SUMMARY
Visceral adipose tissue (VAT) depots are associated with the adverse metabolic consequences of obesity, such as insulin resistance. The developmental origin of VAT depots and the identity and regulation of adipocyte progenitor cells have been active areas of investigation. In recent years, a paradigm of mesothelial cells as a source of VAT adipocyte progenitor cells has emerged based on lineage tracing studies using the Wilms’ tumor gene, Wt1, as a marker for cells of mesothelial origin. Here, we show that Wt1 expression in adipose tissue is not limited to the mesothelium but is also expressed by a distinct preadipocyte population in mice and humans. We identify keratin 19 (Krt19) as a highly specific marker for the adult mouse mesothelium and demonstrate that Krt19-expressing mesothelial cells do not differentiate into visceral adipocytes. These results contradict the assertion that the VAT mesothelium can serve as a source of adipocytes.
Graphical abstract
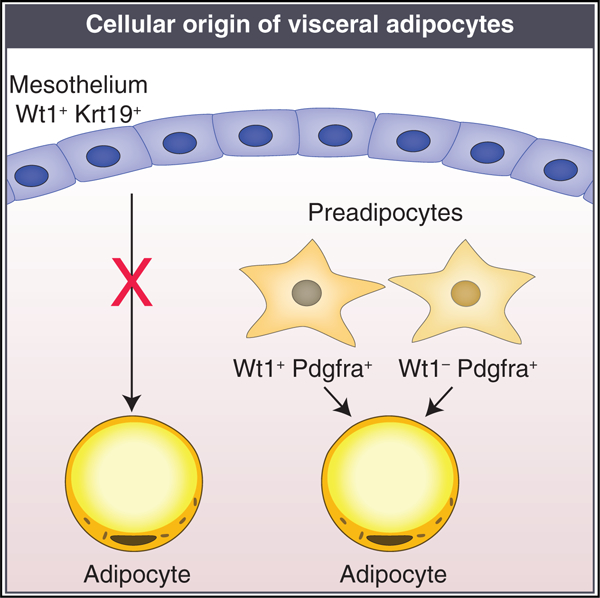
In brief
Mesothelial differentiation into adipocytes has been proposed based on fate mapping experiments using Wt1 as a marker. Westcott et al. show that Wt1 is expressed in stromal preadipocytes in addition to mesothelium and that fate mapping using a specific mesothelial marker, Krt19, does not support adipocyte differentiation.
INTRODUCTION
Adipogenic precursor cell populations have been proposed to arise from the mural cell compartment of the adipose vasculature (Tang et al., 2008), endothelium (Tran et al., 2012), and bone marrow (Rydén et al., 2015) and from a subset of Sca-1+CD24+ (Rodeheffer et al., 2008) or Pdgfra+ (Rodeheffer et al., 2008) stromal cells. In addition, a mesothelial source of visceral adipocytes has been hypothesized based on the established function of WT1 in mesenchymal development (Hastie, 2017), its presence in visceral adipose tissue (VAT) and absence in other white adipose depots (Chau et al., 2011), and its importance in development of tissues such as the epicardium (Zhou et al., 2008). Chau et al. (2014) used fate mapping techniques to demonstrate that Wt1-expressing cells differentiate into mature adipocytes in murine VAT. Importantly, the conclusion that traced adipocytes are of mesothelial origin depends on exclusive expression of Wt1 in mesothelial cells. In this study, we demonstrate that Wt1 is expressed in a distinct preadipocyte population in addition to the mesothelium and that mesothelial cells tracked with a more specific marker do not undergo adipocyte differentiation.
RESULTS
Wt1 is not specific to visceral adipose mesothelium and is also expressed in a distinct preadipocyte population
The advent of single-cell RNA sequencing (scRNA-seq) technology has enabled rigorous interrogation of previously accepted cell-type-specific markers as well as discovery of new ones. We performed scRNA-seq on adult male adipose tissue (Li et al., 2021). We specifically examined the epididymal adipose depot (Figure 1A) and identified distinct mesothelial and preadipocyte populations (Figure 1B). Although established adipocyte progenitor markers such as Pdgfra and Sca-1 are present exclusively in the preadipocyte cluster, and the mesothelial markers Msln and Upk3b are present exclusively in the mesothelial cluster, Wt1 is strongly expressed in both populations (Figures 1C and 1D). Expression of WT1 in mesothelial and preadipocyte clusters has also been noted in human omental single-nucleus RNA-seq data (Figure S1). Within the mouse preadipocyte population, Wt1 expression was not evenly distributed. To further explore the distribution of Wt1 in preadipocytes, we performed subclustering analysis on mouse preadipocytes. Of the four subpopulations identified, one was marked by high Wt1 expression (Figures S2A–S2D). Gene Ontology revealed that this subcluster also expresses a number of inflammatory genes, including those in the tumor necrosis factor (TNF), nuclear factor κB (NF-κB), and interleukin-17 (IL-17) signaling pathways (Figures S2E and S2F).
Figure 1. scRNA-seq of mouse SVF reveals distinct preadipocyte and mesothelial populations that express Wt1.
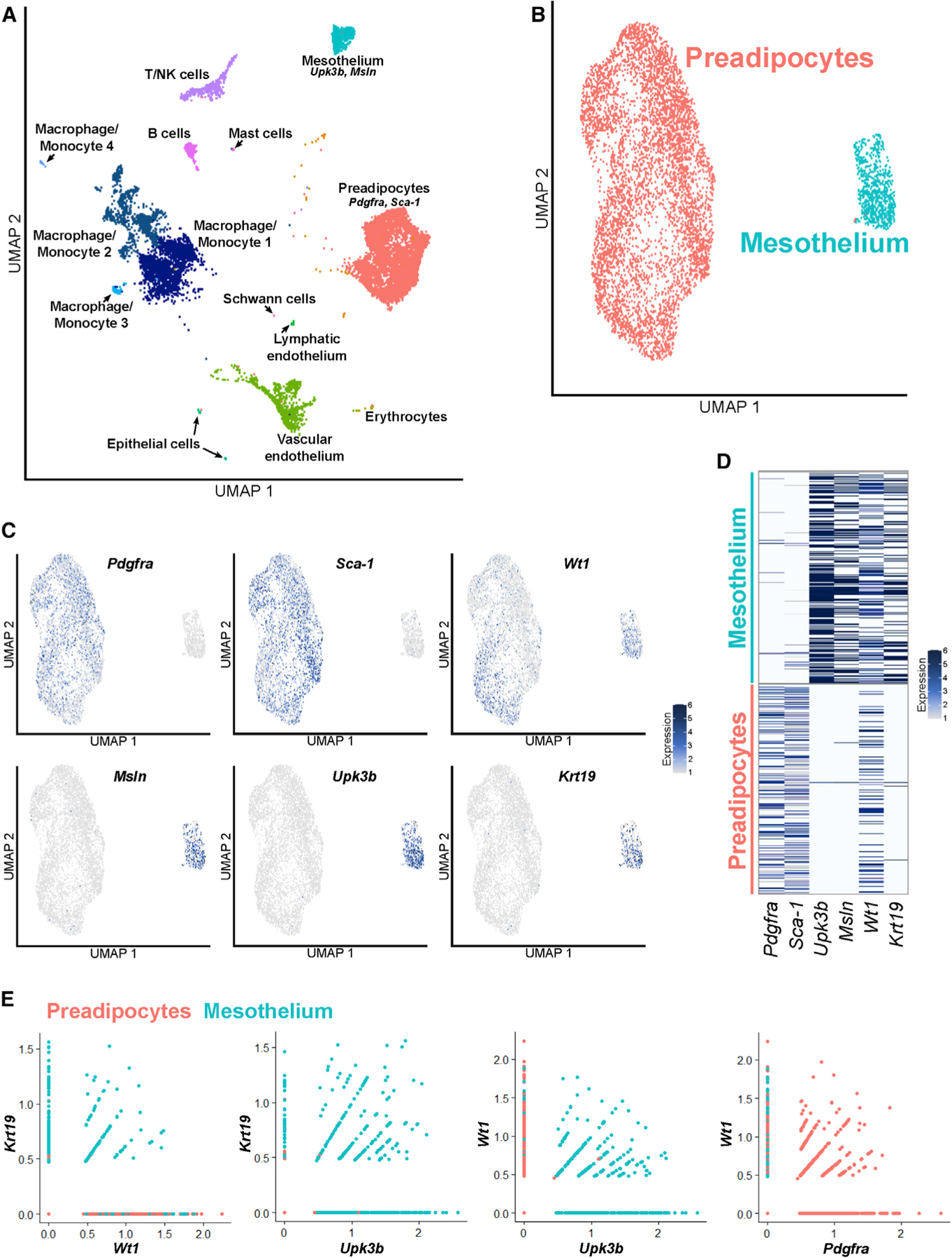
(A) Uniform Manifold Approximation and Projection (UMAP) plot of scRNA-seq performed on the SVF of mouse epididymal fat (eWAT), which identifies all expected populations, including distinct populations of preadipocytes and mesothelial cells.
(B) UMAP plot of the preadipocyte and mesothelial populations.
(C) The preadipocyte population is characterized by expression of Pdgfra and Sca-1, whereas the mesothelium expresses Upk3b and Msln. Although Krt19 expression mirrors that of mesothelial markers, Wt1 is expressed in preadipocytes and mesothelial cells.
(D) Heatmap demonstrates that Wt1 is expressed in cells expressing preadipocyte and mesothelial genes.
(E) Most cells expressing Wt1 but not Krt19 are preadipocytes, whereas virtually all Krt19-expressing cells are mesothelial. A significant subset of Wt1+ cells co-expresses Pdgfra.
In contrast to Wt1, our data indicate that Krt19 is a highly specific marker for the mesothelium in mouse and human (Figures 1C, 1D, and S1). A member of the keratin family, KRT19 is an intermediate filament protein discovered in squamous cell carcinomas and found in epithelioid cells (Wu and Rheinwald, 1981). KRT19 is expressed in human omental mesothelium but absent from subcutaneous fat (Darimont et al., 2008).
Trypsin digestion (Kenny et al., 2007) of intact fat pads was used to enzymatically isolate the mesothelium from perigonadal VAT depots, and qPCR for the mesothelial genes Msln, Upk3b, and Krt19 confirmed high enrichment in the mesothelial fraction (Figure 2A). Wt1, however, was expressed in the mesothelium and the residual stromal vascular fraction (SVF). To isolate Wt1-expressing and Krt19-expressing cells, we obtained tamoxifen-inducible Cre mouse models for Wt1 (Wt1CreERT2; Zhou et al., 2008) and Krt19 (Krt19CreERT; Means et al., 2008) and crossed them to a reporter strain that expresses tdTomato following Cre recombination. We next performed flow cytometry analysis of Wt1CreERT2: and Krt19CreERT:tdTomato+ SVF cells stained for platelet derived growth factor receptor alpha (PDGFRA) and demonstrated that Wt1CreERT2: tdTomato+ cells display extensive co-labeling for PDGFRA but Krt19CreERT:tdTomato+ cells did not (Figure 2B). Likewise, CD45− PDGFRA+ cells sorted from wild-type SVF express Wt1 but not the other, more specific mesothelial markers (Figure 2C). Using our Wt1CreERT2: and Krt19CreERT:tdTomato+ models, we confirmed that trypsin-digested mesothelium is labeled by both models, but only Wt1CreERT2 also labels a substantial proportion of the remaining SVF population (Figure 2D).
Figure 2. Wt1 is not a specific marker for the mesothelium and also labels Pdgfra+ preadipocytes.
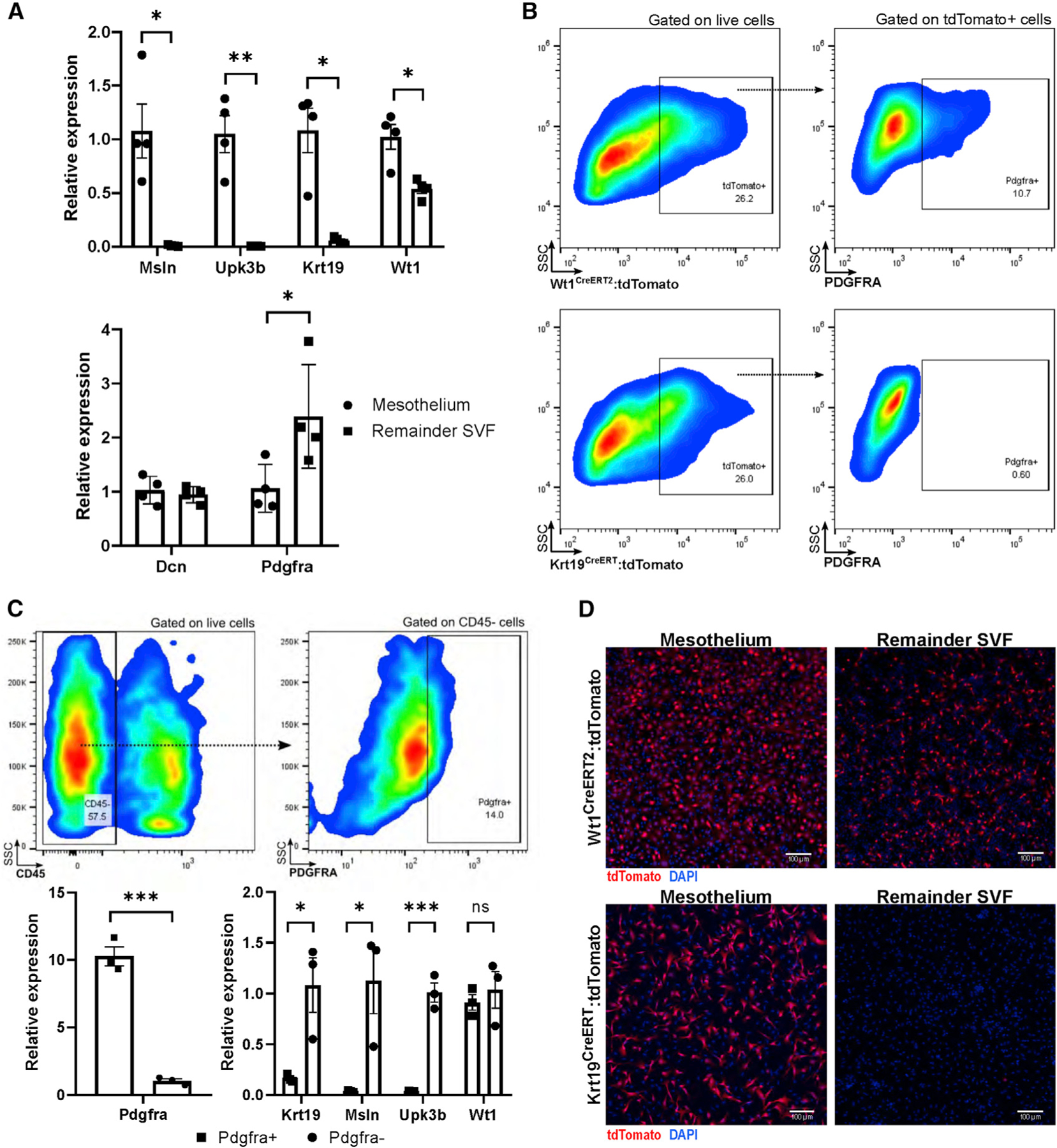
(A) The mesothelium can be digested away from the remainder of the adipose parenchyma with trypsin, a technique that highly enriches for mesothelial markers but not for Wt1, which is also highly expressed in the remainder SVF. The remainder SVF highly expresses Pdgfra, whereas Dcn is expressed by mesothelium and other stromal cells. n = 4 biological replicates of 2 mice each; mean ± SEM; *p < 0.05, **p < 0.01.
(B) Using Wt1CreERT2 and Krt19CreERT models crossed with a tdTomato reporter, flow cytometry reveals that Wt1-expressing cells contain a PDGFRA+ preadipocyte population whereas Krt19-expressing cells do not. Results are representative of two independent experiments.
(C) SVF cells isolated from wild-type mice were gated against CD45 and sorted into PDGFRA+ and PDGFRA− populations; mesothelial gene expression was restricted to PDGFRA− populations, whereas Wt1 was also present in the PDGFRA+ preadipocyte population. n = 3 groups of 2 mice each; mean ± SEM; ns, p R 0.05; *p < 0.05; ***p < 0.001.
(D) Trypsin-digested mesothelium contains a high proportion of tdTomato+ cells in Krt19CreERT and Wt1CreERT2 models, whereas the remainder SVF contains tdTomato+ cells in only Wt1CreERT2 mice. SSC, side scatter. Images are representative of 10 low-powered fields of cells cultured from 4 separate mice.
Cultured mesothelial cells do not undergo adipocyte differentiation in vitro
We next assessed the ability of Wt1+ and Krt19+ cells to undergo adipogenesis ex vivo. Wt1CreERT2: and Krt19CreERT:tdTomato+ mice were injected with tamoxifen, and a week later, VAT was harvested and collagenase digested, and the SVF was cultured in adipocyte differentiation medium. Although many newly formed adipocytes were tdTomato+ when Wt1CreERT2 was used as the driver, virtually no tdTomato+ adipocytes were seen in the Krt19CreERT sample (Figure 3A).
Figure 3. Krt19-expressing mesothelial cells do not differentiate into adipocytes in vitro.
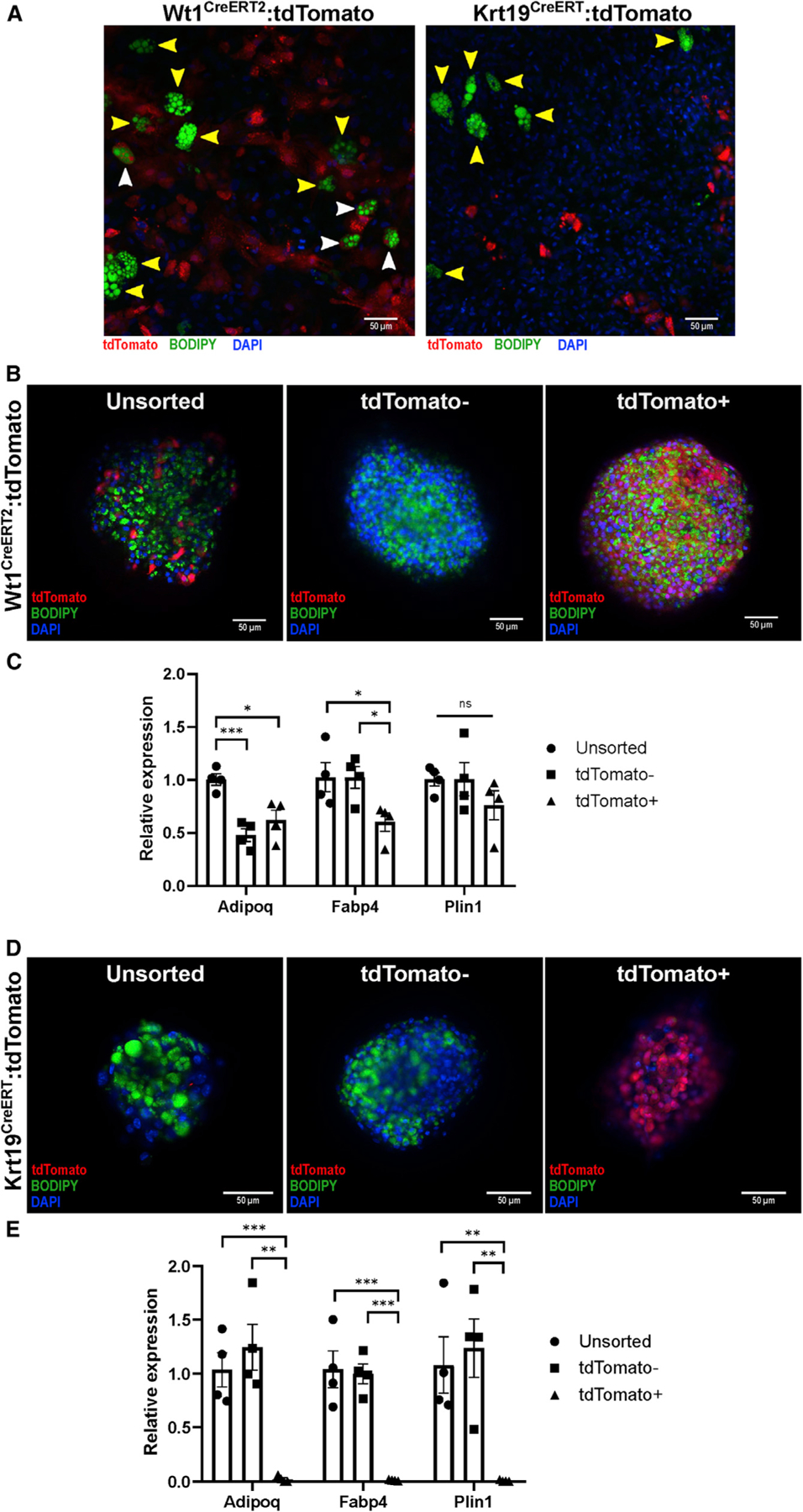
(A) SVF from Wt1CreERT2 and Krt19CreERT mice was isolated and underwent adipogenic differentiation on standard collagen-coated plates. Although adipocytes differentiated from Wt1CreERT2:tdTomato SVF contained tdTomato+ (white arrowheads) and tdTomato− (yellow arrowheads), no tdTomato+ adipocytes were found in the Wt1CreERT2:tdTomato samples. Representative images from 36 low-powered fields are shown.
(B and C) SVF was isolated from Wt1CreERT2:tdTomato mice after tamoxifen labeling, sorted into tdTomato+ and tdTomato− groups, and plated on low-adhesion plates, forming microspheres. After adipogenic differentiation, unsorted, tdTomato+, and tdTomato− cells derived from Wt1CreERT2:tdTomato mice, as demonstrated by BODIPY labeling of lipid droplet formation and qPCR for the adipocyte markers Adipoq, Fabp4, and Plin1. n = 4 microspheres derived from 3 separate mice; mean ± SEM; ns, p R 0.05; *p < 0.05; ***p < 0.001.
(D and E) SVF isolated from Krt19CreERT:tdTomato mice was sorted into tdTomato+ and tdTomato− populations, and although unsorted and tdTomato− cells differentiated normally as shown by BODIPY staining and qPCR, tdTomato+ cells did not undergo adipogenic differentiation. n = 4 microspheres derived from 5 separate mice; mean ± SEM; **p < 0.01, ***p < 0.001.
We next employed a 3D spheroid differentiation model to improve differentiation of visceral adipocytes from their progenitors (Emont et al., 2015), which also has the advantage of down-scaling for use on only a few thousand cells (Spallanzani et al., 2019). Wt1CreERT2: and Krt19CreERT:tdTomato+ mice were injected with tamoxifen, and the VAT SVF was isolated a week later and sorted by flow cytometry into tdTomato+ and tdTomato− populations, which were subsequently plated on an ultra-low attachment surface plate to facilitate spheroid formation and then exposed to adipocyte differentiation medium. Although tdTomato+ cells isolated from Wt1CreERT2 mice differentiated into adipocytes, as demonstrated by staining with 4,4-Difluoro-1,3,5,7,8-Pentamethyl-4-Bora-3a,4a-Diaza-s-Indacene (also known as BODIPY) for lipid accumulation as well as qPCR for the adipocyte-related genes adiponectin (Adipoq), fatty acid-binding protein 4 (Fabp4), and perilipin 1 (Plin1) (Figures 3B and 3C), tdTomato+ cells isolated from Krt19CreERT mice did not accumulate lipid or express adipocyte genes (Figures 3D and 3E).
Krt19-expressing mesothelial cells do not contribute to mature adipocytes in visceral adipose depots in vivo in response to aging or a high-fat diet
After demonstrating that Krt19-expressing mesothelial cells do not undergo adipocyte differentiation in vitro, we sought to confirm these findings in vivo. Wt1CreERT2: and Krt19CreERT mice were crossed with the mTmG reporter model, in which Cre recombination results in a switch from tdTomato to GFP expression, and male and female offspring were labeled with tamoxifen at 6 weeks of age. Littermates were placed on a high-fat diet 1 week after injection or maintained on a standard chow diet. At 15 weeks of age, mice were sacrificed, and gonadal VAT was whole mounted for confocal microscopy (Berry et al., 2014). Although there was GFP labeling of the mesothelial layer in both models (Figures 4A, 4B, and 4D), GFP+ adipocytes were only seen in Wt1CreERT2 mice fed chow or high-fat diet (HFD) (Figures 4A and 4B). Confocal images of 6–10 sections from 3 mice per group were obtained, and the percentage of GFP+ adipocytes was calculated (Figure 4C). Despite counting over 2,200 adipocytes in the Krt19CreERT chow-fed cohort and over 1,600 adipocytes in the HFD-fed Krt19CreERT cohort, no GFP+ adipocytes were identified. Male and female perigonadal depots were studied, with no difference noted between the sexes in the Wt1CreERT2 or Krt19CreERT models (Figure S3A). To further rule out the adipogenic potential of Krt19-lineage cells, we extended the period of high-fat feeding to 18 weeks but still saw no GFP+ adipocytes, and aging chow-fed Krt19CreERT:mtmG mice for 1 year also failed to yield any GFP+ adipocytes (Figures S3B and S3C).
Figure 4. Krt19-lineage mesothelial cells do not differentiate into adipocytes in vivo.
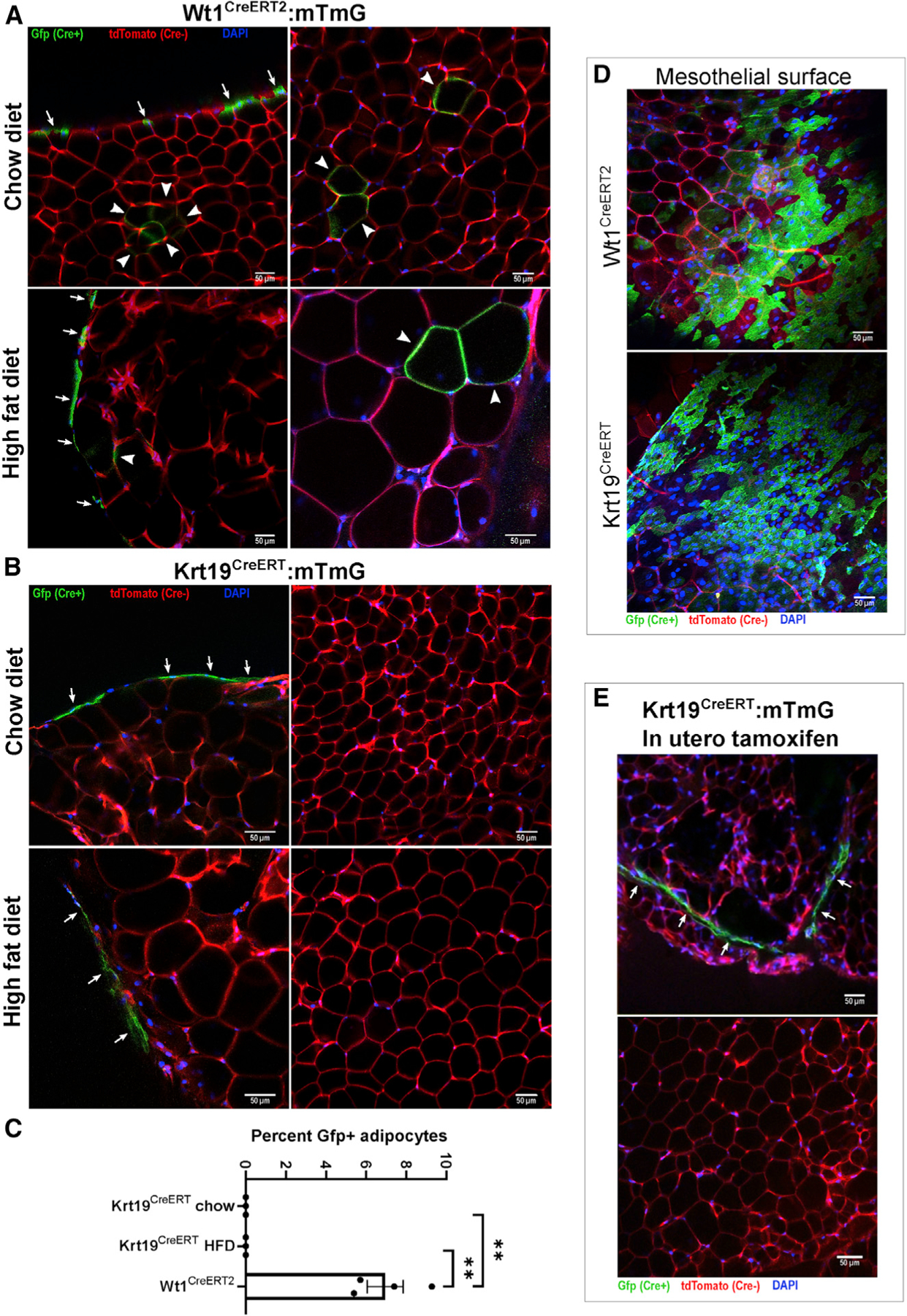
Wt1CreERT2:mTmG and Krt19CreERT:mTmG mice were injected with tamoxifen at 6 weeks of age and sacrificed at 15 weeks of age; mice receiving a high-fat diet did so starting at 7 weeks age. Whole mounts of perigonadal fat were imaged by confocal microscopy.
(A) In Wt1CreERT2:mTmG mice, labeling of the mesothelial layer (arrows) as well as multiple Gfp+ adipocytes (arrowheads) were readily observed. Shown are representative images from 26 low-powered fields obtained from 4 separate mice.
(B) In contrast, Krt19CreERT:mTmG mice only had labeling of the mesothelial layer, and no GFP+ adipocytes were observed. Representative images from 50 low-powered fields were obtained from 6 separate mice.
(C) The percentage of Gfp+ adipocytes was quantified. In the chow and HFD Krt19-CreER groups, over 2,200 and 1,600 adipocytes, respectively, were counted without any Gfp+ adipocytes observed. n = 3–4 mice, 6–10 fields per mouse; mean ± SEM; **p < 0.01.
(D) GFP+ mesothelium was readily identifiable in Wt1CreERT2:mTmG and Krt19CreERT:mTmG models.
(E) To evaluate the possibility that mesothelial cells, which develop in utero, may transition to a preadipocyte identity later in development, Krt19CreERT:mTmG dams were injected with tamoxifen at gestational day E14.5, and offspring were sacrificed at 6 weeks of age. The mesothelium contained GFP+ cells, but no GFP+ adipocytes were identified. Shown are representative images from 15 fields obtained from 2 separate mice.
Data derived from mice labeled with tamoxifen as adults do not preclude the possibility that the fetal mesothelium could give rise to adipose precursors that still express Wt1, even though the adult mesothelium may no longer contribute to the adipocyte pool. In fact, Chau et al. (2014) demonstrated that exposing Wt1CreERT2:mTmG mice to tamoxifen in utero on embryonic day 14.5 (E14.5) resulted in GFP+ adipocytes in the adult mouse (Chau et al., 2014). We therefore repeated this experiment using Krt19CreERT:mTmG mice, exposing them to tamoxifen on day 14.5 of gestation via subcutaneous injection of the dam. This manipulation led to GFP+ cells in the mesothelium, but again we did not identify any mature GFP+ adipocytes in the grown offspring (Figure 4E).
DISCUSSION
Adipose tissue development is a complex process that occurs in a depot-specific manner (Kahn et al., 2019). Although some have suggested that white adipose tissue (WAT) adipocyte progenitors are homogeneous and lack subtypes (Acosta et al., 2017), numerous studies now indicate that there is diversity within the preadipocyte pool (Ferrero et al., 2020). This complexity has led to challenges in identifying gene markers that comprehensively identify adipocyte progenitors, and almost a dozen different Cre lines have been used to perform embryonic adipocyte lineage tracing (Sebo and Rodeheffer, 2019). Beyond developmental preadipocyte commitment, terminal adipocyte differentiation from progenitors is also under the control of various metabolic and hormonal stimuli, including caloric overload (Wang et al., 2013), cold exposure (Wang et al., 2013), ischemia (Zangi et al., 2017), and wound healing (Plikus et al., 2017), adding to the complexity of studying adipocyte maturation. scRNA-seq has provided an important tool to unravel the complexity of adipose tissue and generate hypotheses about its development and the role of different cell types under varying metabolic conditions.
The role of the mesothelium as a source of visceral preadipocytes during development and adulthood is intriguing and plausible, given the variety of sources for adipocytes that have been identified previously. The idea has gained widespread acceptance, and subsequent studies have used Wt1 as a marker for mesothelium-derived preadipocytes (Lee et al., 2019). Chau et al. (2014) demonstrated that Wt1+ cells in visceral fat pads express the preadipocyte marker Sca-1 and argue that this is consistent with mesothelial cells’ capacity to undergo adipocyte differentiation. However, we demonstrate that Sca-1 and Pdgfra are, in fact, not expressed in the mesothelium, whereas Wt1 is expressed in the mesothelium and preadipocyte populations, indicating that Wt1+ mesothelial cells are distinct from Wt1+ preadipocytes. Consistent with our findings, previous studies have demonstrated Wt1 expression in non-mesothelial stromal populations, including integration of multiple mouse VAT scRNA-seq experiments (Burl et al., 2018; Ferrero et al., 2020; Hepler et al., 2018; Rondini and Granneman, 2020). Furthermore, scRNA-seq profiling of mouse eWAT after CL 316,243 treatment demonstrated that Msln+/Upk3b+/Krt19+ stromal cells do not initiate adipogenic programs in response to β3-adrenergic stimulation (Burl et al., 2018). Given the implications of these findings for the specificity of Wt1 as a marker for the mesothelium and, thus, the conclusion that mesothelial cells can differentiate into adipocytes, we performed in vivo lineage tracing using a tamoxifen-inducible Cre model driven by the specific mesothelial transcriptional marker Krt19. These studies demonstrated that mesothelial cells, in fact, do not contribute to VAT adipogenesis in mice under homeostatic conditions or with caloric overload. That Wt1 is expressed in visceral but not subcutaneous depots may indeed suggest distinct embryologic origins among WAT depots, but our findings clarify that the mesothelium does not represent a developmental intermediate for adipocytes.
The physiology of the mesothelium of visceral adipose depots remains under-characterized. Mesothelial cells have been suggested to play a role in adipose tissue inflammation in obesity (Darimont et al., 2008), and a recent study indicated the presence of an inflammatory subset of mesothelial cells that expands in mice after HFD feeding (Sárvári et al., 2021). Wt1-expressing preadipocytes and adipocytes have been described previously to be more highly responsive to TNF-α than other white adipocytes (Lee et al., 2019), and this could be taken to support the idea that inflammatory mesothelial cells may be differentiating into more pro-inflammatory adipocytes. However, given the distinction between Wt1+ mesothelium and Wt1+ preadipocyte pools demonstrated by our data, it is clear that studies of the mesothelium should use a more specific marker than Wt1, and studies of Wt1-expressing cells must recognize the underlying heterogeneity of that population.
Our study also highlights the heterogeneity of stromal preadipocytes, in particular with respect to Wt1 expression. Wt1 is not expressed by subcutaneous or brown preadipocytes and marks varying proportions of visceral depots in mice (Chau et al., 2014; Lee et al., 2019). Adipocytes differentiated in vitro from a Wt1-lineage immortalized preadipocyte clone demonstrated robust c-Jun N-terminal kinase (JNK) phosphorylation after treatment with TNF-α (Lee et al., 2019), which is consistent with our finding that Wt1-expressing preadipocytes also express genes that participate in TNF signaling and other inflammatory pathways. Our Wt1-high preadipocyte cluster also specifically expresses Nr4a1 (Nur77; data not shown), which has been shown previously to inhibit adipocyte differentiation (Chao et al., 2008). This transcriptional heterogeneity suggests functional specialization of preadipocytes which may have implications for adipose tissue development and is worthy of further study.
In addition to elucidating key VAT biology, this study raises important issues with respect to lineage tracing based on transcriptional markers. Identifying a single specific and sensitive marker for lineage tracing can be challenging. For example, aP2-Cre was initially used as an adipocyte-specific-Cre until it was found to be inefficient at labeling all adipocytes, and it also labeled some blood-lineage cells and endothelial cells (Jeffery et al., 2014). Given Wt1’s broad expression during development, it is perhaps not surprising that it lacks specificity for a single cell type within a tissue, consistent with findings in other tissues in which Wt1 is critical for development, such as the heart (Rudat and Kispert, 2012). Intersectional genetics approaches have been developed to address these challenges (Han et al., 2021), and the increasingly widespread application of single-cell and single-nucleus RNA sequencing will hopefully make selection of cell-type-specific markers more precise and more easily interpretable. It should be noted that the lineage tracing of adipocytes can be complicated by the effect of tamoxifen, which has been shown to induce de novo adipogenesis and may maintain nuclear CreERT translocation after even 2 months of washout (Ye et al., 2015). Because our tamoxifen-inducible Krt19CreERT models do not show any evidence of adipogenesis, these caveats do not affect our conclusions but should be taken into account when designing lineage tracing experiments.
STAR★METHODS
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| FITC-conjugated mouse anti-CD150a (PDGFRA) | Thermo Fisher | 11–1401-82; RRID: AB_2572476 |
| Chemicals, peptides, and recombinant proteins | ||
| Tamoxifen | Sigma-Aldrich | T5648 |
| Sunflower seed oil | Sigma-Aldrich | S5007 |
| Insulin | Sigma-Aldrich | I1882 |
| Rosiglitazone | Cayman Chemicals | 71740 |
| 3-Isobutyl-1-methylxanthine | Sigma-Aldrich | I5879 |
| Dexamethasone | Sigma-Aldrich | D4902 |
| Fluoromount-G | Southern Biotech | 0100 |
| SYBR Green PCR master mix | Thermo Fisher | 4312704 |
| Z-fix Concentrate | Anatech Ltd | 171 |
| BODIPY 493/503 (4,4-Difluoro-1,3,5,7,8-Pentamethyl-4-Bora-3a,4a-Diaza-s-Indacene) | Invitrogen | D3922 |
| Hoechst 33342, Trihydrochloride, Trihydrate | Invitrogen | H3570 |
| 0.25% Trypsin, 0.1% EDTA in HBSS w/o Calcium, Magnesium and Sodium Bicarbonate | Corning | 25053CI |
| Collagenase type II | Sigma-Aldrich | C6885 |
| Triton X-100 Surfact-Amps | Thermo Fisher | 28314 |
| Trizol | Thermo Fisher | 15596018 |
| Critical commercial assays | ||
| RNeasy MinElute Cleanup Kit | QIAGEN | 74204 |
| Deposited data | ||
| Mouse adipose SVF single-cell RNA-seq dataset | Single cell atlas portal | https://singlecell.broadinstitute.org/single_cell/study/SCP708/mouse-adipose-stromal-vascular-fraction |
| Experimental models: Organisms/strains | ||
| Mouse: Krt19CreERT: Krt19tm1(cre/ERT)Ggu/J | The Jackson Laboratory | 026925 |
| Mouse: Wt1CreERT2: Wt1tm2(cre/ERT2)Wtp/J | The Jackson Laboratory | 010912 |
| Mouse: mTmG: B6.129(Cg)-Gt(ROSA) 26Sortm4(ACTB-tdTomato,-EGFP)Luo/J | The Jackson Laboratory | 007676 |
| Mouse: tdTomato: B6.Cg-Gt(ROSA) 26Sortm9(CAG-tdTomato)Hze/J | The Jackson Laboratory | 007909 |
| Oligonucleotides | ||
| qPCR primers | See Table S1 | Thermo Fisher |
| Software and algorithms | ||
| FlowJo v 10.7.1 | BD | N/A |
| Seurat v 3.0.3.9036 | Stuart et al., 2019 | N/A |
| Zen Black v 2.3 | Zeiss | N/A |
| Other | ||
| SORP FACSAria III | BD | N/A |
| LSM 880 Upright Laser Scanning Confocal Microscope | Zeiss | N/A |
| Ultra-Low Attachment Surface Plate, 96 well | Corning | 4515 |
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to the lead contact Evan Rosen (erosen@bidmc.harvard.edu).
Materials availability
This study did not generate unique reagents.
Data and code availability
Single-cell RNA-seq data has been deposited to the Single Cell Atlas at the Broad Institute https://singlecell.broadinstitute.org/. Mouse adipose SVF data is available here: https://singlecell.broadinstitute.org/single_cell/study/SCP708/mouse-adipose-stromal-vascular-fraction.
All code to reproduce clustering data may be found on GitHub: https://github.com/rosen-lab/sc-svf-nts.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animals
All animal experiments were performed in accordance with a protocol approved by the BIDMC Institutional Animal Care and Use Committee. Mice were maintained under a 12 hr light/12hr dark cycle at 22°C on chow diet (8664 Harlan Teklad, 6.4% wt/wt fat) unless otherwise specified. Krt19-CreERT (Means et al., 2008) (stock no. 026925), Wt1-CreERT2 (Zhou et al., 2008) (stock no. 010912) lox-stop-lox tdTomato (stock no 007914) and mTmG mice (stock no 007676) were all obtained from Jackson Labs. Tamoxifen-induced Cre labeling was performed at 6 weeks of age with subcutaneous tamoxifen injections (100 mg/kg dissolved in sunflower seed oil) on four consecutive days. For high fat diet experiments mice were injected with tamoxifen at 6 weeks of age as above and either continued on chow diet or switched to high fat diet consisting of 20% calories from protein, 60% from fat, and 20% from carbohydrate (Research Diets, D12492) at 7 weeks of age; mice were sacrificed at 15 weeks of age for confocal imaging. For in utero tamoxifen exposure, the dam was injected with a single dose of tamoxifen (100 mg/kg) subcutaneously on day E14.5 of gestation; offspring were sacrificed at 6 weeks of age for confocal imaging. Both male and female mice were used to perform the presented experiments and similar results were obtained from mice of both sexes.
METHOD DETAILS
SVF isolation and flow cytometry
SVF isolation was performed as described elsewhere (Cho et al., 2014); briefly, mouse gonadal fat depots were washed with PBS on ice and then minced and incubated with collagenase type II (1 mg/ml) (Sigma-Aldrich) and incubated on a shaker water bath incubator at 37°C for 20 minutes with vigorous shaking by hand every 5 minutes. EDTA, pH 8.0 at a concentration of 10 mM was then added for an additional 5 minutes, after which wash media (DMEM+glutamax with 10% bovine calf serum and 1% penicillin-streptomycin) or FACS buffer (PBS with 1mM EDTA, 25 mM HEPES and 1% fetal bovine serum) was added. Cells were filtered through a 100 µm filter and washed, and if flow cytometry was to be performed, incubated with ACK lysis buffer for 5 minutes. SVF was then washed and filtered through 40 µm filter followed by plating on collagen-coated multiwell plates in growth media (DMEM+glutamax, 15% FBS, 1% penicillin-streptomycin) or suspended in FACS buffer for flow cytometry. For flow cytometry, cells were incubated in FITC-conjugated mouse anti-CD150a (PDGFRA) antibody (1:1000 dilution) (ThermoFisher 11–1401-82). Flow cytometric cell sorting and analysis was performed on a SORP FACSAria III (BD). Flow cytometry data was processed using FlowJo version 10.7.1 (BD).
Trypsin mesothelial digestion
Gonadal fat pads were removed intact and washed once in PBS on ice and then transferred to trypsin solution (0.25% trypsin, 0.1% EDTA, Corning diluted 1:1 in PBS) and placed in shaker water bath incubator at 37°C for 30 minutes, with vigorous shaking by hand every several minutes. Intact adipose depots were then removed with forceps and minced and processed as described above (‘‘remainder SVF’’). Wash media was added to the remaining solution which contained mesothelial cells and cells were washed twice prior to plating. For qPCR studies, cells from individual animals were plated on separate wells of a collagen-coated 12-well plate for 1 hour, washed with PBS three times, and lysed in Trizol (Thermo Fisher). For confocal imaging, mesothelial cells from two mice were plated per well of a BioCoat collagen-coated 4-well culture slide (Corning) for approximately 16 hours prior to fixation with 4% para-formaldehyde. After fixation, cells were permeabilized with Triton X-100, stained with Hoechst, and coverslips mounted with Fluoromount-G (Southern Biotech).
In vitro adipocyte differentiation assays
For standard in vitro differentiation assays, SVF was plated on collagen-coated 12-well plates in growth media for 2 days and then switched to differentiation induction media containing DMEM+glutamax, 10% FBS, 1% penicillin-streptomycin, 0.5 µg/ml insulin, 5 µM dexamethasone, 1 µM rosiglitazone, and 0.5 mM 3-isobutyl-1-methylxanthine for two days followed by maintenance media (DMEM+glutamax, 10% FBS, 1% penicillin-streptomycin, 0.5 µg/ml insulin) for an additional seven days prior to imaging, with media changes every 2 days. Cells were fixed with a formaldehyde-containing fixative Z-fix and stained with BODIPY to label neutral lipid accumulation that is consistent with adipocyte differentiation, and Hoechst for nuclear labeling. For microsphere differentiation, cells were flow-sorted as above, washed in wash media, and plated on collagen-coated multi-well plates for 16 hours. Cells were then suspended with trypsin and counted prior to plating approximately 5,000 cells per well into a 96-well ultra-low attachment surface plate (Corning, cat# 4515). Pre-plating the cells appeared to improve viability and spheroid formation compared to directly sorting into the ultra-low attachment surface plate. Cells were cultured in growth media for 2 days to allow spheroid formation, followed by 3 days in differentiation induction media as described above, followed by 5 days in maintenance media with insulin alone, with media change after 3 days in maintenance media. A subset of spheroids were then washed in PBS, fixed with Z-fix, stained with BODIPY and Hoechst, and placed onto slides for confocal imaging, while the remaining spheroids were washed and lysed in Trizol for RNA purification and qPCR.
Quantitative PCR
Cells were collected in Trizol as above. Total RNA was isolated using RNeasy MinElute Cleanup Kit (QIAGEN) using manufacturer’s instructions. Up to 1 µg of RNA was reverse-transcribed using High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher). SYBR Green PCR Master Mix (Thermo Fisher) and gene primers (Table S1) were used to perform qPCR on QuantStudio 6 Flex Real-Time PCR system (Thermo Fisher) and normalized to housekeeping gene Tbp. Results are presented as fold change (2−ΔCT).
Confocal microscopy
All confocal images were obtained using a Zeiss LSM 880 Upright Laser Scanning Confocal Microscope using a 10X or 20X objective. Images were processed using Zen Black 2.3 software. Multi well slides and microspheres were prepared as above. Whole mount images were obtained by dividing gonadal depot into several small sections, washing in PBS once, fixing with Z-fix and Hoechst for 20 minutes, washing with PBS three times, and mounting on a slide with Fluoromount-G.
QUANTIFICATION AND STATISTICAL ANALYSIS
Single-cell RNA sequencing & clustering
Drop-seq was performed on Collagenase-digested adipose tissue SVF derived from 10-week old wild-type male C57BL/6 mice using procedures described in detail previously (Campbell et al., 2017; Li et al., 2021; Macosko et al., 2015). Raw sequencing data were filtered for quality and trimmed of adaptor sequences and poly(A) tails using Drop-seq tools (https://github.com/broadinstitute/Drop-seq), then aligned to the mouse genome (GRCm38/mm10) with STAR (Dobin et al., 2013) v2.6.1e. Digital expression matrices were generated from uniquely-mapped reads, including intronic and exonic features from the GENCODE M16 mouse genome annotation. Doublet cell populations were identified and removed with Scrublet (Wolock et al., 2019) using a doublet score threshold of 0.25. Clustering and further analyses were performed with the Seurat (v3.0.3.9036) software package (Satija et al., 2015; Stuart et al., 2019). First, we removed genes expressed in fewer than 2 cells, and cells with fewer than 400 UMIs or with greater than 10% mitochondrial content. Data were normalized and scaled and variable features detected using SCTransform (Hafemeister and Satija, 2019). Cells were then clustered using the top 50 PCs among the variable gene set. UMAP (Becht et al., 2018) projections were generated from these same PCs for visualization. For subclustering, preadipocytes were subset from the larger dataset, PCA was rerun on the subset data, and UMAP projections and clusters were calculated using the top 20 PCs. Human adipose single-nucleus RNA-seq data were processed similarly to the above. Reads were mapped to the GRCh38/hg38 human genome assembly and counts generated from mappings to GENCODE 27 genome annotation (introns and exons).
Gene Ontology analysis
Markers were calculated for each preadipocyte subcluster using a non-parametric Wilcoxon rank sum test, markers were then filtered to retain only those with an adjusted p value < 0.05 and evaluated for enrichment in KEGG pathways containing under 300 genes using clusterProfiler (Yu et al., 2012).
Statistical analysis
Statistics were performed on GraphPad Prism version 8. Paired two-tailed t tests were performed on mesothelium-remainder SVF qPCR samples, and unpaired two-tailed t tests were performed on microsphere qPCR samples and in vivo adipocyte counts.
Supplementary Material
Highlights.
Wt1 marks mesothelial cells and preadipocytes in visceral adipose tissue
Krt19 is a specific mesothelial marker
Wt1+ cells, but not Krt19+ cells, are adipocyte progenitors in mouse visceral fat
ACKNOWLEDGMENTS
We gratefully acknowledge the Boston Nutrition Obesity Research Center Functional Genomics and Bioinformatics Core (NIH P30DK046200) for expertise regarding single-cell RNA sequencing analysis and the Beth Israel Deaconess Medical Center Flow Cytometry and Confocal Imaging Cores for assistance with flow sorting and confocal microscopy studies, respectively. This work was supported by funding from the NIH (RC2DK116691). G.P.W. received funding from NIH grant T32DK007516, administrative supplement RC2DK116691–03S1, and the Endocrine Fellows Foundation. M.P.E. is supported by NIH grant F32DK124914. We thank all members of the Rosen lab for thoughtful and constructive discussions.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2021.109388.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Acosta JR, Joost S, Karlsson K, Ehrlund A, Li X, Aouadi M, Kasper M, Arner P, Rydén M, and Laurencikiene J (2017). Single cell transcriptomics suggest that human adipocyte progenitor cells constitute a homogeneous cell population. Stem Cell Res. Ther 8, 250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becht E, McInnes L, Healy J, Dutertre C-A, Kwok IWH, Ng LG, Ginhoux F, and Newell EW (2018). Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol 37, 38–44. [DOI] [PubMed] [Google Scholar]
- Berry R, Church CD, Gericke MT, Jeffery E, Colman L, and Rodeheffer MS (2014). Imaging of adipose tissue. Methods Enzymol 537, 47–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burl RB, Ramseyer VD, Rondini EA, Pique-Regi R, Lee Y-H, and Granneman JG (2018). Deconstructing Adipogenesis Induced by b3-Adrenergic Receptor Activation with Single-Cell Expression Profiling. Cell Metab 28, 300–309.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell JN, Macosko EZ, Fenselau H, Pers TH, Lyubetskaya A, Tenen D, Goldman M, Verstegen AMJ, Resch JM, McCarroll SA, et al. (2017). A molecular census of arcuate hypothalamus and median eminence cell types. Nat. Neurosci 20, 484–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao LC, Bensinger SJ, Villanueva CJ, Wroblewski K, and Tontonoz P (2008). Inhibition of adipocyte differentiation by Nur77, Nurr1, and Nor1. Mol. Endocrinol 22, 2596–2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chau Y-Y, Brownstein D, Mjoseng H, Lee W-C, Buza-Vidas N, Nerlov C, Jacobsen SE, Perry P, Berry R, Thornburn A, et al. (2011). Acute multiple organ failure in adult mice deleted for the developmental regulator Wt1. PLoS Genet 7, e1002404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chau Y-Y, Bandiera R, Serrels A, Martínez-Estrada OM, Qing W, Lee M, Slight J, Thornburn A, Berry R, McHaffie S, et al. (2014). Visceral and subcutaneous fat have different origins and evidence supports a mesothelial source. Nat. Cell Biol 16, 367–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho KW, Morris DL, and Lumeng CN (2014). Flow cytometry analyses of adipose tissue macrophages. Methods Enzymol 537, 297–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darimont C, Avanti O, Blancher F, Wagniere S, Mansourian R, Zbinden I, Leone-Vautravers P, Fuerholz A, Giusti V, and Macé K (2008). Contribution of mesothelial cells in the expression of inflammatory-related factors in omental adipose tissue of obese subjects. Int. J. Obes 32, 112–120. [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emont MP, Yu H, Jun H, Hong X, Maganti N, Stegemann JP, and Wu J (2015). Using a 3D Culture System to Differentiate Visceral Adipocytes In Vitro. Endocrinology 156, 4761–4768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrero R, Rainer P, and Deplancke B (2020). Toward a Consensus View of Mammalian Adipocyte Stem and Progenitor Cell Heterogeneity. Trends Cell Biol 30, 937–950. [DOI] [PubMed] [Google Scholar]
- Hafemeister C, and Satija R (2019). Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol 20, 296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X, Zhang Z, He L, Zhu H, Li Y, Pu W, Han M, Zhao H, Liu K, Li Y, et al. (2021). A suite of new Dre recombinase drivers markedly expands the ability to perform intersectional genetic targeting. Cell Stem Cell 28, 1160–1176.e7. [DOI] [PubMed] [Google Scholar]
- Hastie ND (2017). Wilms’ tumour 1 (WT1) in development, homeostasis and disease. Development 144, 2862–2872. [DOI] [PubMed] [Google Scholar]
- Hepler C, Shan B, Zhang Q, Henry GH, Shao M, Vishvanath L, Ghaben AL, Mobley AB, Strand D, Hon GC, and Gupta RK (2018). Identification of functionally distinct fibro-inflammatory and adipogenic stromal subpopulations in visceral adipose tissue of adult mice. eLife 7, e39636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffery E, Berry R, Church CD, Yu S, Shook BA, Horsley V, Rosen ED, and Rodeheffer MS (2014). Characterization of Cre recombinase models for the study of adipose tissue. Adipocyte 3, 206–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn CR, Wang G, and Lee KY (2019). Altered adipose tissue and adipocyte function in the pathogenesis of metabolic syndrome. J. Clin. Invest 129, 3990–4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenny HA, Krausz T, Yamada SD, and Lengyel E (2007). Use of a novel 3D culture model to elucidate the role of mesothelial cells, fibroblasts and extra-cellular matrices on adhesion and invasion of ovarian cancer cells to the omentum. Int. J. Cancer 121, 1463–1472. [DOI] [PubMed] [Google Scholar]
- Lee KY, Luong Q, Sharma R, Dreyfuss JM, Ussar S, and Kahn CR (2019). Developmental and functional heterogeneity of white adipocytes within a single fat depot. EMBO J 38, e99291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Li E, Czepielewski RS, Chi J, Guo X, Han Y-H, Wang D, Wang L, Hu B, Dawes B, et al. (2021). Neurotensin is an anti-thermogenic peptide produced by lymphatic endothelial cells. Cell Metab Published online May 19, 2021. 10.1016/j.cmet.2021.04.019. [DOI] [PMC free article] [PubMed]
- Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, et al. (2015). Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell 161, 1202–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Means AL, Xu Y, Zhao A, Ray KC, and Gu G (2008). A CK19CreERT Knockin Mouse Line Allows for Conditional DNA Recombination in Epithelial Cells in Multiple Endodermal Organs. Genesis 46, 318–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plikus MV, Guerrero-Juarez CF, Ito M, Li YR, Dedhia PH, Zheng Y, Shao M, Gay DL, Ramos R, Hsi T-C, et al. (2017). Regeneration of fat cells from myofibroblasts during wound healing. Science 355, 748–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodeheffer MS, Birsoy K, and Friedman JM (2008). Identification of white adipocyte progenitor cells in vivo. Cell 135, 240–249. [DOI] [PubMed] [Google Scholar]
- Rondini EA, and Granneman JG (2020). Single cell approaches to address adipose tissue stromal cell heterogeneity. Biochem. J 477, 583–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudat C, and Kispert A (2012). Wt1 and epicardial fate mapping. Circ. Res 111, 165–169. [DOI] [PubMed] [Google Scholar]
- Rydén M, Uzunel M, Hård JL, Borgström E, Mold JE, Arner E, Mejhert N, Andersson DP, Widlund Y, Hassan M, et al. (2015). Transplanted Bone Marrow-Derived Cells Contribute to Human Adipogenesis. Cell Metab 22, 408–417. [DOI] [PubMed] [Google Scholar]
- Sárvári AK, Van Hauwaert EL, Markussen LK, Gammelmark E, Marcher A-B, Ebbesen MF, Nielsen R, Brewer JR, Madsen JGS, and Mandrup S (2021). Plasticity of Epididymal Adipose Tissue in Response to Diet-Induced Obesity at Single-Nucleus Resolution. Cell Metab 33, 437–453.e5. [DOI] [PubMed] [Google Scholar]
- Satija R, Farrell JA, Gennert D, Schier AF, and Regev A (2015). Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol 33, 495–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebo ZL, and Rodeheffer MS (2019). Assembling the adipose organ: adipocyte lineage segregation and adipogenesis in vivo. Development 146, dev172098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spallanzani RG, Zemmour D, Xiao T, Jayewickreme T, Li C, Bryce PJ, Benoist C, and Mathis D (2019). Distinct immunocyte-promoting and adipocyte-generating stromal components coordinate adipose tissue immune and metabolic tenors. Sci. Immunol 4, eaaw3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM 3rd, Hao Y, Stoeckius M, Smibert P, and Satija R (2019). Comprehensive Integration of Single-Cell Data. Cell 177, 1888–1902.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Zeve D, Suh JM, Bosnakovski D, Kyba M, Hammer RE, Tallquist MD, and Graff JM (2008). White fat progenitor cells reside in the adipose vasculature. Science 322, 583–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran K-V, Gealekman O, Frontini A, Zingaretti MC, Morroni M, Giordano A, Smorlesi A, Perugini J, De Matteis R, Sbarbati A, et al. (2012). The vascular endothelium of the adipose tissue gives rise to both white and brown fat cells. Cell Metab 15, 222–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang QA, Tao C, Gupta RK, and Scherer PE (2013). Tracking adipogenesis during white adipose tissue development, expansion and regeneration. Nat. Med 19, 1338–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolock SL, Lopez R, and Klein AM (2019). Scrublet: Computational Identification of Cell Doublets in Single-Cell Transcriptomic Data. Cell Syst 8, 281–291.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu YJ, and Rheinwald JG (1981). A new small (40 kd) keratin filament protein made by some cultured human squamous cell carcinomas. Cell 25, 627–635. [DOI] [PubMed] [Google Scholar]
- Ye R, Wang QA, Tao C, Vishvanath L, Shao M, McDonald JG, Gupta RK, and Scherer PE (2015). Impact of tamoxifen on adipocyte lineage tracing: Inducer of adipogenesis and prolonged nuclear translocation of Cre recombinase. Mol. Metab 4, 771–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G, Wang L-G, Han Y, and He Q-Y (2012). clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16, 284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zangi L, Oliveira MS, Ye LY, Ma Q, Sultana N, Hadas Y, Chepurko E, Später D, Zhou B, Chew WL, et al. (2017). Insulin-Like Growth Factor 1 Receptor-Dependent Pathway Drives Epicardial Adipose Tissue Formation After Myocardial Injury. Circulation 135, 59–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B, Ma Q, Rajagopal S, Wu SM, Domian I, Rivera-Feliciano J, Jiang D, von Gise A, Ikeda S, Chien KR, and Pu WT (2008). Epicardial progenitors contribute to the cardiomyocyte lineage in the developing heart. Nature 454, 109–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Single-cell RNA-seq data has been deposited to the Single Cell Atlas at the Broad Institute https://singlecell.broadinstitute.org/. Mouse adipose SVF data is available here: https://singlecell.broadinstitute.org/single_cell/study/SCP708/mouse-adipose-stromal-vascular-fraction.
All code to reproduce clustering data may be found on GitHub: https://github.com/rosen-lab/sc-svf-nts.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


