Abstract
The use of peptide amphiphiles (PAs) is becoming increasingly popular, not only because of their unique self-assembly properties but also due to the versatility of designs, allowing biological responsiveness, biocompatibility, and easy synthesis, which could potentially contribute to new drug design and disease treatment concepts. Oligonucleotides, another major functional bio-macromolecule class, have been introduced recently as new functional building blocks into PAs, further enriching the tools available for the fabrication of bio-functional PAs. Taking advantage of this, in the present work, two nucleic base-linked (adenine, A and thymine, T) RGD-rich peptide amphiphiles (NPAs) containing the fluorophores naphthalimide and rhodamine (Nph-A and Rh-T) were designed and synthesized. The two NPAs exhibit distinctive assembly behaviours with spherical (Rh-T) and fibrous (Nph-A) morphologies, and mixing Nph-A with Rh-T leads to a densely crosslinked colloidal network (Nph-A/Rh-T) via mutually promoted supramolecular polymerization via nucleation-growth assembly. Because of the RGD-rich sequences in the crosslinked network, further research on in situ targeted cancer cell (MDA-MB-231) encapsulation via RGD–integrin recognition was performed, and the modulation of cell behaviours (e.g., cell viability and migration) was demonstrated using both confocal laser scanning microscopy (CLSM) imaging and a scratch wound healing assay.
A cross-linking of peptide–nucleic base amphiphiles leads to a dense colloidal network that can perform targeted cancer cell encapsulation in situ.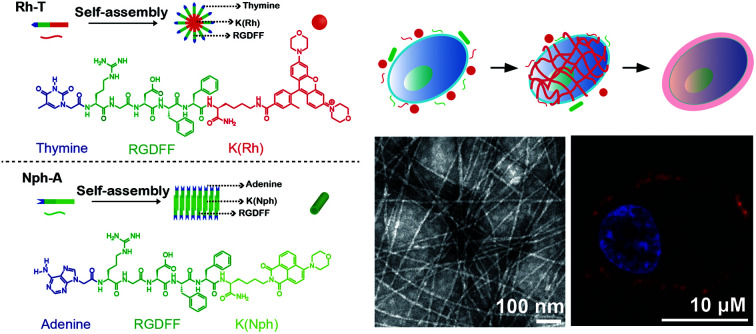
Introduction
Peptide amphiphiles (PAs) are remarkably versatile and useful as building blocks for the construction of complex supramolecular structures using a bottom-up model.1 To date, diversified supramolecular assembly strategies have been reported2 and proven to be promising and powerful methods for biomedical materials with applications as antimicrobial agents,3 in cancer therapy,4 and in regenerative medicine.5 Among the available assembly methods, the nucleation-growth mechanism is considered as one of the most promising strategies, because it allows the control of the self-assembly shape and the function of the peptides. In 2012, two PAs were designed to explore the growth mechanism of peptide amphiphiles in the presence of early formed spherical micelles.6 More recently, Cui et al. took advantage of a small hydrophobic molecule, paclitaxel (PTX), as an effective promoter to induce nanofibre formation and elongation of a peptide–PTX conjugate.7 Based on this mechanism, many attempts have been made to design stimuli-responsive peptide sequences and expand the scope of PA self-assembly in bio-applications. For example, peptides can be modified with enzyme-activated substrates that can perform spontaneous intracellular self-assembly upon interaction with specific enzymes, thus leading to cyto- or organelle-dysfunction.8,9 This in situ nanodrug preparation strategy is now widely used as an efficient, targeted theranostic approach for cancer cells,10,11 and can be applied to other tumour-related stimuli like micro-environments (e.g., hypoxia, pH)12,13 and small molecule metabolites (e.g., ATP).14 Moreover, the enzyme-activated intracellular colloidal formation can also act as a “nanoreactor” to facilitate in situ drug release and overcome the obstacle of inefficient dosing due to the relatively low concentration of drug uptake.15
Recently, nucleic bases have been introduced as new functional building blocks into the PAs,16 due to their diverse stimuli-responsiveness (e.g., high affinity with oligonucleotides,17 multiple hydrogen bonding in duplexes/triplexes,18 metal ion coordination,19 pH/photo-responsiveness20) as well as good biocompatibility and biodegradability.21 The design and fabrication of nucleic based tethered PAs are thus enriched. Inspired by this, herein two nucleic base-linked (adenine, A and thymine, T) peptide amphiphiles (NPAs) containing the integrin-targeted RGD sequences were designed and synthesized (Fig. 1a). The two NPAs self-assemble into either spherical or fibrous nanostructures, whereas the mixture of these two NPAs (1 : 1) leads to a distinctive, mutually promoted supramolecular polymerization by a nucleation-growth assembly, resulting in the formation of a well-structured, densely crosslinked colloidal network. Taking advantages of the crosslinking peptide network and the integrin-targeted feature of the RGD sequences, an in situ “bottom-up” encapsulation of the targeted cancer cells (e.g., MDA-MB-231, a triple negative breast cancer cell line that overexpresses integrin receptors) upon easy mixing of NPAs is achieved in this work (Fig. 1b). Different from the previously mentioned intracellular self-assembly approach, the NPA “capsule” directly interacts with receptors on the targeted MDA-MB-231 cytomembrane and may disturb the cellular mass and signal exchanges (e.g., nutrient, metabolite or waste) between the cell and the surrounding micro-environment, thus imposing a negative effect on the cell stability and activity.22 As a result, the viability and migration capability of the MDA-MB-231 cell line used are significantly affected by the crosslinked NPAs co-assembled on the cytomembrane, and thus, providing a new strategy for targeted cancer therapy.
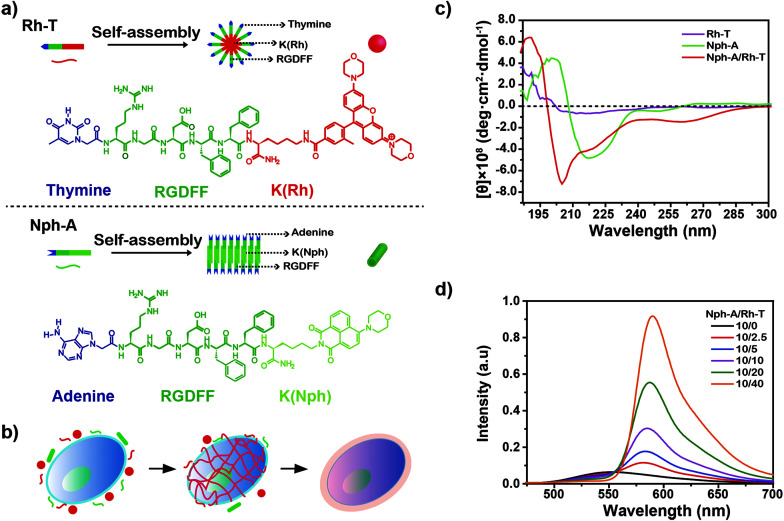
Fig. 1. (a) The molecular structures of peptides Rh-T and Nph-A and an illustration of the assembly of the peptides. (b) An illustration of cell encapsulation using the crosslinked Nph-A/Rh-T colloidal network on the cytomembrane. (c) Circular dichroism (CD) spectra of Nph-A (10.0 μM), Rh-T (10.0 μM), and Nph-A/Rh-T (10 μM, 1 : 1). (d) Fluorescence spectra of Nph-A/Rh-T formed with different ratios of Nph-A and Rh-T in TBS (50 mM Tris, pH 7.4, 25 °C).
Results and discussion
Molecular design
The two NPAs, adenine–RGDFFK–Nph (Nph-A) and thymine–RGDFFK–Rh (Rh-T), were designed and contained three regions. Region 1 is the nucleic base (A or T), which can enhance the intermolecular interactions with the help of hydrogen bonding. Region 2 is a short peptide (RGDFF), of which the RGD sequence is expected to increase the uptake of the peptides by the cancer cells via a mutual recognition mechanism with the integrin receptor. The diphenylalanine peptide (l-Phe-l-Phe, FF) is generally considered as the simplest peptide building block for self-assembly, and which has been demonstrated to self-assemble into well-ordered tubular structures with a long persistence length (∼100 μm) by a combination of hydrogen bonding and π–π stacking of the aromatic residues.23 Region 3 is the fluorophore of naphthalimide or rhodamine (Nph/Rh) with the lysine (K) link. The different geometry and π–π stacking interactions of the fluorophores together with the hydrogen-bonding of the peptide sequence FF determine the self-assembly types of the peptide amphiphiles Nph-A and Rh-T (Fig. 1a and S1–S3, ESI†).24 According to the reported general procedure, both peptides, Nph-A and Rh-T, were synthesized using a manual Fmoc solid-phase peptide synthesis (SPPS) method, and were isolated by preparative HPLC on a reversed-phase C18 column with a 98.3%, and 98.5% purity and a 19.8%, and 20.6% in yield, respectively. The structures of both compounds were confirmed by analytical HPLC and mass spectrometry (Fig. S4–S6, ESI†).
Self-assembly of Nph-A and Rh-T
To investigate the self-assembly behaviour of the designed NPAs, Nph-A and Rh-T were all pre-treated in hexafluoroisopropanol (HFIP) to eliminate any pre-existing aggregates.25 All HFIP treated samples were vacuum dried using a rotary evaporator, followed by addition of a Tris buffered saline buffer solution (TBS) to promote the formation of discrete nanostructures. After 2 h of incubation, all the assembled nanostructures were imaged by transmission electron microscopy (TEM) to characterize their morphologies and dimensions. As shown in Fig. S7a (ESI†), Nph-A revealed a fibrous structure with uniform diameters of 10–15 nm (Fig. S8a, ESI†) and lengths reaching the micrometer-scale. In contrast, peptide Rh-T showed a spherical shaped aggregation with diameters of 20–40 nm (Fig. S7b and S8b, ESI†). The self-assembly behaviours of the peptides were further examined by dynamic light scattering (DLS, Fig. S9a, ESI†). The Nph-A and Rh-T formed different sized aggregations with diameters of 426 nm and 58 nm in a monomodal size distribution in TBS (50 mM Tris, pH 7.4, 25 °C), respectively. The differences in diameters and assembly structures might be due to the different hydrophobic groups/fluorophores (Region 3) linked to the peptide sequences. According to a molecular simulation study by Velichko26 and Manandhar,27 the assembly of a classic β-sheet-containing peptide amphiphile in aqueous solution can undergo two distinct pathways for the formation of supramolecular polymers, depending on the strength of hydrogen bonding relative to the interactions between the hydrophobic units. In the regime where hydrophobic interactions dominate, micellar aggregates are first formed, followed by the formation of hydrogen bonding in the periphery that eventually results in supramolecular polymeric growth. Thus, the extended π system and rigid planar structures of the 1,8-naphthalimide derivatives were beneficial to forming a co-facial structure, which leads to the nanostructure of the nanofibers. However, rhodamine's nonplanar configuration resulted in a different assembly morphology of the nanosphere.28
The secondary structures of the Nph-A and Rh-T assemblies were further characterized using circular dichroism (CD) and Fourier transform infrared (FTIR) spectroscopies. The CD spectrum of Nph-A showed a maximum peak at 200 nm and a minimum peak at 217 nm in TBS, which was characteristic of the formation of a twisted β-sheet structure (Fig. 1c).29,30 Whereas the spherical Rh-T assembly did not display characteristic signals on the CD spectra, indicating its isotropic feature. The FTIR spectrum of Nph-A showed a strong band at 1637 cm−1 and a weak signal at 1550–1600 cm−1, which are typical signals of an antiparallel β-sheet.31 In contrast, Rh-T showed a broad peak at 1647 cm−1, which is due to amide I absorption and the sharp peak at 1200–1250 cm−1 indicated a randomly coiled structure (Fig. S9b, ESI†).32
Crosslinking of Nph-A and Rh-T
Although individual Nph-A self-assembles into nanofibers and Rh-T self-assembles into spherical form, interestingly, the equivalent mixture of Nph-A and Rh-T (10 μM for each) unexpectedly led to a formation of a crosslinked fibril network with a uniform diameter of 8 nm and lengths reaching the micrometer-scale (Fig. S7c and S8c, ESI†). The DLS results showed a higher degree of NPA aggregation after the mixing of Nph-A and Rh-T. A double-modal size distribution appeared, which was consistent with the TEM results, indicating a possible crosslinking between Nph-A and Rh-T (Fig. S9a, ESI†). From the CD spectra, it can be assumed that the mixture assembled into highly oriented chiral structures via a stronger β-sheet formation that led to both an increased positive peak at 190 nm and a negative peak at 205 nm. The negative peak at 218 nm could be attributed to the π–π stacking of the aromatic side chains FF and to the distortion of the β-sheets, which was also much stronger than the Nph-A assembly alone (Fig. 1c).33 A weak peak at around 260–280 nm revealed the existence of the helical stacking of adenine and thymine. Fluorescence emission spectroscopy was also performed to check the Förster resonance energy transfer (FRET) between the naphthalimide in Nph-A and the rhodamine in Rh-T. The Nph-A and Rh-T showed remarkable fluorescence at 527 nm and 589 nm in TBS, respectively. The fluorescence intensity of the Nph-A/Rh-T mixture (from 4 : 1 to 1 : 4) showed a decrease at 527 nm and a subsequent increase at 589 nm (Fig. 1d), which demonstrated the efficient FRET between the naphthalimide in Nph-A and the rhodamine in Rh-T. This result provides more evidence for the crosslinking of Nph-A and Rh-T because the efficient FRET can be attributed to the shortened distance between the two fluorophores. The freeze-dried Nph-A/Rh-T were also characterized by scanning electron microscopy (SEM) and porous microstructures were observed, a typical hydrogel-like structure previously reported for other supramolecular hydrogels (Fig. S10, ESI†),34,35 demonstrating again the crosslinking feature of Nph-A/Rh-T.
It was speculated that the peptide self-assembly as well as hydrogen bonding from the nucleic bases might induce the crosslinking of Nph-A/Rh-T. The two aromatic peptides were blended to form a hydrophobic area, together with phenylalanine. At the same time, adenine and thymine formed an additional hydrogen bonding based on the complementary pairing, further strengthening the crosslinking between the peptide monomers. As previously reported, Shimada et al. observed the formation of spherical micelles as a precursor morphology prior to their elongation into long filamentous structures.36 In this research, it is hypothesized that the crosslinking was promoted by enhanced hydrogen binding between two NPAs. The assembled Rh-T spherical micelles as the precursor morphology promoted the extension of the assembled Nph-A, which lead to the formation of the fiber network.
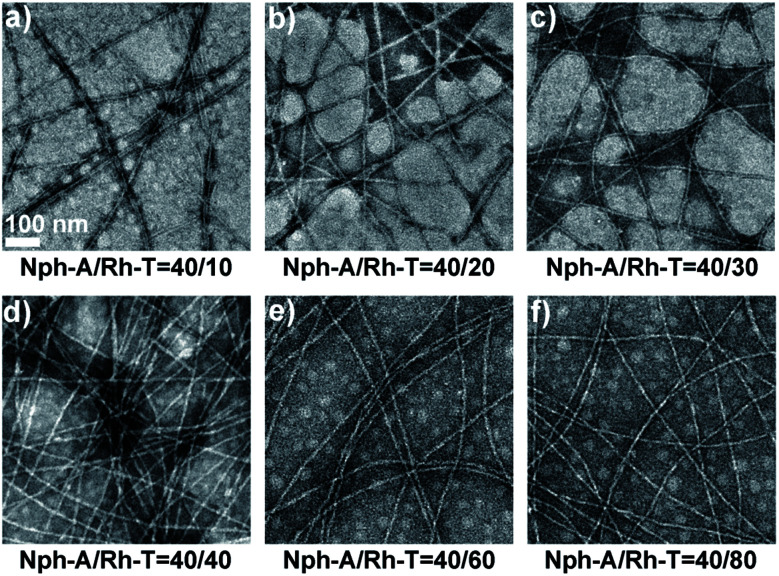
In order to investigate the role of nucleic bases in supramolecular self-assembly, a control peptide was further synthesized with thymine replacing adenine as Nph-T (Fig. S3, ESI†). The TEM image shows that Nph-T itself forms nanofibers with diameters of 14–16 nm and lengths in the micron range, similar to that of Nph-A (Fig. S7d, ESI†). Yet, no crosslinked network was observed when mixing Nph-T with Rh-T (molar ratio of 1 : 1, 10 μM, Fig. S7e, ESI†). To determine the optimized assembly ratio, a series of Nph-A/Rh-T mixtures with ratios of 4 : 1 to 1 : 2 were prepared (aged for 2 h) for examining the crosslinking performances of these two NPAs. At all ratios, the TEM images exhibited a long fiber network with diameters of ca. 6–8 nm. As the amount of Rh-T increased, the density of the fiber network increased and a densely crosslinked fiber network was observed when the ratio was increased to 1 : 1 (Fig. 2a–d). The morphology and network density did not vary obviously when the amount of Rh-T was further increased (Fig. 2e and f). Therefore, an equal ratio of the Nph-A/Rh-T mixture was selected for further applications in in situ cell encapsulation.
Fig. 2. Transmission electron microscopy (TEM) images of the assemblies of mixtures with different molar ratios of Nph-A/Rh-T: (a) 4 : 1, (b) 2 : 1, (c) 4 : 3, (d) 1 : 1, (e) 2 : 3, and (f) 1 : 2 (Nph-A = 10 μM). Scale bar = 100 nm.
Cellular uptake and encapsulation
As an important member of the integrin family, the adhesion molecule αvβ3 has been widely investigated for use in tumour imaging and therapy because of its pivotal roles in tumour proliferation and metastasis. In particular, due to its upregulated expression in proliferating tumour cells compared to normal cells with a low αvβ3 expression, integrin αvβ3 has been identified as an ideal therapeutic target for tumour therapy. The peptide ligands containing the RGD motif, which shows a strong binding affinity and selectivity to integrin αvβ3, have been employed as tumour-homing ligands for integrin targeted therapeutic applications.
In this work, it was assumed that the densely crosslinked network of Nph-A/Rh-T together with the RGD-rich feature might form a peptide “cocoon” that encapsulated the αvβ3-overexpressed tumour cells. To verify the hypothesis, the in situ cell encapsulation performances of the Nph-A/Rh-T network were investigated by confocal laser scanning microscopy (CLSM) after incubation with two cell lines of MDA-MB-231 (human breast cancer cell that overexpress the integrin αvβ3 (ref. 37)) and HeLa (human cervical carcinoma that has limited integrin αvβ3 expression38). After incubation with RGD-rich Rh-T for 2.5 h, both αvβ3-positive MDA-MB-231 cells and αvβ3-negative HeLa cell lines exhibited a red fluorescence in cytoplasm, which was probably due to the internationalization of the nanoparticle-sized Rh-T assembly through the lysosomal pathway (Fig. S11a–d and S12a–d, ESI†).39 Both cell lines treated with Nph-A showed a negligible green fluorescence, which might be attributed to the fibrous morphology of Nph-A that was unfavorable for cell uptake (Fig. S11e–h and S12e–h, ESI†).
Interestingly, the MDA-MB-231 cells incubated with Nph-A/Rh-T exhibited an obviously enhanced red fluorescence of rhodamine along the outline of the cells rather than in the cytoplasm (Fig. S11i–l, ESI†), when compared with use of Rh-T alone. This might indicate the possible interactions between the overexpressed integrin αvβ3 on the cytomembrane of MDA-MB-231 cells and the RGD-rich Nph-A/Rh-T networks that lead to their encapsulation on the cell surface. Conversely, no obvious sign of cell encapsulation was detected in HeLa cells, as the red rhodamine fluorescence was mainly observed in the cytoplasm (Fig. S12i–l, ESI†), similar to the case shown in Fig. S12a–d (ESI†). The sharp difference of the encapsulation performance on HeLa cells might be attributed to the limited expression of the integrin αvβ3 receptor on the HeLa cell membrane.
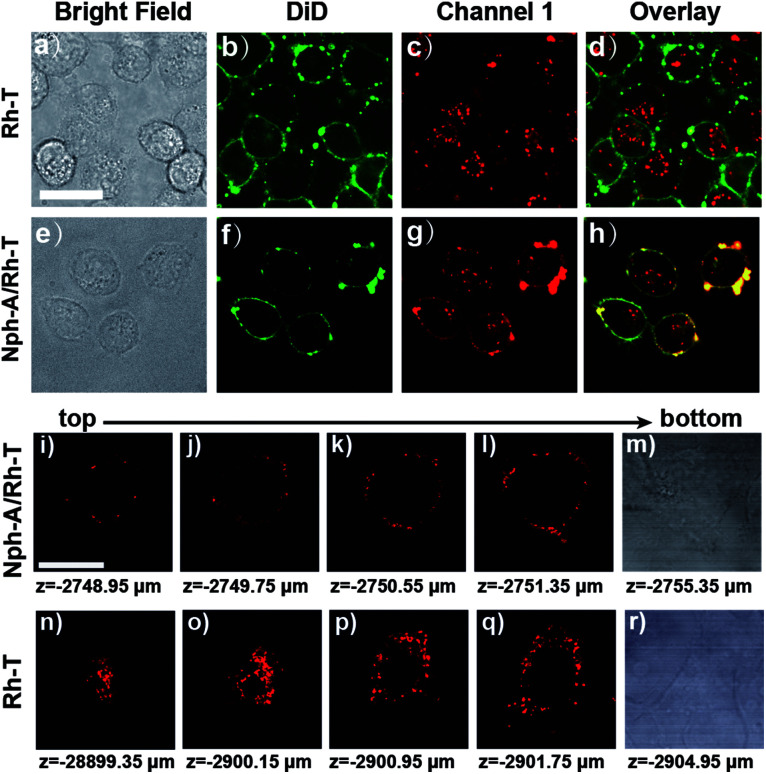
To further identify the Nph-A/Rh-T network with RGD sequence formed encapsulation on the cell surface of the MDA-MB-231 cells, the cytomembrane was stained with DiD (DiIC18(5), Biotium), a commercially available membrane-staining dye, and performed the colocalization imaging of the co-stained cell surface (Fig. 3 and S13, ESI†). As shown in Fig. 3e–h and S13e–h (ESI†), the cells treated with Nph-A/Rh-T exhibited red rhodamine fluorescence that overlapped well with the fluorescence of the cytomembrane stain DiD, indicating the formation of crosslinked peptide capsules on the cell surface. Conversely, colocalization of fluorescence between rhodamine and DiD was not observed when the cells were incubated with Rh-T alone (Fig. 3a–d and S13a–d, ESI†) or non-crosslinked Nph-T/Rh-T (Fig. S13i–l, ESI†), further proving the crucial role of the crosslinked Nph-A/Rh-T network on the efficient cell encapsulation.
Fig. 3. Confocal fluorescence images of (a–d) MDA-MB-231 cells incubated with Rh-T (10 μM) and stained with DiD (20 μM); and (e–h) MDA-MB-231 cells incubated with Nph-A/Rh-T (1 : 1, Nph-A = 10 μM and Rh-T = 10 μM) and stained with DiD (20 μM). 3D confocal images of channel 1 (red) of the MDA-MB-231 cells incubated with (i–m) Nph-A/Rh-T (1 : 1, Nph-A = 10 μM and Rh-T = 10 μM) and (n–r) Rh-T (= 20 μM). Scanned from bottom to top. Channel DiD: excitation: 640 nm, emission collected: 663–738 nm. Channel 1 for Rh-T: excitation: 561 nm, emission collected: 570–620 nm. Scale bar: 10 μm.
Next, the 3D confocal imaging of MDA-MB-231 cell with Nph-A/Rh-T was performed. The bright red circle of fluorescence, whose diameter gradually shrank when the cell was scanned from bottom to top, showed the spatial encapsulation of the MDA-MB-231 cell (Fig. 3i–m). As a control experiment, cells were cultured with Rh-T alone (Fig. 3n–r) and a bright mass of red fluorescence with an unclear outline was observed instead. These results demonstrated that the Nph-A/Rh-T network was able to specifically attach onto the cell surface and it prevented itself from being endocytosed into the cytoplasm.
Cell viability and migration assays
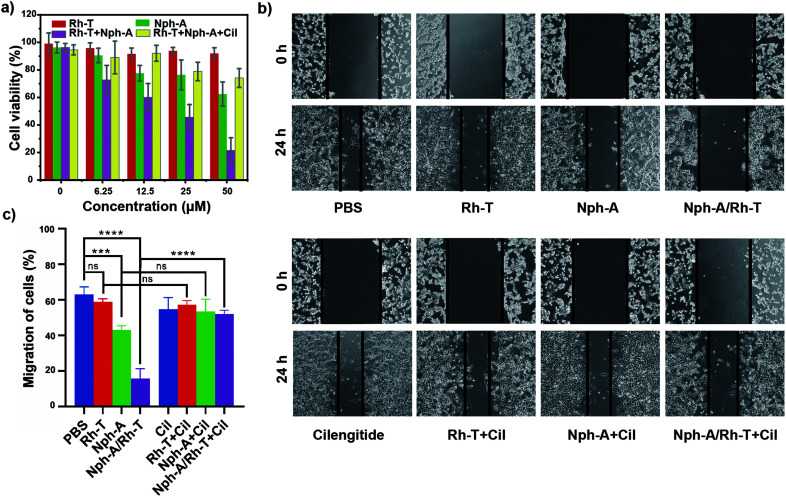
To examine whether the cell encapsulation with the Nph-A/Rh-T network could affect the cell behaviours of αvβ3-positive MDA-MB-231, e.g., viability and migration, the CCK-8 and scratch wound healing assays were investigated. Firstly, as revealed by the CCK-8 assay, the MDA-MB-231 cells cultured with Rh-T (0–50 μM) showed good viabilities of over 90% after 24 h incubation (Fig. 4a). The MDA-MB-231 cells cultured with Nph-A (0–50 μM) also showed good viability, although a little lower than that of Rh-T at high concentrations. This difference might be attributed to the potential cytotoxicity of the fibrous Nph-A assembly attached to the cytomembrane in high concentrations.22 These results provide evidence that neither Rh-T or Nph-A would affect the cell viability. The crosslinked Nph-A/Rh-T, however, exhibited a significant impact on the viability of the MDA-MB-231 cells viability (45.6%) when reaching a concentration of 12.5 μM. It was assumed that the encapsulation of MDA-MB-231 cells via RGD-αvβ3 recognition of the crosslinking networks composed of Nph-A/Rh-T could hamper materials (nutrients and metabolites) and signal transportation between the cells and the surrounding micro-environment, which leads to the decreased cell viability. To further confirm that the RGD-αvβ3 recognition is responsible for the observed cell inhibition, the MDA-MB-231 cells were pre-treated with the αvβ3 inhibitor (cilengitide, Cil) before incubation with Nph-A/Rh-T. In sharp contrast, the cells successively treated with Cil and Nph-A/Rh-T showed limited cell inhibition even at a high Nph-A/Rh-T concentrations, indicating that the interaction between the αvβ3 and RGD-containing peptides facilitated the cell encapsulation by αvβ3 recognition (Fig. 4a). Moreover, control experiments were also performed on the HeLa cell line, which had limited αvβ3 expression. The CCK-8 assay exhibited similar cell viability (80–90%) upon treatment with Rh-T, Nph-A and Nph-A/Rh-T, in different concentrations (Fig. S14a, ESI†), indicating the negligible inhibition effects of the Nph-A/Rh-T crosslinked network on the HeLa cells, on account of inefficient encapsulation.
Fig. 4. (a) Results of the CCK-8 assay of MDA-MB-231 cells incubated with Rh-T, Nph-A, and Nph-A/Rh-T (1 : 1) at various concentrations (0 μM, 6.25 μM, 12.5 μM, 25 μM, and 50 μM). (b) Results of the scratch wound healing assays of MDA-MB-231 cells with Rh-T (12.5 μM), Nph-A (12.5 μM), and Nph-A/Rh-T (1 : 1, 12.5 μM) with and without cilengitide (Cil, 40 μM). (c) Quantification of the migration rates of cells (%). ns ≥ 0.05, ***p < 0.001, ****p < 0.0001.
The inhibitory action on cell migration was further evaluated using a scratch wound healing assay. As shown in Fig. 4b, the scratched cell “wound” was photographed at 0 and 24 h before and after various treatments. It was clearly seen that the control MDA-MB-231 cells had a nearly healed “wound” after 24 h incubation, whereas the wound closure was inhibited by different amounts after Rh-T, Nph-A and Nph-A/Rh-T treatments for 24 h, and Nph-A/Rh-T showed the highest inhibition. However, pre-treatment of the cells by an αvβ3 inhibitor (cilengitide, Cil) before the treatments showed similar wound closures after 24 h, suggesting that αvβ3 recognition played a crucial role in cell migration. To quantify the inhibition of cell migration, the wound area was determined and the average migration rate of the cells was calculated.40 As shown in Fig. 4c, the cell migration speed of the control group was 63.0%. After treatments for 24 h, the cell migration speed of the Rh-T, Nph-A and Nph-A/Rh-T groups decreased to 59.0%, 51.5%, and 15.7%, respectively, and the cells pre-treated with Cil showed no significant difference to the control cells. The same experiments were also carried out using HeLa cell lines (Fig. S14b and c, ESI†). As expected, no evident inhibitory effects were discovered when treating the HeLa cells with Rh-T (26.7%), Nph-A (26.6%) and Nph-A/Rh-T (27.6%) for 24 h, when compared to the control group (26.6%). These results demonstrated that the cell-encapsulation with a RGD-rich Nph-A/Rh-T network could inhibit the proliferation and migration of tumour cells via αvβ3 recognition.
Conclusions
In summary, two nucleic base-linked peptide amphiphiles (NPAs) that displayed different assembly performances were designed and synthesized. Peptide Nph-A with adenine as the N-terminal aggregates into a twisted ribbon structure due to β-sheet formation. Peptide Rh-T with thymine as the N-terminal induces isotropic molecular packing, resulting in spherical micelles. Interestingly, the crosslinking of Nph-A and Rh-T leads to the formation of a densely crosslinked colloidal network stabilized via hydrogen bonding between adenine and thymine. Taking advantage of the RGD-rich peptide crosslinked network, it was further applied to targeted cancer cell encapsulation via RGD–integrin recognition, and the manipulation of cell behaviours (e.g., cell migration) was demonstrated. It is expected that this design strategy will provide a general approach for controlling/modulating the biological behaviours and processes of targeting cells via self-assembly-facilitated cell surface engineering, which could one day lead to diverse applications in cancer theranostics and single cell studies.
Data availability
Further details of the experiments are available in the ESI.
Author contributions
Y. Zhou, D. Yao and J. Zhang designed the study. Y. Zhou, D. Yao, P. Qiu performed the experiments. Y. Zhou, D. Yao and J. Zhang analyzed the data. Y. Song, Y. Zhu and H. Pan made contribution to the discussions during the work. Y. Zhou, D. Yao and J. Zhang prepared the manuscript. J. Wu and J. Zhang concepted the project and supervised this research. All authors provided feedback on the paper.
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
The authors acknowledge financial support from the NSFC (21878086, 21788102) and the Shanghai Rising-Star Program (19QA1402500 to JZ). The project was supported by the Shanghai Municipal Science and Technology Major Project (Grant No. 2018SHZDZX03), the International Cooperation Program of Shanghai Science and Technology Committee (17520750100), and the Fundamental Research Funds for the Central Universities (222201717003).
Electronic supplementary information (ESI) available. See DOI: 10.1039/d1sc02995a
References
- Percec V. Ungar G. Peterca M. Science. 2006;313:55–56. doi: 10.1126/science.1129512. [DOI] [PubMed] [Google Scholar]
- He Z. F. Jiang W. Schalley C. A. Chem. Soc. Rev. 2015;44:779–789. doi: 10.1039/C4CS00305E. [DOI] [PubMed] [Google Scholar]
- Laverty G. McCloskey A. P. Gilmore B. F. Jones D. S. Zhou J. Xu B. Biomacromolecules. 2014;15:3429–3439. doi: 10.1021/bm500981y. [DOI] [PubMed] [Google Scholar]
- Kuang Y. Xu B. Angew. Chem., Int. Ed. 2013;52:6944–6948. doi: 10.1002/anie.201302658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pena B. Martinelli V. Jeong M. Bosi S. Lapasin R. Taylor M. R. G. Long C. S. Shandas R. Park D. Mestroni L. Biomacromolecules. 2016;17:1593–1601. doi: 10.1021/acs.biomac.5b01734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada T. Sakamoto N. Motokawa R. Koizumi S. Tirrell M. J. Phys. Chem. B. 2012;116:240–243. doi: 10.1021/jp209105z. [DOI] [PubMed] [Google Scholar]
- Su H. Zhang W. J. Wang H. Wang F. H. Cui H. G. J. Am. Chem. Soc. 2019;141:11997–12004. doi: 10.1021/jacs.9b04730. [DOI] [PubMed] [Google Scholar]
- Chandra Saha P. Das R. S. Chatterjee T. Bhattacharyya M. Guha S. Bioconjugate Chem. 2020;31:1301–1306. doi: 10.1021/acs.bioconjchem.0c00153. [DOI] [PubMed] [Google Scholar]
- Jeena M. T. Palanikumar L. Go E. M. Kim I. Kang M. G. Lee S. Park S. Choi H. Kim C. Jin S. M. Bae S. C. Rhee H. W. Lee E. Kwak S. K. Ryu J. H. Nat. Commun. 2017;8:26. doi: 10.1038/s41467-017-00047-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du W. Hu X. Wei W. Liang G. Bioconjugate Chem. 2018;29:826–837. doi: 10.1021/acs.bioconjchem.7b00798. [DOI] [PubMed] [Google Scholar]
- Feng Z. Han X. Wang H. Tang T. Xu B. Chem. 2019;5:2442–2449. doi: 10.1016/j.chempr.2019.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. Y. Shi K. J. Sabet Z. F. Fu W. J. Zhou H. Xu S. X. Liu T. You M. Cao M. J. Xu M. Z. Cui X. J. Hu B. Liu Y. Chen C. Y. Sci. Adv. 2019;5:eaax0937. doi: 10.1126/sciadv.aax0937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M. Ning Y. Chen J. Duan X. Song N. Ding D. Su X. Yu Z. Nano Lett. 2019;19:7965–7976. doi: 10.1021/acs.nanolett.9b03136. [DOI] [PubMed] [Google Scholar]
- Wang H. Feng Z. Qin Y. Wang J. Xu B. Angew. Chem., Int. Ed. 2018;57:4931–4935. doi: 10.1002/anie.201712834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An H. W. Li L. L. Wang Y. Wang Z. Hou D. Lin Y. X. Qiao S. L. Wang M. D. Yang C. Cong Y. Ma Y. Zhao X. X. Cai Q. Chen W. T. Lu C. Q. Xu W. Wang H. Zhao Y. Nat. Commun. 2019;10:4861. doi: 10.1038/s41467-019-12848-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel R. Sarma S. Shukla A. Parmar P. Goswami D. Saraf M. Mol. Biol. Rep. 2020;47:8113–8131. doi: 10.1007/s11033-020-05819-3. [DOI] [PubMed] [Google Scholar]
- Guler M. O. Pokorski J. K. Appella D. H. Stupp S. I. Bioconjugate Chem. 2005;16:501–503. doi: 10.1021/bc050053b. [DOI] [PubMed] [Google Scholar]
- Brinke G. T., Bouteiller L., Ikkala O. and Binder W., Hydrogen Bonded Polymers, Springer-Verlag, Berlin, 2007, vol. 207, pp. 113–177 [Google Scholar]
- Lin Y. Y. Pashuck E. T. Thomas M. R. Amdursky N. Wang S. T. Chow L. W. Stevens M. M. Angew. Chem., Int. Ed. 2017;56:2361–2365. doi: 10.1002/anie.201610976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger O. Adler-Abramovich L. Levy-Sakin M. Grunwald A. Liebes-Peer Y. Bachar M. Buzhansky L. Mossou E. Forsyth V. T. Schwartz T. Ebenstein Y. Frolow F. Shimon L. J. Patolsky F. Gazit E. Nat. Nanotechnol. 2015;10:353–360. doi: 10.1038/nnano.2015.27. [DOI] [PubMed] [Google Scholar]
- Yuan D. Du X. W. Shi J. F. Zhou N. Zhou J. Xu B. Angew. Chem., Int. Ed. 2015;54:5705–5708. doi: 10.1002/anie.201412448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang Y. Shi J. F. Li J. Yuan D. Alberti K. A. Xu Q. B. Xu B. Angew. Chem., Int. Ed. 2014;53:8104–8107. doi: 10.1002/anie.201402216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X. H. Zhu P. L. Li J. B. Chem. Soc. Rev. 2010;39:1877–1890. doi: 10.1039/B915765B. [DOI] [PubMed] [Google Scholar]
- Yeh M.-Y. Huang C.-T. Lai T.-S. Che F.-Y. Chu N.-T. Tseng D. T.-H. Hung S.-C. Lin H.-C. Langmuir. 2016;32:7630–7638. doi: 10.1021/acs.langmuir.6b01809. [DOI] [PubMed] [Google Scholar]
- Kumar M. Kumar N. Bhalla V. Singh H. Sharma P. R. Kaur T. Org. Lett. 2011;13:1422–1425. doi: 10.1021/ol2001073. [DOI] [PubMed] [Google Scholar]
- Velichko Y. S. Stupp S. I. de la Cruz M. O. J. Phys. Chem. B. 2008;112:2326–2334. doi: 10.1021/jp074420n. [DOI] [PubMed] [Google Scholar]
- Manandhar A. Kang M. Chakraborty K. Tang P. K. Loverde S. M. Org. Biomol. Chem. 2017;15:7993–8005. doi: 10.1039/C7OB01290J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbeloa F. L. Ojeda P. R. Arbeloa I. L. J. Lumin. 1989;44:105–112. doi: 10.1016/0022-2313(89)90027-6. [DOI] [Google Scholar]
- Lara C. Reynolds N. P. Berryman J. T. Xu A. Q. Zhang A. F. Mezzenga R. J. Am. Chem. Soc. 2014;136:4732–4739. doi: 10.1021/ja500445z. [DOI] [PubMed] [Google Scholar]
- Ranjbar B. Gill P. Chem. Biol. Drug Des. 2009;74:101–120. doi: 10.1111/j.1747-0285.2009.00847.x. [DOI] [PubMed] [Google Scholar]
- Bandekar J. Krimm S. Biopolymers. 1988;27:909–921. doi: 10.1002/bip.360270603. [DOI] [Google Scholar]
- Lazarev Y. A. Grishkovsky B. A. Khromova T. B. Biopolymers. 1985;24:1449–1478. doi: 10.1002/bip.360240804. [DOI] [PubMed] [Google Scholar]
- Bowerman C. J. Liyanage W. Federation A. J. Nilsson B. L. Biomacromolecules. 2011;12:2735–2745. doi: 10.1021/bm200510k. [DOI] [PubMed] [Google Scholar]
- Li Z. Y. Davidson-Rozenfeld G. Vázquez-González M. Fadeev M. Zhang J. J. Tian H. Willner I. J. Am. Chem. Soc. 2018;140:17691–17701. doi: 10.1021/jacs.8b10481. [DOI] [PubMed] [Google Scholar]
- Li Z. Y. Liu Y. Y. Li Y. J. Wang W. H. Song Y. Y. Zhang J. J. Tian H. Angew. Chem., Int. Ed. 2021;60:5157–5161. doi: 10.1002/anie.202013011. [DOI] [PubMed] [Google Scholar]
- Shimada T. Sakamoto N. Motokawa R. Koizumi S. Tirrell M. J. Phys. Chem. B. 2012;116:240–243. doi: 10.1021/jp209105z. [DOI] [PubMed] [Google Scholar]
- Liu Z. Chen W. Li Y. H. Xu Q. Anal. Chem. 2016;88:11955–11962. doi: 10.1021/acs.analchem.6b03927. [DOI] [PubMed] [Google Scholar]
- Bolzati C. Salvarese N. Carpanese D. Seraglia R. Melendez-Alafort L. Rosato A. Capasso D. Saviano M. Del Gatto A. Comegna D. Zaccaro L. J. Med. Chem. 2018;61:9596–9610. doi: 10.1021/acs.jmedchem.8b01075. [DOI] [PubMed] [Google Scholar]
- Hu Q. Q. Li H. Wang L. H. Gu H. Z. Fan C. H. Chem. Rev. 2019;119:6459–6506. doi: 10.1021/acs.chemrev.7b00663. [DOI] [PubMed] [Google Scholar]
- Grada A. Otero-Vinas M. Prieto-Castrillo F. Obagi Z. Falanga V. J. Invest. Dermatol. 2017;137:e11–e16. doi: 10.1016/j.jid.2016.11.020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Further details of the experiments are available in the ESI.