Abstract
Objective:
Accurate diagnosis is particularly challenging in Parkinson’s disease (PD), multiple system atrophy (MSAp), and progressive supranuclear palsy (PSP). We compare the utility of 3 promising biomarkers to differentiate disease state and explain disease severity in parkinsonism: the Automated Imaging Differentiation in Parkinsonism (AID-P), the Magnetic Resonance Parkinsonism Index (MRPI), and plasma-based neurofilament light chain protein (NfL).
Methods:
For each biomarker, the area under the curve (AUC) of receiver operating characteristic curves were quantified for PD versus MSAp/PSP and MSAp versus PSP and statistically compared. Unique combinations of variables were also assessed. Furthermore, each measures association with disease severity was determined using stepwise multiple regression.
Results:
For PD versus MSAp/PSP, AID-P (AUC, 0.900) measures had higher AUC compared with NfL (AUC, 0.747) and MRPI (AUC, 0.669), P < 0.05. For MSAp versus PSP, AID-P (AUC, 0.889), and MRPI (AUC, 0.824) measures were greater than NfL (AUC, 0.537), P < 0.05. We then combined measures to determine if any unique combination provided enhanced accuracy and found that no combination performed better than the AID-P alone in differentiating parkinsonisms. Furthermore, we found that the AID-P demonstrated the highest association with the MDS-UPDRS (Radj2–AID-P, 26.58%; NfL,15.12%; MRPI, 12.90%).
Conclusions:
Compared with MRPI and NfL, AID-P provides the best overall differentiation of PD versus MSAp/PSP. Both AID-P and MRPI are effective in differentiating MSAp versus PSP. Furthermore, combining biomarkers did not improve classification of disease state compared with using AID-P alone. The findings demonstrate in the current sample that the AID-P and MRPI are robust biomarkers for PD, MSAp, and PSP.
Keywords: MRI, multiple system atrophy, neurofilament light, Parkinson’s disease, progressive supranuclear palsy
Parkinson’s disease (PD), multiple system atrophy parkinsonian variant (MSAp), and progressive supranuclear palsy (PSP) are difficult to distinguish in the early stages because of their shared clinical manifestations.1-3 Diagnostic accuracy in early PD (< 5 years duration) has been estimated at only 58%, and more than half of the misdiagnosed patients have either multiple system atrophy (MSA) or PSP.1,2 These 3 parkinsonisms initially present with similar motor and nonmotor features and all share nigrostriatal degeneration; however, MSAp and PSP have more widespread neuropathology and neurodegeneration.3,4 Moreover, accurate diagnosis has important implications for correct prognosis and inclusion in clinical trials, and a biomarker to differentiate these parkinsonisms is critical.
Ongoing efforts to directly compare promising biomarkers to assist with diagnosis among these neurode-generative parkinsonisms have so far been limited. Plasma-based neurofilament light chain (NfL) protein is a potential biomarker for distinguishing PD from MSAp and PSP with good diagnostic accuracy, but it has performed poorly when distinguishing MSAp from PSP.5 In comparison, the Magnetic Resonance Parkinsonism Index (MRPI) is a magnetic resonance imaging (MRI)–based measure incorporating pons, midbrain, and cerebellar peduncle ratios to distinguish PSP from other forms of parkinsonism; however, it cannot distinguish PD from MSA.6-8 It is possible that combining NfL and MRPI could provide a stepwise biomarker for PD, MSAp, and PSP. Recently, the Automated Imaging Differentiation in Parkinsonism (AID-P) has been developed and uses diffusion MRI across numerous tracts and regions to distinguish PD from MSAp/PSP and MSAp from PSP and could therefore address the limitations of NfL and MRPI measures.9 To date no study has ever combined or compared these potentially useful biomarkers in the same cohort. Thus, the purpose of this study was to evaluate the AID-P, MRPI, and NfL measures in differentiating PD from atypical parkinsonism as well as between MSAp and PSP in the same cohort and to determine how well these measures associate with disease severity. Furthermore, we sought to determine if any combination of these measures was robust at differentiating between parkinsonisms.
Methods
Participants
Participants included 39 PD, 17 MSAp, and 16 PSP patients (Table 1). All participants were referred from the University of Florida Normal Fixel Institute for Neurological Disease and were diagnosed by a movement disorder specialist according to established criteria.10-12 Disease severity was assessed using Part III of the Movement Disorders Society Unified Parkinson’s Disease Rating Scale (MDS-UPDRS III).13 All patients with MSA had the parkinsonian subtype. Patients with PSP were also evaluated using the 2017 MDS crtieria to determine subtype, and 11 patients with PSP had the Richardson’s syndrome subtype and the remaining 5 had parkinsonian PSP.14 We grouped patients with PSP with the Richardson’s syndrome subtype and patients with parkinsonian PSP as one group. The Montreal Cognitive Assessment was used to assess cognitive function.15 Disease duration was defined as the time since the current diagnosis was stable for 36 months on average for all groups (Table 1). Although most patients were taking medication, all testing was performed 12 to 14 hours after overnight withdrawal of antiparkinsonian medication. The total levodopa equivalent daily dose16 and other clinical measures are shown for each group in Table 1. The study was approved by the institutional review board at the University of Florida, and participant informed consent was obtained.
TABLE 1.
Demographics and clinical data
| PD | MSAp | PSP | |
|---|---|---|---|
| Sample size | 38 | 17 | 16 |
| Age, y | 64.84 (9.03) | 67.71 (9.12) | 70.94 (7.77) |
| Sex, male/female | 29/9 | 12/5 | 7/9 |
| Disease duration, mos. | 48.13 (29.44) | 55.47 (35.39) | 35.94 (32.14) |
| Total LEDD | 693.32 (259.09)b,c | 448.82 (570.93)a | 429.69 (364.83)a |
| Hoehn & Yahr stage | 1.92 (0.49)b,c | 3.35 (1.00)a,c | 3.06 (1.18)a,b |
| MDS-UPDRS III | 28.13 (11.88)b,c | 44.53 (14.45)a | 41.81 (13.87)a |
| MoCA | 25.50 (2.87)b,c | 21.18 (4.73)a | 21.19 (5.38)a |
| NfL | 13.30 (6.29)b,c | 27.08 (17.81)a | 26.05 (14.88)a |
| MRPI | 10.66 (2.60)c | 10.85 (4.90)c | 18.77 (7.01)a,b |
Abbreviations: PD, Parkinson’s disease; MSAp, multiple system atrophy parkinsonian variant; PSP, progressive supranuclear palsy; LEDD, levodopa equivalent daily dose; MDS-UPDRS III, Part III of the Movement Disorders Society Unified Parkinson’s Disease Rating Scale; MoCA, Montreal Cognitive Assessment; NfL, neurofilament light chain protein; MRPI, Magnetic Resonance Parkinsonism Index.
P < 0.05 versus PD.
P < 0.05 versus MSAp.
P < 0.05 versus PSP.
Structural T1 Acquisition and Preprocessing
T1-weighted images (resolution: 1 mm isotropic, repetition time = 8.2 milliseconds, echo time = 3.7 mm, flip angle = 8°, field of view = 240 mm × 240 mm) were acquired in 170 axial slices using a 3T magnetic resonance scanner (Philips Achieva; Philips Healthcare, Andover, MA) and a 32-channel quadrature volume head coil. The MRPI was calculated in an automated manner by incorporating the pons (P), midbrain (M), middle cerebellar peduncle (MCP), and superior cerebellar peduncle (SCP) areas into a single metric: (P/M)* (MCP/SCP).17
Biochemical Collection and Analysis
Plasma NfL concentrations were measured with ultrasensitive single molecule array technology. Samples were measured using the commercially available NF-light digital immunoassay kit (Quanterix, Lexington, MA). Prior to analyses, plasma samples were thawed at room temperature (1 cycle), mixed thoroughly, and centrifuged at 14,000g for 3 minutes. The supernatant was loaded onto a Quanterix HD-1 Analyzer with a 1:4 specified dilution. Measures were completed in duplicate, and the technician was blinded to the clinical information.
Diffusion MRI Acquisition and Preprocessing
Diffusion MRI images (64 directions; b values = 0/1000 s/mm2; resolution: 2 × 2 × 2 mm) were collected using the 3T magnetic resonance (Philips Achieva) and a 32-channel quadrature volume head coil. Data were preprocessed to obtain free-water (FW) and FW-corrected fractional anisotropy (FAT) maps and transferred to Montreal Neurological Institute space using the Automated Imaging Differentiation in Parkinsonism (AID-P).9 This pipeline calculates the FW and FAT in 17 regions of interest and 43 tracts of interest and inputs these values into PD versus MSAp/PSP and MSAp versus PSP machine-learning models to output a diagnostic probability for each patient. The training dataset for the AID-P is the cohort from the original publication, which includes 406 PD, 70 MSAp, and 103 PSP patients from 17 international sites.18
Statistical Analysis
All statistical analyses were performed in R (version 3.5.1; R Foundation for Statistical Computing, Vienna, Austria). Patient demographics, clinical variables, MRPI, and NfL concentrations were compared with an analysis of variance or Welch’s analysis of variance in the case of nonparametric variables as determined by Levene’s test. First, we conducted receiver operating characteristic (ROC) analyses to determine the diagnostic accuracy of the AID-P, MRPI, and NfL measures. For the AID-P, the current sample served as a test sample using model weights already established in prior work.18 PD versus MSAp/PSP and MSAp versus PSP diagnostic probabilities were obtained by inputting the subject space diffusion MRI maps into the AID-P algorithm. We used the outputted probabilities from these analyses as inputs into the ROC analysis to determine area under the curve (AUC). For MRPI and NfL, the measure of interest as well as age and sex were entered into a binary logistic regression analysis to obtain probabilities for each individual, and thus the models were fit in the current study sample. The AUC for each combination of variables was statistically compared using Delong’s test.19 Next, AID-P, MRPI, and NfL probabilities were assessed to determine if they predict the MDS-UPDRS III using forward stepwise multiple regression analysis. For the AID-P, measures that are the top 10 measures in both the PD versus MSAp/PSP and MSAp versus PSP machine-learning models were included (superior cerebellar peduncle FW, dentate nucleus FW, subthalamo-pallidal tract FW, descending supplemental motor area tract FW, ventral premotor cortex transcallosal tract FW).9 Age and sex covariates were entered into the AID-P model. For NfL and MRPI, the variable of interest was inputted in addition to age and sex covariates. Finally, we determined if different multimodal combinations of variables (AID-P, MRPI, NfL) were superior than unimodal metrics in differentiating PD versus MSAp/PSP and MSAp versus PSP. There were 4 different combinations tested: (1) AID-P + NfL + MRPI, (2) AID-P + NfL, (3) AID-P + MRPI, and (4) NfL + MRPI. These multimodal models were compared with the most significant unimodal models using Delong’s test.
Results
Between-Group Differences
There were no between-group differences in age (P = 0.07), sex distribution (P = 0.06), or disease duration (P = 0.21) (Table 1). Total levodopa equivalent daily dose was significantly different between groups (P = 0.023), in which the MSAp and PSP groups had a higher levodopa equivalent daily dose than the PD group. There was a significant between-group difference in the Hoehn & Yahr stage (P < 0.001), in which the MSAp and PSP groups had higher scores than the PD group, and the MSAp group also exhibited higher scores compared with the PSP group. The MSAp and PSP groups had higher MDS-UPDRS III (P < 0.001) than the PD group. Furthermore, the MSAp and PSP groups had lower Montreal Cognitive Assessment scores compared with the PD group (P < 0.001). Between-group differences were also evaluated in the NfL and MRPI measures, and both measures demonstrated a significant effect (P < 0.001). For NfL, the PD group had lower values compared with MSAp and PSP. For the MRPI, the PSP group had higher values compared with PD and MSAp.
Diagnostic Accuracy of the AID-P, MRPI, and NfL
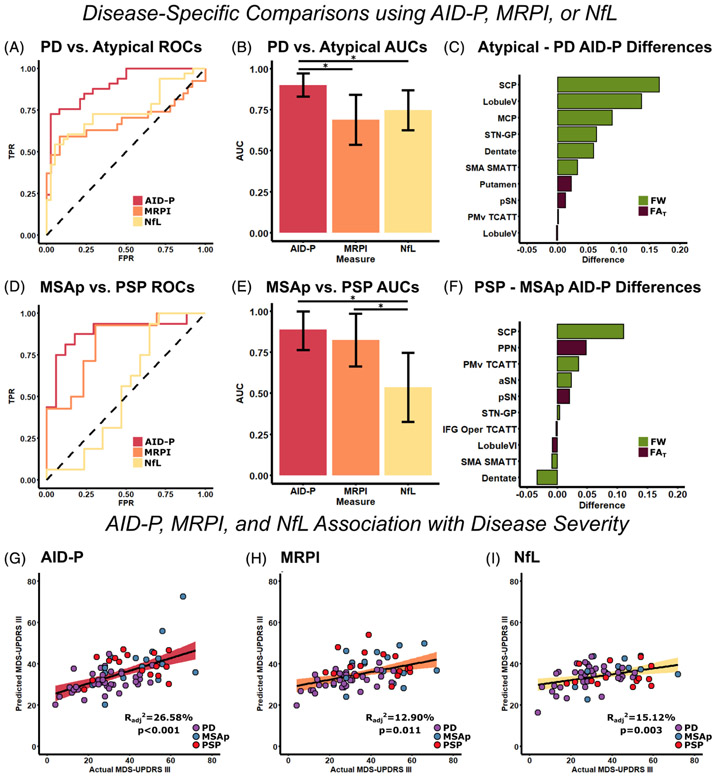
To evaluate the ability of AID-P, MRPI, and NfL to differentiate different forms of parkinsonism, 2 disease-specific models (PD vs. MSAp/PSP and MSAp vs. PSP) were used. For the AID-P, FAT and FW values were directly input into previously developed machine-learning models to obtain group probabilities for each individual. For MRPI and NfL, binary logistic regression was conducted (with age and sex covariates) to obtain group probabilities for each individual. These probabilities were entered into ROC curve analyses to obtain AUCs. For PD versus MSAp/PSP, the AID-P (AUC, 0.900; 95% CI, 0.830–0.971) measures were higher (P < 0.05) than the MRPI (AUC, 0.689; 95% CI, 0.537–0.842) and NfL (AUC, 0.747; 95% CI, 0.626–0.869) measures. For MSAp versus PSP, the AID-P (AUC, 0.889; 95% CI, 0.765–1.00) and MRPI (AUC, 0.824; 95% CI, 0.664–0.985) measures were higher (P < 0.05) than the NfL (AUC, 0.537; 95% CI, 0.326–0.747) measures. All AUC, sensitivity, and specificity values are reported in Table 2. The ROC curves for each biomarker (AID-P, MRPI, NfL) for each analysis are shown in Figure 1A,D with the corresponding AUC values and Delong’s test shown in Figure 1B,E. The FAT and FW differences in the AID-P are shown for PD versus MSAp/PSP and MSAp versus PSP in Figure 1C,F, respectively. As the MRPI is particularly accurate in distinguishing PSP from PD/MSAp, we conducted AID-P, MRPI, and NfL ROC analyses for this comparison. The AID-P (AUC, 0.864; 95% CI, 0.720–1), MRPI (AUC, 0.886; 95% CI, 0.786–0.987), and NfL (AUC, 0.761; 95% CI, 0.629–0.894) measures all performed with similar accuracies (all P > 0.05).
TABLE 2.
Receiver operating characteristic performance measures
| PD vs. Atypical |
MSAp vs. PSP |
|||||
|---|---|---|---|---|---|---|
| Combination of Measures |
AUC | Sensitivity | Specificity | AUC | Sensitivity | Specificity |
| AID-P | 0.900 (0.830–0.971) | 72.72 | 97.37 | 0.889 (0.765–1) | 87.50 | 82.35 |
| MRPI | 0.689 (0.537–0.842) | 59.26 | 91.67 | 0.824 (0.664–0.985) | 92.86 | 69.23 |
| NfL | 0.747 (0.626–0.869) | 54.55 | 94.74 | 0.537 (0.326–0.747) | 100 | 29.41 |
| AID-P + NfL + MRPI | 0.931 (0.870–0.992) | 88.89 | 86.11 | 0.907 (0.764–1) | 85.71 | 92.31 |
| AID-P + NfL | 0.923 (0.865–0.982) | 75.76 | 94.74 | 0.897 (0.774–1) | 87.50 | 88.24 |
| AID-P + MRPI | 0.894 (0.814–0.974) | 74.07 | 94.44 | 0.907 (0.772–1) | 78.60 | 100 |
| NfL + MRPI | 0.824 (0.706–0.943) | 81.48 | 80.56 | 0.824 (0.665–0.983) | 85.71 | 69.23 |
Abbreviations: PD, Parkinson’s disease; MSAp, multiple system atrophy parkinsonian variant; PSP, progressive supranuclear palsy; AUC, area under the curve; AID-P, Automated Imaging Differentiation in Parkinsonism; MRPI, Magnetic Resonance Parkinsonism Index; NfL, neurofilament light chain protein.
FIG. 1.
Unimodal prediction of disease state and disease severity using AID-P, MRPI, and NfL. ROC curves were created for the AID-P (red), MRPI (orange), and NFL (yellow) measures for PD versus MSAp/PSP (A), and the AUC value for each curve was quantified (B). The FW (green) and FAT (purple) difference between MSAp/PSP and PD was quantified for the top 10 contributors to the AID-P PD versus MSAp/PSP machine-learning model (C). The same analyses were repeated for MSAp versus PSP (D–F). Stepwise regression was conducted to determine the best set of variables to predict disease severity for AID-P (G, red), MRPI (H, orange), and NfL (I, yellow).*Significance at P < 0.05 and error bars represent 95% confidence intervals. AID-P, Automated Imaging Differentiation in Parkinsonism; aSN, anterior substantia nigra; AUCs, areas under the curve; FAT, FW-corrected fractional anisotropy; FW, free-water; IFG, inferior frontal gyrus; MSAp, multiple system atrophy; MDS-UPDRS III, Part III of the Movement Disorders Society Unified Parkinson’s Disease Rating Scale; MRPI, Magnetic Resonance Parkinsonism Index; MSAp, multiple system atrophy parkinsonian variant; NfL, neurofilament light chain protein; PD, Parkinson’s disease; pSN, posterior substantia nigra; PSP, progressive supranuclear palsy; ROCs, receiver operating characteristics; SMA, supplementary motor area; SMATT, sensorimotor area tract template; STN-GP, subthalamic nucleus-globus pallidus; TCATT, transcallosal tract template; TPR, true positive rate.
AID-P, MRPI, and NfL Association with Disease Severity
The AID-P, MRPI, and NfL measures were used in a forward stepwise multiple regression analysis to determine their association with the MDS-UPDRS III in patients with parkinsonism. All measures were significantly associated with disease severity. The final AID-P model included dentate nucleus FW, subthalamo-pallidal tract FW, descending supplemental motor area tract FW, and age. For the MRPI and NfL models, age and sex covariates were included in the final models. Figure 1G-I shows the predicted versus actual MDS-UPDRS III plots for the AID-P, MRPI, and NfL measures.
Multimodal Diagnostic Accuracy in Parkinsonism
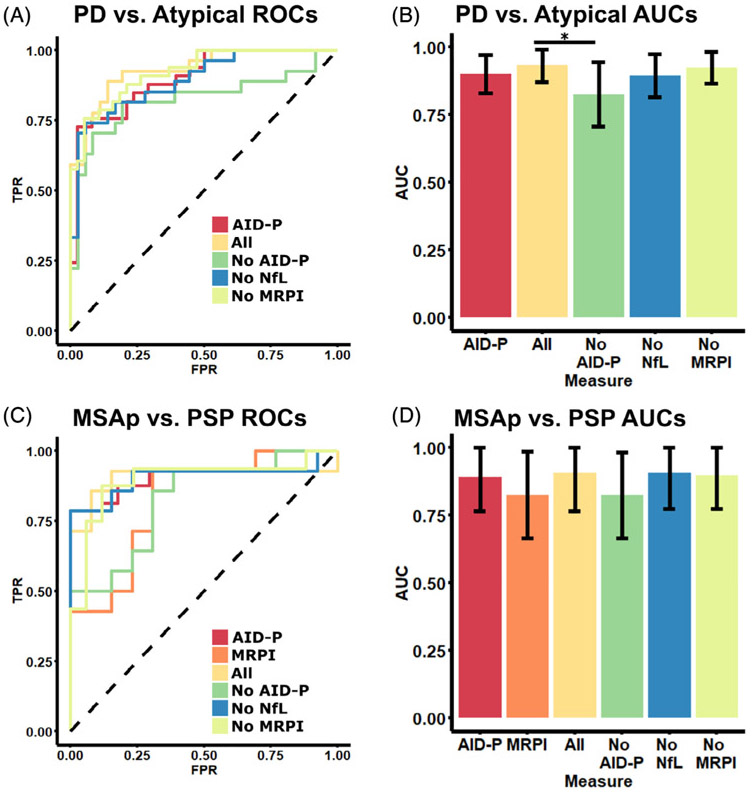
The most significant unimodal diagnostic models were compared to all possible multimodal models to determine if the use of several metrics was useful in differentiating between subtypes of parkinsonism. There was a total of 4 different multimodal models that were built for the PD versus MSAp/PSP and MSAp versus PSP comparisons: (1) AID-P + NfL + MRPI, (2) AID-P + NfL, (3) AID-P + MRPI, and (4) NfL + MRPI. These multimodal comparisons were compared with the highest performing unimodal models. For PD versus MSAp/PSP, the best performing model (ie, AID-P) was compared with the 4 multimodal models. The ROC curves and AUCs with 95% confidence intervals for these models are shown in Table 2 and presented graphically in Figure 2A,B. No multimodal model performed better than the AID-P model; however, the model with all 3 metrics (Fig. 2B, yellow) performed better (P < 0.05) than the model with only MRPI and NfL (ie, “no AID-P”; Fig. 2B, green). For MSAp versus PSP, the AID-P and MRPI models performed similarly and were therefore compared with the multimodal models. All multimodal models performed similarly to the unimodal models (all P > 0.05).
FIG. 2.
Multimodal classification of disease state using AID-P, MRPI, and NfL. The best-performing models from the unimodal analyses were compared to multimodal classification. For PD versus MSAp/PSP (A,B), the best unimodal performance was found for the AID-P (red). The AID-P measure was compared with a model with all metrics (yellow, AID-P, MRPI, and NfL). Each measure was then iteratively removed from the “All” model to create 3 different bimodal models (green, no AIDP; blue, no NfL; green-yellow, no MRPI). This was repeated for the MSAp versus PSP (C,D) comparison in which the unimodal MRPI model was included as it performed similarly to the AID-P model. *Significance at P < 0.05 and error bars represent 95% confidence intervals. AID-P, Automated Imaging Differentiation in Parkinsonism; AUCs, areas under the curve; FPR, false positive rate; MRPI, Magnetic Resonance Parkinsonism Index; MSAp, multiple system atrophy parkinsonian variant; NfL, neurofilament light chain protein; PD, Parkinson’s disease; PSP, progressive supranuclear palsy; ROCs, receiver operating characteristics; TPR, true positive rate.
Discussion
We assessed the AID-P, MRPI, and NfL in their ability to associate with parkinsonian state. The findings indicate that AID-P outperformed both the MRPI and NfL in differentiating PD from atypical parkinsonism (MSAp/PSP) and was better than NfL for differentiating between MSAp and PSP. Furthermore, the MRPI differentiated MSAp from PSP equally well, but did poorly for PD versus atypical parkinsonism. When comparing these unimodal models to predict parkinsonian state with models that combined biomarkers, we found that overall, there were no differences between the AID-P and any multimodal model in differentiating parkinsonisms. In other words, combining NfL and MRPI measures did not improve diagnostic accuracy compared with AID-P. In addition, although all biomarkers were predictive of disease severity, this study found that the AID-P is more strongly associated with the MDS-UPDRS III. This is the first study to compare these 3 biomarkers of parkinsonism in the same cohort.
A major benefit of neuroimaging measures is that they allow for the examination of pathologically relevant regions of interest in the brain and therefore allow us to target vulnerable networks in specific forms of parkinsonism. Although blood-based biomarkers that can index neurodegeneration may be easily accessible and less expensive compared with neuroimaging biomarkers, they are less specific to these brain networks. In this case, NfL in the neuronal cytoplasm plays a particularly vital role in maintaining neuronal structure20 and is consequently elevated in disorders with neuronal damage that result in the leaking of NfL into cerebrospinal fluid and plasma.21 Moreover, NfL is elevated in the presence of neurodegeneration in general, and does not allow us to probe specific networks. Several studies have measured cerebrospinal fluid and plasma-based NfL and determined its relationship with disease severity in parkinsonism.5,22,23 Here, we replicate these findings by showing that the MDS-UPDRS III is associated with plasma-based NfL in this parkinsonism cohort. Studies have also sought to determine if NfL is capable of differentiating disease state in parkinsonism, and both cerebrospinal fluid and plasma-based NfL have shown some promise in differentiating PD from MSAp/PSP. However, NfL has a low to moderate AUC when differentiating MSAp from PSP,5,20,22,23 and these findings confirm this prior observation.
An established neuroimaging metric is the MRPI, which is calculated using T1-weighted imaging to obtain spatial information from the middle cerebellar peduncle, superior cerebellar peduncle, midbrain, and pons to calculate an overall index. This metric has shown a strong ability to differentiate PSP from other parkinsonisms, but there has yet to be evidence that this metric can differentiate PD from MSA.6-8 In contrast, recent research has used advanced diffusion MRI metrics, in conjunction with machine learning, to create the AID-P.9 Specifically, advanced neuroimaging techniques were used to partition each voxel into a fluid component (ie, FW) and tissue component (ie, FAT) within 17 regions of interest and 43 neuronal tracts of interest. The AID-P was developed using a robust machine-learning algorithm in hundreds of patients from 17 different international sites. AID-P was capable of differentiating PD from MSA/PSP and MSA from PSP with high diagnostic accuracy (PD vs. MSA/PSP AUC, 0.955; MSA vs. PSP AUC, 0.926).9 In the present study, AID-P, MRPI, and NfL were directly compared in their ability to distinguish between different forms of parkinsonism. AID-P was found to have higher AUC compared with MRPI and NfL measures for differentiating PD from MSAp/PSP. AID-P and MRPI both had higher AUC compared with NfL in differentiating MSAp from PSP. Furthermore, we found that the AID-P had stronger associations with parkinsonism disease severity than NfL and MRPI. A potential reason that the AID-P had a higher AUC for PD versus MSAp/PSP is that it captures a more widespread window of neurodegeneration across multiple vulnerable brain networks associated with parkinsonism and thus may be more sensitive to disease state.
Although the current study directly compared these biomarkers for the first time, there are clear limitations in this article. First, these comparisons were conducted in a single cohort, and it is possible that not all findings would translate to other cohorts. Second, the sample size is a limiting factor for both the imaging and biofluid markers in this study, particularly for the PSP and MSAp cohorts. Third, although age and sex differences were not statistically significant between groups, they did approach significance and thus were added as covariates in the models. Fourth, although we demonstrate that the AID-P, MRPI, and NfL measures are associated with disease severity, this was a cross-sectional study and results should not be interpreted as longitudinal. Finally, although patients were tested following overnight withdrawal from antiparkinson medication, it is possible that medications with a longer half-life could have still influenced motor symptoms assessed by the MDS-UPDRS.
Conclusion
In conclusion, the present study evaluated 3 promising biomarkers, the AID-P, MRPI, and NfL, in a cohort of patients who were diagnosed by movement disorders specialists and followed over time. At the time of imaging, the patients on average had a disease duration of 3 years or more, suggesting that the clinical diagnosis was stable. The findings show that AID-P provides the best overall differentiation of PD versus MSAp/PSP, and both AID-P and MRPI are effective in differentiating MSAp versus PSP. Furthermore, no combination of these biomarkers performed better than the AID-P alone in differentiating PD from atypical parkinsonism. This is the first study to directly compare these 3 biomarkers, and the findings in the current sample demonstrate that the AID-P, using non-invasive diffusion MRI, provides diagnostic utility in parkinsonism.
Funding agencies:
National Institutes of Health Grants U01 NS102038 and R01 NS052318.
Footnotes
Relevant conflicts of interests/financial disclosures: D.E.V., M.S.O., and N.R.M. have National Institutes of Health grants related to this work. A.J. has stock options in Quanterix that performs the neurofilament analysis.
References
- 1.Adler CH, Beach TG, Hentz JG, et al. Low clinical diagnostic accuracy of early vs advanced Parkinson disease: clinicopathologic study. Neurology 2014;83(5):406–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beach TG, Adler CH. Importance of low diagnostic accuracy for early Parkinson’s disease. Mov Disord 2018;33(10):1551–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dickson DW. Parkinson’s disease and parkinsonism: neuropathology. Cold Spring Harb Perspect Med 2012;2(8). 10.1101/cshperspect.a009258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Planetta PJ, Ofori E, Pasternak O, et al. Free-water imaging in Parkinson’s disease and atypical parkinsonism. Brain 2016;139(Pt 2):495–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hansson O, Janelidze S, Hall S, et al. Blood-based NfL: a biomarker for differential diagnosis of parkinsonian disorder. Neurology 2017;88(10):930–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morelli M, Arabia G, Salsone M, et al. Accuracy of magnetic resonance parkinsonism index for differentiation of progressive supranuclear palsy from probable or possible Parkinson disease. Mov Disord 2011;26(3):527–533. [DOI] [PubMed] [Google Scholar]
- 7.Quattrone A, Nicoletti G, Messina D, et al. MR imaging index for differentiation of progressive supranuclear palsy from Parkinson disease and the Parkinson variant of multiple system atrophy. Radiology 2008;246(1):214–221. [DOI] [PubMed] [Google Scholar]
- 8.Constantinides VC, Paraskevas GP, Velonakis G, Toulas P, Stamboulis E, Kapaki E. MRI Planimetry and Magnetic Resonance Parkinsonism Index in the differential diagnosis of patients with parkinsonism. Am J Neuroradiol 2018;39(6):1047–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Archer DB, Bricker JT, Chu WT, et al. Development and validation of the automated imaging differentiation in parkinsonism (AID-P): a multicentre machine learning study. Lancet Digital Health 2019;1(5):e222–e231. [DOI] [PubMed] [Google Scholar]
- 10.Gilman S, Wenning GK, Low PA, et al. Second consensus statement on the diagnosis of multiple system atrophy. Neurology 2008;71(9):670–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Litvan I, Agid Y, Calne D, et al. Clinical research criteria for the diagnosis of progressive supranuclear palsy (Steele-Richardson-Olszewski syndrome): report of the NINDS-SPSP international workshop. Neurology 1996;47(1):1–9. [DOI] [PubMed] [Google Scholar]
- 12.Hughes AJ, Ben-Shlomo Y, Daniel SE, Lees AJ. What features improve the accuracy of clinical diagnosis in Parkinson’s disease: a clinicopathologic study. 1992. Neurology 2001;57(10 suppl 3):S34–S38. [PubMed] [Google Scholar]
- 13.Goetz CG, Fahn S, Martinez-Martin P, et al. Movement Disorder Society-sponsored revision of the Unified Parkinson’s Disease Rating Scale (MDS-UPDRS): process, format, and clinimetric testing plan. Mov Disord 2007;22(1):41–47. [DOI] [PubMed] [Google Scholar]
- 14.Hoglinger GU, Respondek G, Stamelou M, et al. Clinical diagnosis of progressive supranuclear palsy: the Movement Disorder Society criteria. Mov Disord 2017;32(6):853–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nasreddine ZS, Phillips NA, Bedirian V, et al. The Montreal Cognitive Assessment, MoCA: a brief screening tool for mild cognitive impairment. J Am Geriatr Soc 2005;53(4):695–699. [DOI] [PubMed] [Google Scholar]
- 16.Tomlinson CL, Stowe R, Patel S, Rick C, Gray R, Clarke CE. Systematic review of levodopa dose equivalency reporting in Parkinson’s disease. Mov Disord 2010;25(15):2649–2653. [DOI] [PubMed] [Google Scholar]
- 17.Nigro S, Arabia G, Antonini A, et al. Magnetic Resonance Parkinsonism Index: diagnostic accuracy of a fully automated algorithm in comparison with the manual measurement in a large Italian multicentre study in patients with progressive supranuclear palsy. Eur Radiol 2017;27(6):2665–2675. [DOI] [PubMed] [Google Scholar]
- 18.Archer DB, Bricker JT, Chu WT, et al. Development and validation of the automated imaging differentiation in parkinsonism (AID-P): a multicentre machine learning study. Lancet Digital Health 2019;1(5):e222–e231. [DOI] [PubMed] [Google Scholar]
- 19.DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics 1988;44(3):837–845. [PubMed] [Google Scholar]
- 20.Abdo WF, Bloem BR, Van Geel WJ, Esselink RA, Verbeek MM. CSF neurofilament light chain and tau differentiate multiple system atrophy from Parkinson’s disease. Neurobiol Aging 2007;28(5):742–747. [DOI] [PubMed] [Google Scholar]
- 21.Holmberg B, Rosengren L, Karlsson JE, Johnels B. Increased cerebrospinal fluid levels of neurofilament protein in progressive supranuclear palsy and multiple-system atrophy compared with Parkinson’s disease. Mov Disord 1998;13(1):70–77. [DOI] [PubMed] [Google Scholar]
- 22.Magdalinou NK, Paterson RW, Schott JM, et al. A panel of nine cerebrospinal fluid biomarkers may identify patients with atypical parkinsonian syndromes. J Neurol Neurosurg Psychiatry 2015;86(11):1240–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hall S, Ohrfelt A, Constantinescu R, et al. Accuracy of a panel of 5 cerebrospinal fluid biomarkers in the differential diagnosis of patients with dementia and/or parkinsonian disorders. Arch Neurol 2012;69(11):1445–1452. [DOI] [PubMed] [Google Scholar]