Abstract
The genus Macaca serves as an ideal research model for speciation and introgressive gene flow due to its short period of diversification (about five million years ago) and rapid radiation of constituent species. To understand evolutionary gene flow in macaques, we sequenced four whole genomes (two M. arctoides and two M. thibetana) and combined them with publicly available macaque genome data for genome-wide analyses. We analyzed 14 individuals from nine Macaca species covering all Asian macaque species groups and detected extensive gene flow signals, with the strongest signals between the fascicularis and silenus species groups. Notably, we detected bidirectional gene flow between M. fascicularis and M. nemestrina. The estimated proportion of the genome inherited via gene flow between the two species was 6.19%. However, the introgression signals found among studied island species, such as Sulawesi macaques and M. fuscata, and other species were largely attributed to the genomic similarity of closely related species or ancestral introgression. Furthermore, gene flow signals varied in individuals of the same species (M. arctoides, M. fascicularis, M. mulatta, M. nemestrina and M. thibetana), suggesting very recent gene flow after the populations split. Pairwise sequentially Markovian coalescence (PSMC) analysis showed all macaques experienced a bottleneck five million years ago, after which different species exhibited different fluctuations in demographic history trajectories, implying they have experienced complicated environmental variation and climate change. These results should help improve our understanding of the complicated evolutionary history of macaques, particularly introgressive gene flow.
Keywords: Macaca, Whole genome, Introgression, Gene flow, Demographic history
INTRODUCTION
Natural hybridization between closely related species plays an important role in evolution and is now recognized as common in animals, including primates (Arnold & Meyer, 2006; Baiz et al., 2020; Cortés-Ortiz et al., 2007; Fan et al., 2018; Gante et al., 2016; Kuhlwilm et al., 2019; Martin & Jiggins, 2017; Rogers et al., 2019; Zhang et al., 2017). Hybridization is an important source of genetic variation during the evolutionary process and can introduce adaptive variants to recipient populations and even drive new species formation (Figueiro et al., 2017; Martin & Jiggins, 2017; Zinner et al., 2011). The footprints left by hybridization in genomes allow the detection of ancient hybridization and gene flow, with sequencing technology enabling research on whole-genome data. The ever-growing database of whole genomes from multiple species and populations can now be used to address important evolutionary questions, such as introgressive hybridization, incomplete lineage sorting (ILS), and speciation-with-gene-flow (Árnason et al., 2018).
At present, the genus Macaca is comprised of 23 extant species. Macaques are the most widely distributed non-human primates and occupy a wide range of habitats (Ito et al., 2018; Roos et al., 2019). Except for M. sylvanus, which is distributed in Africa (Algeria and Morocco), all other species are found in Asia (see IUCN Red List). Historically, macaques dispersed into Eurasia during the late Miocene, roughly coinciding with the sea level drop associated with the Messinian Salinity Crisis (Abbott et al., 2013; Roos et al., 2019). The oldest putative Macaca fossils in Europe and Asia date to 5.9–5.3 million years ago (Ma) and 4.9–3.6 Ma, respectively (Roos et al., 2019). After reaching Asia, Macaca experienced rapid speciation and radiation due to climate and environmental change during the late Pliocene and Pleistocene (Abbott et al., 2013; Roos et al., 2019; Woodruff, 2010). Macaca can be classified into four or seven species groups (Delson, 1980; Fooden, 1976; Groves, 2001; Roos et al., 2014, 2019). The seven species group classification includes three new species groups, i.e., arctoides, mulatta, and sulawesi (Roos et al., 2014), by separating M. arctoides, M. mulatta+M. fuscata and all Sulawesi macaque species (respectively) from three (sinica, fascicularis, and silenus) of the four species groups (Delson, 1980). In this study, we used the four species group classification (sylvanus, silenus, fascicularis, and sinica) proposed by Delson (1980). Our samples covered all Asian groups of macaques, including silenus (M. nemestrina, M. nigra, and M. tonkeana), fascicularis (M. fascicularis, M. fuscata, and M. mulatta), and sinica (M. arctoides, M. assamensis, and M. thibetana).
Hybridization has been previously reported in Macaca, e.g., between M. fuscata and M. cyclopis (Hamada et al., 2012), M. mulatta and M. fascicularis (Hamada et al., 2016; Ito et al., 2020; Rovie-Ryan et al., 2021), M. thibetana and Chinese M. mulatta (Fan et al., 2014), M. arctoides and fascicularis species group (Fan et al., 2018; Jiang et al., 2016; Li et al., 2009; Tosi et al., 2000, 2003b), and between Sulawesi macaques (Ciani et al., 1989). Active tectonics and changing climate and sea levels during the Pliocene and Pleistocene and the periodic formation of land bridges in Southeast Asia significantly influenced Macaca evolution and allowed possible hybridization or introgression among species/species groups (Meijaard, 2003). In addition, macaques have an almost identical chromosome karyotype, a factor that may facilitate hybridization among species and the production of viable hybrid offspring (Hamada et al., 2012; Yang & Shi, 1994). In recent years, there has been a considerable increase in whole-genome-based studies on hybridization in macaques. For example, Fan et al. (2014) reported on hybridization between M. thibetana and M. mulatta based on whole-genome sequencing analysis of M. thibetana. Fan et al. (2018) identified an introgression event in the evolutionary history of M. arctoides by genome-wide analysis. Osada et al. (2021) reported on sex-biased admixture between the fascicularis and sinica species groups based on whole-genome analysis of 17 macaques. Vanderpool et al. (2020) performed phylogenomic analysis of primates, including three species of macaques, and found introgression between M. nemestrina and M. fascicularis. Ito et al. (2020) suggested potential gene flow between M. fascicularis and M. mulatta following secondary contact after a period of isolation, and that the migration rate from M. mulatta to M. fascicularis may be slightly higher than in the opposite direction.
However, most field observations and molecular-based analyses of Macaca hybridization have focused on the most recently diverged species groups of fascicularis and sinica, with gene flow between the more basal silenus group and other groups less studied. Moreover, genus-wide hybridization or gene flow studies of Macaca are scarce, particularly with whole-genome data. Here, we included three species from the silenus group and presented high-coverage whole-genome data of four macaques (two M. arctoides and two M. thibetana). Combined with other publicly available data, our study included 14 individuals from nine Macaca species, covering all species groups of Asian macaques. We estimated local evolutionary histories, reconstructed a multispecies coalescent phylogenetic tree, and explored ancient or ongoing introgressive gene flow in macaques based on whole-genome DNA sequences. Furthermore, we also analyzed the genetic diversity and demographic history of macaques.
MATERIALS AND METHODS
Ethics statement
All samples were collected following the Regulations for the Implementation of the Protection of Terrestrial Wildlife of China (State Council Decree [1992] No. 13). Throughout the procedures, all animals received humane care to avoid suffering and to ensure animal welfare. All animal protocols involved in this study were reviewed and approved by the Ethics Committee of the College of Life Science, Sichuan University, China (Permit No.: 20200327009).
Samples and sequencing
The genomes of two M. arctoides and two M. thibetana were sequenced using Illumina HiSeq 2500 technology with a 150 bp paired-end strategy. DNA was isolated from blood using a commercially available DNeasy Blood and Tissue Kit (Qiagen, Germany). Sequencing libraries were prepared with an insert size of 300 bp. The two M. arctoides individuals were from the Yunnan Province and Guangxi Zhuang Autonomous Region of China, respectively. The two M. thibetana monkeys were from the Anhui Province and Guangxi Zhuang Autonomous Region of China, respectively (Table 1). In addition, we downloaded 11 genome sequence data from the NCBI SRA database. In total, we used whole-genome data from 14 individuals belonging to Macaca and one individual of Papio anubis in our study. The 15 whole-genome sequences covered all species groups of Asian macaques and included 10 species: i.e., M. arctoides (stump-tailed macaque), M. thibetana (Tibetan macaque), M. assamensis (Assamese macaque), M. nemestrina (southern pig-tailed macaque), M. nigra (black-crested macaque), M. tonkeana (Tonkean macaque), M. fascicularis (crab-eating macaque), M. mulatta (rhesus macaque), M. fuscata (Japanese macaque), and P. anubis (olive baboon). Detailed information on the 15 genomes and sequencing data is given in Table 1 and the “Detailed information on samples” section in the Supplementary Materials.
Table 1. Information on samples and genomic data.
| Scientific name | Common name | Sample ID | NCBI accession No. | Sex | Sample origin | Sequencing platform | No. of bases (Gb) | Depth* |
| For individual macaque genome samples, scientific name, common name, sample ID, NCBI accession No., sex, sample origin, sequencing platform, No. of bases, and sequencing depth are shown. New whole-genome sequences of this study are marked in bold. *: Calculations were based on total length of M. mulatta genome assembly Mmul_8.0.1, 3 236 224 332 bp. | ||||||||
| M. arctoides | Stump-tailed macaque | Marc_R02 | This study, SAMN15194901 | Male | Yunnan, China | Illumina | 152.0 | ~47.0× |
| Marc_R19 | This study, SAMN15194902 | Female | Guangxi, China | Illumina | 112.2 | ~34.7× | ||
| M. assamensis | Assamese macaque | Mass | SRR2981114 | Male | Yunnan, China | Illumina | 154.0 | ~47.6× |
| M. fascicularis | Crab-eating macaque | Mfas_1 | SRR8194877 | Female | Unknown (in captivity in China) | Illumina | 92.1 | ~28.5× |
| Mfas_Mau | ERS629711 | Male | Mauritius | Illumina | 61.6 | ~19.0× | ||
| M. fuscata | Japanese macaque | Mfus | DRR002234 | Unknown | Japan | Illumina | 142.5 | ~44.0× |
| M. mulatta | Rhesus macaque | Mmul_Chi | SRR1944102 | Female | China | Illumina | 144.5 | ~44.7× |
| Mmul_Ind | SRR1952166 | Female | India | Illumina | 131.3 | ~40.6× | ||
| M. nemestrina | Southern pig-tailed macaque | Mnem_1 | SRR1698391, SRR1698394, SRR1698403, SRR1698405 | Female | Unknown | Illumina | 152.7 | ~47.2× |
| Mnem_2 | SRR5947292 | Male | Borneo, Malaysia | Illumina | 140.7 | ~43.5× | ||
| M. nigra | Black crested macaque | Mnig | SRR5947294 | Female | Sulawesi, Indonesia | Illumina | 135.8 | ~42.0× |
| M. thibetana | Tibetan macaque | Mthi_HT1 | This study, SAMN15194903 | Female | Anhui, China | Illumina | 93.7 | ~29.0× |
| Mthi_R25 | This study, SAMN15194904 | Female | Guangxi, China | Illumina | 113.9 | ~35.2× | ||
| M. tonkeana | Tonkean macaque | Mton | SRR5947293 | Male | Sulawesi, Indonesia | Illumina | 135.4 | ~41.8× |
| P. anubis | Olive baboon | Panu | SRR8723580 | Female | Unknown | Illumina | 137.9 | ~42.6× |
Single nucleotide variant (SNV) calling
Quality control of reads was performed using fastp (v.0.20.0) (Chen et al., 2018) and Trim Galore (http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/, v.0.6.1). Firstly, low-quality bases were trimmed at both ends of the reads using fastp with parameters: fastp --detect_adapter_for_pe -5 -3. Adapter sequences were then cut using Trim Galore with parameters: trim_galore -q 20 --phred33 --stringency 3 -e 0.1 --length 50 --max_n 0 --trim-n -j 4 -paired. Filtered reads were mapped to the M. mulatta genome Mmul_8.0.1 (NCBI Assembly ID GCA_000772875.3) with Bowtie2 (v.2.3.5.1) (Langmead & Salzberg, 2012). As a new Mmul_10 reference genome is now available, genome-wide analyses based on the new reference may be slightly different from the present study. From the mapped reads, SNVs were called using the Genome Analysis Toolkit (GATK, v.4.1.2.0) (DePristo et al., 2011) following the GATK germline short variant discovery workflow (https://software.broadinstitute.org/gatk/best-practices/workflow?id=11145). Repetitive sequences of the Mmul_8.0.1 assembly were annotated using RepeatMasker (v.open-4.0.9) (Tarailo-Graovac & Chen, 2009). The SNV sites were filtered out when one or more of the following criteria were met: overlap with repetitive sequences, DP<3.0, QD<2.0, SOR>3.0, FS>60.0, MQ<40.0, MQRankSum<–12.5, and ReadPosRankSum<–8.0. Statistics of SNVs were calculated using bcftools (v.1.9) (Li, 2011) (Table 2).
Table 2. SNV information for each analyzed macaque.
| Sample | Filtered | Downsampled (~15×) | ||||||||
| SNVs | Homo | Het | Callable sites | Het | Heterozygosity | Ts | Tv | Ts/Tv | ||
| For individual macaque genome samples, short ID, total number of single nucleotide variants (SNVs), number of heterozygous SNVs (Het), number of homozygous SNVs (Homo), and transitions/transversions (Ts/Tv) are shown. | ||||||||||
| Marc_R02 | 7 271 275 | 5 383 371 | 1 887 904 | 2 555 381 332 | 3 782 713 | 0.001 480 | 10 682 013 | 4 949 053 | 2.16 | |
| Marc_R19 | 7 000 609 | 5 675 953 | 1 324 656 | 2 553 354 858 | 2 644 171 | 0.001 036 | 10 334 343 | 4 762 614 | 2.17 | |
| Mass | 7 926 468 | 4 560 039 | 3 366 429 | 2 519 264 130 | 6 121 131 | 0.002 430 | 10 831 112 | 5 023 843 | 2.16 | |
| Mfas_1 | 6 909 052 | 3 101 413 | 3 807 639 | 2 562 592 812 | 7 612 441 | 0.002 971 | 10 058 278 | 4 638 571 | 2.17 | |
| Mfas_Mau | 6 773 377 | 3 895 560 | 2 877 817 | 2 549 849 925 | 5 881 316 | 0.002 307 | 10 190 936 | 4 659 300 | 2.19 | |
| Mfus | 4 805 372 | 3 403 338 | 1 402 034 | 2 562 072 275 | 2 619 778 | 0.001 023 | 7 074 787 | 3 279 216 | 2.16 | |
| Mmul_Chi | 5 192 005 | 2 071 981 | 3 120 024 | 2 556 851 619 | 5 748 608 | 0.002 248 | 7 168 259 | 3 273 752 | 2.19 | |
| Mmul_Ind | 4 058 527 | 1 370 638 | 2 687 889 | 2 550 406 218 | 5 072 952 | 0.001 989 | 5 723 495 | 2 602 782 | 2.20 | |
| Mnem_1 | 8 733 029 | 4 860 252 | 3 872 777 | 2 559 986 484 | 7 267 811 | 0.002 839 | 12 211 436 | 5 571 807 | 2.19 | |
| Mnem_2 | 8 568 074 | 4 907 725 | 3 660 349 | 2 557 580 297 | 7 210 573 | 0.002 819 | 12 310 845 | 5 636 798 | 2.18 | |
| Mnig | 7 743 384 | 6 043 224 | 1 700 160 | 2 553 652 584 | 3 353 047 | 0.001 313 | 11 259 320 | 5 250 264 | 2.14 | |
| Mthi_HT1 | 6 133 336 | 5 691 120 | 442 216 | 2 488 747 849 | 903 396 | 0.000 363 | 9 401 283 | 4 277 321 | 2.20 | |
| Mthi_R25 | 6 858 249 | 5 921 203 | 937 046 | 2 552 204 059 | 1 886 287 | 0.000 739 | 10 137 409 | 4 730 651 | 2.14 | |
| Mton | 8 229 873 | 5 597 616 | 2 632 257 | 2 555 184 660 | 5 168 771 | 0.002 023 | 11 872 755 | 5 501 580 | 2.16 | |
| Panu | 17 507 271 | 15 461 142 | 2 046 129 | 2 529 072 236 | 3 620 049 | 0.001 431 | 25 935 871 | 11 112 178 | 2.33 | |
Phylogenetic reconstruction
The SNVs of all samples were merged using bcftools (v.1.9) with the parameter -0. The SNVs from every non-overlapping 100 kb genome fragment were concatenated separately. Heterozygous SNVs were represented according to the degenerate base naming rules of the International Union of Pure and Applied Chemistry (IUPAC). We called the concatenated sequences “SNV-fragment”. In total, 26 633 SNV-fragments were obtained for phylogenetic analysis. For each SNV-fragment, a phylogenetic tree was computed using IQ-TREE (v.1.6.10) (Hoang et al., 2018; Kalyaanamoorthy et al., 2017; Nguyen et al., 2015) with parameters: -st DNA -bb 1000 -m MFP+ASC. Branches of the SNV-fragment trees with support values lower than 30% were contracted using Newick utilities (v.1.6) (Junier & Zdobnov, 2010). Based on all SNV-fragment trees, ASTRAL (v.5.6.3) (Zhang et al., 2018) and MP-EST (v.2.0) (Liu et al., 2010) were used to estimate the species tree with default parameters. The species tree was rooted with P. anubis. These two programs generated the same phylogenetic topology. We tested multiple genome fragment sizes (30, 50, 70, and 100 kb). All genome fragment sizes led to consistent results.
Divergence time estimation
We randomly selected one of the first 1 000 SNVs and selected one every 1 000 SNVs with this SNV as the starting point. We then concatenated these SNVs to form an alignment to estimate divergence time. The final alignment length was 28 498 bp. To reduce the consumption of computing resources and time, the first 20 000 sites of the alignment were used to estimate divergence time with SNAPP (Bryant et al., 2012) in BEAST (v2.6.0) (Bouckaert et al., 2014) under the following parameters: (1) four age constraints, which were derived from our unpublished genome assembly-based divergence time estimations of Macaca by MCMCTree (Yang, 2007) (Supplementary Table S1); (2) chain length of 1 000 000 MCMC iterations; and (3) species tree generated from ASTRAL as a guide tree. An ascertainment-bias correction was applied to the divergence time estimations because of no constant site in the SNV data (Bryant et al., 2012; Lewis, 2001). We performed divergence time estimation three times to confirm similar results (see Stange et al. (2018) for detailed methods of divergence time estimation). The topology generated from SNAPP was used for subsequent analyses.
Gene flow analyses
The D statistic (Green et al., 2010) utilizes the number of biallelic sites that meet the ABBA or BABA site pattern in a four-taxon phylogeny, and requires a phylogenetic topology following (((P1, P2), P3), O) with P1 to P3 being ingroups and O being the outgroup. We applied the D statistic to all asymmetric four-taxon phylogenies that could be extracted from the species tree with P. anubis as an outgroup using Dsuite (v.0.2 r17) (Malinsky et al., 2021). This resulted in 364 gene flow analyses. Four-taxon phylogenies with corrected P<0.001 were retained. Thefd statistic quantifies the proportion of the genome affected by introgression and utilizes four-taxon phylogeny equaling the D statistic (Martin et al., 2015). The fd statistic of the retained four-taxon phylogenies was calculated using Dsuite. Gene flow direction can be estimated by a derivation of the D statistic, i.e., DFOIL analysis (Pease & Hahn, 2015). DFOIL analysis requires a symmetric five-taxon phylogeny with a specific topology. Therefore, not all combinations of five taxa could be analyzed. All symmetric five-taxon phylogenies (with P. anubis as the outgroup) that fit DFOIL analysis were extracted and DFOIL analyses were finished using ExDFOIL (Lambert et al., 2019).
We found that all species pairs between the fascicularis and silenus species groups exhibited very strong gene flow signals. Therefore, further analyses were undertaken to determine if these signals reflected hybridization between the examined extant species after all dichotomous divergence events. We calculated the fd statistic for all species pairs based on non-overlapping 30 kb windows. The script for calculating fd based on sliding windows was deposited in GitHub (https://github.com/sy-snake/Sliding_window_D_and_fd). We selected pairs from the M. fascicularis-silenus group species (M. nemestrina, M. nigra, and M. tonkeana) and M. nemestrina-fascicularis group species (M. fascicularis, M. fuscata, and M. mulatta) as target pairs. We then selected the top 5% of windows with the strongest fd value of target species pairs for subsequent analyses. Of these windows, we discarded any that met the following criteria: (1) appeared in more than 60% of all species pairs; (2) window coverage was larger than two times or less than one third of the sample’s average coverage; and (3) window heterozygosity was larger than two times the sample’s whole-genome heterozygosity. The overlapping ratios of the remaining windows of target pairs from the M. fascicularis-silenus group species and M. nemestrina-fascicularis group species were calculated to determine whether these strong hybridization signals were due to genomic similarity of closely related species or hybridization between checked species after all dichotomous divergence events. If we assume that the genome fragments exchanged during hybridization are random, then these windows should not be shared at a high level among species pairs. If this ratio is high, we can assume that strong hybridization signals are largely due to the genomic similarity of closely related species.
We also checked genes (including upstream 5 kb) located in the top 5% of windows of species pair M. fascicularis-M. nemestrina using bedtools with parameters: bedtools intersect -wo -f 0.5 -F 0.5 -e. Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO)-based enrichment analyses of these genes were performed using g:Profiler (Reimand et al., 2007). We set M. mulatta as the background organism, statistical domain scope to “all known genes”, significance threshold to “g:SCS thread”, and remaining parameters to default.
Inference of reticulate phylogenies
PhyloNetworks (Solís-Lemus et al., 2017) uses maximum pseudolikelihood methods to estimate phylogenetic networks. Here, we used the “snaq!” program in PhyloNetworks and applied different maximum numbers of hybrid nodes (hmax: 0-6) using the 26 633 SNV-fragment trees as input for 100 iterations. Each network generated from PhyloNetworks has a network score, with lower scores indicating better networks. Similarly, PhyloNet (Wen et al., 2018) was specifically developed to reconstruct reticulate phylogenies from a set of gene trees. We used the “InferNetwork_MPL” method with 1 000 runs and six as the maximum number of reticulations, yielding five optimal networks to analyze the 26 633 SNV-fragment trees. We ran the program multiple times with a different maximum number of reticulations (0–6). When the maximum number of reticulations was set to five, the number of reticulations in the phylogenetic networks no longer increased. We set the maximum number of reticulations to six to ensure that the number of reticulations did not increase. Consensus networks of the 26 633 SNV-fragment trees were generated using phangorn (v.2.5.5) (Schliep, 2011) with different proportions.
Demographic history
The demographic histories of the 14 Macaca individuals were inferred from genome sequences using the pairwise sequentially Markovian coalescence model (PSMC) (Li & Durbin, 2011). Only autosomes of reference assembly Mmul_8.0.1 were used in the demographic history analyses. Input files for PSMC were generated using bcftools mpileup and vcfutils with the minimum and maximum depth of coverage thresholds set to one third and two times the sample’s average depth, respectively. PSMC was run for 25 iterations, with a N0-scaled maximum coalescent time of 15, initial θ/ρ ratio of 5, and time intervals of 64 parameterized as “1×6+58×1”. Bootstrapping was performed for 100 runs. PSMC plots were scaled with a mutation rate of μ=0.58×10-8 per bp per generation (Wang et al., 2020) and a generation time of g=10. According to published reports, the reproductive age of M. mulatta ranges from about four years old to late teens, resulting in a median parent birth age of about 10 years (Kubisch et al., 2012; Xue et al., 2016). Therefore, we used 10 as the generation time.
Genome-wide heterozygosity
Genetic diversity was assessed using heterozygosity of each individual. To exclude potential effects of different sequencing coverage of each macaque, the BAM files were downsampled to ~15× using GATK. Heterozygosity was estimated based on the downsampled BAM files. SNVs of autosomes were called using bcftools with the parameter “bcftools mpileup -C50 -Ou | bcftools call -m”. Heterozygosity of individuals was calculated as the number of heterozygous sites divided by the total number of callable sites across the whole genome. In addition, we performed a calculation of heterozygosity from genomic non-overlapping 1 Mb windows.
RESULTS
Genome sequencing and SNV calling
Sequencing of genomic DNA from the two M. arctoides and two M. thibetana using Illumina technology yielded 93.7–152.0 Gb of data. Combined with the 11 whole-genome sequences downloaded from the NCBI SRA database, a total of 15 whole-genome sequences were used for subsequent analyses. Reference genome mapping of the 15 whole-genome sequences against the M. mulatta genome assembly Mmul_8.0.1 yielded genome coverages of 17.8–43.0×. Table 1 and Supplementary Table S2 detail the sequencing and mapping data and provide accession numbers of the included species. RepeatMasker (Tarailo-Graovac & Chen, 2009) identified 51.95% of repetitive sequences in the M. mulatta (reference) genome. Of these, 12.75% and 18.94% were short and long interspersed elements (SINE and LINEs), respectively (Supplementary Table S3). The repetitive sequence regions were excluded in SNV calling. After a series of filters, we identified 4.06–8.73 million SNVs in the 14 macaque genomes. Detailed information on the SNVs is shown in Table 2. Concatenating SNVs from every non-overlapping 100 kb genome fragment yielded 26 633 alignments, totaling 28 497 813 bp in length for each sample. These alignments were used for subsequent evolutionary analyses.
Local and genome-wide evolutionary histories of macaques
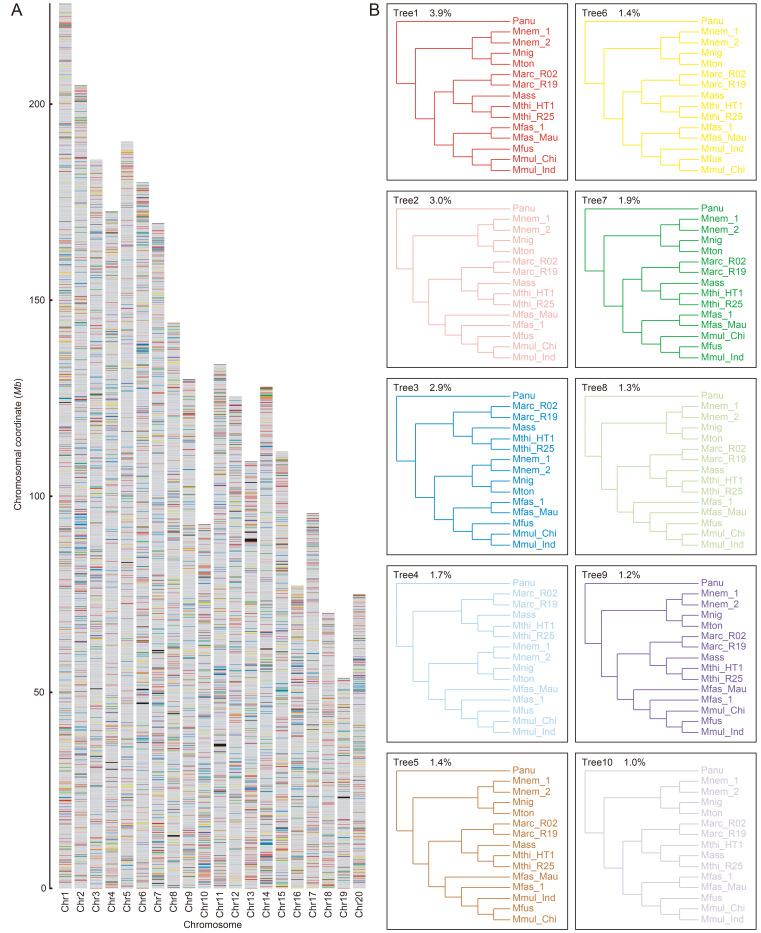
Model testing suggested different models for different SNV-fragments. The 26 633 SNV-fragment trees were then constructed based on SNV-fragment-specified models. The distributions of SNV-fragment trees across genomes at the individual level and species group level are shown in Figure 1 and Supplementary Figure S1. The local evolutionary histories of the macaques showed many conflicts (Figure 1), indicating different genomic areas had different evolutionary signals across the macaque genome, which implies complex evolutionary histories of macaques. Various programs were used to reconstruct the macaque species tree based on the 26 633 SNV-fragment trees, which generated similar results except for the position of Mfas_1 (M. fascicularis specimen from China) (Figure 2A, B). Our general topology conforms to previous nuclear marker analyses of the phylogenetic relationships within Macaca (Fan et al., 2018; Jiang et al., 2016; Li et al., 2009). Here, the macaques clustered into three branches, corresponding to the fascicularis species group (M. mulatta, M. fuscata, and M. fascicularis), sinica species group (M. thibetana, M. assamensis, and M. arctoides), and silenus species group (M. nemestrina, M. nigra, and M. tonkeana) (Figure 2C). With respect to the position of Mfas_1, it was either grouped by itself at the basal node of M. mulatta and M. fuscata by ASTRAL and MP-EST (Figure 2A) or grouped with Mfas_Mau (M. fascicularis specimen from Mauritius) by SNAPP (Figure 2B). The estimated quartet scores for these two clusters were 0.4208 (ASTRAL and MP-EST) and 0.4047 (SNAPP), which are too close to describe the position of Mfas_1 accurately. A similar phylogenetic inconsistency was also observed following CONSENSE (Felsenstein, 1989) analysis of the SNV-fragment trees. Although a majority-rule consensus tree (Supplementary Figure S2) confirmed the topology of the ASTRAL and MP-EST species tree (Figure 2A), the separate grouping of the two M. fascicularis specimens occurred 9 959 times, accounting for 37.39% of the 26 633 trees (Supplementary Table S4).
Figure 1.
Local evolutionary history of macaques at individual level
A: Each bar represents a chromosome, in terms of M. mulatta genome. Colored bands represent tree topologies of each 100 kb window. Colors correspond to topologies in (B). B: Ten most common trees. Values at top left corner are percentage of all 100 kb windows that recover that topology. Colors correspond to colored bands in (A), with gray and black regions showing other topologies and missing data.
Figure 2.
MSC (multispecies coalescent) tree
All node support values are 100%. A: MSC species tree generated from ASTRAL and MP-EST based on 26 633 SNV-fragments. B: Species tree with divergence time. Topology and divergence time were both estimated in SNAPP based on 20 000 SNV sites. This tree shows a conflict of Mfas_1 position with (A). Numbers on nodes indicate divergence time. Purple bars on nodes represent 95% confidence interval of divergence time. C: MSC tree at species level.
Divergence time was estimated based on 20 000 SNV sites (Figure 2B). The estimated divergence time of baboons and macaques was ~10.38 (95% CI: 9.30–11.51) Ma, congruent with previous reports (Abbott et al., 2013; Roos et al., 2019). The silenus and fascicularis/sinica species groups diverged ~1.95 (95% CI: 1.74–2.17) Ma, and the fascicularis and sinica species groups diverged ~1.56 (95% CI: 1.39–1.73) Ma.
Consensus network analysis of the SNV-fragment trees yielded cuboid structures of connecting alternative branches, thus indicating reticulate phylogenetic relationships among macaques (Figure 3). Cuboid structures mainly occurred in the fascicularis and sinica species groups at a threshold of 11%, thus indicating phylogenetic conflicts within/between the two species groups and potential interspecific gene flow. There was no cuboid structure in the net center, suggesting that the evolutionary relationship of ancestral macaques is relatively clear. At lower thresholds, the phylogenetic signal became more complex, and a cuboid structure appeared in the center of the net, thus implying additional phylogenetic conflict (Supplementary Figure S3).
Figure 3.
Consensus network of 26 633 SNV-fragment trees with 11% threshold
Cuboid structures mainly occur in fascicularis and sinica species groups, indicating phylogenetic conflicts in these two species groups and potential interspecific gene flow. There is no cuboid structure in the net center, indicating that the evolutionary relationship of ancestral macaques is relatively clear.
Gene flow analyses
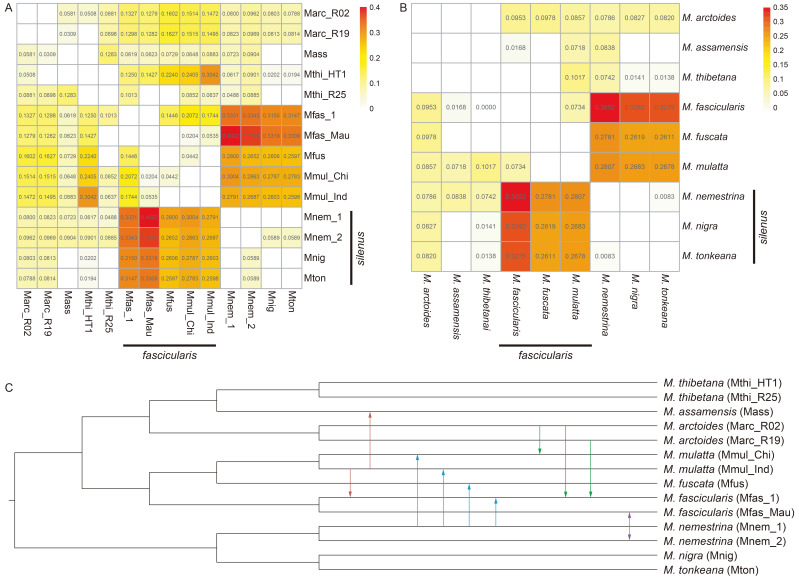
Both D and DFOIL analyses identified substantial gene flow signals among macaques (Figure 4; Supplementary Tables S5, S6). The gene flow signals detected by the D statistic (corrected P<0.001) at the individual and species level are shown inFigure 4A, B, respectively. Most species pairs showed significant gene flow signals, with strong signals detected in all species pairs between the fascicularis species group (M. fascicularis, M. fuscata, M. mulatta) and silenus species group (M. nemestrina, M. nigra, M. tonkeana). DFOIL analyses identified the direction of gene flow signals between the 11 individual pairs (Supplementary Table S6). Ten of these signals were significant according to both D and DFOIL statistical analyses. We mapped the direction of these gene flow signals in the species tree (Figure 4C). Unidirectional gene flow signals were found from M. mulatta to M. assamensis and M. fascicularis, from M. nemestrina to M. fuscata and M. mulatta, and from M. arctoides to M. mulatta and M. fascicularis. A bidirectional gene flow signal was detected between Mauritian M. fascicularis and Bornean M. nemestrina.
Figure 4.
Gene flow signals in macaques
A: Gene flow identified by D statistic (corrected P<0.001) at individual level. Numbers in grids representD values, with higher values indicating higher introgression level. Grids without numbers indicate that no gene flow signal was detected. Redder colors indicate higher D statistics. Maximum D value of each individual pair was used for drawing. B: Gene flow identified by D statistic (corrected P<0.001) at species level. The meanings of color and number in grid are the same as that in (A). MaximumD value of each species pair was used for drawing. C: Species tree of macaques with directional gene flow signals. Only signals confirmed by both D and DFOIL statistics are shown here. Arrows indicate direction of gene flow.
Different introgression signals were observed in individuals from different populations of the same species. For instance, the Anhui M. thibetana (Mthi_HT1) had an introgression signal with M. fuscata, but the Guangxi M. thibetana (Mthi_R25) did not have this signal. Different individuals of M. fascicularis and M. nemestrina also showed different introgression signals (Figure 4A, C). Mfas_1 (a captive breed specimen collected from China, M. fascicularis) showed introgression signals with the Guangxi M. thibetana (Mthi_R25) and M. fuscata specimens, while the Mauritian M. fascicularis specimen (Mfas_Mau) did not show these signals (Figure 4A). Mnem_2 (specimen from Borneo, M. nemestrina) showed a significant introgression signal with M. nigra and M. tonkeana, but Mnem_1 (specimen of M. nemestrina) did not (Figure 4A).
The fraction of the genomes shared through gene flow was estimated using fd (Martin et al., 2015) (Figure 5). The M. fascicularis and M. nemestrina species pair generated the highest fd value (0.061 9). In addition, the fd values of species pairs between the fascicularis and silenus species groups (0.023 9–0.061 9) were higher than that of the other species pairs. The M. fascicularis and M. mulatta species pair also generated a relatively high fd value (0.024 0).
Figure 5.
Proportion of genome shared through gene flow estimated by fd statistic at (A) individual level and (B) species level
Number in grid is value of fd. Grids without numbers indicate that no gene flow signal was detected. The fd values of species pairs between fascicularis and silenus species groups were higher than that of other species pairs, with M. fascicularis and M. nemestrina species pair showing highest fd value. In addition, M. fascicularis and M. mulatta species pair showed a high fd value.
In addition to analyses based on site patterns, gene flow was investigated by topology-based analyses using PhyloNetworks (Solís-Lemus et al., 2017) and PhyloNet (Wen et al., 2018). Consistent with the D and DFOIL results, gene flow signals were identified between the fascicularis and silenus species groups (Figure 6; Supplementary Figures S4, S5). Different maximum numbers of reticulations (0–6) were set when estimating networks using PhyloNetworks. We generated seven phylogenetic networks, each with a network score (lower score indicates better network). Networks with the maximum number of reticulations set to six had the lowest score (Figure 6B). However, networks with five or six reticulations (Supplementary Figure S6) could not be re-rooted by P. anubis due to hybrid edges on the P. anubis branch. Therefore, we only presented phylogenetic networks with four reticulations, which had the lowest score next to networks with five and six reticulations. PhyloNetworks identified gene flow signals between the following pairs: fascicularis and silenus species group, M. fuscata and Chinese M. mulatta, two individuals of M. fascicularis, and unsampled species and Anhui M. thibetana (Figure 6A). PhyloNetworks also determined the proportion of genes inherited via gene flow between these pairs (Figure 6A). For the ancestral lineage of the fascicularis species group, 39.5% of genes were from an ancestor of the silenus species group. Furthermore, 33.2% of M. fuscata genes were from Chinese M. mulatta and 23.2% of Anhui M. thibetana genes were from an unsampled species. In addition, PhyloNet identified several gene flow signals between the fascicularis and sinica (M. arctoides, M. assamensis, M. thibetana) species groups (Supplementary Figures S4, S5).
Figure 6.
Rooted network of macaques
A: Rooted network of macaques with four reticulations inferred by PhyloNetworks based on 26 633 SNV-fragments. Hybrid edges are annotated with their inheritance values, which measure proportion of genes inherited via gene flow. B: Network scores based on different maximum number of reticulations (0–6). We estimated networks with a different maximum number of reticulations, and each generated a network score, with lower scores indicating better networks. However, networks with five and six reticulations could not be re-rooted by P. anubis due to hybrid edges on the P. anubis branch. Therefore, only the network with four reticulations is shown here.
Given that strong introgression signals appeared in all species pairs between the fascicularis and silenus species groups, we compared the top 5% of windows showing the strongest introgression signals of several species pairs to determine if these signals reflected hybridization between the examined extant species after all dichotomous divergence events. These windows showed similar genomic distributions among the species pairs (Figure 7; Supplementary Figure S7). An example distribution of these windows across chromosome 18 is shown in Figure 7. In total, the proportion of shared top 5% of windows was 60.14%–67.93% for the M. fascicularis-silenus group species pairs and 45.84%–47.33% for the M. nemestrina-fascicularis group species pairs. The windows specific to each species pair only accounted for 8.70%–21.43% (Table 3).
Figure 7.
Distribution of top 5% of windows with strongest introgression signals according to fd across chromosome 18
Species of species pairs belong to fascicularis and silenus species groups. Results showed that top 5% of windows with strongest introgression signals for different species pairs had similar genomic distributions.
Table 3. Statistics on top 5% of windows with strongest introgression signals based on fd for species pairs between fascicularis and silenus species groups .
| Species pair | No. of top 5% windows | No. of shared windows | Percentage of shared windows | No. of specific windows | Percentage of specific window | |
| For M. fascicularis-silenus group species pairs, 60.14%–67.93% of top 5% of windows were shared, and this proportion for M. nemestrina-fascicularis group species pairs was 45.84%–47.33%. Windows specific to each species pair only accounted for 8.70%–21.43%. | ||||||
| M. fascicularis-silenus group species |
M. fascicularis-
M. nemestrina |
2 920 | 1 756 | 60.14 | 378 | 12.95 |
|
M. fascicularis-
M. nigra |
2 585 | 67.93 | 225 | 8.70 | ||
|
M. fascicularis-
M. tonkeana |
2 814 | 62.40 | 257 | 9.13 | ||
| M. nemestrina-fascicularis group species |
M. fascicularis-
M. nemestrina |
2 920 | 1 382 | 47.33 | 378 | 12.95 |
|
M. fuscata-
M. nemestrina |
3 015 | 45.84 | 646 | 21.43 | ||
|
M. mulatta-
M. nemestrina |
2 999 | 46.08 | 446 | 14.87 | ||
There were 893 genes located in the top 5% of autosomal windows of the M. fascicularis-M. nemestrina species pair. Enrichment analysis (Supplementary Tables S7) showed that there were no enriched terms related to fertilization or reproduction. This implies that introgression may not have contributed to the formation of reproductive isolation.
Genetic diversity and demographic history
Genetic diversity was assessed using individual heterozygosity. Genome-wide heterozygosity varied considerably among macaques, ranging from 0.000 363 to 0.002 971 (Table 2; Supplementary Figure 8A). Average heterozygosity was lowest for M. thibetana (0.000 551) and highest for M. nemestrina (0.002 829).
Figure 8.
Genome-wide heterozygosity and demographic history
A: Genome-wide heterozygosity estimated from non-overlapping 1 Mb windows. B–D: Historical effective population sizes (Ne) of fascicularis, silenus, and sinica species groups. X-axis represents time; Y-axis represents Ne. Plots were scaled using a mutation rate (μ) of 0.58×10-8 bp-1 generation-1 and a generation time (g) of 10.
The historical effective population sizes (Ne) of macaques were modeled from the distribution of heterozygous sites across the genome using PSMC analysis (Li & Durbin, 2011) (Figure 8B–D; Supplementary Figure S8). The ancestral effective population sizes for all macaques were almost the same and all experienced a bottleneck ~5 Ma. After that, the Ne of most macaques increased gradually, although that of M. nigra remained relatively stable. The M. thibetana population was largest ~0.80 Ma, after which it decreased. The M. arctoides population was largest ~2 Ma and declined thereafter. The variation trend of the M. assamensis population was similar to that of M. thibetana, but more gradual. The M. mulatta population increased after 5 Ma until 0.13 Ma, but there were two slight declines during the increasing period. The M. fascicularis population reached its largest size ~1.30 Ma, and then declined. The M. fuscata population remained small after 5 Ma, with small fluctuations. The M. nemestrina population increased from 5 Ma until 0.20 Ma. The M. tonkeana population remained small from 5 Ma to 1.70 Ma, and then increased to its peak at ~0.13 Ma.
DISCUSSION
Phylogeny of macaques
Our genome-wide-based multispecies coalescent phylogenetic tree shows a topology consistent with previous research based on nuclear markers and genome-wide data (Deinard & Smith, 2001; Fan et al., 2018; Jiang et al., 2016; Li et al., 2009). The reconstructed species tree (Figure 2C) supported the classification of both the four and seven species groups, the differences simply being different hierarchical classifications of the macaques. The two M. fascicularis specimens had inconsistent positions in the phylogenetic trees when we used different software (Figure 2A, B). The quartet scores supporting the two phylogenetic topologies were very close (0.420 8 and 0.404 7). CONSENSE analysis separately grouped the two M. fascicularis specimens in 37.39% of the 26 633 trees. These results imply that distinct genetic backgrounds exist between the two M. fascicularis. Mfas_1 was a captive bred individual from China not within its natural range (Ito et al., 2020; Osada et al., 2021; Trask et al., 2013) and may have been introduced from nearby countries such as Vietnam and Thailand. As reported previously, unidirectional introgression hybridization from Chinese M. mulatta to Indochinese M. fascicularis has played an important role in the formation of the Indochinese M. fascicularis genome (Stevison & Kohn, 2009; Tosi & Coke, 2007; Yan et al., 2011). The Mfas_1 genome may carry fragments originally from M. mulatta and the mosaic genome may result in a closer phylogenetic relationship between Mfas_1 and M. mulatta than its conspecific Mfas_Mau (Figure 2A).
It should be noted that our samples included not only wild-caught macaques but also captive-bred and unknown-origin samples (e.g., Mfas_1 and Mnem_1). Therefore, certain results regarding these two samples may not necessarily be conclusive. Further studies that include only wild-caught Macaca individuals are necessary to confirm our results.
Reticulate evolution in macaques
Phylogenetic inconsistencies among different markers are signals of complex evolutionary history and should not be ignored when analyzing genome fragments longer than 25 kb (Kumar et al., 2017; Nakhleh, 2013). There are many possible reasons for phylogenetic conflict, such as mutation, drift, selection, ILS, and hybridization (Nakhleh, 2013). Our genome-wide analyses showed that the local evolutionary histories of various genomic regions in the macaques were significantly different (Figure 1). Coalescent-based analyses of whole-genome sequences and network analyses also indicated that the macaque genomes were characterized by contradictory genealogies (Figures 2, 3, 6). Consensus network and gene flow analyses also suggested a reticulate evolutionary history of macaques (Figures 3–6). Phylogenetic networks provide a better model than trees for capturing reticulate evolutionary history (Nakhleh, 2013). Macaque evolution appears to be a process of gradual divergence with complex gene flow. Therefore, the divergence of macaques is best understood as a phylogenetic network. This may be a reason why the evolution of macaques has been reconstructed differently in published studies (Fan et al., 2017; Jiang et al., 2016; Li et al., 2009; Li & Zhang, 2005).
Reticulate evolutionary relationships have also been found in other mammals (Árnason et al., 2018; Figueiro et al., 2017; Kumar et al., 2017), as well as in birds (Zhang et al., 2017), fish (Gante et al., 2016), amphibians (Pabijan et al., 2017), reptiles (Barley et al., 2019), and insects (Edelman et al., 2019). Therefore, speciation with genetic exchange is common in animals, and should be viewed as a selective process that maintains species divergence even under gene flow (Kumar et al., 2017; Lexer & Widmer, 2008; Nosil, 2008).
Although we used a variety of software that consider ILS in gene flow analysis, ILS is an important factor that can lead to incongruences between gene trees or between gene and species trees and may not be completely ruled out. If several speciation events have occurred in one lineage within a relatively short time, different alleles may eventually fix in different descendant lineages, leading to incongruences as mentioned above. ILS is an important factor in evolutionary history but was unexamined in this study. Future studies including ILS analysis are necessary to improve our understanding of the evolutionary history of Macaca.
Signals of gene flow
Various macaque hybridization studies based on different molecular markers (e.g., autosome, mtDNA, Y chromosome, and genome-wide) and morphological characteristics have been published (Evans et al., 2017; Fan et al., 2014, 2018; Hamada et al., 2016; Ito et al., 2020; Jiang et al., 2016; Matsudaira et al., 2018; Osada et al., 2021; Rovie-Ryan et al., 2021; Tosi et al., 2000, 2003a, 2003b; Vanderpool et al., 2020; Yan et al., 2011), which suggest complicated gene flow patterns in macaques. However, most previous studies have focused on intragroup patterns or on the fascicularis and sinica species groups, e.g., between M. mulatta and M. fascicularis (Hamada et al., 2016), and proto-M. fascicularis aurea and sinica species group (Matsudaira et al., 2018). However, there is limited information about gene flow between the silenus group and other species within the genus Macaca.
Here, we analyzed more Macaca species, including three species from the silenus group, and found strong introgression signals in all species pairs between the fascicularis and silenus groups. In particular, D analysis confirmed the strongest introgression signals between M. nemestrina and M. fascicularis (Figure 4B). These two species have a wide geographical distribution overlap (Ang et al., 2020; Eudey et al., 2020; Roos et al., 2014), and natural hybridization events have been reported by field observation (Bernsteil, 1966). A recent study on the reference genomes of M. nemestrina, M. fascicularis, and M. mulatta also indicated strong gene flow signals among them (Vanderpool et al., 2020), consistent with our results. However, the previous study included only three macaque species, whereas our genome-wide analyses were based on multiple species and population samples from silenus and fascicularis species groups, which indicated that M. nemestrina-M. fascicularis showed the strongest introgression signal among all species pairs (Figure 4B). The signal between M. nemestrina and M. fascicularis is most likely caused by ancient hybridization events between the two species after their dichotomous divergence. We further estimated that the proportion of genome inherited via gene flow between the two species was 6.19% (fd statistic; Figure 5A).
We also noted that species pairs between the fascicularis and silenus species groups, namely M. fascicularis-M. nigra/M. tonkeana and M. fuscata/M. mulatta-M. nemestrina, showed slightly weaker introgression signals than M. nemestrina-M. fascicularis. Other species pairs from these two groups, namely M. nigra-M. fuscata/M. mulatta and M. tonkeana-M. fuscata/M. mulatta, showed the weakest introgression signals (Figure 4B). These findings indicate weaker introgression signals between species pairs that have more distant phylogenetic relationships with M. nemestrina or M. fascicularis. Furthermore, considering the non-overlapping distribution of species in other species pairs, excluding M. nemestrina-M. fascicularis, we suspect that introgression signals between the fascicularis and silenus species groups may be largely due to the similarity of genomes of closely related species, especially for insular species such as M. tonkeana, M. nigra, and M. fuscata, for instance, introgression signals between the two Sulawesi macaques (M. tonkeana and M. nigra) and the fascicularis species group. The two Sulawesi macaques are restricted to Sulawesi island and have no current distribution overlap with other macaques. However, previous studies have suggested that Bornean M. nemestrina expanded eastwards and reached Sulawesi, most likely by natural rafting, and diverged into numerous distinct Sulawesi species about 1.9–2.0 Ma (Meijaard, 2003; Ziegler et al., 2007). Sulawesi macaques thus share many genomic similarities with Bornean M. nemestrina. Owing to the strong introgression between M. nemestrina and M. fascicularis, the introgression signals detected between the fascicularis group and two Sulawesi macaques are most likely attributed to the genomic similarity of the closely related species. Similar to the Sulawesi macaques, introgression signals between M. fuscata and the silenus species group are probably due to the genomic similarity of M. fuscata to the Chinese M. mulatta, which both belong to the fascicularis species group (Chu et al., 2007). In addition, approximately 60.14%–67.93% of the top 5% of windows for the M. fascicularis-silenus group species pairs, and 45.84%–47.33% for the M. nemestrina-fascicularis group species pairs were shared (Table 3). These high ratios of shared windows confirmed that the introgression signals detected between Sulawesi macaques and the fascicularis group species, and between M. fuscata and the silenus group species are most likely due to genomic similarity, rather than hybridization after all dichotomous divergence events.
However, genomic similarity does not seem to be the cause of the introgression signal between Mauritian M. fascicularis (Mfas_Mau) and Bornean M. nemestrina (Mnem_2), which showed a bidirectional introgression signal, as detected by DFOIL (Figure 4C). Mauritius is over 6 600 km from Borneo and naturally occurring introgression between these species seems impossible, implying that the introgression signal between them is most likely from an ancient hybridization event. Macaca fascicularis was introduced to Mauritius by the Portuguese in the 16th century (Tosi & Coke, 2007), with possible origin from Sumatra, Java, or Peninsular Malaysia (Tosi & Coke, 2007). Therefore, the gene flow signal between Mauritian M. fascicularis and Bornean M. nemestrina mirrors the gene flow between Bornean M. nemestrina and M. fascicularis from Sumatra, Java, or Peninsular Malaysia. Sumatra, Java, Peninsular Malaysia, and Borneo were connected during most of the Pleistocene due to lower sea levels (Louys & Turner, 2012). The connection of these islands provided avenues for hybridization. As the bidirectional signal was only detected in one of the two M. nemestrina, there must have been a hybridization event after divergence of the two populations, i.e., after 0.33 Ma according to our divergence time estimation (Figure 2B).
It should be noted that there may be another explanation for the strong hybridization signals between the fascicularis and silenus species groups. For example, introgression may have occurred between ancestral lineages of these two species groups, and these signals were eventually retained in different descendant lineages. This explanation is consistent with the PhyloNetworks findings (Figure 6). The observed hybridization pattern may result in a similar distribution of hybridization signals across genomes among species pairs, consistent with Figure 7. It is also possible that ancestral hybridization and hybridization after divergence both occurred, which would make it difficult to determine all hybridization events correctly (Vanderpool et al., 2020). Studies with more species, larger sample sizes, and modeling analyses will allow detection of a more complex evolutionary history and clarification of any separate hybridization events.
Our study also detected introgression signals congruous with previous studies. Hybridization between M. fascicularis and M. mulatta has been extensively reported and there is a postulated hybrid zone (Hamada et al., 2016; Ito et al., 2020; Yan et al., 2011). Here, D analysis also detected a significant signal between these species. Similar to previously reported gene flow between M. thibetana and Chinese M. mulatta (Fan et al., 2014), our results also suggested a significant gene flow signal between these two lineages. Additionally, we detected a significant introgression signal between M. thibetana and Indian M. mulatta. Given the non-overlapping geographical distributions of these two lineages, the introgression signal is probably a result of genomic similarity between the Chinese and Indian M. mulatta or ancestral introgression.
Previous research has suggested that ~30% of the Vietnamese M. fascicularis genome is of Chinese M. mulatta origin, and ~8.84% of the M. thibetana genome contains putative introgression regions when compared to Chinese M. mulatta (Fan et al., 2014; Yan et al., 2011). According to our estimations, the proportion of introgression reached 5.93% for M. fascicularis and Chinese M. mulatta, and 3.53% for M. thibetana and M. mulatta (Figure 5A). As fd is a conservative estimator, the proportion of genome shared through gene flow may be underestimated (Martin et al., 2015). Although the amount of introgression varied between the fd statistic and other estimators, we still obtained the approximate proportions of introgression (Martin et al., 2015), which allow us to compare the degree of hybridization between different species.
Recent introgression signals in different populations
In our study, individuals from five species originated from different populations, which allows for the assessment of introgression after the divergence of different populations. Surprisingly, introgression signals varied in the different populations of all five species (Figure 4A, C). This phenomenon has also been reported in M. mulatta, M. fascicularis, and M. arctoides (Fan et al., 2018; Osada et al., 2021; Rovie-Ryan et al., 2021). These results suggest that gene flow occurred after the split of populations, and thus may be very recent. Although morphological characteristics, genital structure, and sexual behavior differ significantly among macaque species, the chromosome karyotypes of macaques are very similar, and hybrid offspring of Macaca are fertile (Bunlungsup et al., 2017; Ciani et al., 1989; Evans et al., 2017; Hamada et al., 2012; Yang & Shi, 1994). The results of our research and previous studies suggest that gene flow among macaques is an ongoing process, and there is incomplete reproductive isolation. Therefore, hybridization occurs between different sympatric macaques.
Genetic diversity and demographic history of macaques
The heterozygosity of M. thibetana was the lowest among macaques, with a similar level as the endangered snub-nosed monkeys (0.015%–0.068%) (Zhou et al., 2016). Previous study also reported a lower genome-wide heterozygosity of M. thibetana (0.0898%) than of M. fascicularis (0.3004%–0.3179%) and M. mulatta (0.2612%–0.2617%) (Fan et al., 2018). The heterozygosity of the other macaques is higher than that of humans (0.08%–0.12%) and great apes (0.065%–0.178%) (Prado-Martinez et al., 2013). The high heterozygosity of macaques is consistent with their widespread distribution (Roos et al., 2019).
We reconstructed the demographic histories of nine species of Macaca using PSMC. Fluctuation in demographic history is usually considered to be from adaptations to changes in the environment and climate. The varied and complicated fluctuation patterns in the different macaque species suggested that they experienced different environmental variation and climate change. According to our study, all macaques went through a bottleneck ~5 Ma. This bottleneck may indicate the ancestral population of all Asian macaques when they first arrived in Eurasia (Roos et al., 2019). The bottleneck may be due to the long-term cooling, drying, and even formation of the ephemeral Northern Hemisphere glaciations between 6.0 and 5.5 Ma during the late Miocene (Holbourn et al., 2018). Subsequent increases in population sizes may have been in response to the warm and wet early Pliocene climates (Sniderman et al., 2016). A similar bottleneck has also been identified in other studies (Fan et al., 2014, 2018; Liu et al., 2018). After the bottleneck, macaques underwent differentiation and radiation. Inconsistent demographic historical dynamics were produced when adapting to different environments and climate. Overall, these findings strongly indicate that the population sizes of macaques varied differently and considerably during the Pliocene and Pleistocene, with successive fluctuations likely resulting in different genetic diversity between these lineages.
CONCLUSIONS
Our genome-wide analyses covered all species groups of Asian macaques. We found that complex introgressive gene flow played an important role in the evolutionary history of Macaca, and the evolution of macaques should be more accurately understood as phylogenetic networks rather than trees. Introgression signals between the fascicularis and silenus species groups were the strongest of all signals detected. In particular, bidirectional gene flow occurred between Mauritian M. fascicularis and Bornean M. nemestrina. However, introgression signals between the island species (e.g., Sulawesi macaques and M. fuscata) and other species may be attributed to genomic similarities between closely related species or ancestral introgression. Different introgression signals in different populations of the same species suggest that gene flow in macaques is likely to be an ongoing process. In conclusion, our study provides comprehensive insight into gene flow in macaques from a genome-wide perspective. However, the contribution of high-level introgression to adaptive history in macaque species remains unclear and is worthy of further study with more extensive population sampling.
SUPPLEMENTARY DATA
Supplementary data to this article can be found online.
COMPETING INTERESTS
The authors declare that they have no competing interests.
AUTHORS’ CONTRIBUTIONS
J.L. and Z.X.F. designed the research. J.L. and Y.S. collected the samples. Y.S. and C.J. performed the experiments. Y.S., J.L., K.H.L., and H.Q. contributed to data analysis. Y.S., Z.X.F., and J.L. wrote the manuscript. M.P. revised the manuscript and improved the language. All authors read and approved the final version of the manuscript.
ACKNOWLEDGEMENTS
We thank Song Wang at Nanning Zoo and Zhan-Long He at the Chinese Academy of Medical Sciences for their support in sample collection.
Funding Statement
This work was supported by the National Natural Science Foundation of China (31530068, 31770415) and Fundamental Research Funds for the Central Universities (SCU2021D006)
References
- 1.Abbott R, Albach D, Ansell S, Arntzen JW, Baird SJE, Bierne N, et al Hybridization and speciation. Journal of Evolutionary Biology. 2013;26(2):229–246. doi: 10.1111/j.1420-9101.2012.02599.x. [DOI] [PubMed] [Google Scholar]
- 2.Ang A, Boonratana R, Choudhury A, Supriatna J. 2020. Macaca nemestrina. The IUCN Red List of Threatened Species 2020: e.T12555A181324867.
- 3.Árnason U, Lammers F, Kumar V, Nilsson MA, Janke A Whole-genome sequencing of the blue whale and other rorquals finds signatures for introgressive gene flow. Science Advances. 2018;4(4):eaap9873. doi: 10.1126/sciadv.aap9873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arnold ML, Meyer A Natural hybridization in primates: one evolutionary mechanism. Zoology (Jena) 2006;109(4):261–276. doi: 10.1016/j.zool.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 5.Baiz MD, Tucker PK, Mueller JL, Cortés-Ortiz L X-linked signature of reproductive isolation in humans is mirrored in a howler monkey hybrid zone. Journal of Heredity. 2020;111(5):419–428. doi: 10.1093/jhered/esaa021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barley AJ, de Oca ANM, Reeder TW, Manríquez-Morán NL, Monroy JCA, Hernández-Gallegos O, et al Complex patterns of hybridization and introgression across evolutionary timescales in Mexican whiptail lizards (Aspidoscelis) . Molecular Phylogenetics and Evolution. 2019;132:284–295. doi: 10.1016/j.ympev.2018.12.016. [DOI] [PubMed] [Google Scholar]
- 7.Bernsteil IS Naturally occurring primate hybrid. Science. 1966;154(3756):1559–1560. doi: 10.1126/science.154.3756.1559. [DOI] [PubMed] [Google Scholar]
- 8.Bouckaert R, Heled J, Kühnert D, Vaughan T, Wu CH, Xie D, et al BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Computational Biology. 2014;10(4):e1003537. doi: 10.1371/journal.pcbi.1003537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bryant D, Bouckaert R, Felsenstein J, Rosenberg NA, RoyChoudhury A Inferring species trees directly from biallelic genetic markers: bypassing gene trees in a full coalescent analysis. Molecular Biology and Evolution. 2012;29(8):1917–1932. doi: 10.1093/molbev/mss086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bunlungsup S, Kanthaswamy S, Oldt RF, Smith DG, Houghton P, Hamada Y, et al Genetic analysis of samples from wild populations opens new perspectives on hybridization between long-tailed (Macaca fascicularis) and rhesus macaques (Macaca mulatta) . American Journal of Primatology. 2017;79(12):e22726. doi: 10.1002/ajp.22726. [DOI] [PubMed] [Google Scholar]
- 11.Chen SF, Zhou YQ, Chen YR, Gu J Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34(17):i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chu JH, Lin YS, Wu HY Evolution and dispersal of three closely related macaque species, Macaca mulatta, M. cyclopis, and M. fuscata, in the Eastern Asia . Molecular Phylogenetics and Evolution. 2007;43(2):418–429. doi: 10.1016/j.ympev.2006.11.022. [DOI] [PubMed] [Google Scholar]
- 13.Ciani AC, Stanyon R, Scheffrahn W, Sampurno B Evidence of gene flow between Sulawesi macaques. American Journal of Primatology. 1989;17(4):257–270. doi: 10.1002/ajp.1350170402. [DOI] [PubMed] [Google Scholar]
- 14.Cortés-Ortiz L, Duda Jr TF, Canales-Espinosa D, García-Orduña F, Rodríguez-Luna E, Bermingham E Hybridization in large-bodied new world primates. Genetics. 2007;176(4):2421–2425. doi: 10.1534/genetics.107.074278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deinard A, Smith DG Phylogenetic relationships among the macaques: evidence from the nuclear locus NRAMP1 . Journal of Human Evolution. 2001;41(1):45–59. doi: 10.1006/jhev.2001.0480. [DOI] [PubMed] [Google Scholar]
- 16.Delson E. 1980. Fossil macaques, phyletic relationships and a scenario of deployment. In: Lindburg DG. The Macaques: Studies in Ecology, Behavior and Evolution. New York: Van Nostrand Reinhold.
- 17.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, et al A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nature Genetics. 2011;43(5):491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Edelman NB, Frandsen PB, Miyagi M, Clavijo B, Davey J, Dikow RB, et al Genomic architecture and introgression shape a butterfly radiation. Science. 2019;366(6465):594–599. doi: 10.1126/science.aaw2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eudey A, Kumar A, Singh M, Boonratana R. 2020. Macaca fascicularis (Errata Version Published in 2021). The IUCN Red List of Threatened Species 2020: e.T12551A195354635.
- 20.Evans BJ, Tosi AJ, Zeng K, Dushoff J, Corvelo A, Melnick DJ Speciation over the edge: gene flow among non-human primate species across a formidable biogeographic barrier. Royal Society Open Science. 2017;4(10):170351. doi: 10.1098/rsos.170351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fan PF, Liu Y, Zhang ZC, Zhao C, Li C, Liu WL, et al Phylogenetic position of the white-cheeked macaque (Macaca leucogenys), a newly described primate from southeastern Tibet . Molecular Phylogenetics and Evolution. 2017;107:80–89. doi: 10.1016/j.ympev.2016.10.012. [DOI] [PubMed] [Google Scholar]
- 22.Fan ZX, Zhao G, Li P, Osada N, Xing JC, Yi Y, et al Whole-genome sequencing of Tibetan macaque (Macaca thibetana) provides new insight into the macaque evolutionary history . Molecular Biology and Evolution. 2014;31(6):1475–1489. doi: 10.1093/molbev/msu104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fan ZX, Zhou AB, Osada N, Yu JQ, Jiang J, Li P, et al Ancient hybridization and admixture in macaques (genus Macaca) inferred from whole genome sequences . Molecular Phylogenetics and Evolution. 2018;127:376–386. doi: 10.1016/j.ympev.2018.03.038. [DOI] [PubMed] [Google Scholar]
- 24.Felsenstein J PHYLIP - phylogeny inference package (Version 3.2) Cladistics. 1989;5(2):164–166. [Google Scholar]
- 25.Figueiro HV, Li G, Trindade FJ, Assis J, Pais F, Fernandes G, et al Genome-wide signatures of complex introgression and adaptive evolution in the big cats. Science Advances. 2017;3(7):e1700299. doi: 10.1126/sciadv.1700299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fooden J Provisional classification and key to living species of macaques (primates: Macaca) . Folia Primatologica. 1976;25(2–3):225–236. doi: 10.1159/000155715. [DOI] [PubMed] [Google Scholar]
- 27.Gante HF, Matschiner M, Malmstrom M, Jakobsen KS, Jentoft S, Salzburger W Genomics of speciation and introgression in princess cichlid fishes from Lake Tanganyika. Molecular Ecology. 2016;25(24):6143–6161. doi: 10.1111/mec.13767. [DOI] [PubMed] [Google Scholar]
- 28.Green RE, Krause J, Briggs AW, Maricic T, Stenzel U, Kircher M, et al A draft sequence of the neandertal genome. Science. 2010;328(5979):710–722. doi: 10.1126/science.1188021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Groves C. 2001. Primate Taxonomy. Washington: Smithsonian Institution Press.
- 30.Hamada Y, San AM, Malaivijitnond S Assessment of the hybridization between rhesus (Macaca mulatta) and long-tailed macaques (M. fascicularis) based on morphological characters . American Journal of Physical Anthropology. 2016;159(2):189–198. doi: 10.1002/ajpa.22862. [DOI] [PubMed] [Google Scholar]
- 31.Hamada Y, Yamamoto A, Kunimatsu Y, Tojima S, Mouri T, Kawamoto Y Variability of tail length in hybrids of the Japanese macaque (Macaca fuscata) and the taiwanese macaque (Macaca cyclopis) . Primates. 2012;53(4):397–411. doi: 10.1007/s10329-012-0317-3. [DOI] [PubMed] [Google Scholar]
- 32.Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS UFBoot2: improving the ultrafast bootstrap approximation. Molecular Biology and Evolution. 2018;35(2):518–522. doi: 10.1093/molbev/msx281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Holbourn AE, Kuhnt W, Clemens SC, Kochhann KGD, Jöhnck J, Lübbers J, et al Late Miocene climate cooling and intensification of southeast Asian winter monsoon. Nature Communications. 2018;9(1):1584. doi: 10.1038/s41467-018-03950-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ito T, Kanthaswamy S, Bunlungsup S, Oldt RF, Houghton P, Hamada Y, et al Secondary contact and genomic admixture between rhesus and long-tailed macaques in the Indochina Peninsula. Journal of Evolutionary Biology. 2020;33(9):1164–1179. doi: 10.1111/jeb.13681. [DOI] [PubMed] [Google Scholar]
- 35.Ito T, Lee YJ, Nishimura TD, Tanaka M, Woo JY, Takai M Phylogenetic relationship of a fossil macaque (Macaca cf. robusta) from the Korean peninsula to Extant species of macaques based on zygomaxillary morphology . Journal of Human Evolution. 2018;119:1–13. doi: 10.1016/j.jhevol.2018.02.002. [DOI] [PubMed] [Google Scholar]
- 36.Jiang J, Yu JQ, Li J, Li P, Fan ZX, Niu LL, et al Mitochondrial genome and nuclear markers provide new insight into the evolutionary history of macaques. PLoS One. 2016;11(5):e0154665. doi: 10.1371/journal.pone.0154665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Junier T, Zdobnov EM The newick utilities: high-throughput phylogenetic tree processing in the UNIX shell. Bioinformatics. 2010;26(13):1669–1670. doi: 10.1093/bioinformatics/btq243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS ModelFinder: fast model selection for accurate phylogenetic estimates. Nature Methods. 2017;14(6):587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kubisch HM, Falkenstein KP, Deroche CB, Franke DE Reproductive efficiency of captive Chinese- and Indian-origin rhesus macaque (Macaca mulatta) females . American Journal of Primatology. 2012;74(2):174–184. doi: 10.1002/ajp.21019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kuhlwilm M, Han S, Sousa VC, Excoffier L, Marques-Bonet T Ancient admixture from an extinct ape lineage into bonobos. Nature Ecology & Evolution. 2019;3(6):957–965. doi: 10.1038/s41559-019-0881-7. [DOI] [PubMed] [Google Scholar]
- 41.Kumar V, Lammers F, Bidon T, Pfenninger M, Kolter L, Nilsson MA, et al The evolutionary history of bears is characterized by gene flow across species. Scientific Reports. 2017;7:46487. doi: 10.1038/srep46487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lambert SM, Streicher JW, Fisher-Reid MC, de la Cruz FRM, Martinez-Méndez N, García-Vázquez UO, et al Inferring introgression using RADseq and DFOIL: Power and pitfalls revealed in a case study of spiny lizards (Sceloporus) . Molecular Ecology Resources. 2019;19(4):818–837. doi: 10.1111/1755-0998.12972. [DOI] [PubMed] [Google Scholar]
- 43.Langmead B, Salzberg SL Fast gapped-read alignment with bowtie 2. Nature Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lewis PO A likelihood approach to estimating phylogeny from discrete morphological character data. Systematic Biology. 2001;50(6):913–925. doi: 10.1080/106351501753462876. [DOI] [PubMed] [Google Scholar]
- 45.Lexer C, Widmer A Review. The genic view of plant speciation: recent progress and emerging questions. Philosophical transactions of the Royal Society B. 2008;363(1506):3023–3036. doi: 10.1098/rstb.2008.0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li H A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics. 2011;27(21):2987–2993. doi: 10.1093/bioinformatics/btr509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li H, Durbin R Inference of human population history from individual whole-genome sequences. Nature. 2011;475(7357):493–496. doi: 10.1038/nature10231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li J, Han K, Xing JC, Kim HS, Rogers J, Ryder OA, et al Phylogeny of the macaques (Cercopithecidae: Macaca) based on Alu elements . Gene. 2009;448(2):242–249. doi: 10.1016/j.gene.2009.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li QQ, Zhang YP Phylogenetic relationships of the macaques (Cercopithecidae: Macaca), Inferred from mitochondrial DNA sequences . Biochemical Genetics. 2005;43(7–8):375–386. doi: 10.1007/s10528-005-6777-z. [DOI] [PubMed] [Google Scholar]
- 50.Liu L, Yu LL, Edwards SV A maximum pseudo-likelihood approach for estimating species trees under the coalescent model. BMC Evolutionary Biology. 2010;10:302. doi: 10.1186/1471-2148-10-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu ZJ, Tan XX, Orozco-terWengel P, Zhou XM, Zhang LY, Tian SL, et al Population genomics of wild Chinese rhesus macaques reveals a dynamic demographic history and local adaptation, with implications for biomedical research. GigaScience. 2018;7(9):giy106. doi: 10.1093/gigascience/giy106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Louys J, Turner A Environment, preferred habitats and potential refugia for Pleistocene Homo in southeast Asia . Comptes Rendus Palevol. 2012;11(2–3):203–211. [Google Scholar]
- 53.Malinsky M, Matschiner M, Svardal H Dsuite - fast D-statistics and related admixture evidence from VCF files . Molecular Ecology Resources. 2021;21(2):584–595. doi: 10.1111/1755-0998.13265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Martin SH, Davey JW, Jiggins CD Evaluating the use of ABBA-BABA statistics to locate introgressed loci. Molecular Biology and Evolution. 2015;32(1):244–257. doi: 10.1093/molbev/msu269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Martin SH, Jiggins CD Interpreting the genomic landscape of introgression. Current Opinion in Genetics & Development. 2017;47:69–74. doi: 10.1016/j.gde.2017.08.007. [DOI] [PubMed] [Google Scholar]
- 56.Matsudaira K, Hamada Y, Bunlungsup S, Ishida T, San AM, Malaivijitnond S Whole mitochondrial genomic and Y-chromosomal phylogenies of burmese long-tailed macaque (Macaca fascicularis aurea) suggest ancient hybridization between fascicularis and sinica species groups . Journal of Heredity. 2018;109(4):360–371. doi: 10.1093/jhered/esx108. [DOI] [PubMed] [Google Scholar]
- 57.Meijaard E Mammals of south-east Asian islands and their Late Pleistocene environments. Journal of Biogeography. 2003;30(8):1245–1257. doi: 10.1046/j.1365-2699.2003.00890.x. [DOI] [Google Scholar]
- 58.Nakhleh L Computational approaches to species phylogeny inference and gene tree reconciliation. Trends in Ecology & Evolution. 2013;28(12):719–728. doi: 10.1016/j.tree.2013.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Molecular Biology and Evolution. 2015;32(1):268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nosil P Speciation with gene flow could be common. Molecular Ecology. 2008;17(9):2103–2106. doi: 10.1111/j.1365-294X.2008.03715.x. [DOI] [PubMed] [Google Scholar]
- 61.Osada N, Matsudaira K, Hamada Y, Malaivijitnond S Testing sex-biased admixture origin of macaque species using autosomal and x-chromosomal genomic sequences. Genome Biology and Evolution. 2021;13(1):evaa209. doi: 10.1093/gbe/evaa209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pabijan M, Zieliński P, Dudek K, Stuglik M, Babik W Isolation and gene flow in a speciation continuum in newts. Molecular Phylogenetics and Evolution. 2017;116:1–12. doi: 10.1016/j.ympev.2017.08.003. [DOI] [PubMed] [Google Scholar]
- 63.Pease JB, Hahn MW Detection and polarization of introgression in a five-taxon phylogeny. Systematic Biology. 2015;64(4):651–662. doi: 10.1093/sysbio/syv023. [DOI] [PubMed] [Google Scholar]
- 64.Prado-Martinez J, Sudmant PH, Kidd JM, Li H, Kelley JL, Lorente-Galdos B, et al Great ape genetic diversity and population history. Nature. 2013;499(7459):471–475. doi: 10.1038/nature12228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Reimand J, Kull M, Peterson H, Hansen J, Vilo J g:Profiler—a web-based toolset for functional profiling of gene lists from large-scale experiments. Nucleic Acids Research. 2007;35(S2):W193–W200. doi: 10.1093/nar/gkm226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rogers J, Raveendran M, Harris RA, Mailund T, Leppälä K, Athanasiadis G, et al The comparative genomics and complex population history of Papio baboons . Science Advances. 2019;5(1):eaau6947. doi: 10.1126/sciadv.aau6947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Roos C, Boonratana R, Supriatna J, Fellowes JR, Groves CP, Nash SD, et al An updated taxonomy and conservation status review of Asian primates. Asian Primates Journal. 2014;4(1):2–38. [Google Scholar]
- 68.Roos C, Kothe M, Alba DM, Delson E, Zinner D The radiation of macaques out of Africa: evidence from mitogenome divergence times and the fossil record. Journal of Human Evolution. 2019;133:114–132. doi: 10.1016/j.jhevol.2019.05.017. [DOI] [PubMed] [Google Scholar]
- 69.Rovie-Ryan JJ, Khan FAA, Abdullah MT Evolutionary pattern of Macaca fascicularis in southeast Asia inferred using y-chromosomal gene . BMC Ecology and Evolution. 2021;21(1):26. doi: 10.1186/s12862-021-01757-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schliep KP Phangorn: phylogenetic analysis in R. Bioinformatics. 2011;27(4):592–593. doi: 10.1093/bioinformatics/btq706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sniderman JMK, Woodhead JD, Hellstrom J, Jordan GJ, Drysdale RN, Tyler JJ, et al Pliocene reversal of late Neogene aridification. Proceedings of the National Academy of Sciences of the United States of America. 2016;113(8):1999–2004. doi: 10.1073/pnas.1520188113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Solís-Lemus C, Bastide P, Ané C PhyloNetworks: a package for phylogenetic networks. Molecular Biology and Evolution. 2017;34(12):3292–3298. doi: 10.1093/molbev/msx235. [DOI] [PubMed] [Google Scholar]
- 73.Stange M, Sánchez-Villagra MR, Salzburger W, Matschiner M Bayesian divergence-time estimation with genome-wide single-nucleotide polymorphism data of sea catfishes (Ariidae) supports Miocene closure of the Panamanian isthmus. Systematic Biology. 2018;67(4):681–699. doi: 10.1093/sysbio/syy006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Stevison LS, Kohn MH Divergence population genetic analysis of hybridization between rhesus and Cynomolgus macaques. Molecular Ecology. 2009;18(11):2457–2475. doi: 10.1111/j.1365-294X.2009.04212.x. [DOI] [PubMed] [Google Scholar]
- 75.Tarailo-Graovac M, Chen NS Using repeatmasker to identify repetitive elements in genomic sequences. Current Protocols in Bioinformatics. 2009;25(1):4–10. doi: 10.1002/0471250953.bi0410s25. [DOI] [PubMed] [Google Scholar]
- 76.Tosi AJ, Coke CS Comparative phylogenetics offer new insights into the biogeographic history of Macaca fascicularis and the origin of the mauritian macaques . Molecular Phylogenetics and Evolution. 2007;42(2):498–504. doi: 10.1016/j.ympev.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 77.Tosi AJ, Disotell TR, Morales JC, Melnick DJ Cercopithecine y-chromosome data provide a test of competing morphological evolutionary hypotheses. Molecular Phylogenetics and Evolution. 2003a;27(3):510–521. doi: 10.1016/S1055-7903(03)00024-1. [DOI] [PubMed] [Google Scholar]
- 78.Tosi AJ, Morales JC, Melnick DJ Comparison of Y chromosome and mtDNA phylogenies leads to unique inferences of macaque evolutionary history. Molecular Phylogenetics and Evolution. 2000;17(2):133–144. doi: 10.1006/mpev.2000.0834. [DOI] [PubMed] [Google Scholar]
- 79.Tosi AJ, Morales JC, Melnick DJ Paternal, maternal, and biparental molecular markers provide unique windows onto the evolutionary history of macaque monkeys. Evolution. 2003b;57(6):1419–1435. doi: 10.1111/j.0014-3820.2003.tb00349.x. [DOI] [PubMed] [Google Scholar]
- 80.Trask JAS, Garnica WT, Smith DG, Houghton P, Lerche N, Kanthaswamy S Single-nucleotide polymorphisms reveal patterns of allele sharing across the species boundary between rhesus (Macaca mulatta) and cynomolgus (M. fascicularis) macaques . American Journal of Primatology. 2013;75(2):135–144. doi: 10.1002/ajp.22091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Vanderpool D, Minh BQ, Lanfear R, Hughes D, Murali S, Harris RA, et al Primate phylogenomics uncovers multiple rapid radiations and ancient interspecific introgression. PLoS Biology. 2020;18(12):e3000954. doi: 10.1371/journal.pbio.3000954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang RJ, Thomas GWC, Raveendran M, Harris RA, Doddapaneni H, Muzny DM, et al Paternal age in rhesus macaques is positively associated with germline mutation accumulation but not with measures of offspring sociability. Genome Research. 2020;30(6):826–834. doi: 10.1101/gr.255174.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wen DQ, Yu Y, Zhu JF, Nakhleh L Inferring phylogenetic networks using PhyloNet. Systematic Biology. 2018;67(4):735–740. doi: 10.1093/sysbio/syy015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Woodruff DS Biogeography and conservation in southeast Asia: how 2.7 million years of repeated environmental fluctuations affect today’s patterns and the future of the remaining refugial-phase biodiversity. Biodiversity and Conservation. 2010;19(4):919–941. doi: 10.1007/s10531-010-9783-3. [DOI] [Google Scholar]
- 85.Xue C, Raveendran M, Harris RA, Fawcett GL, Liu XM, White S, et al The population genomics of rhesus macaques (Macaca mulatta) based on whole-genome sequences . Genome Research. 2016;26(12):1651–1662. doi: 10.1101/gr.204255.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yan GM, Zhang GJ, Fang XD, Zhang YF, Li C, Ling F, et al Genome sequencing and comparison of two nonhuman primate animal models, the cynomolgus and Chinese rhesus macaques. Nature Biotechnology. 2011;29(11):1019–1023. doi: 10.1038/nbt.1992. [DOI] [PubMed] [Google Scholar]
- 87.Yang FT, Shi LM Studies of the miotic chromosomes, meiosis and spermatogenesis of a macaque hybrid. Acta Genetica Sinica. 1994;21(1):24–29. [PubMed] [Google Scholar]
- 88.Yang ZH PAML 4: phylogenetic analysis by maximum likelihood. Molecular Biology and Evolution. 2007;24(8):1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 89.Zhang C, Rabiee M, Sayyari E, Mirarab S ASTRAL-III: polynomial time species tree reconstruction from partially resolved gene trees. BMC Bioinformatics. 2018;19(S6):153. doi: 10.1186/s12859-018-2129-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhang DZ, Song G, Gao B, Cheng YL, Qu YH, Wu SY, et al Genomic differentiation and patterns of gene flow between two long-tailed tit species (Aegithalos) . Molecular Ecology. 2017;26(23):6654–6665. doi: 10.1111/mec.14383. [DOI] [PubMed] [Google Scholar]
- 91.Zhou XM, Meng XH, Liu ZJ, Chang J, Wang BS, Li MZ, et al Population genomics reveals low genetic diversity and adaptation to hypoxia in snub-nosed monkeys. Molecular Biology and Evolution. 2016;33(10):2670–2681. doi: 10.1093/molbev/msw150. [DOI] [PubMed] [Google Scholar]
- 92.Ziegler T, Abegg C, Meijaard E, Perwitasari-Farajallah D, Walter L, Hodges JK, et al Molecular phylogeny and evolutionary history of southeast Asian macaques forming the M. silenus group . Molecular Phylogenetics and Evolution. 2007;42(3):807–816. doi: 10.1016/j.ympev.2006.11.015. [DOI] [PubMed] [Google Scholar]
- 93.Zinner D, Arnold ML, Roos C The strange blood: natural hybridization in primates. Evolutionary Anthropology: Issues, News, and Reviews. 2011;20(3):96–103. doi: 10.1002/evan.20301. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data to this article can be found online.