Abstract
Radiogenomics is a new field of medical science that integrates two omics, radiomics and genomics, and may bring a major paradigm shift in traditional personalized medicine strategies that require tumor tissue samples. In addition, the acquisition of the data does not require special imaging equipment or special imaging conditions, and it is possible to use image information from computed tomography, magnetic resonance imaging, positron emission tomography‐computed tomography in clinical practice, so the versatility and cost‐effectiveness of radiogenomics are expected. So far, the field of radiogenomics has developed, especially in the fields of brain tumors and breast cancer, but recently, reports of radiogenomic research on gastroenterological cancer are increasing. This review provides an overview of radiogenomic research methods and summarizes the current radiogenomic research in gastroenterological cancer. In addition, the application of artificial intelligence is considered to be indispensable for the integrated analysis of enormous omics information in the future, and the future direction of this research, including the latest technologies, will be discussed.
Keywords: artificial intelligence, gastroenterological cancer, imaging, personalized medicine, radiogenomics
This review provides an overview of radiogenomic research methods and summarizes the current radiogenomic research in gastrointestinal cancer. Radiogenomics may bring a major paradigm shift in traditional personalized medicine strategies that require tumor tissue samples. Radiogenomics have developed, especially in the field of brain tumors and breast cancer, but reports of radiogenomic research on gastrointestinal cancer are comparatively limited.
1. INTRODUCTION
For a long time, attempts have been made to diagnose disease using imaging tests. For example, the clinical TNM classification derived using computed tomography (CT) and magnetic resonance imaging (MRI) predicts resectability and prognosis of tumors. So far, the following three methods have been tried for improving the accuracy of the prediction: (1) improving the accuracy of the equipment, which can be described as an attempt to obtain an image closer to the pathological image; (2) devising the appropriate imaging method, which is an attempt to obtain new biomarkers by performing an imaging method designed to mimic biological phenomena, such as reflect blood flow information and cell density; and (3) devising the analysis procedure for the acquired image. However, methods (1) and (2) are expensive and can only be practiced in a limited number of facilities, especially in the initial stage. In contrast, method (3) is easy to generalize and has the advantage that cases can be collected retrospectively.
Radiomics is one of the current goals of method (3). Radiomics is a research field that extracts several tens of thousands of image features from images using mathematical methods, such as morphology/histogram/texture analysis, and compares them with other clinical information. For example, the average apparent diffusion coefficient (ADC) value in MRI and the maximum standardized uptake value (SUV) in fluorodeoxyglucose‐positron emission tomography (FDG‐PET) are one of the image features, and radiomics extracts many such values for analysis. Furthermore, radiogenomics is a research field that integrates and analyzes two different sets of omics information and explores their correlation: radiomics, which deals with image features, and genomics, which deals with genome and gene expression analyses. Studies on radiogenomics have been actively reported in recent years. 1 , 2
Predicting genomic information using images from routine practice is the expected result of radiogenomics. In recent years, when a diagnostic model is constructed using a machine learning algorithm from a large number of image features and genetic information, it is expected to be more accurate than conventional analysis and exceeds human diagnostic ability in some fields. 3 This is because general images from CT and MRI correlate with the histopathology results.
Conversely, precision medicine, which is a so‐called tailor‐made medical treatment that analyzes and selects the optimal treatment method for each patient based on their genetic information, has been proposed, and expectations are rising. However, it is considered that examining and providing individual treatment methods for each patient has various problems leading to soaring medical costs. In addition, genetic retrieval is invasive, expensive, and time‐consuming. In addition, in order to provide precision medicine to all cancer patients, there are many problems that need to be solved, such as capital investment for analysis and human resource development. Moreover, in the reports so far, there remains a big question regarding the cost‐effectiveness of patients who can benefit from current precision medicine, which is a low percentage of all the cases tested.
Radiogenomics, which uses images used in daily clinical practice, is easy to use as a method for practicing precision medicine. The field of radiogenomics is currently relatively well‐reported, especially for brain tumors and breast cancer. In contrast, for studies on gastroenterological cancer, there are only a few reports examining the relationship between protein expression and imaging findings, and many of these studies lack quantitative evaluation of images and do not fully reflect radiogenomics. 4 Recently, studies on radiogenomics targeting hepatocellular carcinoma (HCC) and colorectal cancer (CRC) have been reported, and advanced studies have been reported targeting esophageal cancer and pancreatic cancer. 5 , 6 In this review, the research methodology of radiogenomics will be introduced, reports focusing on gastroenterological cancer will be summarized, and future changes that may be introduced to the medical field will be explained.
2. THE WORKFLOW OF RADIOGENOMICS
Radiogenomics first involves image acquisition from CT and MRI that are also used in ordinary medical care. This is followed by tumor segmentation, feature extraction, predictive modeling, and model validation (Figure 1). The details of each procedure are described below.
FIGURE 1.
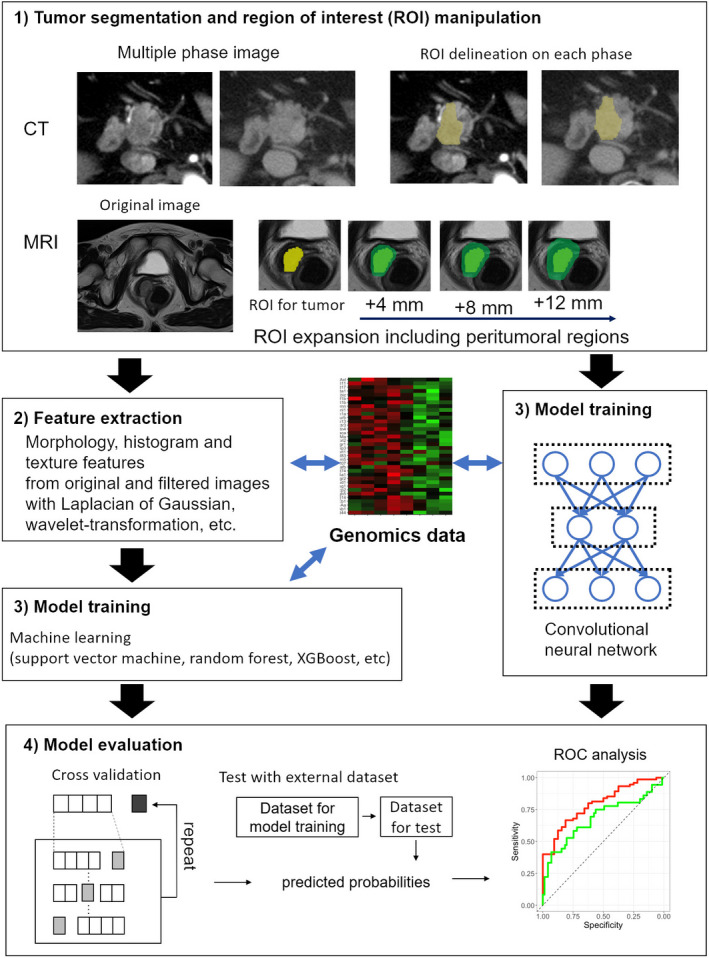
The workflow of radiogenomics
2.1. Image acquisition
One of the merits of radiogenomics is that it does not require a special imaging method. In the current clinical oncology field, various imaging modalities such as CT, MRI, FDG‐PET, and even ultrasound are used, and the diagnostic assessment is made by directly visualizing the underlying anatomical or physiological characteristics of each tumor in an individual patient. 7 , 8 Radiogenomics needs to be further developed so that it can acquire as much objective data as possible. In addition, different images can be acquired when the same tumor is visualized depending on the imaging machine, imaging parameters, image reconstruction parameters, etc. Therefore, standardization of data is important, and it is desirable to unify the image device and the image data acquisition method when acquiring an image.
2.2. Tumor segmentation
The segmentation of areas of interest is primarily performed by diagnostic radiologists. However, there is no consistent way to outline a tumor. In addition, it is possible that there will be a significant difference while drawing the same tumor contour depending on the doctor who evaluates it. Therefore, unified segmentation methods and common recognition among evaluators are required, at least within the same study. It should also be performed by a diagnostic radiologist who is familiar with radiogenomic analysis. 9 In contrast, bias may be weakened using different regions of interest (ROI) generated by different evaluators when using the radiomics method. 10 In the future, segmentation using artificial intelligence (AI) will be implemented, which may solve the problems of inter‐rater reliability. However, a robust method of auto‐segmentation has not yet been established. It has also been suggested that accuracy varies depending on the target of setting the ROI. Particularly, it has been reported that the predictive ability can be increased by including the tumor margin and surroundings. 6 , 11 , 12 , 13 , 14 Pathologically, it is often accompanied by reactive changes, such as microscopic infiltration, inflammatory cell infiltration, and fibrosis around the tumor, and radiogenomics seems to evaluate the same reactive changes.
2.3. Imaging feature extraction
In this step, quantitative data are extracted from the image, which is the core step in radiogenomics. Features related to tumor shape, histogram, and texture were calculated from the image. In addition, the number of features can be increased by applying various filters to the original image. Over 1000 features can be extracted as needed. Several commonly used radiogenomic analysis software programs are available as open source software. 15 , 16
2.4. Predictive model training
The classical radiogenomics approach is used for confirming the correlation between genetic data and extracted imaging features and for comparing the levels of imaging features with and without gene expression. In recent years, machine learning has been used as a technique for handling a large number of extracted imaging features.
2.5. Model evaluation
Machine learning models, including convolutional neural networks (CNNs), require parameter adjustment for model construction. The data required for model training are referred to as the training dataset, and the data required for parameter adjustment during training are referred to as the validation dataset. Furthermore, new data that evaluate the performance of the built model are called a test dataset. If the training/validation dataset and test dataset are not separated, overfitting will yield excessively good results. Ideally, the test dataset should have data from different facilities, but in practice, it is often difficult. In this case, cross‐validation is performed. In cross‐validation, model construction and performance tests are repeated by dividing the data on hand and rearranging the number of times the training/validation dataset and test dataset was divided; this prevents overfitting. 17 , 18
3. GENOME‐LEVEL ANALYSIS
In the previous report, there is currently no strict distinction between the terms "radiomics" and "radiogenomics." Generally, radiomics evaluates clinical outcomes, such as prognosis and prediction of susceptibility to treatments such as anticancer drugs and radiation, whereas radiogenomics evaluates mutations or expressions in molecular biological factors, such as genomes, genes, and proteins. Originally, the term omics refers to the whole study of "‐ome" or "objects" of a particular property. Therefore, genomics refers to the study of the genome, transcriptomics to the study of the transcriptome, and epigenomics to the study of the epigenome. 19 Radiogenomics is originally a fusion research of radiomics, which represents the research of comprehensive image features and genomics, making it a comprehensive genome research, and there is a possibility that new research directions other than the research described here may emerge. The term radiogenomics is sometimes used in research for genotyping, which indicates radiosensitivity, but it is not common at present. 20 , 21 , 22 Therefore, the literature review and the selection of articles need to be performed accurately.
3.1. Search for gene or genomic abnormalities
There have been several reports on genomic mutations that can be indicators of treatment responsiveness and resistance to chemotherapy and radiotherapy and can also be biomarkers for predicting prognosis. 23 , 24 , 25 , 26 These are generally assessed using a sequencer. In addition, gene and protein expressions are also used for treatment prediction and prognosis prediction. 27 , 28 , 29 , 30 Recently, these expressions have been evaluated in different carcinomas for predicting the therapeutic effect of immune checkpoint inhibitors and for evaluating the expression of PD‐L1 antigen as a basis for introducing the treatment. 31 , 32 These evaluations were performed using reverse transcription‐polymerase chain reaction (RT‐PCR) or immunostaining. These are key evaluation methods for precision medicine but require biopsy sampling and/or surgical excision. Unfortunately, in the cases of recurrence of metastasis or multiple metastases for which curative treatment is difficult, it is often impractical to collect invasive lesions from each lesion. In such cases, genomic abnormalities and gene/protein expression abnormalities may be predicted using the radiogenomics method.
3.2. Comprehensive gene or genomic search
In recent years, technological innovation has progressed, and the methods for comprehensive evaluation of gene expression abnormalities and genomic mutations have been developed. 33 , 34 , 35 , 36 Thus, it has become possible to obtain molecular information in units of tens of thousands in a few hours to a few days, instead of searching for individual genomic, genetic, and protein information. This also generalizes the term omics. In addition, these comprehensive search techniques are the basic methods of current precision medicine. However, its economic cost is not negligible. 37 The need for a tumor sample and the quality of the sample are also important. 38 , 39 Cancer heterogeneity is also a major problem, and heterogeneity within the same tumor and heterogeneity between tumors within the same individual is a major obstacle for predicting the effects of precision medicine based on sampling. 40
3.3. Examination using database
It is now possible to utilize already analyzed genetic information using open source databases. For example, the Alzheimer's Disease Neuroimaging Initiative (ADNI) database has genome‐wide association study data. In addition, the Cancer Genome Atlas (TCGA) provides genetic data centered on next‐generation sequencers. These data can be downloaded and/or processed using open source software, such as TCGA‐Assembler. These open sources can be used for exploratory research in radiogenomics research and for modeling accuracy testing using external data.
4. RADIOGENOMICS IN GASTROENTEROLOGICAL CANCER
A PubMed search with the keyword radiogenomics yielded more than 400 articles, and the number kept increasing. In addition, many of these articles were related to cancer, which shows that this new medical science field is gaining attention. However, most of the reports are on brain tumors, lung cancers, breast cancers, and ovarian cancers, and the studies on malignant tumors of the digestive system are limited. 2 , 41 , 42 , 43 , 44 , 45 , 46 , 47 The following is a review of previous radiogenomic studies on gastroenterological malignancies.
4.1. Liver cancer
Of the Radiogenomics studies on gastroenterological cancers so far, the most were on hepatocellular carcinoma (HCC) (Table 1). 48 Segal et al tried to reconstruct gene expression profile in HCC using CT images; 78% of the gene expression profile could be reconstructed by combining 28 image features, and cell proliferation, hepatocyte synthesis function, and patient prognosis were clarified, i.e. the phenotype of the HCC genome can be decoded using noninvasive imaging, which allows noninvasive, continuous, and frequent molecular profiling for personalized medicine. 48
TABLE 1.
Overview of radiogenomic literature on hepatocellular carcinoma
| No. | Imaging modality | Molecular of interest | Feature type | Identified features | Reference no. |
|---|---|---|---|---|---|
| 1 | CT | General gene expression | Qualitative | Imaging signature | 46 |
| 2 | CT | 91‐gene signature for microvascular invasion | Qualitative | Imaging signature | 47 |
| 3 | MRI (God‐EOB‐DTPA) | SLCO1B3 | Qualitative | Dynamic contrast | 48 |
| 4 | CT, MRI (God‐EOB‐DTPA) | β‐catenin, OATP1B3 | Qualitative | Diffusion‐weighted imaging | 49 |
Microvascular invasion in HCC is an independent poor predictor of prognosis after resection or liver transplantation. Banergee et al used radiomics for predicting 91 gene expression signatures for microvascular invasion using contrast‐enhanced CT images. 49 Preoperative CT images of 157 patients with HCC who underwent resection (N = 72) or transplantation (N = 85) between 2000 and 2009 at three centers were used for assessing the presence or absence of microvascular invasion, and the ability of radiogenomics to predict microvascular invasion was evaluated. The diagnostic accuracy, sensitivity, and specificity of radiogenomics for predicting microvascular invasion were 89%, 76%, and 94%, respectively. High radiogenomic scores were associated with lower overall survival than lower radiogenomic scores (P < .001; 48 months vs 147 months). From these results, it was concluded that radiogenomics is a noninvasive radiobiomarker that accurately predicts histological venous invasion in HCC surgery candidates.
HCC is mainly visualized as a low‐intensity lesion on contrast‐enhanced MRI using the contrast agent gadolinium ethoxybenzyl diethylenetriamine pentaacetic acid (Gd‐EOB‐DTPA). However, some HCCs are known to be visualized as high‐intensity lesions. Miura et al also conducted a comprehensive genetic analysis for clarifying the clinicopathological and biological characteristics of these high‐intensity HCC lesions. Of the 77 patients, 14 were classified as having high‐intensity HCC lesions. In high‐intensity HCC lesions, the protein induced by vitamin K absence or antagonist‐II (PIVKAII) value was low, and many of these lesions were histologically well‐differentiated. Comprehensive gene expression analysis revealed that SLCO1B3 gene was highly expressed in high‐intensity HCC lesions. 50 In addition, Kitao et al identified image features associated with β‐catenin gene mutations and investigated their relationship and histopathological factors; the study involved 138 patients with HCC who underwent radical resection. HCC with β‐catenin mutation showed a lower median contrast‐to‐noise ratio on diffusion‐weighted images compared to other HCCs. HCC with β‐catenin mutation showed high‐grade differentiation, with a prominent pseudogland‐like pattern and bile production. The characteristic imaging findings were a high enhancement rate in enhanced MRI and a high apparent diffusion coefficient in diffusion‐weighted imaging. There was a significant positive correlation between the expression of β‐catenin, glutamine synthetase (GS), and organic anion transporting polypeptide 1B3 (OATP 1B3) (P < .0001). 51
4.2. Colorectal cancer
There is an interesting report of radiogenomic analysis of liver metastases of colorectal cancer rather than primary lesions, 52 where texture analysis was performed using pretreatment contrast‐enhanced CT images of 77 patients with liver metastases (Table 2). It was clarified that skewness, which is one of the texture parameters, had a negative correlation with the KRAS mutation (P = .02). In a study by Yang et al, contrast‐enhanced CT images of primary lesions were used for investigating whether the radiomic signature could predict KRAS/NRAS/BRAF mutations in colorectal cancer 53 ; 346 image features were extracted from the CT images of the primary lesion in the portal vein layer and the association between gene mutations, clinical background, tumor staging, and histological differentiation were examined using two cohorts: the test cohort and validation cohort. A ReliefF and support vector machine were used for extracting image features, and the extracted image yields were significantly associated with KRAS/NRAS/BRAF mutations (P < .001). In addition, the ROC analysis for predicting KRAS, NRAS, and BRAF mutations showed AUC, sensitivity, and specificity of 0.869, 0.757, and 0.833 in the test cohort, respectively, and 0.829, 0.686, and 0.857 in the validation cohort, respectively.
TABLE 2.
Overview of radiogenomic literature on colorectal cancer
Chen et al tried to distinguish between mutant and wild‐type KRAS genes in patients with colorectal cancers using parameters from various FDG‐PET scans. 54 Patients with colorectal cancer who underwent preoperative PET/CT were included in this study. Several PET/CT‐related parameters were measured, including SUVmax, metabolic tumor volume threshold, total lesion glycolysis, and PET/CT‐based tumor width. The tumor and PET/CT‐related parameters were used for statistical analysis for determining whether KRAS mutation and wild types could be distinguished. The results showed that the multivariate analysis used SUVmax and a threshold level of 40% of maximum TW uptake (TW40%) as the two predictors of KRAS mutations. The association between various gene mutations, including those other than KRAS in patients with colorectal cancer, and parameters derived from PET/CT were also investigated 55 ; 130 colorectal cancer patients underwent PET/CT in this study. Similar to previous studies, several PET/CT‐related parameters were measured, including SUVmax. Using high‐resolution melting methods for analyzing gene mutations, the tumor and PET/CT‐related parameters correlated with parameterized changes in TP53, KRAS, APC, BRAF, and PIK3CA genes. Genetic alterations in TP53, KRAS, and APC were identified in 41 (40%), 34 (33%), and 27 (26%) tumors, respectively. Five and four of the patients showed PIK3CA and BRAF mutations, respectively. The TP53 variant showed a higher SUVmax. The odds ratio was 1.28 (P = .04; 95% confidence interval, 1.01‐1.61). Tumors with KRAS mutations had increased FDG accumulation (TW40%) and odds ratios of 1.15 (P = .001; 95% confidence interval, 1.06‐1.24). The accuracy of TW40% for KRAS was 61%, while the accuracy of SUVmax greater than 10 was 60% for the prediction of TP53 mutation. It has been reported that an increase in SUVmax and TW40% was associated with colorectal cancer with TP53 and KRAS mutations, respectively.
4.3. Others
MicroRNA is a single‐stranded non‐coding RNA and it has recently has been attracting attention as a molecule for regulating gene expression (Table 3). Previously, our group reported that miR‐1246 in serum is a useful biomarker for esophageal cancer. 56 Since then, many groups have reported that miR‐1246 in blood is a useful biomarker for various carcinomas. 57 , 58 , 59 We investigated whether miR‐1246 expression in the serum of patients with esophageal cancer could be predicted using radiogenomics and found that the expression of miR‐1246 in serum can be predicted with a significant probability by combining the image features obtained using six contrast‐enhanced CTs. 5 Using preoperative CT images of pancreatic cancer, Attiyeh et al validated the mutational states of KRAS, TP53, CDKN2A, and SMAD4 genes, which are known as driver genes for pancreatic cancer, using sequencers and immunohistochemistry. Radiological features of the tumor were extracted from preoperative CT scans and were evaluated for predictability of gene mutations. Genomic and immunohistochemical analyses revealed that 16 (46%) of the patients had SMAD4 mutations. A total of 255 image features were extracted from the CT image, and 28 important features were obtained using feature selection. Therefore, it was possible to distinguish between patients with normal SMAD4 gene and with mutant SMAD4 gene using image features. 60 We also examined the expression of p53 and PD‐L1 in more than 100 pancreatic cancer patients using immunostaining. It became clear that all factors were prognostic factors. Furthermore, the AUC, sensitivity, and specificity of p53 and PD‐L1 expression predictions using the image features obtained from contrast CT images were 0.795, 66.7%, 81.3% and 0.683, 41.7%, and 93.0%, respectively. It was shown to be significantly predictable for p53. However, a sufficient predictive model could not be created for PD‐L1, indicating that radiogenomics cannot always replace molecular biological factors. 6
TABLE 3.
Overview of radiogenomic literature on gastrointestinal cancers (other than HCC and colorectal cancer)
5. ARTIFICIAL INTELLIGENCE
AI is at the heart of the fourth industrial revolution and is expected to introduce an innovative data‐driven analytical paradigm and bring significant advances in information processing technology in the context of clinical decision support systems. In radiogenomics, it is expected that the overwhelming information processing ability of AI will be utilized for image processing, analysis of huge amounts of image features, and analysis of molecular biological factors, and in some cases, AI may have already been applied.
5.1. AI applications
There have been few reports on the application of AI in radiogenomics of gastroenterological cancer, brain tumors, and breast cancer. 6
IDH mutations in glioma patients are thought to lead to prolonged survival and influence treatment strategies. In a study aimed at predicting the IDH status of glioma from MRI using AI, preoperative images of 496 patients were collected from multiple centers and divided into training, validation, and test sets, and verification was performed. The images were trained on a neural network of each MR sequence (FLAIR, T2, T1 precontrast, and T1 post‐contrast) and a predictive model was built from the output. An IDH prediction accuracy of 83.0% (AUC = 0.93) and 85.7% (AUC = 0.94) was achieved for the validation and test sets, respectively. The result was epoch‐making as a development of a deep learning method for noninvasively predicting the IDH genotype of glioma. 61 In addition, using MRI data and molecular information obtained retrospectively from the Cancer Imaging Archives, IDH1 mutation status in 259 patients with low‐grade or high‐grade glioma was predicted using CNN. It was shown that the IDH1 mutant state can be identified with a high predictive ability of 94%. The AI approach has been shown to classify gene mutations in glioma, and neural networks have been shown to learn imaging components without prior feature selection or human‐led training. 62 In another report, the performance of deep learning for predicting IDH1 mutational status was validated in a dataset of 151 patients with low‐grade glioma. Tumors were segmented using a modified CNN structure with six convolutional layers and a fully connected layer with 4096 neurons. Image features were obtained by normalizing the information in the last convolution layer of the CNN instead of calculating them from segmented images. Fisher vectors were used for encoding CNN features from image slices of various sizes. The AUC for normal radiomics was 86%, while that for CNN was 92%. CNN can be a powerful method for extracting useful information from medical images. 63
Ha et al challenged the development of a CNN algorithm that can predict the molecular subtype of breast cancer based on MRI images. It was validated in 216 patients with pretreatment MRI and immunohistochemical staining of pathological data. The CNN architecture was designed in 14 layers. They used many types of normalization, including dropouts, L2, feature map dropouts, and transition layers. A class balance holdout set of 40 patients was used as the test set. Seventy‐four cases of luminal A, 106 cases of luminal B, 13 cases of HER2+, and 23 cases of basal breast tumors were examined. The accuracy of the test set was 70%. The ROC of the AUC was 0.853. Sensitivity and specificity were 0.603 and 0.958, and CNN‐based MRI analysis of breast cancer was predictable for the molecular subtype of breast cancer. 64
6. THE FUTURE OF RADIOGENOMICS
There is an expectation to enhance traditional diagnostic imaging and identify clinically relevant imaging features of cancer patients using AI technology for extracting and analyzing more imaging information. In contrast, even if the same object is imaged, the image changes greatly depending on the imaging device and imaging conditions. Unless reproducibility is guaranteed, its use as a quantitative value is limited. The Radiological Society of North America, the world's largest radiological society, launched the Quantitative Imaging Biomarkers Alliance to standardize the quantitative values obtained from images and establish them for use in clinical trials and routine clinical practice. In the future, image reproducibility will be examined by focusing on such organizations. Dedicated open access databases with multimodal data (imaging, genomics, clinical, laboratory) suitable for radiogenomic analysis may also raise the interest of the scientific community in this area.
Radiogenomics, tumor‐selective molecular imaging, and various molecular biology techniques are inevitably converging in the age of AI, and the resulting synergies are enormous for the diagnosis and treatment of cancer, resulting in significant progress in the medical field. Most of the current research focuses on brain tumors, lung cancer, and breast cancer, but we believe that the field of radiogenomics will make a significant contribution to the field of gastroenterological cancer in the future.
7. CONCLUSIONS
Radiogenomic analysis can lead to the identification of new biomarkers based on the association of image signatures with genomic information of the lesion and can minimize the application of invasive methods. The introduction of AI is indispensable for the enormous amount of multimodal information processing, and it is thought that they will contribute to the development of radiogenomics in the future. Radiogenomics is also considered an indispensable tool for diagnosis and treatment in the field of gastroenterological cancer.
DISCLOSURE
Conflict of Interest: The authors declare no conflicts of interest in association with the present study.
Hoshino I, Yokota H. Radiogenomics of gastroenterological cancer: The dawn of personalized medicine with artificial intelligence‐based image analysis. Ann Gastroenterol Surg. 2021;5:427–435. 10.1002/ags3.12437
REFERENCES
- 1. Kuo MD, Gollub J, Sirlin CB, Ooi C, Chen X. Radiogenomic analysis to identify imaging phenotypes associated with drug response gene expression programs in hepatocellular carcinoma. J Vasc Interv Radiol. 2007;18(7):821–31. [DOI] [PubMed] [Google Scholar]
- 2. Diehn M, Nardini C, Wang DS, McGovern S, Jayaraman M, Liangv Y, et al. Identification of noninvasive imaging surrogates for brain tumor gene‐expression modules. Proc Natl Acad Sci U S A. 2008;105(13):5213–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hu W, Yang H, Xu H, Mao Y. Radiomics based on artificial intelligence in liver diseases: where we are? Gastroenterol Rep (Oxf). 2020;8(2):90–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Horvat N, Monti S, Oliveira BC, Rocha CCT, Giancipoli RG, Mannelli L. State of the art in magnetic resonance imaging of hepatocellular carcinoma. Radiol Oncol. 2018;52(4):353–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hoshino I, Yokota H, Ishige F, Iwatate Y, Takeshita N, Nagase H, et al. Radiogenomics predicts the expression of microRNA‐1246 in the serum of esophageal cancer patients. Sci Rep. 2020;10(1):2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Iwatate Y, Hoshino I, Yokota H, Ishige F, Itami M, Mori Y, et al. Radiogenomics for predicting p53 status, PD‐L1 expression, and prognosis with machine learning in pancreatic cancer. Br J Cancer. 2020.;123 (8):1253–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wu J, Tha KK, Xing L, Li R. Radiomics and radiogenomics for precision radiotherapy. J Radiat Res. 2018;59(suppl_1):i25–i31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Park VY, Lee E, Lee HS, Kim HJ, Yoon J, Son J, et al. Combining radiomics with ultrasound‐based risk stratification systems for thyroid nodules: an approach for improving performance. Eur Radiol. 2020. Available from 10.1007/s00330-020-07365-9. [DOI] [PubMed] [Google Scholar]
- 9. Lambin P, Leijenaar RTH, Deist TM, Peerlings J, de Jong EEC, van Timmeren J, et al. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol. 2017;14(12):749–62. [DOI] [PubMed] [Google Scholar]
- 10. Wada T, Yokota H, Horikoshi T, Starkey J, Hattori S, Hashiba J, et al. Diagnostic performance and inter‐operator variability of apparent diffusion coefficient analysis for differentiating pleomorphic adenoma and carcinoma ex pleomorphic adenoma: comparing one‐point measurement and whole‐tumor measurement including radiomics approach. Jpn J Radiol. 2020;38(3):207–14. [DOI] [PubMed] [Google Scholar]
- 11. Takada A, Yokota H, Watanabe Nemoto M, Horikoshi T, Matsushima J, Uno T. A multi‐scanner study of MRI radiomics in uterine cervical cancer: prediction of in‐field tumor control after definitive radiotherapy based on a machine learning method including peritumoral regions. Jpn J Radiol. 2020;38(3):265–73. [DOI] [PubMed] [Google Scholar]
- 12. Liu C, Ding J, Spuhler K, Gao Y, Serrano Sosa M, Moriarty M, et al. Preoperative prediction of sentinel lymph node metastasis in breast cancer by radiomic signatures from dynamic contrast‐enhanced MRI. J Magn Reson Imaging. 2019;49(1):131–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Braman NM, Etesami M, Prasanna P, Dubchuk C, Gilmore H, Tiwari P, et al. Intratumoral and peritumoral radiomics for the pretreatment prediction of pathological complete response to neoadjuvant chemotherapy based on breast DCE‐MRI. Breast Cancer Res. 2017;19(1):57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dou TH, Coroller TP, van Griethuysen JJM, Mak RH, Aerts H. Peritumoral radiomics features predict distant metastasis in locally advanced NSCLC. PLoS One. 2018;13(11):e0206108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Echegaray S, Bakr S, Rubin DL, Napel S. Quantitative Image Feature Engine (QIFE): an Open‐Source, Modular Engine for 3D Quantitative Feature Extraction from Volumetric Medical Images. J Digit Imaging. 2018;31(4):403–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang L, Fried DV, Fave XJ, Hunter LA, Yang J, Court LE. IBEX: an open infrastructure software platform to facilitate collaborative work in radiomics. Med Phys. 2015;42(3):1341–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Geras KJ, Mann RM, Moy L. Artificial Intelligence for Mammography and Digital Breast Tomosynthesis: Current Concepts and Future Perspectives. Radiology. 2019;293(2):246–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chen S, Zhong X, Hu S, Kachelrieß M, Lell M. Automatic multi‐organ segmentation in dual‐energy CT (DECT) with dedicated 3D fully convolutional DECT networks. Med Phys. 2020;47(2):552–62. [DOI] [PubMed] [Google Scholar]
- 19. Story MD, Durante M. Radiogenomics. Med Phys. 2018;45(11):e1111–e1122. [DOI] [PubMed] [Google Scholar]
- 20. Burnet NG, Elliott RM, Dunning A, West CM. Radiosensitivity, radiogenomics and RAPPER. Clin Oncol. 2006;18(7):525–8. [DOI] [PubMed] [Google Scholar]
- 21. Herskind C, Talbot CJ, Kerns SL, Veldwijk MR, Rosenstein BS, West CM. Radiogenomics: A systems biology approach to understanding genetic risk factors for radiotherapy toxicity? Cancer Lett. 2016;382(1):95–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Proud C. Radiogenomics: the promise of personalized treatment in radiation oncology? Clin J Oncol Nurs. 2014;18(2):185–9. [DOI] [PubMed] [Google Scholar]
- 23. George B, Chowdhury SM, Hart A, Sircar A, Singh SK, Nath UK, et al. Ibrutinib Resistance Mechanisms and Treatment Strategies for B‐Cell lymphomas. Cancers. 2020;12(5).1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Baraibar I, Mezquita L, Gil‐Bazo I, Planchard D. Novel drugs targeting EGFR and HER2 exon 20 mutations in metastatic NSCLC. Crit Rev Oncol Hematol. 2020;148:102906. [DOI] [PubMed] [Google Scholar]
- 25. El Dika I, Ilson DH. Current and future therapies for targeting HER2 mutations in gastrointestinal cancer. Expert Rev Anticancer Ther. 2018;18(11):1085–92. [DOI] [PubMed] [Google Scholar]
- 26. Linardou H, Dahabreh IJ, Kanaloupiti D, Siannis F, Bafaloukos D, Kosmidis P, et al. Assessment of somatic k‐RAS mutations as a mechanism associated with resistance to EGFR‐targeted agents: a systematic review and meta‐analysis of studies in advanced non‐small‐cell lung cancer and metastatic colorectal cancer. Lancet Oncol. 2008;9(10):962–72. [DOI] [PubMed] [Google Scholar]
- 27. Liang R, Chen X, Chen L, Wan F, Chen K, Sun Y, et al. STAT3 signaling in ovarian cancer: a potential therapeutic target. J Cancer. 2020;11(4):837–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pegram MD, Miles D, Tsui CK, Zong Y. HER2‐Overexpressing/Amplified Breast Cancer as a Testing Ground for Antibody‐Drug Conjugate Drug Development in Solid Tumors. Clin Cancer Res. 2020;26(4):775–86. [DOI] [PubMed] [Google Scholar]
- 29. Kwon Y, Kim M, Jung HS, Kim Y, Jeoung D. Targeting Autophagy for Overcoming Resistance to Anti‐EGFR Treatments. Cancers. 2019;11(9).1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kuwano M, Shibata T, Watari K, Ono M. Oncogenic Y‐box binding protein‐1 as an effective therapeutic target in drug‐resistant cancer. Cancer Sci. 2019;110(5):1536–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Borghaei H, Paz‐Ares L, Horn L, Spigel DR, Steins M, Ready NE, et al. Nivolumab versus docetaxel in advanced nonsquamous non‐small‐cell lung cancer. N Engl J Med. 2015;373(17):1627–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Brahmer J, Reckamp KL, Baas P, Crinò L, Eberhardt WE, Poddubskaya E, et al. Nivolumab versus docetaxel in advanced squamous‐cell non‐small‐cell lung cancer. N Engl J Med. 2015;373(2):123–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Reis‐Filho JS, Pusztai L. Gene expression profiling in breast cancer: classification, prognostication, and prediction. Lancet. 2011;378(9805):1812–23. [DOI] [PubMed] [Google Scholar]
- 34. Ntzani EE, Ioannidis JP. Predictive ability of DNA microarrays for cancer outcomes and correlates: an empirical assessment. Lancet. 2003;362(9394):1439–44. [DOI] [PubMed] [Google Scholar]
- 35. Hiley CT, Le Quesne J, Santis G, Sharpe R, de Castro DG, Middleton G, et al. Challenges in molecular testing in non‐small‐cell lung cancer patients with advanced disease. Lancet. 2016;388(10048):1002–11. [DOI] [PubMed] [Google Scholar]
- 36. Nangalia J, Campbell PJ. Genome Sequencing during a Patient's Journey through Cancer. N Engl J Med. 2019;381(22):2145–56. [DOI] [PubMed] [Google Scholar]
- 37. Bennette CS, Gallego CJ, Burke W, Jarvik GP, Veenstra DL. The cost‐effectiveness of returning incidental findings from next‐generation genomic sequencing. Genet Med. 2015;17(7):587–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Franczak C, Dubouis L, Gilson P, Husson M, Rouyer M, Demange J, et al. Integrated routine workflow using next‐generation sequencing and a fully‐automated platform for the detection of KRAS, NRAS and BRAF mutations in formalin‐fixed paraffin embedded samples with poor DNA quality in patients with colorectal carcinoma. PLoS One. 2019;14(2):e0212801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nagahashi M, Shimada Y, Ichikawa H, Nakagawa S, Sato N, Kaneko K, et al. Formalin‐fixed paraffin‐embedded sample conditions for deep next generation sequencing. J Surg Res. 2017;220:125–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gerlinger M, Rowan AJ, Horswell S, Math M, Larkin J, Endesfelder D, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366(10):883–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zinn PO, Mahajan B, Sathyan P, Singh SK, Majumder S, Jolesz FA, et al. Radiogenomic mapping of edema/cellular invasion MRI‐phenotypes in glioblastoma multiforme. PLoS One. 2011;6(10):e25451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mazurowski MA, Clark K, Czarnek NM, Shamsesfandabadi P, Peters KB, Saha A. Radiogenomics of lower‐grade glioma: algorithmically‐assessed tumor shape is associated with tumor genomic subtypes and patient outcomes in a multi‐institutional study with The Cancer Genome Atlas data. J Neurooncol. 2017;133(1):27–35. [DOI] [PubMed] [Google Scholar]
- 43. Gevaert O, Xu J, Hoang CD, Xu Y, Quon A, Rubin DL, et al. Non‐small cell lung cancer: identifying prognostic imaging biomarkers by leveraging public gene expression microarray data–methods and preliminary results. Radiology. 2012;264(2):387–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rizzo S, Petrella F, Buscarino V, De Maria F, Raimondi S, Barberis M, et al. CT Radiogenomic Characterization of EGFR, K‐RAS, and ALK Mutations in Non‐Small Cell Lung Cancer. Eur Radiol. 2016;26(1):32–42. [DOI] [PubMed] [Google Scholar]
- 45. Yamamoto S, Maki DD, Korn RL, Kuo MD. Radiogenomic analysis of breast cancer using MRI: a preliminary study to define the landscape. AJR Am J Roentgenol. 2012;199(3):654–63. [DOI] [PubMed] [Google Scholar]
- 46. Mazurowski MA, Zhang J, Grimm LJ, Yoon SC, Silber JI. Radiogenomic analysis of breast cancer: luminal B molecular subtype is associated with enhancement dynamics at MR imaging. Radiology. 2014;273(2):365–72. [DOI] [PubMed] [Google Scholar]
- 47. Winterhoff B, Hamidi H, Wang C, Kalli KR, Fridley BL, Dering J, et al. Molecular classification of high grade endometrioid and clear cell ovarian cancer using TCGA gene expression signatures. Gynecol Oncol. 2016;141(1):95–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Segal E, Sirlin CB, Ooi C, Adler AS, Gollub J, Chen X, et al. Decoding global gene expression programs in liver cancer by noninvasive imaging. Nat Biotechnol. 2007;25(6):675–80. [DOI] [PubMed] [Google Scholar]
- 49. Banerjee S, Wang DS, Kim HJ, Sirlin CB, Chan MG, Korn RL, et al. A computed tomography radiogenomic biomarker predicts microvascular invasion and clinical outcomes in hepatocellular carcinoma. Hepatology. 2015;62(3):792–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Miura T, Ban D, Tanaka S, Mogushi K, Kudo A, Matsumura S, et al. Distinct clinicopathological phenotype of hepatocellular carcinoma with ethoxybenzyl‐magnetic resonance imaging hyperintensity: association with gene expression signature. Am J Surg. 2015;210(3):561–9. [DOI] [PubMed] [Google Scholar]
- 51. Kitao A, Matsui O, Yoneda N, Kozaka K, Kobayashi S, Sanada J, et al. Hepatocellular Carcinoma with β‐Catenin Mutation: Imaging and Pathologic Characteristics. Radiology. 2015;275(3):708–17. [DOI] [PubMed] [Google Scholar]
- 52. Lubner MG, Stabo N, Lubner SJ, del Rio JAM, Song C, Halberg RB, et al. CT textural analysis of hepatic metastatic colorectal cancer: pre‐treatment tumor heterogeneity correlates with pathology and clinical outcomes. Abdom Imaging. 2015;40(7):2331–7. [DOI] [PubMed] [Google Scholar]
- 53. Yang L, Dong D, Fang M, Zhu Y, Zang Y, Liu Z, et al. Can CT‐based radiomics signature predict KRAS/NRAS/BRAF mutations in colorectal cancer? Eur Radiol. 2018;28(5):2058–67. [DOI] [PubMed] [Google Scholar]
- 54. Chen SW, Chiang HC, Chen WT, Hsieh TC, Yen KY, Chiang SF, et al. Correlation between PET/CT parameters and KRAS expression in colorectal cancer. Clin Nucl Med. 2014;39(8):685–9. [DOI] [PubMed] [Google Scholar]
- 55. Chen SW, Lin CY, Ho CM, Chang YS, Yang SF, Kao CH, et al. Genetic Alterations in Colorectal Cancer Have Different Patterns on 18F‐FDG PET/CT. Clin Nucl Med. 2015;40(8):621–6. [DOI] [PubMed] [Google Scholar]
- 56. Takeshita N, Hoshino I, Mori M, Akutsu Y, Hanari N, Yoneyama Y, et al. Serum microRNA expression profile: miR‐1246 as a novel diagnostic and prognostic biomarker for oesophageal squamous cell carcinoma. Br J Cancer. 2013;108(3):644–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Shi Y, Wang Z, Zhu X, Chen L, Ma Y, Wang J, et al. Exosomal miR‐1246 in serum as a potential biomarker for early diagnosis of gastric cancer. Int J Clin Oncol. 2020;25(1):89–99. [DOI] [PubMed] [Google Scholar]
- 58. Huang D, Qu D. Early diagnostic and prognostic value of serum exosomal miR‐1246 in non‐small cell lung cancer. Int J Clin Exp Pathol. 2020;13(7):1601–7. [PMC free article] [PubMed] [Google Scholar]
- 59. Wei C, Li Y, Huang K, Li G, He M. Exosomal miR‐1246 in body fluids is a potential biomarker for gastrointestinal cancer. Biomark Med. 2018;12(10):1185–96. [DOI] [PubMed] [Google Scholar]
- 60. Attiyeh MA, Chakraborty J, McIntyre CA, Kappagantula R, Chou Y, Askan G, et al. CT radiomics associations with genotype and stromal content in pancreatic ductal adenocarcinoma. Abdom Radiol (NY). 2019;44(9):3148–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Chang K, Bai HX, Zhou H, Su C, Bi WL, Agbodza E, et al. Residual Convolutional Neural Network for the Determination of IDH Status in Low‐ and High‐Grade Gliomas from MR Imaging. Clin Cancer Res. 2018;24(5):1073–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Chang P, Grinband J, Weinberg BD, Bardis M, Khy M, Cadena G, et al. Deep‐Learning Convolutional Neural Networks Accurately Classify Genetic Mutations in Gliomas. AJNR Am J Neuroradiol. 2018;39(7):1201–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Li Z, Wang Y, Yu J, Guo Y, Cao W. Deep Learning based Radiomics (DLR) and its usage in noninvasive IDH1 prediction for low grade glioma. Sci Rep. 2017;7(1):5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ha R, Mutasa S, Karcich J, Gupta N, Pascual Van Sant E, Nemer J, et al. Predicting Breast Cancer Molecular Subtype with MRI Dataset Utilizing Convolutional Neural Network Algorithm. J Digit Imaging. 2019;32(2):276–82. [DOI] [PMC free article] [PubMed] [Google Scholar]


