
Different sources of the Cas9 nuclease. a The Cas9 DNA plasmid (either two different plasmids or a single plasmid) is delivered to the cell nucleus, followed by undergoing transcription in the process. b The Cas9 mRNA is delivered to the cytoplasm to be translated to produce Cas9 protein. c The RNP (Cas9-sgRNA complex) is delivered to the nucleus and can directly target the desired genes in the cell
Having no need for the DNA to be transcribed, delivery via mRNA can be a beneficial method compared to the DNA plasmid delivery [63]. This process is also advantageous since there is no need for the CRISPR components to cross the nuclear membrane; only a direct delivery to the cytoplasm is enough for the ribosomes to express the delivered mRNA. However, secured delivery and rapid expression of mRNA is essential due to unstable nature of mRNA to RNases in physiological environment. In fact, stability of mRNA compromises the gene editing efficiency, making this method somewhat still challenging to be translated in clinics. A number of studies has been carried out to increase the stability and expression of the mRNA by chemical modification of nucleic acid molecules [67–69].
Lastly, the ribonucleoprotein (RNP) complex is the simplest and most efficient means of delivery with higher gene editing efficiency since additional expression is not required and nuclear localization is the only localization process required [61, 63]. Many studies are now focusing on this format since it offers many benefits such as the sgRNA being protected by the RNP complex from any degradative enzymes and the complex having a relatively short half-life which cuts the chances for off-target mutations inside the cell [70]. However, like the other sources of Cas9 protein, delivering the RNP complex is still inundated with impediments such as the highly negatively-charged nature of sgRNA and the proper encapsulation of the largely-sized Cas9 nuclease.
Delivery strategies for CRISPR-based gene editing: conventional approach
Physical internalization
Microinjection and electroporation have been widely used as intuitive physical means to directly internalize the CRISPR components (Fig. 4). As a long-established strategy of mechanical transfection, the microinjection uses the combination of a microscope and a micrometer-scale needle that effectively pierces through the cell membrane. The direct delivery of the CRISPR/Cas construct into the cell, effectively bypassing the barriers such as extracellular matrix and cell membrane, makes microinjection a powerful method, and this consequently has been used in many research areas [71–73]. The cargo size, which is considered one of the important limiting factors that should be addressed in delivery systems, is also not an issue when using this technique. In addition, this technique confers that manual injection provides a way to better control the dosage of the cargoes that are being delivered onto the cells [60, 61]. However, there are certainly major drawbacks when using this specific delivery method. Firstly, when delivering the Cas9 in DNA plasmid format, there is a high chance that a random integration of the vector into the chromosome will happen. Secondly, the delivery of Cas9 (in mRNA format) and sgRNA seems to be an arduous task since injecting the CRISPR/Cas construct happens not in one location, but in two different locations which are nucleus and cytoplasm, and so doing it with two different microinjections is impractical [71]. Finally, the use of microscope renders the overall delivery useless when used in in vivo setting [60].
Fig. 4.
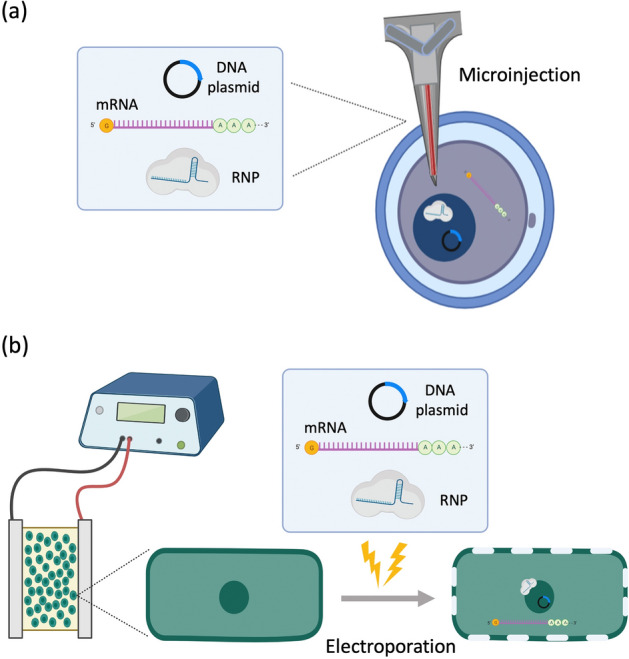
CRISPR-Cas9 system delivery by physical methods. a Microinjection: The cargoes are delivered using a microscopic needle which can penetrate through the cell membrane and nuclear membrane. b Electroporation: The cell membrane is perturbed through the use of voltage; the presence of pores allows the cargoes to be delivered onto their designated sites
Electroporation, on the other hand, employs an optimized voltage with which electrical pulses are sent through and consequently disrupt the cell membrane. This creates temporary pores which can then be used to deliver the system into the cells. One of the advantages this technique offers is that it does not discriminate among diverse types of cells, and so those cells that are originally difficult to control can now accept the CRISPR components that are being delivered; one typical example includes the use of tube electroporation that effectively delivered the CRISPR/Cas9 construct into human stem and primary cells with high efficiency and low cytotoxicity [74]. A different approach towards electroporation has been also performed, so-called zygote electroporation of nuclease (ZEN), which successfully delivered CRISPR-Cas9 material into the mouse zygotes [75]. The technique has been also known to a safe method since the presence of any chemical or viral components is not needed to carry out its function. Like microinjection, however, electroporation is not suitable as a delivery strategy in in vivo and clinical settings due to mammalian cells being quite sensitive to high voltages [60].
Viral vectors
Viruses have been dealt as essential elements in the fields of molecular biology and biotechnology as they are easy to manipulate and can serve as a vector for delivery of genetic information to understand the cellular dynamics and functions. They are generally considered as natural vectors for genetic information that led them to being used in many different platforms nowadays particularly in gene therapy (Fig. 5) [76]. Adeno-associated viruses (AAVs), which are mainly characterized by having a linear, single-stranded DNA (ssDNA) genome, have become a mainstay in biological research due to their significant features. These viruses can infect a variety of cells, including human cells, without inducing severe immune response in return. In this regard, AAVs have been broadly used to deliver CRISPR-Cas9 components into cells both in vitro and in vivo. However, long-term insertional mutagenesis is still a noticeable obstacle to be adopted in clinics despite of the efforts engineering the AAV-based gene delivery system not to integrate into the genome of the target organism [77, 78]. Moreover, small packaging size of the vector which cannot fully accommodate the large Cas9 gene, sgRNA, fluorescent tags and other reporters is another challenge faced in AAV-based delivery methods.
Fig. 5.
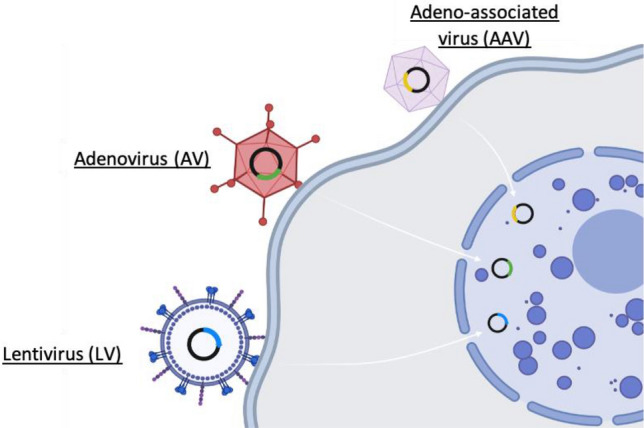
CRISPR-Cas9 system delivery using viral vectors. Adeno-associated virus (AAV), adenovirus (AVs), and lentivirus (LV) deliver the Cas9 protein in a plasmid DNA format; these viruses can infect a wide array of cells and exhibit a high gene editing capacity
In addition to AAVs, adenoviruses (AVs) and lentiviruses (LVs) have been also used to deliver the CRISPR-Cas9 components into cells. The AVs and LVs have a better cloning capacity, which make delivery of long engineered Cas9 proteins, gRNAs, tags and fusion proteins in a single viral vector. Moreover, it has been reported that LVs have a highly efficient transduction process in both dividing and non-dividing cells, indicating that they can infect many types of cells. These features of LVs make them suitable, to some extent, as carriers for CRISPR-based gene editing tool [79]. In spite of their high gene editing capacity, their exposure to the host may trigger immune responses which can obstruct the overall delivery of CRISPR-based system into the cells [80]. Also, long term exposure to the active CRISPR system expressed by the viral DNA may also increase the risk of off-target effects [60, 61].
Overall, the applications of CRISPR-based gene editing in translational medicine are still limited due to the lack of proper delivery system, despite of high potential as a gene therapy tool. Alternatively, nanoparticles have attracted great attention as promising candidates for the CRISPR delivery system due to their diverse design to achieve target specificity, immune escape, and efficient intracellular delivery. Tailored design of nanoparticles to have adaptable shape and surface chemistry may enable efficient and safe delivery of CRISPR components in vivo.
Nanoparticle-mediated delivery for CRISPR systems
Lipid-based nanoparticles
Lipid-based nanoparticles (LNPs) are spherical nanostructures typically ranging from 10 to 500 nm in size. Based on the structures, these particles may be subdivided into liposomes, nanostructured lipid carriers (NLCs), and solid lipid nanoparticles (SLNs). Regardless of the type, they have been used as carriers for therapeutic agents in many disease researches due to their versatility, high biocompatibility, biodegradability and protection of payloads [81–83]. Moreover, since nucleic acids cannot easily pass through the cell membrane due to their highly anionic nature, these particles seem to be suited in delivering these molecules by encapsulating them with cationic lipids and protecting them from enzymatic degradation [84, 85]. With the aid of technology and the development of new biological and chemical techniques, these nanoparticles are now being used as carriers in delivering the CRISPR-based tool to hopefully treat many diseases, including the hard-to-treat cancer, in human.
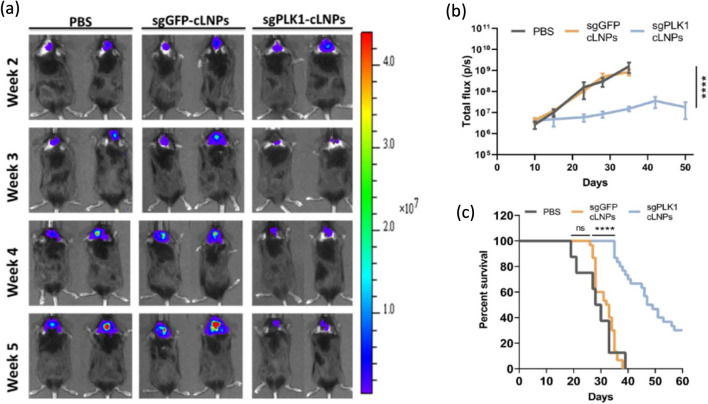
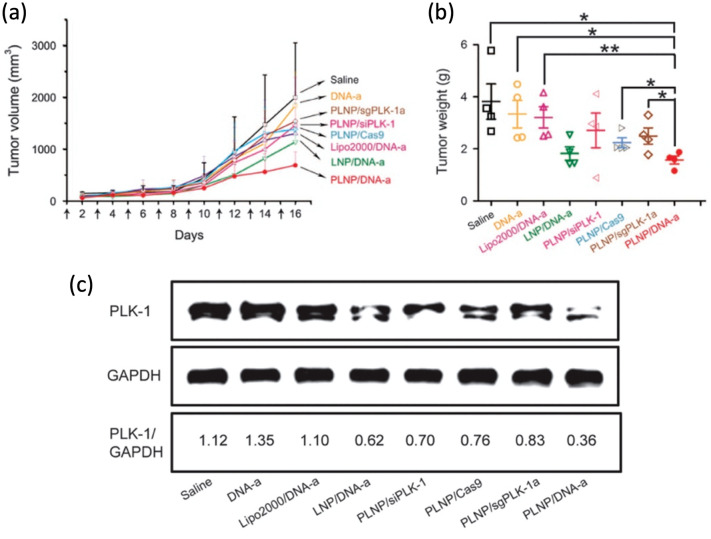
Ionizable amino lipid library can provide a means to screen cationic lipids to construct LNPs which is designed to encapsulate both Cas9 and sgRNA. Recent study has shown that the Cas9 mRNA was modified with 5-methoxyuridine to confer stability and reduce the risks of immunogenicity [18]. Introducing this LNPs with sgRNA targeting PLK1 gene achieved ~ 70% gene editing in aggressive orthotopic glioblastoma model and ~ 80% gene editing in disseminated ovarian tumor model inhibiting tumor growth and improving survival rate (Fig. 6). Cationic LNPs with a core–shell structure containing the protamine, Cas9-sgPLK-1 plasmid DNA and chondroitin sulfate (CS) as a negative core have shown another potential as a powerful CRISPR delivery strategy. The plasmid DNA was condensed in the core with positively charged protamine as a seed with the help of highly negatively charged CS. This core was encapsulated by a cationic lipid shell containing DOTAP, DOEP, cholesterol and polyethylene glycol (PEG) functionalized DSPE to increase stability, solubility, half-life in blood circulation while reducing toxicity and immunogenicity [86, 87]. This core–shell PEG functionalized lipid nanoparticles (PLNPs) showed ~ 47.4% of transfection efficiency and ~ 16.1% of indel mutation in PLK gene target site while commercially available transfection agent (Lipofectamine 2000) showed only ~ 3% transfection efficiency and ~ 1.2% of indel mutation using A375 cell line in vitro. Intratumorally injected PLNPs in melanoma-bearing mice induced ~ 3% of indel mutation in PLK locus indicating successful delivery of CRISPR constructs confirmed by deep sequencing and decreased PLK-1 expression level and tumor size (Fig. 7) [88].
Fig. 6.
Tumor inhibition by sgPLK1-cLNPs. a, b Intratumoral injection of sgPLK1-cLNPs caused a significant tumor growth inhibition in 005 GBM-bearing mice as opposed to that of sgGFP-cLNPs. c The survival curve showed that sgPLK1-cLNPs helped the cancer-bearing mice to have a longer lifespan. Reprinted with permission from American Association for the Advancement of Science [18]
Fig. 7.
a, b Mice treated with the PLNP with core–shell structure showed the lowest tumor volume and tumor weight after 16 days, indicating successful inhibition of the growth of tumors. c Western blotting analysis showed that the PLNP was effective in silencing the PLK-1 protein. Reprinted with permission from Springer Nature [88]
Inorganic nanoparticles
Recently, the use of inorganic materials has become the forefront in drug delivery systems, due to their desirable characteristics and technological significance. As a result, many inorganic nanoparticles have been suggested as nano-carriers for drug delivery since they are generally non-toxic, biocompatible, and more stable than organic materials [89–91]. Moreover, these materials also possess the ability to deliver many therapeutic gene agents such as antisense oligonucleotides, siRNA and mRNA [92, 93]. Extensive investigations have shown safe and efficient delivery of CRISPR/Cas9 system using various types of inorganic nanoparticles both in vitro and in vivo.
Gold nanoparticles
Gold nanoparticles (AuNPs) are comprised of the element gold (Au) whose diameter typically ranges from 1 to 100 nm. Once these gold particles are suspended in water, they are called as “colloidal gold”. Being the most stable metal nanoparticles, the AuNPs are involved in many different applications such as in electronics, sensors and probes, catalysis, and therapeutic agent delivery [94–97]. One of the defining characteristics of AuNPs is that they are considered chemically inert, thus the chances of triggering an immune response upon delivery are very low, making them suitable as candidate for intracellular delivery of the CRISPR-based system.
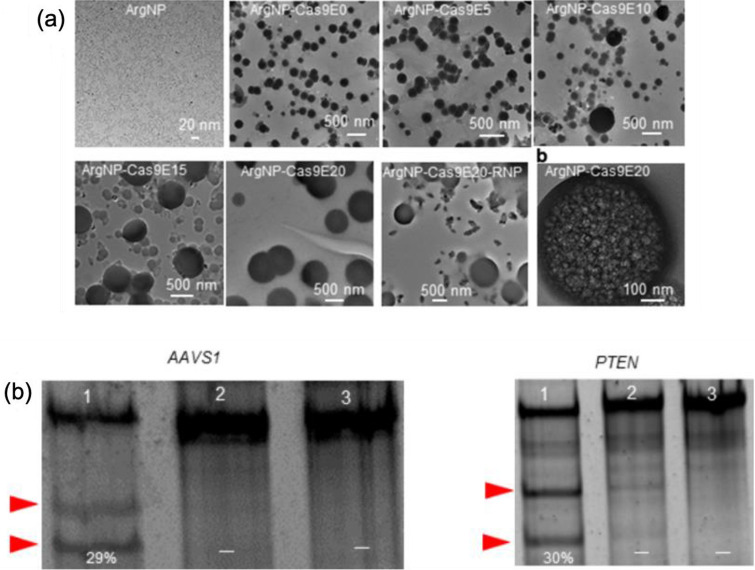
One of the recent advancement in AuNPs-based drug delivery systems has shown that the nanoassemblies between AuNPs (coated with arginine) and Cas9 protein (inserted with glutamate peptide tag, E-tag) and sgRNA reported a ~ 90% delivery efficiency and up to ~ 30% gene editing efficiency upon cellular uptake [98]. The engineering of Cas9 to carry the negatively-charged glutamate tag was crucial in accomplishing a strong self-assembled structure with the cationic arginine-AuNPs; the presence of nuclear localization signal (NLS) also enabled the Cas9 to have a clear target path towards the nucleus, increasing the chances of having a precise gene editing. The nanoassemblies were formed at varying ratios of both Cas9 and AuNPs, and it has been observed that the length of glutamate-tag plays a role both in self-assembly formation between Cas9 and AuNPs and in cytoplasmic delivery (Fig. 8a). Real-time tracking experiment revealed that the Cas9 carrying shorter E-tag (e.g., Cas9E0 and Cas9E5) led to a poor cytoplasmic delivery, by which lack of proper self-assembly with cationic arginine-AuNPs might be the major cause. The mechanism of delivery was also investigated by incubating the HeLa cells, which have been pretreated with endocytosis inhibitors or a cholesterol depletion inducing drug. Cytoplasmic/nuclear Cas9E20 delivery was blocked by the cholesterol depletion inducing drug while it was not affected by endocytosis inhibitors, which clearly suggest that the delivery occurred through a cholesterol-dependent membrane fusion-like process. Lastly, the gene editing capability of the CRISPR-based construct was evaluated using the human AAVS1 gene as the target. The indel analysis results showed that up to 29% of indel efficiency has been achieved. At the same time, the human PTEN gene was also targeted to confirm the efficacy of the method. The results showed that 30% of indel efficiency has been induced in the gene, indicating an efficient genome editing capability (Fig. 8b).
Fig. 8.
a Nanoassemblies between Cas9 and AuNPs: The length of glutamate (E-tag) significantly plays a role in the self-assembly formation between Cas9En (glutamate-tagged Cas9) and ArgNPs (arginine-AuNPs); larger nanoassemblies are observed when the length of E-tag is increased. b Gene editing capability: Approximately 30% of indel efficiency (lane 1) has been observed in the targeted AAVS1 and PTEN gene, indicating an efficient gene editing as opposed to delivering Cas9E15-RNP alone (Lane 2) and cell only control (Lane 3). Arrowheads (red) designate the DNA cleavages. Reprinted with permission from American Chemical Society [98]. (Color figure online)
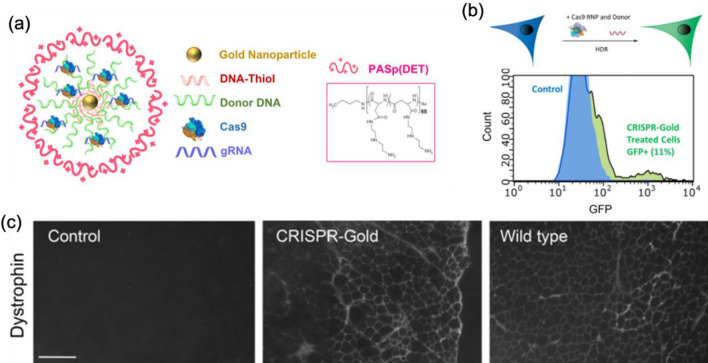
Another form of AuNPs and CRISPR component assembly (CRISPR-Gold) focus on homology-directed repair (HDR) based gene editing by replacing original DNA sequence with a donor DNA sequence co-delivered with Cas9/sgRNA to broaden the scope for the treatment [99]. The CRISPR-Gold is composed of 15 nm gold nanoparticle in the center, conjugated to thiol modified DNA which is hybridized with single stranded donor DNA for HDR. Cas9 RNP binds to this donor DNA and the whole nanoparticle is coated with endosoaml escape polymer PAsp (DET). Cas9 RNPs are loaded with non-specific electrostatic interaction with the AuNPs and affinity to the donor template DNA (Fig. 9a). The CRISPR-Gold targeting CXC34 gene in a number of cell types including human embryonic stem cells (hESC) and induced pluripotent stem cells (hiPSC) showed 3–4% HDR efficiency (Fig. 9b). The nanoparticles are internalized through endocytosis via cavaeolae/raft-dependent pathway and PAsp(DET) disrupts endosome for endosomal escape. It was used to treat Duchenne muscular dystrophy (DMD) model mouse in vivo improving strength and agility in mdx mice by correcting the mutated Dmd allele to its wild type with about 5.4% of correction rate, which is far higher than 0.3% by Cas9 RNP and donor DNA alone (Fig. 9c). This CRISPR-Gold system also treated Fmr1 knockout fragile X syndrome model mouse significantly rescueing the incrased repetitive behaviors by knocking out mGluR5 gene with 14.6% of indel mutation frequency reducing 40–50% of its mRNA and protein expression level [20].
Fig. 9.
a CRISPR-Gold design: The CRISPR-Gold structure is composed of gold nanoparticles as its core, and the affinity of Cas9 RNP with the loaded DNA enabled its adsorption onto the nanoparticles. b Inducing HDR in vitro: The treatment of BFP-HEK cells with CRISPR-Gold converted approximately 11.3% of the cells to GFP expressing cells. c Dystrophin expression by CRISPR-Gold: A single injection of CRISPR-Gold in mdx mice resulted in restoring the dystrophin expression, indicating a successful reversion from mutated gene into its wild-type version. Reprinted with permission from Springer Nature [99]
Iron oxide nanoparticles
Due to their special magnetic properties of magnetite (Fe3O4) and/or maghemite (-Fe2O3), the iron oxide nanoparticles (IONPs) have been extensively used in many biomedical applications such as sensors, tags, imaging, and drug delivery systems and have been applied in many major clinical diagnosis including magnetic resonance image (MRI) [100–102]. Following the success of IONPs as drug delivery system, the IONPs was used as a carrier to deliver CRISPR/Cas9 construct into porcine fetal fibroblasts (PFFs) [103]. The combination of physicochemical properties and external force, termed as magnetofection, enables magnetic field to concentrate and deposit the CRISPR-based material, which is enclosed in the magnetic nanoparticles, on the cell that is being transfected. A comparison between magnetofection-induced CRISPR/Cas9 and classic lipofection has been performed to confirm whether the former technique improves the delivery of CRISPR/Cas9 constructs into cells. Using the genomic cleavage detection assay, it has been observed that the cell lines treated with CRISPR/Cas9 had much intensified cleaved bands, an indication that magnetofection was more successful in inducing DNA cleavage at porcine H11 locus compared to lipofection. Furthermore, the TIDE web tool analysis showed that the indel frequencies were much higher in magnetofection, 15.3% for 10 pg DNA/cell and 27.6% for 20 pg DNA/cell, compared to lipofection which only had 4.3% and 7.4%, respectively (Fig. 10). Overall, the combined effects of magnetic nanoparticles, which have iron oxide material as the core, and gradient magnetic field proved to be an effective method in delivering CRISPR/Cas9 into cells and in inducing genome editing afterwards.
Fig. 10.
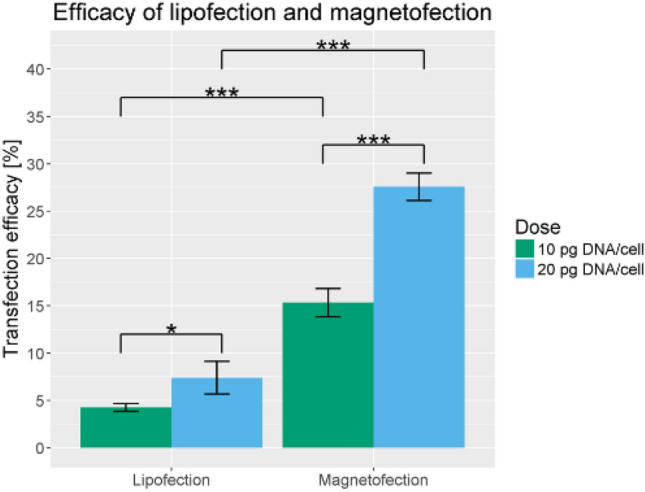
Comparison between magnetofection and lipofection. The plot shows that magnetofection, both in 10 pg DNA/cell and 20 pg DNA/cell plasmid concentrations, resulted in a higher transfection efficiency as opposed to lipofection. From M. Reprinted with permission from Springer Nature [103]
Mesoporous silica nanoparticles
Mesoporous silica nanoparticles (MSNs) have a systematic framework composed of silicon oxide, which serves as the foundation of large networks of pores, whose diameter typically ranges from 2 to 20 nm. The MSNs are able to effectively deliver a number of therapeutic regimens such as drugs, nucleic acids, and proteins which are helpful for blocking the advancement of diseases. As a template matrix, silica nanoparticles are commonly prepared using the Stober process, a sol–gel technique where a precursor tetraethylorthosilicate (TEOS) reacts via hydrolysis and condensation to form a new phase (sol) in water-alcohol solution with ammonia as a catalyst. Afterwards, the nanoparticles suspended within the sol condense into the gel phase. The rate of hydrolysis and ocndensation of silica and interaction between template and silica decides size and morphology of MSNs. The base triethanolamine can be used as a substitute for the basicity provided by ammonia to synthesize MSNs, although it is also reported that triethanolamine has the ability to inhibit the growth of the nanoparticles [104]. The use of cationic surfactants including cetyltrimethylammonium bromide or cetyltrimethylammonium chloride as a template has been shown to direct the silicate source and make them condensed around the micelles originally formed by these surfactants to form ordered silica structures.
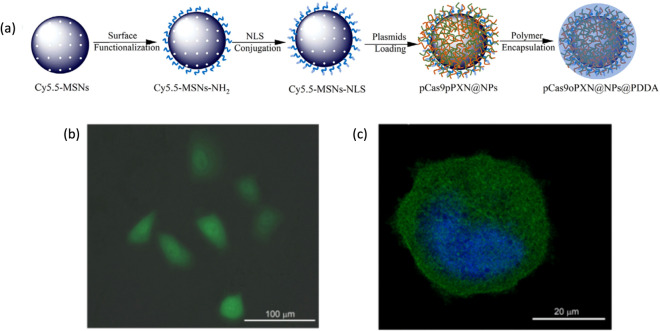
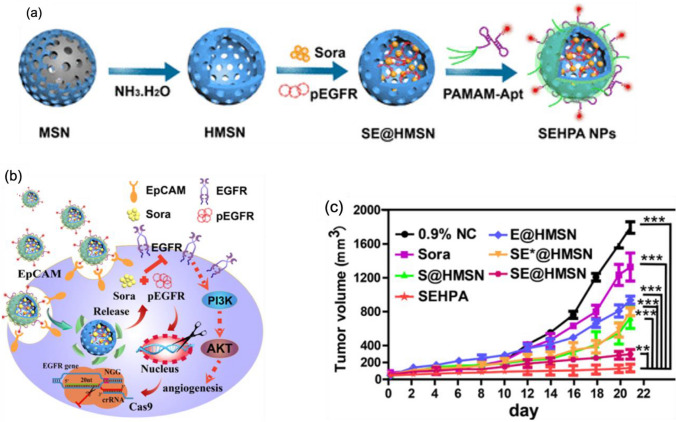
Monodispersed spherical MSNs were used for delivery of plasmids for expression of CRISPR/Cas9 sgRNA complex and donor template for HDR mediated knock in of GFP into PXN gene [105]. MSN was functionalized with positively charged amine group for strong electrostatic interaction with negatively charged plasmid DNAs. Additionally, a nuclear localization signal was attached to the MSNs for better nuclear localization of plasmids following endosomal escape. The MSNs were then encapsulated with positively charged pH-responsive polymer (poly(dimethyldiallylammonium chloride), PDDA using microfluidic co-flow focusing nanoprecipitation method. The PDDA coating protects the plasmids from environment, its positive charge enhances cellular internalization and its pH responsive degradation facilitates endosomal escape, which is essential for nanocarriers to prevent lysosomal degradation of payloads. This nanoparticle showed successful GFP knock-in into PXN gene proven by expression of GFP from the transfected U2OS cell and colocalization with staining against PXN protein (Fig. 11). In another study, it has been reported that constructing core–shell hollow mesoporous silica nanoparticles (HMSNs) as carriers for both chemotherapy drugs and gene therapy approaches was successful in inducing a synergistic inhibition on EGFR signaling, with the nanocomplex showing at least more than 60% gene editing efficiency [106]. The drug used is Sorafenib, which inhibits a number of targets present in hepatocellular carcinoma (HCC), and the gene editing approach is based on delivering EGFR-targeting sgRNA via DNA plasmids (Fig. 11a). Several components have also been added onto the HMSNs to make the carriers become more ideal in exerting its gene-drug co-delivery system effects. Such components include dendrimer polyamidoamine, which can both prevent the leakage of drugs and allow the release in the tumor tissue, and modified DNA aptamer, which has the ability to recognize the epithelial cell adhesion molecules found in HCC (Fig. 12b, c). In vivo experiments showed that the nanoparticles had a resounding therapeutic effect once injected into the tumor-bearing model mice, resulting from the decrease of EGFR protein expression in tumor tissue. This led to inhibition of the growth of tumor as shown in the low volume present in the tumor. Overall, the synergistic effects of gene-drug co-delivery accompanied by the nanosystem provided an alternative way for cancer treatment more efficiently.
Fig. 11.
a Preparation of the MSNs for CRISPR/Cas9 delivery. The plasmids are adsorbed to the nanoparticles well due to their strong interaction with the functionalized amine moieties found in the surface of the particles. b Intense green signal of GFP is observed after 7 days of incubation with the MSNs-based nanocarrier, indicating a successful gene editing. Reprinted with permission from Springer Nature [105]
Fig. 12.
a Schematic illustration of the SEHPA (SE@HMSN/P-Apt: Sora- and pEGFR-loaded HMNS/PAMAM-Aptamer) nanoparticle preparation. b The downregulation of EGFR/PI3K/Akt pathway is caused by the synergistic effects of Sora and pEGFR in SEHPA NPs. c SEHPA nanoparticles inhibited the growth of tumor, indicating a successful anticancer effect. Reprinted with permission from American Chemical Society [106]
Concluding remarks
In the recent events following the discoveries made in the field of molecular biology, genome editing has been at the forefront of many research areas due to its ability of modifying DNA sequences at specific sites, unlike the early genetic engineering techniques which make use of randomly inserted genetic materials into a certain host’s genome. Among the many versatile genome editing tools, the CRISPR/Cas9 system is a revolutionary and effective method that researchers and scientists use to alter any specific DNA sequences in a certain organism’s genome: this is achieved by bringing a specific kind of nuclease, and a guide RNA, to cut specific targeted sites in the organism’s genome. The efficiency, engineering feasibility, and design simplicity of CRISPR/Cas9 system definitely stood out among the previously engineered nucleases like TALEN, ZFN, and meganucleases. Moreover, the accumulated works of researchers who put their time and effort in understanding the CRISPR-based genomic tool have shed light on its potential therapeutic benefits. Such benefits include, but are not limited to, deleting specific genes in the genome, adding therapeutic genes, and correcting mutated genes back to their wild-type versions which seem to be helpful in treating many kinds of diseases.
In the same process, a number of delivery systems have been developed to confirm the efficacy and to maximize the power of the CRISPR-based genome editing tool when it comes to gene editing; these delivery strategies have been used in many studies and have been proven to be effective to some extent, considering that each strategy has also its own drawbacks (Table 1). Safety concerns are of the utmost importance when choosing which strategy can be used to deliver the CRISPR/Cas9 constructs particularly in in vivo setting, and because of this, the shift from viral vectors to non-viral vectors nowadays has palpably increased at a faster rate. Nanoparticles, a prime example of non-viral vectors, have continuously gained momentum in the research field due to their desirable characteristics; these characteristics, including nanoscale size, specific targeting, ease of size tunability and surface functionalization and modification, little to no immune response, and low cost, make them very suitable candidates in delivering CRISPR/Cas9 constructs into a wide range of cells. Furthermore, the range and potential for what could be improved and/or added to this tool, for the purpose of intensifying its effectiveness, are limitless: for instance, the research study involving the combination of CRISPR-based constructs, delivered by mesoporous silica nanoparticles (MSNs), and the Sorafenib drug proved to be marked with superiority in delivering a synergistic inhibition of EGFR expression in cancer cell lines; another study tackled the effective delivery of CRISPR/Cas9 constructs, enclosed in iron oxide nanoparticles, using external magnetic force. As pointed out in the above sections, the therapeutic potential of CRISPR-based genome editing tool is satisfactory with which changing of the genes responsible for the occurrence of certain kinds of diseases becomes the central focus of each work.
Table 1.
Different strategies for CRISPR/Cas9 delivery are shown below, with each strategy having its own advantages and limitation
| Delivery Strategy | Definition and/or Use | Source of Cas9 Protein | Application | Limitations | Text References | |
|---|---|---|---|---|---|---|
| Physical Methods | Microinjection | Use of a glass micropipette to inject a liquid substance at a microscopic | DNA plasmid; mRNA (Cas9 + sgRNA); protein (RNP) | in vitro, ex vivo | Not possible on in vivo experiment | 46–47 |
| Electroporation | Use of electrical pulse to create temporary pores in cell membranes through which substances can pass into cells | DNA plasmid; mRNA (Cas9 + sgRNA); protein (RNP) | in vitro, ex vivo | Difficult to perform in in vivo experiment | 46–47 | |
| Viral Vectors | Adeno-associated viruses [AAVs] | Vectors for genetic information; has a high gene editing capacity | DNA plasmid | in vivo | Cloning capacity is limited | 46–47, 65 |
| Adenoviruses [AVs] | Vectors for genetic information; has a high gene editing capacity | DNA plasmid | in vitro, ex vivo | May cause off-target effects | 46–47 | |
| Lentiviruses [LVs] | Vectors for genetic information; has a high gene editing capacity | DNA plasmid | in vitro, ex vivo | May cause off-target effects | 46–47 | |
| Non-viral Vectors | Lipid-based Nanoparticles | Composed of lipids; size is of spherical form and size ranges from 10 to 1000 nm | DNA plasmid; mRNA (Cas9 + sgRNA) | in vitro, in vivo | Efficiency depends on cell types | 73, 76 |
| Gold Nanoparticles [AuNPs] | Composed of the element gold; size ranges from 1 to 100 nm | Protein (RNP) | in vitro, in vivo | Efficiency depends on cell types | 98, 99 | |
| Iron Oxide Nanoparticles [IONPs] | Composed of magnetic compounds (magnetite/maghemite); size ranges from 1 to 100 nm | DNA plasmid | in vitro, in vivo | Efficiency depends on cell types | 103 | |
| Mesoporous Silica Nanoparticles [MSNs] | Comprised of silicon oxide (SiO2); pore diameter ranges from 2 to 50 nm | DNA plasmid | in vitro, in vivo | Efficiency depends on cell types | 105, 106 | |
In summary, up-to-date research revolving around nanoparticles as carriers for CRISPR-based genome editing tool opens up a new era, and the fact that these nanoparticles are continuously compromised by a number of different factors is a reasonable issue that can be address upon further work. For instance, the final goal for various components of a certain nanoparticle system and whether they pose cytotoxic effects after delivery still remain an elusive topic for most researchers. With how technology progresses every second, however, it is only a matter of time before several large improvements will be made to these non-viral vectors. The outlook we have is certainly positive, and we firmly believe that in few years later, CRISPR/Cas9-loaded nanoparticles will be at their fullest potential, leading the framework in clinical trials for the treatment of many kinds of diseases and for the benefit of mankind.
Acknowledgements
This work was supported by the Basic Science Research Program (No. 2020R1C1C1006081 and 2020R1A4A3078645) through the National Research Foundation of Korea funded by the Korea government (MSIT) and by the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI) and Korea Dementia Research Center (KDRC), funded by the Ministry of Health & Welfare and Ministry of Science and ICT, Republic of Korea (HU20C0094).
Declarations
Conflict of interest
The authors declare that they do not have any conflicts of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Komor AC, Badran AH, Liu DR. CRISPR-based technologies for the manipulation of eukaryotic genomes. Cell. 2017;168:20–36. doi: 10.1016/j.cell.2016.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Konermann S, Brigham MD, Trevino AE, Joung J, Abudayyeh OO, Barcena C, Hsu PD, Habib N, Gootenberg JS, Nishimasu H, Nureki O, Zhang F. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 2015;517:583–U332. doi: 10.1038/nature14136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gilbert LA, Horlbeck MA, Adamson B, Villalta JE, Chen Y, Whitehead EH, Guimaraes C, Panning B, Ploegh HL, Bassik MC, Qi LS, Kampmann M, Weissman JS. Genome-scale CRISPR-mediated control of gene repression and activation. Cell. 2014;159:647–661. doi: 10.1016/j.cell.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Findlay GM, Boyle EA, Hause RJ, Klein JC, Shendure J. Saturation editing of genomic regions by multiplex homology-directed repair. Nature. 2014;513:120–123. doi: 10.1038/nature13695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelsen TS, Heckl D, Ebert BL, Root DE, Doench JG, Zhang F. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Komor AC, Badran AH, Liu DR. CRISPR-based technologies for the manipulation of eukaryotic genomes (vol 168, pg 20, 2017) Cell. 2017;169:559–559. doi: 10.1016/j.cell.2017.04.005. [DOI] [PubMed] [Google Scholar]
- 7.Kaiser J. Virus used in gene therapies may pose cancer risk, dog study hints. Science. 2020 doi: 10.1126/science.aba7696. [DOI] [Google Scholar]
- 8.Vora S, Tuttle M, Cheng J, Church G. Next stop for the CRISPR revolution: RNA-guided epigenetic regulators. Febs J. 2016;283:3181–3193. doi: 10.1111/febs.13768. [DOI] [PubMed] [Google Scholar]
- 9.Mali P, Yang LH, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, Li YQ, Fine EJ, Wu XB, Shalem O, Cradick TJ, Marraffini LA, Bao G, Zhang F. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A Programmable Dual-RNA-Guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Foo J, Michor F. Evolution of acquired resistance to anti-cancer therapy. J Theor Biol. 2014;355:10–20. doi: 10.1016/j.jtbi.2014.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yin H, Kauffman KJ, Anderson DG. Delivery technologies for genome editing. Nat Rev Drug Discovery. 2017;16:387–399. doi: 10.1038/nrd.2016.280. [DOI] [PubMed] [Google Scholar]
- 14.LaFountaine JS, Fathe K, Smyth HDC. Delivery and therapeutic applications of gene editing technologies ZFNs, TALENs, and CRISPR/Cas9. Int J Pharmaceut. 2015;494:180–194. doi: 10.1016/j.ijpharm.2015.08.029. [DOI] [PubMed] [Google Scholar]
- 15.Yin H, Xue W, Chen SD, Bogorad RL, Benedetti E, Grompe M, Koteliansky V, Sharp PA, Jacks T, Anderson DG. Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype (vol 32, pg 551, 2014) Nat Biotechnol. 2014;32:952–952. doi: 10.1038/nbt0914-952d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nelson CE, Wu YY, Gemberling MP, Oliver ML, Waller MA, Bohning JD, Robinson-Hamm JN, Bulaklak K, Rivera RMC, Collier JH, Asokan A, Gersbach CA. Long-term evaluation of AAV-CRISPR genome editing for Duchenne muscular dystrophy. Nat Med. 2019;25:427–432. doi: 10.1038/s41591-019-0344-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hanlon KS, Kleinstiver BP, Garcia SP, Zaborowski MP, Volak A, Spirig SE, Muller A, Sousa AA, Tsai SQ, Bengtsson NE, Loov C, Ingelsson M, Chamberlain JS, Corey DP, Aryee MJ, Joung JK, Breakefield XO, Maguire CA, Gyorgy B. High levels of AAV vector integration into CRISPR-induced DNA breaks. Nat Commun. 2019;10:1–11. doi: 10.1038/s41467-019-12449-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rosenblum D, Gutkin A, Kedmi R, Ramishetti S, Veiga N, Jacobi AM, Schubert MS, Friedmann-Morvinski D, Cohen ZR, Behlke MA, Lieberman J, Peer D. CRISPR-Cas9 genome editing using targeted lipid nanoparticles for cancer therapy. Sci Adv. 2020;6:eabc9450. doi: 10.1126/sciadv.abc9450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheng Q, Wei T, Farbiak L, Johnson LT, Dilliard SA, Siegwart DJ. Selective organ targeting (SORT) nanoparticles for tissue-specific mRNA delivery and CRISPR-Cas gene editing. Nat Nanotechnol. 2020;15:313–320. doi: 10.1038/s41565-020-0669-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee B, Lee K, Panda S, Gonzales-Rojas R, Chong A, Bugay V, Park HM, Brenner R, Murthy N, Lee HY. Nanoparticle delivery of CRISPR into the brain rescues a mouse model of fragile X syndrome from exaggerated repetitive behaviours. Nat Biomed Eng. 2018;2:497–507. doi: 10.1038/s41551-018-0252-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carlson-Stevermer J, Abdeen AA, Kohlenberg L, Goedland M, Molugu K, Lou M, Saha K. Assembly of CRISPR ribonucleoproteins with biotinylated oligonucleotides via an RNA aptamer for precise gene editing. Nat Commun. 2017;8:1–13. doi: 10.1038/s41467-017-01875-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alsaiari SK, Patil S, Alyami M, Alamoudi KO, Aleisa FA, Merzaban JS, Li M, Khashab NM. Endosomal escape and delivery of CRISPR/cas9 genome editing machinery enabled by nanoscale zeolitic imidazolate framework. J Am Chem Soc. 2018;140:143–146. doi: 10.1021/jacs.7b11754. [DOI] [PubMed] [Google Scholar]
- 23.Rouet P, Smih F, Jasin M. Introduction of double-strand breaks into the genome of mouse cells by expression of a rare-cutting endonuclease. Mol Cell Biol. 1994;14:8096–8106. doi: 10.1128/mcb.14.12.8096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O'Driscoll M, Jeggo PA. The role of double-strand break repair - insights from human genetics. Nat Rev Genet. 2006;7:45–54. doi: 10.1038/nrg1746. [DOI] [PubMed] [Google Scholar]
- 25.Guo T, Feng YL, Xiao JJ, Liu Q, Sun XN, Xiang JF, Kong N, Liu SC, Chen GQ, Wang Y, Dong MM, Cai Z, Lin H, Cai XJ, Xie AY. Harnessing accurate non-homologous end joining for efficient precise deletion in CRISPR/Cas9-mediated genome editing. Genome Biol. 2018;19:170. doi: 10.1186/s13059-018-1518-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yan MY, Li SS, Ding XY, Guo XP, Jin Q, Sun YC. A CRISPR-assisted nonhomologous end-joining strategy for efficient genome editing in mycobacterium tuberculosis. MBio. 2020;11:e02364–e2419. doi: 10.1128/mBio.02364-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jasin M, Haber JE. The democratization of gene editing: Insights from site-specific cleavage and double-strand break repair. DNA Repair. 2016;44:6–16. doi: 10.1016/j.dnarep.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ryu S-M, Hur JW, Kim K. Evolution of CRISPR towards accurate and efficient mammal genome engineering. BMB Rep. 2019;52:475–481. doi: 10.5483/BMBRep.2019.52.8.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sansbury BM, Hewes AM, Kmiec EB. Understanding the diversity of genetic outcomes from CRISPR-Cas generated homology-directed repair. Commun Biol. 2019;2:458. doi: 10.1038/s42003-019-0705-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Souza N. Primer: genome editing with engineered nucleases. Nat Methods. 2012;9:27–27. doi: 10.1038/nmeth.1848. [DOI] [PubMed] [Google Scholar]
- 31.Epinat JC, Arnould S, Chames P, Rochaix P, Desfontaines D, Puzin C, Patin A, Zanghellini A, Paques F, Lacroix E. A novel engineered meganuclease induces homologous recombination in yeast and mammalian cells. Nucleic Acids Res. 2003;31:2952–2962. doi: 10.1093/nar/gkg375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chapdelaine P, Pichavant C, Rousseau J, Paques F, Tremblay JP. Meganucleases can restore the reading frame of a mutated dystrophin. Gene Ther. 2010;17:846–858. doi: 10.1038/gt.2010.26. [DOI] [PubMed] [Google Scholar]
- 33.Carlson DF, Fahrenkrug SC, Hackett PB. Targeting DNA with fingers and TALENs. Mol Ther-Nucl Acids. 2012;1:e3. doi: 10.1038/mtna.2011.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khan SH. Genome-editing technologies: concept, pros, and cons of various genome-editing techniques and bioethical concerns for clinical application. Mol Ther Nucleic Acids. 2019;16:326–334. doi: 10.1016/j.omtn.2019.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li H, Yang Y, Hong W, Huang M, Wu M, Zhao X. Applications of genome editing technology in the targeted therapy of human diseases: mechanisms, advances and prospects. Signal Transduct Target Ther. 2020;5:1. doi: 10.1038/s41392-019-0089-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Song GY, Jia ML, Chen K, Kong XC, Khattak B, Xie CX, Li AL, Mao L. CRISPR/Cas9: A powerful tool for crop genome editing. Crop J. 2016;4:75–82. doi: 10.1016/j.cj.2015.12.002. [DOI] [Google Scholar]
- 37.Stoddard BL. Homing endonucleases from mobile group I introns: discovery to genome engineering. Mobile DNA-Uk. 2014;5:1–16. doi: 10.1186/1759-8753-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Holkers M, Maggio I, Liu J, Janssen JM, Miselli F, Mussolino C, Recchia A, Cathomen T, Goncalves MA. Differential integrity of TALE nuclease genes following adenoviral and lentiviral vector gene transfer into human cells. Nucleic Acids Res. 2013;41:e63. doi: 10.1093/nar/gks1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hockemeyer D, Soldner F, Beard C, Gao Q, Mitalipova M, DeKelver RC, Katibah GE, Amora R, Boydston EA, Zeitler B, Meng X, Miller JC, Zhang L, Rebar EJ, Gregory PD, Urnov FD, Jaenisch R. Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nat Biotechnol. 2009;27:851–857. doi: 10.1038/nbt.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hockemeyer D, Wang H, Kiani S, Lai CS, Gao Q, Cassady JP, Cost GJ, Zhang L, Santiago Y, Miller JC, Zeitler B, Cherone JM, Meng X, Hinkley SJ, Rebar EJ, Gregory PD, Urnov FD, Jaenisch R. Genetic engineering of human pluripotent cells using TALE nucleases. Nat Biotechnol. 2011;29:731–734. doi: 10.1038/nbt.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pattanayak V, Ramirez CL, Joung JK, Liu DR. Revealing off-target cleavage specificities of zinc-finger nucleases by in vitro selection. Nat Methods. 2011;8:765–770. doi: 10.1038/nmeth.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rahimi H, Salehiabar M, Charmi J, Barsbay M, Ghaffarlou M, Roohi Razlighi M, Davaran S, Khalilov R, Sugiyama M, Nosrati H, Kaboli S, Danafar H, Webster TJ. Harnessing nanoparticles for the efficient delivery of the CRISPR/Cas9 system. Nano Today. 2020;34:100895. doi: 10.1016/j.nantod.2020.100895. [DOI] [Google Scholar]
- 43.Mojica FJM, Diez-Villasenor C, Garcia-Martinez J, Soria E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J Mol Evol. 2005;60:174–182. doi: 10.1007/s00239-004-0046-3. [DOI] [PubMed] [Google Scholar]
- 44.Rath D, Amlinger L, Rath A, Lundgren M. The CRISPR-Cas immune system: biology, mechanisms and applications. Biochimie. 2015;117:119–128. doi: 10.1016/j.biochi.2015.03.025. [DOI] [PubMed] [Google Scholar]
- 45.Hille F, Charpentier E. CRISPR-Cas: biology, mechanisms and relevance. Philos T R Soc B. 2016;371:20150496. doi: 10.1098/rstb.2015.0496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kick L, Kirchner M, Schneider S. CRISPR-Cas9: From a bacterial immune system to genome-edited human cells in clinical trials. Bioengineered. 2017;8:280–286. doi: 10.1080/21655979.2017.1299834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Makarova KS, Haft DH, Barrangou R, Brouns SJ, Charpentier E, Horvath P, Moineau S, Mojica FJ, Wolf YI, Yakunin AF, van der Oost J, Koonin EV. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol. 2011;9:467–477. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Burstein D, Harrington LB, Strutt SC, Probst AJ, Anantharaman K, Thomas BC, Doudna JA, Banfield JF. New CRISPR-Cas systems from uncultivated microbes. Nature. 2017;542:237–241. doi: 10.1038/nature21059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shmakov S, Smargon A, Scott D, Cox D, Pyzocha N, Yan W, Abudayyeh OO, Gootenberg JS, Makarova KS, Wolf YI, Severinov K, Zhang F, Koonin EV. Diversity and evolution of class 2 CRISPR-Cas systems. Nat Rev Microbiol. 2017;15:169–182. doi: 10.1038/nrmicro.2016.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schwank G, Koo BK, Sasselli V, Dekkers JF, Heo I, Demircan T, Sasaki N, Boymans S, Cuppen E, van der Ent CK, Nieuwenhuis EES, Beekman JM, Clevers H. Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients. Cell Stem Cell. 2013;13:653–658. doi: 10.1016/j.stem.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 51.Sanchez-Rivera FJ, Jacks T. Applications of the CRISPR-Cas9 system in cancer biology. Nat Rev Cancer. 2015;15:387–395. doi: 10.1038/nrc3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zuckermann M, Hovestadt V, Knobbe-Thomsen CB, Zapatka M, Northcott PA, Schramm K, Belic J, Jones DTW, Tschida B, Moriarity B, Largaespada D, Roussel MF, Korshunov A, Reifenberger G, Pfister SM, Lichter P, Kawauchi D, Gronych J. Somatic CRISPR/Cas9-mediated tumour suppressor disruption enables versatile brain tumour modelling. Nat Commun. 2015;6:1–9. doi: 10.1038/ncomms8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dever DP, Bak RO, Reinisch A, Camarena J, Washington G, Nicolas CE, Pavel-Dinu M, Saxena N, Wilkens AB, Mantri S, Uchida N, Hendel A, Narla A, Majeti R, Weinberg KI, Porteus MH. CRISPR/Cas9 beta-globin gene targeting in human haematopoietic stem cells. Nature. 2016;539:384–389. doi: 10.1038/nature20134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ou ZH, Niu XH, He WY, Chen YC, Song B, Xian YX, Fan D, Tang DL, Sun XF. The combination of CRISPR/Cas9 and iPSC technologies in the gene therapy of human beta-thalassemia in mice. Sci Rep-Uk. 2016;6:1–13. doi: 10.1038/s41598-016-0001-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pankowicz FP, Barzi M, Legras X, Hubert L, Mi T, Tomolonis JA, Ravishankar M, Sun Q, Yang DN, Borowiak M, Sumazin P, Elsea SH, Bissig-Choisat B, Bissig KD. Reprogramming metabolic pathways in vivo with CRISPR/Cas9 genome editing to treat hereditary tyrosinaemia. Nat Commun. 2016;7:1–6. doi: 10.1038/ncomms12642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Halim D, Wilson MP, Oliver D, Brosens E, Verheij JBGM, Han Y, Nanda V, Lyu Q, Doukas M, Stoop H, Brouwer RWW, van IJcken WFJ, Slivano OJ, Burns AJ, Christie CK, Bentley KLD, Brooks AS, Tibboel D, Xu SW, Jin ZG, Djuwantono T, Yan W, Alves MM, Hofstra RMW, Miano JM. Loss of LMOD1 impairs smooth muscle cytocontractility and causes megacystis microcolon intestinal hypoperistalsis syndrome in humans and mice. P Natl Acad Sci USA. 2017;114:E2739–E2747. doi: 10.1073/pnas.1620507114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rodriguez-Rodriguez DR, Ramirez-Solis R, Garza-Elizondo MA, Garza-Rodriguez MD, Barrera-Saldana HA. Genome editing: A perspective on the application of CRISPR/Cas9 to study human diseases (Review) Int J Mol Med. 2019;43:1559–1574. doi: 10.3892/ijmm.2019.4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016;533:420–424. doi: 10.1038/nature17946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Anzalone AV, Randolph PB, Davis JR, Sousa AA, Koblan LW, Levy JM, Chen PJ, Wilson C, Newby GA, Raguram A, Liu DR. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature. 2019;576:149–157. doi: 10.1038/s41586-019-1711-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lino CA, Harper JC, Carney JP, Timlin JA. Delivering CRISPR: a review of the challenges and approaches. Drug Deliv. 2018;25:1234–1257. doi: 10.1080/10717544.2018.1474964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yip BH. Recent Advances in CRISPR/Cas9 delivery strategies. Biomolecules. 2020;10:839. doi: 10.3390/biom10060839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Williams PD, Kingston PA. Plasmid-mediated gene therapy for cardiovascular disease. Cardiovasc Res. 2011;91:565–576. doi: 10.1093/cvr/cvr197. [DOI] [PubMed] [Google Scholar]
- 63.Liang XQ, Potter J, Kumar S, Zou YF, Quintanilla R, Sridharan M, Carte J, Chen W, Roark N, Ranganathan S, Ravinder N, Chesnut JD. Rapid and highly efficient mammalian cell engineering via Cas9 protein transfection. J Biotechnol. 2015;208:44–53. doi: 10.1016/j.jbiotec.2015.04.024. [DOI] [PubMed] [Google Scholar]
- 64.Tabebordbar M, Zhu KX, Cheng JKW, Chew WL, Widrick JJ, Yan WX, Maesner C, Wu EY, Xiao R, Ran FA, Cong L, Zhang F, Vandenberghe LH, Church GM, Wagers AJ. In vivo gene editing in dystrophic mouse muscle and muscle stem cells. Science. 2016;351:407–411. doi: 10.1126/science.aad5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schmeer M, Buchholz T, Schleef M. Plasmid DNA manufacturing for indirect and direct clinical applications. Hum Gene Ther. 2017;28:856–861. doi: 10.1089/hum.2017.159. [DOI] [PubMed] [Google Scholar]
- 66.Fu Y, Foden JA, Khayter C, Maeder ML, Reyon D, Joung JK, Sander JD. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol. 2013;31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hendel A, Bak RO, Clark JT, Kennedy AB, Ryan DE, Roy S, Steinfeld I, Lunstad BD, Kaiser RJ, Wilkens AB, Bacchetta R, Tsalenko A, Dellinger D, Bruhn L, Porteus MH. Chemically modified guide RNAs enhance CRISPR-Cas genome editing in human primary cells. Nat Biotechnol. 2015;33:985–U232. doi: 10.1038/nbt.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rahdar M, McMahon MA, Prakash TP, Swayze EE, Bennett CF, Cleveland DW. Synthetic CRISPR RNA-Cas9-guided genome editing in human cells. P Natl Acad Sci USA. 2015;112:E7110–E7117. doi: 10.1073/pnas.1520883112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li B, Zhao WY, Luo X, Zhang XF, Li CL, Zeng CX, Dong YZ. Engineering CRISPR-Cpf1 crRNAs and mRNAs to maximize genome editing efficiency. Nat Biomed Eng. 2017;1:1–10. doi: 10.1038/s41551-016-0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shalaby K, Aouida M, El-Agnaf O. Tissue-specific delivery of CRISPR therapeutics: strategies and mechanisms of non-viral vectors. Int J Mol Sci. 2020;21:7353. doi: 10.3390/ijms21197353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Horii T, Arai Y, Yamazaki M, Morita S, Kimura M, Itoh M, Abe Y, Hatada I. Validation of microinjection methods for generating knockout mice by CRISPR/Cas-mediated genome engineering. Sci Rep. 2014;4:4513. doi: 10.1038/srep04513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vilarino M, Rashid ST, Suchy FP, McNabb BR, van der Meulen T, Fine EJ, Ahsan S, Mursaliyev N, Sebastiano V, Diab SS, Huising MO, Nakauchi H, Ross PJ. CRISPR/Cas9 microinjection in oocytes disables pancreas development in sheep. Sci Rep-Uk. 2017;7:1–10. doi: 10.1038/s41598-016-0028-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Elaswad A, Khalil K, Cline D, Page-McCaw P, Chen WB, Michel M, Cone R, Dunham R. Microinjection of CRISPR/Cas9 Protein into Channel Catfish, Ictalurus punctatus, Embryos for Gene Editing. Jove-J Vis Exp. 2018;131:e56275. doi: 10.3791/56275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Xu XY, Gao DB, Wang P, Chen J, Ruan JX, Xu J, Xia XF. Efficient homology-directed gene editing by CRISPR/Cas9 in human stem and primary cells using tube electroporation. Sci Rep-Uk. 2018;8:1–11. doi: 10.1038/s41598-018-30227-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Qin W, Dion SL, Kutny PM, Zhang Y, Cheng AW, Jillette NL, Malhotra A, Geurts AM, Chen YG, Wang H. Efficient CRISPR/Cas9-mediated genome editing in mice by zygote electroporation of nuclease. Genetics. 2015;200:423–430. doi: 10.1534/genetics.115.176594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bulcha JT, Wang Y, Ma H, Tai PWL, Gao GP. Viral vector platforms within the gene therapy landscape. Signal Transduct Target. 2021;6:1–24. doi: 10.1038/s41392-020-00451-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Carter BJ. Adeno-associated virus and the development of adeno-associated virus vectors: A historical perspective. Mol Ther. 2004;10:981–989. doi: 10.1016/j.ymthe.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 78.Daya S, Berns KI. Gene therapy using adeno-associated virus vectors. Clin Microbiol Rev. 2008;21:583–593. doi: 10.1128/CMR.00008-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kotterman MA, Chalberg TW, Schaffer DV. Viral vectors for gene therapy: translational and clinical outlook. Annu Rev Biomed Eng. 2015;17:63–89. doi: 10.1146/annurev-bioeng-071813-104938. [DOI] [PubMed] [Google Scholar]
- 80.Pistello M, Antonelli G. Integration of the viral genome into the host cell genome: a double-edged sword. Clin Microbiol Infec. 2016;22:296–298. doi: 10.1016/j.cmi.2016.01.022. [DOI] [PubMed] [Google Scholar]
- 81.Rostami E, Kashanian S, Azandaryani AH, Faramarzi H, Dolatabadi JEN, Omidfar K. Drug targeting using solid lipid nanoparticles. Chem Phys Lipids. 2014;181:56–61. doi: 10.1016/j.chemphyslip.2014.03.006. [DOI] [PubMed] [Google Scholar]
- 82.Montoto SS, Muraca G, Ruiz ME. Solid Lipid Nanoparticles for Drug Delivery: Pharmacological and Biopharmaceutical Aspects. Front Mol Biosci. 2020;7:319. doi: 10.3389/fmolb.2020.587997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mitchell MJ, Billingsley MM, Haley RM, Wechsler ME, Peppas NA, Langer R. Engineering precision nanoparticles for drug delivery. Nat Rev Drug Discov. 2021;20:101–124. doi: 10.1038/s41573-020-0090-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kulkarni JA, Cullis PR, van der Meel R. Lipid nanoparticles enabling gene therapies: from concepts to clinical utility. Nucleic Acid Ther. 2018;28:146–157. doi: 10.1089/nat.2018.0721. [DOI] [PubMed] [Google Scholar]
- 85.Samaridou E, Heyes J, Lutwyche P. Lipid nanoparticles for nucleic acid delivery: Current perspectives. Adv Drug Deliver Rev. 2020;154:37–63. doi: 10.1016/j.addr.2020.06.002. [DOI] [PubMed] [Google Scholar]
- 86.Immordino ML, Dosio F, Cattel L. Stealth liposomes: review of the basic science, rationale, and clinical applications, existing and potential. Int J Nanomed. 2006;1:297–315. doi: 10.2217/17435889.1.3.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Romberg B, Hennink WE, Storm G. Sheddable coatings for long-circulating nanoparticles. Pharm Res. 2008;25:55–71. doi: 10.1007/s11095-007-9348-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhang LM, Wang P, Feng Q, Wang NX, Chen ZT, Huang YY, Zheng WF, Jiang XY. Lipid nanoparticle-mediated efficient delivery of CRISPR/Cas9 for tumor therapy. Npg Asia Mater. 2017;9:e441–e441. doi: 10.1038/am.2017.185. [DOI] [Google Scholar]
- 89.Singh RK, Kim HW. Inorganic nanobiomaterial drug carriers for medicine. Tissue Eng Regen Med. 2013;10:296–309. doi: 10.1007/s13770-013-1092-y. [DOI] [Google Scholar]
- 90.Chen SZ, Hao XH, Liang XJ, Zhang Q, Zhang CM, Zhou GQ, Shen SG, Jia G, Zhang JC. Inorganic nanomaterials as carriers for drug delivery. J Biomed Nanotechnol. 2016;12:1–27. doi: 10.1166/jbn.2016.2122. [DOI] [PubMed] [Google Scholar]
- 91.Patra JK, Das G, Fraceto LF, Campos EVR, Rodriguez-Torres MDP, Acosta-Torres LS, Diaz-Torres LA, Grillo R, Swamy MK, Sharma S, Habtemariam S, Shin HS. Nano based drug delivery systems: recent developments and future prospects. J Nanobiotechnol. 2018;16:1–33. doi: 10.1186/s12951-018-0392-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kim T, Hyeon T. Applications of inorganic nanoparticles as therapeutic agents. Nanotechnology. 2014;25:012001. doi: 10.1088/0957-4484/25/1/012001. [DOI] [PubMed] [Google Scholar]
- 93.Wang FL, Li CY, Cheng J, Yuan ZQ. Recent advances on inorganic nanoparticle-based cancer therapeutic agents. Int J Env Res Pub He. 2016;13:1182. doi: 10.3390/ijerph13121182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Daniel MC, Astruc D. Gold nanoparticles: Assembly, supramolecular chemistry, quantum-size-related properties, and applications toward biology, catalysis, and nanotechnology. Chem Rev. 2004;104:293–346. doi: 10.1021/cr030698+. [DOI] [PubMed] [Google Scholar]
- 95.Zhang GM. Functional gold nanoparticles for sensing applications. Nanotechnol Rev. 2013;2:269–288. doi: 10.1515/ntrev-2012-0088. [DOI] [Google Scholar]
- 96.Elahi N, Kamali M, Baghersad MH. Recent biomedical applications of gold nanoparticles: a review. Talanta. 2018;184:537–556. doi: 10.1016/j.talanta.2018.02.088. [DOI] [PubMed] [Google Scholar]
- 97.Hu XP, Zhang YT, Ding TT, Liu J, Zhao H. Multifunctional Gold Nanoparticles: A Novel Nanomaterial for Various Medical Applications and Biological Activities. Front Bioeng Biotech. 2020;8:990. doi: 10.3389/fbioe.2020.00990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mout R, Ray M, Yesilbag Tonga G, Lee YW, Tay T, Sasaki K, Rotello VM. Direct cytosolic delivery of CRISPR/Cas9-ribonucleoprotein for efficient gene editing. ACS Nano. 2017;11:2452–2458. doi: 10.1021/acsnano.6b07600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lee K, Conboy M, Park HM, Jiang F, Kim HJ, Dewitt MA, Mackley VA, Chang K, Rao A, Skinner C, Shobha T, Mehdipour M, Liu H, Huang WC, Lan F, Bray NL, Li S, Corn JE, Kataoka K, Doudna JA, Conboy I, Murthy N. Nanoparticle delivery of Cas9 ribonucleoprotein and donor DNA in vivo induces homology-directed DNA repair. Nat Biomed Eng. 2017;1:889–901. doi: 10.1038/s41551-017-0137-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Laurent S, Forge D, Port M, Roch A, Robic C, Elst LV, Muller RN. Magnetic iron oxide nanoparticles: Synthesis, stabilization, vectorization, physicochemical characterizations, and biological applications. Chem Rev. 2008;108:2064–2110. doi: 10.1021/cr068445e. [DOI] [PubMed] [Google Scholar]
- 101.Yu ZB, Peng C, Luo Y, Zhu JZ, Chen C, Shen MW, Shi XY. Poly(gamma-glutamic acid)-stabilized iron oxide nanoparticles: synthesis, characterization and applications for MR imaging of tumors. Rsc Adv. 2015;5:76700–76707. doi: 10.1039/C5RA15814A. [DOI] [Google Scholar]
- 102.Luo S, Ma C, Zhu MQ, Ju WN, Yang Y, Wang X. Application of Iron Oxide Nanoparticles in the Diagnosis and Treatment of Neurodegenerative Diseases With Emphasis on Alzheimer's Disease. Front Cell Neurosci. 2020;14:21. doi: 10.3389/fncel.2020.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hryhorowicz M, Grzeskowiak B, Mazurkiewicz N, Sledzinski P, Lipinski D, Slomski R. Improved delivery of CRISPR/Cas9 system using magnetic nanoparticles into porcine fibroblast. Mol Biotechnol. 2019;61:173–180. doi: 10.1007/s12033-018-0145-9. [DOI] [PubMed] [Google Scholar]
- 104.Wu SH, Mou CY, Lin HP. Synthesis of mesoporous silica nanoparticles. Chem Soc Rev. 2013;42:3862–3875. doi: 10.1039/c3cs35405a. [DOI] [PubMed] [Google Scholar]
- 105.Xu X, Koivisto O, Liu C, Zhou J, Miihkinen M, Jacquemet G, Wang D, Rosenholm JM, Shu Y, Zhang H. Effective delivery of the CRISPR/Cas9 system enabled by functionalized mesoporous silica nanoparticles for GFP-tagged paxillin knock-in. Adv Therapeutics. 2021;4:2000072. doi: 10.1002/adtp.202000072. [DOI] [Google Scholar]
- 106.Zhang BC, Luo BY, Zou JJ, Wu PY, Jiang JL, Le JQ, Zhao RR, Chen L, Shao JW. Co-delivery of sorafenib and CRISPR/Cas9 based on targeted core-shell hollow mesoporous organosilica nanoparticles for synergistic HCC therapy. ACS Appl Mater Interfaces. 2020;12:57362–57372. doi: 10.1021/acsami.0c17660. [DOI] [PubMed] [Google Scholar]