Graphical abstract
Keywords: anti-viral, tetravalent, synthetic, neutralizing, RBD-binding
Abstract
Neutralizing antibodies (nAbs) hold promise as therapeutics against COVID-19. Here, we describe protein engineering and modular design principles that have led to the development of synthetic bivalent and tetravalent nAbs against SARS-CoV-2. The best nAb targets the host receptor binding site of the viral S-protein and tetravalent versions block entry with a potency exceeding bivalent nAbs by an order of magnitude. Structural studies show that both the bivalent and tetravalent nAbs can make multivalent interactions with a single S-protein trimer, consistent with the avidity and potency of these molecules. Significantly, we show that the tetravalent nAbs show increased tolerance to potential virus escape mutants and an emerging variant of concern. Bivalent and tetravalent nAbs can be produced at large-scale and are as stable and specific as approved antibody drugs. Our results provide a general framework for enhancing antiviral therapies against COVID-19 and related viral threats, and our strategy can be applied to virtually any antibody drug.
Introduction
As of July 17, 2021, the ongoing COVID-19 viral pandemic has tallied more than 188,000,000 confirmed cases and caused over 4,000,000 deaths (www.who.int). Moreover, the highly infectious nature of the disease has imposed severe global economic hardship due to the need for social distancing and lockdown measures. A number of repurposed drugs have shown only limited or uncertain efficacy against COVID-19.1, 2 Although licensed vaccines are now being applied,3 their effectiveness across demographics, availability worldwide, and resistance to adoption by the public remains an unknown. Almost certainly, COVID-19 will remain a serious human health concern for the foreseeable future and there is an urgent need for the development of therapeutics capable of treating patients and those at high risk for infection and/or with a poor prognosis.
Several lines of evidence suggest that SARS-CoV-2 neutralizing antibodies (nAbs) that bind directly to the virus spike glycoprotein and inhibit entry into host cells have therapeutic potential. First, many infected individuals either remain asymptomatic or recover rapidly with only minimal symptoms, and plasma from these convalescent patients usually contains nAbs.4, 5 Second, transfer of plasma containing nAbs from convalescent patients to symptomatic patients has been beneficial in some cases.6, 7, 8 Third, recombinant nAbs that inhibit the interaction between SARS-CoV-2 and host cells confer protection in cell-based assays and animal models,9, 10 and efficacy has been observed for nAbs targeting related coronaviruses SARS-CoV11, 12, 13 and MERS.14 Consequently, a number of nAbs have entered clinical trials as post-infection treatment of COVID-19 associated with SARS-CoV-2 (Clinicaltrials.gov - NCT04452318, NCT04497987), with some (Bamlanivimab, Casirivimab, and Imdevimab) receiving Emergency Use Authorization for treatment of mildly ill subjects.
SARS-CoV-2 virions contain 25–100 glycosylated spike (S) proteins that protrude from the viral membrane.15, 16 The S-protein binds to the host cell receptor, angiotensin-converting enzyme 2 (ACE2), to mediate viral entry.17 The S-protein is a homotrimer and each of its three receptor binding domains (RBD) can be found in either the “up” or the “down” conformation, the former required for ACE2 binding. The most potent nAbs against both SARS-CoV-2 and SARS-CoV bind to the RBD and sterically block its interaction with ACE2.18, 19, 20, 21, 22 Consequently, we focused our efforts on developing nAbs that bound to the RBD and competed with ACE2.
To date, all clinically advanced candidate nAbs against SARS-CoV-2 infection have been derived by cloning from B cells of recovered COVID-19 patients or from other natural sources.10, 18, 20, 21, 22, 23, 24 Here, we applied an alternative strategy using in vitro selection with phage-displayed libraries of synthetic Abs built on a single human IgG framework derived from a clinically validated drug, trastuzumab.25 This approach enabled rapid production of high affinity nAbs with drug-like properties ready for pre-clinical assessment. Moreover, the use of a highly stable framework enabled facile and modular design of ultra-high-affinity nAbs in tetravalent formats that retained favorable drug-like properties while exhibiting neutralization potencies that greatly exceeded those of the bivalent IgG format. Our tetravalent platform provides a general approach for rapidly improving the potency of virtually any nAb targeting pathogen-related, receptor-binding proteins, including SARS-CoV-2 and related coronaviruses and while improving existing COVID-19 nAb drugs, can be adapted in response to resistant mutations or to future viral threats.
Results
Engineering of anti-RBD Fabs and IgGs
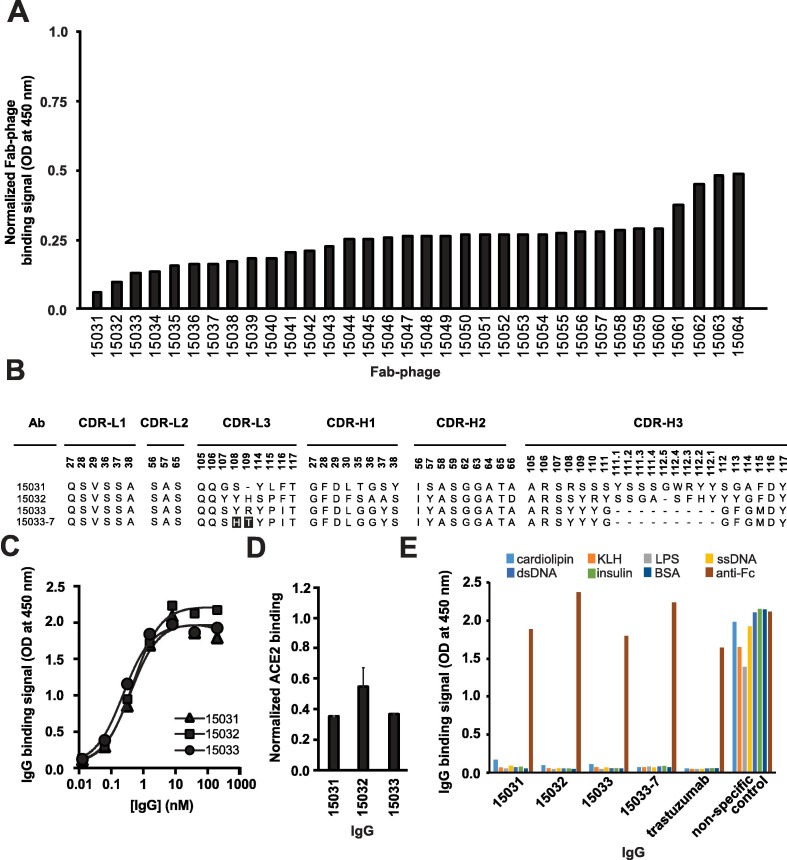
Using a phage-displayed human antigen-binding fragment (Fab) library similar to library F,26 we performed four rounds of selection for binding to the biotinylated RBD of the S-protein of SARS-CoV-2 immobilized on streptavidin-coated plates. Screening of 384 Fab-phage clones revealed 348 that bound to the RBD but not to streptavidin. The Fab-phage were screened by ELISA and those exhibiting > 50% loss of binding to the RBD in the presence of 200 nM ACE2 were sequenced, resulting in 34 unique clones (Figure 1 (A)) that were converted into the full-length human IgG1 format for purification and functional characterization.
Figure 1.
Characterization of anti-RBD Abs by ELISA. (A) Binding of unique Fab-phage clones to immobilized RBD blocked by solution-phase ACE2. Signal was normalized to the signal in the absence of ACE2. (B) CDR sequences of Abs for which the binding to RBD was strongly blocked by ACE2. Positions are numbered according to the IMGT nomenclature.46 Sequences in 15033-7 that differ from 15033 are shaded black. (C) Serial dilutions of IgG binding to immobilized S-protein trimer. The EC50 values derived from the curves are shown in Table 1 and values are representative of 2 independent experiments. (D) Binding of biotinylated ACE2 to immobilized S-protein blocked by solution-phase IgG. Signal was normalized to the signal in the presence of a non-binding control IgG and error bars show the standard error of the mean of duplicate samples. (E) Assessment of non-specific binding of IgGs to immobilized antigens or a goat anti-human Fc Ab (positive control).
To determine relative binding strength, ELISAs were performed with serial dilutions of IgG protein binding to streptavidin-captured, biotinylated S-protein trimer. These assays showed that three IgGs bound with sub-nanomolar EC50 values (Figure 1(B), (C) and Table 1 ). Each IgG also partially blocked the binding of biotinylated ACE2 to immobilized S-protein (Figure 1(D)). Moreover, similar to the clinical benchmark IgG trastuzumab, these three IgGs did not bind to seven immobilized, heterologous biomolecules whose promiscuous interaction with some IgGs can be predictive of poor pharmacokinetics in vivo (Figure 1(E)).27, 28 We also used biolayer interferometry (BLI) to measure binding kinetics and to determine avidities more accurately. All three antibodies exhibited sub-nanomolar KD apparent (Table 1), in agreement with estimates determined by ELISA (Figure 1(C)). Among these, IgG 15033 exhibited the highest avidity, which was mainly due to a two- or seven-fold higher on-rate than IgG 15031 or 15032, respectively. Based on the binding kinetics, we focused further efforts on Ab 15033.
Table 1.
Affinity, potency and biophysical characteristics of nAbs.
| Ab | Format | BLI analysisa |
Virus inhibitionb |
SEC analysisc |
TM2d(oC) | Yielde (mg/L) | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| kon (105M-1s−1) | koff (10-5s−1) | Apparent KD (pM) | IC50 (pM) | IC50 (ng/mL) | Monomer Peak (%) | Elution volume (mL) | ||||
| Trastuzumab | IgG | NA | NA | NA | NA | NA | >95 | 9.2 | 79.5 | 303 |
| 15033 | IgG | 17 | 50 | 300 | 3260 | 489 | 91 | 9.3 | 81 | 207 |
| 15033-7 | IgG | 18 | 7 | 39 | 550 | 83 | 95 | 9.1 | 81 | 165 |
| 15033 | Fab-IgG | 46 | 1.5 | 3 | 170 | 43 | >95 | 8.2 | 87 | 214 |
| 15033-7 | Fab-IgG | 40 | 0.8 | 2 | 60 | 15 | 93 | 8 | 85.5 | 194 |
| 15033 | IgG-Fab | 36 | 1.9 | 5 | 160 | 41 | >95 | 8.2 | 87 | 166 |
| 15033-7 | IgG-Fab | 30 | <0.1 | <1 | 37 | 9.3 | 93 | 8.1 | 85 | 118 |
Calculated by assay of serial dilutions of nAb binding to immobilized S-trimer, as shown in Figure 3(D).
Calculated from assays shown in Figure 5(A).
Calculated from analysis shown in Figure 3(B).
Calculated from analysis shown in Figure S3.
Calculated from proteins produced by transient transfection of Expi293F cells followed by affinity purification with Protein A sepharose, as shown in Figure 3(C).
We took advantage of the modular design principles of our synthetic Ab library to improve the affinity of Ab 15033. The naïve library was constructed with tailored diversification of key positions in all three heavy chain complementarity-determining regions (CDRs) and the third CDR of the light chain (CDR-L3). We reasoned that the already high affinity of Ab 15033 could be further improved by recombining the heavy chain with a library of light chains with naïve diversity in CDR-L3. Following selection for RBD binding, the light chain library yielded numerous variants, of which 17 were purified in the IgG format and analyzed by BLI. Several of the variant light chains resulted in IgGs with improved binding kinetics compared with IgG 15033, and in particular, IgG 15033-7 (Figure 1(B)) exhibited significantly improved avidity (KD apparent = 300 pM for 15033 or 39 pM for 15033-7) due to an off-rate that was an order of magnitude slower (Table 1).
Structural analysis of Fab:RBD and Fab:S-protein complexes
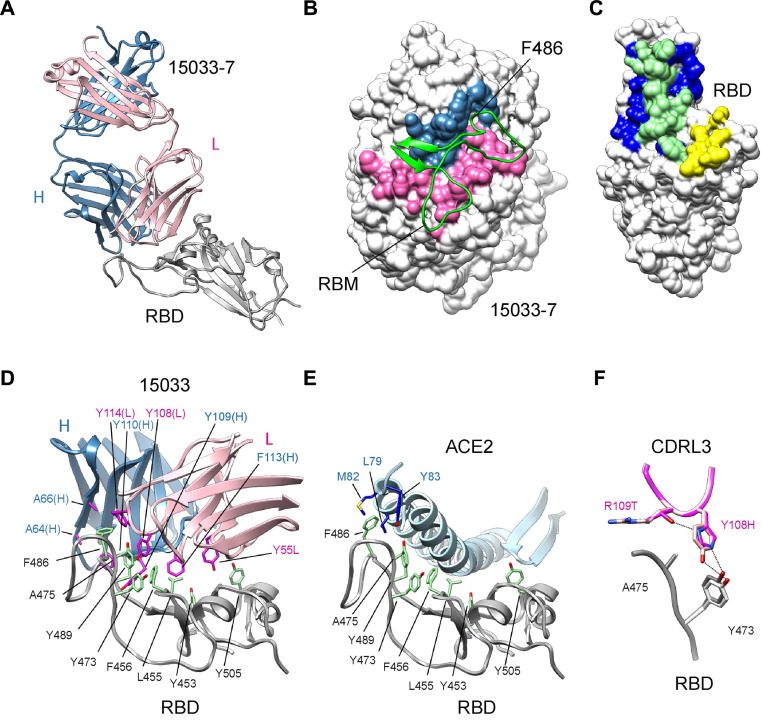
To rationalize the molecular basis for the differences between nAbs 15033 and 15033-7, and confirm blockade of ACE2 binding, we first solved the X-ray crystal structures of Fabs in complex with the SARS-CoV-2 RBD at 3.2 and 3.0 Å resolution, respectively (Figure 2 (A), Table S1). As expected, backbone superposition showed that the two complexes were essentially identical (RMSD = 0.17 Å). The binding of Fab 15033-7 to the RBD resulted in an interface with 1130 and 1112 Å2 of buried surface area on the RBD and Fab, respectively. Of the surface area buried on the Fab, 59% comes from the light chain and 41% from the heavy chain, with the Fab paratope centered on the receptor binding motif of the RBD (Figure 2(B)). The contributions of individual CDR loops to RBD binding are detailed in Table S2. Comparison of the Fab and ACE2 footprints on the RBD revealed that they overlap extensively, with 69% of the ACE2 footprint covered by that of the Fab footprint (Figure 2(C)). It follows that direct steric hindrance explains the ability of 15033 and 15033-7 to block the RBD-ACE2 interaction (Figure 1(D)).
Figure 2.
X-ray crystallographic analysis of Fab 15033-7 in complex with the RBD. (A) Ribbon diagram Fab 15033-7 in complex with the RBD. The RBD is shown in white and the light and heavy chains of the Fab are shown in pink and blue, respectively. (B) Fab 15033-7 (surface representation) and its interaction with the receptor binding motif (residues Leu452 to Ser494, green ribbons) of the RBD. The paratope defined by the Fab-RBD complex is colored blue (heavy chain) and pink (light chain) and the remainder of the Fab is colored white. (C) Surface representation of the RBD colored to show the Fab footprint (blue and green), the ACE2 footprint (yellow and green) and their significant overlap (green). (D) Non-polar residues (magenta) on Fab 15033 (light chain, pink; heavy chain, blue) make key interactions with a stretch of exposed non-polar residues (green) on the RBD (white). (E) The same non-polar residues (green) on the RBD (white) are critical for binding to ACE2 (light blue). RBD residue Phe486 makes key interactions in both the Fab and ACE2 complexes. (F) Fabs 15033 (light pink) and 15033-7 (dark pink) differ by only two residues, both found in CDR-L3. In 15033-7, His108 forms an additional hydrogen bond with Thr109, an interaction that stabilizes the local conformation of the CDR-L3 loop.
Fabs 15033 and 15033-7 recognize a patch of surface-exposed non-polar residues on the RBD using several non-polar residues in both their heavy and light chain CDRs (Figure 2(D)). The significance of this interaction mode is made clear by a comparison with the ACE2-RBD complex (Figure 2(E)). The same patch of non-polar RBD residues mediates the interaction with ACE2, a reflection of their importance in both the Fab-RBD and ACE2-RBD complexes. The RBD of SARS-CoV-2 binds to ACE2 with high affinity and exploiting this surface on the RBD may explain why Ab 15033 emerged as the most potent Ab from our initial screens. RBD residue Phe486 is particularly noteworthy as its side chain is completely buried in a pocket between CDRs H2, H3 and L3.
Abs 15033 and 15033-7 differ at only two positions (15033, Tyr108L/Arg109L; 15033-7, His108L/Thr109L). In the RBD-15033 complex, Tyr108L makes van der Waals interactions with RBD residues Tyr473 and Ala475, as well as a weak hydrogen bond to the side chain of Tyr473 (Figure 2(F)). In the higher affinity RBD-15033-7 complex, the side chain of the equivalent residue, His108L, makes similar interactions. However, its side chain also makes an additional intramolecular hydrogen bond to the corresponding residue, Thr109L, the other residue that differs between 15033 and 15033-7. Taken together, these residue changes likely stabilize the 15033-7 CDR-L3 loop conformation with a concomitant improvement in both the van der Waals and hydrogen bond interactions with RBD residues Tyr473 and Ala475.
Using electron cryo-microscopy (cryo-EM), we also determined the structure of Fab 15033-7 in complex with the S-protein trimer (Table S3). This analysis resulted in the identification of at least 4 different complexes with either two or three Fabs bound to each S-protein trimer (Figures 3, S1, S2). For all but one of the Fabs in these complexes, interpretable density for the entire Fab was observed, a result of Fab-Fab interactions that served to immobilize the Fab and the RBD to which it was bound.
Figure 3.
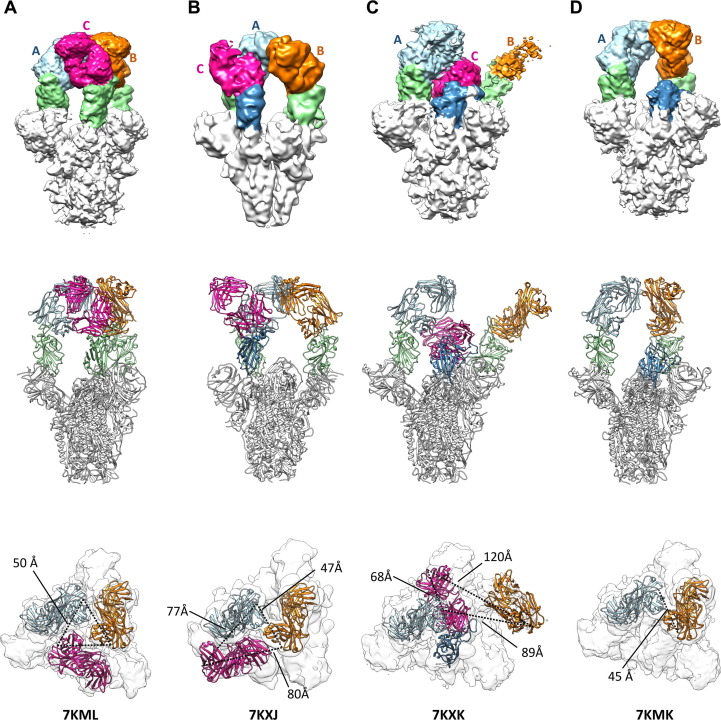
Cryo-EM analysis of Fab 15033-7 in complex with the trimeric S-protein ectodomain. Four Fab-S-protein complexes were observed in the cryo-EM maps: (A) the 3-Fab-bound, 3-“up”, C3 symmetric structure; (B) the 3-Fab-bound, 3-“up”, asymmetric structure; C) the 3-Fab-bound, 2-“up”-1-“down” structure; (D) the 2-Fab-bound, 2-“up” structure. Upper row: side view of the EM maps in surface representation; middle row: side view of the model in ribbon representation; lower row: top view of the EM maps with the Fabs in ribbon representation and S-protein in surface representation. The dotted lines indicate the distances between the C-terminal ends of the Fab heavy chains. Light blue, unit A Fab; orange, unit B Fab; magenta, unit C Fab; green/blue, RBD. In (C) and (D) the RBDs (blue) are in the “down” position. In (B) the RBD (blue) and its bound Fab (unit C) is rotated, breaking the C3-symmetry observed in (A). In (C) the density for the B unit Fab is weak. The distances involving it were derived from the best fit of a Fab (orange) to the density (also see Figures S1 and S2).
In two of the complexes with three Fabs bound (Figure 3 (A), (B)), all three RBDs were found in the “up” conformation. The Fabs were relatively well-ordered in both complexes, as adjacent Fabs contacted each other around the three-fold rotation axis describing the S-protein trimer. Although three-fold symmetry was observed in one of the complexes (Figures 3(A), S1(A)), in the other, one of the Fab-RBD units made unique Fab-Fab interactions that broke the symmetry (Figures 3(B), S1(B)).
In the third complex with three Fabs bound, two of the RBDs were in the “up” conformation and the third RBD was in the “down” conformation (Figures 3(C), S1(C)). To accommodate the latter, one of the other Fab-RBD units was pushed away from the 3-fold rotation axis where it was unable to make Fab-Fab contacts. As such, the density for the Fab was relatively weak, an indication of motion/disorder. The remaining Fab-RBD unit stacked over the one in the “down” conformation, making extensive and yet again different Fab-Fab contacts. This led to a well-packed arrangement where the Fab bound to the “down” RBD was sandwiched between the two “up” RBDs.
In the complex with 2 Fabs bound (Figures 3(D), S1(D)), the bound RBDs were in the “up” conformation and the Fab-Fab interactions were very similar to those found in the symmetrical complex with three Fabs bound (Figures 3(A), S1(A)). The structure showed that the unbound RBD, which was in the “down” conformation, could not sterically accommodate a Fab and further affirmed that one of the two “up” Fab-RBD units must move away from the 3-fold rotation axis if binding to it was to occur.
Analysis of the distances between the C-termini of the heavy chains of the Fabs in these complexes showed that they range from 45 to 120 Å (Figure 3, lower row). In some cases, they are oriented such that two of them, with some repositioning of the Fab-RBD unit, could be linked to a single IgG molecule. Indeed, as described below, negative stain electron microscopy (nsEM) confirmed simultaneous binding of both Fab arms of a single IgG 15033-7 molecule to the S-protein trimer.
Taken together, our structural analysis showed that Fab 15033/15033-7 blocks ACE2 binding to the RBD by direct steric hindrance and that simultaneous binding of both IgG arms to the S-protein trimer likely enhances potency through avidity effects. It also showed that Fab 15033/15033-7 can make four conformationally distinct complexes with the S-protein trimer through interactions involving the RBD in either the “up” or “down” conformation, a property also shared by some other ACE2-blocking Fabs.29 While it was not immediately apparent why these Fabs were able to recognize both “up” and “down” positions, such flexibility bodes well for efforts aimed at the design of improved antibody-based therapeutics. Finally, it showed that different Fab-Fab interaction modes served to define and stabilize the complexes observed.
Engineering of tetravalent nAbs with enhanced avidities
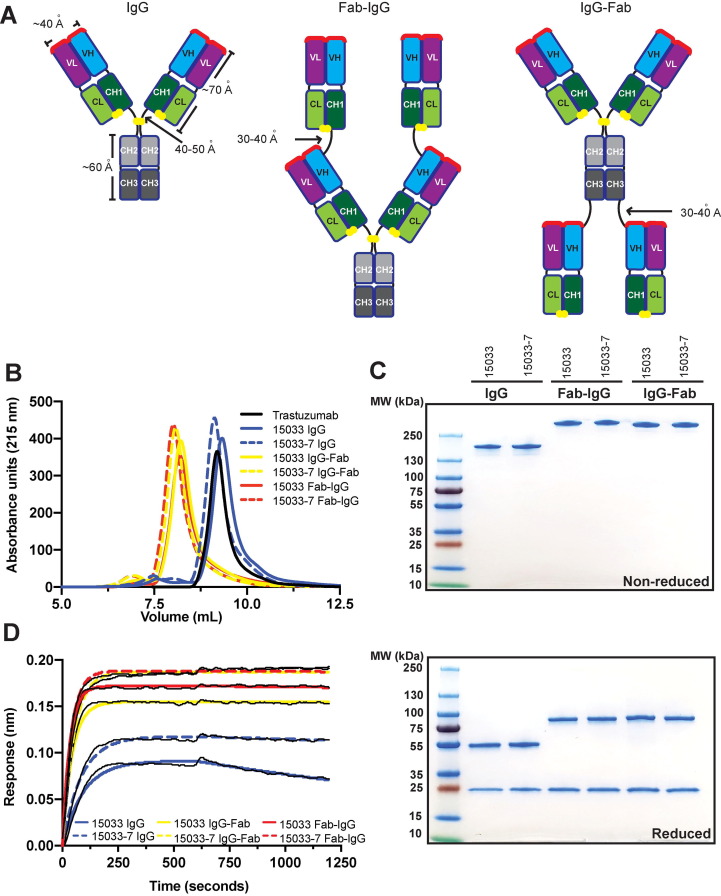
Building on these observations, we explored whether we could further enhance the interactions of these nAbs with the S-protein trimer using modular design strategies. We generated tetravalent versions of 15033 and 15033-7 by fusing additional copies of the Fab to either the N- or C- terminus of the IgG heavy chain to construct molecules that we termed Fab-IgG or IgG-Fab, respectively (Figure 4 (A)).
Figure 4.
Design and characterization of tetravalent nAbs. (A) Schematic of the bivalent IgG format, and the tetravalent Fab-IgG and IgG-Fab formats. Paratopes are shown in red, linkers are shown in black, and disulfide bonds are shown as yellow spheres. (B) Analytical gel filtration SEC of nAbs. (C) SDS-PAGE analysis of nAbs under non-reducing (top) or reducing conditions (bottom). (D) BLI sensor traces for nAbs (6.7 nM) binding to immobilized S-protein trimer.
Our ultimate goal is to produce nAbs that can be used to counter SARS-CoV-2 infections either as a therapeutic and/or as a prophylactic. Aside from high affinity and specificity, effective nAb drugs must also possess favorable properties including high yields from recombinant expression in mammalian cells, high thermodynamic stability, and the lack of aggregation and excessive hydrophobic surface area. To examine these properties, we produced IgGs 15033 and 15033-7, and their Fab-IgG and IgG-Fab counterparts, by transient expression in Expi293F cells. All six proteins were expressed in high yield (160–200 mg/L) and showed high thermostability with CH3/Fab melting temperatures exceeding even that of the trastuzumab Fab (81–87 °C vs 79.5 °C, Table 1, Figure S3). Size exclusion chromatography revealed that each IgG eluted as a predominantly (91 to > 95%) monodisperse peak with elution volumes nearly identical to trastuzumab (Figure 4(B) and Table 1). Tetravalent molecules eluted as single peaks in advance of trastuzumab, consistent with their larger molecular weights. We also showed that the IgG and tetravalent versions of both 15033 and 15033-7 could be purified to near homogeneity by protein-A affinity chromatography as evidenced by SDS-PAGE (Figure 4(C)). Taken together, these analyses demonstrated that the IgGs and their tetravalent derivatives possess excellent biophysical properties that will facilitate drug development and production at large scale.
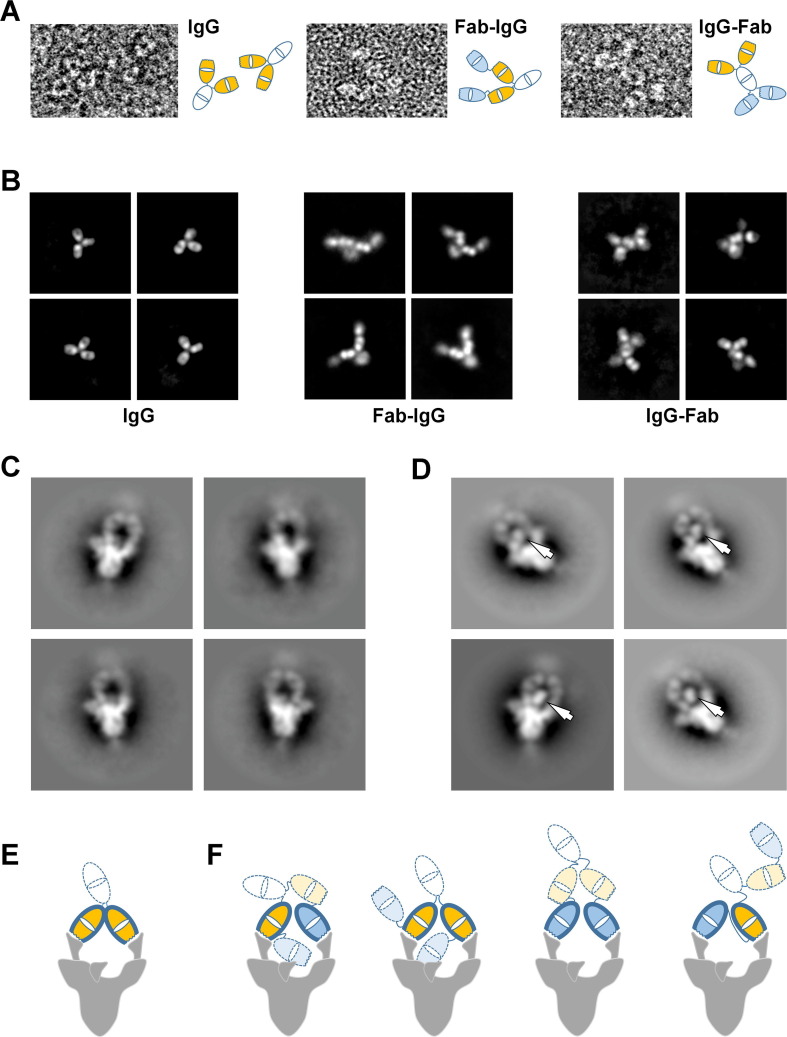
Importantly, the tetravalent Abs exhibited greatly reduced off-rates compared with their bivalent IgG counterparts, with KD apparent for the S-protein trimer in the low single-digit picomolar range as measured by BLI (Figure 4(D), Table 1). Negative stain micrographs (Figure 5 (A)) and 2D class averages of IgG 15033-7 and its tetravalent formats (Figure 5(B)) provided insights in to modular structure of these antibodies. nsEM of IgG 15033-7, in complex with the S-protein trimer, showed that the two arms of a single IgG bound the S-protein trimer in a pincer-like fashion (Figure 5(C),(E)). Fab-IgG 15033-7 also makes pincer-like interactions with a single S-protein trimer (Figure 5(D),(F), Figures S6, S7), but the complexes revealed additional density that differed from those observed for the complex of the S-protein trimer with the IgG. In particular, we observed density consistent with that from an additional Fab in the Fab-IgG complexes, suggesting that all three of the S-protein RBDs are bound by Fabs from a single tetravalent nAb molecule, Fab-IgG in this instance. Given the range of complexes observed in our cryo-EM analysis, it is possible that the Fab-IgG, with four available Fabs, can engage all three RBDs within a single S-protein molecule (Figure 5(F)). Further analysis will be required to fully establish whether this occurs, potentially explaining the decreased off-rates shown by the tetravalent proteins due to avidity. Nevertheless, it is clear that both the Fab-IgG and IgG-Fab possess an increased potential for multivalent interactions with one or more S-protein trimers. The latter mode of interaction may be possible on the viral surface and likely contributes to enhanced potency observed for the tetravalent IgGs, as shown below using a cell-based infectivity assay.
Figure 5.
Negative stain electron microscopy analysis of nAbs and their interaction with the S-protein trimer. (A) Examples of the 15033-7 IgG (left), Fab-IgG (middle) and IgG-Fab (right) molecules observed in the negative stain micrographs. Schematic interpretations are shown to the right of each image. (B) Selected negative stain electron microscopy 2D class averages for the 15033-7 IgG (left), Fab-IgG (middle) and IgG-Fab (right). (C) Negative stain electron microscopy 2D class averages of IgG 15033-7 in complex with the S-protein trimer. (D) Negative stain electron microscopy 2D class averages of Fab-IgG 15033-7 in complex with the S-protein trimer. The arrows indicate Fab-sized densities not observed in the IgG complex in (C). (E) Tentative schematic of the complex in (C) showing a trimer with 2 RBDs “up” and one RBD “down”. (F) Schematics of the four possible ways that the Fab-IgG can bind to the two “up” RBDs of a trimer that has two RBDs “up” and one RBD “down”. In (E) and (F), the following color scheme was used: white, Fc; orange, the Fabs of a native IgG; blue, the Fabs added to the native IgG. Wavy lines indicate the locations of the CDRs. The Fabs with heavy outline are making specific interactions through their CDRs. As shown by our Cryo-EM structures, the RBD in the “down” conformation is accessible for specific interaction with one of the remaining Fabs (orange or blue with light dotted outline) shown in (F). This would lead to a trivalent interaction with the S-protein trimer and may explain the additional Fab-sized densities observed in (D).
Inhibition of SARS-CoV-2 infection in cell-based assays
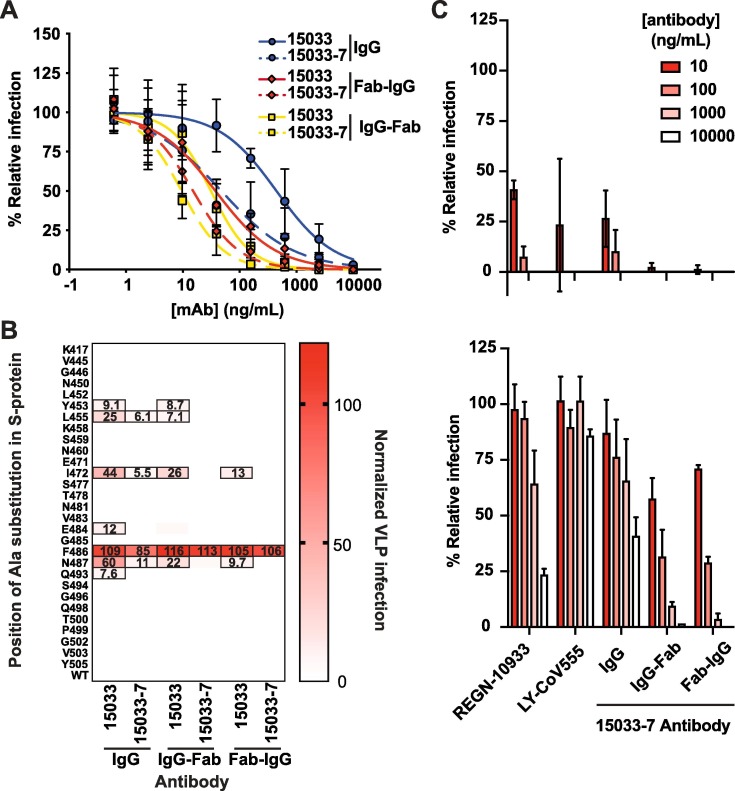
We assessed the effects of the Abs on virus infection in an assay that measured the infection of ACE2-expressing Vero E6 cells with the SARS-CoV-2 strain 2019n-CoV/USA_WA1/2020. All three high affinity nAbs (15031, 15032 and 15033) from the naïve library (Figure 1) exhibited dose-dependent neutralization of SARS-CoV-2 infection, confirming their inhibitory capacity (Figure S4). Consistent with their affinities, IgG 15033 was the most potent with an IC50 of 3.3 nM, and its potency was confirmed with the observation of strong neutralization of a second SARS-CoV-2 strain (2019-nCoV/Italy-INMI1). Neutralization of the second strain was consistent with that of the first strain, and for simplicity, only one set of data are shown.
The tetravalent Fab-IgG and IgG-Fab versions of 15033 exhibited improved potencies with IC50 values of 170 and 160 pM, respectively (Figure 6 (A) and Table 1). The optimized IgG 15033-7 also exhibited high neutralizing potency with an IC50 of 550 pM and its tetravalent Fab-IgG and IgG-Fab versions exhibited the best potencies of all the molecules tested with IC50 values of 60 and 37 pM, respectively. Taken together, these results support the ability of naïve synthetic Ab libraries to yield highly potent nAbs with drug-like properties. Moreover, optimization via engineering of tetravalent formats can produce drug-like molecules with ultra-high potencies, exceeding those of bivalent IgGs.
Figure 6.
Neutralization of SARS-CoV-2 and pseudotyped VLPs. (A) Neutralization of SARS-CoV-2 strain 2019 n-CoV/USA_WA1/2020 by bivalent and tetravalent nAbs (also see Figure S4). The virus was pre-treated with serial dilutions of nAb and infection of ACE2-expressing Vero E6 cells was measured relative to untreated control. Samples were run in triplicate and results are representative of two independent experiments. Error bars indicate standard error of the mean. (B) Neutralization of a panel of pseudotyped VLPs displaying SARS-CoV-2 S-proteins with single alanine mutations in or near the ACE2-binding site of the S-protein RBD (also see Figure S5). The VLPs were treated with 50 nM of the indicated nAb and uptake by ACE2-expressing HEK-293 cells was measured in duplicate, and results are representative of two independent experiments. The heat map shows uptake normalized to uptake in the absence of nAb. Boxed cells indicate VLP mutations that represent potential escape vulnerabilities for a given nAb, as defined by > 5% uptake with nAb treatment compared with untreated control (the percent uptake is shown in each cell). (C) Ab-mediated neutralization of isogenic strains of the D614G (upper panel) and B.1.351 (lower panel) SARS-CoV-2 variants, as assessed in the focus reduction neutralization assay at the indicated Ab concentrations relative to assays conducted in the absence of Ab.
Resistance to potential viral escape mutants
To explore the sensitivity of our nAbs to potential escape mutants, we generated HIV-gag-based, lentivirus-like particles (VLPs) pseudotyped with the SARS-CoV-2 S-protein. We confirmed ACE2-dependent infection of HEK-293 cells stably over-expressing exogenous ACE2 by pseudotyped VLPs, and that infection was inhibited by either Fc-tagged RBD (RBD-Fc) or IgG 15033 (data not shown). Using this system, we generated a panel of 44 pseudotyped VLP variants (Figure S5(A)), each containing a single alanine substitution at an RBD position within or close to the ACE2-binding site. Fifteen of these VLP variants exhibited a > 4-fold reduction in infection compared with the wild-type (wt) VLP, suggesting that the substituted residues contributed favorably to interactions between the RBD and ACE2, and were excluded from further analysis. The remaining 29 VLP variants infected with high efficiency, suggesting that these are positions where residue changes could abrogate antibody binding without affecting the ACE2 interaction.
Using the panel of 29 infective VLP variants, we measured VLP infection after treatment with the various nAbs (Figure 6(B)). We defined as escape mutants those VLP variants for which entry in the presence of 50 nM nAb was > 5% of the entry in the absence of the nAb. Based on this definition, we found that 7 of the variants were able to escape from IgG 15033 (Figures 6(B), S5(A)), whereas only 4 could escape from IgG 15033-7. We also found that 15033 in tetravalent Ab format neutralized more variants than it did as an IgG, and that, remarkably, tetravalent Ab 15033-7 strongly neutralized all of the variants but one (Phe486). As discussed above, RBD residue Phe486 is found in the non-polar interface between the RBD and the Fab and it makes numerous contacts with CDRs H2, H3 and L3, sitting deep in a pocket formed by these contacts (Figure S5(B),(C)). As such, it represents a bona fide positional vulnerability where mutant viruses still capable of binding ACE2 could evade neutralization by nAb 15033-7. Comprehensive maps of escape mutations confirm the vulnerability of other nAbs to this mutation, but also suggest strategies by which escape can be overcome with nAb cocktails or bi-specific nAbs.30 Overall, these results showed that for IgGs, increasing the affinity of the Fab-RBD interaction increased neutralizing potency and resistance to mutation, and moreover, that tetravalent presentation of the Fab provides even greater potency and resistance to potential escape mutants.
Resistance to an emerging variant of concern
Recently, variants of SARS-CoV2 with enhanced transmissibility have emerged31, 32, 33 and have been shown to possess mutations within the RBD that could potentially reduce the efficacy of neutralizing antibodies that target this region of the S-protein.34, 35 In particular, the B.1.351 variant possesses three mutations within the RBD (K417N / E484K / N501Y), which have been shown to increase resistance to therapeutic antibodies, convalescent plasma and vaccine sera.34, 35 Notably, the E484K mutation alone reduces the inhibitory activity of many neutralizing antibodies that target the RBD.34
To assess and compare the in vitro neutralization activities of 15033-7 IgG, its tetravalent formats, and approved therapeutic nAbs (REGN10933 and LY-CoV555) - versus the dominant D614G and emerging B.1.351 variants - we conducted cell-based neutralization assays (described above) using isogenic S-protein variants in the background of the Washington strain of the virus.34 These assays clearly showed that all three IgGs possessed similar potencies against the D614G variant, while both IgGs 15033-7 and REGN10933 suffered significant loss of potency against the B.1.351 variant, and neutralization by Ly-CoV555 IgG was completely abrogated (Figure 6(C)). In contrast with the bivalent IgGs, the tetravalent 15033-7 nAbs showed enhanced potency against the D614G variant and were also reasonably potent against the B.1.351 variant. This confirms a key point: improved potency obtained through avidity effects upon conversion to tetravalent formats renders nAbs less sensitive to mutations that have emerged since isolation of the original lead nAbs. This further suggests that tetravalent nAbs may also offer enhanced efficacy against variants that have yet to emerge, highlighting a major advantage of this approach.
Discussion
SARS-CoV-2 has wreaked havoc on global health and the economy, highlighting the need for drug development technologies to combat not only COVID-19, but emerging infectious diseases in general. In this context, we have used synthetic antibody engineering to rapidly develop unique formats of human nAbs as therapeutic candidates. Our nAb formats include natural bivalent IgGs as well as ultra-potent tetravalent molecules arising from a modular design strategy that adds two additional Fabs to the canonical IgG molecule. Importantly, we have shown that enhanced avidities and potencies characteristic of the tetravalent nAbs are achieved without compromising the favorable properties (e.g. yield, solubility, and stability) that make IgG molecules ideal drugs. Moreover, we showed that tetravalent nAbs exhibit enhanced potency against emerging variants of concern and potential virus escape mutants that may yet emerge, a property that highlights the utility of these molecules as therapeutics against SARS-CoV-2 and other viruses capable of mutational escape.
Structural analyses showed that IgG and Fab-IgG 15033-7 make pincer-like interactions with a single SARS-CoV-2 S-protein trimer, an observation providing insight into the basis for the avidity observed for these molecules. It also provides strong evidence that the tetravalent Fab-IgG can engage all three RBDs of the S-protein trimer, and that the RBD can be bound by a Fab in either the “up” or “down” conformation. We did not optimize the tetravalent formats; their very high avidities arose naturally from the geometry of the Fab-RBD complex, the spatial relationships between the Fabs, the flexibility of the RBDs on the S-protein trimer, and fortuitous Fab-Fab interactions that clearly stabilized the complexes observed. We and others have generated many additional Fabs that can inhibit the virus but employ different epitopes to do so, and incorporating these Fabs into tetravalent frameworks, including bispecific formats targeting two distinct epitopes, has the potential to generate a vast array of ultra-high affinity nAbs with minimal further effort. Taken together, these observations underscore the enormous potential of our modular design approach for the development of novel and highly effective nAb therapeutics, including the potential to further optimize even currently approved COVID-19 nAb therapeutics.
COVID-19 has also exposed the need for drug development to respond to emerging viral threats in real time. In this regard, isolation of nAbs from the B cells of infected individuals has emerged as a rapid approach to obtaining leads for drug development. Indeed, several reports have shown that these technologies can deliver drug-grade therapeutic nAbs for manufacturing and subsequent clinical trials in approximately six months.18, 36 Further highlighting the urgent need and the rapid speed at which drug development has proceeded, these IgG-based nAbs have received emergency-use approval from the US FDA as both cocktail and single agent, and these extraordinarily rapid drug development timelines set a benchmark for alternative technologies. The potencies of other anti-SARS CoV-2 nAbs obtained in similar fashion range from nanomolar37 to picomolar,38 but those that have been awarded approval for emergency use have potencies in the middle of this range.18, 36 Thus, it is worth noting that antibodies with higher in vitro potencies may further improve in vivo efficacy, and conversion to tetravalent formats may enhance potency of nAbs in general.
With our platform, we show that synthetic in vitro Ab engineering is comparable to B cell cloning, in terms of both speed and potency of drug development.18, 39 Our project was initiated at the beginning of April 2020 with the identification of our first naïve Ab leads. Within a month, we validated lead IgG molecules as neutralizing agents in cell-based assays with authentic virus. In parallel, we initiated further selections to optimize the Fab 15033 paratope, that yielded our best lead nAb 15033-7; both Fabs were then reformatted as tetravalent molecules (Fab-IgG and IgG-Fab) to further enhance potency. Though others have now shown enhanced potency of SARS-CoV-2 neutralization with oligomers of Ab variable domains40 and synthetic proteins,41 insofar as our format closely resembles natural Abs, they can also be manufactured at large scale, possess long half-lives and likely exhibit low immunogenicity, all of which are required of effective drugs. Indeed, by the beginning of October, we established manufacturing-grade stable mammalian cells capable of producing multi-gram quantities of drug-grade nAbs from a litre of culture, in both the IgG and tetravalent formats. Thus, synthetic Ab engineering technologies can match the six-month lead-to-manufacture timelines established by methods based on the cloning of natural nAbs.
Our synthetic engineering technologies offer exquisite control over Ab design, and by introducing tailored diversity into an optimized IgG framework, ensure that candidate therapeutics possess biophysical properties that are ideal for drug development. Facilitated by the use of highly stable frameworks, we have demonstrated the rapid construction of complex tetravalent formats that enhance potency while retaining favorable drug-like properties. To minimize risk of immunogenicity, we chose as a framework the highly validated drug trastuzumab, which has been well tolerated with minimal immunogenicity in thousands of breast cancer patients,42 and we incorporated synthetic diversity that was designed to mimic natural diversity at only the most hypervariable positions in the CDRs.26 Perhaps most importantly, synthetic methods do not rely on infected patients (i.e. natural repertoires) as a source of drug leads. By its very nature, B cell cloning is a reactive technology that can be implemented only after a viral outbreak, placing limitations on drug development response times. With synthetic in vitro methods, drug development can be undertaken in a proactive manner, enabling development and stockpiling of potential therapeutics in advance of outbreaks. Indeed, large-scale surveillance and sequencing efforts have provided unprecedented access to the genomes of numerous SARS-CoV-2-related viruses and other pathogens with the potential to cross species barriers and infect humans.43, 44 With our approach, it is feasible to develop - in advance and in parallel - synthetic Abs against hundreds of antigens,45 and working within a collaborative international network, we have initiated efforts with this aim.
Methods
Cells
Mammalian cells were maintained in humidified environments at 37 °C in 5% CO2 in the indicated media. Vero E6 (ATCC), HEK293T (ATCC) and HEK293T cells stably overexpressing ACE2 were maintained at 37 °C in 5% CO2 in DMEM containing 10% (vol/vol) FBS. Expi293F cells (ThermoFisher) were maintained at 37 °C in 8% CO2 in Expi293F expression media (ThermoFisher).
Protein production
The previously reported piggyBac transposase-based expression plasmid PB-T-PAF47 containing a CMV promotor (PB-CMV) and a woodchuck hepatitis virus posttranscriptional regulatory element (WPRE) was used for large-scale transient expression. cDNA encoding the SARS-CoV-2 S-protein ectodomain trimer (residues 1–1211), followed by a foldon trimerization motif,48 a 6xHis tag and an AviTag biotinylation motif49 was cloned in to the PB-CMV vector using standard molecular biology techniques, and residues 682–685 (RRAR) and 986–987 (KV) were mutated to SSAS or PP sequences, respectively, to remove the furin cleavage site on the SARS-CoV-2 spike protein or to stabilize the pre-fusion form of the spike,50 respectively. The SARS-CoV-2 receptor binding domain (RBD, residues 328–528), the soluble human ACE2 construct (residues 19–615), and the SARS-CoV RBD (residues 315–514), each followed by a 6xHis tag and an AviTag, were similarly cloned into the same vector. For expression, PB-CMV expression constructs were mixed with Opti-MEM media (Gibco) containing 293fectin reagent (Thermo Fisher) and the mixture was incubated for 5 min before addition to the shaker flask containing 106 Freestyle 293-F cells/mL grown in suspension in Freestyle 293 expression media (Thermo Fisher). Expression was allowed to continue for 6 days before purification.
Protein purification and in vitro biotinylation
Expressed proteins were harvested from expression medium by binding to Ni-NTA affinity resin followed by elution with 1X PBS containing 300 mM imidazole and 0.1% (v/v) protease inhibitor cocktail (Sigma), then further purified by size-exclusion chromatography. For the RBDs and ACE2, a Superdex 200 Increase (GE healthcare) column was used. For the S-protein ectodomain, a Superose 6 Increase (GE healthcare) column was used. Purified proteins were site-specifically biotinylated in a reaction with 200 µM biotin, 500 µM ATP, 500 µM MgCl2, 30 µg/mL BirA, 0.1% (v/v) protease inhibitor cocktail and not more than 100 µM of the protein-AviTag substrate. The reactions were incubated at 30 °C for 2 h and biotinylated proteins were then purified by size-exclusion chromatography.
Phage display selections
A synthetic, phage-displayed antibody library26 was selected for binding to SARS-CoV-2 RBD in solution. In each round, phage library was first depleted on neutravidin immobilized in wells of a 96-well Maxisorp plate from a 2 µg/mL solution incubated with shaking overnight at 4 °C, then incubated with 50 nM biotinylated RBD in solution for two hours at RT. Protein-phage complexes were captured in wells coated with neutravidin as above for 15 min at RT. After washing with 1X PBS pH 7.4 containing 0.05% Tween, phage were eluted for 5 min with 0.1 M HCl and then neutralized with 1 M Tris pH 8.0. Eluted phage were amplified, purified, and selected for binding to target for a total of 5 rounds, after which, individual phage clones were subjected to DNA sequencing, as described.26
Enzyme-linked immunosorbent assays
For ELISAs, plates were coated with neutravidin, as above, then blocked with PBS, 0.2% BSA for 1 h. Biotinylated target protein was captured from solution by incubation in neutravidin-coated and BSA-blocked wells for 15 min with shaking at RT, and subsequently, phage or Ab was added and allowed to bind for 30 min. Plates were washed, incubated with an appropriate secondary antibody, and developed with TMB substrate as described.51
Construction of genes encoding tetravalent Abs
DNA fragments encoding heavy chain Fab regions (VH-CH1; terminating at hinge residue Thr10, IMGT numbering46 were amplified by PCR from the IgG expression constructs. Tetravalent Ab constructs were generated by fusing these fragments with their respective IgG heavy chain in the pSCSTa mammalian expression vector using Gibson assembly (New England Biolabs, Ipswich, MA). Fab-IgG constructs were arranged by fusing a heavy chain Fab domain to the N-terminus of the IgG using a S(G4S)3 linker. IgG-Fab constructs were arranged by fusing a heavy chain Fab domain to the C-terminus using a G(G4S)2GGGTG linker. For both formats, the Fc region terminated at Gly129 (IMGT numbering46).
Ab production and purification
IgG and tetravalent Abs were produced in Expi293F cells (ThermoFisher) by transient transfection, by diluting heavy and light chain construct DNA in OptiMem serum-free media (Gibco) before the addition of and incubation with FectoPro (Polyplus Transfection) for 10 min. For IgG expression, equivalent amounts of plasmids encoding heavy chain or light chain were transfected, whereas for tetravalent formats, a ratio of 2:1 light chain to heavy chain plasmids was used. Following addition of the DNA complex to Expi293F cells and a 5-day expression period, Abs were purified using rProtein A Sepharose (GE Healthcare), then buffer exchanged and concentrated using Amicon Ultra-15 Centrifugal Filter devices (Millipore). IgGs were stored in PBS (Gibco), and tetravalent Abs were stored in 10 mM L-Histidine, 0.9% sucrose, 140 mM NaCl, pH 6.0.
Size exclusion chromatography
Protein samples (50 μg) were injected onto a TSKgel BioAssist G3SWxl column (Tosoh) fitted with a guard column using an NGC chromatography system and a C96 autosampler (Biorad). The column was preequilibrated in a PBS mobile phase and protein retention was monitored by absorbance at 215 nm during a 1.5 CV isocratic elution in PBS.
Biolayer interferometry
The binding kinetics and estimation of apparent affinity (KD) of Abs binding to the S-protein were determined by BLI with an Octet HTX instrument (ForteBio) at 1000 rpm and 25 °C. Biotinylated S-protein was first captured on streptavidin biosensors from a 2 µg/mL solution to achieve a binding response of 0.4–0.6 nm and unoccupied sites were quenched with 100 µg/mL biotin. Abs were diluted with assay buffer (PBS, 1% BSA, 0.05% Tween 20) and 67 nM of an unrelated biotinylated protein of similar size was used as negative control. Following equilibration with assay buffer, loaded biosensors were dipped for 600 s into wells containing 3-fold serial dilutions of each Ab starting at 67 nM, and subsequently, were transferred back into assay buffer for 600 s. Binding response data were corrected by subtraction of response from a reference and were fitted with a 1:1 binding model using ForteBio’s Octet Systems software 9.0.
Differential scanning fluorimetry
Thermostabilities of Abs were determined by differential scanning fluorimetry using Sypro Orange, as described,52 with a 1 µM solution of Ab and temperature range from 25-100 °C in 0.5 °C increments.
Generation of pseudotyped VLPs
HEK-293 cells (ATCC) were seeded in a 6-well plate at 0.3 × 106 cells/well in DMEM (ThermoFisher) supplemented with 10% FBS and 1% penicillin–streptomycin (Gibco) and grown overnight at 37 °C with 5% CO2. HEK-293 cells were then co-transfected with 1 µg of pNL4-3.luc.R-E- plasmid (luciferase expressing HIV-1 with defective envelop protein) (NIH AIDS Reagent Program) and 0.06 μg of CMV-promoter driven plasmid encoding wt or mutant S-protein using Lipofectamine™ 2000 transfection reagent (ThermoFisher). Pseudotyped VLPs were harvested by collecting supernatant 48 h after transfection and were filter sterilized (0.44 µm, Millipore Sigma, Cat. No. SLHA033SS).
Infection assays with pseudotyped VLPs
HEK-293 cells stably over-expressing full-length human ACE2 protein were seeded in 96-well white polystyrene microplates (Corning, Cat. No. CLS3610) at 0.03 × 106 cells/well in DMEM (10% FBS and 1% penicillin–streptomycin) and were grown overnight at 37 °C with 5% CO2. Pseudotyped VLPs were mixed with Ab, incubated at room temperature for 10 min, and added to the cells. The cells were incubated at 37 °C with 5% CO2, the medium was replaced with fresh DMEM (10% FBS and 1% penicillin–streptomycin) after 6 h, and again every 24 h up to 72 h. To measure the luciferase signal (VLP entry), DMEM was removed and cells were replaced in DPBS (ThermoFisher) and mixed with an equal volume of ONE-Glo™ EX Luciferase Assay System (Promega). Relative luciferase units were measured using a BioTek Synergy Neo plate reader (BioTek Instruments Inc.). The data were analyzed by GraphPad Prism Version 8.4.3 (GraphPad Software, LLC).
SARS-CoV-2 focus reduction neutralization assay
SARS-CoV-2 strain 2019 n-CoV/USA_WA1/2020 was obtained from the Centers for Disease Control (USA) and Prevention. The chimeric, infectious SARS-CoV-2 variant was generated with the B.1.351 S-protein in the 2019 n-CoV/USA_WA1/2020 background.34 Virus stocks were produced in Vero CCL81 cells (ATCC) and titrated by focus-forming assay on Vero E6 cells.53 Serial dilutions of mAbs were incubated with 102 focus-forming units (FFU) of SARS-CoV-2 for 1 h at 37 °C. MAb-virus complexes were added to Vero E6 cell monolayers in 96-well plates and incubated at 37 °C for 1 h. Cells were overlaid with 1% (w/v) methylcellulose in MEM supplemented with 2% FBS. Plates were harvested after 30 h by removing overlays and were fixed with 4% PFA in PBS for 20 min at RT. Plates were washed and sequentially incubated with 1 µg/mL of CR302254 anti-S-protein antibody and HRP-conjugated goat anti-human IgG in PBS, 0.1% saponin, 0.1% BSA. SARS-CoV-2-infected cell foci were visualized using TrueBlue peroxidase substrate (KPL) and were quantitated on an ImmunoSpot microanalyzer (Cellular Technologies). Data were processed using Prism software (GraphPad Prism 8.0).
X-ray crystallography
The SARS-CoV-2 RBD (residues 328–528) was expressed with a C-terminal 6xHis tag from HEK293F GnT1-minus cells. The Fabs were expressed from HEK293F cells. The RBD was purified from the expression media by metal-affinity chromatography using Ni-NTA beads (Qiagen). The Fab fragments were purified from the expression media using rProtein A Sepharose Fast Flow beads (GE healthcare). The RBD was treated with endoglycosidase H55 and Carboxypeptidase A (Sigma-Aldrich), to remove the N-glycans and the C-terminal 6xHis tag. The RBD and Fabs were further purified by ion-exchange and hydrophobic interaction chromatography. Before crystallization, the RBD-Fab complexes were purified using size-exclusion chromatography on a Superdex 200 Increase column (GE Healthcare). For both Fab 15033 and Fab 15033-7, the optimized crystallization conditions contained 1.2–1.6 M (NH4)2SO4 and 8–18% glycerol. X-ray diffraction data were collected at 100 K on beamline 08IB-1 at the Canadian Light Source. The diffraction data were integrated and scaled using the XDS package.56 The structures were solved by molecular replacement using the program Phaser.57 A homology model of the VH-VL region was produced by the SysImm Repertoire Builder server (https://sysimm.org/rep_builder).58 The CDR loops were then deleted from this VH-VL model. The constant (CH1-CL) region search model and the RBD search model were both derived from PDB entry 6W41.54 The three models were used to solve the structure by molecular replacement. The atomic models were built using Coot59 and refined with Phenix.refine.60
Negative stain electron microscopy
3 µL of the S-protein ectodomain were applied to the surface of a glow-discharged (60 s, 15 mA), carbon-coated copper mesh grid. The grid was washed with water and stained with 3 µL of 2% uranyl formate. Micrographs were acquired on a Talos L120C 120 kV electron microscope equipped with a Ceta 16 M CMOS camera. Particle selection and two-dimensional classification were carried out using cryoSPARC v2. Contrast transfer function (CTF) estimation was performed using GCTF.61
Cryo-EM
Four additional substitutions for proline residues (817P/A892P/A899P/A942P) were introduced to the S-2P SARS-CoV-2 S-protein ectodomain to stabilize the prefusion conformation.62 Fab 15033-7 was mixed with the SARS-CoV-2 spike ectodomain at a 3:1 molar ratio. 3 µL of the Fab-spike mixture containing 0.4 mg/mL total protein were applied to a glow-discharged (60 s, 15 mA) C-flat 2/2 carbon holey grid (CF-2/2-4C, Electron Microscopy Science). Plunge vitrification was performed using a Thermo Fisher Scientific Vitrobot Mark IV instrument. The grids were blotted for 2.5 s at 100% humidity and 277 K before being plunge-frozen in liquid ethane cooled to 90 K. Specimen screening and optimization were performed using a Talos L120C 120 kV electron microscope equipped with a Ceta 16 M CMOS camera.
High-resolution data were collected on a Thermo Fisher Scientific Titan Krios G3 300 kV microscope equipped with a Falcon 4 Direct Electron Detector. The data were collected at 75000x nominal magnification, resulting in a calibrated pixel size of 1.03 Å. Each movie was recorded in counting mode with a 10 s exposure and saved in 30 fractions. The total exposure was 38 electrons per Å2. The data were collected with a 1.0–2.2 µm defocus setting. A total of ∼ 6431 movies were collected for the final high-resolution dataset.
Full-frame motion and local motion corrections of the cryo-EM movies were performed using implementations of the alignframes_lmbfgs algorithm63 in cryoSPARC v2.64 CTF parameters were estimated using GCTF.61 Particle selection was initially performed using a Gaussian blob picker, then by Topaz neural network picking,65 both in cryoSPARC v2. Two-dimensional classification of the particle images was performed using cryoSPARC v2. The particle images corresponding to Fab-S-protein complexes in the different conformations were separated by performing heterogenous refinement in cryoSPARC v2. Initial map generation and homogenous, heterogenous and non-uniform three-dimensional refinements were performed in cryoSPARC v2. For the map with three RBDs in the up conformation, the three-dimensional refinements were carried out with either C1 or C3 symmetry imposed. The maps generated with C3 symmetry were used for model building. A previously reported SARS-CoV-2 spike structure, PDB entry 6VXX,66 and the 15033-7:RBD complex reported here, were first docked into the map using UCSF Chimera67 and then manually modified using Coot.68 The HV-LV and HC1-LC domains of the Fab were docked individually to account for elbow angle differences (SI Figure ZFH). The resulting model was refined with C3 symmetry against the cryo-EM map using Rosetta69 and Phenix.70 The resulting C3-symmetric model was used as starting model to build the other three structures, which were also refined against the corresponding cryo-EM maps using Rosetta and Phenix. The atomic models were validated using Molprobity71 and the comprehensive validation (cryo-EM) tool in Phenix.70 To improve the interpretability of the Fab-RBD interaction, local refinement of the Fab-RBD unit was performed in cryoSPARC v2 using a mask encompassing one of the Fab-RBD units in the C3-symmetric structure. Moderate resolution improvement for the Fab-RBD unit was achieved (Figure S2(H)).
Data deposition
The crystal structures of the RBD-Fab 15033 and RBD-Fab 15033-7 complexes were deposited in the PDB with accession codes 7KLG and 7KLH, respectively. The S-protein-Fab 15033-7 cryo-EM maps and structures were deposited to EMDB and PDB with the following accession codes: EMD-22925/PDB:7KMK (2 Fab bound, 2-“up”-1-“down”), EMD-22926/PDB:7KML (3 Fab bound, 3-“up”, C3-symmetric), EMD-23064/PDB:7KXJ (3 Fab bound, 3-“up” asymmetric), EMD-23065/PDB:7KXK (3 Fab bound, 2-“up”, 1-“down”).
Significance statement
These results demonstrate that novel antibody formats, generated using simple protein engineering techniques based on principles of modularity, can be used to rapidly enhance antiviral therapies against COVID-19 in terms of potency and resistance to mutations, without compromising the drug-like properties of an antibody that are required for in vivo use. It also provides confirmation that phage-display platforms are comparable to the dominant approach of single B-cell sequencing for isolation of drug-grade antibodies in both terms of speed of development and the quality of antibodies obtained. Insofar as this strategy can be applied to virtually any antibody drug and to other viruses, it should have broad appeal to drug developers, protein engineers, bio-defense officials, virologists and more.
CRediT authorship contribution statement
Shane Miersch: Conceptualization, Methodology, Investigation, Supervision, Writing – original draft, Writing – review & editing, Visualization, Validation, Project administration. Zhijie Li: Methodology, Investigation, Data curation, Visualization, Formal analysis, Validation, Writing – original draft, Writing – review & editing. Reza Saberianfar: Methodology, Investigation. Mart Ustav: Conceptualization, Methodology, Investigation. James Brett Case: Investigation, Validation. Levi Blazer: Conceptualization, Investigation. Chao Chen: Investigation. Wei Ye: Investigation. Alevtina Pavlenco: Investigation. Maryna Gorelik: Investigation. Julia Garcia Perez: Investigation. Suryasree Subramania: Investigation. Serena Singh: Investigation. Lynda Ploder: Investigation. Safder Ganaie: Investigation. Rita E. Chen: Investigation. Daisy W. Leung: Investigation. Pier Paolo Pandolfi: Investigation. Giuseppe Novelli: Investigation. Giulia Matusali: Investigation. Francesca Colavita: Investigation. Maria R. Capobianchi: Investigation. Suresh Jain: Funding acquisition. J.B. Gupta: Funding acquisition, Supervision, Writing – review & editing. Gaya K. Amarasinghe: Supervision, Writing – review & editing. Michael S. Diamond: Supervision, Writing – review & editing. James Rini: Conceptualization, Writing – review & editing, Supervision, Funding acquisition. Sachdev S. Sidhu: Conceptualization, Writing – original draft, Supervision, Funding acquisition.
Acknowledgments
Acknowledgements
This study was supported in part by contracts and grants from NIH (R01 AI157155) and the Defense Advanced Research Project Agency (HR001117S0019) to MSD, NIH grants P01AI120943 and R01AI123926 to GKA, and an NIH grant R01AI107056 to DWL. This study was also supported in part by a CIHR operating grant (COVID-19 Rapid Research Funding #OV3-170649) to JR and SSS, and by the Lazio Region (Italy, DGR n. 653 29 September 2020) and the Rome Foundation (Italy, Prot 317A/I). Additional support was generously provided by FAST grants #2161 to GKA and SSS and #2189 to SSS, from Emergent Ventures through the Thistledown Foundation (Canada) and the Mercatus Center at George Mason University. We are grateful to Daniele Lapa (INMI), for his contribution in determining the neutralizing power of the antibodies tested in Rome. We are also grateful to Carlo Tomino for his valuable help in preparing for the regulatory aspects of monoclonal antibodies in Italy.
Declaration of Competing Interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: MSD is a consultant for Inbios, Vir Biotechnology, NGM Biopharmaceuticals, Carnival Corporation and on the Scientific Advisory Boards of Moderna and Immunome. The Diamond laboratory has received unrelated funding support in sponsored research agreements from Moderna, Vir Biotechnology, and Emergent BioSolutions. SSS, SJ, SM, MU, JBG and PPP are shareholders in Virna Therapeutics.
Edited by M.F. Summers
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jmb.2021.167177.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.Beigel J.H., Tomashek K.M., Dodd L.E., Mehta A.K., Zingman B.S., Kalil A.C., Hohmann E., Chu H.Y., Luetkemeyer A., Kline S., de Castilla D.L., Finberg R.W., Dierberg K., Tapson V., Hsieh L., Patterson T.F., Paredes R., Sweeney D.A., Short W.R., Touloumi G., Lye D.C., Ohmagari N., Oh M., Ruiz-Palacios G.M., Benfield T., Fätkenheuer G., Kortepeter M.G., Atmar R.L., Creech C.B., Lundgren J., Babiker A.G., Pett S., Neaton J.D., Burgess T.H., Bonnett T., Green M., Makowski M., Osinusi A., Nayak S., Lane H.C. Remdesivir for the Treatment of Covid-19 — Preliminary Report. N. Engl. J. Med. 2020 doi: 10.1056/nejmoa2007764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boulware D.R., Pullen M.F., Bangdiwala A.S., Pastick K.A., Lofgren S.M., Okafor E.C., Skipper C.P., Nascene A.A., Nicol M.R., Abassi M., Engen N.W., Cheng M.P., LaBar D., Lother S.A., MacKenzie L.J., Drobot G., Marten N., Zarychanski R., Kelly L.E., Schwartz I.S., McDonald E.G., Rajasingham R., Lee T.C., Hullsiek K.H. A randomized trial of hydroxychloroquine as postexposure prophylaxis for covid-19. N. Engl. J. Med. 2020;383:517–525. doi: 10.1056/NEJMoa2016638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Poland G.A., Ovsyannikova I.G., Kennedy R.B. SARS-CoV-2 immunity: review and applications to phase 3 vaccine candidates. The Lancet. 2020;396:1595–1606. doi: 10.1016/S0140-6736(20)32137-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Robbiani D.F., Gaebler C., Muecksch F., Lorenzi J.C.C., Wang Z., Cho A., Agudelo M., Barnes C.O., Gazumyan A., Finkin S., Hägglöf T., Oliveira T.Y., Viant C., Hurley A., Hoffmann H.H., Millard K.G., Kost R.G., Cipolla M., Gordon K., Bianchini F., Chen S.T., Ramos V., Patel R., Dizon J., Shimeliovich I., Mendoza P., Hartweger H., Nogueira L., Pack M., Horowitz J., Schmidt F., Weisblum Y., Michailidis E., Ashbrook A.W., Waltari E., Pak J.E., Huey-Tubman K.E., Koranda N., Hoffman P.R., West A.P., Rice C.M., Hatziioannou T., Bjorkman P.J., Bieniasz P.D., Caskey M., Nussenzweig M.C. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature. 2020;584:437–442. doi: 10.1038/s41586-020-2456-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Long Q.X., Tang X.J., Shi Q.L., Li Q., Deng H.J., Yuan J., Hu J.L., Xu W., Zhang Y., Lv F.J., Su K., Zhang F., Gong J., Wu B., Liu X.M., Li J.J., Qiu J.F., Chen J., Huang A.L. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat. Med. 2020;26:1200–1204. doi: 10.1038/s41591-020-0965-6. [DOI] [PubMed] [Google Scholar]
- 6.Li L., Zhang W., Hu Y., Tong X., Zheng S., Yang J., Kong Y., Ren L., Wei Q., Mei H., Hu C., Tao C., Yang R., Wang J., Yu Y., Guo Y., Wu X., Xu Z., Zeng L., Xiong N., Chen L., Wang J., Man N., Liu Y., Xu H., Deng E., Zhang X., Li C., Wang C., Su S., Zhang L., Wang J., Wu Y., Liu Z. Effect of convalescent plasma therapy on time to clinical improvement in patients with severe and life-threatening COVID-19: A randomized clinical trial. JAMA. 2020 doi: 10.1001/jama.2020.10044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shen C., Wang Z., Zhao F., Yang Y., Li J., Yuan J., Wang F., Li D., Yang M., Xing L., Wei J., Xiao H., Yang Y., Qu J., Qing L., Chen L., Xu Z., Peng L., Li Y., Zheng H., Chen F., Huang K., Jiang Y., Liu D., Zhang Z., Liu Y., Liu L. Treatment of 5 critically ill patients with COVID-19 with convalescent plasma. JAMA – J. Am. Med. Assoc. 2020;323:1582–1589. doi: 10.1001/jama.2020.4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duan K., Liu B., Li C., Zhang H., Yu T., Qu J., Zhou M., Chen L., Meng S., Hu Y., Peng C., Yuan M., Huang J., Wang Z., Yu J., Gao X., Wang D., Yu X., Li L., Zhang J., Wu X., Li B., Xu Y., Chen W., Peng Y., Hu Y., Lin L., Liu X., Huang S., Zhou Z., Zhang L., Wang Y., Zhang Z., Deng K., Xia Z., Gong Q., Zhang W., Zheng X., Liu Y., Yang H., Zhou D., Yu D., Hou J., Shi Z., Chen S., Chen Z., Zhang X., Yang X. Effectiveness of convalescent plasma therapy in severe COVID-19 patients. PNAS. 2020;117:9490–9496. doi: 10.1073/pnas.2004168117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alsoussi W.B., Turner J.S., Case J.B., Zhao H., Schmitz A.J., Zhou J.Q., Chen R.E., Lei T., Rizk A.A., McIntire K.M., Winkler E.S., Fox J.M., Kafai N.M., Thackray L.B., Hassan A.O., Amanat F., Krammer F., Watson C.T., Kleinstein S.H., Fremont D.H., Diamond M.S., Ellebedy A.H. A potently neutralizing antibody protects mice against SARS-CoV-2 infection. J. Immunol. 2020;205:915–922. doi: 10.4049/jimmunol.2000583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shi R., Shan C., Duan X., Chen Z., Liu P., Song J., Song T., Bi X., Han C., Wu L., Gao G., Hu X., Zhang Y., Tong Z., Huang W., Liu W.J., Wu G., Zhang B., Wang L., Qi J., Feng H., Wang F.S., Wang Q., Gao G.F., Yuan Z., Yan J. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature. 2020;584:120–124. doi: 10.1038/s41586-020-2381-y. [DOI] [PubMed] [Google Scholar]
- 11.Sui J., Li W., Murakami A., Tamin A., Matthews L.J., Wong S.K., Moore M.J., Tallarico A.S.C., Olurinde M., Choe H., Anderson L.J., Bellini W.J., Farzan M., Marasco W.A. Potent neutralization of severe acute respiratory syndrome (SARS) coronavirus by a human mAb to S1 protein that blocks receptor association. PNAS. 2004;101:2536–2541. doi: 10.1073/pnas.0307140101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu Z., Chakraborti S., He Y., Roberts A., Sheahan T., Xiao D., Hensley L.E., Prabakaran P., Rockx B., Sidorov I.A., Corti O., Vogel L., Feng Y., Kim J.O., Wang L.F., Baric R., Lanzavecchia A., Curtis K.M., Nabel G.J., Subbarao K., Jiang S., Dimitrov D.S. Potent cross-reactive neutralization of SARS coronavirus isolates by human monoclonal antibodies. PNAS. 2007;104:12123–12128. doi: 10.1073/pnas.0701000104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ter Meulen J., Bakker A.B.H., Van Den Brink E.N., Weverling G.J., Martina B.E.E., Haagmans B.L., Kuiken T., De Kruif J., Preiser W., Spaan W., Gelderblom H.R., Goudsmit J., Osterhaus A.D.M.E. Human monoclonal antibody as prophylaxis for SARS coronavirus infection in ferrets. Lancet. 2004;363:2139–2141. doi: 10.1016/S0140-6736(04)16506-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Corti D., Zhao J., Pedotti M., Simonelli L., Agnihothram S., Fett C., Fernandez-Rodriguez B., Foglierini M., Agatic G., Vanzetta F., Gopal R., Langrish C.J., Barrett N.A., Sallusto F., Baric R.S., Varani L., Zambon M., Perlman S., Lanzavecchia A. Prophylactic and postexposure efficacy of a potent human monoclonal antibody against MERS coronavirus. PNAS. 2015;112:10473–10478. doi: 10.1073/pnas.1510199112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klein S., Cortese M., Winter S.L., Wachsmuth-Melm M., Neufeldt C.J., Cerikan B., Stanifer M.L., Boulant S., Bartenschlager R., Chlanda P. SARS-CoV-2 structure and replication characterized by in situ cryo-electron tomography. BioRxiv. 2020 doi: 10.1101/2020.06.23.167064. 2020.06.23.167064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ke Z., Oton J., Qu K., Cortese M., Zila V., McKeane L., Nakane T., Zivanov J., Neufeldt C.J., Cerikan B., Lu J.M., Peukes J., Xiong X., Kräusslich H.G., Scheres S.H.W., Bartenschlager R., Briggs J.A.G. Structures and distributions of SARS-CoV-2 spike proteins on intact virions. Nature. 2020:1–7. doi: 10.1038/s41586-020-2665-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoffmann M., Kleine-Weber H., Schroeder S., Mü M.A., Drosten C., Pö S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hansen J., Baum A., Pascal K.E., Russo V., Giordano S., Wloga E., Fulton B.O., Yan Y., Koon K., Patel K., Chung K.M., Hermann A., Ullman E., Cruz J., Rafique A., Huang T., Fairhurst J., Libertiny C., Malbec M., Lee W.Y., Welsh R., Farr G., Pennington S., Deshpande D., Cheng J., Watty A., Bouffard P., Babb R., Levenkova N., Chen C., Zhang B., Hernandez A.R., Saotome K., Zhou Y., Franklin M., Sivapalasingam S., Lye D.C., Weston S., Logue J., Haupt R., Frieman M., Chen G., Olson W., Murphy A.J., Stahl N., Yancopoulos G.D., Kyratsous C.A. Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science (New York, N.Y.) 2020;369:1010–1014. doi: 10.1126/science.abd0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.D. Pinto, Y.-J. Park, M. Beltramello, A.C. Walls, M.A. Tortorici, S. Bianchi, S. Jaconi, K. Culap, F. Zatta, A. De Marco, A. Peter, B. Guarino, R. Spreafico, E. Cameroni, J.B. Case, R.E. Chen, C. Havenar-Daughton, G. Snell, A. Telenti, H.W. Virgin, A. Lanzavecchia, M.S. Diamond, K. Fink, D. Veesler, D. Corti, Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody, (n.d.). https://doi.org/10.1038/s41586-020.
- 20.Cao Y., Su B., Guo X., Sun W., Deng Y., Bao L., Zhu Q., Zhang X., Zheng Y., Geng C., Chai X., He R., Li X., Lv Q., Zhu H., Deng W., Xu Y., Wang Y., Qiao L., Tan Y., Song L., Wang G., Du X., Gao N., Liu J., Xiao J., dong Su X., Du Z., Feng Y., Qin C., Qin C., Jin R., Xie X.S. Potent neutralizing antibodies against SARS-CoV-2 identified by high-throughput single-cell sequencing of convalescent patients’ B cells. Cell. 2020;182 doi: 10.1016/j.cell.2020.05.025. 73-84.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rogers T.F., Zhao F., Huang D., Beutler N., Burns A., He W.T., Limbo O., Smith C., Song G., Woehl J., Yang L., Abbott R.K., Callaghan S., Garcia E., Hurtado J., Parren M., Peng L., Ramirez S., Ricketts J., Ricciardi M.J., Rawlings S.A., Wu N.C., Yuan M., Smith D.M., Nemazee D., Teijaro J.R., Voss J.E., Wilson I.A., Andrabi R., Briney B., Landais E., Sok D., Jardine J.G., Burton D.R. Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science. 2020;369:956–963. doi: 10.1126/science.abc7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wan J., Xing S., Ding L., Wang Y., Gu C., Wu Y., Rong B., Li C., Wang S., Chen K., He C., Zhu D., Yuan S., Qiu C., Zhao C., Nie L., Gao Z., Jiao J., Zhang X., Wang X., Ying T., Wang H., Xie Y., Lu Y., Xu J., Lan F. Human-IgG-neutralizing monoclonal antibodies block the SARS-CoV-2 infection. Cell Rep. 2020;32 doi: 10.1016/j.celrep.2020.107918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Noy-Porat T., Makdasi E., Alcalay R., Mechaly A., Levy Y., Bercovich-Kinori A., Zauberman A., Tamir H., Yahalom-Ronen Y., Israeli M., Epstein E., Achdout H., Melamed S., Chitlaru T., Weiss S., Peretz E., Rosen O., Paran N., Yitzhaki S., Shapira S.C., Israely T., Mazor O., Rosenfeld R. A panel of human neutralizing mAbs targeting SARS-CoV-2 spike at multiple epitopes. Nat. Commun. 2020;11:1–7. doi: 10.1038/s41467-020-18159-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wec A.Z., Wrapp D., Herbert A.S., Maurer D.P., Haslwanter D., Sakharkar M., Jangra R.K., Dieterle M.E., Lilov A., Huang D., Tse L.V., Johnson N.V., Hsieh C.L., Wang N., Nett J.H., Champney E., Burnina I., Brown M., Lin S., Sinclair M., Johnson C., Pudi S., Bortz R., Wirchnianski A.S., Laudermilch E., Florez C., Fels J.M., O’Brien C.M., Graham B.S., Nemazee D., Burton D.R., Baric R.S., Voss J.E., Chandran K., Dye J.M., McLellan J.S., Walker L.M. Broad neutralization of SARS-related viruses by human monoclonal antibodies. Science. 2020;369:731–736. doi: 10.1126/science.abc7424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cobleigh M.A., Vogel C.L., Tripathy D., Robert N.J., Scholl S., Fehrenbacher L., Wolter J.M., Paton V., Shak S., Lieberman G., Slamon D.J. Multinational study of the efficacy and safety of humanized anti-HER2 monoclonal antibody in women who have HER2-overexpressing metastatic breast cancer that has progressed after chemotherapy for metastatic disease. J. Clin. Oncol. 1999;17:2639–2648. doi: 10.1200/jco.1999.17.9.2639. [DOI] [PubMed] [Google Scholar]
- 26.Persson H., Ye W., Wernimont A., Adams J.J., Koide A., Koide S., Lam R., Sidhu S.S. CDR-H3 diversity is not required for antigen recognition by synthetic antibodies. J. Mol. Biol. 2013;425:803–811. doi: 10.1016/j.jmb.2012.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mouquet H., Scheid J.F., Zoller M.J., Krogsgaard M., Ott R.G., Shukair S., Artyomov M.N., Pietzsch J., Connors M., Pereyra F., Walker B.D., Ho D.D., Wilson P.C., Seaman M.S., Eisen H.N., Chakraborty A.K., Hope T.J., Ravetch J.V., Wardemann H., Nussenzweig M.C. Polyreactivity increases the apparent affinity of anti-HIV antibodies by heteroligation. Nature. 2010;467:591–595. doi: 10.1038/nature09385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jain T., Sun T., Durand S., Hall A., Houston N.R., Nett J.H., Sharkey B., Bobrowicz B., Caffry I., Yu Y., Cao Y., Lynaugh H., Brown M., Baruah H., Gray L.T., Krauland E.M., Xu Y., Vásquez M., Wittrup K.D. Biophysical properties of the clinical-stage antibody landscape. PNAS. 2017;114:944–949. doi: 10.1073/pnas.1616408114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barnes C.O., Jette C.A., Abernathy M.E., Dam K.M.A., Esswein S.R., Gristick H.B., Malyutin A.G., Sharaf N.G., Huey-Tubman K.E., Lee Y.E., Robbiani D.F., Nussenzweig M.C., West A.P., Bjorkman P.J. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature. 2020 doi: 10.1038/s41586-020-2852-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Greaney A.J., Starr T.N., Gilchuk P., Zost S.J., Binshtein E., Loes A.N., Hilton S.K., Huddleston J., Eguia R., Crawford K.H.D., Dingens A.S., Nargi R.S., Sutton R.E., Suryadevara N., Rothlauf P.W., Liu Z., Whelan S.P.J., Carnahan R.H., Crowe J.E., Bloom J.D. Complete mapping of mutations to the SARS-CoV-2 spike receptor-binding domain that escape antibody recognition. Cell Host Microbe. 2020 doi: 10.1016/j.chom.2020.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tegally H., Wilkinson E., Giovanetti M., Iranzadeh A., Fonseca V., Giandhari J., Doolabh D., Pillay S., San E.J., Msomi N., Mlisana K., von Gottberg A., Walaza S., Allam M., Ismail A., Mohale T., Glass A.J., Engelbrecht S., Van Zyl G., Preiser W., Petruccione F., Sigal A., Hardie D., Marais G., Hsiao M., Korsman S., Davies M.-A., Tyers L., Mudau I., York D., Maslo C., Goedhals D., Abrahams S., Laguda-Akingba O., Alisoltani-Dehkordi A., Godzik A., Wibmer C.K., Sewell B.T., Lourenço J., Alcantara L.C.J., Kosakovsky Pond S.L., Weaver S., Martin D., Lessells R.J., Bhiman J.N., Williamson C., de Oliveira T. Emergence of a SARS-CoV-2 variant of concern with mutations in spike glycoprotein. Nature. 2021:1–8. doi: 10.1038/s41586-021-03402-9. [DOI] [PubMed] [Google Scholar]
- 32.E. Volz, S. Mishra, M. Chand, J.C. Barrett, R. Johnson, S. Hopkins, A. Gandy, A. Rambaut, N.M. Ferguson, Transmission of SARS-CoV-2 Lineage B.1.1.7 in England: Insights from linking epidemiological and genetic data, MedRxiv. (2021) 2020.12.30.20249034. https://doi.org/10.1101/2020.12.30.20249034.
- 33.N.R. Faria, T.A. Mellan, C. Whittaker, I.M. Claro, D. da S. Candido, S. Mishra, M.A.E. Crispim, F.C. Sales, I. Hawryluk, J.T. McCrone, R.J.G. Hulswit, L.A.M. Franco, M.S. Ramundo, J.G. de Jesus, P.S. Andrade, T.M. Coletti, G.M. Ferreira, C.A.M. Silva, E.R. Manuli, R.H.M. Pereira, P.S. Peixoto, M.U. Kraemer, N. Gaburo, C. da C. Camilo, H. Hoeltgebaum, W.M. Souza, E.C. Rocha, L.M. de Souza, M.C. de Pinho, L.J.T. Araujo, F.S. V Malta, A.B. de Lima, J. do P. Silva, D.A.G. Zauli, A.C. de S. Ferreira, R.P. Schnekenberg, D.J. Laydon, P.G.T. Walker, H.M. Schlüter, A.L.P. Dos Santos, M.S. Vidal, V.S. Del Caro, R.M.F. Filho, H.M. Dos Santos, R.S. Aguiar, J.L.P. Modena, B. Nelson, J.A. Hay, M. Monod, X. Miscouridou, H. Coupland, R. Sonabend, M. Vollmer, A. Gandy, M.A. Suchard, T.A. Bowden, S.L.K. Pond, C.-H. Wu, O. Ratmann, N.M. Ferguson, C. Dye, N.J. Loman, P. Lemey, A. Rambaut, N.A. Fraiji, M. do P.S.S. Carvalho, O.G. Pybus, S. Flaxman, S. Bhatt, E.C. Sabino, Genomics and epidemiology of a novel SARS-CoV-2 lineage in Manaus, Brazil., MedRxiv : The Preprint Server for Health Sciences. (2021) 2021.02.26.21252554. https://doi.org/10.1101/2021.02.26.21252554.
- 34.Chen R.E., Zhang X., Case J.B., Winkler E.S., Liu Y., VanBlargan L.A., Liu J., Errico J.M., Xie X., Suryadevara N., Gilchuk P., Zost S.J., Tahan S., Droit L., Turner J.S., Kim W., Schmitz A.J., Thapa M., Wang D., Boon A.C.M., Presti R.M., O’Halloran J.A., Kim A.H.J., Deepak P., Pinto D., Fremont D.H., Crowe J.E., Corti D., Virgin H.W., Ellebedy A.H., Shi P.-Y., Diamond M.S. Resistance of SARS-CoV-2 variants to neutralization by monoclonal and serum-derived polyclonal antibodies. Nat. Med. 2021:1–10. doi: 10.1038/s41591-021-01294-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.P. Wang, L. Liu, S. Iketani, Y. Luo, Y. Guo, M. Wang, J. Yu, B. Zhang, P.D. Kwong, B.S. Graham, J.R. Mascola, J.Y. Chang, M.T. Yin, M. Sobieszczyk, C.A. Kyratsous, L. Shapiro, Z. Sheng, M.S. Nair, Y. Huang, D.D. Ho, Increased Resistance of SARS-CoV-2 Variants B.1.351 and B.1.1.7 to Antibody Neutralization., BioRxiv : The Preprint Server for Biology. (2021). https://doi.org/10.1101/2021.01.25.428137.
- 36.B.E. Jones, P.L. Brown-Augsburger, K.S. Corbett, K. Westendorf, J. Davies, T.P. Cujec, C.M. Wiethoff, J.L. Blackbourne, B.A. Heinz, D. Foster, R.E. Higgs, D. Balasubramaniam, L. Wang, R. Bidshahri, L. Kraft, Y. Hwang, S. Žentelis, K.R. Jepson, R. Goya, M.A. Smith, D.W. Collins, S.J. Hinshaw, S.A. Tycho, D. Pellacani, P. Xiang, K. Muthuraman, S. Sobhanifar, M.H. Piper, F.J. Triana, J. Hendle, A. Pustilnik, A.C. Adams, S.J. Berens, R.S. Baric, D.R. Martinez, R.W. Cross, T.W. Geisbert, V. Borisevich, O. Abiona, H.M. Belli, M. De Vries, A. Mohamed, M. Dittmann, M. Samanovic, M.J. Mulligan, J.A. Goldsmith, C.-L. Hsieh, N. V Johnson, D. Wrapp, J.S. Mclellan, B.C. Barnhart, B.S. Graham, J.R. Mascola, C.L. Hansen, E. Falconer, Title: LY-CoV555, a rapidly isolated potent neutralizing antibody, provides protection in a non-human primate model of SARS-CoV-2 infection, (n.d.). https://doi.org/10.1101/2020.09.30.318972.
- 37.Wu Y., Wang F., Shen C., Peng W., Li D., Zhao C., Li Z., Li S., Bi Y., Yang Y., Gong Y., Xiao H., Fan Z., Tan S., Wu G., Tan W., Lu X., Fan C., Wang Q., Liu Y., Zhang C., Qi J., Gao G.F., Gao F., Liu L. A noncompeting pair of human neutralizing antibodies block COVID-19 virus binding to its receptor ACE2. Science. 2020:eabc2241. doi: 10.1126/science.abc2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu L., Wang P., Nair M.S., Yu J., Rapp M., Wang Q., Luo Y., Chan J.F.W., Sahi V., Figueroa A., Guo X.V., Cerutti G., Bimela J., Gorman J., Zhou T., Chen Z., Yuen K.Y., Kwong P.D., Sodroski J.G., Yin M.T., Sheng Z., Huang Y., Shapiro L., Ho D.D. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature. 2020;584:450–456. doi: 10.1038/s41586-020-2571-7. [DOI] [PubMed] [Google Scholar]
- 39.Zost S.J., Gilchuk P., Chen R.E., Case J.B., Reidy J.X., Trivette A., Nargi R.S., Sutton R.E., Suryadevara N., Chen E.C., Binshtein E., Shrihari S., Ostrowski M., Chu H.Y., Didier J.E., MacRenaris K.W., Jones T., Day S., Myers L., Eun-Hyung Lee F., Nguyen D.C., Sanz I., Martinez D.R., Rothlauf P.W., Bloyet L.M., Whelan S.P.J., Baric R.S., Thackray L.B., Diamond M.S., Carnahan R.H., Crowe J.E. Rapid isolation and profiling of a diverse panel of human monoclonal antibodies targeting the SARS-CoV-2 spike protein. Nat. Med. 2020;26:1422–1427. doi: 10.1038/s41591-020-0998-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schoof M., Faust B., Saunders R.A., Sangwan S., Rezelj V., Hoppe N., Boone M., Billesbølle C.B., Puchades C., Azumaya C.M., Kratochvil H.T., Zimanyi M., Deshpande I., Liang J., Dickinson S., Nguyen H.C., Chio C.M., Merz G.E., Thompson M.C., Diwanji D., Schaefer K., Anand A.A., Dobzinski N., Zha B.S., Simoneau C.R., Leon K., White K.M., Chio U.S., Gupta M., Jin M., Li F., Liu Y., Zhang K., Bulkley D., Sun M., Smith A.M., Rizo A.N., Moss F., Brilot A.F., Pourmal S., Trenker R., Pospiech T., Gupta S., Barsi-Rhyne B., Belyy V., Barile-Hill A.W., Nock S., Liu Y., Krogan N.J., Ralston C.Y., Swaney D.L., García-Sastre A., Ott M., Vignuzzi M., Walter P., Manglik A. An ultrapotent synthetic nanobody neutralizes SARS-CoV-2 by stabilizing inactive Spike. Science. 2020:eabe3255. doi: 10.1126/science.abe3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cao L., Goreshnik I., Coventry B., Case J.B., Miller L., Kozodoy L., Chen R.E., Carter L., Walls A.C., Park Y.J., Strauch E.M., Stewart L., Diamond M.S., Veesler D., Baker D. De novo design of picomolar SARS-CoV-2 miniprotein inhibitors. Science. 2020;370:426–431. doi: 10.1126/science.abd9909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kilany L.A.A., Gaber A.A.S., Aboulwafa M.M., Zedan H.H. Trastuzumab immunogenicity development in patients’ sera and in laboratory animals. BMC Immunol. 2021;22 doi: 10.1186/S12865-021-00405-Z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Daszak P., Olival K.J., Li H. A strategy to prevent future epidemics similar to the 2019-nCoV outbreak. Biosafety and Health. 2020;2:6–8. doi: 10.1016/j.bsheal.2020.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shi Z., Hu Z. A review of studies on animal reservoirs of the SARS coronavirus. Virus Res. 2008;133:74–87. doi: 10.1016/j.virusres.2007.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hornsby M., Paduch M., Miersch S., Sääf A., Matsuguchi T., Lee B., Wypisniak K., Doak A., King D., Usatyuk S., Perry K., Lu V., Thomas W., Luke J., Goodman J., Hoey R.J., Lai D., Griffin C., Li Z., Vizeacoumar F.J., Dong D., Campbell E., Anderson S., Zhong N., Gräslund S., Koide S., Moffat J., Sidhu S., Kossiakoff A., Wells J. A high through-put platform for recombinant antibodies to folded proteins. Mol. Cell. Proteomics. 2015;14:2833–2847. doi: 10.1074/mcp.O115.052209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lefranc M.P., Pommié C., Ruiz M., Giudicelli V., Foulquier E., Truong L., Thouvenin-Contet V., Lefranc G. IMGT unique numbering for immunoglobulin and T cell receptor variable domains and Ig superfamily V-like domains. Dev. Comp. Immunol. 2003;27:55–77. doi: 10.1016/S0145-305X(02)00039-3. [DOI] [PubMed] [Google Scholar]
- 47.Li Z., Michael I.P., Zhou D., Nagy A., Rini J.M. Simple piggyBac transposon-based mammalian cell expression system for inducible protein production. PNAS. 2013;110:5004–5009. doi: 10.1073/pnas.1218620110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tao Y., Strelkov S.V., Mesyanzhinov V.V., Rossmann M.G. Structure of bacteriophage T4 fibritin: a segmented coiled coil and the role of the C-terminal domain. Structure. 1997;5:789–798. doi: 10.1016/S0969-2126(97)00233-5. [DOI] [PubMed] [Google Scholar]
- 49.Fairhead M., Howarth M. Site-specific biotinylation of purified proteins using BirA. Methods Mol. Biol. 2015;1266:171–184. doi: 10.1007/978-1-4939-2272-7_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pallesen J., Wang N., Corbett K.S., Wrapp D., Kirchdoerfer R.N., Turner H.L., Cottrell C.A., Becker M.M., Wang L., Shi W., Kong W.P., Andres E.L., Kettenbach A.N., Denison M.R., Chappell J.D., Graham B.S., Ward A.B., McLellan J.S. Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen. PNAS. 2017;114:E7348–E7357. doi: 10.1073/pnas.1707304114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Miersch S., Maruthachalam B.V., Geyer C.R., Sidhu S.S. Structure-directed and tailored diversity synthetic antibody libraries yield novel anti-EGFR antagonists. ACS Chem. Biol. 2017;12:1381–1389. doi: 10.1021/acschembio.6b00990. [DOI] [PubMed] [Google Scholar]
- 52.Niedziela-Majka A., Kan E., Weissburg P., Mehra U., Sellers S., Sakowicz R. High-throughput screening of formulations to optimize the thermal stability of a therapeutic monoclonal antibody. J. Biomol. Screen. 2015;20:552–559. doi: 10.1177/1087057114557781. [DOI] [PubMed] [Google Scholar]
- 53.Case J.B., Rothlauf P.W., Chen R.E., Liu Z., Zhao H., Kim A.S., Bloyet L.M., Zeng Q., Tahan S., Droit L., Ilagan M.X.G., Tartell M.A., Amarasinghe G., Henderson J.P., Miersch S., Ustav M., Sidhu S., Virgin H.W., Wang D., Ding S., Corti D., Theel E.S., Fremont D.H., Diamond M.S., Whelan S.P.J. Neutralizing antibody and soluble ACE2 inhibition of a replication-competent VSV-SARS-CoV-2 and a clinical isolate of SARS-CoV-2. Cell Host Microbe. 2020;28:475–485.e5. doi: 10.1016/j.chom.2020.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yuan M., Wu N.C., Zhu X., Lee C.C.D., So R.T.Y., Lv H., Mok C.K.P., Wilson I.A. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science (New York, N.Y.) 2020;368:630–633. doi: 10.1126/science.abb7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Robbins P.W., Trimble’+d R.B., Wirth D.F., Hering C., Maley F., Maleycp8 G.F., Das R., Gibsonh B.W., Royalh N., Biemann K. Primary structure of the streptomyces enzyme endo-@-N-acetylglucosaminidase H*. The J. Biol. Chem. 1984 [PubMed] [Google Scholar]
- 56.Kabsch W. XDS. Acta Crystallographica. Section D, Biol. Crystallography. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schritt D., Li S., Rozewicki J., Katoh K., Yamashita K., Volkmuth W., Cavet G., Standley D.M. Repertoire builder: High-throughput structural modeling of B and T cell receptors. Mol. Syst. Des. Eng. 2019;4:761–768. doi: 10.1039/c9me00020h. [DOI] [Google Scholar]
- 59.Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Afonine P.V., Grosse-Kunstleve R.W., Echols N., Headd J.J., Moriarty N.W., Mustyakimov M., Terwilliger T.C., Urzhumtsev A., Zwart P.H., Adams P.D. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 2012;68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang K. Gctf: Real-time CTF determination and correction. J. Struct. Biol. 2016;193:1–12. doi: 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hsieh C.L., Goldsmith J.A., Schaub J.M., DiVenere A.M., Kuo H.C., Javanmardi K., Le K.C., Wrapp D., Lee A.G., Liu Y., Chou C.W., Byrne P.O., Hjorth C.K., Johnson N.V., Ludes-Meyers J., Nguyen A.W., Park J., Wang N., Amengor D., Lavinder J.J., Ippolito G.C., Maynard J.A., Finkelstein I.J., McLellan J.S. Structure-based design of prefusion-stabilized SARS-CoV-2 spikes. Science. 2020;369:1501–1505. doi: 10.1126/SCIENCE.ABD0826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rubinstein J.L., Brubaker M.A. Alignment of cryo-EM movies of individual particles by optimization of image translations. J. Struct. Biol. 2015;192:188–195. doi: 10.1016/j.jsb.2015.08.007. [DOI] [PubMed] [Google Scholar]
- 64.Punjani A., Rubinstein J.L., Fleet D.J., Brubaker M.A. CryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods. 2017;14:290–296. doi: 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- 65.Bepler T., Morin A., Rapp M., Brasch J., Shapiro L., Noble A.J., Berger B. Positive-unlabeled convolutional neural networks for particle picking in cryo-electron micrographs. Nat. Methods. 2019;16:1153–1160. doi: 10.1038/s41592-019-0575-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera - A visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 68.Emsley P., Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 69.Wang R.Y.R., Song Y., Barad B.A., Cheng Y., Fraser J.S., DiMaio F. Automated structure refinement of macromolecular assemblies from cryo-EM maps using Rosetta. ELife. 2016;5 doi: 10.7554/eLife.17219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liebschner D., Afonine P.V., Baker M.L., Bunkoczi G., Chen V.B., Croll T.I., Hintze B., Hung L.W., Jain S., McCoy A.J., Moriarty N.W., Oeffner R.D., Poon B.K., Prisant M.G., Read R.J., Richardson J.S., Richardson D.C., Sammito M.D., Sobolev O.V., Stockwell D.H., Terwilliger T.C., Urzhumtsev A.G., Videau L.L., Williams C.J., Adams P.D. Macromolecular structure determination using X-rays, neutrons and electrons: Recent developments in Phenix. Acta Crystallographica Section D: Struct. Biol. 2019;75:861–877. doi: 10.1107/S2059798319011471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Williams C.J., Headd J.J., Moriarty N.W., Prisant M.G., Videau L.L., Deis L.N., Verma V., Keedy D.A., Hintze B.J., Chen V.B., Jain S., Lewis S.M., Arendall W.B., Snoeyink J., Adams P.D., Lovell S.C., Richardson J.S., Richardson D.C. MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci. 2018;27:293–315. doi: 10.1002/pro.3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.