Introduction
The retinal pigment epithelium (RPE) is a monolayer of pigmented cells in the retina, which is a part of the blood/retina barrier. The RPE cells are responsible for absorption of energy from scattered light, and transportation of metal ions, water, and metabolic end products between the subretinal space and the bloodstream thereby facilitating delivery of nutrients to photoreceptors (1). Importantly, the retinal chromophore of rod and cone visual pigments is constantly exchanged between photoreceptors and the RPE to maintain photoreceptor excitability in a process referred as the visual, or retinoid cycle (2, 3). In addition, a portion of the photoreceptor outer segments are shed every day and phagocytosed by the RPE (4). Failure in any of these complex functions can lead to retinal degenerations, visual function loss, and even blindness. Numerous genes in the RPE cells are involved in the biological functions in the visual cycle. Mutations of these genes could lead to dysfunctions of the RPE, resulting in inherited retinal degeneration (IRD) (5–8). Gene replacement therapy has been promising for the treatment of monogenic retinal disorders by replacing a mutated gene with a copy of healthy gene into the diseased cells and then relying on the endogenous protein translational machinery to generate normal protein and restore the lost function (9, 10). Gene therapy using adeno-associated virus (AAV) has been approved for the treatment of Lebers’ congenital amaurosis type 2 (LCA2) (11, 12). Unfortunately, AAV has a limited cargo capacity and cannot be directly expanded for the treatment of retinal diseases caused by the mutations of large genes (13). In addition, it is costly to manufacture AAV based gene therapy. There is an unmet clinical need for the design and development of safe and efficient gene delivery platforms for specific delivery of therapeutic genes in targeted cells.
Previously, we developed a smart pH-sensitive amino lipid (1-aminoethyl)iminobis[N-(oleoylcysteinyl-1-amino-ethyl)propionamide) (ECO), which self-assembles with the gene cargo into stable nanoparticles formulations, facilitates a pH-sensitive amphiphilic endosomal escape process followed by a reductive cytosolic release (PERC effect) of nucleic acids, and achieves efficient intracellular delivery of genetic materials without size limitations (14–20). We also created all-trans-retinylamine modified ECO/pDNA nanoparticles for specific gene delivery to the RPE to treat LCA2. By targeting interphotoreceptor retinoid-binding protein (IRBP), enhanced gene expression in the RPE and a rescuing effect were observed in an LCA mouse model (14). However, due to the poor stability of all-trans-retinylamine and consequent decrease of transfection efficiency, the use of a targeting ligand with high stability is needed to modify the ECO-based gene delivery system with a PEG spacer; this could significantly improve the delivery efficiency and therapeutic efficacy of the system (21, 22).
ACU4429 (Emixustat) is a stable analogue of all-trans-retinylamine, and binds to the retinal pigment epithelium 65 protein (RPE65) to inhibit the retinoid cycle. Developed as a small-molecule visual cycle modulator, it prevents accumulation of toxic byproducts resulted from the visual cycle in the RPE (23–28). More importantly, ACU4429 has high chemical stability and binds to the RPE through the same mechanism as all-trans-retinylamine, which is a promising ligand to target the RPE. Polyethylene glycol (PEG) spacers are commonly involved in targeting ligand designs, due to the advantages of minimizing non-specific cellular uptake and increasing biocompatibility (29, 30). However, PEGylation compromises the efficiency of cellular uptake and endosomal escape properties of non-viral gene delivery systems. Previously, a pH-sensitive hydrazone linker was introduced in the PEG spacer in targeted ECO/siRNA nanoparticles. The pH-sensitive PEG shedding due to acid-catalyzed hydrolysis of the hydrazone linker in the endosomes significantly enhanced the endosomal escape of ECO/siRNA nanoparticles and resulted in an excellent therapeutic effect for cancer RNAi therapy (22).
In this work, ACU4429 was introduced as a targeting ligand to achieve specific and efficient gene delivery to the RPE by conjugating the ECO-based gene delivery platform with a PEG spacer and a pH-sensitive hydrazone linker (ACU-PEG-HZ). ACU4429 targeted dual pH-sensitive ECO/pDNA nanoparticles (ACU-PEG-HZ-ECO/pDNA) were prepared by self-assembly of the ACU-PEG-HZ ligand with ECO and plasmid DNA. A plasmid DNA expressing ABCA4 protein was used as a large therapeutic gene to test the specificity and efficacy of the targeted nanoparticles. The ABCA4 protein is the flippase located in photoreceptor outer segments that helps the elimination of retinaldehyde, a toxic photoproduct from the visual cycle (31, 32). Mutations in the ABCA4 gene can cause Stargart disease (STGD) and cone-rod dystrophies (33–35). A recent study has shown that ABCA4 is detected in the RPE of mice, and genetically modified mice with ABCA4 expression only in the RPE, and not in photoreceptor cells, demonstrated a partial rescue effect of photoreceptor degeneration in Abca4−/−mice, suggesting that gene therapy to enhance ABCA4 expression in the RPE could prevent photoreceptor degeneration in Abca4−/− mice and possibly in patients with ABCA4 mutations (36). RPE-specific delivery and gene expression of ACU-PEG-HZ-ECO/pDNA nanoparticles was assessed both in vitro and in vivo with ABCA4 plasmids of different promoters.
Results
The ACU4429 targeting ligand with a PEG spacer and a hydrazone linker (ACU-PEG-HZ-MAL) was synthesized as described in Figure 1A. An excess of ACU4429 was first reacted with a 3.4 kD HS-PEG-NHS to give ACU-PEG-SH. A hydrazide group was then introduced to the ACU-PEG-SH by reacting with an excess of EMCH to form ACU-PEG-CO-NHNH2, which then reacted with N-4-acetylphenyl maleimide to give the targeting ligand with a PEG spacer and a HZ linker ACU4429-PEG-HZ-MAL. The maleimido group was introduced for conjugating to one of the thiol groups on ECO. The intermediates and ACU-PEG-HZ-MAL were purified by precipitation and characterized by MALDI-TOF mass spectrometry (Figure 1B). The molecular peak shifts in the spectra toward higher molecular weight indicated formation of desired products during the synthesis of ACU-PEG-HZ-MAL at each step.
Figure 1.
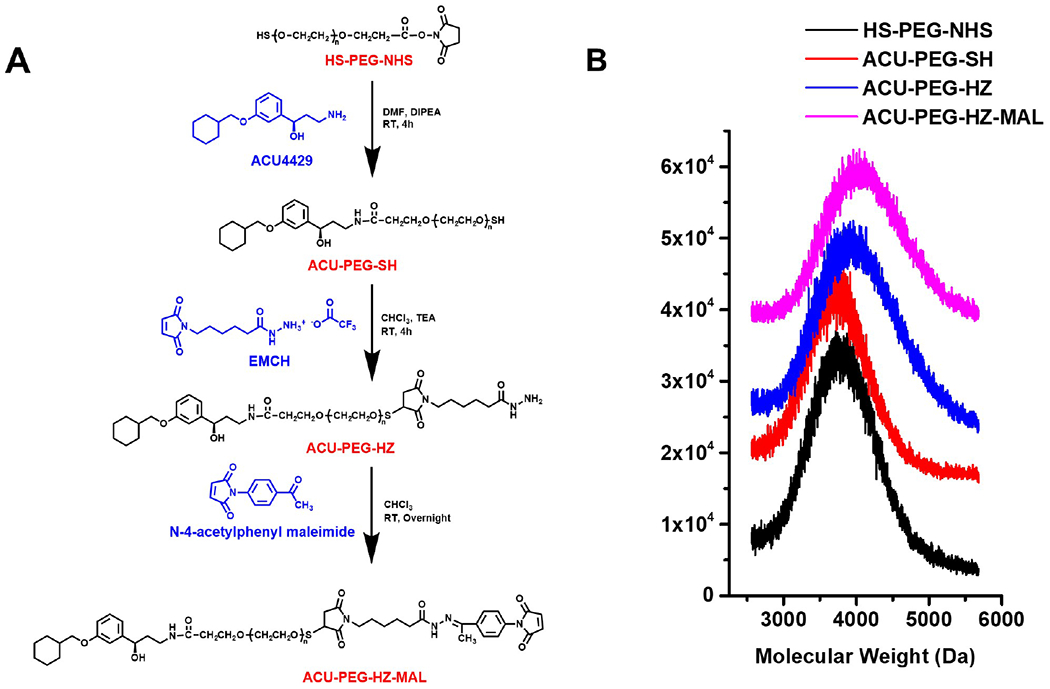
Design of ACU-PEG-HZ-MAL Targeting Ligand. (A) Synthetic route of ACU-PEG-HZ-MAL targeting ligand. (B) MALDI-TOF mass spectra of ACU-PEG-HZ-MAL and intermediates.
To formulate targeted ECO/pABCA4 nanoparticles, ACU-PEG-HZ-MAL targeting ligand (2.5 mol-%) was first mixed and reacted with ECO molecules in aqueous solution for 30 min. The targeted nanoparticles then formed by self-assembly when pABCA4 was added to the solution (Figure 2A). ECO/pABCA4 nanoparticles (no ligand) and PEG-ECO/pABCA4 (non-targeting ligand 3.4 kD PEG chain conjugated) nanoparticles were similarly prepared as controls. The nanoparticles were characterized using dynamic light scattering. All nanoparticles had similar sizes and size distribution with a diameter of 195.91±12.93 nm for ECO/pABCA4, 199.54±0.74 nm for PEG-ECO/pABCA4 and 195.13±5.74 nm for ACU-PEG-HZ-ECO/pABCA4 (Figure 2B,C). DLS zeta potential measurements showed that the nanoparticles had positive surface charge, with 39.05±0.64 mV for ECO/pABCA4 and 33.51±0.71 mV for PEG-ECO/pABCA4 and 33.83±0.40 mV for ACU-PEG-HZ-ECO/pABCA4 nanoparticles (Figure 2D,E). To determine the best N/P (amine/phosphate) ratio for effective intracellular gene delivery, transfections in ARPE-19 cells were performed using ECO/pCMV-GFP nanoparticles formulated at N/P ratio 6, 8, 10 and 12. Confocal images of GFP expression (Figure 2F) demonstrated increased GFP expression with N/P ratios. However, potential increased cytotoxicity was observed when the N/P ratio was 12. We selected N/P ratio of 10 for the following evaluations, due to a potential higher transfection efficiency of delivering large genes.
Figure 2.
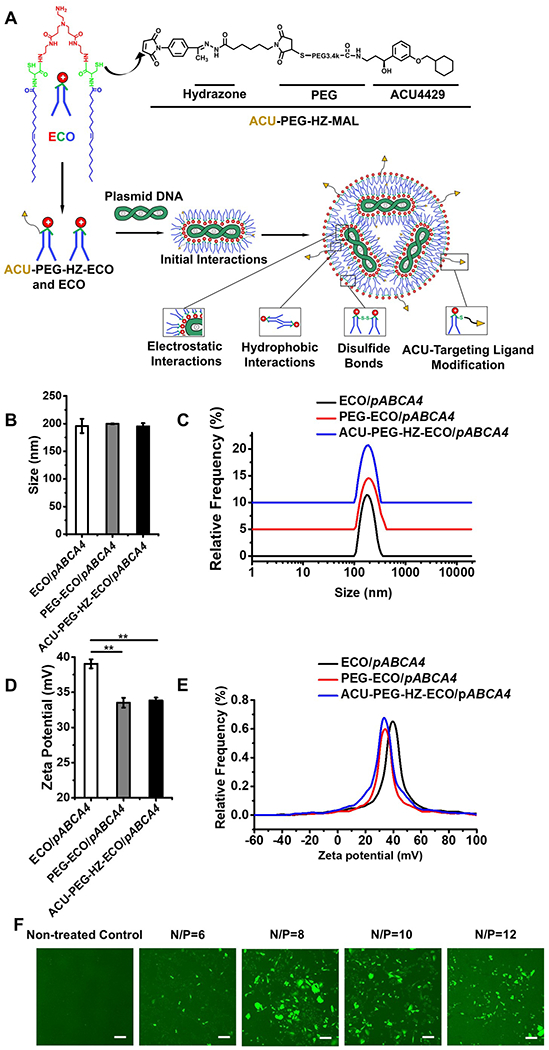
Formulation and characterization of ACU4429 modified ECO/pABCA4 nanoparticles. (A) Scheme of ACU-PEG-HZ-ECO/pDNA nanoparticle formulation, (B) nanoparticle sizes, (C) size distributions, (D) zeta potentials, (E) zeta potential distributions, and (F) confocal images of GFP expression 24 h after transfection in ARPE-19 cells under 10 % serum transfection media using ECO/pCMV-GFP nanoparticles of N/P ratios 6, 8, 10 and 12. (n = 3, error bars = ± SD, **P < 0.05. Scale bars represent 20 μm.).
To evaluate the stability and encapsulation of ECO/pABCA4 and ACU4429 modified ECO/pABCA4 nanoparticles, an agarose gel electrophoresis was performed. The complete encapsulation of pABCA4 (16 kb) was observed in the nanoparticles with no free DNA smear in the gels using the standard nanoparticle formulation procedure (Figure 3A). The bands on the top of the gel of ECO/pABCA4 and modified ECO/pABCA4 nanoparticles demonstrated limited mobility of encapsulated pABCA4, indicating good stability of these nanoparticles. However, if ECO and pABCA4 were mixed shortly only by pipetting, DNA was not completely encapsulated and a free pABCA4 smear was observed (black arrow). Controls of free ECO and PEG ligands didn’t show any bands in the gel. The introduction of the ACU-4429 targeting ligand with a PEG spacer produced only slight reduction of zeta potential that did not affect the size and encapsulation of ECO/pABCA4 nanoparticles. To evaluate cytotoxicity of ECO/pABCA4 and ACU4429 modified ECO/pABCA4 nanoparticles, a CCK-8 assay was performed in ARPE-19 cells (Figure 3B). No significant cytotoxicity was observed for ECO/pABCA4 and modified ECO/pABCA4 nanoparticles compared with the non-treated control. Free ECO, PEG ligands and combinations of both also didn’t demonstrated significant cytotoxicity (p < 0.05).
Figure 3.
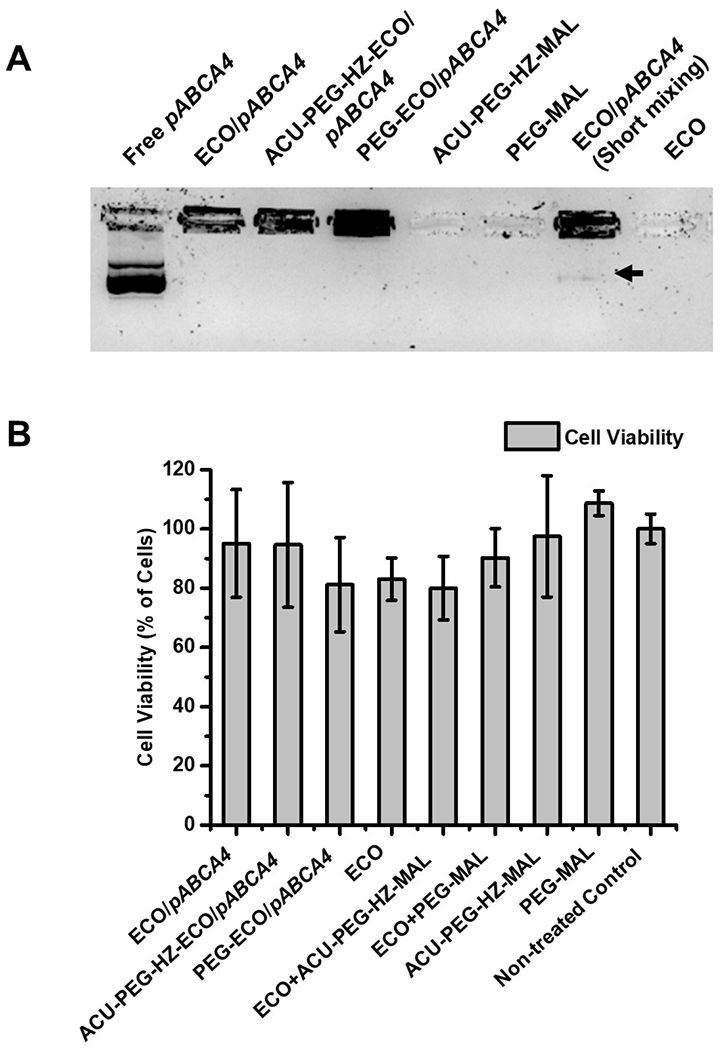
Stability, encapsulation and cytotoxicity of ECO/pABCA4 and ACU4429 modified ECO/pABCA4 nanoparticles. (A) Agarose gel electrophoresis of ECO/pABCA4, PEG-ECO/pABCA4 and ACU-PEG-HZ-ECO/pABCA4 nanoparticles with PEG ligands as controls (The arrow indicated the DNA smear of free pABCA4 not encapsulated by short pipette mixing of ECO and pABCA4.). (B) Viability of ARPE-19 cells treated with ECO/pABCA4 and ACU4429 modified ECO/pABCA4 nanoparticles, and free ECO, PEG ligands and non-treated ARPE cells as control (significance analysis was done using one-way ANOVA, p< 0.05).
To evaluate the intracellular transfection efficiency of ACU-PEG-HZ-ECO/pDNA nanoparticles, ARPE-19 cells were transfected with ACU-PEG-HZ-ECO/Cy3-pDNA nanoparticles at N/P = 10, with the non-targeted PEG-ECO/Cy3-pDNA nanoparticles as a control. Fluorescence confocal microscopy clearly revealed the differences in intracellular uptake and distribution of the fluorescently labeled DNA between these nanoparticle formulations. The ACU4429 targeted nanoparticles with the hydrazone linker demonstrated stronger cellular uptake at an early time point with a dispersed cytoplasmic DNA distribution at later time points (Figure 4A). At 1 h, both nanoparticles interacted with the cell membrane and began internalization. At 2 h, both nanoparticles were internalized by the cells. It seems that the targeted nanoparticles had higher cellular uptake than the non-targeted nanoparticles. At later time points, especially at 24 h, a highly dispersed distribution of the labeled DNA was observed in the cytoplasm of the cells treated with ACU-PEG-HZ-ECO/Cy3-pDNA nanoparticles, while only spotted intracellular distribution was observed with PEG-ECO/Cy3-pDNA nanoparticles (Figure 4A). The intracellular transfections of both nanoparticle systems were also quantified using a flow cytometry analysis (Figure 4B), which was consistent with the confocal observations. ACU-PEG-HZ-ECO/Cy3-pDNA nanoparticles demonstrated higher mean fluorescence intensities for all the time points. These results suggest excellent endosomal escape ability of ACU-PEG-HZ-ECO/Cy3-pDNA nanoparticles. The excellent endosomal escape ability was translated to high gene expression efficiency in the ARPE-19 cells transfected by ACU-PEG-HZ-ECO/pABCA4 nanoparticles (Figure 4C). Significantly higher ABCA4 mRNA expression was observed for ACU-PEG-HZ-ECO/pABCA4 nanoparticles than PEG-ECO/pABCA4 nanoparticles 48 h after transfection. Taken together, the targeted smart ACU-PEG-HZ-ECO/pDNA nanoparticles improved endosomal escape and cytosolic DNA delivery efficiency, and resulted in higher levels of gene expression.
Figure 4.
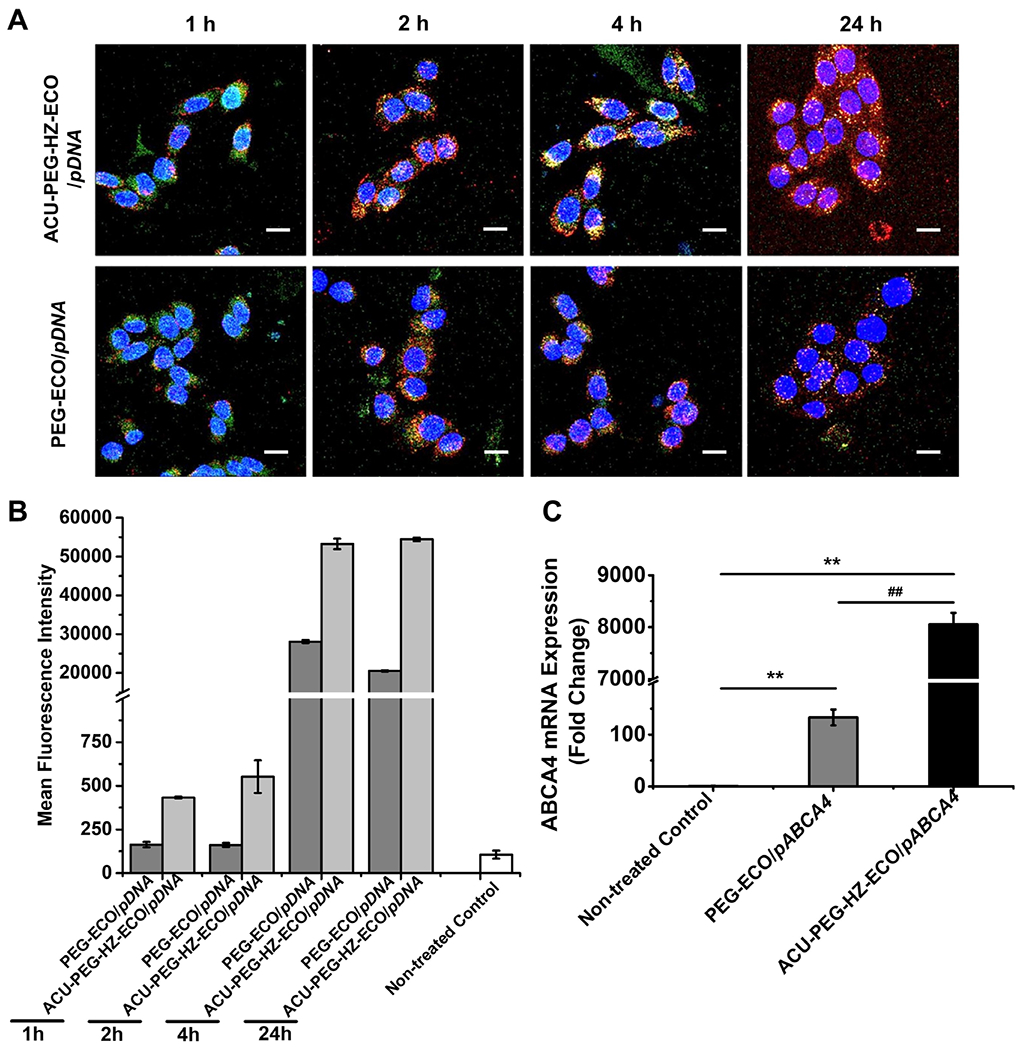
In vitro transfection efficiency of ACU-4429-PEG-HZ-ECO/pDNA nanoparticles. (A) Confocal fluorescence images (B) Flow cytometry measurements of the cytosolic delivery of ACU-PEG-HZ-ECO/Cy3-pDNA and PEG-ECO/Cy3-pDNA nanoparticles at an N/P ratio of 10 in ARPE-19 cells. DNA plasmid was labeled with Cy3 (red), nuclei with Hoechst 33342 (blue), and Late endosomes with LysoTracker Green (green). Flow cytometry recorded mean fluorescence intensities of Cy3 positive ARPE-19 cells at each time point. (C) qRT-PCR of ABCA4 mRNA expression at 48 h in ARPE-19 cells transfected with ACU-PEG-HZ-ECO/pCMV-ABCA4 and PEG-ECO/pCMV-ABCA4 nanoparticles (n = 3, error bars = ± SD, **, ##P < 0.05. Scale bars represent 20 μm).
To evaluate the in vivo targeting efficiency, ACU-PEG-HZ-ECO/Cy3-pDNA nanoparticles were injected into the subretinal space in Abca4−/− mice. ACU-PEG-HZ-ECO/Cy3-pDNA demonstrated excellent targeting efficiency in the subretinal space, where abundant IRBP is expressed (Figure 5A). ACU-PEG-HZ-ECO/Cy3-pDNA nanoparticles remained in the interphotoreceptor matrix (IPM), demonstrated by the red signals in the subretinal space. In comparison, the non-targeted ECO/Cy3-pDNA nanoparticles diffused in both the RPE and photoreceptors.
Figure 5.
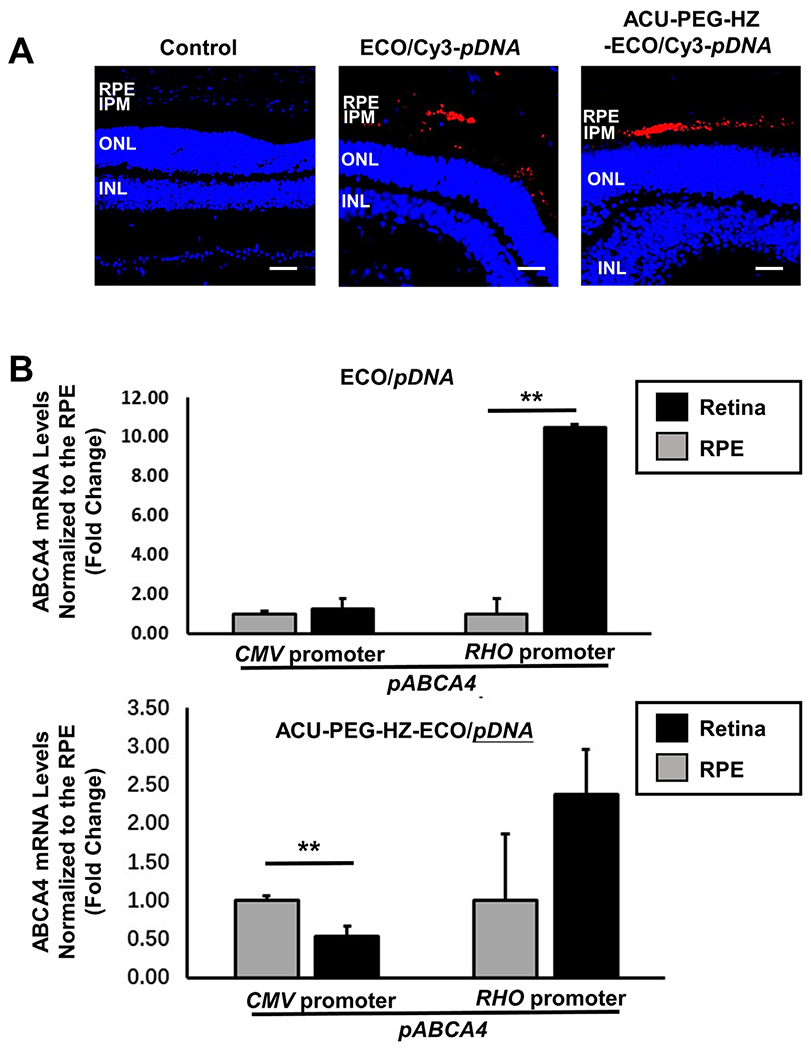
In vivo evaluation of the targeted ACU-PEG-HZ-ECO/pDNA nanoparticles for gene delivery to the RPE. (A) The distribution of ECO/Cy3-pDNA nanoparticles and ACU-PEG-HZ-ECO/Cy3-pDNA nanoparticles in the subretinal space of Abca4−/− mice 4 days after subretinal injection (Control: PBS injection; IPM: interphotoreceptor matrix; ONL: outer nuclear layer; INL: inner nuclear layer). (B) qRT-PCR analysis of ABCA4 mRNA expression in Abca4−/− mice 7 days after subretinal injection of ECO/pCMV-ABCA4 and ECO/pRHO-ABCA4 nanoparticles modified with PEG and ACU-PEG-HZ (n = 3, error bars = ± SD, **P < 0.05. Scale bars represent 50 μm).
Binding of ACU-PEG-HZ-ECO/pDNA nanoparticles to IRBP in the IPM resulted in enhanced RPE specific gene expression after subretinal injections of the nanoparticles. Both ACU-PEG-HZ-ECO/pABCA4 and PEG-ECO/pABCA4 nanoparticles were formulated with a photoreceptor-specific RHO promoter or a common CMV promoter by self-assembly. The nanoparticles (1 μL) were injected into the subretinal space of Abca4−/− mice at a dose of 200 ng DNA. For non-targeted ECO/pCMV-ABCA4 nanoparticles, similar ABCA4 mRNA expression in the retina and the RPE with the common promoter was demonstrated, while significantly higher ABCA4 mRNA expression in the retina was achieved with PEG-ECO/pRHO-ABCA4 by using the specific RHO promoter (Figure 5B). For the ACU4429 targeted nanoparticles, more ABCA4 mRNA expression was observed in the RPE than the retina for ACU-PEG-HZ-ECO/pCMV-ABCA4 because of enhanced RPE uptake mediated by ACU4429. Due to the photoreceptor specificity of the RHO promoter in conjunction with the targeting effect of ACU-4429 to the RPE and low gene expression with pRHO-ABCA4 in the RPE cells, no statistical difference between ABCA4 mRNA expression in the retina and the RPE was found with ACU-PEG-HZ-ECO/pRHO-ABCA4. Taken together, the results indicated that the stable retinylamine analogue ACU4429 modified ECO/pDNA nanoparticles effectively facilitated delivery of therapeutic DNA and induced gene expression in the RPE.
Discussion
The retinal pigment epithelium is one of the most important layers in the retina, and responsible for crucial functions in the visual pathway (8). Mutations in the genes associated with this pathway can cause disparate retinal diseases (6). In order to treat the related genetic disorders, RPE specific gene delivery is critical for specific transgene expression to achieve effective gene therapy with acceptable safety (37). ACU4429 has shown excellent RPE targeting efficiency due to its specific binding to IRBP. In the retina, the space between rod outer segments and the RPE is filled with the interphotoreceptor matrix, where IRBP is the major protein that facilitates selective transportations of all-trans-retinol from the photoreceptors to the RPE. This property has been explored as an effective targeting strategy for specific delivery to the RPE, where all-trans-retinylamine has been used as a targeting ligand (14). However, all-trans-retinylamine is light sensitive and has shown poor stability, which could diminish its targeting efficiency. ACU-4429 is a synthetic small molecule with a similar structure to all-trans-retinylamine. It is highly stable and is used for inhibiting the isomerohydrolase reaction of the all-trans-retinoids. We have shown here that modification of ECO/pDNA nanoparticles with ACU4429, a stable all-trans-retinoid analogue, can also enhance gene delivery to the RPE through binding to IRBPs. It is speculated that after injection of the ACU-PEG-HZ-ECO/pDNA nanoparticles into the subretinal space, ACU4429 would quickly bind to IRBPs, which would then transport the nanoparticles near the apical surface of the RPE and facilitate the internalization of the nanoparticles by the RPE (Figure 6). Consequently, more Cy3-pDNA and expression of pABCA4 plasmid was observed in the RPE than the retina when the ECO/pDNA nanoparticles were modified with ACU4429 as a targeting ligand.
Figure 6.
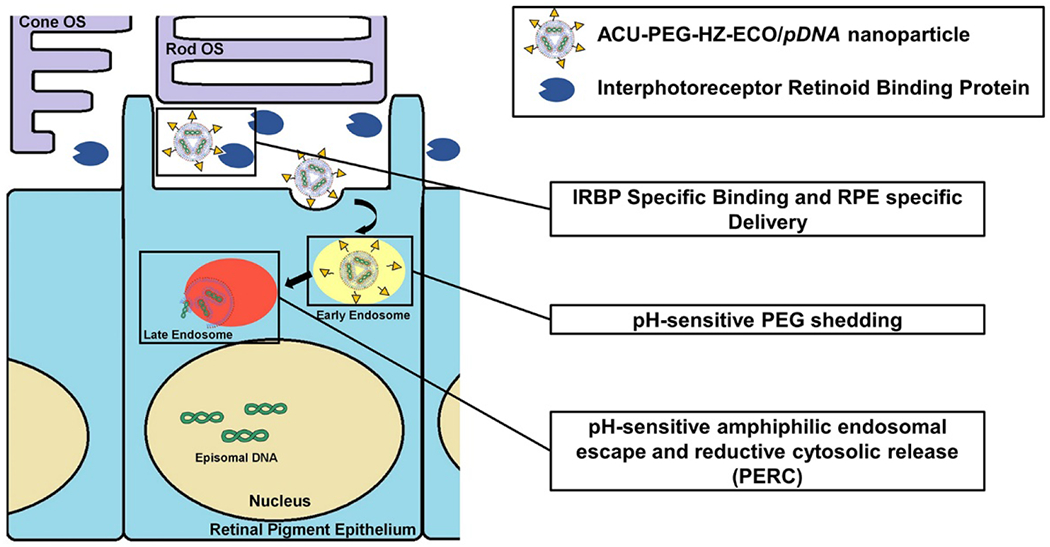
The targeting mechanism of ACU-PEG-HZ-ECO/pDNA nanoparticles for gene delivery in the RPE. Once injected into the subretinal space, ACU-PEG-HZ-ECO/pDNA nanoparticles bind to interphotoreceptor retinoid binding protein (IRBP) in the interphotoreceptor matrix. IRBP binding helps transport the nanoparticles to the targeted RPE cells. Following endocytosis, the PEG layer of the targeted nanoparticles sheds off due to the acid-catalyzed hydrolysis of the hydrazone linker in the acidic endosome. The ECO/pDNA nanoparticles then escape from the endosomal entrapment and then release the therapeutic plasmid DNA through the PERC mechanism.
PEGylation is a common strategy in the design of target gene delivery platforms to minimize non-specific cellular uptake and potential toxic side effects (29). However, PEGylation compromises the efficiency of endosomal escape and cytosolic release of the gene cargo. In order to overcome this obstacle, a pH-sensitive hydrazone linker was used to conjugate the targeting agent with the PEG spacer to shed the PEG layer through a pH-sensitive hydrolysis of hydrazone once targeted nanoparticles were in acidic endosomes. The core ECO/pDNA nanoparticles would be exposed to enhance endosomal escape and cytosolic release of the DNA cargo through the PERC effect of ECO. The enhanced amphiphilicity of ECO due to the protonation of the amino head groups of ECO in the acidic endosomes (pH = 5–6) would destabilize the endosomal membrane to facilitate the escape of the ECO/pDNA nanoparticles into cytosol. The reductive environment of the cytosol would reduce the disulfide bonds in the nanoparticles, lead to dissociation of the ECO/pDNA formulations and release the DNA payloads (14–17, 19). In contrast, the regular PEGylated ECO/pDNA nanoparticles had limited ability to escape from the endosomal entrapment. The results in this work have shown that the strategy of using a pH-sensitive hydrazone linker facilitated endosomal escape and cytosolic release of the DNA payload of the nanoparticles and significantly enhanced the gene expression of the ACU-PEG-HZ-ECO/pDNA as compared to PEG-ECO/pDNA nanoparticles.
Recently, ABCA4 expression was detected in the mouse RPE, comprising 1% of its level in photoreceptors of the neural retina. Expression of ABCA4 in the RPE has a partial response in the ocular Abca4−/− phenotype (36). Delivery and expression of ABCA4 gene in the RPE cells have the potential to prevent the progression of Stargardt disease. We used ABCA4 plasmids with a common CMV promoter and a rod photoreceptor specific RHO promoter to demonstrate the ability of ACU4429 targeted ECO/pDNA nanoparticles for specific delivery of a large ABCA4 gene in the RPE. More significant expression of ABCA4 was observed for the targeted nanoparticles with the CMV promoter in the RPE than in the retina, while the targeted nanoparticles with the RHO promoter had lower relative ABCA4 expression in the retina than the non-targeted nanoparticles. The results have validated the hypothesis that the targeted ECO/pDNA nanoparticles are effective for specific delivery of therapeutic genes into the RPE in vivo. ACU4429 modified ECO/pDNA nanoparticles have the promise to be used as non-viral system to deliver therapeutic genes of any sizes to the RPE for gene therapy of monogenetic visual disorders.
Conclusion
In conclusion, the ACU4429 modified dual pH-sensitive lipid ECO/pDNA nanopartics were readily prepared with self-assembly of the targeting agent with a pH-sensitive cleavable PEG spacer, ECO and pDNA. The sheddable PEG spacer is effective to improve cytosolic gene delivery and gene expression by enhancing endosomal escape of the core ECO/pDNA nanoparticles. The smart ECO/pDNA nanoparticles also mediated excellent RPE specific gene expression in mice. Therefore, after ACU4429-PEG-hydrazone modification, the pH-sensitive non-viral ECO/pDNA nanoparticles can be promising as a safe and efficient platform for targeted delivery of gene therapeutics to the RPE, and treatment for monogenic visual disorders.
MATERIALS AND METHODS
Reagents
All reagents ordered from vendors were directly used without extra purification unless they were otherwise detailed in this section. Organic solvents such as Methylene chloride (DCM), acetonitrile (ACN) chloroform, and methanol were ordered from Thermo Fisher Scientific (Hampton, NH). NHS-PEG-SH (MW 3400) was purchased from Nanocs lnc (New York, NY). The synthesis of Lipid ECO followed the procedures reported previously (38). For cell culture, penicillin fetal bovine serum, and streptomycin were purchased from Invitrogen (Carlsbad, CA). The ABCA4 plasmid (pCMV-ABCA4) was kindly gifted by Dr. Robert S. Molday (University of British Columbia), which included human ABCA4 cDNA sequence of full-length (NCBI Accession # NM_000350.2) on a pCEP4 backbone. pRHO-ABCA4 was prepared as previously described (20). Cy3-pDNA was purchased from Mirus Bio (catalog number MIR7904). ACU-4429 was a gift from Dr. Krzysztof Palczewski (University of California, Irvine).
Cell Culture:
ARPE-19 (ATCC, Manassas, Virginia) cells were passaged and maintained in a Dulbecco’s modified Eagle’s medium containing fetal bovine serum (10%), streptomycin (100 μg/mL), and penicillin (100 units/mL). Cells were kept in a humidified incubator at 37 ºC and 5% CO2.
Animal:
Pigmented Abca4−/− knockout mice were obtained as described previously (39) and maintained with mixed backgrounds of 129Sv/Ev or C57BL/6. Animals were housed and bred in the Animal Resource Center at CWRU. All procedures followed approved protocols by the CWRU Institutional Animal Care and Use Committee (IACUC#2014–0053), which were also in compliance with recommendations from the Association for Research for Vision and Ophthalmology and the American Veterinary Medical Association Panel on Euthanasia.
Synthesis of ACU-PEG-HZ-MAL:
ACU-PEG-HZ-MAL was prepared following multi-step reactions. ACU4429 (25 mg) was dissolved in DMF (5 mL) and 1 mL of NHS-PEG3400-SH (70.4 mg) in DMF was added drop-wise to the mixture, followed by the addition of DIPEA (100 μL). After stirring at room temperature for 4 hours, the mixture was added dropwise to diethyl ether (3X volume excess) to remove unreacted ACU4429. ACU-PEG3400-SH was obtained as precipitate in ether. To fully reduce any disulfide bond present in the synthesized ACU-PEG-SH, dithiothreitol (100 mg) was added to a solution of the synthesized ligand and stirred overnight. A desalting spin column (1.8K MWCO) (Thermo Fisher Scientific, Waltham, MA) was used to remove free dithiothreitol. The product was lyophilized, and further characterized by MALDI-TOF mass spectrometry (Mw increase of 160 was observed). ACU-PEG-Hydrazide was synthesized by the reaction of ACU-PEG-SH (25.2 mg) and N-ε-maleimidocaproic acid hydrazide (4.25 mg) in the presence of triethylamine (4.39 μL) mixed in a 5 mL chloroform solution for 4 hours at room temperature. ACU-PEG-Hydrazide was purified after centrifuge using a spin column (1.8K MWCO). The product was lyophilized, and characterized by MALDI-TOF mass spectrometry. Finally, ACU-PEG-Hydrazide (12 mg) was reacted with N-4-acetylphenyl maleimide (1.2 mg) in DMF overnight at room temperature. The final product ACU-PEG-HZ-MAL was purified through ether precipitation and obtained after lyophilization, which was characterized by MALDI-TOF mass spectrometry with a peak molecular weight of 4,000 Da.
Preparation of ECO/pDNA and ACU-PEG-HZ-ECO/pDNA Nanoparticles:
ECO (2.5 mM) in ethanol and ACU-PEG-HZ-MAL targeting ligand (0.4 mM in water) (2.5 mol % of ECO) was first mixed and reacted in aqueous solution for 30 min. Plasmid DNA (0.5 mg/mL) aqueous solution at predetermined volume from the N/P ratio (amine to phosphate ratio) of 10 were added to the ECO and ACU-PEG-HZ-MAL mixture, and vortexed for 30 min at room temperature to give ACU-PEG-HZ-ECO/pDNA nanoparticles. An ECO stock solution (25 mM) of was used to formulate nanoparticles for in vivo experiments. Encapsulations of pDNA by lipid ECO in nanoparticle formulations were characterized by an agarose gel electrophoresis method. Agarose gel (0.7%) in TBE buffer was performed at 100 V for 30 min.
Dynamic Light Scattering:
The sizes and zeta potentials was characterized for nanoparticle formulations of ECO/pDNA (N/P=10), PEG-ECO/pDNA (N/P=10) and ACU-PEG-HZ-ECO/pDNA (N/P = 10) using a dynamic light scattering method with an Anton Paar Litesizer 500 (Anton Paar USA Inc, Ashland, VA). Each sample was analyzed three times at 20 ºC.
In Vitro Transfection:
Transfections were performed on 12-well plates, where ARPE-19 cells were seeded at a concentration of 4 × 104 cells/well. Cells were allowed to grow for 24 h before transfection. Nanoparticles at pDNA concentration of 1 μg/mL in DMEM with 10% serum were added to ARPE-19 cells and incubated for 8 h at 37 ºC. The media containing nanoparticles was then replaced with fresh DMEM (10% serum). ARPE-19 cells were further incubated for an additional 48 h. Expression of ABCA4 was evaluated by qRT-PCR at mRNA level.
Transfection of ECO/pCMV-GFP nanoparticles of different N/P ratios were also conducted similarly in ARPE-19 cells, where DMEM (10% serum) and a pDNA concentration of 1 μg/mL were also used. The transfection was performed in 12-well plate with ARPE-19 cell concentration of 4 × 104 cells per well. The nanoparticles were formulated under N/P ratios of 6, 8, 10 and 12 and were then incubated with ARPE-19 cells as previously described. After 24 h, fluorescence images of GFP expression were acquired using an Olympus FV1000 confocal microscope.
Cytotoxicity:
Cytotoxicity of ECO/pABCA4 and ACU4429 modified ECO/pABCA4 nanoparticles was investigated using a CCK-8 assay (Dojindo Molecular Technologies, Inc, Washington, D.C.). Cell viability was evaluated using ARPE-19 cells on 96-well plates, where cells were seeded at a concentration of 1 × 104 cells per well. ARPE-19 cells were incubated with ECO/pABCA4 or ACU4429 modified ECO/pABCA4 nanoparticles at DNA concentrations of 1 μg/mL in DMEM (10% serum) for 8 h at 37 °C, after which, nanoparticle containing DMEM was replaced with fresh DMEM (10% serum). The cells were allowed to grow until 48 h and washed with PBS. The CCK-8 reagent was added to each well followed by an incubated of 1.5 h at 37°C. The absorbance at 450 nm was recorded using a plate reader. Cell viability was characterized by normalizing to the absorption of non-treated control. Free ECO, PEG ligands and combinations were used as controls. The amount of each molecule is the same as what was used in each formulation.
qRT-PCR:
For cells, a scraper was used for cell lysis and homogenization. For animal retinal tissues, eye samples were harvested from mice and dissected in a PBS buffer to separate neural retina and the RPE. Both tissues were homogenized separately using a glass rod in a 1.5 mL tube loaded with 0.6 mL of the lysis buffer. The RNA extractions for cells and tissue samples were conducted using a QIAGEN RNeasy kit following the manufacturer’s instructions. cDNAs were synthesized from mRNA transcripts using a QIAGEN miScriptII reverse transcriptase kit (Germantown, MD). The qRT-PCR analysis was performed in a Mastercycler instrument (Eppendorf, Hauppauge, NY) using a SYBR Green Master mix (AB Biosciences, Allston, MA). Fold changes of mRNA levels were determined by normalization to 18S. Eyes injected with PBS (SHAM) were used as controls.
Intracellular Uptake:
Intracellular uptake was first evaluated using microscopic method using glass-bottom micro-well dishes, where ARPE-19 cells were seeded at a concentration of 4 × 104 cells/dish and grew for 24 h at 37 ºC. Then, the cell nuclei were stained with Hoechst 33342 (4 μg/mL) (Invitrogen) and endosomes/lysosomes with LysoTracker Green (100 mM) (Life Technologies, Carlsbad, CA). ARPE-19 cells were then incubated with ACU-PEG-HZ-ECO/Cy3-pDNA or PEG-ECO/Cy3-pDNA (N/P = 10) nanoparticles in DMEM (10% serum) for 1 h, 2h, 4 h and 24 h (where nanoparticle containing media was replaced by fresh DMEM at 4 h and cells were further incubated for 24 h). After incubation, cells were washed with PBS three times and then fixed in 0.5 mL of PBS containing 4% paraformaldehyde (4% PFA). Samples were imaged for cy3 fluorescence using an Olympus FV1000 confocal microscope (Olympus, Center Valley, PA). Quantitative analysis was performed using a flow cytometry method. To prepare samples, cells were washed with PBS and then harvested after treatment with trypsin (0.25%, 0.26 mmol EDTA) (Invitrogen, Carlsbad, CA). The cell pellets were obtained by centrifugation (1500 rpm, 5 min) and resuspended in 4% PFA (0.5 mL). The cell suspensions were finally forced through a cell strainer (35 μm) (BD Biosciences, Franklin Lakes, NJ). Cy3 positive ARPE-19 cells were quantified by measurements of the fluorescence intensities for more than 3,000 cells from each sample using a BD FACSCalibur flow cytometer (BD Biosciences, Franklin Lakes, NJ).
In Vivo Subretinal Transfection with PEG-ECO/pDNA and ACU-PEG-HZ-ECO/pDNA Nanoparticles:
Subretinal injection was performed as previously described (40) (41). The nanoparticle solution (1 µL) was injected by a pump with a steady speed of 150 nL/sec into the mouse eye. Successful administration was confirmed by bleb formation in the subretinal space. A total of 200 ng of Cy3-labeled plasmid or pABCA4 (CMV or RHO promoters) was delivered. The contralateral eye injected with 1 µL of PBS was used as control. At least three days after the subretinal administration, examinations using OCT were performed to ensure the heal of retinal structure of the injection site with minimal retinal detachment.
Immunohistochemistry:
Histological samples were prepared as previously described (14). The eyes were harvested from mice after subretinal injections, washed with PBS, and fixed in 4% PFA for 2 h. Then eye cups were prepared and further fixed in 4% PFA overnight. The eyecups were then immersed in Tissue-Tek optimal cutting temperature compound (OCT) (Sakura, Torrance, CA) solutions containing 20% sucrose through a gradual sucrose gradient. After incubation in 20% sucrose/OCT overnight, the eye cups were imbedded in cryomolds and flash frozen in OCT. Histological slides (10 μm-thick) were collected under frozen conditions. The slides were then warmed to room temperature and further washed with Tris buffer containing 1% tween 20 (TBST) buffer. Followed by an antigen retrieval (20 min) in citrate buffer (10 mM, pH 6.0) using a pressure cooker, the slides were ready for staining. DAPI was used to label cell nuclei. Fluorescence images of the distribution of Cy3-labeled pDNA and DAPI were collected using an Olympus FV1000 confocal microscope (Olympus).
Statistical Analysis:
Experiments were performed in triplicate and the number of animals is listed in the figure captions. Experimental data are presented as averages with standard deviations. Statistical analysis was performed with one-way ANOVA and two-tailed Student’s t-tests. A 95% confidence interval was used and P ≤ 0.05 was accepted as statistically significant.
Key points for patent application in this section:
Formulations of PEG-ECO/pABCA4 and stable retinoid analogue targeted ACU4429 ACU-PEG-HZ-ECO/pABCA4 nanoparticles with the pH-sensitive hydrazone linker (HZ) for gene therapy of Stargardt disease.
The ACU-PEG-HZ-ECO/pABCA4 nanoparticles provide specific delivery of ABCA4 gene into the retinal pigment epithelium to express ABCA4 gene for treating Stargardt disease.
Acknowledgements
This project was supported by the Gund-Harrington Scholars Award from the Harrington Discovery Institute and by the Foundation Fighting Blindness. Further support was obtained through grants from the NIH (NEI R24-EY-024864 and NEI R24-EY-027283), the Canadian Institute for Advanced Research (CIFAR), and the Alcon Research Institute (ARI). Z.R.L. is an M. Frank Rudy and Margaret Domiter Rudy Professor of Biomedical Engineering. KP is the Irving H. Leopold Chair of Ophthalmology.
Footnotes
Conflict of interest
The authors have declared that no conflict of interest exists.
Contributor Information
Da Sun, Department of Biomedical Engineering, Case Western Reserve University, Cleveland, OH 44106.
Rebecca M. Schur, Department of Biomedical Engineering, Case Western Reserve University, Cleveland, OH 44106
Avery E. Sears, Department of Pharmacology, Case Western Reserve University, Cleveland, OH 44106
Song-Qi Gao, Department of Biomedical Engineering, Case Western Reserve University, Cleveland, OH 44106.
Wenyu Sun, Department of Biomedical Engineering, Case Western Reserve University, Cleveland, OH 44106.
Amirreza Naderi, Department of Biomedical Engineering, Case Western Reserve University, Cleveland, OH 44106.
Timothy Kern, Department of Ophthalmology, University of California Irvine, Irvine, CA 92697-7600.
Krzysztof Palczewski, Department of Ophthalmology, University of California Irvine, Irvine, CA 92697-7600.
Zheng-Rong Lu, Department of Biomedical Engineering, Case Western Reserve University, Cleveland, OH 44106.
Reference
- (1).Strauss O (2005) The Retinal Pigment Epithelium in Visual Function. Physiological Reviews 85, 845–881. [DOI] [PubMed] [Google Scholar]
- (2).Kiser PD, Golczak M, and Palczewski K (2014) Chemistry of the Retinoid (Visual) Cycle. Chemical Reviews 114, 194–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Kiser PD, Golczak M, Maeda A, and Palczewski K (2012) Key enzymes of the retinoid (visual) cycle in vertebrate retina. Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids 1821, 137–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Mazzoni F, Safa H, and Finnemann SC (2014) Understanding photoreceptor outer segment phagocytosis: Use and utility of RPE cells in culture. Experimental Eye Research 126, 51–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Thompson DA, and Gal A (2003) Vitamin A metabolism in the retinal pigment epithelium: genes, mutations, and diseases. Progress in Retinal and Eye Research 22, 683–703. [DOI] [PubMed] [Google Scholar]
- (6).Gu SM, Thompson DA, Srikumari CRS, Lorenz B, Finckh U, Nicoletti A, Murthy KR, Rathmann M, Kumaramanickavel G, Denton MJ, and Gal A (1997) Mutations in RPE65 cause autosomal recessive childhood–onset severe retinal dystrophy. Nature Genetics 17, 194–197. [DOI] [PubMed] [Google Scholar]
- (7).Bird A (1996) Age-related macular disease. The British journal of ophthalmology 80, 2–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Bok D (1993) The retinal pigment epithelium: a versatile partner in vision. Journal of Cell Science 1993, 189–195. [DOI] [PubMed] [Google Scholar]
- (9).Bainbridge JWB, Tan MH, and Ali RR (2006) Gene therapy progress and prospects: the eye. Gene Therapy 13, 1191–1197. [DOI] [PubMed] [Google Scholar]
- (10).Scholl HPN, Strauss RW, Singh MS, Dalkara D, Roska B, Picaud S, and Sahel J-A (2016) Emerging therapies for inherited retinal degeneration. Science Translational Medicine 8, 368rv6-368rv6. [DOI] [PubMed] [Google Scholar]
- (11).Schimmer J, and Breazzano S (2015) Investor Outlook: Focus on Upcoming LCA2 Gene Therapy Phase III Results. Human Gene Therapy Clinical Development 26, 144–149. [DOI] [PubMed] [Google Scholar]
- (12).Simonelli F, Maguire AM, Testa F, Pierce EA, Mingozzi F, Bennicelli JL, Rossi S, Marshall K, Banfi S, Surace EM, Sun J, Redmond TM, Zhu X, Shindler KS, Ying GS, Ziviello C, Acerra C, Wright JF, McDonnell JW, High KA, Bennett J, and Auricchio A (2010) Gene therapy for leber’s congenital amaurosis is safe and effective through 1.5 years after vector administration. Molecular Therapy 18, 643–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Rodrigues GA, Shalaev E, Karami TK, Cunningham J, Slater NKH, and Rivers HM (2018) Pharmaceutical Development of AAV-Based Gene Therapy Products for the Eye. Pharmaceutical Research 36, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Sun D, Sahu B, Gao S, Schur RM, Vaidya AM, Maeda A, Palczewski K, and Lu Z-R (2017) Targeted Multifunctional Lipid ECO Plasmid DNA Nanoparticles as Efficient Non-viral Gene Therapy for Leber’s Congenital Amaurosis. Molecular Therapy - Nucleic Acids 7, 42–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Sun D, Maeno H, Gujrati M, Schur R, Maeda A, Maeda T, Palczewski K, and Lu Z-R (2015) Self-Assembly of a Multifunctional Lipid With Core–Shell Dendrimer DNA Nanoparticles Enhanced Efficient Gene Delivery at Low Charge Ratios into RPE Cells. Macromolecular Bioscience 15, 1663–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Sun D, Sun Z, Jiang H, Vaidya AM, Xin R, Ayat NR, Schilb AL, Qiao PL, Han Z, Naderi A, and Lu Z-R (2019) Synthesis and Evaluation of pH-Sensitive Multifunctional Lipids for Efficient Delivery of CRISPR/Cas9 in Gene Editing. Bioconjugate Chemistry 30, 667–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Vaidya AM, Sun Z, Ayat N, Schilb A, Liu X, Jiang H, Sun D, Scheidt J, Qian V, He S, Gilmore H, Schiemann WP, and Lu Z-R (2019) Systemic Delivery of Tumor-Targeting siRNA Nanoparticles against an Oncogenic LncRNA Facilitates Effective Triple-Negative Breast Cancer Therapy. Bioconjugate Chemistry 30, 907–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Dinculescu A, Glushakova L, Min S-H, and Hauswirth WW (2005) Adeno-Associated Virus-Vectored Gene Therapy for Retinal Disease. Human Gene Therapy 16, 649–663. [DOI] [PubMed] [Google Scholar]
- (19).Ayat NR, Sun Z, Sun D, Yin M, Hall RC, Vaidya AM, Liu X, Schilb AL, Scheidt JH, and Lu Z-R (2019) Formulation of Biocompatible Targeted ECO/siRNA Nanoparticles with Long-Term Stability for Clinical Translation of RNAi. Nucleic Acid Therapeutics 29, 195–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Sun D, Schur RM, Sears AE, Gao S-Q, Vaidya A, Sun W, Maeda A, Kern T, Palczewski K, and Lu Z-R (2020) Non-viral Gene Therapy for Stargardt Disease with ECO/pRHO-ABCA4 Self-Assembled Nanoparticles. Molecular Therapy 28, 293–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Dawson MI (2017) Chemistry and Biology of Synthetic Retinoids: 0, CRC Press. [Google Scholar]
- (22).Gujrati M, Vaidya AM, Mack M, Snyder D, Malamas A, and Lu ZR (2016) Targeted Dual pH‐Sensitive Lipid ECO/siRNA Self‐Assembly Nanoparticles Facilitate In Vivo Cytosolic sieIF4E Delivery and Overcome Paclitaxel Resistance in Breast Cancer Therapy. Advanced healthcare materials 5, 2882–2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Kubota R, Al-Fayoumi S, Mallikaarjun S, Patil S, Bavik C, and Chandler JW (2014) Phase 1, dose-ranging study of emixustat hydrochloride (ACU-4429), a novel visual cycle modulator, in healthy volunteers. Retina 34, 603–609. [DOI] [PubMed] [Google Scholar]
- (24).Zhang J, Kiser PD, Badiee M, Palczewska G, Dong Z, Golczak M, Tochtrop GP, and Palczewski K (2015) Molecular pharmacodynamics of emixustat in protection against retinal degeneration. The Journal of clinical investigation 125, 2781–2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Jack LS, Sadiq MA, Do DV, and Nguyen QD (2016) Emixustat and lampalizumab: potential therapeutic options for geographic atrophy, in Retinal Pharmacotherapeutics pp 302–309, Karger Publishers. [DOI] [PubMed] [Google Scholar]
- (26).Rosenfeld PJ, Dugel PU, Holz FG, Heier JS, Pearlman JA, Novack RL, Csaky KG, Koester JM, Gregory JK, and Kubota R (2018) Emixustat hydrochloride for geographic atrophy secondary to age-related macular degeneration: a randomized clinical trial. Ophthalmology 125, 1556–1567. [DOI] [PubMed] [Google Scholar]
- (27).Fitzsimmons ME, Sun G, Kuksa V, and Reid MJ (2018) Disposition, profiling and identification of emixustat and its metabolites in humans. Xenobiotica 48, 592–604. [DOI] [PubMed] [Google Scholar]
- (28).Dugel PU, Novack RL, Csaky KG, Richmond PP, Birch DG, and Kubota R (2015) Phase II, randomized, placebo-controlled, 90-day study of emixustat HCl in geographic atrophy associated with dry age-related macular degeneration. Retina (Philadelphia, Pa.) 35, 1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Lee M, and Kim SW (2005) Polyethylene Glycol-Conjugated Copolymers for Plasmid DNA Delivery. Pharmaceutical Research 22, 1–10. [DOI] [PubMed] [Google Scholar]
- (30).Kawano K, and Maitani Y (2011) Effects of Polyethylene Glycol Spacer Length and Ligand Density on Folate Receptor Targeting of Liposomal Doxorubicin In Vitro. Journal of Drug Delivery 2011,1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Quazi F, Lenevich S, and Molday RS (2012) ABCA4 is an N-retinylidene-phosphatidylethanolamine and phosphatidylethanolamine importer. Nature Communications 3, 925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Molday RS (2007) ATP-binding cassette transporter ABCA4: Molecular properties and role in vision and macular degeneration. Journal of Bioenergetics and Biomembranes 39, 507–517. [DOI] [PubMed] [Google Scholar]
- (33).Maugeri A, Klevering BJ, Rohrschneider K, Blankenagel A, Brunner HG, Deutman AF, Hoyng CB, and Cremers FPM (2000) Mutations in the ABCA4 (ABCR) Gene Are the Major Cause of Autosomal Recessive Cone-Rod Dystrophy. The American Journal of Human Genetics 67, 960–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Klevering BJ, Deutman AF, Maugeri A, Cremers FPM, and Hoyng CB (2005) The spectrum of retinal phenotypes caused by mutations in the ABCA4 gene. Graefe’s Archive for Clinical and Experimental Ophthalmology 243, 90–100. [DOI] [PubMed] [Google Scholar]
- (35).Koenekoop RK (2003) The gene for Stargardt disease, ABCA4, is a major retinal gene: a mini-review. Ophthalmic Genetics 24, 75–80. [DOI] [PubMed] [Google Scholar]
- (36).Lenis TL, Hu J, Ng SY, Jiang Z, Sarfare S, Lloyd MB, Esposito NJ, Samuel W, Jaworski C, Bok D, Finnemann SC, Radeke MJ, Redmond TM, Travis GH, and Radu RA (2018) Expression of ABCA4 in the retinal pigment epithelium and its implications for Stargardt macular degeneration. Proceedings of the National Academy of Sciences 115, E11120–E11127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Yin H, Kanasty RL, Eltoukhy AA, Vegas AJ, Dorkin JR, and Anderson DG (2014) Non-viral vectors for gene-based therapy. Nature Reviews Genetics 15, 541. [DOI] [PubMed] [Google Scholar]
- (38).Malamas AS, Gujrati M, Kummitha CM, Xu R, and Lu Z-R (2013) Design and evaluation of new pH-sensitive amphiphilic cationic lipids for siRNA delivery. Journal of Controlled Release 171, 296–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Maeda A, Maeda T, Golczak M, and Palczewski K (2008) Retinopathy in Mice Induced by Disrupted All-trans-retinal Clearance. Journal of Biological Chemistry 283, 26684–26693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Timmers AM, Zhang H, Squitieri A, and Gonzalez-Pola C (2001) Subretinal injections in rodent eyes: effects on electrophysiology and histology of rat retina. Mol. Vis. 7, 131–7. [PubMed] [Google Scholar]
- (41).Johnson CJ, Berglin L, Chrenek MA, Redmond TM, Boatright JH, and Nickerson JM (2008) Technical brief: subretinal injection and electroporation into adult mouse eyes. Mol. Vis. 14, 2211–26. [PMC free article] [PubMed] [Google Scholar]


