Abstract
BACKGROUND
The coronavirus disease 2019 (COVID-19) pandemic presents a significant challenge to the medical profession, increasing in the presence of microbial co-infection. Bacterial and Fungal co-infections increase the risk of morbidity and mortality in patients with COVID-19.
AIM
To study the bacterial profile in patients with COVID-19 who needed admission to receive treatment in the main centres concerned with managing COVID-19 disease in the Kingdom of Bahrain.
METHODS
The study was a retrospective observational analysis of the bacterial profile and the bacterial resistance in patients with confirmed COVID-19 disease who needed admission to receive treatment in the main centres assigned to manage patients with COVID-19 disease in the Kingdom of Bahrain from February to October 2020. We used the electronic patients’ records and the microbiology laboratory data to identify patients’ demographics, clinical data, microbial profile, hospital or community-acquired, and the outcomes.
RESULTS
The study included 1380 patients admitted with confirmed COVID-19 disease during the study period. 51% were admitted from February to June, and 49% were admitted from July to October 2020, with a recurrence rate was 0.36%. There was a significant increase in bacterial and fungal co-infection in the second period compared to the first period. The most common isolated organisms were the gram-negative bacteria (mainly Klebsiella pneumoniae, Pseudomonas aeruginosa, multi-drug resistant Acinetobacter baumannii, and Escherichia coli), the gram-positive bacteria (mainly coagulase negative Staphylococci, Enterococcus faecium, Enterococcus faecalis, Staphylococcus aureus) and fungaemia (Candida galabrata, Candida tropicalis, Candida albicans, Aspergillus fumigatus, Candida parapsilosis, Aspergillus niger). The hospital-acquired infection formed 73.8%, 61.6%, 100% gram-negative, gram-positive and fungaemia. Most of the hospital-acquired infection occurred in the second period with a higher death rate than community-acquired infections.
CONCLUSION
Bacterial and fungal co-infections in patients admitted with confirmed COVID-19 disease pose higher morbidity and mortality risks than those without co-infections. We should perform every effort to minimize these risks.
Keywords: COVID-19, Bacterial co-infection, Fungi, Hospital-acquired infection, Kingdom of Bahrain
Core Tip: Coronavirus pandemic presents a significant challenge to the medical profession. Bacterial and fungal co-infections are common complications of viral infections with increasing morbidity and mortality. We observed a significant increase in the number of bacterial and fungal co-infection over the study period. In addition, gram-negative infections carry a higher risk of morbidity and mortality.
INTRODUCTION
The coronavirus disease 2019 (COVID-19) pandemic, which began with the first reported case in December 2019 in China, led to a Public Health Emergency worldwide, including in Bahrain. This pandemic presents a significant challenge to the medical profession, especially with the contradicting data about the origin of the virus[1-3].
Bacterial co-infection is a common complication of viral infections with increasing morbidity and mortality in conjunction with more burden on healthcare resources. Serious bacterial infections may be missed when all attention focuses on COVID-19. Therefore, recognition of co-infection in patients with COVID-19 is of utmost importance. It enables us to implement the appropriate management and proper control of antibiotic use, with effective delivery of antimicrobial stewardship[4]. There are different reports about the prevalence of bacterial co-infection with COVID-19 assuming less bacterial co-infection with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) than influenza and other viral diseases[5]. On the other hand, some opinions based on the previous experience with the severe acute respiratory syndrome (SARS) outbreak in 2003 and the Middle East Respiratory Syndrome outbreak in 2012 suggest underestimation of bacterial co-infections in COVID-19 because of non-discriminatory use of antibiotics or the limitation of the overwhelmed clinical examinations in healthcare systems during the pandemic[6]. Bacteria can promote viral capability by augmenting virion stability, promoting viral infection of eukaryotic cells, and increasing co-infection rates. At the same time, virus binding of bacteria can also impact bacterial biology, including bacterial adherence to eukaryotic cells[7].
Bacterial co-infections in patients with COVID-19 are especially important when they require intensive care, including invasive mechanical ventilation support. For example, bacterial co-infections occurred in more than a third of children requiring invasive ventilation for bronchiolitis and were associated with more extended pediatric intensive care unit stay and mechanical ventilation[8]. Furthermore, patients admitted to intensive care unit (ICU) with prolonged illness/intubation have more frequent detection of multidrug-resistant gram-negative pathogens, likely reflecting hospital-acquired infection[9]. Therefore, it is vital to consider (investigate and empirically treat) bacterial co-infection when assessing these patients. Unfortunately, there is no consensus about treating patients with COVID-19 disease, which differs from one setting to another and from one country to another. Therefore, experts suggest not to use prophylactic antibiotics as a routine in patients with COVID-19, especially at the early stage or for non-intubated patients and recommend close monitoring of the signs of secondary infection, especially in critically ill patients who have been admitted to ICU for more than 48 h[10]. Furthermore, considering the long-term impact of the antimicrobial resistance development due to the unnecessary usage of antimicrobial agents, we should know the common bacterial and fungal infections that could complicate COVID-19, and know their expected antibiogram, and strictly monitor the rate of development of resistant bacterial strains[11]. Unfortunately, there are not enough data about the bacterial co-infections in patients admitted with COVID-19 disease. Therefore, we aimed to study the microbiological profile and the bacterial antibiogram in patients with COVID-19 who needed admission to receive treatment in the main centres concerned with managing COVID-19 disease in the Kingdom of Bahrain.
MATERIALS AND METHODS
Study design and setting
The study was a retrospective observational analysis of the microbiological profile of the patients admitted with confirmed COVID-19 disease to the different Ministry of Health (MOH) COVID isolation and treatment centres in the Kingdom of Bahrain for nine months period from February 2020 to October 2020. Inpatients with confirmed SARS-CoV-2 infection who had clinical suspicion of sepsis and/or bacterial co-infection were included in the study. Data were extracted and reviewed from the inpatients’ electronic health medical records from all MOH inpatients. The demographics, clinical data, microbiological profile, and outcomes of included patients were extracted, and the data were tabulated using the Microsoft Excel database.
Definitions
According to the national guidelines, the patients were stratified and allocated to specific COVID-19 Care centres into mild, moderate, and severe. The severe cases were assigned to the tertiary care centres with advanced care facilities. The medications differed according to the severity of the case and the presence of criteria of suspected sepsis.
Inpatients with the clinical suspension of sepsis/bacterial co-infection: COVID inpatients suspected clinically to have bacterial co-infection as decided by their treating physician during their clinical care, and septic workup were collected and sent to the microbiology laboratory.
Community-acquired infection: When clinical suspicion of sepsis/bacterial co-infection and the clinical samples for microbiology testing were collected from patients at the time of admission or within the initial 48 h from admission to COVID-19 facility.
Hospital-acquired infection: When clinical suspicion of sepsis/bacterial co-infection and the clinical samples for microbiology testing were collected after 48 h from the time of admission to COVID-19 facility.
Clinical isolates: The first bacterial pathogen growth for each patient from any clinical specimen was counted as a clinical isolate. Isolates were considered duplicate and not considered if identified from the same patient with the same organism and antimicrobial profile.
Laboratory technique
All the patients confirmed to have COVID-19 disease by positive testing using real-time reverse transcriptase-polymerase chain reaction for nasopharyngeal, sputum, endotracheal aspiration, or bronchoalveolar lavage samples. Clinical samples such as blood culture, sputum culture, stool culture, endotracheal aspirate or bronchoalveolar lavage culture were ordered according to the clinical indications when bacterial co-infection was suspected. These samples were cultured with the relevant media (nutritive, differential and/or selective), atmospheres and duration. The phenotypic detection was done using MALDI-TOF MS (Bruker Daltonics, Germany). Antimicrobial Susceptibility Testing was performed using BD Phoenix (BD Diagnostics, Baltimore, MD, United States) and interpreted according to the Clinical Laboratory Standards Institute[12]. We followed the trend of antibacterial sensitivity to evaluate the antimicrobial resistance.
Data analysis
All data were anonymized and collated on Excel 2017 (Microsoft, Redmond, WA, United States). We used TexaSoft, WINKS SDA Software 2011 (Sixth Edition, Cedar Hill, TX, United States) to perform the statistical analysis. We computed the percentages and frequencies for different categorical variables, and a cross-tabulation was computed between every two categorical variables. Finally, the Chi-Squared test determined whether there were significant relationships between every two categorical variables. We considered a P value of less than 0.05 as statistically significant. A biomedical statistician performed the statistical review of the study.
Ethical approval
The study was approved by the National COVID-19 Research Team and Secondary Care Research Committee of Salmaniya Medical Complex, Ministry of Health, the Kingdom of Bahrain. However, the study had no ethical consideration as it was a retrospective non-interventional study with no exposure to any patient data.
RESULTS
Table 1 showed the demographics of the included inpatients. The study included 1380 patients admitted with confirmed COVID-19 disease and had clinical suspicion of sepsis during the study period from February to October 2020, with a Male: Female ratio of 0.9, mean age of 50.2 ± 18.1 years, and 73% of them were Bahraini. The death rate was 11.5% for all the admitted patients during the study period. 51% of inpatients with clinical suspicion of sepsis were admitted from February to June, and 49% were admitted from July to October 2020. Five patients had confirmed recurrences (0.36%), all five patients recovered. From those admitted patients with confirmed COVID-19 diseases and clinical suspicion of sepsis, 261 patients (19%) had confirmed bacterial and fungal co-infections, 75% of them were Bahraini with a mean age of 58.5 ± 18.7 years, Male: Female ratio of 0.8, and a death rate of 42.5%. Two of these patients had a recurrence, and both survived. The remaining 1119 admitted patients (81%) had negative bacterial and fungal culture. Their mean age was 48.4 ± 17.6 years, with a male: female ratio of 0.9; 73% of them were Bahraini with a death rate of 4.3%. The group with confirmed bacterial and fungal co-infections had a significantly higher age (P < 0.0001) and rate of death (P < 0.0001) than the group without confirmed bacterial or fungal co-infection.
Table 1.
Comparison patients’ demographic for total admitted patients with/without Bacterial or fungal coinfections
|
|
Total admitted patients (COVID with clinical suspicion of sepsis)
|
Patients without coinfection (negative bacterial culture)
|
Patients with coinfection (positive bacterial culture)
|
P
value
|
| n (%) | 1380 | 1119 (81.1) | 261 (18.9) | < 0.0001 |
| Male/female | 0.92 | 0.87 | 1.13 | > 0.05 |
| Bahraini/non-Bahraini | 2.80 | 2.70 | 3.10 | > 0.05 |
| Mean age (yr) ± SD | 50.2 ± 18.1 | 48.4 ± 17.6 | 58.5 ± 18.7 | < 0.0001 |
| Death | 159 (11.5%) | 48 (4.30%) | 111 (42.5%) | < 0.0001 |
| Recurrences | 5 (0.36%) | 3 (0.27%) | 2 (0.77%) | > 0.05 |
COVID: Corona virus disease; SD: Standard deviation.
Table 2 showed the demographics of the patients with gram-positive, gram-negative bacteria, fungal and mixed infections. There were no significant differences between the number, age, gender, and nationality between the gram-positive and gram-negative bacteria. However, gram-negative infection occurred in older age and has a significantly higher death rate and more hospital-acquired infection rates than gram-positive bacteria. All the gram-negative isolates were detected from the centres allocated for the severe cases. Moreover, mixed infections occurred in less than a quarter of cases, with significantly higher age and death rate than other types of co-infections. All cases of mixed infections were hospital-acquired. We also observed that the number of patients with bacterial or fungal infection was significantly higher in the July-to-October period (P < 0.0001) with higher mean age (P < 0.01) compared to the first period of the study between February to June. In addition, the number of co-infections with gram-negative bacteria was significantly higher (P < 0.0001) in the July to October period than that of the February-to- June. The same also was observed in fungal co-infections. The number of mixed co-infections was also significantly higher in the July-to-October period (P < 0.01).
Table 2.
Comparison patients’ demographics and microbial profile for patients with gram-positive and gram-negative Bacteria and mixed infections
|
|
Gram + ve coinfection
|
Gram-ve coinfection
|
Mixed coinfection
|
Candida
|
P
value1
|
P
value2
|
P
value3
|
| n (%) | 136 (54) | 115 (46) | 82 (23.8) | 115 (46) | > 0.05 | < 0.0001 | < 0.0001 |
| Male/female | 0.82 | 0.67 | 0.74 | 0.88 | > 0.05 | > 0.05 | > 0.05 |
| Bahraini/non-Bahraini | 2.50 | 3.10 | 1.90 | 2.10 | > 0.05 | > 0.05 | > 0.05 |
| Mean age (yr) ± SD | 57.7 ± 18.2 | 60 ± 18.2 | 64.3 ± 14.3 | 63.4 ± 16.4 | > 0.05 | < 0.01a | < 0.05 |
| Death | 39 (28.7%) | 61 (53%) | 62 (75.6%) | 81 (70.4%) | < 0.0001 | < 0.0001 | < 0.01 |
| HA infection | 78 (57.3%) | 86 (75%) | 79 (96%) | 97 (84.3%) | < 0.001 | < 0.0001 | < 0.0001 |
Comparison between gram + ve and gram-ve coinfection.
Comparison between gram + ve and mixed coinfections.
Comparison between gram-ve and mixed coinfections. HA: Hospital acquired; SD: Standard deviation.
Table 3 showed the microbiological profile in patients with confirmed COVID-19 disease in the whole study period with a total of 472 isolates from 261 admitted patients. The gram-negative bacteria were isolated from 34.7% [59% showed Multidrug-resistant (MDR) strains], and gram-positive isolates were isolated from 34.7% of the patients (53% showed MDR strains). In comparison, fungal infections were isolated from 32% of the patient, 25% were isolated from the blood (Fungaemia). There was no significant difference in the isolates number in the two study periods, from February to June and July to October. However, the percentage of gram-negative isolates increased from 26.8% in the first period to 73% in the second period (P < 0.0001) and the percentage of MDR among gram-negative strains increased from 41% in the first period to 65.8% in the second period (P < 0.01). Thus, the MDR gram-negative strains isolated in the second period formed 81.4% of the total MDR strains isolated throughout the study (P < 0.0001). The most common gram-negative strains isolated through the study were Klebsiella pneumoniae (K. pneumoniae) followed by Pseudomonas aeruginosa (P. aeruginosa), then MDR Acinetobacter baumannii (A. baumannii), Escherichia coli (E. coli), Stenotrophomonas maltophilia (S. maltophilia), and Enterobacter cloacae (E. cloacae).
Table 3.
Microbiological profile in the admitted patients with confirmed coronavirus disease 2019 during the study period (472 isolates)
|
Type of the organism
|
Number
|
% of MDR
|
||||
| Gram Negative Isolates (164) | Klebsiella pneumoniae | Total | 39 | 97.4 | ||
| ESBL | 11 | |||||
| CRE | 27 | |||||
| Pseudomonas aeruginosa | Total | 38 | 26.3 | |||
| CRP | 8 | |||||
| MDR | 2 | |||||
| Acinetobacter baumannii (MDR) | 36 | 100 | ||||
| Escherichia coli | Total | 28 | 68 | |||
| ESBL | 11 | |||||
| CRE | 8 | |||||
| Stenotrophomonas maltophilia | 12 | 0 | ||||
| Enterobacter cloacae | Total | 2 | 100 | |||
| CRE | 2 | |||||
| Other | 9 | 0 | ||||
| Total G-ve isolates | 164 | 59 | ||||
| Total G-ve MDR strains | 97 | |||||
| Gram positive isolates (164) | Coagulase negative Staphylococci (CoNS) | Staphylococcus hominis | Total | 31 | 58 | |
| MRCoNS | 18 | |||||
| Staphylococcus epidermidis | Total | 25 | 78.6 | |||
| MRCoNS | 22 | |||||
| Staphylococcus heemolyticus | MRCoNS | 18 | 100 | |||
| Staphylococcus capitis | Total | 10 | 50 | |||
| MRCoNS | 5 | |||||
| Staphylococcus pettenkoferi | MRCoNS | 1 | 100 | |||
| Total CoNS | Total | 85 | 75.3 | |||
| MRCoNS | 64 | |||||
| Enterococcus faecium | Total | 24 | 16.6 | |||
| VRE | 3 | |||||
| HLGR | 1 | |||||
| Enterococcus faecalis | Total | 20 | 5.0 | |||
| HLGR | 1 | |||||
| Staphylococcus aureus | Total | 15 | 53.3 | |||
| MRSA | 8 | |||||
| Others | 20 | |||||
| Total G + ve isolates | 164 | 47 | ||||
| Total G + ve MDR Strains | 77 | |||||
| Fungal isolates(144) | Fungaemia | Candida galabrata | 11 | |||
| Candida tropicalis | 9 | |||||
| Candida albicans | 7 | |||||
| Aspergillus fumigatus | 3 | |||||
| Candida parapsilosis | 3 | |||||
| Aspergillus niger | 3 | |||||
| Total | 36 | |||||
| Candida species | 108 | |||||
| Total fungal isolates | 144 | |||||
| Total number of the microbial isolates | 472 | 36.9 | ||||
| Total number of mdr bacterial strains | 174 | |||||
CRE: Carbapenem-resistant Enterobacteriaceae; ESBL: Extended spectrum beta-lactamase; HLGR: High level aminoglycoside resistance; MDR: Multidrug-resistant; MRCoNS: Methicillin-resistant coagulase-negative Staphylococci; MRSA: Methicillin-resistant Staphylococcus aureus; VRE: Vancomycin-resistant enterococci; Other gram-negative bacteria: Citrobacter freundii, Salmonella species, Pantoea species, Proteus mirabilis, Serratia marcescens, Elizabethkingia meningoseptica; Other gram-positive bacteria: Streptococcus agalactiae (Strep. Group B), Corynebacterium afermentans, Bacillus licheniformis, Leuconostoc mesenteroides, Staphylococcus caprae, Staphylococcus lugdunensis, Staphylococcus warneri, Streptococcus parasanguinis, Gemella sanguinis, Micrococcus luteus, Propionibacterium acnes, Rhodococcus erythropolis, Aerococcus viridans, Staphylococcus gallinarum.
On the other hand, the gram-positive bacteria showed a significant increase in the total number of isolates in the second period but no significant difference in the number of total MDR strains or the number of coagulase-negative Staphylococci in the two study periods. Moreover, there was a significant increase in the number of methicillin-resistant coagulase-negative Staphylococci (MRCoNS) in the second period compared with the first periods. The most common gram-positive strains isolated throughout the study were Staphylococcus hominis (S. hominis) (MRCoNS), followed by Staphylococcus epidermidis (S. epidermidis) (CoNS), Enterococcus faecium (E. faecium), Enterococcus faecalis (E. faecalis), and Staphylococcus aureus (S. aureus). In addition, the rate of fungaemia was significantly higher in the second period (6-fold increase) compared to the first period (P < 0.0001).
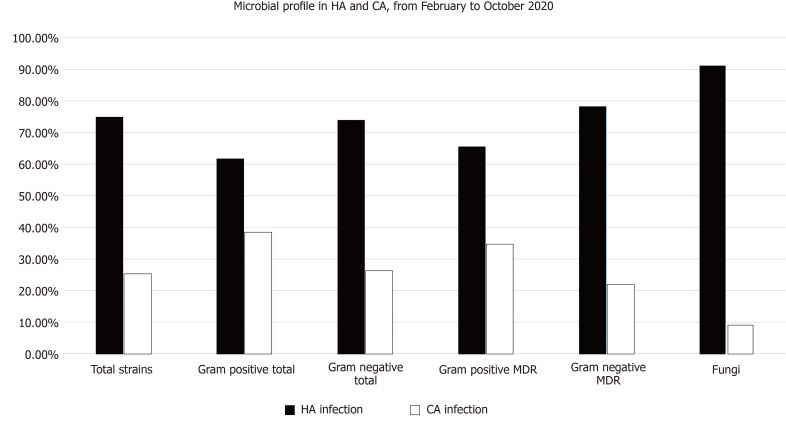
Table 4 and Figure 1 showed a comparison between the community and hospital-acquired infections (HAI) and their microbiologic profile in patients with confirmed COVID-19 disease with a total of 472 isolates during the whole study periods. Hospital-acquired infections formed 70% of the total infections. Those patients with HAI had a significantly higher mean of age (P < 0.01) than those of CAI. In addition, the percentage of the gram-negative isolates, including the MDR strains, were significantly higher in the HAI than CAI. The most common gram-negative strains were K. pneumoniae, followed by MDR A. baumannii, P. aeruginosa, E. coli, and S. maltophilia. At the same time, the total number of gram-positive isolates, including the MDR strains, were significantly higher in patients with HAI compared to patients with CAI (P < 0.0001). The most common gram-positive strains were S. epidermidis (CoNS), followed by E. faecium, E. faecalis, Staphylococcus haemolyticus (CoNS), S. hominis (CoNS), and S. aureus, as shown in the table. All isolates with fungaemia were obtained from patients with HAI. No cases with fungaemia were recorded from CAI.
Table 4.
Comparison between the community and hospital acquired infections and their microbiologic profile from coronavirus disease 2019 confirmed patients (total isolates 472)
|
Character
|
HA infection
|
CA infection
|
P
value
|
||||
| Patient number (total 261) 22 patients has both HA and CA | 185 (70.8%) | 98 (37.5%) | < 0.0001 | ||||
| Age | 60.8 ± 16.8 | 54. ± 20.6 | < 0.01 | ||||
| Male: Female | 0.85 | 0.80 | > 0.05 | ||||
| Bharani | 137 (74%) | 71 (72.4%) | > 0.05 | ||||
| Death | 102 (55%) | 23 (23.5%) | < 0.0001 | ||||
| Gram negative isolates (164) | Klebsiella pneumoniae | Total | 30 | 9 | |||
| ESBL | 11 | 0 | |||||
| CRE | 15 | 2 | |||||
| Acinetobacter baumannii (MDR) | 29 | 7 | |||||
| Pseudomonas aeruginosa | Total | 28 | 10 | ||||
| CRP | 6 | 2 | |||||
| MDR | 2 | 2 | |||||
| Escherichia coli | Total | 13 | 15 | ||||
| ESBL | 6 | 6 | |||||
| CRE | 5 | 2 | |||||
| Stenotrophomonas maltophilia | 11 | 1 | |||||
| Enterobacter cloacae | Total | 2 | 0 | ||||
| CRE | 1 | 0 | |||||
| Others | 8 | 1 | |||||
| Total number of the G-ve isolates (164) | 121 (73.8%) | 43 (26.2%) | < 0.0001 | ||||
| Number of G-ve resistant Strains | 75 (62%) | 21(48.8%) | > 0.05 | ||||
| % from total Resistant strains (96) | 78.1% | 21.9% | < 0.0001 | ||||
| Gram positive isolates (164) | Coagulase negative Staphylococci (CoNS) | Staphylococcus epidermidis | Total | 19 | 6 | ||
| MRCoNS | 18 | 4 | |||||
| Staphylococcus haemolyticus | MRCoNS | 14 | 4 | ||||
| Staphylococcus hominis | Total | 12 | 19 | ||||
| MRCoNS | 6 | 13 | |||||
| Staphylococcus capitis | Total | 5 | 5 | ||||
| MRCoNS | 5 | 0 | |||||
| Staphylococcus pettenkoferi | MRCoNS | 1 | 0 | ||||
| Total CoNS | Total | 41 (40.6%) | 31 (49.2%) | > 0.05 | |||
| MRCoNS | 37 (90%) | 21 (67.7%) | < 0.05 | ||||
| Enterococcus faecium | Total | 17 | 7 | ||||
| VRE | 2 | 1 | |||||
| HLGR | 1 | 0 | |||||
| Enterococcus faecalis | Total | 16 | 4 | ||||
| HLGR | 1 | 0 | |||||
| Staphylococcus aureus | Total | 8 | 8 | ||||
| MRSA | 3 | 5 | |||||
| Others | 9 | 10 | |||||
| Total number of the G + ve isolates (164) | 101 (61.6%) | 63 (38.4%) | < 0.0001 | ||||
| Number of G + ve resistant strains | 51 (50.5%) | 27 (42.9%) | > 0.05 | ||||
| % from total Resistant strains (78) | 65.4% | 34.6% | < 0.0001 | ||||
| Fungal isolates (144) | Candida galabrata | 11 | 0 | ||||
| Candida albicans | 7 | 0 | |||||
| Candida tropicalis | 9 | 0 | |||||
| Candida parapsilosis | 3 | 0 | |||||
| Aspergillus fumigatus | 3 | 0 | |||||
| Aspergillus niger | 3 | 0 | |||||
| Total | 36 | 0 | |||||
| Candida species | 95 | 13 | |||||
| Total fungal isolates (144) | 131 (91%) | 13 (9%) | < 0.0001 | ||||
| Total microbial isolates (472) | 353 (74.8%) | 119 (25.2%) | < 0.0001 | ||||
| Total number of resistant bacterial strains | 126 (35.7%) | 48 (40.3%) | > 0.05 | ||||
| Percentage from total resistant bacterial strains (174) | 72.4% | 27.6% | < 0.0001 | ||||
CA: Community Acquired; CRE: Carbapenem-resistant Enterobacteriaceae; ESBL: Extended spectrum beta-lactamase; HA: Hospital acquired; HLGR: High level aminoglycoside resistance; MDR: Multidrug-resistant; MRCoNS: Methicillin-resistant coagulase-negative Staphylococci; MRSA: Methicillin-resistant Staphylococcus aureus; VRE: Vancomycin-resistant enterococci; Other gram-negative bacteria: Citrobacter freundii, Salmonella species, Pantoea species, Proteus mirabilis, Serratia marcescens, Elizabethkingia meningoseptica; Other gram-positive bacteria: Streptococcus agalactiae (Strep. Group B), Corynebacterium afermentans, Bacillus licheniformis, Leuconostoc mesenteroides, Staphylococcus caprae, Staphylococcus lugdunensis, Staphylococcus warneri, Streptococcus parasanguinis, Gemella sanguinis, Micrococcus luteus, Propionibacterium acnes, Rhodococcus erythropolis, Aerococcus viridans, Staphylococcus gallinarum.
Figure 1.
Microbial profile in hospital-acquired infections and community-acquired infections from February to October 2020. MDR: Multidrug-resistant; HA: Hospital-acquired; CA: Community-acquired.
DISCUSSION
Microbial co-infections are commonly identified in viral respiratory infections. They are key reasons for difficult diagnosis, poor prognosis, increased morbidity and mortality, and greater use of healthcare resources. The prevalence and characteristic of bacterial co-infection in patients with confirmed COVID-19 disease are not well studied, especially in the Kingdom of Bahrain, with a broad knowledge gap. Bacterial co-infection could occur before admission of the patient to the hospital (Community-acquired) or could complicate the course of the illness as a secondary infection (Hospital-acquired). Our observational study identified a rate of 19% of bacterial co-infection through the study with increased rates of laboratory-confirmed bacterial and fungal co-infections in patients admitted with confirmed COVID-19 disease during the second period compared to the first period of the study despite that the total number of the admitted patients remained nearly the same. A study by Garcia-Vidal et al[13] had similar results to our results in the first period. They found an incidence of 7.2% of bacterial co-infection in their study, which conducted between February and April 2020[13]. Zhang et al[14] showed that the severely affected patients with COVID-19 disease had a significantly higher rate of bacterial (25.5%) and fungal (10.9%) co-infections. At the same time, a meta-analysis by Lansbury et al[5] indicated that about 7% of hospitalized patients with COVID-19 disease had bacterial co-infections, which increased to 14% in studies that only included ICU patients.
Nevertheless, this meta-analysis had a lower rate than that observed in our study, as it analyzed data from the earliest cases of the SARS-CoV-2 pandemic, which could differ from the current situation. Another metanalysis by Langford et al[15] showed that the overall proportion of COVID-19 patients with bacterial infection was 6.9% and increased to 8.1% of critically ill patients. The increased rate of bacterial co-infection in the second period in our study is related to the change in the admission criteria in the second period of the study to be more selective for the sick patients with medical comorbidities that need hospital management and allowing asymptomatic and mildly symptomatic patients to be managed at home. The death rate reached 42.5% in patients with bacterial co-infection than the patients without (4.3%). This high rate of death in the presence of microbial co-infection was also reported in a previous study in China which showed that 96% of patients with confirmed COVID-19 disease and secondary bacterial infections died. About half of the non-survivors experienced a secondary infection[16].
In the current study, there was a high incidence of gram-negative bacteria in patients who need hospitalization with increased mortality rates. Most of the gram-negative bacterial co-infections were hospital-acquired (75%). Consequently, every effort should be made to minimize this risk. Multi-drug resistant strains were present in more than half of the gram-negative bacterial isolates. This point should be considered during the management till the results of the antibiotic sensitivity are achieved. Being male and older than 60 years carries a higher risk for gram-negative as well as mixed co-infections. There was also a marked increase in the rate of gram-negative bacteria in the second period of the study, notably K. pneumoniae, followed by P. aeruginosa, MDR A. baumannii, E. coli, S. maltophilia, and E. cloacae, K. pneumoniae and P. aeruginosa were attributed to respiratory, then blood and urine-sourced infections. The MDR rate among the gram-negative bacteria was 65.8% in the second period and 41% in the first period of the study. This agreed with the work of Kokkoris et al[17], who reported an increase in the gram-negative blood-stream infections identified in ICU-admitted patients with confirmed COVID-19 disease, primarily due to MDR pathogens. A similar study in Egypt showed that MDR K. pneumoniae and A. baumannii were the predominant gram-negative bacteria that carried different resistance-associated genes[18]. The improper use of antibiotics could be implicated in increasing the resistance frequency. Many studies showed that antimicrobials were being administered at a high rate in patients with COVID-19 disease even in the presence of a low number of confirmed bacterial infection[19].
In the present study, the rate of co-infection with gram-positive bacteria in admitted patients was 11.8%. The most common isolated organisms were coagulase-negative Staphylococci (S. hominis, S. epidermidis, Staphylococcus heemolyticus, and others), forming 52.5% of total gram-positive isolates, followed by E. faecium, E. faecalis, and S. aureus with 47% of them were MDR strains. There was a significant increase in gram-positive bacteria in the second period than the first period of the study (P < 0.05). However, the resistance rate non-significantly decreased in the second period compared with the first period (P > 0.05). This observation agreed with the work of Sepulveda et al[20], who found that coagulase-negative staphylococcus species accounted for 59.7% of all positive cultures among patients with COVID-19 disease in New York City. Hughes et al[21] also found that coagulase-negative Staphylococcus species were the most common organisms isolated from the blood culture, followed by Acinetobacter species. Thus, infection with SARS-CoV-2 may reduce the patient’s immunity and increase the risk of bacterial infections. In a retrospective study in Wuhan, China, 19 patients in the ICU with confirmed COVID-19 disease had markedly reduced CD4 and CD8 T-cells[22]. This immune compromise increases the risk of co-infection with both viruses and bacteria, increases the risk of bacterial resistance, and the requirements of the patients to extended courses of IV antibiotic therapy[23].
In the current study, we observed the presence of fungaemia in about 10% of microbial co-infection. The most common fungi isolated were Candida galabrata, Candida tropicalis, Candida albicans, and Aspergillus fumigatus. The death rate in our patients who had fungal co-infection was very high (70.4%). This finding agreed with the study done in Upper Egypt by Ramadan et al[18], who found that Candida albicans and Candida glabrata were the most common fungal isolates. Patients hospitalised for COVID-19 are at risk for HAIs, with fungaemia; bloodstream infections caused by Candida or aspergillus. Invasive fungal infections add more prudent to the already immune-compromised patients with COVID-19 disease, especially diagnostic tools’ limitations and the critical clinical settings that put these patients at additional risk. Fungal infections resistant to antifungal treatment have also been described in patients with severe COVID-19. Early diagnosis and monitoring for Candida infections and antifungal resistant infections are essential to reduce death from COVID-19 in patients with severe COVID-19[24,25]. Mixed infections in the current study had a very high death rate, representing a significant threat to the patients with COVID-19 and necessitate aggressive treatment. To avoid missing these types of severe infection, patients should be recruited on admission to intensive care units and sampled longitudinally throughout the disease course using culture-independent techniques capable of identifying complex mixed infections[26].
In the current study, the HAI was about 71% of the total bacterial, and fungal infections in patients admitted with COVID-19 disease. The death rate in HAI was 55% compared to 23.5% in community-acquired infection. The age in HAI was also higher than in CAI. Older age is a significant risk factor to have HAI in patients with COVID-19[27]. Intrahospital and interhospital clonal transmission of bacteria could be a factor for HAI. Rational utilization of antibiotics and steroids to treat patients with COVID-19 is essential in preventing nosocomial infection. We should give particular attention to diabetic patients and patients with invasive devices[28]. HAI is a risk factor to have resistant strains. The percentage of resistant strains in HAI reached 62% in gram-negative and 50.5% in gram-positive isolates in the current study.
Antimicrobial resistance is a global problem, especially among gram-negative pathogens. The current study showed a high resistance pattern in bacterial co-infection in patients with COVID-19. In the gram-negative bacteria, about 28% of K. pneumoniae were extended spectrum beta-lactamase (ESBL), and 69% were CRE. All A. baumannii strains were MDR. About 39% of E. coli were ESBL, and 22% were CRE. In P. aeruginosa, 21% were CRP, while 8% were MDR. In gram-positive isolates, 75% of coagulase-negative Staphylococci and 53% of S. aureus were Methicillin-resistant. Antibiotic resistance is a critical reason for the failure of antibiotic therapy. At the same time, COVID-19 disease can exacerbate antibiotic resistance[29]. This increased resistance results from the interplay of different factors, including the micro-organisms, patients, and hospital environment, including the antibiotic use and the infection control practices. Increasing antibiotic resistance is also caused by improper antibiotic prescription and transmission of resistant bacterial strains within the hospitals by cross colonisation of patients via the hands of healthcare staff and subsequent spread between hospitals by transfer of the colonised patients[30]. Strategies to control antibiotic resistance in hospitals include multidisciplinary cooperation in implementing local policies on the use of antibiotics and infection control measures, timely detection with adequate microbiology laboratory standards and reporting of the antibiotic-resistant strains, improved surveillance, and aggressive control of transmission of epidemic resistant bacteria. We should integrate the antimicrobial stewardship activities into the pandemic response across the broader health system[31].
Limitation of the study
Despite being a multicentre study, it had some limitations. Being a retrospective study reduces control over multiple confounders and data collection. We did not study the mechanism of bacterial resistance due to lack of time and the workload during the pandemic. We also included only the infections that were documented by culture and, therefore, some episodes may be missing, and viral co-infection was not included. Finally, this study was done in the Kingdom of Bahrain, with its own unique local epidemiologic effects on antimicrobial resistance, limiting the generalisability of the findings.
CONCLUSION
Bacterial and fungal co-infections are common and place a significant threat to the patient with COVID-19 disease. At the same time, COVID-19 disease increases the risk of bacterial and fungal co-infections. We observed a high death rate in patients with hospital-acquired gram-negative co-infections. At the same time, older age was noted, especially in HAI. In addition, bacterial resistance was a significant problem in bacterial co-infection. Therefore, we should perform every effort to prevent microbial co-infections to minimize both morbidity and mortality.
ARTICLE HIGHLIGHTS
Research background
The coronavirus (COVID-19) pandemic presents a significant challenge to health worldwide. Bacterial and Fungal co-infections increase the risk of morbidity and mortality in patients with COVID-19, in conjunction with more burden on healthcare resources.
Research motivation
With the increasing risk of mortality among patients with COVID-19, there is a solid need to study the different factors that could increase or decrease this risk. Therefore, recognition of co-infection in patients with COVID-19 is of utmost importance. It enables us to implement the appropriate management and proper control of antibiotic use, with effective delivery of antimicrobial stewardship. Therefore, the centres that provide care for patients with COVID-19 in the kingdom of Bahrain participated in the current research.
Research objectives
We aimed to study the microbiological profile and the bacterial antibiogram in patients with COVID-19 who needed admission to receive treatment in the main centres concerned with managing COVID-19 disease in the Kingdom of Bahrain.
Research methods
The study was a retrospective observational analysis of the microbiological profile of the patients admitted with confirmed COVID-19 disease to the different Ministry of Health COVID isolation and treatment centres in the Kingdom of Bahrain for nine months period from February 2020 to October 2020.
Research results
There was a significant increase in the number of bacterial and fungal co-infection over the study period. The most common isolated organisms were the gram-negative bacteria (mainly Klebsiella pneumoniae, Pseudomonas aeruginosa, multi-drug resistant Acinetobacter baumannii, and Escherichia coli), the gram-positive bacteria (mainly coagulase negative Staphylococci, Enterococcus faecium, Enterococcus faecalis, Staphylococcus aureus) and fungaemia (Candida galabrata, Candida tropicalis, Candida albicans, Aspergillus fumigatus, Candida parapsilosis, Aspergillus niger). The hospital-acquired infection formed 73.8%, 61.6%, 100% gram-negative, gram-positive, and fungaemia. Most of the hospital-acquired infection occurred in the second period with a higher death rate than community-acquired infections.
Research conclusions
Bacterial and fungal co-infections in patients admitted with confirmed COVID-19 disease pose higher morbidity and mortality risks than those without co-infections. Therefore, we should perform every effort to minimize these risks.
Research perspectives
We need to study bacterial resistance mechanisms among the patients infected with COVID-19 and have co-infection with resistant bacterial strains. We also need to study viral co-infection and its effects on morbidity and mortality. Finally, we should compare our data with the data from other countries to generalize the obtained results.
ACKNOWLEDGEMENTS
The authors thank the anonymous reviewers who provided the manuscript with their valuable comments.
Footnotes
Institutional review board statement: The study was approved by the National COVID-19 Research Team and Secondary Care Research Committee of Salmaniya Medical Complex, Ministry of Health, the Kingdom of Bahrain.
Informed consent statement: The study had no ethical consideration as it was a retrospective non-interventional study with no exposure to any patient data.
Conflict-of-interest statement: The authors declare no conflict of interests for this article.
Manuscript source: Invited manuscript
Peer-review started: January 16, 2021
First decision: May 5, 2021
Article in press: May 19, 2021
Specialty type: Infectious diseases
Country/Territory of origin: Bahrain
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Zhang FY S-Editor: Zhang L L-Editor: A P-Editor: Xing YX
Contributor Information
Nermin Kamal Saeed, Medical Microbiology Section, Pathology Department, Salmaniya Medical Complex, Manama 00000, Bahrain; Microbiology Department, Royal College of Surgeons in Ireland - Bahrain, Manama 00000, Bahrain.
Safaa Al-Khawaja, Infection Disease Unit, Department of Internal Medicine, Salmaniya Medical Complex, Ministry of Health, Kingdom of Bahrain, Manama 00000, Bahrain; Department of Infectious Disease, Arabian Gulf University, Manama 00000, Bahrain.
Jameela Alsalman, Infection Disease Unit, Department of Internal Medicine, Salmaniya Medical Complex, Ministry of Health, Kingdom of Bahrain, Manama 00000, Bahrain; Department of Infectious Disease, Arabian Gulf University, Manama 00000, Bahrain.
Safiya Almusawi, Medical Microbiology Section, Pathology Department, Salmaniya Medical Complex, Manama 00000, Bahrain; Microbiology Department, Royal College of Surgeons in Ireland - Bahrain, Manama 00000, Bahrain.
Noor Ahmed Albalooshi, Medical Microbiology Section, Pathology Department, Salmaniya Medical Complex, Manama 00000, Bahrain.
Mohammed Al-Biltagi, Department of Pediatrics, University Medical Center, King Abdulla Medical City, Arabian Gulf University, Manama 00000, Bahrain; Department of Pediatrics, Faculty of Medicine, Tanta University, Tanta 000000, Al Gharbia, Egypt. mbelrem@hotmail.com.
Data sharing statement
The data that support the findings of this study are available from the corresponding author, [Al-Biltagi M], upon reasonable request.
References
- 1.Lai CC, Wang CY, Hsueh PR. Co-infections among patients with COVID-19: The need for combination therapy with non-anti-SARS-CoV-2 agents? J Microbiol Immunol Infect . 2020;53:505–512. doi: 10.1016/j.jmii.2020.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carrat F, Figoni J, Henny J, Desenclos JC, Kab S, de Lamballerie X, Zins M. Evidence of early circulation of SARS-CoV-2 in France: findings from the population-based "CONSTANCES" cohort. Eur J Epidemiol . 2021;36:219–222. doi: 10.1007/s10654-020-00716-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amendola A, Bianchi S, Gori M, Colzani D, Canuti M, Borghi E, Raviglione MC, Zuccotti GV, Tanzi E. Evidence of SARS-CoV-2 RNA in an Oropharyngeal Swab Specimen, Milan, Italy, Early December 2019. Emerg Infect Dis . 2021;27:648–650. doi: 10.3201/eid2702.204632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scott E. Androgen deprivation with or without radiation therapy for clinically node-positive prostate cancer. Lin CC, Gray PJ, Jemal A, Efstathiou JA, Surveillance and Health Services Research Program, Intramural Research, American Cancer Society, Atlanta, GA (CCL, AJ); Department of Radiation Oncology, Massachusetts General Hospital, Harvard Medical School, Boston, MA (PJG, JAE). J Natl Cancer Inst. 2015 May 9;107(7). pii: djv119. [Print 2015 Jul]. doi: 10.1093/jnci/djv119. Urol Oncol . 2017;35:122–123. doi: 10.1016/j.urolonc.2016.12.020. [DOI] [PubMed] [Google Scholar]
- 5.Lansbury L, Lim B, Baskaran V, Lim WS. Co-infections in people with COVID-19: a systematic review and meta-analysis. J Infect . 2020;81:266–275. doi: 10.1016/j.jinf.2020.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang CY, Chan KG. Underestimation of co-infections in COVID-19 due to non-discriminatory use of antibiotics. J Infect . 2020;81:e29–e30. doi: 10.1016/j.jinf.2020.06.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Neu U, Mainou BA. Virus interactions with bacteria: Partners in the infectious dance. PLoS Pathog . 2020;16:e1008234. doi: 10.1371/journal.ppat.1008234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wiegers HMG, van Nijen L, van Woensel JBM, Bem RA, de Jong MD, Calis JCJ. Bacterial co-infection of the respiratory tract in ventilated children with bronchiolitis; a retrospective cohort study. BMC Infect Dis . 2019;19:938. doi: 10.1186/s12879-019-4468-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peleg AY, Hooper DC. Hospital-acquired infections due to gram-negative bacteria. N Engl J Med . 2010;362:1804–1813. doi: 10.1056/NEJMra0904124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shang Y, Pan C, Yang X, Zhong M, Shang X, Wu Z, Yu Z, Zhang W, Zhong Q, Zheng X, Sang L, Jiang L, Zhang J, Xiong W, Liu J, Chen D. Management of critically ill patients with COVID-19 in ICU: statement from front-line intensive care experts in Wuhan, China. Ann Intensive Care . 2020;10:73. doi: 10.1186/s13613-020-00689-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rawson TM, Moore LSP, Zhu N, Ranganathan N, Skolimowska K, Gilchrist M, Satta G, Cooke G, Holmes A. Bacterial and Fungal Coinfection in Individuals With Coronavirus: A Rapid Review To Support COVID-19 Antimicrobial Prescribing. Clin Infect Dis . 2020;71:2459–2468. doi: 10.1093/cid/ciaa530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing, 30th ed. Melvin P. Weinstein: Clinical and Laboratory Standards Institute, 2020. Available from: https://clsi.org/standards/products/microbiology/documents/m100/
- 13.Garcia-Vidal C, Sanjuan G, Moreno-García E, Puerta-Alcalde P, Garcia-Pouton N, Chumbita M, Fernandez-Pittol M, Pitart C, Inciarte A, Bodro M, Morata L, Ambrosioni J, Grafia I, Meira F, Macaya I, Cardozo C, Casals C, Tellez A, Castro P, Marco F, García F, Mensa J, Martínez JA, Soriano A COVID-19 Researchers Group. Incidence of co-infections and superinfections in hospitalized patients with COVID-19: a retrospective cohort study. Clin Microbiol Infect . 2021;27:83–88. doi: 10.1016/j.cmi.2020.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang G, Hu C, Luo L, Fang F, Chen Y, Li J, Peng Z, Pan H. Clinical features and short-term outcomes of 221 patients with COVID-19 in Wuhan, China. J Clin Virol . 2020;127:104364. doi: 10.1016/j.jcv.2020.104364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Langford BJ, So M, Raybardhan S, Leung V, Westwood D, MacFadden DR, Soucy JR, Daneman N. Bacterial co-infection and secondary infection in patients with COVID-19: a living rapid review and meta-analysis. Clin Microbiol Infect . 2020;26:1622–1629. doi: 10.1016/j.cmi.2020.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou F, Yu T, Du R, Fan G, Liu Y, Liu Z, Xiang J, Wang Y, Song B, Gu X, Guan L, Wei Y, Li H, Wu X, Xu J, Tu S, Zhang Y, Chen H, Cao B. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet . 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kokkoris S, Papachatzakis I, Gavrielatou E, Ntaidou T, Ischaki E, Malachias S, Vrettou C, Nichlos C, Kanavou A, Zervakis D, Perivolioti E, Ranellou K, Argyropoulou A, Zakynthinos S, Kotanidou A, Routsi C. ICU-acquired bloodstream infections in critically ill patients with COVID-19. J Hosp Infect . 2021;107:95–97. doi: 10.1016/j.jhin.2020.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ramadan HK, Mahmoud MA, Aburahma MZ, Elkhawaga AA, El-Mokhtar MA, Sayed IM, Hosni A, Hassany SM, Medhat MA. Predictors of Severity and Co-Infection Resistance Profile in COVID-19 Patients: First Report from Upper Egypt. Infect Drug Resist . 2020;13:3409–3422. doi: 10.2147/IDR.S272605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rothe K, Feihl S, Schneider J, Wallnöfer F, Wurst M, Lukas M, Treiber M, Lahmer T, Heim M, Dommasch M, Waschulzik B, Zink A, Querbach C, Busch DH, Schmid RM, Schneider G, Spinner CD. Rates of bacterial co-infections and antimicrobial use in COVID-19 patients: a retrospective cohort study in light of antibiotic stewardship. Eur J Clin Microbiol Infect Dis . 2021;40:859–869. doi: 10.1007/s10096-020-04063-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sepulveda J, Westblade LF, Whittier S, Satlin MJ, Greendyke WG, Aaron JG, Zucker J, Dietz D, Sobieszczyk M, Choi JJ, Liu D, Russell S, Connelly C, Green DA. Bacteremia and Blood Culture Utilization during COVID-19 Surge in New York City. J Clin Microbiol . 2020;58 doi: 10.1128/JCM.00875-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hughes S, Troise O, Donaldson H, Mughal N, Moore LSP. Bacterial and fungal coinfection among hospitalized patients with COVID-19: a retrospective cohort study in a UK secondary-care setting. Clin Microbiol Infect . 2020;26:1395–1399. doi: 10.1016/j.cmi.2020.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Diao B, Wang C, Tan Y, Chen X, Liu Y, Ning L, Chen L, Li M, Wang G, Yuan Z, Feng Z, Zhang Y, Wu Y, Chen Y. Reduction and Functional Exhaustion of T Cells in Patients With Coronavirus Disease 2019 (COVID-19) Front Immunol . 2020;11:827. doi: 10.3389/fimmu.2020.00827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ganji A, Farahani I, Khansarinejad B, Ghazavi A, Mosayebi G. Increased expression of CD8 marker on T-cells in COVID-19 patients. Blood Cells Mol Dis . 2020;83:102437. doi: 10.1016/j.bcmd.2020.102437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Posteraro B, Torelli R, Vella A, Leone PM, De Angelis G, De Carolis E, Ventura G, Sanguinetti M, Fantoni M. Pan-Echinocandin-Resistant Candida glabrata Bloodstream Infection Complicating COVID-19: A Fatal Case Report. J Fungi (Basel) . 2020;6 doi: 10.3390/jof6030163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gold Medal: E. W. Meijer / Nagoya Silver Medal: H. Suga / Prelog Medal and Lectureship: S. B. H. Kent / Tajima Prize: X. Hu. Angew Chem Int Ed Engl . 2018;57:619. doi: 10.1002/anie.201711956. [DOI] [PubMed] [Google Scholar]
- 26.Cox MJ, Loman N, Bogaert D, O'Grady J. Co-infections: potentially lethal and unexplored in COVID-19. Lancet Microbe . 2020;1:e11. doi: 10.1016/S2666-5247(20)30009-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carter B, Collins JT, Barlow-Pay F, Rickard F, Bruce E, Verduri A, Quinn TJ, Mitchell E, Price A, Vilches-Moraga A, Stechman MJ, Short R, Einarsson A, Braude P, Moug S, Myint PK, Hewitt J, Pearce L, McCarthy K COPE Study Collaborators. Nosocomial COVID-19 infection: examining the risk of mortality. The COPE-Nosocomial Study (COVID in Older PEople) J Hosp Infect . 2020;106:376–384. doi: 10.1016/j.jhin.2020.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karterud S. The valence theory of Bion and the significance of (DSM-III) diagnoses for inpatient group behavior. Acta Psychiatr Scand . 1988;78:462–470. doi: 10.1111/j.1600-0447.1988.tb06368.x. [DOI] [PubMed] [Google Scholar]
- 29.Strathdee SA, Davies SC, Marcelin JR. Confronting antimicrobial resistance beyond the COVID-19 pandemic and the 2020 US election. Lancet . 2020;396:1050–1053. doi: 10.1016/S0140-6736(20)32063-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Struelens MJ. The epidemiology of antimicrobial resistance in hospital acquired infections: problems and possible solutions. BMJ . 1998;317:652–654. doi: 10.1136/bmj.317.7159.652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Almagor J, Temkin E, Benenson I, Fallach N, Carmeli Y DRIVE-AB consortium. The impact of antibiotic use on transmission of resistant bacteria in hospitals: Insights from an agent-based model. PLoS One . 2018;13:e0197111. doi: 10.1371/journal.pone.0197111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author, [Al-Biltagi M], upon reasonable request.