Abstract
Surgical resection of the liver is the standard treatment for colorectal liver metastases, but 70% of patients still experience recurrence, resulting in limited survival. Molecular biomarkers promise guidance within the selection process of individualized treatment and provide better prognostic forecasting of recurrence and response to treatment. Presently, most investigated biomarkers include mutations of KRAS, BRAF, TP53, PIK3CA, APC, expression of Ki‐67, and microsatellite instability. As some colorectal cancer tumors exhibit more than one molecular target, in line with a rising number of potential biomarkers, the complexity of their clinical implementation is rising steadily. Therefore, it is important to approach new insights into molecular biomarkers with explicit caution to their clinical applicability and significance, as there are contradictory results arising from multiple available studies and meta‐analyses. This review helps to shed light on the complexity of promising biomarkers in both the prognosis and diagnosis of colorectal liver metastases.
Keywords: biomarker, BRAF, colorectal liver metastases, diagnostic, heterogeneity, KRAS, liver resection, mutations, prognostic, RAS
Biomarkers for diagnosis and prognosis in colorectal liver metastases.

1. INTRODUCTION
Colorectal cancer (CRC) has 1.8 million cases globally and is the third most common cancer and ranks second by mortality. The incidence is rising due to socioeconomic developments, exposome changes, and the rise in age. 1 In all, 65% of CRC patients will develop metastases and 40% of those occur in the liver. Currently, the majority of CRC patients are assigned to curative surgery followed by adjuvant chemotherapy, which is predominantly determined by clinicopathologic features like primary tumor stage, carcinoembryonic antigen (CEA) level, nodal status, number and size of liver metastases, resection margin status, and the interval between primary tumor diagnosis and liver metastasis. 2 , 3 Notably, centers offering multidisciplinary treatment approaches including pathologists, radiologists, oncologists, and colorectal as well as liver surgeons show better survival rates than general hospitals or nonspecialized centers. 4 Over 50% of CRC patients will develop colorectal liver metastasis (CLM) and complete surgical removal still offers the best chance for long‐term survival. 5 Nonetheless, one‐third of CLM patients still succumb because of recurrent disease in the liver, which affects two‐thirds of patients after resection. 6 , 7 Even modern multidisciplinary approaches that entail, e.g., two‐stage hepatectomies after portal vein embolization, radiation, multiple ablation techniques for CLM (radiofrequency and microwave ablation), and expanding surgical techniques did not change this situation significantly until now. Currently, perioperative systematic therapy is suggested; however, a large randomized controlled trial by Nordlinger et al involving 364 patients showed no improvement in 5‐year overall survival (OS) (51% vs 48%; P = .34) in patients treated with hepatic resection and perioperative therapy with FOLFOX4. 8
The molecular factors contributing to disease recurrence are still unknown, while histological factors such as tumor grade and differentiation or lymphovascular invasion are the only help in assessing the risk. 6 The molecular events leading to tumor metastasis is immensely complex, and thus a poorly understood process. Mutations at the initial site of colorectal carcinogenesis are critical events in the metastatic process. The events leading to cancer cell invasion, adaptation, and colonization of the hepatic parenchyma, called the colorectal cancer invasion‐metastasis cascade, involve various molecular pathways, with inter‐ and intracellular interactions with potential clinical interest. 9
From a surgical point of view, there are essentially three predominant clinical scenarios in which validated biomarkers could provide evidence‐based guidance in the future:
Primarily resectable CLM
For primary resectable CLM, perioperative, individualized therapy must eliminate the micrometastatic disease to prevent recurrent CLM or extrahepatic disease recurrence.
Primarily unresectable CLM
Here the primary aim of perioperative therapy is to convert the extent and localization of CLM to a resectable stage of disease. Risk prediction of recurrence and prognosis is of special importance here, since the operative risk must be weighed against expected survival.
Synchronous CLM with the primary CRC in situ
The timing and extent of surgical resection is an ongoing discussion in this situation. The question of whether to resect the primary tumor or CLM first is still unanswered.
Hepatic resection is both the major aim and challenge in the treatment process of CLM. The challenge itself lies in the need for a stratified selection of patients feasible for surgery, which is still mainly based on personal decision‐making by the surgeons. This clinical complexity is possibly also expressed in the wide ranges of the 5‐year OS after CLM (25%–58%). 10 , 11 Due to the genetically heterogeneous nature of colorectal cancer with CLM, the diagnosis, prognosis, and selection for treatment are, therefore, challenging for surgeons. 6 An individualized therapy based on molecular profiling of the genetic landscape is highly required to improve survival rates and avoid an unnecessary burden on the patient as well as the healthcare system. Unfortunately, to date no biomarker has been translated into clinical practice to guide either exclusion of patients from surgery or for the timing of surgery.
The molecular biomarker's potential to evaluate tumor biology by noninvasive methods are promising (see Figure 1), but the definite effectiveness in clinical settings remains unclear. This review aims to provide an overview of the most emergent biomarkers in the multidisciplinary treatment approach of CLM to illustrate evidence‐based treatment opportunities after molecular stratification, focusing especially on the clinical application.
FIGURE 1.
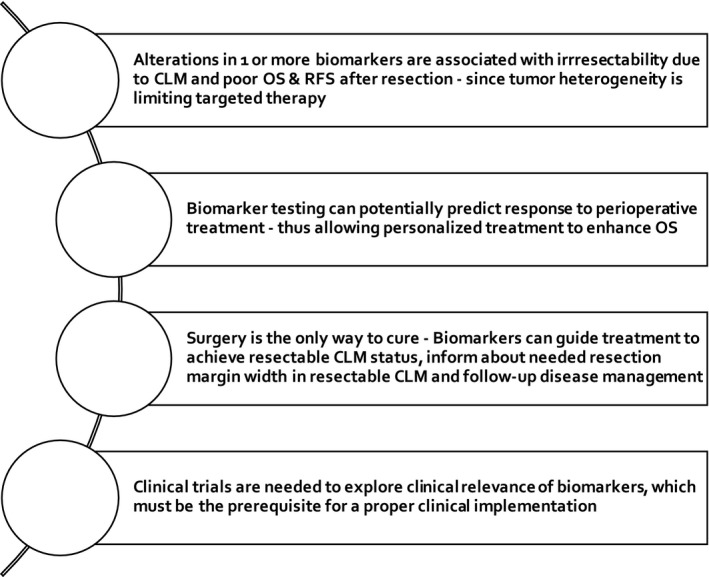
Highlights: Biomarkers and their clinical importance. CLM, colorectal liver metastases; OS, overall survival; RFS, recurrence‐free survival
2. METHODS
This review summarizes and assesses research findings in the English written literature published between 1999 and 2020, with a special focus on the most up‐to‐date research findings using the PubMed database. Keywords and cross‐references of prior published literature including reviews and meta‐analyses were used. Search strings included colorectal, liver, metastasis, biomarker, RAF, KRAS, BRAF, resection in various AND combinations.
2.1. Sidedness of the primary tumor
When making a treatment decision for the side of the primary tumor in the colon is a factor to consider since tumors show different molecular features. Overall, patients with left‐sided tumors have better survival with fewer metastases and are more often suited for curative resection. 12 Taniguchi et al 13 showed that right‐sided tumors are more likely to expose mutations of KRAS and BRAF, which correlate with unresectable liver metastases status, resistance to anti‐EGFR (epidermal growth factor receptor) antibody treatment, and negative prognostic outcome after liver resection. BRAF mutations occur in 32.3% of cases on the right‐sided colon. Additionally, right‐sided tumors with wildtype RAS show a high prevalence (17.2%) of PIK3CA mutations. Thus, testing for those markers in right‐sided tumors can be recommended. 14
2.2. Heterogeneity of primary tumor and metastases
Even though initially carrying identical mutations, further mutations result in genetic heterogeneity between the primary tumor and metastases (intertumor heterogeneity). Thus, primary tumor and distant metastases must be considered genetically heterogeneous. 15
A high degree of primary CRC heterogeneity (intratumor heterogeneity), including mutations in APC, TP53, and KRAS is associated with liver metastasis and poor response to therapy and survival. Tumor heterogeneity helps metastatic CRC to develop collectively through the parallel spread of multiple clonal subpopulations via vascular invasion. 16
Moreover, a high level of intermetastatic genomic heterogeneity in the same patient (intrapatient heterogeneity) might cause worse outcome factors after hepatic resection, including chemoresistance. 17
2.3. Somatic gene alterations in CLM
Alterations in genes like KRAS, P53, BRAF, and PIK3CA frequently influence tumor behavior in CLM. Here we look at the latest research performed around these genes in CLM, and lastly into the crosstalk of the genes that are believed to explain the inconclusive and contradicting study results.
2.3.1. RAS Proto‐oncogenes
Rat sarcoma viral oncogene homologs (RAS) are oncogenes assessed in CLM typically in codons 12, 13, 61, and 146. Oncogenes are mutated or upregulated forms of proto‐oncogenes, typically causing gain‐of‐function and uncontrolled cell proliferation. Mutations in KRAS, NRAS, HRAS, proto‐oncogenes of the RAS family lead to an active MAPK pathway, which results in resistance to preoperative systemic treatment with the EGFR inhibitor (cetuximab). 18 , 19 Thus, RAS status is a validated predictor of response to chemotherapy targeting EGFR. 19 , 20 , 21 RAS mutations, from either primary tumor or liver metastases, are the most researched and known negative prognostic biomarkers for patients with CLM resection associated with poor OS. 22 Brudvik et al 18 showed in their systematic review a 3‐year OS of 52% in RAS mutated patients, while wildtype patients reached 81%. Prospective studies confirmed the relationship between RAS mutations and poor outcomes after resection of CLM. 18 For that reason, testing of RAS mutational status in combination with other clinical‐pathological factors can be advised to estimate tumor response to chemotherapy. Still, conflicting results in the literature mirror the complexity of the topic. Kawaguchi et al 23 found that RAS mutation status alone is not adequate for the prognosis after resection of the liver, but multiple somatic mutations in the genes RAS, TP53, and SMAD4 were associated with worse prognosis than a single mutation after resection of CLM and patients with unresectable colorectal metastases.
2.3.2. KRAS
Kirsten rat sarcoma viral oncogene homolog (KRAS) is an oncogene belonging to the GTPase RAS superfamily and triggers pathways involved in cell proliferation and differentiation as a growth signal transducer downstream of the EGFR. Thus, when over‐active, caused by alteration, KRAS is involved in neoplastic transformation as an early event in colorectal carcinogenesis. KRAS is found to be mutated in 30%–45% of CRC, 25%–52% of CLM tumors 20 and has a high concordance between primary tumor and liver metastases. 24 In patients undergoing liver resection for CLM, KRAS has been shown to be a significant predictor of OS and recurrence‐free survival (RFS). 5 , 21 , 25 , 26 , 27 , 28 For instance, in one study, using a large national multicenter web‐based database including 622 patients, KRAS mutation was an independent predictor of death or recurrence in CLM, expressed as RFS, which was 22% at 5 years for mutant KRAS (mt‐KRAS) and 33% at 5 years for wildtype KRAS (wt‐KRAS) (P =.0053; hazard ratio [HR]: 1.42). Both wt‐KRAS and mt‐KRAS patients received pre‐ and postoperative systemic treatment. 29
To achieve a microscopically margin‐negative resection (RO) in multiple bilobar metastases surgery, meaning no tumor tissue remains in the resection margin, a large proportion of normal parenchyma needs to be resected. This leads to an increased risk of liver failure after surgery and potential morbidity and mortality. 4 Here, the resection margin status is of relevance in wt‐KRAS but shows no prognostic relevance in mt‐KRAS. 30 There is a positive correlation between mt‐KRAS and high incidence of micrometastases, resulting in insufficient safety resection margins. Moreover, patients who received preoperative chemotherapy are associated with a lower incidence of micrometastases and were more likely to receive surgical resection after tumor shrinkage. Thus, a wider resection margin might be recommended in patients with mt‐KRAS to achieve RO resection. 31
Brunsell et al 22 showed that KRAS mutations in multiple resected CLMs were of individual prognostic significance of OS in addition to being a predictive marker for the effect of anti‐EGFR therapy in CLM independent of the systemic treatment.
In this study, most patients with primarily resectable CLM received adjuvant, neoadjuvant, and oxaliplatin‐based chemotherapy. Other patients received conversion treatment including targeted agents to achieve resectable CLM status. In patients with primarily resectable liver metastases, no clinical benefit of anti‐EGFR antibodies has been recognized. 22
2.3.3. BRAF
The v‐raf murine sarcoma b‐viral oncogene (BRAF) gene is another member of the RAS family and plays, as a protein kinase, a crucial role in the mitogen‐activated protein kinase APK/ERK signaling pathway. BRAF activation is associated with cell differentiation, migration, angiogenesis, and proliferation. BRAF gene products act downstream of KRAS in the MAPK signaling pathway. A valine‐to‐glutamic acid amino acid in codon 600 (V600E) substitutions leads to the abnormal activation of the MEK–ERK pathway. BRAF V600E is found to be the most important alteration—making up 90% of all BRAF mutations and an indicator of aggressive disease. 32
BRAF mutation (mt‐BRAF) is found in 15%–35% of patients with resectable CLM and correlates with worse OS and RFS after liver resection. 20 Because only a few mt‐BRAF patients are eligible for liver surgery, clinical studies with resectable CLM are sparsely available, as these patients are usually treated by palliative chemotherapy. The newest evidence states that BRAF mutations do not increase the risk of recurrence after resection of CLM resected patients; however, if recurrence occurs mt‐BRAF is associated with poor survival. 33 However, mt‐BRAF has also been shown to be a biomarker in determining the response to anti‐EGFR antibodies. 34 , 35
BRAF and PIK3CA mutation is found to be more prevalent in right‐sided CRC and is shown in the resistance to anti‐EGFR therapy. 13 , 32
2.3.4. Ki‐67
Ki‐67, a nuclear non‐histone protein, is found in all active phases of the cell cycle and drives cellular proliferation. Increased expression over 50% is linked to a negative prognosis, shown in a lower median survival after hepatic resection. High Ki‐67 expression has been reported in 19.5%–62% of patients with CLM. A study on liver metastases samples of 124 patients with resected CLM revealed that high Ki‐67 expression may be an even stronger predictor of prognosis than KRAS; interestingly, patients with high Ki‐67 expression were more likely to present with synchronous CLM, high tumor burden, and preoperative CEA >200 ng/mL and less likely to undergo curative‐intent resection. On the contrary, a retrospective analysis with 98 liver resection patients reported conflicting results, where Ki‐67 overexpression was a positive prognostic factor of survival. 36 The explanation is believed to be found in the study design: the later study did include subjects that were chemotherapy‐naive, had multiple metastases, and a short time interval to metastasis, while the other did not differentiate. 34
2.4. Multiple alterations
Variable and conflicting results in available studies are most likely to be based on the above‐mentioned complexity and under‐researched interactions of multiple genes, their alteration status, and the different individual cancer‐related pathways. However, mutations occurring in multiple genes are likely to be associated with negative OS and RFS in most studies.
KRAS mutations together with mutations in NRAS (Neuroblastoma RAS viral oncogene homolog) (exons 2, 3, and 4), another proto‐oncogene, are predicting the limited treatment efficacy of anti‐EGFR monoclonal antibodies (mAb) cetuximab and panitumumab due to resistance. The clinical practice guidelines in Europe and the USA involve indications for RAS testing (KRAS and NRAS mutations) before the use of anti‐EGFR agents. 37 , 38 Co‐alterations in RAS and BRAF are observed rarely in colorectal liver metastases, with only 0.05%. A systematic review covering 11 publications found that coaltered RAS and BRAF show different genetic signatures, suggested in molecular profiling; however, no study has proven the role in metastatic disease so far. 39
TP53, together with RAS alteration, is also associated with worse survival. In one study by Kawaguchi et al 23 , including 485 patients, OS and RFS after CLM resection were worse in patients with co‐alteration in RAS and TP53 than in patients with one alteration of the two genes and patients with no alteration. Consequently, the authors suggested that even after the 2 years of recurrence‐free CLM resection, patients with co‐alteration should receive a different, RFS‐focused disease management, considering repeat resection and/or chemotherapy, determining surveillance frequency and intensity, and scheduling clinical surveillance more frequently and intensely. A meta‐analysis of seven trials with 1403 patients concluded that both KRAS and BRAF were negative prognostic factors in patients with hepatic resection of CLM compared to wildtype. Together with other clinical‐pathological factors, assessment of biomarkers could potentially help to predict recurrence and survival after surgical resection. Yet the particular studies had different selection criteria and samples from either primary tumor or liver metastases, and thus proper interpretation is limited. 27
2.4.1. APC & PIK3CA
Adenomatous polyposis coli (APC) is a tumor suppressor gene inducing apoptosis. Loss of function mutation is stimulating the formation of tumors and appears in 47.4% and 48.7% of CLM. However, regarding the prognosis for CLM liver resection, APC together with PIK3CA mutations have been shown to result in lower OS and RFS compared to single mutations only. It is important to note here that more studies are needed to confirm this statement. 34
Phosphatidylinositol‐4, 5‐bisphosphate 3‐kinase, catalytic subunit alpha (PIK3CA) regulates cell proliferation. Studies found a mutation rate of 13.4%–20.9% in CLM patients, 80% of mutations are in exons 9 and 20, whose concomitant existence has been reported to have been linked to poor prognosis. Furthermore, a poor prognostic outcome has been reported for PIK3CA mutation in combination with wt‐Kras status. 40 Taken together, those biomarkers are currently not suitable for clinical use, since more research is badly needed.
2.4.2. Microsatellite instability
Deficient DNA mismatch repair (dMMR) leads to microsatellite instability (MSI) and is associated with less metastatic potential and thus a better prognosis of survival. MSI‐high tumors occur in only 5% of metastatic colorectal cancer, but correspond in 34.4% of cases with a simultaneous presence of BRAF V600E mutations. The MSI‐high and BRAF V600E mutant combination shows a better 5‐year survival (73%) compared to the microsatellite stable phenotype combined with BRAF V600E wildtype (65%). MSI‐high with BRAF V600E wildtype shows the best prognosis of a 75% 5‐year survival. The data suggest therefore a combined testing for MSI & BRAF. 41 The use of immunotherapy for patients with MSI‐high metastatic colorectal cancer as well as preoperative therapy before CLM resection has been studied in clinical trials. 32 The National Cancer Institute recommended a panel of five microsatellite loci for evaluating MSI status in either high‐frequency MSI (MSI‐H) and low‐frequency MSI (MSIL). MSI‐H CRC is mostly located in the right‐sided colon and defined by mucinous features, poor differentiation, and lymphocytic invasion. MSI‐H shows less risk of distant recurrence, while MSI‐L patients show a lower OS than patients with MSI‐H. Immune checkpoint inhibitors (ICI), also known as checkpoint immunotherapy, proved to be effective in up to 50% of the 13%–16% CLM patients who exhibit dMMR or MSI‐H. Therefore, testing for microsatellite instability can predict prognosis and response to ICI therapy. 42
3. DISCUSSION
In this review we aimed to focus on the challenge of resectable and unresectable CLM disease management with a focus on the complexity and potential clinical impact of molecular biomarkers. Prediction of the risk of recurrence after resection of CLM is of special importance for informed decision making intended for the right individualized treatment plan that enables greater survival chances, less risk, and cost‐effectiveness. Moreover, emerging noninvasive molecular biomarkers could help to predict response to various chemotherapeutics and anti‐EGFR antibodies, detect, predict, or decrease recurrence after curative resection earlier, and minimize treatment toxicity. Further benefits include enabling effective complements to first‐line standard therapies and individualized second‐line therapies and give a better prognosis of survival (see Table 1).
TABLE 1.
Biomarkers clinical impact for diagnostic, prognostic, and therapy in patients with CLM
| Biomarker | Prevalence (CLM) | Diagnostic | Prognostic/prediction | Therapeutic |
|---|---|---|---|---|
| RAS | 30% left‐sided,48.2% right‐sided | Testing RAS + other clinico‐pathological factors | Poor OS, 52% vs 81% wt‐RAS after resection; earlier metastatic disease; EGFR antibodies response prediction; worse preoperative chemotherapy response | Wider cross‐resection margins of 15 mm suggested and a wider ablation margin |
| BRAF | 5%–11% | Testing to predict anti EGFR response & prognosis | Poor OS + RFS after resection; no benefit of anti EGFR antibodies | Wider safety margins needed to achieve Ro resection; resection asap after first‐line therapy response |
| KRAS | 29.5% left‐sided; 46.9% right‐ sided | Testing to predict anti‐EGFR response & prognosis | Poor OS + RFS after resection; resistance to anti‐EGFR antibodies | Multidisciplinary treatment after curative resection of CLM |
| Ki‐67 | 19.5%–62% | Less often undergoing resection due to poor clinic‐pathological factors | expression over 50% associated with lower median survival after hepatic resection | Multidisciplinary treatment before and after curative resection of CLM |
| APC & PIK3CA double mutation | 42%–73% & 6.7%–28% | Testing | predicts poor response to preoperative chemotherapy and poor survival in patients with CLM | Personalized treatment to achieve resectable CLM |
Further studies and sound clinical trials are needed to develop a cost‐efficient and clinically effective molecular biomarker screening plan for the fast application of research data into clinical practice for patients with CLM. The complexity of the genetic mutations and the resulting variability of the tumor behavior make this a challenging task. Most identified biomarkers, apart from KRAS and BRAF, did not prove an effectiveness in the majority of available studies because of the molecular heterogeneity of tumors and metastases, and underpowered study design. Likewise, publication bias is an important factor to consider before making a conclusive interpretation. Existing, partly overlapping meta‐analyses also exhibit a significant heterogeneity of included and excluded studies—urging for caution when interpreting the results.
The current body of evidence clearly shows an urgent need for prospective trials as well as prognostic and diagnostic studies to accomplish reproducible results of clinically relevant gene signatures in the treatment of patients with CLM. This claim is underpinned by the fact that even the role of the best‐explored entities such as KRAS and BRAF mutations remains debatable in the clinical real‐world situation. In other words, there is still not enough solid evidence that KRAS or BRAF mutated patients are less likely to obtain a benefit from standard chemotherapeutic strategies. While most data from nonrandomized, retrospective studies, report KRAS status as a potential application as prognosis biomarkers in CRC management, 44 , 45 other studies do not. 21 , 46 Nevertheless, KRAS status has a strong potential for clinical implementation as a prognostic biomarker other than prediction to an anti‐EGFR response. A large meta‐analysis by Brudvik et al 5 combined data of 14 distinct studies researching KRAS status outcome after resection of CLM. This analysis showed an increased risk of recurrence (hazard ratio [HR] 1⁄4 1.89, 95% confidence interval [CI] 1.54e2.32) and worse OS (HR 1⁄4 2.24, 95% CI 1.76e2.85) in mt‐KRAS vs wt‐KRAS after resection. 5
While surgery of the liver is considered today still the only potentially curative treatment for CLM, the risks of an extensive liver resection should be outweighed by the benefits of this surgery. Biomarkers could help to make those decisions easier and more effective by tailoring the therapy to molecular profiling and to avoid risky treatment without a real survival benefit. Thus, especially a metastatic liver disease bears the potential to benefit from these novel biomarkers in diagnosis, prognosis, and stratifying treatment approaches, although there is more knowledge needed from clinical research for validating specific biomarkers. Last, biomarkers were studied in CLM patients, as well as the comparable high frequency in liver metastases, and thus comparable sufficient subjects could be enrolled in the studies. Hence, the implication of biomarkers as prognostic and diagnostic tools remains promising but needs proper further investigation.
CONFLICT OF INTEREST
The authors declare no conflicts of interest for this article.
ACKNOWLEDGMENTS
The authors thank Tanja Iken, affiliate of the Department of General and Visceral Surgery, Medical Center — University of Freiburg, Faculty of Medicine, University of Freiburg, Hugstetter Straße 55, 79106, Freiburg im Breisgau, Germany for providing medical writing support.
Diener MK, Fichtner‐Feigl S. Biomarkers in colorectal liver metastases: Rising complexity and unknown clinical significance?. Ann Gastroenterol Surg. 2021;5:477–483. 10.1002/ags3.12454
REFERENCES
- 1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Cancer J. Clin. 2018;68:394–424. [DOI] [PubMed] [Google Scholar]
- 2. McCullough A. Comprehensive molecular characterization of human colon and rectal cancer. Yearb. Pathol. Lab. Med. 2013;2013:295–6. [Google Scholar]
- 3. Tejpar S, Bertagnolli M, Bosman F, Lenz H, Garraway L, Waldman F, et al. Prognostic and predictive biomarkers in resected colon cancer: current status and future perspectives for integrating genomics into biomarker discovery. Oncologist. 2010;15:390–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. O’Connor JM, Loria FS. Towards precision medicine in colorectal cancer liver metastases. Hepatic Oncol. 2020;7:HEP29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Andreatos N, Ronnekleiv‐kelly S, Margonis GA, Sasaki K, Gani F, Amini N, et al. From bench to bedside: clinical implications of KRAS status in patients with colorectal liver metastasis. Surg Oncol. 2016;25(3):332–8. [DOI] [PubMed] [Google Scholar]
- 6. Zarour L, Anand S, Billingsley K, Bisson W, Cercek A, Clarke M, et al. Colorectal cancer liver metastasis: evolving paradigms and future directions. Cell Mol Gastroenterol Hepatol. 2017;3(2):163–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fong Y, Fortner J, Sun R, Brennan M, Blumgart L. Clinical score for predicting recurrence after hepatic resection for metastatic colorectal cancer. Ann Surg. 1999;230(3):309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nordlinger B, Sorbye H, Glimelius B, Poston G, Schlag P, Rougier P, et al. Perioperative FOLFOX4 chemotherapy and surgery versus surgery alone for resectable liver metastases from colorectal cancer (EORTC 40983): long‐term results of a randomised, controlled, phase 3 trial. Lancet Oncol. 2013;14(12):1208–15. [DOI] [PubMed] [Google Scholar]
- 9. Paschos KA, Majeed AW, Bird NC. Natural history of hepatic metastases from colorectal cancer – Pathobiological pathways with clinical significance. World J Gastroenterol. 2014;20(14):3719–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Brudvik KW, Shindoh J. Limitations of molecular biomarkers in patients with resectable colorectal liver metastases. Chinese Clin Oncol. 2019;8(5):1–7. [DOI] [PubMed] [Google Scholar]
- 11. Bredt LC, Rachid AF. Predictors of recurrence after a first hepatectomy for colorectal cancer liver metastases: a retrospective analysis. World J Surg Oncol. 2014;12(1):1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Engstrand J, Nilsson H, Strömberg C, Jonas E, Freedman J. Colorectal cancer liver metastases – a population‐based study on incidence, management and survival. BMC Cancer. 2018;18:78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Taniguchi H, Uehara K, Nakayama G, Nakayama H, Aiba T, Hattori N, et al. Tumor location is associated with the prevalence of Braf and Pik3ca mutations in patients with wild‐type ras colorectal cancer: a prospective multi‐center cohort study in Japan. Transl Oncol. 2020;13(7):100786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rehman AH, Jones RP, Poston G. Prognostic and predictive markers in liver limited stage IV colorectal cancer. Eur J Surg Oncol. 2019;45(12):pp. 2251–2256.13. [DOI] [PubMed] [Google Scholar]
- 15. Filip S, Vymetalkova V, Petera J, Vodickova L, Kubecek O, John S, et al. Distant metastasis in colorectal cancer patients—do we have new predicting clinicopathological and molecular biomarkers? A comprehensive review. Int J Mol Sci. 2020;21(15):5255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Joung J, Oh B, Hong H, Al‐Khalidi H, Al‐Alem F, Lee H, et al. Tumor heterogeneity predicts metastatic potential in colorectal cancer. Clin Cancer Res. 2017;23(23):7209–16. [DOI] [PubMed] [Google Scholar]
- 17. Molinari C, Marisi G, Passardi A, Matteucci L, De Maio G, Ulivi P. Heterogeneity in colorectal cancer: a challenge for personalized medicine? Int. J. Mol. Sci. 2018;19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Brudvik KW, Jones RP, Giuliante F, Shindoh J, Passot G, Chung MH, et al. RAS Mutation clinical risk score to predict survival after resection of colorectal liver metastases. Ann Surg. 2019;269(1):120–6. [DOI] [PubMed] [Google Scholar]
- 19. Misale S, Yaeger R, Hobor S, Scala E, Janakiraman M, Liska D, et al. Emergence of KRAS mutations and acquired resistance to anti‐EGFR therapy in colorectal cancer. Nature. 2012;486(7404):532–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tsilimigras D, Ntanasis‐Stathopoulos I, Bagante F, Moris D, Cloyd J, Spartalis E, et al. Clinical significance and prognostic relevance of KRAS, BRAF, PI3K and TP53 genetic mutation analysis for resectable and unresectable colorectal liver metastases: A systematic review of the current evidence. Surg Oncol. 2018;27(2):280–8. [DOI] [PubMed] [Google Scholar]
- 21. Garcia‐Carbonero N, Martinez‐Useros J, Li W, Orta A, Perez N, Carames C, et al. KRAS and BRAF mutations as prognostic and predictive biomarkers for standard chemotherapy response in metastatic colorectal cancer: a single institutional study. Cells. 2020;9(1):219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Brunsell TH, Sveen A, Bjørnbeth BA, Røsok BI, Danielsen SA, Brudvik KW, et al. High concordance and negative prognostic impact of RAS/BRAF/PIK3CA mutations in multiple resected colorectal liver metastases. Clin Colorectal Cancer. 2020;19(1):e26–47. [DOI] [PubMed] [Google Scholar]
- 23. Kawaguchi Y, Kopetz S, Newhook T, De Bellis M, Chun Y, Tzeng C, et al. Mutation status of RAS, TP53, and SMAD4 is superior to mutation status of RAS alone for predicting prognosis after resection of colorectal liver metastases. Clin Cancer Res. 2019;25(19):5843–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Knijn N, Mekenkamp L, Klomp M, Vink‐Börger M, Tol J, Teerenstra S, et al. KRAS mutation analysis: a comparison between primary tumors and matched liver metastases in 305 colorectal cancer patients. Br J Cancer. 2011;104(6):1020–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Polivka J, Windrichova J, Pesta M, Houfkova K, Rezackova H, Macanova T, et al. The level of preoperative plasma kras mutations and cea predict survival of patients undergoing surgery for colorectal cancer liver metastases. Cancers (Basel). 2020;12(9):1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Linnekamp J, Wang X, Medema J, Vermeulen L. Colorectal cancer heterogeneity and targeted therapy: a case for molecular disease subtypes. Can Res. 2015;75(2):245–9. [DOI] [PubMed] [Google Scholar]
- 27. Passiglia F, Bronte G, Bazan V, Galvano A, Vincenzi B, Russo A. Can KRAS and BRAF mutations limit the benefit of liver resection in metastatic colorectal cancer patients? A systematic review and meta‐analysis. Crit Rev Oncol Hematol. 2016;99:150–7. [DOI] [PubMed] [Google Scholar]
- 28. Allievi N, Goffredo P, Utria AF, Pisano M, Poiasina E, Lucianetti A, et al. The association of KRAS mutation with primary tumor location and survival in patients undergoing resection of colorectal cancers and synchronous liver metastases. Chinese Clin Oncol. 2019;8(5):1–7. [DOI] [PubMed] [Google Scholar]
- 29. O´Connor J, Sanchez Loria F, Ardiles V, Grondona J, Sanchez P, Andriani O, et al. Prognostic impact of K‐RAS mutational status and primary tumor location in patients undergoing resection for colorectal cancer liver metastases: an update. Future Oncol. 2019;15(27):3149–57. [DOI] [PubMed] [Google Scholar]
- 30. Hatta A, Pathanki A, Hodson J, Sutcliffe R, Marudanayagam R, Roberts K, et al. The effects of resection margin and KRAS status on outcomes after resection of colorectal liver metastases. HPB. 2021;23(1):90–8. [DOI] [PubMed] [Google Scholar]
- 31. Zhang Q, Peng J, Ye M, Weng W, Tan C, Ni S, et al. KRAS mutation predicted more mirometastases and closer resection margins in patients with colorectal cancer liver metastases. Ann Surg Oncol. 2019;27(4):1164–73. [DOI] [PubMed] [Google Scholar]
- 32. Yamashita S, Chun YS, Kopetz SE, Vauthey JN. Biomarkers in colorectal liver metastases. Br J Surg. 2018;105(6):618–27. [DOI] [PubMed] [Google Scholar]
- 33. Bachet JB, Moreno‐Lopez N, Vigano L, Marchese U, Gelli M, Raoux L, et al. BRAF mutation is not associated with an increased risk of recurrence in patients undergoing resection of colorectal liver metastases. Br J Surg. 2019;106(9):1237–47. [DOI] [PubMed] [Google Scholar]
- 34. Barbon C, Margonis GA, Andreatos N, Rezaee N, Sasaki K, Buettner S, et al. Colorectal liver metastases: does the future of precision medicine lie in genetic testing? J Gastrointest Surg. 2018;22(7):1286–96. [DOI] [PubMed] [Google Scholar]
- 35. Tie J, Desai J. Targeting BRAF mutant metastatic colorectal cancer: clinical implications and emerging therapeutic strategies. Target Oncol. 2015;10(2):179–88. [DOI] [PubMed] [Google Scholar]
- 36. Taniguchi H, Uehara K, Nakayama G, Nakayama H, Aiba T. Translational oncology tumor location is associated with the prevalence of Braf and Pik3ca mutations in patients with wild‐type ras colorectal cancer: a prospective multi‐center cohort study in Japan. Transl Oncol. 2020;13(7):100786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ivanecz A, Kavalar R, Palfy M, Pivec V, Sremec M, Horvat M, et al. Can we improve the clinical risk score? The prognostic value of p53, Ki‐67 and thymidylate synthase in patients undergoing radical resection of colorectal liver metastases. HPB. 2014;16(3):235–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Van Cutsem E, Cervantes A, Adam R, Sobrero A, Van Krieken JH, Aderka D, et al. ESMO consensus guidelines for the management of patients with metastatic colorectal cancer. Ann Oncol. 2016;27(8):1386–422. [DOI] [PubMed] [Google Scholar]
- 39. Allegra C, Rumble R, Hamilton S, Mangu P, Roach N, Hantel A, et al. Extended RAS gene mutation testing in metastatic colorectal carcinoma to predict response to anti‐epidermal growth factor receptor monoclonal antibody therapy: American Society of Clinical Oncology Provisional Clinical Opinion Update 2015. J Clin Oncol. 2016;34(2):179–85. [DOI] [PubMed] [Google Scholar]
- 40. Afrăsânie V‐A, Gafton B, Marinca MV, Alexa‐Stratulat T, Miron L, Rusu C, et al. The coexistence of RAS and BRAF mutations in metastatic colorectal cancer: a case report and systematic literature review. J Gastrointest Liver Dis. 2020;29(2):251–6. [DOI] [PubMed] [Google Scholar]
- 41. Lech G, Słotwiński R, Słodkowski M, Krasnodębski IW. 2016 Colorectal cancer : global view Colorectal cancer tumour markers and biomarkers : recent therapeutic advances. World J Gastroenterol. 2016;22(5):1745–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Torshizi Esfahani A, Seyedna SY, Nazemalhosseini Mojarad E, Majd A, Asadzadeh AH. MSI‐L/EMAST is a predictive biomarker for metastasis in colorectal cancer patients. J Cell Physiol. 2019;234(8):13128–36. [DOI] [PubMed] [Google Scholar]
- 43. Tanaka M, Omura K, Watanabe Y, Oda Y, Nakanishi I. Prognostic factors of colorectal cancer: K‐ras mutation, overexpression of the p53 protein, and cell proliferative activity. J Surg Oncol. 1994;57(1):57–64. [DOI] [PubMed] [Google Scholar]
- 44. Tran B, Kopetz S, Tie J, Gibbs P, Jiang Z‐Q, Lieu CH, et al. Impact of BRAF mutation and microsatellite instability on the pattern of metastatic spread and prognosis in metastatic colorectal cancer. Cancer. 2011;117(20):4623–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pino M, Chung D. The chromosomal instability pathway in colon cancer. Gastroenterology. 2010;138(6):2059–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chouhan H, Sammour T, L. Thomas M, W. Moore J. Prognostic significance of BRAF mutation alone and in combination with microsatellite instability in stage III colon cancer. Asia Pac J Clin Oncol. 2018;15(1):69–74. [DOI] [PubMed] [Google Scholar]


