Summary
Activation of the STAT5 transcription factor downstream of the Interleukin-2 receptor (IL-2R) induces expression of Foxp3, a critical step in the differentiation of regulatory T (Treg) cells. Due to the pleiotropic effects of IL-2R signaling, it is unclear how STAT5 acts directly on the Foxp3 locus to promote its expression. Here, we report that IL-2 – STAT5 signaling converged on an enhancer (CNS0) during Foxp3 induction. CNS0 facilitated the IL-2 dependent CD25+Foxp3− precursor to Treg cell transition in the thymus. Its deficiency resulted in impaired Treg cell generation in neonates, which was partially mitigated with age. While the thymic Treg cell paucity caused by CNS0 deficiency did not result in autoimmunity on its own, it exacerbated autoimmune manifestations caused by disruption of the Aire gene. Thus, CNS0 enhancer activity ensures robust Treg cell differentiation early in postnatal life and cooperatively with other tolerance mechanisms minimizes autoimmunity.
Keywords: Regulatory T cells, Foxp3, enhancer, immune tolerance
Introduction
Regulatory T (Treg) cells play an essential role in establishing and maintaining immune tolerance (Josefowicz et al., 2012a; Sakaguchi et al., 2020). The majority of Treg cells express the high affinity subunit (CD25) of the receptor for interleukin (IL)-2, or IL-2R, which signals by inducing the activation of the transcription factor (TF) STAT5. IL-2 plays a central role in multiple aspects of Treg cell biology. Treg cell differentiation requires IL-2 – STAT5, in addition T cell receptor (TCR), signaling to induce Foxp3 expression (Burchill et al., 2003, 2007; Fontenot et al., 2005a, 2005b). Foxp3 serves as the Treg cell lineage specifying TF and its continued expression in differentiated Treg cells is required for Treg cell lineage stability and function (Fontenot et al., 2005b; Gavin et al., 2007; Lin et al., 2007; Williams and Rudensky, 2007). Additionally, in mature Treg cells, IL-2R signaling supports their persistence and immunosuppressive ability (Chinen et al., 2016; Fan et al., 2018; Liu et al., 2015; Toomer et al., 2019; Yao et al., 2007).
CD25+Foxp3− CD4 single positive (SP) thymocytes begin to emerge on day 1 after birth (P1), but then abruptly plateau. This is in contrast with a slower accumulation of Foxp3 expressing CD25+ CD4SP thymocytes, which begins at P3 (Fontenot et al., 2005c). The CD25+Foxp3− CD4SP thymocyte subset has been shown to be enriched for precursors of thymic Treg cells based on their overlapping TCR repertoires and ability to express Foxp3 in response to IL-2 alone (Lio and Hsieh, 2008). These observations support a “two-step” model of Treg cell differentiation where relatively strong TCR stimulation induces CD25 expression in precursor cells and thereby makes them receptive to subsequent IL-2 stimulation that ultimately induces Foxp3 expression (Burchill et al., 2008; Lio and Hsieh, 2008). Consistent with this model, IL-2 produced by self-reactive CD4SP thymocytes serves as a limiting “niche” factor scaling Treg cell differentiation in the thymus (Hemmers et al., 2019; Owen et al., 2018).
The fact that IL-2 – STAT5 signaling induces and sustains Foxp3 expression suggests that this pathway directly acts on the Foxp3 locus. However, given the pleiotropic transcriptional effects of IL-2 signaling on hundreds of targets (Chinen et al., 2016), it remains poorly understood to what extent the IL-2 – STAT5 pathway acts directly on the Foxp3 locus and thereby shapes the Treg cell population. Previously, we and others have described an intronic enhancer in the Foxp3 gene, CNS2 (Conserved Non-coding Sequence 2), which is bound by STAT5 and is required for heritable maintenance of Foxp3 expression in dividing mature Treg cells but dispensable for its induction (Feng et al., 2014; Li et al., 2014; Zheng et al., 2010; Zorn et al., 2006). The latter is consistent with the observations that CNS2 is methylated, inactive, and inaccessible prior to Foxp3 expression (Zheng et al., 2010). We therefore reasoned that a distinct cis-regulatory element must be targeted by IL-2 – STAT5 signaling to induce Foxp3 expression in precursor cells during Treg cell lineage commitment, before TET-induced DNA demethylation activates CNS2 (Yue et al., 2016).
Herein, we identified such a STAT5 binding enhancer and explored its biological functions. This conserved regulatory element proved to overlap substantially with previously identified SATB1 (designated CNS0) and MLL4 binding sites (designated −8.5kb MLL4) (Kitagawa et al., 2017; Placek et al., 2017). Although a role for SATB1 in thymic Treg cell differentiation has been demonstrated, the importance of SATB1 binding to this region remains untested. Given the genome-wide binding of SATB1, it is unknown which Foxp3 enhancers, if any, account for SATB1 function (Kitagawa et al., 2017). Likewise, MLL4 binding to this region has been shown to promote in vitro Treg cell polarization, yet its role in vivo remains less clearly defined (Placek et al., 2017). Using a mouse genetic model, we observed that deletion of CNS0 resulted in a selective impairment in thymic Treg cell generation and a subsequent decrease in peripheral Treg cell numbers, most noticeably during the perinatal period. With age, this defect was mitigated, likely due to peripheral Treg cell expansion. While deletion of this enhancer resulted in a persistent minor increase in immune tone without overt autoimmunity, it markedly enhanced autoimmunity when combined with Aire deficiency. Our results suggest that this IL-2 – STAT5 response element ensures robust Treg cell differentiation early in postnatal life and minimizes autoimmune disease by acting cooperatively with other tolerance mechanisms.
Results
Identification and characterization of the Foxp3 enhancer CNS0
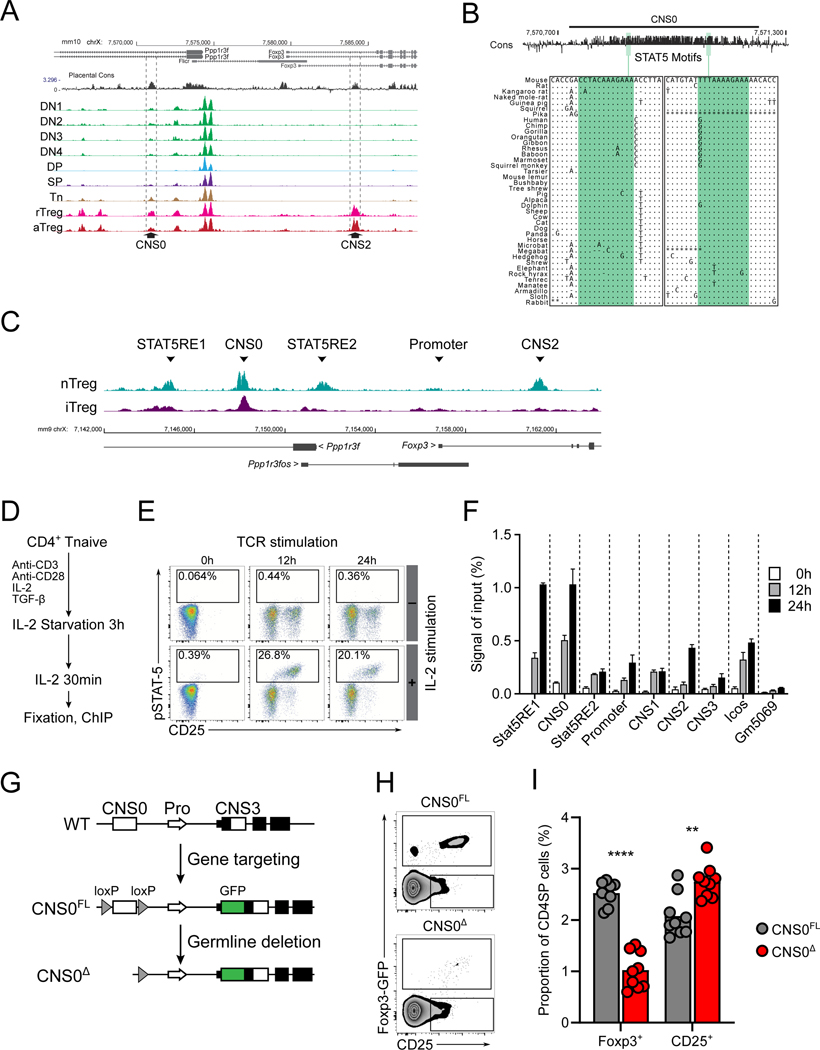
To identify any cis-regulatory elements that promote IL-2 dependent induction of Foxp3 expression during Treg cell lineage commitment, we searched for open chromatin regions (OCRs) in proximity to the Foxp3 gene that would be accessible in T cells with the potential to express Foxp3. ATAC-seq analysis of these cells from the thymus and secondary lymphoid organs (SLO) and of peripheral resting and activated Treg cells revealed an OCR in an intron of the Ppp1r3f gene upstream of Foxp3, accessible in both Treg and precursor cells and demonstrating sequence conservation (Figure 1A). This region seemed to coincide with the SATB1 binding site, termed CNS0, described previously, and proved to overlap substantially with the targeted CNS0 deletion described by the same group in an accompanying study (Kawakami et al., this issue) (Kitagawa et al., 2017). Therefore, we refer to this region as CNS0. This enhancer contained at least two STAT5 motifs, which were significantly conserved across different mammalian species (Figure 1B) (FIMO p-value: 0.00032 and 0.000451; phyloP conservation p-value: < 0.00001 and < 0.00001 for the 5’ and 3’ motifs, respectively).
Figure 1. Identification and deletion of CNS0.
A. Placental sequence conservation and ATAC-seq tracks of indicated populations from Foxp3GFP mice. Double negative thymocytes (DN): CD4−CD8−; Double positive thymocytes (DP): CD4+CD8+; CD4 Single Positive thymocytes (SP): CD4+CD8−TCRβ+Foxp3-GFP− from thymus; and naïve CD4 T cells (Tn): CD4+TCRβ+CD62L+CD44−Foxp3-GFP−; resting Treg cells (rTreg): CD4+TCRβ+CD62L+CD44loFoxp3-GFP+; activated Treg cells (aTreg): CD4+TCRβ+CD62L−CD44hiFoxp3-GFP+ from SLO. CNS2 and CNS0 are indicated by dashed lines.
B. Conservation and positions of STAT5 motifs within CNS0. Multiple sequence alignment of the motifs with other placental mammals are shown. “.” indicates conserved base, “-” indicates deletion, and “*” indicates missing data.
C. Tracks for STAT5 ChIP-seq showing the region surrounding the Foxp3 locus for ex vivo Treg cells (nTreg) and in vitro induced Treg cells (iTreg) stimulated with IL-2. Indicated peaks: CNS2, CNS0, the Foxp3 promoter, the Ppp1r3f promoter (STAT5RE2), and a non-conserved region (STAT5RE1). RE, response element. Data represent 3 biological replicates.
D-F. Schematic of ChIP-qPCR experiment (D) and representative plots showing phosphorylation of STAT5 upon IL-2 stimulation of CD4+ Tn cells after 0, 12, and 24 hours of activation (E). Normalized ChIP-qPCR signals by input DNA for indicated regions after IL-2 stimulation at different time points after activation (F). Icos and Gm5069 refer to gene promoter regions. See STAR Methods for details. Plot shows means and SD, n = 3 per group. Data represent 2 independent experiments.
G. Strategy for generating CNS0FL and CNS0Δ animals. CNS0 was defined as chr X: 7,570,674 – 7,571,274 (mm10). See STAR Methods for details.
H-I. Representative plots (H) and summarized data (I) of the frequencies of Foxp3-GFP+ (Foxp3+) and CD25+Foxp3-GFP− (CD25+) among CD4+CD8−TCRβ+ cells from thymuses of 5- to 18-week-old CNS0FL or CNS0KO male littermates. Each point indicates 1 mouse and data are pooled from 3 independent experiments.
Unpaired t-tests with correction for multiple comparisons using the Holm-Sidak method. p <0.01 **, <0.0001 ****. </P/>See also Figure S1.
Analysis of STAT5 binding with ChIP-seq of in vitro generated Treg (iTreg) cells or ex vivo isolated Treg (nTreg) cells after IL-2 stimulation revealed that CNS0 was bound by STAT5, suggesting that it is a bona fide IL-2 – STAT5 response element (Figure 1C). Accessibility of CNS0 in precursors suggested that STA5 binding to CNS0 might precede Foxp3 expression, and therefore this element may play a role in Foxp3 induction. We tested this possibility by performing ChIP-qPCR with T cells stimulated in vitro under Treg cell inducing conditions (Figure 1D). Consistent with a role of CNS0 in Foxp3 induction, this region was bound by STAT5 as early as 12 hours post activation (Figures 1E and 1F). This was in contrast to the Foxp3 promoter and other known enhancers, which showed weak STAT5 binding even 24 hours after stimulation (Figure 1F).
To investigate the role for CNS0 in regulating Foxp3 expression and Treg cell differentiation, we generated conditional CNS0FL mice harboring a GFP-Foxp3 fusion reporter and a loxP-flanked CNS0 region. Germline cre mediated recombination of this allele generated CNS0Δ mice (Figure 1G). Since these mice encoded a GFP-Foxp3 fusion protein known to modulate Treg cell function (Darce et al., 2012), CNS0FL or Foxp3GFP animals carrying an identical GFP fusion allele were consistently used as wild type controls. To exclude the possibility that loss of CNS0 caused widespread perturbation of chromatin accessibility at the Foxp3 locus, we performed ATAC-seq analysis of CNS0FL Cd4-cre+ mice and corresponding littermate controls (Cd4-cre−). Beyond the expected loss of CNS0, we observed no changes in chromatin accessibility at all other sites in and around the Foxp3 locus (Figure S1A).
STAT5 binding at CNS0 during in vitro Foxp3 induction suggested that CNS0 may play a role in Treg cell differentiation. Consistent with this notion, frequencies of Foxp3+ cells among CD4SP thymocytes, particularly recently generated CD73−Foxp3+ thymocytes, were reduced in CNS0Δ mice relative to littermate controls (Owen et al., 2019) (Figures 1H, 1I, S1B, and S1C). Thymic Treg cell differentiation is thought to proceed in a two-step manner: TCR dependent expression of CD25 followed by IL-2 mediated induction of Foxp3 (Burchill et al., 2008; Lio and Hsieh, 2008). Thus, we tested whether the observed decrease in thymic Treg cells was accompanied by an increase in CD25+Foxp3− precursor cells. Indeed, this subset was expanded in CNS0Δ animals, suggesting a selective block in the IL-2-dependent transition from pre-Treg to Treg cells. Importantly, thymic T cell development was otherwise unperturbed in CNS0Δ mice, including no changes in the abundance of CD122+GITR+ CD4SPs that gives rise to CD25+Foxp3− Treg cell precursors (Figures S1D and S1E). This process is facilitated by TCR signaling through c-Rel, rather than common γ-chain cytokines (Schuster et al., 2017). Thus, these data support the hypothesis that CNS0 promotes the induction of Foxp3 expression through the IL-2 – STAT5 axis.
Thymic Treg cell differentiation is severely compromised in CNS0-deficient mice in early postnatal life
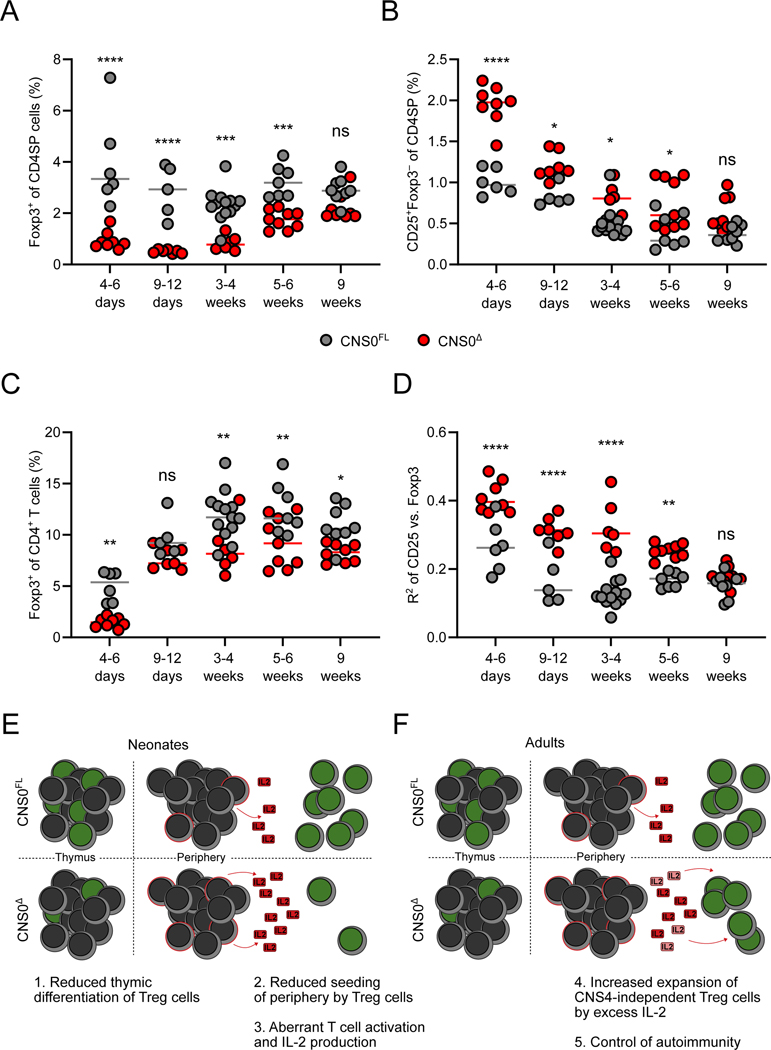
Thymic Treg cell differentiation in adult animals contributes minimally to the peripheral Treg cell pool, in contrast to the neonatal period, when Treg cells seed and thereafter self-renew in SLOs and non-lymphoid tissues (Rubtsov et al., 2010; Yang et al., 2015). Additionally, in adult mice CD25+Foxp3− CD4SP thymocytes are a heterogeneous population, comprising pre-Treg cells, autoreactive conventional T cells and their precursors, and thymocytes undergoing negative selection (Hemmers et al., 2019; Owen et al., 2019). On the other hand, a recent study has demonstrated that specifically in neonates, the majority of Treg cells arise from CD25+Foxp3− precursors (Bending et al., 2018). Thus, we sought to explore the effects of CNS0 deficiency on Treg cell generation in the neonatal thymus. Our analysis of mice across a range of ages revealed very few Treg cells in the thymuses of neonatal CNS0Δ mice, while pre-Treg cells were over-represented in comparison to littermate controls (Figures 2A and 2B). This neonatal defect was even more pronounced for CD73− Treg cells, consistent with an impact of CNS0-deficiency on recently differentiated, rather than mature, thymic Treg cells (Figure S2A). This early block in the second step of Treg cell differentiation progressively diminished as mice became adults, reaching a milder steady-state defect by week 9 of life.
Figure 2. CNS0 deficiency impairs Treg cell differentiation.
A-D. Frequencies of Foxp3-GFP+ (A) and CD25+Foxp3-GFP− cells (B) among CD4+CD8− TCRβ+ cells from thymuses, and Foxp3-GFP+ among CD4+TCRβ+ cell from spleens (C) of CNS0Δ and CNS0FL male littermates of indicated ages. R2 for linear regression of log-transformed fluorescence intensities of CD25 and Foxp3-GFP among CD25+Foxp3-GFP+ CD4SP thymocytes (D). Each point indicates 1 mouse and data are pooled from 5 independent experiments, including a single experiment where mice of both genotypes from all ages were analyzed.
E, F. Model of the effect of CNS0 deficiency on Treg cell induction and expansion in neonatal (E) and adult (F) mice. Cells with green nucleuses represent Treg cells and those with red membranes represent antigen experienced Tconv cells. Faded IL-2 indicates its consumption by Treg cells.
Two-way ANOVA with Sidak test to correct for multiple comparisons. p >0.05 ns, <0.05 *, <0.01 **, <0.001 ***, <0.0001 ****.
See also Figure S2.
Consistent with an early deficit in thymic Treg cell production, Treg cells were nearly absent from the spleen of neonatal mice (Figure 2C). This peripheral deficiency was also mitigated with age, but more rapidly than in the thymus. We confirmed that this dynamic also occurred in non-lymphoid tissues (Figure S2B). We also noticed that there was a stronger correlation between Foxp3 protein and surface CD25 expression in thymic Treg cells in CNS0Δ mice as compared to littermate controls (Figure 2D and S2C), reflecting a paucity of Foxp3hiCD25lo cells. This suggested that during the postnatal burst of thymic Treg cell differentiation, only precursor cells with higher CD25 expression were becoming mature Foxp3+ Treg cells. Given that agonist TCR signaling results in CD25 expression in thymic Treg cell precursors preceding Foxp3 expression, we reasoned that strong TCR stimulation was primarily driving Foxp3 induction in CNS0Δ mice, when the IL-2 dependent component was compromised. To test this notion, we analyzed Il2 haploinsufficient CNS0Δ and CNS0FL mice. We found that thymic CD73− Treg cells were similarly reduced between CNS0-sufficient and -deficient Il2+/− animals as for Il2+/+ mice (Figure S2D). This was despite reduced production of IL-2 in the Il2+/− groups, which also resulted in lower CD25 expression by Treg cells (Figures S2E and S2F). Therefore, in the presence of CNS0, Treg cell differentiation was unperturbed by reduced IL-2 availability stemming from Il2-haploinsufficiency. This suggests that CNS0 confers on the Foxp3 locus high sensitivity to STAT5 mediated transcriptional activation. Conversely, Treg cell differentiation in Il2+/− mice lacking CNS0 was not exacerbated by reduced IL-2 availability. This latter point is consistent with, but does not prove, a lack of a CNS0-independent role for IL-2 signaling in Foxp3 induction. Our observations support the notion that CNS0 confers responsiveness of the Foxp3 gene to scarce IL-2 amounts during Treg cell differentiation in the thymus and suggest that IL-2 facilitates Foxp3 induction in the thymus by acting foremost through CNS0.
The results so far were consistent with a scenario wherein a neonatal paucity of Treg cells in CNS0Δ mice, which stemmed from defective Treg cell differentiation in the thymus, resulted in inadequate early postnatal seeding of SLOs and peripheral tissues by Treg cells (Figure 2E). This led to transient T cell activation and a subsequent increase in the availability of IL-2 due to a combination of its increased production by conventional T cells and diminished consumption by Treg cells (Figure 2E). As CNS0Δ mice age, this excess IL-2 may promote expansion of the numerically diminished Treg cell population and their differentiation into suppressive activated Treg cells (Figure 2F). In support of this model, we observed increased proliferation of Treg cells in mice at an age immediately prior to when Treg cell frequencies began to normalize (Figures S2G). CD25 surface expression is controlled by multiple signals; however, our observation of progressively reduced surface CD25 expression by T cells in Il2+/− demonstrate that CD25 surface expression by mature T cells can reflect in vivo IL-2 availability (Figures S2E, S2F). Therefore, our model is further supported by the fact that surface CD25 expression by both Foxp3+ Treg and activated Foxp3−CD44+CD4+ T cells was increased at an age immediately preceding the increased Treg cell proliferation (Figures S2H and S2I). Taken together, these results suggest that a relative enrichment for high affinity TCRs and transiently increased IL-2 availability may compensate in part for the Treg cell developmental defect caused by CNS0 deficiency.
Acquisition of Foxp3 expression is dependent on IL-2 signaling through CNS0
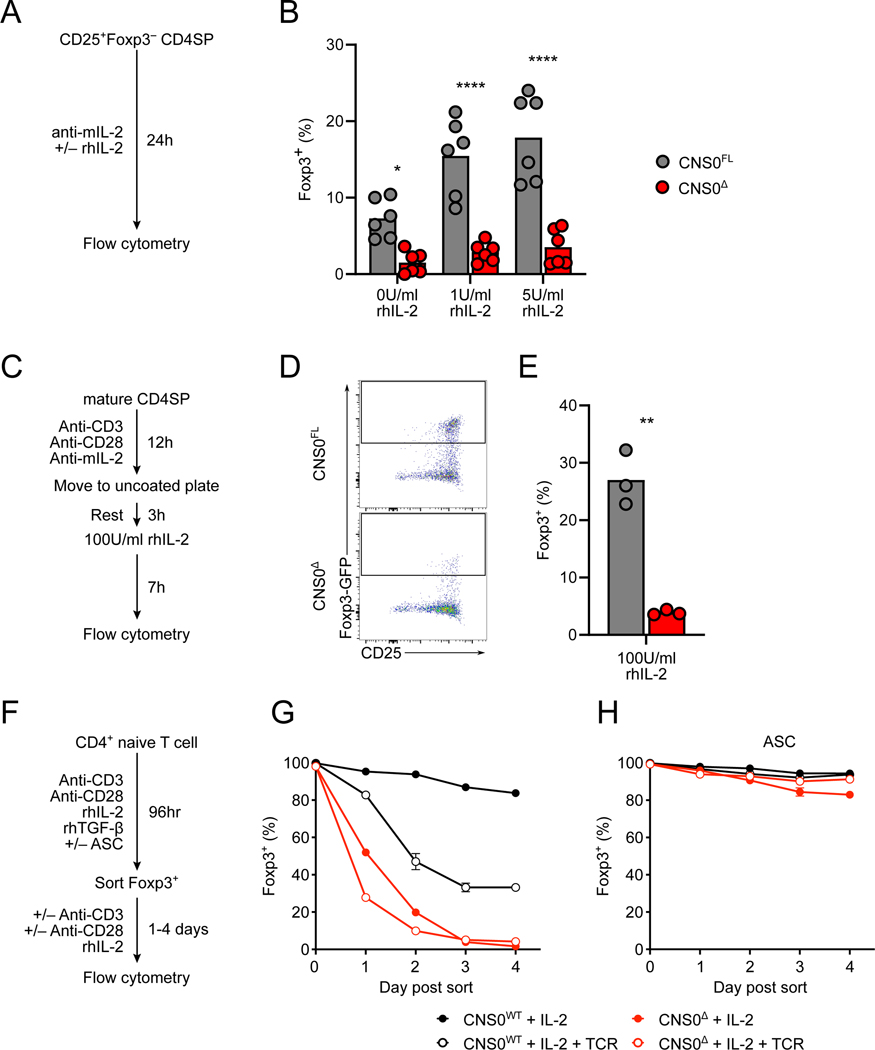
Next, we set out to directly test the proposed role of CNS0 in enabling the IL-2-dependent step of Foxp3 induction and Treg cell differentiation using an in vitro model of their two-step thymic differentiation (Hemmers et al., 2019; Lio and Hsieh, 2008). Sorted CD25+Foxp3− CD4SP thymocytes from CNS0Δ mice failed to induce Foxp3 expression in response to IL-2, in contrast to those from CNS0FL mice (Figures 3A and 3B). This observation supported the notion that the accumulation of CD25+Foxp3− CD4SP thymocytes in CNS0Δ mice was due to failed expression of Foxp3 in response to IL-2 stimulation by CD25+ Treg cell precursors. Given the heterogeneous composition of the CD25+Foxp3− CD4SP thymocyte population, we also modeled the entire two-step differentiation process in vitro (Figure 3C). While a substantial fraction of CNS0-sufficient CD4SP thymocytes expressed Foxp3 in response to this treatment, very few CNS0-deficient CD4SP cells did (Figures 3D and 3E). Thus, during Treg cell differentiation IL-2 – STAT5 signaling requires CNS0 to induce Foxp3 expression.
Figure 3. IL-2 dependent role of CNS0 in initiating and sustaining Foxp3 expression.
A. Schematic of experiments shown in (B). See STAR Methods for details.
B. Frequencies of Foxp3-GFP+ cells among Treg cell precursors (CD4+CD8−Foxp3-GFP− CD25+TCRβ+) sorted from the thymuses of 8-week-old CNS0FL or CNS0Δ male littermates incubated ex vivo with indicated amounts of IL-2 for 24 hours. Each point indicates 1 mouse and data are pooled from 2 independent experiments.
C. Schematic of experiments shown in (D-E). See STAR Methods for details.
D, E. Representative plots (D) and summarized data (E) of the frequencies of Foxp3-GFP+ cells among ex vivo stimulated mature CD4SP (CD4+CD8−CD73−Foxp3-GFP−CD25− TCRβhiCD24lo) cells sorted from the thymuses of 5- to 6-week-old CNS0FL or CNS0Δ male littermates. Each point indicates 1 mouse and data are representative of 3 independent experiments.
F. Schematic of experiments shown in (G, H). See STAR Methods for details.
G, H. Summarized data of the frequencies of Foxp3 expression by in vitro induced Treg cells after additional 4 days of culture. Treg cells were generated from Tn cells (CD4+Foxp3-GFP− CD62L+CD44−TCRβ+) in the absence (G) or presence (H) of sodium ascorbate (ASC). Plots show means and SD, n = 3 per group. Data represent >3 independent experiments.
</P/>Unpaired t-tests with correction for multiple comparisons using the Holm-Sidak method. p <0.05 *, <0.01 **, <0.0001 ****.
See also Figure S3.
These observations suggest a division of labor between CNS2 and CNS0, the two principal STAT5 responsive elements for the Foxp3 locus: the former drives the IL-2 dependent induction of Foxp3 expression during Treg cell differentiation, while the latter maintains Foxp3 expression in an IL-2 dependent manner post-Foxp3 induction. However, a granular temporal analysis of nascent thymic Treg cells has shown that CNS2 only becomes demethylated and therefore functional after an initial brief period of sustained Foxp3 expression (Bending et al., 2018; Herppich et al., 2019). This leaves a time window during which Foxp3 expression needs to be sustained before CNS2 becomes active, raising the possibility that in addition to promoting Foxp3 induction, CNS0 might play a role in maintaining Foxp3 expression during this transitional state. To explore this possibility, we turned to analysis of in vitro generated iTreg cells, where CNS2 remains methylated and inactive (Zheng et al., 2010). In these cultures, CNS0-deficient naïve CD4+ T (Tn) cells also displayed diminished ability to induce Foxp3 expression in response to IL-2 (Figures S3A and S3B). To test the potential contribution of CNS0 to the maintenance of Foxp3 expression during Treg cell differentiation prior to CNS2 demethylation, we generated Foxp3+ iTreg cells, then sorted them on day 4 and further cultured them in the presence of IL-2 with or without TCR re-stimulation (Figure 3F). CNS0-deficient cells lost Foxp3 expression to a greater extent than control cells, with or without TCR re-stimulation (Figure 3G). To test whether CNS0 still contributed to the stability of Foxp3 expression when CNS2 was active, we treated differentiating iTreg cells with sodium ascorbate (ASC), which increases TET enzymatic activity (Blaschke et al., 2013; Yin et al., 2013). This treatment resulted in efficient demethylation of CNS2 in both CNS0-deficient and -sufficient Foxp3+ cells (Figure S3C), consistent with previous studies (Nikolouli et al., 2017; Sasidharan Nair et al., 2016; Yue et al., 2016). In this setting, cells of neither genotype lost Foxp3 expression regardless of TCR re-stimulation (Figure 3H). Importantly, the ASC induced stabilization of Foxp3 expression in iTreg cells was CNS2-dependent as shown using CNS2-deficient Tn cells in a similar assay (Figure S3D). This finding suggests that in addition to promoting the induction of Foxp3 expression, CNS0 stabilizes the expression of recently induced Foxp3 before CNS2 becomes active and takes on the function of maintaining Foxp3 expression in differentiated Treg cells. In further support of a division of labor between CNS0 and CNS2, when we induced CNS0 deletion after adoptive transfer of Treg cells, we observed that in contrast to CNS2, CNS0 was dispensable for the maintenance of Foxp3 expression in mature Treg cells (Figure S3E).
A genomic region, apparently partially overlapping with CNS0, has been characterized as an MLL4-bound site and proposed to assist in the deposition of histone H3K4 mono-methylation (H3K4me1) at multiple regions within the Foxp3 locus (Placek et al., 2017). We performed H3K4me1 ChIP-qPCR in order to determine whether loss of CNS0 affects histone methylation in the Foxp3 locus and found reduced H3K4me1 at the Foxp3 promoter, but not CNS3 in the absence of CNS0 in Tn cells (Figure S3H). These results raise the possibility that IL-2 – STAT5 signaling through CNS0 might promote Foxp3 induction in part through directing MLL4 activity to the Foxp3 promoter.
Compensatory mechanisms support a functional Treg cell population in the absence of CNS0
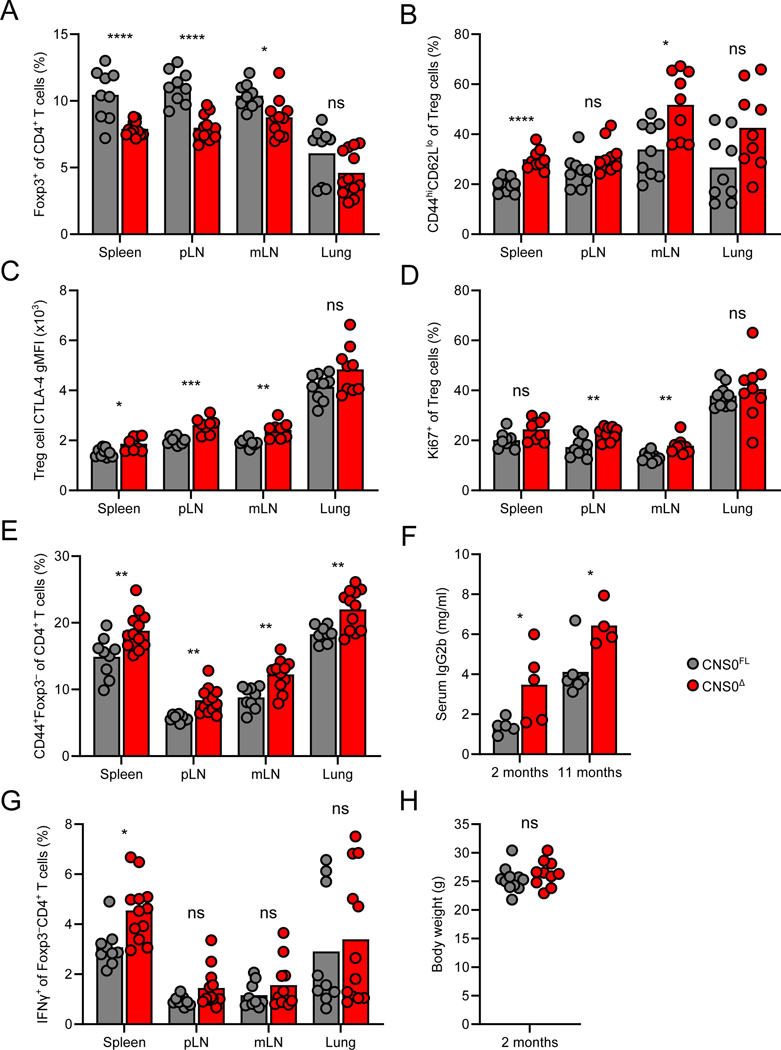
Flow cytometric analyses of adult CNS0Δ animals revealed overall reduced frequencies of Treg cells, yet higher proportions of activated Treg cells in SLOs and non-lymphoid tissues in comparison to littermate controls (Figures 4A and 4B). Furthermore, CNS0-deficient Treg cells expressed higher amounts of CTLA-4 and were more proliferative (Figures 4C and 4D). We also observed an increase in antigen-experienced conventional CD4+ T cells, as well as elevated titers of serum IgG2b, an indication of previous or ongoing B cell activation (Figures 4E and 4F). However, production of IFNγ and other pro-inflammatory cytokines by T cells in CNS0-deficient mice was only modestly increased or even unchanged (Figures 4G, S4A, and data not shown). Overall, these mice were outwardly healthy with no differences in body weights relative to controls (Figure 4H). Even aged CNS0Δ animals did not show increased immune activation or histological indications of tissue pathology (Figures S4B-E and Table S1). Collectively, these data further support our model that the near absence of Treg cells in CNS0-deficient neonates results in transient immune activation, which is then counteracted by subsequent expansion and heightened activation of Treg cells, resulting in sufficient long-term prevention of pathology. Indeed, we observed that CNS0-deficient Treg cells were more efficient on a per cell basis at suppressing proliferation of conventional T cells in vitro than control Treg cells, consistent with the ability of CNS0-deficient Treg cells to suppress autoimmunity in vivo despite being numerically diminished (Figures S4F and S4G).
Figure 4. Immune perturbation but no overt pathology in adult CNS0-deficient mice.
A, E, G. Frequencies of Foxp3-GFP+ cells (A) and CD44hiFoxp3-GFP− (E) among CD4+TCRβ+ cells, and IFNγ+ cells among Foxp3-GFP−CD4+TCRβ+ cells (G) in indicated tissues (pLN: pooled brachial, axillary, and inguinal lymph nodes; mLN: pooled mesenteric lymph nodes) from 8- to 10-week-old CNS0Δ and CNS0FL male littermates. Each point indicates 1 mouse and data are pooled from 2 independent experiments.
B-D. Frequencies of CD44hiCD62L− cells (B), CTLA-4 gMFI, (C) and frequencies of Ki67+ cells (D) among Treg cells in indicated tissues from 8- to 10-week-old CNS0Δ and CNS0FL male littermates. n=9 per group.
F. Serum IgG2b quantification by ELISA of CNS0Δ and CNS0FL male littermates of indicated ages. Each point indicates 1 mouse. Data represent 2 independent experiments.
H. Body weight of 8- to 10-week-old CNS0Δ and CNS0FL male littermates.
Unpaired t-tests with correction for multiple comparisons using the Holm-Sidak method. p >0.05 ns, <0.05 *, <0.01 **, <0.001 ***, <0.0001 ****.
See also Figure S4.
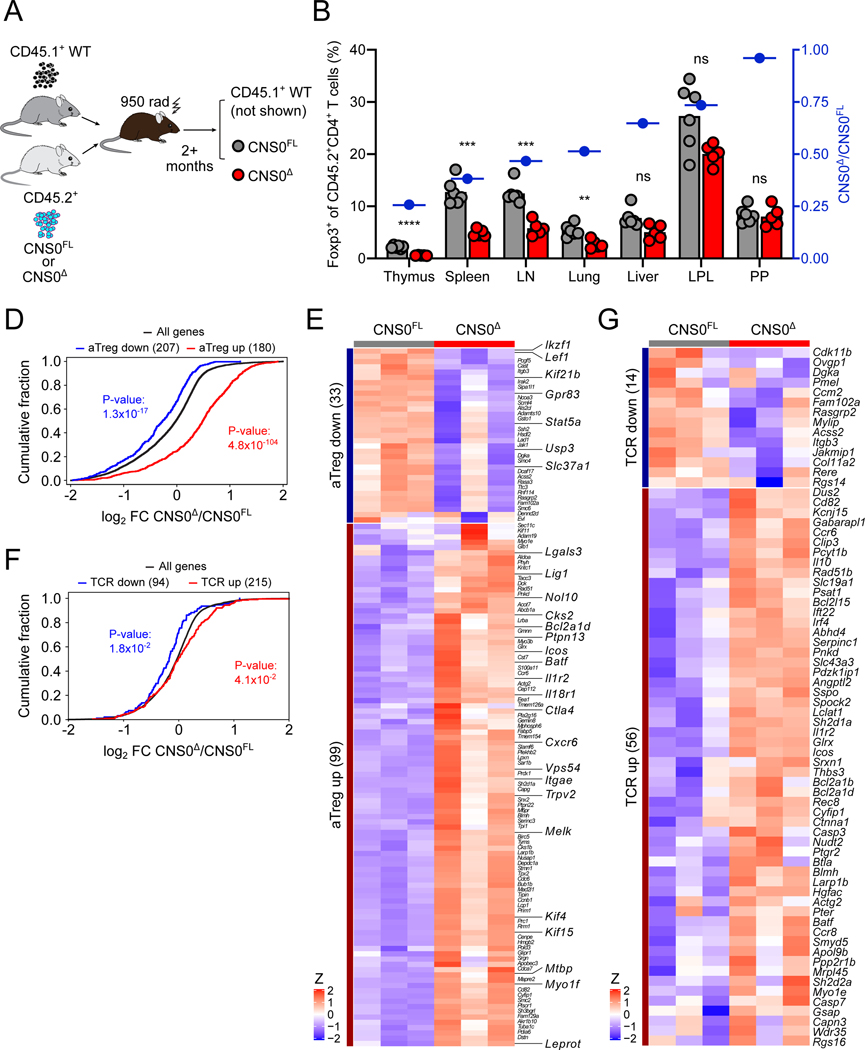
The increased activation of Treg cells in CNS0Δ mice (Figures 4B-D) can be a consequence of: (1) a response to the mildly increased immune tone; (2) stronger TCR or common γ-chain cytokine signaling required to drive CNS0-independent Treg cell differentiation; or (3) increased activation of the few Treg cells exiting the thymus as they expand to fill peripheral niches (Figures 2E and 2F). To distinguish among these possibilities, we first generated mixed bone marrow chimeric mice, where control (WT) CD45.1 bone marrow cells were co-transferred along with either CNS0Δ or CNS0FL bone marrow cells into lethally irradiated recipients (Figure 5A). In this setting where any aberrant immune activation was suppressed by the presence of WT CD45.1 Treg cells in both groups, we still detected increased activation of CNS0Δ Treg cells (Figure S5A). We also observed increased expression of CTLA-4 by and increased proliferation of CNS0Δ Treg cells (Figures S5B and S5C). These findings suggest that the elevated activation status of CNS0-deficient Treg cells was due to their intrinsic properties rather than a consequence of the increased immune tone. While we found that CNS0-deficient Treg cells were greatly underrepresented in the thymuses of the mixed bone marrow chimeric mice, it was noteworthy that this defect was rescued to varying degrees in the SLOs peripheral tissues (Figures 5B).
Figure 5. Increased cell-intrinsic activation partially compensates for CNS0-deficiency.
A. Schematic of the generation of bone marrow chimeric (BMC) mice analyzed in (B, S5A-C). See STAR Methods for details.
B. Frequencies of Foxp3-GFP+ Treg cells among CD45.2+CD4+ T cells in indicated tissues (LN: pooled brachial, axillary, and inguinal lymph nodes; LPL: small intestine lamina propria; PP: Peyer’s patches) of CNS0FL or CNS0Δ BMC mice. Blue symbols indicate ratios of CNS0Δ to CNS0FL Treg cell frequencies. Each point indicates 1 mouse, and data are representative of 2 independent experiments.
D, E. RNA-seq of CNS0Δ versus CNS0FL Treg cells sorted from the thymuses of 8-week-old CNS0Δ/WT and CNS0FL/WT female littermates.
Cumulative distribution function plot of gene expression (log2 fold change, FC). All genes (black) and genes with increased (red) or decreased (blue) expresison in activated vs. resting Treg cells. p, one-tailed Kolmogorov-Smirnov test.
E. Heatmap showing row Z-score normalized gene expression. Genes are restricted to those contained in the indicated gene sets (activated vs resting Treg) and significantly differentially expressed (adjusted p-value <0.05) between genotypes.
F, G. RNA-seq of Treg cells sorted from the thymuses of 8-week-old CNS0FL and CNS0Δ male littermates.
F. Cumulative distribution function plot of differentially expressed genes (log2 FC). All genes (black) and genes with increased (red) or decreased (blue) expresison in TCR-bearing vs. TCR-deleted Treg cells. p, one-tailed Kolmogorov-Smirnov test.
G. Heatmap showing row Z-score normalized gene expression. Genes are restricted to those contained in the indicated gene sets (TCR-dependent genes) and significantly differentially expressed (adjusted p-value <0.05) between genotypes.
(B) Unpaired t-tests with correction for multiple comparisons using the Holm-Sidak method. p >0.05 ns, <0.01 **, <0.001 ***, <0.0001 ****.
(D, F) p, by one-tailed Kolmogorov-Smirnov test.
See also Figure S5.
These results suggested that the increased activation of CNS0KO Treg cells compensated in the periphery for their reduced thymic generation. To gain insight into the nature of this cell-intrinsic compensation, we performed RNA-seq analysis of thymic Treg cells from female CNS0Δ/WT and CNS0FL/WT heterozygotes, which both harbor WT Treg cells in addition to CNS0-deficient and -sufficient populations due to random X chromosome inactivation. We observed overall increased expression of genes associated with Treg cell activation in the CNS0-deficient Treg cells, including those linked to their suppressive function such as Ctla4, Il18r1, Icos, and Il1r2 (Figures 5D and 5E). The heightened activation of thymic CNS0deficient Treg cells raised the possibility that high affinity TCRs, which afford stronger stimulation and increased activation, might allow a small number of precursor cells to differentiate into Treg cells in the absence of CNS0. Consistent with this idea, RNA-seq analysis of thymic Treg cell transcriptomes from male CNS0Δ and CNS0FL mice showed overall higher expression of TCR-dependent genes in the former, including significantly increased expression of genes associated with T cell activation and immunosuppression such as Il10 and Icos, Irf4, and Batf (Figures 5F and 5G). Additionally, increased expression of several chemokine receptors was consistent with an enhanced ability of Treg cells from CNS0-deficient mice to migrate to inflammatory sites and non-lymphoid tissues in order to maintain immune tolerance when numerically disadvantaged upon exiting the thymus (Figures 5F and 5G).
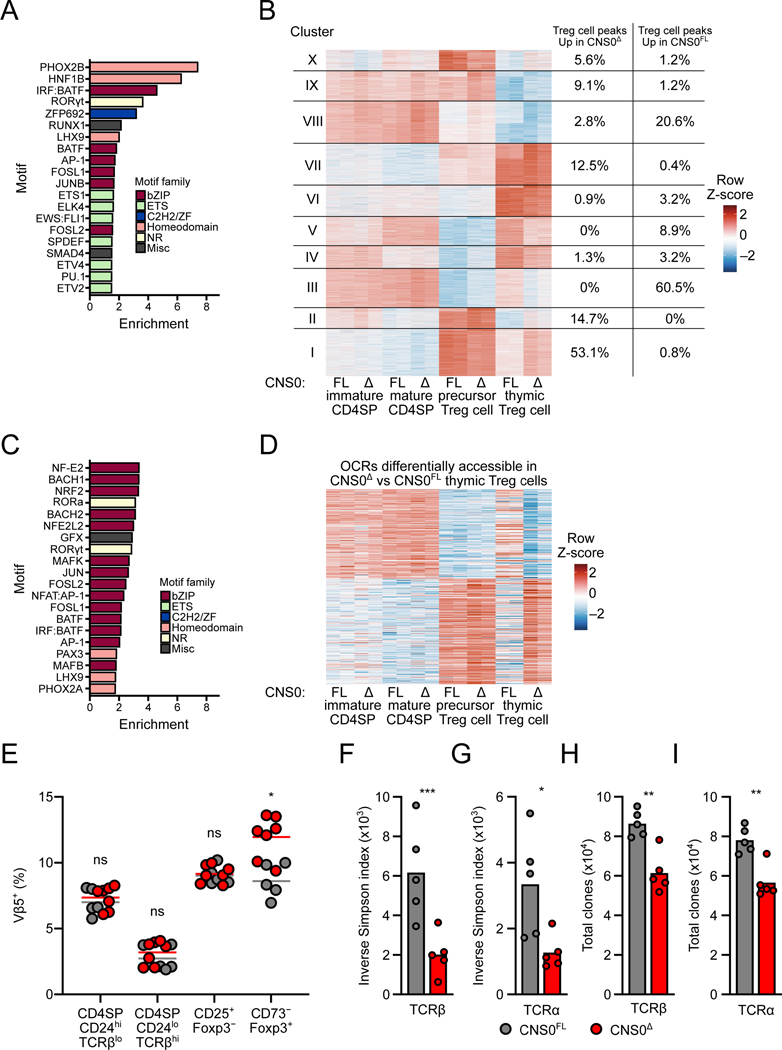
Additionally, we supplemented and analyzed our ATAC-seq data of various CNS0Δ and control thymocyte subsets to identify OCRs differentially accessible according to genotype (Hemmers et al., 2019). This analysis revealed limited CNS0 dependent chromatin perturbation in thymic Foxp3+ Treg cells and CD25+ Treg precursors and confirmed no changes at other OCRs in and around the Foxp3 locus (Figures S6A and S6B). Consistent with the flow cytometric analysis (Figures S1D and S1E), there were minimal alterations in the other CD4SP subsets (Figures S6C and S6D). Motif enrichment analysis revealed enrichment of homeodomain and bZIP motifs in OCRs with increased accessibility in CNS0-deficient Treg cells (Figure 6A). TFs from the bZIP family are known mediators of TCR signaling, as are several homeodomain TFs (Gaud et al., 2018; Kang et al., 1992; Moore et al., 2018). K-means clustering of OCRs differentially accessible in any pairwise comparisons revealed a set of OCRs with greater accessibility in CNS0Δ Treg cells, including a majority of those differentially accessible (cluster I, Figure 6B and S7A, see Table S2 for underlying data). Consistent with the previous analysis, this cluster was enriched for bZIP motifs (Figure 6C). These OCRs were most accessible in CD25+Foxp3− Treg cell precursors of either genotype (Figure 6B and S7A), further supporting the notion that these regions were responsive to TCR signaling, as these cells are believed to have recently experienced agonist TCR signals (Bending et al., 2018; Hemmers et al., 2019; Lio and Hsieh, 2008; Owen et al., 2019; Schuster et al., 2017). A similar trend was also observed for OCRs differentially accessible between CNS0-deficient and -sufficient thymic Treg cells: regions more accessible in CNS0Δ Treg cells overlapped with those most accessible in CD25+Foxp3− cells of either genotype (Figure 6D, S7B, and S7C). We found no motifs significantly enriched (p < 0.05) in OCRs with reduced accessibility in CNS0Δ Treg cells and only weak enrichment of MADS and Zinc Finger family motifs in the corresponding cluster (cluster III, Figure 6B and data not shown).
Figure 6. Increased TCR signaling drives thymic Treg cell differentiation in the absence of CNS0.
A-D. ATAC-seq of indicated populations sorted from the thymuses of 16-week-old CNS0FL and CNS0Δ male littermates. See STAR Methods for details.
A. Enrichment (presence in target versus in background) of known TF motifs, determined by HOMER, in summit-centered peaks of OCRs significantly more accessible in CNS0Δ Treg cells. Top 20 are shown, all with q <0.05 and ranked by enrichment.
B. K-means clustering of OCRs with significant differential accessibility in any pairwise comparison. Columns show individual samples and data are normalized by Z-scores per row. Right-hand columns indicate proportion of OCRs significantly more accessible in CNS0Δ Treg cells (left) or CNS0FL Treg cells (right) within each cluster.
C. Enrichment (presence in target versus in background) of known TF motifs, determined by HOMER, in summit-centered peaks of OCRs in k-means cluster I. Top 20 are shown, all with q <0.05 and ranked by enrichment. Coloring indicates families of DNA-binding domains.
D. K-means clustering of OCRs with significant differential accessibility between CNS0Δ and CNS0FL Treg cells. Columns show individual samples and data are normalized by Z-scores per row.
E. Frequencies of Vβ5+ cells among indicated populations from the thymuses of 6- to 10-week-old CNS0FL and CNS0Δ male littermates. Each point indicates 1 mouse. Data are pooled from 2 experiments.
F-H. Inverse Simpson index indicating the diversity intensity (F, G) and total unique TCR clones (H, I) of TCRβ (F, H) or TCRα (G, I) chain sequences from splenic Treg cells isolated from 6-week-old CNS0FL and CNS0Δ male mice. Each point indicates 1 mouse and data are from a single experiment.
Unpaired t-tests with correction for multiple comparisons using the Holm-Sidak method. p >0.05 ns, <0.05 *, <0.01 **, <0.001 ***.
See also Figures S6 and S7.
Collectively, the ATAC- and RNA-seq analyses suggested that CNS0 deficiency limits Treg cell differentiation to precursors receiving robust TCR stimulation. Endogenous mammary tumor virus-8 and −9 encoded superantigens (vSAG-8 and vSAG-9) exhibit reactivity with Vβ5 containing TCRs (Scherer et al., 1993). As a result, these vSAGs induce partial deletion of Vβ5+ CD4SP thymocytes while also promoting Vβ5+ Treg cell differentiation in C57BL/6 (B6) mice, which display increased Vβ5 utilization by thymic Treg cells in comparison to conventional CD4SP cells (Pacholczyk et al., 2002; Romagnoli et al., 2002). Therefore, relative frequencies of Vβ5+ Treg cells can reflect Treg cell selection driven by a strong, abundant TCR agonist. We observed an increase in thymic Vβ5+ CD73− Treg cells in CNS0-deficient versus -sufficient littermates, but no changes in the usage of non-reactive Vβs (Figures 6E, S6E, and S6F). These results support the above notion that the Treg cell precursors differentiating into mature Treg cells in the absence of CNS0 are skewed towards those that received stronger TCR agonist signaling. This, and the heightened proliferation of Treg cells from CNS0-deficient mice suggest that the CNS0Δ Treg cells are clonally expanded. To test this, we sequenced TCRα and TCRβ chains of peripheral Treg cells from CNS0Δ and CNS0FL animals. This analysis revealed reduced diversity intensity of both TCR chains for CNS0-deficient Treg cells, as measured by inverse Simpson index, suggesting their clonal expansion (Figures 6F and 6G). Accordingly, total unique TCRα and TCRβ clones were also reduced for CNS0Δ Treg cells (Figures 6H and 6I). Together, these results suggest that in the absence of CNS0, some thymic Treg cell precursors, which express strongly self-reactive TCRs, bypass the IL-2-dependent phase of Foxp3 induction and give rise to functionally competent Treg cells. This heightened TCR self-reactivity of CNS0-deficient Treg cells likely accounts for their high CD25 expression, which in turn supports their expansion. However, this results in a less diverse Treg cell population with increased clonal expansion, as suggested by our model (Figure 2F).
CNS0 deficiency increases susceptibility to autoimmune disease
While these compensatory Treg cell-intrinsic mechanisms likely account for the overall health of CNS0-deficient mice on the B6 genetic background, which is resistant to autoimmunity, the majority of common pathologies, including autoimmune and inflammatory diseases, are subject to complex polygenic control. Thus, it remains possible that CNS0 deficiency could markedly exacerbate autoimmune disease when combined with impairment in other distinct genetic mechanisms of self-tolerance.
AIRE is a transcriptional regulator expressed by medullary thymic epithelial cells to enable presentation of tissue-restricted antigens (TRAs), which subsequently induces negative selection that inactivates TRA-reactive T cells and promotes the differentiation of TRA-specific Treg cells (Proekt et al., 2017). Loss-of-function mutations of Aire result in the human autoimmune syndrome APECED (APS-1) (Björses et al., 1998). Similarly, Aire deficiency in mice leads to autoimmune inflammation affecting multiple organs (Anderson et al., 2002). In both humans and mice, Aire has also been recognized as a major autoimmune susceptibility gene: hypomorphic mutations in Aire in combination with other genetic lesions precipitate autoimmune pathologies (Proekt et al., 2017).
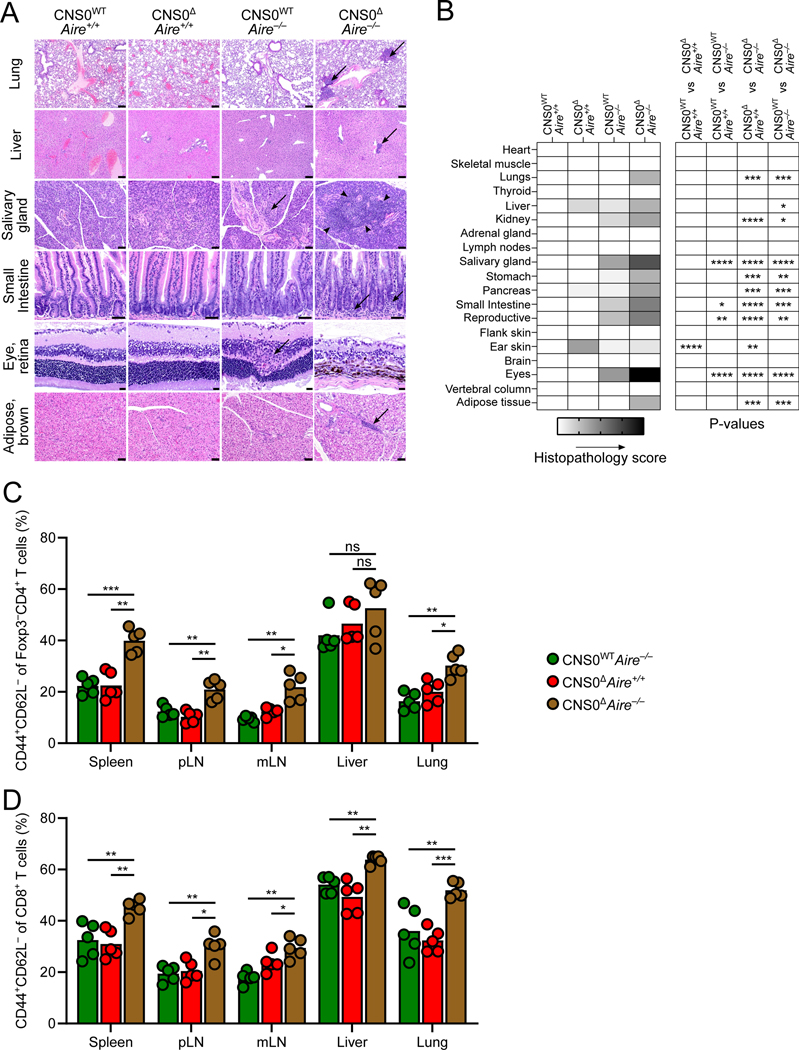
Therefore, we combined CNS0 and Aire deficiency to uncover the role of CNS0-dependent Treg cell development in an autoimmune background. We analyzed inflammation and tissue damage by histopathology in four groups of mice: CNS0WTAire+/+, CNS0ΔAire+/+, CNS0WTAire−/−, and CNS0ΔAire−/−, (termed WT, CNS0Δ, Aire−/−, and Δ/Δ, respectively), all harboring the same Foxp3GFP fusion protein reporter. We observed that the tissue-specific autoimmunity in the Aire−/− background was significantly exacerbated in the absence of CNS0 (Figures 7A and 7B). Histopathological assessment revealed increased inflammatory infiltration in lungs, livers, salivary glands, intestines, and adipose tissue, as well as cell loss and retinal atrophy in the eyes of Δ/Δ mice, relative to Aire−/− and CNS0Δ controls (Figure 7A). Organs known to be affected by Aire-deficiency, such as salivary gland, intestine, reproductive organs, and eye exhibited increased pathology when combined with CNS0-deficiency (Figure 7B, compare WT vs Aire−/− against Aire−/− vs Δ/Δ). Additionally, tissues with no overt pathology in CNS0-sufficient Aire−/− mice, such as lung, stomach, and adipose tissue exhibited inflammatory lesions (Figure 7B). This was accompanied by increased T cell activation in Δ/Δ mice relative to Aire−/− and CNS0Δ animals (Figures 7C and 7D). These results indicate that IL-2 – STAT5 – CNS0 dependent Treg cell differentiation confers robust immunosuppressive capacity to mitigate breaks in organ-specific tolerance.
Figure 7. Genetic perturbation of Aire exacerbates autoimmunity in CNS0-deficient animals.
A. Representative images of hematoxylin and eosin stained slides. Arrows indicate aggregates of inflammatory cells and arrowheads circumscribe the inflammatory regions. Scale bars indicate 100 μm (lung, liver), 50 μm (salivary gland, small intestine, and adipose tissue), or 20 μm (eye).
B. Histopathological scoring. Shading indicates mean score for each group (0–4) and p values denote two-way ANOVA with Sidak test to correct for multiple comparisons.
C, D. Frequencies of CD44+CD62L− cells among CD4+Foxp3-GFP− T cells (C) and CD44+CD62L− cells among CD8+ T cells (D).
Unpaired t-tests with correction for multiple comparisons using the Holm-Sidak method. p >0.05 ns or blank (B), <0.05 *, <0.01 **, <0.001 ***, <0.0001 ****.
Discussion
Treg cells were originally described as a subset of CD4+ T cells with high CD25 expression, which proved essential for their differentiation and function (Chinen et al., 2016; Fan et al., 2018; Furtado et al., 2002; Malek et al., 2002; Panduro et al., 2016; Sakaguchi et al., 1995; Toomer et al., 2019). Both have been subsequently shown to depend upon sustained expression of Foxp3 (Fontenot et al., 2003; Gavin et al., 2007; Hori et al., 2003; Khattri et al., 2003; Lin et al., 2007; Williams and Rudensky, 2007). Although previous work has demonstrated pleiotropic roles of IL-2 – STAT5 signaling in Treg cell biology, its direct function in Foxp3 induction during Treg cell differentiation remains poorly understood (Burchill et al., 2003, 2007; Feng et al., 2014; Fontenot et al., 2005a; Li et al., 2014; Yao et al., 2007; Zorn et al., 2006).
We identified a distal enhancer, CNS0, located in an intron of the Ppp1r3f gene upstream of the Foxp3 locus, as the key IL-2 – STAT5 response element acting during Foxp3 induction in Treg cell precursors. CNS0 is conserved across mammalian species, features conserved STAT5 binding motifs, and is accessible in Treg cell precursors. CNS0 was also the first site bound by STAT5 in the vicinity of the Foxp3 gene during in vitro Treg cell induction. Notably, a previously characterized c-Rel binding Foxp3 enhancer, CNS3, facilitates Foxp3 induction particularly at suboptimal TCR signal strengths (Feng et al., 2015; Zheng et al., 2010). Thus, CNS3 and CNS0 might collectively provide a means for the two principal signals driving Treg cell generation—agonist TCR signaling, followed by IL-2 stimulation—to act directly on the Foxp3 locus. It is also possible that CNS3 and CNS0 enable two distinct but interwoven Treg cell differentiation trajectories, differentially reliant on sustained TCR or IL-2 signaling, respectively.
Previously, Sakaguchi’s group suggested that this region participated in chromatin looping during thymic Treg cell differentiation on the basis of SATB1 binding in pooled Treg and pre-Treg cells (Kitagawa et al., 2017). Although a functional role for CNS0 in short range chromatin looping has not been tested in that study or herein, it is possible that SATB1 may assist STAT5 in Foxp3 induction by enabling interactions between this region and the Foxp3 promoter. Another group described an overlapping region as a binding site for MLL4 and suggested that chromatin looping across this region, CNS3, and the Foxp3 promoter caused local enrichment of H3K4me1 at these sites (Placek et al., 2017). However, in contrast to our results, animals lacking this site, did not have significantly reduced thymic Treg cell frequencies, probably because of the age of the animals at the time of analysis, the markers used to identify thymic Treg cells, the precise region targeted, or some combination thereof. Although we detected H3K4me1 at CNS0 and reduced H3K4me1 at the Foxp3 promoter in CNS0-deficient Tn cells, CNS0 deficiency had no measurable effect on chromatin accessibility at the Foxp3 locus in Treg cells or their precursors and no effect on H3K4me1 at CNS3. Therefore, as with SATB1, further experiments are required to assess the potential epistatic relationship between STAT5 and MLL4 activities at CNS0 in Foxp3 induction and Treg cell differentiation. Additionally, CNS0 is proximal to the previously described lncRNA Flicr, which was suggested to negatively regulate Foxp3 expression when IL-2 is limiting (Zemmour et al., 2017). As we observed reduced expression of Flicr in the absence of CNS0 (data not shown), its proposed function as a negative regulator of Foxp3 seems inconsistent with the function of CNS0 in promoting IL-2 dependent Treg cell differentiation.
CNS0 deficiency resulted in impaired IL-2 dependent induction of Foxp3 expression and thymic Treg cell differentiation in the neonatal period. This led to transient immune activation, which was thereafter reined in by an activated Treg cell population that had numerically recovered in adult animals. However, in the context of an additional genetic perturbation—Aire deficiency—lack of CNS0 exacerbated and expanded autoimmune pathology. A key function of Aire appears to be the generation of TRA-specific Treg cells, particularly during the neonatal period, which are important for the long-term prevention of tissue-specific autoimmunity (Aschenbrenner et al., 2007; Guerau-de-Arellano et al., 2009; Malchow et al., 2013; Perry et al., 2014; Yang et al., 2015). Given that the most pronounced Treg cell paucity in CNS0-deficient mice is observed during the same time window, it is possible that the Treg cells arising independently of CNS0 comprise at least to some extent those selected by Aire-dependent antigens. Therefore, these two genetic lesions would impede generation of complementary populations of Treg cells, leading to a severe Treg cell paucity at a crucial developmental time. On the other hand, Aire deficiency also compromises Treg cell-independent elimination of pathogenic TRA-reactive T cell clones, reflected by the compounded disease in Foxp3 and Aire double deficient mice (Chen et al., 2005). It is therefore also possible that CNS0 is particularly important for controlling the pathogenic T cells by supporting the diversification of the Treg cell TCR repertoire.
More generally, there is a parallel between these results and our recent findings from studies of CNS1, a Foxp3 intronic enhancer specifically involved in extrathymic Treg cell differentiation (Campbell et al., 2018; Josefowicz et al., 2012b; Zheng et al., 2010). In that case, lack of CNS1 resulted in delayed accumulation of Treg cells in the colon in response to microbial colonization and transient aberrant type II immune activation with lingering perturbations in the absence of ongoing inflammation. In both cases, acute and localized Treg cell paucity results in transient immune activation, which leads to pathology under extenuating circumstances, but is otherwise controlled. Nonetheless, the evolutionary conservation of these regulatory elements suggests that such specific circumstances generate selective pressure over time and that organismal fitness depends on the context-specific regulation and optimization of Foxp3 expression that these genetic elements afford.
Numerous studies demonstrate that IL-2 plays at least two principal roles in Treg cell biology: it is crucial for Treg cell differentiation in the thymus and also promotes the survival and function of peripheral Treg cells (Chinen et al., 2016; Fan et al., 2018; Feng et al., 2014; Hemmers et al., 2019; Li et al., 2014; Liu et al., 2015; Owen et al., 2018; Toomer et al., 2019). Here, we show that these distinct modes are separately controlled through a temporally regulated switch from CNS0 to CNS2 during the transition from precursors to fully differentiated Treg cells. In this vein, there are several examples of early- and late-acting enhancers controlling expression of the same gene during the differentiation of immune cells (Bagadia et al., 2019; Ellmeier et al., 1998; Hernández-Munain et al., 1999). We found that CNS0 was dispensable in fully differentiated Treg cells, while playing important roles during Treg cell lineage commitment. CNS0 thereby mirrors the functionality of CNS2 as the latter is required for the maintenance of Foxp3 expression in mature Treg cells but has no effect on Foxp3 induction during Treg cell lineage commitment (Feng et al., 2014; Li et al., 2014; Zheng et al., 2010). The role of CNS0 in sustaining early Foxp3 expression became redundant upon demethylation and consequent activation of CNS2. Furthermore, stability of Foxp3 expression in mature Treg cells was independent of CNS0, and the progressive loss of Foxp3 expression by dividing CNS2-deficient cells suggests that CNS0 fails to compensate for CNS2 deficiency (Feng et al., 2014; Li et al., 2014). Thus, the integration of IL-2 – STAT5 signaling at the Foxp3 locus by CNS0 followed by a switch to CNS2 exemplifies temporal enhancer partitioning that anticipates changing external stimuli in a manner that meets shifting biological needs.
Limitations of the study
The mechanistic basis of IL-2 – STAT5 – CNS0 dependent activation of Foxp3 transcription and subsequent Treg cell differentiation remains to be fully defined. In one scenario, CNS0 might drive expression of a long non-coding transcript, whose invasion into the Foxp3 locus allows for the resolution of R-loops formed by bi-directional transcripts, which are found at several Foxp3 cis-regulatory elements. Alternatively, SATB1 may link CNS0 to the Foxp3 promoter, enabling IL-2 – STAT5 dependent transcription of Foxp3 (Kitagawa et al., 2017; Placek et al., 2017). Likewise, our studies have not addressed the specific mechanisms underlying the exacerbated and expanded autoimmune disease resulting from combined CNS0 and Aire deficiencies relative to Aire deficiency alone. Potential disorders due to immune dysregulation resulting from the severe neonatal paucity of Treg cells in CNS0-deficient mice were also unexplored.
STAR Methods
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Alexander Rudensky (rudenska@mskcc.org)
Materials Availability
There are restrictions to the availability of the CNS0FL and CNS0Δ mouse lines generated in this study due to the need for an MTA with Memorial Sloan Kettering Cancer Center. This study did not generate additional new unique reagents.
Data and Code Availability
The ATAC-seq, RNA-seq, ChIP-seq, and TCR-seq data sets generated during this study are available at GEO under accession GSE164118. Other data and code are available from the corresponding authors upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
Foxp3GFP (B6.129-Foxp3tm2Ayr/J; RRID: IMSR_APB:3598), CNS2Δ (B6.129-Foxp3tm6.2Ayr/J; RRID: MGI:4430238), and Foxp3DTR (B6.129(Cg)-Foxp3tm3(DTR/GFP)Ayr/J; RRID: IMSR_JAX:016958) have been previously described and were maintained in house (Fontenot et al., 2005b; Kim et al., 2007; Zheng et al., 2010). Aire−/− (B6.129S2-Airetm1.1Doi/J; RRID: IMSR_JAX:004743), Ndor1UBC-creER (B6.Cg-Ndor1Tg(UBC-cre/ERT2)1Ejb/1J; RRID: IMSR_JAX:007001), TcrbΔTcrdΔ (B6.129P2-Tcrbtm1MomTcrdtm1Mom/J; RRID: IMSR_JAX:002122), and CD45.1 (B6.SJL-PtprcaPepcb/BoyJ; RRID: IMSR_JAX:002014) have been previously described, were purchased from Jackson Laboratories and then maintained in house (Anderson et al., 2002; Mombaerts et al., 1992; Ruzankina et al., 2007; Shen et al., 1985). Il2−/− mice arose from a germline recombination of previously-described Il2fl/fl (B6-Il2tm1.1Kasm/J, MGI:5449713) mice and were maintained in house (Popmihajlov et al., 2012). All mice were housed at the Research Animal Resource Center for Memorial Sloan Kettering Cancer Center and Weill Cornell Medicine and the Animal Resources Center at St. Jude Children’s Research Hospital. All studies conducted at MSKCC were under protocol 08–10-023 and approved by the Sloan Kettering Institute Institutional Animal Care and Use Committee. Experiments conducted at St. Jude Children’s Research Hospital were approved by the Institutional Animal Care and Use Committee (612). All animals used in this study had no previous history of experimentation and were naïve at the time of analysis. Sex and age are indicated and justified in text and figure legends. Generally, male mice were used because of the inability to generate CNS0Δ/Δ and CNS0FL/FL female littermate controls due to CNS0 and Foxp3 being X-linked.
METHOD DETAILS
Generation of CNS0FL and CNS0Δ mice
To generate conditional CNS0 (CNS0FL) mice, bacterial artificial chromosome (BAC) RP23–143D8 was used to insert GFP sequence at the N-terminus of Foxp3 with recombineering (Sharan et al., 2009). A DTA sequence was integrated at the 5’ end of CNS0 for negative selection of engineered embryonic stem (ES) cells. CNS0 was defined as chr X: 7,570,674 – 7,571,274 (mm10) for the purposes of targeting. CNS0 sequence was then replaced with loxP-CNS0-FRT-Neo-FRT-loxP to generate the donor DNA. The resulting BAC construct was linearized with PI-SceI and electroporated into C57BL/6-derived ES cells to select for clones bearing expected homologous recombination with negative and positive selection followed by Southern blotting (CRISPR and Genome Editing Center, the Rockefeller University). Positive ES cell clones were further validated by genotyping PCR and Sanger sequencing before being injected into blastocysts (Mouse Genetic Core, MSKCC). Founders were established by crossing the chimeras with wildtype C57BL/6 mice. They were then confirmed by Southern blotting and crossed to flpe transgenic mice to delete FRT-Neo-FRT, resulting in CNS0FL mice. Deletion of Neo was validated by genotyping PCR. To generate germline deficient CNS0Δ mice, CNS0FL mice were crossed to CMV-cre (Mouse Genetic Core, MSKCC) and pups were genotyped by PCR.
STAT5 ChIP
STAT5 ChIP experiments were performed as previously described (Feng et al., 2014). Specifically, iTreg or nTreg cells were sorted and starved for 4 hours in complete RPMI (1x RPMI 1640 with 5% Fetal Bovine Serum (FBS), 10 mM HEPES buffer (ThermoFisher, 25–060-CI), 1% penicillin/streptomycin (ThermoFisher, 30–002-CI), 2 mM L-glutamine (ThermoFisher, 25–005-CI), 1 mM Sodium Pyruvate (ThermoFisher, 25–000-Cl), 1x MEM Nonessential Amino Acids (ThermoFisher, 25–025-Cl)). Cells were then stimulated with 500 U/mL recombinant human IL-2 (NIH) for 30 minutes. Cells were harvested, washed twice with PBS at room temperature (RT), resuspended at 5×106 cells/mL in 1x PBS supplemented with 1 mM MgCl2, and treated with 2 mM disuccinimidyl glutarate (DSG) cross-linker at RT for 30 minutes on a rotator. Cells were then pelleted via centrifugation, washed twice with 1x PBS at RT, and fixed with 1% formaldehyde (ThermoFisher) at RT for 5 minutes. Fixation was quenched by addition of glycine to a final concentration of 125 mM. Cells were then pelleted down, washed once with cold 1x PBS, and frozen at −80°C or immediately processed. Chromatin was sonicated with truChIP Chromatin Shearing Kit with Focused Ultrasonicator M220 following the manufacturer’s instructions. Chromatin was sheared to 400–800 bp and 10% of samples were aliquoted as input control. In each ChIP reaction, 5 μL of rabbit anti-STAT5 or control rabbit IgG were added to the lysis/binding buffer (50 mM HEPES pH 8.0, 1 mM EDTA, 1% Triton X-100, 0.1% sodium deoxycholate, 0.1% SDS, 140 mM NaCl, 1 mM PMSF, protease inhibitor cocktail) to precipitate the chromatin. After an overnight incubation, protein A and protein G magnetic beads were added to capture the antibody-chromatin complexes. After an extensive wash, DNA was released from the beads with proteinase K followed by phenol:chloroform:isoamyl alcohol extraction and 2-propanol precipitation in the presence of GlycoBlue Coprecipitant. DNA pellets were dissolved in 1x TE buffer (10 mM Tris HCl pH 8.0, 1 mM EDTA) for qPCR or deep sequencing.
To quantify the precipitated DNA, qPCR was performed with locus- or region-specific primers (See Table S4) and PowerUp SYBR Green Master Mix and CFX384 Real Time System. Relative enrichment of the targets was calculated by normalizing the signals of the precipitated DNA to those of the input samples. To perform ChIP-Seq, libraries were prepared with precipitated DNA using KAPA HyperPrep Kit. The library DNA was enriched by size selection with AMPure XP beads and quantified by NEBNext Library Quant Kit. Indexed samples were pooled together for paired end sequencing with 100 cycles on a HiSeq 4000 or HiSeq 2500 (Illumina). Approximately 40 million reads per sample were sequenced for each sample.
H3K4me1 ChIP
5 × 105 naïve CD4+ T cells (CD25−Foxp3-GFP−CD4+CD8−TCRβ+CD62L+CD44−) cells were sorted from pooled spleens and lymph nodes after CD4+ cell enrichment using the Dynabeads™ FlowComp™ Mouse CD4 Kit according to the manufacturer’s instructions. Cells were then crosslinked in 1 mL complete RPMI containing 1% formaldehyde (Sigma-Aldrich, F8775) for 5 minutes at RT. Crosslinking reaction was quenched by adding 50 μL of 2.5 M glycine solution (Sigma-Aldrich, G7126) and cells were washed once with 1x PBS. Then, cell pellets were resuspended in 1 mL Cell Lysis Buffer (25 mM HEPES, 1.5 mM MgCl2 (Sigma-Aldrich, M8266), 10 mM KCl (Sigma-Aldrich, P3911), 0.1% NP-40 (ThermoFisher, 85124)) with freshly added DTT (Sigma-Aldrich, D9779) at a final concentration of 1mM and EDTA-free protease inhibitors and incubated on ice for 10 minutes. Nuclei were collected by centrifugation and resuspended in 100 μL micrococcal nuclease buffer. Chromatin was fragmented by adding 1,200 gel units of Micrococcal Nuclease to each sample, which was incubated at 37 °C for 1 minute before addition of 10 μL 0.5 M EDTA (Sigma-Aldrich, E4884) to stop the reaction. Nuclei were pelleted, resuspended in 200 μL Nuclear Lysis Buffer (50 mM HEPES, 1 mM EDTA, 1% Triton X-100 (Fisher, BP151–500), 0.1% Na-DOC (Sigma-Aldrich, D5670), and freshly added protease inhibitors) containing 140 mM NaCl (Fisher, S671) and 1% SDS (Sigma-Aldrich, L4390), and sonicated on a Bioruptor Twin instrument (diagenode) for 10 cycles (30 s on, 30 s off) on High to disrupt the nuclear membrane. Samples were centrifuged at 13,000 g for 10 minutes at 4 °C and the chromatin-containing supernatant was collected, transferred to a different tube, and diluted 10-fold with Nuclear Lysis Buffer containing 140 mM NaCl to reach a final SDS concentration of 0.1%. 5% of the sonicated chromatin was kept separately for later use as input control. 1 μg anti-H3K4me1 antibody was added to each sample, which was rotated at 4 °C overnight. On the next day, 20 μL Protein A Dynabeads (Invitrogen, 10001D), washed and resuspended in Nuclear Lysis Buffer containing 140 mM NaCl and 0.1% SDS, was added to each sample and incubated at 4 °C for 2 hours with rocking and tilting. Beads were then collected with a microcentrifuge tube magnet after a short spin and washed twice with each of the following buffers, with every wash taking 5 min at 4 °C with rocking and tilting: 1) 1 mL Nuclear Lysis buffer containing 140 mM NaCl and 0.1% SDS, 2) 1 mL Nuclear Lysis buffer containing 500 mM NaCl and 0.1% SDS, 3) 1 mL ChIP Wash Buffer (20 mM Tris-HCl (Sigma-Aldrich, T3253), 140 mM NaCl, 1 mM EDTA, 250 mM LiCl (Sigma-Aldrich, L9650), 0.5% Na-DOC, 0.5% NP-40), and after transferred into a new tube, 4) 500 μL TE Buffer (10 mM Tris-HCl, 1 mM EDTA). To reverse the crosslinking, 300 μL Elution Buffer (20 mM Tris (Sigma-Aldrich, T6066), 50 mM NaCl, 5 mM EDTA, 1% SDS) containing 50 μg/mL proteinase K was added to the beads and the input, and the samples were incubated overnight at 65 °C with shaking at 1300 RPM on a Thermomixer R (Eppendorf). The supernatant was then collected and the beads were resuspended in another 200 μL Elution Buffer and incubated as before for 5 min. The two supernatant fractions were pooled, and the immunoprecipitated DNA was extracted using the MinElute Reaction Cleanup Kit according to manufacturer’s instructions. Relative abundance of immunoprecipitated DNA fragments were assessed by qPCR [5 μL Power SYBR Green PCR Master Mix, 2 μL of 0.5 μM Forward primer, 2 μL of 0.5 μM Reverse primer, 1 μL DNA] on a QuantStudio 6 Flex instrument with region specific primers (See Table S4). H3K4me1 signal intensity was expressed as relative fold enrichment by normalizing the fold enrichment (immunoprecipitation/input) of each region to that of the control region (Gm5069 promoter).
Sequence conservation analysis
STAT5 motifs were discovered in the CNS0 sequence (chrX:7570674–7571274) from the mm10 genome (Mouse Genome Sequencing Consortium et al., 2002) using FIMO (Grant et al., 2011) with the options --thresh 0.1 and using mouse Stat5a and Stat5b motifs from CisBP (Weirauch et al., 2014). Motifs with a p-value < 0.01 were used in subsequent analysis. Conservation of these motifs were accessed by phyloP (Pollard et al., 2010) with the options: --msa-format MAF --method SPH --features [bed file with CNS0 STAT5 motifs] --mode CON and using a model for conservation of all placental mammal species, as well as corresponding multiple sequence alignments downloaded from the UCSC Genome Browser (Kent et al., 2002). Sequences of STAT5 motifs and flanking sequence were extracted from multiple sequence alignments using maf_parse and msa_view tools from the PHAST package (Pollard et al., 2010; Siepel et al., 2005).
Lymphocyte isolation for flow cytometry
For thymus and secondary lymphoid organs, organs were dissected post-euthanasia and placed in 1 mL Wash buffer (1x RPMI 1640 with 2% FBS, 10 mM HEPES buffer, 1% penicillin/streptomycin, 2 mM L-glutamine). Tissues were then mechanically disrupted with the back end of a syringe plunger, and then passed through a 100 μm nylon mesh. For non-lymphoid tissues (Liver, Lung), tissues were placed in 5 mL snap-cap tubes (Eppendorf 0030119401) in 3 mL Wash buffer supplemented with 0.2 U/mL collagenase A and 1 U/mL DNase I, along with three ¼ inch ceramic beads (MP Biomedicals, 116540424-CF) and shaken horizontally at 250 RPM for 45 minutes at 37°C. Digested samples were then passed through a 100 μm strainer and centrifuged to remove collagenase solution. Samples were then treated with 1x ACK (155 mM ammonium chloride, 10 mM potassium bicarbonate, 100 nM EDTA pH 7.2) to lyse red blood cells, and then washed by centrifugation in 40% Percoll™ (ThermoFisher, 45–001-747) in 1x PBS to remove debris and enrich for lymphocytes. To isolate lymphocytes from small intestines, Peyer’s patches were removed and the remaining tissue was cut into 5 mm fragments and washed with 1x PBS supplemented with 5% newborn calf serum. Intraepithelial lymphocytes (IEL) were released by incubating the tissue with 1x PBS, 5% newborn calf serum, and 5 mM EDTA at 37°C with shaking for 20 minutes. After EDTA was removed, lamina propria lymphocytes (LPL) were released by digesting the tissue with 125 U/mL or 1 mg/mL collagenase type 4 and 0.25 mg/mL DNase I in DMEM, 10% newborn calf serum, and 15 mM HEPES at 37°C with shaking for 45 minutes. Digested tissue was then filtered through a 100 μm strainer, pelleted by centrifugation, and fractionated by two layers of Percoll™, 42% and 70%, in 1x PBS and 5% newborn calf serum with centrifugation. The resulting cells between the two layers of Percoll™ were pelleted and resuspended in complete RPMI for downstream analyses. All samples were washed by centrifugation in 5–10 mL Wash buffer.
Flow cytometry
Cells were stained in 96-well V-bottom plates. All centrifugations were carried out at 900x g for 2 minutes. Cell were first stained with a viability dye, and anti-mouse CD16/32 to block Fc binding in 100 μl in 1x PBS at 4°C for 10 minutes. Cells were then washed by addition of 100 μl staining buffer (1x PBS with 0.1% (w/v) BSA (VWR, 97061–422), 0.1% NaN3, 2 mM EDTA, 10 mM HEPES) followed by centrifugation. Cells were stained with fluorophore-conjugated antibodies in 100 μl staining buffer at 4°C for 25 minutes. For staining of nuclear antigens, eBioscience™ Foxp3 / Transcription Factor Staining Buffer Set was used according to the manufacturer’s instructions, adjusted for 96-well staining (100 μl for fixation, 100 μl for intracellular staining, 200 μl for washing). For staining for cytokine production, cells were incubated in complete RPMI supplemented with 50 ng/mL phorbol 12-myristate 13-acetate and 500 ng/mL ionomycin with 1 μg/mL brefeldin A and 2 μM monensin (all from Millipore Sigma) to inhibit ER and Golgi transport for 4 hours at 37°C with 5% CO2. Cells were stained for cytokines by using the Cytofix/Cytoperm™ Fixation/Permeablization Kit according to the manufacturer’s instructions, adjusted for 96-well staining (100 μl for fixation, 100 μl for intracellular staining, 200 μl for washing). Samples were acquired on an LSR II (BD Biosciences) or an Aurora (Cytek) and analyzed by using FlowJo software. To aid acquisition on the Aurora, samples were treated with 40 U/mL DNase I for 10 minutes at RT before acquisition. All sorting was carried out on an Aria II (BD Biosciences). See Table S3 for full antibody usage information.
Flow cytometric identification of thymocyte subsets
For Figures S1D and S1E, thymocyte subsets were identified as follows. All cells were gated as CD45+ZombieNIR−CD90+CD11b−MHC-II−CD19−, with doublets excluded by FSC and SSC. γδ T cells: TCRγδ+; NKT cells: CD1d-Tet+; DN1: TCRγδ−CD1d-Tet−NK1.1−CD4−CD8α− CD44+CD25−; DN2: TCRγδ−CD1d-Tet−NK1.1−CD4−CD8α−CD44+CD25+; DN3: TCRγδ−CD1d-Tet−NK1.1−CD4−CD8α−CD44−CD25+; DN4: TCRγδ−CD1d-Tet−NK1.1−CD4−CD8α−CD44−CD25−; DP TCR−: TCRγδ−CD1d-Tet−NK1.1−CD4+CD8α+CD5loTCRβ−; DP TCR+: TCRγδ−CD1d-Tet− NK1.1−CD4+CD8α+CD5hiTCRβ+; CD8SP: TCRγδ−CD1d-Tet−NK1.1−CD4−CD8α+TCRβ+; CD4SP: TCRγδ−CD1d-Tet−NK1.1−CD4+CD8α−TCRβ+CD25−Foxp3-GFP−; CD122+: TCRγδ−CD1d-Tet−NK1.1−CD4+CD8α−TCRβ+CD25−Foxp3-GFP−GITRhiCD122+; CD25+: TCRγδ−CD1dTet−NK1.1−CD4+CD8α−TCRβ+CD25+Foxp3-GFP−; CD73− Treg: TCRγδ−CD1d-Tet−NK1.1−CD4+CD8α−TCRβ+CD73−Foxp3-GFP+
Calculating Foxp3 vs. CD25 MFI R2
Scaled MFI values for compensated parameters of thymic Treg cells were exported from FlowJo using the “Export/Concatenate” feature. Values were log-transformed and fit with a linear regression in R (R Core Team, 2020).
Foxp3 induction in CD4SP thymocytes
Mature CD4 single positive (SP) thymocytes (CD25−Foxp3-GFP−CD4+CD8−TCRβhiCD24lo) were sorted from thymuses after CD8+ cell depletion using the Dynabeads™ FlowComp™ Mouse CD8 Kit according to the manufacturer’s instructions, keeping the unbound fraction. 96-well flat-bottom plates (tissue culture treated, USA Scientific 5665–5180) were coated with 1 μg/mL anti-mouse CD3 and 1 μg/mL anti-mouse CD28 antibodies in 200 μl 1x PBS at 37°C for more than 2 hours and then washed. Cells were incubated on these plates for 12 hours in complete RPMI in the presence of 5 μg/mL each anti-mouse IL-2 antibodies. Cells were then transferred to uncoated V-bottom plates, in fresh Complete RPMI, with anti-mouse IL-2 antibodies for 3 hours. Recombinant human IL-2 was added at a final concentration of 100 U/mL and cells were incubated for an additional 7 hours before staining and analysis.
Foxp3 induction in CD25+CD4SP thymocytes
CD25+Foxp3− thymocytes (CD25+Foxp3-GFP−CD4+CD8−TCRβ+) were sorted from thymuses after CD8+ cell depletion using the Dynabeads™ FlowComp™ Mouse CD8 Kit according to the manufacturer’s instructions, keeping the unbound fraction. Cells were incubated in 96-well U-bottom plates in complete RPMI in the presence of 5 μg/mL each anti-mouse IL-2 antibody and varying concentrations of recombinant human IL-2 for 24 hours before staining and analysis.
Foxp3 induction and stability assay in naïve CD4+ T cells
Naïve CD4+ T cells (CD25−Foxp3-GFP−CD4+CD8−TCRβ+CD62L+CD44−) cells were sorted from pooled spleens and lymph nodes after CD4+ cell enrichment using the Dynabeads™ FlowComp™ Mouse CD4 Kit according to the manufacturer’s instructions. Tissue culture treated 96-well flat-bottom plates (USA Scientific) were coated with 1 μg/mL anti-mouse CD3 and 1 μg/mL anti-mouse CD28 antibodies (BioXCell) in 200 μL 1x PBS at 37°C for 2 hours and then washed once with 1x PBS. Cells were cultured on these plates for 2–4 days with 1 ng/ml recombinant human TGF-β and varying concentrations of recombinant human IL-2 before staining and analysis. To assay the stability of Foxp3 expression, naïve CD4+ T cells were cultured in Treg cell induction conditions with or without 0.25 mM ascorbic acid-2-phosphate (Sigma) for 4 days. Foxp3-GFP+ cells were then sorted and cultured on new plates coated with or without 1 μg/mL anti-CD3 and 1 μg/mL anti-CD28 antibodies (BioXCell) in the presence of 100 U/mL recombinant IL-2 for 1–4 days. At the end of culture, cells were harvested and stained with a viability dye (BioLegend) followed by cell fixation and Foxp3 staining.
In vitro Treg cell suppression assay
CD4+Foxp3-GFP+ Treg cells were FACS sorted from pooled spleens and lymph nodes. CD4+CD25−CD44−CD62L+ naïve T cells were sorted from the spleens of male CD45.1 mice and stained with CellTrace™ CFSE according to the manufacturer’s manuals. Antigen-presenting cells were prepared from the splenocytes of male CD45.1 mice by depleting CD90.2+ T cells and lysing red blood cells followed by lethal irradiation (20 Gy). Treg cell suppression assay was conducted in 96-well plates with each well containing 200 μL complete RPMI-1640 supplemented with 1 μg/mL anti-CD3 antibody, 1 × 105 antigen-presenting cells, and 4 × 104 CD4 naïve T cells. Treg cells were then added at different ratios to CD4 naïve T cells. Cells were harvested 3 days later for flow cytometric analysis after being stained for viability dye (Tonbo Bioscience) and antibodies against CD4 and CD45.1.
TCR sequencing
Bulk TCR sequencing was performed as previously described (Dash et al., 2015; Feng et al., 2015). Briefly, CD4+Foxp3-GFP+ Treg cells were FACS sorted from the spleens. Total RNA was extracted by TRIzol™ reagent according to the manufacturer’s instructions. cDNA was synthesized by SMARTScribe Reverse Transcriptase with primers targeting mouse TCRα (AGTCAAAGTCGGTGAAC) or TCRβ (ATCTCTGCTTTTGATG) chains and template switch adapter (AAGCAGUGGTAUCAACGCAGAGUNNNNUNNNNUNNNNUCTTrGrGrGrG). The TCRα and TCRβ chains were further amplified with a two-step protocol: First, a mixture of primers (5’-CACTCTATCCGACAAGCAGTGGTATCAACGCAG-3’ and 5’-CACTCTATCCGACAAGCAGT-3’) bound at the template switch adapter and primers targeting the constant regions of TCRα or TCRβ chains (TCRα GCTGTCCTGAGACCGAGGAT or TCRβ ATGGCTCAAACAAGGAGACC) were used. Second, indexing primer (N)2− 4(XXXXX)CAGTGGTATCAACGCAGAG annealing on the template switch adapter was used together with TCRα 3’ indexing primer (N)2–4(XXXXX)CAGGTTCTGGGTTCTGGATGT or TCRβ 3’ indexing primer (N)2–4(XXXXX)AGTCACATTTCTCAGATCCT. Sequencing adapters were then added using the Kapa HyperPrep Kit. Samples were pooled at the same molar ratios and sequenced on a NovaSeq 6000 instrument with at least 20x coverage relative to the numbers of Treg cells used. To analyze TCR diversity, bulk TCR sequence data were de-multiplexed by using the MIGEC V1.2.9 software (Shugay et al., 2014). VDJ gene assignments were calculated and filtered with MIGEC software. VDJtools v1.2.1 software (Shugay et al., 2015) was applied for filtering non-functional TCR and decontaminate samples. To estimate TCR diversity, we used the immunarch package (Nazarov et al., 2020) to explore the T cell repertoires.
Serum Immunoglobulin ELISAs
Serum IgG2b titers were measured using the SBA Clonotyping System-HRP according to the manufacturer’s instructions. ELISAs were read at OD 450 on a Synergy HTX instrument.
Histopathological analysis
Tissue samples were fixed in 10% neutral buffered formalin and processed for hematoxylin and eosin staining. Stained slides were scored for tissue inflammation as previously described (Chaudhry et al., 2009).
Bone marrow chimera generation
Mixed bone marrow chimeric mice were generated as previously described (Feng et al., 2014). Briefly, recipient mice were irradiated (9.5 Gy) 1 day before transfer of 10×106 bone marrow cells from CD45.1 wildtype and CD45.2 CNS0FL or CNS0Δ mice mixed at a 1:1 ratio. After bone marrow transfer, the recipient mice were administrated with 2 mg/ml neomycin (Millipore Sigma) in drinking water for 3 weeks and analyzed 8 to 10 weeks later.
Adoptive transfer for Treg cell stability
Treg cells (Foxp3-GFP+CD4+CD8−TCRβ+) cells were sorted from pooled spleens and lymph nodes after CD4+ cell enrichment using the Dynabeads™ FlowComp™ Mouse CD4 Kit according to the manufacturer’s instructions. Donors were CNS0FL/YFoxp3GFP/YNdor1UBCcreER/+ (CNS0-deletion) or CNS0WT/YFoxp3GFP/YNdor1UBCcreER/+ (control) littermates. Bulk T cells were isolated from pooled spleen and lymph nodes of CD45.1 Foxp3DTR mice by CD90+ cell enrichment using the Dynabeads™ FlowComp™ Mouse Pan T Kit according to the manufacturer’s instructions. Treg cells of either genotype were mixed at a 1:10 ratio with Foxp3DTR T cells and transferred into TcrbΔTcrdΔ mice, with each mouse receiving ~ 1.8 × 105 Treg cells and 1.78 × 106 bulk T cells. To deplete Treg cells derived from CD45.1 Foxp3DTR mice, recipients were treated on days 1, 4, 9, 14, 19, 24, 29, and 34 post-transfer with 250 ng diphtheria toxin by intraperitoneal injection in 200 μL sterile 1x PBS. To induce creER activity (and CNS0 deletion in experimental group), recipients were treated on days 7, 10, and 13 post-transfer with 8 mg tamoxifen by oral gavage in 200 μL sterile corn oil (Sigma-Aldrich, C8267). Tamoxifen treatment was delayed 1 week to spare killing of proliferating CreER+ Treg cells (Kurachi et al., 2019). Mice were analyzed for Foxp3 expression by Treg cells by flow cytometry on day 35 post-transfer.
RNA-sequencing
CD4+Foxp3-GFP+ Treg cells were FACS sorted from the thymuses of 8-week-old female CNS0FL/WT and CNS0Δ/WT or male CNS0FL and CNS0Δ littermates. Peripheral resting (CD44loCD62Lhi) and activated (CD44hiCD62Llo) Treg cells were sorted from the spleen and lymph nodes of 6- to 8-week-old male CNS0FL mice. Sorted Treg cells were lysed with the TRIzol™ reagent. RNA was extracted, sequencing libraries were constructed and sequenced on the HiSeq 2000 platform by the IGO core at MSKCC. Reads processing, mapping, and differential gene expression analysis were performed as previously described (Feng et al., 2015). Activation related transcriptional signatures in Treg cells were determined by the differences between read counts of peripheral activated versus resting Treg cells from CNS0FL mice with fold-change and Benjamini-Hochberg corrected p values (FDR < 0.001). TCR-dependent signatures were used from (Levine et al., 2017). One-tailed Kolmogorov-Smirnov test was used to determine the significance between the distributions of signature genes and all expressed genes.
Sample preparation for ATAC-sequencing
Cells used for ATAC sequencing (Figure 1) were FACS sorted from male Foxp3GFP mice by the following markers: DN1: CD4−CD8−CD44+CD25−, DN2: CD4−CD8−CD44+CD25+, DN3: CD4−CD8−CD44−CD25+, DN4: CD4−CD8−CD44−CD25−, DP: CD4+CD8+, CD4SP: CD4+CD8− CD44−CD25−, (thymus); Tn: CD4+TCRβ+Foxp3-GFP−CD25−CD44loCD62Lhi, rTreg: CD4+Foxp3-GFP+CD44loCD62Lhi, and aTreg: CD4+Foxp3-GFP+CD44hiCD62Llo (SLOs). To isolate DN thymocytes, cells were first depleted using biotinylated antibodies against CD19, CD11b, CD11c, CD4, CD8, TCRβ, TCRγδ, Ter119, and NK1.1 with streptavidin coupled Dynabeads™. For Figure S1, Treg cells (CD4+Foxp3-GFP+) were sorted from the SLOs of 8-week-old Cd4-cretg+CNS0FL and Cd4-cretg−CNS0FL littermates. For Figure 6, thymuses were dissociated with a syringe plunger and passed through a 100 μm strainer. To enrich for CD4SP cells, single cell suspensions were depleted of CD8+ cells using the Dynabeads™ FlowComp™ Mouse CD8 kit according to the manufacturer’s instructions, and collecting the unbound fraction. Cells were then sorted into DNA Lo-Bind (Eppendorf, 022431021) tubes as follows: Treg cells: Foxp3-GFP+CD4+CD8−TCRβ+; preTreg cells: CD25+Foxp3-GFP− CD4+CD8−TCRβ+; mature CD4SP cells: CD25−Foxp3-GFP−CD4+CD8−TCRβhiCD24lo; semi-mature CD4SP cells: CD25−Foxp3-GFP−CD4+CD8−TCRβloCD24hi. There were 2 biological replicates per cell population with 40,000 to 50,000 cells. Immediately after sorting cells were processed to generate ATAC libraries as previously described with minor modifications (Buenrostro et al., 2015). Briefly, cells were pelleted in a fixed rotor benchtop centrifuge at 500x g for 5 minutes at 4°C. Cells were then washed in 1 mL cold 1x PBS and pelleted again. Supernatant was aspirated and cells were resuspended in 50 μL ice-cold cell lysis buffer (10 mM Tris-Cl pH 7.4; 10 mM NaCl; 3 mM MgCl2; 0.1% NP-40) to disrupt plasma membranes. Nuclei were pelleted at 1000x g for 10 minutes. Supernatant was aspirated and nuclei were resuspended in 50 μL transposition reaction mixture (Illumina Tagment Kit: 25 μL TD buffer; 2.5 μL TDE1; 22.5 μL ddH2O). Samples were incubated in a ThermoMixer at 1100 RPM for 45 minutes at 42°C. DNA was then purified usng a MinElute Reaction Cleanup Kit, according to the manufacturer’s instructions. DNA was eluted in 10 μL buffer EB. Libraries were then barcoded and amplified with NEBNext® High-Fidelity Master Mix and primers from (Buenrostro et al., 2013) (50 μL reaction with 10 μL DNA and 2.5 μL of 25 μM primers – 1 cycle of 5 minutes at 72°C, 30 seconds at 98°C; 5 cycles of 10 seconds at 98°C, 20 seconds at 63°C, 1 minute at 72°C). A qPCR on the product determined that an additional 7 cycles (10 seconds at 98°C, 20 seconds at 63°C, 1 minutes at 72°C) were required. Library was purified and size selected with AMPure XP beads: 45 μL of PCR product was incubated with 18 μL beads and supernatant was collected (beads bound larger >2000 bp fragments). Supernatant (63 μL) was then incubated with an additional 63 μL of beads for 5 minutes, supernatant was removed, beads were washed 2x with 75% ethanol, and DNA was eluted into 50 μL H2O by incubating for 2 minutes. Samples were QC-checked and quantified on an Agilent BioAnalyzer by the IGO core at MSKCC before pooling and sequencing on an Illumina HiSeq 3000.
ATAC-sequencing data processing
Fastq files were processed with Trimmomatic (Bolger et al., 2014) to remove adapter sequences and unpaired reads, with the following options: ILLUMINACLIP:TruSeq3-PE.fa:2:30:10 LEADING:3 TRAILING:3 SLIDINGWINDOW:4:15 MINLEN:36. Reads were then aligned to the mouse mm10 genome assembly using STAR (Dobin et al., 2013) and the following options: --runMode alignReads --readFilesCommand zcat --outSAMtype BAM SortedByCoordinate --outBAMcompression 6 --outFilterMultimapNmax 1 --outFilterMismatchNoverLmax 0.06 --outFilterMatchNminOverLread 0.35 --outFilterMatchNmin 30 --alignIntronMax 1 --alignEndsType Local. Duplicate reads were then removed by the following commands using samtools (Li et al., 2009): (1) samtools sort -@ 4 -n (2) samtools fixmate -rm (3) samtools sort -@ 4 (4) samtools markdup -l 1500 -r -d 100 -s (5) samtools sort -@ 4 -n. Peak calling was performed per celltype*genotype using Genrich (Gaspar, 2020) with the following options: -j -d 25 -g 5 -e chrM -v -q 0.01 -a 280. A peak atlas was created by merging peaks across all celltype*genotype groups using a custom R script. Counting reads against the peak atlas was performed using the summarizeOverlaps function of the R package GenomicAlignments (Lawrence et al., 2013). Differential accessibility analysis was performed using DESeq2 (Love et al., 2014). K-means clustering was performed using R and included all peaks showing differential accessibility in any pairwise comparison of celltype*genotype groups. Peaks (150 bp around summit) differentially accessible between CNS0Δ and CNS0FL Treg cells or belonging to clusters I and III were then scanned for known motif enrichment using the script findMotifsGenome.pl from HOMER (Heinz et al., 2010) using the mm10 genome assembly, all clustered peaks as background, and with options: -size given -mask -p 2. For track display, alignments were converted to bigWig files, scaled for number of paired, uniquely aligned, non-duplicate reads, with the following commands using bedtools v2.26.0 (Quinlan and Hall, 2010), and UCSC utility wigToBigWig (Kent et al., 2010), and mm10 chromosome sizes downloaded from UCSC (mm10.genome): (1) bedtools genomecov -bg -scale -g mm10.genome (2) wigToBigWig -clip mm10.genome. All pertinent R scripts available upon request.
Bisulfite sequencing
CD4+ naïve T cells were sorted and cultured in Treg cell induction conditions. 250 μM ascorbic acid 2-phostate was added to cell culture 48 hours later. Foxp3-GFP+ iTreg cells were sorted after 0, 8, 24, and 48 hours of ASC treatment. Genomic DNA from 0.5×106 sorted cells was prepared by proteinase K digestion followed by Phenol:Chloroform:Isoamyl Alcohol extraction and 2-propanol precipitation. 1 μg of DNA was used for processing with the EpiTect Bisulfite Kit. CNS0, promoter, CNS1, CNS2, and CNS3 were amplified by EpiTaq™ HS polymerase with the primers indicated in Table S4. PCR products were separated by agarose gel electrophoresis and retrieved by NucleoSpin Gel and PCR clean-up Kit. DNA fragments from the same sample were pooled together at equimolar ratios and barcoded. Sequencing libraries were prepared with TruSeq DNA PCR-Free LT Kit according to the manufacture’s manual. Samples were sequenced on the MiSeq platform (Illumina) with 1 × 105 reads for every DNA fragment in each sample. Sequencing reads were then aligned to the mouse reference genome with BSMAP2.74 (Xi and Li, 2009). The methylation ratio for each CpG site was extracted using the script methratio.py from BSMAP2.74.
QUANTIFICATION AND STATSTICAL ANALYSIS
Data were analyzed and statistical tests were performed by using GraphPad Prism and R, and plotted using GraphPad Prism and ggplot2 (Wickham, 2009). Statistical details for each experiment can be found in the text and figure legends. Unless otherwise noted, significance was defined as α = 0.05. No specific methods were used to determine strategies for randomization, sample size estimation, and no data or subjects were excluded post hoc. No specific methods were used to determine whether the data met the assumptions of the statistical tests used.
Supplementary Material
List and sequences of primers used for qPCR and library construction. Related to STAR Methods.
List of antibodies and reagents used for flow cytometry. Related to STAR Methods.
Pathological evaluation of multiple tissues from 10- to 11- month old CNS0FL and CNS0Δ male littermates. Related to Figure 4.
Peaks included in ATAC-seq hierarchical clustering, along with coordinates, log2 FC and adjusted p-values. Related to Figures 6, S6, and S7.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Biotinylated Goat anti-mouse Areg polyclonal | R&D Systems | Cat# BAF989; RRID: AB_2060662 |
| Brilliant Violet 750 anti-mouse CD11b | BioLegend | Cat# 101267; RRID: AB_2810328, Clone: M1/70 |
| eFluor 450 anti-mouse CD11c | ThermoFisher | Cat# 48-0114-82; RRID: AB_1548654, Clone: N418 |
| PE-eFluor610 anti-mouse CD11c | ThermoFisher | Cat# 61-0114-82; RRID: AB_2574530, Clone: N418 |
| PE anti-mouse CD122 | BioLegend | Cat# 105906; RRID: AB_2125736, Clone: 5H4 |
| Brilliant Violet 421 anti-mouse CD127 | BioLegend | Cat# 135023; RRID: AB_10897948, Clone: A7R34 |
| Brilliant Violet 785 anti-mouse CD127 | BioLegend | Cat# 135037; RRID: AB_2565269, Clone: A7R34 |
| PE anti-mouse CD127 | TONBO biosciences | Cat# 50-1271-U100; RRID: AB_2621780, Clone: A7R34 |
| PE-Cy5 anti-mouse CD19 | BioLegend | Cat# 115510; RRID: AB_313645, Clone: 6D5 |
| eFluor 450 anti-mouse CD218 | ThermoFisher | Cat# 48-5183-82; RRID: AB_2574069, Clone: P3TUNIA |
| Brilliant Violet 605 anti-mouse CD24 | BioLegend | Cat# 101827; RRID: AB_2563464, Clone: M1/69 |
| eFluor 450 anti-mouse CD24 | ThermoFisher | Cat# 48-0242-82; RRID: AB_1311169, Clone: M1/69 |
| APC anti-mouse CD25 | ThermoFisher | Cat# 17-0251-82; RRID: AB_469366, Clone: PC61.5 |
| Brilliant UltraViolet 395 anti-mouse CD25 | BD Biosciences | Cat# 564022; RRID: AB_2722574, Clone: PC61.5 |
| PE anti-mouse CD25 | ThermoFisher | Cat# 12-0251-83; RRID: AB_465608, Clone: PC61.5 |
| PE-Cy5.5 anti-mouse CD25 | ThermoFisher | Cat# 35-0251-82; RRID: AB_11218898, Clone: PC61.5 |
| PE-Cy7 anti-mouse CD25 | ThermoFisher | Cat# 25-0251-82; RRID: AB_469608, Clone: PC61.5 |
| Brilliant Violet 421 anti-mouse CD304 | BioLegend | Cat# 145209; RRID: AB_2562358, Clone: 3E12 |
| PE-eFluor610 anti-mouse CD304 | ThermoFisher | Cat# 61-3041-82; RRID: AB_2574600, Clone: 3DS304M |
| Brilliant Violet 605 anti-mouse CD3ε | BioLegend | Cat# 100237; RRID: AB_2562039, Clone: 17A2 |
| Brilliant Violet 650 anti-mouse CD3ε | BioLegend | Cat# 100229; RRID: AB_11204249, Clone: 17A2 |
| PE-Cy7 anti-mouse CD3ε | TONBO biosciences | Cat# 60-0031-U100; RRID: AB_2621824, Clone: 145-2C11 |
| AlexaFluor 647 anti-mouse CD4 | BioLegend | Cat# 100530; RRID: AB_389325, Clone: RM4-5 |
| AlexaFluor 700 anti-mouse CD4 | BioLegend | Cat# 116022; RRID: AB_2715958, Clone: RM4-4 |
| Brilliant UltraViolet 496 anti-mouse CD4 | BD Biosciences | Cat# 564667; RRID: AB_2722549, Clone: GK1.5 |
| Brilliant Violet 510 anti-mouse CD4 | BioLegend | Cat# 100553; RRID: AB_2561388, Clone: RM4-5 |
| Brilliant Violet 605 anti-mouse CD4 | BioLegend | Cat# 100548; RRID: AB_2563054, Clone: RM4-5 |
| PerCP-Cy5.5 anti-mouse CD4 | BioLegend | Cat# 116012; RRID: AB_2563023, Clone: RM4-4 |
| Brilliant Violet 650 anti-mouse CD44 | BioLegend | Cat# 103049; RRID: AB_2562600, Clone: IM7 |
| Brilliant Violet 711 anti-mouse CD44 | BD Biosciences | Cat# 563971; RRID: AB_2738518, Clone: IM7 |
| AlexaFluor 700 anti-mouse CD45 | ThermoFisher | Cat# 56-0451-82; RRID: AB_891454, Clone: 30-F11 |
| Brilliant Violet 510 anti-mouse CD45 | BioLegend | Cat# 103137; RRID: AB_2561392, Clone: 30-F11 |
| Brilliant Violet 570 anti-mouse CD45 | BioLegend | Cat# 103136; RRID: AB_2562612, Clone: 30-F11 |
| Brilliant Violet 750 anti-mouse CD45 | BioLegend | Cat# 103157; RRID: AB_2734155, Clone: 30-F11 |
| Brilliant Violet 711 anti-mouse CD49a | BD Biosciences | Cat# 564863; RRID: AB_2738987, Clone: Ha31/8 |
| Brilliant Violet 711 anti-mouse CD5 | BioLegend | Cat# 100639; RRID: AB_2810322, Clone: 53-7.3 |
| Brilliant UltraViolet 737 anti-mouse CD62L | BD Biosciences | Cat# 565213; RRID: AB_2721774, Clone: MEL-14 |
| Brilliant Violet 510 anti-mouse CD62L | BioLegend | Cat# 104441; RRID: AB_2561537, Clone: MEL-14 |
| Brilliant Violet 570 anti-mouse CD62L | BioLegend | Cat# 104433; RRID: AB_10900262, Clone: MEL-14 |
| Brilliant Violet 605 anti-mouse CD62L | BioLegend | Cat# 104438; RRID: AB_2563058, Clone: MEL-14 |
| PE anti-mouse CD62L | ThermoFisher | Cat# 12-0621-83; RRID: AB_465722, Clone: MEL-14 |
| APC anti-mouse CD64 | BioLegend | Cat# 139306; RRID: AB_11219391, Clone: X54-5/7.1 |
| APC anti-mouse CD69 | ThermoFisher | Cat# 17-0691-82; RRID: AB_1210795, Clone: H1.2F3 |
| PE-Cy7 anti-mouse CD69 | ThermoFisher | Cat# 25-0691-82; RRID: AB_469637, Clone: H1.2F3 |
| Brilliant Violet 605 anti-mouse CD73 | BioLegend | Cat# 127215; RRID: AB_2561528, Clone: TY/11.8 |
| eFluor 450 anti-mouse CD73 | ThermoFisher | Cat# 48-0731-82; RRID: AB_1272196, Clone: TY/11.8 |
| PerCP-eFluor 710 anti-mouse CD73 | ThermoFisher | Cat# 46-0731-82; RRID: AB_10853356, Clone: TY/11.8 |
| AlexaFluor 532 anti-mouse CD8α | ThermoFisher | Cat# 58-0081-82; RRID: AB_11220469, Clone: 53-6.7 |
| AlexaFluor 700 anti-mouse CD8α | BD Biosciences | Cat# 557959; RRID: AB_396959, Clone: 53-6.7 |
| Brilliant Violet 510 anti-mouse CD8α | BioLegend | Cat# 100752; RRID: AB_2563057, Clone: 53-6.7 |
| Brilliant Violet 711 anti-mouse CD8α | BioLegend | Cat# 100759; RRID: AB_2563510, Clone: 53–6.7 |
| PE-Cy5 anti-mouse CD8α | ThermoFisher | Cat# 15-0081-83; RRID: AB_468707, Clone: 53–6.7 |
| AlexaFluor 700 anti-mouse CD8β.2 | BioLegend | Cat# 126618; RRID: AB_2563949, Clone: YTS156.7.7 |
| AlexaFluor 700 anti-mouse CD90.2 | BioLegend | Cat# 105320; RRID: AB_493725, Clone: 30-H12 |
| Brilliant Violet 786 anti-mouse CD90.2 | BD Biosciences | Cat# 564365; RRID: AB_2734760, Clone: 30-H12 |
| APC anti-mouse Egr2 | ThermoFisher | Cat# 17-6691-82; RRID: AB_11151502, Clone: erongr2 |
| PE anti-mouse Egr2 | ThermoFisher | Cat# 12-6691-82; RRID: AB_10717804, Clone: erongr2 |
| PE-Cy7 anti-mouse Eomes | ThermoFisher | Cat# 25-4875-82; RRID: AB_2573454, Clone: Dan11mag |
| FITC anti-mouse Foxp3 | ThermoFisher | Cat# 11-5773-82; RRID: AB_465243, Clone: FJK-16s |
| eFluor 660 anti-mouse Gata-3 | ThermoFisher | Cat# 50-9966-42; RRID: AB_10596663, Clone: TWAJ |
| efluor450 anti-mouse GITR | ThermoFisher | Cat# 48-5874-82; RRID: AB_1944394, Clone: DTA-1 |
| FITC anti-mouse GITR | ThermoFisher | Cat# 11-5874-82; RRID: AB_465286, Clone: DTA-1 |
| PerCP-Cy5.5 anti-mouse GM-CSF | BioLegend | Cat# 505410; RRID: AB_2562376, Clone: MP1-22E9 |
| AlexaFluor 532 anti-mouse Gr-1 | ThermoFisher | Cat# 58-5931-82; RRID: AB_11220477, Clone: RB6-8C5 |
| APC anti-mouse IFNγ | ThermoFisher | Cat# 17-7311-82; RRID: AB_469504, Clone: XMG1.2 |
| Brilliant UltraViolet 737 anti-mouse IFNγ | BD Biosciences | Cat# 564693; RRID: AB_2722494, Clone: XMG1.2 |
| PE anti-mouse IFNγ | TONBO biosciences | Cat# 50-7311-U100; RRID: AB_2621810, Clone: XMG1.2 |
| AlexaFluor 647 anti-mouse IgD | ThermoFisher | Cat# 51-5993-82; RRID: AB_837118, Clone: 11-26 |
| PE anti-mouse IL-13 | ThermoFisher | Cat# 12-7133-82; RRID: AB_763559, Clone: eBio13A |
| PE-Cy7 anti-mouse IL-13 | ThermoFisher | Cat# 25-7133-82; RRID: AB_2573530, Clone: ebio13A |
| Brilliant UltraViolet 395 anti-mouse IL-17 | BD Biosciences | Cat# 565246; RRID: AB_2722575, Clone: TC11-18H10 |
| eFluor 450 anti-mouse IL-17 | ThermoFisher | Cat# 48-7177-82; RRID: AB_11149503, Clone: eBio17B7 |
| AlexaFluor 700 anti-mouse IL-2 | BioLegend | Cat# 503818; RRID: AB_528931, Clone: JES6-5H4 |
| Brilliant Violet 510 anti-mouse IL-2 | BioLegend | Cat# 503833; RRID: AB_2562977, Clone: JES6-5H4 |
| PE anti-mouse IL-2 | ThermoFisher | Cat# 12-7021-82; RRID: AB_466150, Clone: JES6-5H4 |
| PerCP-eFluor 710 anti-mouse IL-22 | ThermoFisher | Cat# 46-7221-82; RRID: AB_10598646, Clone: 1H8PWSR |
| APC anti-mouse IL-4 | ThermoFisher | Cat# 17-7041-82; RRID: AB_469494, Clone: 11B11 |
| PE-Cy7 anti-mouse IL-4 | ThermoFisher | Cat# 25-7042-82; RRID: AB_469674, Clone: BVD6-24G2 |
| eFluor 450 anti-mouse Ki-67 | ThermoFisher | Cat# 48-5698-82; RRID: AB_11149124, Clone: SoIA15 |
| PE anti-mouse Ki-67 | ThermoFisher | Cat# 12-5698-82; RRID: AB_11150954, Clone: SolA15 |
| PE-eFluor610 anti-mouse Ki-67 | ThermoFisher | Cat# 61-5698-82; RRID: AB_2574620, Clone: SolA15 |
| PerCP-eFluor 710 anti-mouse Ki-67 | ThermoFisher | Cat# 46-5698-82; RRID: AB_11040981, Clone: SolA15 |
| Brilliant Violet 711 anti-mouse KLRG1 | BD Biosciences | Cat# 564014; RRID: AB_2738542, Clone: 2F1 |
| PE-eFluor610 anti-mouse KLRG1 | ThermoFisher | Cat# 61-5893-82; RRID: AB_2574630, Clone: 2F1 |
| APC anti-mouse Lag-3 | BioLegend | Cat# 125210; RRID: AB_10639727, Clone: C9B7W |
| Brilliant Violet 421 anti-mouse LAP | BioLegend | Cat# 141408; RRID: AB_2650898, Clone: TW7-16B4 |
| Brilliant Violet 711 anti-mouse Ly-6C | BioLegend | Cat# 128037; RRID: AB_2562630, Clone: HK1.4 |
| Brilliant Violet 480 anti-mouse MHCII | BD Biosciences | Cat# 566086; RRID: AB_2869739, Clone: M5/114.15.2 |
| APC-eFluor 780 anti-mouse NK1.1 | ThermoFisher | Cat# 47-5941-82; RRID: AB_2735070, Clone: PK136 |
| Brilliant Violet 510 anti-mouse NK1.1 | BioLegend | Cat# 108738; RRID: AB_2562217, Clone: PK136 |
| eFluor 450 anti-mouse NK1.1 | ThermoFisher | Cat# 48-5941-82; RRID: AB_2043877, Clone: PK136 |
| PE-Cy7 anti-mouse NK1.1 | ThermoFisher | Cat# 25-5941-82; RRID: AB_469665, Clone: PK136 |
| eFluor 660 anti-mouse NKp46 | ThermoFisher | Cat# 50-3351-82; RRID: AB_10598664, Clone: 29A1.4 |
| Brilliant Violet 785 anti-mouse PD1 | BioLegend | Cat# 135225; RRID: AB_2563680, Clone: 29F.1A12 |
| Brilliant Violet 421 anti-mouse RORγt | BD Horizon | Cat# 562894; RRID: AB_2687545, Clone: Q31-378 |
| Brilliant Violet 421 anti-mouse SiglecF | BD Biosciences | Cat# 562681; RRID: AB_2722581, Clone: E50-2440 |
| PE anti-mouse T-bet | ThermoFisher | Cat# 12-5825-82; RRID: AB_925761, Clone: 4B10 |
| APC anti-mouse TCRβ | ThermoFisher | Cat# 17-5961-82; RRID: AB_469481, Clone: H57-597 |
| APC-eFluor 780 anti-mouse TCRβ | ThermoFisher | Cat# 47-5961-82; RRID: AB_1272173, Clone: H57-597 |
| PE-eFluor610 anti-mouse TCRβ | ThermoFisher | Cat# 61-5961-82; RRID: AB_2574644, Clone: H57-597 |
| PerCP-Cy5.5 anti-mouse TCRβ | BioLegend | Cat# 109227; RRID: AB_1575176, Clone: H57-597 |
| Brilliant Violet 421 anti-mouse TCRγδ | BioLegend | Cat# 118119; RRID: AB_10896753, Clone: GL3 |
| PerCP-eFluor 710 anti-mouse TCRγδ | ThermoFisher | Cat# 46-5711-82; RRID: AB_2016707, Clone: GL3 |
| Brilliant Violet 605 anti-mouse TNFα | BioLegend | Cat# 506329; RRID: AB_11123912, Clone: MP6-XT22 |
| PE-Cy7 anti-mouse TCR Vβ5.1, 5.2 | BioLegend | Cat# 139508; RRID: AB_2566021, Clone: MR9-4 |
| PerCP-eFluor 710 anti-mouse TCR Vβ6 | ThermoFisher | Cat# 46-5795-82; RRID: AB_11150054, Clone: RR4-7 |
| AlexaFluor 647 anti-mouse TCR Vβ8.1, 8.2 | BioLegend | Cat# 118414; RRID: AB_1186096, Clone: KJ16-133.18 |
| redFluor™ 710 Anti-Mouse CD45.2 | TONBO biosciences | Cat# 80-0454-U100; RRID: AB_2621988, Clone: 104 |
| Rabbit anti-STAT5 monoclonal | Cell Signaling Technologies | Cat# 94205S; RRID: AB_2737403, Clone: D206Y |
| Rabbit anti-H3Kme1 polyclonal | Abcam | Cat# ab8895; RRID: AB_306847 |
| Hamster anti-mouse CD3ε monoclonal | BioXCell | Cat# BE0001-1; RRID: AB_1107634, Clone: 145-2C11 |
| Hamster anti-mouse CD28 monoclonal | BioXCell | Cat# BE0015-1; RRID: AB_1107624, Clone: 37.51 |
| Rat anti-mouse IL-2 monoclonal | BioXCell | Cat# BE0043; RRID: AB_1107702, Clone: JES6-1A12 |
| Rat anti-mouse IL-2 monoclonal | BioXCell | Cat# BE0043-1; RRID: AB_1107705, Clone: S4B6-1 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Recombinant human Interleukin-2 | NIH-NCI-FNLCRBRB | Cat# BULK Ro 23-6019 |
| DSG (disuccinimidyl glutarate) | ThermoFisher | Cat# 20593 |
| cOmplete™ Protease Inhibitor Cocktail | Millipore Sigma | Cat# 11697498001 |
| cOmplete™ EDTA-free Protease Inhibitor Cocktail | Millipore Sigma | Cat# 11836170001 |
| Collagenase A | Millipore Sigma | Cat# 11088793001 |
| Dnase I | Millipore Sigma | Cat# 10104159001 |
| Collagenase, Type IV | Worthington | Cat# LS004189 |
| Recombinant human TGF-β1 | R&D Systems | Cat# 240-B-002 |
| Recombinant human IL-2 | PeproTech | Cat# AF-200-02 |
| Recombinant mouse IL-4 | PeproTech | Cat# 214-14 |
| Recombinant mouse IL-6 | PeproTech | Cat# 216-16 |
| Diphtheria Toxin, Unnicked, from Corynebacterium diphtheriae | list labs | Cat# 150 |
| Tamoxifen | Millipore Sigma | Cat# T5648 |
| Proteinase K, recombinant, PCR grade | Millipore Sigma | Cat# 3115887001 |
| Uracil-DNA Glycosylase | New England Biolabs |
Cat# M0280 |
| Micrococcal Nuclease | New England Biolabs | Cat# M0247S |
| Rnasin® Ribonuclease Inhibitor, recombinant | Promega | Cat# 2511 |
| PBS57-loaded CD1d Tetramer (APCconjugated) | NIH Tetramer Core Facility |
https://tetramer.yerkes.emory.edu/ |
| Ghost Dye™ Violet 510 | Tonbo Bioscience | Cat# 13-0870-T500 |
| Ghost Dye™ Red 780 | Tonbo Bioscience | Cat# 13-0865-T500 |
| Zombie NIR™ Fixable Viability Kit | BioLegend | Cat# 423106 |
| CellTrace™ CFSE Cell Proliferation Kit | ThermoFisher | Cat# C34554 |
| TRIzol™ Reagent | ThermoFisher | Cat# 15596018 |
| Critical Commercial Assays | ||
| truChIP Chromatin Shearing Kit with Formaldehyde | Covaris | Cat# 520154 |
| Pierce Protein A/G Magnetic Beads | ThermoFisher | Cat# 88803 |
| Dynabeads™ Protein A for Immunoprecipitation | ThermoFisher | Cat # 10001D |
| GlycoBlue™ Coprecipitant | ThermoFisher | Cat# AM9516 |
| PowerUp™ SYBR™ Green Master Mix | ThermoFisher | Cat# A25741 |
| KAPA HyperPrep Kit | Roche | Cat# 7962347001 |
| AMPure XP PCR Purification | Beckman Coulter | Cat# A63881 |
| NEBNext® Library Quant Kit for Illumina® | New England Biolabs |
Cat# E7630S |
| Q5® Hot Start High-Fidelity 2X Master Mix | New England Biolabs |
Cat# M0494 |
| eBioscience™ Foxp3 / Transcription Factor Staining Buffer Set | ThermoFisher | Cat# 00-5523-00 |
| Cytofix/Cytoperm™ Fixation/Permeablization Kit |
BD Biosciences | Cat# 554714 |
| Dynabeads™ FlowComp™ Mouse CD8 Kit | ThermoFisher | Cat# 11462D |
| Dynabeads™ FlowComp™ Mouse CD4 Kit | ThermoFisher | Cat# 11461D |
| Dynabeads™ FlowComp™ Mouse Pan T (CD90.2) Kit | ThermoFisher | Cat# 11465D |
| SMARTScribe™ Reverse Transcriptase | TaKaRa | Cat# 639538 |
| SBA Clonotyping System-HRP | SouthernBiotech | Cat#5300-05 |
| SMARTer® PCR cDNA Synthesis Kit | TaKaRa | Cat# 634926 |
| Illumina Tagment DNA TDE1 Enzyme and Buffer Kit |
Illumina | Cat# 20034197 |
| MinElute Reaction Cleanup Kit | QIAGEN | Cat# 28204 |
| NEBNext® High-Fidelity 2X PCR Master Mix | New England Biolabs |
Cat# M0541L |
| EpiTect Bisulfite Kit | QIAGEN | Cat# 59104 |
| EpiTaq™ HS | Takara | Cat# R110B |
| NucleoSpin Gel and PCR Clean-up Kit | MACHEREYNAGEL | Cat# 740609.250 |
| TruSeq DNA PCR-Free LT Library Prep Kit | Illumina | Cat# 20015962 |
| Deposited Data | ||
| Cis-BP Database | (Weirauch et al., 2014) | http://cisbp.ccbr.utoronto.ca/ |
| C57BL/6 genome assembly (mm10) | (Mouse Genome Sequencing Consortium et al., 2002) | http://genome.ucsc.edu/ |
| C57BL/6 genome assembly (mm9) | (Mouse Genome Sequencing Consortium et al., 2002) | http://genome.ucsc.edu/ |
| UCSC Genome Browser | (Kent et al., 2002) | http://genome.ucsc.edu/ |
| ATAC-seq (Figure 1A) | This study | Accession: GSE164118 |
| ATAC-seq (Figure S1A) | This study | Accession: GSE164118 |
| STAT5 ChIP-seq | This study | Accession: GSE164118 |
| RNA-Seq | This study | Accession: GSE164118 |
| ATAC-seq (Figures 6, S6, and S7) | This study, (Hemmers et al., 2019) |
Accession: GSE164118 |
| TCR-seq | This study | Accession: GSE164118 |
| Experimental Models: Organisms/Strains | ||
| Mouse: Foxp3GFP: B6.129-Foxp3tm2Ayr/J | (Fontenot et al., 2005) |
RRID: IMSR_APB:3598 |
| Mouse: Foxp3DTR: B6.129(Cg)- Foxp3tm3(DTR/GFP)Ayr/J |
(Kim et al., 2007) | RRID: IMSR_JAX:016958 |
| Mouse: Aire−/−: B6.129S2-Airetm1.1Doi/J | (Anderson et al., 2002) | RRID: IMSR_JAX:004743 |
| Mouse: Ndor1UBC-creER: B6.Cg-Ndor1Tg(UBCcre/ERT2)1Ejb/1J | (Ruzankina et al., 2007) | RRID: IMSR_JAX:007001 |
| Mouse: TcrbΔTcrdΔ (B6.129P2Tcrbtm1MomTcrdtm1Mom/J) | (Mombaerts et al., 1992) | RRID: IMSR_JAX:002122 |
| Mouse: CD45.1: B6.SJL-PtprcaPepcb/BoyJ | (Shen et al., 1985) | RRID: IMSR_JAX:002014 |
| Mouse: CNS0FL | This study | N/A |
| Mouse: CNS0Δ | This study | N/A |
| Mouse: Il2−/− | This study | N/A |
| Oligonucleotides | ||
| Primers for STAT5 ChIP, see Table S4 | This study | N/A |
| Primers for Bisulfite sequencing, see Table S4 | This study | N/A |
| Primers for TCR sequencing, see Table S4 | This study | N/A |
| Primers for ATAC-sequencing libraries, see Table S4 | (Buenrostro et al., 2013) | N/A |
| Primers for H3K4me1 ChIP, see Table S4 | This study | N/A |
| All oligonucleotides | Integrated DNA Technologies | https://www.idtdna.com/ |
| Software and Algorithms | ||
| FIMO | (Grant et al., 2011) | http://meme-suite.org/doc/fimo.html |
| PHAST | (Pollard et al., 2010; Siepel et al., 2005) |
http://compgen.cshl.edu/phast/ |
| FlowJo (v10) | BD Biosciences | https://www.flowjo.com/solutions/flowjo |
| R | (R Core Team, 2020) | https://cran.r-project.org/ |
| MIGEC | (Shugay et al., 2014) | https://milaboratory.com/software/migec/ |
| VDJTools | (Shugay et al., 2015) | https://milaboratory.com/software/vdjtools/ |
| immunarch | (Nazarov et al., 2020) | https://immunarch.com/ |
| Trimmomatic | (Bolger et al., 2014) | http://www.usadellab.org/cms/?page=trimmomatic |
| STAR | (Dobin et al., 2013) | https://github.com/alexdobin/STAR |
| samtools | (Li et al., 2009) | http://www.htslib.org/ |
| Genrich | (Gaspar, 2020) | https://github.com/jsh58/Genrich |
| GenomicAlignments (R package) | (Lawrence et al., 2013) | https://doi.org/doi:10.18129/B9.bioc.GenomicAlignments |
| DESeq2 (R Package) | (Love et al., 2014) | https://doi.org/doi:10.18129/B9.bioc.DESeq2 |
| HOMER | (Heinz et al., 2010) | http://homer.ucsd.edu/homer/motif/ |
| bedtools | (Quinlan and Hall, 2010) | https://github.com/arq5x/bedtools2 |
| wigToBigWig | (Kent et al., 2010) | https://hgdownload.cse.ucsc.edu/admin/exe/ |
| BSMAP | (Xi and Li, 2009) | https://code.google.com/archive/p/bsmap/ |
| ggplot2 (R package) | (Wickham, 2009) | https://ggplot2.tidyverse.org/ |
| Prism v9 | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| Other | ||
| Instrument: M220 Focused-ultrasonicator | Covaris | Cat# 500295 |
| Instrument: CFX384 Touch Real-Time PCR Detection System | BIO-RAD | Cat# 1855485 |
| Instrument: QuantStudio™ 6 Flex RealTime PCR System | ThermoFisher | Cat# 4485691 |
| Instrument: Aurora | Cytek | https://cytekbio.com/pages/aurora |
| Instrument: LSR II | BD Biosciences | https://www.bdbiosciences.com/enus/instruments |
| Instrument: Aria II | BD Biosciences | https://www.bdbiosciences.com/enus/instruments |
| Instrument: Synergy HTX | BioTek | Cat# S1A |
| Instrument: Bioruptor® Twin | diagenode | Cat # UCD-400 |
Acknowledgments
We thank J. Verter and A. Bravo for help with animal husbandry, Y. Zhong for guidance on ATAC-seq analysis, and S. Schattgen for assistance with TCR sequencing. We thank S. Hemmers for generating the Il2+/− mice. We thank the Memorial Sloan Kettering Integrated Genomics Core facility for performing sequencing. We thank A. Mendoza and S. Hemmers for critical reading of the manuscript and all the members of the Rudensky laboratory for input. This work was supported by the NCI Cancer Center Support Grants P30 CA008748 (MSKCC) and P30 CA021765 (St. Jude), NIH grants R37 AI034206 (A.Y.R.) and R01 AI153138 (Y. Feng), the Ludwig Center at the Memorial Sloan Kettering Cancer Center (A.Y.R.), and the American Lebanese Syrian Associated Charities (Y. Feng). A.Y.R. is an investigator with the Howard Hughes Medical Institute. S.D. was supported by the General Atlantic Fellowship. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
Declaration of Interests
A.Y.R. is an SAB member and a co-founder of Sonoma Biotherapeutics. All other authors declare no competing interests.
REFERENCES
- Anderson MS, Venanzi ES, Klein L, Chen Z, Berzins SP, Turley SJ, von Boehmer H, Bronson R, Dierich A, Benoist C, et al. (2002). Projection of an immunological self shadow within the thymus by the aire protein. Science 298, 1395–1401. [DOI] [PubMed] [Google Scholar]
- Aschenbrenner K, D’Cruz LM, Vollmann EH, Hinterberger M, Emmerich J, Swee LK, Rolink A, and Klein L (2007). Selection of Foxp3+ regulatory T cells specific for self antigen expressed and presented by Aire+ medullary thymic epithelial cells. Nat. Immunol. 8, 351–358. [DOI] [PubMed] [Google Scholar]
- Bagadia P, Huang X, Liu T-T, Durai V, Grajales-Reyes GE, Nitschké M, Modrusan Z, Granja JM, Satpathy AT, Briseño CG, et al. (2019). An Nfil3-Zeb2-Id2 pathway imposes Irf8 enhancer switching during cDC1 development. Nat. Immunol. 20, 1174–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bending D, Prieto Martín P, Paduraru A, Ducker C, Marzaganov E, Laviron M, Kitano S, Miyachi H, Crompton T, and Ono M (2018). A timer for analyzing temporally dynamic changes in transcription during differentiation in vivo. J. Cell Biol. 217, 2931–2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Björses P, Aaltonen J, Horelli-Kuitunen N, Yaspo ML, and Peltonen L (1998). Gene defect behind APECED: a new clue to autoimmunity. Hum. Mol. Genet. 7, 1547–1553. [DOI] [PubMed] [Google Scholar]
- Blaschke K, Ebata KT, Karimi MM, Zepeda-Martínez JA, Goyal P, Mahapatra S, Tam A, Laird DJ, Hirst M, Rao A, et al. (2013). Vitamin C induces Tet-dependent DNA demethylation and a blastocyst-like state in ES cells. Nature 500, 222–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, and Usadel B (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Giresi PG, Zaba LC, Chang HY, and Greenleaf WJ (2013). Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Wu B, Chang HY, and Greenleaf WJ (2015). ATAC-seq: A Method for Assaying Chromatin Accessibility Genome-Wide. Curr Protoc Mol Biol 109, 21.29.1–21.29.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burchill MA, Goetz CA, Prlic M, O’Neil JJ, Harmon IR, Bensinger SJ, Turka LA, Brennan P, Jameson SC, and Farrar MA (2003). Distinct effects of STAT5 activation on CD4+ and CD8+ T cell homeostasis: development of CD4+CD25+ regulatory T cells versus CD8+ memory T cells. J Immunol 171, 5853–5864. [DOI] [PubMed] [Google Scholar]
- Burchill MA, Yang J, Vogtenhuber C, Blazar BR, and Farrar MA (2007). IL-2 receptor beta-dependent STAT5 activation is required for the development of Foxp3+ regulatory T cells. J Immunol 178, 280–290. [DOI] [PubMed] [Google Scholar]
- Burchill MA, Yang J, Vang KB, Moon JJ, Chu HH, Lio C-WJ, Vegoe AL, Hsieh C-S, Jenkins MK, and Farrar MA (2008). Linked T cell receptor and cytokine signaling govern the development of the regulatory T cell repertoire. Immunity 28, 112–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell C, Dikiy S, Bhattarai SK, Chinen T, Matheis F, Calafiore M, Hoyos B, Hanash A, Mucida D, Bucci V, et al. (2018). Extrathymically Generated Regulatory T Cells Establish a Niche for Intestinal Border-Dwelling Bacteria and Affect Physiologic Metabolite Balance. Immunity 48, 1245–1257.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhry A, Rudra D, Treuting P, Samstein RM, Liang Y, Kas A, and Rudensky AY(2009). CD4+ regulatory T cells control TH17 responses in a Stat3-dependent manner. Science 326, 986–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Benoist C, and Mathis D (2005). How defects in central tolerance impinge on a deficiency in regulatory T cells. Proc Natl Acad Sci U S A 102, 14735–14740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinen T, Kannan AK, Levine AG, Fan X, Klein U, Zheng Y, Gasteiger G, Feng Y, Fontenot JD, and Rudensky AY (2016). An essential role for the IL-2 receptor in Treg cell function. Nat. Immunol. 17, 1322–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darce J, Rudra D, Li L, Nishio J, Cipolletta D, Rudensky AY, Mathis D, and Benoist C (2012). An N-terminal mutation of the Foxp3 transcription factor alleviates arthritis but exacerbates diabetes. Immunity 36, 731–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dash P, Wang GC, and Thomas PG (2015). Single-Cell Analysis of T-Cell Receptor αβ Repertoire. Methods Mol Biol 1343, 181–197. [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellmeier W, Sunshine MJ, Losos K, and Littman DR (1998). Multiple developmental stage-specific enhancers regulate CD8 expression in developing thymocytes and in thymus-independent T cells. Immunity 9, 485–496. [DOI] [PubMed] [Google Scholar]
- Fan MY, Low JS, Tanimine N, Finn KK, Priyadharshini B, Germana SK, Kaech SM, and Turka LA (2018). Differential Roles of IL-2 Signaling in Developing versus Mature Tregs. Cell Rep 25, 1204–1213.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Arvey A, Chinen T, van der Veeken J, Gasteiger G, and Rudensky AY (2014). Control of the inheritance of regulatory T cell identity by a cis element in the Foxp3 locus. Cell 158, 749–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, van der Veeken J, Shugay M, Putintseva EV, Osmanbeyoglu HU, Dikiy S, Hoyos BE, Moltedo B, Hemmers S, Treuting P, et al. (2015). A mechanism for expansion of regulatory T-cell repertoire and its role in self-tolerance. Nature 528, 132–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontenot JD, Gavin MA, and Rudensky AY (2003). Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat. Immunol. 4, 330–336. [DOI] [PubMed] [Google Scholar]
- Fontenot JD, Rasmussen JP, Gavin MA, and Rudensky AY (2005a). A function for interleukin 2 in Foxp3-expressing regulatory T cells. Nat. Immunol. 6, 1142–1151. [DOI] [PubMed] [Google Scholar]
- Fontenot JD, Rasmussen JP, Williams LM, Dooley JL, Farr AG, and Rudensky AY (2005b). Regulatory T cell lineage specification by the forkhead transcription factor foxp3. Immunity 22, 329–341. [DOI] [PubMed] [Google Scholar]
- Fontenot JD, Dooley JL, Farr AG, and Rudensky AY (2005. c). Developmental regulation of Foxp3 expression during ontogeny. J. Exp. Med. 202, 901–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furtado GC, Curotto de Lafaille MA, Kutchukhidze N, and Lafaille JJ (2002). Interleukin 2 signaling is required for CD4(+) regulatory T cell function. J. Exp. Med. 196, 851–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspar JM (2020). jsh58/Genrich. [Google Scholar]
- Gaud G, Lesourne R, and Love PE (2018). Regulatory mechanisms in T cell receptor signalling. Nat Rev Immunol 18, 485–497. [DOI] [PubMed] [Google Scholar]
- Gavin MA, Rasmussen JP, Fontenot JD, Vasta V, Manganiello VC, Beavo JA, and Rudensky AY (2007). Foxp3-dependent programme of regulatory T-cell differentiation. Nature 445, 771–775. [DOI] [PubMed] [Google Scholar]
- Grant CE, Bailey TL, and Noble WS (2011). FIMO: scanning for occurrences of a given motif. Bioinformatics 27, 1017–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerau-de-Arellano M, Martinic M, Benoist C, and Mathis D (2009). Neonatal tolerance revisited: a perinatal window for Aire control of autoimmunity. J Exp Med 206, 1245–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, and Glass CK (2010). Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmers S, Schizas M, Azizi E, Dikiy S, Zhong Y, Feng Y, Altan-Bonnet G, and Rudensky AY (2019). IL-2 production by self-reactive CD4 thymocytes scales regulatory T cell generation in the thymus. J. Exp. Med. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández-Munain C, Sleckman BP, and Krangel MS (1999). A developmental switch from TCR delta enhancer to TCR alpha enhancer function during thymocyte maturation. Immunity 10, 723–733. [DOI] [PubMed] [Google Scholar]
- Herppich S, Toker A, Pietzsch B, Kitagawa Y, Ohkura N, Miyao T, Floess S, Hori S, Sakaguchi S, and Huehn J (2019). Dynamic Imprinting of the Treg Cell-Specific Epigenetic Signature in Developing Thymic Regulatory T Cells. Front Immunol 10, 2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori S, Nomura T, and Sakaguchi S (2003). Control of regulatory T cell development by the transcription factor Foxp3. Science 299, 1057–1061. [DOI] [PubMed] [Google Scholar]
- Josefowicz SZ, Lu L-F, and Rudensky AY (2012a). Regulatory T cells: mechanisms of differentiation and function. Annu. Rev. Immunol. 30, 531–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josefowicz SZ, Niec RE, Kim HY, Treuting P, Chinen T, Zheng Y, Umetsu DT, and Rudensky AY (2012b). Extrathymically generated regulatory T cells control mucosal TH2 inflammation. Nature 482, 395–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang SM, Tsang W, Doll S, Scherle P, Ko HS, Tran AC, Lenardo MJ, and Staudt LM (1992). Induction of the POU domain transcription factor Oct-2 during T-cell activation by cognate antigen. Mol Cell Biol 12, 3149–3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, and Haussler D (2002). The human genome browser at UCSC. Genome Res 12, 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Zweig AS, Barber G, Hinrichs AS, and Karolchik D (2010). BigWig and BigBed: enabling browsing of large distributed datasets. Bioinformatics 26, 2204–2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khattri R, Cox T, Yasayko S-A, and Ramsdell F (2003). An essential role for Scurfin in CD4+CD25+ T regulatory cells. Nat. Immunol. 4, 337–342. [DOI] [PubMed] [Google Scholar]
- Kim JM, Rasmussen JP, and Rudensky AY (2007). Regulatory T cells prevent catastrophic autoimmunity throughout the lifespan of mice. Nat. Immunol. 8, 191–197. [DOI] [PubMed] [Google Scholar]
- Kitagawa Y, Ohkura N, Kidani Y, Vandenbon A, Hirota K, Kawakami R, Yasuda K, Motooka D, Nakamura S, Kondo M, et al. (2017). Guidance of regulatory T cell development by Satb1-dependent super-enhancer establishment. Nat. Immunol. 18, 173–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurachi M, Ngiow SF, Kurachi J, Chen Z, and Wherry EJ (2019). Hidden Caveat of Inducible Cre Recombinase. Immunity 51, 591–592. [DOI] [PubMed] [Google Scholar]
- Lawrence M, Huber W, Pagès H, Aboyoun P, Carlson M, Gentleman R, Morgan MT, and Carey VJ (2013). Software for computing and annotating genomic ranges. PLoS Comput. Biol. 9, e1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine AG, Hemmers S, Baptista AP, Schizas M, Faire MB, Moltedo B, Konopacki C, Schmidt-Supprian M, Germain RN, Treuting PM, et al. (2017). Suppression of lethal autoimmunity by regulatory T cells with a single TCR specificity. J. Exp. Med. 214, 609–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, and 1000 Genome Project Data Processing Subgroup (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Liang Y, LeBlanc M, Benner C, and Zheng Y (2014). Function of a Foxp3 cis-element in protecting regulatory T cell identity. Cell 158, 734–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W, Haribhai D, Relland LM, Truong N, Carlson MR, Williams CB, and Chatila TA (2007). Regulatory T cell development in the absence of functional Foxp3. Nat. Immunol. 8, 359–368. [DOI] [PubMed] [Google Scholar]
- Lio C-WJ, and Hsieh C-S (2008). A two-step process for thymic regulatory T cell development. Immunity 28, 100–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Gerner MY, Van Panhuys N, Levine AG, Rudensky AY, and Germain RN (2015). Immune homeostasis enforced by co-localized effector and regulatory T cells. Nature 528, 225–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malchow S, Leventhal DS, Nishi S, Fischer BI, Shen L, Paner GP, Amit AS, Kang C, Geddes JE, Allison JP, et al. (2013). Aire-dependent thymic development of tumorassociated regulatory T cells. Science 339, 1219–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malek TR, Yu A, Vincek V, Scibelli P, and Kong L (2002). CD4 regulatory T cells prevent lethal autoimmunity in IL-2Rbeta-deficient mice. Implications for the nonredundant function of IL-2. Immunity 17, 167–178. [DOI] [PubMed] [Google Scholar]
- Mombaerts P, Clarke AR, Rudnicki MA, Iacomini J, Itohara S, Lafaille JJ, Wang L, Ichikawa Y, Jaenisch R, and Hooper ML (1992). Mutations in T-cell antigen receptor genes alpha and beta block thymocyte development at different stages. Nature 360, 225–231. [DOI] [PubMed] [Google Scholar]
- Moore MJ, Blachere NE, Fak JJ, Park CY, Sawicka K, Parveen S, Zucker-Scharff I, Moltedo B, Rudensky AY, and Darnell RB (2018). ZFP36 RNA-binding proteins restrain T cell activation and anti-viral immunity. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouse Genome Sequencing Consortium, Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, et al. (2002). Initial sequencing and comparative analysis of the mouse genome. Nature 420, 520–562. [DOI] [PubMed] [Google Scholar]
- Nazarov VI, Tsvetkov VO, Rumynskiy E, Lorenc A, Moore DJ, Greiff V, and ImmunoMind (2020). immunarch: Bioinformatics Analysis of T-Cell and B-Cell Immune Repertoires. [Google Scholar]
- Nikolouli E, Hardtke-Wolenski M, Hapke M, Beckstette M, Geffers R, Floess S, Jaeckel E, and Huehn J (2017). Alloantigen-Induced Regulatory T Cells Generated in Presence of Vitamin C Display Enhanced Stability of Foxp3 Expression and Promote Skin Allograft Acceptance. Front Immunol 8, 748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen DL, Mahmud SA, Vang KB, Kelly RM, Blazar BR, Smith KA, and Farrar MA (2018). Identification of Cellular Sources of IL-2 Needed for Regulatory T Cell Development and Homeostasis. J Immunol 200, 3926–3933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen DL, Mahmud SA, Sjaastad LE, Williams JB, Spanier JA, Simeonov DR, Ruscher R, Huang W, Proekt I, Miller CN, et al. (2019). Thymic regulatory T cells arise via two distinct developmental programs. Nat. Immunol. 20, 195–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacholczyk R, Kraj P, and Ignatowicz L (2002). Peptide specificity of thymic selection of CD4+CD25+ T cells. J Immunol 168, 613–620. [DOI] [PubMed] [Google Scholar]
- Panduro M, Benoist C, and Mathis D (2016). Tissue Tregs. Annu. Rev. Immunol. 34, 609–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry JSA, Lio C-WJ, Kau AL, Nutsch K, Yang Z, Gordon JI, Murphy KM, and Hsieh C-S (2014). Distinct contributions of Aire and antigen-presenting-cell subsets to the generation of self-tolerance in the thymus. Immunity 41, 414–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Placek K, Hu G, Cui K, Zhang D, Ding Y, Lee J-E, Jang Y, Wang C, Konkel JE, Song J, et al. (2017). MLL4 prepares the enhancer landscape for Foxp3 induction via chromatin looping. Nat. Immunol. 18, 1035–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard KS, Hubisz MJ, Rosenbloom KR, and Siepel A (2010). Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res 20, 110–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popmihajlov Z, Xu D, Morgan H, Milligan Z, and Smith KA (2012). Conditional IL-2 Gene Deletion: Consequences for T Cell Proliferation. Front Immunol 3, 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proekt I, Miller CN, Lionakis MS, and Anderson MS (2017). Insights into immune tolerance from AIRE deficiency. Curr. Opin. Immunol. 49, 71–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, and Hall IM (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team (2020). R: A language and environment for statistical computing (Vienna, Austria: R Foundation for Statistical Computing; ). [Google Scholar]
- Romagnoli P, Hudrisier D, and van Meerwijk JPM (2002). Preferential recognition of self antigens despite normal thymic deletion of CD4(+)CD25(+) regulatory T cells. J Immunol 168, 1644–1648. [DOI] [PubMed] [Google Scholar]
- Rubtsov YP, Niec RE, Josefowicz S, Li L, Darce J, Mathis D, Benoist C, and Rudensky AY (2010). Stability of the regulatory T cell lineage in vivo. Science 329, 1667–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzankina Y, Pinzon-Guzman C, Asare A, Ong T, Pontano L, Cotsarelis G, Zediak VP, Velez M, Bhandoola A, and Brown EJ (2007). Deletion of the developmentally essential gene ATR in adult mice leads to age-related phenotypes and stem cell loss. Cell Stem Cell 1, 113–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi S, Sakaguchi N, Asano M, Itoh M, and Toda M (1995). Immunologic self-tolerance maintained by activated T cells expressing IL-2 receptor alpha-chains (CD25). Breakdown of a single mechanism of self-tolerance causes various autoimmune diseases. J. Immunol. 155, 1151–1164. [PubMed] [Google Scholar]
- Sakaguchi S, Mikami N, Wing JB, Tanaka A, Ichiyama K, and Ohkura N (2020). Regulatory T Cells and Human Disease. Annu. Rev. Immunol. 38, 541–566. [DOI] [PubMed] [Google Scholar]
- Sasidharan Nair V, Song MH, and Oh KI (2016). Vitamin C Facilitates Demethylation of the Foxp3 Enhancer in a Tet-Dependent Manner. J Immunol 196, 2119–2131. [DOI] [PubMed] [Google Scholar]
- Scherer MT, Ignatowicz L, Winslow GM, Kappler JW, and Marrack P (1993). Superantigens: bacterial and viral proteins that manipulate the immune system. Annu Rev Cell Biol 9, 101–128. [DOI] [PubMed] [Google Scholar]
- Schuster M, Plaza-Sirvent C, Matthies A-M, Heise U, Jeron A, Bruder D, Visekruna A, Huehn J, and Schmitz I (2017). c-REL and IκBNS Govern Common and Independent Steps of Regulatory T Cell Development from Novel CD122-Expressing Pre-Precursors. J Immunol 199, 920–930. [DOI] [PubMed] [Google Scholar]
- Sharan SK, Thomason LC, Kuznetsov SG, and Court DL (2009). Recombineering: a homologous recombination-based method of genetic engineering. Nat Protoc 4, 206–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen FW, Saga Y, Litman G, Freeman G, Tung JS, Cantor H, and Boyse EA (1985). Cloning of Ly-5 cDNA. Proc Natl Acad Sci U S A 82, 7360–7363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shugay M, Britanova OV, Merzlyak EM, Turchaninova MA, Mamedov IZ, Tuganbaev TR, Bolotin DA, Staroverov DB, Putintseva EV, Plevova K, et al. (2014). Towards error-free profiling of immune repertoires. Nat Methods 11, 653–655. [DOI] [PubMed] [Google Scholar]
- Shugay M, Bagaev DV, Turchaninova MA, Bolotin DA, Britanova OV, Putintseva EV, Pogorelyy MV, Nazarov VI, Zvyagin IV, Kirgizova VI, et al. (2015). VDJtools: Unifying Post-analysis of T Cell Receptor Repertoires. PLoS Comput Biol 11, e1004503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S, et al. (2005). Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res 15, 1034–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toomer KH, Lui JB, Altman NH, Ban Y, Chen X, and Malek TR (2019). Essential and non-overlapping IL-2Rα-dependent processes for thymic development and peripheral homeostasis of regulatory T cells. Nat Commun 10, 1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weirauch MT, Yang A, Albu M, Cote AG, Montenegro-Montero A, Drewe P, Najafabadi HS, Lambert SA, Mann I, Cook K, et al. (2014). Determination and inference of eukaryotic transcription factor sequence specificity. Cell 158, 1431–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H (2009). ggplot2: Elegant Graphics for Data Analysis (New York: Springer-Verlag; ). [Google Scholar]
- Williams LM, and Rudensky AY (2007). Maintenance of the Foxp3-dependent developmental program in mature regulatory T cells requires continued expression of Foxp3. Nat. Immunol. 8, 277–284. [DOI] [PubMed] [Google Scholar]
- Xi Y, and Li W (2009). BSMAP: whole genome bisulfite sequence MAPping program. BMC Bioinformatics 10, 232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Fujikado N, Kolodin D, Benoist C, and Mathis D (2015). Immune tolerance. Regulatory T cells generated early in life play a distinct role in maintaining self-tolerance. Science 348, 589–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Z, Kanno Y, Kerenyi M, Stephens G, Durant L, Watford WT, Laurence A, Robinson GW, Shevach EM, Moriggl R, et al. (2007). Nonredundant roles for Stat5a/b in directly regulating Foxp3. Blood 109, 4368–4375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin R, Mao S-Q, Zhao B, Chong Z, Yang Y, Zhao C, Zhang D, Huang H, Gao J, Li Z, et al. (2013). Ascorbic acid enhances Tet-mediated 5-methylcytosine oxidation and promotes DNA demethylation in mammals. J. Am. Chem. Soc. 135, 10396–10403. [DOI] [PubMed] [Google Scholar]
- Yue X, Trifari S, Äijö T, Tsagaratou A, Pastor WA, Zepeda-Martínez JA, Lio C-WJ, Li X, Huang Y, Vijayanand P, et al. (2016). Control of Foxp3 stability through modulation of TET activity. J Exp Med 213, 377–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemmour D, Pratama A, Loughhead SM, Mathis D, and Benoist C (2017). Flicr, a long noncoding RNA, modulates Foxp3 expression and autoimmunity. Proc. Natl. Acad. Sci. U.S.A. 114, E3472–E3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Josefowicz S, Chaudhry A, Peng XP, Forbush K, and Rudensky AY (2010). Role of conserved non-coding DNA elements in the Foxp3 gene in regulatory T-cell fate. Nature 463, 808–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorn E, Nelson EA, Mohseni M, Porcheray F, Kim H, Litsa D, Bellucci R, Raderschall E, Canning C, Soiffer RJ, et al. (2006). IL-2 regulates FOXP3 expression in human CD4+CD25+ regulatory T cells through a STAT-dependent mechanism and induces the expansion of these cells in vivo. Blood 108, 1571–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List and sequences of primers used for qPCR and library construction. Related to STAR Methods.
List of antibodies and reagents used for flow cytometry. Related to STAR Methods.
Pathological evaluation of multiple tissues from 10- to 11- month old CNS0FL and CNS0Δ male littermates. Related to Figure 4.
Peaks included in ATAC-seq hierarchical clustering, along with coordinates, log2 FC and adjusted p-values. Related to Figures 6, S6, and S7.
Data Availability Statement
The ATAC-seq, RNA-seq, ChIP-seq, and TCR-seq data sets generated during this study are available at GEO under accession GSE164118. Other data and code are available from the corresponding authors upon request.