Abstract
Neurodegenerative diseases (NDs) are a heterogeneous group of aging-associated disorders characterized by the disruption of cellular proteostasis machinery and the misfolding of distinct protein species to form toxic aggregates in neurons. The increasing prevalence of NDs represents a growing healthcare burden worldwide, a concern compounded by the fact that few, if any, treatments exist to target the underlying cause of these diseases. Consequently, the application of a high-throughput, physiologically relevant model system to studies dissecting the molecular mechanisms governing ND pathology is crucial for identifying novel avenues for the development of targeted therapeutics. The nematode Caenorhabditis elegans (C. elegans) has emerged as a powerful tool for the study of disease mechanisms due to its ease of genetic manipulation and swift cultivation, while providing a whole-animal system amendable to numerous molecular and biochemical techniques. To date, numerous C. elegans models have been generated for a variety of NDs, allowing for the large-scale in vivo study of protein-conformation disorders. Furthermore, the comparatively low barriers to entry in the development of transgenic worm models have facilitated the modeling of rare or “orphan” NDs, thereby providing unparalleled insight into the shared mechanisms underlying these pathologies. In this review, we summarize findings from a comprehensive collection of C. elegans neurodegenerative disease models of varying prevalence to emphasize shared mechanisms of proteotoxicity, and highlight the utility of these models in elucidating the molecular basis of ND pathologies.
1. Introduction
Aging is a universal and highly-conserved process that results in the accumulation of physiological damage over time and increasing incidences of disease-associated complications. As advancements in medical technology have afforded a drastic increase in the average human lifespan, a proper mechanistic understanding of aging-associated diseases represents a crucial area of discovery that is necessary for addressing the growing burden of our aging population. Of particular concern are neurodegenerative diseases (NDs), such as Alzheimer’s disease (AD), Parkinson’s disease (PD), and amyotrophic lateral sclerosis (ALS), among others. Despite increasing incidence of NDs in the general population, treatments for the majority of these disorders remain elusive due to several factors. First is their diverse etiologies – while some NDs, such as Huntington’s disease (HD) and spinocerebellar ataxias (SCAs) are genetic in their origin, the vast majority of NDs have no clear singular cause. NDs are also heterogenous in their physical manifestations, ranging from impairment in memory and cognitive processes to progressive interference in motor functions [2]. Furthermore, symptom onset may vary drastically between individuals afflicted by the same ND – for example, cases of ALS may first present with either limb-onset or bulbar-onset symptoms, which differentially predict disease course and survival outcomes [3].
Despite these seemingly vast differences in pathogenicity, NDs are uniquely characterized by a shared hallmark of misfolded protein species, leading to aggregation and deposition of large, misfolded protein aggregates. Due to this defining feature, these NDs are often referred to as “proteinopathies”. Examples of ND-associated proteins discussed here include amyloid-beta (Aβ), tau, huntingtin (Htt), superoxide dismutase (SOD-1), alpha-synuclein (α-syn), and prion protein (PrPSc), among others. In affected cells, the misfolding of theses protein species are thought to induce “gain-of-function” toxicity characterized by failed or improper protein trafficking, premature degradation, or activation of cell stress responses [4-6]. Despite comprehensive knowledge of protein species implicated in NDs, and the identification of patient mutations leading to enhanced susceptibility to aggregation, our understanding of how to harness this knowledge in the context of disease-modifying therapies remains limited. Consequently, research probing the molecular mechanisms underlying aggregation of these toxic species in vivo requires a robust and highly manipulatable model system.
In this review, we provide a comprehensive overview of findings from currently available C. elegans models of common neurodegenerative proteinopathies, such as AD, PD, HD, and ALS, and touch on models of lesser-known NDs, such as Machado-Joseph disease (MJD), prion diseases, and various tauopathies. We further highlight how key mechanistic findings from these models have shed light on how highly conserved longevity pathways and shared protein aggregation hallmarks influence proteotoxicity across the compendium of NDs.
1.1. Caenorhabditis elegans as a model organism for the study of protein misfolding diseases
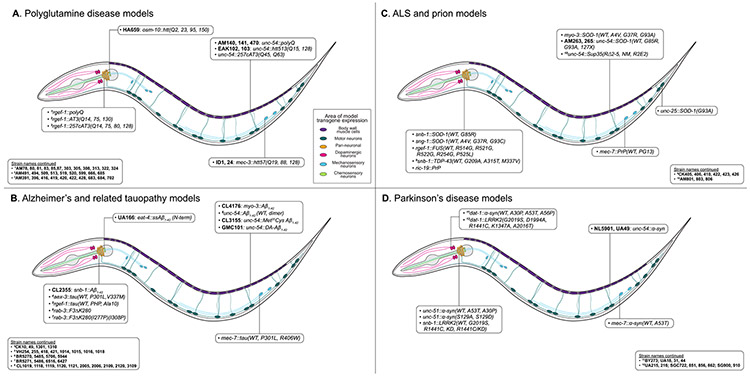
The nematode Caenorhabditis elegans (C. elegans) was first introduced by Sydney Brenner in the mid-1960s as a model organism to study development and neurobiology [1]. Two of his earlier postdoctoral fellows, John E. Sulston and H. Robert Horvitz,went on to use C. elegans to elucidate the fundamental regulation of cell growth and controlled cell death. For their discoveries, Brenner, Sulston, and Horvitz were awarded the 2002 Nobel Prize in Physiology. Nowadays, C. elegans is used to study a vast array of biological processes, including apoptosis, gene regulation, cell metabolism, cell fate regulation, embryogenesis, stress signaling, and aging [7]. As a model organism, the worm provides numerous advantages. It is remarkably simple to culture in the laboratory on a diet of E. coli, and rapidly develops within 3 days post-hatching into fertile adult animals at 20°C. A single adult self-fertilizing hermaphrodite can produce approximately 300 offspring, rendering it amendable to the large-scale growth of thousands of animals for high-throughput screening [8]. Furthermore, C. elegans is transparent, and expression of fluorescent transgenes readily allows for the live study of cellular processes. In the context of disease research, the worm is highly amenable to genetic manipulation and features extensive sequence homology with mammalian genes, making it a valuable system for studying human disease [9]. Both RNA interference (RNAi) [10] and clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 technology [11] are well established in C. elegans, enabling the use of knock-down (RNAi), knock-out, and knock-in approaches (both CRISPR/Cas9) to study the role of individual genes and gene clusters. More recently, C. elegans became the first organism to have every neuron and its’ connections – termed “connectome” – mapped [12]. The sum of these features renders C. elegans an ideal model organism for probing the molecular basis of protein aggregation in NDs. Maps of transgene expression in C. elegans for each of the NDs discussed here are shown in Fig. 1, and a comprehensive summary of these models is listed in Table 1.
Fig. 1.
Anatomical map of ND model transgene expression.
Areas of transgene expression in C. elegans ND models. (A) Polyglutamine disease models. (B) Alzheimer’s and tauopathy models. (C) ALS and prion disease models. (D) Parkinson’s disease models.
Table 1.
Strain, phenotype, and availability information for C. elegans neurodegenerative disease models.
| NDs | Model | Strain name(s) | Expression in C. elegans |
Phenotypes | References | CGC? |
|---|---|---|---|---|---|---|
| polyQ | Is[unc-54::polyQ(19, 82)::gfp/yfp/cfp] | n.a.* | Constitutive muscle | Length-dependent aggregate formation; development delays, motility defects. | (Satyal et al., 2000) | No |
| rmIs[unc-54::polyQ(0, 19, 29, 33, 35, 37, 40, 44, 64, 82)::yfp] | AM140, 141, 470 | Constitutive muscle | Stark aggregation threshold at Q35-40 accompanied by motility defects; Threshold is dynamic and influenced by animal longevity. | (Morley et al., 2002) | No | |
| rmEx[rgef-1::polyQ(0, 19, 35, 40, 67, 86)::cfp/yfp] | AM78, 80, 81, 83, 85, 87; AM303, 305, 308, 313, 322, 324 | Constitutive pan-neuronal | PolyQ length-dependent aggregation and neuronal dysfunction: dysfunctional thrashing, pharyngeal pumping; erratic defecation cycle. | (Brignull et al., 2006) | Yes | |
| HD | rtIs11[osm-10::gfp + osm-10::htt(Q2, 23, 95, 150) + dpy-20(+)] | n.a. | Chemosensory neurons | Q150 expression induces ASH neuronal degeneration, but not death. | (Faber et al., 1999) | Yes |
| Is[mec-3::htt57(Q19, 88, 128)::gfp; mec-3::htt57(Q19, 88, 128)::cfp + mec-3::yfp] | ID1, ID24 | Mechanosensory neurons | Substantial posterior touch insensitivity and anterior Mec phenotype. Significant aggregate deposition and PLM axon abnormalities. | (Parker et al., 2001) | No | |
| rtIs11[osm-10::gfp + osm-10::htt(Q150) + dpy-20(+)] | HA659 | Chemosensory neurons | Genetic screen identified polyQ enhancer-1 (PQE-1); Loss of PQE-1 strongly enhanced neurodegeneration, while overexpression has protective effect | (Faber et al., 2002) | Yes | |
| Is[unc-54::htt513(Q15, Q128)::yfp] | EAK102, 103 | Constitutive muscle | Length-dependent aggregation and toxicity; Htt513(Q128) animals show motor dysfunction and modest decrease in longevity. | (Lee et al., 2017) | Yes | |
| MJD | Full-length ATXN-3 lines: Is[rgef-1::(AT3q14, AT3q75, AT3q130)::yfp] | AM491, 494, 509, 513, 519, 520, 599, 666, 685 | Constitutive pan-neuronal | PolyQ length-dependent aggregation, motor dysfunction; Ventral and dorsal nerve cord neurons highly affected. | (Teixeira-Castro et al., 2011) | No |
| C-terminal ATXN-3 lines: Is[rgef-1::(257cAT3q14, 257cAT3q75, 257cAT3q80, 257cAT3q128)::yfp] | AM391, 396, 416, 419, 420, 422, 428, 683, 684, 702 | Constitutive pan-neuronal | Similar aggregation profile to full-length ATXN-3 with more severe motor phenotype. | (Teixeira-Castro et al., 2011) | No | |
| Is[unc-54::257cAT3(Q45, Q63)::yfp] | n.a. | Constitutive muscle | PolyQ length-dependent aggregation and toxicity not significantly affected by aging. | (Christie et al., 2014) | No | |
| AD | Is[unc-54::Aβ1–42] (WT) or dimer Is[Aβ1–42] | CL1019, 1118, 1119, 1120, 1121, 2005, 2006, 2109, 2120, 3109 | Constitutive muscle | Progressive age-dependent paralysis with formation of amyloid deposits. | (Link, C.D., 1995) | Yes |
| Is[unc-54::Met35Cys Aβ1–42] | CL3115 | Constitutive muscle | No formation of amyloid deposits and no increases in oxidative stress. | (Yatin et al., 1999) | No | |
| smg-1(cc546); Is[snb-1::Aβ1–42 + mtl-2::gfp] | CL2241, 2355 | Inducible pan-neuronal | Defective chemotaxis, formation of amyloid deposits, and serotonin hypersensitivity. | (Wu et al., 2006) | Yes | |
| 3::Aβ1–42::let UTR) + (rol-6(su1006)] | CL4176 | Inducible body-wall muscle | Rapid paralysis; oxidative stress which precedes Aβ fibril formation. | (Drake et al., 2003) | Yes | |
| dvIs100[CL354(unc-54::DA-Aβ1–42) + CL26(mtl-2::gfp)] | GMC101 | Constitutive muscle | Severe paralysis following 48hr exposure to 25°C temperature shift. | (McColl et al., 2012) | Yes | |
| Is[eat-4::ssAβ1–42(N-term) + eat-4::gfp + myo-2::mCherry] | UA166 | Glutamatergic neurons | Age-dependent loss of glutamatergic neurons. | (Treusch et al., 2011) | No | |
| Tauopathies/FTDP-17 | Is[aex-3::4R1N hTau (WT, V337M, P301L) + myo-2::gfp] | CK10, 49, 1301, 1310 | Constitutive pan-neuronal | V337M and P301L mutants show age-dependent uncoordinated phenotype, insoluble tau fibrils, and reduced lifespan. | (Kraemer et al., 2003) | No |
| tmIs[mec-7::tau WT(0N4R, 0N3R) + rol-6(su1006)] | n.a. | Mechanosensory neurons | Mild progressive impairment in touch response; Little detectable tau accumulation in PLM tail neuron. | (Miyasaka et al., 2005) | No | |
| tmIs[mec-7::tau(P301L, R406W) + rol-6(su1006)] | n.a. | Mechanosensory neurons | Severe progressive impairment in touch response, significant tau accumulation in PLM tail neuron. | (Miyasaka et al., 2005) | No | |
| pha-1(e2123ts); Ex[rgef-1::Tau352(WT, PHP, Ala10) + pha-1(+)] | VH254, 255, 418, 421, 1014, 1015, 1016, 1018 | Constitutive pan-neuronal | Progressive motor dysfunction and neurodegeneration. PHP tau induces abnormal motor neuron development; Ala10 worms show early motor impairments and shortened lifespan. | (Brandt et al., 2009) | Yes | |
| Pro-aggregant lines: Is[rab-3::F3ΔK280 + myo-2::mCherry] | BR5270, 5485, 5706, 5944 | Constitutive pan-neuronal | Severe locomotive impairment at day 1 of adulthood; Accelerated aggregate formation; Severe developmental defects in nervous system; impairments in presynaptic transmission. | (Fatouros et al., 2012) | Yes | |
| Anti-aggregant lines: Is[rab-3::F3ΔK280(I277P)(I308P) + myo-2::mCherry] | BR5271, 5486, 6427, 6516 | Constitutive pan-neuronal | No overt locomotive impairment; minimal effects on neurodevelopment. | (Fatouros et al., 2012) | Yes | |
| ALS | ALS Is[hsp-16.2::sod-1 (WT, A4V, G37R, G93A) + myo-3::sod-1(WT, A4V)::gfp + rol-6(su1006)] | n.a. | Heat-shock inducible muscles | Oxidative stress-induced aggregate formation. | (Oeda et al., 2001) | No |
| Is[snb-1::sod-1(WT, G85R)::yfp] | n.a. | Constitutive pan-neuronal | G85R mutants exhibit severe motor dysfunction accompanied by both soluble oligomers and insoluble aggregate deposits. | (Wang et al., 2009) | No | |
| Is[sng-1::sod-1(WT, A4V, G37R, G93C)::gfp] | n.a. | Constitutive pan-neuronal | Mutant homodimers show increased aggregate formation compared to heterodimers, but G85R heterodimers are more toxic in functional assays. | (Witan et al., 2008) | No | |
| Is[unc-54::sod-1(WT, G85R, G93A, G127insTGGGstop)::yfp] | AM263, 265 | Constitutive muscle | SOD-1 mutants show morphologically heterogenous aggregates with varied biophysical properties and mild motility defects | (Gidalevitz et al., 2009) | Yes | |
| Is[unc-25::sod-1(G93A)::gfp] | n.a. | GABAergic motor neurons | G93A SOD-1 animals show progressive motor dysfunction, aggregate formation, and axonal guidance defects. | (Li et al., 2013) | No | |
| lin-15(n765ts); [rgef-1::fus(WT, R514G, R521G, R522G, R524S, P525L) + pab-1::mCherry; lin-15(+)] | n.a. | Constitutive pan-neuronal | Cytoplasmic FUS aggregates; R522G, P525L, FUS513 and FUS501: reduced lifespan. P525L, FUS513 and FUS501: partial or complete paralysis. | (Murakami et al., 2012) | No | |
| Is[snb-1::tdp-43 (WT, G290A, A315T, M337V) + snb-1::gfp] | CK405, 406, 410, 422, 423, 426 | Constitutive pan-neuronal | TDP-43 mutants exhibited severe motor dysfunction. Motor neuron degeneration accompanied by hyperphosphorylation, ubiquitination, and truncated TDP-43 in aggregates. | (Liachko et al., 2010) | No | |
| Is[unc-25::snb-1::gfp] + Ex[snb-1::tdp-43; regf-1::DsRed2; unc122::rfp] | CL1681, 1682, 2609 | Constitutive pan-neuronal | Uncoordinated motor phenotype; abnormal motor neuron synapses. | (Ash et al., 2010) | No | |
| PD | Is[dat-1::α-syn (WT, A53T) + dat-1::gfp] | BY273; UA18, 31, 44 | Dopaminergic neurons | Neuronal loss and dendritic breaks shown in both WT and A53T animals. | (Lakso et al., 2003) | No |
| Is[unc-54::α-syn::yfp + unc-119(+)] | NL5901 | Constitutive muscle | Age-dependent inclusion formation. | (van Ham et al., 2008) | Yes | |
| Is[unc-54::α-syn::gfp + rol-6(su1006)] | UA49 | Constitutive muscle | Misfolding and accumulation of α-synuclein aggregates. | (Hamamichi et al., 2008) | No | |
| Is[unc-51::α-syn(WT, A53T, A30P) + unc-51::egfp] | n.a. | Constitutive pan-neuronal | No motor dysfunction or developmental defects; RNAi knock-down of AP-2 complex subunits induced severe growth/motor abnormalities. | (Kuwahara et al., 2008) | No | |
| Is[mec-7::α-syn(WT, A53T) + rol-6(su1006)] | n.a. | Mechanosensory neurons | Moderate impairments in touch response, impaired neuromuscular transmission. | (Kuwahara et al., 2008) | No | |
| Is[dat-1::α-syn(A30P, A53T, A56P) + dat- 1::mCherry] | n.a. | Dopaminergic neurons | A56P and TP: Pronounced neurodegeneration, DA loss, and significant impairment in DA-dependent behavior; A30P and A53T: Moderate decrease in DA levels, DA-dependent behavioral impairments. | (Karpinar et al., 2009) | No | |
| unc-51::α-syn(S129A, S129D) + unc-51::egfp | n.a. | Constitutive pan-neuronal | Severe motor dysfunction beginning in early development, stunted growth, and synaptic abnormalities. | (Kuwahara et al., 2012) | No | |
| lin-15(n765ts); wlzIs1-7[snb-1::lrrk2(WT, G2019S, R1441C, KD, R1441C/KD) + mec-4::gfp; lin-15(+)] | n.a. | Constitutive pan-neuronal | WT LRRK2 protects against mitochondrial dysfunction. G2019S mutants increased vulnerability of DA neurons to mitochondrial stress. | (Saha et al., 2009) | Yes | |
| baIn20[dat-1::lrrk2(G2019S) + dat-1::gfp] | UA118 | Dopaminergic neurons | Progressive degeneration of DA neurons, significant depletion of dopamine accompanied by locomotor dysfunction and behavioral deficits. G2019S mutants have more rapid progression of disease phenotype. | (Liu et al., 2011) | No | |
| BY250; baEx129[dat-1::lrrk2(G2019S/D1994A)] | UA215, 216 | Dopaminergic neurons | Progressive degeneration of DA neurons accompanied by behavioral deficits, motor dysfunction, and DA depletion; G2019S strain more severe. | (Liu et al., 2011) | No | |
| lin-15(n765ts)X; Is[dat-1::lrrk2(WT, R1441C, G2019S, K1347A) + dat-1::gfp + lin-15(+)] | SGC722, 851, 856, 862 | Dopaminergic neurons | Progressive degeneration of DA neurons and DA loss; behavioral and motor deficits. | (Yao et al., 2013) | No | |
| lin-15(n765ts)X; cwrEx900[dat-1::gfp, dat-1::lrrk2(R1441C/A2016T), lin-15(+)] | SG900, 910 | Dopaminergic neurons | Double mutants show similar profile to R1441C and G2019S LRRK2 models; DAergic defects and neurodegeneration. | (Yao et al., 2013) | No | |
| Prion | cgIs51-53[ric-19::prp + ric-19::gfp] | n.a. | Constitutive pan-neuronal | High PrP levels alter morphology; significant motor impairment and reduction in lifespan. | (Park and Li, 2011) | No |
| lin-15(n765ts); Is[mec-7::prp(WT, PG13) + str-1::gfp; mec-7::gfp + lin-15(+)] | n.a. | Mechanosensory neurons | PG13-PrP mutants show progressive loss of tail-touch response without neuronal death. | (Bizat et al., 2010) | No | |
| rmIs319[unc-54::sup35(RΔ2—5, NM, R2E2)::yfp] | AM801, 803, 806 | Constitutive muscle | Severe disruption of mitochondrial integrity, embryonic/larval arrest, developmental delays, tissue defects, loss of proteostasis. | (Nussbaum-Krammer et al., 2013) | No |
Genetic nomenclature of C. elegans ND models, expression pattern, observable phenotypes, and availability of strain(s) through the Caenorhabditis Genetics Center (CGC).
Denotes that strain nomenclature was not provided by the creators of the model(s) in the primary literature.
1.1.1. Polyglutamine expansion disease models
Polyglutamine (polyQ) repeat expansion diseases are a group of at least 9 inherited neurodegenerative disorders, including Huntington’s disease (HD), spinal and bulbar muscular atrophy (SBMA), dentatorubropallidoluysian atrophy (DRPLA), and six forms of spinocerebellar ataxia (SCA1, 2, 3, 6, 7, and 17) [13]. The molecular basis of these pathologies lies in the aberrant expansion of CAG codon repeats, which encode the amino acid glutamine (Q), in distinct, yet unrelated proteins. In healthy individuals, polyQ tracts are around 35–50 repeats, depending on the affected protein. However, in those carrying a polyQ mutation, the resulting mutant protein may contain more than 40 repeats, reaching over 100 Qs in length in some conditions [14].
1.1.2. Generic polyglutamine (polyQ) models
A popular approach to modeling CAG repeat diseases in C. elegans is to fuse plain polyQ repeats to a fluorescent reporter protein. Given that expanded polyQ tracts alone are sufficient to recapitulate many of the hallmarks of polyglutamine aggregation, the majority of described models are limited to expression of the polyQ tract without inclusion of additional protein sequence context [15]. A 2000 study reported the characterization of transgenic C. elegans expressing polyQ tracts of physiological (Q19) or pathological (Q82) lengths fused to GFP or CFP fluorescent proteins in body-wall muscle cells [16]. In agreement with other models of polyQ expression in neurons, expression of Q82, but not Q19 led to the formation of discrete aggregates, demonstrating that the length-dependent toxicity of polyQ proteins can be recapitulated in C. elegans models through expression in non-neuronal cell types. Using these models, Satyal et al. found that 1) the expression of toxic polyQ repeats altered the global proteostasis environment of the cell, 2) Q82 expression sequestered non-toxic Q19 into aggregates, and 3) Q82 accumulation induced mislocalization of the nuclear protein HRP1 to the cytosol.
To investigate the molecular mechanisms underlying polyQ aggregation at near-threshold lengths, Morley et al. generated C. elegans lines in which polyQ tracts of varying lengths (Q0, Q19, Q29, Q33, Q35, Q40, Q44, Q64, Q82) fused to YFP were expressed in body wall muscle cells [17]. Young adult (day 3–4) animals expressing polyQ::YFP transgenes with 35 or fewer Qs showed diffuse distribution of the protein, while those longer than Q40 exhibited distinct puncta characteristic of protein aggregates. Q40-expressing animals showed a heterogenous distribution of diffuse fluorescence alongside foci. Expression of Q82::YFP resulted in an approximately 10-fold reduction in motility compared to control animals, while Q40::YFP worms demonstrated an intermediate phenotype with high variability. As worms aged, an inverse relationship between increasing aggregate numbers and decreasing motility emerged, and near-threshold animals (e.g. Q35::YFP) showed marked aggregate accumulation at 4–5 days of age, suggesting that the threshold for polyQ toxicity is dynamic and modulated by factors such as organismal age. Furthermore, animals expressing Q82 in an age-1(hx5476) longevity-enhanced mutant background showed a stark reduction of aggregate formation in embryos compared to Q82 in the wild-type background, with decreases in aggregation of 30–50% during larval stages. This trend of reduced phenotypic severity continued until days 4–5 of adulthood, indicating that lifespan-enhancing mutations may delay in the onset of polyQ toxicity.
Brignull et al., generated complementary strains with pan-neuronal expression of a range of polyQ proteins fused to either YFP or CFP (cyan fluorescent protein) [18]. Here, Q19 worms showed diffuse polyQ expression, while Q86 animals exhibited discrete aggregates as early as the first larval stage, as confirmed via fluorescence recovery after photobleaching (FRAP). Neuronal polyQ expression recapitulated Q-length-dependent threshold toxicity, in accordance with aggregation properties observed in body-wall muscle strains. Förster resonance energy transfer (FRET) followed by FRAP further revealed the presence of two distinct, dynamic populations of diffuse or aggregated Q40. Taken together, these data characterized polyQ dynamics in an in vivo, pan-neuronal setting and established FRAP and FRET as complementary techniques to garner information on the biophysical properties protein aggregates in a living system.
1.1.3. Huntington’s disease (HD) models
Huntington’s disease (HD) is the most common polyglutamine expansion disease, with a prevalence of approximately 1 in 10,000 individuals in the United States and Europe [19]. At the molecular level, HD is caused by an autosomal dominant mutation leading to a polyQ expansion in exon 1 of the huntingtin (Htt) protein. Molecular genetics analyses of HD patients indicate that manifestation of the disease occurs with more than 35–40 CAG repeats, and a strong inverse correlation between repeat length and age-of-onset has been well documented [20,21]. Disease onset usually occurs between 30 and 50 years of age, often beginning with physical symptoms manifesting as jerky, random, and uncontrollable movements, termed “chorea” [22]. As the disease progresses, patients experience worsening motor symptoms, as well as decreases in cognitive performance and neuropsychiatric changes.
The first transgenic C. elegans model of huntingtin toxicity was described in 1999, in which an N-terminal huntingtin fragment containing varying Q lengths (Q2, 23, 95, 150) was expressed in nematode ASH chemosensory neurons [23]. ASH neurons expressing Htn-Q150 demonstrated progressive degeneration and accumulation of aggregates. These findings were further validated by a later independent study utilizing C. elegans expressing N-terminal huntingtin with either physiological (Q19) or expanded (Q88 or Q128) Qs in mechanosensory neurons [24]. Here, polyQ toxicity was length-dependent such that Q128 animals demonstrated a significant mechanosensory defective phenotype at the tail, accompanied by aggregated polyQ deposits and morphological abnormalities.
The C. elegans model expressing Htn-Q150 in ASH neurons was later utilized in both chemical mutagenesis screens for mutations enhancing neuronal degeneration [25], and in a small-molecule screens to identify pharmacological suppressors of neuronal death [26]. The mutagenesis screen identified a loss-of-function mutation in a gene designated polyQ enhancer-1 (pqe-1). Loss of functional pqe-1 in Htn-Q150 animals, but not in those expressing physiological Q tract lengths (Htn-Q2, Htn-Q23), significantly accelerated degeneration and death of ASH neurons, without any other overt phenotypes. The pharmacological screen identified rotenone, an inhibitor of complex I of the mitochondrial electron transport chain, as a strong dose-dependent suppressor of ASH neuronal loss in Htn-Q150-expressing animals. Rotenone further mitigated ASH neuron death in L1 larval nematodes, validating its utility independent of effects on growth or development [26].
A more recent HD model strain expresses a Htt513(Q15, Q128):YFP transgene in body wall muscle cells [15]. The N-terminus of Htt is cleaved by caspase-3 at amino acid position 513 (Htt513) [27]. The resulting fragment, which contains the expanded polyQ repeat domain, is proposed to trigger neuronal death through initiation of an apoptotic pathway [28,29]. In C. elegans, muscular expression recapitulates the length-dependent toxicity associated with polyQ expansions and also demonstrates shortened lifespan and motor defects.
1.1.4. Machado-Joseph disease (MJD) models
While HD is the most common polyglutamine expansion disease, transgenic C. elegans have also been developed as models of other, less common disorders in this class of NDs. At least 6 autosomal dominant forms of spinocerebellar ataxia (SCA1, 2, 3, 6, 7, and 17) are caused by CAG repeat expansions, the most common of these being SCA3, or Machado-Joseph disease (MJD). MJD is caused by a CAG repeat expansion in exon 10 of the ATXN3 gene, which encodes ataxin-3 (AT3), a ubiquitously expressed protein hypothesized to play a role in protein quality control pathways [30]. Notably, the threshold for polyQ repeats is not identical between HD, MJD, and other polyQ expansion diseases. While HD is caused by 37–40 or more CAG repeats, pathological polyQ expansions in AT3 range from 61 to 87 repeat units [31].
In C. elegans, expression of full-length and C-terminal mutant AT3 in neurons induces a canonical polyQ-length dependent aggregation phenotype, recapitulating studies expressing polyQ-only fragments in vivo [32]. Interestingly, however, this phenomenon was accompanied by a neuronal sub-type specificity, such that ventral and dorsal nerve-cord neurons were highly affected, and some lateral interneurons appeared resistant, shedding light on how protein context might modulate polyQ-induced proteotoxicity [32]. Furthermore, reduction of insulin/insulin-like growth-factor signaling (IIS) and activation of heat shock protein 1 (HSP-1) chaperone activity reduced pathogenesis in this model, supporting the established link between cellular aging and ND proteotoxicity. The expression of the C-terminal domain of AT3 with various Q lengths in body-wall muscle cells also induced polyQ-length dependent aggregation and toxicity, resulting in motor impairments [33]. However, in contrast to previous reports, neither organismal age nor heat-shock induced proteotoxic stress modulated AT3 aggregation in this model [33]. The cause for these differences remains to be addressed.
Taken as a whole, C. elegans models of disease-relevant Htt proteotoxicity validate previous studies of polyQ tracts in vitro and in vivo and are promising tools for high-throughput studies to modulate aggregate toxicity in more disease-focused contexts. Results obtained from studies utilizing generic polyQ tract and disease-specific (HD, MJD) models highlighted several strongly conserved principles which underlie polyQ toxicity, such as threshold-dependent aggregation and the impact of organismal age on aggregation propensity.
1.2. Amyotrophic lateral sclerosis (ALS)
Amyotrophic lateral sclerosis (ALS) is the most common motor neuron disease (MND) and is characterized by progressive neuronal death in the motor cortex, brainstem, corticospinal tracts, and spinal cord [3]. Patients present with either limb onset (80% of cases) or bulbar onset (20% of cases) of symptoms, with each of the two sub-types having differential disease outcomes. Limb onset cases are characterized by distal limb weakness and spasticity, while bulbar cases generally begin with swallowing and speaking difficulties [34]. The disease is invariably fatal as patients experience progressive paralysis, and death generally occurs 3–5 years after symptom onset due to respiratory failure [3]. The vast majority of cases of ALS are sporadic with no clear underlying etiology; however, approximately 10% of cases are linked to known genetic mutations, referred to as familial ALS (fALS) [35]. Of this 10%, approximately 20% of fALS cases arise from mutations in the gene coding for superoxide dismutase 1 (SOD-1), while 4–5% of cases are attributed to mutations in the RNA-binding proteins TDP-43 and FUS [34].
1.2.1. SOD-1 models
Cu/Zn superoxide dismutase 1 (SOD-1) is a ubiquitously expressed cytosolic enzyme that, under physiological conditions, catalyzes the conversion of superoxide anions into hydrogen peroxide [36]. It has been hypothesized that mutations in SOD-1 result in cell death via a toxic gain-of-function effect, though the exact mechanisms underlying this are yet to be fully understood. Over 100 disease-causing point mutations in the gene sod-1 have been mapped to date [37], 70 of which have been linked to fALS [35]. The vast majority of disease-relevant SOD-1 mutations are autosomal dominant. Mutant SOD-1 can be classified into two groups: Wild-type-like proteins, which exhibit metal ion binding and dismutase activity, and mutants with impaired metal binding which show significantly reduced dismutase activity [38]. The current prevailing model of SOD-1 toxicity proposes that SOD-1 mutations result in cell death via a toxic gain-of-function effect [39,40], though the precise mechanisms underlying this process are yet to be fully understood.
A number of C. elegans-based studies have implicated an association between neuronal oxidative stress and SOD-1 mutant-linked fALS [41]. While the expression of WT SOD-1 in body-wall muscle cells was well tolerated, the presence of mutant SOD-1 (A4VV, G37R, and G93A) led to increased sensitivity to oxidative damage and a significantly decreased rate of degradation of mutant SOD-1, promoting the formation of A4V mutant SOD-1 aggregates [42]. SOD-1 (G85R) formed aggregates when expressed in neurons, triggering severe locomotor defects [43]. Notably, the muscular expression of wild-type or mutant SOD-1::YFP fusion proteins only induced mild cellular toxicity, despite the appearance of SOD-1 aggregates of varying biophysical composition [44]. To study interactions between WT and mutant SOD-1, Witan et al. created C. elegans strains with pan-neuronal expression of genetically engineered SOD-1/SOD-1(G85R) heterodimers [45]. Aggregation of these heterodimers was significantly increased compared to SOD-1 homodimers, but decreased in comparison to SOD-1(G85R) homodimers. Animals expressing SOD-1(WT)/SOD-1(G85R) heterodimers also demonstrated significantly impaired locomotion compared to SOD-1 and SOD-1(G85R) expressing animals, indicating that interactions between wild-type and mutant SOD-1 are more harmful to neuronal function than the presence of SOD-1(G85R) homodimers. These findings were confirmed in in vivo mouse studies, which showed that co-expression of SOD-1 and SOD-1(G85R) accelerates disease [46,47]. In another study, worms expressing SOD-1(G93A) in GABAergic motor neurons in a daf-2(e1370) mutant background showed significant reductions in SOD-1 aggregation and motor dysfunction [48]. Similarly, animals with pan-neuronal expression of SOD-1(G85R) in a daf-2(e1370) mutant background ameliorated SOD-1 aggregation and associated locomotive deficits in a daf-16 dependent manner [49], corroborating findings from other C. elegans ND models suggesting a global role for longevity-enhancing mutations in attenuating proteotoxicity.
1.2.2. TDP-43 models
The TAR DNA-binding protein 43 (TDP-43) is a ubiquitously expressed, highly conserved DNA and RNA binding protein. Under physiological conditions, it is predominantly localized to the nucleus, where it assists in transcriptional repression, alternative mRNA splicing, and mRNA stability [50]. However, in disease states, TDP-43 becomes mislocalized to the cytoplasm and is identified in ubiquitinated neuronal inclusions in patients with ALS and frontotemporal dementia (FTD) [51]. Numerous mutations in TARDBP, which encodes TDP-43, have been linked to cases of fALS and FTD, predominantly in exon 6 of the gene. Interestingly, mutations in TDP-43 have also been identified in cases of sporadic ALS (sALS), and several known mutations have demonstrated overlap between sALS and fALS, as well as between ALS and FTD. This suggests a more global role for TDP-43 dysfunction in the pathology of ALS and related NDs [52]. However, the molecular mechanisms underlying TDP-43-mediated neurotoxicity are unclear, and it is unknown whether disease results from an increase, decrease, or change in TDP-43 function and activity [53].
When expressed in C. elegans neurons, full-length human TDP-43 (hTDP-43) triggers the development of abnormal motor synapses and motor dysfunction, resulting in a prominent uncoordinated (unc) phenotype [53]. Deletion of either the RNA recognition domain (RRM1/2), or the C-terminus blocked TDP-43 neurotoxicity in vivo, yet these species still formed nuclear inclusions. However, fusion of TDP-43 with the C-terminus of TDP-1, the structurally conserved C. elegans TDP-43 ortholog [54], restored toxicity, demonstrating that overexpression of full-length TDP-43 is sufficient to induce neurotoxicity in vivo resulting from the protein’s aberrant nuclear activity. Furthermore, the presence of fALS-linked TDP-43 mutations (G290A, A315T, or M337V) exacerbated motor impairment, resulting in partial or complete paralysis [55]. In contrast, TDP-43 containing mutations that abolish pathological phosphorylation of serine residues 409 and 410 [51,56,57] demonstrated marked amelioration of motor dysfunction evidenced by reduced motor abnormalities and increased responsiveness to stimuli. Similar motor impairments were observed in C. elegans strains with pan-neuronal expression of WT and two ALS-linked TDP-43 mutants (Q331K, M337V) fused to the fluorescent reporter, YFP [58]. Exposure of these animals to elevated temperatures (25 °C) exacerbated TDP-43 accumulation, suggesting a relationship between proteostasis stress, TDP-43 misfolding, and resulting neurotoxicity. In all cases, organismal aging exacerbated TDP-43 phenotypes, in-line with data from other C. elegans studies which accurately recapitulate the aging hallmarks of NDs. At the molecular level, WT and mutant TDP-43 localized in the nuclear bodies of neurons to form insoluble aggregates. Notably, expression of neuronal WT TDP-43 in a daf-2(e1370) mutant background significantly suppressed locomotor deficits and insoluble TDP-43 aggregate formation, following similar reports of daf-2 mediated rescue of proteotoxicity. Taken together, these findings highlight the utility of C. elegans as a model for dissecting the molecular basis of TDP-43 misfolding and toxicity.
1.2.3. Fused in sarcoma/translocated in sarcoma (FUS/TLS) models
At least 13 different mutations in the FUS gene, which encodes the RNA-binding protein fused in sarcoma/translocated in sarcoma (FUS/TLS) have been linked to fALS and sporadic forms of frontotemporal dementia (FTD) [59]. FUS is, like TDP-43, multi-functional and expressed almost exclusively in the nucleus under physiological conditions [60]. However, in patients with disease-causing FUS mutations, abnormal accumulations of the protein are present in the brain and spinal cord [61]. Interestingly, the majority of known FUS mutations are located at its C-terminus, which is hypothesized to result in disruption of the protein’s normal nuclear localization sequence (NLS) [62]. The molecular mechanisms underlying FUS-related pathology in ALS/FTD are unclear; however, commonalities between disease-causing FUS and TDP-43 mutations may indicate that they exert their aberrant effects through similar mechanisms [63].
C. elegans models for dissecting the molecular events behind FUS pathology in fALS were first described in 2012 [64]. Wild-type and four different FUS missense mutations associated with varying disease severity were expressed in C. elegans under the control of an endogenous, pan-neuronal promoter [65]. Here, expression of mutant FUS, but not WT-FUS, led to mislocalization of FUS to the cytoplasm, progressive motor dysfunction, and reduced lifespan [64]. Importantly, the reported clinical severity of missense mutations was directly correlated with the severity of pathology observed in the C. elegans strains, lending supporting evidence to the relevance of studying ALS-related proteinopathy in nematodes. Co-expression of wild-type FUS was not sufficient to rescue mutant FUS toxicity phenotypes, though WT-FUS was predominantly cytoplasmic and was not recruited to mutant nuclear inclusions. Overexpression of WT-FUS in vivo was not able to rescue mutant FUS disease phenotypes, though FUS was predominantly cytoplasmic and was not recruited to mutant nuclear inclusions [64]. These data suggest that mutations in FUS linked to ALS likely induce neurotoxicity through a toxic dominant gain-of-function mechanism.
1.3. Alzheimer’s disease (AD) and related tauopathies
Alzheimer’s disease is the most common neurodegenerative disease worldwide, and accounts for over 80% of dementia cases diagnosed in the elderly [66,67]. Often beginning with mild impairments in learning and memory, the disease progresses to behavioral and neuropsychiatric changes and a steady decline in the ability to perform basic activities of daily living [68]. At a molecular level, AD is characterized by neuronal loss in the neocortex, accompanied by the presence of amyloid-β (Aβ) plaques and intracellular neurofibrillary tangles (NFTs) composed of hyper-phosphorylated tau protein [69,70]. Aβ is a small peptide (36–43 amino acids) that is generated by the step-wise proteolysis of amyloid precursor protein (APP) by β- and γ-secretases [71]. The amyloid cascade hypothesis proposes that the abnormal accumulation of Aβ serves as the initial trigger for AD pathology [72,73]. This model is supported by the identification of mutations in the amyloid precursor protein (APP) [74], presenilin 1 (PSEN1) [75], and presenilin 2 (PSEN2) [76] genes linked to early-onset familial forms of AD. However, the overwhelming failure of therapeutics aimed at reducing Aβ levels in the brain suggests the involvement of additional key factors in AD pathology [77]. Of particular interest is tau, a protein that binds to and stabilizes microtubules to support the neuronal cytoskeleton [78]. In AD, however, tau becomes abnormally hyperphosphorylated, leading to its aggregation and eventual formation of NFTs [79]. The spread of pathological tau accumulations strongly correlates with AD progression [80], and tau pathology is evident at earlier stages of disease than Aβ accumulation [81]. Furthermore, tau pathology is not exclusive to AD – other NDs exhibiting primary tau pathology include Pick’s disease, corticobasal degeneration, and frontotemporal dementia and parkinsonism linked to chromosome 17 (FTDP-17) [82]. Such diseases are broadly referred to as “tauopathies”. In this section, we highlight findings from both C. elegans models of classical amyloid hypothesis-driven proteotoxicity, and those recapitulating primary tauopathies such as FTDP-17.
1.3.1. Amyloid-beta (Aβ) models
Early C. elegans models of Aβ aggregation were based on the expression of Aβ1-42 peptide in body-wall muscle cells [83]. The resulting strains exhibited amyloid deposits and an age-dependent paralysis phenotype, establishing that expression of human Aβ in C. elegans reliably recapitulates the abnormal structural phenotypes associated with toxic protein aggregates. Furthermore, a role for insulin/insulin growth factor-1-like signaling (IIS), a key longevity pathway, in ameliorating Aβ toxicity was uncovered using these early models, which demonstrated increased longevity and decreased aggregation-induced paralysis upon RNAi-induced knock-down of daf-2 [84]. Work by Florez-McClure et al. further revealed that decreased daf-2 signaling promotes the autophagic degradation of Aβ by accelerating the maturation of autophagosomes into autolysosomes [85], suggesting a direct link between conserved longevity pathways and key cellular processes, such as autophagy, which become dysregulated in various NDs.
However, some of these initial C. elegans models were later found to express a truncated Aβ3-42 product, rather than the Aβ1-42 peptide [86]. More recent C. elegans strains express a modified Aβ1-42 version in which the original Aβ signal peptide was altered to contain two additional N-terminal residues (Asp-Ala), facilitating cleavage of the proper 1–42 peptide [87]. These animals, which express Aβ1-42 in body-wall muscle cells, demonstrated accumulation of amyloid inclusions accompanied by age-progressive paralysis. Exposure of worms to 25 °C triggered a severe, fully-penetrant paralysis within 48hrs. The robustness of this phenotype has subsequently been exploited in high-throughput screenings of therapeutic compounds in a whole-animal setting.
A sizeable body of evidence has implicated a role for oxidative stress and free-radicals in Aβ pathogenicity [88]. In particular, the Aβ1-42 Met35Cys residue has been proposed to be critical for free-radical formation in AD models [89]. This hypothesis was tested by assessing protein carbonyl levels in C. elegans strains expressing wild-type human Aβ1-42 peptide or Aβ with a Met35Cys substitution [90]. Interestingly, while wild-type Aβ1-42 lines showed carbonyl levels of 176% relative to vector-only controls, those with the Aβ Met35Cys substitution showed no demonstrable increase in carbonyls, suggesting a critical role for the Met35Cys residue in Aβ oxidative stress. These results were later confirmed utilizing a C. elegans model in which Aβ expression in muscle cells was induced upon shifting animals from 16 °C to 23 °C incubation, a transcriptional switch controlled by the temperature-sensitive smg-1 system [91,92]. Notably, increased oxidative stress levels occurred without the presence of Aβ fibrils, suggesting that Aβ toxicity is mediated by pre-fibrillar species.
While the majority of C. elegans Aβ models express the peptide in body-wall muscle, one inducible pan-neuronal model has been described in the literature [93]. This strain demonstrated formation of amyloid deposits accompanied by deficits in chemotaxis behavior and serotonin (5-HT) hypersensitivity, extending findings of Aβ pathology in muscle models to neuronal impairment.
1.3.2. Tau models
Several transgenic C. elegans models developed for the study of tau pathology have utilized the genetic mutations found in patients with FTDP-17. This disorder is a hereditary form of frontotemporal dementia (FTD) caused by mutations in the microtubule-associated protein tau (MAPT) gene, located on chromosome 17. To date, 53 causative MAPT mutations have been identified across approximately 150 families, though patients with the same mutation may vary drastically in their clinical presentations [94,95]. Due to the complex and, at times, conflicting literature on the role of tau in NDs, the genetic basis underlying FTDP-17 pathogenesis serves as a unique tool for probing the molecular basis of tau aggregation and toxicity.
The first set of transgenic C. elegans tau pathology models consisted of worms expressing either wild-type (WT) or FTDP-17 mutant (V337M or P301L) tau throughout the nervous system [96]. The resulting animals demonstrated a progressive unc phenotype accompanied by the accumulation of insoluble tau deposits, structural abnormalities in neurons, neurodegeneration, and cell death. Notably, this pathology was seen in all three tau forms, but with increased severity in those expressing FTDP-17 mutations. The expression of tau in mechanosensory neurons only modestly decreased touch response, while the presence of tau mutants (P301L and R406W) in these neurons led to severe age-dependent impairments, corroborating findings of increased severity associated with FTDP-17 tau mutants [97]. Electron microscopy (EM) of affected neurons revealed neurodegenerative patterns that have also been reported in tau models in Drosophila [98] and mice [99,100]. As such, C. elegans models can accurately recapitulate aggregation-induced neurotoxicity observed in higher-order animal systems, further validating their utility in the study of ND pathology.
Much controversy exists as to whether large tau neurofibrillary tangles (NFTs) are the primary instigators of cellular toxicity, or if these inclusions instead represent a protective mechanism intended to sequester toxic oligomers away from interfering with functional cellular machinery. To address this question, several studies have utilized C. elegans models expressing an aggregation enhancing FTDP-17 mutant, F3ΔK280, and an engineered “anti-aggregation” form of this construct containing I277P and I308P (F3ΔK280-PP) mutations in tau’s hexapeptide motifs [101-103]. These isoleucine-to-proline point mutations disrupt β-sheet formation, hindering the peptide’s ability to aggregate. Consequently, while expression of pro-aggregant F3ΔK280 tau is highly cytotoxic, expression of the anti-aggregant species at comparable levels is not [104]. Pan-neuronal co-expression of the pro-aggregation F3ΔK280 fragment with full-length tau V337M resulted in significant proteotoxicity, evidenced by uncoordinated locomotion from day 1 of adulthood, axonal defects in motor neurons, and impaired axonal transport of mitochondria, recapitulating primary aspects of tau pathology described in mammalian models [96,105]. Treatment with the novel tau aggregation inhibitor cmp16, which belongs to the aminothienopyridazine (ATPZ) class of compounds, ameliorated motility impairments and slowed the accumulation of neuronal abnormalities. In contrast, F3ΔK280-PP-expressing animals showed only mild phenotypes, with significantly reduced morphological abnormalities.
Another tau model strain expresses WT human tau (hTau) and pseudohyperphosphorylated (PHP; 10 Ser/Thr residues replaced with Glu) tau in all neurons [106]. Worms expressing either hTau or PHP tau demonstrated uncoordinated locomotive phenotypes, but only those expressing PHP tau showed defects in motor neuron development.
Taken together, this work established a novel in vivo model of tau aggregation dynamics and positioned the worm as a powerful tool for physiologically relevant, high-throughput screens. Still, a significant limitation of current C. elegans-based tau studies is a lack of transgenic models beyond those utilizing exceedingly rare FTDP-17 mutations. Development of novel wild-type tau strains to investigate the impact of MAPT splice variants (e.g. 3R/4R ratio) on neuronal health, for example, may significantly expand the utility of tau findings from C. elegans. Notably, a recent study has implicated the low-density lipoprotein receptor-related protein 1 (LRP1) as a key regulator of tau uptake and spread [107]. A C. elegans tau model utilizing human LRP1 or its worm ortholog (lrp-1) [108] may provide a window through which the molecular mechanisms underlying this process in vivo can be examined in unprecedented detail.
1.4. Parkinson’s disease (PD) models
Parkinson’s disease (PD) is the second most common ND, affecting approximately 2–3% of the population over 65 years of age [109]. Clinically, PD is characterized by the presence of four features – resting tremors, muscle rigidity, slowness of movement (bradykinesia), and postural instability [110]. As the disease progresses, patients experience increasing motor dysfunction and may also develop non-motor symptoms, such as mood disturbances, cognitive impairments, and dementia. At a cellular level, PD is caused by the selective death of dopaminergic neurons in the substantia nigra pars compacta, a region of the basal ganglia critical for the planning of motor functions and goal-directed behaviors. This widespread neuronal death is accompanied by the presence of intracellular Lewy bodies – dense aggregates primarily composed of the protein α-synuclein. Similar to other NDs, the majority of PD cases are sporadic, though heritable forms of the disease linked to specific genetic mutations account for an estimated 5–10% of cases. These include rare, dominant mutations in the SCNA gene encoding α-synuclein [111], and mutations in the leucine-rich repeat kinase 2 (LRRK2) [112], genetic variants of which can also act of risk factors for sporadic PD in the general population [113].
1.4.1. α-synuclein models
α-synuclein (α-syn) is a small (14 kDa) protein that, under physiological conditions, localizes at pre-synaptic terminals, and is thought to play a role in the regulation of neurotransmitter release and synaptic function [114,115]. The first transgenic C. elegans model to study of α-synuclein aggregation was based on the overexpression of WT α-syn and its A53T mutant form in all neurons, dopaminergic (DA) neurons, or motor neurons, respectively [116]. Interestingly, motor deficits were found in animals expressing α-syn pan-neuronally or in motor neurons, but not when expression was limited to DA neurons. However, pan-neuronal and DA expression of WT and A53T α-syn led to a significant loss (~30%) of neurons and dendritic breaks detectable in early larval stages, recapitulating the cell-type specificity of synucleinopathies targeting dopaminergic neurons. Knock-down of 4 genes related to the endocytic pathway, in worms with pan-neuronal expression of WT, A53T, or A30P mutant α-syn [117] induced profound growth and motor abnormalities. Notably, apa-2 knock-down promoted accumulation of phosphorylated α-syn in neuron cell bodies, mimicking synucleinopathy. Thus, endocytic pathway dysfunction may play a role in α-synuclein neurotoxicity. Two C. elegans models expressing either α-syn::GFP [118] or α-syn::YFP [119] in body-wall muscle cells have further been utilized for RNAi-based screens of genetic modulators of α-syn aggregation. Work with these models led to the identification of novel gene targets which protected against α-synuclein toxicity, and highlighted the impact of both vesicle trafficking and protein quality control systems in modulating α-syn aggregation.
To examine whether oligomeric α-syn or large α-syn inclusions function as the primary instigators of toxicity in synucleinopathies and related NDs, Karpinar et al. detailed the characterization of transgenic C. elegans lines expressing several α-syn variants with differing aggregation dynamics [120]. These included worms expressing WT or the engineered mutant α-syn (A30P/A56P/A76P), which shows lower aggregation propensity, in body-wall muscle cells, as well as strains expressing WT, α-syn (A30P/A56P/A76P), α-syn (A56P), and two disease-relevant α-syn mutants (A30P, A53T), in dopaminergic neurons. Strikingly, mutations that hinder fibrillization were associated with reductions in dopamine (DA) levels and impairments in DA-dependent behaviors (e.g. food response). This relationship tracked across all C. elegans lines investigated, as well as in 3 other validated PD model systems (HEK293T cells, rat primary neurons, and Drosophila). Taken together, these data indicate an association between an increase in soluble α-syn oligomers and increased neurotoxicity, supporting the hypothesis that these intermediate species are primary instigators of α-syn toxicity.
1.4.2. LRRK2 models
Mutations in the leucine-rich repeat kinase 2 (LRRK2) gene are the most frequent genetic cause of PD, accounting for approximately 4% of familial PD cases, and 1% of sporadic PD diagnoses worldwide [121]. LRRK2 is a member of the ROCO superfamily, characterized by the presence of a Ras-like G-domain (Roc), a C-terminal of Roc domain (COR), and a kinase domain [122]. The exact physiological function of LRRK2 is not known, though it has been linked to an array of diverse cellular functions, including cytoskeletal maintenance, vesicular trafficking, neurite outgrowth, and autophagy [123]. To date, six LRRK2 mutations have been linked to PD pathogenesis, the most common of these being the G2019S mutation, located in the protein’s kinase domain.
The first transgenic C. elegans models of LRRK2-mediated PD pathology were based on strains with pan-neuronal expression of WT, mutant, and kinase-dead (KD) human LRRK2 variants [124]. These included the G2019S and R1441C mutants and the KD and 1441C/KD kinase-dead variants. LRRK2 expression led to rapid loss of dopaminergic markers beginning in early adulthood; this phenotype was more pronounced in animals expressing LRRK2 (G2019S). Notably, the expression of wild-type, but not mutant LRRK2 in neurons significantly reduced toxicity of the mitochondrial complex I inhibitor, rotenone, and the mitochondrial toxin, paraquat, suggesting a role for LRRK2 in modulating mitochondrial function. Treatment of C. elegans strains expressing LRRK2 G2019S or R1441C with four promising LRRK2 kinase inhibitors (LRRK2-IN1, TTT-3002, GW5074, and sorafineb) significantly rescued dopaminergic behavioral deficits and neuron loss [125-128]. Strikingly, for LRRK2 and TTT-3002, this effect persisted even when treatment commenced after symptoms appeared, suggesting that LRRK2 kinase inhibition can reverse dopaminergic degeneration. Taken as a whole, C. elegans models of LRRK2-linked PD pathology served as critical steps to elucidating LRRK2 inhibition as a potential pharmaceutical target for the treatment of PD.
1.5. Prion disease models
Prion diseases are a group of rare, fatal neurodegenerative diseases affecting animals and humans. Uniquely, these disorders are caused by an infectious misfolded protein known as a prion (PrP), from the term “protein-only, infectious particle” [129]. The normal cellular prion protein (PrPC) is a small cell-surface glycoprotein rich in α-helical content. Upon exposure to an infectious prion species, PrPC adopts a pathogenic conformation with high β-sheet character, (PrPSc) [130]. According to the “seeding-nucleation” model, PrPSc catalyzes the conversion of PrPC monomers, leading to the formation of large PrPSc fibrils, which are prone to fragmentation, allowing them to break off and migrate to incite further prion conversion [131,132]. Most prion disease cases are sporadic in origin, the most common being sporadic Cruetzfeldt-Jakob disease (sCJD), which accounts for 85% of all CJD cases [133]. However, they can also arise from heritable genetic mutations or acquired through contact with infected materials, such as in cases of “Mad Cow” disease caused by consumption of contaminated meat products from cattle suffering from Bovine Spongiform Encephalopathy (BSE). Autosomal dominant mutations in the PRNP gene encoding for PrPC have been linked to inherited forms of prion disease, such as familial Cruetzfeldt-Jakob disease (fCJD), fatal familial insomnia (FFI), and Gerstmann-Sträussler-Scheinker syndrome (GSS). Despite their rarity, prion diseases have piqued interest due to the unique infectious nature of the PrPSc agent and its structural likeness to misfolded protein aggregates implicated in other NDs. Consequently, research dissecting the molecular basis of prion pathogenesis has important implications for our broader understanding of the role of protein aggregation in NDs.
To date, five independent transgenic C. elegans prion models have been reported in the literature. A complementary pair of model strains are based on the expression of either human WT PrP or PG13-PrP – a mutant form linked to early-onset Gerstmann-Sträussler-Scheinker syndrome (GSS) [134] – in mechanosensory neurons [135]. Here, worms expressing PG13-PrP, but not WT PrP, demonstrated a progressive loss of touch, indicative of PLM neuronal dysfunction. Furthermore, animals expressing PG13-PrP in a src-2 (a C. elegans Fyn-related kinase ortholog) null background showed a significant reduction in neuronal dysfunction induced by mutant PrP, suggesting a role for Fyn-related kinase signaling in mediating prion neurotoxicity. Another PrP model relies on pan-neuronal expression of the endogenous mouse PrP [136]. Here, molecules of murine PrP were N-glycosylated, GPI-anchored, and found on neuronal plasma membranes, consistent with mammalian studies of PrP expression. Examining multiple lines with a spectrum of PrP-expression levels, it was found that those expressing high levels of PrP demonstrated significant growth impairments, abnormal body morphology, striking reductions in lifespan, and a progressive uncoordinated phenotype. In comparison, none of these phenotypes were observed in lines expressing low-levels of PrP. To assess the potential anti-apoptotic role of PrPC, murine BAX was expressed in dopaminergic neurons, which are required for normal food-sensing behavior in C. elegans [137]. While animals expressing only BAX exhibited compromised food-sensing ability, those crossed with low-expressingric-19::PrP worms demonstrated normal food-sensing behavior, and microscopic analysis revealed a rescue of BAX-induced cell death in those expressing PrPC. As such, PrPC expressing C. elegans recapitulated neuropathological hallmarks of mammalian prion disease, and demonstrated the first evidence of PrP’s anti-BAX activity in vivo.
To study the cellular pathways underlying prion transmission while avoiding the complications of utilizing infectious mammalian prions, Nussbaum-Kramer et al. generated a transgenic C. elegans models expressing three versions of the yeast prion domain of Sup35 fused to a YFP tag in body-wall muscle cells [138]. These included the full-length wild-type domain (NM), a deletion of the oligorepeat region (RΔ2-5), and a domain containing an expansion of the oligorepeat region (R2E2). Here, aggregate formation progressed proportionally to the length of the oligorepeat region, such that R2E2 formed inclusions more rapidly and in higher numbers than NM, while RΔ2-5 remained primarily soluble. Furthermore, R2E2 aggregates were found to be targeted for lysosomal degradation, comprising a pool of vesicles with abnormal tubular morphology. These tubular vesicles were found to spread to non-expressing cells by vesicular transport, and, strikingly, were capable of inducing the aggregation of previously soluble RΔ2-5. As such, expression of the yeast Sup35 prion in a multicellular system and its adaptability to transport mechanisms shed light on the potential utility of this system for the study of common molecular pathways used by misfolded proteins to disseminate toxicity.
1.6. Advantages and limitations of using C. elegans to study neurodegeneration
C. elegans is a crucial animal model in the aging field and in the study of aging-associated diseases. Numerous longevity pathways were first discovered and characterized in C. elegans, such as the insulin-like receptor gene, daf-2. Throughout the study of worm ND models in long-lived mutant backgrounds, extensive evidence suggests a role for this and other highly conserved pathways in the global attenuation of proteotoxicity induced by numerous disease-relevant protein aggregates. Due to their genetic tractability, cost-effectiveness, and naturally short lifespan (~30 days), C. elegans models are uniquely poised to serve as an ideal model to investigate how organismal age contributes to pathological protein aggregation at the molecular level. Furthermore, an large percentage of C. elegans genes are highly conserved, and proteomics analyses suggest that only 11% or less of the C. elegans proteome contains nematode-specific genes [139], further bolstering the idea that discoveries made in worm ND models will yield tractable information relevant to human disease. One particularly compelling advantage of C. elegans is their unique application as an in vivo system to large-scale screening techniques, such as RNA interference (RNAi). With the generation of tissue and even neuron-specific RNAi sensitive strains [140] C. elegans offers an unique platform to elucidate the impact of individual genes on the regulation of neuronal function and health.
The simple architecture of C. elegans also comes along with its own limitations: many defined tissue/organ systems critically affected in NDs, in particular the CNS and brain, are absent in worms. NDs are multifactorial diseases, and a growing body of literature supports a role for co-morbid processes, such as inflammation, in driving their pathology. Given the worm’s lack of an adaptive immune system, C. elegans models are not particularly suitable to studies of this topic. Furthermore, despite their rapid growth, the worm’s small size in comparison to other model organisms can render the task of obtaining sufficient quantities of animals for biochemical studies difficult [141]. Nevertheless, C. elegans serves as an in vivo model amendable to facile transgenic manipulation, vast microscopy approaches, and the direct linking of biochemical read-outs with quantifiable behaviors renders it a key tool in the context of NDs and other aging-associated disorders.
2. Summary
C. elegans ND models serve as powerful tools for studying the molecular pathology governing aggregate formation and disease progression. Strikingly, worm models of NDs recapitulate many key features of protein aggregation in humans, and the presence of worm orthologs for proteins of interest to human disease provides further evidence that the molecular and biochemical insights gained from these models may serve as critical foundations for furthering our understanding of these diseases. Future experimental aims may focus on utilizing these models for identifying key regulators of global proteostasis, such as chaperone machinery, as targets for modulating aggregate-associated toxicity and neurodegeneration.
At a glance:
Neurodegenerative diseases (NDs) are among the most devastating and least understood aging-associated disorders, and pose a substantial and growing burden on healthcare systems worldwide.
Despite decades of intensive research, few, if any, treatments exist for these disorders. The availability of high-throughput model systems for manipulation are crucial to dissecting the molecular pathways regulating protein aggregation in vivo and identifying potential targets for intervention.
First put forward in the 1970s1 as a model for the study of neurodevelopment and developmental cell biology, the nematode Caenorhabditis elegans (C. elegans) has emerged as a powerful tool for examining the molecular basis of ND pathology.
C. elegans models of polyglutamine expansion diseases point to abnormal CAG repeats present in Huntington’s disease and related disorders as disruptors of global proteostasis and define age and threshold toxicity as key modulators of aggregate formation.
Models of ALS-linked patient mutations in RNA binding proteins TDP-43 and FUS hypothesize toxic gain-of-function as a dominant feature of familial and sporadic ALS pathologies.
Acknowledgments
MCT is supported by the Azheimer’s Association (AARG-19-615132) and Brightfocus Foundation (A2019157S). KVP is supported by the NIH Cellular and Molecular Biology Training Grant T32-GM007315.
Table of definitions for abbreviations
- C. elegans
specific nomenclature, and technical terms used in the manuscript
- NDs
Neurodegenerative diseases
- AD
Alzheimer’s disease
- PD
Parkinson’s disease
- ALS
Amyotrophic lateral sclerosis
- HD
Huntington’s disease
- SCA
Spinocerebellar ataxia
- RNAi
RNA interference
- SBMA
Spinal and bulbar muscular atrophy
- DRPLA
Dentatorubropallidoluysian atrophy
- G/C/YFP
Green/cyan/yellow fluorescent protein
- FRAP
Fluorescence recovery after photobleaching
- FRET
Förster resonance energy transfer
- Htt
Huntingtin protein
- ASH
Amphid neurons, single
- MJD
Machado-Joseph disease
- AT3
Ataxin-3
- IIS
Insulin/insulin-like growth factor signaling
- MND
Motor neuron disease
- fALS
Familial amyotrophic lateral sclerosis
- SOD-1
Cu/Zn superoxide dismutase 1
- TDP-43
TAR DNA-binding protein 43
- sALS
Sporadic amyotrophic lateral sclerosis
- Unc
Uncoordinated
- FUS/TLS
Fused in sarcoma/translocated in sarcoma
- NFT
Neurofibrillary tangles
- FTDP-17
Frontotemporal dementia and parkinsonism linked to chromosome 17
- FTD
Frontotemporal dementia
- MAPT
Microtubule-associated protein tau
- ATPZ
Aminothienopyridazine
- LRRK2
Leucine-rich repeat kinase 2
- sCJD
Sporadic Cruetzfeldt-Jakob disease
- fCJD
Familial Cruetzfeldt-Jakob disease
- BSE
Bovine spongiform encephalopathy
- FFI
Fatal familial insomnia
- GSS
Gerstmann-Sträussler-Scheinker syndrome
Footnotes
Declaration of Competing Interest
The authors declare that there is no conflicts of interest.
References
- [1].Brenner S, The genetics of Caenorhabditis elegans, Genetics 77 (1974) 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Gitler AD, Dhillon P, Shorter J, Neurodegenerative disease: models, mechanisms, and a new hope, Dis. Model. Mech 10 (2017) 499–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Wijesekera LC, Leigh PN, Amyotrophic lateral sclerosis, Orphanet J. Rare Dis 4 (2009) 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Kakizuka A, Protein precipitation: a common etiology in neurodegenerative disorders? Trends Genet. TIG 14 (1998) 396–402. [DOI] [PubMed] [Google Scholar]
- [5].Chiti F, Dobson CM, Protein misfolding, functional amyloid, and human disease, Annu. Rev. Biochem 75 (2006) 333–366. [DOI] [PubMed] [Google Scholar]
- [6].Prahlad V, Morimoto RI, Integrating the stress response: lessons for neurodegenerative diseases from C. elegans, Trends Cell Biol. 19 (2009) 52–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].C. elegans II, Cold Spring Harbor Laboratory Press, 1997. [Google Scholar]
- [8].Kaletta T, Hengartner MO, Finding function in novel targets: C. elegans as a model organism, Nat. Rev. Drug Discov 5 (2006) 387–398. [DOI] [PubMed] [Google Scholar]
- [9].Corsi AK, A biochemist’s guide to C. elegans, Anal. Biochem 359 (2006) 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Fire A, et al. , Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans, Nature 391 (1998) 806–811. [DOI] [PubMed] [Google Scholar]
- [11].Dickinson DJ, Ward JD, Reiner DJ, Goldstein B, Engineering the Caenorhabditis elegans genome using Cas9-triggered homologous recombination, Nat. Methods 10 (2013) 1028–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Cook SJ, et al. , Whole-animal connectomes of both Caenorhabditis elegans sexes, Nature 571 (2019) 63–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Lieberman AP, Shakkottai VG, Albin RL, Polyglutamine repeats in neurodegenerative diseases, Annu. Rev. Pathol 14 (2019) 1–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Cohen-Carmon D, Meshorer E, Polyglutamine (polyQ) disorders, Nucleus 3 (2012) 433–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Lee AL, Ung HM, Sands LP, Kikis EA, A new Caenorhabditis elegans model of human huntingtin 513 aggregation and toxicity in body wall muscles, PloS One 12 (2017), e0173644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Satyal SH, et al. , Polyglutamine aggregates alter protein folding homeostasis in Caenorhabditis elegans, Proc. Natl. Acad. Sci. U. S. A 97 (2000) 5750–5755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Morley JF, Brignull HR, Weyers JJ, Morimoto RI, The threshold for polyglutamine-expansion protein aggregation and cellular toxicity is dynamic and influenced by aging in Caenorhabditis elegans, Proc. Natl. Acad. Sci. U. S. A 99 (2002) 10417–10422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Brignull HR, Moore FE, Tang SJ, Morimoto RI, Polyglutamine proteins at the pathogenic threshold display neuron-specific aggregation in a pan-neuronal Caenorhabditis elegans model, J. Neurosci. Off. J. Soc. Neurosci 26 (2006) 7597–7606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Ward JM, La Spada AR, The expanding world of stem cell modeling of Huntington’s disease: creating tools with a promising future, Genome Med. 4 (2012) 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. The Huntington’s Disease Collaborative Research Group, Cell 72 (1993) 971–983. [DOI] [PubMed] [Google Scholar]
- [21].Duyao M, et al. , Trinucleotide repeat length instability and age of onset in Huntington’s disease, Nat. Genet 4 (1993) 387–392. [DOI] [PubMed] [Google Scholar]
- [22].Dayalu P, Albin RL, Huntington disease: pathogenesis and treatment, Neurol. Clin 33 (2015) 101–114. [DOI] [PubMed] [Google Scholar]
- [23].Faber PW, Alter JR, MacDonald ME, Hart AC, Polyglutamine-mediated dysfunction and apoptotic death of a Caenorhabditis elegans sensory neuron, Proc. Natl. Acad. Sci. U. S. A 96 (1999) 179–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Parker JA, et al. , Expanded polyglutamines in Caenorhabditis elegans cause axonal abnormalities and severe dysfunction of PLM mechanosensory neurons without cell death, Proc. Natl. Acad. Sci. U. S. A 98 (2001) 13318–13323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Faber PW, Voisine C, King DC, Bates EA, Hart AC, Glutamine/proline-rich PQE-1 proteins protect Caenorhabditis elegans neurons from huntingtin polyglutamine neurotoxicity, Proc. Natl. Acad. Sci. U. S. A 99 (2002) 17131–17136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Varma H, Cheng R, Voisine C, Hart AC, Stockwell BR, Inhibitors of metabolism rescue cell death in Huntington’s disease models, Proc. Natl. Acad. Sci. U. S. A 104 (2007) 14525–14530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Wellington CL, et al. , Caspase cleavage of gene products associated with triplet expansion disorders generates truncated fragments containing the polyglutamine tract, J. Biol. Chem 273 (1998) 9158–9167. [DOI] [PubMed] [Google Scholar]
- [28].Butterworth NJ, et al. , Trinucleotide (CAG) repeat length is positively correlated with the degree of DNA fragmentation in Huntington’s disease striatum, Neuroscience 87 (1998) 49–53. [DOI] [PubMed] [Google Scholar]
- [29].Sánchez I, et al. , Caspase-8 is required for cell death induced by expanded polyglutamine repeats, Neuron 22 (1999) 623–633. [DOI] [PubMed] [Google Scholar]
- [30].Bettencourt C, Lima M, Joseph Machado, Disease: from first descriptions to new perspectives, Orphanet J. Rare Dis 6 (2011) 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Maciel P, et al. , Improvement in the molecular diagnosis of Machado-Joseph disease, Arch. Neurol 58 (2001) 1821–1827. [DOI] [PubMed] [Google Scholar]
- [32].Teixeira-Castro A, et al. , Neuron-specific proteotoxicity of mutant ataxin-3 in C. elegans: rescue by the DAF-16 and HSF-1 pathways, Hum. Mol. Genet 20 (2011) 2996–3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Christie NTM, Lee AL, Fay HG, Gray AA, Kikis EA, Novel polyglutamine model uncouples proteotoxicity from aging, PloS One 9 (2014), e96835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Chen S, Sayana P, Zhang X, Le W, Genetics of amyotrophic lateral sclerosis: an update, Mol. Neurodegener 8 (2013) 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Cudkowicz ME, et al. , Epidemiology of mutations in superoxide dismutase in amyotrophic lateral sclerosis, Ann. Neurol 41 (1997) 210–221. [DOI] [PubMed] [Google Scholar]
- [36].Fridovich I, Superoxide dismutases, Adv. Enzymol. Relat. Area Mol. Biol 58 (1986) 61–97. [DOI] [PubMed] [Google Scholar]
- [37].Banci L, et al. , SOD1 and amyotrophic lateral sclerosis: mutations and oligomerization, PloS One 3 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Valentine JS, Doucette PA, Zittin Potter S, Copper-zinc superoxide dismutase and amyotrophic lateral sclerosis, Annu. Rev. Biochem 74 (2005) 563–593. [DOI] [PubMed] [Google Scholar]
- [39].Bruijn LI, et al. , ALS-linked SOD1 mutant G85R mediates damage to astrocytes and promotes rapidly progressive disease with SOD1-containing inclusions, Neuron 18 (1997) 327–338. [DOI] [PubMed] [Google Scholar]
- [40].Bruijn LI, et al. , Aggregation and motor neuron toxicity of an ALS-linked SOD1 mutant independent from wild-type SOD1, Science 281 (1998) 1851–1854. [DOI] [PubMed] [Google Scholar]
- [41].Ferrante RJ, et al. , Evidence of increased oxidative damage in both sporadic and familial amyotrophic lateral sclerosis, J. Neurochem 69 (1997) 2064–2074. [DOI] [PubMed] [Google Scholar]
- [42].Oeda T, et al. , Oxidative stress causes abnormal accumulation of familial amyotrophic lateral sclerosis-related mutant SOD1 in transgenic Caenorhabditis elegans, Hum. Mol. Genet 10 (2001) 2013–2023. [DOI] [PubMed] [Google Scholar]
- [43].Wang J, et al. , An ALS-linked mutant SOD1 produces a locomotor defect associated with aggregation and synaptic dysfunction when expressed in neurons of Caenorhabditis elegans, PLoS Genet. 5 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Gidalevitz T, Krupinski T, Garcia S, Morimoto RI, Destabilizing protein polymorphisms in the genetic background direct phenotypic expression of mutant SOD1 toxicity, PLoS Genet. 5 (2009), e1000399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Witan H, et al. , Heterodimer formation of wild-type and amyotrophic lateral sclerosis-causing mutant Cu/Zn-superoxide dismutase induces toxicity independent of protein aggregation, Hum. Mol. Genet 17 (2008) 1373–1385. [DOI] [PubMed] [Google Scholar]
- [46].Deng H-X, et al. , Conversion to the amyotrophic lateral sclerosis phenotype is associated with intermolecular linked insoluble aggregates of SOD1 in mitochondria, Proc. Natl. Acad. Sci. U. S. A 103 (2006) 7142–7147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Fukada K, et al. , Stabilization of mutant Cu/Zn superoxide dismutase (SOD1) protein by coexpressed wild SOD1 protein accelerates the disease progression in familial amyotrophic lateral sclerosis mice, Eur. J. Neurosci 14 (2001) 2032–2036. [DOI] [PubMed] [Google Scholar]
- [48].Li J, Huang K, Le W, Establishing a novel C. elegans model to investigate the role of autophagy in amyotrophic lateral sclerosis, Acta Pharmacol. Sin 34 (2013) 644–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Boccitto M, Lamitina T, Kalb RG, Daf-2 signaling modifies mutant SOD1 toxicity in C. elegans, PloS One 7 (2012), e33494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Mackenzie IRA, Rademakers R, The role of TDP-43 in amyotrophic lateral sclerosis and frontotemporal dementia, Curr. Opin. Neurol 21 (2008) 693–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Neumann M, et al. , Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis, Science 314 (2006) 130–133. [DOI] [PubMed] [Google Scholar]
- [52].Arai T, et al. , TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis, Biochem. Biophys. Res. Commun 351 (2006) 602–611. [DOI] [PubMed] [Google Scholar]
- [53].Ash PEA, et al. , Neurotoxic effects of TDP-43 overexpression in C. elegans, Hum. Mol. Genet 19 (2010) 3206–3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Ayala YM, et al. , Human, Drosophila, and C.elegans TDP43: nucleic acid binding properties and splicing regulatory function, J. Mol. Biol 348 (2005) 575–588. [DOI] [PubMed] [Google Scholar]
- [55].Liachko NF, Guthrie CR, Kraemer BC, Phosphorylation promotes neurotoxicity in a Caenorhabditis elegans model of TDP-43 proteinopathy, J. Neurosci. Off. J. Soc. Neurosci 30 (2010) 16208–16219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Hasegawa M, et al. , Phosphorylated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis, Ann. Neurol 64 (2008) 60–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Inukai Y, et al. , Abnormal phosphorylation of ser409/410 of TDP-43 in FTLD-U and ALS, FEBS Lett. 582 (2008) 2899–2904. [DOI] [PubMed] [Google Scholar]
- [58].Zhang T, Mullane PC, Periz G, Wang J TDP-43 neurotoxicity and protein aggregation modulated by heat shock factor and insulin/IGF-1 signaling, Hum. Mol. Genet 20 (2011) 1952–1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Kwiatkowski TJ, et al. , Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis, Science 323 (2009) 1205–1208. [DOI] [PubMed] [Google Scholar]
- [60].Lagier-Tourenne C, Polymenidou M, Cleveland DW, TDP-43 and FUS/TLS: emerging roles in RNA processing and neurodegeneration, Hum. Mol. Genet 19 (2010) R46–R64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Deng H-X, et al. , FUS-immunoreactive inclusions are a common feature in sporadic and non-SOD1 familial amyotrophic lateral sclerosis, Ann. Neurol 67 (2010) 739–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Dormann D, et al. , ALS-associated fused in sarcoma (FUS) mutations disrupt Transportin-mediated nuclear import, EMBO J. 29 (2010) 2841–2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Dormann D, Haass C, TDP-43 and FUS: a nuclear affair, Trends Neurosci. 34 (2011) 339–348. [DOI] [PubMed] [Google Scholar]
- [64].Murakami T, et al. , ALS mutations in FUS cause neuronal dysfunction and death in Caenorhabditis elegans by a dominant gain-of-function mechanism, Hum. Mol. Genet 21 (2012) 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Vance C, et al. , Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6, Science 323 (2009) 1208–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Kumar A, Singh A, Ekavali NULL, A review on Alzheimer’s disease pathophysiology and its management: an update, Pharmacol. Rep. PR 67 (2015) 195–203. [DOI] [PubMed] [Google Scholar]
- [67].Jagust W, Vulnerable neural systems and the borderland of brain aging and neurodegeneration, Neuron 77 (2013) 219–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Förstl H, Kurz A, Clinical features of Alzheimer’s disease, Eur. Arch. Psychiatr. Clin. Neurosci 249 (1999) 288–290. [DOI] [PubMed] [Google Scholar]
- [69].Kosik KS, Joachim CL, Selkoe DJ, Microtubule-associated protein tau (tau) is a major antigenic component of paired helical filaments in Alzheimer disease, Proc. Natl. Acad. Sci. U. S. A 83 (1986) 4044–4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Vinters HV, Emerging concepts in Alzheimer’s disease, Annu. Rev. Pathol 10 (2015) 291–319. [DOI] [PubMed] [Google Scholar]
- [71].Nunan J, Small DH, Regulation of APP cleavage by alpha-, beta- and gamma-secretases, FEBS Lett. 483 (2000) 6–10. [DOI] [PubMed] [Google Scholar]
- [72].Hardy J, Allsop D, Amyloid deposition as the central event in the aetiology of Alzheimer’s disease, Trends Pharmacol. Sci 12 (1991) 383–388. [DOI] [PubMed] [Google Scholar]
- [73].Hardy JA, Higgins GA, Alzheimer’s disease: the amyloid cascade hypothesis, Science 256 (1992) 184–185. [DOI] [PubMed] [Google Scholar]
- [74].Goate A, et al. , Segregation of a missense mutation in the amyloid precursor protein gene with familial Alzheimer’s disease, Nature 349 (1991) 704–706. [DOI] [PubMed] [Google Scholar]
- [75].Levy-Lahad E, et al. , Candidate gene for the chromosome 1 familial Alzheimer’s disease locus, Science 269 (1995) 973–977. [DOI] [PubMed] [Google Scholar]
- [76].Sherrington R, et al. , Cloning of a gene bearing missense mutations in early-onset familial Alzheimer’s disease, Nature 375 (1995) 754–760. [DOI] [PubMed] [Google Scholar]
- [77].Mehta D, Jackson R, Paul G, Shi J, Sabbagh M, Why do trials for Alzheimer’s disease drugs keep failing? A discontinued drug perspective for 2010–2015, Expet Opin. Invest. Drugs 26 (2017) 735–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Weingarten MD, Lockwood AH, Hwo SY, Kirschner MW, A protein factor essential for microtubule assembly, Proc. Natl. Acad. Sci. U. S. A 72 (1975) 1858–1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Grundke-Iqbal I, et al. , Abnormal phosphorylation of the microtubule-associated protein tau (tau) in Alzheimer cytoskeletal pathology, Proc. Natl. Acad. Sci. U. S. A 83 (1986) 4913–4917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Bejanin A, et al. , Tau pathology and neurodegeneration contribute to cognitive impairment in Alzheimer’s disease, Brain J. Neurol 140 (2017) 3286–3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Braak H, Del Tredici K, Are cases with tau pathology occurring in the absence of Aβ deposits part of the AD-related pathological process? Acta Neuropathol. 128 (2014) 767–772. [DOI] [PubMed] [Google Scholar]
- [82].Lee VM, Goedert M, Trojanowski JQ, Neurodegenerative tauopathies, Annu. Rev. Neurosci 24 (2001) 1121–1159. [DOI] [PubMed] [Google Scholar]
- [83].Link CD, Expression of human beta-amyloid peptide in transgenic Caenorhabditis elegans, Proc. Natl. Acad. Sci. U. S. A 92 (1995) 9368–9372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Cohen E, Bieschke J, Perciavalle RM, Kelly JW, Dillin A, Opposing activities protect against age-onset proteotoxicity, Science 313 (2006) 1604–1610. [DOI] [PubMed] [Google Scholar]
- [85].Florez-McClure ML, Hohsfield LA, Fonte G, Bealor MT, Link CD, Decreased insulin-receptor signaling promotes the autophagic degradation of beta-amyloid peptide in C. elegans, Autophagy 3 (2007) 569–580. [DOI] [PubMed] [Google Scholar]
- [86].McColl G, et al. , The Caenorhabditis elegans A beta 1-42 model of Alzheimer disease predominantly expresses A beta 3-42, J. Biol. Chem 284 (2009) 22697–22702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].McColl G, et al. , Utility of an improved model of amyloid-beta (Aβ1-42) toxicity in Caenorhabditis elegans for drug screening for Alzheimer’s disease, Mol. Neurodegener 7 (2012) 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Yatin SM, Varadarajan S, Link CD, Butterfield DA, In vitro and in vivo oxidative stress associated with Alzheimer’s amyloid beta-peptide (1-42), Neurobiol. Aging 20 (1999) 325–330, discussion 339-342. [DOI] [PubMed] [Google Scholar]
- [89].Pike CJ, et al. , Structure-activity analyses of beta-amyloid peptides: contributions of the beta 25-35 region to aggregation and neurotoxicity, J. Neurochem 64 (1995) 253–265. [DOI] [PubMed] [Google Scholar]
- [90].Berlett BS, Stadtman ER, Protein oxidation in aging, disease, and oxidative stress, J. Biol. Chem 272 (1997) 20313–20316. [DOI] [PubMed] [Google Scholar]
- [91].Drake J, Link CD, Butterfield DA, Oxidative stress precedes fibrillar deposition of Alzheimer’s disease amyloid beta-peptide (1-42) in a transgenic Caenorhabditis elegans model, Neurobiol. Aging 24 (2003) 415–420. [DOI] [PubMed] [Google Scholar]
- [92].Mango SE, Stop making nonSense: the C. elegans smg genes, Trends Genet. TIG 17 (2001) 646–653. [DOI] [PubMed] [Google Scholar]
- [93].Wu Y, et al. , Amyloid-beta-induced pathological behaviors are suppressed by Ginkgo biloba extract EGb 761 and ginkgolides in transgenic Caenorhabditis elegans, J. Neurosci. Off. J. Soc. Neurosci 26 (2006) 13102–13113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].van der Zee J, Van Broeckhoven C Dementia, Frontotemporal lobar degeneration-building on breakthroughs, Nat. Rev. Neurol 10 (2013) 70–72, 2014. [DOI] [PubMed] [Google Scholar]
- [95].Ghetti B, et al. , Invited review: frontotemporal dementia caused by microtubule-associated protein tau gene (MAPT) mutations: a chameleon for neuropathology and neuroimaging, Neuropathol. Appl. Neurobiol 41 (2015) 24–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Kraemer BC, et al. , Neurodegeneration and defective neurotransmission in a Caenorhabditis elegans model of tauopathy, Proc. Natl. Acad. Sci. U. S. A 100 (2003) 9980–9985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Miyasaka T, et al. , Progressive neurodegeneration in C. elegans model of tauopathy, Neurobiol. Dis 20 (2005) 372–383. [DOI] [PubMed] [Google Scholar]
- [98].Wittmann CW, et al. , Tauopathy in Drosophila: neurodegeneration without neurofibrillary tangles, Science 293 (2001) 711–714. [DOI] [PubMed] [Google Scholar]
- [99].Ishihara T, et al. , Age-dependent emergence and progression of a tauopathy in transgenic mice overexpressing the shortest human tau isoform, Neuron 24 (1999) 751–762. [DOI] [PubMed] [Google Scholar]
- [100].Lewis J, et al. , Neurofibrillary tangles, amyotrophy and progressive motor disturbance in mice expressing mutant (P301L) tau protein, Nat. Genet 25 (2000) 402–405. [DOI] [PubMed] [Google Scholar]
- [101].von Bergen M, et al. , Mutations of tau protein in frontotemporal dementia promote aggregation of paired helical filaments by enhancing local beta-structure, J. Biol. Chem 276 (2001) 48165–48174. [DOI] [PubMed] [Google Scholar]
- [102].Mocanu M-M, et al. , The potential for beta-structure in the repeat domain of tau protein determines aggregation, synaptic decay, neuronal loss, and coassembly with endogenous Tau in inducible mouse models of tauopathy, J. Neurosci. Off. J. Soc. Neurosci 28 (2008) 737–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Eckermann K, et al. , The beta-propensity of Tau determines aggregation and synaptic loss in inducible mouse models of tauopathy, J. Biol. Chem 282 (2007) 31755–31765. [DOI] [PubMed] [Google Scholar]
- [104].Wang YP, Biernat J, Pickhardt M, Mandelkow E, Mandelkow E-M, Stepwise proteolysis liberates tau fragments that nucleate the Alzheimer-like aggregation of full-length tau in a neuronal cell model, Proc. Natl. Acad. Sci. U. S. A 104 (2007) 10252–10257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Fatouros C, et al. , Inhibition of tau aggregation in a novel Caenorhabditis elegans model of tauopathy mitigates proteotoxicity, Hum. Mol. Genet. 21 (2012) 3587–3603. [DOI] [PubMed] [Google Scholar]
- [106].Brandt R, Gergou A, Wacker I, Fath T, Hutter H, A Caenorhabditis elegans model of tau hyperphosphorylation: induction of developmental defects by transgenic overexpression of Alzheimer’s disease-like modified tau, Neurobiol. Aging 30 (2009) 22–33. [DOI] [PubMed] [Google Scholar]
- [107].Rauch JN, et al. , LRP1 is a master regulator of tau uptake and spread, Nature 580 (2020) 381–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Yochem J, Greenwald I, A gene for a low density lipoprotein receptor-related protein in the nematode Caenorhabditis elegans, Proc. Natl. Acad. Sci. U. S. A 90 (1993) 4572–4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Poewe W, et al. , Parkinson disease, Nat. Rev. Dis. Primer 3 (2017) 1–21. [DOI] [PubMed] [Google Scholar]
- [110].Jankovic J, Parkinson’s disease: clinical features and diagnosis, J. Neurol. Neurosurg. Psychiatry 79 (2008) 368–376. [DOI] [PubMed] [Google Scholar]
- [111].Gasser T, Molecular pathogenesis of Parkinson disease: insights from genetic studies, Expet Rev. Mol. Med 11 (2009) e22. [DOI] [PubMed] [Google Scholar]
- [112].Paisán-Ruzíz C, et al. , Cloning of the gene containing mutations that cause PARK8-linked Parkinson’s disease, Neuron 44 (2004) 595–600. [DOI] [PubMed] [Google Scholar]
- [113].Satake W, et al. , Genome-wide association study identifies common variants at four loci as genetic risk factors for Parkinson’s disease, Nat. Genet 41 (2009) 1303–1307. [DOI] [PubMed] [Google Scholar]
- [114].Iwai A, et al. , The precursor protein of non-A beta component of Alzheimer’s disease amyloid is a presynaptic protein of the central nervous system, Neuron 14 (1995) 467–475. [DOI] [PubMed] [Google Scholar]
- [115].Zhang L, et al. , Semi-quantitative analysis of alpha-synuclein in subcellular pools of rat brain neurons: an immunogold electron microscopic study using a C-terminal specific monoclonal antibody, Brain Res. 1244 (2008) 40–52. [DOI] [PubMed] [Google Scholar]
- [116].Lakso M, et al. , Dopaminergic neuronal loss and motor deficits in Caenorhabditis elegans overexpressing human alphα-synuclein, J. Neurochem 86 (2003) 165–172. [DOI] [PubMed] [Google Scholar]
- [117].Kuwahara T, et al. , A systematic RNAi screen reveals involvement of endocytic pathway in neuronal dysfunction in alphα-synuclein transgenic C. elegans, Hum. Mol. Genet 17 (2008) 2997–3009. [DOI] [PubMed] [Google Scholar]
- [118].Hamamichi S, et al. , Hypothesis-based RNAi screening identifies neuro-protective genes in a Parkinson’s disease model, Proc. Natl. Acad. Sci. U. S. A 105 (2008) 728–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].van Ham TJ, et al. , C. elegans model identifies genetic modifiers of alpha-synuclein inclusion formation during aging, PLoS Genet. 4 (2008), e1000027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Karpinar DP, et al. , Pre-fibrillar alpha-synuclein variants with impaired beta-structure increase neurotoxicity in Parkinson’s disease models, EMBO J. 28 (2009) 3256–3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Xiong Y, Dawson TM, Dawson VL, Models of LRRK2 associated Parkinson’s disease, Adv. Neurobiol 14 (2017) 163–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Bosgraaf L, Van Haastert PJM, Roc, a Ras/GTPase domain in complex proteins, Biochim. Biophys. Acta 1643 (2003) 5–10. [DOI] [PubMed] [Google Scholar]
- [123].Rideout HJ, Stefanis L, The neurobiology of LRRK2 and its role in the pathogenesis of Parkinson’s disease, Neurochem. Res 39 (2014) 576–592. [DOI] [PubMed] [Google Scholar]
- [124].Saha S, et al. , LRRK2 modulates vulnerability to mitochondrial dysfunction in Caenorhabditis elegans, J. Neurosci. Off. J. Soc. Neurosci 29 (2009) 9210–9218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Yao C, et al. , Kinase inhibitors arrest neurodegeneration in cell and C. elegans models of LRRK2 toxicity, Hum. Mol. Genet 22 (2013) 328–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Deng X, et al. , Characterization of a selective inhibitor of the Parkinson’s disease kinase LRRK2, Nat. Chem. Biol 7 (2011) 203–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Lee BD, et al. , Inhibitors of leucine-rich repeat kinase-2 protect against models of Parkinson’s disease, Nat. Med 16 (2010) 998–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Liu Z, et al. , Inhibitors of LRRK2 kinase attenuate neurodegeneration and Parkinson-like phenotypes in Caenorhabditis elegans and Drosophila Parkinson’s disease models, Hum. Mol. Genet 20 (2011) 3933–3942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Prusiner SB, Prions. Sci. Am 251 (1984) 50–59. [DOI] [PubMed] [Google Scholar]
- [130].Pan KM, et al. , Conversion of alpha-helices into beta-sheets features in the formation of the scrapie prion proteins, Proc. Natl. Acad. Sci. U. S. A 90 (1993) 10962–10966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Collinge J, Clarke AR, A general model of prion strains and their pathogenicity, Science 318 (2007) 930–936. [DOI] [PubMed] [Google Scholar]
- [132].Gillam JE, MacPhee CE, Modelling amyloid fibril formation kinetics: mechanisms of nucleation and growth, J. Phys. Condens. Matter Inst. Phys. J 25 (2013) 373101. [DOI] [PubMed] [Google Scholar]
- [133].Mead S, et al. , Balancing selection at the prion protein gene consistent with prehistoric kurulike epidemics, Science 300 (2003) 640–643. [DOI] [PubMed] [Google Scholar]
- [134].Laplanche JL, et al. , Prominent psychiatric features and early onset in an inherited prion disease with a new insertional mutation in the prion protein gene, Brain J. Neurol 122 (Pt 12) (1999) 2375–2386. [DOI] [PubMed] [Google Scholar]
- [135].Bizat N, et al. , Neuron dysfunction is induced by prion protein with an insertional mutation via a Fyn kinase and reversed by sirtuin activation in Caenorhabditis elegans, J. Neurosci. Off. J. Soc. Neurosci 30 (2010) 5394–5403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Park K-W, Li L, Prion protein in Caenorhabditis elegans: distinct models of anti-BAX and neuropathology, Prion 5 (2011) 28–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Kindt KS, et al. , Dopamine mediates context-dependent modulation of sensory plasticity in C. elegans, Neuron 55 (2007) 662–676. [DOI] [PubMed] [Google Scholar]
- [138].Nussbaum-Krammer CI, Park K-W, Li L, Melki R, Morimoto RI, Spreading of a prion domain from cell-to-cell by vesicular transport in Caenorhabditis elegans, PLoS Genet. 9 (2013), e1003351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Lai C-H, Chou C-Y, Ch’ang L-Y, Liu C-S, Lin W, Identification of novel human genes evolutionarily conserved in Caenorhabditis elegans by comparative proteomics, Genome Res. 10 (2000) 703–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Firnhaber C, Hammarlund M, Neuron-specific feeding RNAi in C. elegans and its use in a screen for essential genes required for GABA neuron function, PLoS Genet. 9 (2013), e1003921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Johnson TE, Advantages and disadvantages of Caenorhabditis elegans for aging research, Exp. Gerontol 38 (2003) 1329–1332. [DOI] [PubMed] [Google Scholar]