Single-cell RNA sequencing (scRNA-seq) is useful for exploring cell heterogeneity. For large animals, however, little is known regarding spermatogonial stem cell (SSC) self-renewal regulation, especially in dairy goats. In this study, we described a high-resolution scRNA-seq atlas derived from a dairy goat. We identified six somatic cell and five spermatogenic cell subtypes. During spermatogenesis, genes with significantly changed expression were mainly enriched in the Notch, TGF-β, and Hippo signaling pathways as well as the signaling pathway involved in the regulation of stem cell pluripotency. We detected and screened specific candidate marker genes (TKTL1 and AES) for spermatogonia. Our study provides new insights into goat spermatogenesis and the development of testicular somatic cells.
The testis is the most complex organ of the transcriptome and expresses more than 80% of the protein-coding genes in humans and other species (Melé et al., 2015; Soumillon et al., 2013). The complexity of the testicular transcriptome is closely related to spermatogenesis, which is a complex and continuous process that occurs asynchronously in the seminiferous tubules. There are usually 4–5 waves of spermatogenesis in the seminiferous tubules of adult mammals, which makes the separation and molecular characterization of the various substages of spermatogenesis difficult (Oakberg, 1956). In recent years, our understanding of spermatogenesis has relied on histological evaluation and expression of marker genes (de Rooij, 2017; Gaysinskaya & Bortvin, 2015; Mays-Hoopes et al., 1995). The most common methods for analyzing and obtaining different cell types include fluorescence-activated cell sorting (FACS) and STA-PUT (Liu et al., 2015), especially for large animals such as livestock (Chen et al., 2017; Du et al., 2021; Shah et al., 2018). However, FACS is hampered by a lack of specific markers, and can only isolate limited subtypes of enriched male germ cells. These limitations have been problematic for deciphering the molecular hallmarks of the various substages of spermatogenesis, and for elucidating the molecular basis and transcriptional dynamics of the mitotic-to-meiotic switch in mammals. However, the advancement of single-cell RNA sequencing (scRNA-seq) technology has expanded our understanding of heterogeneous tissues, especially mammalian embryos and reproductive organs (Shami et al., 2020; Xiang et al., 2020; Yan et al., 2013).
scRNA-seq is an unbiased method that can sequence thousands of cells in a single experiment, as well as obtain thousands of individual features per cell and provide ultra-high-resolution transcriptomes of animal tissues and organs (Choi & Kim, 2019). After cluster analysis, data can be ordered according to the cell development pathway to form continuous, dynamic, and heterogeneous differentiation process. Currently, scRNA-seq has been widely used in the study of mammalian spermatogenesis (Lau et al., 2020; Shami et al., 2020; Tan et al., 2020).
Guo first used scRNA-seq to analyze human spermatogonial stem cells rich in SSEA4+ and c-KIT+, and obtained new candidate spermatogonial stem cell marker genes, including TCF7, PIWIL2, and BMPR1A/B (Guo et al., 2017). Subsequently, scRNA-seq was applied to analyze the entire testicular tissue in mice and humans, with cells divided into germ cell and somatic cell clusters. In mice, four germ cell clusters and seven somatic cell clusters were defined (Green et al., 2018). In humans, testis cells were subdivided into eight germ cell clusters, and five somatic cell clusters (Guo et al., 2018). Furthermore, given that the composition and proportion of cells in the testis vary with age, scRNA-seq has also been used to analyze and compare testicular tissues in different periods and species (Guo et al., 2021; Hermann et al., 2018; Lau et al., 2020; Liao et al., 2019; Shami et al., 2020; Tan et al., 2020).
Different from the first wave of spermatogenesis in postnatal mice (Oakberg, 1957), humans and large animals (monkeys, pigs, cattle, and sheep) have puberty. And the testes of large prepubertal animals usually do not have whole spermatogenesis. Although the process of spermatogenesis is similar in mammals, it is still difficult to translate knowledge generated in mice to large animals due to the existence of puberty. Existing research has mainly focused on higher primates, and there is a lack of research and data on large domestic animals (Lau et al., 2020; Shami et al., 2020). In addition, although scientists have made considerable efforts to understand processes through the isolation and analysis of specific germ cells, much remains unknown.
Here, we used the 10x Genomics Platform to perform scRNA-seq on the testis of a pre-sexual young Guanzhong dairy goat. Germ and somatic cells were grouped according to the marker genes in sheep, mice, humans, and monkeys, and six somatic cell groups and ten germ cell subgroups were determined. Our research revealed specific marker genes for the development of mammalian puberty somatic cells and spermatogenic cell subtypes in dairy goats.
Guanzhong dairy goat testis tissue was separated into single cells for scRNA-seq. The single-cell library was constructed using the ChromiumTM Controller and ChromiumTM Single Cell 3' Reagent v2 Kit (10x Genomics, USA). scRNA-seq was performed using the Illumina Hiseq 4000 platform (BGI-Shenzhen, China). After removing low-quality reads, Cell Ranger v3.0.2 was used for gene expression quantification and cell-type identification based on cell barcodes and unique molecular identifier (UMI) information of reads in every single cell. The matrix data file generated from the 10-fold genomics alignment was used as input in the Seurat package in R for subsequent analysis, including cell type identification, principal component analysis (PCA), and t-Stochastic-Neighbor Embedding (t-SNE) dimensionality reduction to visualize single-cell clustering. Several clusters from the t-SNE plot were used for pseudotime analysis with Monocle 2. Through data filtering, normalization, cell classification and clustering, key gene selection, dimensionality reduction, and sorting, the pseudotime trajectory of germ cells in spermatogenesis was obtained. Functional enrichment of marker genes within each cluster was then investigated by Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses.
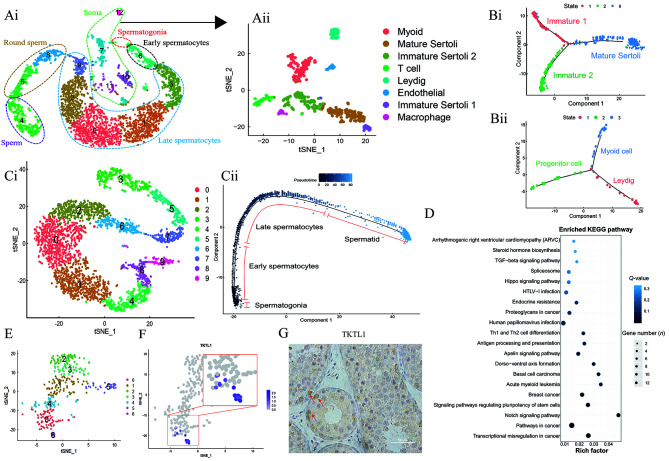
We sequenced 11 753 cells and selected 3 890 cells in the range of 200 to 6 000 feature RNAs. These cells were divided into 13 clusters, including four clusters of somatic cells and nine clusters of germ cells (Figure 1Ai). The expression patterns of known marker genes and GO analysis assigned the nine germ cell types to spermatogonia (UTF1, FOXO1), early spermatocytes (SYCP2, SYCP3), late spermatocytes (ACR, LCA5), round sperm (TEX29, ACRV1), and sperm (PRM1, TNP2) in the germ cell (DDX4) cluster (Supplementary Figure S1).
Figure 1.
Single-cell RNA sequencing reveals atlas of dairy goat testis cells
Ai: t-SNE cluster containing 3 890 cells (cells are colored by 13 broad cell types). Aii: t-SNE diagram of somatic cells in testis of dairy goat (cells are colored by eight broad cell types). Bi: Pseudotime trajectory analysis of Sertoli cells. Bii: Pseudotime trajectory analysis of Leydig and Myoid cells. Ci: t-SNE image of testicular germ cells in dairy goats (cells are colored by 10 broad cell types). Cii: Pseudotime trajectory analysis of germ cell clusters. Shades of blue indicate sorting according to pseudotime value. D: KEGG analysis of main differentially expressed genes during spermatogenesis. E: t-SNE diagram of re-clustering of spermatocytes and spermatogonia in dairy goat testes (cells are colored by seven broad cell types). F: Expression distribution of TKTL1 in spermatogonia clusters. G: Immunohistochemical staining for TKTL1 expression in dairy goat. Red arrows indicate spermatogonia. Scale bar: 50 μm.
The process of spermatogenesis in mammals requires the support of a specialized microenvironment (i.e., niche) composed of multiple somatic cell types (Morrison and Spradling, 2008; Scadden, 2006). Here, the somatic cell clusters (Clusters 7, 10, 11, and 12) were re-clustered and subdivided into eight somatic cell clusters (Figure 1Aii). Clustering analysis of these somatic cells revealed six major cell types. All clusters were identified as known cell types based on previously reported cell type-specific marker genes: i.e., Sertoli cells (SOX9), Leydig cells (INSL3), endothelial cells (HES1), myoid cells (ACTA2), macrophages (CD83), and T cells (NKG7) (Supplementary Figure S2).
Guo et al. (2020) identified two types of immature Sertoli cells in pubertal testes, which eventually develop into the same type of mature Sertoli cells, and myoid and Leydig cells, which develop and differentiate from the same progenitor. However, this phenomenon does not exist in mice (Bellvé et al., 1977; Tharmalingam et al., 2018). To explore whether this condition is prevalent in adolescent dairy goat testes, we analyzed and examined the Sertoli cell population (Clusters 1, 2, and 6 in Figure 1Aii), Leydig cell population (Cluster 4 in Figure 1Aii), and myoid cell population (Cluster 0 in Figure 1Aii). Cluster analysis defined three Sertoli clusters, and Monocle pseudotime analysis divided the cells of these clusters into three different states. As the peseudotiming progresses, the two initial states were merged into the same state, which we defined respectively as Immature 1, Immature 2, and Mature Sertoli (Figure 1Bi). Through correlation analysis and identification, Cluster 6 corresponded to Mature Sertoli and was rich in ribosomal protein genes (Supplementary Figure S2). Unlike Guo et al. (2020), cluster analysis did not initially place the Leydig and myoid cells into one large cluster, but instead into two separate clusters. However, Monocle pseudotime analysis showed that the Leydig and myoid cells are differentiated from common progenitor cells (Figure 1Bii), as also reported in the result of Guo et al. (2020).
To study each germ cell type and subtype during spermatogenesis in dairy goat, we re-clustered the germ cell clusters (Clusters 0, 1, 2, 3, 4, 5, 6, 8, and 9 in Figure 1Ai). Cell types were determined based on known marker genes: Clusters 8 and 9 were spermatogonia and early spermatocytes, Clusters 1, 2, 4, and 6 were late spermatocytes, Clusters 5 and 7 were round spermatids, and Cluster 3 was sperm cells (Figure 1Ci). Monocle pseudotime analysis verified our conclusions. The germ cell clusters were arranged from left to right to form a continuous curve (Figure 1Cii). Monocle was used to analyze differentially expressed genes during spermatogenesis. During spermatogenesis, genes showing significant changes in expression were mainly enriched in the Notch, Hippo, and TGF-β signaling pathways, as well as the signaling pathway regulating the pluripotency of stem cells (Figure 1D). Genes specifically expressed at the beginning of spermatogenesis were mainly enriched in chromatin binding, DNA binding, positive regulation of transcription, and enhancer sequence-specific DNA binding. With progressing spermatogenesis, late spermatocytes mainly expressed genes related to adenosine triphosphate (ATP) binding, nuclear acid-binding, spermatogenesis, and male meiosis. At the end of spermatogenesis, sperm cells specifically expressed genes related to sperm motility, cell differentiation, and cell fate determination (Supplementary Figure S3).
In mammalian testes, the proportion of stem cells is particularly low. In rodents, for example, stem cells account for only ~0.03% of testicular germ cells (Tagelenbosch & de Rooij, 1993). In the current study, only ~10 cells were defined as spermatogonia. As such, it was not possible to classify them individually. So we chose germ-cell Clusters 8 and 9 (Figure 1Ci) for re-clustering and grouping and used known marker genes in humans and mice (SOHLH1, UTF1, ZBTB16, and FOXO1) to identify Cluster 6 as a spermatogonia cluster (Figure 1E; Supplementary Figure S4). However, many marker genes common in humans and mice, including SALL4, PIWIL4, TSPAN33, and GFRA1 (Fayomi & Orwig, 2018; Lovelace et al., 2016; Wang et al., 2018; Wei et al., 2021a, b), were barely distributed in the dairy goat spermatogonia clusters. This may be due to the small proportion of undifferentiated spermatogonia in the spermatogonia clusters obtained in our study. We performed functional enrichment analysis of spermatogonia clusters, and results showed that compared with early spermatocytes, spermatogonia marker genes were significantly enriched in protein binding, cytoplasm, and extracellular exosome-related pathways. Related pathways such as ubiquitin-binding and regulation of cell migration were also enriched (Supplementary Figure S4).
To determine the specific marker genes of spermatogonia in dairy goat, we further analyzed the specific expression genes of the spermatogonia clusters. We selected AES and TKTL1 as candidate marker genes for spermatogonia in dairy goat. Transketolase 1 (TKTL1) plays an important role in the pentose phosphate pathway (PPP) branch (Zheng & Li, 2018). Furthermore, AES, also known as Groucho-related gene 5 (GRG5), is a multifunctional protein with functions in stem cell biology and can be used as a stem cell and as an indicator of neural fate (Chanoumidou et al., 2018). We analyzed the location and expression of these two genes in the dairy goat germ cells (Supplementary Figure S4) and their location in the spermatogonia cluster (Figure 1F). Rolland found that the TKTL1 protein is expressed throughout the maturation process of germ cells, with spermatogonia exhibiting the strongest labeling (Rolland et al., 2013). Our results showed that AES expression was highest in the heart tissue, with moderate expression in the testis (Supplementary Figure S5), whereas TKTL1 expression was highest in the testis tissue and localized in the spermatogonia (Figure 4F), confirming that TKTL1 is a specific marker gene for dairy goat spermatogonia. These results were further verified using sheep data (Figure S5).
In our research, the dairy goat testicular cells were divided into six somatic cell (i.e., Sertoli, Leydig, endothelial, myoid, macrophage, and T cells) and 10 germ cell subgroups. Among these cells, Monocle was used to analyze the trajectory of the Sertoli cells. Results showed that both immature and mature Sertoli cells existed in the testes of the 100-day-old dairy goat, and mature Sertoli cells developed from two immature Sertoli cells. Although cluster analysis divided Leydig and myoid cells into two independent clusters, the pseudotime trajectory showed that these two types of cells were differentiated from the same progenitor cell. Our results confirmed that, unlike mice, the Sertoli, Leydig, and myoid cells had similar developmental trajectories and fates in pubertal dairy goat testes.
We identified five types of germ cells, namely spermatogonia, early spermatocytes, late spermatocytes, round sperm, and sperm. Differential analysis was carried out according to the gene expression of each type of cell, and the characteristic genes specifically expressed by the cell cluster were obtained. The discovery of new characteristic genes will provide important molecular markers for future research on male goat reproduction. The pseudotime differentiation trajectory of the germ cells was successfully constructed based on Monocle analysis. During spermatogenesis, genes with significant changes in expression level were mainly enriched in the Notch, Hippo, and TGF-β signaling pathways, as well as the signaling pathway that regulates stem cell pluripotency. Our studies found that the related pathways of spermatogenesis in dairy goats are roughly similar to those of humans and mice. We also screened and identified specific genes expressed in Guanzhong dairy goat spermatogonia, i.e., TKTL1 and AES, and verified the expression of TKTL1 and AES in sheep (Yang et al., 2021).
In summary, we applied scRNA-seq technology to analyze the process of spermatogenesis in Guanzhong dairy goats at the single-cell level for the first time, revealing the somatic development of dairy goats during puberty. The results of this study should enrich our understanding of the spermatogenesis of Guanzhong dairy goats and provide theoretical and technical support for dairy goat breeding research.
SUPPLEMENTARY DATA
Supplementary data to this article can be found online.
COMPETING INTERESTS
The authors declare that they have no competing interests.
AUTHORS’ CONTRIBUTIONS
X.W.Y., X.M.D., Q.Y.S., M.F.Z., D.H.Y., W.J.X., and N.L. performed the experiments. Y.D.W., T.T.L., W.B.C., C.L.B., X.L.L., G.P.L. S.P., and M.Z.L. analyzed the data. J.L.H. designed the project. X.W.Y and X.M.D wrote the manuscript, J.L.H. revised the manuscript. All authors read and approved the final version of the manuscript.
Funding Statement
This work was supported by the National Natural Science Foundation of China (31572399, 32072806, 32072815, 32002246, 61772431, 62072377), and Program of Shaanxi Province Science and Technology Innovation Team (2019TD-036), Program of State Key Lab of Reproductive Regulation & Breeding of Grassland Livestock (SKL-OT-201801), Program of State Key Laboratory of Respiratory Disease (SKLRD-OP-202114), Science and Technology Major Project of Inner Mongolia Autonomous Region of China (ZDZX2018065), the First-class University and Academic Program from Northwest A&F University (Z1010221003) and Financial aid for basic operation fee of Central University(Z1090219146)
Contributor Information
Sha Peng, Email: pengshacxh@nwsuaf.edu.cn.
Ming-Zhi Liao, Email: liaomz@nwsuaf.edu.cn.
Jin-Lian Hua, Email: jinlianhua@nwsuaf.edu.cn.
References
- 1.Bellvé AR, Cavicchia JC, Millette CF, O'Brien DA, Bhatnagar YM, Dym M Spermatogenic cells of the prepuberal mouse: isolation and morphological characterization. Journal of Cell Biology. 1977;74(1):68–85. doi: 10.1083/jcb.74.1.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chanoumidou K, Hadjimichael C, Athanasouli P, Ahlenius H, Klonizakis A, Nikolaou C, et al Groucho related gene 5 (GRG5) is involved in embryonic and neural stem cell state decisions. Scientific Reports. 2018;8(1):13790. doi: 10.1038/s41598-018-31696-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen XX, Che DX, Zhang PF, Li XL, Yuan QQ, Liu TT, et al Profiling of miRNAs in porcine germ cells during spermatogenesis. Reproduction. 2017;154(6):789–798. doi: 10.1530/REP-17-0441. [DOI] [PubMed] [Google Scholar]
- 4.Choi YH, Kim JK Dissecting cellular heterogeneity using single-cell RNA sequencing. Molecules and Cells. 2019;42(3):189–199. doi: 10.14348/molcells.2019.2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Rooij DG The nature and dynamics of spermatogonial stem cells. Development. 2017;144(17):3022–3030. doi: 10.1242/dev.146571. [DOI] [PubMed] [Google Scholar]
- 6.Du XM, Wu SY, Wei YD, Yu XW, Ma FL, Zhai YX, et al PAX7 promotes CD49f-positive dairy goat spermatogonial stem cells' self-renewal . Journal of Cellular Physiology. 2021;236(2):1481–1493. doi: 10.1002/jcp.29954. [DOI] [PubMed] [Google Scholar]
- 7.Fayomi AP, Orwig KE Spermatogonial stem cells and spermatogenesis in mice, monkeys and men. Stem Cell Research. 2018;29:207–214. doi: 10.1016/j.scr.2018.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gaysinskaya V, Bortvin A Flow cytometry of murine spermatocytes. Current Protocols in Cytometry. 2015;72(1):7.44.1–7.44.24. doi: 10.1002/0471142956.cy0744s72. [DOI] [PubMed] [Google Scholar]
- 9.Green CD, Ma QY, Manske GL, Shami AN, Zheng XN, Marini S, et al A comprehensive roadmap of murine spermatogenesis defined by single-cell RNA-Seq. Developmental Cell. 2018;46(5):651–667. doi: 10.1016/j.devcel.2018.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guo JT, Grow EJ, Mlcochova H, Maher GJ, Lindskog C, Nie XC, et al The adult human testis transcriptional cell atlas. Cell Research. 2018;28(12):1141–1157. doi: 10.1038/s41422-018-0099-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo JT, Grow EJ, Yi CL, Mlcochova H, Maher GJ, Lindskog C, et al Chromatin and single-cell RNA-Seq profiling reveal dynamic signaling and metabolic transitions during human spermatogonial stem cell development. Cell Stem Cell. 2017;21(4):533–546. doi: 10.1016/j.stem.2017.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guo JT, Nie XC, Giebler M, Mlcochova H, Wang YQ, Grow EJ, et al The dynamic transcriptional cell atlas of testis development during human puberty. Cell Stem Cell. 2020;26(2):262–276. doi: 10.1016/j.stem.2019.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo JT, Sosa E, Chitiashvili T, Nie XC, Rojas EJ, Oliver E, et al Single-cell analysis of the developing human testis reveals somatic niche cell specification and fetal germline stem cell establishment. Cell Stem Cell. 2021;28(4):764–778. doi: 10.1016/j.stem.2020.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hermann BP, Cheng KR, Singh A, Roa-De La Cruz L, Mutoji KN, Chen IC, et al The mammalian spermatogenesis single-cell transcriptome, from spermatogonial stem cells to spermatids. Cell Reports. 2018;25(6):1650–1667. doi: 10.1016/j.celrep.2018.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lau X, Munusamy P, Ng MJ, Sangrithi M Single-cell RNA sequencing of the Cynomolgus macaque testis reveals conserved transcriptional profiles during mammalian spermatogenesis . Developmental Cell. 2020;54(4):548–566. doi: 10.1016/j.devcel.2020.07.018. [DOI] [PubMed] [Google Scholar]
- 16.Liao JY, Ng SH, Luk AC, Suen HC, Qian Y, Lee AWT, et al Revealing cellular and molecular transitions in neonatal germ cell differentiation using single cell RNA sequencing. Development. 2019;146(6):dev174953. doi: 10.1242/dev.174953. [DOI] [PubMed] [Google Scholar]
- 17.Liu Y, Niu MH, Yao CC, Hai YN, Yuan QQ, Liu Y, et al Fractionation of human spermatogenic cells using STA-PUT gravity sedimentation and their miRNA profiling. Scientific Reports. 2015;5(1):8084. doi: 10.1038/srep08084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lovelace DL, Gao Z, Mutoji K, Song YC, Ruan JH, Hermann BP The regulatory repertoire of PLZF and SALL4 in undifferentiated spermatogonia. Development. 2016;143(11):1893–1906. doi: 10.1242/dev.132761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mays-Hoopes LL, Bolen J, Riggs AD, Singer-Sam J Preparation of spermatogonia, spermatocytes, and round spermatids for analysis of gene expression using fluorescence-activated cell sorting. Biology of Reproduction. 1995;53(5):1003–1011. doi: 10.1095/biolreprod53.5.1003. [DOI] [PubMed] [Google Scholar]
- 20.Melé M, Ferreira PG, Reverter F, DeLuca DS, Monlong J, Sammeth M, et al The human transcriptome across tissues and individuals. Science. 2015;348(6235):660–665. doi: 10.1126/science.aaa0355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morrison SJ, Spradling AC Stem cells and niches: mechanisms that promote stem cell maintenance throughout life. Cell. 2008;132(4):598–611. doi: 10.1016/j.cell.2008.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oakberg EF A description of spermiogenesis in the mouse and its use in analysis of the cycle of the seminiferous epithelium and germ cell renewal. American Journal of Anatomy. 1956;99(3):391–413. doi: 10.1002/aja.1000990303. [DOI] [PubMed] [Google Scholar]
- 23.Oakberg EF Duration of spermatogenesis in the mouse. Nature. 1957;180(4595):1137–1138. doi: 10.1038/1801137a0. [DOI] [PubMed] [Google Scholar]
- 24.Rolland AD, Lavigne R, Dauly C, Calvel P, Kervarrec C, Freour T, et al Identification of genital tract markers in the human seminal plasma using an integrative genomics approach. Human Reproduction. 2013;28(1):199–209. doi: 10.1093/humrep/des360. [DOI] [PubMed] [Google Scholar]
- 25.Scadden DT The stem-cell niche as an entity of action. Nature. 2006;441(7097):1075–1079. doi: 10.1038/nature04957. [DOI] [PubMed] [Google Scholar]
- 26.Shah MA, Xu CF, Wu SX, Zhao WS, Luo H, Yi CP, et al Isolation and characterization of spermatogenic cells from cattle, yak and cattleyak. Animal Reproduction Science. 2018;193:182–190. doi: 10.1016/j.anireprosci.2018.04.067. [DOI] [PubMed] [Google Scholar]
- 27.Shami AN, Zheng XN, Munyoki SK, Ma QY, Manske GL, Green CD, et al Single-cell RNA sequencing of human, macaque, and mouse testes uncovers conserved and divergent features of mammalian spermatogenesis. Developmental Cell. 2020;54(4):529–547. doi: 10.1016/j.devcel.2020.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Soumillon M, Necsulea A, Weier M, Brawand D, Zhang XL, Gu HC, et al Cellular source and mechanisms of high transcriptome complexity in the mammalian testis. Cell Reports. 2013;3(6):2179–2190. doi: 10.1016/j.celrep.2013.05.031. [DOI] [PubMed] [Google Scholar]
- 29.Tagelenbosch RAJ, de Rooij DG A quantitative study of spermatogonial multiplication and stem cell renewal in the C3H/101 F1 hybrid mouse . Mutation Research/Fundamental and Molecular Mechanisms of Mutagenesis. 1993;290(2):193–200. doi: 10.1016/0027-5107(93)90159-D. [DOI] [PubMed] [Google Scholar]
- 30.Tan K, Song HW, Wilkinson MF Single-cell RNAseq analysis of testicular germ and somatic cell development during the perinatal period. Development. 2020;147(3):dev183251. doi: 10.1242/dev.183251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tharmalingam MD, Jorgensen A, Mitchell RT Experimental models of testicular development and function using human tissue and cells. Molecular and Cellular Endocrinology. 2018;468:95–110. doi: 10.1016/j.mce.2017.12.011. [DOI] [PubMed] [Google Scholar]
- 32.Wang M, Liu XX, Chang G, Chen YD, An G, Yan LY, et al Single-Cell RNA sequencing analysis reveals sequential cell fate transition during human spermatogenesis. Cell Stem Cell. 2018;23(4):599–614. doi: 10.1016/j.stem.2018.08.007. [DOI] [PubMed] [Google Scholar]
- 33.Wei YD, Du XM, Yang DH, Ma FL, Yu XW, Zhang MF, et al Dmrt1 regulates the immune response by repressing the TLR4 signaling pathway in goat male germline stem cells. Zoological research. 2021;42(1):14–27. doi: 10.24272/j.issn.2095-8137.2020.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wei YD, Yang DH, Du XM, Yu XW, Zhang MF, Tang FR, et al Interaction between DMRT1 and PLZF protein regulates self-renewal and proliferation in male germline stem cells. Molecular and cellular biochemistry. 2021;476(2):1123–1134. doi: 10.1007/s11010-020-03977-3. [DOI] [PubMed] [Google Scholar]
- 35.Xiang LF, Yin Y, Zheng Y, Ma YP, Li YG, Zhao ZG, et al A developmental landscape of 3D-cultured human pre-gastrulation embryos. Nature. 2020;577(7791):537–542. doi: 10.1038/s41586-019-1875-y. [DOI] [PubMed] [Google Scholar]
- 36.Yan LY, Yang MY, Guo HS, Yang L, Wu J, Li R, et al Single-cell RNA-Seq profiling of human preimplantation embryos and embryonic stem cells. Nature Structural & Molecular Biology. 2013;20(9):1131–1139. doi: 10.1038/nsmb.2660. [DOI] [PubMed] [Google Scholar]
- 37.Yang H, Ma JY, Wan Z, Wang Q, Wang ZB, Zhao J, et al Characterization of sheep spermatogenesis through single-cell RNA sequencing. FASEB Journal. 2021;35(2):e21187. doi: 10.1096/fj.202001035RRR. [DOI] [PubMed] [Google Scholar]
- 38.Zheng X, Li HX TKTL1 modulates the response of paclitaxel-resistant human ovarian cancer cells to paclitaxel. Biochemical and Biophysical Research Communications. 2018;503(2):572–579. doi: 10.1016/j.bbrc.2018.06.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data to this article can be found online.