Abstract
ATP-binding cassette sub-family G member 2 (ABCG2) is a homodimeric ATP-binding cassette (ABC) transporter that not only has a key role in helping cancer cells to evade the cytotoxic effects of chemotherapy, but also in protecting organisms from multiple xeno- and endobiotics. Structural studies indicate that substrate and inhibitor (ligands) binding to ABCG2 can be differentiated quantitatively by the number of amino acid contacts, with inhibitors displaying more contacts. Although binding is the obligate initial step in the transport cycle, there is no empirical evidence for one amino acid being primarily responsible for ligand binding. By mutagenesis and biochemical studies, we demonstrated that the phylogenetically conserved amino acid residue, F439, was critical for both transport and the binding of multiple substrates and inhibitors. Structural modeling implied that the π-π interactions from each F439 monomer mediated the binding of a surprisingly diverse array of structurally unrelated substrates and inhibitors and that this symmetrical π-π interaction “clamps” the ligand into the binding pocket. Key molecular features of diverse ABCG2 ligands using the π-π clamp along with structural studies created a pharmacophore model. These novel findings have important therapeutic implications because key properties of ligands interacting with ABCG2 have been disovered. Furthermore, mechanistic insights have been revealed by demonstrating that for ABCG2 a single amino acid is essential for engaging and initiating transport of multiple drugs and xenobiotics.
Keywords: BCRP, cancer, CETSA, drug, thermal shift assay, kinase inhibitor, transporter
1 |. INTRODUCTION
ATP-binding cassette (ABC) transporters have key roles in the energy-dependent movement of molecules across biological membranes, including important chemotherapeutics. ATP-binding cassette sub-family G member 2 (ABCG2), also known as breast cancer resistance protein (BCRP), a ubiquitously expressed transporter, has an important physiological role in creating a barrier to penetration of many cytotoxins into cells. For instance, ABCG2 protects hematopoietic progenitors from cytotoxins, both endogenous and chemotherapeutics, and has key protective roles in the blood-brain and blood-testis barriers.1–3 Further, genetic variation in ABCG2 contributes to the susceptibility to toxicity from endogenous substances, for example, uric acid.4–6 ABCG2 dysfunction is linked to hyperuricemia, which can result in gout, kidney disease, and hypertension, all of which are secondary to the impaired export of uric acid.4,5 ABCG2, like its functional homologs ABCB1 (P-glycoprotein) and ABCC1 (MRP1), has a major protective role against many chemotherapeutics, effecting the pharmacokinetics of many commonly used drugs, and its expression correlates with a poor prognosis and treatment outcome of certain cancers (eg, acute myeloid leukemia7). ABCG2 has broad substrate specificity, with a seeming preference for ligands (defined here as either substrates and/or inhibitors) that are polycyclic, hydrophobic compounds.8 Among the many medicines and xenobiotics, the therapeutically important kinase inhibitors (KIs) interact with ABCG2 as both substrate and inhibitor (see Table S1) producing important drug interactions.9 Despite recent cryo-electron microscopy studies on the substrate and inhibitor binding pocket of ABCG2,10,11 there is no knowledge or empirical evidence, in either cells or native cell membranes, for ligands preferring any one amino acid to mediate binding to ABCG2.
One of the crucial unanswered questions concerning multidrug transporters, such as ABCG2, is: Is there a specific amino acid that is key to substrate and inhibitor binding to ABCG2? The cryo-electron microscopy structures of ABCG2 are unclear as to whether there is a single amino acid that promotes binding to the binding pocket.10,11 At present, it appears that inhibitors can be differentiated from substrates by both their greater size and sheer number and location of potential contacts with ABCG2.11 For ABCG2, an analog of the inhibitor, Ko143 interacted with 16 residues distributed among transmembrane helices (TMH) 1, 2, and 5,10 whereas a substrate, estrone-3-sulfate, interacted with fewer residues. The general interpretation was that the total number of contacts enabled tight binding of inhibitors (eg, tariquidar) compared to substrates (eg, estrone-sulfate). Thus, inhibitors, with more contacts were hypothesized to create a “wedge” that blocked substrate transport. Accordingly, one might infer that disruption of inhibitor binding to ABCG2 would require the loss of more molecular contacts than that of a substrate, and that substrates and inhibitors might not depend on the the same contacts to bind to ABCG2.
Unexpectedly, for ABCG2, our studies discovered that in a native cellular environment, the substitution of a highly conserved amino-acid caused the loss of substrate transport and inhibitor binding. Our studies reveal a key feature of the amino acid F439, where, through π-π stacking, it appears to form a molecular “clamp” in the ABCG2 binding pocket. This conserved feature might be important to other ABCG family members (eg, ABCG5/8). Importantly, this substitution of alanine for phenylalanine at position 439 results in the loss of ABCG2 interaction with multiple ligands, suggesting that this amino acid interaction is a key initiating step for ABCG2 transport and inhibitor interaction in cells.
2 |. MATERIALS AND METHODS
2.1 |. Chemicals
Fumitremorgin C (FTC) and Ko143 were obtained from Enzo Life Sciences. Hoechst 33342 was obtained from Life Technologies. Pheophorbide a (PhA) was obtained from Frontier Scientific Inc PhA was dissolved in DMSO and aliquots protected from light at −20°C. Other compounds were stored at −20°C at 10 mM in DMSO.
2.2 |. Plasmid construction and site-directed mutagenesis
All site-directed mutations were generated in the MSCV-ABCG2-IRES-GFP expression plasmid using QuikChange II XL site-directed mutagenesis kit (Agilent Technologies, Santa Clara, CA). The primers used in this study are indicated in the Table 1.
TABLE 1.
Oligonucleotide primers used to generate ABCG2 mutationsa
| Mutants | Forward primer (5′->3′) | Reverse primer (5′->3′) |
|---|---|---|
| N436A | ctcttcttcctgacgaccGCccagtgtttcagcagtgt | acactgctgaaacactggGCggtcgtcaggaagaagag |
| F439A | tgacgaccaaccagtgtGCcagcagtgtttcagccg | cggctgaaacactgctgGCacactggttggtcgtca |
| F439W | tgacgaccaaccagtgttGGagcagtgtttcagccg | cggctgaaacactgctCCaacactggttggtcgtca |
| F439Y | tgacgaccaaccagtgttAcagcagtgtttcagccg | cggctgaaacactgctgTaacactggttggtcgtca |
Capital letters indicate substitution.
2.3 |. Cell culture and transfection
Mouse embryonic fibroblast (MEF) cell line was derived from Abcg2−/− knockout (KO) mice. The Abcg2-KO mice were obtained from the Sorrentino lab.2 The Abcg2-KO MEFs were engineered to express human ABCG2 (Uniprot: Q9UNQ0). Cells were cultured at 37°C with 5% of CO2. Cells were grown in Dulbecco’s Modified Essential Medium (D-MEM) (Lonza, Basel, Switzerland) supplemented with 10% of fetal bovine serum (Gibco, Grand Island, NY), penicillin (100 U/mL), streptomycin (100 μg/mL) (Gibco, Grand Island, NY), and l-glutamine (2 mM) (Gibco, Grand Island, NY). The Abcg2-KO MEF cell line was transduced with the retroviruses harboring cDNAs encoding either the mutant ABCG2 N436A, F439A, F439W, or F439Y.
2.4 |. Transport assays
The intracellular accumulation of fluorescent PhA, a specific ABCG2 substrate,12,13 or Hoechst 33342 was used to determine the function of ABCG2. 10 μM of Fumitremogin C (FTC), a specific inhibitor of ABCG2 activity,14 was used as a positive control. Cells were seeded into a 96-well cell culture plates at 10 000 cells/well and PhA or Hoechst 33342 (0.5 μM of final concentration) and either FTC or the test ligand added to the culture medium followed by an incubation for 1 hour (PhA) or 2 hours (Hoechst 33342) in the dark. Thereafter, the medium was removed, cells were washed twice with ice-cold PBS and the PhA fluorescence was then quantitatively determined with a Synergy H4 microplate reader (BioTek, Winooski, VT, USA) at an excitation wavelength of 405 nm (PhA) or 355 nm (Hoechst 33342) and an emission wavelength of 667 nm (PhA) or 460 nm (Hoechst 33342). The data were normalized by the proportion accumulated relative (%) to FTC inhibition (see Figure S1 for FTC dose-response curve). The accumulation of Hoechst 33342 was normalized to protein concentration as determined using the BCA Protein Assay Kit (Pierce, Rockford, IL, USA).
2.5 |. Cellular thermal shift assay
The cellular thermal shift assay (CETSA)15–17 was adapted for ABCG2 as follows: To establish the melting temperature (Tm) for ABCG2, MEFs lacking Abcg2 (KO), but harboring human ABCG2 were gently detached with a weak trypsin solution (0.05% of trypsin). After this, the cells were washed with PBS three times followed by resuspension in PBS supplemented with protease inhibitors (complete Tablets-Mini EASYpack, Roche, Basel, Switzerland). A portion of the cell suspension was reserved for protein determination by the BCA protein assay. The resuspended cells were then heated in a PCR machine for 3 minutes at various temperatures (59–75°C), followed by a 3 min incubation at room temperature. Samples were then treated with ice-cold PBS supplemented with NP-40 to a final concentration of 0.8%, snap-frozen in liquid nitrogen, and then placed into a thermoshaker at 25°C to initiate thawing before being transferred to ice until the entire contents were thawed. This freeze-thaw cycle was repeated twice before ultracentrifugation to precipitate the denatured protein (20 minutes at 4°C and 100 000 g) and the soluble fractions were subjected to immunoblot analysis using an ABCG2 antibody. Based on the ABCG2 thermal denaturation curve, we empirically selected 67°C as the temperature to interrogate the ability of ligands to stabilize wildtype (WT) ABCG2. To determine the relative thermal stabilization of ABCG2 by ligands, cells were incubated with ligand for 60 minutes at 37°C before trypsin treatment. The immunoblot data were normalized with respect to the specific ABCG2 inhibitor, Ko143 and the signal intensity reported as % of Ko143 (Figure 1B). In isothermal dose-response experiments, the signal intensity was normalized to the intensity of the highest concentration.
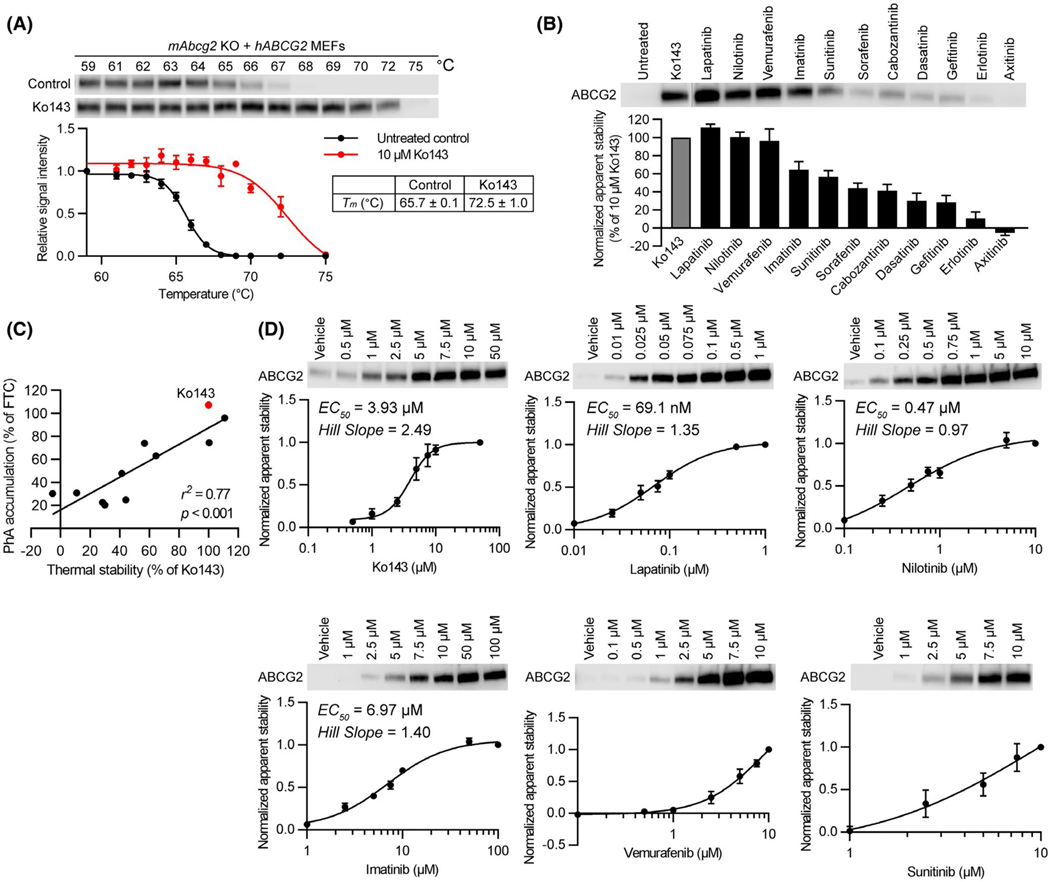
FIGURE 1.
Relationship between kinase inhibitor-binding to ABCG2 and pheophorbide a (PhA) transport inhibition. A, CETSA-melting curve of ABCG2 and ABCG2 CETSA curve shift upon ligand binding. Western blot for ABCG2 is shown in the inset. mAbcg2-KO MEFs expressing human ABCG2 were incubated with 10 μM of Ko143 (n = 3, red) or 0.1% of DMSO (untreated control) (n = 7, black) for 60 min at 37°C. Samples were heated for 3 min at various temperatures (59–75°C). The signal intensity was normalized to the intensity of the 59°C sample. B, CETSA was performed to measure the binding ability of kinase inhibitors to ABCG2 in mAbcg2-KO MEFs expressing human ABCG2. mAbcg2-KO MEFs expressing human ABCG2 were incubated with 10 μM of kinase inhibitor or 10 μM of Ko143 (positive control) for 60 min at 37°C. Samples were heated for 3 min at 67°C. The immunoblot data were normalized with respect to Ko143 and the signal intensity reported as % of Ko143. Each bar represents the mean + SE (n = 5–7). C, Scatter plot showing significant positive correlation between kinase inhibitor binding to ABCG2 and transport inhibition. The calculated correlation coefficient (r2) between thermal stability in 10 μM of kinase inhibitor or Ko143 treantment (Figure 1B) and PhA accumulation in 1 μM of kinase inhibitor or Ko143 treantment (PhA transport inhibition, Figure S1) was 0.77 (P < .001). D, mAbcg2-KO hABCG2 expressing MEFs were incubated with various concentrations of Ko143 or kinase inhibitor (Lapatinib, Nilotinib, Imatinib, Vemurafenib, or Sunitinib) for 60 min at 37°C. Dose-dependent stabilization of ABCG2 was assessed after heating samples at 67°C for 3 min. Each bar represents the mean ± SE (n = 3–5)
2.6 |. Isolation of plasma membranes
MEF cell lines were pelleted and washed with 1x PBS before suspension in a hypotonic buffer (0.5 mM of Tris (7.4), 0.1 mM of EDTA and protease inhibitors) and freezing. After gentle thawing, cells were pelleted by centrifugation, suspended in 2.5 mL of hypotonic buffer and spun for 40 minutes in an SW32Ti swinging bucket rotor at 100 000 g. Pellets were removed and resuspended into Homogenization Buffer (250 mM of sucrose, 50 mM of Tris, 250 μM of CaCl2, and protease inhibitors) and homogenized with 50 strokes of a tight dounce pestle. Following centrifugation at 500 g for 5 minutes, the pellets were then resuspended in Homogenization Buffer and homogenized again before an additional 500 g centrifugation step. The supernatants of each spin were pooled together and layered over a 5 mL 35% of sucrose pad formed in 50 mM of Tris (pH 7.4) and centrifuged in the SW32Ti rotor for 1.5 hours at 100 000 g. The interface was carefully removed and diluted with Dilution Buffer (50 mM of Tris, (pH 7.4), 25 mM of sucrose) and the crude membranes were pelleted at 100 000 g. Pellets were resuspended in 25 mM of Tris (pH 7.4) and incubated overnight at 4°C on ice. These pellets were then passed 10x through a 27 gauge needle before protein calculation. The membranes were typically usually used immediately or stored at −80°C for one thaw cycle.
2.7 |. Membrane thermal shift
Human ABCG2-expressing membrane vesicles, from HEK293 cells, were purchased from Solvo Biotechnology (Budapest, Hungary) or prepared from MEFs. Thermal shift assay was adapted according to Ashok et al.18 Briefly, membrane vesicles (5 μg protein/20 μL final reaction volume) were heated in a PCR machine for 3 minutes at various temperatures (37–70°C) to establish a thermal denaturation curve. Samples were then treated with ice-cold PBS supplemented with NP-40 to a final concentration of 0.8%, snap-frozen in liquid nitrogen, and then placed into a thermoshaker at 25°C to initiate thawing before being transferred to ice after the entire contents were thawed. After this, the freeze-thaw cycle was repeated and ultracentrifugation performed to precipitate the denatured protein (20 minutes at 4°C and 100 000 g). The supernatant was subjected to immunoblot analysis using an ABCG2 antibody (Enzo Life Sciences Inc, BXP53, 1:1000). Based on an extrapolation from the thermal denaturation curve that was predicted to produce 99% loss of protein in the supernatant, 65°C was slected for wildtype, 67°C for N436A, 61°C for F439A, 65°C for F439Y, and 67°C for F439W. For ABCG2-expressing HEK293 membrane vesicles, 62°C was selected based on the ABCG2 thermal denaturation curve. To assess the ability of ligands to thermally stabilize ABCG2, membranes were incubated with ligand for 60 minutes or with the non-hydrolyzable ATP analog, AMP-PNP for 30 minutes at 37°C. The signal intensity of the heated sample was normalized to the signal intensity of the unheated samples (WT and mutants) and the signal intensity reported as % of WT-ABCG2.
2.8 |. Immunoblot analysis
Cell lysate was subjected to SDS-PAGE, and proteins were transferred to a nitrocellulose membrane. Immunoblot analysis was performed using primary antibody against ABCG2 (Enzo Life Sciences Inc, BXP53, 1:1000) and was subjected to further incubation with HRP-conjugated secondary antibodies raised against rat IgG (Jackson ImmunoResearch Laboratories Inc, West Grove, PA) as previously described.19 Chemiluminescence signal was detected using LICOR Odyssey system. The bands were analyzed using LI-COR Image Studio Lite Ver 5.2 software for quantification of western blot signals.
2.9 |. Pharmacophore model
A global pharmacophore model was created in molecular operating environment (MOE) by aligning all 20 ABCG2 ligands using the Flexible Alignment application with default parameters.20 The best alignment, as determined by the “Grand Alignment Score,” was used to create a consensus pharmacophore model, the “Global Pharmacophore Model.”
2.10 |. In silico ligand modeling
Docking experiments were performed on the inward facing electron microscopy (EM) structure of ABCG2 (PDB: 6ETI) using MOE 2019v01. The protein structure was prepared by protonating at physiological pH (7.4) followed by partial charge calculations using default parameters in MOE. Site finder implemented in the software was employed to identify residues in the TMDs (transmembrane domains) which were then manually selected to restrict the binding cavity to the solvent accessible residues inside the binding cavity. Ligands were drawn in MOE, which were then stochastically minimized for generating 3D conformations. Partial charges on ligands were calculated using the Gastegier PEOE method implemented in the software. The docking experiment employed the alpha triangle placement method along with London-dG as scoring function. A total of 100 poses (without duplication) were generated for each docked ligand. Each pose was then minimized and rescored using the GBVI/WSA-dG scoring function.
2.11 |. Cytotoxicity assays
CellTiter-Glo luminescent cell viability assay was used to determine the cytotoxicity of mitoxantrone, topotecan, BisPOM-PMEA, and cladribine using the MEFs lacking Abcg2 (KO) but harboring the human ABCG2-WT, ABCG2-F439A, or vector control. Cells were seeded into a 96-well cell culture plates at 1000 cells/well and cytotoxic drug was added to the culture medium followed by an incubation for 3 days.
2.12 |. Data analysis and curve fitting
All data analysis and curve fitting were carried out using GraphPad Prism (v. 8.1. 2) (Prism; La Jolla, CA). Melting temperature curve (Tm) or dose-response curve (EC50) were fit with the nonlinear regression with three parameters (1) or four parameters (variable slope) (2) and are shown in each figure according to the best-fit model as determined by r2 value.
| (1) |
| (2) |
Top and Bottom are plateaus in the units of the Y axis.
Data shown are representative of mean ± SE of at least three independent experiments. Apparent EC50 and IC50 values were calculated from dose-response data using nonlinear regression analysis.
3 |. RESULTS
3.1 |. CETSA reveals KIs bind with differential affinity to ABCG2
The cellular thermal shift assay (CETSA) was developed to assist in the identification of cancer chemotherapeutic protein targets.17,21 The guiding principle is that a target protein will unfold and precipitate, upon heating, whereas a protein engaged with a ligand will generally require a higher temperature to unfold and precipitate.15–17,21 To validate this approach, we constructed a thermal denaturation curve for human ABCG2 (expressed in abcg2 knockout mouse embryo fibroblasts (MEFs) to eliminate dimerization with the endogenous ABCG2) that revealed a Tm of 65.7°C. Upon treatment of cells with the well-known ABCG2 inhibitor, Ko143, the Tm was elevated almost 7 to 72.5°C (Figure 1A), a shift in the Tm of similar magnitude by Ko143 binding to purified ABCG2 was reported.10
To determine if there was a relationship between ligand induced thermal stability and transport, eleven KIs, many reported as substrates and inhibitors (Table S1), were evaluated for their ability to inhibit the transport of the ABCG2-specific substrate, PhA.12 The KIs showed dose-dependent inhibition, and a wide range of potencies (Figure S1). To determine if these KIs thermally stabilized ABCG2, cells were incubated with 10 μM of each KI followed by an incubation at 67°C. Some KIs induced a strong thermal shift while others produced either a weak or minimal thermal shift, suggesting either little or no ability to bind ABCG2 and provide thermal stability (Figure 1B). We then determined if there was a relationship between the CETSA measured ABCG2 thermal stabilization (expressed as a proportion (%) of Ko143 induced stabilization) and transport inhibition by comparing the % inhibition by the KI relative to that produced by FTC (see Figure S1). Among many of the KIs, the relationship between KI thermal stabilization of ABCG2 and transport inhibition was strong with a calculated correlation coefficient (r2) between ABCG2 thermal stability and transport inhibition of 0.77 (P < .001 (Figure 1C). Vemurafenib showed strong binding to ABCG2, but weak transport inhibitory effect, however, even with inclusion of this outlier the correlation was still strong (r2 = .58, P = .004).
We next hypothesized that the potency of a KI to produce an ABCG2 thermal shift might be related to their potency of ABCG2 transport inhibition. As a control, Ko143 was used because it has a known binding site on ABCG2.10,11 Ko143 displayed dose-dependent stabilization of ABCG2 with an EC50 of 3.93 μM and a Hill coefficient of 2.49 (Figure 1D), consistent with the ABCG2 structure displaying two potential Ko143 analogs in the binding pocket.10 Next, the KIs were tested. Lapatinib displayed the lowest EC50 for binding of about 69 nM, whereas sunitinib was the least potent. Among the KIs, most showed a Hill slope slightly greater than one. Importantly, a strong correlation was observed between the KI thermal stabilization of ABCG2 (Figure 1B) and IC50 for transport inhibition (Figure S1) (r2 = .88, P = .002) (Figure S2).
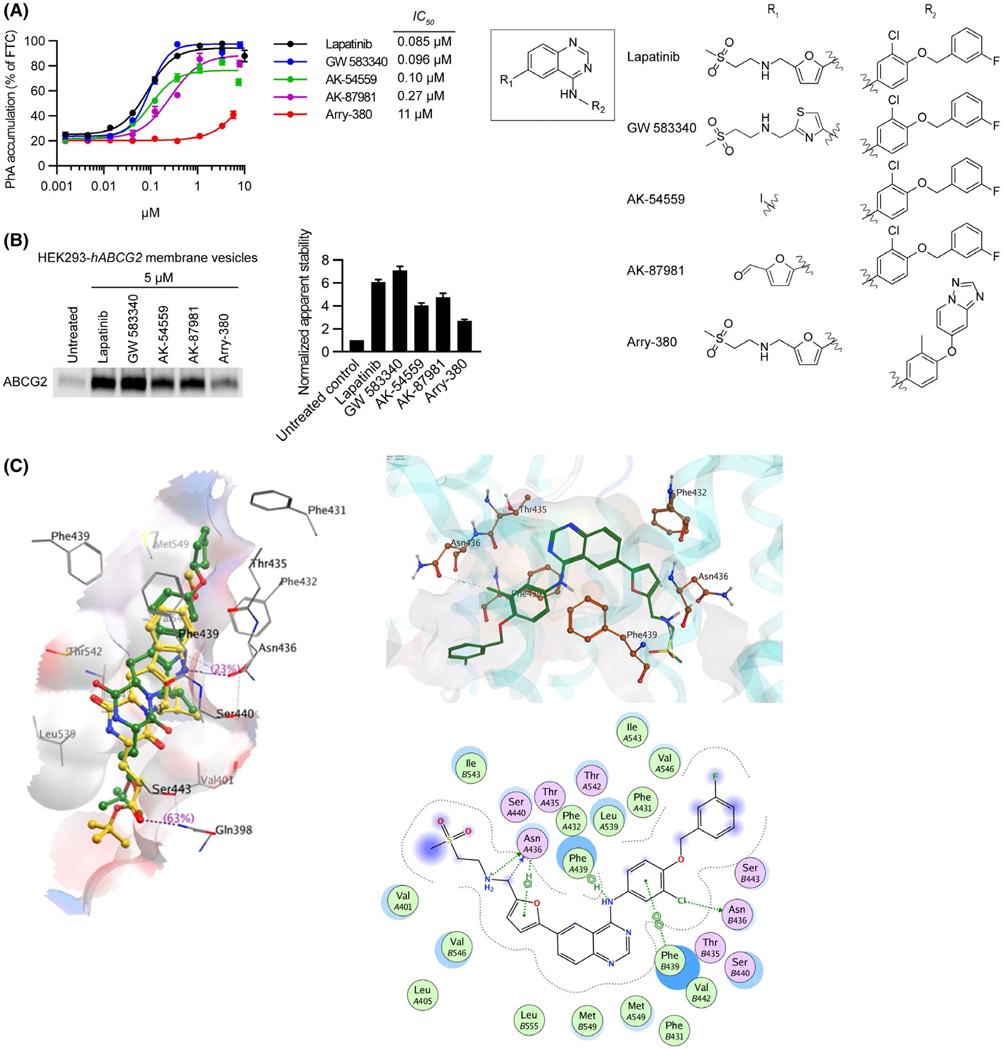
3.2 |. Lapatinib analogs and structural model of binding
Because Lapatinib was the most potent in inducing a thermal shift among the KIs tested, we investigated if it had molecular features that were important determinants for interaction with ABCG2 (Figure 2). Four Lapatinib analogs were available to evaluate for their ability to inhibit ABCG2 mediated transport. Three of these had IC50 values similar to Lapatinib (IC50 = 0.085 μM), while the fourth, Arry-380 was 130-fold lower at 11 μM (Figure 2A). These studies were then extended to assess the ability of these ligands to thermally stabilize ABCG2 by the membrane thermal shift assay. Lapatinib and GW583340 were most effective in stabilizing ABCG2 against thermal denaturation followed by AK-87981 and AK-54559. Paralleling the transport inhibition data, the least potent stabilizer was Arry-380 (Figure 2B). Structure-based docking experiments using ABCG2 (PDB: 6ETI) predicted the most likely binding poses of the KIs used in this study. We noted that a single amino acid, F439, was predicted to be involved in binding via π-π stacking to all of the drugs. F439 is close to the two-fold symmetry axis of this dimeric transporter, and a F439 residue from each monomer participates in ligand binding. The top docking pose of Ko143 was consistent with the cryo-EM structure that would allow the binding of two molecules. In contrast, most of the other KIs, such as Lapatinib, had poses, where only one drug molecule was threaded in between the two F439 residues. Since none of the KIs have symmetry, the two F439 residues appear to form a “clamp” via π-π interaction with the aromatic groups within the various drugs. These observations helped to elucidate the nature of the ABCG2 pharmacophore (generated separately below). Although F439 was identified as a universal interacting partner with all the KIs examined, the N436 was also predicted to hydrogen-bond with many of the drugs.
FIGURE 2.
Lapitinib analogs reveal important residues for interacting with ABCG2. A, 0.5 μM of PhA was used as substrate and Fumitremorgin C (FTC) (positive control), Lapatinib, or Lapitinib analogs were used as inhibitors to measure inhibitory effect (IC50) of ABCG2. mAbcg2-KO MEFs expressing human ABCG2 were incubated with PhA and inhibitor for 60 min. Each bar represents the mean ± SE (n = 4). Molecular structures of Lapatinib and its analogs (right). Core structure is shown at the middle, and the individual R1 and R2 constituents of Lapatinib and its analogs GW 583340, AK-54559, AK-87981, and Arry-380 are shown right the core structure. B, HEK293-hABCG2 membrane vesicles were incubated with 5 μM of Lapatinib or Lapatinib analogs for 60 min at 37°C. Samples were heated for 3 min at 62°C. Representative western blot is shown on the left side and the densitometry quantification data from three independent experiments is shown on the right side. C, Diagrammatic representation of Ko143 analog mz29 and lapatinib in the ABCG2 binding cavity. Left panel: the binding of Ko143 (yellow) and mz29 (green) with PDB 6ETI. Right panel: 3D (top side) and 2D (bottom side) representation of Lapatinib, a tyrosine kinase inhibitor, within the ABCG2 binding cavity
3.3 |. Highly conserved F439 is required to confer ABCG2 thermal stability for multiple ligands
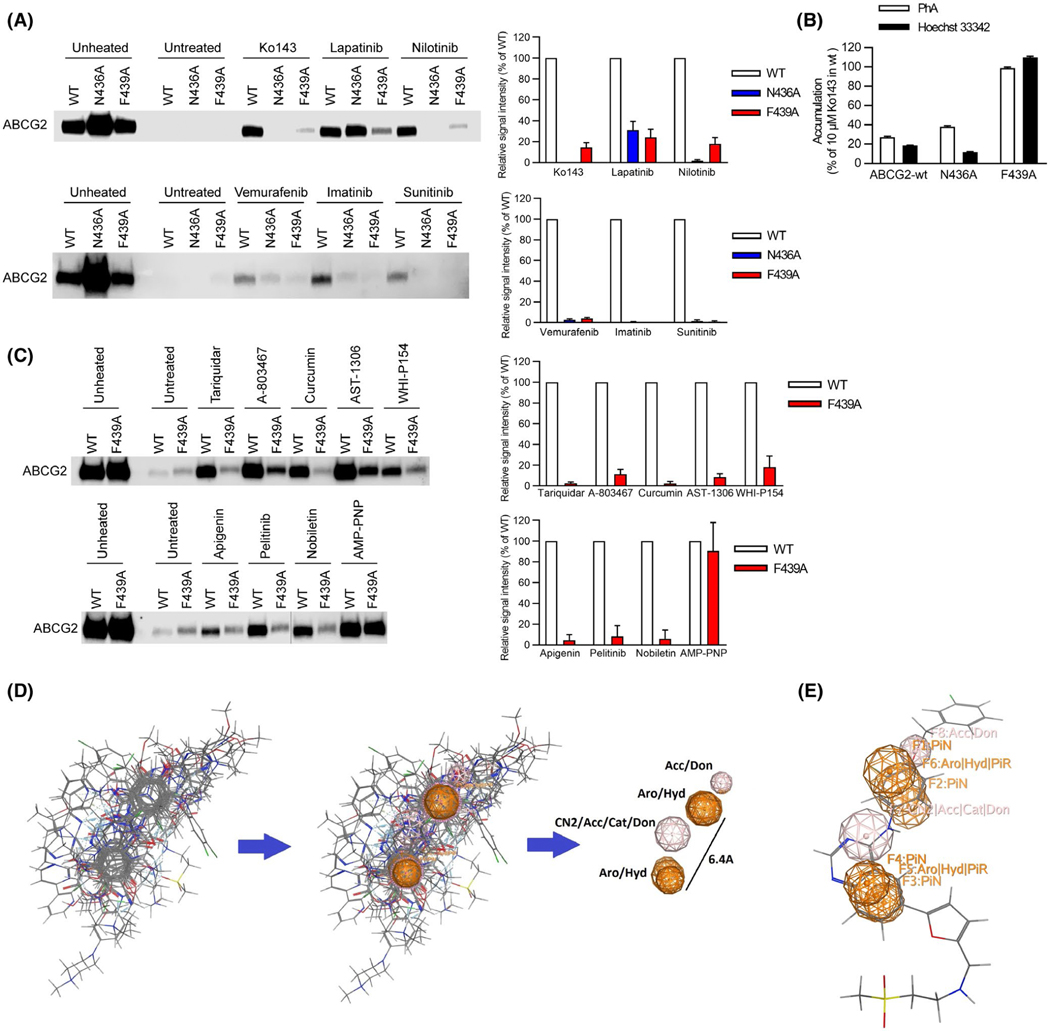
Based on the Lapatinib modeling studies (Figure 2C), which suggested that residues N436 and F439 frequently contribute to ligand interactions with ABCG2, we hypothesized that they might play a key role for KIs interacting with ABCG2. To investigate this, we developed ABCG2 retroviral expression vectors harboring site-directed mutants that converted either F439 to alanine or N436 to alanine. These were then transduced into Abcg2-knockout MEFs. Subsequently, membranes were prepared. These membranes display the same properties as cells in that ATP and its non-hydrolyzable analog (AMP-PNP) and stabilize ABCG2 against thermal denaturation (Figure 3B, Figure S4A-D). The Tm for F439A (Tm =57°C) was lower than WT-ABCG2 (WT Tm =59°C), whereas the Tm for N436A was higher at 62°C (see Figure S3B). To test the impact of these mutations on KI stabilization of ABCG2, the denaturation temperature we selected was 65°C for wildtype, 67°C for N436A, and 61°C for the F439A mutant. These denaturation temperatures, predicted to precipitate greater than 99% of the ABCG2 compared to unheated samples, were selected after interpolatation from their respective denaturation curves. Each membrane preparation was assessed for their level of ABCG2 expression in the absence of thermal denaturation (to account for any variation not only in ABCG2 expression of the mutants, but also between each membrane preparation), and then when heated to a denaturation temperature, where 99% of ABCG2 precipitated (labeled “untreated”). Both ABCG2-F439A and N436A strongly reduced Ko143 stabilization of ABCG2. The N436A affected the thermal stabilization of ABCG2 by many KIs, but Lapatinib partially stabilized N436A, and none of the KIs were capable of stabilizing the F439A-ABCG2 mutant to the same extent as WT-ABCG2 (Figure 3A). Importantly, unlike a previous report11 the N436A-ABCG2 mutant retained PhA and Hoechst 33342 transport activity similar to WT-ABCG2. In contrast, the F439A-ABCG2 mutant was incapable of transporting PhA and Hoechst 33342 (Figure 3B). These results suggest that F439 is key for the binding of these ligands to ABCG2.
FIGURE 3.
A key amino acid residue in the binding pocket is required for binding to ABCG2. A, Role of N436 and F439 in kinase inhibitor binding assessed by thermal shift. Representative western blots are shown on the left side and the densitometry quantification data from three to four independent experiments are shown on the right side. Membrane vesicles prepared from mAbcg2-KO MEFs expressing human ABCG2 (WT, N436A, and F439A) were incubated with 10 μM of kinase inhibitor for 60 min at 37°C. Samples were heated for 3 min at 65°C for WT-ABCG2 (white), 67°C for N436A (blue), and 61°C for F439A mutant (red). B, N436A retains transport of Hoechst 33342 (black) and PhA (white), whereas F439A has lost transport ability. Each bar represents the mean + SE (n = 8). C, F439 contribution to binding of various ligands to ABCG2. Representative western blots are shown on the left side and the densitometry quantification data from three independent experiments are shown on the right side. Membrane vesicles prepared from mAbcg2-KO MEFs expressing human ABCG2 (WT and F439A) were incubated with 10 μM of ABCG2 ligand for 60 min or 1 mM of AMP-PNP for 30 min at 37°C. Samples were heated for 3 min at 65°C for WT-ABCG2 (white) and 61°C for F439A mutant (red). D, Best flexible alignment of all ABCG2 ligands after a 48 hr alignment run in MOE, was used to compute the consensus pharmacophore model that had two aromatic features at 6.4Å to each other. E, Docked Lapatinib molecule mapped onto the consensus pharmacophore model, showing the connection between the docking model and the consensus pharmacophore model. Don = H-bond donor; Acc = H-bond acceptor; PiN = Ring normal; Aro = Aromatic; PiR = pi-Ring; Hyd = Hydrophobic; Cat = Cation (use MOE reference)
The strong broad effect of F439A on KI induced thermal stabilization of ABCG2 led us to more generally evaluate additional ABCG2 interacting ligands of diverse molecular features (AST-1306, Tariquidar, Nobiletin, Apgenin, Pelitinib, WHI-P154, Curcumin, A-803467) (Figure 3C). As a positive control for ABCG2 binding, the non-hydrolyzable ATP analog, AMP-PMP, stabilized both WT-ABCG2 and F439A, similarly, indicating nucleotide binding was unimpaired by the F439A substitution (Figure 3C). Thus, while WT-ABCG2 was stabilized by every additional ligand, none were capable of inducing strong thermal stabilization of the F439A-ABCG2 (Figure 3C).
We investigated the ligands for common features that might be affected by the F439 mutation. This was accomplished by flexibly aligning the compounds in MOE and extracting features from the alignment to generate a consensus pharmacophore (Figure 3D). The consensus “global” pharmacophore model had two aromatic/hydrophobic centers separated by 6.4Å which indicated that the aromatic centers will, on average be separated by approximately a two carbon spacer. The model also contained two hydrogen bond donor/hydrogen bond acceptor centers. A docked Lapatinib molecule is overlaid on this consensus pharmacophore to illustrate the key features of the model (Figure 3E). This model likely accounts for Arry-380 displaying both weaker ABCG2 binding and inhibition compared to Lapatinib (see Figure 2A,B), by suggesting that the aromatic benzene ring of Lapatinib is important for binding and that the heterocyclic ring of Arry-380 might clash, thereby restricting its binding. We were able to map 12 out of the 20 ABCG2 ligands (Table 2) directly onto its pharmacophoric features. The model also mapped 13 of the 19 docked poses (best pose from each of the 19 docked ABCG2 ligands), with the correspondence indicating that both the docking and the pharmacophore model captured the essential features of these ABCG2 ligands (list is in Table S2).
TABLE 2.
Ligands used to develop the global ABCG2 pharmacophore model
| Ko143 | Lapatinib | Nilotinib | Vemurafenib | Imatinib |
|---|---|---|---|---|
| Sunitinib | Sorafenib | Gefitinib | Tariquidar | Curcumin |
| AST-1306 | WHI-P154 | Apigenin | Pelitinib | Nobiletin |
| Hoechst 33342 | Pheophorbide a | Mitoxantrone | Topotecan | 4-[(2-phenylquinazolin-4-yl)amino]benzonitrile |
3.4 |. F439 mutation affects substrate export with conservative amino acid substitutions restoring function
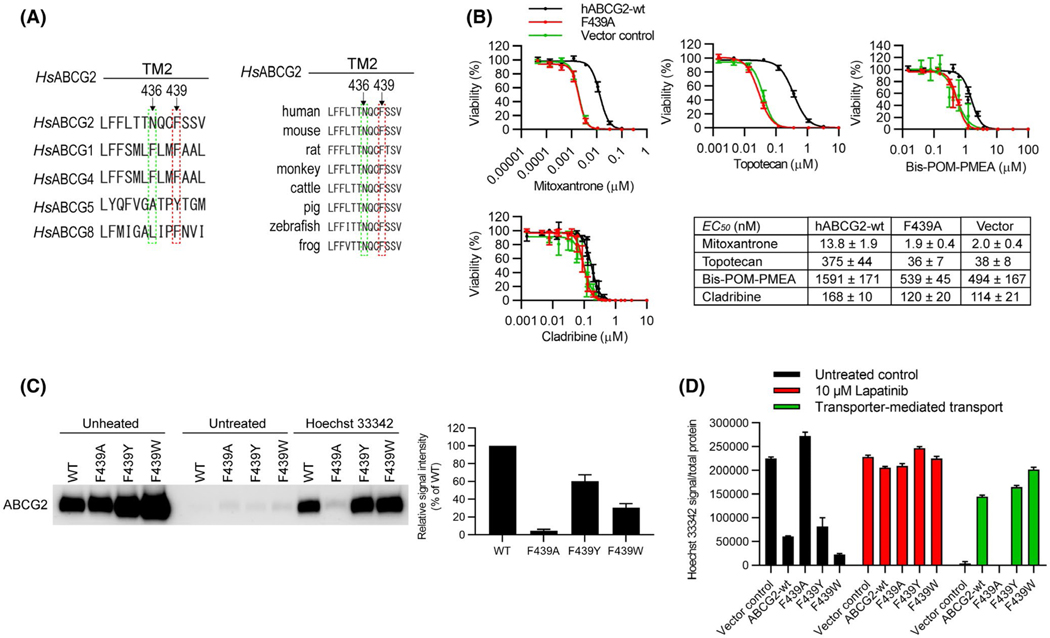
Among the ABCG family members, the position analogous to F439 in ABCG2 harbors an aromatic amino acid (Figure 4A, left) and as the sequence alignment shows, the amino acid F439 is conserved in ABCG2 orthologs from frogs to humans (Figure 4A, right). To further investigate the role of the amino acid F439 in substrate transport, we determined whether the F439A-ABCG2 was capable of conferring resistance (a proxy for transport) to a variety of cytotoxic drugs known to be exported by ABCG2.22–24 Unlike WT-ABCG2, cells expressing F439A-ABCG2 were as sensitive as cells lacking ABCG2 to the cytotoxic effects of structurally and mechanistically diverse cytotoxic drugs, such as mitoxantrone, topotecan, Bis-POM-PMEA, and cladribine (Figure 4B) suggesting F439A-ABCG2 is incapable of transporting these drugs.
FIGURE 4.
Mutation of the conserved binding residues rescues ABCG2 ligand-binding and transport. A, The human ABCG2 sequence containing the residues N436 and F439 was aligned with the corresponding sequences in its ABCG2 homologs (left) and orthologs (right). The residue F439 in ABCG2 are either conserved or substituted by other hydrophobic residue Y (ABCG5) in the other G-subfamily ABC transporters. The residue N436 in ABCG2 is not conserved in the other G-subfamily ABC transporters (left). The amino acid F439 is conserved in ABCG2 orthologs from frogs to humans (right). B, The cytotoxic effects (EC50 values) were determined by CellTiter-Glo luminescent cell viability assay of cytotoxic drugs Mitoxantrone, Topotecan, Bis-POM-PMEA, and Cladribine using the mAbcg2-KO MEFs expressing human ABCG2-WT (black), ABCG2-F439A (red), or vector control (green). Each bar represents the mean ± SE (n = 5–10). C, MEF-hABCG2-WT, F439A, F439Y, or F439W membrane vesicles were incubated with 10 μM of Hoechst 33342 for 60 min at 37°C. Representative western blot is shown on the left side and the densitometry quantification data from four independent experiments is shown on the right side. D, 0.5 μM of Hoechst 33342 was used as ABCG2 substrate and 10 μM of Lapatinib was used as ABCG2 inhibitor to measure transport activity of ABCG2-WT, F439A, F439Y, and F439W. The cells were incubated with Hoechst 33342 (and Lapatinib) for 120 min. Each bar represents the mean + SE (n = 8)
Because binding is the obligate initial step in the transport cycle and the aromatic acid residue is conserved among ABCG family members at positions analogous to F439 (Figure 4A), we tested if the substrate, Hoechst 33342 stabilized either ABCG2 or F439A-ABCG2. Importantly, based on ABCG5,25 we created F439W-ABCG2 and F439Y-ABCG2 by substituting phenylalanine with either tyrosine or tryptophan in a human ABCG2 retroviral expression plasmids which were then subsequently transduced into Abcg2-knockout MEFs. We determined that Hoechst 33342 stabilized WT-ABCG2, F439W-ABCG2, and F439Y-ABCG2 but not F439A-ABCG2 (Figure 4C). We then investigated the role of an aromatic residue at this position in Hoechst 33342 transport. The uptake of Hoechst 33342 was determined for WT-ABCG2 (ie, F439), F439A-ABCG2, F439W-ABCG2, and F439Y-ABCG2 in the absence (black bar) or presence (red bar) of Lapatinib with the difference in uptake between the two conditions representing ABCG2 mediated transport (green bar) (Figure 4D). As expected, F439A-ABCG2 displayed no net ABCG2 mediated transport, whereas F439W-ABCG2 and F439Y-ABCG2 had transport activity similar to WT-ABCG2. As F439 is near the dimer interface, we tested the possibility that F439A-ABCG2 might be impaired in dimerization and compared it to WT-ABCG2, F439W-ABCG2, and F439Y-ABCG2. Non-denaturing gel electrophoresis revealed that F439A-ABCG2 formed a dimer like WT-ABCG2, F439W-ABCG2, and F439Y-ABCG2 (Figure S3A).
4 |. Discussion
For the drug and xenobiotic transporting ABC transporter, ABCG2, recent cryo-electron microscopy structures have provided valuable snapshots of some substrates and inhibitors in the binding pocket (also termed cavity one), that apparently use the same residues to interact with ABCG2. It was unknown if one or more residues were required for binding to ABCG2. We selected some of these binding pocket residues for further studies and demonstrated, surprisingly, that a single highly conserved amino acid, F439 was essential for binding all the ligands tested. Importantly, F439 was also crucial for ABCG2 mediated substrate transport. Based on the structures of the ligands requiring F439 a unique pharmacophore model was developed that has the capability of predicting numerous drugs that interact with ABCG2. Further, in silico docking of ligands to a structure of the ABCG2 homodimer (Figure 5A-E), suggested that the two F439 aromatic rings “clamp” ligands in the binding pocket (Figure 5F). Importantly, conversion of this phenylalanine to amino acids with similar planar, aromatic side-chain rings, that is, tryptophan or tyrosine (which is also found in ABCG5 and essential for cholesterol transport25), retained binding, and transport activity comparable to wildtype ABCG2 (Figure 4C,D) which suggests that this aromatic “clamp” may be a key feature for ligand engagement by other ABCG family members.
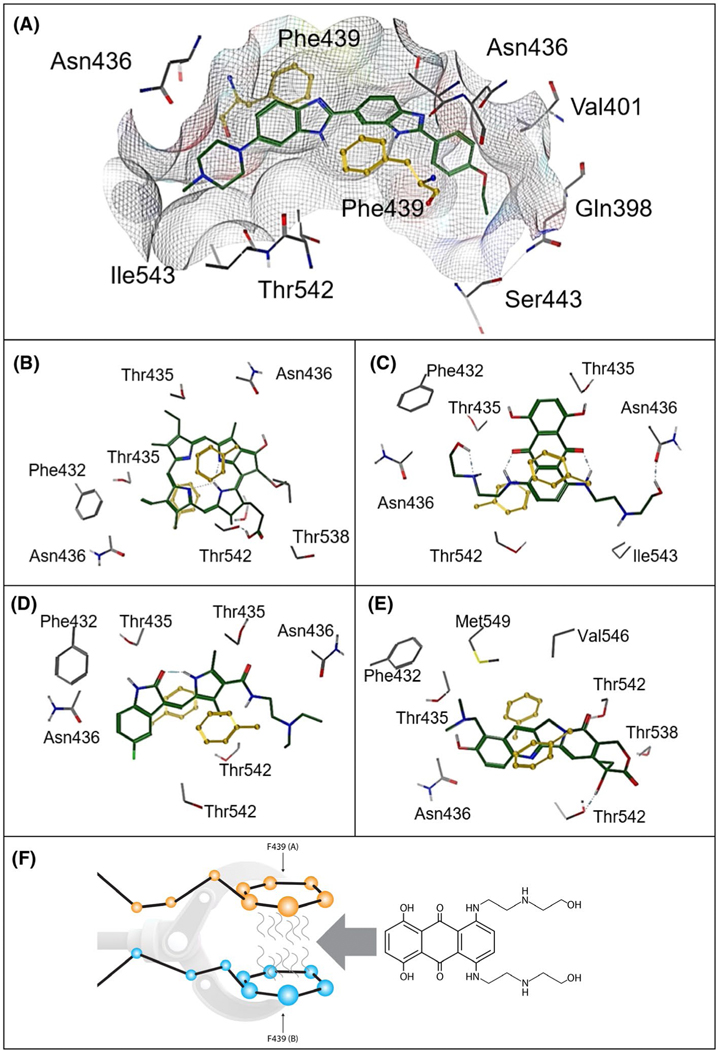
FIGURE 5.
Diagrammatic representation of structurally different ligands in the ABCG2 binding cavity and a depiction of the F439 interaction. A, Hoeschst 33342. B, Pheophorbide a. C, Mitoxantrone. D, Topotecan. E, Sunitinib. F439 (yellow) provides an anchoring point for ABCG2 ligands (A-E). F, “A” and “B” indicate F439 has been donated from separate ABCG2 monomers. The molecule that will interdigitate between the two F439 is the ABCG2 substrate and chemotherapeutic agent, mitoxantrone
Our studies do not exclude the possibility that other residues in the binding pocket impact how ligands interact with ABCG2. Instead, we show that many structurally diverse ligands require F439 for optimal binding to ABCG2. This is consistent with the findings of Manolaridis et al11 demonstrating F439A could not transport estrone-3-sulfate. However, our findings constrast with their findings, where N436A which was shown incapable of estrone-3-sulfate transport. Our studies demonstrate that N436A is nearly as capable of transporting PhA and Hoechst 33342 as WT-ABCG2. Thus, while N436A appears to have impaired interaction with some ligands, it is unlike F439A which has no demonstrable binding or transport activity which we speculate might be due to impaired interaction with a membrane constituent like cholesterol,26,27 an important modulator of ABCG2 activity.28 Perhaps the increased thermal stabilization by the alanine substitution at N436 alters ABCG2 conformation to preserve the interaction with certain ligands and not others. Based on the findings from G-protein coupled receptors (GPCRs), transmembrane alanine substitutions generally produced thermostabilization.29,30 Indeed, this is the basis of a structural approach that has resulted in the structures of many GPCRs.
Studies of ABCG2 ligand binding have suggested that more than one drug/ligand binding pocket exists31,32 and analysis of the outward- and inward-facing conformations of the ABCG2 homodimer supports this interpretation, with both an internal binding site and an extracellular surface binding site that also spans the two-fold symmetry axis, but residues near the extracellular surface.10,26 Multiple binding pockets may occur in WT-ABCG2 and in the drug-selected, gain-of-function variant, R482G-ABCG2, that reportedly has more than one drug binding site.33 Multiple binding pockets may be a general property of membrane transporters as multidrug pumps interact with ligands through both surface binding sites and buried sites (eg, AcrAB/TolC).34,35 Consistent with the structural model, cellular studies have predicted that mitoxantrone binds ABCG2 through two asymmetric binding sites, a surface exposed site and a buried, second binding site.31,33,36 Other studies, have also proposed that ABCG2 drug binding occurs through at least two binding sites.32 From the ABCG2 structure, one might infer that these two binding sites are so called “cavity one” and “cavity two”.26 It is likely that cavity one and two have different affinity for ligands. Importantly, our studies do not preclude the existence of two distinct ligand binding sites in ABCG2, but instead suggest that the ABCG2 binding site in cavity one (harboring F439) is the preferred site, and the one required for maximal ABCG2 interaction with a broad array of ligands, both substrates and inhibitors. Furthermore, based upon our transport studies with Hoechst 33342 (and the drug resistance studies (Figure 4B) as a proxy for transport), we propose that the substrate translocation pathway first requires binding to cavity one and that this is mediated by an obligate interaction with F439 which is sequentially followed by the opening of the “leucine plug”,26,37 and substrate movement into cavity two, ultimately leading to substrate release. It is notable that inhibitors also require F439A for binding to cavity one. This suggests that inhibitors, such as Ko143 and tariquidar compete directly with substrates to prevent their initial binding, regardless of whether they ultimately become “wedged” by additional contacts, as was proposed,11 will require future kinetic and affinity binding studies to elucidate such an important relationship.
The identification of a key phylogenetically conserved amino acid, essential to ABCG2 interaction with a diverse array ligands was indispensible to the creation of structural models depicting the interaction of multiple substrates and inhibitors (Figure 5). This empirical knowledge was essential to the creation of unique pharmacophore and docking models. Our global pharmacophore model can be utilized for virtual screens to identify new and novel substrates and inhibitors of ABCG2 which will be invaluable in prioritizing the development of drugs that avoid an interaction with ABCG2. Indeed, although defects in ABCG2 are important causes of gout, due to defective uric acid transport, the ability of ABCG2 to transport uric acid4–6,38,39 seems couterintuitive as it is a small, soluble molecule compared to the large hydrophobic drugs such as the KIs that seem more frequently thought of as ABCG2 substrates. However, when tested, our docking and pharmacophore model suggest that the F439 clamp can π-π stack with the uric acid heterocyclic rings (Figure S6). Indeed, uric acid was predicted by the top poses to interact with ABCG2 in a location between the two F439 residues. In total, these findings not only provide a novel basis for ABCG2 interacting with multiple ligands (one that was not obvious or easily inferred from the recent structural findings), but also provides mechanistic insights that suggest that in a native cellular environment, substrates must first interact with a single-amino acid, F439, which appears to be a pre-requisite to initiating transport. This is in contrast to N436A, which is still capable of transport. These findings have therapeutic important implications toward the efforts to predict and identify drugs that can evade ABCG2 which are particulary important for drugs attempting to penetrate into tumors or cross the blood-brain barrier.3,40–42
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by NIH grants R01CA194057, R01 Act CA194206 (JS), R35-GM118041 (TC), P30CA21745, CA21865, CA96832, and by ALSAC. We thank Diana Zhou for her excellent assistance with some of the experiments contained herein. We thank the members of the Schuetz labs for constructive comments and edits.
Funding information
NIH, ALSAC, Grant/Award Number: R01CA194057, R35-GM118041, P30CA21745, CA21865, CA96832
Abbreviations:
- ABCG2
ATP-binding cassette sub-family G member 2
- CETSA
cellular thermal shift assay
- EC50
half maximal effective concentration
- EM
electron microscopy
- FTC
fumitremorgin C
- GPCR
G-protein coupled receptor
- IC50,
half maximal inhibitory concentration
- KI
kinase inhibitor
- MEF
mouse embryonic fibroblast
- MOE
molecular operating environment
- PhA
pheophorbide
- Tm,
melting temperature
- TMDs
transmembrane domains
- TMH
transmembrane helices
Footnotes
SUPPORTING INFORMATION
Additional supporting information may be found online in the Supporting Information section.
CONFLICT OF INTEREST
The authors declare no conflicts of interest.
REFERENCES
- 1.Zhou S, Schuetz JD, Bunting KD, et al. The ABC transporter Bcrp1/ABCG2 is expressed in a wide variety of stem cells and is a molecular determinant of the side-population phenotype. Nat Med. 2001;7:1028–1034. [DOI] [PubMed] [Google Scholar]
- 2.Zhou S, Morris JJ, Barnes Y, Lan L, Schuetz JD, Sorrentino BP. Bcrp1 gene expression is required for normal numbers of side population stem cells in mice, and confers relative protection to mitoxantrone in hematopoietic cells in vivo. Proc Natl Acad Sci. 2002;99:12339–12344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Enokizono J, Kusuhara H, Ose A, Schinkel AH, Sugiyama Y. Quantitative investigation of the role of breast cancer resistance protein (Bcrp/Abcg2) in limiting brain and testis penetration of xenobiotic compounds. Drug Metab Dispos. 2008;36:995–1002. [DOI] [PubMed] [Google Scholar]
- 4.Ichida K, Matsuo H, Takada T, et al. Decreased extra-renal urate excretion is a common cause of hyperuricemia. Nat. Commun. 2012;3:764–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dehghan A, Köttgen A, Yang Q, et al. Association of three genetic loci with uric acid concentration and risk of gout: a genome-wide association study. Lancet. 2008;372:1953–1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hosomi A, Nakanishi T, Fujita T, Tamai I. Extra-renal elimination of uric acid via intestinal efflux transporter BCRP/ABCG2. PLoS One. 2012;7:e30456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fukuda Y, Wang Y, Lian S, et al. Upregulated heme biosynthesis, an exploitable vulnerability in MYCN-driven leukemogenesis. JCI Insight. 2017;2:e92409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shukla S, Kouanda A, Silverton L, Talele TT, Ambudkar SV. Pharmacophore modeling of nilotinib as an inhibitor of ATP-binding cassette drug transporters and BCR-ABL kinase using a three-dimensional quantitative structure-activity relationship approach. Mol Pharm. 2014;11:2313–2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hussaarts KGAM, Veerman GDM, Jansman FGA, van Gelder T, Mathijssen RHJ, van Leeuwen RWF. Clinically relevant drug interactions with multikinase inhibitors: a review. Ther Adv Med Oncol. 2019;11:1758835918818347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jackson SM, Manolaridis I, Kowal J, et al. Structural basis of small-molecule inhibition of human multidrug transporter ABCG2. Nat Struct Mol Biol. 2018;25:333–340. [DOI] [PubMed] [Google Scholar]
- 11.Manolaridis I, Jackson SM, Taylor NMI, Kowal J, Stahlberg H, Locher KP. Cryo-EM structures of a human ABCG2 mutant trapped in ATP-bound and substrate-bound states. Nature. 2018;563:426–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Robey RW, Steadman K, Polgar O, et al. Pheophorbide a is a specific probe for ABCG2 function and inhibition. Cancer Res. 2004;64:1242–1246. [DOI] [PubMed] [Google Scholar]
- 13.Yasuda K, Ganguly S, Schuetz EG. Pheophorbide A: fluorescent bcrp substrate to measure oral drug-drug interactions in real-time in vivo. Drug Metab Dispos. 2018;46:1725–1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Allen JD, van Loevezijn A, Lakhai JM, et al. Potent and specific inhibition of the breast cancer resistance protein multidrug transporter in vitro and in mouse intestine by a novel analogue of fumitremorgin C. Mol Cancer Ther. 2002;1:417–425. [PubMed] [Google Scholar]
- 15.Reinhard FBM, Eberhard D, Werner T, et al. Thermal proteome profiling monitors ligand interactions with cellular membrane proteins. Nat Methods. 2015;12:1129–1131. [DOI] [PubMed] [Google Scholar]
- 16.Molina DM, Jafari R, Ignatushchenko M, et al. Monitoring drug target engagement in cells and tissues using the cellular thermal shift assay. Science. 2013;341:84–87. [DOI] [PubMed] [Google Scholar]
- 17.Martinez Molina D, Nordlund P. The cellular thermal shift assay: a novel biophysical assay for in situ drug target engagement and mechanistic biomarker studies. Annu Rev Pharmacol Toxicol. 2015;56:141–161. [DOI] [PubMed] [Google Scholar]
- 18.Ashok Y, Nanekar R, Jaakola VP. Defining thermostability of membrane proteins by western blotting. Protein Eng Des Sel. 2015;28:539–542. [DOI] [PubMed] [Google Scholar]
- 19.Fukuda Y, Cheong PL, Lynch J, et al. The severity of hereditary porphyria is modulated by the porphyrin exporter and Lan antigen ABCB6. Nat Commun. 2016;7:12353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gasteiger J, Marsili M. Iterative partial equalization of orbital electronegativity-a rapid access to atomic charges. Tetrahedron. 1980;36:3219–3228. [Google Scholar]
- 21.Jafari R, Almqvist H, Axelsson H, et al. The cellular thermal shift assay for evaluating drug target interactions in cells. Nat Protoc. 2014;9:2100–2122. [DOI] [PubMed] [Google Scholar]
- 22.Takenaka K, Morgan JA, Scheffer GL, et al. Substrate overlap between Mrp4 and Abcg2/Bcrp affects purine analogue drug cytotoxicity and tissue distribution. Cancer Res. 2007; 67:6965–6972. [DOI] [PubMed] [Google Scholar]
- 23.Nagai S, Takenaka K, Nachagari D, et al. Deoxycytidine kinase modulates the impact of the ABC transporter ABCG2 on clofarabine cytotoxicity. Cancer Res. 2011;71:1781–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fukuda Y, Schuetz JD. ABC transporters and their role in nucleoside and nucleotide drug resistance. Biochem Pharmacol; 2012;83:1073–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee JY, Kinch LN, Borek DM, et al. Crystal structure of the human sterol transporter ABCG5/ABCG8. Nature. 2016;533:561–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taylor NMI, Manolaridis I, Jackson SM, Kowal J, Stahlberg H, Locher KP. Structure of the human multidrug transporter ABCG2. Nature. 2017;546:504–509. [DOI] [PubMed] [Google Scholar]
- 27.Jackson SM, Manolaridis I, Kowal J, et al. Structural basis of small-molecule inhibition of human multidrug transporter ABCG2. Nat Struct Mol Biol. 2018;25:333–340. [DOI] [PubMed] [Google Scholar]
- 28.Pál A, Méhn D, Molnár E, et al. Cholesterol potentiates ABCG2 activity in a heterologous expression system: improved in vitro model to study function of human ABCG2. J Pharmacol Exp Ther. 2007;321:1085–1094. [DOI] [PubMed] [Google Scholar]
- 29.Vaidehi N, Grisshammer R, Tate CG. How can mutations thermostabilize G-protein-coupled receptors? Trends Pharmacol Sci. 2016;37:37–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Serrano-Vega MJ, Magnani F, Shibata Y, Tate CG.Conformational thermostabilization of the 1-adrenergic receptor in a detergent-resistant form; 2008;105:877–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cox MH, Kapoor P, Briggs DA, Kerr ID. Residues contributing to drug transport by ABCG2 are localised to multiple drug-binding pockets. Biochem J. 2018;105:877–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ferreira RJ, Bonito CA, Cordeiro MNDS, Ferreira MJU, Dos Santos DJVA. Structure-function relationships in ABCG2: insights from molecular dynamics simulations and molecular docking studies. Sci Rep. 2017;7:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clark R, Kerr ID, Callaghan R. Multiple drugbinding sites on the R482G isoform of the ABCG2 transporter. Br J Pharmacol. 2006;149:506–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Elkins CA, Mullis LB. Substrate competition studies using whole-cell accumulation assays with the major tripartite multidrug efflux pumps of Escherichia coli. Antimicrob Agents Chemother; 2007;51:923–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Z, Fan G, Hryc CF, et al. An allosteric transport mechanism for the AcrAB-TolC multidrug efflux pump. eLife. 2017;6:e24905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Haider AJ, Cox MH, Jones N, et al. Identification of residues in ABCG2 affecting protein trafficking and drug transport, using co-evolutionary analysis of ABCG sequences. Biosci Rep. 2015;6:e24905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Khunweeraphong N, Stockner T, Kuchler K. The structure of the human ABC transporter ABCG2 reveals a novel mechanism for drug extrusion. Sci Rep. 2017;7:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.So A, Thorens B. Uric acid transport and disease. J Clin Invest. 2010;120:1791–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matsuo H, Takada T, Ichida K, et al. Common defects of ABCG2, a high-capacity urate exporter, cause gout: a function-based genetic analysis in a Japanese population. Sci. Transl. Med. 2009;120:1791–1799 [DOI] [PubMed] [Google Scholar]
- 40.Morfouace M, Cheepala S, Jackson S, et al. ABCG2 transporter expression impacts group 3 medulloblastoma response to chemotherapy. Cancer Res. 2015; 75:3879–3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Robey RW, Pluchino KM, Hall MD, Fojo AT, Bates SE, Gottesman MM. Revisiting the role of ABC transporters in multidrug-resistant cancer. Nat Rev Cancer. 2018;18:452–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wijaya J, Fukuda Y, Schuetz JD. Obstacles to brain tumor therapy: Key ABC transporters. Int J Mol Sci. 2017;18:2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.