Abstract
Pathogenic streptococcal species are responsible for a broad spectrum of human diseases ranging from non-invasive and localized infections to more aggressive and life-threatening diseases, which cause great economic losses worldwide. Streptococci possess a dozen two-component systems (TCSs) that play important roles in the response to different environmental changes and adjust the expression of multiple genes to successfully colonize and infect host cells. In this review, we discuss the progress in the study of a conserved TCS named CiaRH in pathogenic or opportunistic streptococci including Streptococcus pneumoniae, Streptococcus pyogenes, Streptococcus agalactiae, Streptococcus mutans, Streptococcus gordonii, Streptococcus sanguinis, and Streptococcus suis, focusing on the function and regulatory networks of CiaRH, which will provide a promising strategy for the exploration of novel antistreptococcal therapies. This review highlights the important role of CiaRH and provides an important basis for the development of antistreptococcal drugs and vaccines.
Keywords: two-component system CiaRH, histidine kinase CiaH, response regulator CiaR, streptococci, regulatory network
Introduction
Streptococcus is a genus of Gram-positive bacteria that can colonize humans and animals, and is the dominant species in the host oral cavity and upper respiratory tract (Patenge et al., 2013). Most streptococci are non-pathogenic and live in benign and commensal relationships with their hosts. However, several of them are pathogenic including Streptococcus pneumoniae, Streptococcus pyogenes (also known as group A Streptococcus, GAS), Streptococcus agalactiae (also known as group B Streptococcus, GBS), Streptococcus mutans, Streptococcus gordonii, Streptococcus sanguinis, and Streptococcus suis, which can cause a wide range of infectious diseases, such as pneumonia, meningitis, septicemia, scarlet fever, toxic shock, protophilic, endocarditis, dental caries, and streptococcal toxic shock syndrome (van der Poll and Opal, 2009; Ralph and Carapetis, 2013; Jiang et al., 2019). These pathogenic streptococcal species are responsible for substantial mortality and morbidity and pose a major threat to human health.
Streptococci have evolved several mechanisms to respond to environmental changes allowing bacterial survival and infection of host cells. One of them, two-component systems (TCSs), is widely distributed in bacteria and is found in some archaea, fungi, plants, and lower eukaryotes; however, they are absent in humans and other mammals (Wuichet et al., 2010; Capra and Laub, 2012; Galperin et al., 2018). In bacteria, a typical TCS consists of two different proteins: a histidine kinase (HK) and a cognate response regulator (RR). First, the HK receives external stimuli and autophosphorylates at a histidine (His) residue. Then, the phosphoryl group is transferred from the His to an aspartic acid (Asp) residue of the RR. Finally, the RR is activated by this phosphorylation and triggers an appropriate cellular response by interacting with the regulatory regions of the related target genes (Gao and Stock, 2009). Most bacteria possess more than 10 TCSs that regulate multiple cellular functions such as metabolism, virulence, stress response, biofilm formation, antibiotic resistance, and competence. Therefore, TCSs are considered promising targets for the development of novel antibacterial drugs (Gotoh et al., 2010).
Based on literature report and the P2CS (prokaryotic TCSs) database1, S. pneumoniae TIGR4 and R6 encode 13 TCSs (Throup et al., 2000); S. mutans UA159, S. sanguinis SK36, and S. pyogenes MGAS5005 contain 14 TCSs (Smith and Spatafora, 2012; Buckley et al., 2018); both S. gordonii str. Challis substr. CH1 and S. suis 2 possess 15 TCSs (Zheng et al., 2018); and S. agalactiae 2603V/R possesses 17 TCSs (Tettelin et al., 2002). Moreover, there is strain-to-strain variation in the number of TCS in these streptococcal species, for example, S. agalactiae 2603V/R possesses 17 TCSs, while S. agalactiae SA20-06 possesses 13 TCSs. One of the best characterized TCSs present in many streptococcal species is CiaRH, which is composed of the HK CiaH and the RR CiaR. In this review, we present an overview of the function and regulatory network of CiaRH in streptococci.
The Homology of CiaRH in Streptococci
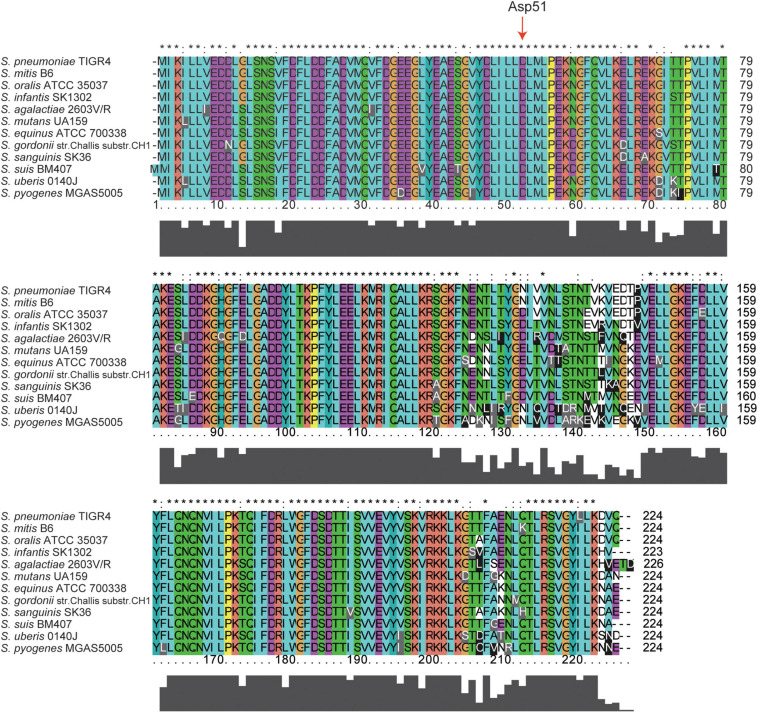
Amino acid sequence alignment analysis revealed that CiaH proteins share between 49 and 97% identical residues and between 66 and 98% similar residues (Table 1). CiaR proteins are much more conserved in streptococci, and the amino acid identities of CiaR proteins range from 85 to 99%, while amino acid similarities range from 91 to 100% (Table 1). These data suggested that CiaRH systems, especially CiaR proteins, are highly conserved in streptococci. Furthermore, almost all Asp residues (D) including Asp51, which receives the phosphoryl group from the His of CiaH (Guenzi et al., 1994), are invariant in all streptococcal CiaR proteins (Figure 1). Collectively, these data indicated that the CiaRH system, especially the CiaR protein, is highly conserved in streptococcal species.
TABLE 1.
Amino acid identities and similarities of CiaR and CiaH proteins of various streptococci to S. pneumoniae TIGR4 CiaR and CiaH proteins.
| Bacteria | Identities with CiaR | Similarities with CiaR | Identities with CiaH | Similarities with CiaH |
| S. pneumoniae TIGR4/R6/D39 | 100%/100%/100% | 100%/100%/100% | 100%/100%/100% | 100%/100%/100% |
| S. mutans UA159/NN2025/LJ23 | 88%/88%/88% | 92%/92%/92% | 56%/56%/56% | 73%/73%/73% |
| S. gordonii str. Challis substr. CH1/BCA22/DD07 | 90%/90%/91% | 95%/95%/95% | 66%/66%/66% | 80%/80%/80% |
| S. sanguinis SK36/SK49/SK355 | 89%/89%/88% | 94%/94%/95% | 61%/60%/60% | 73%/74%/72% |
| S. pyogenes MGAS5005/10270/10750 | 85%/85%/85% | 91%/91%/91% | 51%/51%51% | 71%/71%/71% |
| S. agalactiae 2603V/R/H36B/A909 | 88%/88%/88% | 95%/95%/95% | 52%/52%/52% | 72%/72%/72% |
| S. suis BM407/SC84/05HAS68 | 88%/88%/88% | 94%/94%/94% | 49%/49%/49% | 66%/66%/66% |
| Streptococcus mitis B6/S022-V7-A3/ATCC 6249 | 99%/99%/97% | 100%/100%/99% | 97%/95%/84% | 98%/98%/93% |
| Streptococcus infantis SK1302/SPAR10/ATCC 700779 | 94%/94%/94% | 97%/97%/97% | 88%/81%/81% | 94%/89%/89% |
| Streptococcus uberis 0140J/NCTC3858/4672 | 85%/85%/85% | 91%/91%/91% | 51%/51%/50% | 69%/70%/69% |
| Streptococcus oralis ATCC 35037/subsp. Tigurinus 1366/subsp. Tigurinus AZ_3a | 98%/98%/97% | 99%/99%/99% | 84%/84%84% | 93%/92%93% |
| Streptococcus equinus ATCC 700338/NCTC8140/10386 | 87%/88%/86% | 93%/92%/92% | 51%/52%/52% | 69%/68%/68% |
FIGURE 1.
Multiple amino acid sequence alignment of CiaR across 12 streptococcal species. Homologous sequences with 91–100% similarities. The 12 streptococcal species, namely, S. pneumoniae TIGR4, S. mitis B6, S. oralis ATCC 35037, S. infantis SK1302, S. agalactiae 2603V/R, S. mutans UA159, S. equinus ATCC 700338, S. gordonii str. Challis substr. CH1, S. sanguinis SK36, S. suis BM407, S. uberis 0140J, and S. pyogenes MGAS5005. The residues labeled with * represent the invariant residues. The height in the bar graph represents the conservation of amino acids.
Phosphorylation of the CiaRH System
Under a variety of growth conditions, the CiaRH system appears to be constitutively active in wild-type streptococci. Both CiaH and CiaR proteins contain a two-domain structure: a receiver domain and an effector domain. Commonly, CiaH senses environmental stimuli via receiver domain and autophosphorylates the appropriate His residue in its effector domain after binding ATP, subsequently transferring the phosphoryl group to a conserved Asp51 in the receiver domain of CiaR, thus activating CiaR to elicit a cellular response by changing the downstream gene expression (Guenzi et al., 1994). Although CiaH is necessary for sensing most signals, not all Cia input signals are CiaH-dependent, and CiaR can obtain phosphoryl groups independently of CiaH (Marx et al., 2014).
In S. pneumoniae, depending on the growth medium, deletion of ciaH weakly elicited the CiaR-dependent promoter activities or strongly inhibited them, suggesting that CiaH possesses kinase or phosphatase activities to phosphorylate or dephosphorylate CiaR, leading to the reversible regulation of gene expression (Halfmann et al., 2011). In the absence of CiaH, CiaR appears to be highly active (Halfmann et al., 2011). A subsequent study found that in the absence of CiaH, CiaR can obtain a phosphoryl group from acetyl phosphate (AcP) to become activated (Marx et al., 2014). Additionally, AckA, an AcP synthesis-related acetate kinase, can bind to CiaR and negatively regulate the AcP-dependent phosphorylation of CiaR (Marx et al., 2014; Kaiser et al., 2020). In addition to TCS kinases, a great number of proteins have been reported to control RRs, and the phosphorylation of serine/threonine/tyrosine and even the acetylation of RRs also affect RR-mediated regulation (Mitrophanov and Groisman, 2008; Kalantari et al., 2015; Janczarek et al., 2018). In addition to the phosphoryl groups donated by CiaH and AcP to CiaR, there may be other phosphodonors for CiaR, other phosphoreceptors for CiaH, or other post-translational modifications such as phosphorylation of serine/threonine/tyrosine residues of CiaR, which warrants further exploration.
Major Functions of CiaRH in Streptococci
Competence Development
In bacteria, competence is a transient physiological state that permits cells to uptake and integrates exogenous DNA into the bacterial genome, which plays a key role in virulence, biofilm formation, and antibiotic resistance (Claverys et al., 2006; Charpentier et al., 2012). Natural competence was originally discovered in S. pneumoniae and was later demonstrated in many bacteria such as in S. mutans but also in S. suis (Avery et al., 1979; Perry and Kuramitsu, 1981; Claverys et al., 2006). In streptococci, natural competence for genetic transformation can be stimulated by competence-stimulating peptide (CSP) or sigX-inducing peptide (XIP) and initiated by the com locus (Cheng et al., 1997; Alloing et al., 1998; Shanker et al., 2016). In Mitis and Anginosus Streptococcus groups, CSP is the product of the comC gene and is exported by the ComAB transporter; after that, extracellular CSP is sensed by the ComDE TCS (e.g., S. pneumoniae) (Havarstein et al., 1996; Cheng et al., 1997; Alloing et al., 1998; Shanker et al., 2016). In contrast, in all other groups of Streptococcus including S. mutans, S. pyogenes, S. agalactiae, and S. suis, XIP is encoded by comS, transported by the oligopeptide permease (Opp) transporter, and sensed by ComR to induce the expression of sigX, then initiates the competence (Mashburn-Warren et al., 2010; Shanker et al., 2016).
In S. pneumoniae, several studies have suggested that CiaRH negatively regulates competence development (Guenzi et al., 1994; Zahner et al., 1996; Echenique et al., 2000; Dagkessamanskaia et al., 2004). The ciaHC306 mutant represents CiaRH in the constitutively active state, which leads to a competence deficiency in S. pneumoniae; moreover, the addition of exogenous cannot stimulate the competence of the ciaHC306 mutant (Guenzi et al., 1994; Zahner et al., 1996). Echenique et al. (2000) found that ciaRH negatively regulates comCDE transcription and then modulates the competence stimulated by O2. Conversely, the inactivation of ciaR leads to competence induction. In addition, CiaRH is a pre-requisite for discontinuing the competent state of cells dependent on CiaR (Dagkessamanskaia et al., 2004). Interestingly, data on the contribution of CiaH and CiaR to competence are different. In S. mutans, CiaRH has been shown to be involved in the competence process, and competence development is influenced by CiaH but not CiaR (Qi et al., 2004; Ahn et al., 2006).
Biofilm Formation
Biofilms are structured communities composed of one or multiple bacterial species that elicit many human infectious diseases, including dental caries (Maier, 2021). Twenty-five species of oral streptococci inhabit the human oral cavity, accounting for approximately 20% of the total oral bacteria (Nicolas and Lavoie, 2011). Dental caries is a classic biofilm-associated condition that is induced by diet (sugars) and microbiota–matrix interactions that occur on tooth surfaces, resulting in destruction of mineralized tooth tissue (Bowen et al., 2018; Chen et al., 2019). Among these oral streptococci, S. mutans is considered the most important cariogenic bacteria contributing to the formation of dental caries, and S. sanguinis and S. gordonii have been proposed to initiate oral biofilm formation (Kolenbrander et al., 2010; Bowen et al., 2018).
In contrast to the ciaRH operon in S. pneumoniae, the ciaRH operon in S. mutans contains three genes–ciaR, ciaH, and ciaX–that comprise a three-component signal transduction system (He et al., 2008). The CiaX contains a calcium-binding domain and ciaXRH operon expression is repressed by calcium through CiaX, and the inactivation of ciaH, ciaR, or ciaX results in attenuated biofilm formation (Qi et al., 2004; He et al., 2008). In addition to S. mutans, the CiaRH system and its cognate signal play a key role in biofilm development in other oral streptococci, e.g., S. sanguinis and S. gordonii (Kolenbrander et al., 2010; Davey et al., 2016a; Ota et al., 2018; Zhu et al., 2018). Biofilm formation and expression of CiaRH in thiol-disulfide oxidoreductase (SdbA)-deficient mutants are drastically increased, and further experiments validated that the effect of SdbA on biofilm formation is controlled by the CiaRH and ComDE TCSs in S. gordonii (Davey et al., 2016a,b).
CiaRH has also been proposed to be the key regulator of biofilm formation in S. sanguinis (Zhu B. et al., 2017; Ota et al., 2018; Zhu et al., 2018). Biofilms in ciaR deletion mutants are fragile, and the transcription of arginine biosynthesis genes (argR, argB, argC, argG, argH, and argJ) is increased (Zhu B. et al., 2017). Double deletion of ciaR and argB restored the biofilm formation ability that was lost in the ciaR mutant, indicating that ciaR influences biofilm formation by regulating an arginine biosynthesis pathway, especially regulation of the argB gene (Zhu B. et al., 2017). Moreover, CiaRH negatively regulated the expression of the type IV pilus retraction ATPase PilT and biofilm formation by controlling csRNA1-1 and csRNA1-2, two of the six known cia-dependent small RNAs (csRNAs) in S. sanguinis (Ota et al., 2018). While CiaR is regulated by the transcription factor brpL, mutation of brpL activates CiaR, which results in increased glucan production, cell aggregation, and biofilm formation (Zhu et al., 2018).
Similar effects of CiaRH on biofilm formation were also found in S. pneumoniae and S. pyogenes. Deletion of ciaR/H significantly decreased biofilm formation in vivo, which resulted in the inhibition of nasopharynx colonization by S. pneumoniae (Blanchette-Cain et al., 2013). In S. pyogenes, transcriptome analysis revealed that the superantigen SpeA suppresses biofilm formation, and this process is mediated by the CiaRH TCS (Babbar et al., 2019). Doubtlessly, inactivation of CiaRH can attenuate the biofilm-forming capacity in streptococci; therefore, targeting CiaRH may be an antibiofilm strategy for the development of antistreptococcal therapies.
Antibiotic Resistance and Stress Tolerance
Antibiotic resistance in bacteria is increasing worldwide and becoming a serious threat to global human health (Doernberg et al., 2017; Doi et al., 2017). Many TCSs play crucial roles in antibiotic resistance by regulating genes to modify the antibiotic or its target, and biosynthesize efflux pumps to extrude the antibiotic (Huang et al., 2020). Competence and biofilm development are implicated in antibiotic resistance, and CiaRH regulates competence and biofilm development processes. Therefore, CiaRH also indirectly participates in antibiotic resistance.
In S. pneumoniae, CiaRH was originally reported to be involved in cefotaxime susceptibility, and the ciaHC306 mutant was resistant to cefotaxime (Guenzi et al., 1994; Giammarinaro et al., 1999). Currently, it is known that CiaRH also participates in cycloserine, bacitracin, vancomycin, and penicillin resistance (Haas et al., 2005; Mascher et al., 2006; Rogers et al., 2007; El Khoury et al., 2017). cDNA microarray analysis revealed that vancomycin can induce the expression of CiaRH in S. pneumoniae TIGR4 (a vancomycin-sensitive strain) but not in Tupelo (a vancomycin-tolerant strain) (Haas et al., 2005). Furthermore, CiaRH in the active state results in resistance to cycloserine, bacitracin, and vancomycin, while bacteria with CiaRH in the inactive state are hypersusceptible to these antibiotics (Mascher et al., 2006). Both DNA microarrays and transcriptome analysis revealed that CiaRH was induced when S. pneumoniae strains were exposed to penicillin (Rogers et al., 2007; El Khoury et al., 2017).
Much less is known about the contribution of CiaRH to antibiotic resistance in oral streptococci. To colonize and thrive on teeth, S. mutans expresses a set of genes to resist several antibacterial factors, including cationic antimicrobial peptides (AMPs) derived from the host. A report by Mazda et al. (2012) described that ciaRH regulated the teichoic acid biosynthesis operon dlt to resist AMPs in S. mutans biofilm cells.
In addition to participating in antibiotic resistance, CiaRH also has a central role in the regulation of stress tolerance, such as oxidative stress and acid tolerance (Ibrahim et al., 2004; Cortes et al., 2015). In the absence of ciaR, S. pneumoniae was more sensitive to oxidative stress, and this phenomenon was restored by complementation with high-temperature requirement A protein (HtrA) (Ibrahim et al., 2004). Cortes et al. (2015) provided evidence that CiaRH and ComDE TCSs participate in acid tolerance to help pneumococcus survive in acidic environments (e.g., in pneumocytes).
Likewise, several reports revealed that CiaRH is associated with acid tolerance in S. mutans (Qi et al., 2004; Zhu W. et al., 2017). The ciaH gene greatly reduced the growth rate of S. mutans under acidic culture conditions (Qi et al., 2004). A recent study revealed that sRNA133474 (a small non-coding RNA) negatively regulated the mRNA expression of ciaR, liaR, and covR to contribute to the acid tolerance of S. mutans in dental caries lesions (Zhu W. et al., 2017). The CiaRH of S. gordonii also contributes to acid tolerance, and the growth of ciaRH, comDE, or vicRK mutants was slower than that of the wild-type strain under acidified media (Liu and Burne, 2009).
In S. agalactiae, ΔciaR mutant exhibited a lower survival rate than the wild-type strain when exposed to cationic AMPs, lysozyme, and ROS (Quach et al., 2009). In S. suis, DNA microarray data showed that acidic stress conditions can stimulate the expression of ciaR/H genes, indicating that CiaRH may play an important role in protecting S. suis against acidic stress or in adapting to these conditions (Yan et al., 2011).
In S. pyogenes, the natural competence is initiated by ComRS and SigX, but not by ComDE and CiaRH (Shanker et al., 2016). Thus, antibiotic resistance between S. pyogenes wild-type and ciaH mutant strains was not significantly different (Riani et al., 2007).
Bacteriocin Production
Bacteriocins, proteins or peptides with pronounced antimicrobial activity, are produced by certain bacteria to inhibit the growth of neighboring bacteria belonging to the same or different species (Negash and Tsehai, 2020; Gradisteanu Pircalabioru et al., 2021). In S. mutans, S. gordonii, and S. pneumoniae, the CiaRH system and its cognate signal play a key role in bacteriocin production (Qi et al., 2004; Dawid et al., 2009; Davey et al., 2016b). Mutacins are bacteriocins produced by S. mutans that exhibit antimicrobial effects on closely related streptococcal species and on other bacteria in dental plaques (Hamada and Ooshima, 1975). The deletion of ciaH, but not of ciaR mutant, repressed mutacin I production (Qi et al., 2004). Interestingly, data on the contribution of CiaRH to bacteriocin production in S. gordonii and S. pneumoniae are in opposition to those in S. mutans. In S. pneumoniae, bacteriocin (also named pneumocin MN) production was negatively regulated by CiaH, deletion of ciaH induced the production of pneumocin MN, and activation of ciaH diminished the production of pneumocin MN (Dawid et al., 2009). Similarly, in S. gordonii, CiaRH was activated, while the production of bacteriocin was inhibited in the ΔsdbA mutant, and loss-of-function CiaRH in the ΔsdbA mutant could restore bacteriocin production (Davey et al., 2016b).
Virulence and Pathogenesis
Over the past decade, a large number of studies have demonstrated that CiaRH also has important roles in the virulence and pathogenesis of streptococci. During the infection process, bacteria can adhere to, invade, and colonize the host organism with the help of virulence factors, leading to related infectious diseases. In S. pneumoniae, CiaRH controls the expression of htrA, which is connected to virulence. In a murine model of pneumonia, disruption of ciaRH or htrA significantly reduced pneumococcal colonization in nasopharyngeal tissue (Sebert et al., 2002). A further study showed that supplementation of the S. pneumoniae ΔciaR mutant with HtrA can recover virulence (Ibrahim et al., 2004). In the process of colonization, sialic acids (Sias) serve as carbon sources for S. pneumoniae and are consumed by the sialidase NanA (Vimr et al., 2004). Recently, a study reported that CiaR sensed N-acetylneuraminic acid (Neu5Ac, the most abundant Sias in humans) to result in an increase in htrA and other genes involved in sialic acid metabolism and ROS tolerance, contributing to increased pneumococcal virulence (Hentrich et al., 2016). In addition, CiaR stimulated the Plic1P1 promoter activity of choline metabolism-related lic loci and promoted pneumococcal colonization in the host (Johnston et al., 2016).
Streptococcus agalactiae is a major opportunistic pathogen that asymptomatically colonizes the vaginal and gastrointestinal tracts of healthy human adults, including pregnant women; however, this species causes invasive diseases, including meningitis, pneumonia, and sepsis in newborns (Doran and Nizet, 2004). To cause meningitis, S. agalactiae must be able to invade and survive within brain microvascular endothelial cells and then breach the blood–brain barrier (BBB) to enter the central nervous system (CNS). CiaRH is regarded as a key factor involved in intracellular survival and the ultimate disease progression of meningitis. Indeed, S. agalactiae strains lacking the ciaR gene exhibited impaired survival in neutrophils, murine macrophages, and human brain microvascular endothelial cells (hBMECs), and a competitive infection assay in a mouse model demonstrated that the ΔciaR mutant had a lower survival advantage in the bloodstream and brain than in the wild-type strain (Quach et al., 2009). Furthermore, mutants in CiaR-regulated genes SAN_2180 and SAN_0039 (Δ2180 and Δ0039) were determined to have a lower intracellular survival rate in hBMECs and a lower survival advantage in the bloodstream and brain in a mouse infection model. These results observed in Δ2180 and Δ0039 were similar to those in ΔciaR, implying that S. agalactiae may modulate SAN_2180 and SAN_0039 gene expression through CiaRH to promote intracellular survival and virulence (Mu et al., 2016). Recently, a study conducted by Spencer et al. (2019) compared the transcriptomes of cas9 (CRISPR-associated protein-9) and ciaR mutants (Δcas9 and ΔciaR) and determined that the RR CiaR was regulated by Cas9 to contribute to vaginal colonization and persistence of S. agalactiae.
Streptococcus suis is a highly invasive pathogen that causes infectious diseases in both swine and humans, resulting in manifestations such as meningitis, septicemia, pneumonia, endocarditis, arthritis, and septic shock (Lun et al., 2007; Wertheim et al., 2009). Li et al. (2011) reported that CiaRH contributes to the virulence of S. suis, and the ciaRH deletion mutant (ΔciaRH) exhibited lower adherence to epithelial cells (Hep-2 and PIEC) and higher susceptibility toward killing by RAW2647 macrophages than the wild-type strain. Moreover, in vivo, ΔciaRH was attenuated in murine and pig animal models of infection, but showed increased survival rates in mice and pigs and reduced bacterial loads in specific organs. Similarly, the ΔciaRH strain also showed an increased survival rate in the zebrafish larval model compared with the wild-type strain (Zaccaria et al., 2016). Recently, because ciaR was significantly increased when S. suis was stimulated with human polymorphonuclear leukocytes (PMNs), CiaRH has been proposed to be a critical regulatory system for resistance to phagocytosis by human PMNs (Chang et al., 2018).
Overall, these data indicated that CiaRH potentially promotes virulence and pathogenesis including the survival rate, adherence, and colonization capabilities of S. pneumoniae, S. agalactiae, and S. suis. However, less is known about the virulence and pathogenesis of CiaRH in S. mutans, although it was shown that CiaRH was induced by Candida albicans to enhance the S. mutans biofilm accumulation, thereby regulating the virulence of S. mutans (He et al., 2017).
Cell Wall Biosynthesis and Autolysis
Teichoic acid is an important component of the Gram-positive cell wall that plays vital roles in autolysis, drug resistance, biofilm formation, and virulence (Wu et al., 2020). The two operons dltABCD and licABC are engaged in teichoic acid biosynthesis and are required for D-alanylation and choline metabolism of teichoic acid in streptococci, respectively. Several studies have shown that CiaRH controls the expression of dltABCD and licABC in S. pneumoniae (Mascher et al., 2003; Dagkessamanskaia et al., 2004; Halfmann et al., 2007; Slager et al., 2019). Autolysis is a programmed cell death process triggered by autolysin that causes cell wall self-digestion. The CiaRH system in the defective state accelerated the autolysis of pneumococci, while the CiaRH system in the active state prevented autolysis (Giammarinaro et al., 1999; Lange et al., 1999). As mentioned above, activation of the CiaRH system resulted in resistance to cell wall inhibitors, while deletion of the CiaRH system resulted in hypersusceptibility to cell wall inhibitors. Taken together, these results suggest that the CiaRH system in S. pneumoniae induces the expression of dltABCD and licABC to confer resistance to cell wall inhibitors and autolysis.
In addition, a similar effect of CiaRH on dltABCD and autolysin was found in S. mutans, which also results in antibiotic resistance and stress tolerance (Wu et al., 2010; Mazda et al., 2012). However, whether CiaRH affects cell wall biosynthesis and autolysis in other streptococci remains to be tested.
Regulatory Network of CiaRH in Streptococci
As mentioned above, the CiaRH TCS affects streptococcal competence development, biofilm formation, antibiotic resistance, stress tolerance, bacteriocin production, virulence, pathogenesis, cell wall biosynthesis, and autolysis. However, some biological functions of the CiaRH TCS in streptococci are the opposite, and the regulatory network of CiaRH is different among streptococcal species.
Regulatory Network of CiaRH in S. pneumoniae
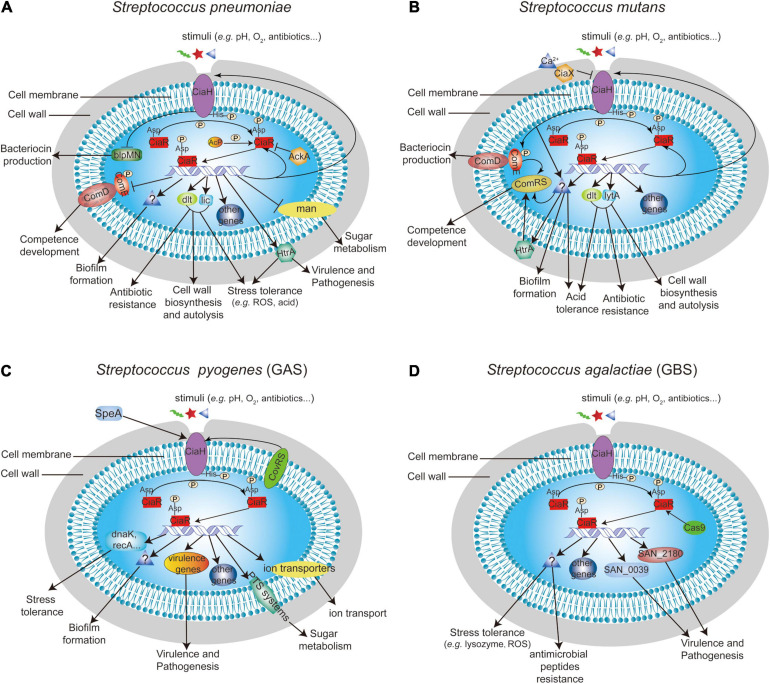
The global gene expression effect of CiaRH in S. pneumoniae has been reported in four genomic/profiling-related manuscripts. The authors of these studies demonstrated that CiaRH controls the expression of more than 70 genes, and most genes were positively regulated by CiaRH (Mascher et al., 2003; Dagkessamanskaia et al., 2004; Halfmann et al., 2007; Slager et al., 2019; Figure 2A). In 2003, a study conducted by Mascher et al. (2003) revealed that CiaRH regulates the mannose phosphotransferase system (manLMN), phosphorylcholine system (licABC), Acetyl esterase (axe), ABC transporter (cyl), amylomaltase and maltose phosphorylase (malPM), teichoic acid biosynthesis system (dltABCD), serine protease (htrA-spo0J), conserved hypothetical protein (spr0782), extracellular protein (spr0931) operons, and 39 genes from 20 competence regulons, including comCDE. In addition to these genes, CiaRH also controls the expression of the DNA biosynthesis-related gene dnaA-dnaN and TCS03 (hk03-rr03) in S. pneumoniae (Dagkessamanskaia et al., 2004). It is worth noting that CiaRH can affect the expression of ciaRH, thus forming a feedback loop (Dagkessamanskaia et al., 2004). A further work showed that CiaR can bind to TTTAAG-N5-TTTAAG, a repeat sequence in the promoters of regulated genes, and thereby directly regulate the expression levels of 24 genes from 15 promoters (Halfmann et al., 2007). Except for manLMN, which was negatively controlled by CiaR, 21 genes, including five small non-coding csRNAs (ccnABCDE), were positively regulated by CiaR, and these genes were associated with choline modification of teichoic acids, sugar metabolism, stress responses, chromosome segregation, and protease maturation (Halfmann et al., 2007). A recent transcriptome study demonstrated that 38 genes distributed over 18 operons were directly or indirectly regulated by CiaR (Slager et al., 2019). Among these genes, six translation-related genes (rimP, nusA, SPV_0480, SPV_0481, infB, and rbfA), 23S rRNA [uracil(1939)-C(5)]-methyltransferase (rlmCD), and a novel uncharacterized non-coding RNA (srf-21) were found to be CiaR-regulated genes for the first time (Slager et al., 2019).
FIGURE 2.
Summary of the known function of CiaRH and its regulatory network in (A) S. pneumoniae, (B) S. mutans, (C) S. pyogenes, and (D) S. agalactiae.
Regulatory Network of CiaRH in Oral Streptococci
The regulatory network of CiaRH has also been reported in oral streptococci, including S. mutans, S. sanguinis, and S. gordonii. A total of 100 genes encoded on the S. mutans chromosome were regulated by CiaRH, including its own gene ciaXRH (Wu et al., 2010; Figure 2B). Moreover, eight Cia regulons were identified, including the ciaXRH operon itself, which is directly regulated by CiaR. The consensus sequence upstream of the putative −10 regions of these genes where the CiaR protein was bound to the promoters was NTTAAG-n5-WTTAAG (Wu et al., 2010). In addition, CiaRH can lead to an increase in the expression of the teichoic acid biosynthesis operon dlt in S. mutans biofilm cells (Mazda et al., 2012).
In S. sanguinis, CiaRH exerts its regulatory effect by mediating the expression of arginine biosynthesis genes, arginine/histidine permease genes, and csRNAs (Zhu B. et al., 2017; Ota et al., 2018). Less is known about the regulatory network of CiaRH in S. gordonii. Loss of SdbA can activate the CiaRH system, thereby negatively regulating the Com system to reduce bacteriocin production (Davey et al., 2016b).
Regulatory Network of CiaRH in S. pyogenes
Streptococcus pyogenes is an exclusive human pathogen that causes diseases ranging from pharyngitis, impetigo, abscesses, cellulitis, sepsis, necrotizing fasciitis, and streptococcal toxic shock syndrome to acute post-streptococcal glomerulonephritis, acute rheumatic fever, and rheumatic heart disease (Carapetis et al., 2005; Walker et al., 2014). In this bacterium, CiaRH was regulated by the CovRS TCS and was associated with virulence (Graham et al., 2002), and CiaRH sensed SpeA to modulate biofilm formation (Babbar et al., 2019; Figure 2C). In addition, a study conducted by Riani et al. (2007) established transcriptome analyses between the wild-type strain and the ciaH mutant, which revealed that CiaH influences the transcription of 132 genes (63 genes were upregulated and 69 genes were downregulated in the ciaH mutant compared with the wild-type strain), including genes that encode divalent cation and other ion transporters, PTS systems, ribosomal proteins, virulence proteins, stress response proteins, and hypothetical and phage proteins (Figure 2C). However, unlike CiaRH in S. pneumoniae, only six stress response genes of 132 genes were activated in the ciaH mutant of S. pyogenes, and no cell wall turnover genes showed changes between wild-type and ciaH mutant strains (Riani et al., 2007).
Regulatory Network of CiaRH in S. agalactiae
The effect of CiaRH on bacterial virulence, survival, and colonization has been well studied in S. agalactiae. Microarray data coupled with quantitative RT-PCR data revealed that CiaR positively regulated the expression of several genes, including hypothetical protein (SAN_2180) and peptidases (SAN_0039, SAN_0058, and SAN_0059) (Quach et al., 2009). Further studies showed that the SAN_2180 and SAN_0039 proteins were modulated by CiaR to enhance the virulence of S. agalactiae in mice (Mu et al., 2016; Figure 2D). Moreover, CiaR was regulated by Cas9 and contributed to S. agalactiae vaginal colonization and persistence (Spencer et al., 2019; Figure 2D).
Concluding Remarks and Future Perspectives
Multidrug-resistant bacteria, including multidrug-resistant streptococci, have increased in prevalence and become a major threat to global public health. Novel drugs with modes of action different from conventional antibiotics are urgently needed to combat bacterial infections caused by these pathogens. Because TCSs are widely distributed in bacteria but do not exist in humans, targeting TCSs has been explored as a novel strategy to combat bacterial infections, especially drug-resistant bacterial infections (Worthington et al., 2013; Hirakawa et al., 2020; Rajput et al., 2021). The CiaRH TCS composed of CiaH and CiaR is an important signal transduction system that mediates various streptococcal behaviors, including competence development, biofilm formation, antibiotic resistance, stress tolerance, bacteriocin production, virulence, pathogenesis, cell wall biosynthesis, and autolysis. Moreover, the CiaRH system, especially the CiaR protein, is highly conserved in streptococcal species. Therefore, CiaRH may serve as an attractive antibiofilm or antivirulence target to develop new drugs to fight against various drug-resistant streptococcal infections. However, several issues remain to be addressed, e.g., (a) in addition to the known functions, what other functions does CiaRH possess? (b) Can CiaH or CiaR serve as markers to screen cell wall antibiotic-resistant streptococci? (c) Which drugs can target CiaR for the treatment of streptococcal infections? A more complete understanding of the CiaRH system will aid in preventing and controlling streptococcal infections.
Author Contributions
L-YH wrote the manuscript. Y-JL and ZG analyzed the data and provided the visual images. SL and X-YY provided the initial idea, funding, and edited the manuscript. All authors contributed to manuscript revision and read and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding. This work was financially supported by the National Natural Science Foundation of China (81860356, to X-YY; 31860259 to ZG), Guizhou Provincial Natural Science Foundation (QKH-J[2020]1Y352, to X-YY; QKH-ZK[2021]1Y420, to SL; and QKH-[2018]5772-018, to SL), and Excellent Young Talents Fund Program of Zunyi Medical University (18zy-005, to X-YY).
References
- Ahn S. J., Wen Z. T., Burne R. A. (2006). Multilevel control of competence development and stress tolerance in Streptococcus mutans UA159. Infect. Immun. 74 1631–1642. 10.1128/iai.74.3.1631-1642.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alloing G., Martin B., Granadel C., Claverys J. P. (1998). Development of competence in Streptococcus pneumonaie: pheromone autoinduction and control of quorum sensing by the oligopeptide permease. Mol. Microbiol. 29 75–83. 10.1046/j.1365-2958.1998.00904.x [DOI] [PubMed] [Google Scholar]
- Avery O. T., Macleod C. M., Mccarty M. (1979). Studies on the chemical nature of the substance inducing transformation of pneumococcal types. Inductions of transformation by a desoxyribonucleic acid fraction isolated from pneumococcus type III. J. Exp. Med. 149 297–326. 10.1084/jem.149.2.297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babbar A., Barrantes I., Pieper D. H., Itzek A. (2019). Superantigen SpeA attenuates the biofilm forming capacity of Streptococcus pyogenes. J. Microbiol. 57 626–636. 10.1007/s12275-019-8648-z [DOI] [PubMed] [Google Scholar]
- Blanchette-Cain K., Hinojosa C. A., Akula Suresh, Babu R., Lizcano A., Gonzalez-Juarbe N., et al. (2013). Streptococcus pneumoniae biofilm formation is strain dependent, multifactorial, and associated with reduced invasiveness and immunoreactivity during colonization. mBio 4 e745–e713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen W. H., Burne R. A., Wu H., Koo H. (2018). Oral Biofilms: Pathogens, Matrix, and Polymicrobial Interactions in Microenvironments. Trends Microbiol. 26 229–242. 10.1016/j.tim.2017.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley S. J., Timms P., Davies M. R., Mcmillan D. J. (2018). In silico characterisation of the two-component system regulators of Streptococcus pyogenes. PLoS One 13:e0199163. 10.1371/journal.pone.0199163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capra E. J., Laub M. T. (2012). Evolution of two-component signal transduction systems. Annu. Rev. Microbiol. 66 325–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carapetis J. R., Steer A. C., Mulholland E. K., Weber M. (2005). The global burden of group A streptococcal diseases. Lancet Infect. Dis. 5 685–694. 10.1016/s1473-3099(05)70267-x [DOI] [PubMed] [Google Scholar]
- Chang P., Li W., Shi G., Li H., Yang X., Xia Z., et al. (2018). The VraSR regulatory system contributes to virulence in Streptococcus suis via resistance to innate immune defenses. Virulence 9 771–782. 10.1080/21505594.2018.1428519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charpentier X., Polard P., Claverys J. P. (2012). Induction of competence for genetic transformation by antibiotics: convergent evolution of stress responses in distant bacterial species lacking SOS? Curr. Opin. Microbiol. 15 570–576. 10.1016/j.mib.2012.08.001 [DOI] [PubMed] [Google Scholar]
- Chen L., Chakraborty B., Zou J., Burne R. A., Zeng L. (2019). Amino Sugars Modify Antagonistic Interactions between Commensal Oral Streptococci and Streptococcus mutans. Appl. Environ. Microbiol. 85 e370–e319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Q., Campbell E. A., Naughton A. M., Johnson S., Masure H. R. (1997). The com locus controls genetic transformation in Streptococcus pneumoniae. Mol. Microbiol. 23 683–692. 10.1046/j.1365-2958.1997.2481617.x [DOI] [PubMed] [Google Scholar]
- Claverys J. P., Prudhomme M., Martin B. (2006). Induction of competence regulons as a general response to stress in gram-positive bacteria. Annu. Rev. Microbiol. 60 451–475. 10.1146/annurev.micro.60.080805.142139 [DOI] [PubMed] [Google Scholar]
- Cortes P. R., Pinas G. E., Cian M. B., Yandar N., Echenique J. (2015). Stress-triggered signaling affecting survival or suicide of Streptococcus pneumoniae. Int. J. Med. Microbiol. 305 157–169. 10.1016/j.ijmm.2014.12.002 [DOI] [PubMed] [Google Scholar]
- Dagkessamanskaia A., Moscoso M., Henard V., Guiral S., Overweg K., Reuter M., et al. (2004). Interconnection of competence, stress and CiaR regulons in Streptococcus pneumoniae: competence triggers stationary phase autolysis of ciaR mutant cells. Mol. Microbiol. 51 1071–1086. 10.1111/j.1365-2958.2003.03892.x [DOI] [PubMed] [Google Scholar]
- Davey L., Halperin S. A., Lee S. F. (2016a). Mutation of the Streptococcus gordonii Thiol-Disulfide Oxidoreductase SdbA Leads to Enhanced Biofilm Formation Mediated by the CiaRH Two-Component Signaling System. PLoS One 11:e0166656. 10.1371/journal.pone.0166656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey L., Halperin S. A., Lee S. F. (2016b). Mutation of the Thiol-Disulfide Oxidoreductase SdbA Activates the CiaRH Two-Component System, Leading to Bacteriocin Expression Shutdown in Streptococcus gordonii. J. Bacteriol. 198 321–331. 10.1128/jb.00800-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawid S., Sebert M. E., Weiser J. N. (2009). Bacteriocin activity of Streptococcus pneumoniae is controlled by the serine protease HtrA via posttranscriptional regulation. J. Bacteriol. 191 1509–1518. 10.1128/jb.01213-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doernberg S. B., Lodise T. P., Thaden J. T., Munita J. M., Cosgrove S. E., Arias C. A., et al. (2017). Gram-Positive Bacterial Infections: Research Priorities, Accomplishments, and Future Directions of the Antibacterial Resistance Leadership Group. Clin. Infect Dis. 64 S24–S29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi Y., Bonomo R. A., Hooper D. C., Kaye K. S., Johnson J. R., Clancy C. J., et al. (2017). Gram-Negative Bacterial Infections: Research Priorities, Accomplishments, and Future Directions of the Antibacterial Resistance Leadership Group. Clin. Infect Dis. 64 S30–S35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doran K. S., Nizet V. (2004). Molecular pathogenesis of neonatal group B streptococcal infection: no longer in its infancy. Mol. Microbiol. 54 23–31. 10.1111/j.1365-2958.2004.04266.x [DOI] [PubMed] [Google Scholar]
- Echenique J. R., Chapuy-Regaud S., Trombe M. C. (2000). Competence regulation by oxygen in Streptococcus pneumoniae: involvement of ciaRH and comCDE. Mol. Microbiol. 36 688–696. 10.1046/j.1365-2958.2000.01891.x [DOI] [PubMed] [Google Scholar]
- El Khoury J. Y., Boucher N., Bergeron M. G., Leprohon P., Ouellette M. (2017). Penicillin induces alterations in glutamine metabolism in Streptococcus pneumoniae. Sci. Rep. 7:14587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin M. Y., Makarova K. S., Wolf Y. I., Koonin E. V. (2018). Phyletic Distribution and Lineage-Specific Domain Architectures of Archaeal Two-Component Signal Transduction Systems. J. Bacteriol. 200 e681–e617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao R., Stock A. M. (2009). Biological insights from structures of two-component proteins. Annu. Rev. Microbiol. 63 133–154. 10.1146/annurev.micro.091208.073214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giammarinaro P., Sicard M., Gasc A. M. (1999). Genetic and physiological studies of the CiaH-CiaR two-component signal-transducing system involved in cefotaxime resistance and competence of Streptococcus pneumoniae. Microbiology 145 1859–1869. 10.1099/13500872-145-8-1859 [DOI] [PubMed] [Google Scholar]
- Gotoh Y., Eguchi Y., Watanabe T., Okamoto S., Doi A., Utsumi R. (2010). Two-component signal transduction as potential drug targets in pathogenic bacteria. Curr. Opin. Microbiol. 13 232–239. 10.1016/j.mib.2010.01.008 [DOI] [PubMed] [Google Scholar]
- Gradisteanu Pircalabioru G., Popa L. I., Marutescu L., Gheorghe I., Popa M., Czobor Barbu I., et al. (2021). Bacteriocins in the Era of Antibiotic Resistance: Rising to the Challenge. Pharmaceutics 13:196. 10.3390/pharmaceutics13020196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham M. R., Smoot L. M., Migliaccio C. A., Virtaneva K., Sturdevant D. E., Porcella S. F., et al. (2002). Virulence control in group A Streptococcus by a two-component gene regulatory system: global expression profiling and in vivo infection modeling. Proc. Natl. Acad. Sci. U. S. A. 99 13855–13860. 10.1073/pnas.202353699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenzi E., Gasc A. M., Sicard M. A., Hakenbeck R. (1994). A two-component signal-transducing system is involved in competence and penicillin susceptibility in laboratory mutants of Streptococcus pneumoniae. Mol. Microbiol. 12 505–515. 10.1111/j.1365-2958.1994.tb01038.x [DOI] [PubMed] [Google Scholar]
- Haas W., Kaushal D., Sublett J., Obert C., Tuomanen E. I. (2005). Vancomycin stress response in a sensitive and a tolerant strain of Streptococcus pneumoniae. J. Bacteriol. 187 8205–8210. 10.1128/jb.187.23.8205-8210.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfmann A., Kovacs M., Hakenbeck R., Bruckner R. (2007). Identification of the genes directly controlled by the response regulator CiaR in Streptococcus pneumoniae: five out of 15 promoters drive expression of small non-coding RNAs. Mol. Microbiol. 66 110–126. 10.1111/j.1365-2958.2007.05900.x [DOI] [PubMed] [Google Scholar]
- Halfmann A., Schnorpfeil A., Muller M., Marx P., Gunzler U., Hakenbeck R., et al. (2011). Activity of the two-component regulatory system CiaRH in Streptococcus pneumoniae R6. J. Mol. Microbiol. Biotechnol. 20 96–104. 10.1159/000324893 [DOI] [PubMed] [Google Scholar]
- Hamada S., Ooshima T. (1975). Production and properties of bacteriocins (mutacins) from Streptococcus mutans. Arch. Oral Biol. 20 641–648. 10.1016/0003-9969(75)90131-4 [DOI] [PubMed] [Google Scholar]
- Havarstein L. S., Gaustad P., Nes I. F., Morrison D. A. (1996). Identification of the streptococcal competence-pheromone receptor. Mol. Microbiol. 21 863–869. 10.1046/j.1365-2958.1996.521416.x [DOI] [PubMed] [Google Scholar]
- He J., Kim D., Zhou X., Ahn S. J., Burne R. A., Richards V. P., et al. (2017). RNA-Seq Reveals Enhanced Sugar Metabolism in Streptococcus mutans Co-cultured with Candida albicans within Mixed-Species Biofilms. Front. Microbiol. 8:1036. 10.3389/fmicb.2017.01036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X., Wu C., Yarbrough D., Sim L., Niu G., Merritt J., et al. (2008). The cia operon of Streptococcus mutans encodes a unique component required for calcium-mediated autoregulation. Mol. Microbiol. 70 112–126. 10.1111/j.1365-2958.2008.06390.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentrich K., Lofling J., Pathak A., Nizet V., Varki A., Henriques-Normark B. (2016). Streptococcus pneumoniae Senses a Human-like Sialic Acid Profile via the Response Regulator CiaR. Cell Host Microbe 20 307–317. 10.1016/j.chom.2016.07.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirakawa H., Kurushima J., Hashimoto Y., Tomita H. (2020). Progress Overview of Bacterial Two-Component Regulatory Systems as Potential Targets for Antimicrobial Chemotherapy. Antibiotics 9:635. 10.3390/antibiotics9100635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Li C., Song J., Velkov T., Wang L., Zhu Y., et al. (2020). Regulating polymyxin resistance in Gram-negative bacteria: roles of two-component systems PhoPQ and PmrAB. Future Microbiol. 15 445–459. 10.2217/fmb-2019-0322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim Y. M., Kerr A. R., Mccluskey J., Mitchell T. J. (2004). Control of virulence by the two-component system CiaR/H is mediated via HtrA, a major virulence factor of Streptococcus pneumoniae. J. Bacteriol. 186 5258–5266. 10.1128/jb.186.16.5258-5266.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janczarek M., Vinardell J. M., Lipa P., Karas M. (2018). Hanks-Type Serine/Threonine Protein Kinases and Phosphatases in Bacteria: Roles in Signaling and Adaptation to Various Environments. Int. J. Mol. Sci. 19:2872. 10.3390/ijms19102872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Q., Zhou X., Cheng L., Li M. (2019). The Adhesion and Invasion Mechanisms of Streptococci. Curr. Issues Mol. Biol. 32 521–560. 10.21775/cimb.032.521 [DOI] [PubMed] [Google Scholar]
- Johnston C., Hauser C., Hermans P. W., Martin B., Polard P., Bootsma H. J., et al. (2016). Fine-tuning of choline metabolism is important for pneumococcal colonization. Mol. Microbiol. 100 972–988. 10.1111/mmi.13360 [DOI] [PubMed] [Google Scholar]
- Kaiser S., Hoppstadter L. M., Bilici K., Heieck K., Bruckner R. (2020). Control of acetyl phosphate-dependent phosphorylation of the response regulator CiaR by acetate kinase in Streptococcus pneumoniae. Microbiology 166 411–421. 10.1099/mic.0.000894 [DOI] [PubMed] [Google Scholar]
- Kalantari A., Derouiche A., Shi L., Mijakovic I. (2015). Serine/threonine/tyrosine phosphorylation regulates DNA binding of bacterial transcriptional regulators. Microbiology 161 1720–1729. 10.1099/mic.0.000148 [DOI] [PubMed] [Google Scholar]
- Kolenbrander P. E., Palmer R. J., Jr., Periasamy S., Jakubovics N. S. (2010). Oral multispecies biofilm development and the key role of cell-cell distance. Nat. Rev. Microbiol. 8 471–480. 10.1038/nrmicro2381 [DOI] [PubMed] [Google Scholar]
- Lange R., Wagner C., De Saizieu A., Flint N., Molnos J., Stieger M., et al. (1999). Domain organization and molecular characterization of 13 two-component systems identified by genome sequencing of Streptococcus pneumoniae. Gene 237 223–234. 10.1016/s0378-1119(99)00266-8 [DOI] [PubMed] [Google Scholar]
- Li J., Tan C., Zhou Y., Fu S., Hu L., Hu J., et al. (2011). The two-component regulatory system CiaRH contributes to the virulence of Streptococcus suis 2. Vet. Microbiol. 148 99–104. 10.1016/j.vetmic.2010.08.005 [DOI] [PubMed] [Google Scholar]
- Liu Y., Burne R. A. (2009). Multiple two-component systems modulate alkali generation in Streptococcus gordonii in response to environmental stresses. J. Bacteriol. 191 7353–7362. 10.1128/jb.01053-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lun Z. R., Wang Q. P., Chen X. G., Li A. X., Zhu X. Q. (2007). Streptococcus suis: an emerging zoonotic pathogen. Lancet Infect. Dis. 7 201–209. 10.1016/s1473-3099(07)70001-4 [DOI] [PubMed] [Google Scholar]
- Maier B. (2021). How Physical Interactions Shape Bacterial Biofilms. Annu. Rev. Biophys. 50 401–417. 10.1146/annurev-biophys-062920-063646 [DOI] [PubMed] [Google Scholar]
- Marx P., Meiers M., Bruckner R. (2014). Activity of the response regulator CiaR in mutants of Streptococcus pneumoniae R6 altered in acetyl phosphate production. Front. Microbiol. 5:772. 10.3389/fmicb.2014.00772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascher T., Heintz M., Zahner D., Merai M., Hakenbeck R. (2006). The CiaRH system of Streptococcus pneumoniae prevents lysis during stress induced by treatment with cell wall inhibitors and by mutations in pbp2x involved in beta-lactam resistance. J. Bacteriol. 188 1959–1968. 10.1128/jb.188.5.1959-1968.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascher T., Zahner D., Merai M., Balmelle N., De Saizieu A. B., Hakenbeck R. (2003). The Streptococcus pneumoniae cia regulon: CiaR target sites and transcription profile analysis. J. Bacteriol. 185 60–70. 10.1128/jb.185.1.60-70.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashburn-Warren L., Morrison D. A., Federle M. J. (2010). A novel double-tryptophan peptide pheromone controls competence in Streptococcus spp. via an Rgg regulator. Mol. Microbiol. 78 589–606. 10.1111/j.1365-2958.2010.07361.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazda Y., Kawada-Matsuo M., Kanbara K., Oogai Y., Shibata Y., Yamashita Y., et al. (2012). Association of CiaRH with resistance of Streptococcus mutans to antimicrobial peptides in biofilms. Mol. Oral Microbiol. 27 124–135. 10.1111/j.2041-1014.2012.00637.x [DOI] [PubMed] [Google Scholar]
- Mitrophanov A. Y., Groisman E. A. (2008). Signal integration in bacterial two-component regulatory systems. Genes Dev. 22 2601–2611. 10.1101/gad.1700308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu R., Cutting A. S., Del Rosario Y., Villarino N., Stewart L., Weston T. A., et al. (2016). Identification of CiaR Regulated Genes That Promote Group B Streptococcal Virulence and Interaction with Brain Endothelial Cells. PLoS One 11:e0153891. 10.1371/journal.pone.0153891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negash A. W., Tsehai B. A. (2020). Current Applications of Bacteriocin. Int. J. Microbiol. 2020:4374891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas G. G., Lavoie M. C. (2011). [Streptococcus mutans and oral streptococci in dental plaque]. Can. J. Microbiol. 57 1–20. [DOI] [PubMed] [Google Scholar]
- Ota C., Morisaki H., Nakata M., Arimoto T., Fukamachi H., Kataoka H., et al. (2018). Streptococcus sanguinis Noncoding cia-Dependent Small RNAs Negatively Regulate Expression of Type IV Pilus Retraction ATPase PilT and Biofilm Formation. Infect Immun. 86 e894–e817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patenge N., Fiedler T., Kreikemeyer B. (2013). Common regulators of virulence in streptococci. Curr. Top. Microbiol. Immunol. 368 111–153. 10.1007/82_2012_295 [DOI] [PubMed] [Google Scholar]
- Perry D., Kuramitsu H. K. (1981). Genetic transformation of Streptococcus mutans. Infect. Immun. 32 1295–1297. 10.1128/iai.32.3.1295-1297.1981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi F., Merritt J., Lux R., Shi W. (2004). Inactivation of the ciaH Gene in Streptococcus mutans diminishes mutacin production and competence development, alters sucrose-dependent biofilm formation, and reduces stress tolerance. Infect. Immun. 72 4895–4899. 10.1128/iai.72.8.4895-4899.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quach D., Van Sorge N. M., Kristian S. A., Bryan J. D., Shelver D. W., Doran K. S. (2009). The CiaR response regulator in group B Streptococcus promotes intracellular survival and resistance to innate immune defenses. J. Bacteriol. 191 2023–2032. 10.1128/jb.01216-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajput A., Seif Y., Choudhary K. S., Dalldorf C., Poudel S., Monk J. M., et al. (2021). Pangenome Analytics Reveal Two-Component Systems as Conserved Targets in ESKAPEE Pathogens. mSystems 6 e981–e920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralph A. P., Carapetis J. R. (2013). Group a streptococcal diseases and their global burden. Curr. Top Microbiol. Immunol. 368 1–27. 10.1007/82_2012_280 [DOI] [PubMed] [Google Scholar]
- Riani C., Standar K., Srimuang S., Lembke C., Kreikemeyer B., Podbielski A. (2007). Transcriptome analyses extend understanding of Streptococcus pyogenes regulatory mechanisms and behavior toward immunomodulatory substances. Int. J. Med. Microbiol. 297 513–523. 10.1016/j.ijmm.2007.04.005 [DOI] [PubMed] [Google Scholar]
- Rogers P. D., Liu T. T., Barker K. S., Hilliard G. M., English B. K., Thornton J., et al. (2007). Gene expression profiling of the response of Streptococcus pneumoniae to penicillin. J. Antimicrob Chemother. 59 616–626. 10.1093/jac/dkl560 [DOI] [PubMed] [Google Scholar]
- Sebert M. E., Palmer L. M., Rosenberg M., Weiser J. N. (2002). Microarray-based identification of htrA, a Streptococcus pneumoniae gene that is regulated by the CiaRH two-component system and contributes to nasopharyngeal colonization. Infect. Immun. 70 4059–4067. 10.1128/iai.70.8.4059-4067.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanker E., Morrison D. A., Talagas A., Nessler S., Federle M. J., Prehna G. (2016). Pheromone Recognition and Selectivity by ComR Proteins among Streptococcus Species. PLoS Pathog 12:e1005979. 10.1371/journal.ppat.1005979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slager J., Aprianto R., Veening J. W. (2019). Refining the Pneumococcal Competence Regulon by RNA Sequencing. J. Bacteriol. 201 e780–e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E. G., Spatafora G. A. (2012). Gene regulation in S. mutans: complex control in a complex environment. J. Dent Res. 91 133–141. 10.1177/0022034511415415 [DOI] [PubMed] [Google Scholar]
- Spencer B. L., Deng L., Patras K. A., Burcham Z. M., Sanches G. F., Nagao P. E., et al. (2019). Cas9 Contributes to Group B Streptococcal Colonization and Disease. Front. Microbiol. 10:1930. 10.3389/fmicb.2019.01930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tettelin H., Masignani V., Cieslewicz M. J., Eisen J. A., Peterson S., Wessels M. R., et al. (2002). Complete genome sequence and comparative genomic analysis of an emerging human pathogen, serotype V Streptococcus agalactiae. Proc. Natl. Acad. Sci. U. S. A. 99 12391–12396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Throup J. P., Koretke K. K., Bryant A. P., Ingraham K. A., Chalker A. F., Ge Y., et al. (2000). A genomic analysis of two-component signal transduction in Streptococcus pneumoniae. Mol. Microbiol. 35 566–576. 10.1046/j.1365-2958.2000.01725.x [DOI] [PubMed] [Google Scholar]
- van der Poll T., Opal S. M. (2009). Pathogenesis, treatment, and prevention of pneumococcal pneumonia. Lancet 374 1543–1556. 10.1016/s0140-6736(09)61114-4 [DOI] [PubMed] [Google Scholar]
- Vimr E. R., Kalivoda K. A., Deszo E. L., Steenbergen S. M. (2004). Diversity of microbial sialic acid metabolism. Microbiol. Mol. Biol. Rev. 68 132–153. 10.1128/mmbr.68.1.132-153.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker M. J., Barnett T. C., Mcarthur J. D., Cole J. N., Gillen C. M., Henningham A., et al. (2014). Disease manifestations and pathogenic mechanisms of Group A Streptococcus. Clin. Microbiol. Rev. 27 264–301. 10.1128/cmr.00101-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertheim H. F., Nghia H. D., Taylor W., Schultsz C. (2009). Streptococcus suis: an emerging human pathogen. Clin. Infect. Dis. 48 617–625. [DOI] [PubMed] [Google Scholar]
- Worthington R. J., Blackledge M. S., Melander C. (2013). Small-molecule inhibition of bacterial two-component systems to combat antibiotic resistance and virulence. Future Med. Chem. 5 1265–1284. 10.4155/fmc.13.58 [DOI] [PubMed] [Google Scholar]
- Wu C., Ayala E. A., Downey J. S., Merritt J., Goodman S. D., Qi F. (2010). Regulation of ciaXRH operon expression and identification of the CiaR regulon in Streptococcus mutans. J. Bacteriol. 192 4669–4679. 10.1128/jb.00556-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Han J., Gong G., Koffas M. A. G., Zha J. (2020). Wall teichoic acids: Physiology and applications. FEMS Microbiol. Rev. fuaa064. 10.1093/femsre/fuaa064 [Epub Online ahead of print] [DOI] [PubMed] [Google Scholar]
- Wuichet K., Cantwell B. J., Zhulin I. B. (2010). Evolution and phyletic distribution of two-component signal transduction systems. Curr. Opin. Microbiol. 13 219–225. 10.1016/j.mib.2009.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan W., Xiaotao Z., Yuan Y., Hua J., Yuling Z., Yafang T., et al. (2011). DNA microarray analysis of acid-responsive genes of Streptococcus suis serotype 2. Ann. Microbiol. 61 505–510. 10.1007/s13213-010-0165-6 [DOI] [Google Scholar]
- Zaccaria E., Cao R., Wells J. M., Van Baarlen P. (2016). A Zebrafish Larval Model to Assess Virulence of Porcine Streptococcus suis Strains. PLoS One 11:e0151623. 10.1371/journal.pone.0151623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahner D., Grebe T., Guenzi E., Krauss J., Van Der Linden M., Terhune K., et al. (1996). Resistance determinants for beta-lactam antibiotics in laboratory mutants of Streptococcus pneumoniae that are involved in genetic competence. Microb. Drug Resist. 2 187–191. 10.1089/mdr.1996.2.187 [DOI] [PubMed] [Google Scholar]
- Zheng C., Li L., Ge H., Meng H., Li Y., Bei W., et al. (2018). Role of two-component regulatory systems in the virulence of Streptococcus suis. Microbiol. Res. 214 123–128. 10.1016/j.micres.2018.07.002 [DOI] [PubMed] [Google Scholar]
- Zhu B., Ge X., Stone V., Kong X., El-Rami F., Liu Y., et al. (2017). ciaR impacts biofilm formation by regulating an arginine biosynthesis pathway in Streptococcus sanguinis SK36. Sci. Rep. 7:17183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W., Liu S., Zhuang P., Liu J., Wang Y., Lin H. (2017). Characterization of acid-tolerance-associated small RNAs in clinical isolates of Streptococcus mutans: Potential biomarkers for caries prevention. Mol. Med. Rep. 16 9242–9250. 10.3892/mmr.2017.7751 [DOI] [PubMed] [Google Scholar]
- Zhu B., Song L., Kong X., Macleod L. C., Xu P. (2018). A Novel Regulator Modulates Glucan Production, Cell Aggregation and Biofilm Formation in Streptococcus sanguinis SK36. Front. Microbiol. 9:1154. 10.3389/fmicb.2018.01154 [DOI] [PMC free article] [PubMed] [Google Scholar]