Abstract
During a 2018 antimicrobial resistance surveillance of Escherichia coli isolates from diarrheal calves in Xinjiang Province, China, an unexpectedly high prevalence (48.5%) of fosfomycin resistance was observed. This study aimed to reveal the determinants of fosfomycin resistance and the underlying transmission mechanism. Polymerase chain reaction (PCR) screening showed that all fosfomycin-resistant E. coli carried the fosA3 gene. Pulsed-field gel electrophoresis (PFGE) and southern blot hybridization revealed that the 16 fosA3-positive isolates belonged to four different PFGE patterns (i.e., A, B, C, D). The fosA3 genes of 11 clonally related strains (pattern D) were located on the chromosome, while others were carried by plasmids. Whole-genome and long-read sequencing indicated that the pattern D strains were E. coli O101:H9-ST10, and the pattern C, B, and A strains were O101:H9-ST167, O8:H30-ST1431, and O101:H9 with unknown ST, respectively. Among the pattern C strains, the blaCTX-M-14 gene was co-localized with the fosA3 gene on the F18:A-:B1 plasmids. Interestingly, phylogenetic analysis based on core genome single nucleotide polymorphisms (cgSNPs) showed that the O101:H9-ST10 strains were closely related to a Australian-isolated Chroicocephalus-origin E. coli O101:H9-ST10 strain producing CTX-M-14 and FosA3, with a difference of only 11 SNPs. These results indicate possible international dissemination of the high-risk E. coli clone O101:H9-ST10 by migratory birds.
Keywords: Clonal spread, Bovine, fosA3, blaCTX-M-14, O101:H9-ST10, Chroicocephalus
INTRODUCTION
Bacterial infections in domesticated bovines continue to increase year by year (Ruegg, 2017). Diarrhea in calves, which is partly caused by pathogenic Escherichia coli, is one of the three major bovine diseases causing economic loss to cattle producers (Wieler et al., 2007). Antimicrobials are often used to treat calf diarrhea caused by pathogenic E. coli (Constable, 2004). However, due to the abuse and misuse of antimicrobials, antimicrobial resistance (AMR) among bovine E. coli has become an important issue, especially for bacterial disease treatment and public health. Interestingly, in many cases, antibiotic resistance in cattle-origin E. coli is lower than in that originating from pigs or chickens (Ho et al., 2011; Li et al., 2019).
As an old antibiotic used in the treatment of uncomplicated urinary tract infections, fosfomycin has been reintroduced with other antimicrobials for the clinical treatment of multidrug-resistant (MDR) bacteria due to its excellent antimicrobial activity (Bassetti et al., 2019; Falagas et al., 2016). Although fosfomycin is not approved for animal use in China, fosfomycin resistance is widely reported among food animals nationwide. In addition, the plasmid-mediated fosA3 gene is reported to be a major determinant of fosfomycin resistance and is often co-localized with CTX-M β-lactamase genes (He et al., 2013, 2017; Huang et al., 2020). Consequently, the fosA3 gene can be co-selected under the use of β-lactam antibiotics. During AMR surveillance of E. coli from a cattle farm in Xinjiang Province, China, an unexpectedly high prevalence (48.5%) of fosfomycin resistance was observed, which was significantly higher than previously reported rates in bovines (Chan et al., 2014; Wang et al., 2017b). Hence, this study aimed to uncover the determinants of fosfomycin resistance and the underlying transmission mechanism in diarrheal calf-derived E. coli isolates.
MATERIALS AND METHODS
Bacterial strain
A total of 51 fecal samples were collected from diarrheal calves aged less than one month from a farm located in Yili, Xinjiang, China, in May 2018. These calves had been treated with enrofloxacin, ceftiofur, gentamycin, ampicillin, penicillin, florfenicol, colistin, and tulathromycin. The collected samples were enriched in Luria-Bertani (LB) broth at 37 ℃ for 16–18 h. The overnight culture was then incubated on a MacConkey agar plate. One isolate showing E. coli morphology from each sample was further identified using matrix-assisted laser desorption ionization-time of flight-mass spectrometry (MALDI-TOF MS) (Shimadzu-Biotech Corp., Japan).
Antimicrobial susceptibility testing and detection of resistance genes
According to the Clinical Laboratory Standard Institute (CLSI) guidelines (M07-A11), the minimum inhibitory concentrations (MICs) of all E. coli isolates against fosfomycin with 25 mg/L glucose-6-phosphate, beta-lactams (ampicillin, cefoxitin, ceftazidime, cefquinome, cefotaxime, and imipenem), aminoglycosides (amikacin, streptomycin, apramycin, gentamicin, and neomycin), tetracyclines (tetracycline and doxycycline), florfenicol, trimethoprim-sulfamethoxazole, and ciprofloxacin were determined using agar dilution or broth microdilution methods (colistin and tigecycline). The E. coli ATCC 25922 strain was used for quality control. The MICs were interpreted according to the criteria of the CLSI (M100-S30) (for fosfomycin, ampicillin, cefoxitin, ceftazidime, cefotaxime, imipenem, gentamicin, amikacin, tetracycline, doxycycline, trimethoprim-sulfamethoxazole, and ciprofloxacin), EUCAST (http://www.eucast.org) (for colistin and tigecycline), US Food and Drug Administration (FDA) for streptomycin (S, ≤32 mg/L; R, ≥64 mg/L), US National Antimicrobial Resistance Monitoring System (NARMS) for apramycin (S, ≤8 mg/L; R, ≥64 mg/L), and veterinary CLSI (VET06-S1) (for cefquinome, neomycin, and florfenicol).
Polymerase chain reaction (PCR) amplification was used to screen the fosfomycin resistance gene fosA3 and other important antimicrobial resistance genes (ARGs), including the extended-spectrum beta-lactamase gene blaCTX-M-1G/9G, AmpC beta-lactamase gene blaCMY-2, 16S rRNA methyltransferase genes armA and rmtB, florfenicol resistance gene floR, and colistin resistance gene mcr-1, using previously described primers (Supplementary Table S1) (Cao et al., 2020; Chen et al., 2007; Yan et al., 2004). PCR mapping was used to determine the genetic background of fosA3 with known primers (Supplementary Table S1) (Hou et al., 2012). The PCR products were subjected to Sanger sequencing (TsingKe Biological Technology, Beijing, China), and the obtained sequences were ascertained without mutation by NCBI-BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
Pulsed-field gel electrophoresis (PFGE), S1-PFGE, and southern blot hybridization
The clonal relationship of fosA3-positive E. coli isolates was assessed based on a rapid PFGE protocol (Gautom, 1997). Total DNA was digested by the XbaI enzyme (TaKaRa, Japan), embedded in low-melting-point agarose (Bio-Rad, USA), and subjected to PFGE using the CHEF-MAPPER System (Bio-Rad, USA). The electrophoretic conditions were: initial switch time, 2.16 s; final switch time, 63.8 s; running time, 19 h; angle, 120°; gradient, 6.0 V/cm; temperature, 14 °C; ramping factor, linear. The Salmonella enterica serotype Braenderup H9812 was used as a molecular size marker. The gel was dyed with ethidium bromide, visualized using a gel imaging system (Bio-Rad, USA), and analyzed with BioNumerics v6.6 (Applied Maths, Belgium). DNA patterns were interpreted based on proposed criteria (Tenover et al., 1995). The S1-PFGE protocol was the same as that of PFGE, except that total DNA was digested by S1 nuclease (TaKaRa, Japan). The products were subsequently used to perform southern blot hybridization with a digoxigenin-labelled fosA3 DNA probe (Roche, Germany).
Conjugation experiment
Horizontal transmission ability was determined for all fosA3 genes, and sodium azide-resistant E. coli J53 was used as the recipient for conjugation. The transconjugant was selected on a MacConkey agar plate supplemented with 150 mg/L sodium azide, 128 mg/L fosfomycin, and 25 mg/L glucose-6-phosphate. The transconjugant underwent PCR amplification and Sanger sequencing to confirm the transfer of the fosA3 gene.
Whole-genome sequencing analysis
Total genomic DNA, extracted using a Hipure Bacterial DNA Kit (Magen, China), was subjected to whole-genome sequencing by Novogene (Beijing Novogene Bioinformatics Co., Ltd., China) using Illumina platform Novo-PE150 technology and to long-read sequencing using Oxford Nanopore MinION (Oxford Nanopore Technologies, UK). SPAdes v3.8.7 (Bankevich et al., 2012) was used for de novo assembly. Unicycler v0.4.7 (Wick et al., 2017) was used to obtain the assembled genome. Whole-genome sequencing data were analyzed in silico using MLST v2.11 (https://github.com/tseemann/mlst) for multi-locus sequence typing, ABRicate v0.8 (https://github.com/tseemann/abricate) for screening ARGs, plasmid types, and virulence factors, and SeroTypeFinder v2.0 for serotyping (Joensen et al., 2015). Sequence alignment was performed by Easyfig v2.1 (Sullivan et al., 2011). A phylogenetic tree based on core genome single nucleotide polymorphisms (cgSNPs) was constructed using Parsnp v1.5.4 (https://github.com/marbl/parsnp). Snippy v4.6.0 (https://github.com/tseemann/snippy) was used to calculate total SNP quantity.
Nucleotide sequence accession number
The assembled genomes of the E. coli isolates (XJW9B263 and XJW9B277) based on long-read sequencing were submitted to GenBank under accession Nos. CP067399–CP067401 and CP068041–CP068045, respectively. The raw reads (Illumina) of the E. coli isolates (XJW9B298, XJW9B290, XJW9B274, XJW9B277, XJW9B263, and XJW9B285) were deposited in the Genome Sequence Archive (GSA) under accession No. CRA004296.
RESULTS
Overall resistance phenotypes of E. coli isolates
A total of 33 non-duplicate E. coli strains were obtained. The antimicrobial susceptibility results showed that of the 33 E. coli isolates, 29 (87.9%) exhibited resistance to five or more antimicrobials, and most were resistant to critically important antimicrobials (CIAs), including third and fourth generation cephalosporins (n=24) and ciprofloxacin (n=24) (Figure 1). In particular, 16 (48.5%) isolates showed resistance to fosfomycin, as well as cephalosporins and ciprofloxacin.
Figure 1.
Antimicrobial resistance phenotypes of all E. coli isolates
FOS, fosfomycin; AMP, ampicillin; FOX, cefoxidine; CAZ, ceftazidime; CQ, cefquinome; CTX, cefotaxime; IPM, imipenem; AMK, amikacin; STR, streptomycin; APR, apramycin; GEN, gentamicin; NEO, neomycin; TET, tetracycline; DOX, doxycycline; TGC, tigecycline; FFC, florfenicol; CL, colistin; SXT, trimethoprim-sulfamethoxazole; CIP, ciprofloxacin.
Molecular characterization of fosA3-positive E. coli
PCR screening confirmed that all fosfomycin-resistant isolates were positive for fosA3. In total, 93.8% (n=15), 93.8% (n=15), and 25.0% (n=4) of fosA3-positive isolates co-harbored blaCTX-M, floR, and rmtB, respectively, with blaCMY-2, armA, and mcr-1 not detected.
PFGE was successfully performed for all fosA3-carrying isolates, and four different XbaI PFGE patterns (A to D) were observed (Figure 2). Pattern D, which included 11 (68.8%) fosA3-carrying isolates, was dominant, followed by pattern C (n=3), suggesting pandemic pattern D fosA3-carrying E. coli isolates in this cattle farm.
Figure 2.
PFGE profiles, antimicrobial resistance genes, and genetic structure of fosA3-positive E. coli
fosA3 genes in blue and red indicate location on plasmid and chromosome, respectively.
The conjugation results indicated that five fosA3 genes in A-, B-, and C-pattern isolates were successfully transferred to E. coli J53. The S1-PFGE results confirmed that the fosA3 genes in the A- and C-pattern isolates were located on single plasmids of the same size (~138.9 kb), and the fosA3 gene in the B-pattern isolate was located on a single plasmid (~78.2 kb). Southern blot hybridization revealed that the other 11 fosA3 genes that failed in the conjugation experiment were located on the same-size band (~1 135 kb) in all D-pattern isolates, indicating a chromosomal location for the fosA3 gene.
The PCR mapping results demonstrated that all fosA3 genes were flanked by IS26. In total, three common types of IS26 composite transposons were found, including IS26-ΔISEcp1-blaCTX-M-14-ΔIS903-fosA3-orf1-Δorf2-IS26 (n=4), IS26-fosA3-orf1-orf2-Δorf3-IS26 (n=1), and IS26-fosA3-orf1-Δorf2-IS26 (n=11) (Supplementary Figure S1).
Genomic analysis of fosA3-positive E. coli
Whole-genome sequencing was performed on six fosA3-harbouring E. coli isolates with four different PFGE patterns, and the obtained data were analyzed in silico. The sequence type (ST) of the dominant D-pattern fosA3-harbouring E. coli isolates (XJW9B263 and XJW9B285) was ST10. The other isolates were ST167 (XJW9B274 and XJW9B277), ST1431 (XJW9B298), and unknown ST (XJW9B290). The O101:H9 serotype was obviously dominant among the six whole-genome sequencing isolates, except that the serotype of E. coli XJW9B298 (ST1431) was O8:H30. In addition, multiple ARGs were detected among all whole-genome sequencing isolates, three of which carried the F18:A-:B1 plasmid (Supplementary Table S2).
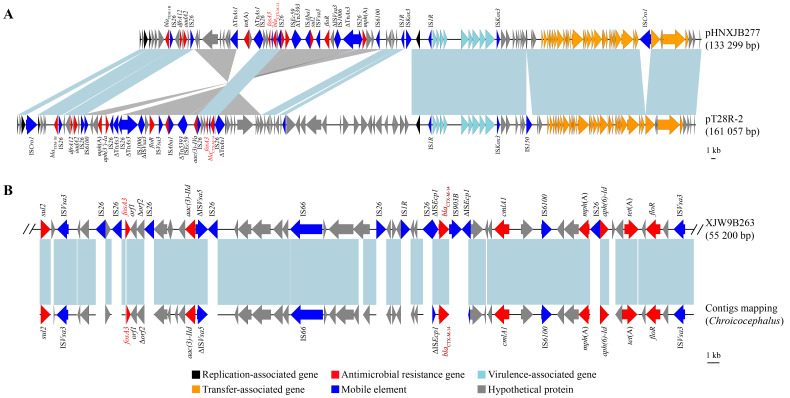
The D-pattern isolate XJW9B263 and C-pattern isolate XJW9B277 were further subjected to long-read sequencing to obtain assembled genomes. Sequence analysis confirmed that fosA3 and blaCTX-M-14 were co-located on the chromosome of XJW9B263 (GenBank: CP067399) and on the 133 299 bp long F18:A-:B1 plasmid pHNXJB277 of XJW9B277 (GenBank accession No.: CP068043).
NCBI-BLAST analysis revealed that pHNXJB277 showed genomic sequence identity with the fosA3-positive F18:A-:B1 plasmid pT28-2R of pet dog origin in Henan Province, China (GenBank accession No.: CP049355.1) (Figure 3A). The backbones of the two F18:A-:B1 plasmids were highly similar, while the multi-resistance region (MRR) varied. Similarly, fosA3 genes co-localized with blaCTX-M-14 on these two plasmids were surrounded by IS26.
Figure 3.
Genetic environment of fosA3 and blaCTX-M-14
A: Complete sequence comparison of two F18:A-:B1 plasmids (pHNXJB277, GenBank accession No.: CP068043; pT28R-2, GenBank accession No.: CP049355.1). B: Incomplete chromosomal sequence of XJW9B263 (GenBank accession No.: CP067399), containing same ARGs as Chroicocephalus-derived isolate (GenBank accession No.: GCA_014156895.1). Blue and gray shadows indicate homologous regions in same and opposite directions, respectively; same color arrows indicate same genes. Branch length is drawn to scale.
Core genome SNP calling of E. coli ST10 and ST167
To explore the origin of pandemic E. coli ST10 and ST167, the assembled contigs of 88 164 E. coli isolates from GenBank were collected to identify STs. A total of 3 444 E. coli isolates of ST10 and 336 E. coli isolates of ST167 were detected and further subjected to cgSNP-based phylogenetic analysis with the pandemic E. coli ST10 (isolates XJW9B263 and XJW9B285) and E. coli ST167 (isolates XJW9B274 and XJW9B277) identified in this cattle farm. Based on our results, all E. coli ST167 isolates from GenBank showed >390 cgSNP differences from XJW9B277, whereas isolates XJW9B274 and XJW9B277 were clonal with only one cgSNP distance. We also calculated the number of cgSNPs in 45 E. coli ST10 isolates that were related to the XJW9B263 clone against the XJW9B263 reference isolate. Isolates XJW9B285 and XJW9B263 from the cattle farm were clonal with no cgSNP difference. However, most of the E. coli ST10 isolates from GenBank showed >100 cgSNP differences from XJW9B263, including isolates originating from humans and animals in China; only seven isolates derived from humans and animals outside China had <100 cgSNP differences from XJW9B263. Of note, an E. coli ST10 strain (GenBank: GCA_014156895.1) isolated from a rectal swab of an Australian silver gull (Chroicocephalus) in 2017 differed from isolate XJW9B263 by just 11 cgSNPs (Figure 4), suggesting a close relationship according to the recommended ≤10 SNP threshold of E. coli (Schürch et al., 2018).
Figure 4.
Core genome SNP-based phylogenetic tree of E. coli ST10 strains
cgSNP indicates total amount of core genome SNPs in E. coli ST10 strains against reference isolate XJW9B263.
Serotypes, ARGs, virulence genes, and plasmids were screened using whole-genome sequences of the original Australian silver gull E. coli isolate (GCA_014156895.1), and further compared with isolate XJW9B263. Results showed that both were E. coli O101:H9-ST10 and carried identical resistance genes (blaCTX-M-14, blaTEM-1B, aph(3'')-Ib, aph(6)-Id, aac(3)-IId, tet(A), cmlA1, floR, mdf(A), mph(A), fosA3, sul2, dfrA14) and plasmids (IncFIB and IncY) (Table 1). XJW9B263 was distinguished from GCA_014156895.1 by only four virulence-associated genes (ecpD, entD, espL4, and espX4), and GCA_014156895.1 additionally carried Col-like replicon Col(MG828). Moreover, the contigs of GCA_014156895.1 successfully matched a partial chromosome of XJW9B263 containing fosA3, blaCTX-M-14, and other ARGs (Figure 3B)
Table 1. Genomic analysis of clonal E. coli O101:H9-ST10 .
| Isolate | XJW9B263 | GCA_014156895.1 |
| a indicates delta fosA3 gene likely truncated by whole-genome sequencing. Virulence genes and plasmids that differ from each other are underlined. | ||
| Collection year | 2018 | 2017 |
| Country | China | Australia |
| Source | Calf | Chroicocephalus |
| Serotype | O101:H9 | O101:H9 |
| Antimicrobial
resistance gene |
blaCTX-M-14, blaTEM-1B, aph(3'')-Ib, aph(6)-Id, aac(3)-IId, tet(A), cmlA1, floR, mdf(A), mph(A), fosA3, sul2, dfrA14 | blaCTX-M-14, blaTEM-1B, aph(3'')-Ib, aph(6)-Id, aac(3)-IId, tet(A), cmlA1, floR, mdf(A), mph(A), fosA3a, sul2, dfrA14 |
| Virulence gene | aslA, ecpA, ecpB, ecpC, ecpD, ecpE, ecpR, entA, entB, entC, entD, entE, entF, entS, espL1, espL4, espX1, espX4, espX5, espY1, fdeC, fepA, fepB, fepC, fepD, fepG, fes, fimB, fimC, fimD, fimE, fimF, fimG, fimH, fimI, ompA | aslA, ecpA, ecpB, ecpC, ecpE, ecpR, entA, entB, entC, entE, entF, entS, espL1, espX1, espX5, espY1, fdeC, fepA, fepB, fepC, fepD, fepG, fes, fimB, fimC, fimD, fimE, fimF, fimG, fimH, fimI, ompA |
| Plasmid | IncFIB, IncY | IncFIB, IncY, Col(MG828) |
DISCUSSION
With the widespread use of antimicrobials among humans and animals, MDR bacteria have emerged (Nikaido, 2009; Schürch et al., 2018), challenging the clinical treatment of bacterial infections. During routine surveillance of antimicrobial resistance in a cattle farm located in Xinjiang, a high prevalence of MDR E. coli isolates of diarrheal calf origin was observed. In particular, a surprisingly high fosfomycin resistance rate (48.5%) was noted, much higher than that of other food animals. For example, fosfomycin resistance rates in chicken-origin E. coli isolates from Guangdong and Northeast China are reported at 27.9% (He et al., 2017) and 27.4% (Jiang et al., 2017), respectively. PCR screening showed that fosA3 genes were present in all fosfomycin-resistant isolates, and thus may mediate fosfomycin resistance. All FosA3-producers also showed resistance to cephalosporins and ciprofloxacin, and almost all fosA3-producers co-harbored blaCTX-M, as commonly described nation- and worldwide (Cunha et al., 2017; Hou et al., 2012; Lupo et al., 2018; Lv et al., 2020; Yang et al., 2014). Regarding this, co-selection by long-term use of cephalosporins and enrofloxacin in this cattle farm may account for the prevalence of fosA3, as reported in a Chinese broiler farm, in which co-selection was considered the hypothetical driving force for the prevalence of plasmid-mediated colistin resistance gene mcr-1 (Cao et al., 2020).
Previous studies have reported that the fosA3 gene in E. coli is not generally spread by clonal transmission, but rather by plasmid-mediated horizontal transmission. Here, however, we found that most fosA3-positive isolates shared similar PFGE profiles, belonging to E. coli O101:H9-ST10 (n=2) and O101:H9-ST167 (n=2), indicating that the spread of fosA3 genes in this cattle farm was likely mediated by vertical clonal transmission.
Serotype O101, which is associated with animal and human diseases, is frequently detected among pathogenic E. coli (Chirila et al., 2017; Mandal et al., 2001; Tan et al., 2012). To the best of our knowledge, however, serotype O101:H9 has only been reported in Shiga toxin-producing E. coli (STEC) from humans with diarrheal disease in Germany (Beutin et al., 2008), in enterotoxigenic E. coli (ETEC) from diarrheal calves in Europe (Contrepois et al., 1998) and children with diarrheal disease in New Caledonia (Begaud et al., 1993), and in E. coli isolated from humans with acute suppurative cholangitis (Sung et al., 1994). In view of the limited number of reports of serotype O101:H9 in China, core genomes of the O101:H9-ST10 and O101:H9-ST167 clones were compared with those of all E. coli isolates submitted in GenBank to explore the origin of the E. coli clones in this cattle farm. Results demonstrated that all GenBank E. coli ST167 isolates were clonally unrelated to the O101:H9-ST167 clone (isolates XJW9B274 and XJW9B277) detected in this study, while core genomes of 45 E. coli ST10 isolates from GenBank were relatively similar to those of the O101:H9-ST10 clone (isolates XJW9B263 and XJW9B285). Further analysis confirmed that most showed >100 cgSNP differences from the O101:H9-ST10 clone; nevertheless, six isolates, all isolated from humans outside of China, including Canada and several European countries (UK, Germany, France, and Estonia), differed from the O101:H9-ST10 clone by <100 cgSNPs. Surprisingly, the core genome sequence of an Australian Chroicocephalus-derived E. coli isolate showed high similarity to the O101:H9-ST10 clone, with just 11 cgSNP differences, and the ARG profiles and plasmid types of both were very similar, indicating a significant clonal relationship. We note that wild birds forage for food at this cattle farm throughout the year, and that cattle feed is often contaminated by bird droppings (Supplementary Figure S2). Considering that the core genome of this clone exhibits greater similarity to foreign isolates, we suspect that the E. coli O101:H9-ST10 clone spreading in this cattle farm originated from foreign wild birds, i.e., the Chroicocephalus-bearing E. coli O101:H9-ST10. In accordance with a migration map of waterbirds worldwide (https://www.eaaflyway.net/), gulls and terns annually traverse the East Asian-Australasian Flyway (EAAF) covering East Asian countries and Australia. Furthermore, Australian Chroicocephalus often mix with the great crested terns (Thalasseus bergii cristatus) that fly to Australia in the south and to Ryukyu Islands and southeastern China in the north (https://birdsoftheworld.org/bow/home). Therefore, although the Australian Chroicocephalus does not migrate to Xinjiang, wild birds foraging for food at the cattle farm may mix with the great crested terns that show an overlapping distribution with the Australian Chroicocephalus, from where they acquire the E. coli O101:H9-ST10 clone (Figure 5).
Figure 5.
Schematic of possible global dissemination of E. coli O101:H9-ST10 from Australian Chroicocephalus to Chinese cattle
Green shadow indicates distribution of great crested tern (Thalasseus bergii cristatus).
Furthermore, though the estimated number of core genome SNPs per year for E. coli is unclear, a cutoff of ≤21 SNPs per genome per year for Klebsiella pneumoniae and a ≤23 SNP threshold for Enterobacteriales of local transmission have been reported (David et al., 2019; Sherry et al., 2019). Therefore, the 11 cgSNP distance between the two E. coli O101:H9-ST10 strains derived from Chroicocephalus and cattle suggests short-term clonal transmission of the high-risk E. coli ST10 from migratory Chroicocephalus birds to the calves.
Although wild birds (gulls) are not exposed to antimicrobials directly, their coastal habitats result in high-level human contact. As such, these birds have been described as reservoirs and vectors of MDR bacteria for the global diffusion of ARGs mediating resistance to CIAs (Mukerji et al., 2019, 2020; Villa et al., 2015; Wang et al., 2017a). To date, Chroicocephalus birds have not been reported in Yili in Xinjiang; however, indirect transmission of MDR bacteria from Chroicocephalus to wild birds that visit this cattle farm is possible. Due to their outdoor breeding, calves may potentially acquire MDR bacteria spread by wild birds that forage or fly over the farm. Thus, greater attention should be paid to wild birds visiting farms and sanitation should be strengthened to slow the potential risk of migratory bird dissemination of MDR bacteria.
CONCLUSIONS
This study described the clonal spread of FosA3- and CTX-M-producing E. coli O101:H9-ST10 among diarrheal calves from a cattle farm in Xinjiang, China. We speculate that the clones originated from migratory (foreign) birds and were transmitted by wild birds foraging on the farm. This is the first direct evidence of migratory birds disseminating bacteria resistant to CIAs across land and countries. These results highlight the need to pay greater attention to the risk of migratory birds spreading MDR microorganisms on a global scale. Moreover, biosafety prevention and control should not only focus on terrestrial pathogen contact, but also pathogens disseminated by birds and/or insects during aerial flight.
DATA AVAILABILITY
The datasets in this study can be found in GenBank under accession Nos. CP067399-CP067401 (XJW9B263) and CP068041-CP068045 (XJW9B277), and in the GSA databank under accession No. CRA004296 (XJW9B298, XJW9B290, XJW9B274, XJW9B277, XJW9B263, and XJW9B285).
SUPPLEMENTARY DATA
Supplementary data to this article can be found online.
COMPETING INTERESTS
The authors declare that they have no competing interests.
AUTHORS’ CONTRIBUTIONS
J.H.L. and J.Y. conceived the research. W.Y.H., X.X.Z., J.Y., G.L.G., M.Y.G., Z.P.C., and L.C.L. collected the data. J.H.L., W.Y.H., X.X.Z., J.Y., L.C.L., F.G.Z., and X.F.S. analyzed and interpreted the data. W.Y.H. drafted the manuscript, J.H.L., J.Y., and X.F.S. revised the report. All authors read and approved the final version of the manuscript.
ACKNOWLEDGEMENTS
We are grateful to Xiao-Jun Yang from the Kunming Institute of Zoology, Chinese Academy of Sciences, for helpful comments on this study.
Funding Statement
This work was supported by the National Natural Science Foundation of China (31625026), International Science and Technology Cooperation Project of Xinjiang Production and Construction Corps (XPCC) (2019BC004), and Innovation Team Project of Guangdong University (2019KCXTD001)
Contributor Information
Jun Yang, Email: junyang@scau.edu.cn.
Jian-Hua Liu, Email: jhliu@scau.edu.cn.
References
- 1.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. Journal of Computational Biology. 2012;19(5):455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bassetti M, Graziano E, Berruti M, Giacobbe DR The role of fosfomycin for multidrug-resistant gram-negative infections. Current Opinion in Infectious Diseases. 2019;32(6):617–625. doi: 10.1097/QCO.0000000000000597. [DOI] [PubMed] [Google Scholar]
- 3.Begaud E, Mondet D, Germani Y Molecular characterization of enterotoxigenic Escherichia coli (ETEC) isolated in New Caledonia (value of potential protective antigens in oral vaccine candidates) . Research in Microbiology. 1993;144(9):721–728. doi: 10.1016/0923-2508(93)90036-2. [DOI] [PubMed] [Google Scholar]
- 4.Beutin L, Kruger U, Krause G, Miko A, Martin A, Strauch E Evaluation of major types of Shiga toxin 2E-producing Escherichia coli bacteria present in food, pigs, and the environment as potential pathogens for humans . Applied and Environmental Microbiology. 2008;74(15):4806–4816. doi: 10.1128/AEM.00623-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cao YP, Lin QQ, He WY, Wang J, Yi MY, Lv LC, et al Co-selection may explain the unexpectedly high prevalence of plasmid-mediated colistin resistance gene mcr-1 in a Chinese broiler farm . Zoological Research. 2020;41(5):569–575. doi: 10.24272/j.issn.2095-8137.2020.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chan J, Lo WU, Chow KH, Lai EL, Law PY, Ho PL Clonal diversity of Escherichia coli isolates carrying plasmid-mediated fosfomycin resistance gene fosA3 from livestock and other animals . Antimicrobial Agents and Chemotherapy. 2014;58(9):5638–5639. doi: 10.1128/AAC.02700-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen L, Chen ZL, Liu JH, Zeng ZL, Ma JY, Jiang HX Emergence of RmtB methylase-producing Escherichia coli and Enterobacter cloacae isolates from pigs in China . Journal of Antimicrobial Chemotherapy. 2007;59(5):880–885. doi: 10.1093/jac/dkm065. [DOI] [PubMed] [Google Scholar]
- 8.Chirila F, Tabaran A, Fit N, Nadas G, Mihaiu M, Tabaran F, et al Concerning increase in antimicrobial resistance in Shiga toxin-producing Escherichia coli isolated from young animals during 1980-2016 . Microbes and Environments. 2017;32(3):252–259. doi: 10.1264/jsme2.ME17023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Constable PD Antimicrobial use in the treatment of calf diarrhea. Journal of Veterinary Internal Medicine. 2004;18(1):8–17. doi: 10.1111/j.1939-1676.2004.tb00129.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Contrepois M, Bertin Y, Pohl P, Picard B, Girardeau JP A study of relationships among F17 a producing enterotoxigenic and non-enterotoxigenic Escherichia coli strains isolated from diarrheic calves . Veterinary Microbiology. 1998;64(1):75–81. doi: 10.1016/S0378-1135(98)00253-3. [DOI] [PubMed] [Google Scholar]
- 11.Cunha MPV, Lincopan N, Cerdeira L, Esposito F, Dropa M, Franco LS, et al Coexistence of CTX-M-2, CTX-M-55, CMY-2, FosA3, and QnrB19 in extraintestinal pathogenic Escherichia coli from poultry in Brazil . Antimicrobial Agents and Chemotherapy. 2017;61(4):e02474–16. doi: 10.1128/AAC.02474-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.David S, Reuter S, Harris SR, Glasner C, Feltwell T, Argimon S, et al Epidemic of carbapenem-resistant Klebsiella pneumoniae in Europe is driven by nosocomial spread . Nature Microbiology. 2019;4(11):1919–1929. doi: 10.1038/s41564-019-0492-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Falagas ME, Vouloumanou EK, Samonis G, Vardakas KZ Fosfomycin. Clinical Microbiology Reviews. 2016;29(2):321–347. doi: 10.1128/CMR.00068-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gautom RK Rapid pulsed-field gel electrophoresis protocol for typing of Escherichia coli O157:H7 and other gram-negative organisms in 1 day . Journal of Clinical Microbiology. 1997;35(11):2977–2980. doi: 10.1128/jcm.35.11.2977-2980.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He D, Liu L, Guo B, Wu S, Chen X, Wang J, et al Chromosomal location of the fosA3 and blaCTX-M genes in Proteus mirabilis and clonal spread of Escherichia coli ST117 carrying fosA3-positive IncHI2/ST3 or F2:A-:B- plasmids in a chicken farm . International Journal of Antimicrobial Agents. 2017;49(4):443–448. doi: 10.1016/j.ijantimicag.2016.12.009. [DOI] [PubMed] [Google Scholar]
- 16.He LY, Partridge SR, Yang XY, Hou JX, Deng YT, Yao QF, et al Complete nucleotide sequence of pHN7A8, an F33:A-:B- type epidemic plasmid carrying blaCTX-M-65, fosA3 and rmtB from China . Journal of Antimicrobial Chemotherapy. 2013;68(1):46–50. doi: 10.1093/jac/dks369. [DOI] [PubMed] [Google Scholar]
- 17.Ho PL, Chow KH, Lai EL, Lo WU, Yeung MK, Chan J, et al Extensive dissemination of CTX-M-producing Escherichia coli with multidrug resistance to 'critically important' antibiotics among food animals in Hong Kong, 2008-10 . Journal of Antimicrobial Chemotherapy. 2011;66(4):765–768. doi: 10.1093/jac/dkq539. [DOI] [PubMed] [Google Scholar]
- 18.Hou J, Huang X, Deng Y, He L, Yang T, Zeng Z, et al Dissemination of the fosfomycin resistance gene fosA3 with CTX-M β-lactamase genes and rmtB carried on IncFII plasmids among Escherichia coli isolates from pets in China . Antimicrobial Agents and Chemotherapy. 2012;56(4):2135–2138. doi: 10.1128/AAC.05104-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang Y, Zeng L, Doi Y, Lv LC, Liu JH Extended-spectrum β-lactamase-producing Escherichia coli . The Lancet Infectious Diseases. 2020;20(4):404–405. doi: 10.1016/S1473-3099(20)30115-8. [DOI] [PubMed] [Google Scholar]
- 20.Jiang W, Men S, Kong LH, Ma SZ, Yang YQ, Wang YX, et al Prevalence of plasmid-mediated fosfomycin resistance gene fosA3 among CTX-M-producing Escherichia coli isolates from chickens in China . Foodborne Pathogens and Disease. 2017;14(4):210–218. doi: 10.1089/fpd.2016.2230. [DOI] [PubMed] [Google Scholar]
- 21.Joensen KG, Tetzschner AMM, Iguchi A, Aarestrup FM, Scheutz F Rapid and easy in silico serotyping of Escherichia coli isolates by use of whole-genome sequencing data . Journal of Clinical Microbiology. 2015;53(8):2410–2426. doi: 10.1128/JCM.00008-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li P, Liu D, Zhang XZ, Tuo HM, Lei CW, Xie XJ, et al Characterization of plasmid-mediated quinolone resistance in gram-negative bacterial strains from animals and humans in China. Microbial Drug Resistance. 2019;25(7):1050–1056. doi: 10.1089/mdr.2018.0405. [DOI] [PubMed] [Google Scholar]
- 23.Lupo A, Saras E, Madec JY, Haenni M Emergence of blaCTX-M-55 associated with fosA, rmtB and mcr gene variants in Escherichia coli from various animal species in France . Journal of Antimicrobial Chemotherapy. 2018;73(4):867–872. doi: 10.1093/jac/dkx489. [DOI] [PubMed] [Google Scholar]
- 24.Lv LC, Huang XY, Wang J, Huang Y, Gao X, Liu YY, et al Multiple plasmid vectors mediate the spread of fosA3 in extended-spectrum-β-lactamase-producing Enterobacterales isolates from retail vegetables in China . mSphere. 2020;5(4):e00507–20. doi: 10.1128/mSphere.00507-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mandal P, Kapil A, Goswami K, Das B, Dwivedi SN Uropathogenic Escherichia coli causing urinary tract infections . Indian Journal of Medical Research. 2001;114:207–211. [PubMed] [Google Scholar]
- 26.Mukerji S, Gunasekera S, Dunlop JN, Stegger M, Jordan D, Laird T, et al Implications of foraging and interspecies interactions of birds for carriage of Escherichia coli strains resistant to critically important antimicrobials . Applied and Environmental Microbiology. 2020;86(20):e01610–20. doi: 10.1128/AEM.01610-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mukerji S, Stegger M, Truswell AV, Laird T, Jordan D, Abraham RJ, et al Resistance to critically important antimicrobials in Australian silver gulls (Chroicocephalus novaehollandiae) and evidence of anthropogenic origins . Journal of Antimicrobial Chemotherapy. 2019;74(9):2566–2574. doi: 10.1093/jac/dkz242. [DOI] [PubMed] [Google Scholar]
- 28.Nikaido H Multidrug resistance in bacteria. Annual Review of Biochemistry. 2009;78:119–146. doi: 10.1146/annurev.biochem.78.082907.145923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ruegg PL A 100-year review: mastitis detection, management, and prevention . Journal of Dairy Science. 2017;100(12):10381–10397. doi: 10.3168/jds.2017-13023. [DOI] [PubMed] [Google Scholar]
- 30.Schürch AC, Arredondo-Alonso S, Willems RJL, Goering RV Whole genome sequencing options for bacterial strain typing and epidemiologic analysis based on single nucleotide polymorphism versus gene-by-gene-based approaches. Clinical Microbiology and Infection. 2018;24(4):350–354. doi: 10.1016/j.cmi.2017.12.016. [DOI] [PubMed] [Google Scholar]
- 31.Sherry NL, Lane CR, Kwong JC, Schultz M, Sait M, Stevens K, et al Genomics for molecular epidemiology and detecting transmission of carbapenemase-producing Enterobacterales in Victoria, Australia, 2012 to 2016 . Journal of Clinical Microbiology. 2019;57(9):e00573–19. doi: 10.1128/JCM.00573-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sullivan MJ, Petty NK, Beatson SA Easyfig: a genome comparison visualizer. Bioinformatics. 2011;27(7):1009–1010. doi: 10.1093/bioinformatics/btr039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sung JY, Shaffer EA, Lam K, Rususka I, Costerton JW Hydrophobic bile salt inhibits bacterial adhesion on biliary stent material. Digestive Diseases and Sciences. 1994;39(5):999–1006. doi: 10.1007/BF02087551. [DOI] [PubMed] [Google Scholar]
- 34.Tan C, Tang XB, Zhang X, Ding Y, Zhao ZQ, Wu B, et al Serotypes and virulence genes of extraintestinal pathogenic Escherichia coli isolates from diseased pigs in China . The Veterinary Journal. 2012;192(3):483–488. doi: 10.1016/j.tvjl.2011.06.038. [DOI] [PubMed] [Google Scholar]
- 35.Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, et al Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. Journal of Clinical Microbiology. 1995;33(9):2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Villa L, Guerra B, Schmoger S, Fischer J, Helmuth R, Zong Z, et al IncA/C plasmid carrying blaNDM-1, blaCMY-16, and fosA3 in a Salmonella enterica serovar Corvallis strain isolated from a migratory wild bird in Germany . Antimicrobial Agents and Chemotherapy. 2015;59(10):6597–6600. doi: 10.1128/AAC.00944-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang J, Ma ZB, Zeng ZL, Yang XW, Huang Y, Liu JH The role of wildlife (wild birds) in the global transmission of antimicrobial resistance genes. Zoological Research. 2017a;38(2):55–80. doi: 10.24272/j.issn.2095-8137.2017.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang XM, Dong ZM, Schwarz S, Zhu Y, Hua X, Zhang YH, et al Plasmids of diverse Inc groups disseminate the fosfomycin resistance gene fosA3 among Escherichia coli isolates from pigs, chickens, and dairy cows in Northeast China . Antimicrobial Agents and Chemotherapy. 2017b;61(9):e00859–17. doi: 10.1128/AAC.00859-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wick RR, Judd LM, Gorrie CL, Holt KE Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Computational Biology. 2017;13(6):e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wieler LH, Sobjinski G, Schlapp T, Failing K, Weiss R, Menge C, et al Longitudinal prevalence study of diarrheagenic Escherichia coli in dairy calves . Berliner und Münchener tierärztliche Wochenschrift. 2007;120(7-8):296–306. [PubMed] [Google Scholar]
- 41.Yan JJ, Hong CY, Ko WC, Chen YJ, Tsai SH, Chuang CL, et al Dissemination of blaCMY-2 among Escherichia coli isolates from food animals, retail ground meats, and humans in southern Taiwan . Antimicrobial Agents and Chemotherapy. 2004;48(4):1353–1356. doi: 10.1128/AAC.48.4.1353-1356.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang XY, Liu WL, Liu YY, Wang J, Lv LC, Chen XJ, et al F33:A-:B-, IncHI2/ST3, and IncI1/ST71 plasmids drive the dissemination of fosA3 and blaCTX-M-55/-14/-65 in Escherichia coli from chickens in China . Frontiers in Microbiology. 2014;5:688. doi: 10.3389/fmicb.2014.00688. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data to this article can be found online.
Data Availability Statement
The datasets in this study can be found in GenBank under accession Nos. CP067399-CP067401 (XJW9B263) and CP068041-CP068045 (XJW9B277), and in the GSA databank under accession No. CRA004296 (XJW9B298, XJW9B290, XJW9B274, XJW9B277, XJW9B263, and XJW9B285).