Abstract
Mutations of PTEN-induced kinase I (PINK1) cause early-onset Parkinson’s disease (PD) with selective neurodegeneration in humans. However, current PINK1 knockout mouse and pig models are unable to recapitulate the typical neurodegenerative phenotypes observed in PD patients. This suggests that generating PINK1 disease models in non-human primates (NHPs) that are close to humans is essential to investigate the unique function of PINK1 in primate brains. Paired single guide RNA (sgRNA)/Cas9-D10A nickases and truncated sgRNA/Cas9, both of which can reduce off-target effects without compromising on-target editing, are two optimized strategies in the CRISPR/Cas9 system for establishing disease animal models. Here, we combined the two strategies and injected Cas9-D10A mRNA and two truncated sgRNAs into one-cell-stage cynomolgus zygotes to target the PINK1 gene. We achieved precise and efficient gene editing of the target site in three newborn cynomolgus monkeys. The frame shift mutations of PINK1 in mutant fibroblasts led to a reduction in mRNA. However, western blotting and immunofluorescence staining confirmed the PINK1 protein levels were comparable to that in wild-type fibroblasts. We further reprogramed mutant fibroblasts into induced pluripotent stem cells (iPSCs), which showed similar ability to differentiate into dopamine (DA) neurons. Taken together, our results showed that co-injection of Cas9-D10A nickase mRNA and sgRNA into one-cell-stage cynomolgus embryos enabled the generation of human disease models in NHPs and target editing by pair truncated sgRNA/Cas9-D10A in PINK1 gene exon 2 did not impact protein expression.
Keywords: Cas9-D10A, Cynomolgus, PINK1
INTRODUCTION
PTEN-induced kinase I (PINK1) was originally identified as a gene up-regulated by overexpression of tumor suppressor PTEN in carcinoma cell lines (Unoki & Nakamura, 2001). Within normal cells, the protein is mainly located in the mitochondria to protect cells from stress-induced mitochondrial dysfunction (Morais et al., 2014; Youle & Van Der Bliek, 2012). Loss of PINK1 function is closely associated with early-onset Parkinson’s disease (PD) (Pickrell & Youle, 2015; Reed et al., 2019), a neurodegenerative disorder characterized by dysfunction of dopaminergic (DA) neurons in the substantia nigra (SN) (Goldstein & Sharabi, 2019; Poewe et al., 2017). However, the pathology of PD caused by PINK1 mutations remains unclear, largely because PINK1 knockout mouse and pig models are unable to recapitulate the typical PD-associated symptoms observed in human patients (Dawson et al., 2010; Nakamura & Edwards, 2007; Zhou et al., 2015). Thus, it is necessary to generate PINK1 mutant animal models in non-human primates (NHPs), which show high similarity to humans, to reveal the unique functions of PINK1 in primate brains.
CRISPR/Cas9-mediated disease models in NHPs with precise genome modifications offer great hope for biomedical research (Chan, 2013; Lasbleiz et al., 2019; Vermilyea & Emborg, 2018). However, the off-target effects of Cas9 on the genomes of monkeys and other species are inevitable and limit widespread application (Zhang et al., 2015). Recent studies have demonstrated that the off-target effects of CRISPR-Cas9 can be reduced by increasing Cas9 specificity, including the use of high-fidelity Cas9 variants (Kleinstiver et al., 2016; Schmid-Burgk et al., 2020), or by reducing the generation of double-strand breaks (DSBs) in off-target sites, including the use of truncated sgRNAs or paired sgRNA/Cas9-D10A nickases (Gopalappa et al., 2018; Ran et al., 2013; Shen et al., 2014). However, some of these methods achieve lower off-target effects at the expense of on-target efficiency. The Cas9 double-nicking approach mediated by Cas9-D10A nickase requires the cooperation of two Cas9 nicking enzymes and a pair of sgRNAs to achieve DSBs. Unlike wild-type (WT) Cas9, which generates DSBs, Cas9-D10A nickase cuts only one DNA strand and generates a single-stranded nick that can be repaired by the high-fidelity base excision repair (BER) pathway (Dianov & Hübscher, 2013), thus minimizing the introduction of indels. Several independent studies have shown that the frequencies of off-target edits generated by paired Cas9-D10A nickase are lower than that with WT nucleases (Gopalappa et al, 2018; Guilinger et al., 2014; Ran et al., 2013). Moreover, paired Cas9-D10A nickase exhibits comparable or significantly higher on-target efficiency than individual WT Cas9 (Gopalappa et al, 2018). Because of its advantages, Cas-D10A has been applied to modify genes in Lactobacillus casei (Song et al., 2017), chicken DF1 cells (Lee et al., 2017), and pigs (Joanna et al., 2018), all showing low or even no off-target effects. However, the gene editing capacity of Cas9-D10A in NHPs and truncated sgRNAs have not yet been reported.
Here, we obtained three PINK1 mutant cynomolgus monkeys via co-injection of Cas9-D10A mRNA and two truncated sgRNAs targeting exon 2 into one-cell-stage cynomolgus embryos. These PINK1 mutant cynomolgus monkeys showed 12.5%–100% editing efficiency on on-target sites, with no indels detected on the off-target sites tested, indicating that pair truncated sgRNA/Cas9-D10A nickase enabled efficient and precise genome modifications in monkeys. The PINK1 mutations did not affect PINK1 protein expression in fibroblasts derived from mutant monkeys compared to WT fibroblasts. Moreover, the DNA alterations of PINK1 exon 2 did not affect the capacity of the induced pluripotent stem cells (iPSCs) reprogrammed from mutant fibroblasts to differentiate into DA neurons. These results suggest that pair truncated sgRNA/Cas9-D10A nickases could be applied to generate highly efficient and precise genome alterations in monkeys and that targeting PINK1 exon 2 alone may not be sufficient to model PD in NHPs.
MATERIALS AND METHODS
Ethics statement
Healthy cynomolgus monkeys (Macaca fascicularis) (2–6 years old, 4–6 kg) were selected for use in this study. All animals were housed at the Yunnan Key Laboratory of Primate Biomedical Research (LPBR), China. All animal procedures were performed following the Association for Assessment and Accreditation of Laboratory Animal Care International (AAALAC) guidelines for the ethical treatment of primates (approval No. KBI K001115033-01).
Preparation of mRNA and sgRNA
The hCas9-D10A plasmids were obtained from Addgene (#41816). The plasmid was linearized with the restriction enzyme PmeI, and mRNA was synthesized and purified using an In Vitro RNA Transcription Kit (Ambion, USA). SgRNA oligos were amplified and transcribed in vitro using the GeneArt Precision gRNA Synthesis Kit (Thermo Fisher Scientific, USA) and purified with the MEGA Clear Kit (Thermo Fisher Scientific, USA) according to the manufacturers’ instructions.
Oocyte collection and in vitro fertilization
Oocyte collection and fertilization were performed as described previously (Niu et al., 2014). In brief, 10 healthy female cynomolgus monkeys aged 5–8 years with regular menstrual cycles were selected as oocyte donors for super ovulation, which was performed by intramuscular injection with rhFSH (recombinant human follitropin alpha, Merck, USA) for 8 days, then rhCG (recombinant human chorionic gonadotropin alpha, Merck, USA) on day 9. Oocytes were collected by laparoscopic follicular aspiration 32–35 h after rhCG administration. Follicular contents were placed in Hepes-buffered Tyrode’s albumin lactate pyruvate (TALP) medium (Gibco, USA) containing 0.3% bovine serum albumin (BSA, Sigma, USA) at 37 °C. Oocytes were stripped of cumulus cells by pipetting after brief exposure (<1 min) to hyaluronidase (0.5 mg/mL, Sigma, USA) in TALP-Hepes to allow visual selection of nuclear mature metaphase II (MII; first polar body present) oocytes. The mature oocytes were subjected to intracytoplasmic sperm injection (ICSI) immediately and then cultured in CMRL-1066 containing 10% fetal bovine serum (FBS) (Gibco, USA) at 37 °C in 5% CO2. Fertilization was confirmed by the presence of the second polar body and two pronuclei.
SgRNA injection, embryo culture, and transplantation
At 6–8 h after ICSI, each zygote was injected with a mixture of Cas9-D10A mRNA (20 ng/mL) and sgRNA (25 ng/mL) to a total volume 5 pL. Microinjections were performed in the cytoplasm of oocytes using a microinjection system under standard conditions. Zygotes were then cultured in chemically defined hamster embryo culture medium-9 (HECM-9) (Gibco, USA) containing 10% FBS (Gibco, USA) at 37 °C in 5% CaO2 to allow embryo development. The culture medium was replaced every other day until the blastocyst stage. Cleaved embryos with high quality at the two-cell to blastocyst stage were transferred into the oviduct of matched recipient monkeys. Twenty-five monkeys were used as surrogate recipients. The earliest pregnancy diagnosis was performed by ultrasonography 20–30 days after embryo transfer. Both clinical pregnancy and number of fetuses were confirmed by fetal cardiac activity and the presence of a yolk sac as detected by ultrasonography.
Off-target analysis
All potential off-target sites with homology to the 23 bp sequence (sgRNA+PAM) were retrieved using a base-by-base scan of the whole cynomolgus genome, allowing ungapped alignments with up to four mismatches in the sgRNA target sequence. In the scan output, potential off-target sites with less than three mismatches in the seed sequence (1 to 7 bases) were selected for polymerase chain reaction (PCR) amplification using umbilical cord genomic DNA as templates. The PCR products were first subjected to T7E1 cleavage assay. The potential off-target sites yielding typical cleavage bands were considered as candidates, and the PCR products of the candidates were then cloned and sequenced to confirm off-target effects.
Western blotting
For western blotting analysis, fibroblasts from the ear were lysed, with PINK1 protein expression then analyzed using GAPDH as an internal loading control. About 40 mg of lysate was mixed with 5×loading buffer, then boiled for 15 min, loaded onto 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) gels, and transferred onto polyvinylidene difluoride (PVDF) membranes (Millipore, USA). The membranes were blocked with 5% non-fat milk for 2 h at room temperature, then incubated with primary antibodies (PINK1, antibody 1-Novus Biologicals, USA; antibody 2-Proteintech, USA; antibody 3-Abcam (ab216144), UK; antibody 4-Abcam (ab23707), UK; GAPDH, Kangcheng, China) overnight at 4 ℃ and subsequently for 1 h at room temperature (25 ℃) with goat anti-rabbit IgG antibody, (H+L) HRP conjugate (Millipore, USA) or goat anti-mouse IgG, (H+L) HRP conjugate (Millipore, USA). The epitope was tested using Pierce ECL Western Blotting Substrate (Thermo Fisher Scientific, USA).
Reprogramming fibroblasts into iPSCs
Fibroblasts were infected by the Sendai virus (Thermo Fisher Scientific). Transfected fibroblasts (1×105 cells per nucleofection) were directly plated into three 10 cm feeder-seeded dishes in Dulbecco’s modified Eagle medium (DMEM) (Thermo Fisher Scientific, USA) with 10% FBS (Hyclone, USA). The fibroblasts were replated 7 days post-infection and cultured in DMEM (Thermo Fisher Scientific, USA) with 10% FBS (Hyclone, USA). The culture medium was changed every other day. On day 12 post-transfection, the medium was replaced with DMEM/F12 (Thermo Fisher Scientific, USA) with 15% KSR containing 10 ng/mL bFGF (Peperotech, USA), 0.1 mmol/L β-Me (STEMCELL Technologies, Canada), NEAA (Thermo Fisher Scientific, USA), and 20% PSGro® Human iPSC/ESC Growth Medium (StemRD, USA). Colonies that morphologically resembled iPSC colonies became visible on day 15 after infection. iPSC lines were cultured on X-ray-inactivated CF-1 mouse embryonic fibroblasts in PSC growth media.
Dopaminergic neuron induction
The iPSCs were digested with collagenase IV (Gibco, USA), and neural induction was induced by switching from ESC growth media to differentiation media in suspension culture (Advance DMEM/F12 (Invitrogen, USA): neurobasal media (Invitrogen, USA) (1:1 mixture) supplemented with 1×N2 (Invitrogen, USA), 1×B27 (Invitrogen, USA), 10 ng/mL bFGF (Millipore, USA), 3 μmol/L CHIR99021 (Cellagen Technology, USA), 5 mmol/L SB431542 (Cellagen Technology, USA), 0.2 mmol/L Compound E (STEMCELL Technologies, Canada), and 0.1 mmol/L LDN193189 (Cellagen Technology, USA). After 6 days, embryoid bodies (EBs) were transferred to 5 mg/mL laminin (Gibco, USA)-coated plates for attachment culture, and the media were switched to neural stem cell (NSC) culture media (neurobasal medium, including B27 (Invitrogen, USA), N2 (Invitrogen, USA), and NEAA (Sigma, USA), 1% Glutmax (Sigma, USA), 3 mmol/L CHIR99021 (Stemcell Technologies, Canada), 5 mmol/L SB431542 (Stemcell Technologies, Canada), 10 ng/mL bFGF (Millipore, USA), and 1 000 U/mL hLIF (Millipore, USA)). To encourage cell propagation, 0.025% trypsin-EDTA (Invitrogen, USA) was used to digest NSC during passage. NESCs were routinely passaged to 1:8 to 1:16 ratios every 3 to 4 days.
For dopaminergic neuron induction, a PSC Dopaminergic Neuron Differentiation Kit (Thermo Fisher Scientific, USA) was used. On day 1, aspirate medium was replaced with 2 mL of complete STEMdiffTM Dopaminergic Neuron Differentiation Medium. After incubation at 37 °C in 5% CO2 for 5–6 days, the medium was changed every other day with warm (37 °C) complete STEMdiffTM Dopaminergic Neuron Differentiation Medium. On day 6/7, cells reached 90%–95% confluence, after which they were seeded on a pre-warmed (37 °C) PLO/laminin-coated (0.05%, 5 μg/mL) (Sigma, USA; Biolaminin, China) dish at a density of 4×104–6×104 cells/cm2 in complete STEMdiffTM Dopaminergic Neuron Differentiation Medium. The cells were then incubated at 37 °C in 5% CO2 for 7 days, with the medium changed every other day with warm (37 °C) complete STEMdiffTM Dopaminergic Neuron Differentiation Medium. On day 13/14, dopaminergic neuronal precursors were seeded onto a pre-warmed (37 °C) cell culture vessel coated with PLO/laminin at a density of 1.5×104–6×104 cells/cm2 in complete STEMdiffTM Dopaminergic Neuron Maturation Medium 1. The cells were distributed evenly, then incubated at 37 °C in 5% CO2 for 5 days, with the medium changed every other day. On day 18/19, the medium was replaced with STEMdiffTM Dopaminergic Neuron Maturation Medium 2 for maturation of dopaminergic neurons for a minimum of 2 weeks, with the medium changed every other day.
Immunostaining
Cells were fixed in 4% paraformaldehyde (DingGuo, China) at room temperature for 15 min, then blocked with phosphate-buffered saline (PBS, Corning, USA) containing 0.2% Triton X-100 (Sigma, USA) and 3% BSA (Sigma, USA) at room temperature for 45 min. The cells were incubated with primary antibodies at 4 ℃ overnight, then with secondary antibodies at room temperature for 1 h. The nuclei were stained with DAPI (Roche, Germany). Antibody details are provided below. The following antibodies were used: anti-OCT4 (1:200; Abcam, UK), Anti-Nanog (1∶300; Abcam, UK), and anti-SOX2 (1∶200; Abcam, UK).
Statistical analyses
All data were analyzed using GraphPad 6.0 software. Welch’s t-test was used to compare differences between two groups.
RESULTS
Pair truncated sgRNA/Cas9-D10A nickases induce efficient genomic editing of PINK1 in cynomolgus embryos
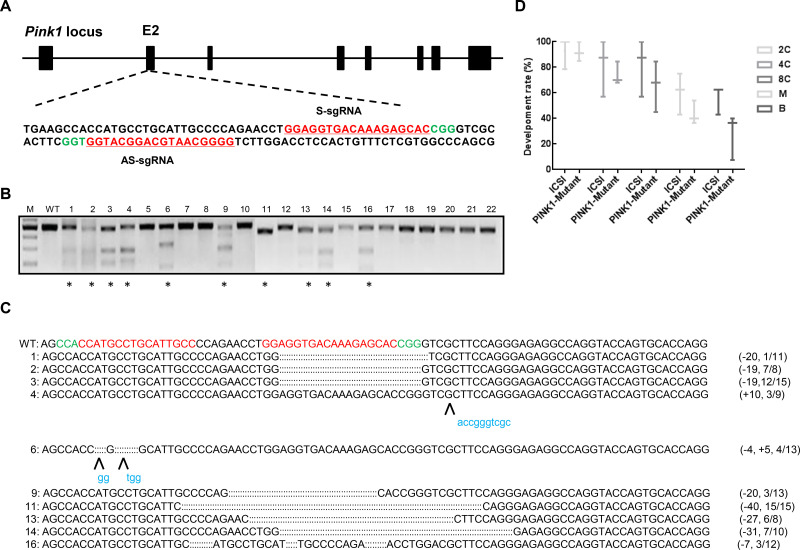
To test whether pair truncated sgRNA/Cas9-D10A works in monkey embryos, we designed two truncated sgRNAs (S-gRNA and AS-gRNA) to target PINK1 exon 2, which encodes the kinase domain essential for PINK1 function (Figure 1A). We injected the 20 ng/mL Cas9-D10A mRNA and 25 ng/mL sgRNA mixture each into one-cell-stage zygotes of cynomolgus monkeys. A total of 22 embryos developed normally into 8-cell or blastocyst stages and were collected and examined for site-specific genome modifications by PCR, T7 endonuclease I (T7E1) cleavage assay, and Sanger sequencing. As shown in Figure 1B, 10 out of 22 (45.5%) embryos carried smaller cleavage bands after T7E1 digestion (Figure 1B). TA cloning followed by Sanger sequencing showed that among the 10 edited embryos, eight contained 7–40 bp deletions with editing efficiencies varying from 9.1%–100%, one harbored a 10 bp insertion, and one carried a 4 bp deletion and 5 bp insertion (Figure 1C). Except for the 27 bp deletion in embryo #13, all indels detected in the edited embryos resulted in frame shift mutations, in principle, enabling the elimination of full-length PINK1 protein production. In addition, to explore whether microinjection of truncated sgRNA/Cas9-D10A RNA impacts monkey embryonic development, we calculated the developmental rates of the 2-cell, 4-cell, 8-cell, morula, and blastocyst stages based on 55 Cas9-D10A injected embryos and 22 counterparts. For all developmental stages tested, the developmental rate of embryos with received a truncated sgRNA/Cas9-D10A injection was comparable to that of the control group, thus demonstrating that microinjection of truncated sgRNA/Cas9-D10A in monkey embryo cells does not affect early embryonic development (Figure 1D).
Figure 1.
Paired Cas9-D10A nickases induce efficient genome editing of PINK1 in cynomolgus monkey embryos
A: Schematic of sgRNAs targeting PINK1 loci. Guide RNA sequences are underlined and highlighted in red. PAM sequences are highlighted in green. B: Cas9-mediated on-target cleavage of PINK1 by T7E1 cleavage assay. PCR products were amplified and subjected to T7E1 digestion. Samples with cleavage bands are marked with an asterisk. M, marker; WT, wild-type. C: Editing profiles of marked samples in (B). Undigested PCR products from (B) were subjected to TA cloning. Single TA clones were selected and analyzed by DNA sequencing. For WT alleles, PAM sequences are highlighted in green and sgRNA sequences are labeled in red. For alleles with indels, deleted bases are replaced with colons and inserted bases are labelled in lower case and highlighted in blue; deletions (-) and insertions (+). D: Developmental rate in Cas9-D10A-injected embryos is comparable to that in ICSI-treated embryos. ICSI, intracytoplasmic sperm injection embryo; PINK1-Mutant, PINK1 sgRNA injected embryo; 2C, 2-cell embryo; 4C, 4-cell embryo; 8C, 8-cell embryo; M, morula; B, blastula.
Pair truncated sgRNA/Cas9-D10A nickases enable one-step generation of PINK1 mutant monkeys
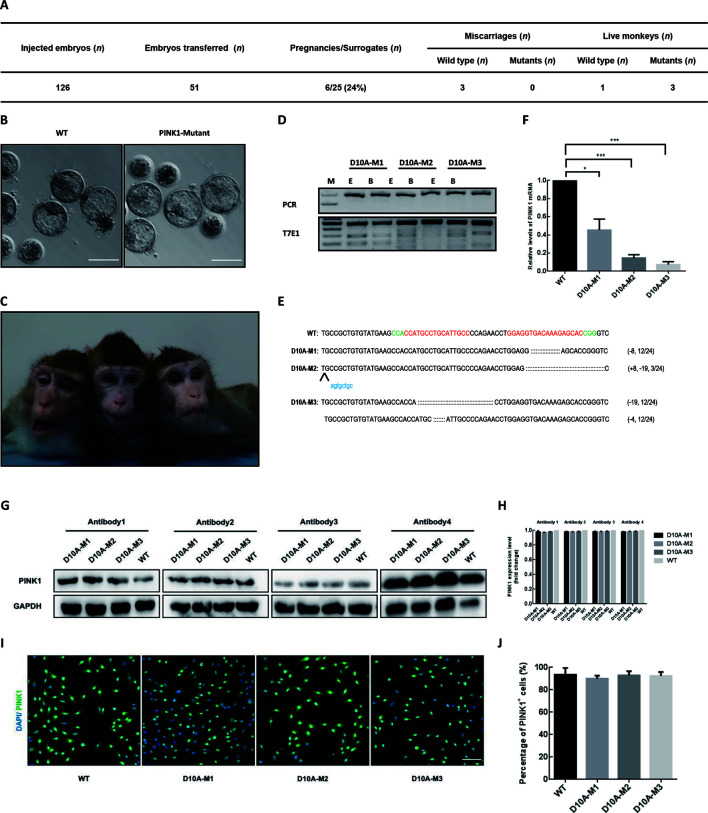
Next, we generated PINK1-modified cynomolgus monkeys using pair truncated sgRNA/Cas9-D10A. In total, 126 fertilized zygotes were microinjected with Cas9-D10A mRNA and sgRNA mixtures as described above. Totally, 51 developed-well Cas9-D10A-injected embryos were transferred into 25 surrogate females, six of which were successfully impregnated. Three of the six completed the pregnancy cycle (~150 days) and successfully gave birth to one or two infants (Figure 2A, C). Based on T7E1 assays of DNA extracted from blood or ear tissues, three newborns were found to carry PINK1 mutations (referred as D10A-M1, -M2, and -M3, respectively), and one was unedited (referred as WT). To further dissect the on-target editing profiles and efficiency, we collected DNA from fibroblasts derived from the mutant monkeys, then amplified the region around the target sites, performed TA cloning assay, and sequenced 24 single clones for each newborn. Sanger sequencing analysis showed that 12 out of 24 10A-M1 clones carried an 8 bp deletion and three out of 24 D10A-M2 clones had an 8 bp insertion and a 19 bp deletion. For the PINK1 mutant monkey D10A-M3, half of the clones (12/24) harbored a 19 bp deletion and the other half harbored a 4 bp deletion, suggesting that D10A-M3 may be a homozygous mutant (Figure 2D, E).
Figure 2.
Paired Cas9-D10A nickases enable one-step generation of PINK1 mutant monkeys
A: Summary of embryos injected, transferred, impregnated, and birthed. B: Representative images of blastocysts developed from zygotes injected with or without Cas9-D10A mRNA and sgRNA. Scale bars: 200 μm. C: Photo of D10A-M1, -M2, and -M3 (left to right) (taken when the monkeys were 3 years old). D: T7E1 cleavage assay of target site containing DNA products amplified from ear or blood tissue of mutant monkeys (D10A-M1, -M2, and -M3). Top panel represents undigested PCR bands and bottom panel represents digested PCR products. E: Ear; B: Blood; M: Marker. E: Editing profiles of mutant monkeys. Regions containing target sites were amplified from mutant fibroblasts and PCR products were subjected to TA cloning. Single TA clones were selected and analyzed by DNA sequencing. For WT allele, PAM sequences are highlighted in green and sgRNA sequences are labeled in red. For alleles with indels, deleted bases are replaced with colons and inserted bases are labelled in lower case and highlighted in blue; deletions (-) and insertions (+). F: RT-qPCR assay on mutant and WT fibroblasts (GAPDH was used for normalization). Compared with WT monkey, all mutant monkeys showed lower PINK1 mRNA expression. ***: P≤0.001; **: P≤0.01; *: P≤0.1. G: Western blotting assay on mutant and WT fibroblasts. H: Relative PINK1 expression levels were calculated using ImageJ 1.8.0 software. Compared with WT monkey, all mutant monkeys exhibited similar PINK1 protein expression. I: Representative images of immunofluorescence staining of mutant and WT fibroblasts. Scale bars: 200 μm. J: Numbers of total cells and PINK1-positive cells were counted using ImageJ 1.8.0 software.
Off-target effects are a major concern for CRISPR/Cas9 applications (Fu et al., 2013; Hsu et al., 2013; Pattanayak et al., 2013). To characterize the potential off-target edits of truncated sgRNA/Cas9-D10A in newborn monkeys, a total of 13 off-target sites predicted by Cas-OFFinder were selected and the regions around all potential off-target loci in mutant fibroblasts underwent PCR amplification, followed by T7E1 assay and/or DNA sequencing. No obvious indels were detected in the 13 potential off-target sites tested (Supplementary Figure S1).
To investigate whether the PINK1 mutations impact the expression levels of the messenger RNA (mRNA) and proteins, reverse-transcription PCR (RT-PCR) and western blotting assays were performed using the fibroblasts derived from PINK1 mutant monkeys and counterparts. The PINK1 mRNA levels in the D10A-M1, -M2, and -M3 monkeys were reduced to 45.3%, 14.7%, and 7.3% that in the WT monkey, respectively (Figure 2F), suggesting that the PINK1 mutations on exon 2 likely induced nonsense-mediated mRNA decay (NMD) by generating a premature stop codon. However, western blotting analysis showed that the expression levels of the PINK1 protein in mutant fibroblasts did not significantly decrease compared to that in WT fibroblasts. To rule out possible antibody effects, we selected four different antibodies for western blotting analysis (Figure 2G, H). Immunofluorescence staining confirmed the presence of PINK1 residual protein in all mutant fibroblasts (Figure 2I, J). To test whether PINK1 protein stability was affected in mutant fibroblasts, we treated D10A-M3 and WT fibroblasts with cycloheximide (CHX), an antibiotic that inhibits protein synthesis. We found that, as treatment time increased, PINK1 protein expression gradually decreased in WT fibroblasts, but did not change significantly in D10A-M3 fibroblasts under the same conditions (Supplementary Figure S2), suggesting that PINK1 protein stability increased in PINK1 mutant fibroblasts. These results indicate that although all PINK1 mutant monkeys carried frame-shift mutations on exon 2, the mutant fibroblasts did not show reduced PINK1 protein expression compared to WT fibroblasts.
PINK1 mutant fibroblast-derived DA neurons did not show specific PD-associated phenotypes
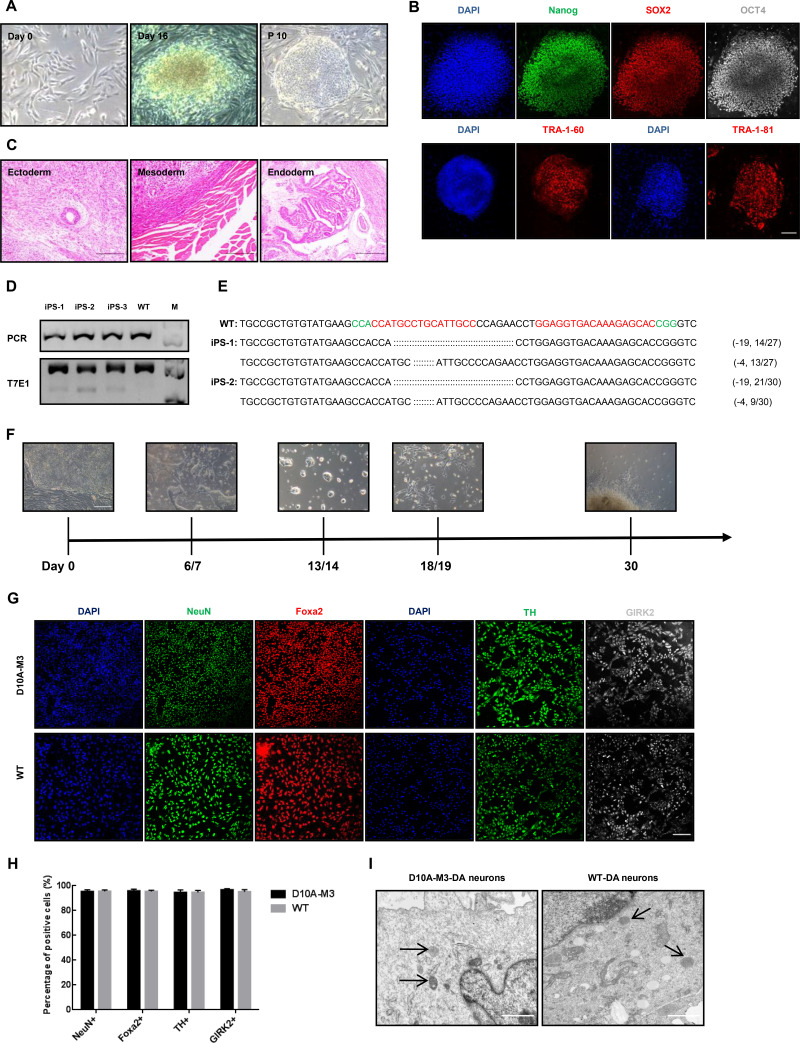
To further explore whether the PINK1 mutations affected function on DA neurons, we reprogrammed the fibroblasts from D10A-M3 and WT monkeys into iPSCs via a Sendai virus-based method (Okita et al., 2007; Takahashi & Yamanaka, 2006) and differentiated the selected iPSC clones into DA neurons in vitro. Three iPSC clones with PINK1 mutations were selected and expanded for differentiation (Figure 3A–E). To identity the types of neuronal cells differentiated from mutant and WT iPSCs, we stained the differentiated cells with specific markers of mature neurons (NeuN, Foxa2, and GIRK2) and DA neurons (TH). Results showed that the proportions of NeuN-, Foxa2-, GIRK2-, and TH-positive cells in mutant iPSC-DA neurons were comparable to that in WT iPSC-DA neurons (Figure 3G, H), indicating that PINK1 mutation did not influence the ability of iPSCs to differentiate into DA neurons. In addition, we compared the mitochondrial morphology of DA neurons induced from D10A-M3 and WT iPSCs under electron microscopy but did not observe any significant differences (Figure 3I).
Figure 3.
PINK1 mutant fibroblast-derived DA neurons did not show any specific PD-associated phenotypes
A: Reprogramming of fibroblasts into iPSCs by Sendai virus. Images represent typical cell phenotypes observed on days 0 and 16 after virus transduction and iPSC phenotypes at passage 10. Scale bars: 500 μm. B: Immunofluorescence staining of pluripotency markers OCT4, Nanog, SOX2, and TRA-1-81/60 in iPSCs. Scale bars: 250 μm. C: Teratoma differentiation of iPSCs in immunodeficient mice (NOD/SCID). Left, ectoderm; middle, mesoderm; right, endoderm. Scale bars: 500 μm. D: T7E1 cleavage assay of target sites amplified from three iPSC clones reprogrammed from D10A-M3 fibroblasts. Top panel represents undigested PCR bands, and bottom panel represents digested PCR products. E: Genotypes of iPSC-1 and iPSC-2. Two iPSCs showing different lengths of DNA fragments. F: Induction process of iPSCs to DA neurons. During induction, different differentiation and maturation media need to be replaced. After 30 days, DA neurons can be obtained. Scale bars: 500 μm. G: Representative images showing immunofluorescence staining of D10A-M3 and WT iPSC-derived DA neuron markers NeuN, Foxa2, TH, and GIRK2. Scale bars: 250 μm. H: Relative expression levels in (G) were calculated via ImageJ 1.8.0 software. No significant differences were observed between the proportion of mature DA neurons in D10A-M3 and WT during induction. I: Mitochondrial morphology of D10A-M3 and WT-derived DA neurons under electron microscopy. Compared with WT monkey, D10A-M3 exhibited similar mitochondrial morphology. Arrows, mitochondria of D10A-M3 and WT-derived neurons Scale bars: 1 μm.
DISCUSSION
This study is the first to report on the application of pair truncated sgRNA/Cas9-D10A nickases in modeling human diseases in NHPs. By injection of paired Cas9-D10A mRNA and two truncated sgRNAs targeting PINK1 exon 2 in one-cell-stage cynomolgus embryos, we successfully obtained three PINK1 mutant cynomolgus monkeys with editing efficiencies varying from 9.1% to 100%. Although frame-shift mutations were detected in the mutant fibroblasts, PINK1 protein expression was not affected when compared to that in WT fibroblasts due to a yet-unknown mechanism. Furthermore, the ability of iPSCs reprogrammed from fibroblasts to differentiate into DA neurons was not impacted by the PINK1 exon 2 mutations, suggesting that PINK1 exon 2 alone may not be a good target for establishing PINK1 knockout animal models.
Similar work was published earlier by Yang et al. (2019), which showed that CRISPR/Cas9 with paired sgRNAs enables the generation of PINK1 knockout rhesus models via inducing a large deletion across exon 2 and 4. We speculate that the large deletions generated in the above study permitted direct RNA splicing from PINK1 exon 1 to exon 5, thus leading to NMD (Brogna & Wen, 2009) and consequently eliminating protein translation. In addition, the PINK1 mutant monkeys (M3 and M4) in Yang et al. (2019) contained small, partial frame-shift deletions on either PINK1 exon 2 or exon 4 and exhibited comparable protein levels to WT monkeys, consistent with our results. These findings could be due to PINK1 WT protein stability or because the capacity of PINK1 WT mRNA translation was enhanced upon the partial loss of PINK1 coding sequences, as verified by western blotting assays (Figure 2G, H). One the other hand, as the commercial antibodies used in our study were designed to recognize the PINK1 carboxyl terminus, we cannot rule out that some yet-unidentified event, such as RNA mis-splicing, may compensate for the damage caused by the frame-shift mutations on PINK1 exon 2, thus producing a protein with a distinct N-terminus but identical C-terminus compared to the WT protein. Due to the presence of the PINK1 protein, all mutant monkeys were born normally and show no symptoms associated with PD, despite now being 6 years of age.
Modeling human genetic diseases in NHPs via CRISPR/Cas9 technology holds great promise for the study of human diseases and the development of corresponding therapeutics. However, this process is challenged by certain genome editing approaches, which are potentially toxic to embryos during microinjection of CRISPR/Cas9 components. We previously successfully generated gene-modified cynomolgus monkeys via co-injection of Cas9 mRNA and sgRNAs into one-cell-stage embryos (Niu et al., 2014), thereby proving Cas9 as an efficient and reliable method for NHP model generation. However, off-target effects, which are of major concern when utilizing CRISPR/Cas9 for gene manipulation, remain unresolved. Double-stranded DNA nicking mediated by Cas9 nickases (D10A or H840A) and truncated sgRNAs for targeting are two optimal ways to minimize the generation of off-target edits without impacting on-target editing efficiency (Fu et al., 2014; Ran et al., 2013). In the current study, we proved that microinjection of Cas9-D10A mRNA and pair truncated sgRNAs into one-cell-stage cynomolgus monkey embryos induced efficient gene modifications, enabled one-step generation of PINK1 mutant monkeys, and did not produce detectable indels in the top 13 potential off-target sites. These results indicate that paired Cas9 may be an efficient and accurate approach to establish human disease models in NHPs.
SUPPLEMENTARY DATA
Supplementary data to this article can be found online.
COMPETING INTERESTS
The authors declare that they have no competing interests.
AUTHORS’ CONTRIBUTIONS
Z.Z.C., Y.Y.N., Q.Y.Y., B.S. C.Z.L. conceptualized, drafted and modified the manuscript. Z.Z.C., J.Y.W., Y.K., X.Y.G. analyzed the data. Z.Z.C., Y.K., D.L.Z., L.Y. collected the data. All authors read and approved the final version of the manuscript.
ACKNOWLEDGEMENTS
We acknowledge the members of the Animal Facility of the Yunnan Key Laboratory of Primate Biomedical Research for excellent animal welfare and husbandry.
Funding Statement
This research was supported by the National Key Research and Development Program (2016YFA0101401 and 2018YFA0801400) and Major Basic Research Project of Science and Technology of Yunnan (2019FY002 and 202001BC070001)
Contributor Information
Bin Shen, Email: binshen@njmu.edu.cn.
Yu-Yu Niu, Email: niuyy@lpbr.cn.
References
- 1.Brogna S, Wen JK Nonsense-mediated mRNA decay (NMD) mechanisms. Nature Structural & Molecular Biology. 2009;16(2):107–113. doi: 10.1038/nsmb.1550. [DOI] [PubMed] [Google Scholar]
- 2.Chan AWS Progress and prospects for genetic modification of nonhuman primate models in biomedical research. ILAR Journal. 2013;54(2):211–223. doi: 10.1093/ilar/ilt035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dawson TM, Ko HS, Dawson VL Genetic animal models of Parkinson's disease. Neuron. 2010;66(5):646–661. doi: 10.1016/j.neuron.2010.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dianov GL, Hübscher U Mammalian base excision repair: the forgotten archangel. Nucleic Acids Research. 2013;41(6):3483–3490. doi: 10.1093/nar/gkt076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fu YF, Sander JD, Reyon D, Cascio VM, Joung JK Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nature Biotechnology. 2014;32(3):279–284. doi: 10.1038/nbt.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goldstein DS, Sharabi Y The heart of PD: lewy body diseases as neurocardiologic disorders. Brain Research. 2019;1702:74–84. doi: 10.1016/j.brainres.2017.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gopalappa R, Suresh B, Ramakrishna S, Kim H Paired D10A Cas9 nickases are sometimes more efficient than individual nucleases for gene disruption. Nucleic Acids Research. 2018;46(12):e71. doi: 10.1093/nar/gky222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guilinger JP, Thompson DB, Liu DR Fusion of catalytically inactive Cas9 to FokI nuclease improves the specificity of genome modification. Nature Biotechnology. 2014;32(6):577–582. doi: 10.1038/nbt.2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V et al. 2013. DNA targeting specificity of RNA-guided Cas9 nucleases. Nature Biotechnology. 31(9):827-32.
- 10.Joanna Z, Magdalena H, Agnieszka NT, Jacek J, Ryszard S, Zdzisław S, et al The production of UL16-binding protein 1 targeted pigs using CRISPR technology. 3 Biotech. 2018;8(1):70. doi: 10.1007/s13205-018-1107-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kleinstiver BP, Pattanayak V, Prew MS, Tsai SQ, Nguyen NT, Zheng ZL, et al High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016;529(7587):490–495. doi: 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lasbleiz C, Mestre-Francés N, Devau G, Luquin MR, Tenenbaum L, Kremer EJ, et al Combining gene transfer and nonhuman primates to better understand and treat parkinson's disease. Frontiers in Molecular Neuroscience. 2019;12:10. doi: 10.3389/fnmol.2019.00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee JH, Kim SW, Park TS Myostatin gene knockout mediated by Cas9-D10A nickase in chicken DF1 cells without off-target effect. Asian-Australasian Journal of Animal Sciences. 2017;30(5):743–748. doi: 10.5713/ajas.16.0695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morais VA, Haddad D, Craessaerts K, De Bock PJ, Swerts J, Vilain S, et al PINK1 loss-of-function mutations affect mitochondrial complex I activity via NdufA10 ubiquinone uncoupling. Science. 2014;344(6180):203–207. doi: 10.1126/science.1249161. [DOI] [PubMed] [Google Scholar]
- 15.Nakamura K, Edwards RH Physiology versus pathology in Parkinson's disease. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(29):11867–11868. doi: 10.1073/pnas.0704254104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Niu YY, Shen B, Cui YQ, Chen YC, Wang JY, Wang L, et al Generation of gene-modified cynomolgus monkey via Cas9/RNA-mediated gene targeting in one-cell embryos. Cell. 2014;156(4):836–843. doi: 10.1016/j.cell.2014.01.027. [DOI] [PubMed] [Google Scholar]
- 17.Okita K, Ichisaka T, Yamanaka S Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448(7151):313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- 18.Pattanayak V, Lin S, Guilinger JP, Ma E, Doudna JA, Liu DR. 2013. High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity.Nature Biotechnology. 31(9):839-43.
- 19.Pickrell AM, Youle RJ The roles of PINK1, parkin, and mitochondrial fidelity in Parkinson's disease. Neuron. 2015;85(2):257–273. doi: 10.1016/j.neuron.2014.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Poewe W, Seppi K, Tanner CM, Halliday GM, Brundin P, Volkmann J, et al Parkinson disease. Nature Reviews Disease Primers. 2017;3(1):17013. doi: 10.1038/nrdp.2017.13. [DOI] [PubMed] [Google Scholar]
- 21.Ran FA, Hsu PD, Lin CY, Gootenberg JS, Konermann S, Trevino AE, et al Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154(6):1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reed X, Bandrés-Ciga S, Blauwendraat C, Cookson MR The role of monogenic genes in idiopathic Parkinson's disease. Neurobiology of Disease. 2019;124:230–239. doi: 10.1016/j.nbd.2018.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schmid-Burgk JL, Gao LY, Li D, Gardner Z, Strecker J, Lash B, et al Highly Parallel Profiling of Cas9 Variant Specificity. Molecular Cell. 2020;78(4):794–800.e8. doi: 10.1016/j.molcel.2020.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shen B, Zhang WS, Zhang J, Zhou JK, Wang JY, Chen L, et al Efficient genome modification by CRISPR-Cas9 nickase with minimal off-target effects. Nature Methods. 2014;11(4):399–402. doi: 10.1038/nmeth.2857. [DOI] [PubMed] [Google Scholar]
- 25.Song X, Huang H, Xiong ZQ, Ai LZ, Yang S CRISPR-Cas9D10A nickase-assisted genome editing in Lactobacillus casei . Applied and Environmental Microbiology. 2017;83(22):e01259-17. doi: 10.1128/aem.01259-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takahashi K, Yamanaka S Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 27.Unoki M, Nakamura Y Growth-suppressive effects of BPOZ and EGR2, two genes involved in the PTEN signaling pathway . Oncogene. 2001;20(33):4457–4465. doi: 10.1038/sj.onc.1204608. [DOI] [PubMed] [Google Scholar]
- 28.Vermilyea SC, Emborg ME The role of nonhuman primate models in the development of cell-based therapies for Parkinson's disease. Journal of Neural Transmission. 2018;125(3):365–384. doi: 10.1007/s00702-017-1708-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang WL, Liu YB, Tu ZC, Xiao C, Yan S, Ma XS, et al CRISPR/Cas9-mediated PINK1 deletion leads to neurodegeneration in rhesus monkeys . Cell Research. 2019;29(4):334–336. doi: 10.1038/s41422-019-0142-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Youle RJ, Van Der Bliek AM Mitochondrial fission, fusion, and stress. Science. 2012;337(6098):1062–1065. doi: 10.1126/science.1219855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang XH, Tee LY, Wang XG, Huang QS, Yang SH Off-target effects in CRISPR/Cas9-mediated genome engineering. Molecular Therapy-Nucleic Acids. 2015;4:e264. doi: 10.1038/mtna.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou XQ, Xin JG, Fan NN, Zou QJ, Huang J, Ouyang Z, et al Generation of CRISPR/Cas9-mediated gene-targeted pigs via somatic cell nuclear transfer. Cellular and Molecular Life Sciences. 2015;72(6):1175–1184. doi: 10.1007/s00018-014-1744-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data to this article can be found online.