The Chinese longsnout catfish (Leiocassis longirostris Günther) is one of the most economically important freshwater fish in China. As wild populations have declined sharply in recent years, it is also a valuable model for research on sexual dimorphism, comparative biology, and conservation. However, the current lack of high-quality chromosome-level genome information for the species hinders the advancement of comparative genomic analysis and evolutionary studies. Therefore, we constructed the first high-quality chromosome-level reference genome for L. longirostris. The total genome was 703.19 Mb, with 389 contigs and contig N50 length of 4.29 Mb. Using high-throughput chromosome conformation capture (Hi-C) data, the genome sequences (685.53 Mb) were scaffolded into 26 chromosomes ranging from 17.36 to 43.97 Mb, resulting in a chromosomal anchoring rate for the genome of 97.44%. In total, 23 708 protein-coding genes were identified in the genome. Phylogenetic analysis indicated that L. longirostris and its closest related species P. fulvidraco diverged approximately 26.6 million years ago. This high-quality reference genome of L. longirostris should pave the way for future genomic comparisons and evolutionary research.
Leiocassis longirostris (also named Jiangtuan) belongs to the family Bagridae, which contains more than 220 species (Ferraris, 2007), and the order Siluriformes. It is a semi-migratory and commercially important freshwater species endemic to China, especially the Huaihe, Liaohe, Minjiang, Yangtze, and Pearl rivers, and the western regions of the Korean Peninsula (Shen et al., 2014; Wang et al., 2006; Zhu et al., 2005). In recent years, wild populations of L. longirostris have experienced a rapid decline due to over-fishing, water pollution, hydropower construction, and other human activities (Liang et al., 2016; Luo et al., 2000; Wang et al., 2006; Xiao & Yang, 2009). Thus, to facilitate conservation and evolutionary research, we constructed the first high-quality chromosome-level reference genome for L. longirostris using BGISEQ-500, Nanopore, and high-throughput chromosome conformation capture (Hi-C) technologies.
One healthy adult female L. longirostris (Figure 1A) collected from a farm at the Sichuan Academy of Agricultural Sciences in Meishan, Sichuan Province, China, was used for genome sequencing. Muscle tissue was collected for DNA extraction after treatment with the anesthetic tricaine MS-222. Genomic DNA for BGISEQ-500 and Nanopore sequencing was isolated using standard chloroform-isoamyl alcohol extraction procedures (Sambrook et al., 1989). DNA quality and quantity were measured using a NanoDrop™ One UV-Vis spectrophotometer (Thermo Fisher Scientific, USA) and Qubit® 3.0 fluorometer (Invitrogen, USA), respectively.
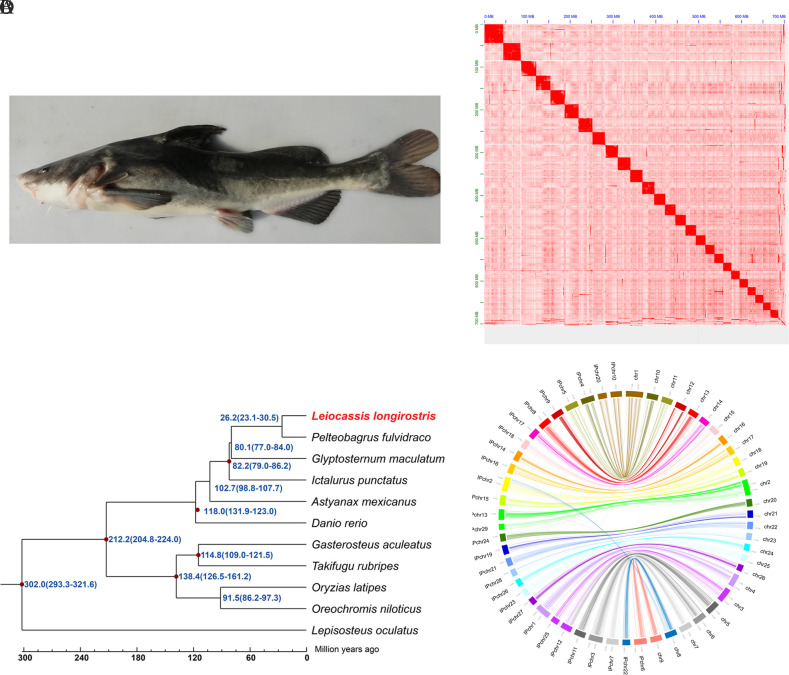
Figure 1.
Genome analysis of L. longirostris
A: Image of L. longirostris used for genome sequencing. B: Hi-C contact map of L. longirostris genome, color bar shows contact density from red (high) to white (low). C: Phylogenetic tree of 11 fish genomes constructed using 3 585 single-copy orthologous genes. D: Genome comparisons between L. longirostris and I. punctatus.
A DNA library (200–400 bp insert size) was constructed following the manufacturer’s instructions as described in previous study (Huang et al., 2017). The library was then sequenced following the BGISEQ-500 protocols (Huang et al., 2017). The short-read data obtained from the BGISEQ-500 platform were filtered using SOAPnuke v1.5.2 (Chen et al., 2018). The adapter sequences were removed from the reads, and paired reads with more than 10% ambiguous or low-quality (Phred score<5) bases were discarded, with BLAST v2.2.31 applied for the evaluation of sample contamination (Altschul et al., 1990). As a result, we obtained a total of 64.11 Gb short reads (Supplementary Table S1). Using Jellyfish v2.2.6 (Marçais & Kingsford, 2011), the K-mer frequency distribution was calculated. The Jellyfish results were subsequently delivered to GenomeScope (Vurture et al., 2017). Using a K-mer size of 17, the K-mer frequency distribution for L. longirostris was obtained (Supplementary Figure S1). As a result, the genome size of L. longirostris was estimated to be 688.99 Mb, with heterozygosity, repeat content, and GC content of 0.35%, 42.53%, and 38.43%, respectively.
For Nanopore sequencing, we prepared a library using a Ligation Sequencing Kit (Oxford Nanopore Technologies, UK, SQK-LSK109) according to the manufacturer’s instructions. The library was sequenced using the Nanopore GridION X5 sequencer (Oxford Nanopore Technologies, UK) with flow cell R9.4 on five flow cells. Base calling was performed using Guppy v2.0.8 with default parameters, and reads were filtered for mean_qscore_template ≥7. NanoPlot v1.0.0 (De Coster et al., 2018) was then used to filter the Nanopore reads. For the construction of the Hi-C library, 1 g of muscle tissue was used to prepare a library according to previously established protocols (Rao et al., 2014). The library was then sequenced on a BGISEQ-500 sequencer (BGI Genomics, China) using 100 bp paired end sequencing.
For transcriptome sequencing, the liver tissues of 15 L. longirostris individuals collected from the same farm were used for RNA extraction with TRIzol reagent (Invitrogen, USA), followed by treatment with DNase I (Invitrogen, USA) to remove genomic DNA. RNA concentration and integrity were measured using a Qubit® RNA Assay Kit and Qubit® 2.0 fluorometer (Life Technologies, USA) and an RNA Nano 6000 Assay Kit with the Agilent Bioanalyzer 2100 system (Agilent Technologies, USA), respectively. Three RNA sequencing libraries (five fish per library) with an insert size of 250–300 bp were prepared using a NEBNext® Ultra™ RNA Library Prep Kit for Illumina® (NEB, USA) following the manufacturer’s protocols, and then sequenced on the Illumina Hiseq X Ten platform (Illumina Inc., USA) as 150 bp paired-end reads. The raw RNA-seq reads were cleaned and assembled as described previously (Ye et al., 2018).
Using the Nanopore sequencing platform, we obtained 43.23 Gb long reads, with an expected average sequencing coverage of 61.48 X for genome assembly (Supplementary Table S1). We then performed de novo genome assembly using Canu v1.8 (Koren et al., 2017) following the correction, trimming, and contig construction steps. After contig assembly, three rounds of contig sequence polishing were performed with cleaned genomic short reads using Pilon v1.23 (Walker et al., 2014). Purge Haplotigs v1.0.3 (Roach et al., 2018) was used to produce an improved and deduplicated assembly. Finally, we obtained the assembled genome of L. longirostris, which was 703.19 Mb in length, with 389 contigs and an N50 contig size of 4.29 Mb. This is a medium-sized genome among other sequenced catfish genomes (Table 1; Supplementary Table S2). We performed genome assembly quality control using the distribution of GC_depth. The GC_depth scatter plots demonstrated a Poisson distribution, indicating that this genome had no significant contamination. The overall GC-content of 39.67% in the L. longirostris genome was slightly higher than that of the walking catfish (Clarias batrachus) (Li et al., 2018) and common carp (Cyprinus carpio) but much lower than that of most teleost genomes (Xu et al., 2014). The completeness of the assembled L. longirostris genome was estimated using BUSCO v3.0.2 (Simão et al., 2015) with the actinopterygii_odb9 database. As a result, 4 293 (93.6 %) of the 4 584 BUSCO genes were completely identified in the genome, including 4 109 (89.6%) single-copy and 184 (4.0%) duplicated genes. These results suggest high genome assembly completeness.
Table 1. Summary of sequenced catfish genomes.
| Species | Family | Sequencing platform | Assembly size (Mb) | Identified genes | Scaffold N50 (Mb) | Contig N50 (kb) | References |
| Longsnout catfish, Leiocassis longirostris | Bagridae | BGISEQ-500, Nanopore, Hi-C | 703.19 | 23 708 | 28.03 | 3 090.00 | This study |
| Yellow catfish, Pelteobagrus fulvidraco | Bagridae | Illumina, PacBio, Hi-C | 732.80 | 24 552 | 25.80 | 1 100.00 | Gong et al., 2018 |
| Illumina, PacBio | 714.00 | 21 562 | 3.65 | 970.00 | Zhang et al., 2018 | ||
| Glyptosternon maculatum | Sisoridae | PacBio, Illumina, 10X Genomics, BioNano | 662.34 | 22 066 | 20.90 | 993.67 | Liu et al., 2018 |
| Channel catfish, Ictalurus punctatus | Ictaluridae | Illumina | 845.40 | 21 556 | 7.25 | 48.50 | Chen et al., 2016 |
| Illumina, PacBio | 783.00 | 26 661 | 7.73 | 77.20 | Liu et al., 2016 | ||
| Giant devil catfish, Bagarius yarrelli | Sisoridae | Illumina, PacBio | 571.00 | 19 027 | 3.10 | 1 600.00 | Jiang et al., 2019 |
| Walking catfish, Clarias batrachus | Clariidae | Illumina | 821.00 | 22 914 | 0.36 | 19.00 | Li et al., 2018 |
| Striped catfish, Pangasianodon hypophthalmus | Pangasiidae | Illumina | 700.00 | 28 600 | 14.29 | 6.00 | Kim et al., 2018 |
For chromosome-level assembly of the L. longirostris genome, Hi-C reads were first filtered using HIC-Pro v2.8.0 (Servant et al., 2015). Juicer v1.5 (Durand et al., 2016a) was then used to analyze the Hi-C datasets, and 3D-DNA v170123 was used to anchor the genome assembly to the chromosomes (Dudchenko et al., 2017) with parameters “-m haploid -s 0 -c 26”. The contact matrix of the L. longirostris contigs was mapped using Juicebox v1.11.08 (Durand et al., 2016b) (Figure 1B). A total of 126.35 Gb clean Hi-C reads were obtained, and 685.53 Mb (97.44% of total genome) genome sequences were successfully scaffolded into 26 pseudochromosomes. The number of chromosome scaffolds is consistent with previous research on karyotypes of L. longirostris (2n=52; Hong & Zhou, 1984). The lengths of chromosomes ranged from 17.36 Mb to 43.97 Mb (Supplementary Table S3). The scaffold N50 of the chromosome-level assembly was 28.03 Mb (Table 1).
For the annotation of repetitive sequences, we used RepeatModeler v1.0.10 (Bao & Eddy, 2002), which employs two complementary computational methods, i.e., RECON v1.08 and RepeatScout v1.0.5 (RepeatScout, RRID:SCR 014653) (Price et al., 2005), to identify repeat element boundaries and family relationships from sequence data. Subsequently, the outputs from the RepeatModeler and RepBase v21.01 library were combined and used for further characterization of transposable elements (TEs), many of which are not repetitive, and other repeats by homology-based methods, including identification with RepeatMasker v4.0.7, rmblast-2.2.28 (RRID:SCR 012954). Using RepBase-based homology and de novo methods, 239.11 Mb (33.99% of total genome) repetitive elements were identified, with DNA transposons (146.40 Mb, 20.81%) being the most abundant type in the genome (Supplementary Table S4-1). The proportion of repetitive elements in L. longirostris is similar to that in the Glyptosternon maculatum genome (33.96%) (Liu et al., 2018) and higher than that of most teleost genomes (Supplementary Table S4-2).
Combined homology-, de novo-, and transcriptome-based methods were used for gene prediction in the genome. The protein sequences of nine fish species, including Danio rerio, Gasterosteus aculeatus, Ictalurus Punctatus, Larimichthys crocea, Oreochromis niloticus, Oryzias latipes, Pangasianodon hypophthalmus, Tachysurus fulvidraco, and Takifugu rubripes, were downloaded from the Ensembl database and mapped onto the assembled L. longirostris genome using BLASTN. Subsequently, GeneWise v2.2.0 (Birney et al., 2004) with default options was used for homologous annotation. For de novo prediction, Augustus v3.1.0 (Stanke & Waack, 2003) was used to predict gene models. In addition, RNA-seq data were aligned to the assembled L. longirostris genome to predict gene coding regions. The gene models were then predicted by combining the above homology-, de novo-, and transcriptome-based information using PASA v2.3.3 (Haas et al., 2003). Various databases, including SwissProt (Boeckmann et al., 2003), Kyoto Encyclopedia of Genes and Genomes (KEGG) (Kanehisa & Goto, 2000), TrEMBL (Boeckmann et al., 2003), InterPro (Zdobnov & Apweiler, 2001), and Gene Ontology (GO) (Ashburner et al., 2000), were used to functionally annotate the predicted protein-coding genes, and GLEAN (Elsik et al., 2007) was used to create a consensus gene set. Finally, a total of 23 708 protein-coding genes were identified in the L. longirostris genome (Supplementary Table S5), of which 21 692, 20 072, 23 114, 21 169, and 16 638 protein-coding genes were annotated in the SwissProt, KEGG, TrEMBL, InterPro, and GO databases, respectively (Supplementary Table S6 and Figure S2). BUSCO was also used to test the completeness of the genome annotation with the actinopterygii_odb9 database, which showed that 92.4% complete and 4.0% fragmented conserved single-copy orthologs were predicted for L. longirostris.
For non-coding RNAs, microRNA (miRNA) and small nuclear RNA (snRNA) were predicted using INFERNAL v1.1 (Nawrocki & Eddy, 2013) and the Rfam database (Kalvari et al., 2018). Transfer RNA (tRNA) and ribosomal RNA (rRNA) were identified using tRNAscan-SE v1.3.1 (Lowe & Eddy, 1997) and RNAmmer v1.2 (Lagesen et al., 2007), respectively. After analysis, 422 miRNAs, 2 118 tRNAs, 1 838 rRNAs, and 1 925 snRNAs were annotated in the L. longirostris genome (Supplementary Table S7).
To identify gene families, protein sequences from the longest transcripts of each gene from L. longirostris and 10 other fish species, including D. rerio, Astyanax mexicanus, G. aculeatus, G. maculatum, I. punctatus, Lepisosteus oculatus, Oreochromis niloticus, Oryzias latipes, Pelteobagrus fulvidraco, and T. rubripes, were aligned using BLASTP with an e-value threshold of 1e-5. OrthoMCL v1.4 (Li et al., 2003) was then used to construct gene families. A total of 19 438 gene families and 3 585 single-copy ortholog families were identified among the 11 species, with 68 gene families specific to L. longirostris (Supplementary Table S8). In addition, 11 729 (89.1%) gene families were shared by the four catfish species, with 301 gene families specific to L. longirostris (Supplementary Figure S3).
To investigate the phylogenetic relationships of L. longirostris with the above 10 fish species, the shared single-copy genes were aligned by MUSCLE v3.8.31 (Edgar, 2004). RAxML v8.2.1163 (Stamatakis, 2014) was then employed to construct a phylogenetic tree with the -m PROTGAMMAAUTO model and 100 bootstrap replicates. MCMCTREE v3.8.31 (Yang, 2007) was used to estimate divergence time based on the “correlated molecular clock” and “HKY85” models. Phylogenetic analysis indicated that L. longirostris and P. fulvidraco, which are both from the family Bagridae, were clustered onto one branch, and L. longirostris was close to the P. fulvidraco, G. maculatum, and I. punctatus clades, which belong to the Siluriformes order. These results are similar to previous phylogenetic analyses based on the mitochondrial genome of L. longirostris (Liu et al., 2019). Our results also showed that L. longirostris diverged ~26.2 million years ago from its closest related species P. fulvidraco (Figure 1C). Furthermore, phylogenetic analysis estimated that I. punctatus diverged from P. fulvidraco around 82.2 million years ago, consistent with the 81.9 million years reported in previous study (Gong et al., 2018). Collinearity analysis of chromosomes between L. longirostris and I. punctatus was performed using LASTZ v1.02.00 (Harris, 2007) with parameters “T=2 C=2 H=2000 Y=3400 L=6000 K=2200”. As a result, all 26 pseudochromosomes of L. longirostris displayed high homology with the corresponding chromosomes of I. punctatus (Figure 1D), suggesting high-quality L. longirostris genome assembly.
In the present study, the first chromosome sequences for L. longirostris were constructed using a combination of BGISEQ-500, Nanopore, and Hi-C technologies. The reference genome exhibited high quality in terms of continuity and completeness. This study should improve our understanding of the L. longirostris genome and provide valuable chromosomal information for genomic comparisons and evolutionary research among important aquaculture species.
DATA AVAILABILITY
The raw genome and RNA sequencing data were deposited in the National Center for Biotechnology Information (NCBI) database under accession No. PRJNA692071.
SUPPLEMENTARY DATA
Supplementary data to this article can be found online.
COMPETING INTERESTS
The authors declare that they have no competing interests.
AUTHORS’ CONTRIBUTIONS
W.P.H., H.L., J.Z., and H.Y. designed the experiments; W.P.H., H.L., J.Z., Z.L., T.S.J., C.H.L., Y.J.Y., M.B.X., and C.W.Z. performed the experiments and analyzed data; W.P.H., G.J.L., H.Y.X., and H.Y. wrote the paper. All authors read and approved the final version of the manuscript.
Funding Statement
This study was supported by the China Agriculture Research System (CARS-46), Fundamental Research Funds for the Central Universities (XDJK2017B008, XDJK2017C035, XDJK2019C025, 5360300098), Natural Science Foundation of Chongqing (cstc2020jcyj-msxmX0438), and National Natural Science Foundation of China (32071651)
Contributor Information
Hui Luo, Email: luohui2629@126.com.
Hua Ye, Email: yhlh2000@126.com.
References
- 1.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ Basic local alignment search tool. Journal of Molecular Biology. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al Gene ontology: tool for the unification of biology. Nature Genetics. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bao ZR, Eddy SR Automated de novo identification of repeat sequence families in sequenced genomes. Genome Research. 2002;12(8):1269–1276. doi: 10.1101/gr.88502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Birney E, Clamp M, Durbin R GeneWise and genomewise. Genome Research. 2004;14(5):988–995. doi: 10.1101/gr.1865504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boeckmann B, Bairoch A, Apweiler R, Blatter MC, Estreicher A, Gasteiger E, et al The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Research. 2003;31(1):365–370. doi: 10.1093/nar/gkg095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen XH, Zhong LQ, Bian C, Xu P, Qiu Y, You XX, et al High-quality genome assembly of channel catfish, Ictalurus punctatus. Gigascience. 2016;5(1):39. doi: 10.1186/s13742-016-0142-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen YX, Chen YS, Shi CM, Huang ZB, Zhang Y, Li SK, et al SOAPnuke: A MapReduce acceleration-supported software for integrated quality control and preprocessing of high-throughput sequencing data. Gigascience. 2018;7(1):gix120. doi: 10.1093/gigascience/gix120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Coster W, D’Hert S, Schultz DT, Cruts M, Van Broeckhoven C NanoPack: Visualizing and processing long-read sequencing data. Bioinformatics. 2018;34(15):2666–2669. doi: 10.1093/bioinformatics/bty149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dudchenko O, Batra SS, Omer AD, Nyquist SK, Hoeger M, Durand NC, et al De novo assembly of the Aedes aegypti genome using Hi-C yields chromosome-length scaffolds . Science. 2017;356(6333):92–95. doi: 10.1126/science.aal3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Durand NC, Robinson JT, Shamim MS, Machol I, Mesirov JP, Lander ES, et al Juicebox provides a visualization system for Hi-C contact maps with unlimited zoom. Cell Systems. 2016b;3(1):99–101. doi: 10.1016/j.cels.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Durand NC, Shamim MS, Machol I, Rao SSP, Huntley MH, Lander ES, et al Juicer provides a one-click system for analyzing loop-resolution Hi-C experiments. Cell Systems. 2016a;3(1):95–98. doi: 10.1016/j.cels.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edgar R MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elsik CG, Mackey AJ, Reese JT, Milshina NV, Roos DS, Weinstock GM Creating a honey bee consensus gene set. Genome Biology. 2007;8(1):R13. doi: 10.1186/gb-2007-8-1-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferraris CJ Checklist of catfishes, recent and fossil (Osteichthyes: Siluriformes), and catalogue of siluriform primary types. Zootaxa. 2007;1418(1):1–628. doi: 10.11646/zootaxa.1418.1.1. [DOI] [Google Scholar]
- 15.Gong GR, Dan C, Xiao SJ, Guo WJ, Huang PP, Xiong Y, et al Chromosomal-level assembly of yellow catfish genome using third-generation DNA sequencing and Hi-C analysis. Gigascience. 2018;7(11):giy120. doi: 10.1093/gigascience/giy120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haas BJ, Delcher AL, Mount SM, Wortman JR, Smith Jr RK, Hannick LI, et al Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies . Nucleic Acids Research. 2003;31(19):5654–5666. doi: 10.1093/nar/gkg770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harris RS. 2007. Improved Pairwise Alignment of Genomic DNA. Ph.D. dissertation, The Pennsylvania State University, Pennsylvania.
- 18.Hong YH, Zhou T Karyotypes of nine species of Chinese Catfishes (Bagridae) Zoological Research. 1984;5(S3):21–28. [Google Scholar]
- 19.Huang J, Liang XM, Xuan YK, Geng CY, Li YX, Lu HR, et al A reference human genome dataset of the BGISEQ-500 sequencer. GigaScience. 2017;6(5):gix024. doi: 10.1093/gigascience/gix024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang WS, Lv YY, Cheng L, Yang KF, Bian C, Wang XA, et al Whole-genome sequencing of the giant devil catfish, Bagarius yarrelli . Genome Biology and Evolution. 2019;11(8):2071–2077. doi: 10.1093/gbe/evz143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kalvari I, Argasinska J, Quinones-Olvera N, Nawrocki EP, Rivas E, Eddy SR, et al Rfam 13. 0: shifting to a genome-centric resource for non-coding RNA families. Nucleic Acids Research. 2018;46(D1):D335–D342. doi: 10.1093/nar/gkx1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kanehisa M, Goto S KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Research. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim O, Nguyen PT, Shoguchi E, Hisata, K, Vo T, Inoue J, et al A draft genome of the striped catfish, Pangasianodon hypophthalmus, for comparative analysis of genes relevant to development and a resource for aquaculture improvement . BMC Genomics. 2018;19(1):733. doi: 10.1186/s12864-018-5079-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, Phillippy AM Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation . Genome Research. 2017;27(5):722–736. doi: 10.1101/gr.215087.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lagesen K, Hallin P, Rodland EA, Staerfeldt HH, Rognes T, Ussery DW RNAmmer: Consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Research. 2007;35(9):3100–3108. doi: 10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li L, Stoeckert Jr CJ, Roos DS OrthoMCL: Identification of ortholog groups for eukaryotic genomes. Genome Research. 2003;13(9):2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li N, Bao LS, Zhou T, Yuan ZH, Liu SK, Dunham R, et al Genome sequence of walking catfish (Clarias batrachus) provides insights into terrestrial adaptation . BMC Genomics. 2018;19(1):952. doi: 10.1186/s12864-018-5355-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liang HW, Guo SS, Luo XZ, Li Z, Zou GW Molecular diagnostic markers of Tachysurus fulvidraco and Leiocassis longirostris and their hybrids . Springerplus. 2016;5(1):2115. doi: 10.1186/s40064-016-3766-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu HP, Liu QY, Chen ZQ, Liu YC, Zhou CW, Liang QQ, et al Draft genome of Glyptosternon maculatum, an endemic fish from Tibet Plateau . GigaScience. 2018;7(9):giy104. doi: 10.1093/gigascience/giy104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu Y, Wu PD, Zhang DZ, Zhang HB, Tang BP, Liu QN, Dai LS Mitochondrial genome of the yellow catfish Pelteobagrus fulvidraco and insights into Bagridae phylogenetics . Genomics. 2019;111(6):1258–1265. doi: 10.1016/j.ygeno.2018.08.005. [DOI] [PubMed] [Google Scholar]
- 31.Liu ZJ, Liu SK, Yao J, Bao LS, Zhang JR, Li Y, et al The channel catfish genome sequence provides insights into the evolution of scale formation in teleosts. Nature Communications. 2016;7:11757. doi: 10.1038/ncomms11757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lowe TM, Eddy SR tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Research. 1997;25(5):955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luo M, Jiang LK, Liu Y, Zhan GQ, Xia SZ Comparative Study on Lsoenzymes in Cleiocassis Longirostris . Chinese Journal of Applied & Environmental Biology. 2000;6(5):447–451. [Google Scholar]
- 34.Marçais G, Kingsford C A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics. 2011;27(6):764–770. doi: 10.1093/bioinformatics/btr011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nawrocki EP, Eddy SR Infernal 1. 1:100-fold faster RNA homology searches. Bioinformatics. 2013;29(22):2933–2935. doi: 10.1093/bioinformatics/btt509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Price AL, Jones NC, Pevzner PA De novo identification of repeat families in large genomes . Bioinformatics. 2005;21(S1):i351–i358. doi: 10.1093/bioinformatics/bti1018. [DOI] [PubMed] [Google Scholar]
- 37.Rao SSP, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, et al A 3D map of the human genome at Kilobase resolution reveals principles of chromatin looping. Cell. 2014;159(7):1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roach MJ, Schmidt SA, Borneman AR Purge Haplotigs: Allelic contig reassignment for third-gen diploid genome assemblies. BMC Bioinformatics. 2018;19(1):460. doi: 10.1186/s12859-018-2485-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular Cloning: A Laboratory Manual. 2nd ed. Cold Spring Harbor: Cold Spring Harbor Laboratory Press.
- 40.Servant N, Varoquaux N, Lajoie BR, Viara E, Chen CJ, Vert JP, et al HiC-Pro: An optimized and flexible pipeline for Hi-C data processing. Genome Biology. 2015;16(1):259. doi: 10.1186/s13059-015-0831-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shen T, He XS, Lei ML, Wang JR, Li XM, Li JM Cloning and structure of a histocompatibility class IIA gene (Lelo-DAA) in Chinese longsnout catfish (Leiocassis longirostris) . Genes & Genomics. 2014;36(6):745–753. [Google Scholar]
- 42.Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31(19):3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- 43.Stamatakis A RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30(9):1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stanke M, Waack S Gene prediction with a hidden Markov model and a new intron submodel. Bioinformatics. 2003;19(S2):ii215–ii225. doi: 10.1093/bioinformatics/btg1080. [DOI] [PubMed] [Google Scholar]
- 45.Vurture GW, Sedlazeck FJ, Nattestad M, Underwood CJ, Fang H, Gurtowski J, et al GenomeScope: fast reference-free genome profiling from short reads. Bioinformatics. 2017;33(14):2202–2204. doi: 10.1093/bioinformatics/btx153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, et al Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One. 2014;9(11):e112963. doi: 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang ZW, Zhou JF, Ye YZ, Wei QW, Wu QJ Genetic structure and low-genetic diversity suggesting the necessity for conservation of the Chinese Longsnout catfish, Leiocassis longirostris (Pisces: Bagriidae) . Environmental Biology of Fishes. 2006;75(4):455–463. doi: 10.1007/s10641-006-0035-z. [DOI] [Google Scholar]
- 48.Xiao MS, Yang G Isolation and characterization of 17 microsatellite loci for the Chinese longsnout catfish (Leiocassis longirostris) . Molecular Ecology Resources. 2009;9(3):1039–1041. doi: 10.1111/j.1755-0998.2009.02554.x. [DOI] [PubMed] [Google Scholar]
- 49.Xu P, Zhang XF, Wang XM, Li JT, Liu GM, Kuang YY, et al Genome sequence and genetic diversity of the common carp, Cyprinus carpio. Nature Genetics. 2014;46(11):1212–1219. doi: 10.1038/ng.3098. [DOI] [PubMed] [Google Scholar]
- 50.Yang ZH PAML 4: phylogenetic analysis by maximum likelihood. Molecular Biology and Evolution. 2007;24(8):1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 51.Ye H, Xiao SJ, Wang XQ, Wang ZY, Zhang ZS, Zhu CK, et al Characterization of Spleen Transcriptome of Schizothorax prenanti during Aeromonas hydrophila Infection . Marine Biotechnology. 2018;20(2):246–256. doi: 10.1007/s10126-018-9801-0. [DOI] [PubMed] [Google Scholar]
- 52.Zhang SY, Li J, Qin Q, Liu W, Bian C, Yi YH, et al Whole-genome sequencing of Chinese yellow catfish provides a valuable genetic resource for high-throughput identification of toxin genes. Toxins. 2018;10:488. doi: 10.3390/toxins10120488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zdobnov EM, Apweiler R InterProScan—An integration platform for the signature-recognition methods in InterPro. Bioinformatics. 2001;17(9):847–848. doi: 10.1093/bioinformatics/17.9.847. [DOI] [PubMed] [Google Scholar]
- 54.Zhu XM, Xie SQ, Lei W, Cui YB, Yang YX, Wootton RJ Compensatory growth in the Chinese longsnout catfish, Leiocassis longirostris following feed deprivation: temporal patterns in growth, nutrient deposition, feed intake and body composition . Aquaculture. 2005;248(1−4):307–314. doi: 10.1016/j.aquaculture.2005.03.006. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data to this article can be found online.
Data Availability Statement
The raw genome and RNA sequencing data were deposited in the National Center for Biotechnology Information (NCBI) database under accession No. PRJNA692071.