Abstract
The coronaviral nonstructural protein 9 (Nsp9) is essential for viral replication; it is the primary substrate of Nsp12’s pseudokinase domain within the viral replication transcription complex, an association that also recruits other components during different stages of RNA reproduction. In the unmodified state, Nsp9 forms an obligate homodimer via an essential GxxxG protein-interaction motif, but its ssRNA-binding mechanism remains unknown. Using structural biological techniques, here we show that a base-mimicking compound identified from a small molecule fragment screen engages Nsp9 via a tetrameric Pi-Pi stacking interaction that induces the formation of a parallel trimer-of-dimers. This oligomerization mechanism allows an interchange of “latching” N-termini, the charges of which contribute to a series of electropositive channels that suggests a potential interface for viral RNA. The identified pyrrolo-pyrimidine compound may also serve as a potential starting point for the development of compounds seeking to probe Nsp9’s role within SARS-CoV-2 replication.
Keywords: SARS-CoV-2, Nsp9, replicase, fragment screen, oligonucleotide binding
Abbreviations: MA, mouse-adapted; Nsp9, nonstructural protein 9; RdRp, RNA-dependent RNA polymerase; RTC, replication/transcription complex; STD, saturation transfer difference
SARS-CoV-2 is the causative agent of the disease COVID-19, a coronavirus whose introduction into the human population and subsequent global spread has inundated health care systems worldwide (1, 2). Coronaviral genomes consist of a large ~30 kb messenger-sense single-stranded RNA that reproduces inside of viral-induced membrane vesicles within host cells (3). Replication occurs as a continuous process alongside a coronaviral-unique discontinuous translation mechanism that also produces subgenomic ER-targeted transcripts (4). Following infection two main classes of coronaviral proteins are translated by host ribosomes: virion structural proteins are encoded by the subgenomic transcripts while nonstructural proteins, or Nsps, are transcribed directly from the genome as parts of an autoproteolytic polyprotein. The Nsps are responsible for replication of SARS-CoV-2 and are considered an obvious point of therapeutic intervention (5). The RNA-dependent RNA polymerase (RdRp) is contained within Nsp12, which in cocomplex with the accessory factors Nsp7 and Nsp8 (6, 7, 8) serves as the core of the viral replication/transcription complex (RTC) (4, 9). In vitro Nsp7, 8 and 12 are sufficient for a basal level of RdRp activity (5); however, replication of the full-length genome and its templating of discontinuous subgenomic transcription is further facilitated by a helicase (Nsp13) (9, 10) and Nsp9 (11, 12).
Coronaviral Nsp9 was originally reported to be an ssRNA-binding protein that forms homodimers in solution with modest affinity for long oligonucleotides (13, 14). A recent cryo-electron microscopy structure of the extended form of the SARS-CoV-2 RTC has also shown that the homodimeric interface of Nsp9, which predominates in solution in vitro (15), can be repurposed to directly interface with the nucleotidyltransferase, or NiRAN, domain of Nsp12 (16). When doing so, the N-terminus of Nsp9 inserts fully into the enzymatic site of the domain to interface with the nucleotide therein while potentially recruiting Nsp10 and Nsp14 (17). This interaction occurs at a distance from the active center of the RNA polymerase domain. NiRAN domains are found throughout the nidoviral family, and their nucleotidylation activity is known to be essential (18), but their precise function is still under study. At least two hypotheses have been proposed for the Nsp9:Nsp12 interaction: Yan et al. (16) suggest a role as nucleotidylation inhibitor during mRNA-cap formation, whereas Slanina et al. (19) propose that Nsp9 itself is the major substrate for the reaction. If the latter modified form of Nsp9 is proven to predominate under physiological conditions, then the posttranscriptionally modified Nsp9 could directly base pair with nucleotides, and earlier RNA-binding studies would need to be revisited. As well as its RTC-associated roles, Nsp9 together with Nsp8 may bind to discrete regions of 7SL RNA to modify host protein trafficking (20).
To aid the characterization of Nsp9COV19, we had previously obtained the structure of its homodimeric form (21). The analogous protein of SARS-CoV-1 shares 98% sequence identity and is known to be essential for viral replication (22), we thus sought to identify compounds with affinity for Nsp9COV19 that could potentially be developed into inhibitors. The integrity of the conserved self-associating GxxxG sequence motif within the C-terminal α-helix of Nsp9 is known to be essential (15). The glycines of this motif occur on the same face of the dimerization helix and closely assemble with their counterparts within the homodimer to yield a tight parallel helical coupling, they represent an obvious surface of the protein to target for inhibition.
This coronaviral Nsp9 dimer is a virally unique fold, which diverges so significantly from other nucleotide binding proteins that potential ssRNA-binding modes cannot be easily deciphered. Herein we identify the residues within Nsp9 COV19 that are chemically perturbed upon ssRNA binding. We then describe the identification of a base-mimetic compound that binds weakly to Nsp9COV19 via a tetrameric π-π stacking mechanism; this induces formation of a hexameric particle that utilizes the same perturbed residues as part of its oligomerization mechanism. The low-affinity compound detailed herein represents a potential starting point for structure-based inhibitor design. Its mode of association with Nsp9COV19 suggests a potential mechanism for single-stranded nucleotide binding within this conserved family of coronaviral proteins.
Results
Identification of residues involved in ssRNA binding by HSQC-NMR
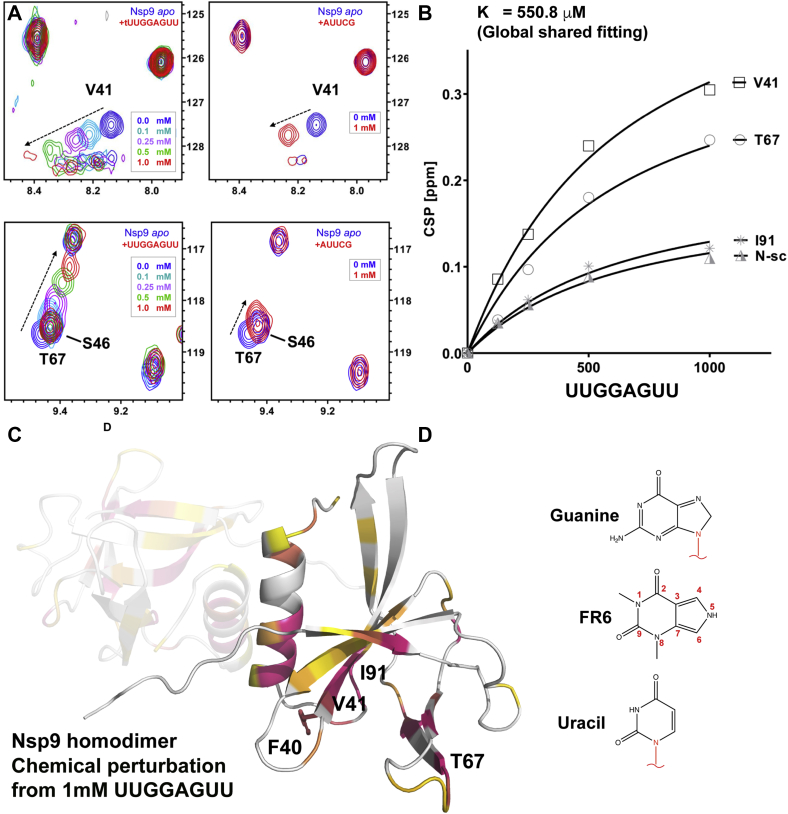
We aimed to identify small-molecule compounds that bind within or near to the ssRNA binding site of Nsp9COV19, but first needed to better characterize the site itself. To do so, we sought to identify the residues involved in oligonucleotide binding using 2D [15N-1H]-HSQC NMR from a uniformly 15N-isotope-labeled protein. Recombinant Nsp9COV19 in isolation yielded well-defined amide cross-peaks with similarities to the previously assigned Nsp9SARS homologue (97% sequence identity) allowing us to tentatively assign the majority of theoretical backbone amide resonances with good confidence (Biological Magnetic Resonance Data Bank ID 6501, Fig. S1A). Our initial assignments agreed with those published elsewhere (23, 24). [15N-1H]-HSQC NMR spectra were then acquired in the presence of short 5 nt and 8 nt ssRNA oligonucleotides. Several amide resonances were observed to undergo concentration-dependent chemical shift perturbations upon titration of 0 to 1000 μM ssRNA with larger shifts observed for the longer oligonucleotide (Fig. 1A and Fig. S1B). Although most peaks remained unperturbed, ssRNA induces large chemical shifts for the backbone amide peaks assigned to Val-41, Thr-67, and Ile-91 (Fig. 1, B and C) as well as a large-scale shift of unassigned sidechain amide peaks (Fig. S1C). Smaller-scale movements were also observed for residues 40, 42, 63, 64, 68, 98, and the point-of closest approach within the GxxxG helices (residues 100–102).
Figure 1.
Identification of residues involved in ssRNA binding and the FR6-binding site in close apposition.A, following peak assignment of the 1H-15N 2D HSQC NMR Nsp9COV19 spectra several peaks are seen to undergo concentration-dependent chemical shift perturbations from their unliganded positions (blue) upon titration of different concentrations of short ssRNA oligomers. B, an approximate KD can thus be obtained for a global shared fitting analysis from the movements of the most perturbed peaks (Val-41, Thr-67, Ile-91, and an unidentified Asn side chain). C, the largest chemical shift perturbations with a white to pink gradient can be mapped onto the crystal structure of Nsp9COV19 solved previously. The positions of the residues used in global fitting are labeled. One subunit of the homodimer is partially transparent. D, chemical structures of guanine, FR6, and uracil.
A global shared fit of the chemically perturbed cross-peaks (Fig. 1B) allowed an estimation of the KD for these short 8 nt ssRNA of ~550 μM (95% confidence between 410 and 750 μM). At the highest ssRNA concentration (red peak in top left panel of Fig. 1A), the resonance also peaks and begins to broaden and reduces in intensity. Thus, two distinct areas of Nsp9 show chemical perturbation in the presence of ssRNA: one corresponding to changes within the homodimer interface and a second site, distinct from the dimer interface that may be involved in nucleotide binding.
Screening for Nsp9COV19 binding compounds via [15N-1H]-HSQC-NMR
After identification of a potential RNA-binding site Nsp9COV19, we next sought to identify small-molecule compounds that might associate at, or nearby, to said site. Our initial stratagem for obtaining such candidates consisted of screening a small fragments library (25) against the recombinant Nsp9COV19 by recording 1D-1H saturation transfer difference NMR on mixtures of five fragments to assess binding. Of 450 fragments, a shortlist of 48 initial hits resulted from this screening process (Fig. S2A). The binding of these fragments was validated by recording [15N-1H]-HSQC NMR for each. Notably several heterocyclic compounds from the initial shortlist held obvious similarities to RNA bases. One such molecule is 1,3-dimethyl-6H-pyrrolo[3,4-d]pyrimidine-2,4-dione, hereafter referred to as FR6 (Fig. 1D). HSQC measurements showed that titration of FR6 with the protein perturbed Phe-40 (Fig. S2B). The 1D 1H spectra of FR6 implied a potential for it to aggregate in solution at high concentrations (>4 mM). Several residues including Phe-40 undergo chemical perturbations upon titration of RNA with Nsp9COV19.
The FR6::Nsp9COV19 cocomplex
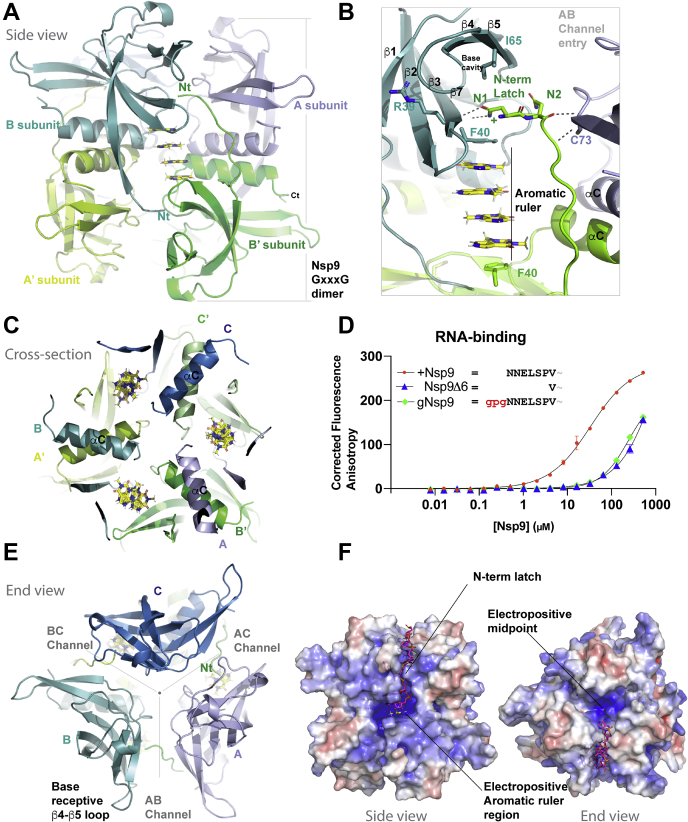
As FR6 is an RNA base-mimetic sans backbone, we theorized it could potentially provide a means to elucidate the molecular interactions that mediate ssRNA binding within Nsp9. In prior studies we had observed a tendency for recombinant Nsp9COV19 crystals produced with extraneous N-terminal tag residues to grow more robustly and diffract to higher resolution than if such residues were removed. We cocomplexed Nsp9COV19 with FR6 in a 5:1 M ratio and identified new crystallization conditions. The FR6::3c-Nsp9COV19 crystals diffracted to 1.58 Å in space group P3221 (see Table S1) with the asymmetric unit containing three molecules of Nsp9COV19 (A–C protomers) (Fig. S2C and Fig. 2A). Clear electron density is observed for all Nsp9COV19 residues, which adopt a largely identical structure to that observed previously: that of a small 7-stranded β-barrel with elongated β1-strand and β6-β7 loop that each protrude to engage opposite faces and ends of the GxxxG dimerization helix. All three protomers have near-identical Cα-backbone conformations except within the β4-β5 loop (residues 57–66). The differences in this loop’s structure within the B-protomer are detailed later.
Figure 2.
The FR6-induced Nsp9COV19hexamer.A, the arrangement within the FR6-bound hexameric form of Nsp9COV19 is displayed in cartoon representation, one ring of protomer subunits uses green hues and the equivalent symmetry-related protomers use blue ones. In this side view, one-third of the hexamer is in the background. B, a closer view of the aromatic ruler region indicating the binding mode of the tetrameric FR6 stacking and the proximity of the N-terminal latch residues and open AB gate entry to the end-cap channel network. C, cross-sectional view of the three π-π stacks within the hexameric form of Nsp9COV19, the αC-dimer helices are labeled for the six subunits. D, fluorescence polarization anisotropy assays using 17mer PolyU to assess ssRNA binding of different recombinant Nsp9 constructs. Constructs correspond to: gNsp9—protein with an additional GPG stub at the N-terminus; +Nsp9—protein with a free backbone amide at Asn-1 and Nsp9. The plot shows corrected anisotropy for each Nsp9COV19 protein concentration; error bars represent the SD from the mean of triplicate measurements. E, top-down view of the Nsp9COV19 hexamer shows the end-cap channel network along with open and closed conformations of β4β5 entry gate. F, representation of the hexamer with the calculated electrostatic surface potential shown in context with the modeled ssRNA fragment. The aromatic ruler and end-cap channel network is labeled.
Within the FR6 complex, crystallographic symmetry operators produce the canonical coronaviral Nsp9 homodimer (14); this protein–protein interaction mechanism accommodates a degree of orientational flexibility about the self-associating GxxxG motifs (21). In the complex, the Cα-backbone structure of the protein aligns most closely to that of gNsp9COV19 form solved previously (pdb: 6WXD, r.m.sd 1.04 Å2), but the respective subunit orientation aligns more closely with that of 3c-Nsp9COV19 (pdb: 6WC1 r.m.sd 1.16 Å2). Overall, Nsp9COV19 retains its structure upon FR6 binding.
FR6 binds at protein proximal and distal sites
Following refinement of the protein residues, extraneous electron density within the omit maps clearly allowed precise positioning of two copies of FR6 per molecule of Nsp9COV19 (Fig. S2D). Binding occurs via a parallel-displaced π-π stacking mechanism with FR6 occupying two distinct sites, one proximal to the protein aligned atop the aromatic ring of Phe-40 (Fig. 2B) and a second distal site parallel to this (Fig. S2E). The proximal binding site is situated between the N-terminus of the GxxxG helix and the nearby β2-β3 loop, a cluster of asparagine and hydrophobic residues serve to interface with the functional groups of the FR6 pyrimidinedione ring. The ligand’s C8 carbonyl group forms a hydrogen bond to the side chain of Asn-98 alongside a water-mediated interaction with the backbone amide of Asn-95. Nearby, the other FR6 carbonyl group hydrogen bonds with the side chain of Asn-33. This proximal binding site engages only one of the ligand’s methylamine groups (via Leu-42 and Leu-94), the other remaining largely solvent exposed.
The electron density of FR6 has potential pseudosymmetry due to similar electron scattering from the carbonyl and methyl groups, hydrogen bonds formed with the protein help orient FR6 within the proximal site, but a lower occupancy form flipped about the FR6 pseudosymmetry axis may also be present at the distal site. FR6 binds at two sites within Nsp9COV19 at the base of the GxxxG helices.
FR6 is a base mimetic with a tetrameric engagement of a Phe-40 aromatic ruler
The pyrimidinedione ring of FR6 bears obvious similarities with that of uracil but also has a heterocyclic structure analogous in form to purine nucleotides. Thus, it is noteworthy that ssRNA and FR6 both result in changes to the local backbone amide environment near Phe-40/Val-41 (Fig. 1A and Fig. S2B). The backbone of Val-41 is overlayed by FR6 (Fig. 2B), and its binding results in two new interactions with its backbone amide. As titration of ssRNA into Nsp9COV19 induced chemical shifts in residues near the FR6-binding site, we next sought to assess whether by acting as a base-proxy whether its association with the protein provided potential information about nucleotide-binding. Although two molecules of FR6 associate with each protein molecule, the pseudosymmetric arrangement of the asymmetric unit’s three protomers converts this into a π-π stack of four aromatic FR6 ligands within a hexameric Nsp9COV19 particle (Fig. 2A). Three copies of the canonical Nsp9 homodimer are seen to align such that their C-terminal α-helices orient toward a central point. The aromatic FR6 stacking has a 14.5° parallel-offset and occurs at each neighboring homodimer interface (Fig. 2, A and B). This is most easily visualized in cross section at the hexamer’s mid-point where the three tetrameric π-π stacks interleave between the αC helical pairings (Fig. 2C); residues within this structural element’s N-termini serve to provide the proximal binding site for each end of the aromatic stacking (Fig. 2B).
The tetrameric π-π ligand stacks are delimited at either side through similar interactions with the side chain of Phe-40 (Fig. 2B), a bookended interface that provides a 4-base aromatic ruler at points throughout the hexamer. While the distal FR6-binding site is relatively open, the tip of the β2-β3 loop serves to cover each end’s protein-proximal site. If nucleotides bind within the same site with bases aligned to FR6, the latter’s solvent-exposed methylamine groups would occupy equivalent positions to that of the β-N1-glycosidic bond. Thus, the necessary presence of the ssRNA phosphate-backbone in physiological substrates is not incompatible with our putative base-binding mode inferred from the FR6 complex. If the observed aromatic ruler within our structure also engages RNA, then artificial tag residues occupy the expected position of the phosphate backbone (Fig. S2F). A free Asn-1 amide may therefore influence nucleotide binding in Nsp9.
ssRNA fluorescence polarization anisotropy
We, and others, have performed fluorescence polarization anisotropy experiments to monitor nucleotide binding to Nsp9COV19 (21). The construct we had used previously retained artificial residues −2Gly-Pro-Gly0 prior to Nsp9’s N-terminal sequence 1NNELSV, a common occurrence with standard recombinant techniques. To determine whether a free amide within Nsp9COV19 influences nucleotide binding, we recloned Nsp9COV19 from its prior form (gNsp9) such that no artificial residues remained after his-tag removal (+Nsp9) along with a second construct lacking the first six residues (Nsp9Δ6). In vitro RNA-binding assays were then repeated with all three constructs, as reported previously gNsp9 displayed relatively weak binding to ssRNA with a Kd of ~200 ± 50 μM, while the fit for the anisotropic data for Nsp9Δ6 provided a Kd within the mM range. In contrast, the ssRNA anisotropy binding data for +Nsp9 suggested a tenfold increase in affinity for this version of the protein (Kd ~ 33 ± 3 μM) (Fig. 2D).
A cyclic exchange of latching Nsp9 N-termini stabilizes hexamerization
The Nsp9 hexamer has two adjacent rings of subunits (blue or green color hues in Fig. 2A) with the canonical homodimer comprised of one from each ring. The homodimer’s αC helices provide a highly stable, yet somewhat dynamic, interring connection. This contrasts with the subunit arrangement around each ring that is less direct and appears stabilized in equal measure by interactions mediated through the aromatic ligands but also utilizes a cyclic exchange of subunit N-termini across neighboring dimers (Fig. 2A). In this latching mechanism, the first two residues of Nsp9 extend to lie between subunits of the opposing end ring (Fig. 2B). In doing so the side chain of Asn-1 forms two new hydrogen bonds to the amide backbone of Val-41. This “latch-engaged” conformation of Asn-1 lies above the benzyl group of Phe-40 further stabilizing the aromatic ruler. Binding of RNA-base like compounds to Nsp9COV19 directly influences Phe-40 and Asn-1 is proximal to the site.
Electropositive channels encircle the surface of the Nsp9COV19 hexamer
If the hexameric form of Nsp9 is also used to bind to RNA, an elongated interaction site would be required capable of accommodating aromatic bases and the phosphoribose backbone. At each endcap of the Nsp9 hexamer, a 6 to 8 Å wide tri-forked channel connects the three FR6 ligand stacks (Fig. 2E). The base of the channel network is created by the residues from the β5 and β6 strands aligning with their counterparts from adjoining end-ring subunits. The first layer of the channel’s walls is then formed by the β3-β4 loop. Access between the endcap channel network and the aromatic ruler occurs over the N-terminal latch (top right Fig. 2B). To assess whether this site could accommodate ssRNA, we made electrostatic calculations for the hexameric particle, which indicate a concentration of positive charge both near the aromatic ruler and throughout the end channel network (Fig. 2F). Within the former, Lys-36, Lys-58, Arg-74, and Asn-1 provide the potential for charge-mediated interactions near to the channel entry gate, while Lys-52, Lys-92, and the dipole moment of a short helical turn (residues 19TQTAC23) present similar contributions within the end channels. Potential hydrophobic sites for base association within the end cap are presented via the side chains of Trp-53, Ile-65, Tyr-66, and Pro-71. Large channels exist within the Nsp9 hexamer that might be able to accommodate oligonucleotides.
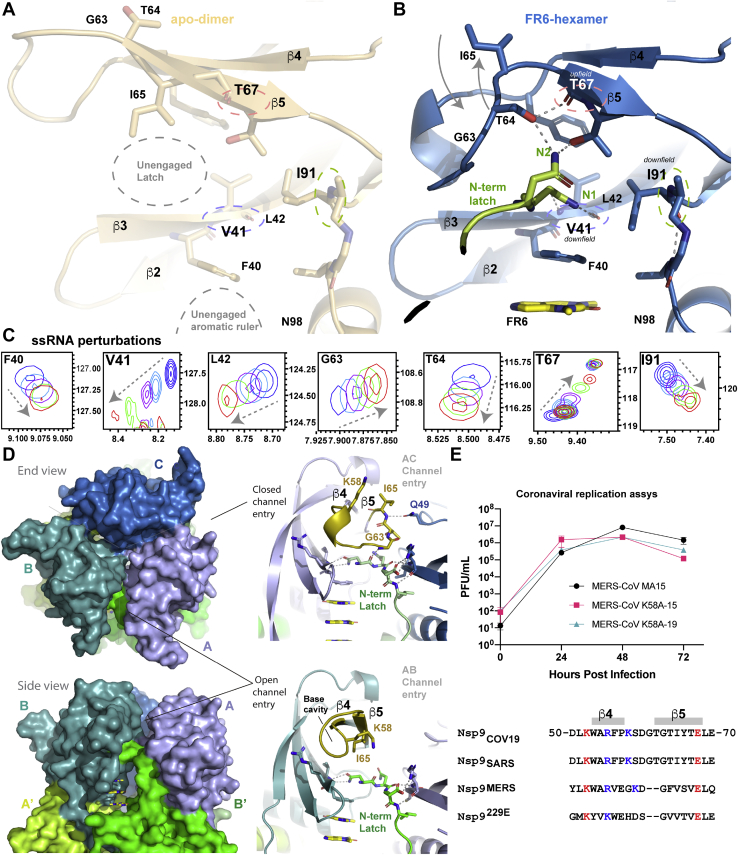
The HSQC peaks perturbed upon addition of ssRNA make up the N-terminal latch
The major peaks displaying chemical perturbation within the HSQC spectra upon addition of ssRNA were Val-41, Thr-67, Ile-91, and unassigned side chain amides. The first three of these residues make up the N-terminal latching mechanism of the FR6-hexamer (Fig. 3, A–C). Self-association of the viral protein results in two new hydrogen bonds between N-1 and the backbone amide of Val-41 upon FR6 binding (Fig. 3B), which would be consistent with its downfield shift (Fig. 3C). In contrast, Thr-67 undergoes an upfield shift potentially arising from a hydroxyl association with its carbonyl group (Fig. 3B). Ile-91 is part of an unusual i + 2 β-turn whose movements may be stabilized when the N-terminal latch associates with its side chain (Fig. 3B). The side chains of Asn-1 and -2 each form new hydrogen bonds upon hexamerization so are potential candidates for the unassigned amides perturbed by ssRNA.
Figure 3.
Chemical shift perturbations observed in HSQC spectrum upon ssRNA binding align with latch-binding residues.A, a structural view of the unengaged latch of Nsp9COV19 within its apo state in which the β4–β5 loop is extended, backbone amides of the three most perturbed peaks upon ssRNA-binding are circled. B, the equivalent region of the protein within the N-latch engaged FR6-hexamer is shown for the C protomer. The new hydrogen bonds formed to V41, T67, and I91 in the hexameric state are highlighted. C, for the residues depicted in panels A and B expanded regions of the 1H-15N 2D HSQC spectra following titration of oligonucleotide into Nsp9COV19 are shown. Coloring is consistent with Figure 1C and the direction of perturbation is highlighted. D, the asymmetric form of the Nsp9COV19 hexamer results in open and closed gates to the end-cap channel network via restructuring of the β4-β5 loop over the N-terminal latches (colored gold). Both states have two hydrogen bonds to Val-41. E, coronaviral replication assay comparing the replication rate of a wild-type MERS-CoV virus with two constructs bearing a K58A mutation up to 72 h post infection using a plaque forming unit assay. Sequence conservation within the β4–β5 loop is shown.
A gating mechanism control entry to the potential nucleotide-binding channels
If the two end-cap channels accommodate nucleotide polymers in the liganded state, then the symmetry of the hexamer will be broken upon binding; that is, all three branches of each tri-forked channel cannot be simultaneously occupied. It is thus noteworthy that within our asymmetric unit, two alternate conformations exist for the β4-β5 loop at the sites of channel entry (residues 57–65, Fig. 3D). That of the B-protomer (bottom Fig. 3D) aligns closely with the predominant form in unliganded Nsp9; in this state Ile-65 is orientated toward the N-terminal latch and together with the alkane core of Arg-39 presents an unoccupied hydrophobic cavity above the latching mechanism (Fig. 2B). This potentially base-receptive form of the β4-β5 loop is present at the AB channel entry gate of the hexamer (Fig. 3D). In contrast, the novel form of the β4-β5 loop occurs within the A and C protomers and makes the other end-cap channel entries more compact. In these positions, the Asn-1 latch is further stabilized by Gly-63 overlaying it (Fig. 3, B and D). In this closed β4-β5 loop conformation, both β-strands are reduced in extent with Ile-65 flipping to present in a solvent-exposed end-cap direction where it forms a hydrogen bond with the side chain of Gln-49 from another end-ring subunit (Fig. 3D).
Positively charged residues line the β4-β5 gate loop (e.g., Lys-58) and are obvious candidates for nucleotide interacting side chains. We had serendipitously made two independent MERS-CoV viral isolates in which Lys-58 (K58) was mutated to alanine. Assuming that the degree of conservation between Nsp9MERS and Nsp9COV19 is consistent with a common functional mechanism, we thus performed a viral replication assay to compare MERS-CoV with a MERS-CoV K58A mutant virus (Fig. 3E). While there was no discernible difference in plaque-forming units at 24 h postinfection, by 72 h an approximately fivefold difference in viral titre was observed for the K58A mutant, indicating a moderate disadvantage in viral replication for the K58A mutant. Therefore, mutation of residues within the β4-β5 loop has discernible impact upon coronaviral replication and merits further studies in infected tissue culture cells and animals.
Coronaviral Nsp9 sequence conservation within the context of the hexamer
Several coronaviral homologs of Nsp9COV19 structures are known in their unliganded states, which share between 35 and 98% sequence identity (13, 14, 21, 26, 27, 28). When their primary sequences are aligned (Fig. S3A) and invariant residues mapped onto the dimeric structure of Nsp9, it is difficult to discern between potential ligand interacting sites and regions of the protein critical for adopting the fold (Fig. S3B). If the same visualization is used within the context of the hexamer (Fig. S3, C and D), residue conservation appears to be concentrated upon N-terminal residues and those making up the base and walls of the end-channel network. Oligomerization thus converts conserved regions of the protein from convex surfaces within the dimer (Fig. S3, A and B) to interdependent concave ones with more obvious potential for ligand interaction (Fig. S3, C and D). Within the FR6-binding site, the hydrophobic nature of Phe-40 is maintained but its aromaticity is not strictly conserved. The N-terminal latch residues also play a dual role in NiRAN domain association.
Discussion
The conserved coronaviral Nsp9 homologues are single-stranded RNA-binding proteins whose unliganded structures had been determined from a number of viral species (14) including SARS-CoV-2 (21). The Nsp9 fold comprises a structure that is clearly distinct from previously studied nucleotide binding proteins making any potential oligonucleotide association mechanism unclear. NMR titration assays indicate several residues within Nsp9 appear to be perturbed upon RNA binding including Phe-40, Val-41, and Ile-91. These residues or those nearby are likely to directly associate with nucleotide. Furthermore, we identify 1,3-dimethyl-6H-pyrrolo[3,4-d]pyrimidine-2,4-dione as a low-affinity ligand that interacts near the putative nucleotide-binding site of Nsp9COV19.
The binding of our purine-like compound induced a hexameric form of Nsp9COV19 that was intriguing but whose relevance is unclear. The residues undergoing chemical perturbation upon oligonucleotide binding map to an intersubunit N-terminal latching mechanism within the hexameric Nsp9 assembly. Moreover, our fluorescence polarization anisotropy assays indicated that a free N-terminus within the protein increased its affinity for oligonucleotide. If a hexameric form of Nsp9 is physiologically relevant, the proximity of the electropositive end-cap channel network to the aromatic ruler suggests potential paths for ssRNA winding. At least two different wrapping topologies are apparent that would be compatible with the presented structure, each utilizing both the aromatic ruler region and the channel network in different ways (Fig. S4). Our experiments support the role of both the proximal and distal FR6 binding sites being occupied upon oligonucleotide binding and leave sufficient room for the phospho-ribose backbone. In the unliganded state residues 1 to 8 are often flexible while in the hexamer they reside next to the aromatic ruler and channel gate inducing oligomerization and oligonucleotide binding. If Nsp9’s association with RNA induces hexamerization in solution, the intersubunit interface at the base of the GxxxG helices is likely to involve a degree of cooperativity during the oligomerization process.
Our potential interpretation of the hexameric Nsp9 assembly is complicated by the recent identification of Asn-1 as the primary substrate for Nsp12’s nucleotidylation activity (19). Such a modification would result in an RNA base posttranscriptionally added to the protein at Asn-1. This modification could potentially also be accommodated within the FR6 proximal site. An alternative explanation for the formation of a hexameric Nsp9 assembly may then be as a storage particle for UMP-modified Nsp9COV19, phosphoramidite P-N bonds are known to be relatively reactive. It is worth noting that UMP modification of Nsp9 may also be linked to its nucleotide-binding role (19). Further work is required to identify how prevalent this form of Nsp9 is within coronaviruses but also which oligomeric state of Nsp9 predominates in solution under different conditions.
Single-stranded nucleotide-binding proteins often provide a number of aromatic-binding pockets, albeit ones that are not as deep or reactive as those normally targeted for drug development. Prior work has gone into developing small molecules that target nucleotide-binding proteins in order to dissect their role in genome replication (29) or as bacterial antibiotic targets (30). The essentiality of Nsp9 for SARS-CoV-2 replication is intriguing (15, 19) and presents a novel opportunity for antiviral development targeting its role within the replication transcription complex (16). The dual-site binding mode of the FR6 fragment to Nsp9COV19 poses challenges with respect to modification of its framework to increase affinity; that is, at present, any modification must be accommodated by two distinct sites. Nonetheless, this compound retains some potential for development with further work required to better define the nucleotide-binding sites of Nsp9COV19. Characterization of the FR6 binding mode helps this effort.
Experimental procedures
Protein production
The maturation of Nsp9 from the Orf1ab polyproteins requires the enzymatic action of the main protease. To produce recombinant Nsp9COV19 for enzymatic assays with a physiological N-terminus, the synthetic gene for Nsp9COV19 was fused to yeast Smt4 sequence between the hexahistidine tag and viral Nsp9 sequence. Protein was produced as before except affinity tag removal was performed using an overnight digestion with Ulp1 protease to yield +Nsp9. Prior to gel filtration purification, cleaved +Nsp9 was separated from the Smt4 sequence through passage over a Hi-trap SP column (GE healthcare), protein eluting under a 0 to 1 M NaCl gradient was then passed over a nickel column and the flow-through subject to gel filtration where it eluted as a single peak. Synthetic genes were ordered for the Nsp9Δ6 mutant described in text.
Crystallization screening also included constructs we had made previously, now renamed gNsp9COV19 and 3c-Nsp9COV19 (21). These retain the artificial residues GPG at the N-termini post cleavage of hexahistidine residues.
Isotope-labeled Nsp9COV19 was purified in an identical manner except that E. coli (DE3) transfected with pET-28-NKI-Nsp9COV19 was grown instead in M9 minimal media containing 15N-labeled ammonium chloride (Merck #299251).
ssRNA oligonucleotides and fluorescence polarization assays
RNA oligonucleotides were purchased from IDT with the following sequences RNA.1(AUUCG) and RNA.2 (UUGGAGUU). Fluorescence anisotropy assays were performed as reported previously (21). Data were measured in triplicate with errors representing the 95% confidence interval obtained from a nonlinear model derived from the program PRISM 8.3.1.
1D-1H ligand saturation transfer difference (STD) NMR spectroscopy
A pilot screen of 90 mixtures containing 450 fragments was screened against 10 μM Nsp9COV19 using 1D-1H STD NMR. NMR experiments were carried out on a Bruker 600 MHz spectrometer equipped with a CryoProbe at 293 K. STD spectra were processed and analyzed by MNova as reported previously (31). A hit rate of 11.8% was observed. The chemical structures of all identified hits are shown in Fig. S2A.
Backbone chemical shift assignments of Nsp9COV19
Chemical shift assignments (1HN,15N,13Cα, and side chain 13Cβ chemical shift assignments) of Nsp9-CoV-1 are available in the Biological Magnetic Resonance Bank (BMRB) with accession numbers 6501. We expressed and purified a uniformly 15N-labeled Nsp9COV19. A 2D TROSY [15N-1H]-HSQC spectrum of Nsp9COV19 was collected in identical buffer and temperature, as reported previously for Nsp9SARS. Overlaid HSQC spectra of Nsp9COV19 and Nsp9SARS are shown in the Fig. S1A. Although the overall chemical shift dispersion pattern between the two spectra is similar, significant chemical shift differences were observed between them for a set of residues. We rationalize that this might be due to the two local residues differences (Nsp9COV19 N34T and Nsp9COV19 H48L) between the two proteins. Backbone amide resonances that show little or no chemical shift perturbations between the spectra are considered for the ligand-binding analysis for Nsp9COV19. 2D HSQC-NMR spectra were processed by Topspin3.6 and analyzed by the software CARA.
Crystallization, data collection, and refinement
3C-Nsp9COV19:FR6 crystals were grown with a three- to five-fold molar excess of compound in the presence of ~2 mM protein. Data were obtained from crystals grown in 2.5 to 3 M sodium malonate and 0.1 M sodium citrate pH 4. Crystals were flash-frozen in liquid nitrogen after being soaked in a cryoprotectant consisting of 1.5 M sodium malonate, 0.1 M sodium citrate 25% v/v glycerol. Diffraction data were collected at the Australian synchrotrons MX1 and MX2 beamlines (32). Data were processed using the program XDS (33), scaled and merged with programs from the CCP4 suite (34). Initial phases were obtained using the molecular replacement program Phaser (35) with trimmed starting model 6W9Q (21). Subsequent rounds of manual building and refinement were performed in the programs COOT (36) and Phenix (37).
MERS-CoV replication assays
Generation of MERS-CoV K58A mutant and replication assays
The Nsp9 K58A mutation was introduced into mouse-adapted (MA) MERS-CoV BAC via a two-step linear lambda red recombination system utilizing an I-SceI homing endonuclease as described in Fehr, 2020 (38). Briefly primers overlapping MERS-CoV Nsp9 containing the K58A mutation and overlap with Kanamycin containing the I-SceI site were used to generate a kanamycin fragment containing overhangs to MERS-CoV Nsp9 that contain the K58A mutation. The fragment was electroporated into GS1783 E. coli cells containing the MA MERS-CoV BAC allowing for homologous recombination. Proper recombination was selected via replicate on LB agar containing kanamycin and chloramphenicol antibiotics. Recombined colonies were treated with arabinose for induction of the I-SceI enzyme. The culture was heat shocked to induce RED recombination effectively removing the fragment containing kanamycin and producing a scarless MA MERS-CoV BAC containing the K58A mutation. Virus was rescued via transfection of Huh-7 cells with 1 to 2 μg of the K58A MERS-CoV BAC and 10 μl lipofectamine 3000 in opti-MEM media. In total, 3 to 4 days post transfection cells and supernatant were collected and virus was propagated on Huh-7 cells to generate stocks and was tittered on Vero81 cells. Growth curves were performed by infecting Huh-7 cells at an MOI of 0.01 of MA MERS-CoV (EMC/2012) or MA MERS-CoV strain bearing the K58A mutation. Virus was collected at 24, 48, and 72 h post infection and freeze thawed. Virus was titered on Vero81 cells. Following adsorption for 1 h, virus inocula were removed and plates were overlaid with 0.6% agarose containing 2% FBS and DMEM supplemented with 10% FBS. After 3 days, agar overlays were removed, fixed with 4% PFA, and stained with 0.1% crystal violet. Viral titers were calculated as pfu/ml. All work with infectious MERS-CoV was performed in a BSL3 laboratory.
Data and code availability
The accession number for the atomic coordinates of the 3C-Nsp9COV19:FR6 and associated structure factors have been deposited at the protein databank (www.rcsb.org) with accession code 7KRI.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare no conflict of interest.
Acknowledgments
Funding for the work originated from the Australian Research Council Centre of Excellence for Advanced Molecular Imaging and the Centre for Fragment-Based Design. This research was undertaken in part using the MX2 beamline at the Australian Synchrotron, part of ANSTO, and made use of the Australian Cancer Research Foundation (ACRF) detector. Additionally, we thank Dr Geoffrey Kong of the Monash Molecular Crystallisation Facility for his assistance with crystallographic screening and optimization. We thank A. Riboldi-Tunnicliffe and R. Williamson for assistance with data collection.
Author contributions
D. R. L., M. J. S., and J. R. conceptualization; D. R. L. and S. A. L. formal analysis; S. P., M. J. S., and J. R. funding acquisition; D. R. L., B. M., S. A. L., and R. N. C. investigation; D. R. L. and B. S. G. project administration; B. S. G., S. P., M. J. S., and J. R. supervision; B. S. G. validation; D. R. L. writing—original draft; D. R. L., B. M., S. A. L., R. N. C., B. S. G., S. P., M. J. S., and J. R. writing—review and editing.
Edited by Craig Cameron
Contributor Information
Dene R. Littler, Email: dene.littler@monash.edu.
Martin J. Scanlon, Email: martin.scanlon@monash.edu.
Jamie Rossjohn, Email: Jamie.rossjohn@monash.edu.
Supporting information
References
- 1.Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y., Yuan M.L., Zhang Y.L., Dai F.H., Liu Y., Wang Q.M. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Jiang R.D. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Snijder E.J., van der Meer Y., Zevenhoven-Dobbe J., Onderwater J.J., van der Meulen J., Koerten H.K., Mommaas A.M. Ultrastructure and origin of membrane vesicles associated with the severe acute respiratory syndrome coronavirus replication complex. J. Virol. 2006;80:5927–5940. doi: 10.1128/JVI.02501-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sawicki S.G., Sawicki D.L. Coronaviruses use discontinuous extension for synthesis of subgenome-length negative strands. Adv. Exp. Med. Biol. 1995;380:499–506. doi: 10.1007/978-1-4615-1899-0_79. [DOI] [PubMed] [Google Scholar]
- 5.Yin W., Mao C., Luan X., Shen D.D., Shen Q., Su H., Wang X., Zhou F., Zhao W., Gao M., Chang S., Xie Y.C., Tian G., Jiang H.W., Tao S.C. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science. 2020;368:1499–1504. doi: 10.1126/science.abc1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sola I., Almazan F., Zuniga S., Enjuanes L. Continuous and discontinuous RNA synthesis in coronaviruses. Annu. Rev. Virol. 2015;2:265–288. doi: 10.1146/annurev-virology-100114-055218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Snijder E.J., Decroly E., Ziebuhr J. The Nonstructural proteins directing coronavirus RNA synthesis and processing. Adv. Virus Res. 2016;96:59–126. doi: 10.1016/bs.aivir.2016.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kirchdoerfer R.N., Ward A.B. Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nat. Commun. 2019;10:2342. doi: 10.1038/s41467-019-10280-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen J., Malone B., Llewellyn E., Grasso M., Shelton P.M.M., Olinares P.D.B., Maruthi K., Eng E.T., Vatandaslar H., Chait B.T., Kapoor T.M., Darst S.A., Campbell E.A. Structural basis for helicase-polymerase coupling in the SARS-CoV-2 replication-transcription complex. Cell. 2020;182:1560–1573.e1513. doi: 10.1016/j.cell.2020.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ivanov K.A., Thiel V., Dobbe J.C., van der Meer Y., Snijder E.J., Ziebuhr J. Multiple enzymatic activities associated with severe acute respiratory syndrome coronavirus helicase. J. Virol. 2004;78:5619–5632. doi: 10.1128/JVI.78.11.5619-5632.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brockway S.M., Clay C.T., Lu X.T., Denison M.R. Characterization of the expression, intracellular localization, and replication complex association of the putative mouse hepatitis virus RNA-dependent RNA polymerase. J. Virol. 2003;77:10515–10527. doi: 10.1128/JVI.77.19.10515-10527.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith E.C., Denison M.R. Coronaviruses as DNA wannabes: A new model for the regulation of RNA virus replication fidelity. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Egloff M.P., Ferron F., Campanacci V., Longhi S., Rancurel C., Dutartre H., Snijder E.J., Gorbalenya A.E., Cambillau C., Canard B. The severe acute respiratory syndrome-coronavirus replicative protein nsp9 is a single-stranded RNA-binding subunit unique in the RNA virus world. Proc. Natl. Acad. Sci. U. S. A. 2004;101:3792–3796. doi: 10.1073/pnas.0307877101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sutton G., Fry E., Carter L., Sainsbury S., Walter T., Nettleship J., Berrow N., Owens R., Gilbert R., Davidson A., Siddell S., Poon L.L., Diprose J., Alderton D., Walsh M. The nsp9 replicase protein of SARS-coronavirus, structure and functional insights. Structure. 2004;12:341–353. doi: 10.1016/j.str.2004.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miknis Z.J., Donaldson E.F., Umland T.C., Rimmer R.A., Baric R.S., Schultz L.W. Severe acute respiratory syndrome coronavirus nsp9 dimerization is essential for efficient viral growth. J. Virol. 2009;83:3007–3018. doi: 10.1128/JVI.01505-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yan L., Ge J., Zheng L., Zhang Y., Gao Y., Wang T., Huang Y., Yang Y., Gao S., Li M., Liu Z., Wang H., Li Y., Chen Y., Guddat L.W. Cryo-EM structure of an extended SARS-CoV-2 replication and transcription complex reveals an intermediate state in cap synthesis. Cell. 2021;184:184–193.e110. doi: 10.1016/j.cell.2020.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yan L., Yang Y., Li M., Zhang Y., Zheng L., Ge J., Huang Y.C., Liu Z., Wang T., Gao S., Zhang R., Huang Y.Y., Guddat L.W., Gao Y., Rao Z. Coupling of N7-methyltransferase and 3'-5' exoribonuclease with SARS-CoV-2 polymerase reveals mechanisms for capping and proofreading. Cell. 2021;184:3474–3485.e3411. doi: 10.1016/j.cell.2021.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lehmann K.C., Gulyaeva A., Zevenhoven-Dobbe J.C., Janssen G.M., Ruben M., Overkleeft H.S., van Veelen P.A., Samborskiy D.V., Kravchenko A.A., Leontovich A.M., Sidorov I.A., Snijder E.J., Posthuma C.C., Gorbalenya A.E. Discovery of an essential nucleotidylating activity associated with a newly delineated conserved domain in the RNA polymerase-containing protein of all nidoviruses. Nucleic Acids Res. 2015;43:8416–8434. doi: 10.1093/nar/gkv838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Slanina H., Madhugiri R., Bylapudi G., Schultheiss K., Karl N., Gulyaeva A., Gorbalenya A.E., Linne U., Ziebuhr J. Coronavirus replication-transcription complex: Vital and selective NMPylation of a conserved site in nsp9 by the NiRAN-RdRp subunit. Proc. Natl. Acad. Sci. U. S. A. 2021;118 doi: 10.1073/pnas.2022310118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Banerjee A.K., Blanco M.R., Bruce E.A., Honson D.D., Chen L.M., Chow A., Bhat P., Ollikainen N., Quinodoz S.A., Loney C., Thai J., Miller Z.D., Lin A.E., Schmidt M.M., Stewart D.G. SARS-CoV-2 disrupts splicing, translation, and protein trafficking to suppress host defenses. Cell. 2020;183:1325–1339.e1321. doi: 10.1016/j.cell.2020.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Littler D.R., Gully B.S., Colson R.N., Rossjohn J. Crystal structure of the SARS-CoV-2 non-structural protein 9, Nsp9. iScience. 2020;23:101258. doi: 10.1016/j.isci.2020.101258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frieman M., Yount B., Agnihothram S., Page C., Donaldson E., Roberts A., Vogel L., Woodruff B., Scorpio D., Subbarao K., Baric R.S. Molecular determinants of severe acute respiratory syndrome coronavirus pathogenesis and virulence in young and aged mouse models of human disease. J. Virol. 2012;86:884–897. doi: 10.1128/JVI.05957-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dudás E.F., Puglisi R., Korn S.M., Alfano C., Bellone M.L., Piaz F.D., Kelly G., Monaca E., Schlundt A., Schwalbe H., Pastore A. Backbone chemical shift spectral assignments of SARS coronavirus-2 non-structural protein nsp9. Biomol. NMR Assign. 2021 doi: 10.1007/s12104-021-10011-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.El-Kamand S., Du Plessis M., Breen N., Johnson L., Beard S., Kwan A.H., Derek R.J., Cubeddu L., Gamsjaeger R. A distinct ssDNA/RNA binding interface in the Nsp9 protein from SARS-CoV-2. Proteins. 2021 doi: 10.1002/prot.26205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taylor A., Doak B.C., Scanlon M.J. Design of a fragment-screening library. Methods Enzymol. 2018;610:97–115. doi: 10.1016/bs.mie.2018.09.018. [DOI] [PubMed] [Google Scholar]
- 26.Chen J., Xu X., Tao H., Li Y., Nan H., Wang Y., Tian M., Chen H. Structural analysis of porcine reproductive and respiratory syndrome virus non-structural protein 7alpha (NSP7alpha) and identification of its interaction with NSP9. Front. Microbiol. 2017;8:853. doi: 10.3389/fmicb.2017.00853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ponnusamy R., Moll R., Weimar T., Mesters J.R., Hilgenfeld R. Variable oligomerization modes in coronavirus non-structural protein 9. J. Mol. Biol. 2008;383:1081–1096. doi: 10.1016/j.jmb.2008.07.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zeng Z., Deng F., Shi K., Ye G., Wang G., Fang L., Xiao S., Fu Z., Peng G. Dimerization of coronavirus nsp9 with diverse modes enhances its nucleic acid binding affinity. J. Virol. 2018;92 doi: 10.1128/JVI.00692-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu D., Bernstein D.A., Satyshur K.A., Keck J.L. Small-molecule tools for dissecting the roles of SSB/protein interactions in genome maintenance. Proc. Natl. Acad. Sci. U. S. A. 2010;107:633–638. doi: 10.1073/pnas.0909191107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Glanzer J.G., Endres J.L., Byrne B.M., Liu S., Bayles K.W., Oakley G.G. Identification of inhibitors for single-stranded DNA-binding proteins in eubacteria. J. Antimicrob. Chemother. 2016;71:3432–3440. doi: 10.1093/jac/dkw340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nebl S., Alwan W.S., Williams M.L., Sharma G., Taylor A., Doak B.C., Wilde K.L., McMahon R.M., Halili M.A., Martin J.L., Capuano B., Fenwick R.B., Mohanty B., Scanlon M.J. NMR fragment screening reveals a novel small molecule binding site near the catalytic surface of the disulfide-dithiol oxidoreductase enzyme DsbA from Burkholderia pseudomallei. J. Biomol. NMR. 2020;74:595–611. doi: 10.1007/s10858-020-00339-5. [DOI] [PubMed] [Google Scholar]
- 32.Aragao D., Aishima J., Cherukuvada H., Clarken R., Clift M., Cowieson N.P., Ericsson D.J., Gee C.L., Macedo S., Mudie N., Panjikar S., Price J.R., Riboldi-Tunnicliffe A., Rostan R., Williamson R. MX2: A high-flux undulator microfocus beamline serving both the chemical and macromolecular crystallography communities at the Australian synchrotron. J. Synchrotron Radiat. 2018;25:885–891. doi: 10.1107/S1600577518003120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kabsch W. XDS. Acta Crystallogr. D Biol. Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Winn M.D., Ballard C.C., Cowtan K.D., Dodson E.J., Emsley P., Evans P.R., Keegan R.M., Krissinel E.B., Leslie A.G., McCoy A., McNicholas S.J., Murshudov G.N., Pannu N.S., Potterton E.A., Powell H.R. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Casanal A., Lohkamp B., Emsley P. Current developments in Coot for macromolecular model building of electron cryo-microscopy and crystallographic data. Protein Sci. 2020;29:1069–1078. doi: 10.1002/pro.3791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liebschner D., Afonine P.V., Baker M.L., Bunkoczi G., Chen V.B., Croll T.I., Hintze B., Hung L.W., Jain S., McCoy A.J., Moriarty N.W., Oeffner R.D., Poon B.K., Prisant M.G., Read R.J. Macromolecular structure determination using X-rays, neutrons and electrons: Recent developments in Phenix. Acta Crystallogr. D Struct. Biol. 2019;75:861–877. doi: 10.1107/S2059798319011471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fehr A.R. Bacterial artificial chromosome-based lambda red recombination with the I-SceI homing endonuclease for genetic alteration of MERS-CoV. Methods Mol. Biol. 2020;2099:53–68. doi: 10.1007/978-1-0716-0211-9_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession number for the atomic coordinates of the 3C-Nsp9COV19:FR6 and associated structure factors have been deposited at the protein databank (www.rcsb.org) with accession code 7KRI.