Abstract
The aim of this study was to develop a rapid and accurate PMA-qPCR method to quantify viable Brochothrix thermosphacta in cold-smoked salmon. B. thermosphacta is one of the main food spoilage bacteria. Among seafood products, cold-smoked salmon is particularly impacted by B. thermosphacta spoilage. Specific and sensitive tools that detect and quantify this bacterium in food products are very useful. The culture method commonly used to quantify B. thermosphacta is time-consuming and can underestimate cells in a viable but not immediately culturable state. We designed a new PCR primer set from the single-copy rpoC gene. QPCR efficiency and specificity were compared with two other published primer sets targeting the rpoC and rpoB genes. The viability dyes PMA or PMAxx were combined with qPCR and compared with these primer sets on viable and dead B. thermosphacta cells in BHI broth and smoked salmon tissue homogenate (SSTH). The three primer sets displayed similar specificity and efficiency. The efficiency of new designed rpoC qPCR on viable B. thermosphacta cells in SSTH was 103.50%, with a linear determination coefficient (r2) of 0.998 and a limit of detection of 4.04 log CFU/g. Using the three primer sets on viable cells, no significant difference was observed between cells treated or untreated with PMA or PMAxx. When dead cells were used, both viability dyes suppressed DNA amplification. Nevertheless, our results did not highlight any difference between PMAxx and PMA in their efficiency to discriminate viable from unviable B. thermosphacta cells in cold-smoked salmon. Thus, this study presents a rapid, specific and efficient rpoC-PMA-qPCR method validated in cold-smoked salmon to quantify viable B. thermosphacta in foods.
Keywords: viable, Brochothrix thermosphacta, spoilage, smoked salmon, PMA, PMAxx-based qPCR, rpoC gene
Introduction
Brochothrix thermosphacta is one of the main food spoilage bacteria, and can cause important economic losses in the food industry. This bacterium can produce off-odors leading to food waste, which moreover contributes to the ecological impact of food spoilage (Remenant et al., 2015; Illikoud et al., 2018a). In beef for example, B. thermosphacta can produce dairy – cheesy and creamy – off-odors (Dainty and Mackey, 1992; Casaburi et al., 2014). Seafood products, whose consumption is constantly increasing worldwide, can also be spoiled by B. thermosphacta, such as cooked and peeled shrimp with the production of strong butter, buttermilk-like, sour, and nauseous off-odors (Mejlholm et al., 2005; Laursen et al., 2006; Jaffrès et al., 2011). In cold-smoked salmon, B. thermosphacta can also produce butter/plastic/rancid, blue-cheese, sour/pungent off-odors (Joffraud et al., 2001, 2006; Stohr et al., 2001). B. thermosphacta has been described as widely disseminated along the food chain, from the raw material to the final product and during storage until use by date. This bacterium has also been isolated from food processing plants (from floors, walls, machines, etc.) which constitute one of the main sources of food contamination during processing (Stackebrandt and Jones, 2006; Nychas et al., 2008).
In the field of microbiological analysis, the non-cultural molecular methods such as quantitative real-time PCR (qPCR) have become essential tools in the last decade to detect and quantify microorganisms, with high precision in complex microbiota, e.g., in food matrices or food processing plants (Kuchta et al., 2014; Franco-Duarte et al., 2019). Quantitative real-time PCR (qPCR) is specific and fast tool to detect and quantify microorganisms, in complex microbiota, e.g., in food matrices or food processing plants (Kuchta et al., 2014). The qPCR detects bacteria in a viable but not immediately culturable state (VBNC), which are underestimates when using culturable microbiological approaches (Postollec et al., 2011). Nevertheless, dead bacteria can be revealed and quantified by qPCR. The qPCR using viability dyes such as propidium monoazide (PMA-qPCR) has been developed. PMA used before DNA extraction can penetrate unviable/dead cells, bind to their DNA and subsequently inhibit the PCR amplification, ensuring a selective quantification of viable bacteria (Elizaquível et al., 2014). PMA-qPCR is largely used nowadays to efficiently detect and quantify viable bacteria in food, e.g., Campylobacter (Josefsen et al., 2010; Castro et al., 2018), Listeria monocytogenes (Pan and Breidt, 2007; Desneux et al., 2015), Salmonella (Liang et al., 2011; Fang et al., 2018), Staphylococcus aureus (Zhang et al., 2015; Dong et al., 2018), Vibrio parahaemolyticus (Zhu et al., 2012; Niu et al., 2018), Escherichia coli O157:H7 (Elizaquível et al., 2012; Zhou et al., 2017), Photobacterium (Macé et al., 2013), and Brochothrix thermosphacta (Pennacchia et al., 2009; Mamlouk et al., 2012). The choice of the targeted phylogenetic marker is determining when such molecular tools are used. The 16S ribosomal RNA gene, commonly used to identify bacterial species, presents biases because multiple copies of this gene are often present in bacterial genomes with possible sequence variations (e.g., genomes of B. thermosphacta strains can contain up to 9 copies of the 16S rRNA gene) (Paoli et al., 2017; Illikoud et al., 2018b). This can lead to mispriming affecting amplification efficiency or quantification reliability. The use of single-copy protein-coding genes such as gyrB (DNA gyrase subunit B) or rpoB and rpoC (RNA polymerase subunit B or C) for bacterial detection and quantification is an alternative to this limitation (Case et al., 2007; Macé et al., 2013; Fougy et al., 2016).
We designed a new B. thermosphacta rpoC gene primer set, and compared its PMA-qPCR efficiency with the efficiency of two previously published rpoB or rpoC gene primer sets. We compared the efficiency of PMA or PMAxx treatments and described a rapid assay to quantify viable B. thermosphacta using PMA-qPCR with the new rpoC primer set. The method was successfully used to quantify B. thermosphacta in artificially contaminated cold-smoked salmon. To our knowledge, this study is a first about the use of qPCR to quantify the spoilage bacterium B. thermosphacta in combination with the use of a viability dye (PMA or PMAxx) and the targeting of a single-copy gene (rpoC or rpoB).
Materials and Methods
Bacterial Strains and B. thermosphacta Enumeration
A total of 26 strains belonging to 12 different bacterial species frequently isolated from various food products, notably cold-smoked salmon (CSS) or from a processing plant were used (Table 1). All strains were recovered from −80°C freezers on their appropriate agar media: Brain Heart Infusion agar (BHI) (VWR Chemicals Prolabo, France); BHI + 2% NaCl (Merck, France); de Man, Rogosa and Sharpe medium (MRS) (Biokar Diagnostics, France); Elliker broth (Biokar Diagnostics, France) supplemented with 1.5% agar (Biokar Diagnostics, France). Each strain was sub-cultured from the colonies on the plates in its corresponding broth and incubated at its optimal temperature growth for 24 to 48 h (Table 1) to obtain bacterial suspensions. B. thermosphacta DSM 20171 in pure culture was enumerated on BHI agar after 48 h incubation at 25°C. B. thermosphacta inoculated in smoked salmon tissue homogenate (SSTH) were enumerated on streptomycin tallous acetate actidione (STAA) agar supplemented with the STAA selective supplement (Oxoid, France), and incubated at 25°C for 48 h. Total viable counts in SSTH samples were estimated on plate count agar (PCA) (Biokar, France) supplemented with 2% NaCl (Merck, France) and incubated at 25°C for 72 h.
TABLE 1.
Bacterial strains used to assess the specificity of the qPCR primers.
| Species | Strains | Isolated from | Incubation temperature (°C) | Growth mediuma |
| Brochothrix thermosphacta | DSM 20171T | Fresh pork sausage | 25 | BHI |
| Brochothrix thermosphacta | Oniris 19/R/663 | Smoked salmon | 25 | BHI |
| Brochothrix thermosphacta | CD 340 (a) | Shrimp | 25 | BHI |
| Brochothrix thermosphacta | EBP 3084 (b) | Salmon | 25 | BHI |
| Brochothrix thermosphacta | TAP 125 (c) | Chicken | 25 | BHI |
| Brochothrix thermosphacta | EBP 3017 (b) | Cod | 25 | BHI |
| Brochothrix campestris | DSM 4712T | Soil | 25 | BHI |
| Carnobacterium maltaromaticum | Oniris 19/R/671 | Smoked salmon factory | 30 | Elliker |
| Carnobacterium divergens | NCDO 2763T | Vacuum-packaged beef | 30 | Elliker |
| Enterococcus faecalis | ATCC 19433T | Unknown | 37 | Elliker |
| Enterococcus faecium | CIP 54.33 | Canned fish | 37 | Elliker |
| Escherichia coli | CIP 53.125 | Human feces | 37 | BHI |
| Companilactobacillus alimentarius | DSM 20181 | Marinated fish product | 30 | MRS |
| Latilactobacillus sakei subsp. sakei | DSM 20017T | Moto starter of sake | 30 | MRS |
| Lactococcus piscium | Oniris 19/R/684 | Smoked salmon | 25 | Elliker |
| Lactococcus raffinolactis | Oniris MIP 2453 | Unknown | 25 | Elliker |
| Listeria monocytogenes | DSM 12464 T | Poultry | 37 | BHI |
| Listeria monocytogenes | CIP 78.35 | Spinal fluid | 37 | BHI |
| Listeria grayi | CIP 68.18T | Feces of chinchilla | 37 | BHI |
| Listeria innocua | CIP 80.11T | Brain of cow | 37 | BHI |
| Photobacterium phosphoreum | CIP 102511T | Unknown | 15 | BHI 2% NaCl |
| Pseudomonas fluorescens | CIP 69.13T | Pre-filter tanks | 30 | BHI |
| Psychrobacter sp. | Oniris 19/R/675 | Smoked salmon factory | 25 | BHI 2% NaCl |
| Serratia liquefaciens | ATCC 27592T | Milk | 37 | BHI |
| Shewanella baltica | Oniris 19/R/670 | Smoked salmon factory | 25 | BHI 2% NaCl |
| Staphylococcus epidermidis | CIP 68.21 | Unknown | 37 | BHI |
DSM, Deutsche Sammlung von Mikroorganismen and Zellkulturen GmbH, Braunschweig, Germany; CIP, Centre de Ressources Biologiques de l’Institut Pasteur, Paris, France; NCDO, National Collection of Dairy Organisms, Shinfield, United Kingdom; ATCC, American Type Culture Collection, Manassas (VA) United States; Oniris, Secalim Bacterial Culture Collection, Nantes, France. T: Type Strain; BHI, Brain Heart Infusion agar (VWR Chemicals Prolabo, France); MRS, de Man Rogosa and Sharpe and Elliker media (Biokar Diagnostics, France). (a) Jaffrès et al. (2009), (b) Chaillou et al. (2015), (c) Illikoud et al. (2019).
Preparation of the Smoked Salmon Tissue Homogenate
Cold-smoked salmon was provided by a French company. B. thermosphacta was not detected in it using standard culture method on STAA, with the STAA selective supplement, and the real-time PCR methods developed in this study. The smoked salmon tissue homogenate (SSTH) was prepared either from packaged smoked salmon slices preserved under vacuum at 4°C (leaving the factory) (SSTHf) or from smoked salmon fillets sent by the French company sampled just after the smoking stage of CSS production (SSTHs). A 20-g portion was aseptically weighed in a sterile stomacher plastic bag equipped with a 63-μm porosity filter (Interscience, France), and 80 mL of sterile water were added to obtain a 5-fold dilution. The sample was then homogenized for 2 min using a stomacher device (Masticator IUL, Spain). The bag-filtered samples were divided into aliquots in sterile vials, and then stored at −80°C until their inoculation by fresh bacterial cultures. In order to limit qPCR inhibition, 1% of bovine serum albumin (BSA) fraction V (Sigma-Aldrich, France) and 4% of polyvinylpyrrolidone (PVP) (Sigma-Aldrich, France) were added to SSTH before DNA extraction (Kreader, 1996; Furet et al., 2009; Schrader et al., 2012).
Viable and Dead Cell Preparation
The efficiency of the PMA and PMAxx treatments was assessed on B. thermosphacta DSM 20171 viable or dead cells in BHI broth or SSTH. The viable cells were obtained from a pre-culture in 10 mL of BHI broth at 25°C for 8 h, followed by a culture in 100 mL of BHI broth for 4 h, in order to obtain an 8 log CFU/mL exponential growth phase culture. The 100% viable cell culture in SSTH was produced as follows: 5 mL of the 8 log CFU/mL exponential growth phase BHI culture were centrifuged at 6,000 x g for 2 min, the supernatant was discarded, and the cells were re-suspended in 5 mL of SSTH (with BSA and PVP). Dead cells were obtained by heating 5 microtubes containing 1 ml of the 8 log CFU/mL exponential growth phase in BHI culture at 90°C for 15 min in a water bath. The suspensions were pooled and centrifuged at 6,000 × g for 2 min. The supernatant was discarded, and the cells were re-suspended in 5 mL of SSTH (with BSA and PVP). Cell viability was checked by plating on STAA agar.
PMA and PMAxx Treatments
PMAxxTM is a new and improved version of viability dye PMA (propidium monoazide) designed by Biotium (Biotium, Inc., Hayward, CA, United States). Like PMA, PMAxxTM is a photo-reactive dye that binds covalently to DNA with high affinity after photolysis with visible light. The PCR cannot amplify this modified DNA. PMA and PMAxxTM dyes are cell membrane-impermeants. PMAxxTM is described to be more effective than PMA to inhibit DNA PCR amplification of dead cell with injured cell membranes, and therefore provides best discrimination between live and dead bacteria (Han et al., 2018). Commercial PMA and PMAxx solutions (20 mM) (VWR, France) were diluted in pure sterile water to obtain 1.5 mM working solutions kept at −20°C in light-tight microtubes. The samples to be treated were separated in 4 sterile microtubes containing each 580 μL of bacterial cell suspensions in BHI or inoculated SSTH. Twenty microliters of PMA working solution were added in 2 microtubes and 20 μL of PMAxx working solution were added in the remaining 2 tubes (50 μM final concentration). The microtubes were placed in the dark at room temperature for 10 min, and mixed occasionally for PMA/PMAxx to penetrate into the dead cells. To photoactivate PMA/PMAxx, the PMA-LiteTM LED Photolysis Device (Biotium) was used as recommended by the PMAxxTM manufacturer (Biotium). The PMA-LiteTM LED Photolysis Device is a thermally stable blue LED light source (LED power: 60 W, output wavelength: 465–475 nm) that provides even illumination to the sides and bottoms of all vials. The microtubes were exposed to blue light for 20 min, to ensure complete cross-linking to the available DNA (free DNA or DNA from unviable cells). The content of the 2 microtubes treated with PMA was pooled and 1 mL of the suspension was conserved for further DNA extraction. The same operation was realized for the microtubes treated with PMAxx. For each cell suspension tested in BHI or in SSTH, one milliliter of viable or dead cell suspension was not treated with PMA or PMAxx and used as an untreated control.
PMA and PMAxx treatments were tested on SSTHs in the presence of mixes of dead and viable B. thermosphacta cell suspensions. Several mix conditions were tested: (i) 5.65 log CFU/g of dead cells mixed with 4.70, 5.70, or 6.70 log CFU/g of viable cells, and (ii) 7.65 log CFU/g of dead cells mixed with 4.70, 5.70, or 6.70 log CFU/g of viable cells. The samples were treated or not with PMA and PMAxx, as mentioned above. Two controls were also included: (i) 5.70 or 7.70 log CFU/g of viable cells treated or not with PMA/PMAxx, and (ii) 5.70 or 7.70 log CFU/g of dead cells treated or not with PMA/PMAxx.
Extraction of Bacterial DNA
Genomic DNA extraction from bacterial cells cultured in BHI broth was performed using the Qiagen DNeasy Blood and Tissue kit (Qiagen, France). One milliliter of culture was centrifuged at 11,000 × g for 10 min at 4°C (Biofuge PimoR, Heraeus). The supernatant was removed. Bacterial DNA extraction was conducted on cell pellets according to the manufacturer’s instructions (Qiagen, France). A Qubit® 2.0 fluorometer using a Qubit® dsDNA BR Assay Kit (Life technologies, Thermo Fisher Scientific, France) was used to quantify the extracted DNA. Extracted DNAs were stored at −20°C until qPCR amplification.
For bacterial DNA extraction in SSTH, one milliliter of SSTH inoculated with B. thermosphacta cells was centrifuged at 11,000 × g for 10 min at 4°C. The supernatant was removed, and the pellet was stored at −20°C until use. DNA extraction was performed with the DNeasy PowerFood Microbial kit (Qiagen, France). Briefly, this kit is designed to isolate high-quality genomic DNA from microorganisms hosted in food. The microbial cell pellet from food was thawed and re-suspended in 450 μL of MBL lysis buffer preheated at 55°C for 10 min. The suspension was transferred to PowerBead tubes, containing beads designed for mechanical microbial-cell lysis, and shaken horizontally for 30 s on a Vortex-Genie 2 device (Scientific industries, United Sates), then on a FastPrep-24 G at 6 m/s for 30 s (MP Biomedicals, France), for mechanical lysis. Each sample was centrifuged at 10,000 × g for 1 min at room temperature. The supernatant was transferred to a microtube, and 100 μL of IRS solution were added. The microtube was incubated on crushed ice for 30 min. The next steps were performed as described in the Qiagen kit instruction manual. DNA samples were stored at −20°C until use. DNA extracts were diluted 1/10 before the qPCR amplification.
Primer Design and Quantitative Real-Time PCR Assay
A new PCR primer set was designed to amplify a rpoC gene DNA fragment specific to B. thermosphacta, excluding other Brochothrix spp. (B. campestris) and closely related species such as Listeria. The in silico primer design was based on multiple alignment of the DNA-dependent RNA polymerase subunit (rpoC) gene sequences of B. thermosphacta and closely related species, available from the GenBank database (release 225.0). The rpoC sequences from B. thermosphacta, B. campestris and the most closely related bacterial species were aligned using the CLC DNA Workbench 6.5 (CLC bio, Aarhus, Denmark) and BioEdit sequence alignment software (Hall, 1999). Primer specificity was tested in silico using the basic local alignment search tool (BLAST) program [National Center for Biotechnology Information (NCBI)] and Primer BLAST (Ye et al., 2012) using Genbank release 225.0.
Two other previously described primer sets targeting the rpoC (Fougy et al., 2016) or rpoB (Illikoud et al., 2019) genes were also used (Table 2). AmplifX 2.0.7 software was used to evaluate the quality of the primers (Jullien, 2019). The annealing temperature was optimized using the temperature gradient test of the Bio-Rad CFX connect real-time PCR detection system (Bio-Rad, France).
TABLE 2.
Description of the three sets of qPCR primers.
| Target gene | Primer pair | Primer sequences (5′- 3′) | Amplicon size (bp) | Tm (°C) | References |
| rpoC | rpoC-126-F | ATACTGTACCAATGGTTGCTC | 126 | 52 | This study |
| rpoC-126-R | CAACAGTGATAACATCAGTTAC | ||||
| rpoC | QSF03-BTH-F | GGACCAGAGGTTATCGAAACATTAACTG | 151 | 56 | Fougy et al. (2016) |
| QSF03-BTH-R | TAATACCAGCAGCAGGAATTGCTT | ||||
| rpoB | rpoB-Fw1 | GCGTGCATTAGGTTTCAGTACA | 394 | 55 | Illikoud et al. (2019) |
| rpoB-Rev1 | TCCAAGACCAGACTCTAATTGCT |
Quantitative real-time PCR amplification was performed using the Sso Advanced universal SYBR Green supermix (Bio-Rad, France). The reaction was performed in a final volume of 12.5 μL containing 1 μM of each primer, 6.25 μL of 2 X Sso Advanced universal SYBR Green Supermix, 2.75 μL of water (molecular biology grade) and 3 μL of DNA extract. DNA concentrations were standardized at 1 ng/μL for the specificity test. The amplification reaction was conducted using a Bio-Rad CFX connect real-time PCR detection system (Bio-Rad, France). The cycling parameters were as follows: initial denaturation step at 95°C for 3 min, 39 cycles of 95°C for 15 s, and 60°C for 30 s. A melting curve from 65 to 95°C was determined after the last amplification cycle and at a temperature transition rate of 0.5°C/3 s. Quantification cycles (Cq) values were automatically obtained by the Biorad CFX Manager software program. All amplification reactions were run in triplicate in three independent assays.
Efficiency of the Quantitative Real-Time PCR
The efficiency of the qPCR was evaluated by using three different sample preparations. The first two were obtained from B. thermosphacta BHI culture: DNA was extracted from a 10-fold serial dilution range of B. thermosphacta BHI culture (7 to 2 log CFU/mL) (BHI culture extract), or from an 8 log CFU/mL of B. thermosphacta BHI suspension. This DNA extract was then serial diluted 10-fold, corresponding to 7 to 2 log CFU/mL (diluted DNA extract). The third sample preparation corresponded to DNA extracted from SSTHf inoculated with B. thermosphacta from 7 to 4 log CFU/mL. The rpoC and rpoB sequences were amplified using the three different species-specific primers used in this study in the qPCR conditions described above. The linear standard curves were generated by plotting the Cq values versus log CFU/mL to determine the analytical efficiency of the qPCR assay. The efficiency of the real-time PCR assay was calculated from the slope (m) of the standard curve according to equation E = 10(–1/m)−1. The qPCR was performed in three independent assays under the same conditions, using three replicates of each template concentration. The linearity of the standard curve was expressed as a coefficient of determination (r2).
Limit of Detection (LOD)
The LOD was established in SSTHf for the two primer sets: rpoC-126-F/R (designed in this study) and rpoB-Fw1/Rev1. Ten replicates of 10-fold serial dilutions of B. thermosphacta inoculated in SSTHf from 7 to 1 log CFU/mL were prepared. DNA extracts from these inoculated samples were used as qPCR templates in triplicate. Each Cq datum corresponding to the amplification of a PCR product whose melting temperature corresponded to that of the studied gene was considered as a positive signal. The LOD was established by comparing the positive signals and the enumeration of cells on STAA agar and analyzed by a Probit model (Kralik and Ricchi, 2017) with R version 3.3.2 (2016-10-31) (The R Foundation for Statistical Computing). The p-value was fixed at 0.05 to detect the gene with 95% probability.
Statistical Analysis
The Shapiro-Wilk’s test was used to check the normality of the data. Student’s t-test or Wilcoxon’s test were applied to determine differences between conditions tested (treated vs. untreated cultures, PMA vs. PMAxx treatments, differences of amplification between the 3 primer sets) on viable or dead B. thermosphacta cells in BHI broth or SSTH. Homoscedasticity was checked on the values of the standard curves obtained in SSTH using Levene’s test. ANOVA was used to control the significant statistical differences between these standard curves. ANOVA, followed by Tukey’s multiple comparison of means test was used to analyze the quantification of viable B. thermosphacta in cold-smoked salmon fillet sampled after the smoking step. The significant threshold of 5% was established for all the statistical tests. The tests were performed using R version 3.3.2 (2016-10-31) (The R Foundation for Statistical Computing) and the XLSTAT add-in software (version 2020) in Microsoft Excel software 2016.
Results
Primer Design
A new PCR primer set was designed to amplify a fragment of the B. thermosphacta rpoC gene encoding a DNA-dependent RNA polymerase subunit. The in silico primers design was based on multiple alignment of B. thermosphacta rpoC gene sequences and on those of closely related species available from the GenBank database. Based on this analysis, a region of the rpoC gene was selected as a target of the primers design to specifically distinguish B. thermosphacta from other related bacterial species examined in this study. The forward primer rpoC-126-F (5′-ATACTGTACCAATGGTTGCTC-3′) matched positions 3005 to 3025, and the reverse primer rpoC-126-R (5′-CAACAGTGATAACATCAGTTAC-3′) matched positions 3109 to 3130 of the B. thermosphacta DSM 20171 type strain rpoC gene (accession number X89231), to amplify a specific 126-bp fragment. Two other previously described primer sets targeting the rpoC or rpoB genes were used. Firstly, the primers QSF03-BTH-F/QSF03-BTH-R targeting the rpoC gene (accession number X89231), with a specific 151-bp amplified fragment (Fougy et al., 2016). The forward primer matched positions 2494 to 2521, and the reverse primer matched positions 2621 to 2644 of the B. thermosphacta DSM 20171 type strain. Secondly, the primers rpoB-Fw1/Rev1 targeting a specific region of B. thermosphacta rpoB gene were used (Illikoud et al., 2019). The forward primer rpoB-Fw1 matched positions 609 to 630, and the reverse primer rpoB-Rev1 matched positions 980 to 1022 of the B. thermosphacta DSM 20171 type strain rpoB gene (Gene ID 29820965), with a specific 394-bp amplified fragment. We selected this later rpoB primer, amplifying a longer fragment, because the PCR inhibition effect by PMA can be dependent on the length of the amplification product (Martin et al., 2013).
In silico studies with BLAST program and Primer BLAST using Genbank release 225.0. demonstrated that the primers developed in the present study (rpoC-126-F/R) and those developed by Fougy et al. (2016) and Illikoud et al. (2019) were B. thermosphacta-specific. Furthermore, the quality assessment of the designed primers using AmplifX 2.0.7. software revealed no hairpin loop, dimer or duplex formation. The hybridization temperature of the primers was optimized using the temperature gradient test of the Bio-Rad CFX connect real-time PCR detection system (Bio-Rad, France). The 3 sets of primers were tested for qPCR specificity and efficiency.
Specificity of the qPCR Assay
The specificity of the qPCR assay was assessed by inclusivity and exclusivity tests using 26 different bacterial strains including six B. thermosphacta strains and 20 non-targeted strains frequently associated with B. thermosphacta in seafood products. DNAs were extracted from pure cultures in the appropriate growth conditions (Table 1). The specificity of the three primer sets was verified using the qPCR Cq values obtained with 3 ng of DNA template (Table 3). All the Cq values of the B. thermosphacta strains were lower than 16.76 ± 0.05 (rpoC QSF03-BTH-F/R, strain CD340). No amplification signal was observed for six non-targeted strains when the rpoB-Fw1/Rev1 primers were used. The lowest Cq value for a non-targeted strain was 29.48 ± 0.18 (Staphylococcus epidermidis CIP 68.21 with the rpoC-126-F/R primers); it was 12.72 Cq above the highest value of the targeted strains, and was considered as the unspecific detection threshold. The primers rpoC QSF03-BTH-F/R produced a single melting peak with Tm values of 81 ± 0.50°C while the rpoB-Fw1/Rev1 and the rpoC-126-F/R primers generated a single melting peak with a Tm value of 82 ± 0.50°C. This confirmed the high specificity levels of the three sets of primers. The three primer sets correctly identified all B. thermosphacta strains, including reference and food-isolate strains. As none of the tested sets gave specific amplification (<29.48 Cq) when DNA from non-targeted strains was used, the three sets were considered as species-specific by q-PCR.
TABLE 3.
Primer specificity (Cq values).
| Strains | rpoC-126-F/R |
QSF03-BTH-F/R |
rpoB-Fw1/Rev1 |
|||
| Cq valuesa | Range of Tm | Cq valuesa | Range of Tm | Cq valuesa | Range of Tm | |
| Brochothrix thermosphacta DSM 20171T | 15.32 ± 0.10 | 82–82.5 | 15.45 ± 0.06 | 81 | 14.67 ± 0.07 | 82 |
| Brochothrix thermosphacta 19/R/663 | 14.42 ± 0.33 | 82–82.5 | 14.63 ± 0.05 | 81 | 13.94 ± 0.07 | 82 |
| Brochothrix thermosphacta CD 340 | 16.43 ± 0.09 | 82–82.5 | 16.76 ± 0.05 | 81–81.5 | 16.43 ± 0.16 | 82–82.5 |
| Brochothrix thermosphacta EBP 3084 | 15.26 ± 0.22 | 82–82.5 | 15.44 ± 0.04 | 81 | 14.60 ± 0.18 | 82 |
| Brochothrix thermosphacta TAP 125 | 15.28 ± 0.18 | 82–82.5 | 15.44 ± 0.08 | 81 | 14.62 ± 0.14 | 82 |
| Brochothrix thermosphacta EBP 3017 | 14.82 ± 0.21 | 82–82.5 | 14.93 ± 0.06 | 81 | 14.00 ± 0.22 | 82 |
| Brochothrix campestris 4712T | 30.49 ± 0.23 | 81.5 | 31.03 ± 0.17 | 80.5–81 | 31.52 ± 0.30 | 82 |
| Carnobacterium maltaromaticum 19/R/671 | 34.60 ± 0.48 | 79.5–81.5 | 36.04 ± 1.10 | 80.5–82 | 35.54* | NA |
| Carnobacterium divergens NCDO 2763T | 35.79 ± 0.47 | 78.5–82.5 | 35.43 ± 0.65 | 80–83.5 | NA | NA |
| Enterococcus faecalis ATCC 19433T | 35.15 ± 0.68 | 79–81 | 35.02 ± 1.14 | 80.5–81 | 36.14 ± 1.39 | 81.5 |
| Enterococcus faecium CIP 54.33 | 34.57 ± 0.79 | 80–81 | 34.37 ± 0.67 | 79.5–80.5 | 34.74 ± 1.35 | 80.5–81 |
| Escherichia coli CIP 53.125 | 31.39 ± 0.60 | 87.5 | 36.88 ± 0.62 | 75.5–81 | NA | NA |
| Companilactobacillus alimentarius DSM 20181 | 34.77 ± 1.09 | 73.5–81.5 | 36.14 ± 0.74 | 80.5–81 | 36.02 ± 0.76 | 82 |
| Latilactobacillus sakei subsp. sakei DSM 20017T | 35.59 ± 0.63 | 79.5–81.5 | 36.12 ± 1.19 | 81–83.5 | NA | NA |
| Lactococcus piscium 19/R/684 | 34.56 ± 0.92 | 79.5–82 | 35.46 ± 1.44 | 80.5–81.5 | 35.05 ± 0.84 | 82 |
| Lactococcus raffinolactis Oniris MIP 2453 | 36.22 ± 0.10 | 79.5–80 | NA | NA | NA | NA |
| Listeria monocytogenes DSM 12464T | 35.61 ± 0.66 | 79.5–83 | 34.12 ± 1.04 | 84–84.5 | 36.51 ± 0.99 | 82 |
| Listeria monocytogenes CIP 78.35 | 35.37 ± 0.49 | 79.5–81.5 | 33.39 ± 1.20 | 84 | 35.53* | NA |
| Listeria grayi CIP 68.18T | 365.09 ± 0.22 | 81.5 | 35.01 ± 1.26 | 80.5–81 | 36.50 ± 0.46 | 82 |
| Listeria innocua CIP 80.11T | 35.71 ± 0.62 | 79.5–83 | 36.32* | 81 | 37.07* | 76 |
| Photobacterium phosphoreum CIP 102511T | 34.59 ± 1.05 | 81.5–83 | 35.31 ± 1.48 | 80.5–81 | 36.47 ± 0.79 | 82 |
| Pseudomonas fluorescens CIP 69.13T | 34.35 ± 0.50 | 91 | NA | NA | NA | NA |
| Psychrobacter sp. 19/R/675 | 35.09 ± 0.40 | 80.5–82.5 | NA | NA | NA | NA |
| Serratia liquefaciens ATCC 27592T | 34.91 ± 0.30 | 82.5 | NA | NA | NA | NA |
| Shewanella baltica 19/R/670 | 32.55 ± 0.38 | 82–87 | NA | NA | NA | NA |
| Staphylococcus epidermidis CIP 68.21 | 29.48 ± 0.18 | 81.5–82 | 29.74 ± 0.22 | 80.5–81 | 29.98 ± 0.25 | 82 |
| NTC | 34.87 ± 1.99 | 72–83 | 38.58* | 76.50 | NA | NA |
aMean and standard deviation of three Cq values (obtained with 3 ng of DNA template). NTC, no template control; NA, no amplification. *no standard deviation calculated (only one Cq available).
qPCR Efficiency Using DNA Extracted From BHI Cultures
The qPCR efficiency of the three primer sets tested on BHI culture extracts and on diluted DNA extracts was calculated from the standard curves plotting the mean Cq values (three replicates) versus log CFU/mL (Table 4). The linear coefficient of determination (r2) ranged from 0.990 (BHI culture extracts; rpoB-Fw1/Rev1) to 1 (diluted DNA extracts; rpoC-126-F/R). Using the primers rpoC-126-F/R (designed for this study), the efficiency levels were 97.45 and 92.58% for diluted DNA extracts and BHI culture extracts, respectively. Using the two other primer set, efficiency was lower and ranged from 89.46 to 92.80%.
TABLE 4.
qPCR efficiency.
| rpoC-126-F/R |
QSF03-BTH-F/R |
rpoB-Fw1/Rev1 |
|||||||
| Equationa y = | Efficiency (%) | r2 | Equationa y = | Efficiency (%) | r2 | Equationa y = | Efficiency (%) | r2 | |
| BHI culture extracts | −3.5136x + 40.558 | 92.58 | 0.997 | −3.5898x + 41.019 | 89.92 | 0.996 | −3.5419x + 42.158 | 91.57 | 0.990 |
| Diluted DNA extracts | −3.3845 + 40.947 | 97.45 | 1 | −3.5074x + 41.542 | 92.80 | 0.998 | −3.6035x + 41.882 | 89.46 | 0.995 |
| Smoked salmon tissue homogenate | −3.2408x + 44.396 | 103.50 | 0.998 | Not determined | −3.5789 + 46.805 | 90.29 | 0.998 | ||
aequations derived from 3 replicates.
qPCR Efficiency and Limit of Detection Using DNA Extracted From Smoked Salmon Tissue Homogenates
The rpoC-126-F/R and rpoB-Fw1/Rev1 primer sets, showing the highest qPCR efficiency on BHI culture extracts, were chosen to evaluate efficiency on DNA extracted from SSTHf. DNA was extracted from SSTHf inoculated with B. thermosphacta DSM 20171 from 7 to 4 log CFU/mL (SSTHf extract). The efficiency of the qPCR amplification using rpoC-126-F/R and rpoB-Fw1/Rev1 (Table 4) was calculated from standard curves (data not shown) generated by plotting the mean Cq values from three replicates vs. log CFU/mL. When the rpoC-126-F/R primers (designed in this study) were used, an efficiency of 103.50% was obtained. Lower efficiency (90.29%) was calculated with the rpoB-Fw1/Rev1 primers. The linear coefficient of determination (r2) was 0.998 with both primer sets.
The limit of detection (LOD) was calculated using a Probit model (Kralik and Ricchi, 2017), only based on SSTHf. It was established at 3.34 log CFU/mL (2.20 × 103 CFU/mL) for the rpoC-126-F/R primers, corresponding to a load of 4.04 log CFU/g of salmon (1.10 × 104 CFU/g), with a 95% probability to detect the gene. For the rpoB-Fw1/Rev1 primers, the LOD was 3.15 log CFU/mL (1.41 × 103 CFU/mL), corresponding to a load of 3.85 log CFU/g of salmon (7.06 × 103 CFU/g).
PMA-qPCR Quantification of Viable B. thermosphacta in BHI Culture
To test the efficiency of the PMA and PMAxx treatments, viable or dead pure-culture B. thermosphacta cells were 10-fold serially diluted in BHI (7 to 3 log CFU/mL), and their DNA was extracted after PMA or PMAxx treatment. The qPCR assays using the three primer sets were used to quantify viable B. thermosphacta. The results were expressed in delta-Cq (i.e., differences in Cq values between PMA/PMAxx-treated cells/untreated dead cells on the one hand, and Cq values of untreated 100% viable cells on the other hand).
When viable cells were treated with PMA or PMAxx, the delta Cq values were low with the three primer sets (Supplementary Data 1). The minimum and maximum delta-Cq values were obtained with rpoB-Fw1/Rev1 (0.01 ± 0.46 vs. 1.34 ± 0.62 for 4 log CFU/mL of viable PMA-treated cells and 5 log CFU/mL of viable PMAxx-treated cells, respectively). These low delta-Cq values indicated no significant difference in the detection and quantification of B. thermosphacta between viable untreated and dye-treated cells. These observations were statistically in accordance with Wilcoxon’s test (p > 0.4). Moreover, there was no significant difference between the delta-Cq values following treatment with the two PMA and PMAxx dyes (p > 0.16) or between the three primer sets (p > 0.18).
When dead cells were used, delta Cq values were higher with PMA/PMAxx-treated cells than with untreated cells. The minimum delta-Cq values were obtained for the 6 log CFU/mL of dead PMA-treated cells with rpoC-126-F/R and rpoC QSF03-BTH-F/R (5.92 ± 0.82 and 5.92 ± 0.83, respectively). The maximum delta-Cq value (8.57 ± 0.61) was obtained with rpoC QSF03-BTH-F/R for the 7 log CFU/mL of dead PMAxx-treated cells. Interestingly, we failed to calculate delta-Cq values for any cell concentration range when rpoB-Fw1/Rev1 was used with PMA/PMAxx-treated cells, indicating that the Cq values were either beyond the unspecific detection threshold (i.e., 29.48) or even none amplification signal was detected. Thus, there was no significant difference between the two rpoC primer sets, while there was a significant difference between the rpoC primer sets and the rpoB primer set (p < 0.005). Finally, there was no significant difference in delta-Cq values between the two PMA/PMAxx dyes (p > 0.16), similarly to viable cells.
Because the delta-Cq values obtained with the 2 sets of rpoC primers were similar and the efficiency of the rpoC-QSF03-BTH-F/R primers was lower, we decided to choose rpoC-126-F/R and rpoB-Fw1/Rev1 for further B. thermosphacta PMA-qPCR quantification in SSTH.
PMA-qPCR Quantification of Viable B. thermosphacta in Smoked Salmon Tissue Homogenates (SSTHf)
We used the rpoC-126-F/R and rpoB-Fw1/Rev1 primer sets to quantify viable B. thermosphacta in SSTHf by qPCR. Suspensions of B. thermosphacta viable or dead cells in SSTHf were 10-fold serially diluted in SSTHf (7 to 4 log CFU/mL), and their DNA was extracted after PMA or PMAxx treatment.
When viable cells were treated with PMA or PMAxx, delta Cq values were low with the two primer sets (Supplementary Data 1), and the highest value was obtained for 5 log CFU/mL of viable PMA-treated cells with the rpoC-126-F/R primer set (1.20 ± 0.41). The delta-Cq values of viable PMA/PMAxx-treated cells were not significantly different from those of untreated viable cells (p > 0.05). Moreover, as previously shown in BHI broth, there was no significant difference in delta-Cq values between the two PMA/PMAxx dyes (p > 0.28) or between the two primer sets (p > 0.36). Interestingly, when dead cells were tested, none of the primer sets allowed determining delta-Cq on PMA- or PMAxx- treated cells because Cq values were beyond the detection limit. Therefore, no quantification was possible after dye treatment. When dead cells were not treated with the dyes, the difference between the Cq values of untreated dead cells vs. untreated viable cells was not significantly different (p > 0.1), even if the delta Cq ranged from 1.68 ± 0.77 to 2.66 ± 1.32.
Quantification of Viable B. thermosphacta in Cold-Smoked Salmon Fillet Sampled After the Smoking Step (SSTHs)
The PMA and PMAxx treatments were tested on SSTHs in the presence of mixes of dead and viable B. thermosphacta cell suspensions. The ability of PMA/PMAxx-qPCR to discriminate viable cells from dead cells in the mixes was evaluated by establishing the relationship between Cq values obtained by qPCR (after PMA/PMAxx treatments or without treatment) and bacterial counts of untreated cells on STAA agar. The results are presented in Figures 1A,B and in Supplementary Data 2. ANOVA and the Tukey’s multiple comparison of means were used to analyze the results. The significance level was set for p-values <0.05.
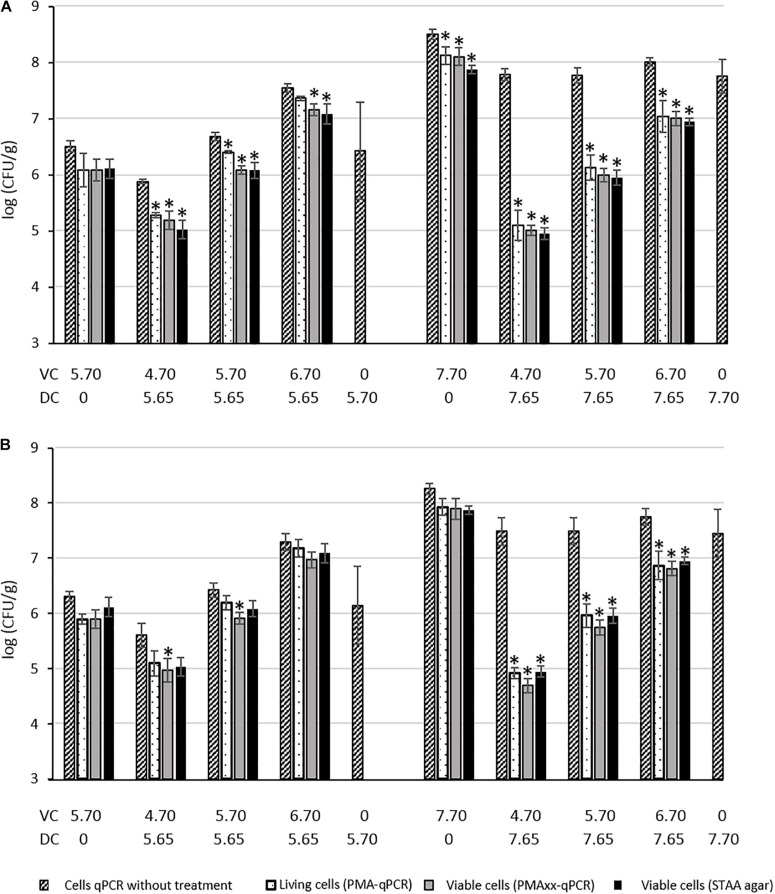
FIGURE 1.
Quantification of dead (DC) and viable (VC) B. thermosphacta cells mixed in smoked salmon tissue homogenate (SSTHs) with plating enumeration on STAA medium, and PMA or PMAxx qPCR using the rpoC-126F/R (A) or rpoB-Fw1/Rev1 (B) primer sets. Results expressed in log CFU/g. *significant difference between quantification results of untreated cells obtained by qPCR method and treated cells quantified by qPCR method or plating method.
For both primer sets, the quantification of 5.70 log CFU/g of PMA/PMAxx-treated viable cells was consistent with the quantification of untreated viable cells, with a non-significant difference (p > 0.05). Likewise, non-significant difference was observed between the quantification of 7.70 log CFU/g of PMA/PMAxx-treated viable cells and the untreated viable cells with rpoB-Fw1/Rev1 but a significant difference was observed with the rpoC-126-F/R primers. As previously shown in SSTHf on 5.70 or 7.70 log CFU/g of dead cells, quantification was only possible with untreated cells, whereas no quantification was possible once the cells were treated with PMA or PMAxx, suggesting that PMA and PMAxx inhibit dead cell DNA amplification.
In the mixes containing 5.65 log CFU/g of dead cells and 4.70 or 5.70 log CFU/g of viable cells, no significant difference was highlighted between PMA-treated and untreated cells using rpoB-Fw1/Rev1 but a significant difference was highlighted between PMAxx-treated and untreated cells. When rpoC-126-F/R was used, a significant difference was observed between PMA/PMAxx-treated and untreated cells in those mixes. No treatment-related difference was noticed in the mixes containing 5.65 log CFU/g of dead cells and 6.70 log CFU/g of viable cells with rpoB-Fw1/Rev1 whereas a significant difference was observed between PMAxx-treated and untreated cells with rpoC-126-F/R. In the mixes containing 7.65 log CFU/g of dead cells and 4.70, 5.70, or 6.70 log CFU/g of viable cells, the efficiency of PMA/PMAxx was more obvious: differences between treated and untreated cells were significant with both primer sets, and viable cells were preferentially amplified and quantified. This shows again the good ability of PMA and PMAxx to inhibit DNA amplification from dead cells.
Discussion
The detection and quantification of unviable cells is admittedly a question of molecular methods targeting the cell DNA directly without culturing steps. This requires methods that discriminate viable cells from unviable cells in biological samples such as food products (Kumar and Ghosh, 2019). In the field of food spoilage, B. thermosphacta is considered as one of the main bacteria, able to spoil many meat and seafood products whatever the packaging (i.e., under air, vacuum, or modified atmosphere) (Illikoud et al., 2018a). B. thermosphacta is even considered as ubiquitous along the food chain because it was also isolated from food factory surfaces (Nychas et al., 2008). Thus, it can be subjected to the environmental and food processing stresses likely to lead to unviable state, making it difficult to detect and quantify it from food products.
We developed an assay aimed at quantifying viable B. thermosphacta using viability dyes (PMA or PMAxx) and the targeting of a single-copy gene (rpoC or rpoB). After designing the primers in silico, we determined their specificity and compared it with that of two other single-copy gene primer sets from previous studies specifically designed to identify B. thermosphacta (Fougy et al., 2016; Illikoud et al., 2019). Using the real-time qPCR inclusivity and exclusivity tests, including DNA extracted and purified from 6 B. thermosphacta strains and 20 non-targeted closely related strains such as B. campestris, we demonstrated the specificity of the three primer sets, without misdetection or misquantification. The difference between targeted and untargeted strains was at least 12.72 Cq. The efficiency of the qPCR assays with the three primer sets was then assessed from the standard curves [Cq = f(CFU (log/mL))]. The most efficient primer set to quantify B. thermosphacta from diluted DNA extracts and BHI culture extracts was rpoC-126-F/R, with satisfactory efficiency levels of 97.45% (r2 = 1) and 92.58% (r2 = 0.997), respectively.
We further investigated the efficiency of PMA and PMAxx treatments to quantify viable B. thermosphacta cells in BHI medium. As described in many previous studies (Zeng et al., 2016), our results showed that a viable dye treatment was clearly relevant to discriminate viable from unviable bacterial cells. The delta-Cq for dead B. thermosphacta cells was higher when PMA or PMAxx treatments were applied than it was for untreated cells, with a difference of at least 3.48 in the delta-Cq values between PMA-treated cells and untreated cells at 6 log CFU/mL when rpoC QSF03-BTH-F/R was used. These results confirm that the number of amplification cycles is higher after dye treatment when it comes to detecting and quantifying dead B. thermosphacta cells rather than viable cells because the amount of amplifiable target DNA is very low when dead cells are treated with dyes. The most important effect of the two dye treatments was also obvious with the rpoB primer set: no delta-Cq values were calculable from dead treated cells, meaning that no amplification was possible in these conditions. In the absence of dye treatment, amplification was still detectable up to 4 log CFU/mL, with a low associated delta-Cq value (1.76 ± 0.70).
Regarding the two rpoC primer sets, they gave quite similar results on viable and dead cells. However, whatever the primer set, there was no significant difference between the PMA and PMAxx treatments, with similar delta-Cq values for the two conditions. Because the delta-Cq values obtained with both rpoC primer sets were quite similar and rpoC-126F/R was slightly more efficient than rpoC-QSF03-BTH-F/R was, we only kept rpoC-126F/R for the further tests on SSTH.
The good efficiency of rpoC-126F/R was confirmed on smoked salmon tissue homogenate (SSTHf), with 103.50% (r2 = 0.998), while rpoB-Fw1/Rev1 showed 90.29% (r2 = 0.998) efficiency. We also determined the limit of detection (LOD) of the qPCR assays on SSTHf, but not on BHI, to be closest to a real food product. The LODs were established at 4.04 and 3.85 log CFU/g of smoked salmon for rpoC-126F/R and rpoB-Fw1/Rev1, respectively. We considered these LODs as relevant according to the food spoilage behavior of B. thermosphacta. In most of the reported cases of food spoilage, the amount of B. thermosphacta in the spoiled foods was above 7 log CFU/g when spoilage was perceived by sensory analysis panelists (Illikoud et al., 2018a). With our rpoC or rpoB qPCR assays, we detected the presence of B. thermosphacta around 3 log before spoilage perception. This could give smoked salmon producers the opportunity to redirect their matrices before spoilage by B. thermosphacta to other transformation processes with a strong stabilization step – e.g., pasteurization – to kill the spoilage agent and limit food losses. The results of the qPCR assays after PMA or PMAxx treatment of viable smoked salmon tissue homogenate were similar to those of viable cells cultured in BHI, with low delta-Cq values and no significant difference between the primer sets. Nevertheless, the effect of the dye treatments on dead cells was more obvious than it was on BHI-cultured cells because no delta-Cq value was calculable on treated dead cells for either primer set. Therefore, no amplification was possible with these primer sets after dye treatment of dead cells. By contrast, amplification was still detectable down to 4 log CFU/mL in untreated cells, with a low associated delta-Cq value (1.74 ± 0.52 and 2.66 ± 1.32 for the rpoC and rpoB primer sets, respectively).
The efficiency of PMA/PMAxx-qPCR to discriminate viable B. thermosphacta cells from dead cells was also confirmed on industrially processed cold-smoked salmon fillets after the smoking step (SSTHs). The DNA amplification of 5.70 or 7.70 log CFU/g of B. thermosphacta dead cells inoculated on SSTHs was inhibited by PMA and PMAxx, whereas qPCR amplification was consistently detectable in the absence of dye treatment. This efficiency was even more obvious in mixed viable/dead-cell conditions, with significantly different quantifications between the treated and the untreated cells with both primer sets. However, we did not find any significant difference between the two primer sets, except for the mixes containing 5.65 log CFU/g of dead cells + 4.70 or 5.70 log CFU/g of PMA/PMAxx-treated viable cells and 5.65 log CFU/g of dead cells + 6.70 log CFU/g of PMAxx-treated viable cells where rpoC-126F/R better discriminated between treated and untreated cells. Moreover, we did not find a better PCR inhibition by PMA or PMAxx with the longer rpoB amplicon (394 bp), as previously described by Martin et al. (2013), compared with the shorter rpoC amplicons (126 or 151 bp). Consequently, our results suggest using rpoC-126-F/R coupled with PMA or PMAxx treatment to specifically quantify B. thermosphacta with good efficiency and reliable discrimination between viable and dead cells in cold-smoked salmon samples.
In the last two decades, many studies have described the interest of molecular methods such as real-time quantitative PCR to detect and quantify a lot of bacterial species in food or other ecosystems instead of the classical culture methods (Postollec et al., 2011; Franco-Duarte et al., 2019), but also the usefulness of viability dyes such as EMA or PMA to discriminate viable cells from unviable or dead cells (Elizaquível et al., 2014; Zeng et al., 2016). Regarding B. thermosphacta, only one study demonstrates the interest of PMA coupled with 16S rDNA-qPCR to detect and quantify this spoilage bacterium in cooked peeled shrimp (Mamlouk et al., 2012). These authors found a better LOD than ours (2.07 log CFU/g of cooked shrimp) with a similar efficiency of 105.3% (r2 = 0.999). Nevertheless, their LOD was not determined from the food matrix, as it is in the present study, but from purified genomic B. thermosphacta DNA serially diluted 10-fold in sterile saline solution and subjected to real-time PCR amplification. Consequently, these conditions reduce the potential presence of PCR inhibitors likely present in food matrices and able to influence on qPCR efficiency, hence a shift of the LOD (Sidstedt et al., 2020).
The new viability dye PMAxx was developed recently. It improves the discrimination between viable and unviable cells (Randazzo et al., 2016). Few studies have been conducted with this new dye so far (Truchado et al., 2020) used it to detect and quantify VBNC L. monocytogenes cells in wash water from processed fruit and vegetables. It has also been used to quantify L. monocytogenes in chocolate liquor, corn flakes, and dry-roasted, shelled pistachios (Ly et al., 2020), quantify viable and non-viable Salmonella from a poultry environment (Zhang et al., 2020), quantify viable Campylobacter in raw milk (Wulsten et al., 2020), detect Clavibacter michiganensis in a viable but non-culturable state in tomato (Han et al., 2018), discriminate between viable and membrane-damaged cells of the plant pathogen Xylella fastidiosa (Sicard et al., 2019) or quantify VBNC Vibrio parahaemolyticus in raw shrimp (Cao et al., 2019). Among all these studies, three of them have compared this new dye with other available dyes, notably EMA or PMA. Two of them have demonstrated a higher suitability of PMAxx than PMA to quantify bacteria in food matrices (Han et al., 2018; Cao et al., 2019). By contrast, another study found PMA more suitable than EMA and PMAxx to detect and quantify viable bacterial cells (Wulsten et al., 2020). Other authors even reported the use of a combination of EMA and PMAxx to reduce the DNA signal from dead cells (with intact and damaged membranes) and viable cells with inactive membranes (Truchado et al., 2020). The use of the EMA-PMAxx combination yielded a more accurate estimation of viable L. monocytogenes cells. Our results did not allow us to clearly distinguish between PMAxx and PMA in their efficiency to discriminate viable B. thermosphacta cells from unviable ones in cold-smoked salmon: they gave similar results in most of the tested conditions.
In conclusion, we developed a rapid, specific and efficient rpoC PMA/PMAxx-qPCR method to detect and quantify B. thermosphacta in pure culture and in cold-smoked salmon samples, with efficient discrimination between viable and dead cells. This qPCR method specifically and quickly enumerates B. thermosphacta (within 3–4 h compared with 48 h for the STAA culture method). Moreover, this PMA/PMAxx-qPCR could be a relevant tool for smoked salmon producers to (i) quickly detect and quantify B. thermosphacta in their products early enough before spoilage is perceived and (ii) limit food losses. Finally, this rpoC PMA/PMAxx-qPCR method could be used to detect and quantify B. thermosphacta in other seafood products and even in other food matrices such as meat products.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author Contributions
AB-A, EJ, HP, and XD conceived and designed the experiments. AB-A contributed to reagents, materials, and analysis tools. AB-A and SS performed the experiments. AB-A, EJ, and HP wrote the manuscript. All authors analyzed the data, contributed to the article, and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thankful to Vincent Gelamur and Julie Courtel from MerAlliance, Thai Union company, for the smoked salmon samples that were kindly provided. We thank the “Pays de la Loire” Region for financial support (Project Number n 2017-06930).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.654178/full#supplementary-material
References
- Cao X., Zhao L., Zhang J., Chen X., Shi L., Fang X., et al. (2019). Detection of viable but nonculturable Vibrio parahaemolyticus in shrimp samples using improved real-time PCR and real-time LAMP methods. Food Control 103 145–152. 10.1016/j.foodcont.2019.04.003 [DOI] [Google Scholar]
- Casaburi A., De Filippis F., Villani F., Ercolini D. (2014). Activities of strains of Brochothrix thermosphacta in vitro and in meat. Food Res. Int. 62 366–374. 10.1016/j.foodres.2014.03.019 [DOI] [Google Scholar]
- Case R. J., Boucher Y., Dahllof I., Holmstrom C., Doolittle W. F., Kjelleberg S. (2007). Use of 16S rRNA and rpoB Genes as Molecular Markers for Microbial Ecology Studies. Appl. Environ. Microbiol. 73 278–288. 10.1128/AEM.01177-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro A. G. S. A., Dorneles E. M. S., Santos E. L. S., Alves T. M., Silva G. R., Figueiredo T. C., et al. (2018). Viability of Campylobacter spp. in frozen and chilled broiler carcasses according to real-time PCR with propidium monoazide pretreatment. Poultry Sci. 97 1706–1711. 10.3382/ps/pey020 [DOI] [PubMed] [Google Scholar]
- Chaillou S., Chaulot-Talmon A., Caekebeke H., Cardinal M., Christieans S., Denis C., et al. (2015). Origin and ecological selection of core and food-specific bacterial communities associated with meat and seafood spoilage. ISME J. 9, 1105–1118. 10.1038/ismej.2014.202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dainty R. H., Mackey B. M. (1992). The relationship between the phenotypic properties of bacteria from chill-stored meat and spoilage processes. J. Appl. Bacteriol. 73 103s–114s. 10.1111/j.1365-2672.1992.tb03630.x [DOI] [PubMed] [Google Scholar]
- Desneux J., Chemaly M., Pourcher A.-M. (2015). Experimental design for the optimization of propidium monoazide treatment to quantify viable and non-viable bacteria in piggery effluents. BMC Microbiol. 15:505–506. 10.1186/s12866-015-0505-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong L., Liu H., Meng L., Xing M., Wang J., Wang C., et al. (2018). Quantitative PCR coupled with sodium dodecyl sulfate and propidium monoazide for detection of viable Staphylococcus aureus in milk. J. Dairy Sci. 101 4936–4943. 10.3168/jds.2017-14087 [DOI] [PubMed] [Google Scholar]
- Elizaquível P., Aznar R., Sánchez G. (2014). Recent developments in the use of viability dyes and quantitative PCR in the food microbiology field. J. Appl. Microbiol. 116 1–13. 10.1111/jam.12365 [DOI] [PubMed] [Google Scholar]
- Elizaquível P., Sánchez G., Selma M. V., Aznar R. (2012). Application of propidium monoazide-qPCR to evaluate the ultrasonic inactivation of Escherichia coli O157:H7 in fresh-cut vegetable wash water. Food Microbiol. 30 316–320. 10.1016/j.fm.2011.10.008 [DOI] [PubMed] [Google Scholar]
- Fang J., Wu Y., Qu D., Ma B., Yu X., Zhang M., et al. (2018). Propidium monoazide real-time loop-mediated isothermal amplification for specific visualization of viable Salmonella in food. Lett. Appl. Microbiol. 67 79–88. 10.1111/lam.12992 [DOI] [PubMed] [Google Scholar]
- Fougy L., Desmonts M.-H., Coeuret G., Fassel C., Hamon E., Hézard B., et al. (2016). Reducing Salt in Raw Pork Sausages Increases Spoilage and Correlates with Reduced Bacterial Diversity. Appl. Environ. Microbiol. 82 3928–3939. 10.1128/AEM.00323-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco-Duarte R., Černáková L., Kadam S., Kaushik K., Salehi B., Bevilacqua A., et al. (2019). Advances in Chemical and Biological Methods to Identify Microorganisms—From Past to Present. Microorganisms 7:microorganisms7050130. 10.3390/microorganisms7050130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furet J.-P., Firmesse O., Gourmelon M., Bridonneau C., Tap J., Mondot S., et al. (2009). Comparative assessment of human and farm animal faecal microbiota using real-time quantitative PCR. FEMS Microbiol. Ecol. 68 351–362. 10.1111/j.1574-6941.2009.00671.x [DOI] [PubMed] [Google Scholar]
- Hall T. A. (1999). BioEdit: A User-Friendly Biological Sequence Alignment Editor and Analysis Program for Windows 95/98/NT. Nucleic Acids Symposium Ser. 41 95–98. 10.14601/Phytopathol_Mediterr-14998u1.29 [DOI] [Google Scholar]
- Han S., Jiang N., Lv Q., Kan Y., Hao J., Li J., et al. (2018). Detection of Clavibacter michiganensis subsp. michiganensis in viable but nonculturable state from tomato seed using improved qPCR. PLoS One 13:e0196525. 10.1371/journal.pone.0196525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illikoud N., Jaffrès E., Zagorec M. (2018a). “Brochothrix thermosphacta,” in Reference Module in Life Sciences, ed. Roitberg B. D. (Amsterdam: Elsevier; ), 10.1016/B978-0-12-809633-8.12106-5 [DOI] [Google Scholar]
- Illikoud N., Klopp C., Roulet A., Bouchez O., Marsaud N., Jaffrès E., et al. (2018b). One complete and three draft genome sequences of four Brochothrix thermosphacta strains, CD 337, TAP 175, BSAS1 3 and EBP 3070. Standards Genomic Sci. 13:0333–z. 10.1186/s40793-018-0333-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illikoud N., Rossero A., Chauvet R., Courcoux P., Pilet M.-F., Charrier T., et al. (2019). Genotypic and phenotypic characterization of the food spoilage bacterium Brochothrix thermosphacta. Food Microbiol. 81 22–31. 10.1016/j.fm.2018.01.015 [DOI] [PubMed] [Google Scholar]
- Jaffrès E., Lalanne V., Macé S., Cornet J., Cardinal M., Sérot T., et al. (2011). Sensory characteristics of spoilage and volatile compounds associated with bacteria isolated from cooked and peeled tropical shrimps using SPME–GC–MS analysis. Int. J. Food Microbiol. 147 195–202. 10.1016/j.ijfoodmicro.2011.04.008 [DOI] [PubMed] [Google Scholar]
- Jaffrès E., Sohier D., Leroi F., Pilet M. F., Prévost H., Joffraud J. J., et al. (2009). Study of the bacterial ecosystem in tropical cooked and peeled shrimps using a polyphasic approach. Int. J. Food Microbiol. 131, 20–29. 10.1016/j.ijfoodmicro.2008.05.017 [DOI] [PubMed] [Google Scholar]
- Joffraud J. J., Leroi F., Roy C., Berdagué J. L. (2001). Characterisation of volatile compounds produced by bacteria isolated from the spoilage flora of cold-smoked salmon. Int. J. Food Microbiol. 66 175–184. 10.1016/S0168-1605(00)00532-8 [DOI] [PubMed] [Google Scholar]
- Joffraud J.-J., Cardinal M., Cornet J., Chasles J.-S., Léon S., Gigout F., et al. (2006). Effect of bacterial interactions on the spoilage of cold-smoked salmon. Int. J. Food Microbiol. 112 51–61. 10.1016/j.ijfoodmicro.2006.05.014 [DOI] [PubMed] [Google Scholar]
- Josefsen M. H., Lofstrom C., Hansen T. B., Christensen L. S., Olsen J. E., Hoorfar J. (2010). Rapid Quantification of Viable Campylobacter Bacteria on Chicken Carcasses, Using Real-Time PCR and Propidium Monoazide Treatment, as a Tool for Quantitative Risk Assessment. Appl. Environ. Microbiol. 76 5097–5104. 10.1128/AEM.00411-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jullien N. (2019). Aix-Marseille Univ, CNRS. Marseille: INP, Inst Neurophysiopathol. [Google Scholar]
- Kralik P., Ricchi M. (2017). A Basic Guide to Real Time PCR in Microbial Diagnostics: Definitions, Parameters, and Everything. Front. Microbiol. 8:00108. 10.3389/fmicb.2017.00108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreader C. A. (1996). Relief of amplification inhibition in PCR with bovine serum albumin or T4 gene 32 protein. Appl. Environ. Microbiol. 62 1102–1106. 10.1128/AEM.62.3.1102-1106.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchta T., Knutsson R., Fiore A., Kudirkiene E., Höhl A., Horvatek Tomic D., et al. (2014). A decade with nucleic acid-based microbiological methods in safety control of foods. Lett. Appl. Microbiol. 59 263–271. 10.1111/lam.12283 [DOI] [PubMed] [Google Scholar]
- Kumar S. S., Ghosh A. R. (2019). Assessment of bacterial viability: a comprehensive review on recent advances and challenges. Microbiology 165 593–610. 10.1099/mic.0.000786 [DOI] [PubMed] [Google Scholar]
- Laursen B. G., Leisner J. J., Dalgaard P. (2006). Carnobacterium Species: Effect of Metabolic Activity and Interaction with Brochothrix thermosphacta on Sensory Characteristics of Modified Atmosphere Packed Shrimp. J. Agricult. Food Chem. 54 3604–3611. 10.1021/jf053017f [DOI] [PubMed] [Google Scholar]
- Liang N., Dong J., Luo L., Li Y. (2011). Detection of Viable Salmonella in Lettuce by Propidium Monoazide Real-Time PCR. J. Food Sci. 76 M234–M237. 10.1111/j.1750-3841.2011.02123.x [DOI] [PubMed] [Google Scholar]
- Ly V., Parreira V. R., Sanchez-Maldonado A. F., Farber J. M. (2020). Survival and Virulence of Listeria monocytogenes during Storage on Chocolate Liquor, Corn Flakes, and Dry-Roasted Shelled Pistachios at 4 and 23°C. J. Food Prot. 83 1852–1862. 10.4315/JFP-20-129 [DOI] [PubMed] [Google Scholar]
- Macé S., Mamlouk K., Chipchakova S., Prévost H., Joffraud J.-J., Dalgaard P., et al. (2013). Development of a Rapid Real-Time PCR Method as a Tool To Quantify Viable Photobacterium phosphoreum Bacteria in Salmon (Salmo salar) Steaks. Appl. Environ. Microbiol. 79 2612–2619. 10.1128/AEM.03677-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamlouk K., Macé S., Guilbaud M., Jaffrès E., Ferchichi M., Prévost H., et al. (2012). Quantification of viable Brochothrix thermosphacta in cooked shrimp and salmon by real-time PCR. Food Microbiol. 30 173–179. 10.1016/j.fm.2011.09.012 [DOI] [PubMed] [Google Scholar]
- Martin B., Raurich S., Garriga M., Aymerich T. (2013). Effect of Amplicon Length in Propidium Monoazide Quantitative PCR for the Enumeration of Viable Cells of Salmonella in Cooked Ham. Food Anal. Methods 6 683–690. 10.1007/s12161-012-9460-0 [DOI] [Google Scholar]
- Mejlholm O., Bøknaes N., Dalgaard P. (2005). Shelf life and safety aspects of chilled cooked and peeled shrimps (Pandalus borealis) in modified atmosphere packaging. J. Appl. Microbiol. 99 66–76. 10.1111/j.1365-2672.2005.02582.x [DOI] [PubMed] [Google Scholar]
- Niu B., Hong B., Zhang Z., Mu L., Malakar P. K., Liu H., et al. (2018). A Novel qPCR Method for Simultaneous Detection and Quantification of Viable Pathogenic and Non-pathogenic Vibrio parahaemolyticus (tlh+, tdh+, and ureR+). Front. Microbiol. 9:01747. 10.3389/fmicb.2018.01747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nychas G.-J. E., Skandamis P. N., Tassou C. C., Koutsoumanis K. P. (2008). Meat spoilage during distribution. Meat Sci. 78 77–89. 10.1016/j.meatsci.2007.06.020 [DOI] [PubMed] [Google Scholar]
- Pan Y., Breidt F. (2007). Enumeration of Viable Listeria monocytogenes Cells by Real-Time PCR with Propidium Monoazide and Ethidium Monoazide in the Presence of Dead Cells. Appl. Environ. Microbiol. 73 8028–8031. 10.1128/AEM.01198-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paoli G. C., Wijey C., Nguyen L.-H., Chen C.-Y., Yan X., Irwin P. L. (2017). Complete Genome Sequences of Two Strains of the Meat Spoilage Bacterium Brochothrix thermosphacta Isolated from Ground Chicken. Genome Announcem. 5 1357–1317. 10.1128/genomeA.01357-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennacchia C., Ercolini D., Villani F. (2009). Development of a Real-Time PCR assay for the specific detection of Brochothrix thermosphacta in fresh and spoiled raw meat. Int. J. Food Microbiol. 134 230–236. 10.1016/j.ijfoodmicro.2009.07.005 [DOI] [PubMed] [Google Scholar]
- Postollec F., Falentin H., Pavan S., Combrisson J., Sohier D. (2011). Recent advances in quantitative PCR (qPCR) applications in food microbiology. Food Microbiol. 28 848–861. 10.1016/j.fm.2011.02.008 [DOI] [PubMed] [Google Scholar]
- Randazzo W., López-Gálvez F., Allende A., Aznar R., Sánchez G. (2016). Evaluation of viability PCR performance for assessing norovirus infectivity in fresh-cut vegetables and irrigation water. Int. J. Food Microbiol. 229 1–6. 10.1016/j.ijfoodmicro.2016.04.010 [DOI] [PubMed] [Google Scholar]
- Remenant B., Jaffrès E., Dousset X., Pilet M.-F., Zagorec M. (2015). Bacterial spoilers of food: Behavior, fitness and functional properties. Food Microbiol. 45 45–53. 10.1016/j.fm.2014.03.009 [DOI] [PubMed] [Google Scholar]
- Schrader C., Schielke A., Ellerbroek L., Johne R. (2012). PCR inhibitors - occurrence, properties and removal. J. Appl. Microbiol. 113 1014–1026. 10.1111/j.1365-2672.2012.05384.x [DOI] [PubMed] [Google Scholar]
- Sicard A., Merfa M. V., Voeltz M., Zeilinger A. R., De La Fuente L., Almeida R. P. P. (2019). Discriminating between viable and membrane-damaged cells of the plant pathogen Xylella fastidiosa. PLoS One 14:e0221119. 10.1371/journal.pone.0221119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidstedt M., Rådström P., Hedman J. (2020). PCR inhibition in qPCR, dPCR and MPS-mechanisms and solutions. Anal. Bioanal. Chem. 412 2009–2023. 10.1007/s00216-020-02490-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stackebrandt E., Jones D. (2006). “The Genus Brochothrix,” in The Prokaryotes, eds Dworkin M., Falkow S., Rosenberg E., Schleifer K.-H., Stackebrandt E. (New York, NY: Springer; ), 477–491. 10.1007/0-387-30744-3_12 [DOI] [Google Scholar]
- Stohr V., Joffraud J. J., Cardinal M., Leroi F. (2001). Spoilage potential and sensory profile associated with bacteria isolated from cold-smoked salmon. Food Res. Int. 34 797–806. 10.1016/S0963-9969(01)00101-6 [DOI] [Google Scholar]
- Truchado P., Gil M. I., Larrosa M., Allende A. (2020). Detection and Quantification Methods for Viable but Non-culturable (VBNC) Cells in Process Wash Water of Fresh-Cut Produce: Industrial Validation. Front. Microbiol. 11:673. 10.3389/fmicb.2020.00673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wulsten I. F., Galeev A., Stingl K. (2020). Underestimated Survival of Campylobacter in Raw Milk Highlighted by Viability Real-Time PCR and Growth Recovery. Front. Microbiol. 11:1107. 10.3389/fmicb.2020.01107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J., Coulouris G., Zaretskaya I., Cutcutache I., Rozen S., Madden T. L. (2012). Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformat. 13:134. 10.1186/1471-2105-13-134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng D., Chen Z., Jiang Y., Xue F., Li B. (2016). Advances and Challenges in Viability Detection of Foodborne Pathogens. Front. Microbiol. 7:1833. 10.3389/fmicb.2016.01833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Khan S., Chousalkar K. K. (2020). Development of PMAxxTM-Based qPCR for the Quantification of Viable and Non-viable Load of Salmonella From Poultry Environment. Front. Microbiol. 11:581201. 10.3389/fmicb.2020.581201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Liu W., Xu H., Aguilar Z. P., Shah N. P., Wei H. (2015). Propidium monoazide combined with real-time PCR for selective detection of viable Staphylococcus aureus in milk powder and meat products. J. Dairy Sci. 98 1625–1633. 10.3168/jds.2014-8938 [DOI] [PubMed] [Google Scholar]
- Zhou B., Liang T., Zhan Z., Liu R., Li F., Xu H. (2017). Rapid and simultaneous quantification of viable Escherichia coli O157:H7 and Salmonella spp. in milk through multiplex real-time PCR. J. Dairy Sci. 100 8804–8813. 10.3168/jds.2017-13362 [DOI] [PubMed] [Google Scholar]
- Zhu R.-G., Li T.-P., Jia Y.-F., Song L.-F. (2012). Quantitative study of viable Vibrio parahaemolyticus cells in raw seafood using propidium monoazide in combination with quantitative PCR. J. Microbiol. Methods 90 262–266. 10.1016/j.mimet.2012.05.019 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.